- 1Department of Plastic Surgery, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China
- 2Department of Plastic Surgery, Huashan Hospital, Fudan University, Shanghai, China
- 3Department of Dermatology, Tongji Hospital, Tongji University School of Medicine, Shanghai, China
Background: Androgen alopecia (AGA), the most common type of alopecia worldwide, has become an important medical and social issue. Accumulating evidence indicates that long noncoding RNAs (lncRNAs) play crucial roles in the progression of various human diseases, including AGA. However, the potential roles of lncRNAs in hair follicle stem cells (HFSCs) and their subsequent relevance for AGA have not been fully elucidated. The current study aimed to explore the function and molecular mechanism of the lncRNA AC010789.1 in AGA progression.
Methods: We investigated the expression levels of AC010789.1 in AGA scalp tissues compared with that in normal tissues and explored the underlying mechanisms using bioinformatics. HFSCs were then isolated from hair follicles of patients with AGA, and an AC010789.1-overexpressing HFSC line was produced and verified. Quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting were performed to verify the molecular mechanisms involved.
Results: AC010789.1 overexpression promoted the proliferation and differentiation of HFSCs. Mechanistically, we demonstrated that AC010789.1 overexpression promotes the biological function of HFSCs by downregulating miR-21-5p and TGF-β1 expression but upregulating the Wnt/β-catenin signaling pathway.
Conclusion: These results reveal that overexpression of AC010789.1 suppresses AGA progression via downregulation of hsa-miR-21-5p and TGF-β1 and promotion of the Wnt/β-catenin signaling pathway, highlighting a potentially promising strategy for AGA treatment.
Introduction
Alopecia, a common disorder occurring worldwide, characterized by hair loss, can be caused by multiple factors, such as heredity, hormonal disorders, immune inflammation, malnutrition, environmental factors, mental disorders, and aging (Ho and Shapiro, 2019). Androgen alopecia (AGA) is the most common type of alopecia and has become an important medical and social issue due to its high incidence; increasingly young onset age; and associated psychological problems, such as depression, anxiety, and emotional disorders (Bas et al., 2015; Molina-Leyva et al., 2016; Katzer et al., 2019; Ding et al., 2020). Currently, finasteride and minoxidil are the only therapeutic drugs approved by the Food and Drug Administration for AGA treatment (Chen et al., 2020a). Finasteride, a specific inhibitor of type II 5α reductase, inhibits the metabolic conversion of testosterone to highly active dihydrotestosterone, reducing the effect of active androgens on hair follicles (Spinucci and Pasquali, 1996). However, finasteride is associated with a risk of sexual dysfunction and depression during treatment (Motofei et al., 2020). Minoxidil treats AGA by predominantly promoting hair growth but can cause side effects, such as contact dermatitis, skin irritation, and dizziness during treatment, and on treatment cessation, alopecia generally recurs (Goren et al., 2017; Jimenez-Cauhe et al., 2019). Hair transplantation is the gold standard for AGA treatment, but the limited number of active hair follicles in the donor site makes it impossible to apply to large areas of baldness (Chouhan et al., 2019a; Chouhan et al., 2019b). Hence, new methods for the treatment of AGA need to be developed urgently.
The three phases of periodic hair follicle growth are resting, growth, and degenerative periods, and cessation of this regeneration cycle is the main mechanism contributing to AGA (Baker and Murray, 2012). Previous studies report that the periodic growth of hair follicles depends on the hair follicle stem cells (HFSCs) located in the bulge area of the hair follicle (Cotsarelis et al., 1990; Fu and Hsu, 2013). HFSCs are a group of adult stem cells with self-renewal ability that specifically express surface markers such as CD34 and CK15 (Morris et al., 2004; Owczarczyk-Saczonek et al., 2018). In normal conditions or during wound repair, HFSCs in the bulge area activate and differentiate into various hair follicle cell types for hair follicle regeneration (Oshima et al., 2001; Yang et al., 2017). However, there is evidence that the scalp hair follicles in patients with AGA are impaired in HFSC activation, thereby preventing their differentiation into hair follicle precursors. Garza et al. (2011) found that, although patients with AGA had a similar quantity of hair follicles in the alopecia scalp area as in nonalopecia areas, the HFSCs in the alopecia areas were in a static state and did not actively differentiate into hair follicle precursors. Thus, activation of HFSC proliferation and differentiation may be an effective breakthrough for the treatment of AGA.
Long noncoding RNAs (lncRNAs), a type of noncoding RNA over 200 nt in length, have attracted considerable interest in recent years. Functional data suggest that they play essential regulatory roles in multiple biological processes, such as cell development, differentiation, disease, subcellular localization, and cell structure maintenance (Yao et al., 2019). Intriguingly, genome analyses comparing AGA scalp tissues with adjacent normal tissues (defined as 5 cm from the margin of the AGA areas with a follicle density >325/cm2) in patients with AGA found a large number of differentially expressed lncRNAs, indicating that the dysregulation of lncRNA expression profiles may be involved in AGA progression (Chew et al., 2016; Bao et al., 2017). Moreover, several lines of evidence demonstrate a novel lncRNA regulatory mechanism in promoting hair follicle regeneration. Zhu et al. (2020) demonstrate that overexpression of lncRNA H19 can directly downregulate the expression of Wnt pathway inhibitors, including DKK1, Kremen2, and sFRP2, which activates Wnt signaling, thereby maintaining the hair follicle regeneration potential of dermal papilla cells (DPCs). Likewise, Lin et al. (2020a); Lin et al. (2020b) found that lncRNA XIST targets miR-424 and PCAT1 targets miR-329 to activate Wnt and hedgehog signaling, respectively, and maintain the regeneration characteristics of DPCs. To date, many molecular mechanisms of DPC-mediated hair follicle regeneration have been investigated. However, the role of the potential connection between lncRNAs and HFSCs in AGA has not been fully elucidated.
The current study aimed to explore the potential function of the lncRNA AC010789.1 in AGA progression. We demonstrate that AC010789.1 expression was downregulated in AGA scalp tissues, and overexpression of AC010789.1 promoted the proliferation and differentiation of HFSCs. Mechanistically, overexpression of AC010789.1 was shown to delay AGA progression by interacting with miR-21 and activating Wnt/β-catenin signaling. Therefore, this study provides insights into the mechanisms underlying AC010789.1 regulation of AGA progression to provide a new theoretical and experimental basis for the prevention and treatment of AGA.
Materials and Methods
Tissue Collection and Ethics Statement
Hair follicle tissues extracted from patients with AGA were obtained from the Shanghai East Hospital affiliated with the Tongji University School of Medicine, and informed consent was obtained from all patients. Ethical approval was obtained from the Shanghai East Hospital Ethics Committee. Tissue specimens were snap-frozen and stored in liquid nitrogen until further use.
Microarray Data Acquisition
Gene expression microarray data sets (GSE84839 and GSE36169) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). The GSE84839 data set was based on the GPL21827 platform (Agilent-079487 Arraystar Human LncRNA microarray V4) and included three pairs of male AGA scalp tissues and adjacent normal tissues. The GSE36169 data set was based on the GPL96 platform (Affymetrix Human Genome U133A Array) and contained AGA and adjacent normal scalp tissues from five individuals. The lncRNA and mRNA expression data in the AGA and adjacent normal tissues were downloaded and used for this study.
Differential Analysis of lncRNA and mRNA Expression
Differential analysis of lncRNA and mRNA expression between AGA and adjacent normal scalp tissues was performed using the GEO2R analysis tool. The platform data were converted using R language software and standardized using the limma array function within the R package (http://www.bioconductor.org/). A p-value < .05, and a base-2 logarithm of fold change (log FC) < −1 or >1 were used as selection criteria to screen differentially expressed lncRNAs and mRNAs (Xiong et al., 2020).
Functional and Pathway Enrichment Analysis
Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/) provides a comprehensive annotation tool to help investigators better clarify the biological function of the submitted genes (Dennis et al., 2003). In this study, DAVID v6.8 was used for gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. GO annotation analysis revealed biological processes (BPs), cellular components (CCs), and molecular functions (MFs) of the genes. Statistical significance was set at p < 0.05. Then, XTalkDB (http://www.xtalkdb.org/home), a database that documents scientific literature supporting crosstalk between pairs of signaling pathways, was used to explore the relationship between pathways.
Integrated Analysis of Interaction Network
GeneMANIA (http://genemania.org/) is a data set that provides a series of functional association information to identify the relation between genes of the submitted set in terms of their genetic interactions, pathways, expression patterns, localization, and protein domain similarity. In this study, protein-protein interaction (PPI) networks were analyzed using the GeneMANIA data set, and Cytoscape (version 3.7.2) was used to visualize the PPI networks.
Isolation and Cultivation of HFSC
Isolation of HFSCs was performed as previously described (Aran et al., 2020). Follicular unit extraction was performed to collect and isolate hair follicle tissues from 20 patients with AGA under a stereomicroscope. A needle was used to separate the hair shaft and papilla, and only the bulge areas were reserved. The bulge areas were treated with dispase II (2.5 mg/ml, Sigma, United States) and collagenase I (1 mg/ml, Gibco, United States) for 60 min, and 0.25% trypsin-ethylene diamine tetra acetic acid (EDTA; Gibco, Grand Island, NY, United States) for 15 min. The obtained cells were cultured in a keratinocyte serum-free medium (K-SFM, Gibco) in a 5% CO2 humidified incubator at 37°C, and the medium was changed every 2–3 days. The third passage (P3) of HFSCs was characterized by immunofluorescence with an anti-K15 antibody (Santa Cruz Biotechnology, TX, United States) and was used for follow-up studies.
Cell Proliferation Assay
Cell Counting Kit 8 (CCK8) assays (Beyotime Biotechnology Company, Jiangsu, China) and 5-ethynyl-2 deoxyuridine (EdU) assays (RiboBio, Guangzhou, China) were performed to assess the cell proliferation ability. For CCK8 assays, cells were seeded at approximately 2×103 cells/well in 96-well plates, and cell attachment was allowed for 12 h. Then, a 10 μl CCK8 test solution was added to each well at 24, 48, 72, and 96 h, and the cells were incubated in a humidified incubator with 5% CO2 at 37°C for 2 h at each time point to evaluate the cell growth viability. The optical density (OD) was measured at 450 nm with a microplate reader (Tecan, Thermo Scientific, United States). For EdU assays, the experiments were performed according to the manufacturer’s instructions (Xiong et al., 2019). Then, the cells were observed under a fluorescence microscope (Zeiss HLA100, Shanghai, China) and analyzed by using ImageJ software (Bethesda, MD, United States). The data shown are representative of three independent experiments.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from tissues or cells using Trizol® Reagent (Life Technologies, United States) and reverse transcribed into cDNA using a cDNA synthesis kit (Thermo Scientific) according to the manufacturer’s instructions. qRT-PCR was performed as previously reported (Xiong et al., 2019). Melt curves were established for the reactions, and the normalized fold expression was calculated using the 2−ΔΔ Ct method. The primer sequences are listed in Table 1.
Western Blotting
The cells from each sample were collected and lysed in radioimmunoprecipitation (RIPA) buffer (Thermo Scientific), followed by centrifugation at 1,200×g for 10 min and subsequent collection of the supernatant. Western blotting was performed as previously described (Xiong et al., 2019). The primary antibodies were GAPDH (1:20,000, Proteintech, China), Lgr5 (1:1,000, Abcam, United States), TGF-β1 (1:1,000, Santa Cruz Biotechnology, United States), and β-catenin (1:1,000, Cell Signaling Technology, United States), and the secondary antibody was horseradish peroxidase-conjugated goat antimouse (1:10,000, Abcam, United States). Protein expression was observed and visualized by chemiluminescence using an Alpha Imager scanner (Tecan, Thermo Scientific).
Statistical Analysis
All data are expressed as the mean ± standard deviation, and statistical significance was determined using Student’s t-test. All statistical analyses were performed using SPSS (version 17.0). Statistical significance was set at p < 0.05.
Results
AC010789.1 Expression is Downregulated in Patients With AGA
To investigate the role of lncRNAs in AGA, we used the gene expression microarray data set GSE84839, which included the data of three pairs of male scalp AGA and adjacent normal tissues. Analysis using the screening criteria of p < 0.05 and |log FC| > 1 revealed a total of 4,939 differentially expressed lncRNAs (4,239 upregulated and 700 downregulated). A volcano map was used to show the distribution of all differentially expressed lncRNAs (Figure 1A). AC010789.1, whose expression was significantly reduced in AGA scalp tissues compared with adjacent normal tissues, was selected for further analysis. To validate the results from the GSE84839 data set, we examined the expression level of AC010789.1 in clinical AGA scalp and adjacent normal tissues. The results showed that AC010789.1 expression was indeed downregulated in the AGA scalp tissues of patients (Figure 1B). These results strongly indicate that AC010789.1 expression is significantly downregulated in AGA scalp tissues and might be an essential predictive factor for patients with AGA.
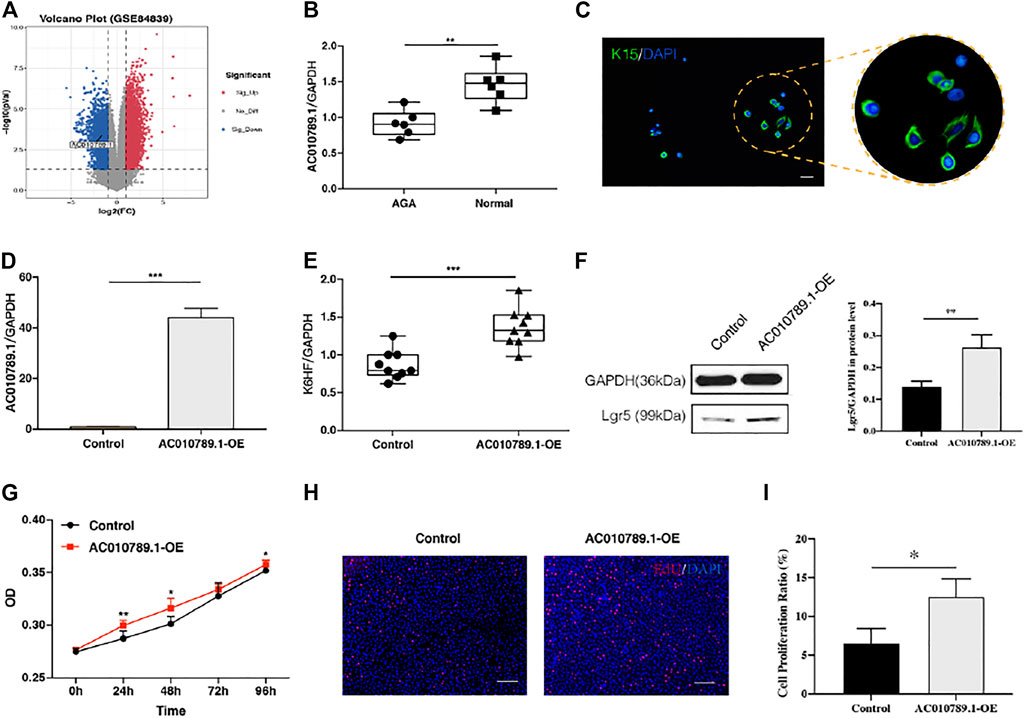
FIGURE 1. AC010789.1 is downregulated in patients with AGA, and its overexpression promotes proliferation and differentiation of HFSCs. (A) Volcano plot of the GSE84839 microarray data set. (B) AC010789.1 was downregulated in scalp tissues compared with adjacent normal tissues of AGA patients. n = 6 (C) The HFSCs highly express K15. HFSCs were stained with green fluorescence, and all nuclei were stained with blue fluorescence. Scale bars indicate 100 μm. (D) The AC010789.1 overexpression HFSC cell lines were successfully constructed. (E–I) Overexpression of AC010789.1 increased cell abilities of differentiation and proliferation. Scale bars indicate 100 μm n = 3 *p < 0.05, **p < 0.01 and ***p < 0.001.
Overexpression of AC010789.1 Promotes Proliferation and Differentiation of HFSCs
HFSCs were isolated from normal hair follicle tissues, and immunofluorescence was performed to detect the hair follicle stem cell marker K15. As shown in Figure 1C, the HFSCs displayed a high expression of K15. To better understand the biological effects of AC010789.1 on AGA development, we constructed an AC010789.1 overexpression plasmid, which was subsequently transfected into HFSCs. The qRT-PCR results in Figure 1D show that AC010789.1 was successfully overexpressed, and the resulting HFSC line (AC010789.1-OE) was used for further study. The qRT-PCR analysis showed that overexpression of AC010789.1 significantly upregulated the expression of HFSC differentiation markers K6HF and Lgr5 (Figures 1E,F). Subsequently, CCK8 and EdU assays revealed that AC010789.1 upregulation also resulted in increased cell proliferation (Figures 1G–I). Taken together, these experiments reveal that AC010789.1 has important functions in regulating the proliferation and differentiation of HFSCs.
AC010789.1 Interacts With miR-21 to Participate in AGA Progression
LncRNAs can act as miRNA sponges to regulate downstream targets. Therefore, we predicted putative candidate AC010789.1-binding miRNAs using RNAhybrid v2.2 and identified hsa-miR-21-5p as a potential candidate (Figure 2A). Next, we demonstrated that the expression level of hsa-miR-21-5p negatively correlated with that of AC010789.1 in AC010789.1-OE samples (Figure 2B). Hence, we considered hsa-miR-21-5p to be a potential target of AC010789.1.
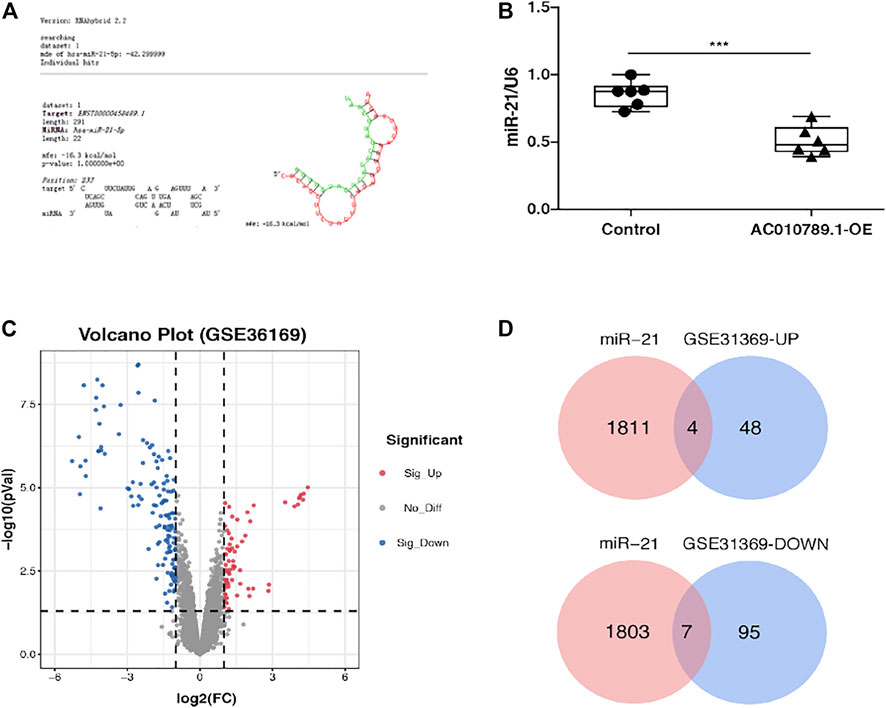
FIGURE 2. AC010789.1 interacts with miR-21 to participate in AGA progress and exploration on the pathogenesis of AGA. (A) RNAhybrid v2.2 showed the putative binding sites of miR-21-5p on AC010789.1. (B) Relative expression of miR-21-5p in AC010789.1-OE. n = 6 (C) Volcano plot of the GSE36169 microarray data set. (D) Venn diagram of upregulated and downregulated DEGs among the mRNA expression profiling sets GSE36169 and hsa-miR-21-5p targeted genes. *p < 0.05, **p < 0.01 and ***p < 0.001.
To further evaluate the mechanism by which AC010789.1 interacts with hsa-miR-21-5p in the pathogenesis of AGA, we analyzed the gene expression microarray data set GSE36169, which contained data of AGA and adjacent normal scalp tissues from five individuals. Analysis using the same screening criteria described above revealed a total of 198 differentially expressed genes (DEGs; 68 upregulated and 300 downregulated), which were represented on a volcano map (Figure 2C). Subsequently, we used the databases miRWalk and miRDB to predict the target genes of hsa-miR-21-5p and Venn diagram software to identify the genes that were common to both sets of analyses (Figure 2D). A total of 11 common genes (four upregulated and nine downregulated) were detected (Table 2).
GO Annotation and KEGG Pathway Enrichment Analysis of DEGs
To further explore the pathogenesis of AGA, we performed GO and KEGG enrichment analyses to further reveal the enrichment status of the DEGs in terms of their MFs, BPs, CCs, and pathways. With regard to BPs (Figures 3A,E), the upregulated DEGs were mainly involved in GO:0050776 (regulation of immune response), GO:0030199 (collagen fibril organization), and GO:0006954 (inflammatory response), and the downregulated DEGs were mainly involved in GO:0008544 (epidermis development), GO:0001942 (hair follicle development), and GO:0007010 (cytoskeleton organization). In terms of CCs (Figures 3B,F), the majority of upregulated DEGs were components of GO:0005576 (extracellular region), GO:0005615 (extracellular space), and GO:0072562 (blood microparticles), and the downregulated DEGs were mainly components of GO:0005882 (intermediate filament), GO:0045095 (keratin filament), and GO:0005615 (extracellular space). For the MFs (Figures 3C,G), the upregulated DEGs were mainly involved in GO:0031720 (haptoglobin binding), GO:0004252 (serine-type endopeptidase activity), and GO:0048407 (platelet-derived growth factor binding), and the downregulated DEGs were mainly involved in GO:0005198 (structural molecule activity), GO:0005509 (calcium ion binding), and GO:0008013 (beta-catenin binding). KEGG pathway analysis was used to explore pathway enrichment of the DEGs (Figures 3D,H). The most upregulated DEGs were significantly enriched in hsa05143 (African trypanosomiasis), hsa05144 (malaria), and hsa05150 (Staphylococcus aureus infection), and the downregulated DEGs were mainly enriched in hsa05144 (malaria), hsa04151 (PI3K-Akt signaling pathway), and hsa04350 (TGF-beta signaling pathway).
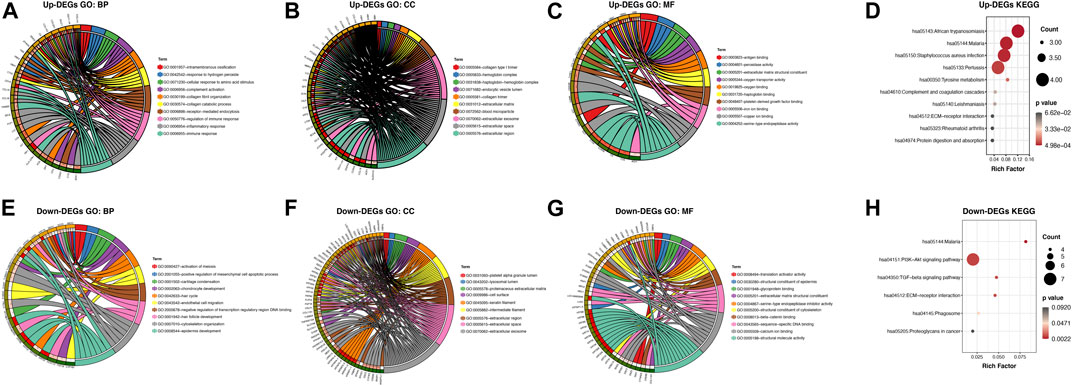
FIGURE 3. GO annotation and KEGG pathway enrichment analysis of the DEGs through the DAVID database. (A,E) The category of “biological process” of upregulated DEGs and downregulated DEGs, respectively. (B,F) The category of “cellular component” of upregulated DEGs and downregulated DEGs, respectively. (C,G) The category of “molecular function” of the predicted TG of upregulated DEGs and downregulated DEGs, respectively. (D,H) The category of “KEGG” of upregulated DEGs and downregulated DEGs, respectively.
Comprehensive Analysis of the Common DEGs
Next, the PPI networks of the common DEGs were analyzed using GeneMANIA, and the 20 most relevant genes were identified. GPM6B, SOX10, MBP, PIM2, CDH19, PMP2, GPM6A, PTPRN, MAG, TNFRSF17, ENPP6, ENPP2, SP2, ITGAV, SPI1, PIGO, CD79A, ENPP7, PIGG, and ENPP5 were primarily associated with the PPI network of the upregulated common genes (Figure 4A), and COL6A1, GABRD, MLLT10, MFAP5, SGK1, RAI14, DDX17, HOXC4, MMP14, ADH1B, VAV2, EFNA5, PLA2G15, RAB31, PCK1, HOXA10, NRP1, ZKSCAN5, MBTPS2, and PRRX1 were primarily associated with the PPI network of the downregulated common genes (Figure 4B). Furthermore, all the common DEGs and their 20 most relevant network genes were analyzed by GO and KEGG enrichment analysis. For BP (Figure 4C), most of the genes were involved in GO:0002040 (sprouting angiogenesis), GO:0035987 (endodermal cell differentiation), and GO:0001525 (angiogenesis). For CC (Figure 4D), the genes were mainly components of GO:0005886 (plasma membrane) and GO:0031012 (extracellular matrix). With regard to MF (Figure 4E), a majority of the genes were involved in GO:0003705 (transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding), GO:0019911 (structural constituent of myelin sheath), and GO:0002020 (protease binding). KEGG pathway enrichment analysis results showed that hsa04151 (PI3K-Akt signaling pathway) and hsa05221 (acute myeloid leukemia) were the most significantly enriched pathways of the genes (Figure 4F).
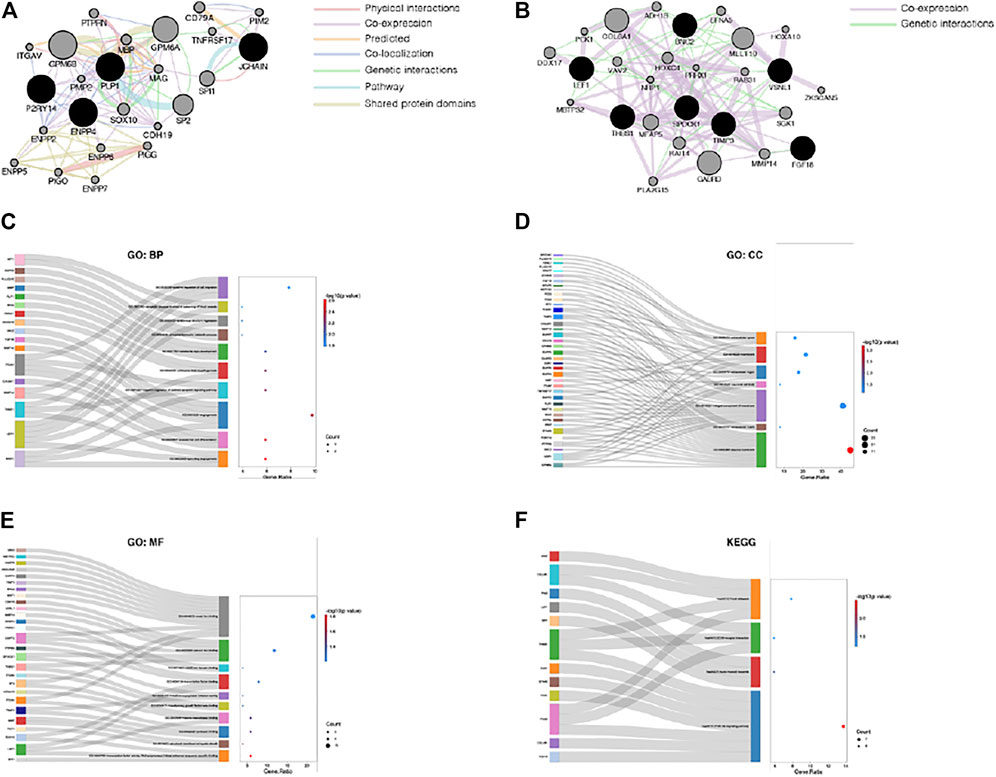
FIGURE 4. Comprehensive analysis of the common DEGs. (A) PPI network of the upregulated common DEGs. (B) PPI network of the downregulated common DEGs. (C) The category of “biological process” of the common DEGs and their most relevant genes. (D) The category of “cellular component” of the common DEGs and their most relevant genes. (E) The category of “molecular function” of the common DEGs and their most relevant genes. (F) The category of “KEGG” of the common DEGs and their most relevant genes.
AC010789.1 Targets the Wnt/β-Catenin Signaling Pathway to Regulate AGA Progression
According to the results of KEGG pathway enrichment analysis, a majority of the enriched pathways were highly associated with Wnt/β-catenin signaling (Figures 5A,B). Thus, we analyzed the mRNA and protein expression levels of several key genes of the Wnt/β-catenin pathway, including DKK-1, TGF-β1, Wnt10b, and β-catenin in AC010789.1-OE. The results showed that the expression of DKK-1 and TGF-β1 was significantly downregulated, whereas the expression of Wnt10b and β-catenin was significantly upregulated in AC010789.1-OE compared with control cells (Figures 5C–F). These data suggest that AC010789.1 may participate in AGA progression by regulating the Wnt/β-catenin pathway.
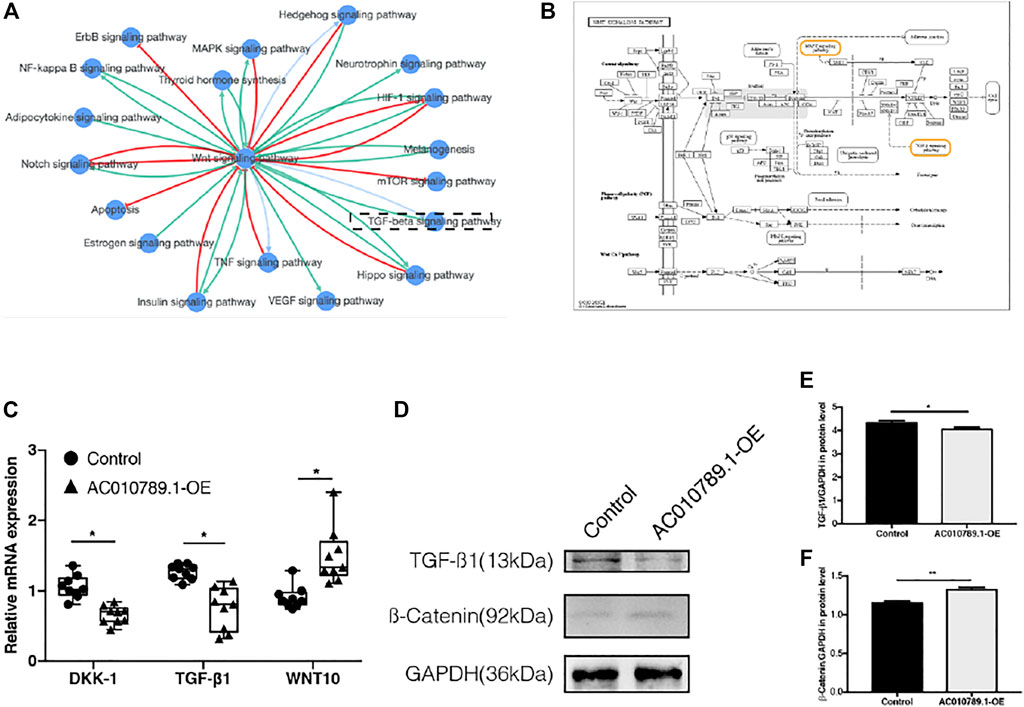
FIGURE 5. AC010789.1 targets with Wnt/β-catenin signaling pathway to participate in AGA progress. (A) The pathway interaction networks with Wnt/β-catenin signaling pathway. (B) KEGG map of Wnt/β-catenin signaling pathway. (C) The mRNA expression levels of DKK-1, TGF-β1, and Wnt10b in AC010789.1-OE. n = 9 (D–F) The protein expression levels of TGF-β1 and β-catenin in AC010789.1-OE. n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001.
Discussion
AGA is an androgen-dependent genetic hair loss disorder characterized by progressive microencapsulation of hair follicles and continuous shortening of the hair follicle growth period (Randall et al., 2000; Rathnayake and Sinclair, 2010). It is currently the most common clinical type of alopecia and can seriously affect a patient’s appearance, mental health, and social behavior. The most common first-line treatments for AGA currently include finasteride and minoxidil, but their application is hindered by limited efficacy, the need for long-term treatment, and inevitable complications. Hair transplant surgery, which involves hair follicle redistribution, is an effective method to improve the appearance of patients; however, as hair follicles cannot be regenerated, AGA patients with large areas of alopecia often have insufficient donor site hair follicles (Rogers, 2015). Therefore, it is necessary to explore the potential molecular mechanisms of AGA onset, progression, and hair follicle regeneration to design more effective treatments.
Several studies indicate that lncRNAs play essential roles in the occurrence and progression of various diseases and that they could be used as new diagnostic and treatment markers. For example, Wang et al. (2018) report that lncHOXA-AS2 promotes the progression of various human tumors by inducing epithelial–mesenchymal transition by directly inhibiting Bax expression, promoting c-Myc and Bcl-2 expression, and activating the Akt-MMP signaling pathway. Accumulating evidence now indicates that the deregulation of the expression of lncRNAs is strongly correlated with the onset and development of AGA (Bao et al., 2017; Zhu et al., 2020). In this study, we used the GSE84839 microarray data set from the GEO database and found that AC010789.1 was expressed to a lower level in AGA scalp tissues than in adjacent normal tissues, which was verified using clinical AGA samples. This suggests that low expression of AC010789.1 is associated with the progression of AGA.
HFSC aging, characterized by a reduction in stemness signatures and a concomitant increase in epidermal commitment, leads to a progressive miniaturization of hair follicles and ultimately, to the hair loss characteristic of AGA (Matsumura et al., 2016). Recently, accumulating evidence indicates that activating HFSCs could be an effective treatment for AGA. Zhang et al. (2020) found that vascular endothelial growth factor significantly reduced 5α-dihydrotestosterone-induced HFSC apoptosis by inhibiting the PI3K-Akt pathway, thereby delaying the progression of AGA. Kubo et al. (2020) used fisetin to induce a telogen-to-anagen transition in hair follicles by inducing the proliferation of HFSCs, thus promoting hair growth. Therefore, we hypothesized that AC010789.1 may promote hair growth by promoting the proliferation and differentiation of HFSCs. In the current study, we successfully isolated K15-positive HFSCs from hair follicles of patients with AGA and constructed an AC010789.1-overexpressing HFSC line. K6HF and Lgr5 are shown to be particularly good markers of hair differentiation and proliferation (Roh et al., 2004; Chen et al., 2020b), and we found that the expression of both marker genes as well as the cell proliferation rate were significantly higher in AC010789.1-OE than in the control group, indicating an important role of AC010789.1 in regulating HFSC functions.
Emerging evidence indicates that a large number of lncRNAs participate in a variety of biological functions by interacting with miRNAs and regulating their target genes (Huang, 2018). In this study, we found that AC010789.1 interacts with miR-21 to participate in the progression of AGA. Many reports show that miR-21 plays an essential role in the regulation of several diseases (Lakhter et al., 2018; Wang et al., 2020). To further explore how the interaction between AC010789.1 and hsa-miR-21-5p regulates the pathogenesis of AGA, the GSE36169 microarray data were downloaded, and the genes targeted by hsa-miR-21-5p were identified. A total of 198 AGA-related DEGs, including 11 common genes intersecting with hsa-miR-21-5p target genes, were identified by differential analysis with adjacent normal controls. GO annotation enrichment analysis was performed to explore the biological functions of the AGA-associated DEGs. The upregulated DEGs were primarily enriched in the BP category of immune and inflammatory responses. Previous studies show that an abundance of immune inflammatory cells in the bulge area of the hair follicle leads to the deregulation of the hair follicle microenvironment, thus impairing the normal function of HFSCs and resulting in alopecia (Wang and Higgins, 2020). Interestingly, the downregulated DEGs were directly related to hair follicle development (GO:0001942) and the hair cycle (GO:0042633). Subsequently, we extended the PPI network of the common genes and performed annotated enrichment analysis. Notably, the most downregulated common genes and their most relevant network genes were enriched in angiogenesis. Vascularization is closely related to hair growth (Gentile and Garcovich, 2019). The vascular system plays a vital role in maintaining the HFSC microenvironment, and angiogenesis helps to increase the blood supply of DPCs and promote hair growth. These findings enhance our understanding of the pathogenesis of AGA and the potential mechanism of interaction between AC010789.1 and miR-21-5p to delay the progression of AGA.
The TGF-β1 and Wnt signaling pathways are the most crucial pathways for maintaining a quiescent niche and regulating the proliferation and differentiation of HFSCs (Yang and Peng, 2010; Ge et al., 2019). Previous studies report that TGF-β1 promotes telogen-to-anagen transition in hair follicles, whereas the transition from anagen to telogen is significantly delayed in the hair follicles of TGF-β1 knockout mice (Foitzik et al., 2000; Daszczuk et al., 2020). The Wnt signaling pathway is the main regulatory pathway of biological development and a key driving factor for stem cells in most tissues (Nusse and Clevers, 2017). In the hair follicle, the Wnt signaling pathway plays a key role in starting the hair follicle cycle by initiating the proliferation response of HFSCs in the bulge area; HFSCs treated with Wnt pathway activator can quickly enter the proliferation period (Greco et al., 2009; Hawkshaw et al., 2020). Moreover, Leirós et al. (2017) found that Wnt pathway inhibitors (DKK-1) impair the differentiation of HFSCs, and the addition of promoters (Wnt10b) can reverse this effect in AGA. In addition, miR-21 is closely related to the Wnt/β-catenin signaling pathway. Previous studies reveal that inhibiting the expression of miR-21 can lead to upregulation of the Wnt/β-catenin pathway, thereby promoting cell activity (Hao et al., 2019; Liu et al., 2019). A recent study also reveals that lncRNA GAS5 competitively combined with miR-21 to regulate the epithelial–mesenchymal transition of human peritoneal mesothelial cells via activation of Wnt/β-catenin signaling (Fan et al., 2021). In this study, we conducted a comprehensive pathway analysis of the DEGs, common genes, and the genes most closely related in the PPI network to help us understand the molecular mechanisms underlying AGA progression. In line with our results, we found that the upregulated DEGs were highly enriched in the TGF-β signaling pathway, and a majority of the pathways were highly correlated with the Wnt/β-catenin signaling pathway. Further analysis showed that AC010789.1 overexpression induced the upregulation of Wnt10b and β-catenin, and downregulation of DKK-1 and TGF-β1. In summary, our results suggest that AC010789.1 regulates Wnt/β-catenin pathway activation, thereby enhancing the proliferation and differentiation of HFSCs and participating in AGA progression.
In summary, our data shows that AC010789.1 overexpression delayed AGA progression through the downregulation of hsa-miR-21-5p and promotion of the Wnt/β-catenin signaling pathway (Figure 6). Our findings provide a novel insight into the mechanism by which AC010789.1 promotes the proliferation and differentiation of HFSCs, which sheds light on the future development of lncRNA-based AGA therapies.
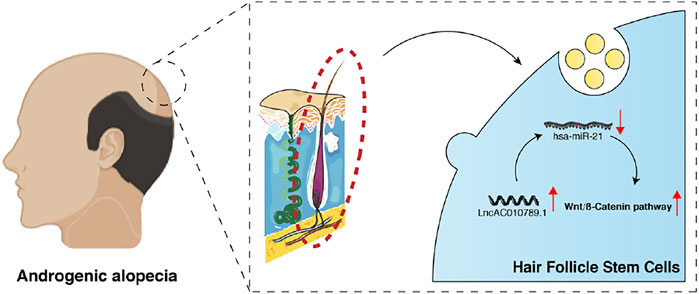
FIGURE 6. A schematic diagram shows how AC010789.1 delays AGA progression by targeting microRNA-21 and the Wnt/β-catenin signaling pathway.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HJ and YL designed the study; JX performed and drafted the experiment; BW and XH revised manuscript, and all authors approved the final version of the manuscript.
Funding
This work was supported by the east hospital affiliated to tongji university introduced talent research startup fund (grant no. DFRC2019008), clinical expansion foundation of PLA (Key Program) (grant no. 18KS1643), tongji hospital affiliated to tongji university excellent talent training program (grant no. HBRC2002), and featured clinical discipline project of shanghai pudong (grant no. PWYts2021-07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aran, S., Zahri, S., Asadi, A., Khaksar, F., and Abdolmaleki, A. (2020). Hair Follicle Stem Cells Differentiation into Bone Cells on Collagen Scaffold. Cell Tissue Bank 21 (2), 181–188. doi:10.1007/s10561-020-09812-9
Baker, R. E., and Murray, P. J. (2012). Understanding Hair Follicle Cycling: a Systems Approach. Curr. Opin. Genet. Dev. 22 (6), 607–612. doi:10.1016/j.gde.2012.11.007
Bao, L., Yu, A., Luo, Y., Tian, T., Dong, Y., Zong, H., et al. (2017). Genomewide Differential Expression Profiling of Long Non-coding RNAs in Androgenetic Alopecia in a Chinese Male Population. J. Eur. Acad. Dermatol. Venereol. 31 (8), 1360–1371. doi:10.1111/jdv.14278
Bas, Y., Seckin, H. Y., Kalkan, G., Takci, Z., Citil, R., Önder, Y., et al. (2015). Prevalence and Types of Androgenetic Alopecia in north Anatolian Population: A Community-Based Study. J. Pak Med. Assoc. 65 (8), 806–809.
Chen, L., Zhang, J., Wang, L., Wang, H., and Chen, B. (2020a). The Efficacy and Safety of Finasteride Combined with Topical Minoxidil for Androgenetic Alopecia: A Systematic Review and Meta-Analysis. Aesth Plast. Surg. 44 (3), 962–970. doi:10.1007/s00266-020-01621-5
Chen, P., Miao, Y., Zhang, F., Huang, J., Chen, Y., Fan, Z., et al. (2020b). Nanoscale Microenvironment Engineering Based on Layer-By-Layer Self-Assembly to Regulate Hair Follicle Stem Cell Fate for Regenerative Medicine. Theranostics 10 (25), 11673–11689. doi:10.7150/thno.48723
Chew, E. G. Y., Tan, J. H. J., Bahta, A. W., Ho, B. S.-Y., Liu, X., Lim, T. C., et al. (2016). Differential Expression between Human Dermal Papilla Cells from Balding and Non-balding Scalps Reveals New Candidate Genes for Androgenetic Alopecia. J. Invest. Dermatol. 136 (8), 1559–1567. doi:10.1016/j.jid.2016.03.032
Chouhan, K., Kota, R. S., Kumar, A., and Gupta, J. (2019a). Assessment of Safe Donor Zone of Scalp and Beard for Follicular Unit Extraction in Indian Men: A Study of 580 Cases. J. Cutan. Aesthet. Surg. 12 (1), 31–35. doi:10.4103/jcas.jcas_142_18
Chouhan, K., Roga, G., Kumar, A., and Gupta, J. (2019b). Approach to Hair Transplantation in Advanced Grade Baldness by Follicular Unit Extraction: A Retrospective Analysis of 820 Cases. J. Cutan. Aesthet. Surg. 12 (4), 215–222. doi:10.4103/jcas.jcas_173_18
Cotsarelis, G., Sun, T.-T., and Lavker, R. M. (1990). Label-retaining Cells Reside in the Bulge Area of Pilosebaceous Unit: Implications for Follicular Stem Cells, Hair Cycle, and Skin Carcinogenesis. Cell 61 (7), 1329–1337. doi:10.1016/0092-8674(90)90696-c
Daszczuk, P., Mazurek, P., Pieczonka, T. D., Olczak, A., Boryń, Ł. M., and Kobielak, K. (2020). An Intrinsic Oscillation of Gene Networks inside Hair Follicle Stem Cells: An Additional Layer that Can Modulate Hair Stem Cell Activities. Front. Cell Dev. Biol. 8, 595178. doi:10.3389/fcell.2020.595178
Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., et al. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4 (5), P3. doi:10.1186/gb-2003-4-5-p3
Ding, Q., Xu, Y.-X., Sun, W.-L., Liu, J.-J., Deng, Y.-Y., Wu, Q.-F., et al. (2020). Early-onset Androgenetic Alopecia in China: a Descriptive Study of a Large Outpatient Cohort. J. Int. Med. Res. 48 (3), 030006051989719. doi:10.1177/0300060519897190
Fan, Y., Zhao, X., Ma, J., and Yang, L. (2021). LncRNA GAS5 Competitively Combined with miR-21 Regulates PTEN and Influences EMT of Peritoneal Mesothelial Cells via Wnt/β-Catenin Signaling Pathway. Front. Physiol. 12, 654951. doi:10.3389/fphys.2021.654951
Foitzik, K., Lindner, G., Mueller‐Roever, S., Maurer, M., Botchkareva, N., Botchkarev, V., et al. (2000). Control of Murine Hair Follicle Regression (Catagen) by TGF‐β1 In Vivo. FASEB j. 14 (5), 752–760. doi:10.1096/fasebj.14.5.752
Fu, J., and Hsu, W. (2013). Epidermal Wnt Controls Hair Follicle Induction by Orchestrating Dynamic Signaling Crosstalk between the Epidermis and Dermis. J. Invest. Dermatol. 133 (4), 890–898. doi:10.1038/jid.2012.407
Garza, L. A., Yang, C.-C., Zhao, T., Blatt, H. B., Lee, M., He, H., et al. (2011). Bald Scalp in Men with Androgenetic Alopecia Retains Hair Follicle Stem Cells but Lacks CD200-Rich and CD34-Positive Hair Follicle Progenitor Cells. J. Clin. Invest. 121 (2), 613–622. doi:10.1172/jci44478
Ge, M., Liu, C., Li, L., Lan, M., Yu, Y., Gu, L., et al. (2019). miR-29a/b1 Inhibits Hair Follicle Stem Cell Lineage Progression by Spatiotemporally Suppressing WNT and BMP Signaling. Cell Rep. 29 (8), 2489–2504. doi:10.1016/j.celrep.2019.10.062
Gentile, P., and Garcovich, S. (2019). Advances in Regenerative Stem Cell Therapy in Androgenic Alopecia and Hair Loss: Wnt Pathway, Growth-Factor, and Mesenchymal Stem Cell Signaling Impact Analysis on Cell Growth and Hair Follicle Development. Cells 8 (5), 466. doi:10.3390/cells8050466
Goren, A., Naccarato, T., Situm, M., Kovacevic, M., Lotti, T., and McCoy, J. (2017). Mechanism of Action of Minoxidil in the Treatment of Androgenetic Alopecia Is Likely Mediated by Mitochondrial Adenosine Triphosphate Synthase-Induced Stem Cell Differentiation. J. Biol. Regul. Homeost Agents 31 (4), 1049–1053.
Greco, V., Chen, T., Rendl, M., Schober, M., Pasolli, H. A., Stokes, N., et al. (2009). A Two-step Mechanism for Stem Cell Activation during Hair Regeneration. Cell Stem Cell 4 (2), 155–169. doi:10.1016/j.stem.2008.12.009
Hao, L., Wang, J., and Liu, N. (2019). Long Noncoding RNA TALNEC2 Regulates Myocardial Ischemic Injury in H9c2 Cells by Regulating miR‐21/PDCD4‐medited Activation of Wnt/β‐catenin Pathway. J. Cell Biochem 120 (8), 12912–12923. doi:10.1002/jcb.28562
Hawkshaw, N. J., Hardman, J. A., Alam, M., Jimenez, F., and Paus, R. (2020). Deciphering the Molecular Morphology of the Human Hair Cycle: Wnt Signalling during the Telogen-Anagen Transformation. Br. J. Dermatol. 182 (5), 1184–1193. doi:10.1111/bjd.18356
Ho, A., and Shapiro, J. (2019). Medical Therapy for Frontal Fibrosing Alopecia: A Review and Clinical Approach. J. Am. Acad. Dermatol. 81 (2), 568–580. doi:10.1016/j.jaad.2019.03.079
Huang, Y. (2018). The Novel Regulatory Role of lncRNA-miRNA-mRNA axis in Cardiovascular Diseases. J. Cell Mol Med 22 (12), 5768–5775. doi:10.1111/jcmm.13866
Jimenez-Cauhe, J., Saceda-Corralo, D., Rodrigues-Barata, R., Hermosa-Gelbard, A., Moreno-Arrones, O. M., Fernandez-Nieto, D., et al. (2019). Effectiveness and Safety of Low-Dose Oral Minoxidil in Male Androgenetic Alopecia. J. Am. Acad. Dermatol. 81 (2), 648–649. doi:10.1016/j.jaad.2019.04.054
Katzer, T., Leite Junior, A., Beck, R., and da Silva, C. (2019). Physiopathology and Current Treatments of Androgenetic Alopecia: Going beyond Androgens and Anti‐androgens. Dermatol. Ther. 32 (5), e13059. doi:10.1111/dth.13059
Kubo, C., Ogawa, M., Uehara, N., and Katakura, Y. (2020). Fisetin Promotes Hair Growth by Augmenting TERT Expression. Front. Cell Dev. Biol. 8, 566617. doi:10.3389/fcell.2020.566617
Lakhter, A. J., Pratt, R. E., Moore, R. E., Doucette, K. K., Maier, B. F., DiMeglio, L. A., et al. (2018). Beta Cell Extracellular Vesicle miR-21-5p Cargo Is Increased in Response to Inflammatory Cytokines and Serves as a Biomarker of Type 1 Diabetes. Diabetologia 61 (5), 1124–1134. doi:10.1007/s00125-018-4559-5
Leirós, G. J., Ceruti, J. M., Castellanos, M. L., Kusinsky, A. G., and Balañá, M. E. (2017). Androgens Modify Wnt Agonists/antagonists Expression Balance in Dermal Papilla Cells Preventing Hair Follicle Stem Cell Differentiation in Androgenetic Alopecia. Mol. Cell Endocrinol. 439, 26–34. doi:10.1016/j.mce.2016.10.018
Lin, B.-J., Lin, G.-Y., Zhu, J.-Y., Yin, G.-Q., Huang, D., and Yan, Y.-Y. (2020a). LncRNA-PCAT1 Maintains Characteristics of Dermal Papilla Cells and Promotes Hair Follicle Regeneration by Regulating miR-329/Wnt10b axis. Exp. Cell Res. 394 (1), 112031. doi:10.1016/j.yexcr.2020.112031
Lin, B.-J., Zhu, J.-Y., Ye, J., Lu, S.-D., Liao, M.-D., Meng, X.-C., et al. (2020b). LncRNA-XIST Promotes Dermal Papilla Induced Hair Follicle Regeneration by Targeting miR-424 to Activate Hedgehog Signaling. Cell Signal. 72, 109623. doi:10.1016/j.cellsig.2020.109623
Liu, X. G., Zhang, Y., Ju, W. F., Li, C. Y., and Mu, Y. C. (2019). MiR-21 Relieves Rheumatoid Arthritis in Rats via Targeting Wnt Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (3 Suppl. l), 96–103. doi:10.26355/eurrev_201908_18635
Matsumura, H., Mohri, Y., Binh, N. T., Morinaga, H., Fukuda, M., Ito, M., et al. (2016). Hair Follicle Aging Is Driven by Transepidermal Elimination of Stem Cells via COL17A1 Proteolysis. Science 351 (6273), aad4395. doi:10.1126/science.aad4395
Molina-Leyva, A., Caparros-Del Moral, I., Gomez-Avivar, P., Alcalde-Alonso, M., and Jimenez-Moleon, J. J. (2016). Psychosocial Impairment as a Possible Cause of Sexual Dysfunction Among Young Men with Mild Androgenetic Alopecia: A Cross-Sectional Crowdsourcing Web-Based Study. Acta Dermatovenerol Croat. 24 (1), 42–48.
Morris, R. J., Liu, Y., Marles, L., Yang, Z., Trempus, C., Li, S., et al. (2004). Capturing and Profiling Adult Hair Follicle Stem Cells. Nat. Biotechnol. 22 (4), 411–417. doi:10.1038/nbt950
Motofei, I. G., Rowland, D. L., Tampa, M., Sarbu, M.-I., Mitran, M.-I., Mitran, C.-I., et al. (2020). Finasteride and Androgenic Alopecia; from Therapeutic Options to Medical Implications. J. Dermatol. Treat. 31 (4), 415–421. doi:10.1080/09546634.2019.1595507
Nusse, R., and Clevers, H. (2017). Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169 (6), 985–999. doi:10.1016/j.cell.2017.05.016
Oshima, H., Rochat, A., Kedzia, C., Kobayashi, K., and Barrandon, Y. (2001). Morphogenesis and Renewal of Hair Follicles from Adult Multipotent Stem Cells. Cell 104 (2), 233–245. doi:10.1016/s0092-8674(01)00208-2
Owczarczyk-Saczonek, A., Krajewska-Włodarczyk, M., Kruszewska, A., Banasiak, Ł., Placek, W., Maksymowicz, W., et al. (2018). Therapeutic Potential of Stem Cells in Follicle Regeneration. Stem Cell Int. 2018, 1–16. doi:10.1155/2018/1049641
Randall, V. A., Hibberts, N. A., Thornton, M. J., Hamada, K., Merrick, A. E., Kato, S., et al. (2000). The Hair Follicle: a Paradoxical Androgen Target Organ. Horm. Res. Paediatr. 54 (5-6), 243–250. doi:10.1159/000053266
Rathnayake, D., and Sinclair, R. (2010). Male Androgenetic Alopecia. Expert Opin. Pharmacother. 11 (8), 1295–1304. doi:10.1517/14656561003752730
Rogers, N. (2015). Hair Transplantation Update. Sem Cutan. Med. Surg. 34 (2), 89–94. doi:10.12788/j.sder.2015.0131
Roh, C., Tao, Q., and Lyle, S. (2004). Dermal Papilla-Induced Hair Differentiation of Adult Epithelial Stem Cells from Human Skin. Physiol. Genomics 19 (2), 207–217. doi:10.1152/physiolgenomics.00134.2004
Spinucci, G., and Pasquali, R. (1996). Finasteride: a New Drug for the Treatment of Male Hirsutism and Androgenetic Alopecia? Clin. Ter 147 (6), 305–315.
Wang, E. C. E., and Higgins, C. A. (2020). Immune Cell Regulation of the Hair Cycle. Exp. Dermatol. 29 (3), 322–333. doi:10.1111/exd.14070
Wang, G., Zhou, Y., Chen, W., Yang, Y., Ye, J., Ou, H., et al. (2020). miR-21-5p Promotes Lung Adenocarcinoma Cell Proliferation, Migration and Invasion via Targeting WWC2. Cbm 28 (4), 549–559. doi:10.3233/cbm-201489
Wang, J., Su, Z., Lu, S., Fu, W., Liu, Z., Jiang, X., et al. (2018). LncRNA HOXA-AS2 and its Molecular Mechanisms in Human Cancer. Clinica Chim. Acta 485, 229–233. doi:10.1016/j.cca.2018.07.004
Xiong, J., Ji, B., Wang, L., Yan, Y., Liu, Z., Fang, S., et al. (2019). Human Adipose-Derived Stem Cells Promote Seawater-Immersed Wound Healing by Activating Skin Stem Cells via the EGFR/MEK/ERK Pathway. Stem Cell Int. 2019, 1–16. doi:10.1155/2019/7135974
Xiong, J., Xue, Y., Xia, Y., Zhao, J., and Wang, Y. (2020). Identification of Key microRNAs of Plasma Extracellular Vesicles and Their Diagnostic and Prognostic Significance in Melanoma. Open Med. (Wars) 15 (1), 464–482. doi:10.1515/med-2020-0111
Yang, H., Adam, R. C., Ge, Y., Hua, Z. L., and Fuchs, E. (2017). Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 169 (3), 483–496. doi:10.1016/j.cell.2017.03.038
Yang, L., and Peng, R. (2010). Unveiling Hair Follicle Stem Cells. Stem Cell Rev Rep 6 (4), 658–664. doi:10.1007/s12015-010-9172-z
Yao, R.-W., Wang, Y., and Chen, L.-L. (2019). Cellular Functions of Long Noncoding RNAs. Nat. Cell Biol 21 (5), 542–551. doi:10.1038/s41556-019-0311-8
Zhang, X., Zhou, D., Ma, T., and Liu, Q. (2020). Vascular Endothelial Growth Factor Protects CD200-Rich and CD34-Positive Hair Follicle Stem Cells against Androgen-Induced Apoptosis through the Phosphoinositide 3-Kinase/Akt Pathway in Patients with Androgenic Alopecia. Ds 46 (3), 358–368. doi:10.1097/dss.0000000000002091
Keywords: androgen alopecia, hair follicle stem cells, AC010789.1, Wnt/β-catenin, TGF-1
Citation: Xiong J, Wu B, Hou Q, Huang X, Jia L, Li Y and Jiang H (2022) Comprehensive Analysis of LncRNA AC010789.1 Delays Androgenic Alopecia Progression by Targeting MicroRNA-21 and the Wnt/β-Catenin Signaling Pathway in Hair Follicle Stem Cells. Front. Genet. 13:782750. doi: 10.3389/fgene.2022.782750
Received: 24 September 2021; Accepted: 11 January 2022;
Published: 15 February 2022.
Edited by:
Jiacan Su, Second Military Medical University, ChinaReviewed by:
Yong Miao, Southern Medical University, ChinaLie Zhu, Shanghai Changzheng Hospital, China
Yingying Jing, Shanghai University, China
Copyright © 2022 Xiong, Wu, Hou, Huang, Jia, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yufei Li, bHVmeWxlZUBob3RtYWlsLmNvbQ==; Hua Jiang, ZG9zamhAMTI2LmNvbQ==
 Jiachao Xiong
Jiachao Xiong Baojin Wu2
Baojin Wu2
