
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 29 March 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.807293
Proteins with a domain of unknown function (DUF) represent a number of gene families that encode functionally uncharacterized proteins in eukaryotes. In particular, members of the DUF1005 family in plants have a 411-amino-acid conserved domain, and this family has not been described previously. In this study, a total of 302 high-confidence DUF1005 family members were identified from 58 plant species, and none were found in the four algae that were selected. Thus, this result showed that DUF1005s might belong to a kind of plant-specific gene family, and this family has not been evolutionarily expanded. Phylogenetic analysis showed that the DUF1005 family genes could be classified into four subgroups in 58 plant species. The earliest group to emerge was Group I, including a total of 100 gene sequences, and this group was present in almost all selected species spanning from mosses to seed plants. Group II and Group III, with 69 and 74 members, respectively, belong to angiosperms. Finally, with 59 members, Group IV was the last batch of genes to emerge, and this group is unique to dicotyledons. Expression pattern analysis of the CiDUF1005, a member of the DUF1005 family from Caragana intermedia, showed that CiDUF1005 genes were differentially regulated under various treatments. Compared to the wild type, transgenic lines with heterologous CiDUF1005 expression in Arabidopsis thaliana had longer primary roots and more lateral roots. These results expanded our knowledge of the evolution of the DUF1005 family in plants and will contribute to elucidating biological functions of the DUF1005 family in the future.
A domain with no confirmed function in the Pfam database is generally called a domain of unknown function (DUF). These domains have two distinct characteristics, including a relatively conserved amino acid sequence and an unknown function (Bateman et al., 2010). In total to date, the Pfam database (version 34) contains 19,179 families, including 6,565 DUF or UPF (uncharacterized protein families), which occupies approximately 34% of all the Pfam families (Mistry et al., 2020). Although the functions of some DUF families have been characterized, a large set of DUF members remains unknown.
Previously published studies have described the vital role of DUF domain-containing proteins in biotic stresses (such as insect pests), abiotic stresses (such as cold, heat, drought, and salt), and plant growth and development. Some DUFs were found to be involved in defense responses and endowed plants with a tolerance to pests and diseases, for example, DUF26 (Wrzaczek et al., 2010; Miyakawa et al., 2014; Lee et al., 2017) and DUF538 (Gholizadeh, 2011; Gholizadeh and Kohnehrouz, 2013; Gholizadeh, 2020). AtRDUF1, AtRDUF2 (Kim et al., 2012; Li et al., 2013), DUF1645 (Cui et al., 2016), and DUF966 (Luo et al., 2014; Zhou et al., 2020) were demonstrated to have a role in the responses of plant to abiotic stress. DUF266 (Parsons et al., 2012), DUF231 (Yuan et al., 2013), DUF246 (Oikawa et al., 2010), DUF1218 (Mewalal et al., 2016), and DUF579 (Urbanowicz et al., 2012) of Arabidopsis thaliana are commonly reported to regulate the development of plant cell walls. Several studies have confirmed that some DUFs, such as DUF640 (Zhao et al., 2004) and DUF827 (Kodama et al., 2011), affect chloroplast development and the growth of plants.
In addition to regulating the plant cell wall and chloroplast, DUF also regulates the development of plant roots, leaves, flowers, and fruits. JA-induced DUF26 functions in rice root development (Jiang et al., 2007), at the same time, and is related to the stomatal density (Arellano-Villagómez et al., 2021). The DUF724 family was highly expressed in Arabidopsis apical meristem tissues (shoots, leaves, roots, etc.) (Cao et al., 2009). In addition, there were many DUFs that regulated root development. These include DUF828 (Prabhakaran Mariyamma, 2016), DUF966 (Shen et al., 2019), DUF668 (Zhao et al., 2021), DUF642 (Gao et al., 2012; Salazar-Iribe et al., 2016), and DUF761 (Zhang et al., 2019). Sugar accumulation 1 (OsSAC1), which consists of two conserved DUFs, namely, DUF4220 and DUF594, causes sugar to accumulate in rice leaves (Zhu et al., 2018). DUF647 regulates the early development of roots through UV-B during root development (Leasure et al., 2009). While DUF640 family members were mainly involved in the regulation of flower development, the DUF784 and DUF1216 (Huang et al., 2008) families were found to play a role in pollen development (Jones-Rhoades et al., 2007).
DUF1005 domain-containing proteins are exclusively present in plant genomes. As of July 2021, a total of 937 genes belonging to the DUF1005 family (Pfam accession: 6219) could be retrieved from 138 species in the Pfam database. Previous studies have shown that DUF1005s were differentially expressed in the transcriptome of cotton, and it was associated with fiber uniformity and fiber elongation (Okubazghi et al., 2017). There were a total of 4 DUF1005s in cucumbers, and they were related to controlling the cucumber lengths (Zhang, 2019). Moreover, proteins with a DUF1005 domain were found in cell wall proteome studies, indicating that they might have functions in cell wall development (Sergeant et al., 2019). The DUF1005-containing protein stigma exposed 1 (SE1) from Vigna radiata has been reported to affect petal and pistil growth during the late development stages in the se1 mutant (Yun et al., 2020). However, few studies have focused on the function of DUF1005 in Caragana intermedia.
C. intermedia is a xerophytic deciduous shrub that belongs to the genus Caragana Fabr. It is mainly distributed in the arid and semiarid desert areas of Inner Mongolia, Ningxia, Shanxi, and northern Shaanxi. It is a native desert shrub with strong drought, salinity, cold resistance, sand-fixing capacity, and high forage value and is a superior xerophytic shrub species that is highly suitable for artificial afforestation in arid desert steppes (Liu et al., 2019). In this study, we identified a gene in C. intermedia that encodes DUF1005, and the morphology and abiotic stress response of CiDUF1005 were further studied in transgenic Arabidopsis. In addition, we performed an analysis of the DUF1005 family in 58 representative plant species, including the identification and evolutionary analysis of DUF1005 family members in 58 representative plant species. The results obtained here should broaden our understanding of the roles of the DUF1005 family and provide a framework for further functional investigation of these genes in plants.
We selected species that are representative of the various stages, based on the progression of plants from moss to plants with seeds in evolution. We identified DUF1005 candidate genes from 58 representative plant species. The protein databases were downloaded from Phytozome12 (Goodstein et al., 2011), ConGenIE (Sundell et al., 2015), and NCBI. Detailed genomic information for the 58 species included in this study is listed in Supplementary Table S1.
To identify the DUF1005 gene family members in the above species, a profile hidden Markov model (HMM) (pHMM) of the DUF1005 (Pfam: PF06219) domain was downloaded from Pfam 31.0 (Mistry et al., 2020). Hmmsearch (HMMer package version 3.1b1) was used to search DUF1005.hmm against the protein sequences from each plant genome (Eddy, 1998). To ensure that the search was reliable, domain hits beyond the gathering threshold (E-value 1e−10) were filtered out before downstream analysis. For gene loci with multiple predicted isoforms, the primary isoform was used if the primary isoform annotation was available; otherwise, the longest protein was used. The Pfam database was employed to confirm the DUF1005 domain in the candidate proteins. The number and completeness of DUF domains were queried on the Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov). According to the 303 protein sequence of DUF1005, we searched homologous proteins in 4 algae with homology comparison in blast+ (Camacho et al., 2009) with above 10−5 e-value.
The compute pI/MW, a tool of the ExPASy server (http://web.expasy.org) was used to calculate the molecular weight (MW) and theoretical isoelectric point (pI) of the DUF proteins (Wiederschain, 2006). The WoLF PSORT program (https://wolfpsort.hgc.jp/) was used to predict protein subcellular localization (Horton et al., 2007).
The amino acid sequences of the DUF1005s identified from 58 plants were subjected to alignment by ClustalW with default settings (Larkin et al., 2007). Phylogenetic analysis was conducted using the maximum likelihood (ML) method. First, the ML estimation using the best-fitting model JTT+F+R6 of sequence evolution was determined by ModelFinder (Kalyaanamoorthy et al., 2017). Then, IQ-TREE (Trifinopoulos et al., 2016) was used to infer the ML tree with 1,000 bootstrap replicates for alignment (Nguyen et al., 2014). All phylogenetic trees were edited and displayed with the online tool iTOL (Letunic and Bork, 2006). MEME software v5.0.5 (http://memesuite.org/tools/meme) was employed to identify conserved motifs with the default parameters (except that the maximum number of motifs was set to 6) (Bailey et al., 2009; Chen et al., 2020). The map was redrawn with Tbtools (Chen et al., 2020).
Total DNA and RNA were isolated using a Plant Genomic DNA Kit (DP305) and RNAprep Pure Plant Kit (DP419) (Tiangen, Beijing, China). The genomic DNA was removed, and the total RNA was converted to cDNA using TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (AT311-02) (TransGen, Beijing, China). We identified a sequence containing the DUF1005 domain from the drought-treated transcriptome database (SRA accession number: SRP121096) of C. intermedia and named it CiDUF1005. To clone the full-length cDNA and gDNA of CiDUF1005 from C. intermedia, PCR was set up with 5× PrimeSTAR Buffer (Mg2+ Plus) (Takara, Dalian, China) of 10 μl, dNTP Mixture (2.5 mM each) of 4 μl, CiDUF1005HA-F/R at 1 μl each, PrimeSTAR HS DNA Polymerase (2.5 U/μl) of 0.5 μl, template cDNA/gDNA of 5 μl, and ddH2O supplemented up to 50 μl. Then, the thermocycler program consisted of 30 cycles at 98°C for 10 s, 70°C for 10 s, and 72°C for 3 min to amplify the target products. The genes were then cloned and sequenced. The GSDS website (Gene Structure Display Server, http://gsds.cbi.pku.edu.cn) was used to analyze the CiDUF1005 structure (Hu et al., 2014). Multiple sequence alignment was carried out using DNAMAN (6.0.3.99) software. Phylogenetic analysis was performed by using MEGA 7 (Kumar et al., 2016) software. The alignment was adjusted manually, while the unrooted phylogenetic trees were constructed by the neighbor-joining method (Sievers et al., 2011).
To produce a recombinant for the constitutive expression of CiDUF1005, SpeI and SacI (Takara, Dalian, China) sites were introduced at the end of the full-length cDNA sequence of CiDUF1005 by PCR amplification, and then, the fragment was digested and subcloned into the pCanG-HA vector (which was kindly provided by Prof. Qi Xie, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China) that was driven by the CaMV35S promoter. The resulting recombinant plasmid was sequenced to confirm that there were no PCR errors. For the plant transformation, the pCanG-HA-CiDUF1005 recombinant vector and empty vector were transferred into Agrobacterium tumefaciens strain GV3101 by electroporation. The Agrobacterium-mediated floral dip method was used for Arabidopsis transformation (Clough and Bent, 1998). Transgenic lines were first selected by kanamycin (25 mg/L), then a total of seven homozygous transgenic lines were obtained, and the expression of CiDUF1005 was verified by qRT–PCR. At the same time, empty vector lines of pCanG-HA were obtained using the same method. Homozygous seeds from the transgenic lines were used for subsequent research. All primers used in this study are listed in Supplementary Table S5.
In this experiment, the Arabidopsis ecotype Col-0 (Columbia-0) was used as the wild type (WT) and for the genetic transformation of CiDUF1005. All seeds from the WT and transgenic lines were kept in darkness for 3 days at 4°C on 0.6% agar medium with half strength (1/2) Murashige and Skoog (MS) after the seeds were sterilized. The seeds were then sown on 1/2 MS medium containing 2.3 g/L of Murashige and Skoog Basal Medium with vitamins (PhytoTech, Lenexa, KS, USA), 1% (w/v) sucrose (Sigma, USA), 0.025% MES (Coolaber, Beijing, China), and 10 g/L of plant agar (Chembase, Nantong, China), and the pH was adjusted to 5.7 with KOH. Plates were cultured for 11 days under long-day conditions (16-h light/8-h dark cycle) at 22°C. Pictures were obtained each day from the first day to the 11th day that the primary root length was measured, and the relative elongation was calculated. Thirty-six seedlings were used to measure root length for each line, and three independent biological replicates were carried out.
Seeds from C. intermedia were collected from Hohhot, Inner Mongolia, China. One-month-old seedlings that were sown in pots containing a soil mixture were used to detect the transcript level of CiDUF1005 under various treatments. For the cold, heat, drought, dehydration, and ABA treatments, we referred to Dr. Xiaomin Han’s method (Han et al., 2015). Three independent biological replicates, each comprising three individual plants, were used for quantitative real-time PCR. Triplicate quantitative assays were performed with each cDNA sample.
For qRT–PCR analysis, the obtained cDNA was diluted with RNase-free water before use. The target genes were expressed with specific primers, and qRT–PCR was performed using SYBR Green I Master Mix (Roche, Basel, Switzerland) and a LightCycler 480 Real Time PCR system (Roche, Switzerland). The thermal cycling program was 95°C for 5 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The melting curves were analyzed at 60°C–95°C after 40 cycles. The relative RNA transcript level was calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). AtEF1α (AT5G60390) and CiEF1α [GenBank No.: KC679842] were used to normalize the Arabidopsis and C. intermedia samples, respectively (Yang et al., 2014). All primers were designed by Primer Premier 5.0 and are listed in Supplementary Table S5. All qRT–PCR assays were carried out with three technical replicates.
The data in this research were subjected to statistical analysis using Microsoft Excel 2016 and SPSS. All data are presented as the mean of three biological replicates ±SD. A statistical analysis was carried out using at least 36 seedlings in each phenotypic experiment. The significant difference between groups was analyzed by Student’s t-test, Duncan’s multiple range tests, and one-way ANOVA. Significant differences were evaluated at p < 0.01 or p < 0.05.
To investigate the number of DUF1005 family members in different species, we conducted a search for DUF1005 across plant lineages, including 58 representative species, by an HMM search. Information on the species and genomes used here is listed in Supplementary Table S1. In the end, 302 full-length DUF1005 were identified from 54 plant species, including the four major plant lineages of bryophytes, lycophytes, angiosperms, and gymnosperms (Figure 1). Then, the protein database was blasted by using the query sequences of the DUF1005 family in 54 plant species and four algae species. The E-value cutoff was set at 1.0e−5 to ensure confidence. We further dissected the algae (Volvox carteri, Chlamydomonas reinhardtii, Ostreococcus lucimarinus, and Micromonas pusilla), and no DUF1005 was found.
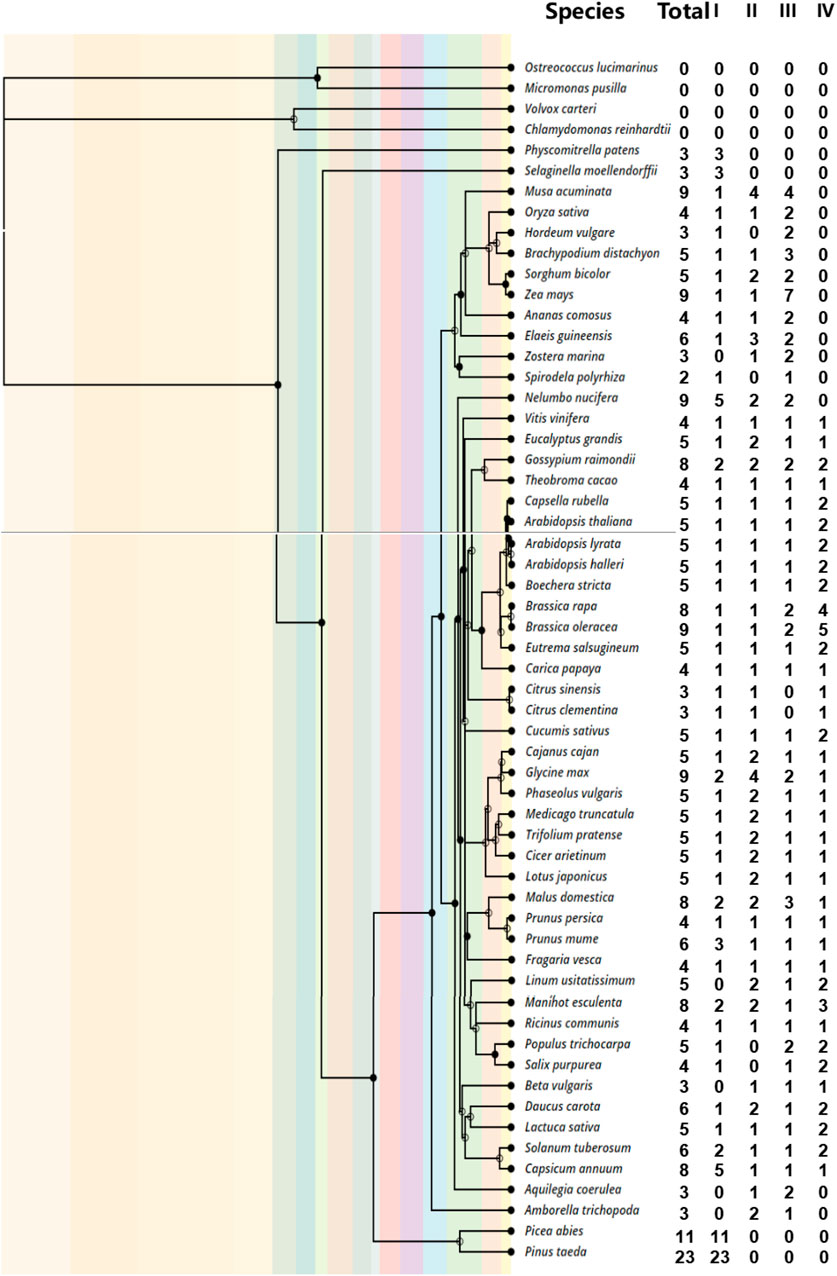
FIGURE 1. The phylogeny of the 58 plants analyzed in this study and the number of DUF1005 genes identified. The order of tree branches and divergence time was derived from the TimeTree database (http://timetree.org/).
According to the number of genes analysis (Figure 1), we found that Spirodela polyrhiza harbored only two DUF1005s, and it ranked last among 54 plant species. In contrast, two gymnosperms, Pinus taeda and P. abies, presented the most DUF1005 members, with 23 and 11, respectively.
The number of DUF1005 family members was relatively small in all plants. Among the other 53 angiosperms except for S. polyrhiza, the number of DUF1005 genes ranged from 3 to 9. For example, there were 3 DUF1005 sequences in 9 species including Physcomitrella patens. Another 9 species, including Oryza sativa, contain 4 genes. There were 6 sequences in 4 species, including Fragaria vesca. There were 8 sequences in 5 species, including Gossypium raimondii. There were 9 sequences in 5 species, including Zea mays. Nineteen species, such as Lotus japonicus, contained 5 DUF1005 sequences in their genomes. Overall, there were few changes in the number of DUF1005 genes among the different plants.
For further verification or detection of the number and completeness of DUF1005 structural domains, we screened out the DUF1005 domain in the NCBI conserved structural domain database (Supplementary Table S2). Most of the identified DUF1005 gene-coding proteins only contained one DUF1005 domain; nevertheless, there were some exceptions. For example, MA_178396g0010 (P. abies) included two deleted DUF1005 domains, PITA_000036082-RA (P. taeda) contained a complete DUF1005 domain and a missing domain, and GSMUA_Achr4P08250_001 (Musa acuminata) contained a complete DUF1005 domain and a PTH2_family_superfamily domain. In addition to the three examples shown above, there were also other proteins with deletions in DUF1005 domains. Among them, the C-terminal domain was deleted in 15 proteins, the N-terminal domain was deleted in 22 proteins, and both ends were deleted in 8 proteins.
To explore the evolutionary relationship of the DUF1005 family in plants, a phylogenetic tree was constructed. IQ-tree software was used to perform a systematic evolutionary analysis of the 302 proteins identified and CiDUF1005 of C. intermedia by the ML method (Figure 2 and Supplementary Figure S1). It is clear that the DUF1005 plant family is divided into four subfamilies, namely, Group I, Group II, Group III, and Group IV (Figure 2 and Supplementary Table S3). The results showed that Group I included a total of 100 genes. Group II and Group III had 69 and 74 genes, respectively. The last group, marked as Group IV, contained 59 genes. The group that emerged the earliest was Group I, and the members of Group I was widely distributed from bryophytes (mosses) to angiosperms (seed plants) except Beta vulgaris, Linum usitatissimum, Aquilegia coerulea, Zostera marina, Amborella trichopoda, and the four algae. Moreover, Group I was ubiquitously distributed in all species, from mosses to seed plants, while Group II and Group III were only found in most angiosperms. The DUF1005 in Group II existed in most angiosperms except G. raimondii, Salix purpurea, Hordeum vulgare, and S. polyrhiza. The DUF1005 of Group III also existed in most angiosperms except Citrus sinensis and Citrus clementina. Obviously, Group IV was the last to emerge, and it is a specific group to dicotyledons in addition to A. coerulea and Nelumbo nucifera. The CiDUF1005 of C. intermedia used in this report belonged to Group III. This implied that the divisions of the four subgroups occur after the gymnosperms to angiosperms.
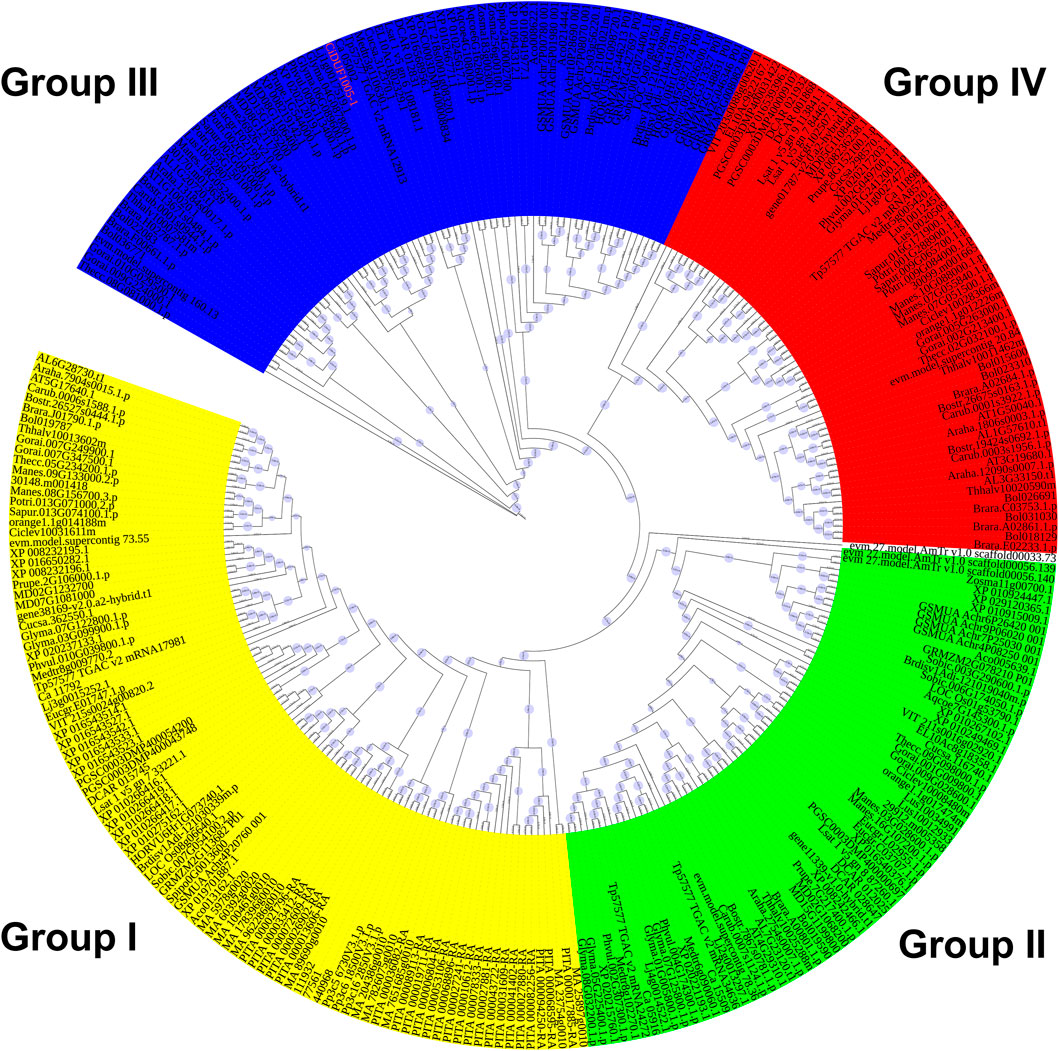
FIGURE 2. Phylogenetic classification of DUF1005 genes in plants. A phylogenetic tree was constructed using the maximum likelihood (ML) method with IQ-tree. The yellow color represents Group I, the green color represents Group II, the blue color represents Group III, and the red color represents Group IV.
Furthermore, we analyzed the protein characteristics of these 303 DUF1005 (including CiDUF1005) sequences. The results showed that DUF1005 proteins varied greatly from 59 amino acids (aa) to 1,656 aa, with an average length of 411 aa (59 aa–1656 aa). All of the DUF1005 candidates and their encoded protein features are listed in Supplementary Table S4. The features of DUF1005 proteins were as follows: the average pI was 8.95 (ranging from 4.5 to 10.88), and 94% of proteins had a pI above 7. Therefore, the pI is in the basic range, and the proteins are rich in basic aa; the average Mw = 44.3 kDa (ranging from 6.25 to 181.45 kDa). The Mw and pI data in Group I and Group III were more scattered, and those in Group II and Group IV were more concentrated (Figure 3A); the average instability index was = 46.7 (ranging from 30.19 to 66.65); the average aliphatic index was = 76.0 (ranging from 61.97 to 119.25); and the average GRAVY was = −0.21 (ranging from −0.667 to 0.346). Thus, DUF1005 is a hydrophilic protein except for some GRAVY > 0 in gymnosperms. The instability index values indicated that DUF1005 was relatively unstable. The subcellular localization prediction results for all 303 DUF1005 proteins indicated that 184 proteins were localized in the chloroplast. Moreover, from Group I to Group IV, the number of aa and the molecular weight continued to increase, and the aliphatic index continued to decrease. Overall, these results indicated that DUF1005 family members were unstable and hydrophilic proteins that were predicted to be located mainly in the chloroplast.
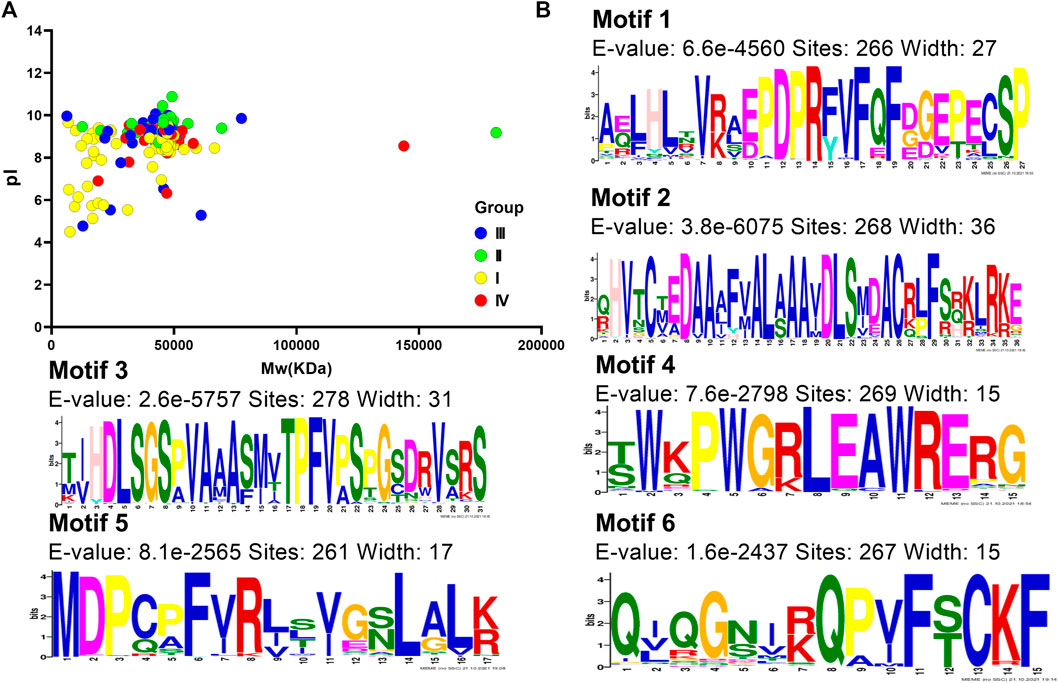
FIGURE 3. DUF1005 protein features. (A) Molecular weight (Mw) and isoelectric point (pI) of DUF1005 genes in plants. The yellow color represents Group I, the green color represents Group II, the red color represents Group III, and the blue color represents Group IV. (B) Sequence logos for the conserved motifs of DUF1005 proteins in plants.
To study the structure of DUF1005 genes, we analyzed their DNA sequences and determined the composition of their introns and exons. The GSDS 2.0 software package was used to map the intron–exon structure of the DUF1005 gene family. The results showed that most of the DUF1005 genes contained two exons (Supplementary Figure S2). Among them, Group I was shown to contain two exons (accounting for 70% of the total number of genes in the subfamily), Group II (65%), Group III (72%), and Group IV (88%). However, Group II contained three exons (20%). The DUF1005 domains of GSMUA_Achr4P08250_001 in Group II had nine exons. Slightly more than half the gymnosperms in Group I genes had three to four exons and had very long introns. The gene structure of the DUF1005 gene family was found to be highly conserved among the various subfamilies, and the number and location of exons were similar among the DUF1005 genes in each subfamily, indicating similar function.
Analyzing motif-representing features, such as DNA-binding sites and protein interaction domains, could help us understand common features of gene family sequences and identify any new conserved motif compositions that might not be recorded in public databases. A total of 6 distinct conserved motifs were found, and the sequence logos for the conserved motifs and their distribution in each protein are illustrated in Figure 3B and Supplementary Figure S3. Most of those containing only motif 3 were located in Group I in gymnosperms. Motif 5 was present in only 261 DUF proteins. Motif 5 was distributed at the N-terminus, and motif 2 was distributed at the C-terminus. Other motifs were found in the middle of the domain. Motif 1 and motif 6 were located nearby, and motifs 3 and 4 stayed close together. Investigations of the amino acid composition indicated that the most frequently occurring amino acid in motifs 3 and 2 were S (Ser) and A (Ala), respectively. Then, these motifs were subjected to SMART online server (http://smart.embl-heidelberg.de/smart/change_mode.pl) for annotation, and the results showed that motif 2 was similar to the DSS1_SEM1 domain (PF05160); both had highly conserved C-T-E-D-A-A-A-F-V-A-L-A-A-A-V-D-L-S amino acid sequences. Furthermore, they were found to be associated with proteasome assembly.
We identified a sequence containing the DUF1005 domain from the drought-treated transcriptome database (SRA accession number: SRP121096) of C. intermedia and named it CiDUF1005. The amplified cDNA and genomic sequence of CiDUF1005 were submitted to GenBank (accession number: MZ074325). The full-length cDNA sequence of CiDUF1005 consisted of 1,399 nucleotides and contained a 1,356-bp open reading frame (ORF). The polypeptide deduced by CiDUF1005 has 451 amino acid residues, and it contains a conserved CiDUF1005 domain with 444 amino acids (Figure 4A). The DUF1005 protein has a predicted theoretical molecular mass of 48.4 kDa and an isoelectric point of 9.25, which were calculated using the ExPASy-Bioinformatics Resource Portal. Comparing the nucleotide sequence of the 1,808-bp genomic DNA with the cloned cDNA showed that CiDUF1005 comprised one intron and two exons, the first exon was 623 bp, the intron was 415 bp, and the second exon was 670 bp in length (Figure 4B).
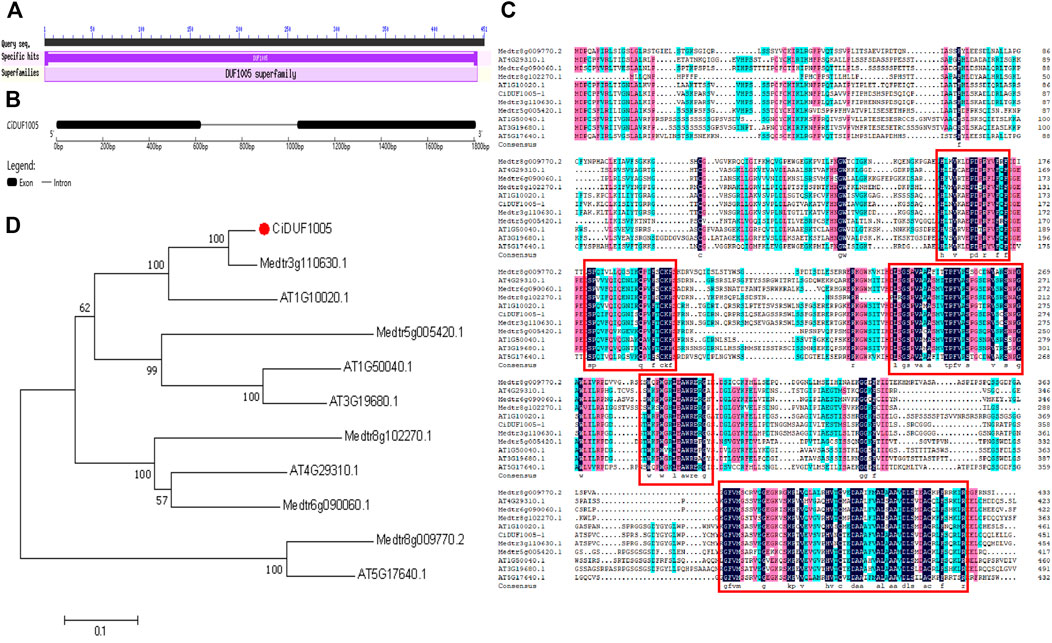
FIGURE 4. Bioinformatics analysis of DUF1005 in plants. (A) The domain of CiDUF1005. (B) The gene structure of CiDUF1005. (C) Multiple sequence alignment of CiDUF1005 and its homologs. The colors of the conserved residues represent different levels of conservation of each column in the alignment. The five basically conserved areas are represented by red rectangles. (D) Phylogenetic tree analysis of CiDUF1005 and its homologs.
The protein sequences of DUF1005 with its homologs from Arabidopsis and Medicago truncatula were compared (Figure 4C). The results showed that the sequence was largely conserved between species and had high homology. Phylogenetic tree analysis showed that CiDUF1005 was closely related to AT1G10020 and Medtr3g110630.1 (Figure 4D). Multiple sequence alignment of protein sequences from three species was performed. Basically, there were 5 primarily conserved regions with high similarity, which is colored in black. In addition, several secondary and tertiary levels of conservation were detected and are colored pink or blue. It is noticeable that the DUF1005 domain covers most of the amino acid residues in all the DUF1005 proteins.
To gain more insight into the role of CiDUF1005 in plant growth and development, we used qRT–PCR to examine the transcription level expression of CiDUF1005 in roots, stems, and leaves (Figure 5). Our results showed that CiDUF1005 was expressed in all tested tissues, but there were no significant differences. We further examined the expression level of CiDUF1005 under conditions such as cold, heat, dehydration, and drought treatments, as well as under the hormone abscisic acid (ABA), which is a plant growth regulator and stress hormone (Zhang et al., 2006; Lata and Prasad, 2011). Our results showed that the transcript of CiDUF1005 could be upregulated within 1 h after cold treatment, reaching a maximum at 12 h. For heat stress, its transcript increased to the maximum level within 1 h and then decreased. Similarly, ABA treatment also rapidly induced a 2-fold increase, followed by a decrease to the basal level. Surprisingly, the expression of CiDUF1005 decreased with both dehydration and drought treatments, and its expression level was reduced by 10-fold under dehydration. The transcript of CiDUF1005 started to decrease continuously after 3 days of drought treatment. This was consistent with the CiDUF1005 trend in the drought-treated transcriptome database (Supplementary Figure S4). Collectively, these results suggested that CiDUF1005 was regulated by abiotic stresses.
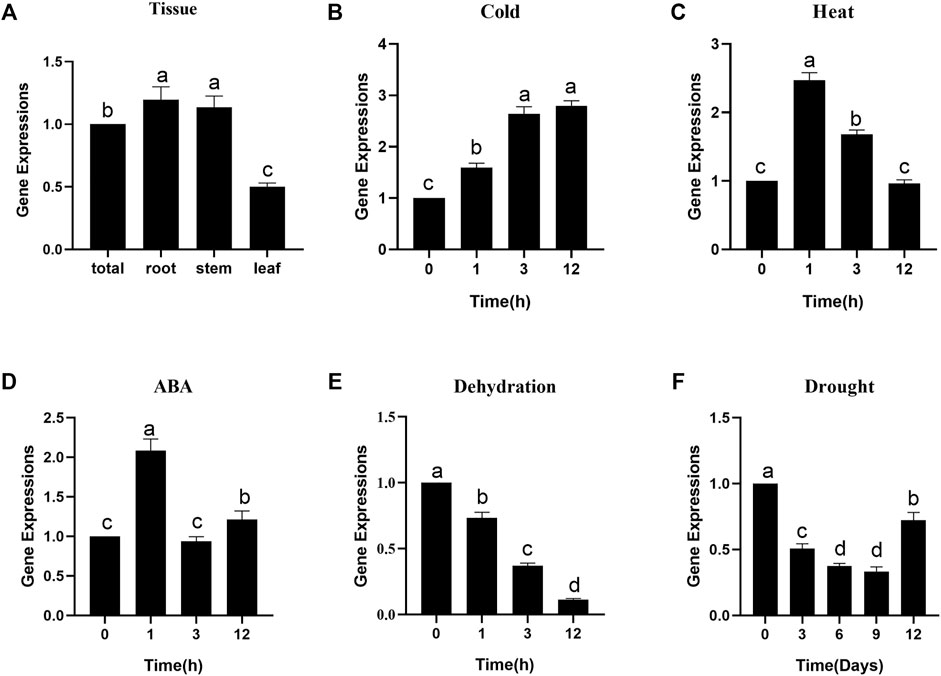
FIGURE 5. The expression pattern of CiDUF1005 in Caragana intermedia. (A) The expression of CiDUF1005 roots, stems, leaves, and whole seedlings (total). (B–F) Pattern of CiDUF1005 expression under different treatments, such as 4°C (B), 42°C (C), and 200 μM of ABA (D), dehydration (E), and drought (F). Different letters indicate significant differences at p < 0.05 according to Duncan’s multiple range test.
To investigate the function of CiDUF1005, we generated homozygous transgenic Arabidopsis with overexpressed CiDUF1005. Among the 7 homozygous transgenic lines, 3 lines with the highest CiDUF1005 expression level were selected for further analysis and were designated OE-4, OE-5, and OE-31 (Supplementary Figure S5). Although no obvious phenotypes between the WT and OE lines were observed during the mature stage, there were significant differences at the seedling stage. In 5-day-old seedlings, the primary roots were much longer in the OE lines than in the WT when grown on 1/2 MS medium (Figures 6A,B). We also used an empty vector transformed Arabidopsis line as a control for comparison with the OE lines, and the results were consistent with the use of the WT, which was used as a control. The primary roots were much longer in the OE lines than in the control line (Supplementary Figure S6). To rule out the possibility that the difference in primary root length was caused by the difference during germination, the growth rate of roots of the OE seedling from the third day to the fifth day was analyzed, and the results showed that the root growth rate of the OE lines increased compared with that of the WT (Figure 6C). The relative rates of the primary root elongation of the three OE lines were 1.4-, 1.43-, and 1.37-fold compared to those of the WT. The evidence suggested that overexpressing CiDUF1005 caused the primary root to grow significantly faster under normal growth conditions.
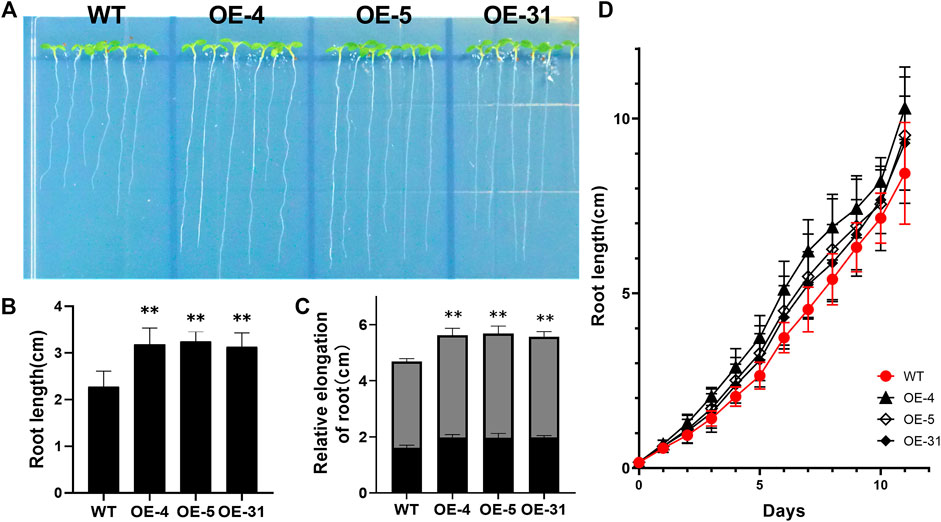
FIGURE 6. Phenotypic differences in the primary root length between the wild type and OE lines. (A) Root morphology of the OE seedlings and wild type. Five-day-old seedlings cultured in 1/2 Murashige and Skoog (MS) were observed. (B) The primary root length of 5-day-old wild type and the transgenic seedlings. (C) The relative elongation of roots after the third (black) and fifth (gray) days. N = 36 ± SD from three independent biological replicates. (D) The primary root length comparison between the OE lines and wild type during 11 days of growth. The error bars represent the means of three technical replicates ±SD. Statistically significant differences from the control group are indicated as **p < 0.01.
To determine at which period CiDUF1005 overexpression promoted root elongation, we measured the root length of the seedling from the first day to the 11th day (Figure 6D and Supplementary Figure S7) after germination. The length of the primary root of the OE lines was longer than that of the WT from the third day. The difference was the largest during the period from the fifth day to the ninth day. After 11 days of growth, the difference in root length decreased gradually between the OE lines and WT (Supplementary Figure S7), while there was no significant difference in adult plants. We also noticed that the OE lines displayed an almost 1.3-fold increase in the number of lateral roots on the 11th day (Figures 7A,B).
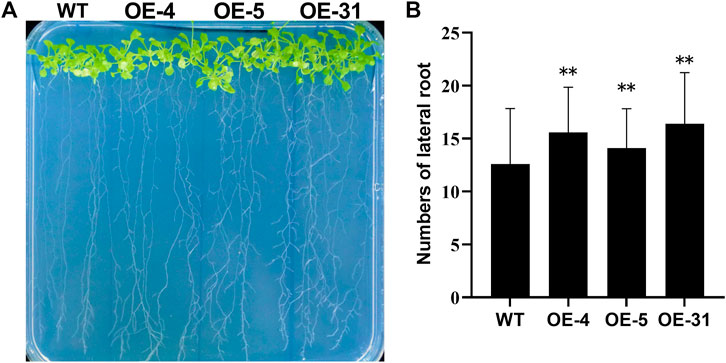
FIGURE 7. Phenotypic differences in the number of lateral roots between the wild type and OE lines. (A) The lateral root morphology of the OE lines and wild-type seedlings. Eleven day-old seedlings cultured in 1/2 Murashige and Skoog (MS) were observed. (B) The number of lateral roots of the 11-day-old wild type and the OE lines. N = 36 ± SD from three independent biological replicates. The error bars represent the means of three technical replicates ±SD. Statistically significant differences are indicated as **p < 0.01.
To further study the responses of CiDUF1005 that were induced by ABA during root development, we measured the root length of CiDUF1005 under ABA treatment. In response to the increasing ABA concentration, the relative elongation of the primary root of OE lines was still longer than that of the WT (Supplementary Figure S8). Additionally, to further investigate the response of CiDUF1005 to drought stress, the primary roots of the WT and OE lines were treated with 350 mM of mannitol. After 5 days of mannitol stress, the OE lines had a longer root length than the WT (Supplementary Figures S9A,B). However, there was no significant difference in the primary root length between the WT and OE lines when measured at day 11 (Supplementary Figures S9C,D).
Taken together, these data strongly confirmed that transgenic CiDUF1005 expression increased the number of lateral roots and promoted primary root elongation during the seedling period.
To understand how CiDUF1005 regulates root development in Arabidopsis, we investigated the expression of ABA-related genes (ARR5, ABF1, ABF2, COR15B, RD29B, RD26, ABI1, ABI5, and RAB18) in OE lines and the WT. In contrast, no clear differences in their expression levels between the WT and OE lines were detected (Supplementary Figure S10). Eventually, we searched the literature and looked for genes that were related to root development in the WT and OE lines. These include GIBBERELLIN 2-OXIDASE2 (GA2ox2) (Rieu et al., 2008; Ubeda-Tomás et al., 2008; Moubayidin et al., 2010) and Expansin-like A (EXLA) (Cosgrove, 2000). The results showed the transcript levels of GA2ox2 (AT1G30040), EXLA1 (AT3G45970), EXLA2 (AT4G38400), and EXLA3 (AT3G45960). GA2ox2, EXLA2, and EXLA3 were significantly lower in the transgenic lines than in the WT (Figure 8). However, there was no difference in the transcript levels of EXLA1 between the WT and transgenic plants. This result suggests that CiDUF1005 might influence primary root elongation by regulating the expression of GA2ox2, EXLA2, and EXLA3, by a currently unknown mechanism.
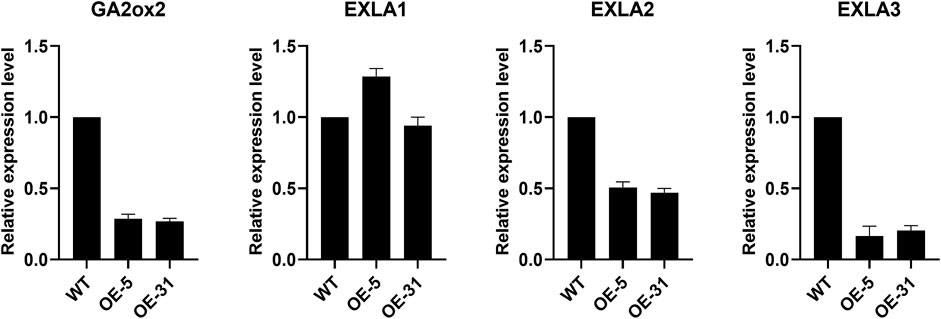
FIGURE 8. Expression levels of the root development genes. Relative expression levels of GA2ox2, EXLA1 EXLA2, and EXLA3 in 5-day-old seedlings. The error bars represent the means of three technical replicates ±SD.
The rapid growth of biological sequence databases has accompanied the development of high-throughput genomic and sequencing techniques (Mudgal et al., 2015). In addition, the fraction of DUF gene families in Pfam entries has increased over the past decade (Finn et al., 2008). Since the last release of Pfam 34.0, it has built 935 new families, killed 15 families, and created 11 new clans. UniProt Reference Proteomes have increased by 21% since Pfam 33.1 and now contain 47 million sequences. The identification of the biological functions of a large number of DUF family genes is a huge challenge. Many studies have shown that genome-wide identification and expression analysis could help researchers understand the origin, diversity, and biological functions of these DUF gene families (Moturu et al., 2018).
As a gene family that encodes proteins with unknown functions, DUF1005 appears to be a plant-specific domain (Batelli et al., 2012). In the present study, we identified 302 DUF1005 family members from 58 different plant species (Figure 1). Most species had 3–9 DUF1005 genes. Although plants were exposed to different environments during evolution, no gene differentiation of DUF1005 occurred, and no special expansion occurred in our analysis. The selected plant species in this study had different genome sizes and different rounds of whole-genome duplication, but they had a similar number of DUF1005. This result indicated that most of the genes in the DUF1005 family were not eliminated by environmental selection; instead, they showed high conservation during evolution. The finding of similar conserved motifs within the DUF1005 family further supports this point. This demonstrates that the domains of all DUF1005 family members support this notion. They remain to be studied in detail from an evolutionary perspective. We did not identify any DUF1005 members in algae strains, indicating that this gene family might originate from the landing of plants (Supplementary Table S3). In addition, no obvious expansion of this family occurred from early plant bryophytes and lycophytes to relatively late-diverging gymnosperms and angiosperms (Supplementary Table S1).
Phylogenetic analysis revealed that 303 DUF1005 were separated into four groups, and their diversification during evolution was revealed (Figure 2 and Supplementary Figure S1). Among these, Group I covered almost all the plant species, Groups II to IV were specific to the angiosperms, and Group IV was dicotyledon-specific. Although the protein sequences of DUF1005 varied greatly in different species and shared low overall sequence similarity, they possessed one highly conserved region (Figure 4).
The DUF1005 gene family in Arabidopsis is composed of 5 genes. The biological function of most DUF1005-encoded proteins remains unknown, which is partly because very few phenotypes related to this family have been uncovered thus far. Among the limited DUF1005 genes with established functions, AT5G17640, also known as Asg1, belonged to the earliest group I. Although Asg1 had no sequence similarities that could suggest its biological function in plant cells, its isolation through functional screening indicated that Asg1 was induced by salt stress in both Solanum tuberosum and Arabidopsis and by ABA in Arabidopsis (Batelli et al., 2012).
Expression analysis showed that CiDUF005 was regulated by various abiotic stresses (Figure 5). To study the abiotic stress response in the OE lines of CiDUF1005, WT and seedlings of the OE lines were subjected to mannitol stress. After 5 days of 350 mM of mannitol stress, except for the significance “**” of OE-4, the OE-5 significance “**” was reduced to “*,” and OE-31 was not significant compared to the untreated condition (Supplementary Figures S9A,B). This result was consistent with previous findings that the expression of CiDUF1005 was decreased under drought treatment (Figure 3 and Supplementary Figure S4). The accumulation of ABA in the root apex was necessary for maintaining elongation in the apical region of the elongation zone (Saab et al., 1992). Further tests are needed to confirm whether the primary root elongation of CiDUF1005 was regulated by ABA signaling, while the OE lines still had longer primary roots than the WT after ABA treatment (Supplementary Figure S8), suggesting that CiDUF1005 was not directly involved in the ABA pathway.
The root system of plants is a primary determinant of growth potential. A root trait contains many factors, such as root length, number, diameter, and root configuration in the soil profile. Root length is one of the most important parameters and primarily reflects a plant’s ability to acquire nutrients from the soil (Teo et al., 1995). However, the underlying mechanisms through which DUF1005 controls root development remain elusive.
Previous studies have shown that DUF1005 might be associated with plant development; for example, VrDUF1005 plays a role in regulating the shape and size of the cell during plant growth (Yun et al., 2020). In our study, CiDUF1005 regulated primary root elongation (Figure 6) and enhanced lateral root formation (Figure 7) during the plant seedling stage.
Therefore, we searched the literature and looked for genes that were related to root development. GA2ox2, which encodes an oxidase that inactivates bioactive C19 gibberellins via 2-oxidation, is a major gibberellin (GA) inactivating pathway in Arabidopsis, and GA delays the switch from cell division to expansion in Arabidopsis roots (Rieu et al., 2008; Ubeda-Tomás et al., 2008; Moubayidin et al., 2010). GA2ox2 expression controls Arabidopsis root meristem cell number, and GA2ox2OE lines formed shorter roots than those of the WT (Li et al., 2017).
Expansin loosened plant cell walls and was involved in cell enlargement and various abiotic stress responses. EXLA is a member of the plant extended protein superfamily. Three expansin-like genes were found in Arabidopsis: EXLA1, EXLA2, and EXLA3 (Cosgrove, 2000). Functional studies have shown that expansins are involved in many developmental processes in plants. Previous research indicated that the roots of exla2 mutants were longer (123%) than the WT roots (Abuqamar et al., 2013). This was consistent with our earlier study, which showed that the transcript levels of EXLA2 were significantly lower in the transgenic plants than in the WT plants (Figure 8). We speculated that CiDUF1005 might regulate root development by influencing the expression of EXLA2 and GA2ox2.
Root length is determined by cell division in the root meristem and cell elongation in the root elongation zone (Beemster et al., 2003). Cell proliferation and division are largely related to mitotic cyclin-dependent kinase activity, such as CCS52A (Vanstraelen et al., 2009). Expansion of root cells in the elongation zone contributes to primary root growth and is largely regulated by plant hormones. It is well established that plant hormones play a key role in root development (Péret et al., 2009). The major plant hormones—ABA, brassinosteroids, cytokinin, ethylene, GA, and auxin (IAA)—are all key regulators of primary root growth (Petricka et al., 2012). Therefore, the detection of hormone content in CiDUF1005 transgenic lines should be helpful to confirm whether DUF1005 regulates root development through hormone signaling pathways, and this will be the main topic in our future research.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Conceived and designed the experiments: QY and GL. Collected public datasets and performed experiments: XT, ZC, XN, and XZ. Analyzed the data: XT, QY, and RW. Wrote the manuscript: XT. Revised the manuscript: YQ and GL. All authors read and approved the final manuscript.
This research was funded by the National Natural Science Foundation of China (No. 32060066), The Major Project of Inner Mongolia Natural Science Foundation (2019ZD05), The Scientific and Technological Projects of Inner Mongolia (2019GG007), The University Scientific and Technological Innovation Team Project of Inner Mongolia (NMGIRT2222), and Postgraduate Innovation Program of Inner Mongolia of China. The funding agencies played no role in the design of the study, data collection, analysis, and interpretation or writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.807293/full#supplementary-material
Abuqamar, S., Ajeb, S., Sham, A., Enan, M. R., and Iratni, R. (2013). A Mutation in Theexpansin-like A2gene Enhances Resistance to Necrotrophic Fungi and Hypersensitivity to Abiotic Stress inArabidopsis Thaliana. Mol. Plant Pathol. 14, 813–827. doi:10.1111/mpp.12049
Arellano-Villagómez, F. C., Guevara-Olvera, L., Zuñiga-Mayo, V. M., E. Cerbantez-Bueno, V., Verdugo-Perales, M., R. Medina, H., et al. (2021). Arabidopsis Cysteine-Rich Receptor-like Protein Kinase CRK33 Affects Stomatal Density and Drought Tolerance. Plant Signal. Behav. 16, 1905335. doi:10.1080/15592324.2021.1905335
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Batelli, G., Massarelli, I., Van Oosten, M., Nurcato, R., Vannini, C., Raimondi, G., et al. (2012). Asg1 Is a Stress-Inducible Gene Which Increases Stomatal Resistance in Salt Stressed Potato. J. Plant Physiol. 169, 1849–1857. doi:10.1016/j.jplph.2012.07.004
Bateman, A., Coggill, P., and Finn, R. D. (2010). DUFs: Families in Search of Function. Acta Cryst. Sect. F 66, 1148–1152. doi:10.1107/S1744309110001685
Beemster, G. T. S., Fiorani, F., and Inzé, D. (2003). Cell Cycle: The Key to Plant Growth Control? Trends Plant Sci. 8, 154–158. doi:10.1016/S1360-1385(03)00046-3
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and Applications. BMC Bioinformatics 10, 421. doi:10.1186/1471-2105-10-421
Cao, X., Yang, K.-Z., Xia, C., Zhang, X.-Q., Chen, L.-Q., and Ye, D. (2009). Characterization of DUF724 Gene Family in Arabidopsis thaliana. Plant Mol. Biol. 72, 61–73. doi:10.1007/s11103-009-9551-5
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Clough, S. J., and Bent, A. F. (1998). Floral Dip: a Simplified Method forAgrobacterium-Mediated Transformation ofArabidopsis Thaliana. Plant J. 16, 735–743. doi:10.1046/j.1365-313x.1998.00343.x
Cosgrove, D. J. (2000). Loosening of Plant Cell walls by Expansins. Nature 407, 321–326. doi:10.1038/35030000
Cui, Y., Wang, M., Zhou, H., Li, M., Huang, L., Yin, X., et al. (2016). OsSGL, a Novel DUF1645 Domain-Containing Protein, Confers Enhanced Drought Tolerance in Transgenic rice and Arabidopsis. Front. Plant Sci. 7, 2001. doi:10.3389/fpls.2016.02001
Eddy, S. R. (1998). Profile Hidden Markov Models. Bioinformatics 14, 755–763. doi:10.1093/bioinformatics/14.9.755
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H.-R., et al. (2008). The Pfam Protein Families Database. Nucleic Acids Res. 36, D281–D288. doi:10.1093/nar/gkm960
Gao, Y., Badejo, A. A., Sawa, Y., and Ishikawa, T. (2012). Analysis of Two L-Galactono-1,4-Lactone-Responsive Genes with Complementary Expression during the Development of Arabidopsis thaliana. Plant Cel Physiol. 53, 592–601. doi:10.1093/pcp/pcs014
Gholizadeh, A., and Kohnehrouz, S. B. (2013). DUF538 Protein Super Family Is Predicted to Be the Potential Homologue of Bactericidal/permeability-Increasing Protein in Plant System. Protein J. 32, 163–171. doi:10.1007/s10930-013-9473-6
Gholizadeh, A. (2011). Heterologous Expression of Stress-Responsive DUF538 Domain Containing Protein and its Morpho-Biochemical Consequences. Protein J. 30, 351–358. doi:10.1007/s10930-011-9338-9
Gholizadeh, A. (2020). Pectin Methylesterase Activity of Plant DUF538 Protein Superfamily. Physiol. Mol. Biol. Plants 26, 829–839. doi:10.1007/s12298-020-00763-9
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2011). Phytozome: A Comparative Platform for green Plant Genomics. Nucleic Acids Res. 40, D1178–D1186. doi:10.1093/nar/gkr944
Han, X., Feng, Z., Xing, D., Yang, Q., Wang, R., Qi, L., et al. (2015). Two NAC Transcription Factors from Caragana Intermedia Altered Salt Tolerance of the Transgenic Arabidopsis. BMC Plant Biol. 15, 208. doi:10.1186/s12870-015-0591-5
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 35, W585–W587. doi:10.1093/nar/gkm259
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2014). GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 31, 1296–1297. doi:10.1093/bioinformatics/btu817
Huang, L., Cao, J., Zhang, A.-H., Zhang, Y.-C., and Ye, Y.-Q. (2008). Characterisation of BcMF10, a Novel Gene Involved in Pollen wall Development of Brassica Rapa Ssp. Chinensis. Funct. Plant Biol. 35, 1194–1204. doi:10.1071/FP08006
Jiang, J., Li, J., Xu, Y., Han, Y., Bai, Y., Zhou, G., et al. (2007). RNAi Knockdown of Oryza Sativa Root Meander Curling Gene Led to Altered Root Development and Coiling Which Were Mediated by Jasmonic Acid Signalling in rice. Plant Cel Environ. 30, 690–699. doi:10.1111/j.1365-3040.2007.01663.x
Jones-Rhoades, M. W., Borevitz, J. O., and Preuss, D. (2007). Genome-Wide Expression Profiling of the Arabidopsis Female Gametophyte Identifies Families of Small, Secreted Proteins. Plos Genet. 3, e171. doi:10.1371/journal.pgen.0030171
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 14, 587–589. doi:10.1038/nmeth.4285
Kim, S. J., Ryu, M. Y., and Kim, W. T. (2012). Suppression of Arabidopsis RING-DUF1117 E3 Ubiquitin Ligases, AtRDUF1 and AtRDUF2, Reduces Tolerance to ABA-Mediated Drought Stress. Biochem. Biophys. Res. Commun. 420, 141–147. doi:10.1016/j.bbrc.2012.02.131
Kodama, Y., Suetsugu, N., and Wada, M. (2011). Novel Protein-Protein Interaction Family Proteins Involved in Chloroplast Movement Response. Plant Signal. Behav. 6, 483–490. doi:10.4161/psb.6.4.14784
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X Version 2.0. Bioinformatics 23, 2947–2948. doi:10.1093/bioinformatics/btm404
Lata, C., and Prasad, M. (2011). Role of DREBs in Regulation of Abiotic Stress Responses in Plants. J. Exp. Bot. 62, 4731–4748. doi:10.1093/jxb/err210
Leasure, C. D., Tong, H., Yuen, G., Hou, X., Sun, X., and He, Z.-H. (2009). ROOT UV-B SENSITIVE2 Acts with ROOT UV-B SENSITIVE1 in a Root Ultraviolet B-Sensing Pathway. Plant Physiol. 150, 1902–1915. doi:10.1104/pp.109.139253
Lee, D. S., Kim, Y. C., Kwon, S. J., Ryu, C.-M., and Park, O. K. (2017). The Arabidopsis Cysteine-Rich Receptor-like Kinase CRK36 Regulates Immunity through Interaction with the Cytoplasmic Kinase BIK1. Front. Plant Sci. 8, 1856. doi:10.3389/fpls.2017.01856
Letunic, I., and Bork, P. (2006). Interactive Tree of Life (iTOL): An Online Tool for Phylogenetic Tree Display and Annotation. Bioinformatics 23, 127–128. doi:10.1093/bioinformatics/btl529
Li, J., Han, Y., Zhao, Q., Li, C., Xie, Q., Chong, K., et al. (2013). The E3 Ligase AtRDUF1 Positively Regulates Salt Stress Responses in Arabidopsis thaliana. PLoS One 8, e71078. doi:10.1371/journal.pone.0071078
Li, H., Torres-Garcia, J., Latrasse, D., Benhamed, M., Schilderink, S., Zhou, W., et al. (2017). Plant-Specific Histone Deacetylases HDT1/2 Regulate GIBBERELLIN 2-OXIDASE2 Expression to Control Arabidopsis Root Meristem Cell Number. Plant Cell 29, 2183–2196. doi:10.1105/tpc.17.00366
Liu, K., Yang, Q., Yang, T., Wu, Y., Wang, G., Yang, F., et al. (2019). Development of Agrobacterium-Mediated Transient Expression System in Caragana Intermedia and Characterization of CiDREB1C in Stress Response. BMC Plant Biol. 19, 237. doi:10.1186/s12870-019-1800-4
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Luo, C., Guo, C., Wang, W., Wang, L., and Chen, L. (2014). Overexpression of a New Stress-Repressive Gene OsDSR2 Encoding a Protein with a DUF966 Domain Increases Salt and Simulated Drought Stress Sensitivities and Reduces ABA Sensitivity in rice. Plant Cel Rep. 33, 323–336. doi:10.1007/s00299-013-1532-0
Mewalal, R., Mizrachi, E., Coetzee, B., Mansfield, S. D., and Myburg, A. A. (2016). The Arabidopsis Domain of Unknown Function 1218 (DUF1218) Containing Proteins, MODIFYING WALL LIGNIN-1 and 2 (At1g31720/MWL-1 and At4g19370/MWL-2) Function Redundantly to Alter Secondary Cell wall Lignin Content. PLoS One 11, e0150254. doi:10.1371/journal.pone.0150254
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2020). Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 49, D412–D419. doi:10.1093/nar/gkaa913
Miyakawa, T., Hatano, K.-i., Miyauchi, Y., Suwa, Y.-i., Sawano, Y., and Tanokura, M. (2014). A Secreted Protein with Plant-Specific Cysteine-Rich Motif Functions as a Mannose-Binding Lectin that Exhibits Antifungal Activity. Plant Physiol. 166, 766–778. doi:10.1104/pp.114.242636
Moturu, T. R., Thula, S., Singh, R. K., Nodzyński, T., Vařeková, R. S., Friml, J., et al. (2018). Molecular Evolution and Diversification of the SMXL Gene Family. J. Exp. Bot. 69, 2367–2378. doi:10.1093/jxb/ery097
Moubayidin, L., Perilli, S., Dello Ioio, R., Di Mambro, R., Costantino, P., and Sabatini, S. (2010). The Rate of Cell Differentiation Controls the Arabidopsis Root Meristem Growth Phase. Curr. Biol. 20, 1138–1143. doi:10.1016/j.cub.2010.05.035
Mudgal, R., Sandhya, S., Chandra, N., and Srinivasan, N. (2015). De-DUFing the DUFs: Deciphering Distant Evolutionary Relationships of Domains of Unknown Function Using Sensitive Homology Detection Methods. Biol. Direct. 10, 38. doi:10.1186/s13062-015-0069-2
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274. doi:10.1093/molbev/msu300
Oikawa, A., Joshi, H. J., Rennie, E. A., Ebert, B., Manisseri, C., Heazlewood, J. L., et al. (2010). An Integrative Approach to the Identification of Arabidopsis and rice Genes Involved in Xylan and Secondary wall Development. PLoS One 5, e15481. doi:10.1371/journal.pone.0015481
Okubazghi, K. W., Li, X.-n., Cai, X.-y., Wang, X.-x., Chen, H.-d., Zhou, Z.-l., et al. (2017). Genome-wide Assessment of Genetic Diversity and Fiber Quality Traits Characterization in Gossypium Hirsutum Races. J. Integr. Agric. 16, 2402–2412. doi:10.1016/S2095-3119(17)61671-X
Parsons, H. T., Christiansen, K., Knierim, B., Carroll, A., Ito, J., Batth, T. S., et al. (2012). Isolation and Proteomic Characterization of the Arabidopsis Golgi Defines Functional and Novel Components Involved in Plant Cell Wall Biosynthesis. Plant Physiol. 159, 12–26. doi:10.1104/pp.111.193151
Péret, B., De Rybel, B., Casimiro, I., Benková, E., Swarup, R., Laplaze, L., et al. (2009). Arabidopsis Lateral Root Development: An Emerging story. Trends Plant Sci. 14, 399–408. doi:10.1016/j.tplants.2009.05.002
Petricka, J. J., Winter, C. M., and Benfey, P. N. (2012). Control of Arabidopsis Root Development. Annu. Rev. Plant Biol. 63, 563–590. doi:10.1146/annurev-arplant-042811-105501
Prabhakaran Mariyamma, N. (2016). Elucidating the Role of the DUF828 Gene Family of Arabidopsis thaliana in Auxin Transport. Dissertation’s Thesis. Canada: University of Lethbridge.
Rieu, I., Eriksson, S., Powers, S. J., Gong, F., Griffiths, J., Woolley, L., et al. (2008). Genetic Analysis Reveals that C19-GA 2-Oxidation Is a Major Gibberellin Inactivation Pathway inArabidopsis. Plant Cell 20, 2420–2436. doi:10.1105/tpc.108.058818
Saab, I. N., Sharp, R. E., and Pritchard, J. (1992). Effect of Inhibition of Abscisic Acid Accumulation on the Spatial Distribution of Elongation in the Primary Root and Mesocotyl of maize at Low Water Potentials. Plant Physiol. 99, 26–33. doi:10.1104/pp.99.1.26
Salazar-Iribe, A., Agredano-Moreno, L. T., Zúñiga-Sánchez, E., Jiménez-Garcia, L. F., and Gamboa-deBuen, A. (2016). The Cell wall DUF642 At2g41800 (TEB) Protein Is Involved in Hypocotyl Cell Elongation. Plant Sci. 253, 206–214. doi:10.1016/j.plantsci.2016.10.007
Sergeant, K., Printz, B., Guerriero, G., Renaut, J., Lutts, S., and Hausman, J. F. (2019). The Dynamics of the Cell wall Proteome of Developing Alfalfa Stems. Biology 8, 60. doi:10.3390/biology8030060
Shen, L., Zhong, T., Wang, L., Zhang, Q., Jin, H., Xu, M., et al. (2019). Characterization the Role of a UFC Homolog, AtAuxRP3, in the Regulation of Arabidopsis Seedling Growth and Stress Response. J. Plant Physiol. 240, 152990. doi:10.1016/j.jplph.2019.152990
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, Scalable Generation of High‐quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Sundell, D., Mannapperuma, C., Netotea, S., Delhomme, N., Lin, Y. C., Sjödin, A., et al. (2015). The Plant Genome Integrative Explorer Resource: PlantGen IE.Org. New Phytol. 208, 1149–1156. doi:10.1111/nph.13557
Teo, Y. H., Beyrouty, C. A., Norman, R. J., and Gbur, E. E. (1995). Nutrient Uptake Relationship to Root Characteristics of rice. Plant Soil 171, 297–302. doi:10.1007/BF00010285
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 44, W232–W235. doi:10.1093/nar/gkw256
Ubeda-Tomás, S., Swarup, R., Coates, J., Swarup, K., Laplaze, L., Beemster, G. T. S., et al. (2008). Root Growth in Arabidopsis Requires Gibberellin/DELLA Signalling in the Endodermis. Nat. Cel Biol. 10, 625–628. doi:10.1038/ncb1726
Urbanowicz, B. R., Pena, M. J., Avci, U., Backe, J., Steet, H. F., Foston, M., et al. (2012). 4-O-methylation of Glucuronic Acid in Arabidopsis Glucuronoxylan Is Catalyzed by a Domain of Unknown Function Family 579 Protein. Proc. Natl. Acad. Sci. 109, 14253–14258. doi:10.1073/pnas.1208097109
Vanstraelen, M., Baloban, M., Da Ines, O., Cultrone, A., Lammens, T., Boudolf, V., et al. (2009). APC/CCCS52Acomplexes Control Meristem Maintenance in theArabidopsisroot. PNAS 106, 11806–11811. doi:10.1073/pnas.0901193106
Wiederschain, G. Y. (2006). The Proteomics Protocols Handbook. Biochemistry (Moscow) 71, 696. doi:10.1134/S0006297906060150
Wrzaczek, M., Brosché, M., Salojärvi, J., Kangasjärvi, S., Idänheimo, N., Mersmann, S., et al. (2010). Transcriptional Regulation of the CRK/DUF26 Group of Receptor-like Protein Kinases by Ozone and Plant Hormones in Arabidopsis. BMC Plant Biol. 10, 95. doi:10.1186/1471-2229-10-95
Yang, Q., Yin, J., Li, G., Qi, L., Yang, F., Wang, R., et al. (2014). Reference Gene Selection for qRT-PCR in Caragana Korshinskii Kom. Under Different Stress Conditions. Mol. Biol. Rep. 41, 2325–2334. doi:10.1007/s11033-014-3086-9
Yuan, Y., Teng, Q., Zhong, R., and Ye, Z.-H. (2013). The Arabidopsis DUF231 Domain-Containing Protein ESK1 Mediates 2-O- and 3-O-Acetylation of Xylosyl Residues in Xylan. Plant Cel Physiol. 54, 1186–1199. doi:10.1093/pcp/pct070
Lin, Y., Laosatit, K., Chen, J., Yuan, X., Wu, R., Amkul, K., et al. (2020). Mapping and Functional Characterization of Stigma Exposed 1, a DUF1005 Gene Controlling Petal and Stigma Cells in Mungbean (Vigna Radiata). Front. Plant Sci. 11, 575922. doi:10.3389/fpls.2020.575922
Zhang, J., Jia, W., Yang, J., and Ismail, A. M. (2006). Role of ABA in Integrating Plant Responses to Drought and Salt Stresses. Field Crops Res. 97, 111–119. doi:10.1016/j.fcr.2005.08.018
Zhang, Y., Zhang, F., and Huang, X. (2019). Characterization of an Arabidopsis thaliana DUF761-Containing Protein with a Potential Role in Development and Defense Responses. Theor. Exp. Plant Physiol. 31, 303–316. doi:10.1007/s40626-019-00146-w
Zhang, Z. (2019). Identification and Functional Analysis of SF2 in Controlling of Fruit Length in Cucumis Sativus L. Dissertation’s Thesis. China: Northwest A&F University.
Zhao, L., Nakazawa, M., Takase, T., Manabe, K., Kobayashi, M., Seki, M., et al. (2004). Overexpression of LSH1, a Member of an Uncharacterised Gene Family, Causes Enhanced Light Regulation of Seedling Development. Plant J. 37, 694–706. doi:10.1111/j.1365-313x.2003.01993.x
Zhao, J., Wang, P., Gao, W., Long, Y., Wang, Y., Geng, S., et al. (2021). Genome-wide Identification of the DUF668 Gene Family in Cotton and Expression Profiling Analysis of GhDUF668 in Gossypium Hirsutum under Adverse Stress. BMC Genomics 22, 1–19. doi:10.1186/s12864-021-07716-w
Zhou, X., Zhu, X., Shao, W., Song, J., Jiang, W., He, Y., et al. (2020). Genome-Wide Mining of Wheat DUF966 Gene Family Provides New Insights into Salt Stress Responses. Front. Plant Sci. 11, 569838. doi:10.3389/fpls.2020.569838
Keywords: DUF1005, family identification, phylogenetic analysis, primary root, Caragana intermedia
Citation: Tian X, Niu X, Chang Z, Zhang X, Wang R, Yang Q and Li G (2022) DUF1005 Family Identification, Evolution Analysis in Plants, and Primary Root Elongation Regulation of CiDUF1005 From Caragana intermedia. Front. Genet. 13:807293. doi: 10.3389/fgene.2022.807293
Received: 02 November 2021; Accepted: 08 February 2022;
Published: 29 March 2022.
Edited by:
Zefeng Yang, Yangzhou University, ChinaCopyright © 2022 Tian, Niu, Chang, Zhang, Wang, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Yang, YXRwX3lhbmdxaUBpbWF1LmVkdS5jbg==; Guojing Li, bGlndW9qaW5nQGltYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.