
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet. , 10 February 2022
Sec. Epigenomics and Epigenetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.760690
This article is part of the Research Topic 10 Years of Frontiers in Genetics: past discoveries, current challenges and future perspectives View all 31 articles
The endosperm is a vital storage tissue in plant seeds. It provides nutrients to the embryos or the seedlings during seed development and germination. Although the genetic information in the endosperm cannot be passed directly to the next generation, its inherited epigenetic marks affect gene expression and its development and, consequently, embryo and seed growth. DNA methylation is a major form of epigenetic modification that can be investigated to understand the epigenome changes during reproductive development. Therefore, it is of great significance to explore the effects of endosperm DNA methylation on crop yield and traits. In this review, we discuss the changes in DNA methylation and the resulting imprinted gene expression levels during plant endosperm development, as well as their effects on seed development.
In angiosperms, the development of seeds requires double fertilization. The egg and central cells independently combine with sperm cells and develop into the embryo and endosperm, respectively, (Bleckmann et al., 2014). The functions of endosperm are mainly to act as the nutrient supplier, to be the mechanical barrier for the embryo, and to be the growth regulator of the embryo during seed development and germination. The endosperm is also a fundamental factor for the seed prosperity of angiosperms (Yan et al., 2014). Additionally, the endosperm is a critical factor in controlling seed viability and dormancy (De Giorgi et al., 2015). The vast majorities of the edible parts of rice, wheat, and corn, which account for approximately 70% of human food consumption, are endosperm tissues, which are rich in starch, protein, vitamins, dietary fiber, and other nutrients needed in the human diet (Kang et al., 2008). Therefore, improving endosperm contents and quality is a significant issue. Regulation of endosperm development involves gene imprinting and epigenetic modifications. DNA methylation is a major epigenetic modification that participates in gene expression, TE silencing, and genome stability during plant development. It is related to epigenetic transgenerational inheritance. Owing to the importance of the endosperm, its DNA methylation and genome imprinting are reviewed here.
This review introduces the methylation and demethylation of plant endosperm and the imprinted gene expression resulting from DNA methylation. We discuss the effects of endosperm DNA methylation on plant development.
DNA methylation, the addition of a methyl (CH3) group at carbon 5 of cytosine by DNA methyltransferases, is a critical epigenetic marker in mammals and plants (Jin et al., 2011). It represents a heritable change in gene expression not encoded by the DNA sequence. DNA methylation is essential for genomic imprinting, transposable element (TE) silencing, gene regulation, genetic evolution, and genomic stability (Zhang et al., 2018). The loss of DNA methyltransferase function can lead to abnormal plant development (Rajkumar et al., 2020). DNA methylation occurs in three sequence contexts in plants: symmetrical CG and CHG sites and asymmetrical CHH (H = C/T/A) sites (Kawakatsu et al., 2017). Different methyltransferases accomplish different DNA methylation patterns through de novo methylation and maintenance of methylation (Law and Jacobsen, 2010). There are three types of DNA methyltransferase in plants: DNA Methyltransferase (MET), Domains Rearranged Methyltransferase (DRM), and plant-specific Chromomethylase (CMT). These methyltransferases perform their duties in de novo and maintenance methylation, and jointly complete the DNA methylation modification in plants (Ashapkin et al., 2016).
De novo methylation refers to the generation of new methylation at sites that have not undergone methylation. The plant-specific RNA-directed DNA methylation (RdDM) pathway catalyzes the de novo methylation of three sequence contexts (Matzke and Mosher, 2014). Asymmetric CHH site methylation can only be maintained through de novo methylation (Zhang et al., 2018). Some repetitive DNA sequences are transcribed by RNA Polymerase IV (Pol IV) to generate single-strand RNAs (ssRNAs). These ssRNAs produce double-strand RNA (dsRNA) by RNA-Dependent RNA Polymerase 2 (RDR2), which is then cut into 24 nt siRNA by DICER-LIKE 3 (DCL3) (Haag and Pikaard, 2011; Matzke and Mosher, 2014; Zhai et al., 2015). In addition, some inverted repeated DNA sequences can also produce dsRNA under the action Pol II and RDR6, which is further cleaved by DCL3 to produce 21 nt siRNA. These 24 nt and 21 nt siRNAs combine with Argonaute 4 (AGO4) to form a siRNA-AGO4 complex, which recruits DRM1 and DRM2 to de novo methylation in the three sites (CG, CHG, CHH) (Matzke and Mosher, 2014). In addition to the RdDM pathway, CMT2 and CMT3 can also catalyze de novo methylation (Law and Jacobsen, 2010). The nucleosome remodeling factor Decrease in DNA Methylation 1 (DDM1) changes nucleosome conformation, binds CMT2 to histone H3 lysine 9 dimethylation (H3K9me2) and mediates the de novo methylation of the adjacent CHG and CHH sites (Kuo et al., 2017).
Maintenance methylation refers to maintaining the methylation form of the original site in the process of DNA replication. The maintenance of CG methylation in plants is completed by the methylation regulator VIMs (Variation in Methylation, VIM1, VIM2, VIM3) protein and DNA methyltransferase MET1. VIMs recognize and bind to the hemimethylated CG site, recruit MET1 to complete CG methylation of the newly synthesized strand, and finally obtain double-stranded DNA methylation of the CG site (Kawashima and Berger, 2014). The maintenance of CHG methylation is mediated by the CMT3-H3K9me2 pathway. CMT3 binds to two H3K9me2 proteins simultaneously and methylates DNA at nearby CHG sites. The methylated CHG DNA recruits Su (var) Homologue 4 (SUVH4), and the deposition of H3K9me2 markers on the nucleosomes surrounding CHG methylated DNA by SUVH4 creates a CHG–H3K9me2 positive feedback loop (Zhang et al., 2018).
In addition, there is an active DNA demethylation process in flowering plants, and this is achieved by DNA glycosylase/lyase through a base excision repair (BER) mechanism. Three types of DNA glycosylases have been found in plants: Demeter (DME), Repressor of silencing1 (ROS1), Demeter-like (DML2 and DML3). Thus, the final methylation level in the genome is determined by the activities of both DNA methyltransferases and demethylases (Gong et al., 2002).
In Arabidopsis, the expression levels of the major DNA methylation enzymes are available at the Arabidopsis RNA-seq database (http://ipf.sustech.edu.cn/pub/athrna/). In wild-type endosperm, the expression of MET1 is low, whereas the expression levels of MET2a and MET2b, which are specifically expressed in central cells, are high. The paternal imprinting genes VIM5 and MET3 are also specifically expressed and highly expressed in the endosperm. Therefore, we speculate that MET2a, MET2b, MET3, and VIM5 may jointly regulate CG methylation in the endosperm (Figure 1), which requires further experimental proof. We profile a simple model based on DME-mediated DNA demethylation in the endosperm (Figure 2). The DME gene is predominantly expressed in the central cell, and DME induces global hypomethylation (Choi et al., 2002; Hsieh et al., 2009). Before fertilization, the central cell and vegetative cell are highly demethylated resulted from the action of DME. DME preferentially targets TE regions (Hsieh et al., 2009; Ibarra et al., 2012). The demethylation of the maternal genome during gametogenesis is also reported in other species—castor bean (Park et al., 2016), rice (Zemach et al., 2010; Park et al., 2016), and maize (Lauria et al., 2004). The vegetative cell produces siRNA into the sperm cells and maintains the sperm cell hypermethylation through the RDdM pathway (Martinez et al., 2016). So the methylation level in the endosperm is much lower than in the embryo after fertilization. The siRNAs produced by the demethylation of the endosperm are transferred to the embryo to maintain the stability of the embryo genome (McCue et al., 2012). The loss of DME function (dme mutant) in endosperm restores CG methylation but unexpectedly further diminishes non-CG methylation, suggesting demethylation in a non-CG context is regulated by a yet unknown DME-independent mechanism (Hsieh et al., 2009; Jullien et al., 2012).
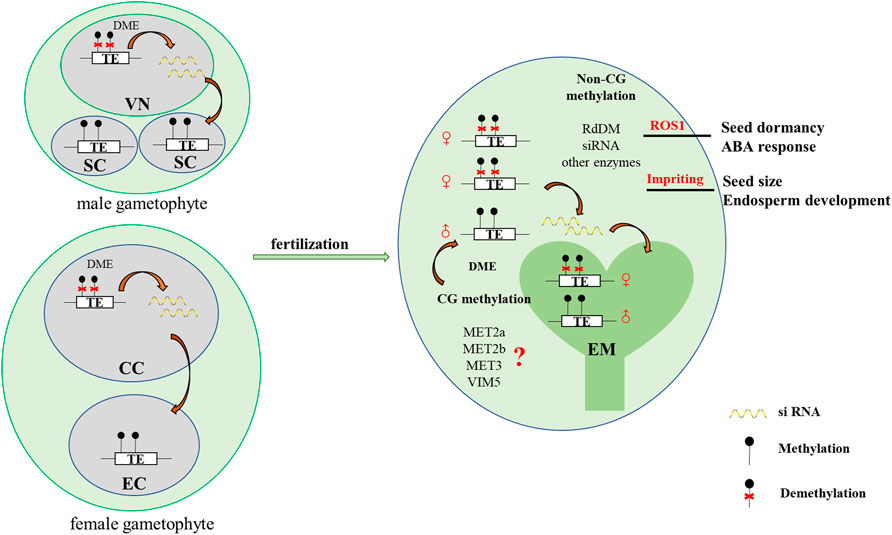
FIGURE 2. A simplified model of DME-mediated DNA demethylation in Arabidopsis endosperm. Before fertilization, the central cell and vegetative cell are highly demethylated resulted from the action of DME. DME preferentially targets TE regions (Gehring et al., 2009; Hsieh et al., 2009; Ibarra et al., 2012). The vegetative cell produces siRNA into the sperm cells and maintains the sperm cell hypermethylation through the RDdM pathway (Martinez et al., 2016). So the methylation level in the endosperm is much lower than in the embryo after fertilization. The siRNAs produced by the demethylation of the endosperm are transferred to the embryo to maintain the stability of the embryo genome. MET2a, MET2b, MET3, and VIM5 may jointly regulate CG methylation in the endosperm. In addition, the differential methylation of the embryo and endosperm leads to imprinting in the endosperm, which may affect endosperm development and control seed size. Additionally, the endosperm demethylase ROS1 regulates seed dormancy. VN: vegetative cell nucleus, SC: sperm cell, CC: central cell, EC: egg cell, EM: embryo.
The endosperm of some plants, such as Arabidopsis, only exist in the early stage of seed development and gradually disappear with seed development (Brown et al., 1999). Most monocotyledons, some dicotyledons, and gymnosperms, have endosperm in their mature seeds, such as castor beans and rice (Greenwood and Bewley, 1982; Brown et al., 1996). The DNA methylation profiles in the endosperm of different plants are significantly different, suggesting that DNA methylation profiles of endosperm are not conserved. By comparing the methylation levels of the endosperm among different plants, it was found that genomic DNA hypomethylation in endosperm relative to the embryo is widespread (Figure 3), especially in dicotyledons (Hsieh et al., 2009; Zemach et al., 2010; Lu et al., 2015; Xu et al., 2016). CG, CHG, and CHH methylation levels were low at 4 days after pollination (DAP), but all three contexts of DNA methylation levels were elevated at 6 days after pollination by DNA methylation sequencing in Arabidopsis endosperm (Pignatta et al., 2014; Moreno-Romero et al., 2016). Hu et al. found that the methylation levels were higher during the early (3–5 DAP) and late stages (13–25 DAP) of endosperm development compared with the middle stage (7–11 DAP) in maize endosperm (Hu et al., 2021). Thus, DNA methylation represents a dynamic process during endosperm development. The DNA methylation changes in the endosperm affect the expression of genes and siRNAs, thereby affecting endosperm formation and seed development (Moore et al., 2013).
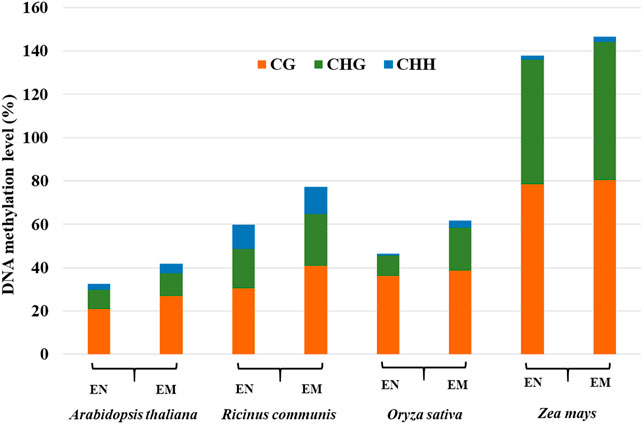
FIGURE 3. DNA methylation levels of CG, CHG, and CHH in the endosperm (EN) and embryo (EM) among different species.
Genomic imprinting is the process of inheriting the epigenetic marking for a particular segment of a chromosome from paternal or maternal alleles (Feil and Berger, 2007). The endosperm is the main organ that undergoes genomic imprinting in flowering plants (Gehring et al., 2011; Hsieh et al., 2011). The epigenetic regulation of genomic imprinting plays an indispensable role in normal endosperm development and seed fertility. The misregulation of imprinted genes affects the sizes of seeds or leads to inviable seeds (Tiwari et al., 2010; Hornslien et al., 2019). The generation of genomic imprinting is mainly caused by the different epigenetic modifications of male and female gametes before fertilization (Batista and Köhler, 2020). When the central cell and sperm cell fuses to form the primordial endosperm nucleus, the differences in epigenetic modification between the male and female genomes result in only one allele being expressed and the other being silenced. The differential loss of DNA methylation in the paternal and maternal alleles produces different chromatin marks in Arabidopsis. For example, the methylated paternal allele can lead to being transcriptionally silent, while the demethylation of maternal allele would become a transcriptionally active state (Kinoshita et al., 2004; Jullien et al., 2006; Tiwari et al., 2008). In Arabidopsis, the expression level of the DNA methyltransferase gene MET1 is low in central cells, whereas the expression level of the demethylase gene DME is high (Huh et al., 2008). Therefore, the central cells maintain a lower DNA methylation level, but the sperm cells maintain a higher DNA methylation level because DME is not expressed (Huh et al., 2008).
RdDM is also critical for silencing of the paternal allele at MEG (maternally expressed imprinted genes) loci; Vu et al. used Col and Cvi to distinguish the parental alleles of Suppressor OF drm1 drm2 cmt3 (SDC) and MOP9.5 (also called AtPI4Kγ3, a type II phosphoinositide 4-kinase), and crossed wild-type ovules with pollen from mutants for RdDM (such as nrpd2a mutant, NRPD2A is the second largest subunit of RNA pol IV and pol V); they observed activation of SDC and MOP9.5 paternal alleles from nrpd2a homozygous plants. Further research found that maternal-specific expression of imprinted genes SDC and MOP9.5 was maintained by MET1. These results suggest that small RNAs have a significant role in setting MEG expression patterns (Vu et al., 2013). PEGs (paternally expressed imprinted genes) can also be hypomethylated at the maternal allele and hypermethylated at the paternal allele (Hsieh et al., 2009; Zhang et al., 2014). So the maternal hypomethylation is essential for the silencing of the maternal allele for many PEGs (Hsieh et al., 2011; Wolff et al., 2011). MEGs are generally more affected by DNA methylation than PEGs (Chen et al., 2018), but the latter is also regulated by histone modification, such as H3K27me3 (Wolff et al., 2011; Zhang et al., 2014). Two other repressive epigenetic marks, H3K9me2 and CHG methylation, also contribute to maternal alleles silencing of PEGs, leading to differential expression of parent-of-origin alleles in the endosperm (Inoue et al., 2017; Moreno-Romero et al., 2019). Silencing of the maternal PHERES 1 (PHE1, a paternally expressed imprinted transcription factor gene) allele depends on the Polycomb Repressive Complex 2 (PRC2), and maternally inherited mutations that encode PRC2 proteins cause biallelic expression of PHE1 (Kohler et al., 2005). The differences in the expression of PRC2 between sperm and central cells resulted in different histone methylation modifications of parental genomes in the endosperm (Luo et al., 2000; Schoft et al., 2011). DNA methylation can prevent H3K27me3 modification and interfere with PRC2 function (Weinhofer et al., 2010; Deleris et al., 2012; Jermann et al., 2014).
Genomic imprinting disruption accompanies endosperm abortion, and the expression of many imprinted genes also changes (Jullien and Berger, 2010; Kradolfer et al., 2013; Florez-Rueda et al., 2016; Tonosaki et al., 2018). Many MEGs affect seed development by regulating endosperm cytogenesis (Niu et al., 2020; Cheng et al., 2021; Tonosaki et al., 2021); whereas most PEGs knock-out mutations generally do not affect normal plant growth and development in Arabidopsis. But PEGs are important for endosperm development in plants, several peg mutants: such as adm (ADMETOS) and peg2 (At1g49290) mutants—can rescue triploid seed abortion (Wolff et al., 2015). And the loss of some PEGs can also lead to serious phenotypic defects. For example, the mutants of PEG1 (Os01g08570, encoding an oxygenase dependent on ketoglutarate and iron), PEG2 (OsFBX365, encoding an F-box domain protein), and PEG3 (OsFBDUF48, encoding a DUF295-domain protein) in rice can reduce starch content and seed fertility (Yuan et al., 2017). The PEGs may be directly involved in regulating reproductive isolation between species. In the endosperm of distant Arabidopsis inter-accession crosses (such as Columbia × Nossen), the expression disorder of PEGs is more significant than that of MEGs (Wolff et al., 2015). In interploidy crosses, some PEGs mutants rescue seed abortion, so they have a dramatically different phenotype than WT (Kradolfer et al., 2013; Wang et al., 2018). Hundreds of possible imprinted genes have been discovered in plants. However, there is still a lack of in-depth research on the biological functions of plant imprinted genes, even though many imprinting genes co-localize with yield-related traits (Yuan et al., 2017). For example, Chen et al. found that the rice grain weight QTL—Grain Weight 2—mainly expressed maternal alleles in the endosperm (Chen et al., 2016; Niu et al., 2020). These studies indicate that both MEGs and PEGs can participate in plant endosperm development.
The DNA methylation of endosperm plays a vital role in regulating seed development and storage material biosynthesis. The removal of imprinted genes can affect endosperm development and lead to seed abortion. In addition, DNA methylation can also regulate endosperm development by regulating the expression of genes and small RNAs. For example, DNA methylation affects starch synthesis in maize endosperm (Hu et al., 2021). DNA methylation also regulates seed size (Rajkumar et al., 2020) and dormancy (Zhu et al., 2018), and it directly affects crop yield and quality. At present, the research on most crops is limited to the regulation of transcription factors, and the research on DNA methylation mainly focuses on model organisms. Although the methylation sequencing of plant endosperm is gradually increasing, the regulatory pathways related to DNA methylation and demethylation in the endosperm are unclear. Therefore, it is recommended to use a combination of methylation sequencing and RNA sequencing (RNA sequencing, single-cell sequencing, small RNA sequencing) to study plant endosperm and establish a complete regulatory network profile. It is of great value to identify the cellular heterogeneity of methylation in plants, but it is still extremely challenging to sequence single-cell DNA methylation in plant endosperm. On the one hand, the presence of seed coat makes it difficult to separate pollution-free endosperm. On the other hand, it is difficult to use bisulfite-transformed DNA fragments by library construction and sequencing for highly methylated and highly repetitive genomes. The regulation of methylation in plant endosperm should be the focus of future research.
DL: Methodology, Investigation, Writing—original draft, Writing—review and editing. JZ: Conceptualization, Methodology, Resources, Funding. MX: Conceptualization, Methodology, Supervision, Writing—review.
This work is supported by the National Key R&D Program of China Grant (2019YFA0903903), and the Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes (2019KSYS006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Ashapkin, V. V., Kutueva, L. I., and Vanyushin, B. F. (2016). Plant DNA Methyltransferase Genes: Multiplicity, Expression, Methylation Patterns. Biochem. Mosc. 81 (2), 141–151. doi:10.1134/S0006297916020085
Batista, R. A., and Köhler, C. (2020). Genomic Imprinting in Plants-Revisiting Existing Models. Genes Dev. 34, 24–36. doi:10.1101/gad.33292410.1101/gad.332924.119
Bleckmann, A., Alter, S., and Dresselhaus, T. (2014). The Beginning of a Seed: Regulatory Mechanisms of Double Fertilization. Front. Plant Sci. 5, 452. doi:10.3389/fpls.2014.00452
Brown, R. C., Lemmon, B. E., Nguyen, H., and Olsen, O.-A. (1999). Development of Endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12 (1), 32–42. doi:10.1007/s004970050169
Brown, R. C., Lemmon, B. E., and Olsen, O.-A. (1996). Development of the Endosperm in rice (Oryza Sativa L.): Cellularization. J. Plant Res. 109 (3), 301–313. doi:10.1007/BF02344477
Chen, C., Begcy, K., Liu, K., Folsom, J. J., Wang, Z., Zhang, C., et al. (2016). Heat Stress Yields a Unique MADS Box Transcription Factor in Determining Seed Size and Thermal Sensitivity. Plant Physiol. 171 (1), 606–622. doi:10.1104/pp.15.01992
Chen, C., Li, T., Zhu, S., Liu, Z., Shi, Z., Zheng, X., et al. (2018). Characterization of Imprinted Genes in Rice Reveals Conservation of Regulation and Imprinting with Other Plant Species. Plant Physiol. 177 (4), 1754–1771. doi:10.1104/pp.17.01621
Cheng, X., Pan, M., E, Z., Zhou, Y., Niu, B., and Chen, C. (2021). The Maternally Expressed Polycomb Group Gene OsEMF2a Is Essential for Endosperm Cellularization and Imprinting in rice. Plant Commun. 2 (1), 100092. doi:10.1016/j.xplc.2020.100092
Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J. J., Goldberg, R. B., et al. (2002). DEMETER, a DNA Glycosylase Domain Protein, Is Required for Endosperm Gene Imprinting and Seed Viability in Arabidopsis. Cell. 110 (1), 33–42. doi:10.1016/s0092-8674(02)00807-3
De Giorgi, J., Piskurewicz, U., Loubery, S., Utz-Pugin, A., Bailly, C., Mène-Saffrané, L., et al. (2015). An Endosperm-Associated Cuticle Is Required for Arabidopsis Seed Viability, Dormancy and Early Control of Germination. Plos Genet. 11 (12), e1005708. doi:10.1371/journal.pgen.1005708
Deleris, A., Stroud, H., Bernatavichute, Y., Johnson, E., Klein, G., Schubert, D., et al. (2012). Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in Arabidopsis thaliana. Plos Genet. 8 (11), e1003062. doi:10.1371/journal.pgen.1003062
Feil, R., and Berger, F. (2007). Convergent Evolution of Genomic Imprinting in Plants and Mammals. Trends Genet. 23 (4), 192–199. doi:10.1016/j.tig.2007.02.004
Florez-Rueda, A. M., Paris, M., Schmidt, A., Widmer, A., Grossniklaus, U., and Städler, T. (2016). Genomic Imprinting in the Endosperm Is Systematically Perturbed in Abortive Hybrid Tomato Seeds. Mol. Biol. Evol. 33 (11), 2935–2946. doi:10.1093/molbev/msw175
Gehring, M., Bubb, K. L., and Henikoff, S. (2009). Extensive Demethylation of Repetitive Elements during Seed Development Underlies Gene Imprinting. Science. 324 (5933), 1447–1451. doi:10.1126/science.1171609
Gehring, M., Missirian, V., and Henikoff, S. (2011). Genomic Analysis of Parent-Of-Origin Allelic Expression in Arabidopsis thaliana Seeds. Plos One. 6 (8), e23687. doi:10.1371/journal.pone.0023687
Gong, Z., Morales-Ruiz, T., Ariza, R. R., Roldán-Arjona, T., David, L., and Zhu, J.-K. (2002). ROS1, a Repressor of Transcriptional Gene Silencing in Arabidopsis, Encodes a DNA Glycosylase/Lyase. Cell. 111 (6), 803–814. doi:10.1016/S0092-8674(02)01133-9
Greenwood, J. S., and Bewley, J. D. (1982). Seed Development in Ricinus communis (castor Bean). I. Descriptive Morphology. Can. J. Bot. 60 (9), 1751–1760. doi:10.1139/b82-222
Haag, J. R., and Pikaard, C. S. (2011). Multisubunit RNA Polymerases IV and V: Purveyors of Non-coding RNA for Plant Gene Silencing. Nat. Rev. Mol. Cell Biol. 12 (8), 483–492. doi:10.1038/nrm3152
Hornslien, K. S., Miller, J. R., and Grini, P. E. (2019). Regulation of Parent-Of-Origin Allelic Expression in the Endosperm. Plant Physiol. 180 (3), 1498–1519. doi:10.1104/pp.19.00320
Hsieh, T.-F., Ibarra, C. A., Silva, P., Zemach, A., Eshed-Williams, L., Fischer, R. L., et al. (2009). Genome-Wide Demethylation of Arabidopsis Endosperm. Science. 324 (5933), 1451–1454. doi:10.1126/science.1172417
Hsieh, T.-F., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M. J., et al. (2011). Regulation of Imprinted Gene Expression in Arabidopsisendosperm. Proc. Natl. Acad. Sci. USA. 108 (5), 1755–1762. doi:10.1073/pnas.1019273108
Hu, Y., Li, Y., Weng, J., Liu, H., Yu, G., Liu, Y., et al. (2021). Coordinated Regulation of Starch Synthesis in Maize Endosperm by microRNAs and DNA Methylation. Plant J. 105 (1), 108–123. doi:10.1111/tpj.15043
Huh, J. H., Bauer, M. J., Hsieh, T.-F., and Fischer, R. L. (2008). Cellular Programming of Plant Gene Imprinting. Cell. 132 (5), 735–744. doi:10.1016/j.cell.2008.02.018
Ibarra, C. A., Feng, X., Schoft, V. K., Hsieh, T.-F., Uzawa, R., Rodrigues, J. A., et al. (2012). Active DNA Demethylation in Plant Companion Cells Reinforces Transposon Methylation in Gametes. Science. 337 (6100), 1360–1364. doi:10.1126/science.1224839
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017). Maternal H3K27me3 Controls DNA Methylation-Independent Imprinting. Nature. 547 (7664), 419–424. doi:10.1038/nature23262
Jermann, P., Hoerner, L., Burger, L., and Schubeler, D. (2014). Short Sequences Can Efficiently Recruit Histone H3 Lysine 27 Trimethylation in the Absence of Enhancer Activity and DNA Methylation. Proc. Natl. Acad. Sci. 111 (33), E3415–E3421. doi:10.1073/pnas.1400672111
Jin, B., Li, Y., and Robertson, K. D. (2011). DNA Methylation: superior or Subordinate in the Epigenetic Hierarchy? Genes & Cancer. 2 (6), 607–617. doi:10.1177/1947601910393957
Jullien, P. E., and Berger, F. (2010). Parental Genome Dosage Imbalance Deregulates Imprinting in Arabidopsis. Plos Genet. 6 (3), e1000885. doi:10.1371/journal.pgen.1000885
Jullien, P. E., Katz, A., Oliva, M., Ohad, N., and Berger, F. (2006). Polycomb Group Complexes Self-Regulate Imprinting of the Polycomb Group Gene MEDEA in Arabidopsis. Curr. Biol. 16 (5), 486–492. doi:10.1016/j.cub.2006.01.020
Jullien, P. E., Susaki, D., Yelagandula, R., Higashiyama, T., and Berger, F. (2012). DNA Methylation Dynamics During Sexual Reproduction in Arabidopsis thaliana. Curr. Biol. 22 (19), 1825–1830. doi:10.1016/j.cub.2012.07.061
Kang, I.-H., Steffen, J. G., Portereiko, M. F., Lloyd, A., and Drews, G. N. (2008). The AGL62 MADS Domain Protein Regulates Cellularization During Endosperm Development inArabidopsis. Plant Cell. 20 (3), 635–647. doi:10.1105/tpc.107.055137
Kawakatsu, T., Nery, J. R., Castanon, R., and Ecker, J. R. (2017). Dynamic DNA Methylation Reconfiguration During Seed Development and Germination. Genome Biol. 18 (1), 171. doi:10.1186/s13059-017-1251-x
Kawashima, T., and Berger, F. (2014). Epigenetic Reprogramming in Plant Sexual Reproduction. Nat. Rev. Genet. 15 (9), 613–624. doi:10.1038/nrg3685
Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S. E., et al. (2004). One-Way Control of FWA Imprinting in Arabidopsis Endosperm by DNA Methylation. Science. 303 (5657), 521–523. doi:10.1126/science.1089835
Köhler, C., Page, D. R., Gagliardini, V., and Grossniklaus, U. (2005). The Arabidopsis thaliana MEDEA Polycomb Group Protein Controls Expression of PHERES1 by Parental Imprinting. Nat. Genet. 37 (1), 28–30. doi:10.1038/ng1495
Kradolfer, D., Wolff, P., Jiang, H., Siretskiy, A., and Köhler, C. (2013). An Imprinted Gene Underlies Postzygotic Reproductive Isolation in Arabidopsis thaliana. Developmental Cell. 26 (5), 525–535. doi:10.1016/j.devcel.2013.08.006
Kuo, H. Y., Jacobsen, E. L., Long, Y., Chen, X., and Zhai, J. (2017). Characteristics and Processing of Pol IV-dependent Transcripts in Arabidopsis. J. Genet. Genomics. 44 (1), 3–6. doi:10.1016/j.jgg.2016.10.009
Lauria, M., Rupe, M., Guo, M., Kranz, E., Pirona, R., Viotti, A., et al. (2004). Extensive Maternal DNA Hypomethylation in the Endosperm of Zea Mays. Plant Cell. 16 (2), 510–522. doi:10.1105/tpc.017780
Law, J. A., and Jacobsen, S. E. (2010). Establishing, Maintaining and Modifying DNA Methylation Patterns in Plants and Animals. Nat. Rev. Genet. 11 (3), 204–220. doi:10.1038/nrg2719
Lu, X., Wang, W., Ren, W., Chai, Z., Guo, W., Chen, R., et al. (2015). Genome-Wide Epigenetic Regulation of Gene Transcription in Maize Seeds. PLoS One. 10 (10), e0139582. doi:10.1371/journal.pone.0139582
Luo, M., Bilodeau, P., Dennis, E. S., Peacock, W. J., and Chaudhury, A. (2000). Expression and Parent-Of-Origin Effects for FIS2, MEA, and FIE in the Endosperm and Embryo of Developing Arabidopsis Seeds. Proc. Natl. Acad. Sci. 97 (19), 10637–10642. doi:10.1073/pnas.170292997
Martínez, G., Panda, K., Köhler, C., and Slotkin, R. K. (2016). Silencing in Sperm Cells Is Directed by RNA Movement from the Surrounding Nurse Cell. Nat. Plants. 2, 16030. doi:10.1038/nplants.2016.30
Matzke, M. A., and Mosher, R. A. (2014). RNA-Directed DNA Methylation: an Epigenetic Pathway of Increasing Complexity. Nat. Rev. Genet. 15 (6), 394–408. doi:10.1038/nrg3683
McCue, A. D., Nuthikattu, S., Reeder, S. H., and Slotkin, R. K. (2012). Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA. Plos Genet. 8 (2), e1002474. doi:10.1371/journal.pgen.1002474
Moore, L. D., Le, T., and Fan, G. (2013). DNA Methylation and its Basic Function. Neuropsychopharmacol. 38 (1), 23–38. doi:10.1038/npp.2012.112
Moreno‐Romero, J., Jiang, H., Santos‐González, J., and Köhler, C. (2016). Parental Epigenetic Asymmetry of PRC 2‐Mediated Histone Modifications in the Arabidopsis Endosperm. EMBO J. 35 (12), 1298–1311. doi:10.15252/embj.201593534
Moreno-Romero, J., Del Toro-De León, G., Yadav, V. K., Santos-González, J., and Köhler, C. (2019). Epigenetic Signatures Associated with Imprinted Paternally Expressed Genes in the Arabidopsis Endosperm. Genome Biol. 20 (1), 41. doi:10.1186/s13059-019-1652-0
Niu, B., Deng, H., Li, T., Sharma, S., Yun, Q., Li, Q., et al. (2020). OsbZIP76 Interacts With OsNF‐YBs and Regulates Endosperm Cellularization in rice (Oryza Sativa). J. Integr. Plant Biol. 62 (12), 1983–1996. doi:10.1111/jipb.12989
Park, K., Kim, M. Y., Vickers, M., Park, J.-S., Hyun, Y., Okamoto, T., et al. (2016). DNA Demethylation Is Initiated in the Central Cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. USA. 113 (52), 15138–15143. doi:10.1073/pnas.1619047114
Pignatta, D., Erdmann, R. M., Scheer, E., Picard, C. L., Bell, G. W., and Gehring, M. (2014). Natural Epigenetic Polymorphisms lead to Intraspecific Variation in Arabidopsis Gene Imprinting. Elife. 3, e03198. doi:10.7554/eLife.03198
Rajkumar, M. S., Gupta, K., Khemka, N. K., Garg, R., and Jain, M. (2020). DNA Methylation Reprogramming During Seed Development and its Functional Relevance in Seed Size/Weight Determination in Chickpea. Commun. Biol. 3 (1), 340. doi:10.1038/s42003-020-1059-1
Schoft, V. K., Chumak, N., Choi, Y., Hannon, M., Garcia-Aguilar, M., Machlicova, A., et al. (2011). Function of the DEMETER DNA Glycosylase in the Arabidopsis thaliana Male Gametophyte. Proc. Natl. Acad. Sci. 108 (19), 8042–8047. doi:10.1073/pnas.1105117108
Tiwari, S., Schulz, R., Ikeda, Y., Dytham, L., Bravo, J., Mathers, L., et al. (2008). Maternally Expressed Pab C-Terminal, a Novel Imprinted Gene Inarabidopsis, Encodes the Conserved C-Terminal Domain of Polyadenylate Binding Proteins. Plant Cell. 20 (9), 2387–2398. doi:10.1105/tpc.108.061929
Tiwari, S., Spielman, M., Schulz, R., Oakey, R. J., Kelsey, G., Salazar, A., et al. (2010). Transcriptional Profiles Underlying Parent-Of-Origin Effects in Seeds of Arabidopsis thaliana. BMC Plant Biol. 10 (1), 72. doi:10.1186/1471-2229-10-72
Tonosaki, K., Ono, A., Kunisada, M., Nishino, M., Nagata, H., Sakamoto, S., et al. (2021). Mutation of the Imprinted Gene OsEMF2a Induces Autonomous Endosperm Development and Delayed Cellularization in rice. Plant Cell. 33 (1), 85–103. doi:10.1093/plcell/koaa006
Tonosaki, K., Sekine, D., Ohnishi, T., Ono, A., Furuumi, H., Kurata, N., et al. (2018). Overcoming the Species Hybridization Barrier by Ploidy Manipulation in the GenusOryza. Plant J. 93 (3), 534–544. doi:10.1111/tpj.13803
Vu, T. M., Nakamura, M., Calarco, J. P., Susaki, D., Lim, P. Q., Kinoshita, T., et al. (2013). RNA-directed DNA Methylation Regulates Parental Genomic Imprinting at Several Loci in Arabidopsis. Development. 140 (14), 2953–2960. doi:10.1242/dev.092981
Wang, G., Jiang, H., Del Toro de León, G., Martinez, G., and Köhler, C. (2018). Sequestration of a Transposon-Derived siRNA by a Target Mimic Imprinted Gene Induces Postzygotic Reproductive Isolation in Arabidopsis. Developmental Cell. 46 (6), 696–705. doi:10.1016/j.devcel.2018.07.014
Weinhofer, I., Hehenberger, E., Roszak, P., Hennig, L., and Köhler, C. (2010). H3K27me3 Profiling of the Endosperm Implies Exclusion of Polycomb Group Protein Targeting by DNA Methylation. Plos Genet. 6 (10), e1001152. doi:10.1371/journal.pgen.1001152
Wolff, P., Jiang, H., Wang, G., Santos-González, J., and Köhler, C. (2015). Paternally Expressed Imprinted Genes Establish Postzygotic Hybridization Barriers in Arabidopsis thaliana. Elife. 4, e10074. doi:10.7554/eLife.10074
Wolff, P., Weinhofer, I., Seguin, J., Roszak, P., Beisel, C., Donoghue, M. T. A., et al. (2011). High-resolution Analysis of Parent-Of-Origin Allelic Expression in the Arabidopsis Endosperm. Plos Genet. 7 (6), e1002126. doi:10.1371/journal.pgen.1002126
Xu, W., Yang, T., Dong, X., Li, D.-Z., and Liu, A. (2016). Genomic DNA Methylation Analyses Reveal the Distinct Profiles in Castor Bean Seeds With Persistent Endosperms. Plant Physiol. 171 (2), 1242–1258. doi:10.1104/pp.16.00056
Yan, D., Duermeyer, L., Leoveanu, C., and Nambara, E. (2014). The Functions of the Endosperm During Seed Germination. Plant Cell Physiol. 55 (9), 1521–1533. doi:10.1093/pcp/pcu089
Yuan, J., Chen, S., Jiao, W., Wang, L., Wang, L., Ye, W., et al. (2017). Both Maternally and Paternally Imprinted Genes Regulate Seed Development in rice. New Phytol. 216 (2), 373–387. doi:10.1111/nph.14510
Zemach, A., Kim, M. Y., Silva, P., Rodrigues, J. A., Dotson, B., Brooks, M. D., et al. (2010). Local DNA Hypomethylation Activates Genes in rice Endosperm. Proc. Natl. Acad. Sci. 107 (43), 18729–18734. doi:10.1073/pnas.1009695107
Zhai, J., Bischof, S., Wang, H., Feng, S., Lee, T.-f., Teng, C., et al. (2015). A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell. 163 (2), 445–455. doi:10.1016/j.cell.2015.09.032
Zhang, H., Lang, Z., and Zhu, J.-K. (2018). Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cell Biol. 19 (8), 489–506. doi:10.1038/s41580-018-0016-z
Zhang, M., Xie, S., Dong, X., Zhao, X., Zeng, B., Chen, J., et al. (2014). Genome-wide High Resolution Parental-specific DNA and Histone Methylation Maps Uncover Patterns of Imprinting Regulation in maize. Genome Res. 24 (1), 167–176. doi:10.1101/gr.155879.113
Keywords: DNA methylation, demethylation, imprinted genes, endosperm, symmetrical methylation, non-symmetrical methylation
Citation: Lu D, Zhai J and Xi M (2022) Regulation of DNA Methylation During Plant Endosperm Development. Front. Genet. 13:760690. doi: 10.3389/fgene.2022.760690
Received: 18 August 2021; Accepted: 17 January 2022;
Published: 10 February 2022.
Edited by:
Joao Batista Teixeira da Rocha, Federal University of Santa Maria, BrazilReviewed by:
Igor Kovalchuk, University of Lethbridge, CanadaCopyright © 2022 Lu, Zhai and Xi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jixian Zhai, emhhaWp4QHN1c3RlY2guZWR1LmNu; Mengli Xi, eGltZW5nbGluamZ1QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.