
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 16 March 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.758304
This article is part of the Research Topic Host-Microbe Interaction and Coevolution View all 11 articles
Understanding how Mycobacterium tuberculosis has evolved into a professional pathogen is helpful in studying its pathogenesis and for designing vaccines. We investigated how the evolutionary adaptation of M. smegmatis mc251 to an important clinical stressor H2O2 allows bacteria to undergo coordinated genetic mutations, resulting in increased pathogenicity. Whole-genome sequencing identified a mutation site in the fur gene, which caused increased expression of katG. Using a Wayne dormancy model, mc251 showed a growth advantage over its parental strain mc2155 in recovering from dormancy under anaerobic conditions. Meanwhile, the high level of KatG in mc251 was accompanied by a low level of ATP, which meant that mc251 is at a low respiratory level. Additionally, the redox-related protein Rv1996 showed different phenotypes in different specific redox states in M. smegmatis mc2155 and mc251, M. bovis BCG, and M. tuberculosis mc27000. In conclusion, our study shows that the same gene presents different phenotypes under different physiological conditions. This may partly explain why M. smegmatis and M. tuberculosis have similar virulence factors and signaling transduction systems such as two-component systems and sigma factors, but due to the different redox states in the corresponding bacteria, M. smegmatis is a nonpathogen, while M. tuberculosis is a pathogen. As mc251 overcomes its shortcomings of rapid removal, it can potentially be developed as a vaccine vector.
Tuberculosis (TB) is caused by the pathogen Mycobacterium tuberculosis and remains a public health threat, resulting in 1.4 million deaths in 2020 (WHO, 2021). The prevalence of multidrug-resistant M. tuberculosis and the rising cases of co-infection with HIV increase this health concern. The World Health Organization (WHO) has estimated that a quarter of the world’s population is infected with M. tuberculosis (WHO, 2021). This latent state may be extended as long as the life of the infected host, but unfortunately, the reactive rate is approximately 5–10% of infected individuals (Flynn and Chan, 2001). TB is treated with chemotherapy, and the latent state of mycobacteria prolongs the time of treatment, which is one of the causes for the development of mycobacterial resistance.
As one of the world’s most successful human pathogens, M. tuberculosis has evolved elegant strategies to escape the immune defensive system of the host. For example, D’Arcy et al. observed that M. tuberculosis-containing phagosomes do not fuse with the lysosome inside infected macrophages (Brown et al., 1969; Armstrong and Hart, 1971). Several studies have indicated that M. tuberculosis being an intracellular pathogen is partially due to its ability to survive and persist in macrophages, in hostile environments with oxidative stress, in low pH, and under starvation and other stresses (Cohen et al., 2018; Nauseef, 2019). When M. tuberculosis infects macrophages, mycobacteria must overcome exogenous reactive oxygen species (ROS), a classical innate defense mechanism against infection (Pieters, 2008). In addition, during latency, M. tuberculosis continues to be exposed to oxidative stress, and thus, the accumulation of mutations caused by oxidative DNA damage is predicted as a potential risk for resistance to antibiotics (Ford et al., 2011). Clinical investigation has shown that cells from TB patients produce less ROS than those of healthy individuals (Jaswal et al., 1992; Kumar et al., 1995). In addition, ROS can also be associated with the treatment of TB (Piccaro et al., 2014; Hu et al., 2021). To avoid ROS attack, an evolved detoxified system is essential for M. tuberculosis survival, persistence, and subsequent reactivation (Piccaro et al., 2014).
As a mycobacterial model of M. tuberculosis, M. smegmatis has contributed to understanding the functions of mycobacteria (Aldridge et al., 2012; Kieser et al., 2015; Gray et al., 2016). The essential genes in M. tuberculosis have a corresponding ortholog gene in M. smegmatis (Dragset et al., 2019; Judd et al., 2021). M. smegmatis mc2155 and M. tuberculosis H37Rv share 2547 mutually orthologous genes (Dragset et al., 2019). At least under certain conditions, M. tuberculosis and M. smegmatis have similar growth mechanisms. The assumed virulence factors identified in M. tuberculosis such as PhoPR and DosR/S/T (Gonzalo-Asensio et al., 2014; Mehra et al., 2015) are also present in M. smegmatis. However, compared to virulent M. tuberculosis, M. smegmatis is a nonpathogenic mycobacterium. M. tuberculosis can persist in the infected host, while the host can quickly remove M. smegmatis (Anes et al., 2003; Anes et al., 2006). Considering that M. tuberculosis with high resistance to hydrogen peroxide (H2O2) persists in the lungs, while M. smegmatis with lower resistance to H2O2 exists in the soil, we hypothesized that the redox state of M. tuberculosis and M. smegmatis may adapt to the corresponding redox environment. The differences between the two bacteria are due to their corresponding degree of resistance to H2O2. To test this hypothesis, we selected a series of H2O2-resistant mutant strains using a clinically important stressor H2O2 and identified the highly H2O2-resistant mycobacterial strain mc251 (Li et al., 2014b). Compared to the wild-type M. smegmatis mc2155 strain, the minimum inhibitory concentration (MIC) of H2O2 was more than 80-fold higher, while the MIC of mc251 to H2O2 was 3.125 mM, similar to that of M. bovis BCG (0.625 mM) (Li et al., 2014a) and M. tuberculosis (0.625 mM), while that of mc2155 was 0.039 mM (Li et al., 2014b). mc251 exhibited a slow growth rate similar to M. tuberculosis.
In this study, we first showed that H2O2-resistant mc251 had a growth advantage both in mice and macrophages compared with wild-type mc2155, which indicated that the higher resistance to H2O2 of mycobacteria is related to higher virulence. Similar to M. tuberculosis that can survive under hypoxia in the lungs and resuscitate under appropriate conditions, mc251 presented a growth advantage to recover from dormancy using the Wayne dormancy model. Furthermore, we showed that Fur mutant carrying the A28V point mutation dysregulated katG levels, which was the main cause for resistance to H2O2 and low levels of ATP. Additionally, the redox-related protein Rv1996, responsible for regulating gene expression, exhibited different phenotypes associated with isoniazid susceptibility in M. smegmatis mc2155 and mc251, M. bovis BCG, and M. tuberculosis mc27000. Thus, the same protein presents different phenotypes under different physiological conditions. Our results suggest that the difference in the corresponding redox status causes the difference in pathogenicity between M. tuberculosis and M. smegmatis.
Previous studies showed that M. smegmatis with the gain of H2O2 resistance, named mc251, improves growth fitness in mycobacteria under stress (Table 1) (Li et al., 2014a; Li et al., 2014b). Prior to selecting H2O2-adapted mycobacterial mutants, we first measured the MIC of wild-type mc2155 to H2O2, which was 0.039 mM. We used 0.0293 mM H2O2 for the initial screening, which was lower than the MIC of mc2155 to H2O2 (Table 1). The whole process was performed as follows: cultures were started from glycerol-frozen stocks and grown to log phase (OD600 of 0.6–0.8). Then, the culture was diluted 1,000 times and grown under 0.0293 mM H2O2 until the OD600 reached logarithmic (log) phase. Cultures were then further diluted 1:1,000, and an additional 0.0293 mM of H2O2 was added to the culture. This process was repeated until the H2O2 concentrations reached 0.4395 mM. In further rounds of culture, H2O2 was added in steps of 0.0879 mM instead of 0.0293 mM until a concentration of 1.5 mM was reached (Supplementary Figure S1; Supplementary Table S1). The actual MIC of H2O2 of the mutant strain, named mc251, selected at 1.5 mM, was 3.125 mM. In a previous study, the mutated mycobacterial strain, mc251, evolved into an M. tuberculosis-like strain that presented slow growth and improved growth fitness under stress (Li et al., 2014b), which H2O2 resistance in mycobacteria linked to virulence. Previous studies including ours have associated isoniazid (INH) with H2O2 resistance (Timmins and Deretic, 2006; Hu et al., 2021). The MIC of isoniazid (INH) for mc251 was dramatically reduced to ∼1% of the MIC for mc2155 (Table 1). To test our hypothesis that the H2O2-resistant strain mc251 would be more persistent in the host than its parental strain mc2155, we performed non-invasive intranasal infections, which can induce respiratory mucosal immune responses and is a promising way of vaccination for respiratory infection diseases. As shown in Figure 1A, intranasal infection of C57BL/6 mice with ∼1 × 107/50 μl and bacterial colony-forming units (CFUs) were counted 1 day after infection. Compared to mc2155 with the percentage of survival in the infected lung of 0.03653 ± 0.01462% (n = 3), the mc251 strain with that of 0.7153 ± 0.2597% (n = 3) had a significantly higher survival percentage (p = 0.0451) (Figure 1B). After infection with live mycobacteria, the bacilli enter alveolar macrophages (AMΦs) and persist within them. To exclude the possibility that the higher CFUs were caused by the larger number of infected bacilli being captured by AMΦs, macrophage-killing assays were explored using the THP-1 cell line. The survival percentage of mutant mc251 is 0.3158 ± 0.1502 (n = 9), while the survival of wild-type mc2155 was 0.01921 ± 0.01223 (n = 6) (p = 0.0003) (Figure 1C). These results indicated that mc251 exhibited enhanced virulence.
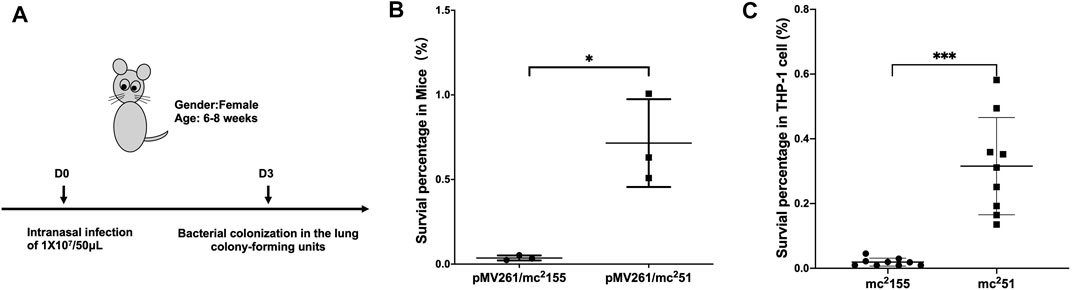
FIGURE 1. M. smegmatis mc251 displayed improved virulence in mice and THP-1 cells. (A) The schematic diagram of the mice infection. C57BL/6 mice were intranasally infected as described in Materials and Methods with ∼1 × 107/50 μl of M. smegmatis mc2155 or mc251. At the 3rd day after infection, the whole lung homogenates were plated to determine bacterial numbers. (B) The H2O2-resistant mutant strain mc251 has a significantly higher bacterial load in the infected lung. The lung burdens 3 days after infection with mc2155 (black circles) or mc251 (black squares) were measured, and the survival percentage was calculated as described in Materials and Methods. Each group consisted of three mice. *p < 0.05. (C) The H2O2-resistant mutant strain mc251 has a survival advantage over wild-type mc2155 in the macrophage-like cell line THP-1. THP-1 cells were infected with mc2155 (black circles) or mc251 (black squares) at a multiplicity of infection of 1 h after infection, THP-1 cell lysates were collected, and the intracellular bacilli were measured, and the survival percentage was calculated as described in Materials and Methods. Each group consisted of nine repeats. ***p < 0.001.
M. tuberculosis has evolved to survive in hypoxic conditions. Over more than 100 years of research, M. tuberculosis has been confirmed to be an obligate aerobic bacterium that cannot replicate under hypoxic conditions. However, M. tuberculosis has incredible survivability in long-term anaerobic environments. Evidence suggests that M. tuberculosis has the ability to reduce the respiratory system to low levels and maintain vitality (Loebel et al., 1933). Due to respiratory depression, ATP is maintained at low levels, which guarantees minimal metabolic activity to ensure membrane integrity under hypoxic conditions. We first compared the survival of mc251 and mc2155 strains under hypoxic conditions. We established a Wayne dormancy model (Wayne and Hayes, 1996), consisting of mycobacterial strain cultures grown in the 7H9 medium to an OD600 of 1.0, which were then transferred to anaerobic tubes containing 1 × 106 cells/ml with a headspace ratio of the culture system of 0.5. Methylene blue (1.5 mg/L) was added as an indicator of oxygen status. The OD600 and CFUs at different indicated times and the color transition time were measured. The indicator of mc251 had become colorless on the 6th day (141 h); on the contrary, the indicator of mc2155 was still in blue at this time (Figure 2A). For mc2155, the indicator became colorless in about 8 days (189 h), indicating that it has entered an anaerobic state. The mc251 entered the anaerobic state faster in the later stages, and the number of viable bacteria in the anaerobic state was significantly higher than mc2155 (Figures 2B,C).
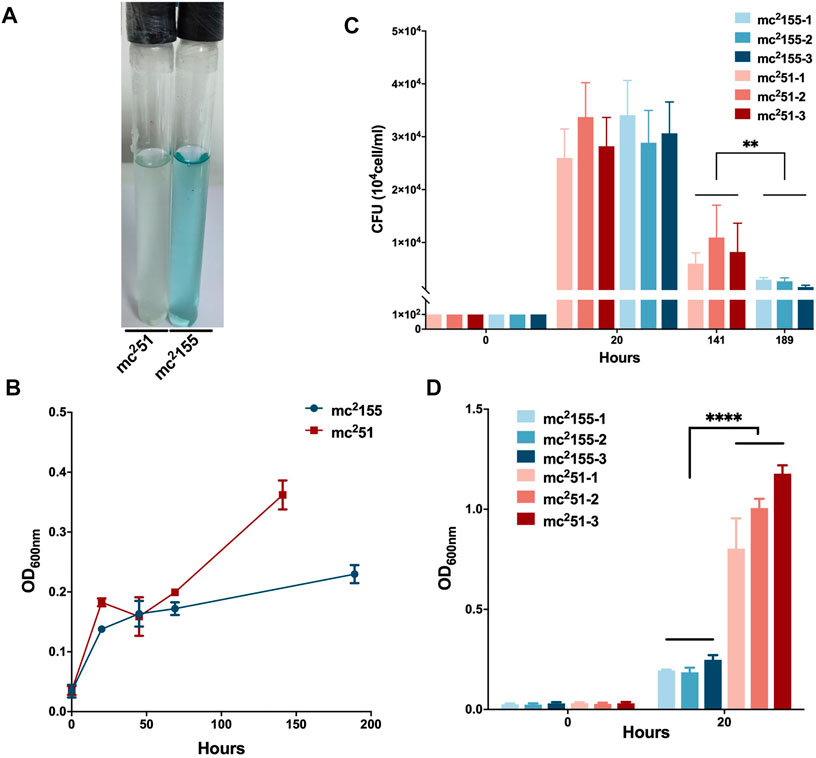
FIGURE 2. Survival in hypoxic environments and resuscitation of M. smegmatis mc2 155 and mc251. (A) The oxygen tension indicator methylene blue of mc251 culture (left) changes to colorless on day 6, while the indicator of mc2155 culture (right) stays blue in the Wayne dormancy model. The cultures were initially inoculated in anaerobic tubes at an OD600 of 0.01 with a headspace ratio of 0.5. The cultures were stirred at 120 rpm. Methylene blue (1.5 mg/L) was used as an oxygen tension indicator. Methylene blue changes from blue to colorless under reducing conditions. Data present results of three biological replicates. Growth rates of strains mc2155 (navy blue) and mc251 (brown) were measured by measuring the OD600 (B) and by determination of CFUs (C) after plating on 7H10. Data are presented as the mean ± standard deviation of three independent replicates. **p < 0.01. (D) The H2O2-resistant mutant strain mc251 shows a growth advantage of recovering from dormancy in an anaerobic state over mc2155. After the indicator methylene blue in the culture of the Wayne dormancy model became colorless, cultures of mc2155 (navy blue) or mc251 (brown) were collected and reinoculated into 7H9 combined with the heart infusion medium at an OD600 of 0.01 and aerobic shaken at 200 rpm. The CFUs were determined 20 h after inoculation. Data are presented as the mean ± standard deviation of three independent replicates. ****p < 0.0001.
For the activation experiment, bacteria cultured in the anaerobic conditions were collected and diluted to 1 × 106 cells/ml in 7H9 and the brain–heart infusion medium and were then grown under aerobic conditions. The three independent mc251 clones were set, and each clone set up three replicates, all of which were grown better than the three independent clones of mc2155 (Figure 2D). Thus, mc251 had the growth advantage of recovering from the dormancy of the anaerobic state over mc2155. We also measured the intracellular ATP levels of mc251 and mc2155. As expected, the abundance of ATP in mc251 (0.1885 ± 0.0481 μM/mg) was lower than that of mc2155 (0.8138 ± 0.1324 μM/mg) (Figure 3B).
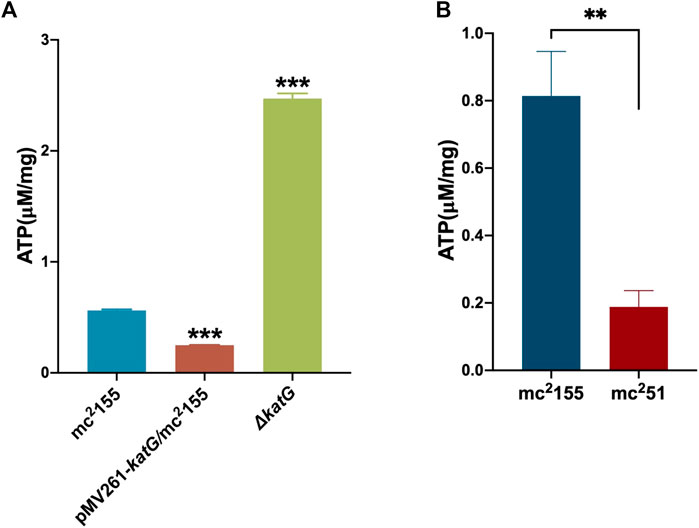
FIGURE 3. ATP content in mc251 is similar to that in pMV261-katG/mc2155. (A) The detection of ATP content in mc2155, ΔkatG, and pMV261-katG/mc2155. The cultures of mc2155 (navy blue), pMV261-katG/mc2155 (brown), and ∆katG (olive drab) at an OD600 of 0.8 were collected. ATP levels were measured in relative light unit (RLU) using a Cytation 3 Cell Imaging Multi-Mode Reader. The corresponding ATP concentrations were calculated according to the ATP standard curve and further converted to μM/mg protein. Data are presented as the mean ± standard deviation of three independent replicates. ***p < 0.001. (B) The intracellular ATP content of mc251 and mc2155. The cultures of mc2155 (navy blue) or mc251 (brown) at an OD600 of 0.8 were collected. ATP levels were measured in relative light unit (RLU) using a Cytation 3 Cell Imaging Multi-Mode Reader. The corresponding ATP concentrations were calculated according to the ATP standard curve and further converted to μM/mg protein. Data are presented as the mean ± standard deviation of three independent replicates. **p < 0.01.
We previously performed whole-genome sequencing to compare differences in mc2155 and mc251 at the genome level. Whole-genome sequencing revealed that there were 29 single-nucleotide polymorphisms (SNPs) in mc251. All 29 SNPs were cloned, and each was transformed into mc2155 strains, and only the fur (msmeg_3460), located upstream of msmeg_3461, encoding KatG (Figure 4A), could restore the resistance phenotype of H2O2 (Li et al., 2014b). The fur-encoded protein Fur negatively regulated katG expression (Pym et al., 2001). The A28V Fur mutation (mFur) in mc251 may also affect the expression of katG. To verify this hypothesis, we examined the binding of mFur to the target DNA (the promoter region of the fur) using electrophoretic mobility shift assays (EMSAs). Compared to wild-type Fur protein, EMSA showed that mFur decreased DNA binding (Figure 4B), which resulted in the katG transcription dysregulation by mFur. In addition, the RNA of mc2155 and mc251 was extracted and quantified. The results showed that compared to the mc2155 strain, the expression of the catalase–peroxidase (KatG) encoding gene katG of the mc251 strain was significantly upregulated to ∼61.82-fold that of the wild-type strain. Taken together, mFur increases the KatG protein level in mc251. KatG is a dual enzyme for catalase and peroxidase, which hydrolyzes ROS (Ng et al., 2004). Thus, the mc251 may maintain ATP at lower levels through KatG, compared to mc2155 levels; that is, the abundance of KatG may affect the mycobacterial redox state and, thus, change the susceptibility to H2O2. We then constructed the ΔkatG (mc2155 with knockout katG) and pMV261-katG/mc2155 (mc2155 with overexpression of katG) strains, and their respective ATP content was tested. As shown in Figure 3A, the KatG level negatively correlated with the ATP level. Furthermore, we constructed the specific site mutant of the fur gene (mfur) in wild-type mc2155 causing an amino acid change of A28V of Fur (Figures 5A,B) by using recombination protein gp61 from Che9c mycobacteriophage (van Kessel et al., 2008), to construct mc2155-mfur (Table 1). As we expected, the Fur mutation at A28V induced high resistance to H2O2 with the MIC of H2O2 in mc2155-mfur being 0.64 mM. We showed that the point mutation of fur dysregulation of katG expression is a major factor leading to the phenotype H2O2 resistance.
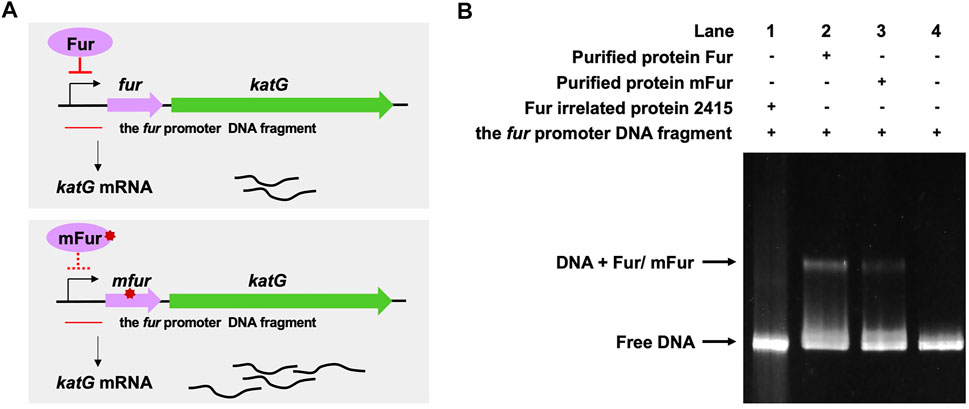
FIGURE 4. A28V Fur mutant protein decreased DNA binding to the fur promoter. (A) Genetic organization of the fur-katG and the schematic diagram of Fur negative regulation of katG (upper panel). Genetic organization of the mfur-katG and the schematic diagram of mFur resulted in derepression of katG with increasing katG mRNA (bottom panel). The red line indicates the fur promoter DNA fragment. (B) Electrophoretic mobility shift assays (EMSAs) of the binding of Fur/mFur protein to the fur promoter DNA fragment. Purified MSMEG_2415 protein (2415), irrelated to Fur, expressed in E. coli, was run in the first lane of a 4–20% polyacrylamide gel. Gel shift caused by Fur (lane 2) and mFur (lane 3) is shown. The image shown is representative of at least three experiments.
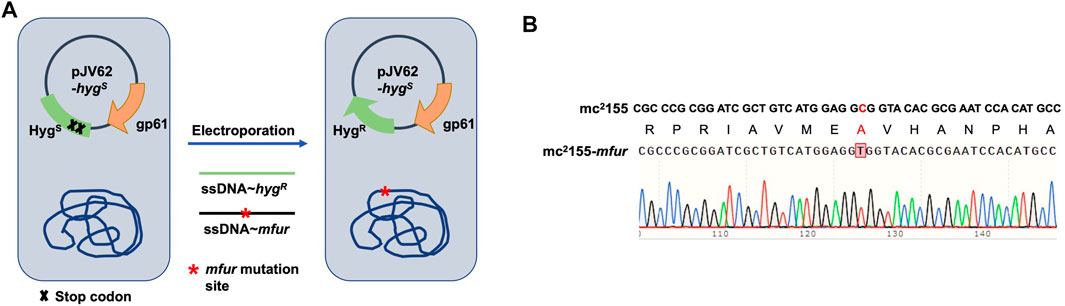
FIGURE 5. Generation of a point mutation furA28V in fur on the chromosome in M. smegmatis. (A) Strategies for recombineering. The site-directed mutagenesis of fur was obtained using Phage Che9c gp61-mediated recombination. Plasmids pJV62-hygs express Che9c gene product gp61, to facilitate single-stranded DNA carrying the point mutation recombination. The clones carrying the mutated site were selected on 7H9 with HygR and KanR. (B) The sequencing result of a point mutation in fur. Sanger sequencing showed the nuclear acid point mutation site c/t, resulting in the protein switch from A to V at site 28th.
As a successful human pathogen, M. tuberculosis has unique respiration properties. M. tuberculosis excretes alkaline supernatants, which is in contrast to other strains that excrete acidic supernatants (Merrill, 1930). The difference between secreted compounds with different acid–base properties suggested that M. tuberculosis has a distinctive redox state. As shown in Table 2, the comparative genomic analysis shows that PhoPR and DosR/S/T, identified as virulence factors of M. tuberculosis, are present in M. smegmatis. The signaling transduction systems such as the two-component systems and the sigma factors of M. tuberculosis are homologous in M. smegmatis (Table 3). Different phenotypes might be due to different redox states. Thus, we considered that the same redox-regulated related protein might perform different functions in different redox states. To test this hypothesis, we examined the biological function of a universal stress protein Rv1996 that increases the expression of KatG, in various mycobacterial strains (Hu et al., 2015). Both previous studies and our studies have linked isoniazid action with redox states, and H2O2 resistance is negatively correlated with INH susceptibility in mycobacteria (Bhaskar et al., 2014; Hu et al., 2015; Vilcheze et al., 2017). We used INH as a chemical probe for monitoring mycobacterial redox states and measured the MICs of INH to the corresponding mycobacterial strains (Figure 6). As predicted, the MICs of INH differed across the tested mycobacterial strains: the MIC of INH in pMV261-rv1996/mc27000 was equal to that of pMV261/mc27000; the MIC of INH in pMV261-rv1996/BCG was lower than that in pMV261/BCG; the MIC of INH in pMV261-rv1996/mc251 was equal to that of pMV261/mc251; and in mc2155, the opposite results were observed with the MIC of INH in pMV261-rv1996/mc2155 being lower than that of INH in pMV261/mc2155 (Figure 6).
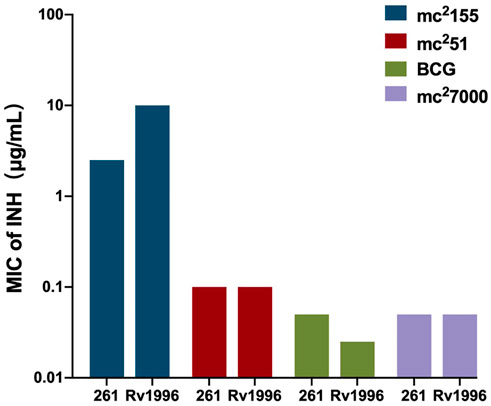
FIGURE 6. Comparison of isoniazid susceptibility in different mycobacterial strains harboring pMV261-rv1996 and the corresponding control strain harboring pMV261.The minimum inhibitory concentrations (MICs) of isoniazid were determined by inoculating each bacterial strain in 7H9 containing serially isoniazid (INH). The values of MIC were recorded. 261 present pMV261. Rv1996 present pMV261-rv1996. The image represents the results of three independent repeats.
Understanding how M. tuberculosis evolved into a professional pathogen is of benefit to the study of its pathogenesis and design of vaccines. The combination of experimental evolution and whole-genome sequencing provides a powerful method for identifying adaptive mutations and elucidating the specific genotype–phenotype relationship (Elena and Lenski, 2003; Lenski, 2017). Historically, the most successful example of continuous selective cultures is M. bovis BCG, the only anti-TB vaccine, which was attenuated after 13 years of continuous in vitro passages of M. bovis BCG. We previously used a similar adaptive evolution strategy to select H2O2-resistant M. smegmatis strains by using a clinically key stressor H2O2 (Li et al., 2014b). Preliminary results showed that the mc251 strain was highly resistant to H2O2 and had greater susceptibility to INH, compared to mc2155. The mc251 phenotype showed an M. tuberculosis-like M. smegmatis phenotype. Altogether, the mutant M. smegmatis mc251 exhibited higher virulence.
The whole-genome sequencing showed the presence of gene mutations in fur, and the mutant Fur resulted in katG levels (Figure 4B). In the Wayne dormancy model, mc251 shows a growth advantage of recovering from dormancy under anaerobic conditions over mc2155 (Figure 2B). In parallel, a high level of katG in mc251 is accompanied by lower ATP levels, which implied mc251 exhibited at a lower level of respiration (Figure 3B). Moreover, we showed that a redox-related protein Rv1996 exhibits a different phenotype under different specific redox states in M. smegmatis mc2155 and mc251, M. bovis BCG, and M. tuberculosis mc27000 (Figure 6). This study indicated that the same genotype presents different phenotypes under different physiological conditions. We at least partially explain why M. smegmatis and M. tuberculosis have similar virulent factors, including a two-component system and sigma factors (Tables 2, 3), but M. smegmatis is a nonpathogen and M. tuberculosis is a pathogen.
M. tuberculosis is a successful human pathogen. It is considered to be derived from the environment (Gutierrez et al., 2005; Wolfe et al., 2007) and has adapted to the immune environment of the human body through long-term evolution. Its successfully established infection is partially attributed to its survival capacity and persistence in macrophages (Podinovskaia et al., 2013). To defend against mycobacterial infection, the host produces ROS, as an important innate defense mechanism. Consequently, M. tuberculosis has evolved a hierarchy and unique antioxidant function and maintains a low level of respiration, manifested by slow growth and persistence in the host. In contrast, M. smegmatis is present in the soil, which is a totally different environment from the host (Zhang and Furman, 2021). In Table 2, we show that M. tuberculosis and M. smegmatis have similar genotypes; however, they show different phenotypes, in terms of INH susceptibility, H2O2 resistance, and virulence. We believe that this striking difference is due to H2O2 resistance. The selected resistance to H2O2 of mc251 shows improved virulence in both the macrophage-killing assay and in an animal model (Figure 1). In fact, several studies have shown that abiotic stress can improve the virulence phenotype of bacterial pathogens (Sundberg et al., 2014; Li et al., 2021). Our study also supports the sit-and-wait hypothesis (Wang et al., 2017), that is, bacterial environmental abiotic stress and virulence evolution. In addition, this study also suggests that we can use mc251 as a model strain, replacing mc2155, to study the regulation of redox homeostasis of M. tuberculosis.
We previously sequenced the whole genome of mc251 strain (Li et al., 2014a) and identified 29 SNPs, compared to mc251. Confirmed with our previous study (Li et al., 2014b), we found that only the fur gene can partially complement the resistant phenotype. This suggested that the fur mutation facilitated elevated H2O2 resistance, although it was not entirely responsible for the high resistance observed in mc251. We also mutated the fur in wild-type mc2155 by genome editing and produced a phenotype similar to mc251, which is highly resistant to H2O2 (Table 1). Large-scale whole-genome sequencing studies on the evolutionary history of tuberculosis also show that key tract mutations at the transcription site will have a critical impact on the particular phenotype (Gagneux, 2018). For example, the change of PhoP in BCG allows infection with bovine pathogenic bacteria capable of infecting humans (Gonzalo-Asensio et al., 2014; Broset et al., 2015). This study reminds us that when designing vaccines, greater attention should be paid to regulators, which may be more efficient targets. M. smegmatis is an effective vaccine for TB and HIV (Sweeney et al., 2011; Kim et al., 2017). The disadvantage of M. smegmatis as a vaccine vector is its transient infection and difficulty to establish a persistent infection and produce adaptive immunity. The M. tuberculosis-like mutant M. smegmatis mc251 may be developed as a vaccine vector.
By comparing the survival of mc2155 and mc251 in the Wayne dormancy animal model, we also found that low respiratory levels are beneficial for survival under anaerobic conditions and resurrection (Figure 2). In the future, we plan to use these strains to compare physiological indicators such as NADH/NAD+, NADPH/NADP+, and ATP, to further understand the mechanisms underlying M. tuberculosis resurrection. This study provides insight into H2O2-resistant mechanisms in mycobacteria and has important implications for linking mycobacteria redox capacity and persistence infection in mice.
The H2O2-resistant Mycobacterium smegmatis strain mc251 was screened in the Mi lab (Li et al., 2014b). Mycobacterium tuberculosis ΔpanCD (named mc27000) (Sambandamurthy et al., 2002) was kindly gifted by J Deng. The M. smegmatis wild-type mc2155, mutant strain mc251, M. bovis BCG Pasteur, and M. tuberculosis mc27000 were cultured in Middlebrook 7H9 (Becton Diskinson Sparks, MD, United States) supplemented with ADS (10% albumin, dextrose, and saline), 0.05% Tween 80 (Sigma, St. Louis, MO, United States), and 0.5% glycerol (Beijing Modern Eastern Fine Chemical Co., Ltd., Beijing, China) for liquid culture and Middlebrook 7H10 (Becton Diskinson Sparks, MD, United States) supplemented with ADS for bacterial colony culture. The colony-forming units (CFUs) of mycobacterial strains were determined by plating serial dilutions of cultures on Middlebrook 7H10 agar plates and incubating at 37°C in an atmosphere of 5% CO2 for the indicated time. For mc27000 culture, panthothenate (24 mg/L) was added. When required, kanamycin (25 mg/L, Amresco, United States) and hygromycin (50 mg/L, Sigma, United States) were added. All bacterial strains used in this study are listed in Table 1.
The susceptibility of isoniazid (INH) or H2O2 of mycobacteria was evaluated using the modified broth microdilution method (Franzblau et al., 1998). In brief, INH or H2O2 was serial diluted using the 7H9 medium. The diluted fold was 1.25- or 2-fold, when required. Then, 40 μl of diluted INH or H2O2 was mixed with 40 μl of mycobacterial suspension with 1 × 107 cells/ml in each well of 96-well microtiter plates and then incubated at 37°C for the indicated days. As an indicator, 0.02% resazurin was added to individual samples, and the color switches from blue to pink were recorded after 4 h. All the experiments were performed in triplicate. The abundance of the cultures was measured using a microplate reader (FLUOstar OPTIMA, BMG Labtech). A difference of two serial dilutions or more indicated a significant difference in the INH or H2O2 susceptibility of bacterial strains.
Female pathogen-free C57BL/6 mice (aged 6–8 weeks) were purchased from Vital River (Beijing, China). For the intranasal infection of M. smegmatis, mice were anesthetized by intraperitoneal injection of pentobarbital sodium (60 mg/kg), and ∼107 CFU/50 µl PBS of mc251 or mc2155 was introduced dropwise through the nostril of each mouse. The bacterial burden throughout the infection was monitored by collecting whole lung tissue at the indicated times after the mice were euthanized, and serial dilutions were then plated on 7H10 agar plates. The dose of infection was confirmed on day one after infection by plating whole lung homogenates from three mice on 7H10 agar. The percentage of survival was calculated as (CFUs after infection/CFUs before infection) × 100%.
Human-derived cell line THP-1 (ATCC TIB-202) was cultured in RPMI medium with 10% fetal bovine serum (FBS, GIBCO, United States). THP-1 cells were activated with 100 ng/ml phorbo-12-myristate-13-acetate (PMA, Sigma, United States) overnight. Infection was carried out at a multiplicity of infection (MOI) of 10 for 1 h at 37°C and 5% CO2 atmosphere. The infected THP-1 cells were washed with RPMI 3 times and then chased for 1 h. The cells with intracellular bacilli were then washed and lysed in sterile cold PBST (PBS with 0.05% Tween 20). Lysates were then vortexed, diluted, and plated on 7H10 agar plates as previously described (Chan et al., 1992). The percentage of survival was calculated as (CFUs after infection/CFUs before infection) × 100%.
Mycobacterial strains were cultured under hypoxic conditions as described by Wayne and Hayes (Wayne and Hayes, 1996). In brief, cultures were initiated at an OD600 of ∼0.01 (1 × 106) and incubated in anaerobic tubes with sealed caps. The headspace ratio of the cultures was 0.5. The cultures were stirred using an 8 mm Teflon stir bar (Fisher Scientific, United States) at 200 rpm. Methylene blue (1.5 mg/L) was used as an oxygen tension indicator. It changes in color from blue to colorless under low oxygen tension. The color transition time was recorded. All experiments were performed in triplicate. Growth was monitored by measuring the OD600 and by determination of CFUs after plating on 7H10.
The indicator methylene blue in the culture became colorless, indicating that the bacteria entered anaerobic conditions. Then, after collecting the bacteria in the anaerobic tube, they were washed with the culture medium or PBS three times and resuspended in a culture medium (7H9 and brain–heart infusion medium), the concentration was adjusted to the same amount of OD600 (OD600 of ∼1.0) and diluted for the CFU count, 1:100 or 1:50 into a fresh culture medium was shaken at 37°C, to monitor the status of the bacteria, and the OD600 was measured. Three independent mycobacterial strain clones were set, and each clone set up three replicates.
The ATP Assay Kit was purchased from Beyotime Biotechnology (Beijing, China). The intracellular ATP assay was performed following the protocol provided by the manufacturer. In brief, the sample measurements were prepared as follows: cultures of indicated mycobacteria were obtained to an OD600 of 0.8. Bacteria were collected by low-temperature centrifugation at the maximum speed, and the pellet was washed with precooled PBS buffer 3 times. A 300 μl volume of ATP detection lysate and 0.5 ml volume of glass beads were added for cell lysis. The lysate obtained was centrifuged at low temperature for 5 min, and the supernatant was placed on ice for later use. The preparation of the standard solution of gradient concentration ATP was performed as follows: the ATP standard solution was serially diluted into 7 concentrations of 10, 3.333, 1.111, 0.37, 0.1234, 0.04115, and 0.01371 μM and stored on ice for later use. The preparation of the working solution for the detection of ATP was performed as follows: an appropriate amount of ATP detection reagent was prepared according to the number of samples, and then, a 90% final volume of ATP detection reagent diluent was added. The prepared working fluid was placed on ice for further use. The determination of the ATP level was performed as follows: 1) the prepared ATP detection working solution was dispensed into 1.5 ml centrifuge tubes, 100 μl per tube, and was allowed to incubate at room temperature for 5 min to allow full reaction of the ATP in the centrifuge tube; 2) during the test, 20 μl of each sample (standard or total protein sample) was added to a 1.5 ml centrifuge tube containing 100 μl of ATP detection working solution and was mixed quickly with a pipette and incubated for 2 s to complete the reaction before using a Cytation 3 Cell Imaging Multi-Mode Reader to determine the relative light unit (RLU); 3) a standard curve was constructed to measure and determine the concentration of the sample by converting the RLU into an ATP concentration; and 4) in order to eliminate the error caused by the difference in the amount of protein during sample preparation, the BCA protein concentration determination kit produced by Beyotime Biotechnology (Beijing, China) was used to determine the protein concentration in the sample. The ATP concentration was converted to μM/mg protein.
The coding regions of fur and mfur were amplified from mc2155 and mc251 genomic DNA and cloned into the Escherichia coli expression vector pET23b (+) (Novagen, Madison, WI, United States) in-frame fused with a C-terminal His6-tag sequence to construct the plasmids pET23b-fur and pET23b-mfur. The final constructs were transformed into BL21 (DE3) for expression, and recombinant Fur/mFur proteins were purified using Ni-NTA agarose (Qiagen, California, United States). The proteins were induced by the addition of 1 mM IPTG at 16°C for 12 h. Protein purification was performed as described previously (Li et al., 2014c). The protocols of the recombinant protein purification are available on request. The recombinant protein MSMEG_2415 was purified as described previously (Li et al., 2014c) and used as a negative control for EMSA, while MSMEG_2415 is irrelated to Fur. The DNA fragment containing the promoter region of fur for gel shift experiments was amplified by PCR with specific primers (forward: 5′-CGTTGGAAAACAACCATTGCAAG-3′, reverse: 5′-CATCCGCAGTTGGGCTTCGAAC-3′). Binding reaction mixtures in 20 μl of binding buffer (20 mM Tris HCl pH 8.0, 1 mM dithiothreitol (DTT), 50 mM KCl, and 5 mM MgCl2) containing 0.15 pmol of the DNA fragment were incubated with purified Fur/mFur protein (0.5 nmol) for 30 min at 30°C. Reaction mixtures were loaded on a 4–20% polyacrylamide gel containing 0.5 × TBE. Gels were run at 70 V at 4°C for 3 h. The gel was stained with Good-view and photographed for the image.
The knockout katG strain was constructed using mycobacteriophage-based specialized transduction (Bardarov et al., 2002; Li et al., 2014c). The upstream and downstream sequences of katG were amplified from M. smegmatis genome DNA. The knockout vector was constructed using phAE159 (Hsu and Jacobs, unpublished data). The mycobacteriophage used for knockout was obtained using MaxPlax packaging extract (Epicentre Biotechnologies, Madison, WI, United States), and a katG knockout strain was obtained by phage transduction, named ∆katG. The KatG overexpression strain was constructed using pMV261 to yield pMV261-katG, and the constructed plasmid was electroporated into mc2155, yielding pMV261-katG/mc2155. The detailed information for construction of all the mycobacterial strains and primers for plasmid construction is available on request.
The single-strand (ss) DNA oligonucleotides used for recombineering were ordered from Genewiz (Suzhou, China). The site-directed mutagenesis of fur was obtained using Phage Che9c gp61-mediated recombination (van Kessel et al., 2008). The detailed information on primers for construction of the fur mycobacterial strain (named mc2155-mfur) is available on request. The coding region containing fur point mutation in the genome (encoding mFur) was amplified and sequenced by Genewiz (Suzhou, China).
The rv1996 gene was amplified and constructed and cloned into pMV261 to yield pMV261-rv1996. The constructed pMV261-rv1996 plasmid was transformed into mycobacterial strains, M. smegmatis mc2155 and mc251, M. bovis BCG Pasteur, and M. tuberculosis mc27000, and the corresponding strains, named pMV261-rv1996/mc2155, pMV261-rv1996/mc251, pMV261-rv1996/BCG, and pMV261-rv1996/mc27000. The empty vector pMV261 was transformed into the corresponding mycobacterial strains, named pMV261/mc2155, pMV261/mc251, pMV261/BCG, and pMV261/mc27000.
Each experiment was carried out at least twice with three–nine mice or samples per group. The CFUs and OD600 were analyzed using an unpaired t-test (Version 8.0 for Windows GraphPad Software). The ATP content was analyzed using ANOVA tests (Version 8.0 for Windows GraphPad Software). ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05.
This study was performed in strict accordance with the recommendations of the Ethics Committee established in the Guide for the Care and Use of Laboratory Animals of the Institute of Microbiology, Chinese Academy of Sciences (IMCAS). The protocol was approved by the Committee on the Ethics of Animal Experiments of the IMCAS. The mice were bred under specific pathogen-free conditions at the IMCAS laboratory animal facility. All animal experiments were conducted under isoflurane anesthesia, and all efforts were made to minimize suffering.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the following: BioProject: PRJNA233977; BioSample: SAMN02951866, https://www.ncbi.nlm.nih.gov/nuccore/JAJD00000000.
KM conceived and designed the experiments; ZJ and ZZ performed the experiments; KM wrote the manuscript; and KM, ZJ, and ZZ revised the manuscript. All authors have read and agreed to the published version of the manuscript.
This work was supported by grants from the Ministry of Science and Technology of China (2018YFC1603900 and 2017YFA0505901 to KM), National Natural Science Foundation of China (31970136 and 32170181 to KM), and International Joint Research Project of the Institute of Medical Science, University of Tokyo (Extension-2019-K3006 to KM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank J. Deng for providing the mycobacterial strain mc27000. They also thank Tong Yin for her help in preparing the experimental materials.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.758304/full#supplementary-material
Supplementary Figure S1 | Evolutionary selection of H2O2-resistant mutations in M. smegmatis Cultures were started from glycerol-frozen stocks and grown to log phase (OD600 of 0.6 - 0.8). Then the cultures were diluted 1:1000 into 5 ml of 7H9 media containing 10% ADS. Hydrogen peroxide (H2O2) was then added to a concentration of 0.0293 mM and cultures were grown until the OD600 reached log phase. Cultures were then further diluted 1:1000 and an additional 0.0293 mM of H2O2 was added to the culture. This process was repeated until the H2O2 concentration reached 0.4395 mM. In further rounds of culture, H2O2was added in steps of 0.0879 mM, instead of 0.0293 mM until an H2O2 concentration of 1.5 mM was reached. To ensure that the H2O2-resistant phenotype was caused by a chromosomal mutation, selected cultures were sub-cultured for 10 passages and then streaked on plates to obtain single colonies. The distinctive single colonies were then inoculated in liquid culture and actual MIC of H2O2 was determined.
Agarwal, N., Woolwine, S. C., Tyagi, S., and Bishai, W. R. (2007). Characterization of the Mycobacterium tuberculosis Sigma Factor SigM by Assessment of Virulence and Identification of SigM-dependent Genes. Infect. Immun. 75, 452–461. doi:10.1128/IAI.01395-06
Aldridge, B. B., Fernandez-Suarez, M., Heller, D., Ambravaneswaran, V., Irimia, D., Toner, M., et al. (2012). Asymmetry and Aging of Mycobacterial Cells lead to Variable Growth and Antibiotic Susceptibility. Science 335, 100–104. doi:10.1126/science.1216166
Anes, E., Kühnel, M. P., Bos, E., Moniz-Pereira, J., Habermann, A., and Griffiths, G. (2003). Selected Lipids Activate Phagosome Actin Assembly and Maturation Resulting in Killing of Pathogenic Mycobacteria. Nat. Cel Biol 5, 793–802. doi:10.1038/ncb1036
Anes, E., Peyron, P., Staali, L., Jordao, L., Gutierrez, M. G., Kress, H., et al. (2006). Dynamic Life and Death Interactions between Mycobacterium Smegmatis and J774 Macrophages. Cell Microbiol 8, 939–960. doi:10.1111/j.1462-5822.2005.00675.x
Armstrong, J. A., and Hart, P. D. A. (1971). Response of Cultured Macrophages to Mycobacterium tuberculosis, with Observations on Fusion of Lysosomes with Phagosomes. J. Exp. Med. 134, 713–740. doi:10.1084/jem.134.3.713
Arora, G., Bothra, A., Prosser, G., Arora, K., and Sajid, A. (2021). Role of post‐translational Modifications in the Acquisition of Drug Resistance in Mycobacterium tuberculosis. FEBS J. 288, 3375–3393. doi:10.1111/febs.15582
Bardarov, S., Bardarov, S., Pavelka, M. S., Sambandamurthy, V., Larsen, M., Tufariello, J., et al. (2002). Specialized Transduction: an Efficient Method for Generating Marked and Unmarked Targeted Gene Disruptions in Mycobacterium tuberculosis, M. Bovis BCG and M. Smegmatis. Microbiology (Reading) 148, 3007–3017. doi:10.1099/00221287-148-10-3007
Bhattacharya, M., Biswas, A., and Das, A. K. (2010). Interaction Analysis of TcrX/Y Two Component System from Mycobacterium tuberculosis. Biochimie 92, 263–272. doi:10.1016/j.biochi.2009.11.009
Boshoff, H. I. M., Myers, T. G., Copp, B. R., McNeil, M. R., Wilson, M. A., and Barry, C. E. (2004). The Transcriptional Responses of Mycobacterium tuberculosis to Inhibitors of Metabolism. J. Biol. Chem. 279, 40174–40184. doi:10.1074/jbc.M406796200
Broset, E., Martín, C., and Gonzalo-Asensio, J. (2015). Evolutionary Landscape of the Mycobacterium tuberculosis Complex from the Viewpoint of PhoPR: Implications for Virulence Regulation and Application to Vaccine Development. mBio 6, e01289–01215. doi:10.1128/mBio.01289-15
Brown, C. A., Draper, P., and Hart, P. D. A. (1969). Mycobacteria and Lysosomes: a Paradox. Nature 221, 658–660. doi:10.1038/221658a0
Calamita, H., Ko, C., Tyagi, S., Yoshimatsu, T., Morrison, N. E., and Bishai, W. R. (2005). The Mycobacterium tuberculosis SigD Sigma Factor Controls the Expression of Ribosome-Associated Gene Products in Stationary Phase and Is Required for Full Virulence. Cel Microbiol 7, 233–244. doi:10.1111/j.1462-5822.2004.00454.x
Chan, J., Xing, Y., Magliozzo, R. S., and Bloom, B. R. (1992). Killing of Virulent Mycobacterium tuberculosis by Reactive Nitrogen Intermediates Produced by Activated Murine Macrophages. J. Exp. Med. 175, 1111–1122. doi:10.1084/jem.175.4.1111
Cohen, S. B., Gern, B. H., Delahaye, J. L., Adams, K. N., Plumlee, C. R., Winkler, J. K., et al. (2018). Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host & Microbe 24, 439–446. doi:10.1016/j.chom.2018.08.001
Dragset, M. S., Ioerger, T. R., Zhang, Y. J., Mærk, M., Ginbot, Z., Sacchettini, J. C., et al. (2019). Genome-wide Phenotypic Profiling Identifies and Categorizes Genes Required for Mycobacterial Low Iron Fitness. Sci. Rep. 9, 11394. doi:10.1038/s41598-019-47905-y
Elena, S. F., and Lenski, R. E. (2003). Evolution Experiments with Microorganisms: The Dynamics and Genetic Bases of Adaptation. Nat. Rev. Genet. 4, 457–469. doi:10.1038/nrg1088
Flynn, J. L., and Chan, J. (2001). Immunology of Tuberculosis. Annu. Rev. Immunol. 19, 93–129. doi:10.1146/annurev.immunol.19.1.93
Fol, M., Chauhan, A., Nair, N. K., Maloney, E., Moomey, M., Jagannath, C., et al. (2006). Modulation of Mycobacterium tuberculosis Proliferation by MtrA, an Essential Two-Component Response Regulator. Mol. Microbiol. 60, 643–657. doi:10.1111/j.1365-2958.2006.05137.x
Ford, C. B., Lin, P. L., Chase, M. R., Shah, R. R., Iartchouk, O., Galagan, J., et al. (2011). Use of Whole Genome Sequencing to Estimate the Mutation Rate of Mycobacterium tuberculosis during Latent Infection. Nat. Genet. 43, 482, 486. doi:10.1038/ng.811
Franzblau, S. G., Witzig, R. S., McLaughlin, J. C., Torres, P., Madico, G., Hernandez, A., et al. (1998). Rapid, Low-Technology MIC Determination with Clinical Mycobacterium tuberculosis Isolates by Using the Microplate Alamar Blue Assay. J. Clin. Microbiol. 36, 362–366. doi:10.1128/Jcm.36.2.362-366.1998
Gagneux, S. (2018). Ecology and Evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 202–213. doi:10.1038/nrmicro.2018.8
Gomez, M., Doukhan, L., Nair, G., and Smith, I. (1998). sigAis an Essential Gene inMycobacterium Smegmatis. Mol. Microbiol. 29, 617–628. doi:10.1046/j.1365-2958.1998.00960.x
Gonzalo-Asensio, J., Malaga, W., Pawlik, A., Astarie-Dequeker, C., Passemar, C., Moreau, F., et al. (2014). Evolutionary History of Tuberculosis Shaped by Conserved Mutations in the PhoPR Virulence Regulator. Proc. Natl. Acad. Sci. 111, 11491–11496. doi:10.1073/pnas.1406693111
Gray, T. A., Clark, R. R., Boucher, N., Lapierre, P., Smith, C., and Derbyshire, K. M. (2016). Intercellular Communication and Conjugation Are Mediated by ESX Secretion Systems in Mycobacteria. Science 354, 347–350. doi:10.1126/science.aag0828
Gutierrez, M. C., Brisse, S., Brosch, R., Fabre, M., Omaïs, B., Marmiesse, M., et al. (2005). Ancient Origin and Gene Mosaicism of the Progenitor of Mycobacterium tuberculosis. Plos Pathog. 1, e5. doi:10.1371/journal.ppat.0010005
Hahn, M.-Y., Raman, S., Anaya, M., and Husson, R. N. (2005). The Mycobacterium tuberculosis Extracytoplasmic-Function Sigma Factor SigL Regulates Polyketide Synthases and Secreted or Membrane Proteins and Is Required for Virulence. J. Bacteriol. 187, 7062–7071. doi:10.1128/JB.187.20.7062-7071.2005
Haydel, S. E., Benjamin, W. H., Dunlap, N. E., and Clark-Curtiss, J. E. (2002). Expression, Autoregulation, and DNA Binding Properties of the Mycobacterium tuberculosis TrcR Response Regulator. J. Bacteriol. 184, 2192–2203. doi:10.1128/Jb.184.8.2192-2203.2002
He, H., and Zahrt, T. C. (2005). Identification and Characterization of a Regulatory Sequence Recognized by Mycobacterium tuberculosis Persistence Regulator MprA. J. Bacteriol. 187, 202–212. doi:10.1128/JB.187.1.202-212.2005
Homerova, D., Halgasova, L., and Kormanec, J. (2008). Cascade of Extracytoplasmic Function Sigma Factors inMycobacterium Tuberculosis: Identification of a σJ-dependent Promoter Upstream ofsigI. FEMS Microbiol. Lett. 280, 120–126. doi:10.1111/j.1574-6968.2007.01054.x
Hu, X., Zhou, X., Yin, T., Chen, K., Hu, Y., Zhu, B., et al. (2021). The Mycobacterial DNA Methyltransferase HsdM Decreases Intrinsic Isoniazid Susceptibility. Antibiotics 10 (11), 1323. doi:10.3390/antibiotics10111323
Hu, X., Li, X., Huang, L., Chan, J., Chen, Y., Deng, H., et al. (2015). Quantitative Proteomics Reveals Novel Insights into Isoniazid Susceptibility in Mycobacteria Mediated by a Universal Stress Protein. J. Proteome Res. 14, 1445–1454. doi:10.1021/pr5011058
James, J. N., Hasan, Z.-u., Ioerger, T. R., Brown, A. C., Personne, Y., Carroll, P., et al. (2012). Deletion of SenX3-RegX3, a Key Two-Component Regulatory System of Mycobacterium Smegmatis, Results in Growth Defects under Phosphate-Limiting Conditions. Microbiology (Reading) 158, 2724–2731. doi:10.1099/mic.0.060319-0
Jaswal, S., Dhand, R., Sethi, A. K., Kohli, K. K., and Ganguly, N. K. (1992). Oxidative Metabolic Status of Blood Monocytes and Alveolar Macrophages in the Spectrum of Human Pulmonary Tuberculosis. Scand. J. Clin. Lab. Invest. 52, 119–128. doi:10.3109/00365519209088775
Judd, J. A., Canestrari, J., Clark, R., Joseph, A., Lapierre, P., Lasek-Nesselquist, E., et al. (2021). A Mycobacterial Systems Resource for the Research Community. mBio 12. doi:10.1128/mBio.02401-20
Kieser, K. J., Boutte, C. C., Kester, J. C., Baer, C. E., Barczak, A. K., Meniche, X., et al. (2015). Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria. Plos Pathog. 11, e1005010. doi:10.1371/journal.ppat.1005010
Kim, B.-J., Gong, J.-R., Kim, G.-N., Kim, B.-R., Lee, S.-Y., Kook, Y.-H., et al. (2017). Recombinant Mycobacterium Smegmatis with a pMyong2 Vector Expressing Human Immunodeficiency Virus Type I Gag Can Induce Enhanced Virus-specific Immune Responses. Sci. Rep. 7, 44776. doi:10.1038/srep44776
Kumar, V., Jindal, S. K., and Ganguly, N. K. (1995). Release of Reactive Oxygen and Nitrogen Intermediates from Monocytes of Patients with Pulmonary Tuberculosis. Scand. J. Clin. Lab. Invest. 55, 163–169. doi:10.3109/00365519509089609
Lee, J.-H., Geiman, D. E., and Bishai, W. R. (2008a). Role of Stress Response Sigma Factor SigG in Mycobacterium tuberculosis. J. Bacteriol. 190, 1128–1133. doi:10.1128/JB.00511-07
Lee, J.-H., Karakousis, P. C., and Bishai, W. R. (2008b). Roles of SigB and SigF in the Mycobacterium tuberculosis Sigma Factor Network. J. Bacteriol. 190, 699–707. doi:10.1128/JB.01273-07
Lenski, R. E. (2017). Experimental Evolution and the Dynamics of Adaptation and Genome Evolution in Microbial Populations. Isme J. 11, 2181–2194. doi:10.1038/ismej.2017.69
Li, F., Xiong, X.-S., Yang, Y.-Y., Wang, J.-J., Wang, M.-M., Tang, J.-W., et al. (2021). Effects of NaCl Concentrations on Growth Patterns, Phenotypes Associated with Virulence, and Energy Metabolism in Escherichia coli BW25113. Front. Microbiol. 12, 705326. doi:10.3389/fmicb.2021.705326
Li, X., Liu, F., Hu, Y., and Mi, K. (2014a). Draft Genome Sequence of Mc 2 51, a Highly Hydrogen Peroxide-Resistant Mycobacterium Smegmatis Mutant Strain. Genome Announc 2. doi:10.1128/genomeA.00092-14
Li, X., Tao, J., Han, J., Hu, X., Chen, Y., Deng, H., et al. (2014b). The Gain of Hydrogen Peroxide Resistance Benefits Growth Fitness in Mycobacteria under Stress. Protein Cell 5, 182–185. doi:10.1007/s13238-014-0024-5
Li, X., Tao, J., Hu, X., Chan, J., Xiao, J., and Mi, K. (2014c). A Bacterial Hemerythrin-like Protein MsmHr Inhibits the SigF-dependent Hydrogen Peroxide Response in Mycobacteria. Front. Microbiol. 5, 800. doi:10.3389/fmicb.2014.00800
Li, Y., Zeng, J., Zhang, H., and He, Z.-G. (2010). The Characterization of Conserved Binding Motifs and Potential Target Genes for M. tuberculosis MtrAB Reveals a Link between the Two-Component System and the Drug Resistance of M. Smegmatis. BMC Microbiol. 10, 242. doi:10.1186/1471-2180-10-242
Loebel, R. O., Shorr, E., and Richardson, H. B. (1933). The Influence of Adverse Conditions upon the Respiratory Metabolism and Growth of Human Tubercle Bacilli. J. Bacteriol. 26, 167–200. doi:10.1128/Jb.26.2.167-200.1933
Mehra, S., Foreman, T. W., Didier, P. J., Ahsan, M. H., Hudock, T. A., Kissee, R., et al. (2015). The DosR Regulon Modulates Adaptive Immunity and Is Essential forMycobacterium tuberculosisPersistence. Am. J. Respir. Crit. Care Med. 191, 1185–1196. doi:10.1164/rccm.201408-1502OC
Merrill, M. H. (1930). Carbohydrate Metabolism of Organisms of the Genus Mycobacterium. J. Bacteriol. 20, 235–286. doi:10.1128/jb.20.4.235-286.1930
Mukherjee, R., and Chatterji, D. (2005). Evaluation of the Role of Sigma B in Mycobacterium Smegmatis. Biochem. Biophysical Res. Commun. 338, 964–972. doi:10.1016/j.bbrc.2005.10.038
Nauseef, W. M. (2019). The Phagocyte NOX2 NADPH Oxidase in Microbial Killing and Cell Signaling. Curr. Opin. Immunol. 60, 130–140. doi:10.1016/j.coi.2019.05.006
Ng, V. H., Cox, J. S., Sousa, A. O., MacMicking, J. D., and McKinney, J. D. (2004). Role of KatG Catalase-Peroxidase in Mycobacterial Pathogenesis: Countering the Phagocyte Oxidative Burst. Mol. Microbiol. 52, 1291–1302. doi:10.1111/j.1365-2958.2004.04078.x
Nowak, E., Panjikar, S., Konarev, P., Svergun, D. I., and Tucker, P. A. (2006). The Structural Basis of Signal Transduction for the Response Regulator PrrA from Mycobacterium tuberculosis. J. Biol. Chem. 281, 9659–9666. doi:10.1074/jbc.M512004200
Parish, T., Smith, D. A., Kendall, S., Casali, N., Bancroft, G. J., and Stoker, N. G. (2003a). Deletion of Two-Component Regulatory Systems Increases the Virulence of Mycobacterium tuberculosis. Infect. Immun. 71, 1134–1140. doi:10.1128/IAI.71.3.1134-1140.2003
Parish, T., Smith, D. A., Roberts, G., Betts, J., and Stoker, N. G. (2003b). The senX3-regX3 Two-Component Regulatory System of Mycobacterium tuberculosis Is Required for Virulence. Microbiology (Reading) 149, 1423–1435. doi:10.1099/mic.0.26245-0
Piccaro, G., Pietraforte, D., Giannoni, F., Mustazzolu, A., and Fattorini, L. (2014). Rifampin Induces Hydroxyl Radical Formation in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 7527–7533. doi:10.1128/AAC.03169-14
Pieters, J. (2008). Mycobacterium tuberculosis and the Macrophage: Maintaining a Balance. Cell Host & Microbe 3, 399–407. doi:10.1016/j.chom.2008.05.006
Plocinska, R., Purushotham, G., Sarva, K., Vadrevu, I. S., Pandeeti, E. V. P., Arora, N., et al. (2012). Septal Localization of the Mycobacterium tuberculosis MtrB Sensor Kinase Promotes MtrA Regulon Expression. J. Biol. Chem. 287, 23887–23899. doi:10.1074/jbc.M112.346544
Podinovskaia, M., Lee, W., Caldwell, S., and Russell, D. G. (2013). Infection of Macrophages withMycobacterium Tuberculosisinduces Global Modifications to Phagosomal Function. Cel Microbiol 15, 843–859. doi:10.1111/cmi.12092
Pym, A. S., Domenech, P., Honore, N., Song, J., Deretic, V., and Cole, S. T. (2001). Regulation of Catalase-Peroxidase (KatG) Expression, Isoniazid Sensitivity and Virulence by furA of Mycobacterium tuberculosis. Mol. Microbiol. 40, 879–889. doi:10.1046/j.1365-2958.2001.02427.x
Raman, S., Hazra, R., Dascher, C. C., and Husson, R. N. (2004). Transcription Regulation by the Mycobacterium tuberculosis Alternative Sigma Factor SigD and its Role in Virulence. J. Bacteriol. 186, 6605–6616. doi:10.1128/Jb.186.19.6605-6616.2004
Raman, S., Puyang, X., Cheng, T.-Y., Young, D. C., Moody, D. B., and Husson, R. N. (2006). Mycobacterium tuberculosis SigM Positively Regulates Esx Secreted Protein and Nonribosomal Peptide Synthetase Genes and Down Regulates Virulence-Associated Surface Lipid Synthesis. J. Bacteriol. 188, 8460–8468. doi:10.1128/JB.01212-06
Rifat, D., and Karakousis, P. C. (2014). Differential Regulation of the Two-Component Regulatory System senX3-regX3 in Mycobacterium tuberculosis. Microbiology (Reading) 160, 1125–1133. doi:10.1099/mic.0.077180-0
Rodrigue, S., Brodeur, J., Jacques, P.-E., Gervais, A. L., Brzezinski, R., and Gaudreau, L. (2007). Identification of Mycobacterial σ Factor Binding Sites by Chromatin Immunoprecipitation Assays. J. Bacteriol. 189, 1505–1513. doi:10.1128/JB.01371-06
Sambandamurthy, V. K., Wang, X., Chen, B., Russell, R. G., Derrick, S., Collins, F. M., et al. (2002). A Pantothenate Auxotroph of Mycobacterium tuberculosis Is Highly Attenuated and Protects Mice against Tuberculosis. Nat. Med. 8, 1171–1174. doi:10.1038/nm765
Schnell, R., Ågren, D., and Schneider, G. (2008). 1.9 Å Structure of the Signal Receiver Domain of the Putative Response Regulator NarL fromMycobacterium Tuberculosis. Acta Cryst. Sect F 64, 1096–1100. doi:10.1107/S1744309108035203
Shrivastava, R., and Das, A. K. (2007). Temperature and Urea Induced Conformational Changes of the Histidine Kinases from Mycobacterium tuberculosis. Int. J. Biol. Macromolecules 41, 154–161. doi:10.1016/j.ijbiomac.2007.01.010
Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T., and Jr, W. R. J. (1990). Isolation and Characterization of Efficient Plasmid Transformation Mutants of Mycobacterium Smegmatis. Mol. Microbiol. 4, 1911–1919. doi:10.1111/j.1365-2958.1990.tb02040.x
Song, T., Song, S.-E., Raman, S., Anaya, M., and Husson, R. N. (2008). Critical Role of a Single Position in the −35 Element for Promoter Recognition by Mycobacterium tuberculosis SigE and SigH. J. Bacteriol. 190, 2227–2230. doi:10.1128/Jb.01642-07
Steyn, A. J. C., Joseph, J., and Bloom, B. R. (2003). Interaction of the Sensor Module of Mycobacterium tuberculosis H37Rv KdpD with Members of the Lpr Family. Mol. Microbiol. 47, 1075–1089. doi:10.1046/j.1365-2958.2003.03356.x
Sun, R., Converse, P. J., Ko, C., Tyagi, S., Morrison, N. E., and Bishai, W. R. (2004). Mycobacterium tuberculosis ECF Sigma Factor sigC Is Required for Lethality in Mice and for the Conditional Expression of a Defined Gene Set. Mol. Microbiol. 52, 25–38. doi:10.1111/j.1365-2958.2003.03958.x
Sundberg, L.-R., Kunttu, H. M. T., and Valtonen, E. (2014). Starvation Can Diversify the Population Structure and Virulence Strategies of an Environmentally Transmitting Fish Pathogen. BMC Microbiol. 14, 67. doi:10.1186/1471-2180-14-67
Sureka, K., Dey, S., Datta, P., Singh, A. K., Dasgupta, A., Rodrigue, S., et al. (2007). Polyphosphate Kinase Is Involved in Stress-Induced mprAB-sigE-Rel Signalling in Mycobacteria. Mol. Microbiol. 65, 261–276. doi:10.1111/j.1365-2958.2007.05814.x
Sweeney, K. A., Dao, D. N., Goldberg, M. F., Hsu, T., Venkataswamy, M. M., Henao-Tamayo, M., et al. (2011). A Recombinant Mycobacterium Smegmatis Induces Potent Bactericidal Immunity against Mycobacterium tuberculosis. Nat. Med. 17, 1261–1268. doi:10.1038/nm.2420
Timmins, G. S., and Deretic, V. (2006). Mechanisms of Action of Isoniazid. Mol. Microbiol. 62, 1220–1227. doi:10.1111/j.1365-2958.2006.05467.x
van Kessel, J. C., Marinelli, L. J., and Hatfull, G. F. (2008). Recombineering Mycobacteria and Their Phages. Nat. Rev. Microbiol. 6, 851–857. doi:10.1038/nrmicro2014
Veyrier, F., Saïd-Salim, B., and Behr, M. A. (2008). Evolution of the Mycobacterial SigK Regulon. J. Bacteriol. 190, 1891–1899. doi:10.1128/JB.01452-07
Walters, S. B., Dubnau, E., Kolesnikova, I., Laval, F., Daffe, M., and Smith, I. (2006). The Mycobacterium tuberculosis PhoPR Two-Component System Regulates Genes Essential for Virulence and Complex Lipid Biosynthesis. Mol. Microbiol. 60, 312–330. doi:10.1111/j.1365-2958.2006.05102.x
Wang, L., Liu, Z., Dai, S., Yan, J., and Wise, M. J. (2017). The Sit-And-Wait Hypothesis in Bacterial Pathogens: A Theoretical Study of Durability and Virulence. Front. Microbiol. 8, 2167. doi:10.3389/fmicb.2017.02167
Wayne, L. G., and Hayes, L. G. (1996). An In Vitro Model for Sequential Study of Shiftdown of Mycobacterium tuberculosis through Two Stages of Nonreplicating Persistence. Infect. Immun. 64, 2062–2069. doi:10.1128/Iai.64.6.2062-2069.1996
Wolfe, N. D., Dunavan, C. P., and Diamond, J. (2007). Origins of Major Human Infectious Diseases. Nature 447, 279–283. doi:10.1038/nature05775
Keywords: mycobacterial pathogenicity, KatG, Fur, H2O2 resistant, TB
Citation: Jiang Z, Zhuang Z and Mi K (2022) Experimental Evolution Reveals Redox State Modulates Mycobacterial Pathogenicity. Front. Genet. 13:758304. doi: 10.3389/fgene.2022.758304
Received: 13 August 2021; Accepted: 10 February 2022;
Published: 16 March 2022.
Edited by:
Wei Huang, Johns Hopkins University, United StatesReviewed by:
Liang Wang, Xuzhou Medical University, ChinaCopyright © 2022 Jiang, Zhuang and Mi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaixia Mi, bWlrQGltLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.