
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 21 November 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1058741
This article is part of the Research Topic Chromosome and Genome Biology of Fungi View all 7 articles
 Callum J. Fraser
Callum J. Fraser Simon K. Whitehall*
Simon K. Whitehall*Heterochromatin is a repressive chromatin state that plays key roles in the functional organisation of eukaryotic genomes. In fungal plant pathogens, effector genes that are required for host colonization tend to be associated with heterochromatic regions of the genome that are enriched with transposable elements. It has been proposed that the heterochromatin environment silences effector genes in the absence of host and dynamic chromatin remodelling facilitates their expression during infection. Here we discuss this model in the context of the key wheat pathogen, Zymoseptoria tritici. We cover progress in understanding the deposition and recognition of heterochromatic histone post translational modifications in Z. tritici and the role that heterochromatin plays in control of genome plasticity and virulence.
Zymoseptoria tritici is responsible for Septoria tritici blotch (STB), a world-wide foliar disease of wheat that is characterised by the appearance of necrotic lesions and in some cases the death of the plant. Yields in an infected crop may be reduced by 30–50% if the disease is not properly controlled via the application of fungicides (Fones and Gurr, 2015). However, the increasing incidence of fungicide resistance means that Z. tritici is a major threat to wheat production (Torriani et al., 2015).
Infection is initiated when Z. tritici spores on the leaf surface germinate and hyphae invade plant tissue through stomata. This is followed by a slow, asymptomatic colonisation of the apoplastic space of the surrounding mesophyll. After approximately 10 days (depending on temperature), there is an abrupt switch from “stealth” infection to necrotrophic growth during which plant cells undergo programmed cell death. The resulting release of nutrients from the dying plant tissue facilitates a rapid increase in fungal biomass and the formation of pycnidia which contain pycnidiospores that are released to infect the leaves of neighbouring plants (Steinberg, 2015).
Plant colonizing fungi produce a repertoire of secreted effector proteins that modulate plant immunity and host cell physiology (Uhse and Djamei, 2018). The expression of effectors is tightly regulated so that they are produced at the appropriate stage and level during infection (Uhse and Djamei, 2018). Accordingly, Z. tritici undergoes several major gene expression reprogramming events during the infection of wheat leaves (Kellner et al., 2014; Rudd et al., 2015). These events include a large up-regulation of secreted proteins approximately 9 days after infection which likely underpins the shift to necrotrophic growth. After approximately 14 days there is a general reduction in the expression of secreted proteins in favour of cell wall and carbohydrate degrading enzymes (Rudd et al., 2015). However, the mechanisms by which these large scale transcriptional reprogramming events are regulated remains to be determined.
The distribution of effector genes in the genomes of filamentous plant pathogens is not random. Instead effector genes, and genes involved in the production of secondary metabolites (SM), tend to be located in clusters within, or close to, regions of the genome that are enriched in repetitive sequences such as transposable elements (TEs) (Dong et al., 2015). An association with TEs is significant because it is increasingly clear that these elements can be utilised by cells as sources of genetic variability that facilitate genome evolution (Colonna Romano and Fanti, 2022). Mobilization of TEs to new sites in the genome may influence the expression of adjacent genes and also their repetitive nature makes them potential substrates for recombination and thus drivers of genomic rearrangements. That TE/repeat-enriched genomic regions are rapidly evolving has given rise to the “two speed genome hypothesis” where specific compartments provide sources of genetic plasticity that drive adaptation to the host species (Dong et al., 2015; Seidl and Thomma, 2017; Fouche et al., 2018).
There is growing evidence indicating that TEs have shaped the compartmentalisation of the Z. tritici genome. Z. tritici has 13 essential core chromosomes and up to 8 conditionally dispensable accessory chromosomes that are gene poor and especially enriched with TEs (Goodwin et al., 2011). Accessory chromosomes are frequently lost during vegetative growth and are prone to rearrangements in meiosis with the breakpoints frequently mapping close to TEs (Wittenberg et al., 2009; Goodwin et al., 2011; Croll et al., 2013; Möller et al., 2019). Core chromosomes from different isolates also exhibit significant structural polymorphism resulting from the insertion or deletion of clusters of TEs (Plissonneau et al., 2016; Plissonneau et al., 2018). The TE-associated chromosomal regions that are present or absent in a strain specific manner (which have been termed accessory, orphan or dispensable regions), are enriched for putative effector genes and account for substantial variations in gene content. Indeed, comparative analysis of the genomes of five Z. tritici isolates identified a core set of 9149 genes and a further 6600 strain-specific accessory genes (Plissonneau et al., 2018).
Host cells employ a variety of defence mechanisms that restrict the activity of TEs to tolerable levels (Goodier, 2016). TEs are often embedded in heterochromatin, a condensed form of chromatin that is typically refractory to transcription and other DNA-dependent processes (Allshire and Madhani, 2018; Marsano and Dimitri, 2022). A key property of heterochromatin is the potential to spread from nucleation sites to cover extended domains (Allshire and Madhani, 2018) and as a result, genes in the vicinity of TEs can be subject to heterochromatic silencing (Choi and Lee, 2020). Indeed, the expression of effector and SM genes is suppressed by heterochromatin in a variety of plant-colonizing fungi (Connolly et al., 2013; Chujo and Scott, 2014; Soyer et al., 2014; Studt et al., 2016). As discussed further below, heterochromatin has also been linked to the pathogenicity of Z. tritici; putative effector genes that are upregulated during the switch to necrotrophic growth are enriched in heterochromatic regions of the genome (Meile et al., 2018; Soyer et al., 2019) and loss of key heterochromatin regulators interferes with the ability of Z. tritici to infect wheat leaves (Möller et al., 2019) (Table 1).
Heterochromatin can be subdivided into two basic forms: constitutive and facultative (Liu et al., 2020). Constitutive heterochromatin is typically associated with highly repetitive regions such as telomeres and centromeres and often plays an important architectural role in chromosomes. Facultative heterochromatin is considered to be a more fluid subclass of heterochromatin which can be remodelled in response to appropriate internal or external signals. For example, in metazoans it is associated with developmentally regulated genes and similarly, facultative heterochromatin regulates the expression of SM genes in some filamentous fungi (Liu et al., 2020; Ridenour et al., 2020).
Heterochromatic regions are characterised by the presence of specific histone post-translational modifications (PTMs). Di- and tri-methylation of histone H3 on lysine 9 (H3K9me2/3) are hallmarks of constitutive heterochromatin while facultative heterochromatin in metazoans is classically marked by histone H3 that is trimethylated on lysine 27 (H3K27me3) (Liu et al., 2020). The genomic locations of heterochromatic PTMs in a Z. tritici reference strain have been mapped using ChIP-seq (Schotanus et al., 2015). H3K9me3 was found to cover around 22% of the genome but only a very small number of protein coding genes were found to be associated with this PTM. Instead H3K9me3 predominantly maps to TEs and to subtelomeric regions. H3K27me3 is also found associated with subtelomeric regions and TEs but, in comparison to H3K9me3, has a broader genomic distribution that encompasses specific genic regions. Furthermore, accessory chromosomes are particularly enriched for H3K27me3. In contrast to some other organisms, pericentomeric regions of Z. tritici chromosomes are not marked with H3K9me3. Indeed, Z. tritici centromeres are quite unusual, being relatively small and lacking extensive AT–rich tracts (Schotanus et al., 2015).
The Z. tritici enzymes responsible for catalysing these modifications are conserved SET [Su (var), Enhancer of zeste, Trithorax] domain lysine methyltransferases (KMTases) (Qian and Zhou, 2006). H3K9me2/3 is mediated by Kmt1 which is a homolog of human Suv39H1/2 (Schizosaccharomyces pombe Clr4, Neurospora crassa DIM5) and H3K27me3 by Kmt6 which is a homolog of Drosophila E(Z) and human EZH2/1 (Möller et al., 2019). The functions of H3K9me2/3 and H3K27me3 in Z. tritici have been investigated through the analysis of mutant strains lacking these KMTases. Deletion of kmt1 results in a severe reduction of fitness in vitro and sensitivity to a range of abiotic stresses (osmotic, oxidative and genotoxic). Furthermore, Δkmt1 strains have severely reduced virulence in wheat infections assays indicating that H3K9me2/3 is important for growth in planta. In contrast, loss of kmt6 (and thus H3K27me3) does not reduce growth in vitro and is associated with only modest reduction in virulence (Möller et al., 2019) (Table 1).
H3K27me3 and H3K9me2/3 modifications play opposing roles in the regulation of genome stability. As outlined above, the H3K27me3-enriched accessory chromosomes are highly unstable and are frequently lost during cell division. Deletion of kmt6 decreases the frequency with which these chromosomes are lost indicating that H3K27me3 promotes their instability (Möller et al., 2019). Increased H3K27me3 is also correlated with an elevated frequency of accessory chromosome loss in Fusarium fujikuroi (Janevska et al., 2018). Analysis of kmt6 mutants has also demonstrated that H3K27me3 promotes the rate of spontaneous mutations (Habig et al., 2021). In contrast, H3K9me2/3 functions to suppress spontaneous mutation rates and genomic instability. Deletion of kmt1 is associated with increased accessory chromosome loss, de-repression of TEs and an elevated incidence of chromosomal rearrangements with break points that often map close to TE clusters (Möller et al., 2019; Habig et al., 2021). The high level of chromosomal instability observed in kmt1 mutants is driven by the widespread re-localisation of H3K27me3 such that it invades regions previously occupied by H3K9me2/3. Accordingly, deletion of kmt6 in a Δkmt1 background suppresses genomic instability (Möller et al., 2019).
In addition to TEs, a large set of protein encoding genes is upregulated in the absence of kmt1, which is consistent with H3K9me2/3 playing an important role in gene silencing. However, the majority of these genes are not directly associated with H3K9me2/3 and the changes in their expression may result, at least in part, from the re-distribution of H3K27me3 that occurs in a kmt1 mutant background (Möller et al., 2019). The majority of H3K27me3-marked genes and TEs located on core chromosomes are not activated by its removal and in contrast to some other filamentous fungi, heterochromatic PTMs do not play a major role in the control of Z. tritici SM genes (Möller et al., 2019; Hassani et al., 2022). However, heterochromatin is implicated in the regulation of effector gene expression and thus the interaction of Z. tritici with its host. Analysis of putative effector genes revealed a significant association with genomic loci that are marked with H3K27me3 or overlapping H3K27me2 and H3K9me3 under axenic growth conditions (Soyer et al., 2019). The regulation of four of these effectors (AvStb6, Avr3D1, QTL_5, and Mycgr3G76589) has been analysed using fluorescent reporter genes (Meile et al., 2020). These loci are silenced under axenic conditions but expression is induced during host colonization. Furthermore, the increase in expression at the switch to necrotrophic growth coincides with a decrease in the associated levels of H3K27me3 at all four genes and all but one also had a reduction in H3K9me3 levels. This is consistent with the active remodelling of heterochromatic domains during infection. Moving these effector genes to an ectopic location results in loss of silencing under axenic conditions and aberrant in planta expression indicating that genomic environment is an important facet of their regulation (Meile et al., 2020).
S. pombe Clr4 and N. crassa DIM-5 H3K9 KMTases function in the context of Cul4 ubiquitin ligase complexes, which are called CLRC (Clr4 methyltransferase complex) and DCDC (DIM-5-7-9 CUL-4 DDB1), respectively (Hong et al., 2005; Horn et al., 2005; Jia et al., 2005; Li et al., 2005; Lewis et al., 2010a; Lewis et al., 2010b). Mutations in the genes encoding CLRC/DCDC subunits result in the loss of H3K9methylation in both organisms. This indicates that the integrity of these complexes is required for the catalytic activity of the KMTase and recent work has shown that CLRC ubiquitylates histone H3 on lysine 14 (H3K14ub), promoting H3K9me by Clr4 (Oya et al.). This H3K14ub-H3K9me “crosstalk” mechanism has not yet been investigated in Z. tritici but homologs of CLRC/DCDC subunits are present (Figure 1). Furthermore, Suv39H1 is associated with CUL4 in human cells (Yang et al., 2015) and knockdown of CUL4 reduces H3K9me levels (Higa et al., 2006). As such, a functional link between CUL4 mediated ubiquitylation and methylation of H3K9 may be broadly conserved.
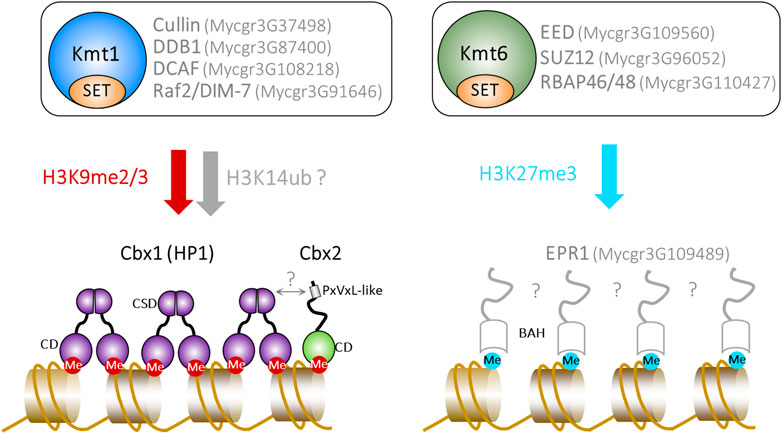
FIGURE 1. The deposition and recognition of heterochromatic histone PTMs in Z. tritici. The top panel shows the predicted composition of Kmt1 (histone H3K9) and Kmt6 (histone H3K27) KMTase complexes. The bottom panel summarises proteins known to be, or predicted to be, involved in the recognition of H3K9me2/3 and H3K27me3. Predicted proteins, activities and interactions are shown in grey. The Ensembl Fungi ID of genes encoding putative proteins is indicated.
Histone PTMs exert their function by acting as docking sites for proteins (termed “readers”) which modify chromatin structure and regulate DNA-dependent processes. The recognition of H3K9me2/3 PTMs is commonly mediated by the Heterochromatin Protein 1 (HP1) family of chromodomain (CD) containing proteins which are present in a variety of eukaryotic organisms including filamentous fungi (Kumar and Kono, 2020). HP1 proteins have a conserved architecture comprised of an N-terminal chromodomain (CD) and a C-terminal chromo-shadow domain (CSD) separated by a flexible hinge region. Recognition of H3K9me2/3 is achieved by the CD. Structural analyses have demonstrated that CDs fold in to a three-stranded β-sheet with an adjacent helix and that methyl-lysine co-ordination is achieved by a “cage” formed by the sidechains of three conserved aromatic residues (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). The CSD mediates homodimerization and provides a docking surface for the interaction of additional proteins with PxVxL and related motifs (Brasher et al., 2000; Cowieson et al., 2000; Huang et al., 2006). Cryo EM analysis has revealed that HP1 dimers are capable of bridging between adjacent nucleosomes (Machida et al., 2018) and further HP1 self-interactions are proposed to facilitate the formation of oligomeric structures which compact nucleosome arrays and provide a platform for the recruitment of additional chromatin regulatory complexes (Canzio et al., 2011; Kumar and Kono, 2020).
Many species have multiple HP1 paralogs (e.g. humans and mice have three, Drosophila has five and S. pombe has two) but filamentous fungi often have a single isoform. The sole HP1 ortholog in Z. tritici, Cbx1, binds to both H3K9me2 and me3 in vitro and is enriched at H3K9me-marked loci in vivo (Fraser et al., 2022). Transcriptomic analysis indicates that Kmt1 and Cbx1 regulate the expression of highly similar sets of protein encoding genes. Surprisingly, loss of cbx1 does not result in a global increase in transcripts derived from TEs and unlike Δkmt1 mutants, Δcbx1 strains are not sensitive to abiotic stresses and exhibit only a moderate reduction in virulence in wheat infection assays (Fraser et al., 2022) (Table 1).
The different phenotypes associated with loss of the HP1 (Cbx1) and the H3K9 methyltransferase (Kmt1) are suggestive of additional H3K9me readers in Z. tritici and indeed a second CD domain protein (Cbx2) that binds to H3K9me has recently been identified (Fraser et al., 2022). In contrast to the conserved HP1 protein, Cbx1, homologs of Cbx2 are restricted to species in just a few fungal families (mainly the Mycosphaerellaceae and Teratosphaeriaceae). Cbx2 is unusual in that it contains two CDs located in the C-terminal region both of which have the conserved “aromatic cage” residues (Fraser et al., 2022). The remaining region lacks any other obvious protein domains and is predicted to be largely unstructured. Mutation of cbx2 alone does not result in marked growth defects either in planta or in vitro. However, combining deletions in cbx1 and cbx2 mimics the sensitivity of Δkmt1 strains to abiotic stresses consistent with the downstream function of H3K9me PTMs being mediated by a combination of these CD proteins (Fraser et al., 2022). Furthermore, Cbx1 and Cbx2 play redundant roles in the silencing of some Kmt1-regulated genes. In vitro analysis of Cbx2 indicates that it shows a strong preference for tri-methylated H3K9 peptides. This is potentially significant as analysis of H3K9me2 and me3 marks in S. pombe have revealed that they demarcate functionally different types of heterochromatin that are associated with different levels of transcriptional silencing (Jih et al., 2017).
While the role of Cbx2 in the establishment and or maintenance of constitutive heterochromatin remains to be determined, it is interesting to note that it shares features with the N. crassa heterochromatin regulator CDP-2 (Honda et al., 2012). Both proteins have CDs located in their C-terminal regions with much of the remaining sequence predicted to be unstructured. Furthermore, alignment of Cbx2 and CDP-2 homologues has revealed a small block of homology in the N-terminal region. CDP-2 is a subunit of a HP1-containing histone deacetylase complex called HCHC which functions in parallel with a HP1-DNA methyltransferase complex (HP1-DIM2) to establish and maintain heterochromatin in N. crassa. The N-terminal region of CDP-2 has a PxVxL-like motif which is required for interaction with the CSD of N. crassa HP1 (Honda et al., 2012). Interestingly, Z. tritici Cbx2 has two sequences in its N-terminal region that conform to a proposed CSD binding motif Φx(V/P)x(L/M/V) (Huang et al., 2006). It will therefore be important to determine whether or not Cbx2 functions in a HDAC complex with the HP1 homolog Cbx1.
DNA cytosine methylation (5 mC) is an additional feature of constitutive heterochromatic loci in many organisms. The deposition of 5 mC has been well studied in N. crassa and is mediated by the DNA methyltransferase (DNMT) DIM-2 which is targeted to H3K9me3-marked genomic loci through a direct interaction with HP1 (Honda and Selker, 2008). While many Z. tritici isolates (including the commonly studied reference strain IPO323), lack functional copies of dim2 and thus 5 mC, several Iranian Z. tritici isolates have been identified which have a functional dim2 allele and high levels of 5 mC at TEs (Möller et al., 2021). Comparison of the genomic patterns of H3K9me3 and 5 mC revealed the co-localization of these modifications suggesting an interaction between Cbx1 (HP1) and Dim2 similar to that observed in N. crassa. In fungi, 5 mC is mainly associated with TEs and is involved in a genome defence mechanism called RIP (Repeat-Induced Point mutation) which is underpinned by the deamination of 5 mC to thymine leading to the accumulation of C to T transitions in repeated elements (Gladyshev, 2017). Accordingly, the presence of functional Dim2 increased mutation rates associated with Z. tritici TEs (Möller et al., 2021).
The deposition and recognition of H3K27me3 in metazoans is mediated by Polycomb repressive complexes 1 and 2 (PRC1 and PRC2), respectively. PRC2 type complexes contain a H3K27 KMTase, together with other core subunits: EED, SUZ12, and RBBP4/7 (Laugesen et al., 2019). A variety of other ‘accessory’ proteins are associated with mammalian PRC2 complexes at substoichiometric levels. The EZH2 KMTase is catalytically inactive in the absence of the other PRC2 subunits and structural studies have revealed that it undergoes a conformational change in the presence of SUZ12 and EED which results in the correct positioning of the methyl donor (S-adenosyl methionine) and the ε-amino group (Jiao and Liu, 2015). PRC1 functions as a reader of H3K27me3 and is thought to bring about transcriptional silencing by directing the compaction of chromatin and the monoubiquitination of histone H2A on lysine 119 (H2AK119Ub) (Piunti and Shilatifard, 2021). Mammals have a variety of PRC1 type complexes which vary in their subunit composition with binding to H3K27me3 being mediated by one of five chromobox (CBX) proteins which are orthologs of the Drosophila CD protein Polycomb (Pc) (Piunti and Shilatifard, 2021).
PRC2 subunits are generally conserved in fungi that mediate H3K27 trimethylation including Z. tritici (Ridenour et al., 2020). An exception is the Cryptococcus neoformans PRC2 complex which apparently lacks a SUZ12 equivalent (Dumesic et al., 2015). Functional analyses of the putative Z. tritici EED (Mycgr3G109560), SUZ12 (Mycgr3G96052) and RBAP46/48 (Mycgr3G110427) subunits are currently lacking. Nonetheless, it appears that Z. tritici possesses a PRC2 complex and that the fundamentals of H3K27me3 deposition are conserved with metazoans (Figure 1). In stark contrast, PRC1 subunits have not been identified in fungi and the mechanisms by which this PTM mediates its biological downstream effects are poorly understood. Despite H3K27me3 being present in a variety of fungal species, to date only one fungal CD protein- Cryptococcus neoformans Ccc1-has been identified that is capable of specifically binding to this PTM (Dumesic et al., 2015). Ccc1 is a component of the PRC2 complex and its loss causes the redistribution of H3K27me3 to regions occupied by H3K9me indicating that Ccc1 restrains PRC2 activity to correct regions of the genome (Dumesic et al., 2015). The degree to which this is a feature of fungal PRC2 complexes remains to be determined as orthologs of Ccc1 have not been identified in other species.
Analysis of a Z. tritici reference genome indicates that it encodes a relatively limited repertoire of CD proteins with the potential to recognise heterochromatic histone PTMs. While the HP1 proteins of some organisms recognise H3K27me3 (Turck et al., 2007; Yale et al., 2016), neither Cbx1 nor Cbx2 show any specificity for H3 peptide tails methylated on K27 in vitro (although an in vivo role cannot be completely discounted) (Fraser et al., 2022). A major role for any of the remaining hypothetical CD proteins (termed Cbx3-Cbx6) in the recognition of H3K27me3 is also unlikely as these proteins lack critical conserved caging aromatic amino acids and/or are encoded by genes on the accessory chromosomes. Some also exhibit similarity to retroviral CD-containing integrases suggesting they are remnants of retrotransposable elements (Fraser et al., 2022).
Recent studies have provided exciting insights into mechanisms of H3K27me3 recognition and silencing independent of PRC1. Using a forward genetic screen in N. crassa, Wiles and colleagues identified EPR-1, a protein with BAH (bromo-adjacent homology) and PHD (plant homeodomain) motifs that is required for transcriptional repression at H3K27me3 marked genes. EPR-1 associates with H3K27me3 marked loci in a manner which is dependent upon the BAH domain (Wiles et al., 2020). Furthermore, the BAH domain of the Fusarium graminearum EPR-1 orthlog, BP1 binds to methylated H3K27 peptides and loss of BP1 de-represses the expression of K27me3-regulated SM genes (Tang et al., 2021). EPR-1 homologs are present in a variety of eukaryotic lineages (Wiles et al., 2020) and indeed BAH-PHD domain proteins have also been linked to PRC2-mediated silencing in plants, which like fungi, lack PRC1 components (Li et al., 2018; Qian et al., 2018; Yang et al., 2018). The BAH domains of human BAHD1 and BAHCC1 have also been shown to function as H3K27me3 readers and BAHD1 is required for optimal silencing of H3K27me3 marked genes (Zhao et al., 2016; Fan et al., 2020; Fan et al., 2021; Xu et al., 2021). Thus BAH mediated recognition of H3K27me3 is conserved across fungi, plants and animals suggesting that BAH represents a more ancient class of Polycomb silencing (Wiles et al., 2020). Characterisation of a putative Z. tritici EPR-1 homolog (Mycgr3G109489) has not yet been reported but it will be important to determine its role in the stability of accessory chromosomes and the control of effector gene expression.
While it has been established that heterochromatic PTMs have an important influence on the genome evolution, effector gene expression and virulence of Z. tritici (Möller et al., 2019; Soyer et al., 2019; Meile et al., 2020; Habig et al., 2021), there are a number of questions surrounding their deposition, maintenance and function that remain to be resolved. Firstly, how are Kmt1 and Kmt6 KMTases targeted to specific regions of the genome? Little is known about the recruitment of Ezh2/Kmt6 type enzymes in fungi and even in relatively well-studied mammalian systems, understanding of PRC2 recruitment is incomplete (Guo et al., 2021). In S. pombe, genomic targeting of Clr4 (H3K9me KMTase) is linked to the RNAi machinery (Martienssen and Moazed, 2015) but no functional link between RNAi and heterochromatin has been established in Z. tritici. Secondly, what are the mechanisms by which constitutive heterochromatin domains are spread and maintained through successive cell divisions in filamentous fungi such as Z. tritici? Unlike their counterparts in metazoans and fission yeast, the Suvar39/Clr4 KMTases of filamentous fungi do not have a recognisable CD in the N-terminal region. This is significant because for human Suv39H1 and S. pombe Clr4 the interaction of the CD with H3K9me2/3 is required for the spreading and maintenance of heterochromatic domains (Zhang et al., 2008; Muller et al., 2016). Thirdly, do all of the phenotypes associated with deletion of kmt1 and kmt6 result from loss of histone modifications? Mammalian Suv39H1 and Ezh2 KMTases both have non-histone substrates (Rodriguez-Paredes and Lyko, 2019; Weirich et al., 2021) and the possibility that this is the case for Z. tritici Kmt1 and Kmt6 needs to be considered. Finally, to what extent does the remodelling of heterochromatin during infection regulate the expression of effector genes (Soyer et al., 2019; Meile et al., 2020) and what are the environmental cues and signalling pathways that control this process? Here it is worth noting that the activity of eukaryotic TEs is often up-regulated in response to environmental stresses (Pappalardo et al., 2021; Ito, 2022) and the colonization of host tissue is a stress-inducing process (Sanchez-Vallet et al., 2018). Indeed, the upregulation of a large number of Z. tritici TE families coincides with the development of symptoms on wheat leaves (Fouche et al., 2020). Therefore, stress-responsive TEs could be an important facet of the controls that shape the chromatin environment of effector genes and thus the interaction of Z. tritici with its host.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
CJF was supported by a PhD studentship from the Newcastle Liverpool Durham BBSRC Doctoral Training Partnership (BB/M011186/1).
We thank Zoë Gardiner for comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allshire, R. C., and Madhani, H. D. (2018). Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19 (4), 229–244. doi:10.1038/nrm.2017.119
Brasher, S. V., Smith, B. O., Fogh, R. H., Nietlispach, D., Thiru, A., Nielsen, P. R., et al. (2000). The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19 (7), 1587–1597. doi:10.1093/emboj/19.7.1587
Canzio, D., Chang, E. Y., Shankar, S., Kuchenbecker, K. M., Simon, M. D., Madhani, H. D., et al. (2011). Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41 (1), 67–81. doi:10.1016/j.molcel.2010.12.016
Choi, J. Y., and Lee, Y. C. G. (2020). Double-edged sword: The evolutionary consequences of the epigenetic silencing of transposable elements. PLoS Genet. 16 (7), e1008872. doi:10.1371/journal.pgen.1008872
Chujo, T., and Scott, B. (2014). Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte-plant symbiosis. Mol. Microbiol. 92 (2), 413–434. doi:10.1111/mmi.12567
Colonna Romano, N., and Fanti, L. (2022). Transposable elements: Major players in shaping genomic and evolutionary patterns. Cells 11 (6), 1048. doi:10.3390/cells11061048
Connolly, L. R., Smith, K. M., and Freitag, M. (2013). The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 9 (10), e1003916. doi:10.1371/journal.pgen.1003916
Cowieson, N. P., Partridge, J. F., Allshire, R. C., and McLaughlin, P. J. (2000). Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 10 (9), 517–525. doi:10.1016/s0960-9822(00)00467-x
Croll, D., Zala, M., and McDonald, B. A. (2013). Breakage-fusion-bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLoS Genet. 9 (6), e1003567. doi:10.1371/journal.pgen.1003567
Dong, S., Raffaele, S., and Kamoun, S. (2015). The two-speed genomes of filamentous pathogens: Waltz with plants. Curr. Opin. Genet. Dev. 35, 57–65. doi:10.1016/j.gde.2015.09.001
Dumesic, P. A., Homer, C. M., Moresco, J. J., Pack, L. R., Shanle, E. K., Coyle, S. M., et al. (2015). Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell 160 (1-2), 204–218. doi:10.1016/j.cell.2014.11.039
Fan, H., Lu, J., Guo, Y., Li, D., Zhang, Z. M., Tsai, Y. H., et al. (2020). BAHCC1 binds H3K27me3 via a conserved BAH module to mediate gene silencing and oncogenesis. Nat. Genet. 52 (12), 1384–1396. doi:10.1038/s41588-020-00729-3
Fan, H., Guo, Y., Tsai, Y. H., Storey, A. J., Kim, A., Gong, W., et al. (2021). A conserved BAH module within mammalian BAHD1 connects H3K27me3 to Polycomb gene silencing. Nucleic Acids Res. 49 (8), 4441–4455. doi:10.1093/nar/gkab210
Fones, H., and Gurr, S. (2015). The impact of Septoria tritici Blotch disease on wheat: An EU perspective. Fungal Genet. Biol. 79, 3–7. doi:10.1016/j.fgb.2015.04.004
Fouche, S., Plissonneau, C., and Croll, D. (2018). The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr. Opin. Microbiol. 46, 34–42. doi:10.1016/j.mib.2018.01.020
Fouche, S., Badet, T., Oggenfuss, U., Plissonneau, C., Francisco, C. S., and Croll, D. (2020). Stress-driven transposable element de-repression dynamics and virulence evolution in a fungal pathogen. Mol. Biol. Evol. 37 (1), 221–239. doi:10.1093/molbev/msz216
Fraser, C. J., Rutherford, J. C., Rudd, J. J., and Whitehall, S. K. (2022). The chromodomain proteins, Cbx1 and Cbx2 have distinct roles in the regulation of heterochromatin and virulence in the fungal wheat pathogen, Zymoseptoria tritici. BioRxiv. doi:10.1101/2022.09.21.508279
Gladyshev, E. (2017). Repeat-induced point mutation and other genome defense mechanisms in fungi. Microbiol. Spectr. 5 (4). doi:10.1128/microbiolspec.FUNK-0042-2017
Goodier, J. L. (2016). Restricting retrotransposons: A review. Mob. DNA 7, 16. doi:10.1186/s13100-016-0070-z
Goodwin, S. B., M'Barek S, B., Dhillon, B., Wittenberg, A. H., Crane, C. F., Hane, J. K., et al. (2011). Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7 (6), e1002070. doi:10.1371/journal.pgen.1002070
Guo, Y., Zhao, S., and Wang, G. G. (2021). Polycomb gene silencing mechanisms: PRC2 chromatin targeting, H3K27me3 'readout', and phase separation-based compaction. Trends Genet. 37 (6), 547–565. doi:10.1016/j.tig.2020.12.006
Habig, M., Lorrain, C., Feurtey, A., Komluski, J., and Stukenbrock, E. H. (2021). Epigenetic modifications affect the rate of spontaneous mutations in a pathogenic fungus. Nat. Commun. 12 (1), 5869. doi:10.1038/s41467-021-26108-y
Hassani, M. A., Oppong-Danquah, E., Feurtey, A., Tasdemir, D., and Stukenbrock, E. H. (2022). Differential regulation and production of secondary metabolites among isolates of the fungal wheat pathogen Zymoseptoria tritici. Appl. Environ. Microbiol. 88 (6), e0229621. doi:10.1128/aem.02296-21
Higa, L. A., Wu, M., Ye, T., Kobayashi, R., Sun, H., and Zhang, H. (2006). CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8 (11), 1277–1283. doi:10.1038/ncb1490
Honda, S., Lewis, Z. A., Shimada, K., Fischle, W., Sack, R., and Selker, E. U. (2012). Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation. Nat. Struct. Mol. Biol. 19 (5), 471–477. S471. doi:10.1038/nsmb.2274
Honda, S., and Selker, E. U. (2008). Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol. Cell. Biol. 28 (19), 6044–6055. doi:10.1128/MCB.00823-08
Hong, E. J., Villen, J., Gerace, E. L., Gygi, S. P., and Moazed, D. (2005). A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2 (3), 106–111. doi:10.4161/rna.2.3.2131
Horn, P. J., Bastie, J. N., and Peterson, C. L. (2005). A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 19 (14), 1705–1714. doi:10.1101/gad.1328005
Huang, Y., Myers, M. P., and Xu, R. M. (2006). Crystal structure of the HP1-EMSY complex reveals an unusual mode of HP1 binding. Structure 14 (4), 703–712. doi:10.1016/j.str.2006.01.007
Ito, H. (2022). Environmental stress and transposons in plants. Genes Genet. Syst., 22-00045. doi:10.1266/ggs.22-00045
Jacobs, S. A., and Khorasanizadeh, S. (2002). Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295 (5562), 2080–2083. doi:10.1126/science.1069473
Janevska, S., Baumann, L., Sieber, C. M. K., Munsterkotter, M., Ulrich, J., Kamper, J., et al. (2018). Elucidation of the two H3K36me3 histone methyltransferases Set2 and Ash1 in Fusarium fujikuroi unravels their different chromosomal targets and a major impact of Ash1 on genome stability. Genetics 208 (1), 153–171. doi:10.1534/genetics.117.1119
Jia, S., Kobayashi, R., and Grewal, S. I. (2005). Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 7 (10), 1007–1013. doi:10.1038/ncb1300
Jiao, L., and Liu, X. (2015). Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350 (6258), aac4383. doi:10.1126/science.aac4383
Jih, G., Iglesias, N., Currie, M. A., Bhanu, N. V., Paulo, J. A., Gygi, S. P., et al. (2017). Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature 547 (7664), 463–467. doi:10.1038/nature23267
Kellner, R., Bhattacharyya, A., Poppe, S., Hsu, T. Y., Brem, R. B., and Stukenbrock, E. H. (2014). Expression profiling of the wheat pathogen Zymoseptoria tritici reveals genomic patterns of transcription and host-specific regulatory programs. Genome Biol. Evol. 6 (6), 1353–1365. doi:10.1093/gbe/evu101
Kumar, A., and Kono, H. (2020). Heterochromatin protein 1 (HP1): Interactions with itself and chromatin components. Biophys. Rev. 12 (2), 387–400. doi:10.1007/s12551-020-00663-y
Laugesen, A., Hojfeldt, J. W., and Helin, K. (2019). Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell 74 (1), 8–18. doi:10.1016/j.molcel.2019.03.011
Lewis, Z. A., Adhvaryu, K. K., Honda, S., Shiver, A. L., Knip, M., Sack, R., et al. (2010a). DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 6 (11), e1001196. doi:10.1371/journal.pgen.1001196
Lewis, Z. A., Adhvaryu, K. K., Honda, S., Shiver, A. L., and Selker, E. U. (2010b). Identification of DIM-7, a protein required to target the DIM-5 H3 methyltransferase to chromatin. Proc. Natl. Acad. Sci. U. S. A. 107 (18), 8310–8315. doi:10.1073/pnas.1000328107
Li, F., Goto, D. B., Zaratiegui, M., Tang, X., Martienssen, R., and Cande, W. Z. (2005). Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr. Biol. 15 (16), 1448–1457. doi:10.1016/j.cub.2005.07.021
Li, Z., Fu, X., Wang, Y., Liu, R., and He, Y. (2018). Polycomb-mediated gene silencing by the BAH-EMF1 complex in plants. Nat. Genet. 50 (9), 1254–1261. doi:10.1038/s41588-018-0190-0
Liu, J., Ali, M., and Zhou, Q. (2020). Establishment and evolution of heterochromatin. Ann. N. Y. Acad. Sci. 1476 (1), 59–77. doi:10.1111/nyas.14303
Machida, S., Takizawa, Y., Ishimaru, M., Sugita, Y., Sekine, S., Nakayama, J. I., et al. (2018). Structural basis of heterochromatin formation by human HP1. Mol. Cell 69 (3), 385–397. doi:10.1016/j.molcel.2017.12.011
Marsano, R. M., and Dimitri, P. (2022). Constitutive heterochromatin in eukaryotic genomes: A mine of transposable elements. Cells 11 (5), 761. doi:10.3390/cells11050761
Martienssen, R., and Moazed, D. (2015). RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 7 (8), a019323. doi:10.1101/cshperspect.a019323
Meile, L., Croll, D., Brunner, P. C., Plissonneau, C., Hartmann, F. E., McDonald, B. A., et al. (2018). A fungal avirulence factor encoded in a highly plastic genomic region triggers partial resistance to septoria tritici blotch. New Phytol. 219 (3), 1048–1061. doi:10.1111/nph.15180
Meile, L., Peter, J., Puccetti, G., Alassimone, J., McDonald, B. A., and Sanchez-Vallet, A. (2020). Chromatin dynamics contribute to the spatiotemporal expression pattern of virulence genes in a fungal plant pathogen. mBio 11. doi:10.1128/mBio.02343-02320
Möller, M., Habig, M., Lorrain, C., Feurtey, A., Haueisen, J., Fagundes, W. C., et al. (2021). Recent loss of the Dim2 DNA methyltransferase decreases mutation rate in repeats and changes evolutionary trajectory in a fungal pathogen. PLoS Genet. 17 (3), e1009448. doi:10.1371/journal.pgen.1009448
Möller, M., Schotanus, K., Soyer, J. L., Haueisen, J., Happ, K., Stralucke, M., et al. (2019). Destabilization of chromosome structure by histone H3 lysine 27 methylation. PLoS Genet. 15 (4), e1008093. doi:10.1371/journal.pgen.1008093
Muller, M. M., Fierz, B., Bittova, L., Liszczak, G., and Muir, T. W. (2016). A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat. Chem. Biol. 12 (3), 188–193. doi:10.1038/nchembio.2008
Nielsen, P. R., Nietlispach, D., Mott, H. R., Callaghan, J., Bannister, A., Kouzarides, T., et al. (2002). Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416 (6876), 103–107. doi:10.1038/nature722
Pappalardo, A. M., Ferrito, V., Biscotti, M. A., Canapa, A., and Capriglione, T. (2021). Transposable elements and stress in vertebrates: An overview. Int. J. Mol. Sci. 22 (4), 1970. doi:10.3390/ijms22041970
Piunti, A., and Shilatifard, A. (2021). The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 22 (5), 326–345. doi:10.1038/s41580-021-00341-1
Plissonneau, C., Sturchler, A., and Croll, D. (2016). The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio 7. doi:10.1128/mBio.01231-16
Plissonneau, C., Hartmann, F. E., and Croll, D. (2018). Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biol. 16 (1), 5. doi:10.1186/s12915-017-0457-4
Qian, C., and Zhou, M. M. (2006). SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell. Mol. Life Sci. 63 (23), 2755–2763. doi:10.1007/s00018-006-6274-5
Qian, S., Lv, X., Scheid, R. N., Lu, L., Yang, Z., Chen, W., et al. (2018). Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL. Nat. Commun. 9 (1), 2425. doi:10.1038/s41467-018-04836-y
Ridenour, J. B., Moller, M., and Freitag, M. (2020). Polycomb repression without bristles: Facultative heterochromatin and genome stability in fungi. Genes (Basel) 11 (6), E638. doi:10.3390/genes11060638
Rodriguez-Paredes, M., and Lyko, F. (2019). The importance of non-histone protein methylation in cancer therapy. Nat. Rev. Mol. Cell Biol. 20 (10), 569–570. doi:10.1038/s41580-019-0147-x
Rudd, J. J., Kanyuka, K., Hassani-Pak, K., Derbyshire, M., Andongabo, A., Devonshire, J., et al. (2015). Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 167 (3), 1158–1185. doi:10.1104/pp.114.255927
Sanchez-Vallet, A., Fouche, S., Fudal, I., Hartmann, F. E., Soyer, J. L., Tellier, A., et al. (2018). The genome biology of effector gene evolution in filamentous plant pathogens. Annu. Rev. Phytopathol. 56, 21–40. doi:10.1146/annurev-phyto-080516-035303
Schotanus, K., Soyer, J. L., Connolly, L. R., Grandaubert, J., Happel, P., Smith, K. M., et al. (2015). Histone modifications rather than the novel regional centromeres of Zymoseptoria tritici distinguish core and accessory chromosomes. Epigenetics Chromatin 8, 41. doi:10.1186/s13072-015-0033-5
Seidl, M. F., and Thomma, B. (2017). Transposable elements direct the coevolution between plants and microbes. Trends Genet. 33 (11), 842–851. doi:10.1016/j.tig.2017.07.003
Soyer, J. L., El Ghalid, M., Glaser, N., Ollivier, B., Linglin, J., Grandaubert, J., et al. (2014). Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans. PLoS Genet. 10 (3), e1004227. doi:10.1371/journal.pgen.1004227
Soyer, J. L., Grandaubert, J., Haueisen, J., Schotanus, K., and Stukenbrock, E. H. (2019). In planta chromatin immunoprecipitation in Zymoseptoria tritici reveals chromatin-based regulation of putative effector gene expression. BioRxiv. doi:10.1101/544627
Steinberg, G. (2015). Cell biology of Zymoseptoria tritici: Pathogen cell organization and wheat infection. Fungal Genet. Biol. 79, 17–23. doi:10.1016/j.fgb.2015.04.002
Studt, L., Rosler, S. M., Burkhardt, I., Arndt, B., Freitag, M., Humpf, H. U., et al. (2016). Knock-down of the methyltransferase Kmt6 relieves H3K27me3 and results in induction of cryptic and otherwise silent secondary metabolite gene clusters in Fusarium fujikuroi. Environ. Microbiol. 18 (11), 4037–4054. doi:10.1111/1462-2920.13427
Tang, G., Yuan, J., Wang, J., Zhang, Y. Z., Xie, S. S., Wang, H., et al. (2021). Fusarium BP1 is a reader of H3K27 methylation. Nucleic Acids Res. 49 (18), 10448–10464. doi:10.1093/nar/gkab844
Torriani, S. F., Melichar, J. P., Mills, C., Pain, N., Sierotzki, H., and Courbot, M. (2015). Zymoseptoria tritici: A major threat to wheat production, integrated approaches to control. Fungal Genet. Biol. 79, 8–12. doi:10.1016/j.fgb.2015.04.010
Turck, F., Roudier, F., Farrona, S., Martin-Magniette, M. L., Guillaume, E., Buisine, N., et al. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3 (6), e86. doi:10.1371/journal.pgen.0030086
Uhse, S., and Djamei, A. (2018). Effectors of plant-colonizing fungi and beyond. PLoS Pathog. 14 (6), e1006992. doi:10.1371/journal.ppat.1006992
Weirich, S., Khella, M. S., and Jeltsch, A. (2021). Structure, activity and function of the Suv39h1 and Suv39h2 protein lysine methyltransferases. Life (Basel) 11 (7), 703. doi:10.3390/life11070703
Wiles, E. T., McNaught, K. J., Kaur, G., Selker, J. M. L., Ormsby, T., Aravind, L., et al. (2020). Evolutionarily ancient BAH-PHD protein mediates Polycomb silencing. Proc. Natl. Acad. Sci. U. S. A. 117 (21), 11614–11623. doi:10.1073/pnas.1918776117
Wittenberg, A. H., van der Lee, T. A., Ben M'barek, S., Ware, S. B., Goodwin, S. B., Kilian, A., et al. (2009). Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS One 4 (6), e5863. doi:10.1371/journal.pone.0005863
Xu, P., Scott, D. C., Xu, B., Yao, Y., Feng, R., Cheng, L., et al. (2021). FBXO11-mediated proteolysis of BAHD1 relieves PRC2-dependent transcriptional repression in erythropoiesis. Blood 137 (2), 155–167. doi:10.1182/blood.2020007809
Yale, K., Tackett, A. J., Neuman, M., Bulley, E., Chait, B. T., and Wiley, E. (2016). Phosphorylation-dependent targeting of Tetrahymena HP1 to condensed chromatin. mSphere 1. doi:10.1128/mSphere.00142-16
Yang, Y., Liu, R., Qiu, R., Zheng, Y., Huang, W., Hu, H., et al. (2015). CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene 34 (1), 104–118. doi:10.1038/onc.2013.522
Yang, Z., Qian, S., Scheid, R. N., Lu, L., Chen, X., Liu, R., et al. (2018). EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat. Genet. 50 (9), 1247–1253. doi:10.1038/s41588-018-0187-8
Zhang, K., Mosch, K., Fischle, W., and Grewal, S. I. (2008). Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15 (4), 381–388. doi:10.1038/nsmb.1406
Keywords: heterochromatin, Zymoseptoria tritici, histone H3 lys 27 trimethylation, histone H3 lys 9 methylation, transposable element, fungal plant pathogen, genome plasticity
Citation: Fraser CJ and Whitehall SK (2022) Heterochromatin in the fungal plant pathogen, Zymoseptoria tritici: Control of transposable elements, genome plasticity and virulence. Front. Genet. 13:1058741. doi: 10.3389/fgene.2022.1058741
Received: 30 September 2022; Accepted: 04 November 2022;
Published: 21 November 2022.
Edited by:
Anna Muszewska, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, PolandReviewed by:
Thierry C. Marcel, INRA Centre Versailles-Grignon, FranceCopyright © 2022 Fraser and Whitehall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simon K. Whitehall, c2ltb24ud2hpdGVoYWxsQG5jbC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.