
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet. , 22 November 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1037091
This article is part of the Research Topic Breaking the Myth: Breeding for Stress Tolerance, Grain Yield, and Quality Traits Simultaneously by Diversifying the Narrow Genetic Base View all 42 articles
Vegetable crops are known as protective foods due to their potential role in a balanced human diet, especially for vegetarians as they are a rich source of vitamins and minerals along with dietary fibers. Many biotic and abiotic stresses threaten the crop growth, yield and quality of these crops. These crops are annual, biennial and perennial in breeding behavior. Traditional breeding strategies pose many challenges in improving economic crop traits. As in most of the cases the large number of backcrosses and stringent selection pressure is required for the introgression of the useful traits into the germplasm, which is time and labour-intensive process. Plant scientists have improved economic traits like yield, quality, biotic stress resistance, abiotic stress tolerance, and improved nutritional quality of crops more precisely and accurately through the use of the revolutionary breeding method known as clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein-9 (Cas9). The high mutation efficiency, less off-target consequences and simplicity of this technique has made it possible to attain novel germplasm resources through gene-directed mutation. It facilitates mutagenic response even in complicated genomes which are difficult to breed using traditional approaches. The revelation of functions of important genes with the advancement of whole-genome sequencing has facilitated the CRISPR-Cas9 editing to mutate the desired target genes. This technology speeds up the creation of new germplasm resources having better agro-economical traits. This review entails a detailed description of CRISPR-Cas9 gene editing technology along with its potential applications in olericulture, challenges faced and future prospects.
The world population is estimated to increase by 10 billion in the next three decades, thereby the demand for food crops is likely to increase by 25–70% (Hunter et al., 2017). Contemporary agriculture will eventually face enormous challenges to produce crops having high yield and better quality which require few inputs (Tilman et al., 2011). Vegetable crops act as protective foods that provide essential nutrients in the human diet due to their richness in vitamins, minerals, dietary fiber and phytochemicals (Dias, 2012). More than 400 g of fruits and vegetables should be consumed daily per person to reduce the risk of cardiovascular diseases. (Bazzano et al., 2002). or cancer (Aune et al., 2017). Like any other food crop, vegetable crops too are prone to many biotic and abiotic stresses (Jaganathan et al., 2018; Boscaiu and Fita 2020) thereby necessitating the development of next-generation architectured crops which are able to sustain adverse environmental stresses (Karkute et al., 2017).
Till now conventional breeding techniques have been extensively used to improve the yield and agronomic performance which is a complex, time-consuming and labor-intensive process (Zhang et al., 2018). It is a selection of improved individuals utilizing genetic variation in the population (Breseghello and Coelho, 2013). It also recombines the desired gene pools resulting in new genotypes or cultivars (Holme et al., 2019). However, when the desirable genetic variation is not available in the gene pool, then it can be generated and used for selection by inducing mutations using various mutagenic agents like non-ionizing radiation i.e. UV rays or ionizing radiation i.e. X and gamma rays, alpha and beta rays, fast and slow neutrons. Some chemicals can also be used as mutagenic agents like ethyl methane sulphonate (EMS), methyl methane sulphonate (MMS), hydrogen fluoride (HF), sodium azide, N-methyl-N-nitrosourea (MNU) and hydroxylamine) (Parry et al., 2009). Mutation breeding has not been extensively utilized in vegetable crop improvement except for a few exceptions being low in its efficiency. Over the last few decades, there have been a large number of significant developments in the molecular biology approaches to improve crop yield and quality. Recently, tremendous progress has been made in genome editing tools like site-directed nucleases (SDNs) which are able to edit the crops at high speed and possess great potential in shaping up the novel genetic makeup of vegetable crops (Tian et al., 2021). Genome editing tools can precisely engineer the genes by either deleting, replacing or inserting specific sequences at the specific targeted location in the target genome to generate novel traits (Corte et al., 2019). Zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated enzymes are the tools for genome editing used to modify plants (Miller et al., 2007). Site-specific double-stranded breaks (DSBs) are enabled by Crispr/Cas which further activate the cellular DNA repair systems (Zhu et al., 2020; Gaj et al., 2013). These DSBs can either be corrected by the non-homologous end joining (NHEJ) pathway or through the homology-directed repair (HDR) pathway (O’Driscoll and Jeggo, 2006; Wang T. et al., 2019a). The use of first-generation technologies like ZFNs and TALENs has been limited due to their adverse mutagenic outcome, low editing efficiency, time-consuming process and labor-intensive selection and screening process (Gaj et al., 2013; Jaganathan et al., 2018). The second-generation genome editing technology i. e. CRISPR/Cas9 is easier to design, and execute and more cost-effective. The use of CRISPR/Cas9 in vegetable crops has substantially expanded gene editing technology and made it possible to create novel genotypes with desired phenotypic features and altered genomic functions at the base pair level (Abdallah et al., 2015; Nunez de Caceres Gonzalez and De la Mora Franco, 2020). We will first go over CRISPR/Cas9 history and development before summarising how it is currently used to modify vegetable crops. Finally, we will talk about the real-world challenges in enhancing vegetable crops with the desired traits.
CRISPR-Cas9 is an advanced genome editing technique that enables scientists to change, add, or remove specific DNA sequences to modify specific regions of the genome. In general, there are three main types (I-III) of CRISPR-Cas systems utilized for target interference (Rouillon et al., 2013). Type II uses its two distinctive nuclease domains, RuvC and HNH, to achieve interference with only a basic effector-module design (Gasiunas et al., 2012). Type II Cas9 from Streptococcus pyogenes (SpCas9) is the most popular CRISPR nuclease employed in CRISPR-Cas technology (Doudna and Charpentier, 2014). The protospacer adjacent motif (PAM) is recognized by the sgRNA-Cas complex, and Cas9 cleaves the target DNA to create a double-strand break (DSB), activating cellular DNA repair processes (Figure 1).
The two essential parts of the CRISPR-Cas9 system that modify DNA are Cas 9 enzyme and guiding RNA (gRNA). The “genetic scissors” known as Cas9 (enzyme), cut the two DNA strands at an exact place in the genome to allow for the addition or deletion of DNA fragments and the guiding RNA (gRNA) is made up of two parts: Crispr RNA (crRNA), a 17–20 nucleotide sequence complementary to the target DNA, and a transactivating Crispr RNA (tracr RNA), which serves as a binding platform for the Cas9 nuclease (Mei et al., 2016) (Figure 1). Each crRNA hybridizes with tracr RNA, and these two RNAs jointly make a complex with the Cas9 nuclease (Deltcheva et al., 2011). The goal of the guide RNA is to find and bind to a target DNA sequence that is complementary to its RNA bases. Double strand breaks, or DSBs, are created when the Cas9 enzyme cuts across both DNA strands at the same location in the DNA sequence as the guide RNA (Jinek et al., 2012; Jiang et al., 2013). The DSBs inflicted by Cas-9 protein are repaired by two mechanisms i.e., non-homologous end-joining (NHEJ) and homology-directed repair (HDR) (Liu et al., 2019) (Figure 2). NHEJ needs enzymes in the repair mechanism in which different DNA segments are joined by excluding a homologous DNA template (Shuman and Glickman, 2007). It is an extraordinarily effective cell repair mechanism that is most often exploited but is prone to errors that can cause minor, spontaneous insertions or deletions. (Yang et al., 2020). The HDR however is quite precise in gene insertion or replacement of DNA segments at the predicted DSB site but requires a large amount of homologous DNA template (Liu et al., 2019; Yang et al., 2020).
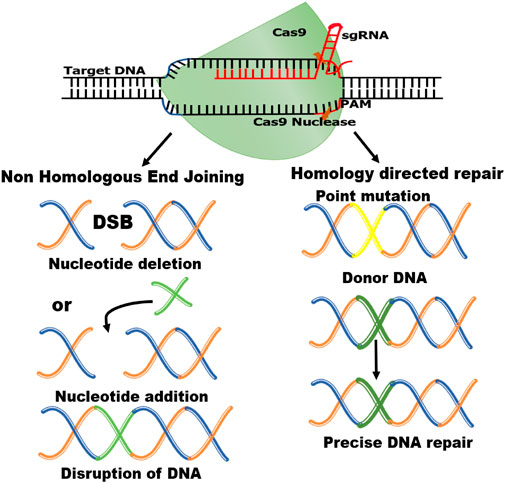
FIGURE 2. Schematic diagram of CRISPR/Cas9 mechanism. Double-strand breaks (DSBs) are induced when Cas9 enzyme and gRNA bind with targeted double-stranded DNA. These DSBs are repaired by Homology-directed repair (HDR) and Non-homologous end-joining (NHEJ) mechanisms.
Several new CRISPR systems have been developed to overcome the limitations of CRISPR/Cas 9 and improve its specificity for more effective genome editing. These cutting-edge technologies are detailed below and could be crucial resources for molecular crop breeding.
Cas protein Cpf1 (also known as Cas12a) is with RNA-guided system being widely used in genome editing (Kim et al., 2017; Moon et al., 2018; Chen et al., 2019). It uses a T-rich PAM sequence to identify the target site, extending the editing sites beyond the G-rich PAM sequences preferred by Cas9. The target site of Cpf1 is located at the distal position and downward the PAM sequence. Cpf1’s guide RNA has 43 base pairs and is shorter than the sgRNA of Cas9 (about 100 bp) (Kim et al., 2017; Chen et al., 2019). Cpf1 generates staggered-ended double-strand breaks, which offers more advantages than Cas9 and improves the effectiveness of the NHEJ-based gene insertion. (Kim et al., 2017; Moon et al., 2018). Cpf1-based genome editing has been reported in rice and soybean (Kim et al., 2017; Xu et al., 2017). Additional research is required to analyze the specificity of Cpf1 in other crops and to enhance the current Cas12a-based applications Schindele and Puchta (2020).
Cas12b prefers T-rich PAM, produces staggered double-strand breaks, and needs both a crRNA and a trans-activating crRNA. Cas12b protein is smaller than Cas9 and Cas12a. Cas12b/C2c1 has been effectively used to carry out multiplex genome editing, induce mutations, and cause deletions at many loci in Arabidopsis (Wu et al., 2020). However, Cas12b shows its optimum activity at higher temperature (Teng et al., 2018), it needs to be altered for making it more useful in crop applications.
Cas13 is a recently identified CRISPR effector which targets specific RNAs in plant cells. This system has high RNA target specificity and efficiency. Cas13 protein belongs to class 2 type VI and contains unique higher eukaryotes and prokaryotes nucleotide-binding domains that are exclusively associated with RNase activity (Wolter and Puchta 2018). Till now, three different Cas13 protein classes, such as Cas13a, Cas13b, and Cas13d, have been used for RNA editing in plants (Schindele et al., 2019), mainly to target RNA for cleavage, for combating RNA viruses (Aman et al., 2018; Wolter and Puchta 2018). It has been demonstrated that CRISPR/LshCas13a system is used to create potyvirus resistance in plants, which suggests that this system can be employed for agricultural and biotechnological applications (Aman et al., 2018).
Cas14a is a highly compact protein, which can be used as an RNA-guided DNA nuclease for target-specific single-stranded DNA (ssDNA) cleavage (Harrington et al., 2018; Khan M. Z. et al., 2019b). Due to its sequence-independent and unrestricted cleavage, it has evolved into an excellent tool for building resistance to economically significant plant ssDNA viruses (Harrington et al., 2018; Khan M. S. S. et al., 2019). Cas14a is only one-third the size of Cas9 and is the smallest working CRISPR system to date. (Harrington et al., 2018). It has the potential to generate resistance against ssDNA viruses that belong to the Geminiviridae and Nanoviridae families (Khan M. S. S. et al., 2019a).
Besides these systems, base editing provides effective, concise, and well-recognized strategies for specific base replacement at the target site without the need for DSBs or donor DNA and independent of homology-directed repair (HDR) (Chen et al., 2019). They are helpful when there is a requirement for desired protein-coding genes to create variations with enhanced economic traits (Li H. et al., 2020c). At present, there are two types of base editors: Cytosine base editors (CBE) (which converts C-G pair to T-A pair) and adenine base editors (ABE) (converts A-T base pair to G-C base pairs (Komor et al., 2016; Gaudelli et al., 2017) Figures 3A,B. Both these editors largely depend upon the availability of PAM sequence as they use DNA binding proteins to induce point mutations at the targeted site (Jin et al., 2019). The major limitation of base editors is their inability to generate precise base edits in point mutations. Research is ongoing to improve the efficiency and role of base editing in random and targeted mutagenesis (Li H. et al., 2020c).
Prime editing, a recent genome-editing tool that induced all kinds of mutations and base substitutions (insertion/deletion) without donor DNA or double-strand breaks. Prime editing uses proteins fused to Cas 9 nickase and prime edited guide RNA with reverse transcriptase to induce mutations. The pegRNA contains not only complimentary sequence as the target sites that directs nCas9 to its target sequence, but also an additional sequence creating desired sequence changes. The Cas9 nicked the PAM containing DNA strand which act as a primer for reverse transcriptase to create extensions in the nicked strand using pegRNA as template and ultimately modifying the target site (Anzalone et al., 2020). PE has been successfully applied in rice and wheat to generate stable edited lines with price gene edits (Butt et al., 2020; Li H. et al., 2020c). It is advantageous over other genome editing tools in crop science with respect to reduced off-target mutations and less requirement of PAM sequences due to RNA template and high-efficiency rates but still experiments are need to be conducted regarding its specificity and potential. The various tools and Databases used for CRISPR/Cas9 mediated genome editing in plant systems are being presented in Tables 1, 2.
As vegetables are susceptible to various abiotic and biotic stresses which reduce optimum production, which highlights the significance of developing resistant/tolerant cultivars. Additionally, in vegetable crops, various quality traits including flavor and nutritional profile, plant architecture, and shelf life can be improved. Various gene editing systems have good potential to improve the quality and yield of vegetables among which CRISPR/Cas is very popular. The major applications of CRISPR/Cas9 in vegetable crops is being discussed below under different sections and the list of various traits modified by CRISPR/Cas is being presented in Table 3.
Some plants lack chlorophyll pigmentation as a result of phytoene desaturase (PDS) gene disruption, which affects the formation of carotenoids and chlorophyll, leading to albino plant phenotypes. There is little information on the albino plant phenotypes in vegetables except for a few publications where albinism has been used to standardize the gene-editing method utilizing CRISPR-Cas. Fully albino plants (pds mutants) were generated in various vegetable crops like tomato, watermelon, melon, cabbage and carrot via editing phytoene desaturase gene through Agrobacterium tumefaciens-mediated transformation (Pan et al., 2016; Tian et al., 2017; Xu J. et al., 2019; Hooghvorst et al., 2019; Ma et al., 2019).
Vegetable crops face many abiotic stresses caused due to temperature, drought, salinity and heat which adversely affect crop productivity. Although traditional breeding techniques are able to combat stresses to certain extent, new innovative technologies like CRISPR-Cas 9 offers the possibility to generate more resilient germplasm in dealing with these stresses (Haque et al., 2018). High temperature is a major stress factor that inhibits the growth and productivity of vegetable crops. It leads to the overproduction of reactive oxygen species (ROS) which causes oxidative damage, ultimately impairing the normal function of plant cells. Highly conserved protein kinases called mitogen-activated protein kinases (MAPKs) are involved in the response to heat stress of vegetable crops (Sharma et al., 2020). Knockout of BRASSINAZOLE RESISTANT 1 (BZR1) impaired the induction of RESPIRATORY BURST OXIDASE HOMOLOG1 (RBOH1) and induced production of H2O2 and heat tolerance in tomato. This exogenous H2O2 recovered the heat tolerance in bzr1 tomato mutant plants (Yin et al., 2018). In addition to this, mutations induced through CRISPR-Cas9 in slmapk3 imparts higher heat stress tolerance in tomato. Slmapk3 mutant also showed less wilting, mild membrane damage, low production of reactive oxygen species and improved antioxidant enzymatic activity under heat stress as reported by Yu et al. (2019). Similarly, in lettuce by using CRISPR/Cas9, LsNCED4 (9-cis-EPOXYCAROTENOIDDIOXYGENASE4) gene, resulting in thermo-inhibition of seed germination was knocked out, which significantly resulted in high-temperature germination in both cultivars (Salinas and Cobham Green), capable of germinating more than 70% at 37°C (Bertier et al., 2018).
One of the most harmful environment factors causing damage to vegetable crops is drought stress. Important signalling molecules that react to drought stress include mitogen-activated protein kinases (MAPKs). In 2017, Wang and his co-workers utilized CRISPR/Cas9) mediated mutagenesis to generate slmapk3 mutants in tomato. In comparison to wild type plants, the slmapk3 mutants had more severe wilting symptoms, greater hydrogen peroxide levels, low antioxidant enzyme activity, and experienced more membrane damage. They concluded that slmapk3 is involved in drought response in tomato by protecting from membrane damage and stimulating transcription of some stress related genes. Chilling stress is the primary obstacle that prevents the growth of some vegetable crops, like tomato, brinjal, and chilli as they are sensitive to severe chilling injury. The highly conserved C-repeat binding factors (CBFs) are involved in regulating cold tolerance. As in tomato, the slcbf mutants were generated using the CRISPR-Cas9 system, but these mutants exhibited severe chilling- injury symptoms as shown by the down-regulation of CBF-related genes as compared to wild-type (WT) plants (Li et al., 2018a). Additionally in both abovementioned studies, the mutants exhibited lower content of proline and protein and more amount of hydrogen peroxide contents and antioxidants than WT plants which further altered hormonal level of plants and reduced the expression of genes. So, there is a need to study the regulatory mechanism of CBF and mitogen-activated protein kinases (MAPKs) genes in order to understand their molecular mechanism.
In addition to this, the SlUVR8 gene was knocked out in tomato to increase tolerance to high UV-B stress using CRISPR-CAS9 gene editing approach by generating sluvr8 mutant lines which confirmed that SlUVR8 plays a significant function in tomato seedling growth and UV-B stress resistance Liu et al. (2020). Excessive concentration of salts within the plant tissues will reduce growth and productivity, as they can affect several pivotal processes, such as germination, photosynthesis, nutrient balance and redox balance, among others (Parihar et al., 2015; Petretto et al., 2019). Recently, the HKT1&2 allele was edited and inserted into tomato Hongkwang cultivar via the CRISPR/Cpf1-mediated homology-directed repair (HDR) mechanism which showed stable inheritance for salt tolerance (Vu et al., 2020). Furthermore, by precise deletion of one or more SlHyPRP1’s functional motifs using CRISPR/Cas9-based multiplexed editing, salt stress-tolerant events in cultivated tomatoes were produced (Tran et al., 2021).
Globally, major losses in the production of vegetable crops are caused by a diverse variety of diseases. A sustainable strategy for supplying the world’s expanding population with food is the development of disease-resistant cultivars. (Thomazella et al., 2016). Traditional plant breeding has been utilized for centuries to develop new varieties, but modern technologies, like genome editing, have the ability to produce improved varieties more quickly, by accurately introducing favourable alleles into locally adapted types (Nekrasov et al., 2017). In tomato, SlDMR6-1 orthologue Solyc03g080190.2 is up-regulated when infected due to Pseudomonas syringae pv. tomato and Phytophthora capsici. The tomato homologue genes were knocked out using CRISPR-Cas9 to cause mutations in DMR6, which resulted in broad-spectrum resistance to Pseudomonas, Phytophthora, and Xanthomonas spp. (Thomazella et al., 2016). Wild-type MILDEW RESISTANT LOCUS O (Mlo) alleles, encode a protein, which provides fungal sensitivity causing powdery mildew disease. In tomato, homozygous loss-of-function of SlMlo1 gene through CRISPR-mediated mutations resulted in resistance to powdery mildew (Nekrasov et al., 2017).
Pseudomonas syringae pv. tomato (Pto) DC3000, a causative agent of tomato bacterial speck disease releases coronatine (COR) which stimulates stomatal opening and encourages the bacterial colonization in the leaves. Ortigosa et al. (2019) developed a tomato genotype resistant to bacterial speck by editing the SlJAZ2 gene (a key co-receptor for coronatine in stomatal guard cells) via the CRISPR/Cas9 system to produce dominating Jasmonate-Zim Domain (JAZ2) repressors (SlJAZ2jas), which prevents coronatine from reopening stomata and provided resistance to PtoDC3000. Jeon et al. (2020) identified biosynthetic gene clusters (ACET1a, ACET1b and Solyc12g100270) in tomato plants required for the production of falcarindiol in response to biotic stress. Mutagenesis through CRISPR revealed the direct role of the cluster in synthesis of falcarindiol which imparts resistance against fungal and bacterial pathogens in tomato. In 2018, through the use of the CRISPR/Cas9 system, tomato plants were made resistant to the tomato yellow leaf curl virus by focusing Through the use of the CRISPR/Cas9 system, tomato plants were made resistant to the tomato yellow leaf curl virus by focusing on the coat protein and replicase sites (Tashkandi et al., 2018).
In order to speed up the breeding of potatoes for resistance to late blight (Phytophthora infestans) and potato virus Y (PVY), CRISPR/Cas has emerged as a substitute and effective method. Targeting P3, CI, Nib, and CP viral genes, Cas13a protein was used to give resistance to three PVY strains (RNA viruses) (Zhan et al., 2019). Similarly, the functional knockouts of StDND1, StCHL1, DMG400000582 (StDMR6-1) and caffeoyl-CoA-O-methyltransferase gene generated potato plants with increased late blight resistance (Hedge et al., 2021; Kieu et al., 2021). Knocking out of Clpsk1 gene, which encodes the PSK precursor, in watermelon to provide increased resistance to Fusarium oxysporum f.sp. niveum (Zhang et al., 2020), whereas, in tomatoes, Solyc08g075770-knockout via CRISPR-Cas9 resulted Fusarium wilt disease sensitivity in the plants (Prihatna et al., 2018). Virus resistance can be induced in cucumber plants by disrupting the function of the recessive eIF4E (eukaryotic translation initiation factor 4E) gene using Cas9/subgenomic RNA (sgRNA) technology (Chandrasekaran et al., 2016). Similarly in Brassica napus, BnWRKY11 and BnWRKY70 were edited with CRISPR/Cas9 vectors and mutations induced in BnWRKY70 generated mutants with enhanced resistance to Sclerotinia spp. (Sun et al., 2018).
Fruit and vegetables (F&V) are highly perishable food products that need advanced post-harvest technologies to maintain their storage stability and extended shelf life (Gallagher and Mahajan., 2011). In tomato, the homology-directed repair (HDR) pathway was used to replace the allele of ALC with the alc gene, resulting in T1 homozygous plants with long shelf life. (Yu et al., 2017). Klap et al. (2017) used CRISPR/Cas9 technology to delete tomato SlAGL6 (SlAGAMOUS-LIKE6) which led to the development of parthenocarpy under high-temperature stress conditions without compromising the weight, fruit shape, or pollen vitality. Other vegetable crops, like the in-demand seedless watermelon or less-seeded fruits, can also use this approach to generate parthenocarpy.
Lycopene is an important plant nutrient with strong antioxidant properties that helps to protect the cells from damage. Lycopene accumulation in the fruit is facilitated by the knockdown of a few genes linked to the carotenoid metabolic pathway. The amount of lycopene was successfully increased to about 5.1 times in genome-edited tomato fruits. These results suggested that the CRISPR/Cas9 system has the potential to greatly increase the amount of lycopene in tomato fruit due to its high effectiveness, infrequent off-target mutations, and stable heredity Li et al. (2018a). The non-proteinogenic amino acid gamma-aminobutyric acid (GABA) has hypotensive properties. Nonaka et al. (2017) To boost GABA accumulation by 7–15 times in tomato fruits, researchers employed CRISPR/CRISPR-associated protein (Cas)9 technology to remove the autoinhibitory domain of SlGAD2 and SlGAD3 and insert a stop codon right before the autoinhibitory domain.
Potato starch quality is important in various food applications. Improved starch quality with full knockout of granule-bound starch synthase (GBSS), starch synthase gene (SS6) and starch-branching enzymes (SBEs) genes SBE1, and SBE2 was reported in potato using CRISPR mediated genome editing (Andersson et al., 2017; Andersson et al., 2018; Kusano et al., 2018; Johansen et al., 2019; Veillet et al., 2019; Sevestre et al., 2020; Zhao et al., 2021). The enzyme polyphenol oxidase (PPO) catalyzes the oxidation of phenolic compounds into highly reactive quinones that cause postharvest browning of cut or bruised fruit (Araji et al., 2014). In the tetraploid potato cultivar Desiree, Gonzalez et al. (2020) investigated the use of the CRISPR/Cas9 system to introduce mutations into the StPPO2 gene. Mutations induced in the four alleles of the StPPO2 gene led to lines with reduced PPO activity (69%) in tubers. CRISPR/Cas has also been utilized in potato for improving traits like carotenoid biosynthesis (Khromov et al., 2018; Banfalvi et al., 2020; Butler et al., 2020) and glycoalkaloids (Nakayasu et al., 2018). Reducing the amount of steroidal glycoalkaloids (SGAs), such as α-solanine and α -chaconine, in tubers is necessary for breeding excellent potatoes since their presence may give potatoes a bitter flavor and have other unfavorable effects on humans. Two SGA-free potato lines were generated by selectively inhibiting a steroid 16-hydroxylase (St16DOX) that is involved in the synthesis of steroidal glycoalkaloids (SGA) in potato (Nakayasu et al., 2018).
Similarly, in brinjal, the three-polyphenol oxidase (PPO) genes SmelPPO4, SmelPPO5, and SmelPPO6 in brinjal were linked to enzymatic browning. To stop the browning of fruit flesh, these three target PPO genes have been eliminated using CRISPR-Cas9-based mutagenesis. (Maioli et al., 2020). It paves the way for the creation of genotypes of eggplant with reduced levels of flesh browning and higher levels of berry polyphenols as this is the first time the CRISPR/Cas9 system has been applied to eggplant for biotechnological uses.
Cucumber gynoecious inbred lines are very important because of their better production yield and cheaper labour cost for crossing. Hu et al. (2017) used CRISPR-Cas9 technique to create Cswip1 mutants by targeting the WPP trp/pro/pro domain Interacting Protein1 (CsWIP1) gene, which encodes a zinc-finger transcription factor. Cswip1 T0 mutants had a gynoecious phenotype with exclusively female flowers and these gynoecious mutants will be beneficial in heterosis breeding to produce high-yielding hybrids.
Similarly, Zhang et al. (2019a) created artificial gynoecious watermelon lines by editing ClWIP1gene using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system. Additionally, SP5G mutations in field tomatoes hasten the blooming process and alter the compact growth habit, resulting in a short flowering interval and an early harvest (Soyk et al., 2017).
Weeds are a significant stress factor that affects the yield and quality of vegetables, and selective herbicides are frequently used to control the growth and development of weeds during cultivation. The herbicide target gene acetolactate synthase (ALS) has been edited by using CRISPR-Cas9 technology in vegetables like tomato, watermelon, soybean, and potato for developing herbicide resistance in plants (Danilo et al., 2019; Tian et al., 2018; Li et al., 2015; Veillet et al., 2019).
Phelipanche aegyptiaca, an obligatory weedy plant parasite, requires the presence of the plant hormone strigolactone (SL) to encourage seed germination. Carotenoid dioxygenase 8 (CCD8), a crucial enzyme in the carotenoid synthesis pathway that generates strigolactone in tomatoes, and More Axillary Growth1 (MAX1), which is involved in strigolactone synthesis, were modified using CRISPR-Cas9 to significantly lower SL content and produce tomato plants resistant to P. aegyptiaca (Bari et al., 2019; Bari et al., 2021).
Recently, Yang et al. (2022) designed and tested the efficiency of sgRNA to use with Crispr/Cas system to edit herbicide-related genes pds (phytoene desaturase), ALS (acetolactate synthase), and EPSPS (5-Enolpyruvylshikimate-3-phosphate synthase) in tomato. The outcomes of the sgRNA efficiency tests confirmed that the transformation process could alter the target locations. They verified that 19 different transgenic tomatoes had adequately been edited by ALS2 P or ALS1 W sgRNAs, and 2 of them carried three base mutations that are likely to change their herbicide resistance.
Genome/gene editing refers to the precise change in either of DNA or RNA sequence of any target organism. This editing can lead to change in a single base pair to completely reorganization of the large genomic region. Sometimes, genes that are not present in the natural gene pool are also introduced into the target individual to generate novel traits. As this technique involve genetic manipulation either by altering genome sequence or by addition of foreign genomic sequence therefore it becomes mandatory to enforce the regulations of the Cartagena Protocol by any country. The Cartagena Protocol on Biosafety set the foundation for regulating the release and international trade of genetically modified organisms. However, there have been differences in the patterns of GM crop cultivation, utilization and legislation. While some nations restrict production and deny consumption, others actively cultivate and consume them (Garcia Ruiz et al., 2018). Some countries regulate the process while others are involved in the regulation of the product (Eckerstorfer et al., 2019; Van Vu et al., 2019).
For instance, in 2018, as per the guide lines of United States Department of Agriculture (USDA) genome editing through CRISPR-Cas 9 is like conventional breeding therefore does not need any regulation under American Regulatory Standards and are exempted from the regulatory frameworks (Waltz, 2016a). This gives advantage in minimizing the time and resources needed for the testing and legislation of the release of the CRISPR edited crops. Growing research output is authentic evidence that CRISPR-Cas edited crops holds significant promise in improving the yield and quality of crops for the consumers across the world.
In the year 2018, Canadian legislation stated that any gene editing technology which produces a novel product must be subjected to further regulatory supervision on toxicity, allergenicity and any effects on other organisms except the target (Smyth, 2017). For example, non-browning apples and non-dark spot potatoes were approved in Canada after a long examination process which ensured that the changes made in these two products were not harmful to the human being.
However, the European Court of Justice (ECJ) has approved many mutagenic crops developed through chemical and physical mutagens (Waltz, 2016b) but considered gene-edited crops under same strict rules as traditional genetically modified (GM) plants. Among the South American countries like Argentina has developed a regulation system as per the guidelines of Cartagena Protocol on Biosafety for the approval of genome-edited products and relies on the case-by-case evaluation with the exemption from the regulation in the absence of transgene (Whelan and Lema, 2015). Chile and Brazil also followed the same regulatory system regarding genome editing as Argentina. Chile has given regulations in 2017 while Brazil in January 2018 (Duensing et al., 2018).
In Australia, the regulatory framework is set by the Gene Technology Act 2000 (GT Act) and GT Regulations 2001 (GT Regulations) with the purpose to protect people’s health, safety and the environment by recognizing threats posed as a consequence of genetic manipulation. The proposed amendments relevant to genome editing would exclude organisms developed with site-directed nucleases (SDN-1) from regulation and stated that organisms developed with SDN2 or SDN-3 are regulated as GMOs (Thysegen, 2019) (Please see Figure 4 depicting mechanism of SDN-1, 2 and 3 types). In the New Zealand, release of genetically modified plants is regulated by the Hazardous Substances and New Organisms (HSNO) Act 1996. The Act generally defines a GMO as any organism whose genome or genetic information has been altered by in vitro methods (Fritsche et al., 2018).
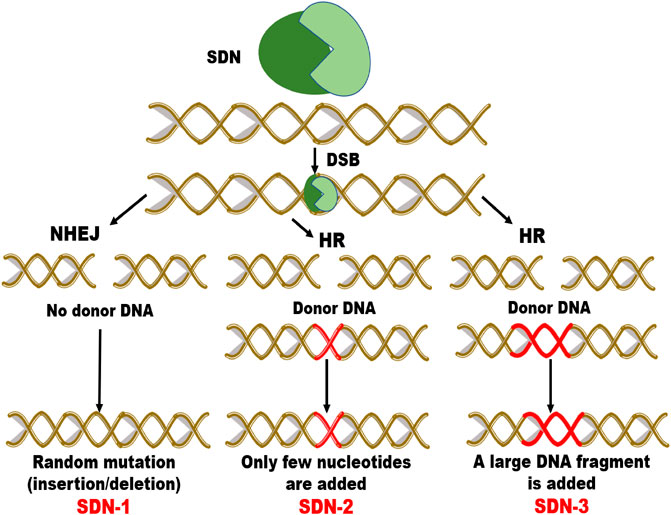
FIGURE 4. Schematic diagram of modification by site directed nuclease (SDN- 1, SDN-2 and SDN-3) types. Double strand break (DSB) is repaired via non-homologous end joining (NHEJ) or homologous recombination (HR). SDN1 results in random insertion/deletion, SDN2 induces addition of few nucleotides and SDN3 inserts a DNA fragment.
In India, all the activities related to the development and use of genetically modified products are regulated as per the ‘‘Rules for the Manufacture/Use/Import/Export and Storage of Hazardous Microorganisms, Genetically Engineered Organisms or Cells’’, 1989 (Rules 1989) which covers new genome editing technologies including CRISPR/Cas9 and notified under the Environment (Protection) Act, 1986, by Genetic Engineering Appraisal Committee (GEAC). (Warrier and Pande 2016). This committee is responsible for granting permits to conduct experimental and large-scale open field trials and also granting approval for the commercial release of genetically altered crops.
The CRISPR/Cas9 system is the latest cutting-edge technology that augments crop improvement by generating high-yielding, better quality, and resistant crop plants to biotic and abiotic stresses crops in a short span of time (Doudna and Charpentier, 2014; Langner et al., 2018). The NHEJ-mediated gene repair creates precise alterations to knock out or change the function of a particular target gene(s) which plays an important role in crop-trait-specific applications, but still there exist many challenges which must be overcome. Foremost is the selection of genes that are to be targeted for mutations and the types of mutation to avoid off-target gene editing. Moreover, it is difficult to carry out genome editing in the target organisms without genome sequencing. Editing a single gene does not result in desired phenotypic changes, because significant agronomic factors are quantitative. To add desired mutant alleles, effective CRISPR-Cas-mediated target site-specific insertion, deletion, and chromosomal recombination procedures can be applied (Zhu et al., 2020).
Once a gene has been identified, the second major challenge is to deliver CRISPR-Cas gene-editing agents into plant cells and the procedure to regenerate the putative edited plants (Chuang et al., 2021; Lino et al., 2018). Actually, it is quite challenging to create a universal and effective genetic transformation and regeneration system for vegetable crops (Niazian et al., 2017). In addition to this, for successful genetic transformation of vegetable crops editing efficiency is to be considered, which is further influenced by various factors, such as the number of sgRNA and GC amount; the expression levels of sgRNA and Cas9; and the secondary structure of the paired sgRNA and target sequence. (Kumlehn et al., 2018; Hu et al., 2019). Since genome editing in vegetable crops has such huge potential, we expect that strategies will be formulated to overcome these challenges in the near future.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdallah, N. A., Channapatna, S. P., and McHughen, A. G. (2015). GM Crops & Food, 6. Taylor & Francis Group, 183–205. doi:10.1080/21645698.2015.1129937
Aman, R., Ali, Z., Butt, H., Mahas, A., Aljedaani, F., Khan, M. Z., et al. (2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 19, 1. doi:10.1186/s13059-017-1381-1
Andersson, M., Turesson, H., Nicolia, A., Falt, A. S., Samuelsson, M., and Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. doi:10.1007/s00299-016-2062-3
Andersson, M., Turesson, H., Olsson, N., Falt, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 164, 378–384. doi:10.1111/ppl.12731
Anzalone, A. Z., Koblan, L. W., and Liu, D. R. (2020). Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844. doi:10.1038/s41587-020-0561-9
Araji, S., Grammer, T. A., Gertzen, R., Anderson, S. D., Mikulic-Petkovsek, M., Veberic, R., et al. (2014). Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 164, 1191–1203. doi:10.1104/pp.113.228593
Aune, D., Giovannucci, E., Boffetta, P., Fadnes, L. T., Keum, N., Norat, T., et al. (2017). Fruit and vegetable intake and the risk of cardiovascular disease, total cancer and all-cause mortality-a systematic review and dose response meta-analysis of prospective studies. Int. J. Epidemiol. 46, 1029–1056. doi:10.1093/ije/dyw319
Bae, S., Park, J., and Kim, J. S. (2014). Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. doi:10.1093/bioinformatics/btu048
Banfalvi, Z., Csakvari, E., Villanyi, V., and Kondrak, M. (2020). Generation of transgene-free PDS mutants in potato by agrobacterium-mediated transformation. BMC Biotechnol. 20, 25. doi:10.1186/s12896-020-00621-2
Bao, H., Ding, Y., Yang, Fei., Zhang, J., Xie, J., Zhao, C., et al. (2022). Gene silencing, knockout and over-expression of a transcription factor ABORTED MICROSPORES (SlAMS) strongly affects pollen viability in tomato (Solanum lycopersicum). BMC Genomics 23, 346. doi:10.1186/s12864-022-08549-x
Bari, V. K., Nassar, J. A., Kheredin, S. M., Gal-On, A., Ron, M., Britt, A., et al. (2019). CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 9, 11438. doi:10.1038/s41598-019-47893-z
Bari, V. K., Nassar, J. A., and Aly, R. (2021). CRISPR/Cas9 mediated mutagenesis of MORE AXILLARY GROWTH 1 in tomato confers resistance to root parasitic weed Phelipanche aegyptiaca. Sci. Rep. 11, 3905. doi:10.1038/s41598-021-82897-8
Bazzano, L. A., He, J., Ogden, L. G., Loria, C. M., Vupputuri, S., Myers, L., et al. (2002). Fruit and vegetable intake and risk of cardiovascular disease in US adults: The first national health and nutrition examination survey epidemiologic follow-up study. Am. J. Clin. Nutr. 76, 93–99. doi:10.1093/ajcn/76.1.93
Beeber, D., and Chain, F. J. J. (2020). crispRdesignR: A versatile guide RNA design package in R for CRISPR/cas9 applications. J. Genomics 8, 62–70. doi:10.7150/jgen.41196
Bertier, L. D., Ron, M., Huo, H., Bradford, K. J., Britt, A. B., and Michelmore, R. W. (2018). High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-Induced modifications of NCED4 in lettuce (Lactuca sativa). G3 (Bethesda) 8, 1513–1521. doi:10.1534/g3.117.300396
Boscaiu, M., and Fita, A. (2020). Physiological and molecular characterization of crop resistance to abiotic stresses. Agronomy 10, 1308. doi:10.3390/agronomy10091308
Breseghello, F., and Coelho, A. S. (2013). Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.) J. Agric. Food Chem. 61, 8277–8286. doi:10.1021/jf305531j
Butler, N. M., Jansky, S. H., and Jiang, J. (2020). First-generation genome editing in potato using hairy root transformation. Plant Biotechnol. J. 18, 2201–2209. doi:10.1111/pbi.13376
Butt, H., Rao, G. S., Sedeek, K., Aman, R., and Mahfouz, M. (2020). Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 18, 2370–2372. doi:10.1111/pbi.13399
Cermak, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High frequency, precise modification of the tomato genome. Genome Biol. 16, 232. doi:10.1186/s13059-015-0796-9
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi:10.1111/mpp.12375
Chen, K., Wang, Y., Zhang, R., Zhang, H., and Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi:10.1146/annurev-arplant-050718-100049
Corte, E.-D., Mahmoud, L. M., Moraes, T. S., Mou, Z., Grosser, J. W., and Dutt, M. (2019). Development of Improved Fruit, Vegetable, and Ornamental Crops Using the CRISPR/Cas9 Genome Editing Technique. Plants 8, 601. doi:10.3390/plants8120601
Chuang, F. Y., Phipps, J. A., Lin, L. F., Hecht, V., Hewitt, W. A., and Wang, Y. P. (2021). Approach for in vivo delivery of CRSIPR/Cas system: a recent update and furture prospect. Cell. Mol. Life Sci. 78, 2683–2708. doi:10.1007/s00018-020-03725-2
D’Ambrosio, C., Giorio, G., and Stigliani, A. L. (2018). Knockout of NPTII marker gene in transgenic tomato plants using the CRISPR/Cas9 system. Transgenic Res. 54, 367–378. doi:10.1007/s11248-018-0079-9
Danilo, B., Perrot, L., Mara, K., Botton, E., Nogue, F., and Mazier, M. (2019). Efficient and transgene-free gene targeting using Agrobacterium-mediated delivery of the CRISPR/Cas9 system in tomato. Plant Cell Rep. 38, 459–462. doi:10.1007/s00299-019-02373-6
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. doi:10.1038/nature09886
Deng, L., Wang, H., Sun, C., Li, Q., Jiang, H., Du, M., et al. (2018). Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J. Genet. Genomics 45, 51–54. doi:10.1016/j.jgg.2017.10.002
Dias, J. (2012). Nutritional quality and health benefits of vegetables: A review. Food Nutr. Sci.Food Nutr. Sci. 3, 1354–1374. doi:10.4236/fns.2012.310179
Ding, F., Wang, M., and Zhang, S. (2018). Sedoheptulose-1, 7-bisphosphatase is involved in methyl jasmonate- and dark-induced leaf senescence in tomato plants. Int. J. Mol. Sci. 19, 3673. doi:10.3390/ijms19113673
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR/Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi:10.1038/nbt.3026
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi:10.1126/science.1258096
Duensing, N., Sprink, T., Parrott, W. A., Fedorova, M., Lema, M. A., Wolt, J. D., et al. (2018). Novel features and considerations for ERA and regulation of crops produced by Genome editing. Front. Bioeng. Biotechnol. 6, 79. doi:10.3389/fbioe.2018.00079
Eckerstorfer, M. F., Engelhard, M., Heissenberger, A., Simon, S., and Teichmann, H. (2019). Plants developed by new genetic modification techniques—Comparison of existing regulatory frameworks in the EU and non-EU countries. Front. Bioeng. Biotechnol. 7, 26. doi:10.3389/fbioe.2019.00026
Filler, H. S., Melamed, B. C., and Levy, A. A. (2017). Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 8, 15605. doi:10.1038/ncomms15605
Fritsche, S., Poovaiah, C., MacRae, E., and Thorlby, G. (2018). A New Zealand perspective on the application and regulation of gene editing. Front. Plant Sci. 9, 1323. doi:10.3389/fpls.2018.01323
Gago, C., Drosou, V., Paschalidis, K., Guerreiro, A., Miguel, G., Antunes, D., et al. (2017). Targeted gene disruption coupled with metabolic screen approach to uncover the LEAFY COTYLEDON1-LIKE4 (L1L4) function in tomato fruit metabolism. Plant Cell Rep. 36, 1065–1082. doi:10.1007/s00299-017-2137-9
Gaj, T., Gersbach, C. A., and Carlos, F. (2013). ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi:10.1016/j.tibtech.2013.04.004
Gallagher, M. J., and Mahajan, P. V. (2011). “Stability and shelf life of fruit and vegetables,” in Food and beverage stability and shelf-life. Editors Kilcast, and Subramaniam (Cambridge, UK: Woodhead Publishing Ltd), 641–656. Chapter 22.
Garcia Ruiz, M. T., Knapp, A. N., and Garcia-Ruiz, H. (2018). Profile of genetically modified plants authorized in Mexico. Gm. Crops Food 9, 152–168. doi:10.1080/21645698.2018.1507601
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 109, E2579–E2586. doi:10.1073/pnas.1208507109
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi:10.1038/nature24644
Gonzalez, M. N., Massa, A. G., Andersson, M., Turesson, H., Olsson, N., Falt, A. S., et al. (2020). Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 9, 1649. doi:10.3389/fpls.2019.01649
Haeussler, M., Schonig, K., Eckert, H., Eschstruth, A., Mianne, J., Renaud, J. B., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. doi:10.1186/s13059-016-1012-2
Haque, E., Taniguchi, H., Hassan, M., Bhowmik, P., Karim, M. R., Smiech, M., et al. (2018). Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front. Plt. Sci. 9, 617. doi:10.3389/fpls.2018.00617d
Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., White, I. P., et al. (2018). Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842. doi:10.1126/science.aav4294
Hedge, N., Joshi, S., Soni, N., and Kushalappa, A. C. (2021). The caffeoyl-CoA O-methyltransferase gene SNP replacement in Russet Burbank potato variety enhances late blight resistance through cell wall reinforcement. Plant Cell Rep. 40, 237–254. doi:10.1007/s00299-020-02629-6
Heigwer, F., Kerr, G., and Boutros, M. (2014). E-CRISP: Fast CRISPR target site identification. Nat. Methods 11, 122–123. doi:10.1038/nmeth.2812
Holme, I. B., Gregersen, P. L., and Brinch-Pedersen, H. (2019). Induced genetic variation in crop plants by random or targeted mutagenesis: Convergence and differences. Front. Plant Sci. 14, 1468. doi:10.3389/fpls.2019.01468
Hooghvorst, I., Lopez-Cristoffanini, C., and Nogues, S. (2019). Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 19, 17077. doi:10.1038/s41598-019-53710-4
Hu, B., Li, D., Liu, X., Qi, J., Gao, D., Zhao, S., et al. (2017). Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant 10, 1575–1578. doi:10.1016/j.molp.2017.09.005
Hu, N., Xian, Z., Li, N., Liu, Y., Huang, W., Yan, F., et al. (2019). Rapid and user-friendly open-source CRISPR/Cas9 system for single- or multi-site editing of tomato genome. Hortic. Res. 6, 7. doi:10.1038/s41438-018-0082-6
Huang, Y., Cao, H., Yang, L., Chen, C., Shabala, L., Xiong, M., et al. (2019). Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae. J. Exp. Bot. 70, 5879–5893. doi:10.1093/jxb/erz328
Hunter, M. C., Smith, R. G., Schipanski, M. E., Atwood, L. W., and Mortensen, D. A. (2017). Agriculture in 2050: Recalibrating targets for sustainable intensification. Bioscience 67, 386–391. doi:10.1093/biosci/bix010
Hwang, G. H., and Bae, S. (2021). Web-Based Base Editing Toolkits: BE-Designer and BE-Analyzer. Methods Mol. Biol. 2189, 81–88. doi:10.1007/978-1-0716-0822-7_7
Hwang, G. H., Park, J., Lim, K., Kim, S., Yu, J., Yu, E., et al. (2018). Web-based design and analysis tools for CRISPR base editing. BMC Bioinforma. 19, 542. doi:10.1186/s12859-018-2585-4
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., Shima, Y., Nakamura, N., et al. (2017). Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 3, 866–874. doi:10.1038/s41477-017-0041-5
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., and Venkataraman, G. (2018). CRISPR for crop improvement: An update review. Front. Plant Sci. 9, 985. doi:10.3389/fpls.2018.00985
Jeon, J. E., Kim, J. G., Fischer, C. R., Mehta, N., Dufour-Schroif, C., Wemmer, K., et al. (2020). A pathogen-responsive gene cluster for highly modified fatty acids in tomato. Cell 180, 176–187. doi:10.1016/j.cell.2019.11.037
Jeong, S. Y., Ahn, H., Ryu, J., Oh, Y., Sivanandhan, G., Won, K. H., et al. (2019). Generation of early-flowering Chinese cabbage (Brassica rapa spp. pekinensis) through CRISPR/Cas9-mediated genome editing. Plant Biotechnol. Rep. 13, 491–499. doi:10.1007/s11816-019-00566-9
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. doi:10.1038/nbt.2508
Jin, S., Zong, Y., Gao, Q., Zhu, Z., Wang, Y., Qin, P., et al. (2019). Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295. doi:10.1126/science.aaw7166
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Johansen, I. E., Liu, Y., Jørgensen, B., Bennett, E. P., Andreasson, E., Nielsen, K. L., et al. (2019). High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 9, 17715. doi:10.1038/s41598-019-54126-w
Jung, Y. J., Lee, G. J., Bae, S., and Kang, K. K. (2018). Reduced ethylene production in tomato fruits upon CRSPR/Cas9-mediated LeMADS-RIN mutagenesis. Hortic. Sci. 36, 396–405. doi:10.12972/kjhst.20180039
Karkute, S. G., Singh, A. K., Gupta, O. P., Singh, P. M., and Singh, B. (2017). CRISPR/Cas9 Mediated Genome Engineering for Improvement of Horticultural Crops. Front Plant Sci. 22, 1635. doi:10.3389/fpls.2017.01635
Kaur, K., Tandon, H., Gupta, A. K., and Kumar, M. (2015). CrisprGE: A central hub of CRISPR/Cas-based genome editing. Database (Oxford) 27, bav055. doi:10.1093/database/bav055
Khan, M. S. S., Basnet, R., Islam, S. A., and Shu, Q. (2019a). Mutational analysis of OsPLDα1 reveals its involvement in phytic acid biosynthesis in rice grains. J. Agric. Food Chem. 41, 11436–11443. doi:10.1021/acs.jafc.9b05052
Khan, M. Z., Haider, S., Mansoor, S., and Amin, I. (2019b). Targeting plant ssDNA viruses with engineered miniature CRISPR-Cas14a. Trends Biotechnol. 37, 800–804. doi:10.1016/j.tibtech.2019.03.015
Khromov, A. V., Gushchin, V. A., Timberbaev, V. I., Kalinina, N. O., Taliansky, M. E., and Makarov, V. V. (2018). Guide RNA design for CRISPR/Cas9-mediated potato genome editing. Dokl. Biochem. Biophys. 479, 90–94. doi:10.1134/s1607672918020084
Kieu, N. P., Lenman, M., Wang, E. S., Petersen, L. B., and Andreasson, E. (2021). Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 11, 4487. doi:10.1038/s41598-021-83972-w
Kim, H., Kim, S. T., Ryu, J., Kang, B. C., Kim, J. S., and Kim, S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8, 14406. doi:10.1038/ncomms14406
Klap, C., Yeshayahou, E., Bolger, A. M., Arazi, T., Gupta, S. K., Shabtai, S., et al. (2017). Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 15, 634–647. doi:10.1111/pbi.12662
Klimek-Chodacka, M., Oleszkiewicz, T., Lowder, L. G., Qi, Y., and Baransk, R. (2018). Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 37, 575–586. doi:10.1007/s00299-018-2252-2
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double stranded DNA cleavage. Nature 533, 420–424. doi:10.1038/nature17946
Kumlehn, J., Pietralla, J., Hensel, G., Pacher, M., and Puchta, H. (2018). The CRISPR/Cas revolution continues: From efficient gene editing for crop breeding to plant synthetic biology. J. Integr. Plant Biol. 60, 1127–1153. doi:10.1111/jipb.12734
Kusano, H., Ohnuma, M., Mutsuro-Aoki, H., Asahi, T., Ichinosawa, D., Onodera, H., et al. (2018). Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci. Rep. 8, 13753. doi:10.1038/s41598-018-32049-2
Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B., and Valen, E. (2016). CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276. doi:10.1093/nar/gkw398
Langner, T., Kamoun, S., and Belhaj, K. (2018). CRISPR Crops: Plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. doi:10.1146/annurev-phyto-080417-050158
Li, Z., Liu, Z. B., Xing, A., Moon, B. P., Koellhoffer, J. P., Huang, L., et al. (2015). Cas9-Guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. doi:10.1104/pp.15.00783
Li, R., Zhang, L., Wang, L., Chen, L., Zhao, R., Sheng, J., et al. (2018a). Reduction of tomato-plant chilling tolerance by CRISPR-Cas9- mediated SlCBF1 mutagenesis. J. Agric. Food Chem. 66, 9042–9051. doi:10.1021/acs.jafc.8b02177
Li, R., Fu, D., Zhu, B., Luo, Y., and Zhu, H. (2018b). CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 94, 513–524. doi:10.1111/tpj.13872
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018c). Lycopene is enriched in tomato fruit by CRISPR/Cas9 mediated multiplex genome editing. Front. Plant Sci. 1-12, 559. doi:10.3389/fpls.2018.00559
Li, R., Li, R., Li, X., Fu, D., Zhu, B., Tian, H., et al. (2018d). Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16, 415–427. doi:10.1111/pbi.12781
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018e). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. doi:10.1038/nbt.4273
Li, R., Liu, C., Zhao, R., Wang, L., Chen, L., Sheng, J., et al. (2019). CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19, 38. doi:10.1186/s12870-018-1627-4
Li, S., Zhu, B., Pirello, J., Xu, C., Zhang, B., Bouzayen, M., et al. (2020a). Roles of RIN and ethylene in tomato fruit ripening and ripening-associated traits. New Phytol. 226, 460–475. doi:10.1111/nph.16362
Li, H., Li, X., Xu, Y., Liu, H., He, M., Tian, X., et al. (2020c). High-efficie.ncy reduction of rice amylose content via CRISPR/Cas9-mediated base editing. Rice Sci. 27, 445–448. doi:10.1016/j.rsci.2020.09.001
Lino, C. A., Harper, J. C., Carney, J. P., and Timlin, J. A. (2018). Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25, 1234–1257. doi:10.1080/10717544.2018.1474964
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., and Chen, L. L. (2017). CRISPR-P 2.0: An improved CRISPR/Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi:10.1016/j.molp.2017.01.003
Liu, M., Rehman, S., Tang, X., Gu, K., Fan, Q., Chen, D., et al. (2019). Methodologies for improving HDR efficiency. Front. Genet. 9, 691–699. doi:10.3389/fgene.2018.00691
Liu, X., Zhang, Q., Yang, G., Zhang, C., Dong, H., Liu, Y., et al. (2020). Pivotal roles of Tomato photoreceptor SlUVR8 in seedling development and UV-B stress tolerance. Biochem. Biophys. Res. Commun. 522, 177–183. doi:10.1016/j.bbrc.2019.11.073
Ma, C., Liu, M., Li, Q., Si, J., Ren, X., and Song, H. (2019). Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 system. Hortic. Plant J. 5, 164–169. doi:10.1016/j.hpj.2019.04.001
Maioli, A., Gianoglio, S., Moglia, A., Acquadro, A., Valentino, D., Milani, A. M., et al. (2020). Simultaneous CRISPR/Cas9 editing of three PPO genes reduces fruit flesh browning in Solanum melongena L. Front. Plant Sci. 11, 607161. doi:10.3389/fpls.2020.607161
Makhotenko, A. V., Khromov, A. V., Snigir, E. A., Makarova, S. S., Makarov, V. V., Suprunova, T. P., et al. (2019). Functional analysis of coilin in virus resistance and stress tolerance of potato Solanum tuberosum using CRISPR-cas9 editing. Dokl. Biochem. Biophys. 484, 88–91. doi:10.1134/S1607672919010241
Mei, Y., Wang, Y., Chen, H., Sun, Z. S., and Da, J. X. (2016). Recent progress in CRISPR/Cas9 technology. J. Genet. Genomics. 43, 63–75. doi:10.1016/j.jgg.2016.01.001
Miller, J. C., tis, M. C., Wang, J. B., Guschin, D. Y., Lee, Y. L., Rupniewski, I., et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25, 778–785. doi:10.1038/nbt1319
Minkenberg, B., Zhang, J., Xie, K., and Yang, Y. (2019). CRISPR-PLANT v2: An online resource for highly specific guide RNA spacers based on improved off-target analysis. Plant Biotechnol. J. 17, 5–8. doi:10.1111/pbi.13025
Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M., and Valen, E. (2014). CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, 401–407. doi:10.1093/nar/gku410
Moon, S. B., Lee, J. M., Kang, J. G., Lee, N. E., Ha, D. I., Kim, D. Y., et al. (2018). Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3’-overhang. Nat. Commun. 9, 3651. doi:10.1038/s41467-018-06129-w
Moreno-Mateos, M. A., Vejnar, C. E., Beaudoin, J. D., Fernandez, J. P., Mis, E. K., Khokha, M. K., et al. (2015). CRISPRscan: Designing highly efficient sgRNAs for CRISPR/Cas9 targeting in vivo. Nat. Methods 12, 982–988. doi:10.1038/nmeth.3543
Naito, Y., Hino, K., Bono, H., and Ui-Tei, K. (2014). CRISPRdirect: Software for designing CRISPR/cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi:10.1093/bioinformatics/btu743
Nakayasu, M., Akiyama, R., Lee, J. H., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. biochem. 131, 70–77. doi:10.1016/j.plaphy.2018.04.026
Nature Plants Editorial (2018). A CRISPR definition of genetic modification. Nat. Plants 4, 233. doi:10.1038/s41477-018-0158-1
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482. doi:10.1038/s41598-017-00578-x
Niazian, M., Sadatnoori, S. A., Galuszka, P., and Mortazavian, S. M. M. (2017). Tissue culture-based Agrobacterium-mediated and in Planta transformation methods. Czech J. Genet. Plant Breed. 53, 133–143. doi:10.17221/177/2016-CJGPB
Nonaka, M., Arai, M., akayama, M., Matsukura, M., and Ezura, M. (2017). Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7, 7057. doi:10.1038/s41598-017-06400-y
Nunez de Caceres Gonzalez, F. F., and De la Mora Franco, D. (2020). “Vegetable crop improvement using CRISPR/Cas9,” in CRISPR/Cas genome editing: Strategies and Potential for crop improvement. Editors A. Bhattacharya, V. Parkhi, and B. Char (Cham: Springer International Publishing), 119–129.
O’Driscoll, M., and Jeggo, P. A. (2006). The role of double-strand break repair—Insights from human genetics. Nat. Rev. Genet. 7, 45–54. doi:10.1038/nrg1746
Oliveros, J. C., Franch, M., Tabas-Madrid, D., San-Leon, D., Montoliu, L., Cubas, P., et al. (2016). Breaking-Cas—Interactive design of guide RNAs for CRISPR-cas experiments for ENSEMBL genomes. Nucleic Acids Res. 44, W267–W271. doi:10.1093/nar/gkw407
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17, 665–673. doi:10.1111/pbi.13006
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6, 24765. doi:10.1038/srep24765
Parihar, P., Singh, S., Singh, R., Singh, V. P., and Prasad, S. M. (2015). Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. Int. 22, 4056–4075. doi:10.1007/s11356-014-3739-1
Park, J., and Bae, S. (2017). Cpf1-database: Web-based genome-wide guide RNA library design for gene knockout screens using CRISPR-cpf1. Bioinformatics 34, 1077–1079. doi:10.1093/bioinformatics/btx695
Park, J., Kim, J. S., and Bae, S. (2016). Cas-database: Web-based genome-wide guide RNA library design for gene knockout screens using CRISPR/Cas9. Bioinformatics 32, 2017–2023. doi:10.1093/bioinformatics/btw103
Parry, M. A. J., Madgwick, P. J., Bayon, C., Tearall, K., Hernandez-Lopez, A., Baudo, M., et al. (2009). Mutation discovery for crop improvement. J. Exp. Bot. 60, 2817–2825. doi:10.1093/jxb/erp189
Paul, H. M., Istanto, D. D., Heldenbrand, J., and Hudson, M. E. (2022). Cropsr: An automated platform for complex genome-wide CRISPR gRNA design and validation. BMC Bioinforma. 23, 74. doi:10.1186/s12859-022-04593-2
Perez, A. R., Pritykin, Y., Vidigal, J. A., Chhangawala, S., Zamparo, L., Leslie, C. S., et al. (2017). GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 35, 347–349. doi:10.1038/nbt.3804
Petretto, G. L., Urgeghe, P. P., Massa, D., and Melito, S. (2019). Effect of salinity (NaCl) on plant growth, nutrient content, and glucosinolate hydrolysis products trends in rocket genotypes. Plant Physiol. biochem. 141, 30–39. doi:10.1016/j.plaphy.2019.05.012
Pliatsika, V., and Rigoutsos, I. (2015). Off-spotter”: Very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/cas guide RNAs. Biol. Direct 10, d4. doi:10.1186/s13062-015-0035-z
Poudel, R., Rodriguez, L. T., Reisch, C. R., and Rivers, A. R. (2022). GuideMaker: Software to design CRISPR-Cas guide RNA pools in non-model genomes. Gigascience 11, giac007–8. doi:10.1093/gigascience/giac007
Pramanik, D., Shelake, R. M., Park, J., Kim, M. J., Hwang, I., Park, Y., et al. (2021). CRISPR/Cas9-Mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int. J. Mol. Sci. 22, 1878. doi:10.3390/ijms22041878
Prihatna, C., Barbetti, M. J., and Barker, S. J. (2018). A novel tomato Fusarium wilt tolerance gene. Front. Microbiol. 9, 1226. doi:10.3389/fmicb.2018.01226
Prykhozhij, S. V., Rajan, V., Gaston, D., and Berman, J. N. (2015). CRISPR multitargeter: A web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS One 10, e0119372. doi:10.1371/journal.pone.0119372
Rodriguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480. doi:10.1016/j.cell.2017.08.030
Roldan, M. V. G., Perilleux, C., Morin, H., Huerga-Fernandez, S., Latrasse, D., Benhamed, M., et al. (2017). Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 7, 4402. doi:10.1038/s41598-017-04556-1
Rouillon, C., Zhou, M., Zhang, J., Politis, A., Beilsten-Edmands, V., Cannone, G., et al. (2013). Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell 52, 124–134. doi:10.1016/j.molcel.2013.08.020
Schindele, P., and Puchta, H. (2020). Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol. J. 8, 1118–1120. doi:10.1111/pbi.13275
Sevestre, F., Facon, M., Fabrice Wattebled, F., and Nicolas Szydlowski, N. (2020). Facilitating gene editing in potato: A single-nucleotide polymorphism (SNP) map of the Solanum tuberosum L. Cv. Desiree genome. Sci. Rep. 10, 2045. doi:10.1038/s41598-020-58985-6
Shao, M., Xu, T., and Chen, C. (2016). The big bang of genome editing technology: Development and application of the CRISPR/CAS9 system in disease animal models. Sci. Press Zool. Res. 37, 191–204. doi:10.13918/j.issn.2095-8137.2016.4.191
Sharma, D., Verma, N., Pandey, C., Verma, D., Bhagat, P., Noryang, S., et al. (2020). “MAP kinase as regulators for stress responses in plants: An overview,” in book: Protein kinases and stress signaling in plants, 369–392. doi:10.1002/9781119541578.ch15
Shuman, S., and Glickman, M. S. (2007). Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 5, 852–861. doi:10.1038/nrmicro1768
Siegner, S. M., Karasu, M. E., Schroder, M. S., Kontarakis, Z., and Corn, J. E. (2021). PnB designer: A web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinforma. 22, 101. doi:10.1186/s12859-021-04034-6
Smyth, S. J. (2017). Canadian regulatory perspectives on genome engineered crops. Gm. Crops Food 8, 35–43. doi:10.1080/21645698.2016.1257468
Soyk, S., Muller, N., Ju Park, S., Schmalenbach, I., Jiang, K., Hayama, R., et al. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49, 162–168. doi:10.1038/ng.3733
Sun, Q., Lin, L., Liu, D., Wu, D., Fang, Y., Wu, J., et al. (2018). CRISPR/Cas9-Mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 19, 2716. doi:10.3390/ijms19092716
Sun, J., Liu, H., Liu, J., Cheng, S., Peng, Y., Zhang, Q., et al. (2019). CRISPR-local: A local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics 35 (14), 2501–2503. doi:10.1093/bioinformatics/bty970
Tashkandi, M., Ali, Z., Aljedaani, F., Shami, A., and Mahfouz, M. M. (2018). Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 13, e1525996. doi:10.1080/15592324.2018.1525996
Teng, F., Cui, T., Feng, G., Guo, L., Xu, K., Gao, Q., et al. (2018). Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 4, 63. doi:10.1038/s41421-018-0069-3
Thomazella, P. d. T., Brail, Q., Dahlbeck, D., and Staskawicz, B. (2016). “CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance,” in Proceedings of the national academy of sciences (Berkeley, CA: bioRxiv), 1–23. doi:10.1101/064824:064824
Thysegen, P. (2019). Clarifying the regulation of genome editing in Australia: Situation for genetically modified organisms. Transgenic Res. 28, 151–159. doi:10.1007/s11248-019-00151-4
Tian, S., Jiang, L., Gao, Q., Zhang, J., Zong, M., Zhang, H., et al. (2017). Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 36, 399–406. doi:10.1007/s00299-016-2089-5
Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., et al. (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 37, 1353–1356. doi:10.1007/s00299-018-2299-0
Tian, S. W., Xing, S. N., and Yong, X. (2021). Advances in CRISPR/Cas9-mediated genome editing on vegetable crops. Vitro Cell. Dev. Biol. -Plant. 4, 672–682. doi:10.1007/s11627-021-10187-z
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U. S. A. 108, 20260–20264. doi:10.1073/pnas.1116437108
Tomilson, L., Yang, Y., Emenecker, R., Smoker, M., Taylor, J., Parkins, S., et al. (2019). Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 17, 132–140. doi:10.1111/pbi.12952
Tran, M. T., Doan, D. T. H., Kim, J., Song, Y. J., Sung, Y. W., Das, S., et al. (2021). CRISPR/Cas9-based precise excision of SlHyPRP1 domain(s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 40, 999–1011. doi:10.1007/s00299-020-02622-z
Ueta, R., Takahito Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., Osakabe, Y., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7, 507. doi:10.1038/s41598-017-00501-4
Van Vu, T., Sung, Y. W., Kim, J., Doan, D. T. H., Tran, M. T., and Kim, J.-Y. (2019). Challenges and perspectives in homology-directed gene targeting in monocot plants. Rice 12, 95. doi:10.1186/s12284-019-0355-1
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M. P., Guyon-Debast, A., Chauvin, J. E., et al. (2019). Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20, 402. doi:10.3390/ijms20020402
Vu, T. V., Sivankalyani, V., Kim, E. J., Doan, D. T. H., Tran, M. T., Kim, J., et al. (2020). Highly efficient homology-directed repair using CRISPR/Cpf1-gemini viral replicon in tomato. Plant Biotechnol. J. 18, 2133–2143. doi:10.1111/pbi.13373
Waltz, E. (2016a). CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 34, 582. doi:10.1038/nbt0616-582
Waltz, E. (2016b). Gene-edited CRISPR mushroom escapes US regulation. Nature 532, 293. doi:10.1038/nature.2016.19754
Wang, L., Chen, L., Li, R., Zhao, R., Yang, M., Sheng, J., et al. (2017). Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 65, 8674–8682. doi:10.1021/acs.jafc.7b02745
Wang, X. L. Y., Chen, S., Tian, H., Fu, D., Zhu, B., Luo, Z. Y., et al. (2018a). Lycopene is enriched in tomato fruit by CRISPR/Cas9-Mediated multiplex genome editing. Front. Plant Sci. 26, 559. doi:10.3389/fpls.2018.00559
Wang, D., Samsul Rizal, N., Yan, C., Allcock, N., Craigon, J., Blanco-Ulate, B., et al. (2018b). Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179, 544–557. doi:10.1104/pp.18.01187
Wang, T., Zhang, H., and Zhu, H. (2019a). CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 6, 77. doi:10.1038/s41438-019-0159-x
Wang, R., Tavano, E. C. D., Lammers, M., Martinelli, A. P., Angenent, G. C., and Maagd, R. A. (2018b). Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 9, 1696. doi:10.1038/s41598-018-38170-6
Warrier, R., and Pande, H. (2016). Genetically engineered plants in the product development pipeline in India. Gm. Crops Food 7, 12–19. doi:10.1080/21645698.2016.1156826
Whelan, A. I., and Lema, M. A. (2015). Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. Gm. Crops Food 6, 253–265. doi:10.1080/21645698.2015.1114698
Wolter, F., and Puchta, H. (2018). The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 94, 767–775. doi:10.1111/tpj.13899
Wong, N., Liu, W., and Wang, X. (2015). Wu-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218. doi:10.1186/s13059-015-0784-0
Wu, F., Qiao, X., Zhao, Y., Zhang, Z., Gao, Y., Shi, L., et al. (2020). Targeted mutagenesis in Arabidopsis thaliana using CRISPR-Cas12b/C2c1. J. Integr. Plant Biol. 11, 1653–1658. doi:10.1111/jipb.12944
Xie, X., Li, F., Tan, X., Zeng, D., Liu, W., Zeng, W., et al. (2022). BEtarget: A versatile web-based tool to design guide RNAs for base editing in plants. Comput. Struct. Biotechnol. J. 20, 4009–4014. doi:10.1016/j.csbj.2022.07.046
Xu, R., Qin, R., Li, H., Li, D., Li, L., Wei, P., et al. (2017). Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 15, 713–717. doi:10.1111/pbi.12669
Xu, J., Kang, B. C., Naing, A. H., Bae, S. J., Kim, J. S., Kim, H., et al. (2019a). CRISPR/Cas9-mediated editing of 1-aminocyclopropane-1-carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol. J. 18, 287–297. doi:10.1111/pbi.13197
Xu, Z. S., Feng, K., and Xiong, A. S. (2019b). CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol. Biotechnol. 61, 191–199. doi:10.1007/s12033-018-00150-6
Yang, Y., Zhu, G., Li, R., Yan, S., Fu, D., Zhu, B., et al. (2017). The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 175, 1690–1702. doi:10.1104/pp.17.01265
Yang, H., Ren, S., Yu, S., Pan, H., Li, T., Ge, S., et al. (2020). Methods favoring homology-directed repair choice in response to CRISPR/cas9 induced-double strand breaks. Int. J. Mol. Sci. 21, 6461. doi:10.3390/ijms21186461
Yang, S. H., Kim, E., Park, H., and Koo, Y. (2022). Selection of the high efficient sgRNA for CRISPR-Cas9 to edit herbicide related genes, PDS, ALS, and EPSPS in tomato. Appl. Biol. Chem. 65, 13. doi:10.1186/s13765-022-00679-w
Yin, Y., Qin, K., Song, X., Zhang, Q., Zhou, Y., Xia, X., et al. (2018). BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 59, 2239–2254. doi:10.1093/pcp/pcy146
Yu, Q. H., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long shelf-life tomato lines. Sci. Rep. 7, 11874. doi:10.1038/s41598-017-12262-1
Yu, W., Wang, L., Zhao, R., Shen, G. J., Zhang, S., Li, R., et al. (2019). Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 19, 354. doi:10.1186/s12870-019-1939-z
Zhan, X., Zhang, F., Zhong, Z., Chen, R., Wang, Y., Chang, L., et al. (2019). Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 17, 1814–1822. doi:10.1111/pbi.13102
Zhang, Y., Karen, M., Ian, D. G., and Caixia, G. (2018). Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210. doi:10.1186/s13059-018-1586-y
Zhang, J., Guo, S., Ji, G., Zhao, H., Sun, H., Ren, Y., et al. (2019a). A unique chromosome translocation disrupting ClWIP1 leads to gynoecy in watermelon. Plant J. 101, 265–277. doi:10.1111/tpj.14537
Zhang, M., Liu, Q., Yang, X., Xu, J., Liu, G., Yao, X., et al. (2020). CRISPR/Cas9-mediated editing of ClPSK1 enhanced watermelon resistance to Fusarium oxysporum. Plant Cell Rep. 39, 589–595. doi:10.1007/s00299-020-02516-0
Zhao, X., Jayarathna, S., Turesson, H., Falt, A-S., Nestor, G., Gonzalez, M. N., et al. (2021). Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci. Rep. 11, 4311. doi:10.1038/s41598-021-83462-z
Zheng, N. Z., Martin, G. B., and Fei, Z. (2019). Plant genome editing database (pged): A call for submission of information about genome-edited plant mutants. Mol. Plant 12, 127–129. doi:10.1016/j.molp.2019.01.001
Zhu, L. J., Holmes, B. R., Aronin, N., and Brodsky, M. H. (2014). CRISPRseek: A bioconductor package to identify target-specific guide RNAs for CRISPR/Cas9 genome-editing systems. PLoS One 9, e108424. doi:10.1371/journal.pone.0108424
Zhu, H., Li, C., and Gao, C. (2020). Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. doi:10.1038/s41580-020-00288-9
Keywords: genome-editing technology, CRISPR-cas application, vegetable crops, advanced, cutting-edge
Citation: Devi R, Chauhan S and Dhillon TS (2022) Genome editing for vegetable crop improvement: Challenges and future prospects. Front. Genet. 13:1037091. doi: 10.3389/fgene.2022.1037091
Received: 05 September 2022; Accepted: 28 October 2022;
Published: 22 November 2022.
Edited by:
Sunil S. Gangurde, University of Georgia, United StatesReviewed by:
Abira Chaudhuri, National Institute of Plant Genome Research (NIPGR), IndiaCopyright © 2022 Devi, Chauhan and Dhillon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruma Devi, cnVtYWRldmlAcGF1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.