
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 03 June 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.812971
This article is part of the Research TopicApplication of Fishes as Biological Models in Genetic StudiesView all 16 articles
Cystatin proteins are known to form a superfamily of cysteine protease inhibitors, which play a key role in protein degradation and are related to different physiological processes, such as development and immunity. Currently, numerous immunoregulatory proteins, such as cystatins, are being used in the control and prevention of diseases in aquaculture. Thus, the objective of this study was to produce recombinant cystatin (rCYST-B) from the red piranha Pygocentrus nattereri and to evaluate its effect on bacterial growth. The gene that encodes cystatin-B was isolated from the spleen of P. nattereri and cloned in an expression system. The protein was produced via a heterologous system involving the yeast Pichia pastoris X-33. The inhibitory activity of purified cystatin-B was evaluated on papain using different concentrations (0–80.0 μg/μL) of rCYST-B. The bacteriostatic action of the protein was evaluated using the Kirby-Bauer method on the growth of Escherichia coli and Bacillus subtilis. rCYST-B showed 100% inhibition at a concentration of 60 μg/μL. Moreover, the bacteriostatic activity of E. coli and B. subtilis showed inhibition of 40.36 and 49.08% compared to the negative control (phosphate buffer), respectively. These results suggest that recombinant CYST-B has biotechnological potential for use in aquaculture.
• Cystatin-B was successfully expressed in methylotrophic yeast, Pichia pastoris X-33.
• Functional analysis of purified rCystatin-B demonstrated papain inhibition activity.
• rCystatin-B efficiently inhibited the growth of E. coli and B. subtilis.
• Recombinant Cystatin-B exhibited potential for therapeutic use in fish.
Aquaculture is a productive sector with accelerated growth, an activity that has become fundamental for the achievement of the Sustainable Development Goals (SDGs) proposed by the United Nations (FAO 2017). According to the Food and Agriculture Organization of the United Nations (FAO) global fisheries production included approximately 179 million tons of fish, of which aquaculture represented production of 82.1 million tons, corresponding to 46% of the total production in 2018 (FAO SOFIA 2020). However, this growth was accompanied by the emergence or re-emergence of several infectious diseases (Pérez-Sánchez et al., 2018; FAO SOFIA 2020), such as acute hepatopancreatic necrosis (AHPND) (OIE 2013), enterocytozoon hepatopenaei (EHP) (Tang et al., 2017), emerging bacteria of the genus Acinetobacter, a potential pathogen in shrimp Penaeus vannamei (Huang et al., 2020) and some parasitic infections such as acanthocephalosis, reported for several groups of fish (Valladão et al., 2019), which have a detrimental influence on their health (Rosny et al., 2016). To overcome this problematic it is necessary to apply good management techniques and to develop new technologies. Biotechnology methods and genetic engineering enable the production of recombinant proteins, which can be used both in the prevention and treatment of diseases, and are considered promising for use in the aquaculture industry.
Cystatins are a large group of proteins that function as protease inhibitors involving cysteine residues in the proteolytic reaction (Berti and Storer 1995). Typically, they are small proteins that inactivate the protease substrate in a specific manner, forming reversible complexes (Turk and Bode 1991). Cysteines play essential roles in the physiology of all living organisms, from protozoa to mammals. In pathogenic microorganisms including bacteria, fungi, and parasites, cysteine protease can act as virulence factor, causing diseases in host organisms (Mottram et al., 2004; Rudenskaya and Pupov 2008). The cystatin superfamily is grouped into three families (Barrett 1986; Magister and Kos 2013), namely, family I (Stefin), family II (cystatins), and family III (kininogen). A fourth family has been reported of invertebrate origin, mainly of nematode parasites (Khaznadji et al., 2005; Li et al., 2010). Family I, composed of Stefin A and B, also known as cystatin A and B, are single-chain polypeptides of approximately 100 residues and molecular weight between 10 and 11 kDa without disulfide bonds or carbohydrate side chains. These are intracellular protease inhibitors present in the cytosol (Barrett 1986; Turk and Bode 1991; Abrahamson et al., 2003). Family 2 cystatins include cystatin C, D, E/M, F, G, S, SA, and SN with a molecular weight between 13–14 kDa and exhibit a signal peptide and two disulfide bridges (Cornwall and Hsia 2003). Family III (kininogens), are large glycoproteins (60–120 kDa) and complex in structure and are found in body fluids, especially in plasma (Barrett 1986; Prunk et al., 2016). All cystatins, regardless of family classification, contain several conserved regions, including an N-terminal glycine segment, a QXVXG sequence that constitutes part of the β-hairpin loop structure, and a proline-tryptophan-containing region that forms a second hairpin loop (Margis et al., 1998; Rzychon and Chmiel 2004). Cystatins play important defensive and regulatory roles during various cellular events. They modulate and stimulate TNF-α and IL-10 synthesis as a defense strategy in response to pathogen infection (Verdot et al., 1999).
In fish, cystatins exhibit protease inhibitory activity and appear to be involved in immune responses against infectious agents (Xiao et al., 2010; Premachandra et al., 2012; Ahn et al., 2013). To explore the functions of fish cystatin, some recombinant cystatins from Keta salmon (Yamashita and Konagaya 1991), trout (Li et al., 2000), and carp (Tzeng et al., 2001) were generated by heterologous expression in Escherichia coli. Although these recombinant proteins have already been developed for fish, the use of a species-specific protein or that obtained from a phylogenetically close species may decrease the side effects, besides potentiating the expected immunoregulatory effect.
Currently, genome sequencing allows the use of genetic information to produce specific proteins for application in aquaculture. However, few species of commercial interest have their genome completely sequenced. One such species is the red piranha Pygocentrus nattereri. The red piranha is a widely distributed carnivorous fish species that is observed in the rivers and lakes of South America (Behr and Signor 2008). In addition to being a popular aquarium fish, piranhas are often sold for human consumption in local markets in the Amazon basin (Duponchelle et al., 2007). Another important characteristic of this species is its similarity phylogenetic with other freshwater species such as tambaqui (Colossoma macropomum of the family Serrasalmidae). Associating the genetic knowledge of species of commercial interest to the development of biotechnologies allows the production of recombinant proteins of industrial interest, such as cystatin, for applications in aquaculture to maintain animal health. Thus, in this study, the main objective was the heterologous production and evaluation of the biological activity of the Cystatin-B derived from the red piranha Pygocentrus nattereri.
Red piranha Pygocentrus nattereri juveniles, weighing approximately 200 g were captured at Janauacá Lake, municipality of Manaquiri - Amazonas, Brazil. The fish were euthanized (n = 13) with eugenol (200 mg/L), and the spleen tissue was harvested, preserved in RNAlater (Ambion, United States) and stored at −80°C.
Total RNA extraction from the red piranha spleen samples was performed using the total RNA extraction protocol using Trizol Reagent ® (Invitrogen, United States), according to the manufacturer’s recommendations. The quality of the extracted RNA was evaluated by 1% (w/v) agarose gel electrophoresis and quantified using a spectrophotometer (BioDrop - Biodrop µLite). Total RNA was treated with the DNAse I (RQ1 RNase-Free DNase - Promega) following the protocol described by the manufacturer. Complementary DNA (cDNA) was generated using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, United States) from 1 µg of total RNA using oligo d (T) primer.
Isolation of the genes encoding cystatin-B was performed by polymerase chain reaction (PCR). Specific primers were previously designed with the Primer-BLAST tool from GenBank (www.ncbi.nlm.nih.gov) based on the sequences XM_017717432 of Pygocentrus nattereri and used in this experiment. The primer sequences are listed as follows: Forward: 5′ CAGCAGGAGAGCAGAAGTTGA 3′ e Reverse 5′ TGTTAGTACGGTTTGTTAAGGGGA 3’. GoTaq® (Promega Corporation. Madison, United States) was used in the reaction, following the manufacturer’s recommendations. PCR cycles were set as follows: initial cycle at 94°C for 2 min, 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 5 min.
After verifying the correct fragment size using electrophoresis in 1% agarose gel, the samples were purified using the PureLink™ PCR Purification Kit® (Invitrogen, Brazil), following the manufacturer’s protocol. The purified PCR products were sequenced using the Sanger method on the Applied Biosystems® Sanger Sequencing 3500 Genetic Analyzer. Automated sequencing was performed by the company ACTGene Molecular Analyses (UFRGS, Brazil). The sequencing result was first analyzed by the quality of the electropherograms obtained using the Chromas software and then aligned using the BLASTn tool (https://blast.ncbi.nlm.nih.gov).
After confirmation by sequencing, protein analyzes were performed based on the predicted protein sequences using the Genebank database. Was used the cystatin-B isoform X1 (XP_017572921.1) protein. Multiple alignment analysis of the amino acid sequences of the CYSTATIN-B protein (named CYST-B herein) was performed using Geneious Prime Software (version 2020.1.2). The signal peptide deduced from the amino acid sequence was predicted by XtalPred (https://xtalpred.godziklab.org) and SignalP 5.0 Software (http://www.cbs.dtu.dk/services/SignalP). The secondary structure of the amino acid sequence of the protein was constructed using PredictProtein (https://predictprotein.org) and Phyre2 system (http://www.sbg.bio.ic.ac.uk/phyre2). The 3D structural model of the protein was established using the Phyre2 system. The phylogenetic tree was constructed based on the amino acid sequence of Cystatin-B, using MEGA X (Kumar et al., 2018) and the Neighbour-joining method with 1000 bootstrap replicates.
Specific primers were re-designed by inserting restriction enzyme sites of XhoI and XbaI (cystBXhoI- For = 5′AACTCGAGCAGCAGCAGGAGAGCAGAAGTTGA 3′ and cystBXbaI-Rev = 5′AATCTAGATGTTAGTTAGTACGGTTTGTTAAGGGGA 3′). The insertion of these sites allowed the cloning of fragments in the pPICZα expression vector (Figure 1). GoTaq® (Promega Corporation. Madison, United States) was used in the reaction, following the manufacturer’s recommendations. PCR cycles were set as follows: initial cycle at 94°C for 2 min, 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, plus a final extension at 72°C for 5 min. Verification of the size of the PCR products was performed using agarose gel electrophoresis (1%). After confirmation, the PCR products were purified using the PureLink PCR Purification Kit (Thermo Fisher Scientific, United States) and cleaved using the enzymes XhoI and XbaI (Promega Corporation, United States), following the protocol described by the manufacturer (Figure 1B). The cleaved fragments were further purified (as described above) for binding to the pPICZα expression vector. The fragment vector was obtained by PCR using the pPICZα plasmid as the template (Figure 1C). The primers pPIC-Forward (XbaI) ATCTAGAACAAAAACTCATCTCAGAGAAGAGG and pPIC-Rever (XhoI) AGCTTCAGCCTCTCTTTTCTCGAGAG were used. PCR conditions were set as follows: an initial cycle of 94°C for 2 min, 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 4 min in addition to a final extension of 72°C for 10 min. The pPIC vector was also cleaved with the same restriction enzymes (XbaI and XhoI) to allow cohesive sequence formation and binding at complementary sites. The binding of the cleaved and purified insert to the vector was performed using the enzyme T7 ligase (Invitrogen, Brazil), following the manufacturer’s recommendations (Figure 1D).
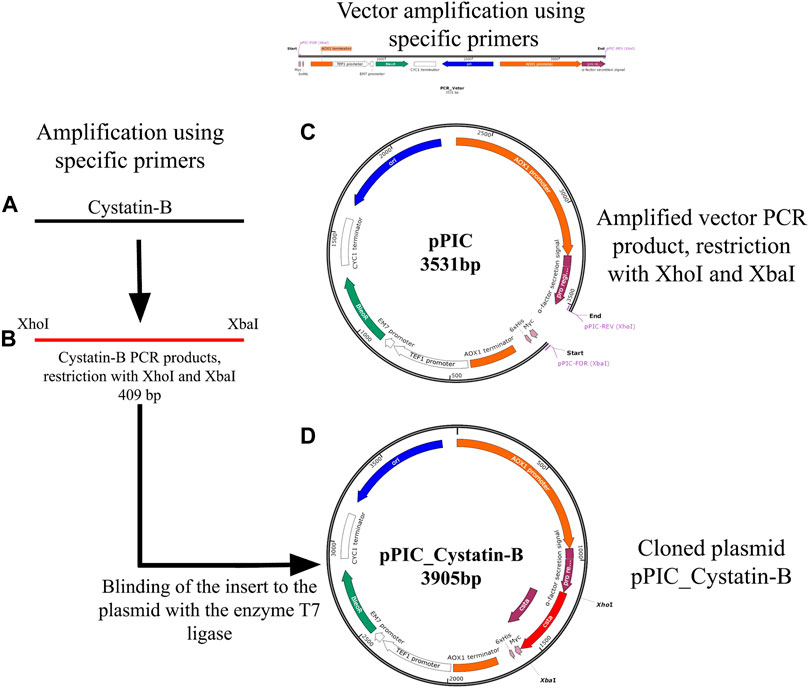
FIGURE 1. Schematic representation of the genetic construct containing cystatin-B (A) Amplification by PCR of the cystatin-B gene of juveniles of Pygocentrus nattereri, using specific primers, designed based on the sequence XM_017717432. (B) Amplified PCR products and cleaved with XhoI and XbaI enzymes. (C). Amplified vector (pPIC) products by PCR, cleaved with XhoI and XbaI. (D). Cloned plasmid pPIC_Cystatin-B (Binding of the insert to the plasmid with the enzyme T7 ligase).
The obtained genetic constructs were used for Escherichia coli TOP10 transformation. The calcium chloride method was used to generate chemically competent cells. Plasmid DNA was extracted using the plasmid DNA kit PureYield ™ Plasmid Miniprep System (Promega). After transformation, bacteria were grown overnight (37°C) in culture plates in LB (Luria-Bertani) solid medium and 50 μg/ml of the selection antibiotic Zeocin™ (Invitrogen, Brazil). E. coli colonies transformed with the cloned plasmid pPIC_cystatin -B were inoculated in LB liquid culture medium containing Zeocin (50 µg/ml−1) with shaking at 225 rpm and a temperature of 37°C. Plasmid DNA extraction was performed using the PureYield™ Plasmid Miniprep System plasmid DNA kit (Invitrogen, Brazil) according to the manufacturer’s recommendations. Plasmids extracted were cleaved with XhoI, XbaI, BamHI, and BgLII and visualized using 1% agarose gel electrophoresis, as well as by PCR using different combinations of primers for specific regions both in the gene of interest and regions of the pPIC_cystatin B plasmid: 5′AOX- (3′ GACTGGTTCCAATTGACAAGC 5′) and 3′AOX -(5′ GCAAATGGCATTCTGACATCC 3′).
Following verification, the plasmid was linearized using the restriction enzyme BamHI, the products were visualized by 1% agarose gel electrophoresis, quantified using a spectrophotometer (BioDrop, Isogen Life Science, Netherlands), and purified. Competent cells were prepared following the procedure in the Easy Select ™ Pichia Expression Kit manual (Thermo Fisher Scientific, United States). The transformation was performed in P. pastoris X33 yeast using the PichiaPink™ Expression System protocol (Thermo Fisher Scientific, United States) with some modifications (was used linearized plasmid DNA). A total of 80 μL of the cells with linearized plasmid DNA were mixed into electroporation cuvettes (0.2 cm) and incubated for 5 min on ice. They were subsequently electroporated (1500 V, 25 μF, 400 Ω). Immediately, 1 ml of ice-cold sorbitol was added to the cuvette, and the contents were transferred to a sterile 1.5 ml Eppendorf-type tube and incubated for 2 h at 30°C without shaking. A total of 100 μL of the transformed cells were plated onto Minimal Dextrose Medium (2% agar; 2% glucose; 4 × 10-5% biotin; yeast nitrogen base YNB 1.34% and 125 μg/ml Zeocin). The plates were incubated at 30°C for a maximum of 4 days until the development of colonies and selection of clones for further induction analyses.
To increase the cell mass the selected clones and the negative control (without plasmid) of P. pastoris were inoculated in 200 ml of Buffered Glycerol Complex Medium (yeast extract 1%; peptone 2%; phosphate buffer pH 6.0 100 mM; YNB 1.34%; biotin 4 × 10-5%; glycerol 1%) and incubated at 30°C with shaking at 250 rpm until the culture showed an OD600 between 2–6, in approximately 18 h. Upon reaching the required OD600, the cultures were centrifuged at 3000 rpm for 10 min at 4°C and resuspended in 300 ml of Buffered Methanol Complex Medium (yeast extract 1%; peptone 2%; phosphate buffer pH 6.0 100 mM; YNB 1.34%; biotin 4 × 10-5%; methane 1%) in 1 L Erlenmeyer flask. The expression of alcohol oxidase enzyme promoter 1 (AOX1) was induced by adding absolute methanol every 24 h to the cell cultures, maintaining a final concentration of 1% (v/v) for 120 h at 30°C with shaking at 250 rpm. Aliquots containing 50 ml of the cultures were collected, centrifuged at 3000 rpm for 10 min at 4°C, and preserved at −80°C for further analysis. The recombinant protein was purified using Ni Sepharose high-Performance nickel-based resin (GE Healthcare, United States) with polyhistidine tail (6xHis). The purified recombinant rCYST-B protein was analyzed via denaturing 15% polyacrylamide gel electrophoresis (SDS-PAGE). Subsequently, the gels were stained with Coomassie blue (0.25%) (Figure 3A), and silver nitrate (20%) (Figure 3B). The concentration of the purified recombinant protein was evaluated according to the method described by Lowry et al. (1951).
The inhibitory effect of the recombinant rCYST-B protein was analyzed using papain as a cysteine protease, performed as described in Xiao et al. (2010) modified for (Yu et al., 2019). The assay was performed using different amounts of rCYST-B proteins (0, 0.1; 0.5; 1.0; 10.0; 15.0; 20.0; 30.0; 60.0 and 80.0 μg), which were incubated with 10 μL of papain (0.1 μg/μL) (Sigma-Aldrich, United States)at 28°C for 30 min. The reaction was initiated by adding 200 μL of Azocasein (0.2% W/V) (Sigma-Aldrich, United States), followed by incubation at 37°C for 2 h. The reaction was inactivated with 200 μL of 10% trichloroacetic acid (TCA). The contents were chilled on ice for 15 min. The precipitate was separated by centrifugation at 15,000 rpm for 5 min. The absorbance was evaluated by spectrophotometry at 450 nm. As a negative control, αGHT protein was used in place of rCYST-B. The relative activity of rCYST-B was determined by: 100x (1–(A440 of rCYST-B)/(A440 control).
The assay for the identification of bacteriostatic action was performed using the Kirby-Bauer method (Bauer et al., 1966). Two types of bacteria (Escherichia coli or Bacillus subtilis) were plated on the surface of Petri dishes with LB medium and incubated at 37°C for 12–16 h. Disks were added with rCYST-B (60 and 80 μg/μL), Spectinomycin 50 ug/uL (positive control), and Potassium Phosphate Buffer 50 mM (negative control). The bacteriostatic action was evaluated qualitatively and quantitatively. The qualitative analysis was related to the presence of inhibition halos around the disk, and the quantitative analysis evaluated the halo diameter (mm), attributed to the bacteriostatic action of the protein. For halo diameter analysis, the plates were photographed (n = 4) and analyzed using ImageJ software.
All experiments were performed in at least triplicates. All values are shown as mean ± standard deviation of the mean. Factorial type ANOVA was applied between the different treatments used to determine the bacteriostatic effect of rCYST-B; significant differences were determined with a p < 0.05. The analyses were performed using GraphPad Prism 8.0 software (GraphPad Software).
All procedures adopted in this study were performed according to the protocols previously approved by the Ethics Committee on Animal Use (CEUA) of Universidade Nilton Lins with approval protocol 015/2017.
The PCR amplified gene products presented a fragment of 409 base pairs (bp) with 100% similarity to the partial sequence available in GeneBank (XM_017717432.2). The sequence of CYST-B showed 104 amino acid residues, with an estimated molecular weight of 11.755 kDa, a theoretical isoelectric point of 6.04, stability index of 33.98, without the presence of a signal peptide. The secondary structure analysis of CYST-B (Figure 2A), showed a structure composed of: loop 49.0%, helix 17.3%, and strand 33.7%. The 3D structure of CYST-B was predicted by Phyre2 (Figure 2B). The tertiary structure of CYST-B was colored using the rainbow command, blue at the N-terminal and red at the C-terminal. The rainbow command colors residues; 99 residues were modeled up to 100.0% confidence and a reliable model was obtained with the following characteristics: (Å): X: 44.068 Y:34.294 Z:30.445. Multiple homologous alignments of the amino acid sequences of CYST-B from P. nattereri (Figure 2C) revealed conserved regions typical of the cystatin superfamily, including two conserved N-terminal glycine residues (G10G11) as well as the glutamine-valine-glycine motif (QXVXG), crucial for the biological activity of the molecule. The motif represented by glutamine-leucine-valine-alanine-glycine (Q52LVAG56) exhibited homology with species such as Colossoma macropomum (GeneBank accession code XP_036414104.1) with 100% identity. In addition, a typical proline-tryptophan (PW) motif was identified at the C-terminus, where the tryptophan residue was replaced by a tyrosine residue (P80Y81). Based on the total amino acid sequence length of CYST-B, phylogenetic tree analysis was performed using MEGA X (Figure 2D). The result indicated a sequence similarity to that of tambaqui Colossoma macropomum, a phylogenetically close species.
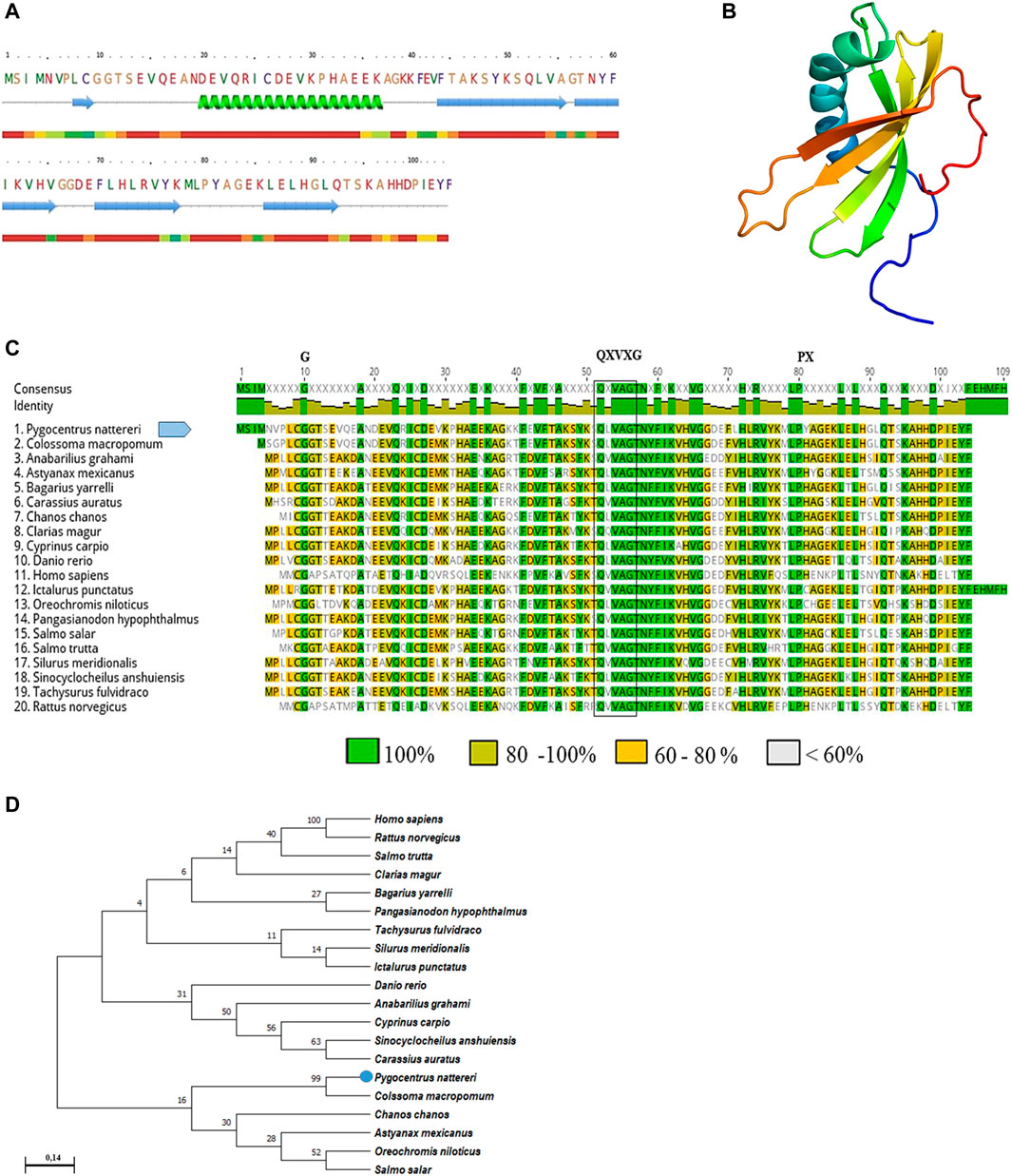
FIGURE 2. Bioinformatic analysis of Pygocentrus nattereri cystatin-B (A). Secondary structure of cystatin-B of P. nattereri generated by PredictProtein and Phyre2 system. fx1 Alpha helix, fx2Beta Strand. (B). The 3D tertiary structure of Cystatin-B generated by the Phyre2 system. Model dimensions (Å): X:44,068 Y:34.294 Z:30.445. (C). Multiple alignments of the cystatin-B amino acid sequence of Pygocentrus nattereri and other sequences. Colossoma macropomum, XP_036414104.1; Anabarilius grahami, ROK23366.1; Astyanax mexicanus, XP_007249105.1; Bagarius yarrelli, TSK98422.1; Carassius auratus, XP_026093937.1; Chanos, XP_030625480.1; Clarias magur, KAF5903616.1; Cyprinus carpio, XP_018952861.1; Danio rerio, NP_001096599.1; Homo sapiens, NP_000091.1; Ictalurus punctatus, XP_017308724.1; Oreochromis niloticus, XP_003443657.1; Pangasianodon hypopthalmus, XP_026793986.1; Salmo salar, XP_014062104.1; Salmo trutta, XP_029625791.1; Silurus meridionalis, KAF7691695.1; Sinocyclocheilus anshuiensis, XP_016296734.1; Tachysurus fulvidraco, XP_027030842.1; Rattus norvegicus, NP_036970.1. Characteristic and conserved residues of glycine (G) and proline (P) are indicated by the letter “G” and “P”, respectively. The characteristic and preserved residues of the QXVXG motif are marked with a box. (D). Phylogenetic tree of cystatin-B and sequences of their counterparts constructed using the neighbor-joining method and 1000 bootstrap replicates. The numbers presented in the branches represent the bootstrap value (%).
SDS-PAGE showed that purified rCYST-B produced a band (Figure 3B, Lanes 3–8) with the expected size of approximately 11.8 kDa. These results confirm the expression of the recombinant rCYST-B protein in P. pastoris X33 cells (Figure 3A), demonstrating a suitable platform for the production of heterologous cystatin proteins.
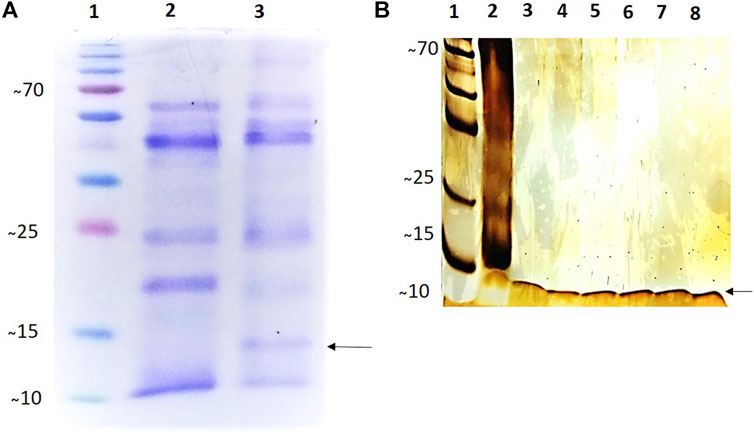
FIGURE 3. Analysis of the expression and purification of rCystatin-B with SDS-PAGE (120 h of induction). (A). Gel stained with 0.25% Coomassie blue; (Lane 1). PMM - Molecular Mass Standard - kDa (Invitrogen). (Lane 2). Lyophilized fraction (crude extract) of Pichia pastoris cell culture without cloned plasmid (negative control) subjected to the induction and expression process; (Lane 3). Transformed and lyophilized fraction of the culture of Pichia pastoris cells subjected to the induction and expression process (inclusion of cloned plasmid: pPIC_Cystatin-B). (B). Gel stained with Silver Nitrate (20%). (Lane 1). PMM - Molecular Mass Standard - kDa (Invitrogen). (Lane 2). Transformed and lyophilized fraction of the culture of Pichia pastoris cells subjected to the induction and expression process (inclusion of cloned plasmid: pPIC_Cystatin-B). (Lanes 3–8). Purified rCystatin-B protein (indicated by the arrow). The recombinant protein was purified using Ni Sepharose High-Performance Nickel based resin.
The in vitro inhibitory activity of the recombinant rCYST-B protein on papain (a cysteine protease), revealed a concentration-dependent inhibition response of the inhibitor, with the increase starting at a concentration of 30 μg/μL and reaching 100% inhibition at the concentration of 60 μg/μL (Figure 4).
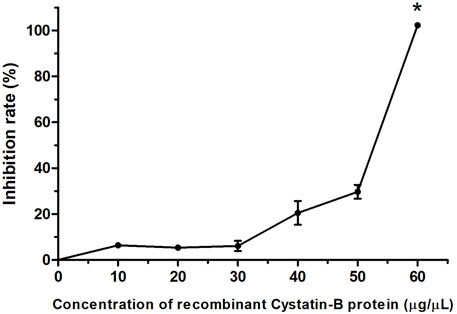
FIGURE 4. Inhibition pattern of rCystatin-B against papain activity. The curve represents the change in residual papain activity along with the addition of rCystatin-B. αGTH (negative control), did not show any inhibitory effect (data not shown). Values are shown as mean ± standard deviation from the mean (n = 3). Asterisc (*) indicate statistical difference (p = 0.003).
The bacteriostatic activity of the recombinant rCYST-B protein on E. coli (Gram-negative bacteria) and B. subtilis (Gram-positive bacteria) demonstrated the presence of inhibition halos (zone of inhibition) at concentrations of 60 and 80 μg/μL of rCYST-B (Figure 5A). The diameter of the inhibition halo at the concentration of 60 μg/μL was 9.66 ± 0.61 mm for E. Coli and 10.50 ± 2.94 mm (p > 0.05) for B. Subtilis (Figure 5B). At the concentration of 80 μg/μL, the halo diameter was 8.64 ± 0.70 mm and 12.41 ± 5.53 mm for E. coli and B. subtilis, respectively (Figure 5C). These values represent a bacteriostatic action in comparison to the negative control (PBS), which did not show a halo of inhibition for any of the replicates. The positive control (spectinomycin antibiotic) showed a halo with a diameter of 23.93 ± 1.42 mm and 21.39 ± 1.83 mm at a concentration of 60 μg/μL for E. coli and B. subtilis, respectively. At the concentration of 80 μg/μL, the halo of inhibition in the positive control was 20.04 ± 2.55 mm and 24.53 ± 3.91 mm for E. coli and B. subtilis, respectively.
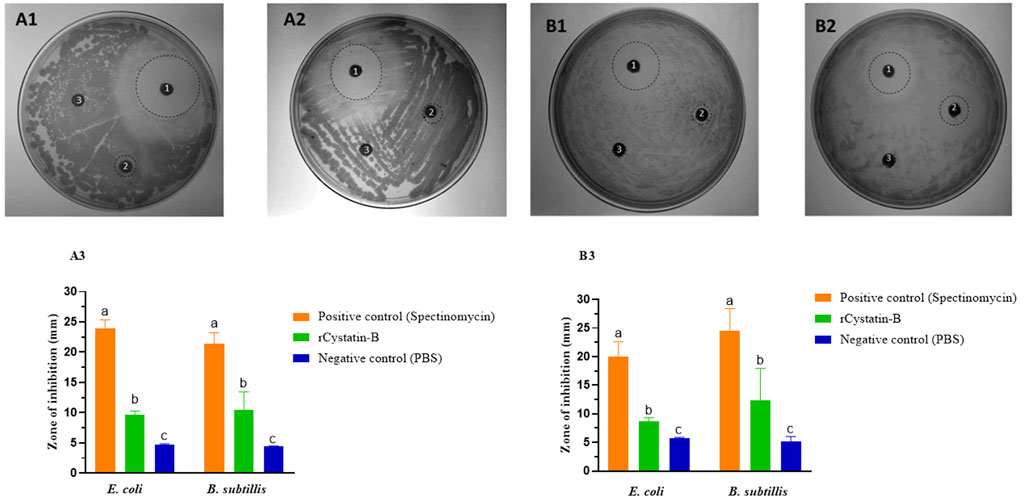
FIGURE 5. Bacteriostatic action of rCystatin-B. (A). 60 μg/μL. (A1). E. coli. (A2). B. subtilis. (A3). Zone of inhibition (diameter mm). (B). 80 µg/µL. (B1). E. coli. (B2). B. subtilis. (B3). Inhibition diameter (mm). The dashed line circles correspond to the observed zone of inhibition. 1). Spectinomycin (positive control). 2) recombinant rCystatin-B protein. 3). PBS (negative control). Values are shown as mean ± standard deviation from the mean (n = 3). Different letters show statistical differences (p < 0.05).
Developing new strategies for disease management and control is one of the current challenges in aquaculture. Biotechnology is considered an important tool for the treatment of emerging diseases associated with aquaculture production. In this study, the objective was to produce a protein with possible immunoregulatory potential for fish. For this, a 409 bp fragment related to the cystatin B gene was isolated from the spleen of the fish Pygocentrus nattereri. This is the first report of the isolation of this gene for this species.
Typically, cystatins in fish are composed of approximately 100 amino acids, varying somewhat according to the species and isoform. For example, in species such as Scophthalmus maximus (Xiao et al., 2010), the gene size of cystatin B was 300 bp length. Ahn et al. (2013) reported the stefin B gene with a size of 297 bp for Paralichthys olivaceus. Cystatin homologs or stefin A have been isolated from species such as Ctenopharyngodon idella with a nucleotide sequence length of 294 bp (Li et al., 2021). Another important feature of cystatins involves the three conserved domains that form the inhibitory sites of interaction with proteases: an N-terminal glycine, a glutamine-X-valine-X-glycine motif, and a C-terminal proline-tryptophan amino acid pair (Bode et al., 1988; Stubbs et al., 1990). In the present study, multiple alignments identified three conserved regions typical of the cystatin superfamily. In rCYST-B, the N-terminal domain containing the conserved glycine residue (G10G11) and the presence of similarity to its homologs from other species were found. The 2 G residues are known to constitute a wedge-shaped border, involved in the inhibition of protease activity, complementary to the active site of papain-like cysteine proteases (Bode et al., 1988; Abrahamson 1993). In addition, it is the domain involved in the interaction between the protease and the inhibitor, which allows it to interact directly with the S3, S2, and S1 substrate-binding regions of the protease (Hall et al., 1995; Bjork et al., 1995). The QXVXG motif, highly conserved in cystatins, was observed in rCYST-B from P. Nattereri. This motif binds to the active site of cysteine proteases, thereby interfering with their interaction with the substrate and inhibiting their activity (Bode et al., 1988; Turk and Bode 1991). The variation in the presence of amino acids proline - tyrosine in the C-terminal region of Cystatin B theoretically does not affect the inhibitory activity, both amino acids have hydrophilic characteristics and very similar isoelectric points. The inhibitory response was observed in the recombinant rCYST-B produced in the present study. Different types of C-terminal variations have been observed in human Stefin B, with replacements by the histidine (H) residue (Bjork et al., 1995) as well as in Acipenser sinensis (Bai et al., 2006) with the replacement by the Leucine (L) residue. Analysis of the phylogenetic tree performed in this study (Figure 2D) confirms the evolutionary relationship and proximity of Cystatin-B to its cystatin homologs as well as the functions among type 1 cystatin (Stefins) (Pemberton 2006).
The estimated molecular weight of rCYST-B was 11.8 kDa, similar results are observed for recombinant cystatin B protein obtained from Ctenopharyngodon idella with 11.48 KDa (Li et al., 2021), Pseudosciaena crocea with 11.4 KDa (Li et al., 2009), Scophthalmus maximus with 11.1 kDa (Xiao et al., 2010), Paralichthys olivaceus with 12.7 KDa (Ahn et al., 2013), reported values for Stefins family cystatins (Ochieng and Chaudhuri 2010).
After bioinformatics analyses, we evaluated the protease inhibition activity of the purified recombinant CYST-B protein. For this, we chose papain as cysteine protease and azocasein as substrate. The rCYST-B from P. Nattereri exhibited concentration-dependent inhibition of papain activity. These results confirmed that the purified recombinant protein was able to maintain its biological functions. The concentration at which 100% inhibition was observed was 60 μg/μL. However, a previous study reported a concentration of 0.5 μg/μL for Stefins B homologs from Oplegnathus fasciatus (Premachandra et al., 2012). Recently, Li et al. (2021), reported a dose-dependent inhibition pattern for the proteolytic activity of papain, cathepsin B, and cathepsin L, showing a decrease in the activity with the increasing amount of Stefin A isolated from Ctenopharyngodon idella. In the case of species such as Crassostrea gigas, the concentration of 50 μg/μL of cystatin A showed an inhibition rate of around 80% (Mao et al., 2018). In contrast, Premachandra et al. (2012), produced and purified a recombinant cystatin B from rock bream Oplegnathus fasciatus and demonstrated 82% inhibition of papain activity at a concentration of 0.5 μg/μL. Even though there is a wide variation in the concentration of cystatin showing 100% inhibition of protease activity, all results suggest that these recombinant proteins may show regulation of exogenous proteases from microorganisms and parasites that infect the host (Scott et al., 2007). These differences in the potential or response of the inhibitory activity of cystatin are supported by the wide spectrum of its structural variation and distribution in different types of cells and tissues (Lefebvre et al., 2008; Xiao et al., 2010); structure characterized by highly conserved regions that form a discrete wedge-shaped structure that blocks the active site of cysteine proteases (Gregory and Maizels, 2008). On the other hand, G11 introduces some flexibility in the N-terminal region of inhibitors, allowing the N-terminal motif to adopt an optimal conformation for enzyme-inhibitor interaction (Hall et al., 1993). The greater inhibitory potential exerted on cysteine proteases has allowed the use of the specificity of Cysteine protease inhibitory activity substrates, as well as different evaluation protocols under in vitro conditions (temperature and enzymatic reaction times) (Bai et al., 2006; Xiao et al., 2010; Wickramasinghe et al., 2020).
In our study, it was evidenced that the protein heterologously produced by P. pastoris was able to inhibit the growth of E. coli and B. subtilis. Inhibition of E. coli and B. subtilis bacterial growth were also observed with Paralichthys olivaceus Cystatin-C (Yu et al., 2019) as well as on Scophthalmus maximus Cystatin-B (Xiao et al., 2010). Previous results confirm the antiviral and antibacterial role of cystatins (Wesierska et al., 2005). In general, proteins with antimicrobial capacity use their own cationic charge to attack and enter in a faster and more effective way through the bacteria membrane, increasing membrane permeability and flow of intracellular contents and subsequently, cell lysis (Izadpanah and Gallo 2005). We believe that this mechanism is used by rCYST-B, however, further research is needed to prove this affirmation. It should be noted that Cystatin-B is involved in immune responses to bacterial and fungal infections and anti-apoptotic processes in the brain (Takahashi et al., 1994; Di Giaimo et al., 2002). They are also expressed at high levels in follicular dendritic cells, which are involved in the immune and inflammatory reaction, present in the germinal centers of secondary lymphoid organs (Rinne et al., 1986). Furthermore, they can prevent apoptosis of B cells containing high-affinity receptors for the antigen present on the surface of follicular dendritic cells (van Eijk and Groot 1999).
Based on the confirmation of the antibacterial potential of rCYST-B, we believe that this protein can be used as an immunoregulatory protein for fish, and also as a food supplement in the fish feed. This hypothesis is based on the fact that recombinant fish cystatins have already been produced and showed potential in preventing fish diseases (Bjorklund et al., 1997). Recombinant cystatins obtained from Keta salmon (Yamashita and Konagaya 1991), trout (Li et al., 2000), and carp (Tzeng et al., 2001) were generated by heterologous expression in Escherichia coli. Even though these recombinant proteins have already been developed for fish, the use of a species-specific protein or that obtained from a phylogenetically close species may decrease the side effects besides potentiating the expected immunoregulatory effect.
The results of this study constitute the first reports on the production of a recombinant rCYST-B protein for a species native to South America. In conclusion, the systematic study of cloning, expression, and characterization of the biological activity of the recombinant cystatin B protein from Pygocentrus nattereri suggests its role as a possible immunoregulatory protein in biological defense against invaders and cellular protection against proteolysis mediated by cysteine proteases.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Ethics Committee on Animal Use (CEUA) of Universidade Nilton Lins with approval protocol 015/2017.
JM Collected the data–Performed the analysis–Discussed the results and wrote to the final manuscript DA Conceived and designed the analysis–Performed the analysis–Discussed the results and contributed to the final manuscript. Supervised the findings of this work.
This work was supported by Brazilian CNPq (National Council for Scientific and Technological Development) (Proc. Number 432071/2018) and FAPEAM (Foundation for Research Support of the State of Amazonas). Special thanks to Professor Luis Fernando Marins for helping with the molecular analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.812971/full#supplementary-material
Abrahamson, M., Alvarez-Fernandez, M., and Nathanson, C.-M. (2003). Cystatins. Biochem. Soc. Symp. 70, 179–199. doi:10.1042/bss0700179
Abrahamson, M. (1993). Cystatins - Protein Inhibitors of Papain-like Cysteine Proteinases Cienc Cult. Sao Paulo) 45, 299.
Ahn, S. J., Bak, H. J., Park, J. H., Kim, S. A., Kim, N. Y., Lee, J. Y., et al. (2013). Olive Flounder (Paralichthys olivaceus) Cystatin B: Cloning, Tissue Distribution, Expression and Inhibitory Profile of Piscine Cystatin B. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 165 (3), 211–218. doi:10.1016/j.cbpb.2013.04.007
Bai, J., Ma, D., Lao, H., Jian, Q., Ye, X., Luo, J., et al. (2006). Molecular Cloning, Sequencing, Expression of Chinese Sturgeon Cystatin in Yeast Pichia pastoris and its Proteinase Inhibitory Activity. J. Biotechnol. 125 (2), 231–241. doi:10.1016/j.jbiotec.2006.02.023
Barrett, A. J. (1986). The Cystatins: a Diverse Superfamily of Cysteine Peptidase Inhibitors. Biomed. Biochim. Acta 45 (11-12), 1363–1374.
Bauer, A. W., Kirby, W. M. M., Sherris, J. C., and Turck, M. (1966). Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathology 45, 493–496. doi:10.1093/ajcp/45.4_ts.493
Behr, E. R., and Signor, C. A. (2008). Distribuição e alimentação de duas espécies simpátricas de piranhas Serrasalmus maculatus e Pygocentrus nattereri (Characidae, Serrasalminae) Do rio Ibicuí, Rio Grande Do Sul, Brasil. Iheringia, Sér. Zool. 98 (4), 501–507. doi:10.1590/S0073-47212008000400014
Berti, P. J., and Storer, A. C. (1995). Alignment/phylogeny of the Papain Superfamily of Cysteine Proteases. J. Mol. Biol. 246, 273–283. doi:10.1006/jmbi.1994.0083
Björk, I., Brieditis, I., and Abrahamson, M. (1995). Probing the Functional Role of the N-Terminal Region of Cystatins by Equilibrium and Kinetic Studies of the Binding of Gly-11 Variants of Recombinant Human Cystatin C to Target Proteinases. Biochem. J. 306, 513–518. doi:10.1042/bj3060513
Björklund, H. V., Johansson, T. R., and Rinne, A. (1997). Rhabdovirus-induced Apoptosis in a Fish Cell Line Is Inhibited by a Human Endogenous Acid Cysteine Proteinase Inhibitor. J. Virol. 71, 5658–5662. doi:10.1128/jvi.71.7.5658-5662.1997
Bode, W., Engh, R., Musil, D., Thiele, U., Huber, R., Karshikov, A., et al. (1988). The 2.0 A X-Ray Crystal Structure of Chicken Egg White Cystatin and its Possible Mode of Interaction with Cysteine Proteinases. EMBO J. 7, 2593–2599. doi:10.1002/j.1460-2075.1988.tb03109.x
Cornwall, G. A., and Hsia, N. (2003). A New Subgroup of the Family 2 Cystatins. Mol. Cell. Endocrinol. 200, 1–8. doi:10.1016/S0303-7207(02)00408-2
Di Giaimo, R., Riccio, M., Santi, S., Galeotti, C., Ambrosetti, D. C., and Melli, M. (2002). New Insights into the Molecular Basis of Progressive Myoclonus Epilepsy: a Multiprotein Complex with Cystatin B. Hum. Mol. Genet. 11, 2941–2950. doi:10.1093/hmg/11.23.2941
Duponchelle, F., Lino, F., Hubert, N., Panfili, J., Renno, J.-F., Baras, E., et al. (2007). Environment-related Life-History Trait Variations of the Red-Bellied Piranha Pygocentrus Nattereri in Two River Basins of the Bolivian Amazon. J. Fish. Biol. 71, 1113–1134. doi:10.1111/j.1095-8649.2007.01583.x
FAO (2017). Iniciativa de Crecimiento Azul. Asociación con los países para lograr los Objetivos de Desarrollo Sostenible. Available at: http://www.fao.org/publications/card/en/c/8801244b-8c61-469e-ab33-7774ae232285/.
Gregory, W., and Maizels, R. (2008). Cystatins from Filarial Parasites: Evolution, Adaptation and Function in the Host-Parasite Relationship☆. Int. J. Biochem. Cell Biol. 40, 1389–1398. doi:10.1016/j.biocel.2007.11.012
Hall, A., Dalbøge, H., Grubb, A., and Abrahamson, M. (1993). Importance of the Evolutionarily Conserved glycine Residue in the N-Terminal Region of Human Cystatin C (Gly-11) for Cysteine Endopeptidase Inhibition. Biochem. J. 291, 123–129. doi:10.1042/bj2910123
Hall, A., Håkansson, K., Mason, R. W., Grubb, A., and Abrahamson, M. (1995). Structural Basis for the Biological Specificity of Cystatin C. J. Biol. Chem. 270, 5115–5121. doi:10.1074/jbc.270.10.5115
Huang, X., Gu, Y., Zhou, H., Xu, L., Cao, H., and Gai, C. (2020). Acinetobacter Venetianus, a Potential Pathogen of Red Leg Disease in Freshwater-Cultured Whiteleg Shrimp Penaeus Vannamei. Aquac. Rep. 18, 100543. doi:10.1016/j.aqrep.2020.100543
Izadpanah, A., and Gallo, R. L. (2005). Antimicrobial Peptides. J. Am. Acad. Dermatology 52, 381–390. doi:10.1016/j.jaad.2004.08.026
Josiah Ochieng, J., and Gautam Chaudhuri, G. (2010). Cystatin Superfamily. J. Health Care Poor Underserved 21, 51–70. doi:10.1353/hpu.0.0257
Khaznadji, E., Collins, P., Dalton, J. P., Bigot, Y., and Moiré, N. (2005). A New Multi-Domain Member of the Cystatin Superfamily Expressed by Fasciola Hepatica. Int. J. Parasitol. 35, 1115–1125. doi:10.1016/j.ijpara.2005.05.001
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lefebvre, C., Vandenbulcke, F., Bocquet, B., Tasiemski, A., Desmons, A., Verstraete, M., et al. (2008). Cathepsin L and Cystatin B Gene Expression Discriminates Immune Cœlomic Cells in the Leech Theromyzon Tessulatum. Dev. Comp. Immunol. 32, 795–807. doi:10.1016/j.dci.2007.11.007
Li, F., An, H., Seymour, T. A., and Barnes, D. W. (2000). Rainbow Trout (Oncorhynchus mykiss) Cystatin C: Expression in Escherichia coli and Properties of the Recombinant Protease Inhibitor. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 125 (4), 493–502. doi:10.1016/S0305-0491(00)00156-5
Li, F., Gai, X., Wang, L., Song, L., Zhang, H., Qiu, L., et al. (2010). Identification and Characterization of a Cystatin Gene from Chinese Mitten Crab Eriocheir Sinensis. Fish Shellfish Immunol. 29, 521–529. doi:10.1016/j.fsi.2010.05.015
Li, R., Li, S., Chen, Z., Jin, Y., Li, S., Li, S., et al. (2021). Grass Carp (Ctenopharyngodon Idella) Stefin A: Systematic Research on its Cloning, Expression, Characterization and Tissue Distribution. Food Chem. 335, 127564. doi:10.1016/j.foodchem.2020.127564
Li, S., Yang, Z., Ao, J., and Chen, X. (2009). Molecular and Functional Characterization of a Novel Stefin Analogue in Large Yellow Croaker (Pseudosciaena Crocea). Dev. Comp. Immunol. 33 (12), 1268–1277. doi:10.1016/j.dci.2009.07.008
Lowry, O., Rosebrough, N., Farr, A. L., and Randall, R. (1951). Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 193, 265–275. doi:10.1016/s0021-9258(19)52451-6
Magister, Š., and Kos, J. (2013). Cystatins in Immune System. J. Cancer 4, 45–56. doi:10.7150/jca.5044
Mao, F., Lin, Y., He, Z., Li, J., Xiang, Z., Zhang, Y., et al. (2018). Dual Roles of Cystatin A in the Immune Defense of the Pacific Oyster, Crassostrea gigas. Fish Shellfish Immunol. 75, 190–197. doi:10.1016/j.fsi.2018.01.041
Margis, R., Reis, E. M., and Villeret, V. (1998). Structural and Phylogenetic Relationships Among Plant and Animal Cystatins. Archives Biochem. Biophysics 359, 24–30. doi:10.1006/abbi.1998.0875
Mottram, J. C., Coombs, G. H., and Alexander, J. (2004). Cysteine Peptidases as Virulence Factors of Leishmania. Curr. Opin. Microbiol. 7, 375–381. doi:10.1016/j.mib.2004.06.010
OIE (2013). Acute Hepatopancreatic Necrosis Disease: A Etiology, Epidemiology, Diagnosis, Prevention and Control References. Retrieved on 11 Nov 2019. Available at: https://www.oie.int/doc/ged/D14023.PDF.
Pemberton, P. A. (2006). PROTEINASE INHIBITORS | Cystatins. 511–517. doi:10.1016/B0-12-370879-6/00329-X
Pérez-Sánchez, T., Mora-Sánchez, B., and Balcázar, J. L. (2018). Biological Approaches for Disease Control in Aquaculture: Advantages, Limitations and Challenges. Trends Microbiol. 26 (11), 896–903. doi:10.1016/j.tim.2018.05.002
Premachandra, H. K. A., Whang, I., Lee, Y.-d., Lee, S., De Zoysa, M., Oh, M.-J., et al. (2012). Cystatin B Homolog from Rock Bream Oplegnathus Fasciatus: Genomic Characterization, Transcriptional Profiling and Protease-Inhibitory Activity of Recombinant Protein. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 163 (1), 138–146. doi:10.1016/j.cbpb.2012.05.012
Prunk, M., Perišić Nanut, M., Sabotič, J., and Kos, J. (2016). Cystatins, Cysteine Peptidase Inhibitors, as Regulators of Immune Cell Cytotoxicity. Period Biol. 118, 9. doi:10.18054/pb.v118i4.4504
Rinne, A., Dorn, A., Järvinen, M., Alavaikko, M., Jokinen, K., and Hopsu-Havu, V. K. (1986). Immunoelectron Microscopical Location of the Acid Cysteine Proteinase Inhibitor in the Lymphatic Tissue of the Tonsils. Acta Histochem. 79 (2), 137–145. doi:10.1016/S0065-1281(86)80072-1
Rosny, H. S., Hossain, M. M. M., Hasan-Uj-Jaman, M., Roy, H. S., Aurpa, I. A., Khondoker, S., et al. (2016). Dietary Supplementation of Garlic (Allium Sativum) to Prevent Acanthocephala Infection in Aquaculture. Int. J. Fish. Aquat. Stud. 4 (3), 188–192. doi:10.22271/fish.2016.v4.i3c.746
Rudenskaya, G. N., and Pupov, D. V. (2008). Cysteine Proteinases of Microorganisms and Viruses. Biochem. Mosc. 73, 1–13. doi:10.1134/s000629790801001x
Rzychon, M., and Chmiel, D. (2004). Stec-Niemczyk, JModes of Inhibition of Cysteine Proteases. Acta Biochim. Pol. 51, 861–873. doi:10.18388/abp.2004_3519
Scott, D. K., Lord, R., Muller, H. K., Malley, R. C., and Woods, G. M. (2007). Proteomics Identifies Enhanced Expression of Stefin A in Neonatal Murine Skin Compared with Adults: Functional Implications. Br. J. Dermatol 156, 1156–1162. doi:10.1111/j.1365-2133.2007.07875.x
Stubbs, M. T., Laber, B., Bode, W., Huber, R., Jerala, R., Lenarcic, B., et al. (1990). The Refined 2.4 A X-Ray Crystal Structure of Recombinant Human Stefin B in Complex with the Cysteine Proteinase Papain: A Novel Type of Proteinase Inhibitor Interaction. EMBO J. 9 (6), 1939–1947. doi:10.1002/j.1460-2075.1990.tb08321.x
Takahashi, M., Tezuka, T., and Katunuma, N. (1994). Inhibition of Growth and Cysteine Proteinase Activity ofStaphylococcus aureusV8 by Phosphorylated Cystatin α in Skin Cornified Envelope. FEBS Lett. 355, 275–278. doi:10.1016/0014-5793(94)01196-6
Tang, K. F. J., Aranguren, L. F., Piamsomboon, P., Han, J. E., Maskaykina, I. Y., and Schmidt, M. M. (2017). Detection of the Microsporidian Enterocytozoon Hepatopenaei (EHP) and Taura Syndrome Virus in Penaeus Vannamei Cultured in Venezuela. Aquaculture 480, 17–21. doi:10.1016/j.aquaculture.2017.07.043
Turk, V., and Bode, W. (1991). The Cystatins: Protein Inhibitors of Cysteine Proteinases. FEBS Lett. 285, 213–219. doi:10.1016/0014-5793(91)80804-C
Tzeng, S.-S., Chen, G.-H., Chung, Y.-C., and Jiang, S.-T. (2001). Expression of Soluble Form Carp (Cyprinus carpio) Ovarian Cystatin in Escherichia coli and its Purification. J. Agric. Food Chem. 49 (9), 4224–4230. doi:10.1021/jf0105135
Valladão, G. M. R., Gallani, S. U., Jerônimo, G. T., and Seixas, A. T. d. (2019). Challenges in the Control of Acanthocephalosis in Aquaculture: Special Emphasis onNeoechinorhynchus Buttnerae. Rev. Aquacult, 1–13. doi:10.1111/raq.12386
van Eijk, M., and de Groot, C. (1999). Germinal Center B Cell Apoptosis Requires Both Caspase and Cathepsin Activity. J. Immunol. 163, 2478–2482.
Verdot, L., Lalmanach, G., Vercruysse, V., Hoebeke, J., Gauthier, F., and Vray, B. (1999). Chicken Cystatin Stimulates Nitric Oxide Release from Interferon-γ-Activated Mouse Peritoneal Macrophages via Cytokine Synthesis. Eur. J. Biochem. 266, 1111–1117. doi:10.1046/j.1432-1327.1999.00964.x
Wesierska, E., Saleh, Y., Trziszka, T., Kopec, W., Siewinski, M., and Korzekwa, K. (2005). Antimicrobial Activity of Chicken Egg White Cystatin. World J. Microbiol. Biotechnol. 21 (1), 59–64. doi:10.1007/s11274-004-1932-y
Wickramasinghe, P. D. S. U., Kwon, H., Elvitigala, D. A. S., Wan, Q., and Lee, J. (2020). Identification and Characterization of Cystatin B from Black Rockfish, Sebastes Schlegelii, Indicating its Potent Immunological Importance. Fish Shellfish Immunol. 104, 497–505. doi:10.1016/j.fsi.2020.05.068
Xiao, P.-p., Hu, Y.-h., and Sun, L. (2010). Scophthalmus maximus Cystatin B Enhances Head Kidney Macrophage-Mediated Bacterial Killing. Dev. Comp. Immunol. 34 (12), 1237–1241. doi:10.1016/j.dci.2010.07.008
Yamashita, M., and Konagaya, S. (1991). A Comparison of Cystatin Activity in the Various Tissues of Chum Salmon Oncorhynchus keta between Feeding and Spawning Migrations. Comp. Biochem. Physiology - Part A Physiology 100 (3), 749–751. doi:10.1016/0300-9629(91)90402-X
Keywords: aquatic diseases, fish, recombinant protein, transgene, stefins
Citation: Ramirez Merlano JA and Almeida DV (2022) Heterologous Production and Evaluation of the Biological Activity of Cystatin-B From the Red Piranha Pygocentrus nattereri. Front. Genet. 13:812971. doi: 10.3389/fgene.2022.812971
Received: 11 November 2021; Accepted: 21 April 2022;
Published: 03 June 2022.
Edited by:
Sandra Isabel Moreno Abril, University of Vigo, SpainReviewed by:
Guilherme Campos Tavares, Federal University of Minas Gerais, BrazilCopyright © 2022 Ramirez Merlano and Almeida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Volcan Almeida, ZGFuaXZvbGNhbkBwcS5jbnBxLmJy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.