- Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague, Prague, Czechia
Arthropod pests are remarkably capable of rapidly adapting to novel forms of environmental stress, including insecticides and climate change. The dynamic interplay between epigenetics and genetics explains the largely unexplored reality underlying rapid climatic adaptation and the development of insecticide resistance in insects. Epigenetic regulation modulates gene expression by methylating DNA and acetylating histones that play an essential role in governing insecticide resistance and adaptation to climate change. This review summarises and discusses the significance of recent advances in epigenetic regulation that facilitate phenotypic plasticity in insects and their symbiotic microbes to cope with selection pressure implied by extensive insecticide applications and climate change. We also discuss how epigenetic changes are passed on to multiple generations through sexual recombination, which remains enigmatic. Finally, we explain how these epigenetic signatures can be utilized to manage insecticide resistance and pest resilience to climate change in Anthropocene.
Highlights
❖ Epigenetics is a rapidly expanding research field addressing previously unknown gene regulation mechanisms.
❖ Epigenetics does not change the DNA sequence but reprograms gene expression in response to environmental stimuli like climate change and insecticide exposure.
❖ How epigenetic changes are passed on to multiple generations remains elusive.
❖ Epigenetic contribution to the rapid evolution of insecticide resistance and climate adaptation is poorly understood.
❖ We provide prospects to tackle pests impeding epigenetic mechanisms.
❖This review will interest molecular biologists, developmental biologists, entomologists, geneticists, and people involved in integrated pest management and readers of this journal.
Introduction
Living organisms are influenced by environmental elements like temperature and light and ecological substances like phytochemicals and toxicants (Skinner, 2014a). Darwinian evolution relies heavily on the environment, and natural selection is a process in which environmental conditions affect the survival or reproductive success of individuals with various phenotypes (Darwin, 1859). The current paradigm in evolutionary biology holds that variations can evolve in response to natural selection due to changes in DNA sequence; however, phenotypic variation is thought to be due to genetic differences and is considered a neo-Darwinian process that the environment could alter through sociobiology-type mechanisms (Baldwin, 1896; Paenke et al., 2007; Laland et al., 2014). In neo-Darwinian evolution, the natural selection process is the primary mechanism through which the environment influences evolution (Olson-Manning et al., 2012; Laland et al., 2014). Combining heredity and evolution through natural selection seemed incorrect since it required an unacceptably high mutation rate to keep the trait variation needed for selection (Huxley, 1940). To answer this, Darwin suggested pangenesis, a sophisticated theory of transmitting environmentally sensitive somatic cells to progeny (Darwin, 1868).
It is not unexpected that climatic changes are altering the physiology, behaviour, abundance, and distribution of many species, given that global temperatures and precipitation patterns are changing quickly and profoundly impact living forms (Franks and Hoffmann, 2012; Good et al., 2016; Richard et al., 2019). The genetic impacts of climate change have not received as much attention as other climate change effects on the environment (Dawson et al., 2011; McMahon et al., 2011). As a result of recent findings in ecological genetics and developments in quantitative genetics and genomics, in our understanding, there are significant gaps in the likelihood and rate of adaptive genetic changes, the roles of regulatory and epigenetic effects, the evolution of plasticity in providing rapid selection responses, and more generally, the genetic architecture of adaptive evolution. Several shreds of evidence suggest that genetically based adaptive evolution traits, including body size, temperature responses, dispersion, and diapause/reproductive timing, may be a result of climate change (Thomas et al., 2001; Franks et al., 2007; Gardner et al., 2011; Hill et al., 2011; Karell et al., 2011; Richard et al., 2021). However, it is unclear whether numerous independently functioning genes govern the evolution of these traits with little influence or by a small number of essential regulatory genes found in genetic and metabolic networks. The exciting idea that heritable epigenetic modifications brought on by climate change could serve as an alternate and more rapid evolution method offers a captivating alternative to selection acting on existing genetic variation (Turner, 2009).
The ability of arthropod pests to develop resistance to insecticides and manage insect infestations is crucial nowadays, and insecticide expenditures in the world are approximately $40 billion each year (Pimentel et al., 1992; Popp, 2011; Bourguet and Guillemaud, 2016). Insecticide exposure in the field resulted in field-evolved resistance and reduced population vulnerability to insecticides (Tabashnik et al., 2014; Olivares-Castro et al., 2021). Individual events of insecticide resistance were better known through case studies, but the overall evolutionary process of insecticide resistance was still poorly understood (Gressel, 2011; Ffrench-Constant, 2013). Aggregating data from insecticide treatments on insect species to determine the trajectory of resistance could provide a road map for new techniques to combat resistance evolution. Observations of insecticide resistance evolution mechanisms in many insect species can provide useful insight into broader trends in resistance development within and between insect species. Most insecticide resistance research has focused on individual insecticide classes with biochemical or genetic mechanisms or the dissemination and growth of resistance in certain fields and species (Ffrench-Constant, 2013; Ffrench-Constant, 2014; Stratonovitch et al., 2014). These methods do not provide insight into the rate of insecticide resistance evolution, but they are critical for understanding how insecticide resistance evolves and spreads. Aggregated data and numerous methodologies were used for examining insecticide resistance development, highlighting larger evolutionary patterns across individual species and offering generalizable insights for pests with limited resources for thorough molecular or genetic studies (Brevik et al., 2018).
Resistance development is influenced by genetic, ecological, and operational factors, and the pace of insecticide resistance evolution varies significantly between species, yet the literature lacks systematic comparisons across geography and insecticide chemistries between species (Georghiou and Taylor, 1977; Council, 1986; Després et al., 2007). Although differences in the rate of evolution of species may explain evolvability, since some species evolve quickly, changes in evolvability differences against insecticides remain unexplained (Kinnison and Hendry, 2001). The evolvability of species is influenced by natural genetic variations such as differing mutation rates, biochemical differences caused by food variations, starting gene frequency differences, the number of generations each year, and population size (Carriére and Tabashnik, 2001; Lee and Gelembiuk, 2008; Hardy et al., 2018).
Insecticides and climate change alter insect development, physiology, and inheritance, and recent studies have focused on a link between insecticide and climate change effects on insects and their epigenetic regulations at the molecular level. Epigenetic mechanisms control gene expression without modifying genetic sequences against different stresses and compensatory mechanisms. This review summarises epigenetic changes from insecticide exposure and climate change on arthropod pests.
Environmental epigenetics
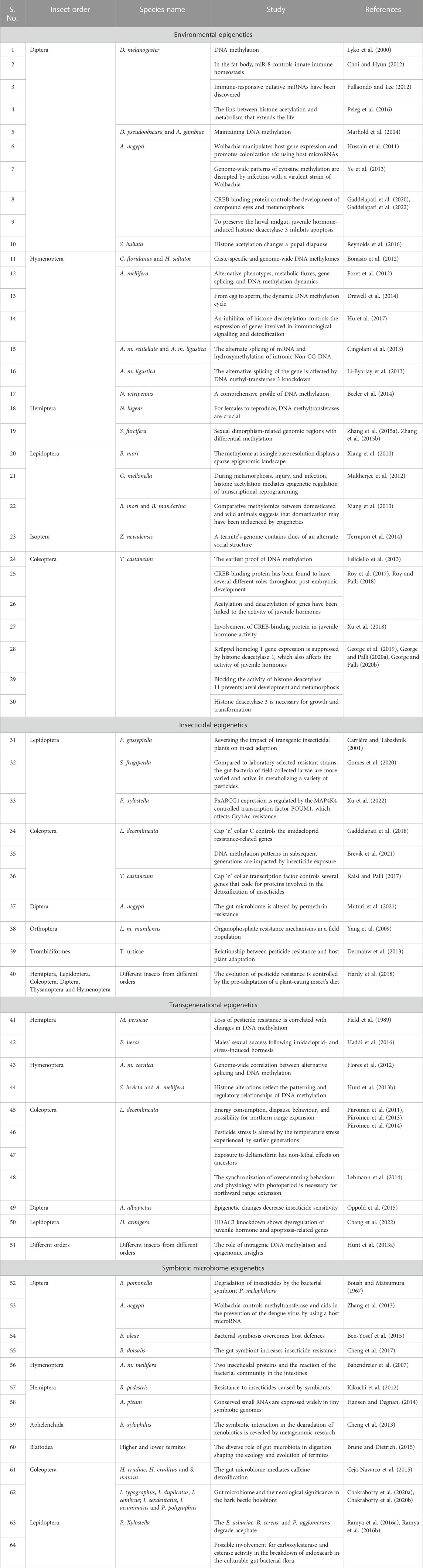
The central idea of modern biology is Charles Darwin’s theory of evolution by natural selection, yet Darwin was unaware of the molecular mechanisms behind this process (Darwin, 1859). A robust neo-Darwinian theory of evolution was made possible by integrating Darwin’s ideas with developments in genetic and molecular sciences during the past century (Olson-Manning et al., 2012). Genetics and mutations are the current main theories for the molecular foundation of evolution, according to which genetic variations caused by chromosomal and random DNA changes directly impact phenotype and phenotypic diversity. Epigenetics is a biological mechanism that can significantly affect genome activity and contribute to phenotypic variance (Figure 1) (Skinner, 2011; Palli, 2021). The word “epigenetics” was first used by Waddington, and the traditional definitions are descriptive without considering the underlying molecular components (Waddington, 1953; Skinner, 2011). Epigenetic information controls how different cellular and organismal phenotypes are produced from the same genome, e.g., how insects can produce phenotypes suited to their habitats (Bonasio et al., 2010; Mukherjee et al., 2015; Glastad et al., 2019). Epigenetics focuses on heritable modifications to the regulation of genes in response to intracellular and extracellular environmental signals (Table 1) (McGuigan et al., 2021). Since many different stimuli can consistently alter gene regulation, epigenetic information can take many other forms as it is generally understood. However, molecular mechanisms that directly influence, modify, or interact with chromatin are often included under molecular epigenetics. Mitotic cell division within individuals and meiotic cell division, which results in progeny birth, can transmit epigenetic information. The development process and how an egg with a single set of genetic instructions can evolve into a multicellular organism made up of several tissues are addressed by intragenerational epigenetic inheritance (Waddington, 1942). The transmission of epigenetic information from a focused individual to offspring is the subject of intergenerational epigenetic inheritance (Heard and Martienssen, 2014). Intergenerational epigenetics, although assumed to be uncommon, is fascinating since it directly influences the course of evolution, and this distinction is crucial. According to current scientific knowledge, epigenetics is “molecular mechanisms around DNA that govern genome activity irrespective of DNA sequence and are mitotically stable” (Skinner, 2011). DNA methylation, histone modifications, chromatin structure, and specific non-coding RNA (ncRNA) are considered epigenetic mechanisms (Figure 1) (Skinner, 2014b). Pre and post-transcriptional gene regulations are the two types of epigenetic mechanisms. Before transcription, DNA methylation and histone acetylation/deacetylation occur, and post-transcriptional gene regulation occurs with small non-coding RNAs/microRNAs (miRNAs) (Figure 1) (Jaenisch and Bird, 2003; Gómez-Díaz et al., 2012). Recent reports also evaluated the influence of coding and non-coding RNAs in forest insect outbreaks (Zhang et al., 2020).

FIGURE 1. Arthropod pest resistance mechanisms emerge in response to environmental and insecticide challenges via epigenetic control. Epigenetic mechanisms include DNA methylation by DNA methyltransferase (DMNT) and histone modifications (acetylation and methylation). Furthermore, non-coding RNAs are also involved in gene expression alteration by modifying DNA or RNA. (snRNA- small nuclear RNA, lncRNA- long non-coding RNA).
In DNA methylation, a methyl group to a cytosine residue in the dinucleotide sequence CpG results in 5-methylcytosine being added, which retains the base-pairing potential of unmodified nucleosides but changes its interaction with regulatory proteins (Figure 1). By methylating even a single CpG site in a promoter region, transcription of downstream genes can be reduced, and gene silencing can occur considerably (Robertson et al., 1995). In arthropods, cytosine methylation is vital in epigenetic gene control (He et al., 2015). DNA methylation has been discovered in several insect orders, including Coleoptera, Diptera, Lepidoptera, Hemiptera, Hymenoptera, Orthoptera, and Odonata (Table 1) (Xiang et al., 2010; Zhang et al., 2015a; Zhang et al., 2015b). DNA methyl-transferases (DMNTs) are enzymes that add a methyl group to specific chromosomal nucleotide bases of DNA and are classified into three families: DNMT1, DNMT2, and DNMT3 (Kausar et al., 2021). Because they are evolutionarily conserved, DNMT1 and DNMT2 are found in a wide range of insects, whereas DNMT3 is found exclusively in a few hymenopteran and hemipteran species, but the honeybee genome has all three DMNTs as well as a duplicated copy of DNMT3 (Glastad et al., 2011; Lyko and Maleszka, 2011). Using genome-wide methylation studies, scientists revealed the most prevalent DNA variation in insects such as Nasonia vitripennis, Bombyx mori, and Apis mellifera (Cingolani et al., 2013; Xiang et al., 2013; Beeler et al., 2014). Bisulfite sequencing can detect specific methylated genes, and CpG methylation was highest in A. mellifera and Tribolium castaneum embryos and decreased in the following stages of development (Feliciello et al., 2013; Drewell et al., 2014). According to genome-wide studies, insects have a significantly lower methylation to unmethylated CpG ratio than vertebrates, and less than 0.5% of the cytosine residues in the CpG dinucleotide are methylated in Drosophila melanogaster (Lyko et al., 2000; Marhold et al., 2004). In comparison to mammalian systems (60%–90%), invertebrates have very modest levels of CpG site methylation (0.36%–20%) (Regev et al., 1998; Suzuki and Bird, 2008). Many studies have found that DNA methylation is frequently restricted to genic regions of the genome (promoters, exons, and introns) rather than intergenic regions (Suzuki and Bird, 2008). Invertebrate insect genomes like hymenopterans (ants, bees, and parasitoid wasps) have lower amounts of DNA methylation than vertebrate genomes because methylation occurs at the promoters of genes and may inhibit transcription and metabolism (Ye et al., 2013; Yan et al., 2015). Several studies have suggested that gene splicing and activation are linked to DNA methylation in insects. Alternative splicing of mRNA transcripts caused by DNA methylation has been linked to behavioural control and caste specialization in eusocial insects (termites, bees, and ants) (Bonasio et al., 2012; Foret et al., 2012; Li-Byarlay et al., 2013; Terrapon et al., 2014). Furthermore, the removal of DNA methylation from the esterase gene of Myzus persicae reduced the transcription of this insecticide-detoxifying enzyme (FIELD, 2000). Recently, Gupta and Nair (2022) observed genetic plasticity in brown planthoppers and clarified the role of epigenomic modifications in converting environmental stimuli into heritable changes that may reduce stress and favor insects. They also showed how 5-azacytidine-mediated disruption DNA methylation influences the expression levels of some stress-responsive genes and highlighted demethylation/methylation as a phenomenon underlying rapid adaptive phenotypes (Gupta and Nair, 2022).
Histones are proteins found in the nuclei of eukaryotic cells that help to construct nucleosomes by packing DNA, and the positive charge of the core histones causes chromatin consistency and regulates transcription factor access (Figure 1) (Adcock and Lee, 2006; Bayarsaihan, 2011). Histones and wrapped DNA are densely packed in the condensed state of chromatin, making it transcriptionally inactive, but transcription factors could attach to DNA in the open chromatin and finally lead to gene expression. The addition or removal of acetyl groups from core histones increases or decreases DNA accessibility and thus promotes or suppresses gene expression, and histone acetylation and deacetylation are reversible epigenetic events that occur before transcription initiation (Marks et al., 2003; Hamon and Cossart, 2008). In insects, many biological and metabolic processes, such as growth regulation, development, metamorphosis, and reproduction, are regulated by juvenile hormones (JH) and ecdysteroids. The transcriptional co-regulators CREB binding protein (CBP) and Trichostatin A (TSA, HDAC inhibitor) activate several transcription factors that govern the expression of genes linked with post-embryonic development in insects. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are enzymes that acetylate and remove acetyl groups from amino acids of histone proteins, respectively. Three of the four classes of HDACs were found in D. melanogaster (Peleg et al., 2016). Our previous research demonstrated the importance of CBP and TSA for the regulation of HAT and HDACs during JH action in the red flour beetle, T. castaneum, and their cell lines and in yellow fever mosquito, Aedes aegypti (Roy et al., 2017; Roy and Palli, 2018; Xu et al., 2018; Gaddelapati et al., 2020). It has been demonstrated that there is a balanced upregulation of HATs and HDACs during insect metamorphosis in Galleria mellonella and Sarcophaga bullata (Mukherjee et al., 2012; Reynolds et al., 2016). In G. mellonella, an early pupal transition occurred in larvae treated with HAT inhibitors (suberonylanilide hydroxamic acid), and intriguingly, injection of HDAC inhibitors (sodium butyrate) delayed pupation. In addition, in the honeybee A. mellifera, sodium butyrate improved insecticide resistance (imidacloprid and microsporidian) by stimulating the production of immune and detoxification genes (Hu et al., 2017). Our previous lab investigated the function of class I and IV HDAC members (HDAC1 and HADC11) in T. castaneum and class I HADC3 in both T. castaneum and A. aegypti, respectively, and showed their effects on the acetylation levels of histones and the expression of genes coding for proteins involved in the regulation of growth, development, and metamorphosis (George et al., 2019; George and Palli, 2020a; George and Palli, 2020b; Gaddelapati et al., 2022).
MicroRNAs (miRNAs) are non-protein-coding short RNAs (18–25 bp) that can epigenetically upregulate or silence their target genes at the post-transcriptional level through either degradation of the target mRNA or translation inhibition (Bartel, 2009). They prevent the translation of mRNAs by base-pairing with untranslated sections or, in rare cases, the coding region (Ambros, 2004; Bartel, 2004; Asgari, 2011). MiRNAs are evolutionarily conserved across eukaryotic species and govern various cellular activities, including immunity, development, differentiation, and death (Bartel and Chen, 2004; Giraldez et al., 2005). Many studies have shown that a single miRNA can regulate hundreds of different target genes and influence more than 30% of animal genes (Bushati and Cohen, 2007; Bartel, 2009; Sato et al., 2011). Seven miRNAs were discovered in D. melanogaster, involved in the control of immunological responses (peptidoglycan receptor proteins), and another miRNA from the fat body was also discovered to influence immunity-related gene translation (Choi and Hyun, 2012; Fullaondo and Lee, 2012). Bacterial and viral infections have been linked to changes in miRNA expression levels in animals (Fassi Fehri et al., 2010). Wolbachia pipientis is an obligatory endosymbiont in a wide variety of invertebrates with the ability to manipulate host reproductive and host insect lifetime (Hussain et al., 2011). In addition, Wolbachia infection may also change the miRNA profile of the A. aegypti by inducing a host miRNA target metalloprotease gene, but, interestingly, inhibiting the miRNA reduces gene expression. To replicate efficiently in the host, the Wolbachia endosymbiont manipulates the host metalloprotease gene via the expression of cellular miRNA (Hussain et al., 2011).
DNA methylation is one example of an epigenetic process that can be imprinted and passed down to generations (Skinner, 2014b). It has been demonstrated that environmental factors encourage the epigenetic transgenerational inheritance of phenotypic variation. With epigenetics, the environment can directly modify phenotypic variation and its subsequent inheritance, e.g., DNA methylation must be removed to produce embryonic stem cells, causing the cell to become pluripotent (Crews et al., 2007; Seisenberger et al., 2013; Skinner, 2014b). Although many environmental influences cannot change the DNA sequence, temperature and nutrition can significantly impact epigenetic processes (Skinner, 2014a). All the examined organisms have highly conserved epigenetic mechanisms (such as DNA methylation) that are modifiable by the environment (Skinner, 2014a). A new molecular mechanism for controlling organism biology is provided by epigenetics. The environment’s capacity to directly affect cell and tissue development and function is essential for the individual’s health and phenotype, except in cases where the epigenetic alterations might be passed down across generations, and this direct environmental epigenetic effect on the individual would probably have little impact on evolution. Epigenetic transgenerational inheritance of disease and phenotypic diversity has been induced by various ecological variables, including temperature, nutrition, and environmental toxins (Skinner, 2014a). The germline transmission of epigenetic information between generations without direct exposure is known as epigenetic transgenerational inheritance (Skinner, 2011). Environmental exposures have imprinted epigenetic marks like DNA methylation during embryonic gonadal sex determination, gametogenesis, or another critical period of germline development (Skinner, 2014a). It has been demonstrated that factors like nutrition, body temperature, stress, and toxins encourage the epigenetic transgenerational inheritance of phenotypic diversity (Anway et al., 2005; Pembrey et al., 2006; Burdge et al., 2011; Song et al., 2013; Skinner, 2014a; Skinner, 2014b). Therefore, the ability of the environment to change the phenotype and alter phenotypic variation, independent of genetics, through this epigenetic mechanism is proposed to be essential for evolution (Anway et al., 2005; Jablonka and Raz, 2009; Day and Bonduriansky, 2011; Kuzawa and Thayer, 2011; Skinner, 2014a). Phenotypic variation is epigenetically transmitted through generations, and the environment can thus promote this process. Darwin cited sexual selection as one of the critical factors in evolution, and a prior study looked at how a hazardous environmental element could encourage the epigenetic transmission of a change in mate preference linked to sexual selection (Darwin, 1859; Crews et al., 2007). Therefore, an environmental element that changed sexual selection was discovered to encourage a long-lasting change in the sperm epigenome passed down through several generations (Crews et al., 2007). These findings suggest that ecological epigenetics may significantly influence evolutionary change (Skinner, 2011).
Epigenetic regulations influencing insecticide resistance
Insecticide resistance has long been a concern affecting crop protection chemicals’ efficacy and usability, as well as firms’ capacity and inclination to invest in innovative crop protection compounds and traits globally (Sparks et al., 2021). In 1984, the Insecticide Resistance Action Committee (IRAC; https://irac-online.org/) was established to offer the crop protection sector a coordinated response to the problem of insecticide resistance and develop a few agrochemical businesses in Europe and the United States into a much bigger group of companies with global representation and an active presence encompassing corporations from more than 20 countries. IRAC also proactively addressed insecticide resistance management (IRM) by making various informational and educational tools (videos, posters, and pamphlets) on insect pests, bioassay methods, modes of insecticide action, and resistance management. The Insecticide Mode of Action (MoA) Classification Scheme, established by IRAC, has evolved from a relatively simple acaricide classification begun in 1998 to a significantly broader scheme that currently covers biologics as well as insecticides and acaricides. Since the discovery of insect resistance to insecticides, a growing number of insects have developed resistance to at least one or more of the insecticides available on the market (Feyereisen et al., 2015). Field-evolved resistance to insecticides is a genetically based reduction in a population’s susceptibility to an insecticide brought on by exposure to the insecticide in the field (Tabashnik et al., 2014). Inherent genetic variations, such as variable mutation rates between species, metabolic variations resulting from dietary variations, and starting gene frequency, may impact evolution (Lee and Gelembiuk, 2008; Hardy et al., 2018). Additionally, the number of generations produced annually and the size of the population may have an impact on the possibility that populations may develop resistance (Carriére and Tabashnik, 2001). The fact that pest species may be subjected to various amounts, chemistries, and rates of insecticides in entirely distinct environmental circumstances makes it challenging to compare the pace of insecticide development. For instance, a pest with a wide distribution may be exposed to a variety of insecticides regularly over several continents, whereas a pest with a more restricted distribution may only be exposed to a single chemical occasionally (Alyokhin et al., 2008; Yang et al., 2009). Arthropods have evolved defence systems in response to host plant allelochemicals (Figure 1). Similarly, they have created a variety of mechanisms, from behavioural to molecular to insecticide resistance (Heckel, 2014). Recent developments also identify transcription factors, internal microbial allies, and mechanisms underlying epigenetic and epitranscriptomics that promote insecticide and host plant resistance (Table 1) (Oppold and Müller, 2017; Gomes et al., 2020; Palli, 2020; Brevik et al., 2021; Muturi et al., 2021; Xu et al., 2022). Arthropods may regularly acquire insecticide resistance not just through novel mutations but also from existing genetic variations, suggesting that phytophagous arthropods have already pre-adapted to resist insecticides (Hawkins et al., 2019; Bras et al., 2022). Thus, if the chemical structure of the insecticides is like the plant defence chemicals, cross-resistance between host plant allelochemicals and insecticides could be a viable mechanism for rapid insecticide resistance development in arthropods (Després et al., 2007). Since the detoxifying mechanisms involved in host plant adaptation and insecticide resistance are similar, changes in host plant use may favour insecticide resistance and vice versa (i.e., pre-adaptation theory). This is supported by comparisons of genetic studies of host plant adaptation and insecticide resistance (Dermauw et al., 2013). Therefore, depending on their evolutionary history and the availability of existing genetic variation, some species may be more likely than others to evolve insecticide resistance (Bras et al., 2022).
In agroecosystems, insecticides are widely used and play a significant selective role in the evolution of insect pests, and the concept of the insecticide treadmill explains how insect pests develop a tolerance to routinely applied insecticides, rendering them useless (STAT, 2019). Based on mutation rates, the rapid gain and loss of resistance appear to happen far more quickly than expected, which raises the possibility that insecticides themselves can speed up mutation or alter the physiological makeup of pest species (Drake et al., 1998; Jablonka and Raz, 2009; Gressel, 2011). One potential justification that has not received much attention is that insecticide resistance may have evolved due to heritable epigenetic changes, which affect gene expression without altering the underlying DNA sequence. Insect pest species that have repeatedly encountered genetic bottlenecks owing to invasion and selection are nevertheless able to adapt relatively quickly, despite having low genetic diversity, which has led to the evolution of insecticide resistance (Gressel, 2011). The same insect pests have developed resistance to all the major classes of insecticides, and it is predicted that they will also develop resistance to new ones (Ffrench-Constant, 2007; Sparks and Nauen, 2015). Extreme genetic bottlenecks also don’t seem to be a barrier to the evolution of insecticide resistance. Insecticide resistance in pests appears inevitable because new phenotypes appear in response to environmental stress at rates that may not be explained by natural selection. Since environmental epigenetic patterning can affect the transgenerational transmission of phenotypic variation, it is crucial to understand how epigenetic processes fit within a neo-Lamarckian paradigm (Skinner, 2015). Xenobiotics and environmental stressors can directly affect the phenotypic responses of organisms to their environment through altering epigenetic changes. The study of epigenetics focuses on how the environment influences inherited alterations in gene expression. DNA methylation, histone changes, and heritable non-coding RNA are a few heritable epigenetic pathways that may be responsible for the transgenerational impacts of insecticides (Vandegehuchte et al., 2010b; Liebers et al., 2014; Niu et al., 2015). A heterodimer of xenobiotic transcription factors, cap n collar C isoform (CncC) and muscular aponeurosis fibromatosis (Maf), mediates cellular defence in invertebrates. These proteins in insects regulate the expression of genes involved in insecticide detoxification. We did RNAi and insecticide bioassays with insecticide detoxification genes in T. castaneum (CYP4G7, CYP4G14, GST-1, ABCA-UB, ABCAA1, ABCA-A1L, and ABCA-9B genes needed CncC for expression) and Colorado potato beetle, L. decemlineata, and discovered that CncC regulates genes coding proteins involved in insecticide detoxification (Kalsi and Palli, 2017; Gaddelapati et al., 2018).
Epigenetic modifications, transgenerational inheritance, and response to xenobiotic stress
Epigenetic changes can be inherited (Vandegehuchte et al., 2010a). Most insect orders exhibit DNA methylation, which is the attachment of a methyl group to the 5-carbon position of the cytosine nucleotide (often the cytosine in CpG dinucleotides), an epigenetic inheritance mechanism that can affect phenotypic diversity (Figure 1; Table 1) (Glastad et al., 2011). In insects, methylation is primarily present in coding areas and is strongly related to gene expression and alternative splicing, which allows a single gene to produce a variety of gene transcripts with different lengths depending on which exons are translated (Flores et al., 2012). Methylation can occur anywhere in the genome, but the effects of DNA methylation differ depending on where it occurs: 1) changes in DNA methylation at the promoter region can affect gene expression in downstream genomic regions; 2) methylation suppresses the gene expression of transposable elements; and 3) gene body methylation can affect gene expression and increase the number of alternative splice variants (Field et al., 1989; Fablet and Vieira, 2011; Glastad et al., 2011; Hunt et al., 2013a). Methylation patterns in insects may change along with their levels of insecticide resistance (Hunt et al., 2013b). The nuclear DNA-wrapped histone proteins can be modified by adding acetyl or methyl groups, altering the regulation of gene expression (Szyf, 2015). Particularly among arthropods, the full ramifications of these alterations are not well understood, and it does seem that some histone changes can be passed down through generations (Holoch and Moazed, 2015). Various non-coding RNA (ncRNA) can be passed down by male or female gametes and sustain DNA methylation and histone modifications and thus influence the chromatin structure, while many recent studies have not looked at heritable RNA (Liebers et al., 2014; Peschansky and Wahlestedt, 2014). A phenotypic response results from a constellation of interrelated effects, including DNA methylation, histone changes, and ncRNAs (Vandegehuchte et al., 2009). It would be ideal to evaluate all three mechanisms simultaneously through concurrent small RNA-seq, bisulfate-treated DNA-seq, and histone modification assays in as many tissues or individuals as possible to fully understand how epigenetic modifications affect transgenerational phenotypic inheritance. To ascertain whether changes in epigenetics and gene expression are consistently different between treatments, it is ideal to sequence numerous generations, and these investigations are severely constrained by the cost of sequencing; however, anticipated decreases in sequencing prices in the future should make these studies possible.
Arthropods exposed to insecticides and other xenobiotic substances may experience alterations in their DNA methylation state, and these epigenetic modifications may last for at least a few generations (Vandegehuchte et al., 2010a; Vandegehuchte et al., 2010b; Poulos et al., 2017). A fungicide exposure causes heritable methylation alterations in mosquitoes and reduces their sensitivity to imidacloprid insecticides (Oppold et al., 2015). Higher rates of spontaneous deamination, which converts methylated cytosines to thymines in methylated areas than in non-methylated areas, can result in higher mutation rates (Baccarelli and Bollati, 2009). Genes that are most elevated in response to insecticide resistance may also be the most likely to experience spontaneous deamination if genes involved with resistance are methylated, which increases expression and increases the likelihood of mutation. Even though it has been demonstrated that DNA methylation and histone modifications frequently co-occur in the genome, it is less known how histone modifications and small RNA affect the epigenetic responses of arthropods to toxins (Szyf, 2015). Histone alterations allow for the epigenetically transmitted transmission of parental hormetic responses to oxidative stress to offspring (Kishimoto et al., 2017). Histone modifications can be altered by various environmental contaminants, including heavy metals, air pollutants, dioxins, and endocrine disruptors, although it is uncertain if these changes are inherited (Rechavi and Lev, 2017). Changes in small RNAs may also be related to the transgenerational inheritance of stress traits since small RNAs have been reported to interact with histone modifications (Piiroinen et al., 2011). A more recent study revealed that juvenile hormone (JH)-related and apoptosis-related genes were dysregulated by HDAC3 knockdown, and it came to the conclusion that the HDAC3 gene is a promising target for controlling H. armigera (Chang et al., 2022).
Implications for transgenerational effects on insect fitness in agroecosystems
We propose that, through epigenetic mechanisms, insecticide use might, directly and indirectly, influence the evolution of insect pests in agroecosystems (Figure 1). The use of insecticides may directly promote the development of beneficial phenotypes that epigenetic changes may suppress (Table 1). Thus, continued insecticide application on populations encountering resistance would function as “natural selection” and systematically boost the prevalence of insecticide-adaptive insect phenotypes. Indirectly, the use of insecticides may maintain stressful surroundings so that hormetic prime insect pests can adapt to stressful situations more readily. For instance, sublethal insecticide exposure might alter adult body size, which may boost insect pests’ resilience to harsh winter circumstances (Alyokhin et al., 2009; Calabrese and Blain, 2011). Additionally, insecticides may improve female fertility or mate-seeking behaviour, resulting in a larger population (Piiroinen et al., 2014; Haddi et al., 2016). Phenotypic characteristics of pest insects that enable them to thrive when exposed to insecticides may also promote global invasions (Alyokhin et al., 2009). The insecticide-treated beetles invest more in fat bodies and have a greater metabolic rate than control ones (Piiroinen et al., 2013). Beetles may be better at detoxifying toxins because of their greater metabolic rates and larger fat bodies and can effectively overwinter using stored fat (Lehmann et al., 2014; Brander et al., 2017).
Contrarily, the intentional use of insecticides in agroecosystems is a component of an active pest management system, wherein insect responses to pressures can positively impact management choices in the future. As highly managed systems, agroecosystems enable more robust experimental control for studies at the field and landscape scales. Along these lines, it would be crucial to understand how individual, population, and species-level epigenetic responses to the same insecticides may vary. Such data would suggest whether epigenetic reactions can be broadly predicted across species and how insecticide resistance might be controlled more effectively. We may be able to understand better the role that epigenetic responses within insects play in the insecticide treadmill by combining new genomic techniques, epigenetic tests, and computationally demanding methodologies.
Microbial influence on host regulatory mechanisms
Nowadays, insect-microbe interactions and the epigenome are among the most intriguing and perplexing fields of insect biological research, involving the symbiotic involvement of bacteria with their host organisms, particularly the gut-insecticide axis (Figure 1) (Gupta and Nair, 2020). Some of the most comprehensive and deep metabolic and ecological connections with gut flora are found in insects (Table 1) (Brune and Dietrich, 2015; Douglas, 2015; Kim et al., 2016; Mukherjee and Dobrindt, 2022). Our group recently expanded current understanding of core gut bacterial communities in Ips bark beetles and their putative functions, such as cellulose degradation, nitrogen fixation, plant xenobiotic compound detoxification, and pathogen inhibition, which could serve as a foundation for future metatranscriptomics, metaproteomics, as well as insect epigenomics research (Chakraborty et al., 2020a; Chakraborty et al., 2020b). In insects, the symbiotic connections mediated by DNA methylation appear to be rare, and only a few examples reveal strong methylation signals. One example is the symbiotic association between Wolbachia, an intracellular bacterium, and the mosquito A. aegypti. Infection with Wolbachia of the mosquito causes changes in host methylation because of impacts on the tRNA methyltransferase DNMT2, as well as immune system modifications that affect antiviral responses (Ye et al., 2013; Zhang et al., 2013). A massive number of non-coding regulatory RNAs, particularly sRNAs in archaea and bacteria, are likely to be more critical in insect-microbe interactions (Babski et al., 2014; Kim et al., 2016). Bacterial sRNAs can target eukaryotic genes, which can be coopted into the RNAi pathway or mimic eukaryotic sRNAs (Kim et al., 2016). Furthermore, bacterial symbionts with drastically reduced genomes have evolved several short RNAs that aid in modulating the expression of important symbiotic genes and regulating core housekeeping operations in their insect hosts (Hansen and Degnan, 2014). While evidence for microbe-mediated direct epigenetic impacts on organismal-level phenotypes in insects is limited, the potential for such regulation is considerable. Some insects (especially social insects) harbor enormous numbers of microbial species, each of which can potentially have some influence on complex regulatory functions (Engel and Moran, 2013). Future follow up studies in this area can reveal more exciting dimensions of inter kingdom interactions during insect symbiosis.
Future perspectives and challenges
According to an emerging theory of environmental epigenetics, environmental exposure to a variety of chemicals can have long-lasting effects that are heritable, and the modifications have a variety of phenotypic impacts that last for generations, ranging from disease etiology to environmental change adaptations (Nilsson et al., 2018; Thiebaut et al., 2019; Sen et al., 2022). These epigenetic modifications can affect developmental bias, phenotypic flexibility, and niche creation, all of which influence the processes of evolution (Figure 1) (Jeremias et al., 2018). It is believed that epigenetic modifications brought on by the environment may have aided in the diversification and evolution of Darwin’s finches (Skinner et al., 2014). In arthropods, DNA methylation can be passed down across generations and affects important gene expression patterns, which may lead to enduring adaptation (Glastad et al., 2014). Additionally, alterations in DNA methylation are linked to modifications in vulnerability to insecticides (Field et al., 1989; Oppold et al., 2015). The rapid emergence of insecticide resistance can be explained in unique ways by understanding these mechanisms, which may also assist in addressing the enigma of insecticide resistance. One potential mechanism by which toxicity exposure reduces overall DNA methylation is due to competition between biological processes (Hunter et al., 2015; Oppold et al., 2015). The availability of methyl groups decides the DNA methylation of genomic DNA, and S-adenosylmethionine provides the methyl groups needed by methyltransferases (DMNT) to methylate DNA (Lee et al., 2009). Homocysteine, which is required as a precursor for S-adenosylmethionine, and glutathione are both antioxidants that conjugate with xenobiotic toxins (Enayati et al., 2005). Detoxification is necessary for the presence of toxins and depletes homocysteine, which may prevent the availability of S-adenosylmethionine for DNA methylation and result in a reduction in DNA methylation in the genome (Lee et al., 2009; Oppold and Müller, 2017). It is possible that the metabolic precursors required to methylate DNA are being depleted in this instance by the biochemical pathways involved in detoxification.
Designing novel chemical tools for insecticide resistance management in pest control requires a thorough understanding of the potential epigenetic mechanisms underlying insecticide resistance. Alternatively, the epigenetic system can most likely be used by insect molecular biologists and biochemists as targets for creating new insecticidal compounds (Jenkins and Muskavitch, 2015). Numerous epigenetic modifiers have been demonstrated to have a role in the control of developmental processes, homeostasis mediation, stimulation of responses to the external environment, and transmission of gene expression patterns through generations (Greer et al., 2011; Cantone and Fisher, 2013). Since epigenetic regulation genes are functionally conserved across all insect species, small molecule antagonists supplied in large quantities may also impact non-target insects (Jenkins and Muskavitch, 2015). Developing new and more effective insecticidal remedies targeting pest species depends on our ability to understand better epigenetic systems, mechanisms, and dynamics among insects. To understand the epigenetic dynamics in the insect gut microbiome related to climate change and insecticide resistance, scientists must concentrate on hologenomic research using cutting-edge laboratory techniques (metagenomes and CRISPR-Cas9 approaches). It will provide insight into how epigenetic modifications control the biomolecular characteristics of both host and microbiome communities (Kim et al., 2016; Gupta and Nair, 2020). Some studies demonstrated that insect resistance to biological (Bacillus thuringiensis and parasitoid wasps) and chemical insecticides (Fenitrothion) could be caused via symbiont-mediated processes (Oliver et al., 2003; Broderick et al., 2006; Kikuchi et al., 2012). However, the development of epigenetic analyses has challenges in establishing molecular techniques (ChIP-seq or ATAC-seq, etc.) to apply genome-wide chromatin dynamics analysis, revealing the epigenomic identity of alternative phenotypes. Instead of focusing on end phenotypes of insects, the studies will need to be focused on crucial stages (and potentially on specific tissues) when and where developmental switches occur and important symbiotic microbes. This will then allow for identifying critical variables that are receptive to environmental and insecticidal signals and will trigger cascades of gene expression alterations that eventually contribute to the formation of various resistance phenotypes. Finally, resistance management can only be effective if all stakeholders, including the crop protection sector, academia, regulators, crop consultants, growers, and other end-users, work together. IRAC was formed to raise resistance awareness among crop protection product developers, coordinate IRM tools and programs, forge alignment on key initiatives, and enable unified communication with other stakeholders. Thus, worries about insecticide resistance and reducing its impact remain paramount for crop protection companies, particularly those involved in discovering and developing new crop protection chemicals. IRAC and its linked IRM standards and programs are concerned with preserving the utility and efficacy of existing and novel crop protection, transgenic plant characteristics, and biological methods.
Conclusion
Due to the widespread use of insecticides for insect pest management and unanticipated climate change, ecosystems can serve as excellent research subjects for studying epigenetic regulators of evolutionary and eco-evolutionary studies. However, the development of resistance also brought about conditions that were challenging to manage. More research on the epigenetic regulation underlying climate change and insecticide resistance evolution is needed to corroborate this pattern. Understanding the ecological and evolutionary consequences of insecticide resistance and climate change in arthropods and their symbiotic microbiomes will shed light on the potential effects of insecticides on multitrophic interactions and ecosystem functioning, which could aid in the development of more sustainable pest management methods (Figure 1; Table 1). To do this, it is critical to integrate a range of methods from molecular and quantitative genetics and genomics with ecological approaches for studying responses to climate change and insecticide resistance. The genetic underpinnings of insecticide resistance adaptation and climate change will be uncovered with recent developments such as genome sequencing, transcription profiling, metagenomics, metatranscriptomics, and bisulfite sequencing. In evolution, a better understanding of the genetic basis of adaptation to climate change and insecticide resistance in natural populations will address several issues, including the number of genes involved in evolutionary changes and their relative importance, the extent to which changes in gene regulation result in evolution, and the role of additional mechanisms, such as epigenetics. Based on existing research, our knowledge of the genetic underpinnings of evolutionary responses to climatic changes and insecticide resistance in arthropods and their symbiotic microorganisms is still limited (Table 1). We believe that the availability of insect genome sequences will remarkably induce epigenetic research on non-model insect pests. Research focused on non-model insects and their symbiotic microbes as holobionts against xenobiotics, insecticides, and climate change in Anthropocene using multi-omics approaches such as genome, transcriptome (coding and non-coding RNAs), metagenome, and metatranscriptome may aid in designing novel pest management strategies by impeding key regulatory mechanisms within pest insects in the postgenomic era.
Author contributions
AR conceptualize the review idea. KM writes the first draft. AR and KM contributed to the figure preparation. Both authors contributed to reviewing and finalizing the draft.
Funding
The review was financed by EVA 4.0, No. CZ.02.1.01/0.0/0.0/16019/0000803 supported by OP RDE.
Acknowledgments
We appreciate the helpful feedback provided by the reviewers and handling editor.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adcock, I., and Lee, K.-Y. (2006). Abnormal histone acetylase and deacetylase expression and function in lung inflammation. Inflamm. Res. 55 (8), 311–321. doi:10.1007/s00011-006-0081-1
Alyokhin, A., Baker, M., Mota-Sanchez, D., Dively, G., and Grafius, E. (2008). Colorado potato beetle resistance to insecticides. Am. J. Potato Res. 85 (6), 395–413. doi:10.1007/s12230-008-9052-0
Alyokhin, A., Guillemette, R., and Choban, R. (2009). Stimulatory and suppressive effects of novaluron on the Colorado potato beetle reproduction. J. Econ. entomology 102 (6), 2078–2083. doi:10.1603/029.102.0609
Ambros, V. (2004). The functions of animal microRNAs. Nature 431 (7006), 350–355. doi:10.1038/nature02871
Anway, M. D., Cupp, A. S., Uzumcu, M., and Skinner, M. K. (2005). Epigenetic transgenerational actions of endocrine disruptors and male fertility. science 308 (5727), 1466–1469. doi:10.1126/science.1108190
Asgari, S. (2011). Role of microRNAs in insect host–microorganism interactions. Front. physiology 2, 48. doi:10.3389/fphys.2011.00048
Babski, J., Maier, L.-K., Heyer, R., Jaschinski, K., Prasse, D., Jäger, D., et al. (2014). Small regulatory RNAs in archaea. RNA Biol. 11 (5), 484–493. doi:10.4161/rna.28452
Babendreier, D., Joller, D., Romeis, J., Bigler, F., and Widmer, F. (2007). Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS Microbiol. Ecol. 59 (3), 600–610. doi:10.1111/j.1574-6941.2006.00249.x
Baccarelli, A., and Bollati, V. (2009). Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 21 (2), 243–251. doi:10.1097/mop.0b013e32832925cc
Bartel, D. P., and Chen, C.-Z. (2004). Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5 (5), 396–400. doi:10.1038/nrg1328
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136 (2), 215–233. doi:10.1016/j.cell.2009.01.002
Bayarsaihan, D. (2011). Epigenetic mechanisms in inflammation. J. Dent. Res. 90 (1), 9–17. doi:10.1177/0022034510378683
Beeler, S. M., Wong, G. T., Zheng, J. M., Bush, E. C., Remnant, E. J., Oldroyd, B. P., et al. (2014). Whole-genome DNA methylation profile of the jewel wasp (Nasonia vitripennis). G3 Genes, Genomes, Genet. 4 (3), 383–388. doi:10.1534/g3.113.008953
Ben-Yosef, M., Pasternak, Z., Jurkevitch, E., and Yuval, B. (2015). Symbiotic bacteria enable olive fly larvae to overcome host defences. R. Soc. open Sci. 2 (7), 150170. doi:10.1098/rsos.150170
Bonasio, R., Li, Q., Lian, J., Mutti, N. S., Jin, L., Zhao, H., et al. (2012). Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 22 (19), 1755–1764. doi:10.1016/j.cub.2012.07.042
Bonasio, R., Tu, S., and Reinberg, D. (2010). Molecular signals of epigenetic states. science 330 (6004), 612–616. doi:10.1126/science.1191078
Bourguet, D., and Guillemaud, T. (2016). The hidden and external costs of pesticide use. Sustain. Agric. Rev. 19, 35–120. doi:10.1007/978-3-319-26777-7_2
Boush, M. G., and Matsumura, F. (1967). Insecticidal degradation by Pseudomonas melophthora, the bacterial symbiote of the apple maggot. J. Econ. Entomology 60 (4), 918–920. doi:10.1093/jee/60.4.918
Brander, S. M., Biales, A. D., and Connon, R. E. (2017). The role of epigenomics in aquatic toxicology. Environ. Toxicol. Chem. 36 (10), 2565–2573. doi:10.1002/etc.3930
Bras, A., Roy, A., Heckel, D. G., Anderson, P., and Karlsson Green, K. (2022). Pesticide resistance in arthropods: Ecology matters too. Ecol. Lett. 25 (8), 1746–1759. doi:10.1111/ele.14030
Brevik, K., Bueno, E. M., McKay, S., Schoville, S. D., and Chen, Y. H. (2021). Insecticide exposure affects intergenerational patterns of DNA methylation in the Colorado potato beetle, Leptinotarsa decemlineata. Evol. Appl. 14 (3), 746–757. doi:10.1111/eva.13153
Brevik, K., Schoville, S. D., Mota-Sanchez, D., and Chen, Y. H. (2018). Pesticide durability and the evolution of resistance: A novel application of survival analysis. Pest Manag. Sci. 74 (8), 1953–1963. doi:10.1002/ps.4899
Broderick, N. A., Raffa, K. F., and Handelsman, J. (2006). Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. 103 (41), 15196–15199. doi:10.1073/pnas.0604865103
Brune, A., and Dietrich, C. (2015). The gut microbiota of termites: Digesting the diversity in the light of ecology and evolution. Annu. Rev. Microbiol. 69, 145–166. doi:10.1146/annurev-micro-092412-155715
Burdge, G. C., Hoile, S. P., Uller, T., Thomas, N. A., Gluckman, P. D., Hanson, M. A., et al. (2011). Progressive, transgenerational changes in offspring phenotype and epigenotype following nutritional transition. PloS one 6 (11), e28282. doi:10.1371/journal.pone.0028282
Bushati, N., and Cohen, S. (2007). microRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205. doi:10.1146/annurev.cellbio.23.090506.123406
Calabrese, E. J., and Blain, R. B. (2011). The hormesis database: The occurrence of hormetic dose responses in the toxicological literature. Regul. Toxicol. Pharmacol. 61 (1), 73–81. doi:10.1016/j.yrtph.2011.06.003
Cantone, I., and Fisher, A. G. (2013). Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 20 (3), 282–289. doi:10.1038/nsmb.2489
Carriére, Y., and Tabashnik, B. (2001). Reversing insect adaptation to transgenic insecticidal plants. Proc. R. Soc. Lond. Ser. B Biol. Sci. 268 (1475), 1475–1480. doi:10.1098/rspb.2001.1689
Ceja-Navarro, J. A., Vega, F. E., Karaoz, U., Hao, Z., Jenkins, S., Lim, H. C., et al. (2015). Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 6 (1), 7618–7619. doi:10.1038/ncomms8618
Chakraborty, A., Ashraf, M. Z., Modlinger, R., Synek, J., Schlyter, F., and Roy, A. (2020a). Unravelling the gut bacteriome of Ips (Coleoptera: Curculionidae: Scolytinae): Identifying core bacterial assemblage and their ecological relevance. Sci. Rep. 10 (1), 18572. doi:10.1038/s41598-020-75203-5
Chakraborty, A., Modlinger, R., Ashraf, M. Z., Synek, J., Schlyter, F., and Roy, A. (2020b). Core mycobiome and their ecological relevance in the gut of five Ips bark beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 11, 568853. doi:10.3389/fmicb.2020.568853
Chang, H., Xu, Z., Li, W., Cai, C., Wang, W., Ge, P., et al. (2022). HDAC3 knockdown dysregulates juvenile hormone and apoptosis-related genes in helicoverpa armigera. Int. J. Mol. Sci. 23 (23), 14820. doi:10.3390/ijms232314820
Cheng, D., Guo, Z., Riegler, M., Xi, Z., Liang, G., and Xu, Y. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5 (1), 13–12. doi:10.1186/s40168-017-0236-z
Cheng, X.-Y., Tian, X.-L., Wang, Y.-S., Lin, R.-M., Mao, Z.-C., Chen, N., et al. (2013). Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci. Rep. 3 (1), 1869. doi:10.1038/srep01869
Choi, I. K., and Hyun, S. (2012). Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev. Comp. Immunol. 37 (1), 50–54. doi:10.1016/j.dci.2011.12.008
Cingolani, P., Cao, X., Khetani, R. S., Chen, C.-C., Coon, M., Sammak, A., et al. (2013). Intronic non-CG DNA hydroxymethylation and alternative mRNA splicing in honey bees. BMC genomics 14 (1), 666. doi:10.1186/1471-2164-14-666
Council, N. R. (1986). Pesticide resistance: Strategies and tactics for management. United States: National Academies Press.
Crews, D., Gore, A. C., Hsu, T. S., Dangleben, N. L., Spinetta, M., Schallert, T., et al. (2007). Transgenerational epigenetic imprints on mate preference. Proc. Natl. Acad. Sci. 104 (14), 5942–5946. doi:10.1073/pnas.0610410104
Darwin, C. (1868). The variation of animals and plants under domestication. Cambridge: Cambridge University Press.
Dawson, T. P., Jackson, S. T., House, J. I., Prentice, I. C., and Mace, G. M. (2011). Beyond predictions: Biodiversity conservation in a changing climate. science 332 (6025), 53–58. doi:10.1126/science.1200303
Day, T., and Bonduriansky, R. (2011). A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am. Nat. 178 (2), E18–E36. doi:10.1086/660911
Dermauw, W., Wybouw, N., Rombauts, S., Menten, B., Vontas, J., Grbić, M., et al. (2013). A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. 110 (2), E113–E122. doi:10.1073/pnas.1213214110
Després, L., David, J.-P., and Gallet, C. (2007). The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 22 (6), 298–307. doi:10.1016/j.tree.2007.02.010
Douglas, A. E. (2015). Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. entomology 60, 17–34. doi:10.1146/annurev-ento-010814-020822
Drake, J. W., Charlesworth, B., Charlesworth, D., and Crow, J. F. (1998). Rates of spontaneous mutation. Genetics 148 (4), 1667–1686. doi:10.1093/genetics/148.4.1667
Drewell, R. A., Bush, E. C., Remnant, E. J., Wong, G. T., Beeler, S. M., Stringham, J. L., et al. (2014). The dynamic DNA methylation cycle from egg to sperm in the honey bee Apis mellifera. Development 141 (13), 2702–2711. doi:10.1242/dev.110163
Enayati, A. A., Ranson, H., and Hemingway, J. (2005). Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14 (1), 3–8. doi:10.1111/j.1365-2583.2004.00529.x
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 37 (5), 699–735. doi:10.1111/1574-6976.12025
Fablet, M., and Vieira, C. (2011). Evolvability, epigenetics and transposable elements. Biomol. Concepts 2, 333–341. doi:10.1515/BMC.2011.035
Fassi Fehri, L., Koch, M., Belogolova, E., Khalil, H., Bolz, C., Kalali, B., et al. (2010). Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PloS one 5 (3), e9500. doi:10.1371/journal.pone.0009500
Feliciello, I., Parazajder, J., Akrap, I., and Ugarković, Đ. (2013). First evidence of DNA methylation in insect Tribolium castaneum: Environmental regulation of DNA methylation within heterochromatin. Epigenetics 8 (5), 534–541. doi:10.4161/epi.24507
Feyereisen, R., Dermauw, W., and Van Leeuwen, T. (2015). Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pesticide Biochem. physiology 121, 61–77. doi:10.1016/j.pestbp.2015.01.004
Ffrench-Constant, R. H. (2014). Insecticide resistance comes of age. Genome Biol. 15, 106. doi:10.1186/gb4162
Ffrench-Constant, R. H. (2013). The molecular genetics of insecticide resistance. Genetics 194 (4), 807–815. doi:10.1534/genetics.112.141895
Ffrench-Constant, R. H. (2007). Which came first: Insecticides or resistance? Trends Genet. 23 (1), 1–4. doi:10.1016/j.tig.2006.11.006
Field, L. M. (2000). Methylation and expression of amplified esterase genes in the aphid Myzus persicae (Sulzer). Biochem. J. 349 (3), 863–868. doi:10.1042/bj3490863
Field, L. M., Devonshire, A. L., Ffrench-Constant, R. H., and Forde, B. G. (1989). Changes in DNA methylation are associated with loss of insecticide resistance in the peach-potato aphid Myzus persicae (Sulz.). FEBS Lett. 243 (2), 323–327. doi:10.1016/0014-5793(89)80154-1
Flores, K., Wolschin, F., Corneveaux, J. J., Allen, A. N., Huentelman, M. J., and Amdam, G. V. (2012). Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC genomics 13 (1), 480–489. doi:10.1186/1471-2164-13-480
Foret, S., Kucharski, R., Pellegrini, M., Feng, S., Jacobsen, S. E., Robinson, G. E., et al. (2012). DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl. Acad. Sci. 109 (13), 4968–4973. doi:10.1073/pnas.1202392109
Franks, S. J., and Hoffmann, A. A. (2012). Genetics of climate change adaptation. Annu. Rev. Genet. 46 (1), 185–208. doi:10.1146/annurev-genet-110711-155511
Franks, S. J., Sim, S., and Weis, A. E. (2007). Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. 104 (4), 1278–1282. doi:10.1073/pnas.0608379104
Fullaondo, A., and Lee, S. Y. (2012). Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev. Comp. Immunol. 36 (2), 267–273. doi:10.1016/j.dci.2011.03.034
Gaddelapati, S. C., Kalsi, M., Roy, A., and Palli, S. R. (2018). Cap'n'collar C regulates genes responsible for imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Biochem. Mol. Biol. 99, 54–62. doi:10.1016/j.ibmb.2018.05.006
Gaddelapati, S. C., Albishi, N. M., Dhandapani, R. K., and Palli, S. R. (2022). Juvenile hormone-induced histone deacetylase 3 suppresses apoptosis to maintain larval midgut in the yellow fever mosquito. Proc. Natl. Acad. Sci. 119 (11), e2118871119. doi:10.1073/pnas.2118871119
Gaddelapati, S. C., Dhandapani, R. K., and Palli, S. R. (2020). CREB-binding protein regulates metamorphosis and compound eye development in the yellow fever mosquito, Aedes aegypti. Biochimica Biophysica Acta (BBA)-Gene Regul. Mech. 1863 (8), 194576. doi:10.1016/j.bbagrm.2020.194576
Gardner, J. L., Peters, A., Kearney, M. R., Joseph, L., and Heinsohn, R. (2011). Declining body size: A third universal response to warming? Trends Ecol. Evol. 26 (6), 285–291. doi:10.1016/j.tree.2011.03.005
George, S., Gaddelapati, S. C., and Palli, S. R. (2019). Histone deacetylase 1 suppresses Krüppel homolog 1 gene expression and influences juvenile hormone action in Tribolium castaneum. Proc. Natl. Acad. Sci. 116 (36), 17759–17764. doi:10.1073/pnas.1909554116
George, S., and Palli, S. R. (2020b). Histone deacetylase 11 knockdown blocks larval development and metamorphosis in the red flour beetle, Tribolium castaneum. Front. Genet. 11, 683. doi:10.3389/fgene.2020.00683
George, S., and Palli, S. R. (2020a). Histone deacetylase 3 is required for development and metamorphosis in the red flour beetle, Tribolium castaneum. BMC genomics 21 (1), 1–14. doi:10.1186/s12864-020-06840-3
Georghiou, G. P., and Taylor, C. E. (1977). Genetic and biological influences in the evolution of insecticide resistance. J. Econ. entomology 70 (3), 319–323. doi:10.1093/jee/70.3.319
Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., et al. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308 (5723), 833–838. doi:10.1126/science.1109020
Glastad, K., Hunt, B. G., Yi, S., and Goodisman, M. (2011). DNA methylation in insects: On the brink of the epigenomic era. Insect Mol. Biol. 20 (5), 553–565. doi:10.1111/j.1365-2583.2011.01092.x
Glastad, K. M., Hunt, B. G., and Goodisman, M. A. (2019). Epigenetics in insects: Genome regulation and the generation of phenotypic diversity. Annu. Rev. Entomol. 64 (185), 185–203. doi:10.1146/annurev-ento-011118-111914
Glastad, K. M., Hunt, B. G., and Goodisman, M. A. (2014). Evolutionary insights into DNA methylation in insects. Curr. Opin. Insect Sci. 1, 25–30. doi:10.1016/j.cois.2014.04.001
Gomes, A. F. F., Omoto, C., and Cônsoli, F. L. (2020). Gut bacteria of field-collected larvae of Spodoptera frugiperda undergo selection and are more diverse and active in metabolizing multiple insecticides than laboratory-selected resistant strains. J. Pest Sci. 93 (2), 833–851. doi:10.1007/s10340-020-01202-0
Gómez-Díaz, E., Jordà, M., Peinado, M. A., and Rivero, A. (2012). Epigenetics of host–pathogen interactions: The road ahead and the road behind. PLoS Pathog. 8 (11), e1003007. doi:10.1371/journal.ppat.1003007
Good, P., Booth, B. B., Chadwick, R., Hawkins, E., Jonko, A., and Lowe, J. A. (2016). Large differences in regional precipitation change between a first and second 2 K of global warming. Nat. Commun. 7 (1), 13667–13668. doi:10.1038/ncomms13667
Greer, E. L., Maures, T. J., Ucar, D., Hauswirth, A. G., Mancini, E., Lim, J. P., et al. (2011). Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479 (7373), 365–371. doi:10.1038/nature10572
Gressel, J. (2011). Low pesticide rates may hasten the evolution of resistance by increasing mutation frequencies. Pest Manag. Sci. 67 (3), 253–257. doi:10.1002/ps.2071
Gupta, A., and Nair, S. (2020). Dynamics of insect–microbiome interaction influence host and microbial symbiont. Front. Microbiol. 11, 1357. doi:10.3389/fmicb.2020.01357
Gupta, A., and Nair, S. (2022). Heritable epigenomic modifications influence stress resilience and rapid adaptations in the Brown planthopper (nilaparvata lugens). Int. J. Mol. Sci. 23 (15), 8728. doi:10.3390/ijms23158728
Haddi, K., Mendes, M. V., Barcellos, M. S., Lino-Neto, J., Freitas, H. L., Guedes, R. N. C., et al. (2016). Sexual success after stress? Imidacloprid-induced hormesis in males of the neotropical stink bug euschistus heros. PloS one 11 (6), e0156616. doi:10.1371/journal.pone.0156616
Hamon, M. A., and Cossart, P. (2008). Histone modifications and chromatin remodeling during bacterial infections. Cell host microbe 4 (2), 100–109. doi:10.1016/j.chom.2008.07.009
Hansen, A. K., and Degnan, P. H. (2014). Widespread expression of conserved small RNAs in small symbiont genomes. ISME J. 8 (12), 2490–2502. doi:10.1038/ismej.2014.121
Hardy, N. B., Peterson, D. A., Ross, L., and Rosenheim, J. A. (2018). Does a plant-eating insect's diet govern the evolution of insecticide resistance? Comparative tests of the pre-adaptation hypothesis. Evol. Appl. 11 (5), 739–747. doi:10.1111/eva.12579
Hawkins, N. J., Bass, C., Dixon, A., and Neve, P. (2019). The evolutionary origins of pesticide resistance. Biol. Rev. 94 (1), 135–155. doi:10.1111/brv.12440
He, Y., Du, Y., Li, J., Liu, P., Wang, Q., and Li, Z. (2015). Analysis of DNA methylation in different tissues of Fenneropenaeus chinensis from the wild population and Huanghai No. 1. Acta Oceanol. Sin. 34 (12), 175–180. doi:10.1007/s13131-015-0765-x
Heard, E., and Martienssen, R. A. (2014). Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 157 (1), 95–109. doi:10.1016/j.cell.2014.02.045
Heckel, D. G. (2014). Insect detoxification and sequestration strategies. Annu. Plant Rev. Insect-Plant Interact. 47, 77–114. doi:10.1002/9781118829783.ch3
Hill, J. K., Griffiths, H. M., and Thomas, C. D. (2011). Climate change and evolutionary adaptations at species' range margins. Annu. Rev. entomology 56, 143–159. doi:10.1146/annurev-ento-120709-144746
Holoch, D., and Moazed, D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16 (2), 71–84. doi:10.1038/nrg3863
Hu, Y.-T., Wu, T.-C., Yang, E.-C., Wu, P.-C., Lin, P.-T., and Wu, Y.-L. (2017). Regulation of genes related to immune signaling and detoxification in Apis mellifera by an inhibitor of histone deacetylation. Sci. Rep. 7 (1), 1–14. doi:10.1038/srep41255
Hunt, B. G., Glastad, K. M., Yi, S. V., and Goodisman, M. A. (2013b). Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol. Evol. 5 (3), 591–598. doi:10.1093/gbe/evt030
Hunt, B. G., Glastad, K. M., Yi, S. V., and Goodisman, M. A. (2013a). The function of intragenic DNA methylation: Insights from insect epigenomes. Integr. Comp. Biol. 53 (2), 319–328. doi:10.1093/icb/ict003
Hunter, R. G., Gagnidze, K., McEwen, B. S., and Pfaff, D. W. (2015). Stress and the dynamic genome: Steroids, epigenetics, and the transposome. Proc. Natl. Acad. Sci. 112 (22), 6828–6833. doi:10.1073/pnas.1411260111
Hussain, M., Frentiu, F. D., Moreira, L. A., O'Neill, S. L., and Asgari, S. (2011). Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc. Natl. Acad. Sci. 108 (22), 9250–9255. doi:10.1073/pnas.1105469108
Jablonka, E., and Raz, G. (2009). Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84 (2), 131–176. doi:10.1086/598822
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (3), 245–254. doi:10.1038/ng1089
Jenkins, A. M., and Muskavitch, M. A. (2015). Evolution of an epigenetic gene ensemble within the genus Anopheles. Genome Biol. Evol. 7 (3), 901–915. doi:10.1093/gbe/evv041
Jeremias, G., Barbosa, J., Marques, S. M., Asselman, J., Gonçalves, F. J., and Pereira, J. L. (2018). Synthesizing the role of epigenetics in the response and adaptation of species to climate change in freshwater ecosystems. Mol. Ecol. 27 (13), 2790–2806. doi:10.1111/mec.14727
Kalsi, M., and Palli, S. R. (2017). Cap n collar transcription factor regulates multiple genes coding for proteins involved in insecticide detoxification in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 90, 43–52. doi:10.1016/j.ibmb.2017.09.009
Karell, P., Ahola, K., Karstinen, T., Valkama, J., and Brommer, J. E. (2011). Climate change drives microevolution in a wild bird. Nat. Commun. 2 (1), 1–7. doi:10.1038/ncomms1213
Kausar, S., Abbas, M. N., and Cui, H. (2021). A review on the DNA methyltransferase family of insects: Aspect and prospects. Int. J. Biol. Macromol. 186, 289–302. doi:10.1016/j.ijbiomac.2021.06.205
Kikuchi, Y., Hayatsu, M., Hosokawa, T., Nagayama, A., Tago, K., and Fukatsu, T. (2012). Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. 109 (22), 8618–8622. doi:10.1073/pnas.1200231109
Kim, D., Thairu, M. W., and Hansen, A. K. (2016). Novel insights into insect-microbe interactions—Role of epigenomics and small RNAs. Front. Plant Sci. 7, 1164. doi:10.3389/fpls.2016.01164
Kinnison, M. T., and Hendry, A. P. (2001). “The pace of modern life II: From rates of contemporary microevolution to pattern and process,” in Microevolution rate, pattern, process (Germany: Springer), 145–164.
Kishimoto, S., Uno, M., Okabe, E., Nono, M., and Nishida, E. (2017). Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Commun. 8 (1), 1–12. doi:10.1038/ncomms14031
Kuzawa, C. W., and Thayer, Z. M. (2011). Timescales of human adaptation: The role of epigenetic processes. Epigenomics 3 (2), 221–234. doi:10.2217/epi.11.11
Laland, K., Uller, T., Feldman, M., Sterelny, K., Müller, G. B., Moczek, A., et al. (2014). Does evolutionary theory need a rethink? Nature 514 (7521), 161–164. doi:10.1038/514161a
Lee, C. E., and Gelembiuk, G. W. (2008). Evolutionary origins of invasive populations. Evol. Appl. 1 (3), 427–448. doi:10.1111/j.1752-4571.2008.00039.x
Lee, D.-H., Jacobs, D. R., and Porta, M. (2009). Hypothesis: A unifying mechanism for nutrition and chemicals as lifelong modulators of DNA hypomethylation. Environ. health Perspect. 117 (12), 1799–1802. doi:10.1289/ehp.0900741
Lehmann, P., Lyytinen, A., Piiroinen, S., and Lindström, L. (2014). Northward range expansion requires synchronization of both overwintering behaviour and physiology with photoperiod in the invasive Colorado potato beetle (Leptinotarsa decemlineata). Oecologia 176 (1), 57–68. doi:10.1007/s00442-014-3009-4
Li-Byarlay, H., Li, Y., Stroud, H., Feng, S., Newman, T. C., Kaneda, M., et al. (2013). RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. 110 (31), 12750–12755. doi:10.1073/pnas.1310735110
Liebers, R., Rassoulzadegan, M., and Lyko, F. (2014). Epigenetic regulation by heritable RNA. PLoS Genet. 10 (4), e1004296. doi:10.1371/journal.pgen.1004296
Lyko, F., and Maleszka, R. (2011). Insects as innovative models for functional studies of DNA methylation. Trends Genet. 27 (4), 127–131. doi:10.1016/j.tig.2011.01.003
Lyko, F., Ramsahoye, B. H., and Jaenisch, R. (2000). DNA methylation in Drosophila melanogaster. Nature 408 (6812), 538–540. doi:10.1038/35046205
Marhold, J., Rothe, N., Pauli, A., Mund, C., Kuehle, K., Brueckner, B., et al. (2004). Conservation of DNA methylation in dipteran insects. Insect Mol. Biol. 13 (2), 117–123. doi:10.1111/j.0962-1075.2004.00466.x
Marks, P. A., Miller, T., and Richon, V. M. (2003). Histone deacetylases. Curr. Opin. Pharmacol. 3 (4), 344–351. doi:10.1016/s1471-4892(03)00084-5
McGuigan, K., Hoffmann, A. A., and Sgrò, C. M. (2021). How is epigenetics predicted to contribute to climate change adaptation? What evidence do we need? Philosophical Trans. R. Soc. B 376 (1826), 20200119. doi:10.1098/rstb.2020.0119
McMahon, S. M., Harrison, S. P., Armbruster, W. S., Bartlein, P. J., Beale, C. M., Edwards, M. E., et al. (2011). Improving assessment and modelling of climate change impacts on global terrestrial biodiversity. Trends Ecol. Evol. 26 (5), 249–259. doi:10.1016/j.tree.2011.02.012
Mukherjee, K., and Dobrindt, U. (2022). The emerging role of epigenetic mechanisms in insect defense against pathogens. Curr. Opin. Insect Sci. 49, 8–14. doi:10.1016/j.cois.2021.10.004
Mukherjee, K., Fischer, R., and Vilcinskas, A. (2012). Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front. zoology 9 (1), 25–12. doi:10.1186/1742-9994-9-25
Mukherjee, K., Twyman, R. M., and Vilcinskas, A. (2015). Insects as models to study the epigenetic basis of disease. Prog. biophysics Mol. Biol. 118 (1-2), 69–78. doi:10.1016/j.pbiomolbio.2015.02.009
Muturi, E. J., Dunlap, C., Smartt, C. T., and Shin, D. (2021). Resistance to permethrin alters the gut microbiota of Aedes aegypti. Sci. Rep. 11 (1), 14406–14408. doi:10.1038/s41598-021-93725-4
Nilsson, E. E., Sadler-Riggleman, I., and Skinner, M. K. (2018). Environmentally induced epigenetic transgenerational inheritance of disease. Environ. Epigenetics 4 (2), dvy016. doi:10.1093/eep/dvy016
Niu, Y., DesMarais, T. L., Tong, Z., Yao, Y., and Costa, M. (2015). Oxidative stress alters global histone modification and DNA methylation. Free Radic. Biol. Med. 82, 22–28. doi:10.1016/j.freeradbiomed.2015.01.028
Olivares-Castro, G., Cáceres-Jensen, L., Guerrero-Bosagna, C., and Villagra, C. (2021). Insect epigenetic mechanisms facing anthropogenic-derived contamination, an overview. Insects 12 (9), 780. doi:10.3390/insects12090780
Oliver, K. M., Russell, J. A., Moran, N. A., and Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. 100 (4), 1803–1807. doi:10.1073/pnas.0335320100
Olson-Manning, C. F., Wagner, M. R., and Mitchell-Olds, T. (2012). Adaptive evolution: Evaluating empirical support for theoretical predictions. Nat. Rev. Genet. 13 (12), 867–877. doi:10.1038/nrg3322
Oppold, A., Kreß, A., Bussche, J. V., Diogo, J., Kuch, U., Oehlmann, J., et al. (2015). Epigenetic alterations and decreasing insecticide sensitivity of the Asian tiger mosquito Aedes albopictus. Ecotoxicol. Environ. Saf. 122, 45–53. doi:10.1016/j.ecoenv.2015.06.036
Oppold, A.-M., and Müller, R. (2017). “Epigenetics: A hidden target of insecticides,” in Advances in insect physiology (Netherlands: Elsevier), 313–324.
Paenke, I., Sendhoff, B., and Kawecki, T. J. (2007). Influence of plasticity and learning on evolution under directional selection. Am. Nat. 170 (2), E47–E58. doi:10.1086/518952
Palli, S. R. (2020). CncC/Maf-mediated xenobiotic response pathway in insects. Archives Insect Biochem. Physiology 104 (2), e21674. doi:10.1002/arch.21674
Palli, S. R. (2021). Epigenetic regulation of post-embryonic development. Curr. Opin. Insect Sci. 43, 63–69. doi:10.1016/j.cois.2020.09.011
Peleg, S., Feller, C., Forne, I., Schiller, E., Sévin, D. C., Schauer, T., et al. (2016). Life span extension by targeting a link between metabolism and histone acetylation in Drosophila. EMBO Rep. 17 (3), 455–469. doi:10.15252/embr.201541132
Pembrey, M. E., Bygren, L. O., Kaati, G., Edvinsson, S., Northstone, K., Sjöström, M., et al. (2006). Sex-specific, male-line transgenerational responses in humans. Eur. J. Hum. Genet. 14 (2), 159–166. doi:10.1038/sj.ejhg.5201538
Peschansky, V. J., and Wahlestedt, C. (2014). Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 9 (1), 3–12. doi:10.4161/epi.27473
Piiroinen, S., Boman, S., Lyytinen, A., Mappes, J., and Lindström, L. (2014). Sublethal effects of deltamethrin exposure of parental generations on physiological traits and overwintering in L eptinotarsa decemlineata. J. Appl. Entomology 138 (1-2), 149–158. doi:10.1111/jen.12088
Piiroinen, S., Ketola, T., Lyytinen, A., and Lindström, L. (2011). Energy use, diapause behaviour and northern range expansion potential in the invasive Colorado potato beetle. Funct. Ecol. 25 (3), 527–536. doi:10.1111/j.1365-2435.2010.01804.x
Piiroinen, S., Lyytinen, A., and Lindström, L. (2013). Stress for invasion success? Temperature stress of preceding generations modifies the response to insecticide stress in an invasive pest insect. Evol. Appl. 6 (2), 313–323. doi:10.1111/eva.12001
Pimentel, D., Acquay, H., Biltonen, M., Rice, P., Silva, M., Nelson, J., et al. (1992). Environmental and economic costs of pesticide use. BioScience 42 (10), 750–760. doi:10.2307/1311994
Popp, J. (2011). Cost-benefit analysis of crop protection measures. J. für Verbraucherschutz und Lebensmittelsicherheit 6 (1), 105–112. doi:10.1007/s00003-011-0677-4
Poulos, R. C., Olivier, J., and Wong, J. W. (2017). The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. Nucleic acids Res. 45 (13), 7786–7795. doi:10.1093/nar/gkx463
Ramya, S. L., Venkatesan, T., Murthy, K. S., Jalali, S. K., and Varghese, A. (2016a). Degradation of acephate by Enterobacter asburiae, Bacillus cereus and Pantoea agglomerans isolated from diamondback moth Plutella xylostella (L), a pest of cruciferous crops. J. Environ. Biol. 37 (4), 611–618.
Ramya, S. L., Venkatesan, T., Srinivasa Murthy, K., Jalali, S. K., and Verghese, A. (2016b). Detection of carboxylesterase and esterase activity in culturable gut bacterial flora isolated from diamondback moth, Plutella xylostella (Linnaeus), from India and its possible role in indoxacarb degradation. Braz. J. Microbiol. 47, 327–336. doi:10.1016/j.bjm.2016.01.012
Rechavi, O., and Lev, I. (2017). Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr. Biol. 27 (14), R720–R730. doi:10.1016/j.cub.2017.05.043
Regev, A., Lamb, M. J., and Jablonka, E. (1998). The role of DNA methylation in invertebrates: Developmental regulation or genome defense? Mol. Biol. Evol. 15 (7), 880–891. doi:10.1093/oxfordjournals.molbev.a025992
Reynolds, J., Bautista-Jimenez, R., and Denlinger, D. (2016). Changes in histone acetylation as potential mediators of pupal diapause in the flesh fly, Sarcophaga bullata. Insect Biochem. Mol. Biol. 76, 29–37. doi:10.1016/j.ibmb.2016.06.012
Richard, G., Jaquiéry, J., and Le Trionnaire, G. (2021). Contribution of epigenetic mechanisms in the regulation of environmentally-induced polyphenism in insects. Insects 12 (7), 649. doi:10.3390/insects12070649
Richard, G., Le Trionnaire, G., Danchin, E., and Sentis, A. (2019). Epigenetics and insect polyphenism: Mechanisms and climate change impacts. Curr. Opin. insect Sci. 35, 138–145. doi:10.1016/j.cois.2019.06.013
Robertson, K. D., Hayward, S. D., Ling, P. D., Samid, D., and Ambinder, R. F. (1995). Transcriptional activation of the epstein-barr virus latency C promoter after 5-azacytidine treatment: Evidence that demethylation at a single CpG site is crucial. Mol. Cell. Biol. 15 (11), 6150–6159. doi:10.1128/mcb.15.11.6150
Roy, A., George, S., and Palli, S. R. (2017). Multiple functions of CREB-binding protein during postembryonic development: Identification of target genes. BMC genomics 18 (1), 1–14. doi:10.1186/s12864-017-4373-3
Roy, A., and Palli, S. R. (2018). Epigenetic modifications acetylation and deacetylation play important roles in juvenile hormone action. BMC genomics 19 (1), 1–15. doi:10.1186/s12864-018-5323-4
Sato, F., Tsuchiya, S., Meltzer, S. J., and Shimizu, K. (2011). MicroRNAs and epigenetics. FEBS J. 278 (10), 1598–1609. doi:10.1111/j.1742-4658.2011.08089.x
Seisenberger, S., Peat, J. R., and Reik, W. (2013). Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr. Opin. Cell Biol. 25 (3), 281–288. doi:10.1016/j.ceb.2013.02.013
Sen, M. K., Hamouzova, K., Košnarová, P., Roy, A., and Soukup, J. (2022). Herbicide resistance in grass weeds: Epigenetic regulation matters too. Front. Plant Sci. 13, 1040958. doi:10.3389/fpls.2022.1040958
Skinner, M. K. (2014a). Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol. Cell. Endocrinol. 398 (1-2), 4–12. doi:10.1016/j.mce.2014.07.019
Skinner, M. K. (2011). Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 6 (7), 838–842. doi:10.4161/epi.6.7.16537
Skinner, M. K. (2015). Environmental epigenetics and a unified theory of the molecular aspects of evolution: A neo-lamarckian concept that facilitates neo-darwinian evolution. Genome Biol. Evol. 7 (5), 1296–1302. doi:10.1093/gbe/evv073
Skinner, M. K. (2014b). Environmental stress and epigenetic transgenerational inheritance. BMC Med. 12 (1), 153–155. doi:10.1186/s12916-014-0153-y
Skinner, M. K., Gurerrero-Bosagna, C., Haque, M. M., Nilsson, E. E., Koop, J. A., Knutie, S. A., et al. (2014). Epigenetics and the evolution of Darwin’s finches. Genome Biol. Evol. 6 (8), 1972–1989. doi:10.1093/gbe/evu158
Song, J., Irwin, J., and Dean, C. (2013). Remembering the prolonged cold of winter. Curr. Biol. 23 (17), R807–R811. doi:10.1016/j.cub.2013.07.027
Sparks, T. C., and Nauen, R. (2015). Irac: Mode of action classification and insecticide resistance management. Pesticide Biochem. physiology 121, 122–128. doi:10.1016/j.pestbp.2014.11.014
Sparks, T. C., Storer, N., Porter, A., Slater, R., and Nauen, R. (2021). Insecticide resistance management and industry: The origins and evolution of the I nsecticide R esistance A ction C ommittee (IRAC) and the mode of action classification scheme. Pest Manag. Sci. 77 (6), 2609–2619. doi:10.1002/ps.6254
Stat, F. (2019). Food and agriculture organization of the united nations. Italy: FAOSTAT Statistical Database.
Stratonovitch, P., Elias, J., Denholm, I., Slater, R., and Semenov, M. A. (2014). An individual-based model of the evolution of pesticide resistance in heterogeneous environments: Control of meligethes aeneus population in oilseed rape crops. PLoS One 9 (12), e115631. doi:10.1371/journal.pone.0115631
Suzuki, M. M., and Bird, A. (2008). DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 9 (6), 465–476. doi:10.1038/nrg2341
Szyf, M. (2015). Nongenetic inheritance and transgenerational epigenetics. Trends Mol. Med. 21 (2), 134–144. doi:10.1016/j.molmed.2014.12.004
Tabashnik, B. E., Mota-Sanchez, D., Whalon, M. E., Hollingworth, R. M., and Carrière, Y. (2014). Defining terms for proactive management of resistance to Bt crops and pesticides. J. Econ. entomology 107 (2), 496–507. doi:10.1603/ec13458
Terrapon, N., Li, C., Robertson, H. M., Ji, L., Meng, X., Booth, W., et al. (2014). Molecular traces of alternative social organization in a termite genome. Nat. Commun. 5 (1), 1–12. doi:10.1038/ncomms4636
Thiebaut, F., Hemerly, A. S., and Ferreira, P. C. G. (2019). A role for epigenetic regulation in the adaptation and stress responses of non-model plants. Front. plant Sci. 10, 246. doi:10.3389/fpls.2019.00246
Thomas, C. D., Bodsworth, E. J., Wilson, R. J., Simmons, A. D., Davies, Z. G., Musche, M., et al. (2001). Ecological and evolutionary processes at expanding range margins. Nature 411 (6837), 577–581. doi:10.1038/35079066
Turner, B. M. (2009). Epigenetic responses to environmental change and their evolutionary implications. Philosophical Trans. R. Soc. B Biol. Sci. 364 (1534), 3403–3418. doi:10.1098/rstb.2009.0125
Vandegehuchte, M. B., De Coninck, D., Vandenbrouck, T., De Coen, W. M., and Janssen, C. R. (2010a). Gene transcription profiles, global DNA methylation and potential transgenerational epigenetic effects related to Zn exposure history in Daphnia magna. Environ. Pollut. 158 (10), 3323–3329. doi:10.1016/j.envpol.2010.07.023
Vandegehuchte, M. B., Kyndt, T., Vanholme, B., Haegeman, A., Gheysen, G., and Janssen, C. R. (2009). Occurrence of DNA methylation in Daphnia magna and influence of multigeneration Cd exposure. Environ. Int. 35 (4), 700–706. doi:10.1016/j.envint.2009.01.002
Vandegehuchte, M. B., Lemière, F., Vanhaecke, L., Berghe, W. V., and Janssen, C. R. (2010b). Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp. Biochem. Physiology Part C Toxicol. Pharmacol. 151 (3), 278–285. doi:10.1016/j.cbpc.2009.11.007
Waddington, C. H. (1953). “Epigenetics and evolution,” in Symposia of the society for experimental biology (Downing St, Cambridge: Company Biologists Ltd Univ Cambridge Dept Zoology), 186–199.
Xiang, H., Li, X., Dai, F., Xu, X., Tan, A., Chen, L., et al. (2013). Comparative methylomics between domesticated and wild silkworms implies possible epigenetic influences on silkworm domestication. BMC genomics 14 (1), 1–11. doi:10.1186/1471-2164-14-646
Xiang, H., Zhu, J., Chen, Q., Dai, F., Li, X., Li, M., et al. (2010). Single base–resolution methylome of the silkworm reveals a sparse epigenomic map. Nat. Biotechnol. 28 (5), 516–520. doi:10.1038/nbt.1626
Xu, J., Roy, A., and Palli, S. R. (2018). CREB-binding protein plays key roles in juvenile hormone action in the red flour beetle, Tribolium Castaneum. Sci. Rep. 8 (1), 1–11. doi:10.1038/s41598-018-19667-6
Xu, L., Qin, J., Fu, W., Wang, S., Wu, Q., Zhou, X., et al. (2022). MAP4K4 controlled transcription factor POUM1 regulates PxABCG1 expression influencing Cry1Ac resistance in Plutella xylostella (L.). Pesticide Biochem. Physiology 182, 105053. doi:10.1016/j.pestbp.2022.105053
Yan, H., Bonasio, R., Simola, D. F., Liebig, J., Berger, S. L., and Reinberg, D. (2015). DNA methylation in social insects: How epigenetics can control behavior and longevity. Annu. Rev. Entomology 60, 435–452. doi:10.1146/annurev-ento-010814-020803
Yang, M., Zhang, J., Zhu, K., Xuan, T., Liu, X., Guo, Y., et al. (2009). Mechanisms of organophosphate resistance in a field population of oriental migratory locust, Locusta migratoria manilensis (Meyen). Archives Insect Biochem. Physiology Publ. Collab. Entomological Soc. Am. 71 (1), 3–15. doi:10.1002/arch.20254
Ye, Y. H., Woolfit, M., Huttley, G. A., Rances, E., Caragata, E. P., Popovici, J., et al. (2013). Infection with a virulent strain of Wolbachia disrupts genome wide-patterns of cytosine methylation in the mosquito Aedes aegypti. PLoS One 8 (6), e66482. doi:10.1371/journal.pone.0066482
Zhang, G., Hussain, M., O’Neill, S. L., and Asgari, S. (2013). Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc. Natl. Acad. Sci. 110 (25), 10276–10281. doi:10.1073/pnas.1303603110
Zhang, J., Xing, Y., Li, Y., Yin, C., Ge, C., and Li, F. (2015a). DNA methyltransferases have an essential role in female fecundity in Brown planthopper, Nilaparvata lugens. Biochem. Biophysical Res. Commun. 464 (1), 83–88. doi:10.1016/j.bbrc.2015.05.114
Zhang, M., Chen, J. L., Liang, S. K., Li, G. H., Wang, F. H., and Ahmad, I. (2015b). Differentially methylated genomic fragments related with sexual dimorphism of rice pests, Sogatella furcifera. Insect Sci. 22 (6), 731–738. doi:10.1111/1744-7917.12179
Keywords: arthropod pests, insecticide resistance, climate change, epigenetic regulations, DNA methylation, histone modifications, symbiotic microbes
Citation: Mogilicherla K and Roy A (2023) Epigenetic regulations as drivers of insecticide resistance and resilience to climate change in arthropod pests. Front. Genet. 13:1044980. doi: 10.3389/fgene.2022.1044980
Received: 15 September 2022; Accepted: 19 December 2022;
Published: 06 January 2023.
Edited by:
Wei Guo, Institute of Zoology (CAS), ChinaReviewed by:
Dong Wei, Southwest University, ChinaDivya Devara, P. V. Narsimha Rao Telangana Veterinary University, India
Jayendra Nath Shukla, Central University of Rajasthan, India
Copyright © 2023 Mogilicherla and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Roy, cm95QGZsZC5jenUuY3o=
 Kanakachari Mogilicherla
Kanakachari Mogilicherla Amit Roy
Amit Roy