
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 19 October 2022
Sec. Computational Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1045244
This article is part of the Research Topic Bioinformatics Analysis of Omics Data for Biomarker Identification in Clinical Research, Volume II View all 53 articles
Background: Infantile hemangiomas (IH) and venous malformations (VM) are the most common types of vascular abnormalities that seriously affect the health of children. Although there is evidence that these two diseases share some common genetic changes, the underlying mechanisms need to be further studied.
Methods: The microarray datasets of IH (GSE127487) and VM (GSE7190) were downloaded from GEO database. Extensive bioinformatics methods were used to investigate the common differentially expressed genes (DEGs) of IH and VM, and to estimate their Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Trough the constructing of protein-protein interaction (PPI) network, gene models and hub genes were obtained by using Cytoscape and STRING. Finally, we analyzed the co-expression and the TF-mRNA-microRNA regulatory network of hub genes.
Results: A total of 144 common DEGs were identified between IH and VM. Functional analysis indicated their important role in cell growth, regulation of vasculature development and regulation of angiogenesis. Five hub genes (CTNNB1, IL6, CD34, IGF2, MAPK11) and two microRNA (has-miR-141-3p, has-miR-150-5p) were significantly differentially expressed between IH and normal control (p < 0.05).
Conclusion: In conclusion, our study investigated the common DEGs and molecular mechanism in IH and VM. Identified hub genes and signaling pathways can regulate both diseases simultaneously. This study provides insight into the crosstalk of IH and VM and obtains several biomarkers relevant to the diagnosis and pathophysiology of vascular abnormalities.
Infantile hemangiomas (IH) are the most common benign tumors in early childhood, with an incidence of 4%–10% (Rodríguez Bandera et al., 2021). Although almost every part of the human body can be involved, hemangiomas are seen more frequently in head and neck (Fernández Faith et al., 2021). The hyper-proliferation of IH can cause significant functional and disfiguring consequences such as ulceration, bleeding, and pain. Vascular malformations affect 3% of the population. Venous malformations (VM) are the most common type representing more than 50% of cases (Gallant et al., 2021). VM grows proportionally with the human body and do not spontaneously involute. It is typically characterized by dilated superficial veins, purple vein bubbles, or blue tinge involving the skin (Kunimoto et al., 2022). Medical interventions or surgical treatments are required by 45% of patients with vascular anomalies (Haggstrom et al., 2006). Based on previous experience, the proper treatment of these diseases requires specialists from multiple disciplines, including plastic surgery, pediatrics, dermatology, radiology and vascular surgery.
IH and VM are generally considered to be two distinct diseases, not only in anatomical, histological, and pathophysiological features, but also in clinical presentation. However, they are both vascular anomalies, representing deleterious mutations in vascular development. The commonality among these lesions is their origin in vascular endothelia (Mahajan et al., 2022). TIE2, encoded by TEK, is an endothelial cell-specific receptor tyrosine kinase that is essential for vessel remodeling and the survival of endothelial cells. Limited studies have identified that somatic mutations in exon 17 of the TEK gene were common changes in vascular tumors and vascular malformations (Ye et al., 2011). Due to the association of IH and VM and some common mutations and immune-related factors, some molecular mechanisms might be involved in the development and progression of IH and VM.
Bioinformatics has developed rapidly in recent years, which can be applied to illustrate large and complicated data sets associated with various diseases. Given the reasonable expectations of the development of sequencing technologies, and the decreasing cost barriers, researchers around the world have contributed a large amount of transcriptomic data. Analysis of mRNA transcriptomics may help uncover the biological processes and underlying mechanisms of disease. However, most research teams mainly focus on a single disease, and then delve into the regulatory mechanisms. The characteristics of the shared changes of the vascular anomalies and their relevance remain less explored. Bioinformatics methods enable us to extract multiple related microarrays for further data mining, which contributes to the reuse of information and the discovery of crosstalk between diseases.
In this study, we tried to identify the common differentially expressed genes (DEGs) between IH and VM patients. Two microarray datasets, GSE7190 and GSE127487, were obtained from GEO database. We analyzed their common DEGs and potential functions. Then, through the construction of PPI network, gene model and hub genes were investigated. Finally, five hub genes were identified and their transcription factor (TF) and microRNA were also traced. The workflow of our study was shown in Figure 1.
GEO (http://www.ncbi.nlm.nih.gov/geo) is a gene expression database created by NCBI, which contains high-throughput gene expression data submitted by research institutes worldwide (Barrett et al., 2013). We used the keywords infantile hemangiomas and venous malformation to search for eligible datasets. Among the inclusion criteria were 1) diagnosis of patients with infantile hemangiomas (IH) and venous malformation (VM), 2) detection of gene level in tissue or blood samples. Exclusion criteria were 1) expression data without normal control, and 2) datasets come from Mus musculus and Rattus norvegicus. Finally, two microarray datasets were eligible: accession numbers GSE127487 (18 infantile hemangiomas (IH) samples and 5 normal samples, Platforms: GPL10558), GSE7190 (2 venous malformation (VM) samples and 2 normal veins samples, Platforms: GPL1708) (Ebenebe et al., 2007; Gomez-Acevedo et al., 2020). For validation, expression profiles of 12 infantile hemangiomas (IH) patients and 4 normal patients were downloaded from GSE69136 dataset (Platforms: GPL19765) (Strub et al., 2016).
Differentially expressed mRNA between infantile hemangiomas (IH) patients and normal skin patients were identified using Limma package of R software. The adj.P.Value <0.05 and LogFC (fold change) > 1.5 or < -1.5 were set as the cutoffs. Used the same procedure to identify differentially expressed genes between venous malformations (VM) and normal veins. Next, the online Venn diagram tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html) was applied to obtain the overlap DEGs among two datasets.
GO (gene ontology) database: the function of genes is divided into three categories: biological process (biological process, BP), cellular components (cellular component, CC), molecular function (molecular function, MF). Using the GO database, we can find out the relationship between the DEGs at the three levels of CC, MF, and BP. Function annotation and Genomes (KEGG) pathways enrichment analysis were performed using DAVID (Huang et al., 2007). P.adjust <0.05 was consider significant.
Protein-protein interaction network was investigated using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING: http://string-db.org, version11.5) (Szklarczyk et al., 2015). It can customize PPI networks, as well as functional characterization of user-uploaded gene/measurement sets. The interactions with a combined score over 0.4 were considered statistically significant. Then, we used Cytoscape to visualize the PPI networks (Saito et al., 2012). The plugin Molecular Complex Detection (MCODE) was applied to identify the module genes that interact most closely with the following criteria: degree cutoff = 2, node score cutoff = 0.2, k-core = 2, max depth = 100 (Bandettini et al., 2012). In addition, GO and KEGG analysis of the model genes were performed.
The plugin Cytohhuba of Cytoscape was applied to identify hub genes in our networks. The top ten core genes in the network are evaluated based on six algorithms (MCC, MNC, degree, closeness, Radiality, EPC). Here, the genes contained in all six algorithms were regarded as hub genes. Subsequently, a co-expression network of hub genes was constructed using GeneMANIA (http://www.genemania.org/), which searches many large, publicly available biological datasets to find related genes (Warde-Farley et al., 2010). These include protein-protein, protein-DNA and genetic interactions, pathways, reactions and phenotypic screening profiles.
In order to better understand the potential regulatory relationship of these hub genes, we established the TF-mRNA-microRNA regulatory network based on TRRUST (http://www.grnpedia.org/trrust) and Mirwalk (http://mirwalk.umm.uni-heidelberg.de/) database. TRRUST is a manually curated database of human and mouse transcriptional factor regulatory networks. Current version of TRRUST contains 8,444 and 6,552 TF-target regulatory relationships of 800 human TFs (Han et al., 2018). The new version of miRWalk stores predicted data obtained with a maschine learning algorithm including experimentally verified miRNA-target interactions (Sticht et al., 2018).
In order to confirm the credibility of our results, hub genes, TFs and microRNA expression was verified. The microarray dataset GSE34989 and GSE7190 were used for hub gene and TFs expression. Additionally, GSE69136 (12 infantile hemangiomas samples (IH) and 4 normal samples) was used for microRNA verification. Student’s t method was used to test the difference and p-value < 0.05 was considered statistically significant.
Two-group comparisons were determined using Student’s t test, and multiple group comparisons were conducted using the analysis of variance t test. All statistical analyses and pictures (Volcano plots, bubble plots, histograms and ROC) were performed and visualized using R software. p-value < 0.05 was considered statistically significant.
After data standardization and filtering, there were 1,093 DEGs (868 upregulated and 225 downregulated) between infantile hemangiomas group (IH) and normal group (Figure 2A). Additionally, 5,740 DEGs (2,992 upregulated and 2,748 downregulated) were identified between venous malformation group (VM) and normal group (Figure 2B). Through the intersection of the Venn diagram, we obtained 144 overlapped DEGs (107 upregulated and 37 downregulated) in GSE127487 and GSE7190 (Figure 2C).
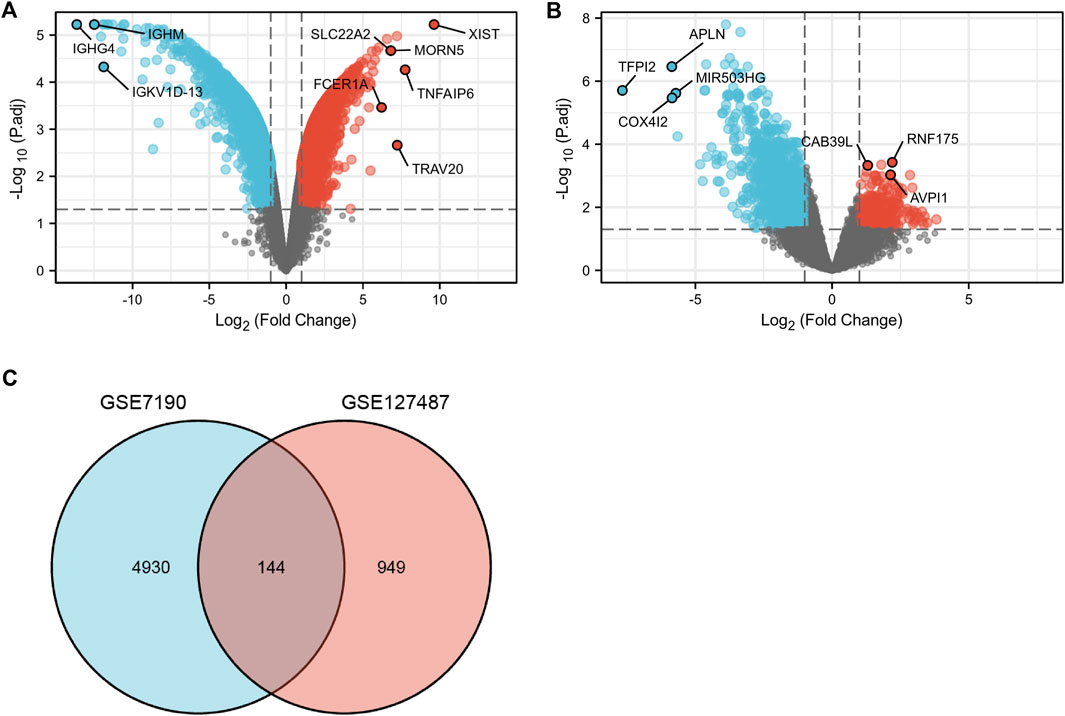
FIGURE 2. The volcano and Venn diagram. (A), the volcano diagram of GSE7190. (B), The volcano diagram of GSE127487. (C), Venn diagram showed an overlap of 144 common DEGs.
Function annotation has been carried out among the 144 common DEGs (p < 0.01). BP category suggested that cell growth (p = 1.02E-05), regulation of vasculature development (p = 1.11E-05) and regulation of angiogenesis (p = 2.08E-05) were important process of the common DEGs (Figures 3A,B). MF results indicated these common DEGs were mostly involved in guanyl-nucleotide exchange factor activity (p = 0.00016) and scavenger receptor activity (p = 0.00049). The microvillus (p = 0.0003), specific granule (p = 0.001), apical part of cell (p = 0.0016) in CC category, indicating these genes play their roles in cell adhesion (Figures 3A,B). In addition, Pertussis (p = 0.00052) and Human cytomegalovirus infection (p = 0.00075) were significant pathways of common DEGs (Figures 3A,B). The GO and KEGG results were summarized in Supplementary Table S1.
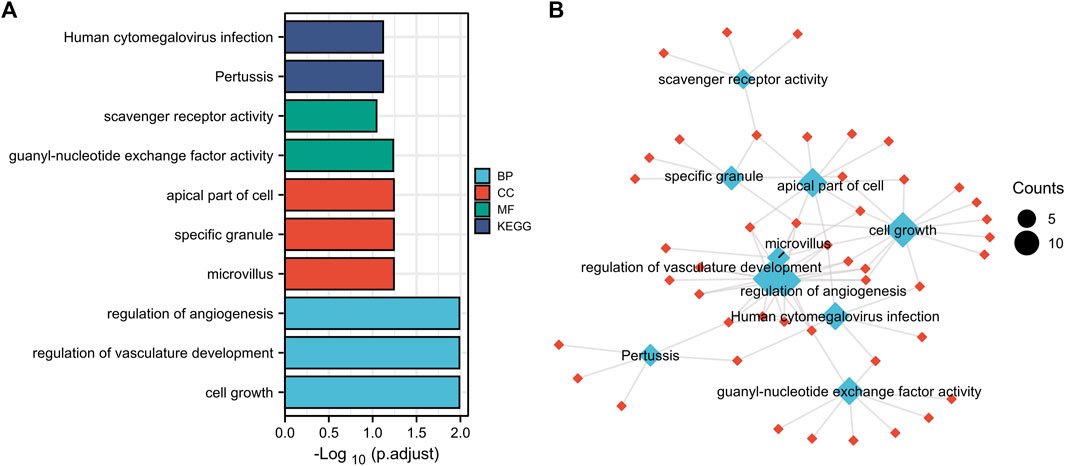
FIGURE 3. Function annotation of 144 common DEGs. (A), GO terms (BP, MF, CC) and KEGG analysis of overlapped common DEGs (p < 0.01). GO, Gene Ontology. BP, biological process, MF, molecular function, CC, cellular component. KEGG, Kyoto Encyclopedia of Genes and Genomes. (B), network of GO and KEGG.
The protein-protein interaction (PPI) network was constructed among 144 common DEGs to explore their potential interactions, including 668 nodes and 13484 edges (Figure 4). It is important to point out that many genes are individual. So, we selected four gene modules that interact most closely in PPI network were obtained through MCODE plugin of Cytoscape, which contained 2,534 edges involving 94 nodes. GO function analysis revealed that these model genes were mainly related to alpha-catenin binding, cell adhesion molecule binding, growth factor activity, synapse pruning, cell differentiation involved in metanephros development, fascia adherens and collagen trimer. KEGG results showed that these genes mainly involved in Focal adhesion, Pertussis and Bacterial invasion of epithelial cells (Figure 5).
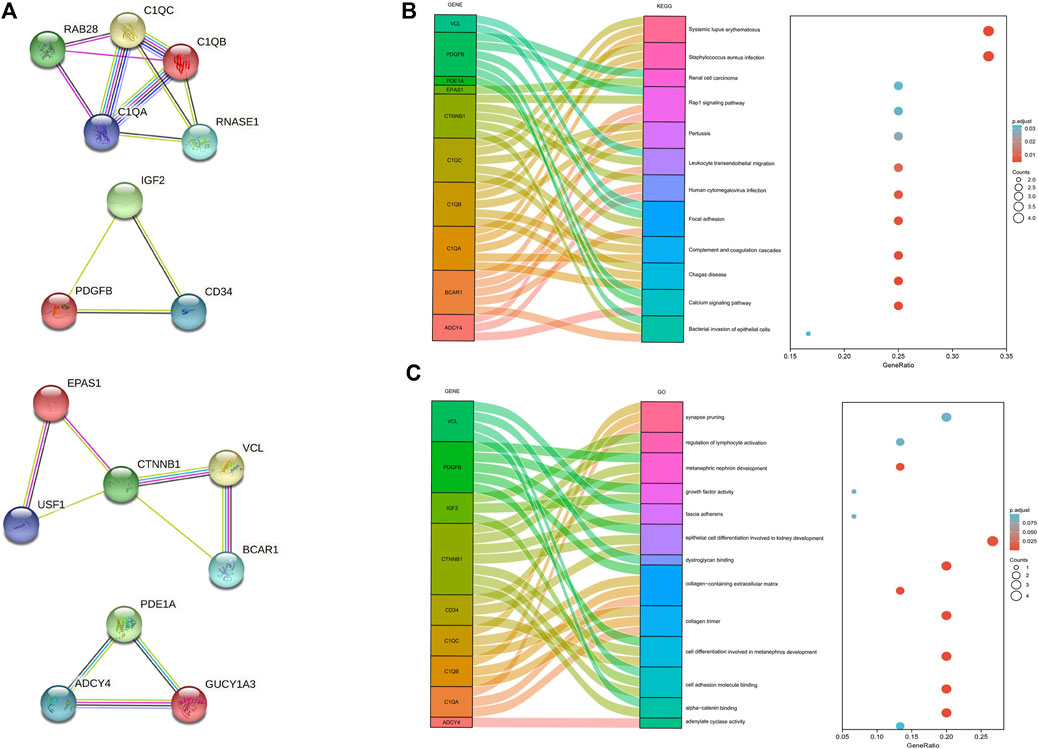
FIGURE 5. Significant gene module and function analysis of the model genes. (A), Four significant gene clustering modules. (B,C), GO and KEGG enrichment analysis of the model genes. The size of the circle represents the number of genes involved, and the abscissa represents the frequency of the genes involved in the term total genes.
By using six algorithms of cytoHubba, we identified the top 10 genes in our network (Figure 6A). After the selection, we found five overlapping hub genes which contained in six algorithms results, including CTNNB1, IL6, CD34, IGF2, MAPK11 (Table 1). Their full name and the function of encoding proteins are shown in Table 2. The co-expression network and functions of hub genes were analyzed using GeneMANIA database (Figure 6B). They showed a complex network with the, physical interactions of 77.6%, co-expression of 8.01%, co-localization of 3.63%, predicted of 5.37% and genetic interactions of 2.87%. Function analysis indicated that these genes mainly involved in insulin-like growth factor binding, leukocyte cell-cell adhesion and positive regulation of cell-cell adhesion (Figure 6C).
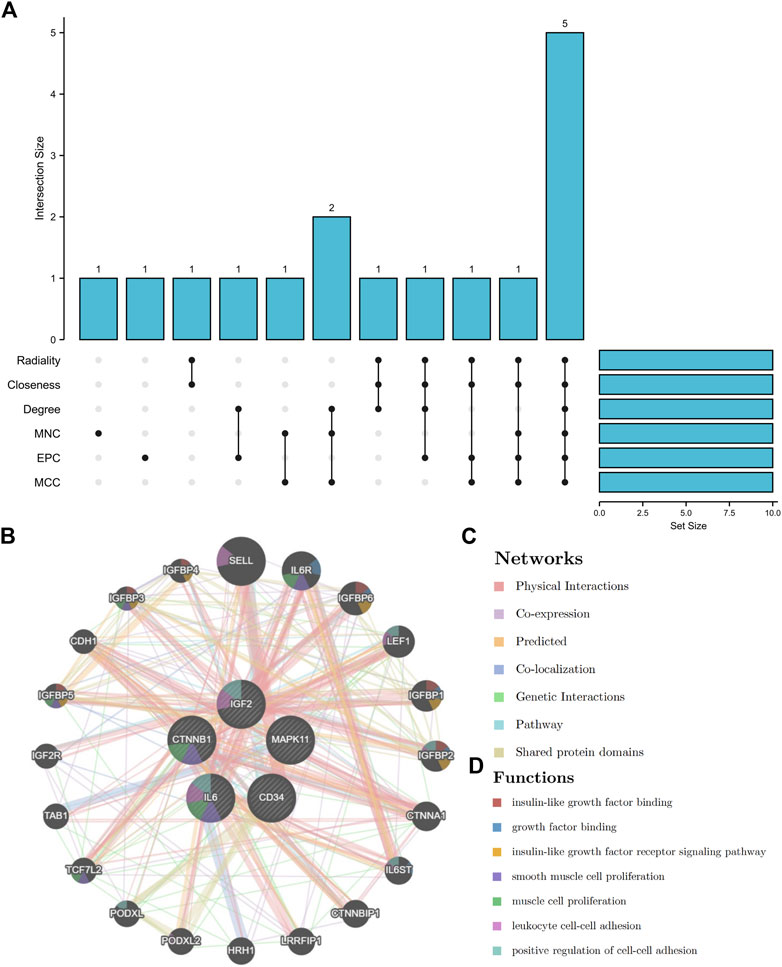
FIGURE 6. The UpSet diagram of hub genes and their co-expression network. (A), six algorithms of cytoHubba selected 5 overlapped hub genes. (B), Hub genes and their co-expression network.
According to the results in TRUUST and Mirwalk, 4 TFs and 7 microRNA have regulatory relationships with these hub genes (Figure 7). CEBPA, EGR2, EGR1 and SP1 were the predicted TFs of IL6 and IGF2 (Qvalue<0.01). In addition, hsa-miR-141-3p can target the 3′UTR of IGF2 (p = 0.01). hsa-miR-150-5p can target the 3′UTR of CTNNB1 (p = 0.01). Moreover, hsa-miR-431-5p, hsa-miR-4319, hsa-miR-129-1-3p, hsa-miR-125b-5p and hsa-miR-129-2-3p were the predicted microRNA of CD34 (p < 0.01).
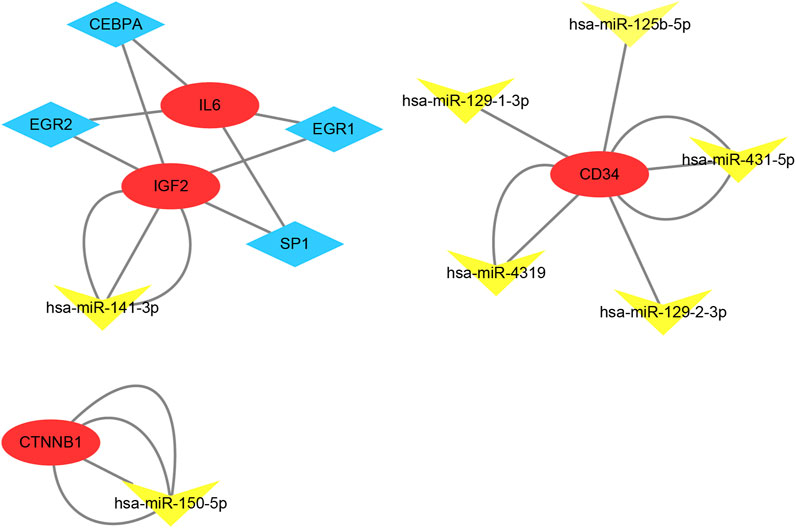
FIGURE 7. TF-mRNA-microRNA regulatory network. Red circles represent hub genes, blue diamonds represent transcription factors and yellow triangles represent microRNA.
Our results showed that CD34, IGF2, MAPK11 were significantly upregulated in IH patients (Figures 8A,C). In contrast, CTNNB1, IL6 were significantly downregulated. Moreover, the expressions of EGR2, EGR1, and SP1 were significantly upregulated in VM, but not significantly in IH. Additionally, has-miR-141-3p and has-miR-150-5p were significantly downregulated in IH (Figure 8E). All of the hub genes (AUC>0.8), has-miR-141-3p (AUC = 1) and has-miR-150-5p (AUC = 0.83) have shown good efficiency in distinguishing diseases from healthy people (Figures 8B,D,F).
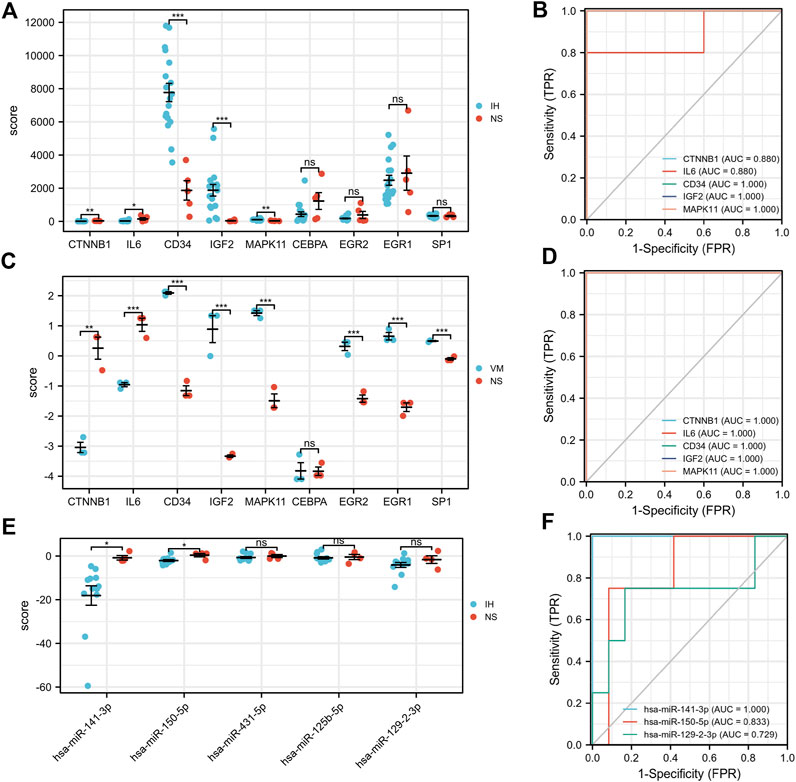
FIGURE 8. Validation of hub genes, TFs and microRNA expression and their ROC curves. *p < 0.05; **p < 0.01; ***p < 0.001.
Evidence suggests that IH and VM share some common pathogenic factors. However, their common differentially expressed mRNAs and the detailed molecular mechanisms remain unclear. In the present study, extensive bioinformatics methods were used to investigate the common DEGs of IH and VM, and to estimate the pathways involved in the hub genes. The P.Value <0.05 and LogFC (fold change) > 1 or < -1 were set as the cutoffs. At this threshold, there were total 989 common DEGs between IH and VM. In order to screen out the most significantly changed genes and narrow the range, we adjusted the threshold to adjP<0.05 and LogFC (fold change) > 1.5 or < -1.5. Then, we obtained 107 overlapping upregulated genes and 37 downregulated genes. Function enrichment analysis indicated that these common DEGs were mainly involved in cell growth, regulation of vasculature development and regulation of angiogenesis. The two most important cells in the course of IH are hemangioma stem cells and hemangioma endothelial cells. Infantile hemangioma is characterized by massive proliferation of hemangioma endothelial cells (Lv et al., 2022). The expression analysis indicates that VM endothelium is misspecified and hyperproliferative, suggesting that VMs are biologically active lesions (Lee et al., 2021). Abnormal proliferation and regulation of vascular endothelial cells can seriously affect vascular development and angiogenesis. Propranolol is a first-line drug for IH treatment and has an antiproliferative and cytotoxic against hemangioma endothelial and stem cells (Ma et al., 2021). These results strongly indicate that the abnormal in vasculature development and angiogenesis are jointly involved in the occurrence and development of these two diseases.
A total of five hub genes (CTNNB1, IL6, CD34, IGF2, MAPK11) were identified by constructing a PPI network. These genes occupy a central position in the PPI network. In addition, these hub genes also play a core role in module 2 and module 3, including CTNNB1, CD34, IGF2. The co-expression and function analysis indicated that these hub genes mainly involved in insulin-like growth factor binding and positive regulation of cell-cell adhesion. Among them, insulin-like growth factor receptor signaling pathway may be the core mechanism affecting vascular abnormalities. IGF2 is required for the continuous expansion of the feto-placental microvasculature in late pregnancy. The angiocrine effects of IGF2 on placental microvasculature expansion are mediated through IGF2R and angiopoietin-Tie2/TEK signaling. Additionally, IGF2 exerts IGF2R-ERK1/2-dependent pro-proliferative and angiogenic effects (Sandovici et al., 2022). CTNNB1 upregulates DLL4 transcription and strongly increases Notch signal in endothelial cells, leading to vascular abnormalities. Reduced Notch signaling in IH cells decreased cell proliferation, migration, and tumor formation (Ma et al., 2021). Many vascular malformations share similar aberrant molecular signaling pathways with cancers and inflammatory disorders (Pang et al., 2020). It is reported that inhibiting the IL-6/STAT3/HIF-1α signaling pathways could suppress IH growth (Maimaiti et al., 2022). These results indicated that the hub genes were involved in several signaling pathways mediated by growth factor and immune gene, which play a critical role in regulating endothelial cells. The regulation of vascular endothelial cell proliferation may be the common mechanism for the treatment of IH and VM. Furthermore, we made an in-depth analysis of the link between the hub gene and these two diseases.
CTNNB1 also named Catenin Beta 1. The protein encoded by CTNNB1 is part of a complex of proteins that constitute adherens junctions (AJs). AJs are necessary for the creation and maintenance of epithelial cell layers by regulating cell growth and adhesion between cells (Liu et al., 2022). Studies have shown that endothelial specific stabilization of Wnt/β-catenin signaling changes the early vascular development of embryos (Corada et al., 2010). β-catenin upregulates Dll4 transcription and strongly increases Notch signaling in the endothelium, resulting in loss of vascular remodeling, intersomatic vascular elongation, branch defect and loss of venous characteristics. Recently, Duan X et al. found that FOXC1A can regulate vascular integrity and brain vascular development through targeting CTNNB1 (Duan et al., 2022). In addition, CTNNB1 was also found to be closely related to a variety of tumor diseases such as liver cancer, gastrointestinal carcinoma and endometrial carcinoma (Ambrozkiewicz et al., 2022; Chao et al., 2022). Although few direct studies have shown the correlation between CTNNB1 and IH. We still insist that CTNNB1 is a potential biomarker strongly associated with the development and progression of IH and VM.
IL6 (interleukin 6) encodes a cytokine that functions in many autoimmune diseases or infections. It is reported that IL6 can regulate macrophage polarization controls atherosclerosis associated vascular intimal hyperplasia (Chen et al., 2022). Dritsoula et al. (2022) demonstrated that IL-6, through STAT3 phosphorylation, activates LRG1 transcription resulting in vascular destabilization. Here, studies have reported a predictive association between IL6 and subsequent cerebral cavernous malformation disease clinical activity (Girard et al., 2018).
IGF2 (insulin-like growth factor 2) is a member of the insulin family, which are involved in human development and growth. It has been detected to be highly expressed during the proliferating phase of IH, but the underlying mechanism is unclear (Yu et al., 2004). The same results can be found in Muller’s experiments that the serum levels of GLUT1, IGF-2, and VEGF-A in IH were significantly higher than those in healthy control (Aydin Köker et al., 2021). IGF2 can stimulate multiple steps of endothelial progenitor cells (EPC) homing in vitro and promote both EPC recruitment and incorporation into the neovascular area, resulting in enhanced angiogenesis (Maeng et al., 2009). Taken together, the utilization of the IGF2 system may facilitate the development of novel therapeutic approaches for IH and VM.
CD34 (hematopoietic progenitor cell antigen 34) plays an important role in the adhesion of stem cells to extracellular matrix or stromal cells. It is related to multiple diseases such as hemangiopericytoma, malignant and neurofibroma. Immunohistochemical staining showed that CD34 was positive in IH lesion cells (Johnson et al., 2018). In vivo experiments, miR-130a inhibition effectively suppressed the tumor growth by reducing the expression of angiogenic markers and the percentage of CD31+ and CD34+ to inhibit angiogenesis (Gao et al., 2017). The deletion of FOXF1 reduces the expression of endothelial genes that are essential for vascular development, such as CD34 (Ren et al., 2014). Therefore, regulating the expression of CD34 may be a potential method for effective treatment of IH and VM.
Mitogen-Activated Protein Kinase 11 (MAPK11) is involved in a variety of cellular processes, including cell proliferation, differentiation and development. Acute H (2) O (2) activation of MAPK11-p38 is the main cause of endothelial dysfunction during pregnancy (Chen et al., 2005). In addition, it has been reported that Ak1-MAPK11-cofilin signal is essential for the proliferation and neovascularization of mouse retinal endothelial cells induced by hypoxia (Kumar et al., 2016). No studies have reported the link between MAPK11 expression and IH and VM. Taken together, we consider that MAPK11 is a novel biomarker closely associated with vascular anomalies.
Finally, it is worth noting that all of the hub genes have shown good efficiency in distinguishing diseases from healthy people (AUC of ROC>0.8). Furthermore, we found that has-miR-141-3p and has-miR-150-5p significantly downregulated in IH patients. In our TF-mRNA-microRNA network, has-miR-141-3p can target 3′UTR of IGF2. Since their expression in IH is exactly opposite, we speculate that the has-miR-141-3p-IGF2 axis is a potential pathway regulating vascular abnormalities. In some cases, history and clinical presentation are not sufficient to diagnose vascular abnormalities, especially in infants and young children. Our results provide additional noninvasive biomarkers to help distinguish between the two diagnoses.
We must acknowledge the limitations in this study. First, the sample size of each group is slightly small. Second, further experimentally validation is required to explore the function and changes of hub genes and signaling pathways. These follow-up steps contribute to a deeper understanding of crosstalk between the two diseases.
In conclusion, our study investigated the common DEGs and molecular mechanism in IH and VM. Base on PPI network analysis, five hub genes (CTNNB1, IL6, CD34, IGF2, MAPK11) were identified. Furthermore, we found that has-miR-141-3p and has-miR-150-5p significantly downregulated in IH patients. The regulation of vascular endothelial cell proliferation may be the common mechanism for the treatment of IH and VM. This study provides insight into the crosstalk of IH and VM and obtains several biomarkers relevant to the diagnosis and pathophysiology of vascular abnormalities.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
BH designed and carried out bioinformatics analyses and drafted the manuscript. PZ and Y-YZ did the statistical analyses and drew the figures; KW collected the data; X-MC and D-JY initial the study. All authors read and approved the final manuscript.
This work was supported by China National Nuclear Corporation Medical Department “Nuclear Medicine Technology Innovation” Project (ZHYLYB2021009). National Natural Science Foundation of China (32071238, 82073477), Natural Science Foundation of Sichuan Province (2020YJ0194).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1045244/full#supplementary-material
GEO, Gene Expression Omnibus; DEGs, differentially expressed genes; PPI, protein−protein interaction; IH, infantile hemangiomas; NS, normal skin; VM, venous malformations; FDR, false discovery rate; FC, fold changes.
Ambrozkiewicz, F., Trailin, A., Červenková, L., Vaclavikova, R., Hanicinec, V., Allah, M., et al. (2022). CTNNB1 mutations, TERT polymorphism and CD8+ cell densities in resected hepatocellular carcinoma are associated with longer time to recurrence. BMC cancer 22 (1), 884. doi:10.1186/s12885-022-09989-0
Aydin Köker, S., Kömüroğlu, A., Köksoy, A., Şiraz, Ü., Tekin, E., and Köker, A. (2021). Evaluation of GLUT1, IGF-2, VEGF, FGF 1, and angiopoietin 2 in infantile hemangioma. Arch. Pediatr. 28 (4), 296–300. doi:10.1016/j.arcped.2021.02.009
Bandettini, W., Kellman, P., Mancini, C., Booker, O., Vasu, S., Leung, S., et al. (2012). MultiContrast delayed enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: A clinical validation study. J. Cardiovasc. Magn. Reson. 14, 83. doi:10.1186/1532-429X-14-83
Barrett, T., Wilhite, S., Ledoux, P., Evangelista, C., Kim, I., Tomashevsky, M., et al. (2013). NCBI geo: Archive for functional genomics data sets--update. Nucleic Acids Res. 41, D991–D995. doi:10.1093/nar/gks1193
Chao, A., Wu, K., Lin, C., Lee, Y., Huang, H., Tang, Y., et al. (2022). Towards less invasive molecular diagnostics for endometrial cancer: Massively parallel sequencing of endometrial lavage specimens in women attending for an office hysteroscopy. J. Mol. Med. 100 (9), 1331–1339. doi:10.1007/s00109-022-02239-7
Chen, D., Li, S., Qian, X., Moon, C., and Zheng, J. (2005). Tyrosine phosphorylation of caveolin 1 by oxidative stress is reversible and dependent on the c-src tyrosine kinase but not mitogen-activated protein kinase pathways in placental artery endothelial cells. Biol. Reprod. 73 (4), 761–772. doi:10.1095/biolreprod.105.040881
Chen, J., Wang, W., Ni, Q., Zhang, L., and Guo, X. (2022). Interleukin 6-regulated macrophage polarization controls atherosclerosis-associated vascular intimal hyperplasia. Front. Immunol. 13, 952164. doi:10.3389/fimmu.2022.952164
Corada, M., Nyqvist, D., Orsenigo, F., Caprini, A., Giampietro, C., Taketo, M., et al. (2010). The Wnt/beta-catenin pathway modulates vascular remodeling and specification by upregulating Dll4/Notch signaling. Dev. Cell 18 (6), 938–949. doi:10.1016/j.devcel.2010.05.006
Dritsoula, A., Dowsett, L., Pilotti, C., O'Connor, M., Moss, S., and Greenwood, J. (2022). Publisher Correction: Angiopathic activity of LRG1 is induced by the IL-6/STAT3 pathway. Sci. Rep. 12 (1), 5347. doi:10.1038/s41598-022-09460-x
Duan, X., Shi, Y., Zhao, S., Yao, L., Sheng, J., and Liu, D. (2022). Foxc1a regulates zebrafish vascular integrity and brain vascular development through targeting amotl2a and ctnnb1. Microvasc. Res. 143, 104400. doi:10.1016/j.mvr.2022.104400
Ebenebe, C., Diehl, S., Bartnick, K., Dörge, H., Becker, J., Schweigerer, L., et al. (2007). Three reactive compartments in venous malformations. Thromb. Haemost. 97 (5), 763–773. doi:10.1160/th07-01-0021
Fernández Faith, E., Shah, S., Witman, P., Harfmann, K., Bradley, F., Blei, F., et al. (2021). Clinical features, prognostic factors, and treatment interventions for ulceration in patients with infantile hemangioma. JAMA Dermatol. 157 (5), 566–572. doi:10.1001/jamadermatol.2021.0469
Gallant, S., Chewning, R., Orbach, D., Trenor, C., and Cunningham, M. (2021). Contemporary management of vascular anomalies of the head and neck-Part 1: Vascular malformations: A review. JAMA Otolaryngol. Head. Neck Surg. 147 (2), 197–206. doi:10.1001/jamaoto.2020.4353
Gao, F., Wang, F., Liu, R., Xue, F., Zhang, J., Xu, G., et al. (2017). Epigenetic silencing of miR-130a ameliorates hemangioma by targeting tissue factor pathway inhibitor 2 through FAK/PI3K/Rac1/mdm2 signaling. Int. J. Oncol. 50 (5), 1821–1831. doi:10.3892/ijo.2017.3943
Girard, R., Zeineddine, H., Koskimäki, J., Fam, M., Cao, Y., Shi, C., et al. (2018). Plasma biomarkers of inflammation and angiogenesis predict cerebral cavernous malformation symptomatic hemorrhage or lesional growth. Circ. Res. 122 (12), 1716–1721. doi:10.1161/CIRCRESAHA.118.312680
Gomez-Acevedo, H., Dai, Y., Strub, G., Shawber, C., Wu, J., and Richter, G. (2020). Identification of putative biomarkers for Infantile Hemangiomas and Propranolol treatment via data integration. Sci. Rep. 10 (1), 3261. doi:10.1038/s41598-020-60025-2
Haggstrom, A., Drolet, B., Baselga, E., Chamlin, S., Garzon, M., Horii, K., et al. (2006). Prospective study of infantile hemangiomas: Clinical characteristics predicting complications and treatment. Pediatrics 118 (3), 882–887. doi:10.1542/peds.2006-0413
Han, H., Cho, J., Lee, S., Yun, A., Kim, H., Bae, D., et al. (2018). TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386. doi:10.1093/nar/gkx1013
Huang, D., Sherman, B., Tan, Q., Collins, J., Alvord, W., Roayaei, J., et al. (2007). The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8 (9), R183. doi:10.1186/gb-2007-8-9-r183
Johnson, E., Davis, D., Tollefson, M., Fritchie, K., and Gibson, L. (2018). Vascular tumors in infants: Case report and review of clinical, histopathologic, and immunohistochemical characteristics of infantile hemangioma, pyogenic granuloma, noninvoluting congenital hemangioma, tufted angioma, and kaposiform hemangioendothelioma. Am. J. Dermatopathol. 40 (4), 231–239. doi:10.1097/DAD.0000000000000983
Kumar, R., Janjanam, J., Singh, N., and Rao, G. (2016). A new role for cofilin in retinal neovascularization. J. Cell Sci. 129 (6), 1234–1249. doi:10.1242/jcs.179382
Kunimoto, K., Yamamoto, Y., and Jinnin, M. (2022). ISSVA classification of vascular anomalies and molecular biology. Int. J. Mol. Sci. 23 (4), 2358. doi:10.3390/ijms23042358
Lee, H., Xu, Y., He, L., Choi, W., Gonzalez, D., Jin, S., et al. (2021). Role of venous endothelial cells in developmental and pathologic angiogenesis. Circulation 144 (16), 1308–1322. doi:10.1161/CIRCULATIONAHA.121.054071
Liu, W., Jiang, X., Li, X., Sun, K., Yang, Y., Yang, M., et al. (2022). LMBR1L regulates the proliferation and migration of endothelial cells through Norrin/β-catenin signaling. J. Cell Sci. 135 (6), jcs259468. doi:10.1242/jcs.259468
Lv, Z., Yang, K., and Wang, Y. (2022). Long non-coding RNA breast cancer-associated transcript 54 sponges microRNA-1269b to suppress the proliferation of hemangioma-derived endothelial cells. Bioengineered 13 (3), 6188–6195. doi:10.1080/21655979.2022.2027064
Ma, X., Lv, K., Wu, L., and Ouyang, T. (2021). Propranolol inhibits stemness of hemangioma through Jagged1. Ann. Transl. Med. 9 (22), 1682. doi:10.21037/atm-21-5563
Maeng, Y., Choi, H., Kwon, J., Park, Y., Choi, K., Min, J., et al. (2009). Endothelial progenitor cell homing: Prominent role of the IGF2-igf2r-PLCbeta2 axis. Blood 113 (1), 233–243. doi:10.1182/blood-2008-06-162891
Mahajan, P., Bergstrom, K., Phung, T., and Metry, D. (2022). The genetics of vascular birthmarks. Clin. Dermatol. doi:10.1016/j.clindermatol.2022.02.006
Maimaiti, A., Aierken, Y., Zhou, L., He, J., Abudureyimu, A., and Li, S. (2022). Inhibiting interleukin-6/signal transducers and activators of transduction-3/hypoxia-inducible factor-1α signaling pathway suppressed the growth of infantile hemangioma. European journal of pediatric surgery. Zeitschrift fur Kinderchirurgie. doi:10.1055/s-0042-1749436
Pang, C., Lim, C., Brookes, J., Tsui, J., and Hamilton, G. (2020). Emerging importance of molecular pathogenesis of vascular malformations in clinical practice and classifications. Vasc. Med. 25 (4), 364–377. doi:10.1177/1358863X20918941
Ren, X., Ustiyan, V., Pradhan, A., Cai, Y., Havrilak, J., Bolte, C., et al. (2014). FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ. Res. 115 (8), 709–720. doi:10.1161/CIRCRESAHA.115.304382
Rodríguez Bandera, A., Sebaratnam, D., Wargon, O., and Wong, L. (2021). Infantile hemangioma. Part 1: Epidemiology, pathogenesis, clinical presentation and assessment. J. Am. Acad. Dermatol. 85 (6), 1379–1392. doi:10.1016/j.jaad.2021.08.019
Saito, R., Smoot, M., Ono, K., Ruscheinski, J., Wang, P., Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nat. Methods 9 (11), 1069–1076. doi:10.1038/nmeth.2212
Sandovici, I., Georgopoulou, A., Pérez-García, V., Hufnagel, A., López-Tello, J., Lam, B., et al. (2022). The imprinted Igf2-Igf2r axis is critical for matching placental microvasculature expansion to fetal growth. Dev. Cell 57 (1), 63–79.e8. e8. doi:10.1016/j.devcel.2021.12.005
Sticht, C., De La Torre, C., Parveen, A., and Gretz, N. (2018). miRWalk: An online resource for prediction of microRNA binding sites. PloS one 13 (10), e0206239. doi:10.1371/journal.pone.0206239
Strub, G., Kirsh, A., Whipple, M., Kuo, W., Keller, R., Kapur, R., et al. (2016). Endothelial and circulating C19MC microRNAs are biomarkers of infantile hemangioma. JCI insight 1 (14), e88856. doi:10.1172/jci.insight.88856
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi:10.1093/nar/gku1003
Warde-Farley, D., Donaldson, S., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220. doi:10.1093/nar/gkq537
Ye, C., Pan, L., Huang, Y., Ye, R., Han, A., Li, S., et al. (2011). Somatic mutations in exon 17 of the TEK gene in vascular tumors and vascular malformations. J. Vasc. Surg. 54 (6), 1760–1768. doi:10.1016/j.jvs.2011.06.098
Keywords: infantile hemangiomas, venous malformation, bioinformatics analysis, differentially expressed genes, microRNA
Citation: Huang B, Zhang P, Zhong Y-Y, Wang K, Chen X-M and Yu D-J (2022) Transcriptional data analysis reveals the association between infantile hemangiomas and venous malformations. Front. Genet. 13:1045244. doi: 10.3389/fgene.2022.1045244
Received: 15 September 2022; Accepted: 10 October 2022;
Published: 19 October 2022.
Edited by:
Shibiao Wan, University of Nebraska Medical Center, United StatesReviewed by:
Rujin Wang, Regeneron Genetic Center, United StatesCopyright © 2022 Huang, Zhang, Zhong, Wang, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dao-Jiang Yu, eWRqNTEwODdAMTYzLmNvbQ==; Xiao-Ming Chen, MjkyMjg4NzZAcXEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.