- The Affiliated Hospital of Chengde Medical University, Chengde, Hebei, China
Background: Primary liver cancer is the sixth most commonly diagnosed cancer and the third leading cause of cancer death worldwide in 2020, and it ranks fifth in global incidence. Liver resection or liver transplantation are the two most prominent surgical procedures for treating primary liver cancer. Both inevitably result in HIRI, causing severe complications for patients and affecting their prognosis and quality of survival. Ferroptosis, a newly discovered mode of cell death, is closely related to HIRI. We used bioinformatics analysis to explore the relationship between the two further.
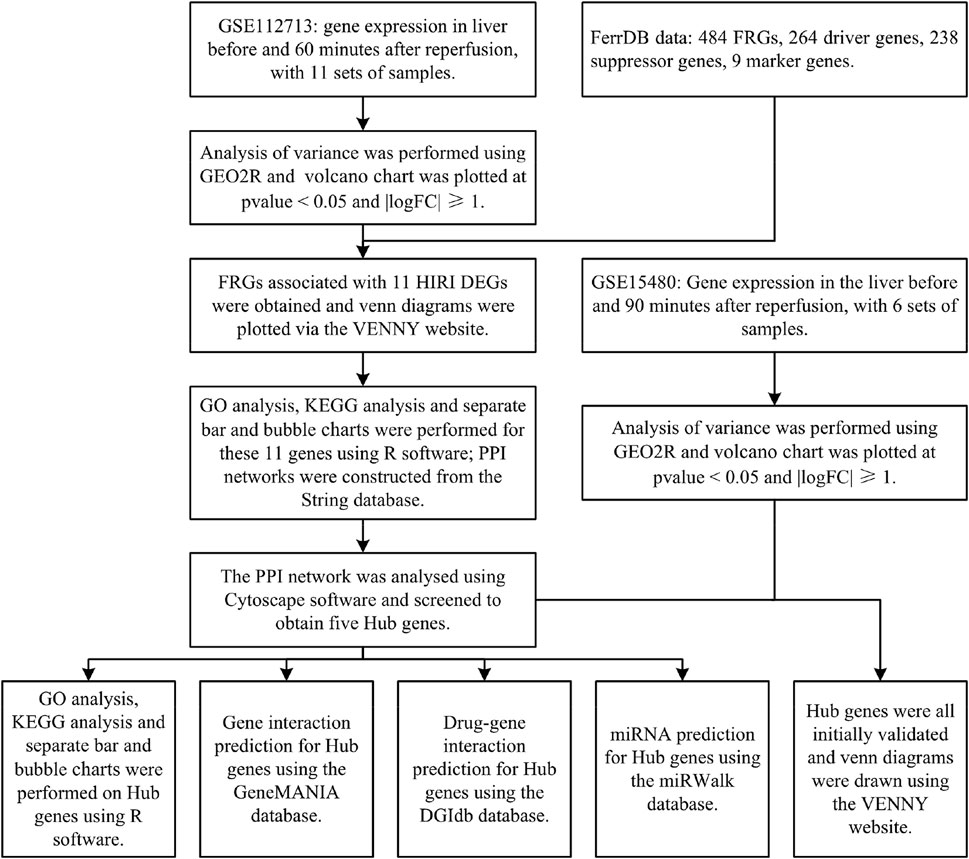
Methods: The GEO database dataset GSE112713 and the FerrDB database data were selected to use bioinformatic analysis methods (difference analysis, FRGs identification, GO analysis, KEGG analysis, PPI network construction and analysis, Hub gene screening with GO analysis and KEGG analysis, intergenic interaction prediction, drug-gene interaction prediction, miRNA prediction) for both for correlation analysis. The GEO database dataset GSE15480 was selected for preliminary validation of the screened Hub genes.
Results: We analysed the dataset GSE112713 for differential gene expression before and after hepatic ischemia-reperfusion and identified by FRGs, yielding 11 genes. These 11 genes were subjected to GO, and KEGG analyses, and PPI networks were constructed and analysed. We also screened these 11 genes again to obtain 5 Hub genes and performed GO analysis, KEGG analysis, intergenic interaction prediction, drug-gene interaction prediction, and miRNA prediction on these 5 Hub genes. Finally, we obtained preliminary validation of all these 5 Hub genes by dataset GSE15480.
Conclusion: There is a close relationship between HIRI and ferroptosis, and inhibition of ferroptosis can potentially be a new approach to mitigate HIRI treatment in the future.
Introduction
Cancer is the leading cause of human mortality and a significant barrier to increasing life expectancy in countries worldwide (Bray F et al., 2021). Primary liver cancer is the sixth most commonly diagnosed cancer and the third leading cause of cancer death worldwide in 2020, with approximately 906,000 new cases and 830,000 deaths (Sung H et al., 2021). In most areas, morbidity and mortality rates are two to three times higher for men than women (Sung H et al., 2021). Primary liver cancer ranks fifth in global incidence and second in male mortality (Sung H et al., 2021). Hepatectomy or liver transplantation are the two main surgical procedures used to treat primary liver cancer (Maki H et al., 2022), but hepatic ischemia-reperfusion injury (HIRI) is inevitable with hepatectomy or liver transplantation. HIRI may lead to liver dysfunction or even failure, causing severe inflammatory and stress reactions in patients. It can seriously affect various organs and systems, affecting their prognosis and quality of life (Nastos C et al., 2014). There are many studies on the pathophysiological mechanisms of HIRI, but they are still inconclusive and controversial. The concept of ferroptosis was first introduced by Dr Brent R. Stockwell in 2012 (Dixon S J et al., 2012). It is an iron-dependent mode of cell death caused by lipid peroxidation and massive accumulation of reactive oxygen radicals. It is a novel form of programmed cell death distinct from apoptosis, cell necrosis, and cell autophagy. Ferroptosis is associated with various biological contexts, from development to ageing, immunity and cancer[6]. Recent studies have shown that ferroptosis is closely related to the pathophysiology of many diseases, with ferroptosis being directly linked to ischemia-reperfusion injury (IRI) in many organs and, of course, HIRI being inextricably linked to ferroptosis (Li J. 2020; Stockwell B R. 2022). However, the relationship between HIRI and ferroptosis has been poorly studied, so we have used bioinformatics analysis to investigate the relationship between HIRI and ferroptosis (Figure 1) and provide a direction and foundation for future research.
Materials and methods
Data sources
The Gene Expression Omnibus (GEO) database is a public gene expression database created by the National Center for Biotechnology Information (NCBI) in the United States. It contains high-throughput gene expression data and microarray gene expression data (Edgar R. 2002). We selected the dataset GSE112713 for bioinformatic analysis of ferroptosis-related genes (FRGs). We selected one data set from GSE112713, a study on normothermic machine perfusion (NMP) inhibits proinflammatory responses in the liver and promotes regeneration: liver transplant patients, transplanted liver donors preserved by conventional refrigerated methods, and liver tissue extracted for microarray gene expression analysis before and 60 min after reperfusion at the end of preservation, for a total of 11 sets of samples (Jassem W et al., 2019).
The FerrDB database is the first in the world to integrate all data from the ferroptosis-related research literature in Pubmed (Zhou N et al., 2020), through which we downloaded FRGs and obtained a total of 484 FRGs, including driver genes 264, suppressor genes 238 and marker genes 9.
Differential analysis of HIRI-related genes and mapping of volcano chart
We performed the differential analysis of gene expression data before and after liver reperfusion for 11 selected samples in the GSE112713 dataset by the GEO2R (Barrett T et al., 2012) online tool in the GEO database. We used the R software (version: 4.2.1) (R Core Team. 2022) ggplot2 package (version: 3.3.6) (H. Wickham. 2016) with p-value<0.05 and |logFC|≥1 for volcano chart plotting.
FRGs identification and mapping of venn diagrams
We took the differentially expressed genes (DEGs) obtained for HIRI and intersected them with FRGs, and plotted them online via the VENNY (version: 2.1) website (Oliveros, 2007) to plot the venn diagram online.
Gene ontology analysis and kyoto encyclopedia of genes and genomes analysis
We took the intersecting genes obtained from the DEGs of HIRI with FRGs obtained using the R software (version: 4.2.1) (R Core Team. 2022) clusterProfiler package (version: 4.4.4) (Wu T et al., 2021), the org.Hs.eg.db package (version: 3.15.0) (Marc C. 2022), the enrichplot package (version: 1.16.2) (Yu G. 2022), and ggplot2 package (version: 3.3.6) (H. Wickham. 2016) for GO analysis and KEGG analysis, and ranked the results of GO analysis and KEGG analysis from smallest to largest with p-value<0.05. The top ten results in GO analysis and the top thirty results in KEGG were selected and plotted as bar graphs and bubble plots respectively.
Building protein-protein interaction networks
The String database (version: 11.5) is one of the most data-rich and widely used databases for studying protein interactions (Szklarczyk D et al., 2019). We put the obtained DEGs of HIRI with the intersecting genes obtained from FRGs through this database for PPI network construction.
Cytoscape software analyses PPI networks and screens for hub genes
Cytoscape software (version: 3.9.1) is software that graphically displays networks and performs analysis and editing (Shannon P et al., 2003). CytoHubba is an APP within this software that screens Hub genes in PPI networks, and we used one of the most widely used MCC algorithms to process PPI networks and screen Hub genes (Chin C-H et al., 2014).
Hub gene GO analysis and KEGG analysis
We performed GO and KEGG analyses on the screened Hub genes using the same methods described above.
Predicting interactions between genes
The GeneMANIA database can help predict interactions between genes (Mostafavi S et al., 2008), and we used this database to predict interactions between genes associated with Hub genes.
Prediction of pharmacogenetic interactions
The DGIdb database (version: 4.2.0) is a drug-gene interaction database that provides information on the association of genes with their known or potential drugs (Freshour S L et al., 2021), which we used to predict the drugs associated with them for Hub genes and visualised them using Cytoscape software (version: 3.9.1) for visualisation.
miRNA prediction
The miRWalk database (version: 2.0) enables the prediction of gene-miRNA interactions (Sticht C et al., 2018). We used this database to make relevant predictions of miRNA expression for Hub genes and used it validated as a screening condition. The results were further screened and finally visualised using Cytoscape software (version: 3.9.1).
Initial validation of the hub gene
We selected dataset GSE15480 from the GEO database to validate Hub genes. Dataset GSE15480 studies the global gene expression in deceased donor liver transplantation with ischemic preconditioning (Raza A et al., 2010). We selected six standards (no IPC) at preimplantation versus six standards (no IPC) at 90 min post-reperfusion for differential analysis by the GEO2R (Barrett T et al., 2012) online tool in the GEO database. We used the R software (version: 4.2.1) (R Core Team. 2022) ggplot2 package (version: 3.3.6) (H. Wickham. 2016) to p-value<0.05 and |logFC|≥1 for volcano chart plotting. Finally, the venn diagram was plotted online with FRGs, genes in Table 1, and Hub genes through the VENNY (version: 2.1) website (Oliveros, 2007) for initial validation of Hub genes.
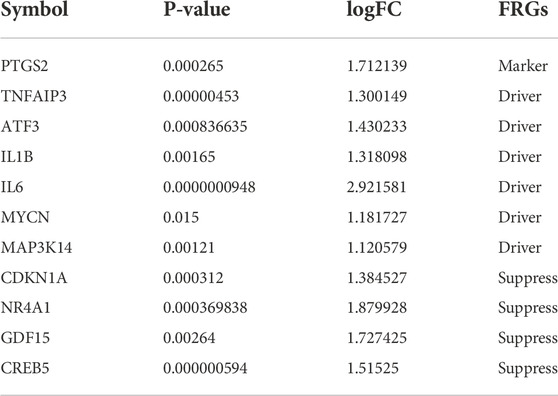
TABLE 1. Symbols of DEGs for 11 HIRI-related FRGs, including the corresponding p-value, logFC, and FRGs.
Results
Differential analysis of HIRI-related genes and mapping of volcano chart
We obtained 166 HIRI-associated DEGs, of which 161 were up-regulated genes and 5 were down-regulated genes and mapped the volcano chart (Figure 2A).
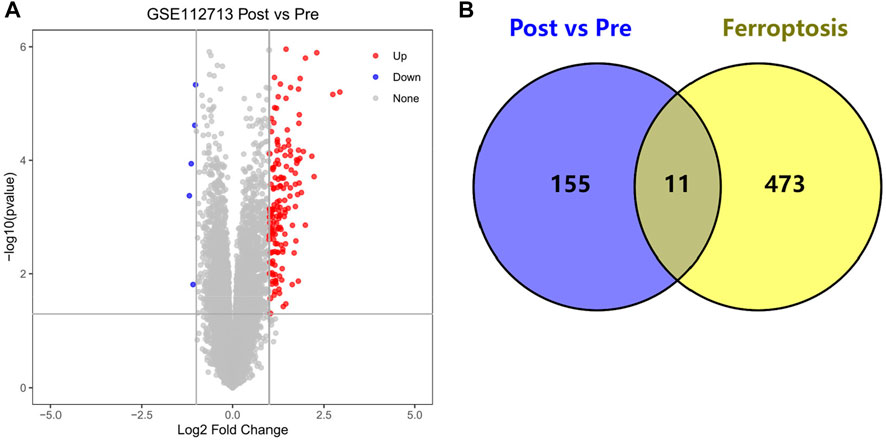
FIGURE 2. (A) Volcano chart mapped by 166 HIRI-related DEGs, (B) Venn chart of 166 “Post vs. Pre” DEGs with 484 “Ferroptosis” genes.
FRGs identification and mapping of venn diagrams
We obtained DEGs for 11 HIRI-associated FRGs (Table 1) and plotted venn chart (Figure 2B).
GO analysis of HIRI-related FRGs
The top 10 results of the GO analysis with the smallest p-value for biological process (BP), cellular component (CC) and molecular function (MF) were selected and plotted in order of bar chart (Figure 3A) and bubble chart (Figure 3B). We can see from the figure that in BP, the central enrichment is in “cellular response to external stimulus”, “response to lipopolysaccharide” and “response to molecule of bacterial origin”; in CC, the central enrichment is in “nuclear membrane” and “endoplasmic reticulum lumen”; in MF, the central enrichment is in “cytokine activity”, “DNA-binding transcription activator activity, RNA polymerase II-specific”, “DNA-binding transcription activator activity”, “receptor ligand activity” and “signaling receptor activator activity".
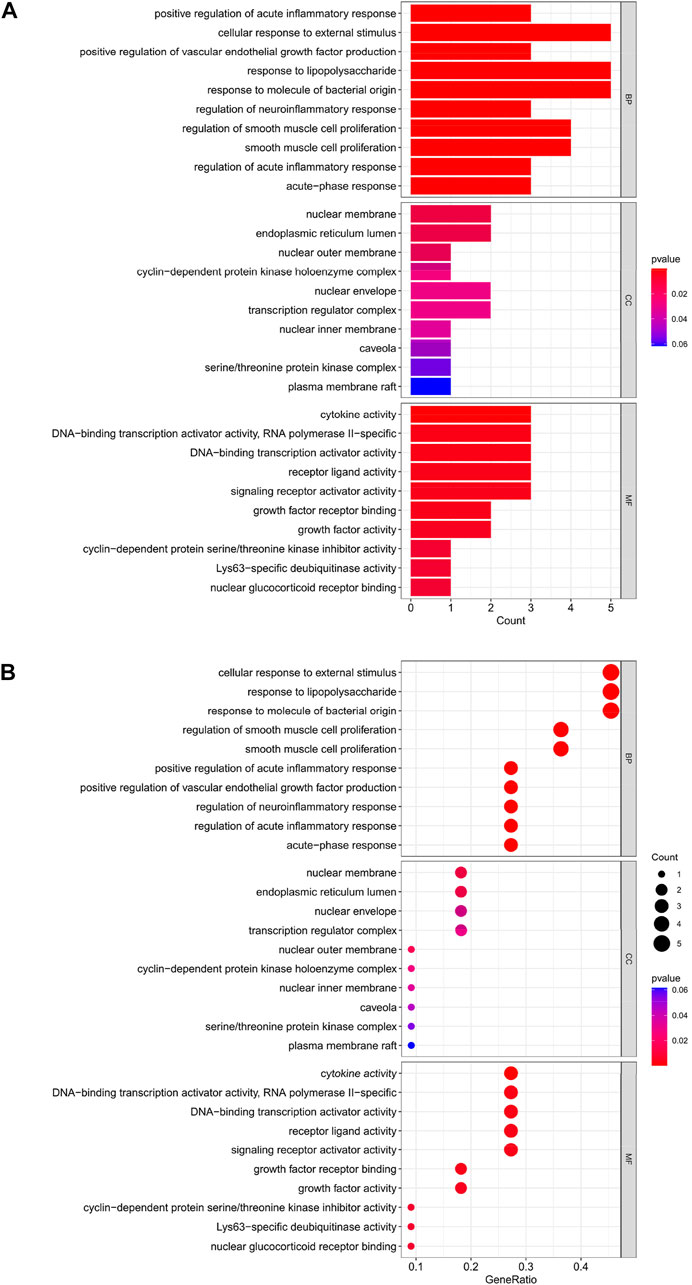
FIGURE 3. (A) Bar chart plotted against the results of GO analysis for the 11 genes in Table 1, (B) Bubble chart plotted against the results of GO analysis for the 11 genes in Table 1.
KEGG analysis of HIRI-associated FRGs
The top 30 analyses with the smallest p-value in the KEGG analysis were selected to plot a bar chart (Figure 4A) with a bubble chart (Figure 4B). The figure shows that there are eight main areas of enrichment, including “TNF signaling pathway” and “Human cytomegalovirus infection".
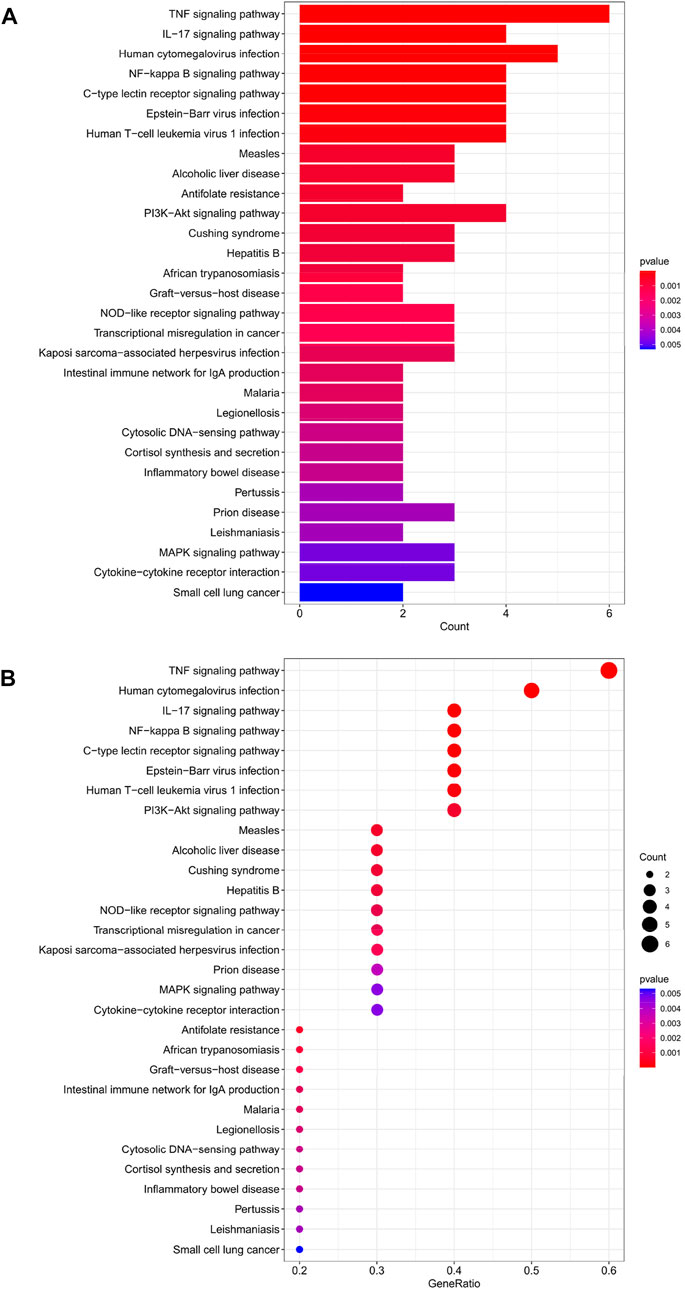
FIGURE 4. (A) Bar chart plotted against the results of KEGG analysis for the 11 genes in Table 1, (B) Bubble chart plotted against the results of KEGG analysis for the 11 genes in Table 1.
HIRI-related FRGs build PPI networks
The PPI network was constructed for the 11 genes in Table 1 (Figure 5A), which has 11 nodes and 22 edges, with the “CREB5” gene not associated with any other gene.
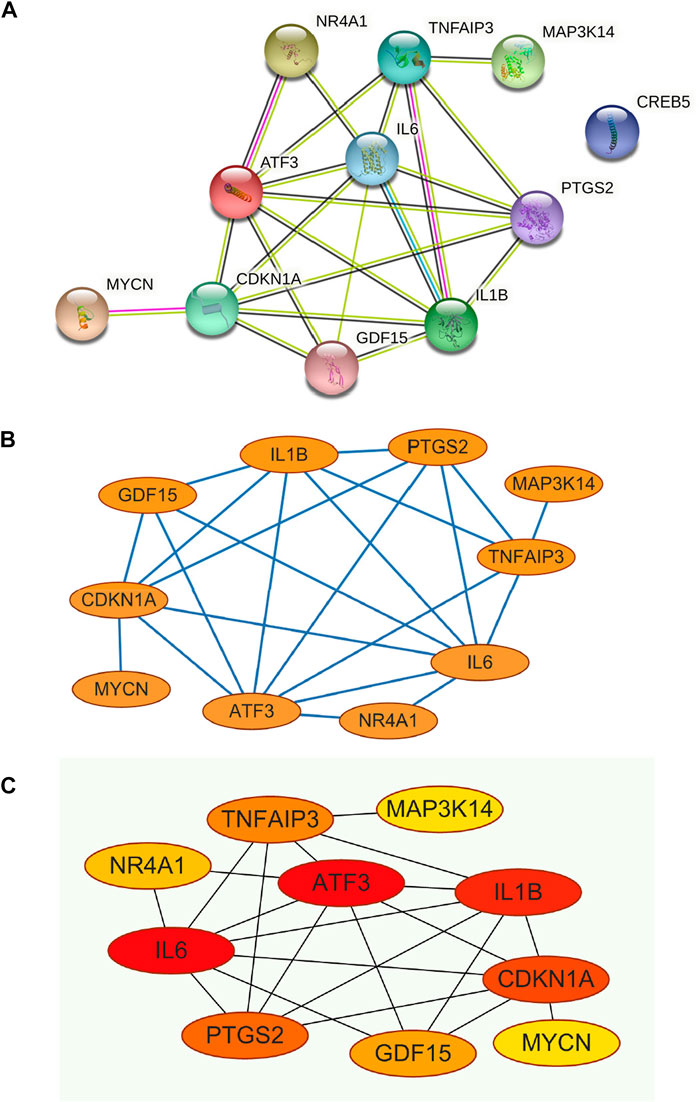
FIGURE 5. (A) Construction of PPI networks for the 11 genes in Table 1, (B) The 11 genes in Table 1 were analysed using Cytoscape software, (C) Analysis of the 11 genes in Table 1 using the MCC algorithm for the CytoHubba function in Cytoscape software yielded Hub Genes.
Cytoscape software analyses PPI networks to screen for hub genes
The constructed PPI network was analysed by the Cytoscape software CytoHubba function (Figure 5B and Figure 5C), and 11 genes were scored using the MCC algorithm (Table 2). The top 5 genes with the highest scores were selected as Hub genes, namely “ATF3”, “IL6”, “IL1B”, “CDKN1A”, and “PTGS2”.
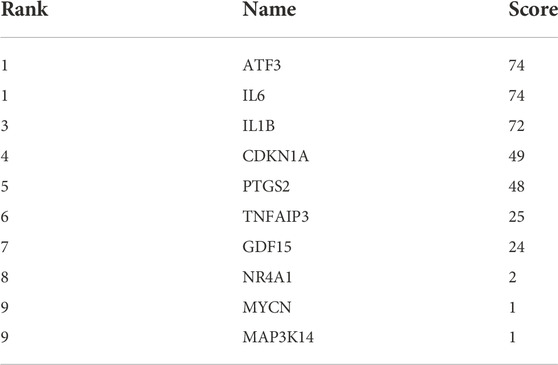
TABLE 2. The 11 genes in Table 1 were scored by applying the MCC algorithm for the CytoHubba function in Cytoscape software.
Hub gene GO analysis
The top 10 results of the GO analysis with the smallest p-value for BP, CC and MF were selected and plotted in turn as a bar chart (Figure 6A) and bubble chart (Figure 6B). In the GO analysis of these 5 Hub genes, ten aspects were enriched in BP, including “cellular response to external stimulus”; in CC, the central enrichment was in 4 areas such as “endoplasmic reticulum lumen”; in MF, it is mainly enriched in “growth factor receptor binding”, “cytokine activity”, “cytokine receptor binding”, “receptor ligand activity”, “signaling receptor activator activity” these five aspects.
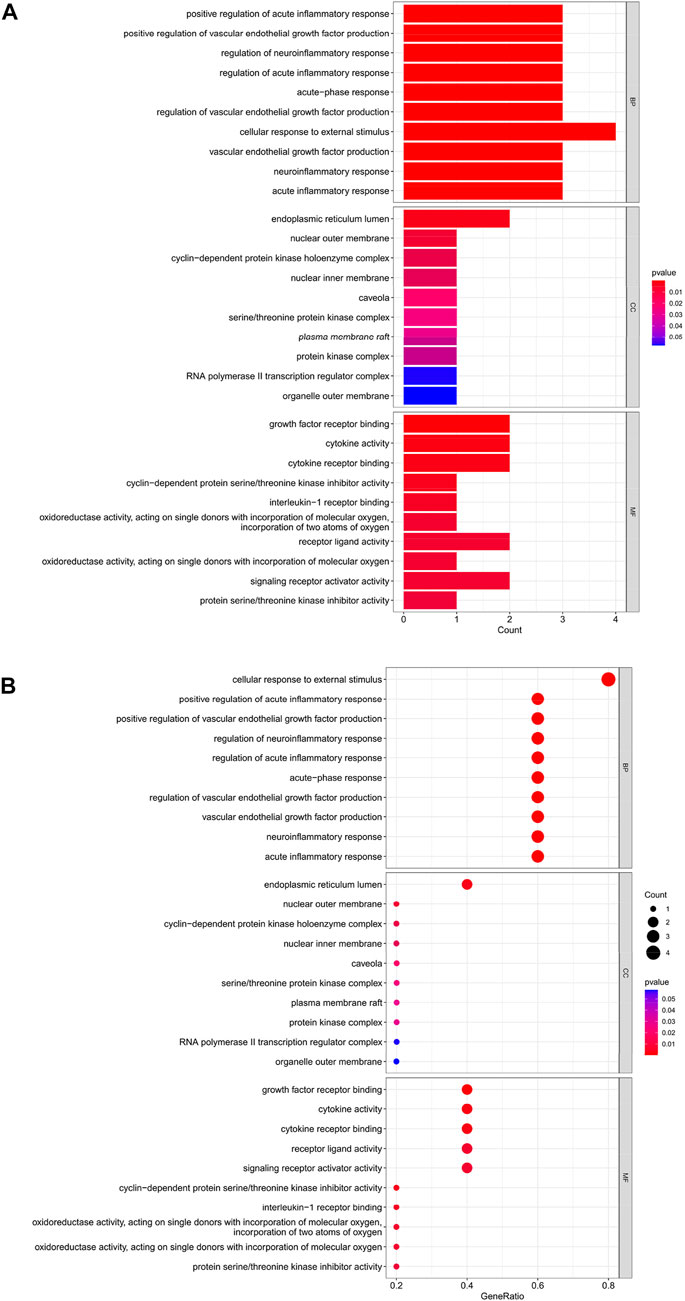
FIGURE 6. (A) Bar chart plotted against the results of GO analysis for the Hub gene, (B) Bubble chart plotted against the results of GO analysis for the Hub gene.
Hub gene KEGG analysis
The top 30 analyses with the smallest p-value in the KEGG analysis were selected to plot a bar chart (Figure 7A) with a bubble chart (Figure 7B). As we can see from the figure, the central enrichment is in six areas, including “Human cytomegalovirus infection".
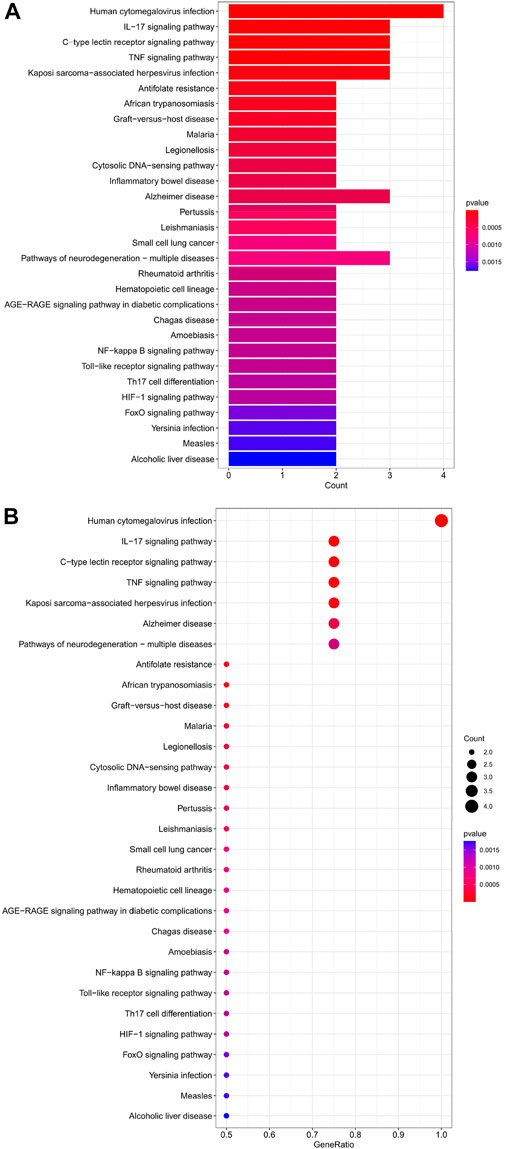
FIGURE 7. (A) Bar chart plotted against the results of KEGG analysis for the Hub gene, (B) Bubble chart plotted against the results of KEGG analysis for the Hub gene.
Predicting interactions between genes
The five Hub genes screened were predicted to interact with each other, and 20 genes were predicted to potentially interact with these five Hub genes (Figure 8A).
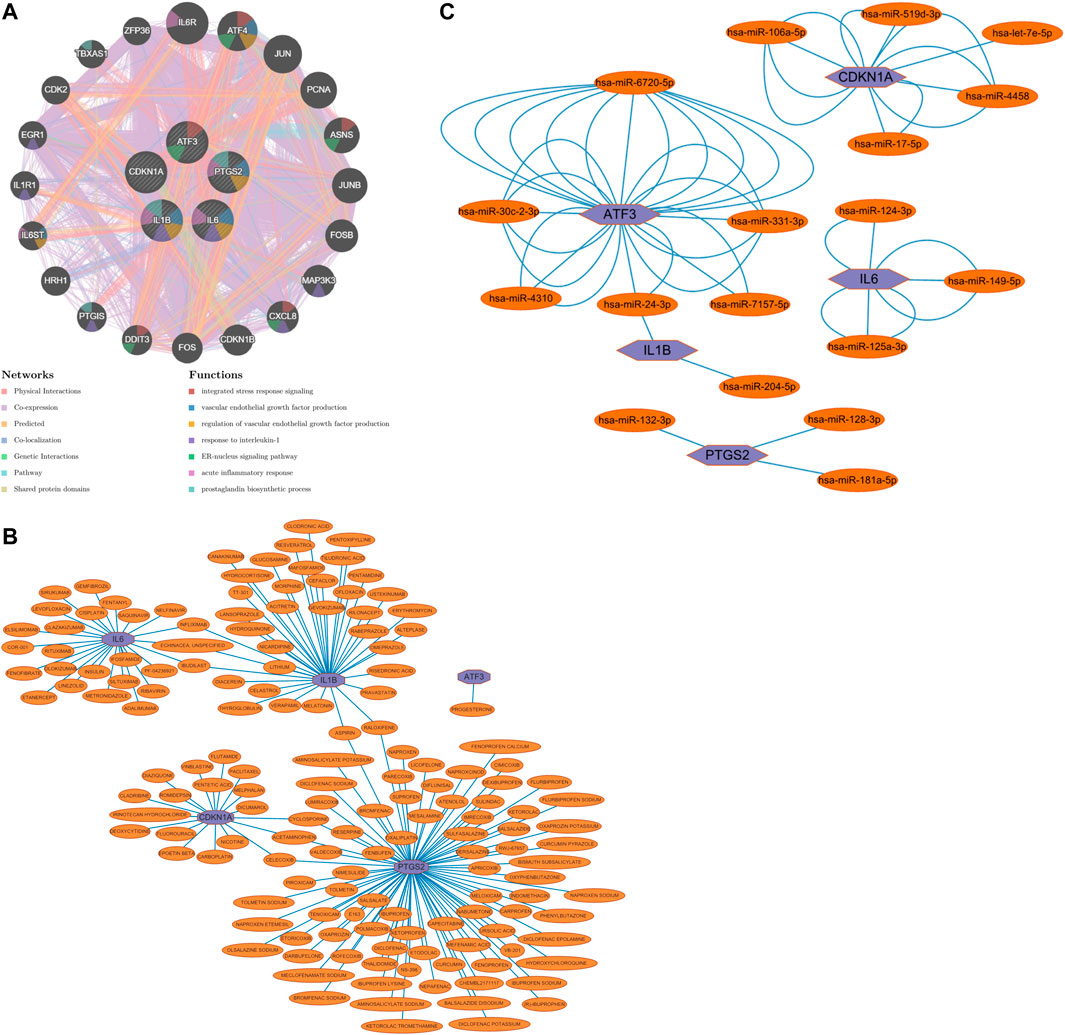
FIGURE 8. (A) Prediction of gene interactions on Hub Gene, (B) Prediction of drug-gene interactions on Hub Gene, (C) The miRNA expression prediction on Hub Gene.
Prediction of drug-gene interactions
Drug-gene interaction predictions for the five Hub genes screened predicted that one could act on ATF3, 25 on IL6, 37 on IL1B, 18 on CDKN1A and 83 on PTGS2; three on both IL6 and IL1B; two on both IL1B and PTGS2; and three on both CDKN1A and PTGS2 (Figure 8B).
miRNA prediction
The five Hub genes screened were subjected to miRNA prediction. After screening the predicted miRNAs, there were six miRNAs potentially expressed by ATF3, two miRNAs potentially expressed by IL1B, three miRNAs potentially expressed by PTGS2, three miRNAs potentially expressed by IL6, and five miRNAs potentially expressed by CDKN1A, with one miRNA potentially expressed in common by ATF3 and IL1B (Figure 8C).
Initial validation of the hub gene
We performed differential analysis on the dataset GSE15480 and obtained a total of 142 DEGs, including 129 up-regulated genes and 13 down-regulated genes, and mapped the volcano chart (Figure 9A). Compared with 484 genes in FRGs, 10 of them were initially validated, namely ATF3, TNFAIP3, IL1B, IL6, PTGS2, JUN, CDKN1A, GJA1, MAP3K14, NR4A1. Compared with 11 genes in Table 1, 8 were initially validated, namely ATF3, TNFAIP3, IL1B, IL6, PTGS2, CDKN1A, MAP3K14, and NR4A1. These were initially validated compared to the five Hub genes screened in Table 2 (Figure 9B).
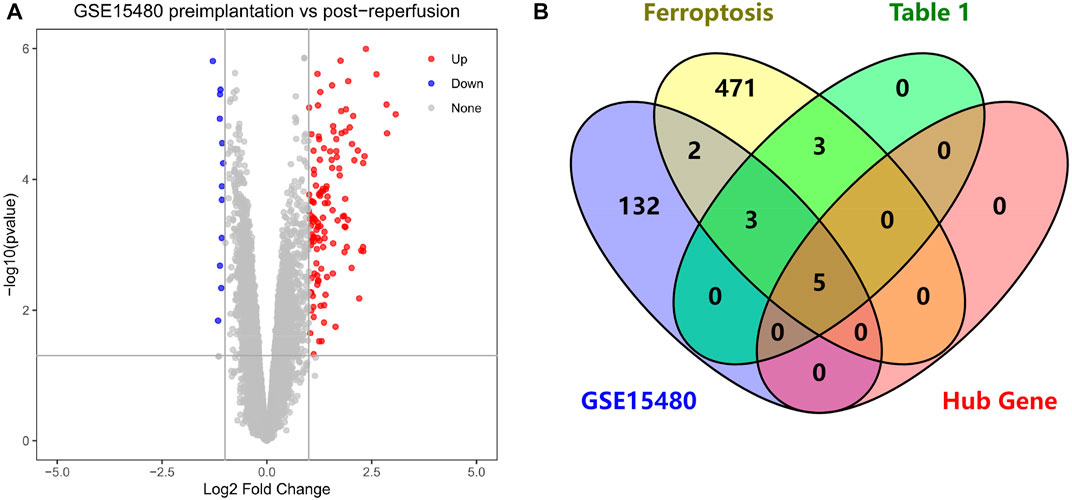
FIGURE 9. (A) Volcano chart mapped by 142 HIRI-related DEGs, (B) Venn chart of “Ferroptosis”, “Table 1”, “GSE15480”, “Hub Gene”.
Discussion
HIRI is a complex pathophysiological process involving multiple factors acting together to exacerbate liver damage, dysfunction and increased structural damage when blood flow is restored after a period of insufficient or interrupted blood flow to the liver (Jaeschke H. 2003; Klune J R et al., 2010). Not only does it affect the physiological functions of the liver, leading to liver dysfunction or even failure in patients, but the severe stress and inflammatory response brought about by HIRI can cause acute dysfunction of other organs such as the heart, kidneys and lungs, and even acute multi-organ failure. HIRI is, therefore, not just a little complication but a severe complication with devastating systemic effects that severely affect patients’ prognosis and quality of survival (Nastos C et al., 2014). Whereas hepatectomy or liver transplantation is currently the predominant surgical procedure for treating primary liver cancer (Maki H et al., 2022), both inevitably result in HIRI. Therefore, the question of how to reduce or even avoid the harmful effects of HIRI has been a hot topic in clinical work and has not been effectively addressed.
The pathophysiological mechanisms of HIRI are currently known in terms of anaerobic metabolism, oxygen-free radical production, Ca2+ overload, Kupffer cell production, activation of the complement system, apoptosis and necrosis (Jaeschke H. 2003; Montalvo-Jave E E et al., 2008; Klune J R et al., 2010). However, many unknown pathophysiological mechanisms exist and have not been confirmed by research. The pathophysiological mechanisms of HIRI have therefore been controversial in clinical practice. Ferroptosis is a newly discovered mode of cell death and has been shown to play an essential role in HIRI (Li J. 2020; Stockwell B R. 2022). As a complement to the pathophysiological mechanisms of HIRI, there are fewer studies in this area, so we explored the relationship between the two in theory employing bioinformatics analysis.
Through our studies, we have found that differences in expression at the gene level occur in the liver before and after reperfusion after a period of ischaemia. We define p-value<0.05 and |logFC|≥1 as DEGs. These DEGs determine the differential expression of proteins. As seen from our plot of the volcano (Figure 2A), the vast majority of these DEGs are up-regulated genes. We, therefore, suggest that the differential proteins expressed by DEGs may be a cause of HIRI. Of these DEGs, 11 of these genes are FRGs (Figure 2B) (Table 1). Most of these FRGs are Driver genes (Zhou N et al., 2020), genes that drive ferroptosis, have a positive regulation of ferroptosis, and have a Mark gene (Zhou N et al., 2020) that does not regulate ferroptosis but which marks the occurrence of ferroptosis, reinforcing the existence of a strong link between ferroptosis and HIRI. In GO analysis of these 11 genes (Figure 3B) with KEGG analysis (Figure 4B), the necessary conditions were provided for us to validate ferroptosis with HIRI further. In the PPI network constructed for these 11 genes (Figure 5A), expression was strongly linked between genes, but the CREB5 gene was not linked to any other gene and could therefore be targeted for elimination in subsequent studies. We analysed the PPI network again using statistical methods (Figure 5B), narrowed it down and screened it again (Figure 5C), and screened for five Hub genes (Table 2). For the five Hub genes GO analysis (Figure 6B) with KEGG analysis in (Figure 7B), which is an essential basis for our following study. In a protein interaction prediction of Hub genes, there were 20 genes with which they interacted that were co-expressed in some way (Figure 8A), which provided a direction for our following study. We applied the study data from the dataset GSE15480 to provide a preliminary validation of our study. As shown in Figure 9B, the FRGs in this dataset overlap with the 11 genes screened in Table 1 and contained all of the Hub genes we screened. This further suggests that ferroptosis plays an essential role in HIRI with these five genes.
Activation transcription factor 3 (ATF3) is a basic leucine zipper (bZIP) DNA binding protein, a member of the ATF/CREB family of transcription factors, and the stress response induces widespread expression of ATF3 (Hai T et al., 1999). Furthermore, HIRI can lead to severe stress reactions in the organism. It has been shown that ATF3 promotes erastin-induced ferroptosis by suppressing system Xc– (Wang L et al., 2020). ATF3 plays an essential role in ferroptosis (Dai C et al., 2020). In HIRI, ATF3 expression is significantly increased, significantly up-regulated and is a very important DEG (Zabala V et al., 2019; Zhong Y et al., 2019). In a comprehensive study of HIRI factor binding proteomics and transcriptomics (Huang S et al., 2019): it was shown that ATF3 is up-regulated by DEGs. This study reveals that ATF3 may be a core factor in HIRI and has potential clinical implications. In our study, ATF3 was the Hub gene, so we speculate that ferroptosis plays an essential role in HIRI in which ATF3 plays an important role.
Interleukin-6 (IL6) is a cytokine with pleiotropic activity that plays an essential role in the acute immune response of the body (Akira S et al., 1993). IL6 has been shown to be required for the induction of ATF3 (Hai T et al., 1999). In contrast, in bioinformatic analysis of ferroptosis in myocardial IRI (Wang Z et al., 2022), IL6 played an essential role in both. In our study, ATF3 and IL6 were screened as Hub genes in the ferroptosis bioinformatic analysis of HIRI; they were intricately linked in the PPI network and intergenic interaction prediction, and ATF3 and IL6 had the same maximum score when scored by the Cytoscape software CytoHubba functional MCC algorithm. We, therefore, hypothesise that ATF3 and IL6 have a central role in the induction of ferroptosis in HIRI. Also, the Hub genes screened in a raw letter analysis of IRI mechanisms in patients undergoing coronary artery bypass grafting (Shen Z et al., 2019) were identical to the Hub genes IL6, IL1B and PTGS2 predicted by our study, and PTGS2 is a Marker gene, and IL6 and IL1B are both Driver genes, thus reinforcing that ferroptosis has an essential role in HIRI.
How to mitigate or avoid the harm caused by HIRI to patients has always been a significant concern in clinical work and is currently a hot topic of research in clinical work. Ferroptosis, one of the pathophysiological mechanisms of HIRI, is beneficial in reducing HIRI by inhibiting the occurrence of ferroptosis. Numerous studies have shown (Kim K M et al., 2020; Yamada N et al., 2020; Liu H et al., 2021; Luo L et al., 2021) that some drugs, such as ferrostatin-1, alpha-tocopherol, deferoxamine, etc. or a low iron diet can reduce HIRI. Therefore, we have also done research in this area and predicted drug-gene interactions for the five Hub genes that were screened (Figure 8B). These predicted drugs could be a reference for future clinical work in targeting HIRI.
MicroRNA (miRNA) is a class of small, endogenous, non-coding RNA molecules of approximately 20–40 nucleotides in length (Saliminejad K et al., 2019). It has a compelling physiological role in regulating gene expression, controlling early development, cell proliferation, apoptosis, cell death, lipid metabolism and cell differentiation, directly affecting tissues, organs and even our entire system[40]. miRNA dysregulation has been demonstrated as a causal factor in disease progression, and miRNAs can be used as biomarkers and therapeutic targets (Paul P et al., 2018). In clinical work, we can use miRNA to monitor organ status, diagnose diseases, treat diseases, etc. Numerous studies have shown that miRNA expression is altered in IRI in organs such as the heart, brain, kidney and intestine and that differential miRNA expression occurs in HIRI. miRNA mediates HIRI, while HIRI can be alleviated by regulating miRNA expression (Cao M et al., 2021; Ingram H et al., 2022; Ye L et al., 2022). We performed miRNA expression prediction for five Hub genes (Figure 8C), further revealing the relationship between ferroptosis and HIRI, which can be used to diagnose the occurrence of ferroptosis in HIRI and may provide a new approach to mitigate HIRI in clinical work. The miRNAs we predicted have the same results as those predicted in the raw letter analysis of IRI mechanisms in coronary artery bypass grafting patients (Shen Z et al., 2019) and different ones. Among them, we predicted has-miR-128-3p, and it has been shown that inhibition of miR-128-3p by Tongxinluo protects human cardiomyocytes from ischemia/reperfusion injury via upregulation of p70s6k1/p-p70s6k1 (Chen G et al., 2017). We predicted hsa-miR-24-3p, and it has been shown that rosuvastatin alleviates ischemia/reperfusion injury in cardiomyocytes by downregulating hsa-miR-24-3p to target up-regulated uncoupling protein 2 (Wei W et al., 2019). We, therefore, hypothesise that there is also some benefit in alleviating ferroptosis in HIRI by regulating miRNA levels.
Although we have done a great deal of work on the relationship between HIRI and ferroptosis, we acknowledge that our study has some limitations. We have only speculated on the link between HIRI and ferroptosis in theory and have used another HIRI-related dataset for preliminary validation of this. We have not further validated our conclusions in animal experiments or clinical trials. Also, our data selection is somewhat limited in that the datasets we selected were all about HIRI in liver transplantation and not about HIRI in liver resection. Therefore, these limitations will also be the focus of our future research efforts to investigate the link between HIRI and ferroptosis in depth and to design and implement rigorous animal or clinical trials to test the theoretical speculations we have obtained. We will apply the results of our research to clinical work to reduce the risks of HIRI, improve the prognosis of patients and enhance their quality of life.
Conclusion
Ferroptosis, a newly discovered mode of cell death, plays an essential role in HIRI. The 11 DEGs and 5 Hub genes of HIRI that we screened by the bioinformatic analysis were closely associated with ferroptosis. Further bioinformatic analysis methods of these genes are beneficial in revealing the potential biological pathways and mechanisms of ferroptosis in HIRI, providing a reference for further in-depth studies. The targeted drugs and miRNA predicted for the five Hubs provide some reference value for clinical work in assessing the diagnosis of ferroptosis in HIRI, the extent of its occurrence and the mitigation of HIRI by means of ferroptosis inhibition. The five Hub genes can be used as potential biomarkers and therapeutic targets for HIRI and also provide some theoretical basis and research directions to explore the relationship between HIRI and ferroptosis further.Oliveros, 2007, R Core Team. R, 2022.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
SS and JL conceived the project and designed the bioinformatics analysis. SS, JX, YG, and JL performed the bioinformatics analysis, and analyzed and interpreted the data. SS drafted the manuscript. SS, JX, YG, and JL revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Government of Hebei Province, China funded the Clinical Medicine Outstanding Talents Project, the Science and Technology Research and Development Program of Chengde City, Hebei Province, China (201904A044).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akira, S., Taga, T., and Kishimoto, T. (1993). Interleukin-6 in biology and medicine. Adv. Immunol. 54, 1–78. doi:10.1016/s0065-2776(08)60532-5
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2012). NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 41 (D1), D991–D995. doi:10.1093/nar/gks1193
Bray, F., Laversanne, M., Weiderpass, E., and Soerjomataram, I. (2021). The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 127 (16), 3029–3030. doi:10.1002/cncr.33587
Cao, M., Song, W., Liang, R., Teng, L., Zhang, M., Zhang, J., et al. (2021). MicroRNA as a potential biomarker and treatment strategy for ischemia-reperfusion injury. Int. J. Genomics 2021, 1–12. doi:10.1155/2021/9098145
Chen, G., Xu, C., Zhang, J., Li, Q., Cui, H. H., Li, X. D., et al. (2017). Inhibition of miR-128-3p by Tongxinluo protects human cardiomyocytes from ischemia/reperfusion injury via upregulation of p70s6k1/p-p70s6k1. Front. Pharmacol. 8, 775. doi:10.3389/fphar.2017.00775
Chin, C-H., Chen, S-H., Wu, H-H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8 (S4), S11. doi:10.1186/1752-0509-8-S4-S11
Dai, C., Chen, X., Li, J., Comish, P., Kang, R., and Tang, D. (2020). Transcription factors in ferroptotic cell death. Cancer Gene Ther. 27 (9), 645–656. doi:10.1038/s41417-020-0170-2
Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., et al. (2012). Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149 (5), 1060–1072. doi:10.1016/j.cell.2012.03.042
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 (1), 207–210. https://www.ncbi.nlm.nih.gov/geo/, doi:10.1093/nar/30.1.207
Freshour, S. L., Kiwala, S., Cotto, K. C., Coffman, A. C., McMichael, J. F., Song, J. J., et al. (2021). Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 49 (D1), D1144–D1151. https://www.dgidb.org/, doi:10.1093/nar/gkaa1084
Hai, T., Wolfgang, C. D., Marsee, D. K., Allen, A. E., and Sivaprasad, U. (1999). ATF3 and stress responses. Gene Expr. 7 (4-6), 321–335.
Huang, S., Ju, W., Zhu, Z., Han, M., Sun, C., Tang, Y., et al. (2019). Comprehensive and combined omics analysis reveals factors of ischemia-reperfusion injury in liver transplantation. Epigenomics 11 (5), 527–542. doi:10.2217/epi-2018-0189
Ingram, H., Dogan, M., Eason, J. D., Kuscu, C., and Kuscu, C. (2022). MicroRNAs: Novel targets in hepatic ischemia–reperfusion injury. Biomedicines 10, 12. doi:10.3390/biomedicines10040791
Jaeschke, H. (2003). Molecular mechanisms of hepatic ischemia-reperfusion injury and preconditioning. Am. J. Physiol. Gastrointest. Liver Physiol. 284 (1), G15–G26. doi:10.1152/ajpgi.00342.2002
Jassem, W., Xystrakis, E., Ghnewa, Y. G., Yuksel, M., Pop, O., Martinez-Llordella, M., et al. (2019). Normothermic machine perfusion (NMP) inhibits proinflammatory responses in the liver and promotes regeneration: Hepatology. Hepatology 70 (2), 682–695. doi:10.1002/hep.30475
Kim, K. M., Cho, S. S., and Ki, S. H. (2020). Emerging roles of ferroptosis in liver pathophysiology. Arch. Pharm. Res. 43 (10), 985–996. doi:10.1007/s12272-020-01273-8
Klune, J. R., and Tsung, A. (2010). Molecular biology of liver ischemia/reperfusion injury: Established mechanisms and recent advancements. Surg. Clin. North Am. 90 (4), 665–677. doi:10.1016/j.suc.2010.04.003
Li, J. (2020). Ferroptosis: Past, present and future. Cell death Dis. 11, 1–13. doi:10.1038/s41419-020-2298-2
Liu, H., and Man, K. (2021). New insights in mechanisms and therapeutics for short- and long-term impacts of hepatic ischemia reperfusion injury post liver transplantation. Int. J. Mol. Sci., 22, 17.
Luo, L., Mo, G., and Huang, D. (2021). Ferroptosis in hepatic ischemia‑reperfusion injury: Regulatory mechanisms and new methods for therapy (Review). Mol. Med. Rep. 23, 225. doi:10.3892/mmr.2021.11864
Maki, H., and Hasegawa, K. (2022). Advances in the surgical treatment of liver cancer. Biosci. Trends 16 (3), 178–188. doi:10.5582/bst.2022.01245
Marc, C. (2022). Org Hs eg db. Genome Wide Annot. Hum. R Package Version 3.15.0[J]. doi:10.18129/B9.bioc.org.Hs.eg.db
Montalvo-Jave, E. E., Escalante-Tattersfield, T., Ortega-Salgado, J. A., Pina, E., and Geller, D. A. (2008). Factors in the pathophysiology of the liver ischemia-reperfusion injury. J. Surg. Res. 147 (1), 153–159. doi:10.1016/j.jss.2007.06.015
Mostafavi, S., Ray, D., Warde-Farley, D., Grouios, C., and Morris, Q. (2008). GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9 (1), S4. http://genemania.org/, doi:10.1186/gb-2008-9-s1-s4
Nastos, C., Kalimeris, K., Papoutsidakis, N., Tasoulis, M. K., Lykoudis, P. M., Theodoraki, K., et al. (2014). Global consequences of liver ischemia/reperfusion injury. Oxidative Med. Cell. Longev. 2014, 1–13. doi:10.1155/2014/906965
Oliveros, J. C. (2007). Venny. An interactive tool for comparing lists with Venn's diagrams. BioinfoGP. https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Paul, P., Chakraborty, A., Sarkar, D., Langthasa, M., Rahman, M., Bari, M., et al. (2018). Interplay between miRNAs and human diseases. J. Cell. Physiol. 233 (3), 2007–2018. doi:10.1002/jcp.25854
R Core Team. R (2022). A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Raza, A., Dikdan, G., Desai, K. K., Shareef, A., Fernandes, H., Aris, V., et al. (2010). Global gene expression profiles of ischemic preconditioning in deceased donor liver transplantation. Liver Transplant. 16 (5), 588. doi:10.1002/lt.22049
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 234 (5), 5451–5465. doi:10.1002/jcp.27486
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shen, Z., Lu, J., Wei, J., Zhao, J., Wang, M., Wang, M., et al. (2019). Investigation of the underlying hub genes and mechanisms of reperfusion injury in patients undergoing coronary artery bypass graft surgery by integrated bioinformatic analyses. Ann. Transl. Med. 7 (22), 664. doi:10.21037/atm.2019.10.43
Sticht, C., De La Torre, C., Parveen, A., and Gretz, N. (2018). miRWalk: An online resource for prediction of microRNA binding sites. PLOS ONE 13 (10), e0206239. http://mirwalk.umm.uni-heidelberg.de/, doi:10.1371/journal.pone.0206239
Stockwell, B. R. (2022). Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 185 (14), 2401–2421. doi:10.1016/j.cell.2022.06.003
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (1), D607–D613. https://cn.string-db.org/, doi:10.1093/nar/gky1131
Wang, L., Liu, Y., Du, T., Yang, H., Lei, L., Guo, M., et al. (2020). ATF3 promotes erastin-induced ferroptosis by suppressing system Xc. Cell Death Differ. 27 (2), 662–675. doi:10.1038/s41418-019-0380-z
Wang, Z., He, Z., Xuan, Q., Zhang, Y., Xu, J., Lin, J., et al. (2022). Analysis of the potential ferroptosis mechanism and multitemporal expression change of central ferroptosis-related genes in cardiac ischemia-reperfusion injury. Front. Physiol. 13, 934901. doi:10.3389/fphys.2022.934901
Wei, W., Peng, J., and Shen, T. (2019). Rosuvastatin alleviates ischemia/reperfusion injury in cardiomyocytes by downregulating hsa-miR-24-3p to target upregulated uncoupling protein 2. Cell. Reprogr. 21 (2), 99–107. doi:10.1089/cell.2018.0039
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2 (3), 100141. doi:10.1016/j.xinn.2021.100141
Yamada, N., Karasawa, T., Wakiya, T., Sadatomo, A., Ito, H., Kamata, R., et al. (2020). Iron overload as a risk factor for hepatic ischemia-reperfusion injury in liver transplantation: Potential role of ferroptosis. Am. J. Transpl. 20 (6), 1606–1618. doi:10.1111/ajt.15773
Ye, L., and Li, S. (2022). Novel targets and therapeutic strategies to protect against hepatic ischemia reperfusion injury. Front. Med. 8, 15.
Yu, G. (2022). Enrichplot: Visualization of functional enrichment result. R package version 1.16.2. Available at: https://yulab-smu.top/biomedical-knowledge-mining-book/.
Zabala, V., Boylan, J. M., Thevenot, P., Frank, A., Senthoor, D., Iyengar, V., et al. (2019). Transcriptional changes during hepatic ischemia-reperfusion in the rat. PLOS ONE 14 (12), e0227038. doi:10.1371/journal.pone.0227038
Zhong, Y., Zhu, Y., Dong, J., Zhou, W., Xiong, C., Xue, M., et al. (2019). Identification of key transcription factors AP-1 and AP-1-dependent miRNAs forming a Co-regulatory network controlling PTEN in liver ischemia/reperfusion injury. BioMed Res. Int. 2019, 1–11. doi:10.1155/2019/8962682
Zhou, N., and FerrDb, Bao J. (2020). A manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database 2020, baaa021. http://www.zhounan.org/ferrdb/current/, doi:10.1093/database/baaa021
Keywords: hepatic ischemia-reperfusion injury, ferroptosis, bioinformatics analysis, prediction of drug-gene interactions, MiRNA prediction
Citation: Sun S, Xue J, Guo Y and Li J (2022) Bioinformatics analysis of genes related to ferroptosis in hepatic ischemia-reperfusion injury. Front. Genet. 13:1072544. doi: 10.3389/fgene.2022.1072544
Received: 17 October 2022; Accepted: 15 November 2022;
Published: 02 December 2022.
Edited by:
Zili Zhang, Nanjing University of Chinese Medicine, ChinaReviewed by:
Yang Wu, Nanjing Medical University, ChinaZhuang Yanshuang, Taizhou Traditional Chinese Medicine Hospital, China
Copyright © 2022 Sun, Xue, Guo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianling Li, bHlqaWFubGluZ0AxNjMuY29t
 Shuo Sun
Shuo Sun Jianming Xue
Jianming Xue Yunfei Guo
Yunfei Guo