- 1College of Life Science, Agriculture and Forestry, Qiqihar University, Qiqihar, China
- 2Heilongjiang Provincial Key Laboratory Resistance Gene Engineering, Qiqihar, China
CIPKs are a subclass of serine/threonine (Ser/Thr) protein kinases. CBLs are ubiquitous Ca2+ sensors that interact with CIPK with the aid of secondary Ca2+ messengers for regulation of growth and development and response to stresses faced by plants. The divergent roles of the CIPK-CBL interaction in plants include responding to environmental stresses (salt, cold, drought, pH, ABA signaling, and ion toxicity), ion homeostasis (K+, NH4+, NO3−, and microelement homeostasis), biotic stress, and plant development. Each member of this gene family produces distinct proteins that help plants adapt to diverse stresses or stimuli by interacting with calcium ion signals. CIPK consists of two structural domains—an N-terminal domain and a C-terminal domain—connected by a junction domain. The N-terminal domain, the site of phosphorylation, is also called the activation domain and kinase domain. The C-terminal, also known as the regulatory domain of CIPK, further comprises NAF/FISL and PPI. CBL comprises four EF domains and conserved PFPF motifs and is the site of binding with the NAF/FISL domain of CIPK to form a CBL-CIPK complex. In addition, we also performed a bibliometric analysis of the CIPK gene family of data extracted from the WoSCC. A total of 95 documents were retrieved, which had been published by 47 sources. The production over time was zigzagged. The top key terms were gene, CIPK, abiotic stress, and gene expression. Beijing Forestry University was the top affiliation, while The Plant Cell was the top source. The genomics and metabolomics of this gene family require more study.
1 The CIPK gene family
CIPKs are a subclass of serine/threonine (Ser/Thr) protein kinases. CBL (calcineurin B-like proteins) are ubiquitous Ca2+ sensors that interact with CIPK (CBL-interacting protein kinases) with the aid of secondary messengers Ca2+ to regulate growth and development, and also respond to various stresses faced by plants (Sanyal et al., 2016). Ca2+ plays a regulatory role in the formation of the CBL-CIPK complex and the enhancement of kinase domain activity (Sánchez-Barrena et al., 2007). The various roles of CIPK-CBL interaction in plants include responding to environmental stress (salt stress, cold stress, drought stress, pH stress, ABA signaling, ion toxicity), ion homeostasis (K+ homeostasis, NH4+ homeostasis, NO3− homeostasis, microelement homeostasis), biotic stress, and plant development (Ding et al., 2022). Each member of this gene family produces distinct proteins that help plants to adapt to diverse stresses or stimuli by interacting with calcium ion signals.
1.1 Structure of the CIPK-CBL complex
CIPK consists of two structural domains: an N-terminal domain and a C-terminal domain, which are connected by a junction domain. The N-terminal domain, the site of phosphorylation, is also called the activation domain and kinase domain (Guo et al., 2001). This domain comprises three conserved amino acids (Ser, Thr, and Tyr) that are crucial for the proper CIPK functioning and activity. CIPK activity is inhibited if any one of the three amino acids is mutated to Asn. The C-terminal is also the regulatory domain of CIPK and further comprises NAF/FISL and PPI (Sanyal et al., 2015). CBL includes four EF domains and conserved PFPF motifs and is the site of binding with the NAF/FISL domain of CIPK to form the CBL-CIPK complex. When the intracellular level of Ca2+ is less, the NAF domain is not bound to the PFPF domain due to the self-inhibitory property of FISL the domain, thus blocking the kinase activity (Batistic et al., 2008). When the Ca2+ level increases, the PFPF motifs of CBL bind to the NAF domain of CIPK due to the conformational change in protein structure and hydrophobic interactions, causing a release of kinase activity and downstream regulation for various stress responses (Figure 1) (Sánchez-Barrena et al., 2007).
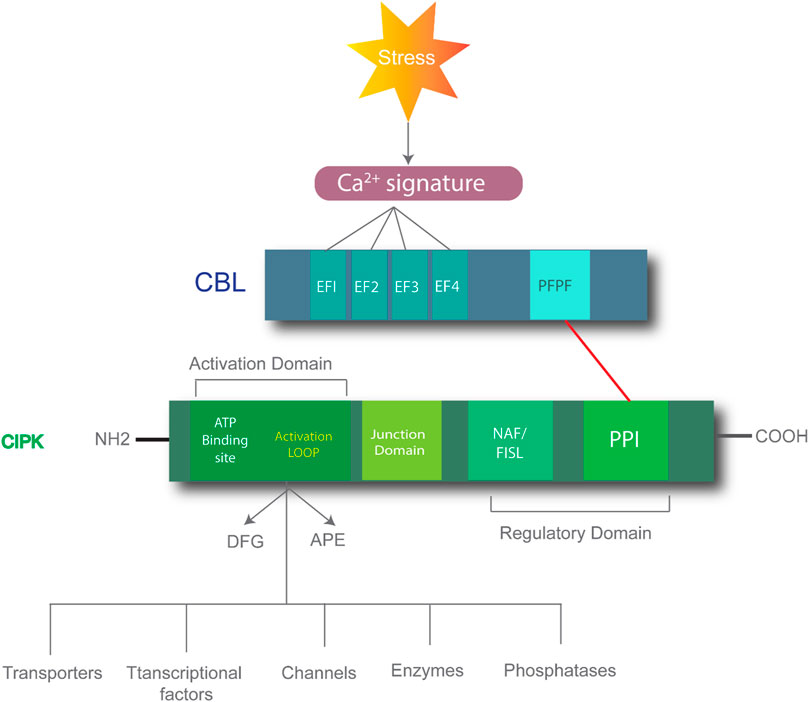
FIGURE 1. Process of CBL-CIPK complex-mediated responses. Stress can induce the calcium ion signatures which bind to EF1-EF4 domains on the CBL. CIPK consists of the N-terminal domain comprising the activation domain and the C-terminal domain comprising the regulatory domain, which includes the NAF/FISL and PPI domains. The red line shows the CBL and CIPK interaction. This complex formation regulates downstream processes.
1.2 How many genes?
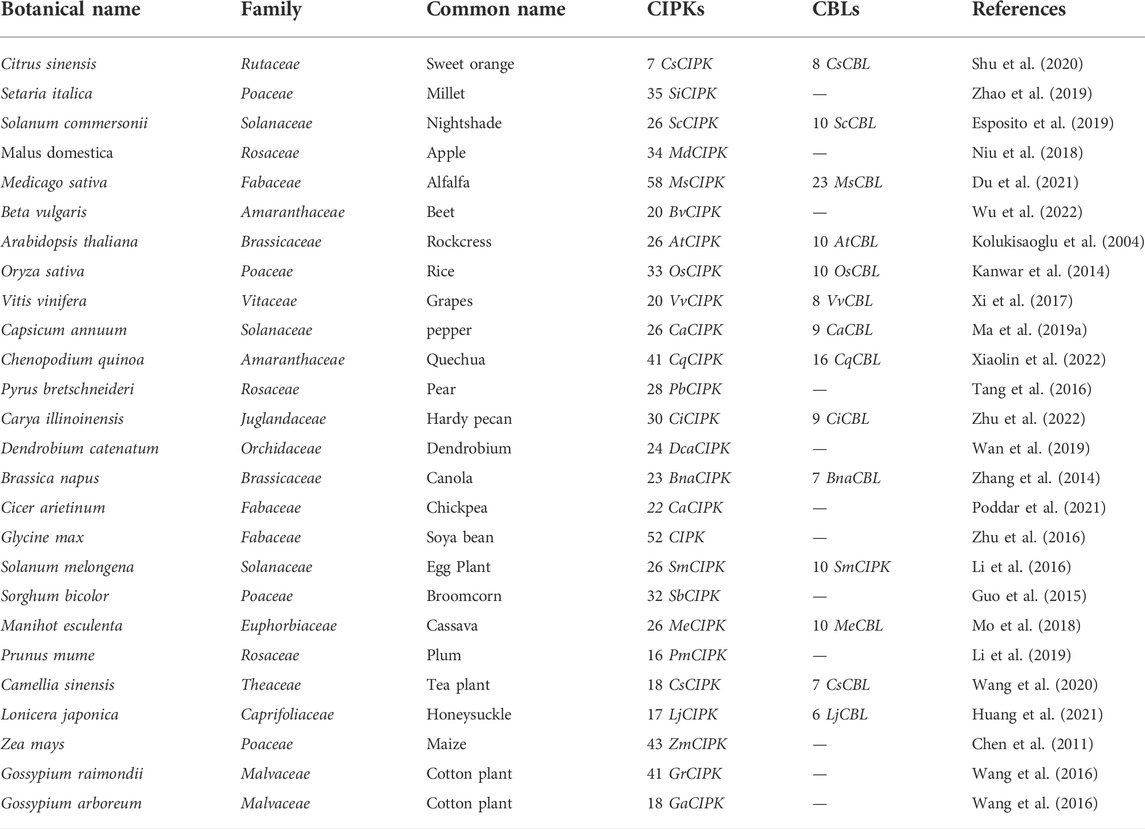
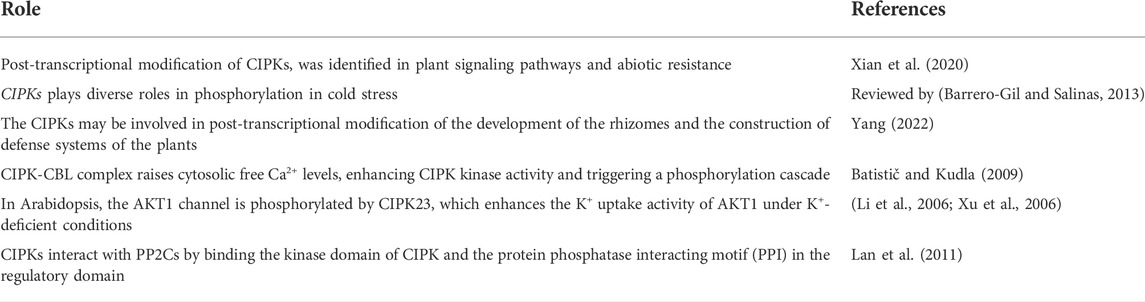
Each plant has a different number of CIPK genes that have been discovered so far. For instance, the total number of CIPK genes in Arabidopsis, Populus, and Cassava are presented in Supplementary Table S1. There are several CBL and CIPK genes found in various plants, some examples are mentioned in Tables 1, 2. The CIPK gene family is further divided into two groups: Those with more introns and those with fewer introns. Additionally, fluorescent labeling reveals that the majority of CIPK proteins are found in the cytoplasm and nucleus. In the cytoplasm and nucleus, the GFP fusion proteins of several CIPKs, including CIPK1, CIPK2, CIPK3, CIPK4, CIPK7, CIPK8, CIPK10, CIPK14, CIPK17, CIPK21, CIPK23, and CIPK24, exhibit significant fluorescence (Mao et al., 2016b). However, the subcellular localization and targeting mechanisms of most CBL-CIPK complexes are yet mysterious.
1.3 Brief introduction to some members of the CIPK gene family
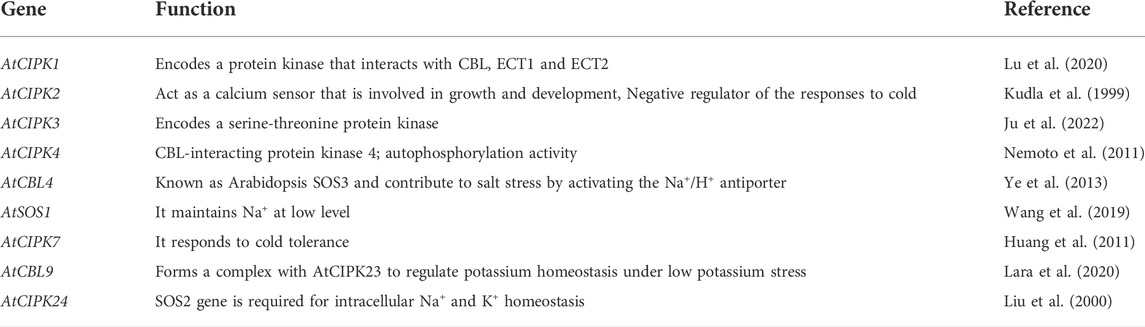
A total of 26 CIPK genes have been identified in Arabidopsis (Kolukisaoglu). Several of these genes and their roles are described in Table 3.
Individual CIPK genes from other plant species have also been reported and characterized in terms of function. For instance, OsCIPK23 overexpression improved rice drought tolerance by increasing the expression levels of drought-related genes (Yang et al., 2008). Drought activated the cotton CIPK gene GhCIPK6, the overexpression of which in Arabidopsis improved drought tolerance (He et al., 2013). These findings imply that the CIPK family plays a significant role in the development of plant stress tolerance.
1.4 Roles of CIPK in transcriptional regulation in plants
The divergent roles of the CIPK-CBL interaction in plants include response to environmental stress (salt stress, cold stress, drought stress, pH stress, ABA signaling, ion toxicity), ion homeostasis (K+, NH4+, NO3−, and microelement homeostasis), biotic stress, and plant development (Ding et al., 2022). We discuss these roles in the following sections.
1.4.1 Environmental stress: salt stress
Elevated salt levels in plants can cause salt toxicity, resulting in halted growth and development. Similarly, extremely low cellular K+ levels can affect plant development. Plants have developed protection mechanisms against salt stress. For instance, to lessen increasing cytoplasmic levels of Na+, plants overcome the threat of salt damage by increasing Na+ discharge, decreasing Na+ intake, and sequestering Na+ into the vacuoles. The first identified CBL-CIPK complex to maintain ion homeostasis is SOS (salt overly sensitive) pathway. High Na+ concentration activates the formation of CBL4/SOS3 and CIPK24/SOS2 complex due to increased concentration of Ca2+ (Yin et al., 2020). This complex activated SOS1, is a plasma membrane anti-porter for Na+/H+ ions. SOS1 helps plants maintain the ion balance by transporting Na+ from the cytoplasm to the extracellular space and controls Na+ transport from root to shoot. CBL10-CIPK24/SOS2 maintains Na+ levels by vacuole sequestration (Shi et al., 2000; Yin et al., 2020). Similarly, CBL1/9-CIPK23 plays a role in maintaining the K+ balance in response to low cellular levels. Low levels of intracellular K+ activate Ca2+, resulting in the formation of the CBL1/9-CIPK23 complex, in which AKTI is located in the plasma membrane, as shown in Figure 2 (Cheong et al., 2007). The CIPK23 kinase domains activate the AKTI channel in the plasma membrane by physically interacting and phosphorylating the channel to initiate K+ uptake into the cytoplasm (Li et al., 2006).
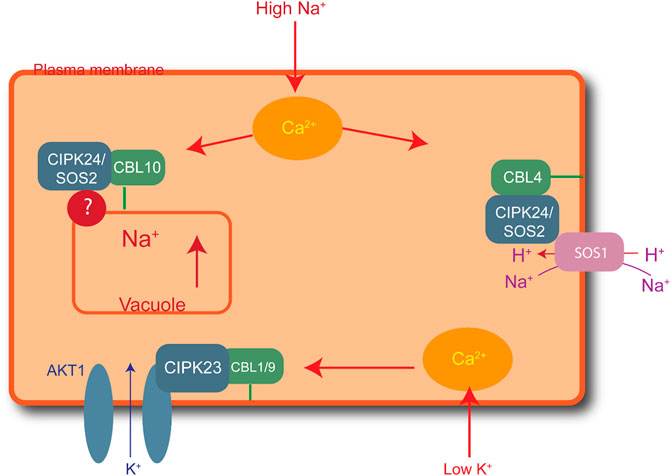
FIGURE 2. Role of CIPK-CBL complexes in managing salt stress in Arabidopsis. CBL4-CIPK24 (SOS2) activates SOS1 for the extracellular transfer of excess sodium ions. Similarly, CBL10-CIPK24 (SOS2) transfers extra sodium ions inside the vacuole, and CBL1/9-CIPK23 plays a role in maintaining a balance of potassium ions by activating AKT1 receptors in the plasma membrane to import potassium ions in low-potassium conditions.
1.4.2 Environmental stress: cold stress
Cold stress can have severe effects on overall plant survival. Cold stress can inhibit plant growth as it reduces membrane fluidity and affects water and nutrient absorption; thus disturbing cellular metabolism and reactions (Chinnusamy et al., 2007). Plants have developed mechanisms to respond to decreasing temperature conditions through various pathways, including the CIPK7-CBL1 complex-mediated approach (Shinozaki et al., 2003). CIPK7-CBL1 may be involved in the response to cold stress by producing sugars, as sugars reduce the risk of cell damage by cold. AtCIPK7 may lead to the phosphorylation of sucrose synthase for sucrose metabolism, as shown in Figure 3 (Huang et al., 2011).
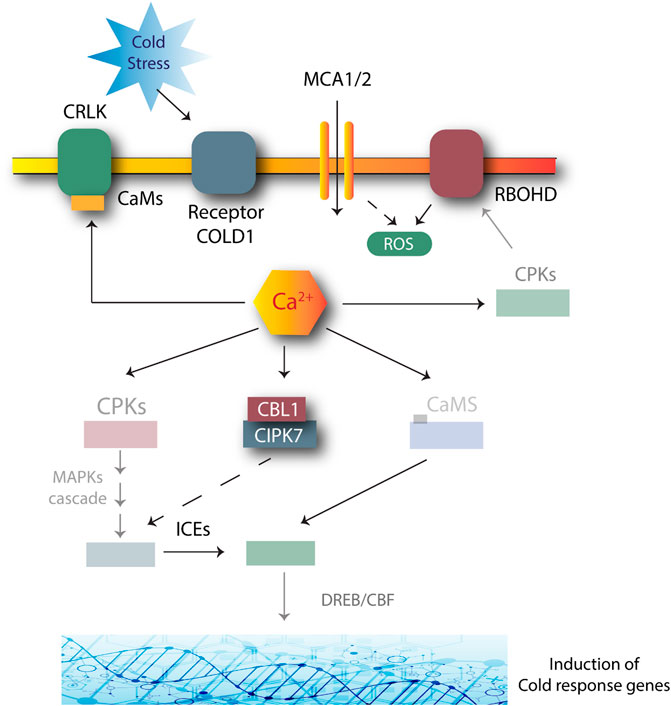
FIGURE 3. Role of CIPK-CBL complexes in managing cold stress in Arabidopsis. Many genes are involved in this response. Cold stress is first perceived by COLD1 receptors. The signals activate the secondary Ca2+ messengers. The transient changes produced in a cell due to Ca2+ activation and AtSRC2 produce ROS due to the NADPH oxidase activity of the AtRBOHF. Increased ROS production positively regulates the Ca2+ channels. CBL1-CIPK plays a role by phosphorylating and activating the MAPK cascades. CRLK1 or CRLK2 interacts with calmodulin (CaM) to further activate the MAPK cascade. The MAPK pathway suppresses the breakdown of ICE1 to tolerate stress. Dashed lines: unclear mechanisms; solid lines: induction.
1.4.3 Environmental stress: drought stress
Drought stress can seriously threaten plant growth through reduced efficiency water use, smaller leaf size, stem elongation, etc. CBL1/9-CIPK23 responds to drought stress in plants by altering the sensitivity of ABA in guard cells. CIPK11 negatively regulates the stress response in coordination with transcriptional factor Di19-3 (Ma X. et al., 2019; Ma Y. et al., 2019).
1.4.4 Environmental stress: ABA signaling
ABA is a vital phytohormone in plants that plays diverse roles, including stomata opening and closing, pathogen defense, seed germination, and plant growth and development. Studies have revealed the role of the CIPK-CBL complex in the regulation of the signal transduction pathway. CIPK3/CBL9 and CIPK15/CBL1 play vital roles in ABA signaling. Experiments have revealed a negative regulatory role of AtCBL9 in ABA signaling during seed germination. A study in plants lacking CBL9 with mutant cbl9 showed the accumulation of higher levels of abscisic acid and hypersensitivity to abscisic acid (Pandey et al., 2004; Pandey et al., 2008). Similarly, to understand the involvement of CIPK15/CBL1 in ABA signaling, plants with mutant CBL1 (cbl1) have been studied. Study results showed that CBL1 is not involved in the regulation of ABA signaling while the interaction of CIPK1 with CBL1 and CBL9 plays a role in the mediation of abscisic acid signaling (D'Angelo et al., 2006). Similarly, CIPK6 loss-of-function lines (cipk6) also showed a higher accumulation of abscisic acid in seedlings, thus demonstrating the role of CIPK6 in ABA signaling (Chen et al., 2013).
1.4.5 Environmental stress: ion toxicity
Cadmium ions can accumulate in plants and are highly toxic to plant growth and development (Cuypers et al., 2010). AtCPK11 plays a role in overcoming cadmium stress by enhancing expression of the ABA signaling pathway (Gu et al., 2021). IRT1 is cadmium transported in roots and the ABA pathway reduces the two crucial transcriptional factors, FIT and bHL039, which regulate IRT1 to block the entry of cadmium and other ions (Fan et al., 2014). Similarly, CIPK23 causes IRT1 degradation to stop the entry of toxic ions into cells (Dubeaux et al., 2018).
1.4.6 Environmental stress: pH stress
CIPK11/CBL2 plays a key role in pH stress management in plants. CIPK11 targets the H+-ATPases AHA2, which is involved in extracellular acidification. CIPK11 (PKS5) negatively regulates H+-ATPase AHA2, as shown in Figure 4 (Fuglsang et al., 2007). The AtCIPK11-AtCBL2 complex phosphorylates H+-ATPase AHA2, preventing it from interacting with the 14-3-3 protein in alkaline conditions (Sanyal et al., 2020).
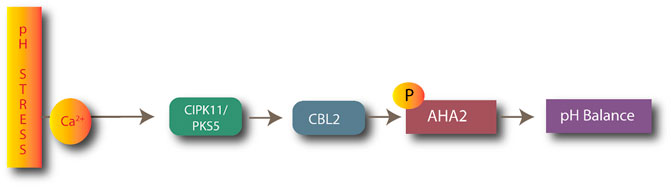
FIGURE 4. Process of maintaining pH balance by CBL2-CIPK11 complexes after activation of AHA ATPases.
1.4.7 Ion homeostasis
Ion homeostasis is a dynamic process for maintaining the physiological processes of plants. The CIPK-CBL complexes play crucial roles in the homeostasis of NO3−, NH4+, K+, and microelements (Mulet et al., 2020). The CBL-CIPK complex allows plant cells to sequester extra Mg2+ into vacuoles, thereby protecting plant cells from high Mg toxicity (Tang et al., 2015). CBL4-CIPK5 complex confers salt, but not drought and chilling, tolerance by regulating ion homeostasis (Huang et al., 2020).
1.4.8 NO3– homeostasis
Nitrate is the chief source of nitrogen for plants and acts as a signal molecule by inducing the expression of nitrate-associated genes, a process called the primary nitrate response. Nitrate induction includes two responses; namely, the high-affinity (km = 30 µM) and low-affinity (km = 0.9 µM) (Liu et al., 2020) stages. The genes of the CIPK family, including CIPK8 and CIPK23, play a crucial role in nitrate homeostasis by regulating the low and high-affinity stages of nitrate induction, respectively. CIPK8 positively regulates the low-affinity response by inducing the expression of the CHL1 (NRT1.1) and NRT2.1 transporters. CIPK23 is a negative regulator of high-affinity response and acts in antagonism with CIPK8 (Zhao et al., 2018). The CBL1/0-CIPK23 complex is located in the plasma membrane, where CIPK23 can convert low-affinity CHL1 to high-affinity by phosphorylation of threonine residues in low-nitrate conditions. Thus, at low nitrate concentrations, higher primary nitrate reactions are prevented. In contrast, high nitrate concentrations inhibit CHL1 phosphorylation by binding to low-affinity sites. This produces high primary nitrate reactions and, ultimately, transports nitrates (Ho et al., 2009).
1.4.9 NH4+ homeostasis
High levels of NH4+ are toxic for plant growth. The CBL1-CIPK23 complex helps maintain ammonium balance by phosphorylating and inactivating the threonine residue of cytosolic AMT1, thus preventing the entry of further ammonium ions (Figure 5) (Straub et al., 2017).
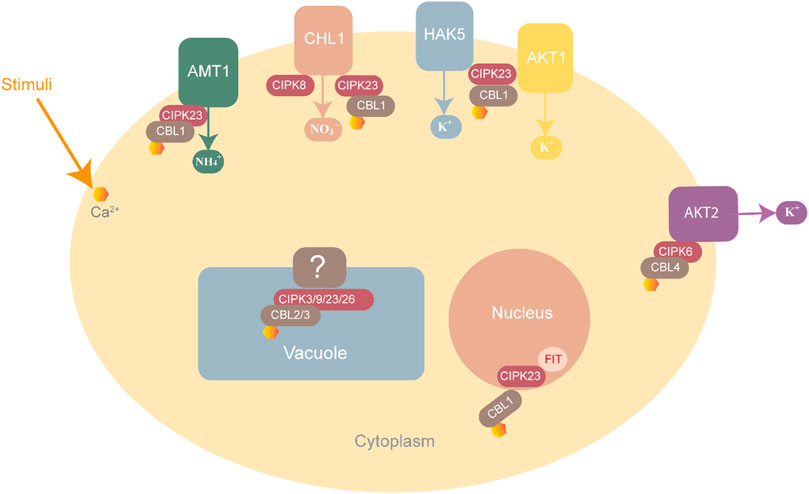
FIGURE 5. CBL-CIPK complexes involved in ion homeostasis. NH4+ homeostasis is maintained through AMT1 inactivation by the CBL-CIPK23 complex to block entry into the cell. K+ homeostasis is maintained by three complexes, CBL1-CIPK23, which interact with HAK5 and AKT1 while CBL4-CIPK6 interacts with AKT2 for K+ uptake into the cell. NO3 homeostasis is maintained by CIPK8 and CIPK23, which interact with CHL1 as positive and negative regulators respectively.
1.4.10 K+ homeostasis
Plants usually suffer at low potassium concentrations. The CBL1/9-CIPK23-AIP1-AKT1 pathway plays a role in maintaining a balance of this ion in plants. CBL1/9 recruits CIPK23 to form an activating complex at the root cells, where CIPK23 activates the AKT1 transporter to increase potassium ion intake by phosphorylating AKT1 (Srivastava et al., 2020). Other than AKT1, CIPK23 can also regulate the potassium ion concentration by interacting with HAK5 (K+ transporter) and KUP4 (K+ regulator) (Srivastava et al., 2020). Many essential microelements in homeostasis are also maintained by CBL-CIPK families. For instance, CBL1/9-CIPK11 and CBL1/9-CIPK23, CIPK7, and CIPK21 play roles in iron homeostasis, as shown in Figure 6 (Ding et al., 2022).
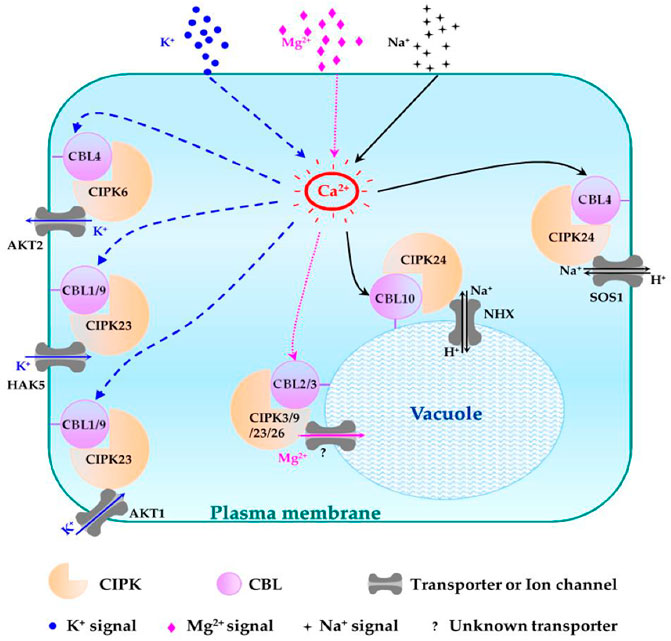
FIGURE 6. Schematic representation of different CBL-CIPK complexes and their functions in regulating Na+, K+, and Mg2+ homeostasis. The regulatory pathways of K+, Mg2+, and Na+ are indicated in blue, pink, and black lines, respectively. Question mark (?): unknown tonoplast-localized transporter (Mao et al., 2016a).
1.4.11 Biotic stress
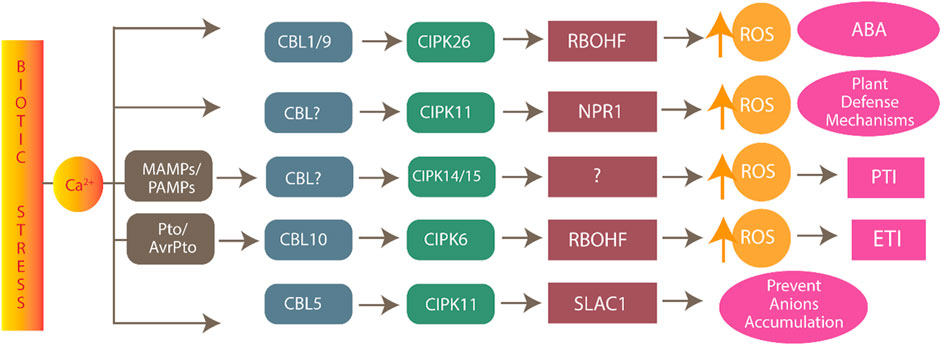
CIPK plays a vital role in protecting plants from biotic stress such as pathogens by directly or indirectly increasing reactive oxygen species (ROS) production (Torres, 2010). ROS are produced by plants through variable defense mechanisms (e.g., PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI)) as a sign that the plant has recognized the pathogen and started to protect itself from biotic stress (Torres, 2010). For instance, CIPK26 protects plants from pathogens via the production of ROS. Similarly, respiratory burst oxidase homolog (RBOH) is also involved in ROS production in plants. The CBL1/9-CIPK26 complex activates RBOH, which further triggers ROS production via the ABA pathway and leads to stomata closure (Fuglsang et al., 2007). Similarly, CBL5-CIPK11 activates other plant defense mechanisms by activating NPR1 (nonexpressor of pathogenesis-related genes), which acts downstream of salicylic acid, which is well known for defense in plants and activates defense genes. The CBL5-CIPK11 complex also reportedly regulates WRKY38 and WRKY62 in disease response (Figure 7) (Tena et al., 2011).
1.4.12 Plant development
CIPK6/12/13/19 plays a vital role in plant development and growth. CIPK12 and CIPK19 are key regulators in the growth of pollen tube tips in plants (Zhou et al., 2015). CIPK13 plays a part in reversible protein phosphorylation, which is essential for maintaining many plant processes such as metabolism, photosynthesis, gene expression, cell development, and structure formation (Schliebner et al., 2008; Schönberg and Baginsky, 2012). CIPK6 is involved in cell division, enhancement of auxin sensitivity, and root growth and development (Tripathi et al., 2009). Finally, CIPK25 aids in the regulation of root meristem development and size (Tripathi et al., 2009).
1.5 CBL-CIPK complex mediates different signaling pathways in plants
As explained above, the CBL-CIPK complex is involved in responses to high salinity, osmotic or drought stress, cold, pH, ABA, K+, nitrate, and other stresses. Crosstalk occurs between the CBL-CIPK complex and other classical pathways such as the CDPK, AMP-activated protein kinase (AMPK), salt overly sensitive (SOS), and reactive oxygen species (ROS) pathways. AtCBL4/SOS3 was the first CBL protein identified in Arabidopsis and was shown to function specifically in the SOS pathway, interacting with AtCIPK24/SOS2 under salinity stress in roots (Liu and Zhu, 1998; Qiu et al., 2002). During germination and seedling development, AtCBL1 reacts to signals involving glucose and gibberellin (GA) (Li et al., 2013). AtCBL1 and AtCBL9 regulate the Arabidopsis NADPH oxidase RbohF after interacting with AtCIPK26 in the ROS pathway (Kimura et al., 2013).
2 Bibliometric analysis
Bibliometrics is the application of statistical techniques to evaluate the scientific content of books, papers, and other publications (Ellegaard and Wallin, 2015). In library and information science, bibliometric techniques are preferably employed. Bibliometrics examines and analyzes scientific metrics and different indicators highly associated with the respective domains (Ellegaard and Wallin, 2015). The earliest bibliometrics studies date back to the late 19th century and changed significantly following the Second World War in the wake of the “periodical crises” and the new technological possibilities provided by computing tools (Wolfram, 2003). The Science Citation Index resulting from Eugene Garfield and Derek John de Solla Price’s citation network analysis became the cornerstone of organized bibliometrics research programs in the early 1960s (Wolfram, 2003). Creating the citation graph, which displays a network of citations between texts, is the foundation of the widely used bibliometric technique known as citation analysis (Hutchins et al., 2019). To examine the influence of their area bibliometric methodologies have been applied to specific publications to identify especially influential papers in certain fields of study (Weingart, 2005). The creation of thesauri, evaluating reader usage, and descriptive linguistics are approaches frequently incorporated in bibliometrics.
2.1 Data sources for bibliometric analysis
Currently, bibliometric analysis can be performed using several databases. However, the development of Scopus and the Web of Science has simplified citation analysis. With certain restrictions, data sources including Dimensions, Google Scholar, and PubMed are also used for data extraction (AlRyalat et al., 2019; Donthu et al., 2021). In 2010, new initiatives in support of open citation data challenged the background of infrastructures for citation data. For example, Web of Science and Scopus have dramatically eased meta-data analysis (Ellegaard, 2018). The Web of Science database provides users access to billions of cited sources in the humanities, social sciences, and life sciences dating back to 1900. This database can be accessed through the website: https://www.webofscience.com. Millions of entries from journals, books, and conference proceedings are included in the Scopus extended abstract and citation database. Scopus can be accessed via the website: https://www.scopus.com/home.uri.
2.1.1 Web of Science
The Web of Science (WoS) is a subscription platform available to institutions and offers access to numerous databases containing reference and citation data from conference proceedings, academic journals, and other publications in a variety of disciplines (AlRyalat et al., 2019). WoS was created by the “Institute for Scientific Information” but is currently held by Clarivate, formerly Thomson Reuters’ Intellectual Property and Science division (Wikipedia, 2022).
2.1.2 Scopus
Elsevier is among the top abstract and citation databases. Scopus was first introduced in 2004 and comprises almost 36,377 titles from approximately 11,678 publishers worldwide (Scopus, 2022). Among these titles, 34,346 are peer-reviewed journals in top-tier subject areas (biological sciences, physical sciences, social sciences, and health sciences). It encompasses certain different sources, for instance, trade journals, book series, and journals (Khudhair et al., 2020). Each year, different parameters such as the h-Index, CiteScore, SCImago Journal Rank, and SNIP numerical quality measures are used to evaluate each journal included in the Scopus database to ensure that it is of a high enough standard (Source Normalized Impact per Paper) (Rankings, 2022). Patent database searches are also incorporated into Scopus searches (Kulkarni et al., 2009).
2.2 Introduction to bibliometric analysis tools
Although several bibliometric databases are available, each was created with a specific objective (Mallig, 2010; Choudhri et al., 2015). Among bibliometric analysis tools, Bibliometrix has the most comprehensive collection of methods and is appropriate for practitioners because of Biblioshiny (Aria and Cuccurullo, 2017). VOSviewer can import and export data from a variety of sources and offers excellent visualization (Van Eck and Waltman, 2017). SciMAT provides the most robust pre-processing and export capabilities (Cobo et al., 2012) and has been used in biomedical sciences (Granada et al., 2020), applied intelligence, and machine learning (Rincon-Patino et al., 2018; López-Robles et al., 2021). Due to the variety of characteristics, researchers must decide on the desired analytical results and select the choice that best serves their objectives.
2.2.1 VOSviewer
VOSviewer (visualization of similarities) is a program for creating and visualizing bibliometric networks. Different types of networks can be developed using VOSviewer. For instance, bibliographic coupling and co-authorship relationships can be shown as attractive networks (Van Eck and Waltman, 2010). VOSviewer allows the detailed analysis of bibliometric maps from various aspects and provides the viewer with the ability to zoom and scroll through the maps to perform detailed examination. To prevent the overlapping of labels, smart labeling algorithms are used. For viewing a moderately large number of items in bibliometric maps, the viewing capability of VOSviewer is remarkable compared to other software (Van Eck and Waltman, 2010). Similarly, VOSviewer can also be used to produce citation and co-citation networks. These networks can be journals, researchers, or individual articles (Van Eck and Waltman, 2010). Data retrieved from the Web of Science, Scopus, Dimensions, Lens, and PubMed databases can be used to create co-authorship, citation-based, and co-occurrence networks (VOSviewer, 2022). Similarly, data downloaded from the APIs of Crossref, Europe PMC, and OpenAlex can also be used to create networks. This program can save screenshots of data as bitmaps and vectors with high resolution (VOSviewer, 2022).
2.2.2 Bibliometrix
All of the primary bibliometric methods of analysis are included in the open-source, scientometric, and bibliometric research tool known as Bibliometrix developed by Aria and Cuccurullo (2017). No coding experience is required to use this software to analyze fields like journals, topics, authors, timespan, etc. (Bibliometrix, 2022). The Bibliometrix package includes a variety of features to extract bibliographic data from databases. The best examples of these databases include SCOPUS, WoS, PubMed, Clarivate Analytics, Digital Science, and Dimensions. Bibliometrics can create data matrices for coupling, scientific collaboration analysis, co-citation, and co-word analysis. It can also be used for structural analysis of new information in data and developments.
2.2.3 Gephi
Gephi is another free program for bibliometric data visualization as massive network graphs (Bastian et al., 2009). To speed exploration, Gephi utilizes a 3-D platform to display graphs in real-time (Gephi, 2021).
3 Bibliometric analyses of the CIPK gene family
A bibliometric analysis of the CIPK gene family was performed. Data were extracted from the Web of Science Core Collection on August 26, 2022. We selected three editions of the core collection; namely, the “Science Citation Index Expanded (SCI-EXPANDED)-1999-present”, “Conference Proceedings Citation Index (AHCI)—2003-present”, and “Emerging Sources Citation Index (ESCI0—2017-present”. The keyword strings CIPK OR CIPK GENE OR CIPK GENE FAMILY were used. Publications were collected from 2003 to 2022. A total of 95 articles were retrieved from the database.
3.1 Information
The main information is provided in Table 4. The time duration was 2003 to 2022. A total of 95 documents published by 47 sources were retrieved.
3.2 Publication analysis of the CIPK gene family
The raw data were analyzed to extract the publications per annum. The data were first published in 2003, with a single article, entitled Isolation and characterization of a novel rice Ca2+-regulated protein kinase gene involved in responses to diverse signals including cold, light, cytokinins, sugars and salts. The production over time zigzagged and no papers were published in 2005. In 2015, the number of publications reached double figures, with a total of 10 articles published. However, the number decreased to five articles in 2016. Since then, a continuous increase was observed (Figure 8).
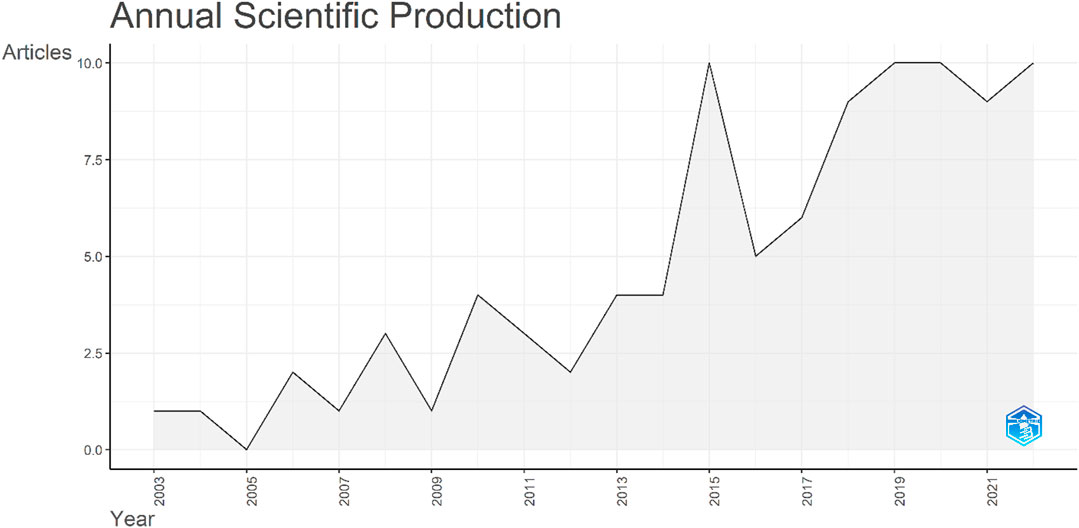
FIGURE 8. Total number of publications on the CIPK gene family over the study period. The x-axis indicates the years while the y-axis indicates the number of publications.
3.3 Relevancy network
The relevancy network was mapped by targeting 1) authors’ affiliations, 2) title terms, and 3) keywords. The top title terms were gene, cipk, family, and identification. The leading keywords were cipk, abiotic stress, gene expression, and salt stress. The top affiliation was Beijing Forestry University (Figure 9).
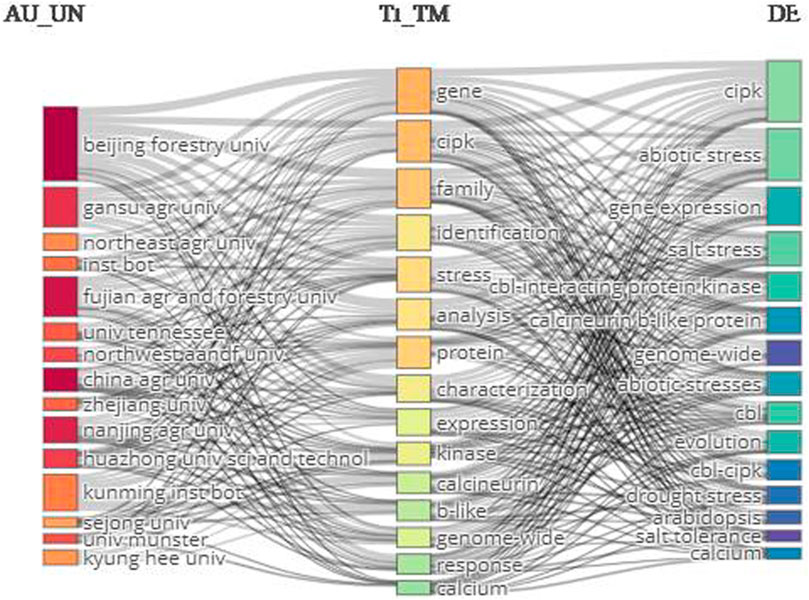
FIGURE 9. Three-field network map. The right-hand, middle, and left-hand sides show the keywords, terms extracted from titles, and author affiliations, respectively.
3.4 Most relevant sources
The retrieved data was analyzed to find the most relevant sources (Figure 10A). Frontiers in Plant Science was the most common journal, with 10 publications. The current IF of the journal is 6.627 and is in category Q1. The most locally cited sources were also identified (Figure 10B), among which Plant Cell was first with 486 citations, followed by Plant Physiology Journal, with 474 citations.
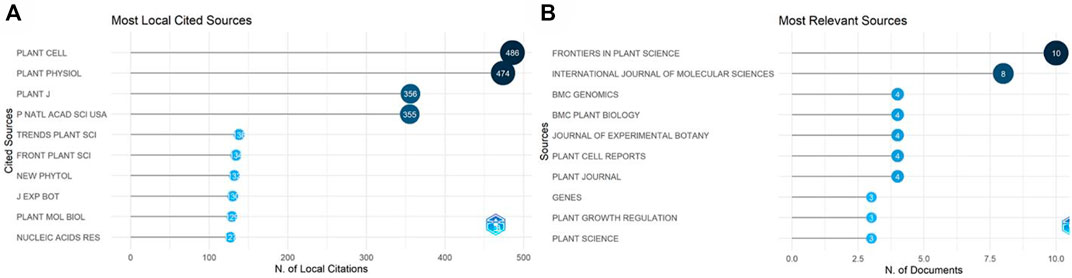
FIGURE 10. Most relevant sources. (A) The most locally cited sources. (B) The most relevant sources according to the number of publications.
3.5 Authors-citation network analysis
We used VOSviewer to map the co-authorship-to-author network (Figure 11A). Similarly, we mapped the network of citations to the authors. A total of 532 authors were included in these publications. We mapped only the linked author network, as shown in Figure 11B. Analysis of the co-occurrence of keywords identified a total of 538 keywords, among which abiotic stress, abscisic acid, and Arabidopsis were the leading terms (Figure 11C).
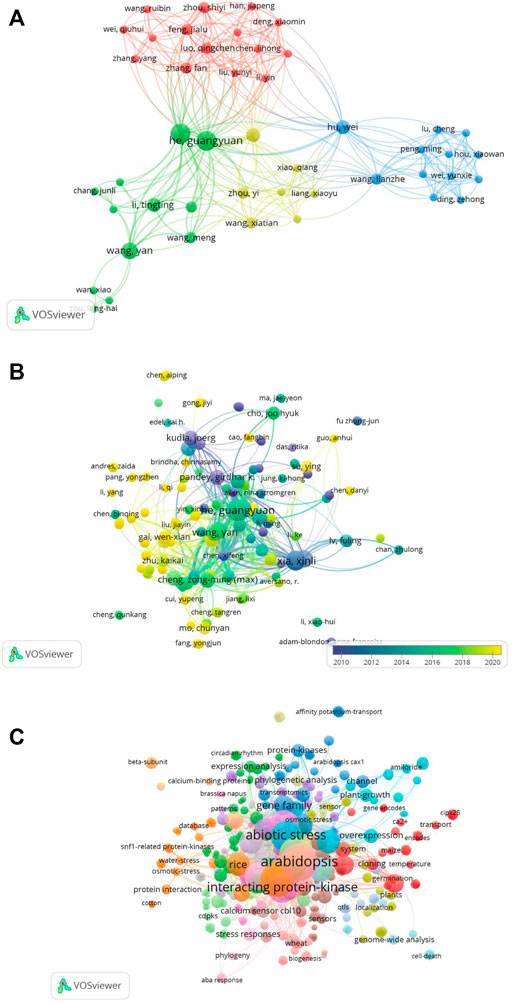
FIGURE 11. (A) Co-authors to authors’ network. Only linked authors are shown. VOSviewer was used to draw the network. (B) Citations to author network map including a total of 532 authors. The overlay visualization of the linked author network is shown. (C) Co-occurrence of keywords. A total of 538 keywords were identified and mapped using VOSviewer. Abiotic stress, abscisic acid, and Arabidopsis were the leading terms.
3.6 Term co-occurrence map
We used VOSviewer to sketch the term co-occurrence map based on the text data. The terms were extracted from the titles and abstracts. A total of 2,856 terms were included. To draw the network map, we set the threshold criterion of minimum occurrence to 3 for key terms and applied “binary” counting. A total of 300 terms met this criterion. The mapped network is shown in Figure 12.
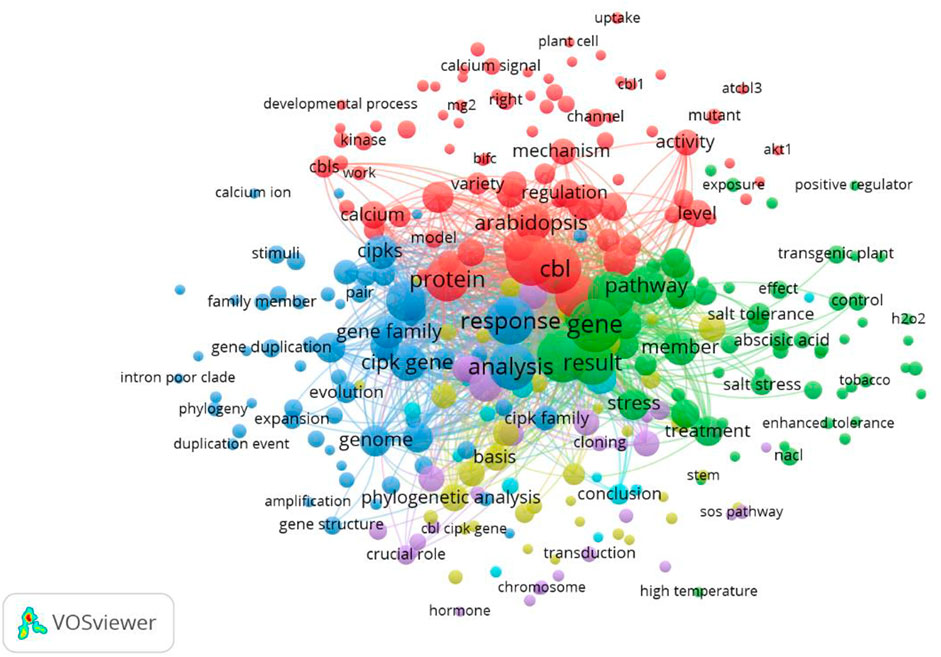
FIGURE 12. Term co-occurrence map. A total of 2,856 terms were extracted from the titles and abstracts. The mapped network is sketched using a minimum occurrence of 3. A total of 300 terms were mapped.
4 What should be discovered in the future?
CIPK proteins play important roles in the Ca2+ signaling pathway and affect plant development, in addition to participating in biotic and abiotic stress responses. CIPKs have been identified and functionally characterized in many crops such as Arabidopsis, rice, maize, and canola (Niu et al., 2018), but less is known about CIPKs in wheat and other economically important crops. Much remains to be studied in this gene family in terms of genomics and metabolomics analysis.
Author contributions
CY designed the study, supervised the project, revised the paper, and provided financial support. JY-f, WY, GY, WQ, and YX drafted the paper, analyzed the data, and applied the software.
Funding
This study was financially supported by the National Natural Science Foundation of China (No. 31501785), Natural Science Foundation of Heilongjiang Province (No. QC2017026), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2018102), and Fundamental Research Funds in Heilongjiang provincial universities (145109518).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1041078/full#supplementary-material
References
Alryalat, S. a. S., Malkawi, L. W., and Momani, S. M. (2019). Comparing bibliometric analysis using PubMed, Scopus, and web of science databases. J. Vis. Exp., e58494. doi:10.3791/58494
Aria, M., and Cuccurullo, C. (2017). bibliometrix: An R-tool for comprehensive science mapping analysis. J. Inf. 11, 959–975. doi:10.1016/j.joi.2017.08.007
Barrero-Gil, J., and Salinas, J. (2013). Post-translational regulation of cold acclimation response. Plant Sci. 205, 48–54. doi:10.1016/j.plantsci.2013.01.008
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: An open source software for exploring and manipulating networks,” in Proceedings of the international AAAI conference on web and social media), 361. Available at: https://ojs.aaai.org/index.php/ICWSM/article/view/13937 (Accessed September 2022).
Batistič, O., and Kudla, J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793, 985–992. doi:10.1016/j.bbamcr.2008.10.006
Batistic, O., Sorek, N., SchüLtke, S., Yalovsky, S., and Kudla, J. R. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell. 20, 1346–1362. doi:10.1105/tpc.108.058123
Chen, L., Wang, Q.-Q., Zhou, L., Ren, F., Li, D.-D., and Li, X.-B. (2013). Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 40, 4759–4767. doi:10.1007/s11033-013-2572-9
Chen, X., Gu, Z., Xin, D., Hao, L., Liu, C., Huang, J., et al. (2011). Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics 38, 77–87. doi:10.1016/j.jcg.2011.01.005
Cheong, Y. H., Pandey, G. K., Grant, J. J., Batistic, O., Li, L., Kim, B. G., et al. (2007). Two calcineurin B‐like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52, 223–239. doi:10.1111/j.1365-313X.2007.03236.x
Chinnusamy, V., Zhu, J., and Zhu, J.-K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi:10.1016/j.tplants.2007.07.002
Choudhri, A. F., Siddiqui, A., Khan, N. R., and Cohen, H. L. (2015). Understanding bibliometric parameters and analysis. Radiographics 35, 736–746. doi:10.1148/rg.2015140036
Cobo, M. J., López‐Herrera, A. G., Herrera‐Viedma, E., and Herrera, F. (2012). SciMAT: A new science mapping analysis software tool. J. Am. Soc. Inf. Sci. Technol. 63, 1609–1630. doi:10.1002/asi.22688
Cuypers, A., Plusquin, M., Remans, T., Jozefczak, M., Keunen, E., Gielen, H., et al. (2010). Cadmium stress: An oxidative challenge. Biometals 23, 927–940. doi:10.1007/s10534-010-9329-x
D'angelo, C., Weinl, S., Batistic, O., Pandey, G. K., Cheong, Y. H., Schültke, S., et al. (2006). Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 48, 857–872. doi:10.1111/j.1365-313X.2006.02921.x
Ding, X., Liu, B., Sun, X., Sun, X., and Zheng, C. (2022). New functions of CIPK gene family are continue to emerging. Mol. Biol. Rep. 49, 6647–6658. doi:10.1007/s11033-022-07255-x
Donthu, N., Kumar, S., Mukherjee, D., Pandey, N., and Lim, W. M. (2021). How to conduct a bibliometric analysis: An overview and guidelines. J. Bus. Res. 133, 285–296. doi:10.1016/j.jbusres.2021.04.070
Du, W., Yang, J., Ma, L., Su, Q., and Pang, Y. (2021). Identification and characterization of abiotic stress responsive CBL-CIPK family genes in Medicago. Int. J. Mol. Sci. 22, 4634. doi:10.3390/ijms22094634
Dubeaux, G., Neveu, J., Zelazny, E., and Vert, G. (2018). Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell. 69, 953–964. doi:10.1016/j.molcel.2018.02.009
Ellegaard, O. (2018). The application of bibliometric analysis: Disciplinary and user aspects. Scientometrics 116, 181–202. doi:10.1007/s11192-018-2765-z
Ellegaard, O., and Wallin, J. A. (2015). The bibliometric analysis of scholarly production: How great is the impact? Scientometrics 105, 1809–1831. doi:10.1007/s11192-015-1645-z
Esposito, S., D'amelia, V., Carputo, D., and Aversano, R. (2019). Genes involved in stress signals: The CBLs-CIPKs network in cold tolerant solanum commersonii. Biol. plant. 63, 699–709. doi:10.32615/bp.2019.072
Fan, S. K., Fang, X. Z., Guan, M. Y., Ye, Y. Q., Lin, X. Y., Du, S. T., et al. (2014). Exogenous abscisic acid application decreases cadmium accumulation in Arabidopsis plants, which is associated with the inhibition of IRT1-mediated cadmium uptake. Front. Plant Sci. 5, 721. doi:10.3389/fpls.2014.00721
Fuglsang, A. T., Guo, Y., Cuin, T. A., Qiu, Q., Song, C., Kristiansen, K. A., et al. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 19, 1617–1634. doi:10.1105/tpc.105.035626
Gephi, G. (2021). Gephi - the open graph viz platform. Available at: https://gephi.org/ (Accessed September 2022).
Gu, S., Wang, X., Bai, J., Wei, T., Sun, M., Zhu, L., et al. (2021). The kinase CIPK11 functions as a positive regulator in cadmium stress response in Arabidopsis. Gene 772, 145372. doi:10.1016/j.gene.2020.145372
Guo, M., Liu, Q., Yu, H., Zhou, T., Zou, J., Zhang, H., et al. (2015). Characterization of alkali stress‐responsive genes of the CIPK family in sweet sorghum [sorghum bicolor (L.) moench]. Crop Sci. 55, 1254–1263. doi:10.2135/cropsci2013.08.0520
Guo, Y., Halfter, U., Ishitani, M., and Zhu, J.-K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 13, 1383–1400. doi:10.1105/tpc.13.6.1383
He, L., Yang, X., Wang, L., Zhu, L., Zhou, T., Deng, J., et al. (2013). Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 435, 209–215. doi:10.1016/j.bbrc.2013.04.080
Granada, U., de, , and Viedma, E. H. (2020). Global trends in coronavirus research at the time of covid-19: A general bibliometric approach and content analysis using SciMAT. Available at: https://digibug.ugr.es/handle/10481/64562 (Accessed September 2022)
Ho, C.-H., Lin, S.-H., Hu, H.-C., and Tsay, Y.-F. (2009). CHL1 functions as a nitrate sensor in plants. Cell. 138, 1184–1194. doi:10.1016/j.cell.2009.07.004
Huang, C., Ding, S., Zhang, H., Du, H., and An, L. (2011). CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci. 181, 57–64. doi:10.1016/j.plantsci.2011.03.011
Huang, L., Li, Z., Fu, Q., Liang, C., Liu, Z., Liu, Q., et al. (2021). Genome-wide identification of CBL-CIPK gene family in honeysuckle (Lonicera japonica Thunb.) and their regulated expression under salt stress. Front. Genet. 12, 751040. doi:10.3389/fgene.2021.751040
Huang, S., Chen, M., Zhao, Y., Wen, X., Guo, Z., and Lu, S. (2020). CBL4-CIPK5 pathway confers salt but not drought and chilling tolerance by regulating ion homeostasis. Environ. Exp. Bot. 179, 104230. doi:10.1016/j.envexpbot.2020.104230
Hutchins, B. I., Baker, K. L., Davis, M. T., Diwersy, M. A., Haque, E., Harriman, R. M., et al. (2019). The NIH open citation collection: A public access, broad coverage resource. PLoS Biol. 17, e3000385. doi:10.1371/journal.pbio.3000385
Ju, C., Zhang, Z., Deng, J., Miao, C., Wang, Z., Wallrad, L., et al. (2022). Ca2+-dependent successive phosphorylation of vacuolar transporter MTP8 by CBL2/3-CIPK3/9/26 and CPK5 is critical for manganese homeostasis in Arabidopsis. Mol. Plant 15, 419–437. doi:10.1016/j.molp.2021.11.012
Kanwar, P., Sanyal, S. K., Tokas, I., Yadav, A. K., Pandey, A., Kapoor, S., et al. (2014). Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell. calcium 56, 81–95. doi:10.1016/j.ceca.2014.05.003
Khudhair, H. Y., Jusoh, D., Bin, A., F Abbas, A., Mardani, A., and Nor, K. M. (2020). A review and bibliometric analysis of service quality and customer satisfaction by using Scopus database. Int. J. Manag. 11. doi:10.34218/IJM.11.8.2020.044
Kimura, S., Kawarazaki, T., Nibori, H., Michikawa, M., Imai, A., Kaya, H., et al. (2013). The CBL-interacting protein kinase CIPK26 is a novel interactor of Arabidopsis NADPH oxidase AtRbohF that negatively modulates its ROS-producing activity in a heterologous expression system. J. Biochem. 153, 191–195. doi:10.1093/jb/mvs132
Kolukisaoglu, W. S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi:10.1104/pp.103.033068
Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. U. S. A. 96, 4718–4723. doi:10.1073/pnas.96.8.4718
Kulkarni, A. V., Aziz, B., Shams, I., and Busse, J. W. (2009). Comparisons of citations in Web of Science, Scopus, and Google Scholar for articles published in general medical journals. Jama 302, 1092–1096. doi:10.1001/jama.2009.1307
Lan, W.-Z., Lee, S.-C., Che, Y.-F., Jiang, Y.-Q., and Luan, S. (2011). Mechanistic analysis of AKT1 regulation by the CBL–CIPK–PP2CA interactions. Mol. Plant 4, 527–536. doi:10.1093/mp/ssr031
Lara, A., Ródenas, R., Andrés, Z., Martínez, V., Quintero, F. J., Nieves-Cordones, M., et al. (2020). Arabidopsis K+ transporter HAK5-mediated high-affinity root K+ uptake is regulated by protein kinases CIPK1 and CIPK9. J. Exp. Bot. 71, 5053–5060. doi:10.1093/jxb/eraa212
Li, J., Jiang, M.-M., Ren, L., Liu, Y., and Chen, H.-Y. (2016). Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genomics 291, 1769–1781. doi:10.1007/s00438-016-1218-8
Li, L., Kim, B.-G., Cheong, Y. H., Pandey, G. K., and Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 103, 12625–12630. doi:10.1073/pnas.0605129103
Li, P., Zheng, T., Li, L., Zhuo, X., Jiang, L., Wang, J., et al. (2019). Identification and comparative analysis of the CIPK gene family and characterization of the cold stress response in the woody plant Prunus mume. PeerJ 7, e6847. doi:10.7717/peerj.6847
Li, Z.-Y., Xu, Z.-S., Chen, Y., He, G.-Y., Yang, G.-X., Chen, M., et al. (2013). A novel role for Arabidopsis CBL1 in affecting plant responses to glucose and gibberellin during germination and seedling development. PLoS One 8, e56412. doi:10.1371/journal.pone.0056412
Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U. S. A. 97, 3730–3734. doi:10.1073/pnas.060034197
Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. doi:10.1126/science.280.5371.1943
Liu, K.-H., Diener, A., Lin, Z., Liu, C., and Sheen, J. (2020). Primary nitrate responses mediated by calcium signalling and diverse protein phosphorylation. J. Exp. Bot. 71, 4428–4441. doi:10.1093/jxb/eraa047
López-Robles, J. R., Cobo, M. J., Gutiérrez-Salcedo, M., Martínez-Sánchez, M., Gamboa-Rosales, N. K., and Herrera-Viedma, E. (2021). 30th anniversary of applied intelligence: A combination of bibliometrics and thematic analysis using SciMAT. Appl. Intell. (Dordr). 51, 6547–6568. doi:10.1007/s10489-021-02584-z
Lu, X., Li, X., Xie, D., Jiang, C., Wang, C., Li, L., et al. (2020). The Ca2+‐regulated protein kinase CIPK1 integrates plant responses to phosphate deficiency in Arabidopsis thaliana. Plant Biol. 22, 753–760. doi:10.1111/plb.13137
Ma, X., Gai, W.-X., Qiao, Y.-M., Ali, M., Wei, A.-M., Luo, D.-X., et al. (2019a). Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC genomics 20, 775–818. doi:10.1186/s12864-019-6125-z
Ma, Y., Cao, J., Chen, Q., He, J., Liu, Z., Wang, J., et al. (2019b). The kinase CIPK11 functions as a negative regulator in drought stress response in Arabidopsis. Int. J. Mol. Sci. 20, E2422. doi:10.3390/ijms20102422
Mallig, N. (2010). A relational database for bibliometric analysis. J. Inf. 4, 564–580. doi:10.1016/j.joi.2010.06.007
Mao, J., Manik, S. M. N., Shi, S., Chao, J., Jin, Y., Wang, Q., et al. (2016a). Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes. 7, 62. doi:10.3390/genes7090062
Mao, J., Manik, S. N., Shi, S., Chao, J., Jin, Y., Wang, Q., et al. (2016b). Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes. 7, 62. doi:10.3390/genes7090062
Mo, C., Wan, S., Xia, Y., Ren, N., Zhou, Y., and Jiang, X. (2018). Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front. Plant Sci. 9, 269. doi:10.3389/fpls.2018.00269
Mulet, J. M., Campos, F., and Yenush, L. (2020). Editorial: Ion Homeostasis in Plant Stress and Development. Front. Plant Sci. 11, 618273. doi:10.3389/fpls.2020.618273
Nemoto, K., Seto, T., Takahashi, H., Nozawa, A., Seki, M., Shinozaki, K., et al. (2011). Autophosphorylation profiling of Arabidopsis protein kinases using the cell-free system. Phytochemistry 72, 1136–1144. doi:10.1016/j.phytochem.2011.02.029
Niu, L., Dong, B., Song, Z., Meng, D., and Fu, Y. (2018). Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple (Malus domestica). Int. J. Mol. Sci. 19, 2131. doi:10.3390/ijms19072131
Pandey, G. K., Cheong, Y. H., Kim, K.-N., Grant, J. J., Li, L., Hung, W., et al. (2004). The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 16, 1912–1924. doi:10.1105/tpc.021311
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B.-G., and Luan, S. (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1, 238–248. doi:10.1093/mp/ssn003
Poddar, N., Singh, A., and Kumar, S. (2021). CBL-interacting protein kinases (CIPKs) in Chickpea: Genome-wide identification, structure and expression analysis under abiotic stresses and development. Available at: https://www.preprints.org/manuscript/202103.0369/v1 (Accessed September 2022).
Qiu, Q.-S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U. S. A. 99, 8436–8441. doi:10.1073/pnas.122224699
Rankings, S. (2022). Scopus journal rankings [Online], 26. Available: https://www.scopus.com/sources (Accessed Sep, 2022).
Rincon-Patino, J., Ramirez-Gonzalez, G., and Corrales, J. C. (2018). Exploring machine learning: A bibliometric general approach using SciMAT. F1000Res. 7, 1210. doi:10.12688/f1000research.15620.1
Sánchez-Barrena, M. J., Fujii, H., Angulo, I., Martínez-Ripoll, M., Zhu, J.-K., and Albert, A. (2007). The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell. 26, 427–435. doi:10.1016/j.molcel.2007.04.013
Sanyal, S. K., Mahiwal, S., Nambiar, D. M., and Pandey, G. K. (2020). CBL–CIPK module-mediated phosphoregulation: Facts and hypothesis. Biochem. J. 477, 853–871. doi:10.1042/BCJ20190339
Sanyal, S. K., Pandey, A., and Pandey, G. K. (2015). The CBL–CIPK signaling module in plants: A mechanistic perspective. Physiol. Plant. 155, 89–108. doi:10.1111/ppl.12344
Sanyal, S., Rao, S., Mishra, L., Sharma, M., and Pandey, G. (2016). Plant stress responses mediated by CBL–CIPK phosphorylation network. Enzymes 40, 31–64. doi:10.1016/bs.enz.2016.08.002
Schliebner, I., Pribil, M., Zuhlke, J., Dietzmann, A., and Leister, D. (2008). A survey of chloroplast protein kinases and phosphatases in Arabidopsis thaliana. Curr. Genomics 9, 184–190. doi:10.2174/138920208784340740
Schönberg, A., and Baginsky, S. (2012). Signal integration by chloroplast phosphorylation networks: An update. Front. Plant Sci. 3, 256. doi:10.3389/fpls.2012.00256
Scopus (2022). Scopus database [online]. Available: https://www.scopus.com/home.uri (Accessed Sep, 2022)
Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U. S. A. 97, 6896–6901. doi:10.1073/pnas.120170197
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi:10.1016/s1369-5266(03)00092-x
Shu, B., Cai, D., Zhang, F., Zhang, D., Liu, C., Wu, Q., et al. (2020). Identifying citrus CBL and CIPK gene families and their expressions in response to drought and arbuscular mycorrhizal fungi colonization. Biol. plant. 64, 773–783. doi:10.32615/bp.2020.123
Srivastava, A. K., Shankar, A., Nalini Chandran, A. K., Sharma, M., Jung, K.-H., Suprasanna, P., et al. (2020). Emerging concepts of potassium homeostasis in plants. J. Exp. Bot. 71, 608–619. doi:10.1093/jxb/erz458
Straub, T., Ludewig, U., and Neuhäuser, B. (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell. 29, 409–422. doi:10.1105/tpc.16.00806
Tang, J., Lin, J., Li, H., Li, X., Yang, Q., Cheng, Z.-M., et al. (2016). Characterization of CIPK family in Asian pear (Pyrus bretschneideri Rehd) and co-expression analysis related to salt and osmotic stress responses. Front. Plant Sci. 7, 1361. doi:10.3389/fpls.2016.01361
Tang, R.-J., Zhao, F.-G., Garcia, V. J., Kleist, T. J., Yang, L., Zhang, H.-X., et al. (2015). Tonoplast CBL–CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 112, 3134–3139. doi:10.1073/pnas.1420944112
Tena, G., Boudsocq, M., and Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529. doi:10.1016/j.pbi.2011.05.006
Torres, M. A. (2010). ROS in biotic interactions. Physiol. Plant. 138, 414–429. doi:10.1111/j.1399-3054.2009.01326.x
Tripathi, V., Parasuraman, B., Laxmi, A., and Chattopadhyay, D. (2009). CIPK6, a CBL‐interacting protein kinase is required for development and salt tolerance in plants. Plant J. 58, 778–790. doi:10.1111/j.1365-313X.2009.03812.x
Van Eck, N. J., and Waltman, L. (2017). Citation-based clustering of publications using CitNetExplorer and VOSviewer. Scientometrics 111, 1053–1070. doi:10.1007/s11192-017-2300-7
Van Eck, N., and Waltman, L. (2010). Software survey: VOSviewer, a computer program for bibliometric mapping. scientometrics 84, 523–538. doi:10.1007/s11192-009-0146-3
VOSviewer (2022). Features of VOSviewer. Available at: https://www.vosviewer.com/features/highlights/ (Accessed September 2022).
Wan, X., Zou, L.-H., Zheng, B.-Q., and Wang, Y. (2019). Circadian regulation of alternative splicing of drought-associated CIPK genes in Dendrobium catenatum (Orchidaceae). Int. J. Mol. Sci. 20, 688. doi:10.3390/ijms20030688
Wang, J., Lu, X., Yin, Z., Mu, M., Zhao, X., Wang, D., et al. (2016). Genome-wide identification and expression analysis of CIPK genes in diploid cottons. Genet. Mol. Res. 15, 15048852. doi:10.4238/gmr15048852
Wang, L., Feng, X., Yao, L., Ding, C., Lei, L., Hao, X., et al. (2020). Characterization of CBL–CIPK signaling complexes and their involvement in cold response in tea plant. Plant Physiol. biochem. 154, 195–203. doi:10.1016/j.plaphy.2020.06.005
Wang, Q., Guan, C., Wang, P., Ma, Q., Bao, A.-K., Zhang, J.-L., et al. (2019). The effect of AtHKT1;1 or AtSOS1 mutation on the expressions of Na⁺ or K⁺ transporter genes and ion homeostasis in Arabidopsis thaliana under salt stress. Int. J. Mol. Sci. 20, 1085. doi:10.3390/ijms20051085
Weingart, P. (2005). Impact of bibliometrics upon the science system: Inadvertent consequences? Scientometrics 62, 117–131. doi:10.1007/s11192-005-0007-7
Wikipedia (2022). Web of science. [Online]. Available: https://en.wikipedia.org/wiki/Web_of_Science (Accessed 09 26, 2022).
Wolfram, D. (2003). Applied informetrics for information retrieval research. Santa Barbara: ABC-CLIO, LLC. Available at: https://www.abc-clio.com/products/f1095c/
Wu, G.-Q., Xie, L.-L., Wang, J.-L., Wang, B.-C., and Li, Z.-Q. (2022). Genome-Wide identification of CIPK genes in sugar beet (beta vulgaris) and their expression under NaCl stress. J. Plant Growth Regul., 1–15. doi:10.1007/s00344-021-10545-6
Xi, Y., Liu, J., Dong, C., and Cheng, Z.-M. (2017). The CBL and CIPK gene family in grapevine (Vitis vinifera): Genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 8, 978. doi:10.3389/fpls.2017.00978
Xian, J., Wang, Y., Niu, K., Ma, H., and Ma, X. (2020). Transcriptional regulation and expression network responding to cadmium stress in a Cd-tolerant perennial grass Poa Pratensis. Chemosphere 250, 126158. doi:10.1016/j.chemosphere.2020.126158
Xiaolin, Z., Baoqiang, W., Xian, W., and Xiaohong, W. (2022). Identification of the CIPK-CBL family gene and functional characterization of CqCIPK14 gene under drought stress in quinoa. BMC genomics 23, 447–518. doi:10.1186/s12864-022-08683-6
Xu, J., Li, H.-D., Chen, L.-Q., Wang, Y., Liu, L.-L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 125, 1347–1360. doi:10.1016/j.cell.2006.06.011
Yang, W., Kong, Z., Omo-Ikerodah, E., Xu, W., Li, Q., and Xue, Y. (2008). Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). J. Genet. Genomics 35, 531–543. doi:10.1016/S1673-8527(08)60073-9
Yang, W. L., Niu, K., Xiang, M., Jia, Z., Ma, H., Wang, Y., et al. (2022). Transcriptional regulation of different rhizome parts reveal the candidate genes that regulate rhizome development in poa pratensis. DNA Cell. Biol. 41, 151–168. doi:10.1089/dna.2021.0337
Ye, J., Zhang, W., and Guo, Y. (2013). Arabidopsis SOS3 plays an important role in salt tolerance by mediating calcium-dependent microfilament reorganization. Plant Cell. Rep. 32, 139–148. doi:10.1007/s00299-012-1348-3
Yin, X., Xia, Y., Xie, Q., Cao, Y., Wang, Z., Hao, G., et al. (2020). The protein kinase complex CBL10–CIPK8–SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 71, 1801–1814. doi:10.1093/jxb/erz549
Zhang, H., Yang, B., Liu, W.-Z., Li, H., Wang, L., Wang, B., et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 14, 8–24. doi:10.1186/1471-2229-14-8
Zhao, J., Yu, A., Du, Y., Wang, G., Li, Y., Zhao, G., et al. (2019). Foxtail millet (Setaria italica (L.) P. Beauv) CIPKs are responsive to ABA and abiotic stresses. PLoS One 14, e0225091. doi:10.1371/journal.pone.0225091
Zhao, L., Liu, F., Crawford, N. M., and Wang, Y. (2018). Molecular regulation of nitrate responses in plants. Int. J. Mol. Sci. 19, 2039. doi:10.3390/ijms19072039
Zhou, L., Lan, W., Chen, B., Fang, W., and Luan, S. (2015). A calcium sensor-regulated protein kinase, calcineurin B-Like protein-interacting protein kinase19, is required for pollen tube growth and polarity. Plant Physiol. 167, 1351–1360. doi:10.1104/pp.114.256065
Zhu, K., Chen, F., Liu, J., Chen, X., Hewezi, T., and Cheng, Z.-M. M. (2016). Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci. Rep. 6, 28225–28312. doi:10.1038/srep28225
Keywords: CIPK, gene family, biotic and abiotic stresses, bibliometric, web of science, protein kinases, transcription regulation
Citation: Yang C, Yi-feng J, Yushu W, Yansong G, Qi W and Xue Y (2022) Diverse roles of the CIPK gene family in transcription regulation and various biotic and abiotic stresses: A literature review and bibliometric study. Front. Genet. 13:1041078. doi: 10.3389/fgene.2022.1041078
Received: 10 September 2022; Accepted: 24 October 2022;
Published: 15 November 2022.
Edited by:
Giuseppe Diego Puglia, National Research Council (CNR), ItalyReviewed by:
Guosheng Xie, Huazhong Agricultural University, ChinaJunliang Yin, Yangtze University, China
Copyright © 2022 Yang, Yi-feng, Yushu, Yansong, Qi and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Yang, MDI2NjNAcXFocnUuZWR1LmNu
 Chen Yang
Chen Yang Jin Yi-feng1,2
Jin Yi-feng1,2