- 1Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou, China
- 2Institute of Horticultural Sciences, University of Agriculture Faisalabad, Faisalabad, Pakistan
- 3Faculty of Agriculture and Veterinary Sciences, Superior University, Lahore, Pakistan
- 4The Key Laboratory for Crop Germplasm Resource of Zhejiang Province, Hangzhou, China
Salicylic acid (SA) is a well-studied phenolic plant hormone that plays an important role in plant defense against the hemi-biothrophic and biothrophic pathogens and depends on the living cells of host for the successful infection. In this study, a pathogenesis test was performed between Vitis davidii and V. vinifera cultivars against grape white rot disease (Coniella diplodiella). V. davidii was found to be resistant against this disease. SA contents were found to be higher in the resistant grape cultivar after different time points. RNA-seq analysis was conducted on susceptible grapevine cultivars after 12, 24, and 48 h of SA application with the hypothesis that SA may induce defense genes in susceptible cultivars. A total of 511 differentially expressed genes (DEGs) were identified from the RNA-seq data, including some important genes, VvWRKY1/2, VvNPR1, VvTGA2, and VvPR1, for the SA defense pathway. DEGs related to phytohormone signal transduction and flavonoid biosynthetic pathways were also upregulated. The quantitative real-time PCR (qRT-PCR) results of the significantly expressed transcripts were found to be consistent with the transcriptome data, with a high correlation between the two analyses. The pathogenesis-related gene 1 (VvPR1), which is an important marker gene for plant defense, was selected for further promoter analysis. The promoter sequence showed that it contains some important cis-elements (W-box, LS7, as-1, and TCA-element) to recruit the transcription factors VvWRKY, VvNPR1, and VvTGA2 to express the VvPR1 gene in response to SA treatment. Furthermore, the VvPR1 promoter was serially deleted into different fragments (−1,837, −1,443, −1,119, −864, −558, −436, and −192 ) bp and constructed vectors with the GUS reporter gene. Deletion analysis revealed that the VvPR1 promoter between −1837 bp to −558 bp induced significant GUS expression with respect to the control. On the basis of these results, the −558 bp region was assumed to be an important part of the VvPR1 promoter, and this region contained the important cis-elements related to SA, such as TCA-element (−1,472 bp), LS7 (−1,428 bp), and as-1 (−520 bp), that recruit the TFs and induce the expression of the VvPR1 gene. This study expanded the available information regarding SA-induced defense in susceptible grapes and recognized the molecular mechanisms through which this defense might be mediated.
Introduction
Grapevine (Vitis spp., family Vitaceae) is a commercially important fruit grown around the globe, and its history extends over 8,000 years (Fischer et al., 2004). Currently, most worldwide production is from the European grapevine (V. vinifera L.), which is the main table grape species in China. Because of the character of the East Asiatic climate (monsoon) with high precipitation and temperature, grapes are vulnerable to different fungal diseases (Wan et al., 2015; Pap et al., 2016; Sapkota et al., 2019). A major fungal disease affecting grapevine is grape white rot (caused by Coniella diplodiella (Speg.) Sacc.), which reduced the grape yield by at least 16.3% in grape-producing regions (Li et al., 2008). White rot disease infects the leaves, berries, and new shoots. The application of antifungal agents is recommended for successful grape production, but those agents have hazardous effects on the environment. At present, many grapevine cultivars are found resistant to grape white rot, especially the wild grape relative, making it an important source for grape white resistance through breeding.
Plants have sophisticated defense mechanisms for pathogen recognition. In the systemic acquired resistance (SAR) pattern recognition receptor (PRR)-triggered immunity, PTI is the first tier of plant immunity. PTI is governed by the recognition of pathogen-associated molecular patterns (PAMPs) and is very effective against most pathogens (Boller and Felix, 2009; Dodds and Rathjen, 2010; Dou and Zhou, 2012). However, to overcome the PTI, pathogens manufacture effector proteins and penetrate them in into the host cell to increase their survival in the host. Then, the plant mediates its second tier of immunity, effector-triggered immunity (ETI), to respond to the effector-triggered susceptibility through their resistance genes by recognizing pathogen effectors (van Loon et al., 2006; Wang et al., 2012; Sawinski et al., 2013). Pathogenesis-related proteins (PRs) are activated by abiotic and biotic factors and play vital roles in plant defense (Sels et al., 2008); the expression of PRs can increase plant resistance following infection by pathogenic bacteria (Wildermuth et al., 2001). A major protein in the PR family is PR1, that responds to disease resistance in plants against the environmental stress (Godiard et al., 1990). Jasmonic acid (JA) and SA perform a vital role in plant SAR through PR1 proteins, different transcription factors, and enzymes that control the expression of PR1 in these pathways (Després et al., 2000; Gu et al., 2002; Després et al., 2003; Lorenzo et al., 2004). It was also found that methyl jasmonate (MeJA) induced the defense genes expression through the MAPK pathway in grapevine (Rahman et al., 2022). PR1 proteins are considered to be antioomycete and antifungal proteins, and the function of PR1 is still enigmatic, in contrast to other PR proteins whose functions have been elucidated (Joshi et al., 2021; Kaur et al., 2022). Recently, tomato and tobacco PR1 protein sterol-binding activity has been found critical for its antimicrobial activity (Gamir et al., 2017). Plant resistance against pathogenic oomycetes and bacteria in tobacco has been found higher from the overexpression of the pepper basic PR1 gene (Sarowar et al., 2005). The promoters of pathogenesis-related genes contained some cis-regulatory elements responsible for their expression upon phytohormones signaling. For example, there are some SA-inducible cis-regulatory elements in promoters of some pathogenesis-related genes in Arabidopsis thaliana PR-1 and tobacco PR-1a, like activation sequence-1 (as-1). In the promoter of PR-1a, tobacco possesses an as-1-like element with inverted TGACG motifs, a binding site for TGA transcription factors (Strompen et al., 1998; Niggeweg et al., 2000; Grüner et al., 2003). The promoters of the 39 BR signaling genes are involved in various regulatory mechanisms and interdependent processes that influence growth, development, and stress response in rice (Ahmar and Gruszka, 2022).
SA is a well-studied phenolic plant hormone that plays an important role in plant defense against the hemi-biothrophic and biothrophic pathogens that depend on the living cells of host for the successful infection. Neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants (Vlot et al., 2009; Raza et al., 2022). SA biosynthesis may activate the resistance genes that initiate SAR through hypersensitive response by accumulating at the place of pathogen attack. They are then distributed to other plant parts as a mobile signal in the form of methyl salicylate to induce the defense response (Zhao et al., 2005). Many plants cannot effectively employ these mechanisms. It was found that exogenous SA application has induced PR proteins (Raskin, 1992), as SA effectiveness has been verified against bacteria (Mohan Babu et al., 2003; Mandal et al., 2013), fungi (He and Wolyn, 2005; Mandal et al., 2009), and viruses (Van Huijsduijnen et al., 1986). SA treatment in tobacco reduced the multiplication of bacteria and disease symptoms against Erwinia carotovora (Palva et al., 1994). The exogenous treatment of SA enhances the resistance of asparagus against Fusarium oxysporum f. sp. asparagi, with increases in the levels of lignifications, phenylalanine ammonia-lyase, and peroxidases (He and Wolyn, 2005). Similarly, in tomato roots, SA application induced the defense response against the F. oxysporum f. sp. Lycopersici and reduced vascular browning symptoms by increasing phenylalanine ammonia-lyase, peroxidases, β-1,3-glucanase, and lignifications (He and Wolyn, 2005; Mandal et al., 2009). PR proteins were also induced by the SA application in grapevine leaves [10]. The PR1 expression induced through exogenous application of SA against the alfalfa mosaic virus (A1MV) (Van Huijsduijnen et al., 1986). The SA master regulator NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1), degraded from oligomer to monomer with the elevation of SA accumulation in the infected cell through NPR3 and NPR4, which results in the effector-triggered cell death; NPR1 also mediated SA resistance in the neighboring cells to promote cell survival (Fu and Dong, 2013; Yan and Dong, 2014). In addition, SA signaling engages a feedback circuit to amplify defense responses that are negatively regulated by EDR1, a MAPKKK (Frye et al., 2001; Xiao et al., 2005).
Previously, researchers analyzed the individual mechanisms for the evaluation of induced resistance involved in stress response. However, these techniques provide limited explanations for the defense mechanisms promoted by the elicitors. To evaluate the elicitors in plant defense, a large number of studies have been conducted on gene expression in response to different phytohormones. A transcriptome study performed on sorghum with exogenous SA treatment induced numerous defense genes, such as several PR genes, the JA pathway, and phenylpropanoid, that exhibit different pattern of synergistic as well as antagonistic effects between SA and JA. Exogenous SA application of on grapevine leaves induced different PR proteins (Renault et al., 1996), but no transcriptome study has been reported on the grapevine leaves in SA application. Our study compared the pathogenesis of grape white rot disease in V. vinifera L. cv. Zaotianmeiguixiang and Chinese wild grape species V. davidii, in which the wild grape cultivar showed resistance against white rot with higher levels of SA. Transcriptome analysis was performed on susceptible grapevine leaves against the SA treatment to investigate the important transcripts responsible for the defense response. This study was proposed to investigate the effect of exogenous SA application on the susceptible grapevine cultivar through the induction of SA marker genes by transcriptome analysis. For further functional analysis, a candidate gene, PR1, was selected for promoter analysis against the SA treatment through transient expression on tobacco leaves.
Materials and methods
Plant material
Grapevine 2-year-old “resistant” V. davidii accession 0940 and “susceptible” V. vinifera (Zaotianmeiguixiang) plants were grown in a greenhouse under controlled conditions (25 ± 5°C, 16-hour light/8-hour dark photoperiod, 65% relative humidity) at the Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences (CAAS), Henan, China. Sand and peat (50/50, v/v) used as potting media, and the plants were watered twice in a week. Tobacco (Nicotiana benthamiana) plants were grown under greenhouse conditions (16-hour light/8-hour dark photoperiod, 65% relative humidity, 25 ± 5 °C) in potting media (vermiculite/perlite/moss, 2/3/5, v/v/v). The N. benthamiana plants were used for the Agrobacterium-mediated transient assay at the sixth-leaf stage.
Pathogenicity test and SA treatment
For the pathogenicity test, the causal organism of white rot (C. diplodiella, strain WR01) was taken from the Institute of Plant Protection, CAAS, and grown at 28°C on potato dextrose agar medium. The plants were inoculated with four mycelium gelose discs (diameter = 2 mm) of C. diplodiella on each leaf using small pins, and the leaf was covered with a plastic bag to retain the moisture throughout the infection period. After 72 h of post-inoculation (hpi), leaf samples were observed. Each treatment had three control replicates and four independent biological infected replicates. The susceptible V. vinifera (Zaotianmeiguixiang) plants were sprayed with 100 μM SA (Sigma-Aldrich Chemicals GmbH, Schnelldorf, Germany) solution containing 0.05% (v/v) Tween 20 treatment, and control plants were sprayed with 0.05% (v/v) Tween 20 (Yang et al., 2019). Three replications were taken for each treatment; each replication contained three leaf samples, and leaf samples were collected after 12, 24, and 48 h of treatment and immediately stored at −80°C before RNA extraction.
Salicylic acid measurement
Grapevine leaves were collected after 12, 48, and 72 h of after the white rot (C. diplodiella, strain WR01) inoculation and kept in liquid nitrogen. SA was measured according to the procedure detailed in Hu et al. (2019). A triple-quadrupole LC/MS system (1290 Infinity II-6470, Agilent Technologies, USA) was used for the measurement of SA content, as explained by Hu et al. (2019). Three independent replicates were used for this experiment.
Total RNA extraction, library construction of mRNA, and data analysis
The total RNA was extracted following the CTAB-pBIOZOL reagent and ethanol precipitation protocol recommended by the manufacturer (Mu et al., 2017). The mRNA was purified using oligo(dt) attached to magnetic beads. The mRNA was fragmented into small fragments with the fragment buffer at the appropriate temperature. The first strand of cDNA was synthesized through reverse transcription with a random hexamer primer; then second-strand cDNA was synthesized. A-Tailing Mix and index adapters were added to the mixture for end repairing of RNA. Previously synthesized cDNA fragments were amplified through PCR, purified by Ampure XP beads, and dissolved in the EB solution. The quality control of the product was validated on the Agilent Technologies 2100 bioanalyzer. For the final library, the double-stranded PCR product was denatured through heating and circularized by the splint oligo sequence. The single strand circular DNA (ssDNA) was considered as the final library. The final library was amplified with phi29 to make DNA nanoball (DNB), which had 300 copies of one molecule. DNBs were loaded into the patterned nanoarray, and pair end 100 bases reads were produced on DNBSeq platform (BGI-Shenzhen, China). This project used SOAPnuke (v1.4.0), the filtering software that was independently developed by BGI. First, the reads containing the unknown base N content greater than 5% and the reads containing the connector (connector contamination) were removed. The low-quality reads (those with a quality value of less than 15, which account for more than 20% of the total number of bases in the reads) were also removed. The filtered “clean reads” are saved in FASTQ format. To compare the RNA-seq reads with the reference genome of Vitis vinifera (http://plants.ensembl.org/Vitis_vinifera/Info/Index?db=core;g=VIT_08s0007g00570;r=8:1482803614830056;t=VIT_08s0007g00570.t01) (accessed on 10 January 2022), hierarchical indexing for spliced alignment of transcripts (HISAT) (v2.1.0) (Kim et al., 2015) software was used. Bowtie2 (v2.2.5) was used to calculate the mapping rate to align clean reads to the reference gene sequence, and RSEM was used to calculate the expression levels of genes and transcripts (Li and Dewey, 2011; Langmead and Salzberg, 2012). Bioinformatics analysis was performed on the successfully mapped clean reads on the reference genome.
Quantitative real-time PCR
A Roche Light Cycler 480 Real-Time PCR system and a Roche Light Cycler 480 SYBR Green I Master were used to run the qRT-PCR. The qRT-PCR conditions were as follows: 95°C for 30 s for denaturation, followed by 40 cycles of 5 s at 95°C, at 55°C for 30 s, and 72°C for 10 s with the primers (Supplementary Table S1). Three biological replicates were used for all of the reactions, and Bio-Rad CFX Manager software was used to determine the threshold cycle (Ct). The qRT-PCR method was used according to the manufacturer’s instructions. The relative quantitative expression level was calculated by the 2−∆∆CT method (Livak and Schmittgen, 2001). The gene expression level of grapevine level was analyzed using VvActin as the reference gene.
Promoter isolation of VvPR1 gene and sequence analysis
The genomic DNA was extracted from the grapevine leaves using a DN 15-Plant DNA Mini Kit according to the manufacturer’s instructions. The DNA concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The primer pair for the promoter of VvPR1 (VIT_03s0088g00810) was designed from the reference sequence of V. vinifera (Supplementary Table S2). A region approximately 1900 base pairs upstream from the coding region was thought to be the putative VvPR1 promoter. The VvPR1 promoter was amplified using the Premix high-fidelity (Takara) enzyme, and the PCR conditions were followed according to Rahman et al. (2022). The PCR product was purified on 1.5% agarose gel, cloned on the pCE2 Blunt vector, and sequenced for the verification of the promoter sequence. The PlantCARE online tool was used to predict the cis regulatory elements in the VvPR1 promoter (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 17 February 2022) (Lescot et al., 2002).
Construction of beta-glucuronidase vectors
The VvPR1 promoter serially deleted into the promoter fragments and cloned them into the pCE2 Blunt vector by designing primers of different lengths from promoter sequence (Supplemetary Table S2). Each forward primer contained the HindIII restriction site at the 5′ end, and the reverse primer contained the BamHI restriction site at the 5′ end. The PCR reaction was performed, and the PCR product was purified on the agarose gel using the gel extraction kit. Meanwhile, the expression vector (pBI-121) was also digested by restriction enzymes (HindIII and BamHI) for 2 hours and then subcloned with the purified PCR products. Seven promoter fragments (−1837 bp to ATG, −1,443 bp to ATG, −1,119 bp to ATG, −864 bp to ATG, −558 bp to ATG, −436 bp to ATG, and −192 bp to ATG) were separately fused into the expression vector pBI-121 with the GUS reporter gene, yielding pBI-121:pPR1 (Figure 1). The strong promoter CaMV35 in the expression vector pBI-121 was used as positive control, and pBI-101 with no promoter was used as a negative control. All recombinant vectors were cloned and propagated in Escherichia coli (DH5α strain). Then, the constructs of promoter/GUS fusion were inserted into the Agrobacterium tumefaciens strain GV3101 by heat shocks.
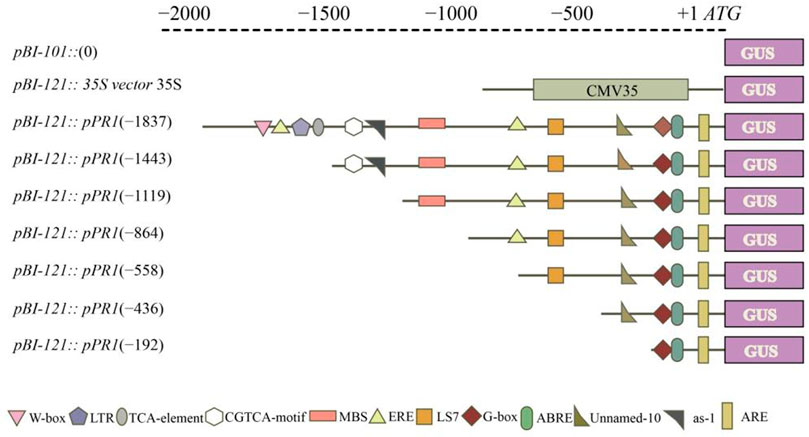
FIGURE 1. Schematic representation of the VvPR1 promoter. Constructs for assaying GUS (β-glucuronidase) expression in tobacco leaves. The constructs of serially deleted promoter fragments of the VdPR1 gene were fused to the GUS reporter gene in the vector pBI-121.
Agrobacterium-mediated transient expression assay with abiotic stress treatment
Agrobacterium was used for the transient expression, as mentioned by Yang et al. (2000). The serially deleted fragments of promoter:GUS harbored by Agrobacterium GV3101 were grown in the LB medium supplemented with the antibiotics rifampicin (60 μg ml−1) and kanamycin (50 μg ml−1). The Agrobacterium strains were cultured in 50 ml of LB broth at 28 °C overnight. The LB broth was centrifuged for 10 min at 6000× g to collect the Agrobacterium cells, which were then resuspended in infiltration media (10 mM MgCl2, 100 µM acetosyringone 10 mM MES, (pH 5.6), (Sigma-Aldrich)) and adjusted to an OD600 of 0.8. A needleless syringe was used to inject the infiltrate of Agrobacterium suspension into tobacco leaves, which were then placed in a moist chamber at 26°C for 24 h and then shifted to the growth room. SA (100 μM, 0.2% Tween-20) and ABA (100 μM, 0.2% Tween-20) treatment was applied to the tobacco leaves harboring the pBI-121:pPR1/GUS, and the control plants were only sprayed with 0.2% Tween-20. SA-treated and control plants were placed in the baskets and covered with the polyethylene-perforated plastic bags (having six holes (1 cm diameter) on each side and 0.03 mm thickness). Three replications were performed for each treatment, and each was also repeated three times for the transient GUS expression on the tobacco leaves. Samples were collected after 24 h of treatment.
GUS activity measurement
Histochemical staining analysis was performed to measure the GUS expression transiently as described by Jefferson et al. (1987). GUS staining solution was prepared as explained by Yu et al. [80]. Tobacco leaf discs were collected and dipped in a GUS staining solution (0.5 mg L−1 5-bromo-4-chloro-3-indolyl-b-D- glucuronic acid, 10 mM Na2EDTA, 100 mM NaH2PO4, 0.1% Triton X-100, and 0.5 Mm K4Fe(CN)6.3H2O (X-Gluc, Sigma-Aldrich, Shanghai, China), pH 7.0) and incubated at 37°C for 24 h. Leaf discs were incubated in 70% ethanol at 37°C to remove the chlorophyll contents for more clear observation and rinsed several times with ethanol. The quantitative GUS assay of promoter transiently expressed in tobacco leaves was measured as described by Jefferson et al. (1987). Microtubes were filled with leaf powder after grinding. The extraction buffer, phosphoric acid buffer (0.5M EDTA, TritonX-100(10%), 2M KPO4 (pH 7.8)), 80% glycerol, and beta mercapto ethanol (1 ml), was added to the microtube and vortexed. The material inside the microcentrifuge tube was centrifuged at 120,00× g for 15 min at 4°C, and the supernatant was transferred to a microcentrifuge tube already placed on ice. The whole fluorogenic reaction was performed at a volume of 1 ml mixed with extraction buffer in 1 mM 4-methylumbelliferyl-h-D-glucuronide (MUG) (Duchefa Biochemie, Haarlem, Netherlands), which also comprised an aliquot of protein extract at a volume of 0.1 ml. The standards of bovine serum albumin (BSA) were used for the quantification of protein extracts as explained by Bradford (1976). The results were found to be similar after measuring GUS three times.
Statistical analysis
Microsoft Excel (2016) was used for values calculation, and Student’s t-test was used for calculating differences among values. The significance levels are shown as follows: * represents p ≤ 0.05, and ** represents p ≤ 0.01. All experiments were repeated three times with three independent biological replicates.
Results
Structure and disease symptoms of grapevine leaves
In this study, the grapevine leaves were inoculated with C. diplodiella, and disease symptoms were found to be higher in V. vinifera than the V. davidii after 72 h of inoculation (Figure 2A). The hypersensitive response (HR) occurred in V. davidii, where the sudden cell death happened at the site of infection and stopped the spreading of the pathogen infection. At the base of the leaf structure, there was no significant difference in leaf thickness between V. davidii and V. vinifera (Table 1). The endogenous SA contents were also measured after C. diplodiella inoculation, and SA contents were found to be higher in V. davidii than V. vinifera after specific time points of inoculation (Figure 2B).
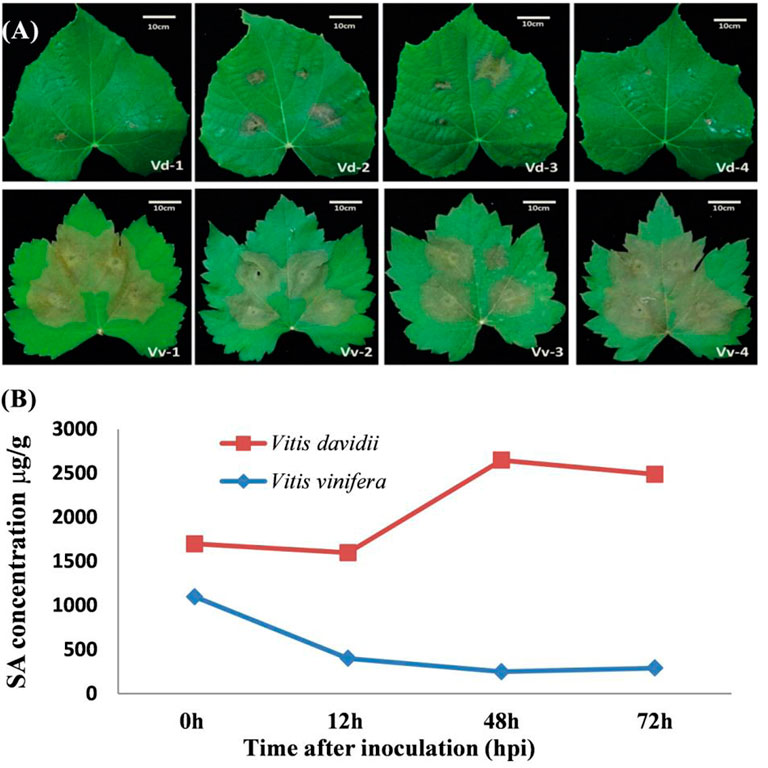
FIGURE 2. (A) Symptoms of Coniella diplodiella infection on the leaf samples of Vitis vinifera Manicure Finger (Vv) and Vitis davidii accession 0940 (Vd). Two-week-old leaf samples were collected at the 3-4 position. Typical hypersensitive response (HR) symptoms were observed in Vd but not in Vv at 72 h post-infection (hpi). There were four replications of each species. (B) Endogenous measurement of SA from the V. davidii and V. vinifera after 0, 12, 48, and 72 h of white rot disease (Coniella diplodiella) inoculation.
Transcriptomic analysis of SA-treated grapevine leaves at different time points
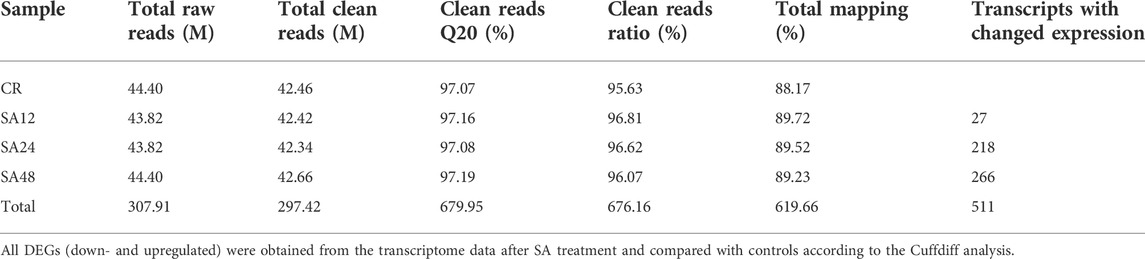
Transcriptome analysis was performed on the grapevine leaves treated with SA after 12 h and 48 h. As most disease attack happened on the aerial parts of the plants, SA treatment was applied to grapevines. An individual leaf sample comprised ≥6.3 Gb data with a Q20 ≥ 97.14% quality score. A total of 57.6 Gb clean data were mapped from twelve leaf samples (Table 2). On average, from each leaf sample, 88.17%–89.72% reads were uniquely mapped and aligned with reference genome V. vinifera L. The expression levels (p ≤ 0.05) of the control and the SA-treated samples were compared on the basis of a Cuffdiff analysis. For differentially expressed genes (DEGs), p-values (0.01) and log2-fold changes (log2FC) ≥ 1 or ≤−1 were used for threshold values after 12 h and 24 h of SA treatment. To identify the DEGs against the SA application after 12, 24, and 48 h, a volcano plot was designed against FC(log2) and −log10(significance) (Figures 3A–C). A Venn diagram was used to show the distribution and representation of DEGs after 12h and 48 h of SA treatment. The Venn diagram represents that 17, 114, and 155 genes were the unique set of genes that were expressed after 12, 24, and 48 h of SA treatment, respectively, and eight genes were coregulated at all time points (Figure 3D). A total of 511 DEGs were identified in which 12, 33, and 44 genes were downregulated, and 15, 183, and 240 genes were upregulated after 12, 24, and 48 h of SA treatments, respectively (Figure 3E).
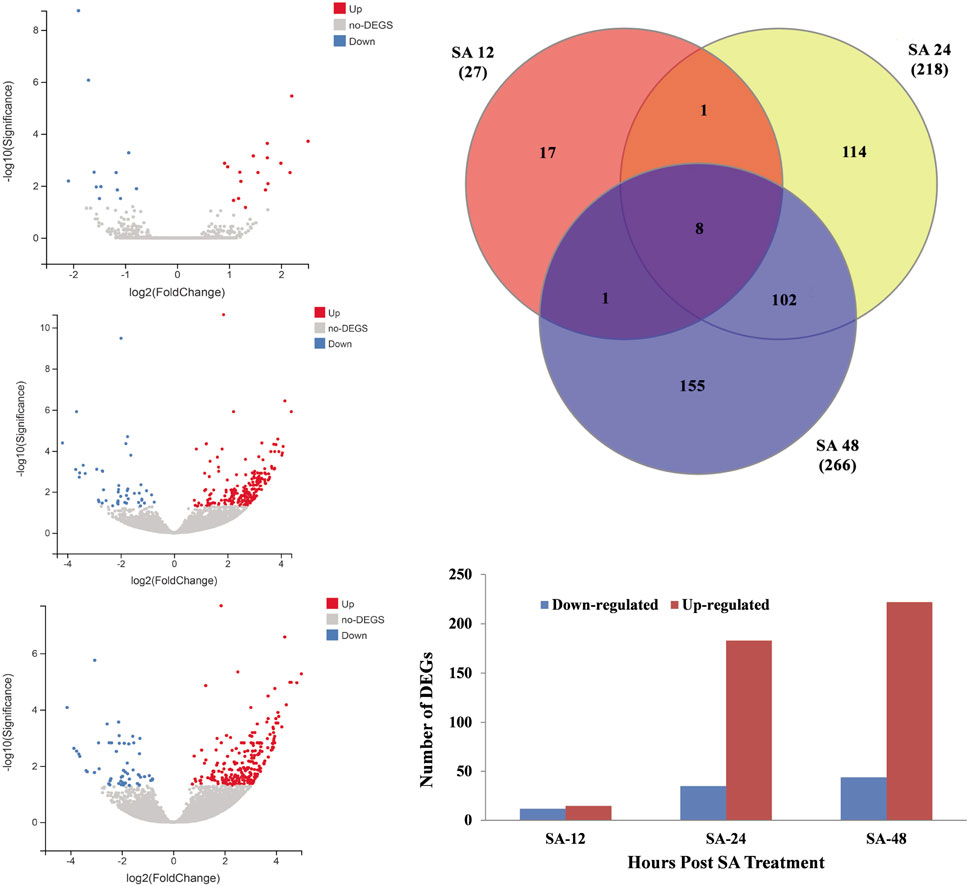
FIGURE 3. Distribution of DEGs from RNA-seq data. (A) Volcano graph of DEGs representing the downregulated genes with blue color and upregulated DEGs with red color after 12 h of SA treatment. (B) Volcano graph of DEGs representing the downregulated genes with blue color and upregulated DEGs with red color after 24 h of SA treatment. (C) Volcano graph of DEGs representing the downregulated genes with blue color and upregulated DEGs with red color after 48 h of SA treatment. (D) Venn diagram analysis of DEGs identified from all time points. (E) Total number of DEGs that were significantly down- or upregulated in response to SA treatment. Log2 FC ≥ 1 or ≤−1 and p < 0.01 FDR.
Gene ontology analysis
Gene ontology (GO) analysis of the DEGs after 12, 24, and 48 h of SA treatment was performed to classify them into three main domains of GO: biological process, cellular component, and molecular function. The biological process category contained nine GO terms, of which cellular process contained a significant number of enriched genes, followed by metabolic process. The cellular component contains three biological terms in which a cellular anatomical entity was enriched with significant DEGs. The molecular function has seven GO terms in which DEGs were significantly enriched in catalytic activity followed by binding (Supplementary Figure S1). More DEGs were enriched in the GO terms and domains after 48 of SA treatment. Three domains from three GO terms (cellular process, cellular anatomic entity, and catalytic activity) had the maximum number of downregulated genes (14, 32, and 25, respectively) and the maximum number of upregulated genes (83, 131, and 127, respectively) genes after the 48 h of SA treatment (Table 3).
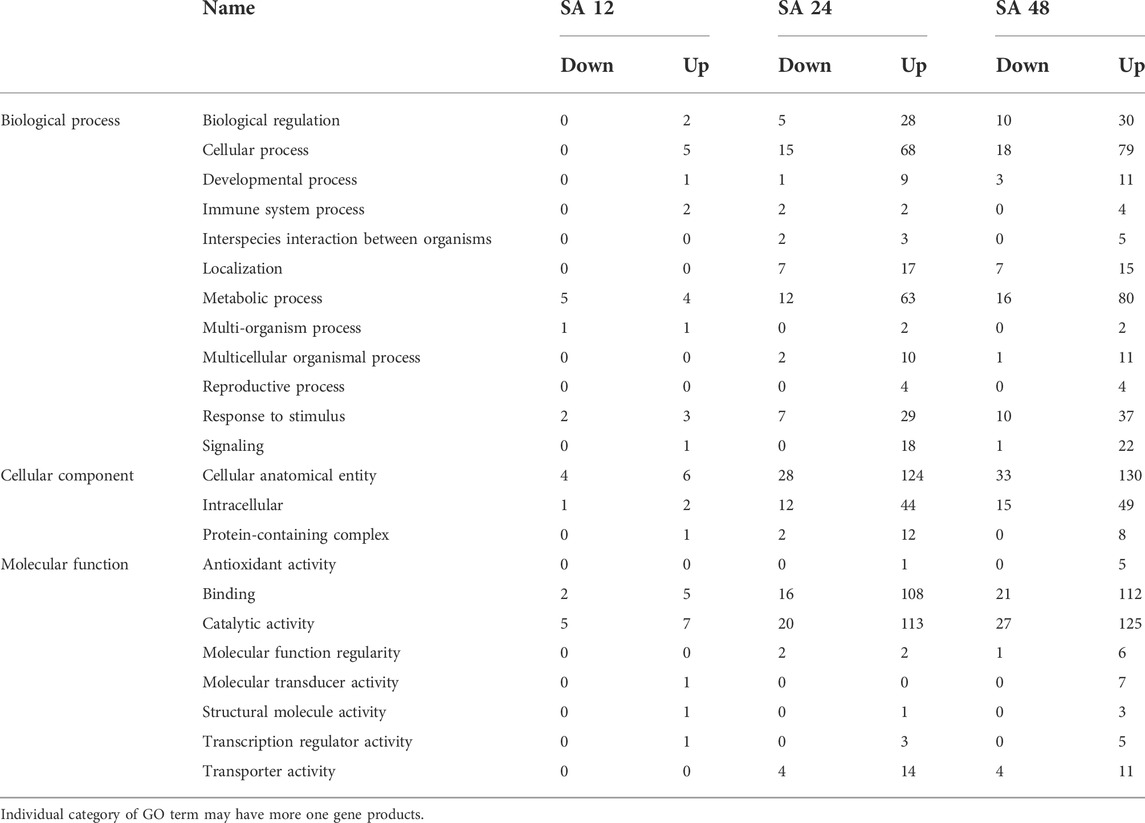
TABLE 3. Functional categorization of DEGs through GO analysis after SA treatment with different time points.
Kyoto Encyclopedia of Genes and Genomes analysis
To understand the biological pathways induced by SA in the grapevine leaves, all DEGs were mapped against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. DEGs mapped on the KEGG database revealed that most significant changes in response to SA treatment were related to the plant immune and defense response. According to KEGG, the metabolic pathway divided into five categories: environmental information processing, cellular processes, organic systems, genetic information processing, and metabolism. The highest number of KEGG pathways induced in grapevine leave were global and overview map (8, 47, and 49), signal transduction (3, 60, and 79), and immune system (4, 39, and 55) after 12, 24, and 48 h of SA treatment (Figure 4). The top 20 KEGG pathways comprised most of the defense and immune related pathways; among them, phenylalanine metabolism, MAPK signaling pathway, Ras signaling pathway, alanine aspartate, glutamate metabolism, and Toll-like receptor signaling pathway that have a crucial role in plant disease resistance (Supplementary Figure S2).
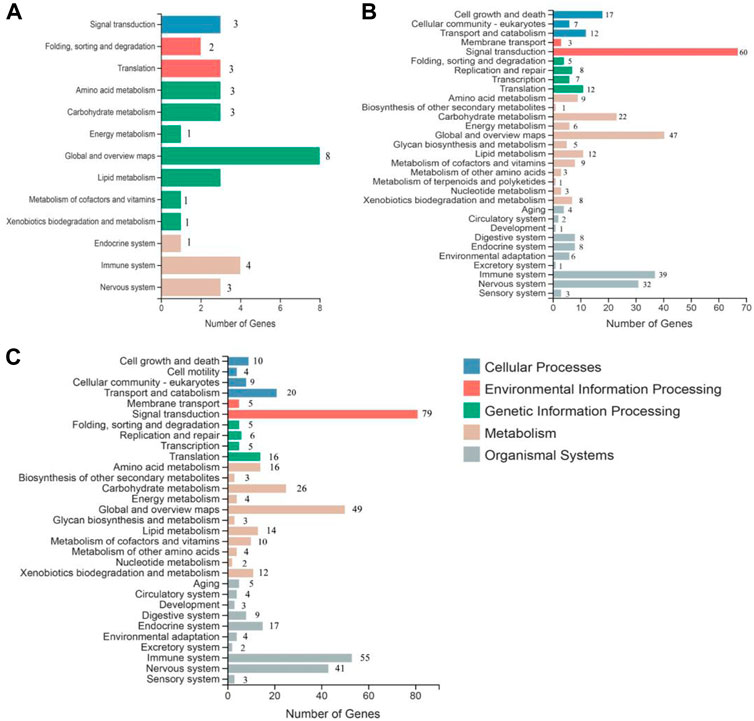
FIGURE 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of significantly expressed transcripts at all time points of SA treatment: (A) 12 h of SA treatment, (B) 24 h SA treatment, and (C) 48 h of SA treatment.
SA plant defense signaling
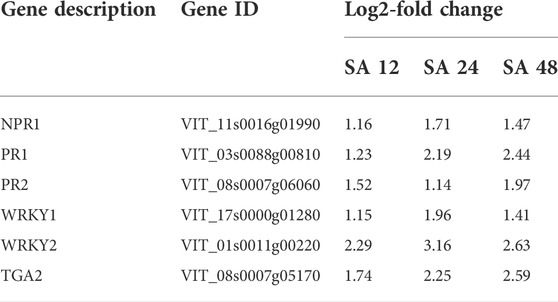
In plant defense, SA helps to encode the proteins related to antimicrobial activities through the induction of pathogenesis-related (PR) genes. To date, 17 PR families have been identified; among them, PR1, PR2, and PR5 are induced by biotrophic and semibiotrophic pathogens. Additionally, the expression of VvPR1 and VvPR2 was upregulated (1.14 to 2.44 fold) by the exogenous application of SA on grapevine leave after different time points (12, 24, and 48). A master regulator of SA-mediated plant defense is VvNPR1 (non-expresser of PR genes 1), and it regulates the VvPR1 gene through the binding with TGA transcription factors. VvNPR1 was upregulated (1.16 to 1.97 fold) at all time points after SA treatment, and VvTGA2 was also identified from the transcriptome data and upregulated (1.74 to 2.59 fold). VvWRKY1 and VvWRKY2, another transcription factor, was also upregulated (1.15 to 3.16 fold) after the 12, 24, and 48 h SA treatment on the grapevine leaves (Table 4; Figure 5).
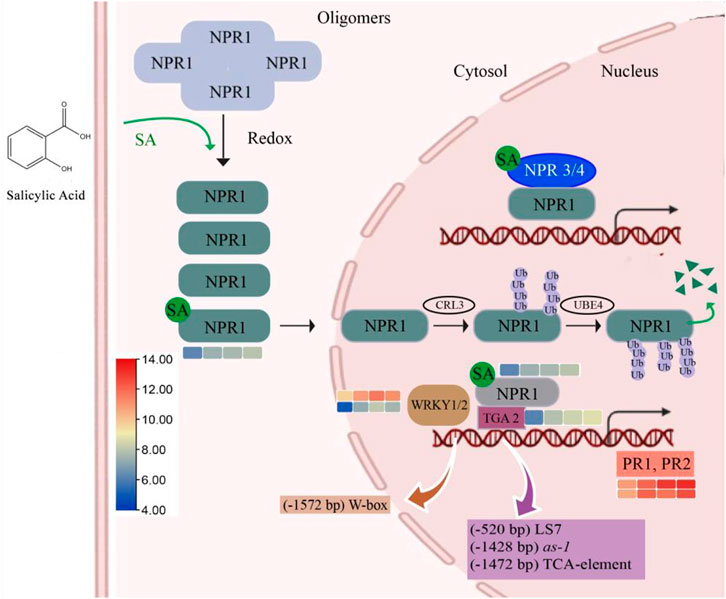
FIGURE 5. SA pathway related to plant defense. A higher SA level can induce the monomerization process of NPR1 and induced NPR1-dependent gene expression through direct interactions with TGA transcription factors. Meanwhile, direct binding with SA derepressed the suppression of NPR3 and NPR4 on SA-induced genes, which further enhanced SA-induced NPR1-dependent gene expression. Efficient turnover of monomeric NPR1 proteins in the nucleus is required for a rate-limited SA-induced gene expression, and this is also dependent on the homeostasis of NPR1-ubiquitination.
Verification of differential gene expression
The validation of the RNA-seq data was performed by selecting random transcripts from significantly expressed transcripts. We selected only nine genes for qPCR, of which five genes (VvPR1, VvPR2, VvTGA2, VvSTB-14, and VvNPR1) were continuously increasing their expression from all time points. The expression of VvWRKY2, VvEDS1, and VvCHI4D increased at 24 h of SA treatment, whereas the expression of VvBAK1 and VvTGA2 decreased at 24 h of SA treatment. The qPCR expression results were found consistent with transcriptome data (Figure 6). The most important plant defense gene, VvPR1, is a DEG that responds to different biotic and abiotic stress conditions, so this gene was selected for further functional validation in response to SA application.
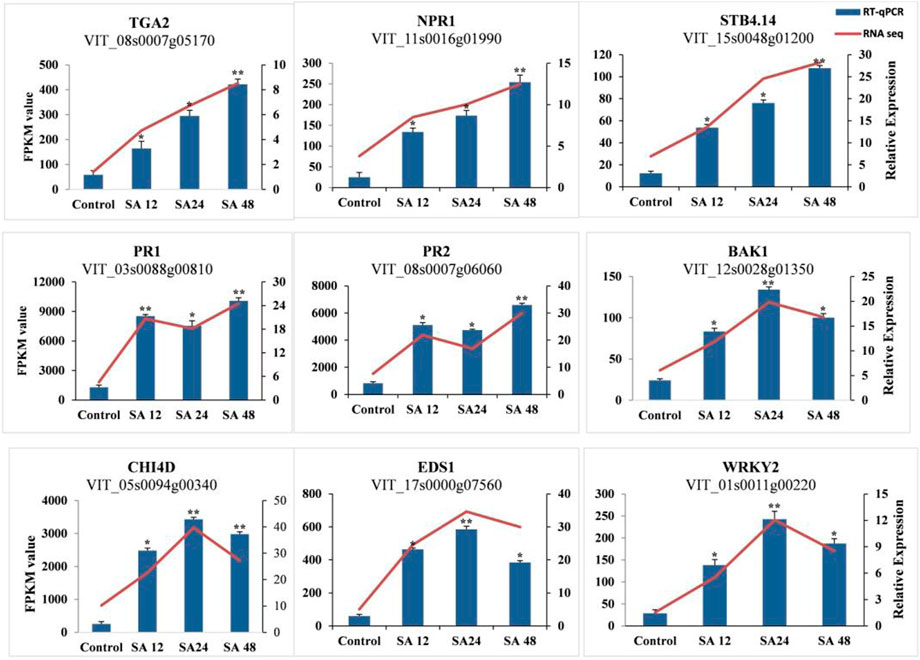
FIGURE 6. Validation of RNA-seq data with RT-qPCR. RT-qPCR of the selected DEGs was used for the verification of RNA-seq data. Error bars indicate the standard error as mean +SD. Significant differences between the control and treated samples are indicated by an asterisk (*). The sign * represents p ≤ 0.05, and ** represents p ≤ 0.01.
VvPR1 promoter sequence analysis
About 1837 bp of VvPR1 upstream from the ATG was considered as a putative promoter and cloned in a pCE2 Blunt vector. The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used for sequence analysis of the VvPR1 promoter and revealed many motifs, sequences, and cis-elements responsible for gene regulation and expression in many eukaryotic promoters (Supplementary Figure S3; Supplementary Table S3). The cis-elements related to defense, hormones, and stress were as found in other plant promoters. The VvPR1 promoter was enriched with CAAT-Box and TATA-box as follows: 1) light-responsive elements (AT1, LS7, AE-box, chs-CMA1a, Box4, and G-box), 2) stress-responsive elements (ARE, LTR, and MBS) respond to low temperature, drought, and anaerobic conditions 3) hormone-responsive elements (ERE, P-box, CGTCA-motif, and ABRE), 4) growth-associated elements (circadian and O2-site) that confer responsiveness to circadian control and zein metabolism regulation. Cis-acting elements (F-box and Unnamed-10) that had unclear functions were also found.
GUS expression of VvPR1 promoter against SA treatment
The serially deleted fragments of the VvPR1 promoter (−1,837 bp, −1,443 bp, −1,119 bp, −864 bp, −558 bp, −436 bp, and −192 bp) were cloned into the binary expression vector pBI121::GUS (Figure 7A). GUS expression was measured through histochemical staining and fluorometric assays in tobacco (N. benthamiana) plants. The above-mentioned constructs were infiltrated into the tobacco leaves and examined after 24 h of SA application by histochemical staining (Figure 7B) and fluorometric assays (Figure 7C). All deletion fragments of the promoter showed significantly higher GUS activity except −436 bp and −192 bp. The full-length promoter (−1,837 bp) exhibited the highest GUS protein activity, followed by the −864 bp, and −558 bp, which had shown significantly higher GUS activity with respect to the control. The −558 bp promoter fragment was found to be the shortest promoter fragment for the expression of the GUS reporter gene under SA treatment.
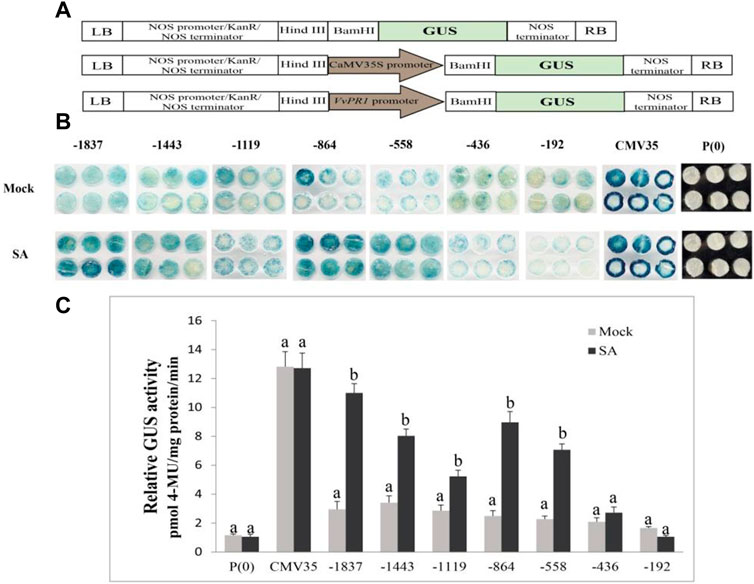
FIGURE 7. Schematic representation for the vector construction, histochemical staining, and fluorometric assay. (A) Promoter-GUS expression constructs showed the schematic structure. P(0); negative control; P(35S); positive control, and VvPR1 promoter in the forward orientation, respectively. (B) Histochemical staining analysis; GUS activity in transiently transformed N. benthamiana leaves with serially deleted VvPR1 promoter fragments (−1837, −1,443, −1,119, −845, −558, −436, and −192) bp against SA. (C) Fluorometric assay of VvPR1 promoter fragments (−1837, −1,443, −1,119, −845, −558, −436, and −192) bp in response to SA in tobacco leaves through transient expression. Different letters on the bars showed a significant difference between SA-treated fragments and the control according to the least significant difference (LSD) test (p < 0.05).
GUS expression of VvPR1 promoter against ABA treatment
The effect of another plant hormone, ABA, on the activation of the VvPR1 promoter was also investigated through a GUS assay in tobacco leaves harboring promoter-GUS chimeric constructs. The GUS expression of VvPR1 promoter fragments from −1837 bp to −192 bp had induced significantly higher GUS activity with respect to the control except for −1,119 bp and −436 bp (Figures 8A,B).
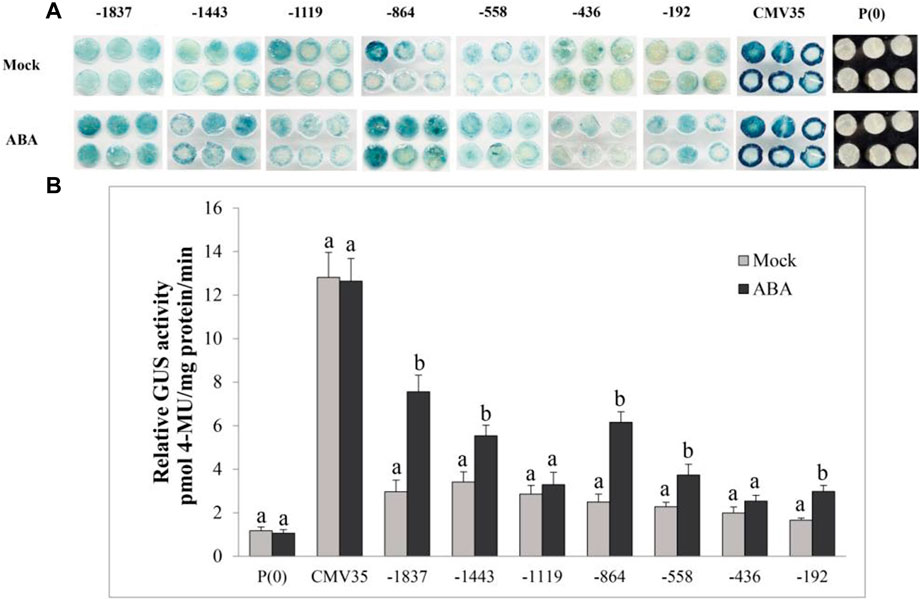
FIGURE 8. Histochemical staining and fluorometric assay of VvPR1 in response to ABA. (A) Histochemical staining analysis; GUS activity in transiently transformed N. benthamiana leaves with serially deleted VvPR1 promoter fragments (−1,837, −1,443, −1,119, −845, −558, −436, and −192) bp against ABA. (B) Fluorometric assay of VvPR1 promoter fragments (−1,837, −1,443, −1,119, −845, −558, −436, and −192) bp in response to SA in tobacco leaves through transient expression. Different letters on the bars showed a significant difference between the SA-treated promoter fragments and the control according to the least significant difference (LSD) test (p < 0.05).
Discussion
Grape white rot disease is the major threat to Vitis species and grapevine cultivation in China. To identify the resistant germplasm for breeding and research purposes, pathogenesis tests were performed on the available germplasm. It was found that Chinese wild grape species V. davidii was resistant to the grape white rot disease (Figure 2) as was previously reported in our laboratory (Zhang et al., 2013; Zhang et al., 2019). Phytohormones such as SA have a key role in plants to respond to different environmental stresses and pathogen attacks (Alazem and Lin, 2015). The best defense-related hormone is known as SA (Ryals et al., 1994; Durrant and Dong, 2004a; Fu and Dong, 2013). When a pathogen attacks a plant, it induces the SA accumulation and the defense response (Vernooij et al., 1994; Sharma and Davis, 1997; Tsuda et al., 2008). In our current study, the amount of SA production in the resistant cultivar (V. davidii) was higher than in the susceptible (V. vinifera). We performed the transcriptome analysis on the susceptible grapevine cultivar treated with exogenous SA, hypothesizing that SA may induce the defense genes in grapevine plants. The RNA-seq analysis was conducted on SA-treated grapevine leaves and 511 DEGs were identified in which 12, 33, and 44 genes were down-regulated, and 15, 183, and 240 genes were upregulated after 12, 24, and 48 h of SA treatment, respectively. A higher number of DEGs were enriched in the GO terms and domains after 48 of SA treatment. Three domains from three GO terms, including cellular process, cellular anatomic entity, and catalytic activity, enriched a maximum number of downregulated and upregulated genes after 48 h of SA treatment on grapevine leaves. The top 20 KEGG pathways comprised most of the defense and immune related pathways; among them, phenylalanine metabolism, MAPK signaling pathway, Ras signaling pathway, Toll-like receptor signaling pathway, and alanine aspartate and glutamate metabolism that have a crucial role in plant disease resistance.
The SA level is elevated during the MTI and PTI response of the plant (Iwai et al., 2007; Garcion et al., 2008; Palmer et al., 2017). SA plays a vital role in the plant’s defense against biotrophic and semi-biotrophic pathogens (Fu and Dong, 2013). Moreover, exogenous SA treatment and its active analogs also induce defense mechanisms in plants against semibiotrophic and biotrophic pathogens (Lu, 2009). In plant defense, SA helps to encode the proteins related to antimicrobial activities through the induction of PR genes. To date, 17 PR families have been identified; among them, PR1, PR2, and PR5 are activated by biotrophic and semibiotrophic pathogens (Stintzi et al., 1993; Hoffmann-Sommergruber, 2000). SA also control the expression of PR1, PR2, and PR5 (Leah et al., 1991; Zhang et al., 2010); they are also used for the SA pathway as a marker. In the current study, PR1 and PR2 genes were upregulated after the SA treatment on the grapevine leaves at all time points. NPR1 is detected through the Arabidopsis mutants with an abolished expression of the PR gene (Cao et al., 1997). NPR1 is known as a master regulator of plant defense through the SA; it controls almost 98% of SA-mediated genes (Wang et al., 2006). In this study, NPR1 was also significantly upregulated from the leaf samples after the SA treatment. SA controls the translocation of NPR1 through the specific redox reactions (Mou et al., 2003). The oligomers of NPR1 formed by the intermolecular disulfide bond are found in the cytoplasm in the absence of infection or SA treatment, but after the SA treatment or infection, intermolecular bonds break, and monomers of NPR1 translocate into the nucleus, where they induce the expression of defense-related genes.
During the plant defense response, NPR1 regulated the expression of PR genes through cofactors known as TGAs because DNA-binding domains are missing on NPR1 (Zhang et al., 1999; Kesarwani et al., 2007). In Arabidopsis, it has been found that TGA2, 3, 5, 6, and 7 show interaction with NPR1 and NPR1 helps to bind TGA transcription factors on the as-1 element in the PR1 promoter region to induce the expression (Després et al., 2000; Després et al., 2003; Johnson et al., 2003). Additionally, VvTGA2 TF was identified from the DEGs after the SA treatment on the grapevine leaves that may bind the promoter of VvPR1 gene with VvNPR1 to activate the plant defense response. NPR3 and NPR4 also bind SA but have been identified as negative regulators of plant defense, in contrast to NPR1, which plays a vital role in SA signaling (Zhang et al., 2006; Fu et al., 2012). NPR1 also works as an SA receptor through SA binding (Wu et al., 2012; Ding et al., 2018). During the plant defense, the NPR1 paralogues, NPR3 and NPR4, are SA receptors that bind SA with different affinities and function as adaptors of the Cullin 3 ubiquitin E3 ligase to mediate NPR1 degradation in an SA-regulated manner (Fu et al., 2012). Meanwhile, Ding et al. claimed that SA-based plant immunity was also accomplished independently by NPR3 and NPR4 (Fu et al., 2012; Ding et al., 2018).
After the RT-PCR of the random DEGs from the RNA-seq data, the VvPR1 gene was selected for further study because PR1 is the SA marker gene for plant defense response. The promoter of the VvPR1 gene was isolated from grapevine, and it was found that the VvPR1 promoter was enriched with CAAT-boxes and TATA-boxes. Other cis-elements, such as stress, hormone, light, growth and development, and associated elements, were also detected from the VvPR1 promoter. In this study, the VvPR1 promoter was enriched with TATA boxes, especially up to −800 bs upstream from the ATG. TATA-boxes are abundant in stress-related genes and are absent in essential genes (Tirosh et al., 2006; Walther et al., 2007). They are important with a variable and rapid gene induction (Newman et al., 2006; Roelofs et al., 2010; Vos et al., 2015). A previous study of Arabidopsis found that the PR-1 promoter contains cis-acting regulatory elements responsible for its induction on exogenous 2,6-dichloroisonicotinic acid (INA) and SA (Lebel et al., 1998). Hormones, different stresses, and pathogens activate the pepper PR1 promoter, possibly by transactivating the CARAV1 and CAZFP1 transcription factors (Hong et al., 2005).
Phytohormones (SA, JA, and ABA) are well-known major signaling components in plant defense signaling networks (Dong, 1998). SA is an important biomolecule in disease resistance that recognizes the pathogen effectors directly or indirectly and induces local resistance and systematic resistance against the biotrophic pathogens (Durrant and Dong, 2004b). In this study, the VvPR1 promoter fragments showed high GUS induction from −1,837 bp to −558 bp against SA. The expression of GUS of the −1,443 bp of VvPR1 promoter increased 2.35-fold with respect to the control under the treatment of SA. In addition, the −1,119, −864, and −558 bp deletion fragments resulted in 1.85-, 3.60-, and 3.11-fold increases in GUS expression by the SA treatment, respectively. This indicated that the minimal cis-regulatory elements important for the molecular response to SA might be present in the 1282-bp region between −1837 and −558 of the VvPR1 promoter. The expression of the VvPR1 promoter upon the SA treatment may be due to the presence of the TCA-element, activation sequence-1 (as-1), and the LS7 cis-acting elements. In this study, the VvPR1 promoter contained the TCA-element, as-1 and LS7 cis-acting elements at -1,472 bp, −1,428 bp, and −520 bp, respectively. Previous studies provide strong evidence that the TCA element is involved in SA signaling and PR-1 promoter induction by providing the site for TGA transcription factors and recruitment of NPR1 (Pastuglia et al., 1997). On the accumulation of SA in the cell, NPR1 monomers translocate in the nucleus (Zhang et al., 1999; Kinkema et al., 2000; Zhou et al., 2000), where they interact with TGA transcription factors and activate the expression of the PR-1 gene and subsequently SAR activation (Zhang et al., 2003; Kesarwani et al., 2007). Activation sequence-1 (as-1) is an SA-inducible cis-element in promoters of some pathogenesis-related genes in Arabidopsis thaliana PR-1 and tobacco PR-1a. In the promoter of PR-1a, tobacco possesses an as-1-like element with inverted TGACG motifs, a binding site for TGA transcription factors (Strompen et al., 1998; Niggeweg et al., 2000; Grüner et al., 2003). Further studies have shown that LS7 is also involved in SA-inducible PR-1 gene expression (Pape et al., 2010).
ABA is an important plant hormone that responds to biotic and abiotic stresses (Ton et al., 2009; Sah et al., 2016; Alazem et al., 2017). On the application of ABA, several transcription factors bind to target genes and respond to the stress (Song et al., 2016). In this study, the GUS protein expression has been induced upon ABA treatment from all deleted fragments of VvPR1 except −1,119 and −436; this indicated that these regions might contain some ABA-repressing elements. The induction of GUS with ABA treatment may be due to the presence of ABRE (a cis-acting element involved in the abscisic acid responsiveness) (Shinozaki and Yamaguchi-Shinozaki, 1997) that is located at −163 bp upstream from the transcription initiation start site. It was also found that ABA promotes proteasome-mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana (Ding et al., 2016) that may express the PR-1 gene under ABA treatment. Overall, the GUS protein expression by the VvPR1 promoter under the treatment of ABA was found to be low compared to SA; this low activation may be due to the presence of the low number of ABRE cis-acting elements in the VvPR1 promoter.
Conclusion
In conclusion, SA levels were found to be higher in resistant grapevine (V. davidii) than in susceptible grapevine (V. vinifera) after a pathogenicity test against white rot disease. After the transcriptome analysis, the GO and KEGG pathway analysis revealed that DEGs include important genes related to the SA defense pathway known as VvPR1. The promoter of the VvPR1 gene was also analyzed through serial deletion of the VvPR1 promoter with the GUS reporter gene. Deletion analysis showed that the region between −1837 bp and −558 bp of the VvPR1 promoter expressed significantly high GUS protein with respect to the control. On the basis of these results, this region was deduced to be an important part of the VvPR1 promoter, which contained the most important cis-elements (TCA-elements, LS7, and as-1) responsible for VvPR1 gene expression in response to SA application (Figure 9). Overall, the current study validated the available information about SA-mediated defense responses in grapevine species that are susceptible to various diseases.
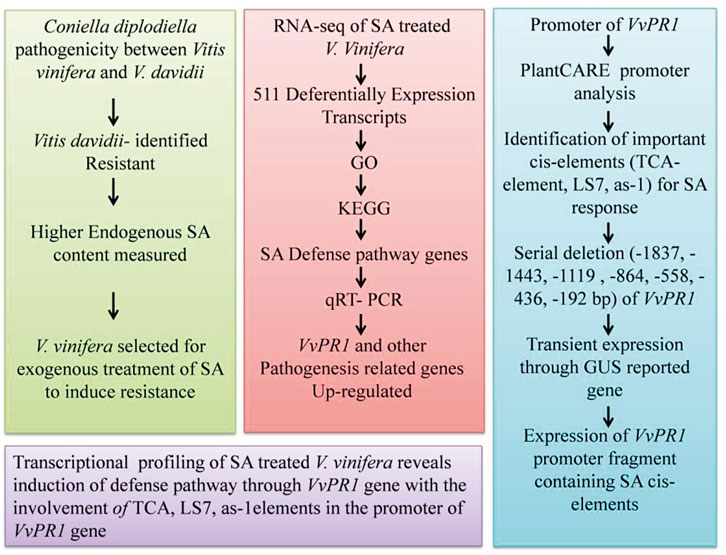
FIGURE 9. Flow chart of study pattern showing the different steps of inducing the pathogenicity of grape white rot, the RNA-seq data analysis, promoter analysis through serial deletion of promoter, and the conclusion.
Data availability statement
The sequencing data has been deposited into the NationalGenomics Data Center (Accession Number: PRJNA845078; link: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA845078.
Author contributions
FR, CL, and YZ conceived and designed the research. FR, RL, IK, LS, and YW performed the experiments. XF, AA, MA, AK, and PL generated the images. FR, JJ, MA, and XF, AA wrote and revised the manuscript. CL, YZ, PL, and XF contribute reagents/materials and interpretation of the results. All authors have read and approved the final draft.
Funding
This work was supported by grants from the National Key Research and Development Program of China (No. 2021YFD1200200), the National Natural Science Foundation of China: 31872057, the China Agriculture Research System: CARS-29, and the Agricultural Science and Technology Innovation Program: CAAS-ASTIP-2017-ZFRI.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1033288/full#supplementary-material
References
Ahmar, S., and Gruszka, D. (2022). In-silico study of brassinosteroid signaling genes in rice provides insight into mechanisms which regulate their expression. Front. Genet. 13, 953458. doi:10.3389/fgene.2022.953458
Alazem, M., He, M.-H., Moffett, P., and Lin, N.-S. (2017). Abscisic acid induces resistance against bamboo mosaic virus through Argonaute 2 and 3. Plant Physiol. 174, 339–355. doi:10.1104/pp.16.00015
Alazem, M., and Lin, N. S. (2015). Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 16, 529–540. doi:10.1111/mpp.12204
Boller, T., and Felix, G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi:10.1146/annurev.arplant.57.032905.105346
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi:10.1006/abio.1976.9999
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi:10.1016/s0092-8674(00)81858-9
Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. doi:10.1105/tpc.012849
Després, C., Delong, C., Glaze, S., Liu, E., and Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. doi:10.2307/3870928
Ding, Y., Dommel, M., and Mou, Z. (2016). Abscisic acid promotes proteasome‐mediated degradation of the transcription coactivator NPR 1 in Arabidopsis thaliana. Plant J. 86, 20–34. doi:10.1111/tpj.13141
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467. e15. doi:10.1016/j.cell.2018.03.044
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi:10.1038/nrg2812
Dong, X. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. doi:10.1016/1369-5266(88)80053-0
Dou, D., and Zhou, J.-M. (2012). Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 12, 484–495. doi:10.1016/j.chom.2012.09.003
Durrant, W. E., and Dong, X. (2004a). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi:10.1146/annurev.phyto.42.040803.140421
Durrant, W. E., and Dong, X. (2004b). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi:10.1146/annurev.phyto.42.040803.140421
Fischer, B., Salakhutdinov, I., Akkurt, M., Eibach, R., Edwards, K., Toepfer, R., et al. (2004). Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor. Appl. Genet. 108, 501–515. doi:10.1007/s00122-003-1445-3
Frye, C. A., Tang, D., and Innes, R. W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. U. S. A. 98, 373–378. doi:10.1073/pnas.011405198
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi:10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi:10.1038/nature11162
Gamir, J., Darwiche, R., Van'T Hof, P., Choudhary, V., Stumpe, M., Schneiter, R., et al. (2017). The sterol‐binding activity of PATHOGENESIS‐RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. doi:10.1111/tpj.13398
Garcion, C., Lohmann, A., Lamodière, E., Catinot, J., Buchala, A., Doermann, P., et al. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. doi:10.1104/pp.108.119420
Godiard, L., Ragueh, F., Froissard, D., Leguay, J.-J., Grosset, J., Chartier, Y., et al. (1990). Analysis of the synthesis of several pathogenesis-related proteins in tobacco leaves infiltrated with water and with compatible and incompatible isolates of Pseudomonas solanacearum. Mol. Plant. Microbe. Interact. 3, 207–213. doi:10.1094/mpmi-3-207
Grüner, R., Strompen, G., Pfitzner, A. J., and Pfitzner, U. M. (2003). Salicylic acid and the hypersensitive response initiate distinct signal transduction pathways in tobacco that converge on the as‐1‐like element of the PR‐1a promoter. Eur. J. Biochem. 270, 4876–4886. doi:10.1046/j.1432-1033.2003.03888.x
Gu, Y.-Q., Wildermuth, M. C., Chakravarthy, S., Loh, Y.-T., Yang, C., He, X., et al. (2002). Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14, 817–831. doi:10.1105/tpc.000794
He, C., and Wolyn, D. (2005). Potential role for salicylic acid in induced resistance of asparagus roots to Fusarium oxysporum f. sp. asparagi. Plant Pathol. 54, 227–232. doi:10.1111/j.1365-3059.2005.01163.x
Hoffmann-sommergruber, K. (2000). Plant allergens and pathogenesis-related proteins. Int. Arch. Allergy Immunol. 122, 155–166. doi:10.1159/000024392
Hong, J. K., Lee, S. C., and Hwang, B. K. (2005). Activation of pepper basic PR-1 gene promoter during defense signaling to pathogen, abiotic and environmental stresses. Gene 356, 169–180. doi:10.1016/j.gene.2005.04.030
Hu, Y., Gao, Y.-R., Yang, L.-S., Wang, W., Wang, Y.-J., and Wen, Y.-Q. (2019). The cytological basis of powdery mildew resistance in wild Chinese Vitis species. Plant Physiol. biochem. 144, 244–253. doi:10.1016/j.plaphy.2019.09.049
Iwai, T., Seo, S., Mitsuhara, I., and Ohashi, Y. (2007). Probenazole-induced accumulation of salicylic acid confers resistance to Magnaporthe grisea in adult rice plants. Plant Cell Physiol. 48, 915–924. doi:10.1093/pcp/pcm062
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi:10.1002/j.1460-2075.1987.tb02730.x
Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15, 1846–1858. doi:10.1105/tpc.012211
Joshi, V., Joshi, N., Vyas, A., and Jadhav, S. (2021). Biocontrol agents and secondary metabolites. Elsevier.Pathogenesis-related proteins: Role in plant defense
Kaur, S., Samota, M. K., Choudhary, M., Choudhary, M., Pandey, A. K., Sharma, A., et al. (2022). How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 28, 485–504. doi:10.1007/s12298-022-01146-y
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. doi:10.1104/pp.106.095299
Kim, D., Langmead, B., and Salzberg, S. L. (2015). Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi:10.1105/tpc.12.12.2339
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi:10.1038/nmeth.1923
Leah, R., Tommerup, H., Svendsen, I., and Mundy, J. (1991). Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 266, 1564–1573. doi:10.1016/s0021-9258(18)52331-0
Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR–1 gene expression in Arabidopsis. Plant J. 16, 223–233.
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Li, B., and Dewey, C. N. (2011). Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinforma. 12, 1–16. doi:10.1186/1471-2105-12-323
Li, D., Wan, Y., Wang, Y., and He, P. (2008). Relatedness of resistance to anthracnose and to white rot in Chinese wild grapes. Vitis 47, 213–215.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. methods 25, 402–408. doi:10.1006/meth.2001.1262
Lorenzo, O., Chico, J. M., Saénchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi:10.1105/tpc.022319
Lu, H. (2009). Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 4, 713–717. doi:10.4161/psb.4.8.9173
Mandal, S., Kar, I., Mukherjee, A. K., and Acharya, P. (2013). Elicitor-induced defense responses in Solanum lycopersicum against Ralstonia solanacearum. The Scientific World Journal.
Mandal, S., Mallick, N., and Mitra, A. (2009). Salicylic acid-induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiol. biochem. 47, 642–649. doi:10.1016/j.plaphy.2009.03.001
Mohan babu, R., Sajeena, A., Vijaya Samundeeswari, A., Sreedhar, A., Vidhyasekaran, P., Seetharaman, K., et al. (2003). Induction of systemic resistance to Xanthomonas oryzae pv. oryzae by salicylic acid in Oryza sativa (L.). J. Plant Dis. Prot. (2006). 110, 419–431. doi:10.1007/bf03356119
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi:10.1016/s0092-8674(03)00429-x
Mu, W., Wei, J., Yang, T., Fan, Y., Cheng, L., Yang, J., et al. (2017). RNA extraction for plant samples using CTAB-pBIOZOL. Protocols.
Newman, J. R., Ghaemmaghami, S., Ihmels, J., Breslow, D. K., Noble, M., Derisi, J. L., et al. (2006). Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846. doi:10.1038/nature04785
Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788. doi:10.1023/a:1006319113205
Palmer, I. A., Shang, Z., and Fu, Z. Q. (2017). Salicylic acid-mediated plant defense: Recent developments, missing links, and future outlook. Front. Biol. (Beijing). 12, 258–270. doi:10.1007/s11515-017-1460-4
Palva, T. K., Hurtig, M., Saindrenan, P., and Palva, E. T. (1994). Salicylic acid induced resistance to Erwinia carotovora subsp. carotovora in tobacco. Mol. Plant. Microbe. Interact. 7, 356–363. doi:10.1094/mpmi-7-0356
Pap, D., Riaz, S., Dry, I. B., Jermakow, A., Tenscher, A. C., Cantu, D., et al. (2016). Identification of two novel powdery mildew resistance loci, Ren6 and Ren7, from the wild Chinese grape species Vitis piasezkii. BMC Plant Biol. 16, 1–19. doi:10.1186/s12870-016-0855-8
Pape, S., Thurow, C., and Gatz, C. (2010). The Arabidopsis PR-1 promoter contains multiple integration sites for the coactivator NPR1 and the repressor SNI1. Plant Physiol. 154, 1805–1818. doi:10.1104/pp.110.165563
Pastuglia, M., Roby, D., Dumas, C., and Cock, J. M. (1997). Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 9, 49–60. doi:10.1105/tpc.9.1.49
Rahman, F. U., Zhang, Y., Khan, I. A., Liu, R., Sun, L., Wu, Y., et al. (2022). The promoter analysis of VvPR1 gene: A candidate gene identified through transcriptional profiling of methyl jasmonate treated grapevine (Vitis vinifera L.). Plants 11, 1540. doi:10.3390/plants11121540
Raskin, I. (1992). Role of salicylic acid in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 439–463. doi:10.1146/annurev.pp.43.060192.002255
Raza, A., Salehi, H., Rahman, M. A., Zahid, Z., Haghjou, M. M., Najafi-Kakavand, S., et al. (2022). Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front. Plant Sci. 13, 3061. doi:10.3389/fpls.2022.961872
Renault, A.-S., Deloire, A., and Bierne, J. (1996). Pathogenesis-related proteins in grapevines induced by salicylic acid and Botrytis cinerea. Vitis 35, 49–52.
Roelofs, D., Morgan, J., and Stürzenbaum, S. (2010). The significance of genome-wide transcriptional regulation in the evolution of stress tolerance. Evol. Ecol. 24, 527–539. doi:10.1007/s10682-009-9345-x
Ryals, J., Uknes, S., and Ward, E. (1994). Systemic acquired resistance. Plant Physiol. 104, 1109–1112. doi:10.1104/pp.104.4.1109
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7, 571. doi:10.3389/fpls.2016.00571
Sapkota, S., Chen, L.-L., Yang, S., Hyma, K. E., Cadle-Davidson, L., and Hwang, C.-F. (2019). Construction of a high-density linkage map and QTL detection of downy mildew resistance in Vitis aestivalis-derived ‘Norton. Theor. Appl. Genet. 132, 137–147. doi:10.1007/s00122-018-3203-6
Sarowar, S., Kim, Y. J., Kim, E. N., Kim, K. D., Hwang, B. K., Islam, R., et al. (2005). Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 24, 216–224. doi:10.1007/s00299-005-0928-x
Sawinski, K., Mersmann, S., Robatzek, S., and Böhmer, M. (2013). Guarding the green: Pathways to stomatal immunity. Mol. Plant. Microbe. Interact. 26, 626–632. doi:10.1094/MPMI-12-12-0288-CR
Sels, J., Mathys, J., De Coninck, B. M., Cammue, B. P., and De Bolle, M. F. (2008). Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. biochem. 46, 941–950. doi:10.1016/j.plaphy.2008.06.011
Sharma, Y. K., and Davis, K. R. (1997). The effects of ozone on antioxidant responses in plants. Free Radic. Biol. Med. 23, 480–488. doi:10.1016/s0891-5849(97)00108-1
Shinozaki, K., and Yamaguchi-shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115, 327–334. doi:10.1104/pp.115.2.327
Song, L., Huang, S.-S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550. doi:10.1126/science.aag1550
Stintzi, A., Heitz, T., Prasad, V., Wiedemann-Merdinoglu, S., Kauffmann, S., Geoffroy, P., et al. (1993). Plant ‘pathogenesis-related’proteins and their role in defense against pathogens. Biochimie 75, 687–706. doi:10.1016/0300-9084(93)90100-7
Strompen, G., Grüner, R., and Pfitzner, U. M. (1998). An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol. 37, 871–883. doi:10.1023/a:1006003916284
Tirosh, I., Weinberger, A., Carmi, M., and Barkai, N. (2006). A genetic signature of interspecies variations in gene expression. Nat. Genet. 38, 830–834. doi:10.1038/ng1819
Ton, J., Flors, V., and Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. doi:10.1016/j.tplants.2009.03.006
Tsuda, K., Sato, M., Glazebrook, J., Cohen, J. D., and Katagiri, F. (2008). Interplay between MAMP‐triggered and SA‐mediated defense responses. Plant J. 53, 763–775. doi:10.1111/j.1365-313X.2007.03369.x
Van huijsduijnen, R. H., Alblas, S., De Rijk, R., and Bol, J. (1986). Induction by salicylic acid of pathogenesis-related proteins and resistance to alfalfa mosaic virus infection in various plant species. J. General Virology 67, 2135–2143. doi:10.1099/0022-1317-67-10-2135
Van loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi:10.1146/annurev.phyto.44.070505.143425
Vernooij, B., Uknes, S., Ward, E., and Ryals, J. (1994). Salicylic acid as a signal molecule in plant-pathogen interactions. Curr. Opin. Cell Biol. 6, 275–279. doi:10.1016/0955-0674(94)90147-3
Vlot, A. C., Dempsey, D. M. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi:10.1146/annurev.phyto.050908.135202
Vos, I. A., Moritz, L., Pieterse, C. M., and Van Wees, S. (2015). Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 6, 639. doi:10.3389/fpls.2015.00639
Walther, D., Brunnemann, R., and Selbig, J. (2007). The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet. 3, e11. doi:10.1371/journal.pgen.0030011
Wan, R., Hou, X., Wang, X., Qu, J., Singer, S. D., Wang, Y., et al. (2015). Resistance evaluation of Chinese wild Vitis genotypes against Botrytis cinerea and different responses of resistant and susceptible hosts to the infection. Front. Plant Sci. 6, 854. doi:10.3389/fpls.2015.00854
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, e123. doi:10.1371/journal.ppat.0020123
Wang, K., Senthil-Kumar, M., Ryu, C.-M., Kang, L., and Mysore, K. S. (2012). Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol. 158, 1789–1802. doi:10.1104/pp.111.189217
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi:10.1038/35107108
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi:10.1016/j.celrep.2012.05.008
Xiao, S., Calis, O., Patrick, E., Zhang, G., Charoenwattana, P., Muskett, P., et al. (2005). The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 42, 95–110. doi:10.1111/j.1365-313X.2005.02356.x
Yan, S., and Dong, X. (2014). Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20, 64–68. doi:10.1016/j.pbi.2014.04.006
Yang, J., Wang, Y., Liu, L., Liu, L., Wang, C., Wang, C., et al. (2019). Effects of exogenous salicylic acid and pH on pathogenicity of biotrophy-associated secreted protein 1 (BAS1)-overexpressing strain, Magnaporthe oryzae. Environ. Sci. Pollut. Res. Int. 26, 13725–13737. doi:10.1007/s11356-018-2532-y
Yang, Y., Li, R., and Qi, M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551. doi:10.1046/j.1365-313x.2000.00760.x
Zhang, Y., Cheng, Y. T., Qu, N., Zhao, Q., Bi, D., and Li, X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. doi:10.1111/j.1365-313X.2006.02903.x
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U. S. A. 96, 6523–6528. doi:10.1073/pnas.96.11.6523
Zhang, Y., Sun, H., Fan, X., Jiang, J., and Liu, C. (2013). Identification and evaluation of resistance of Vitis to grape white rot. J. Fruit Sci. 30, 191–196.
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. doi:10.1105/tpc.014894
Zhang, Y., Xu, S., Ding, P., Wang, D., Cheng, Y. T., He, J., et al. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. U. S. A. 107, 18220–18225. doi:10.1073/pnas.1005225107
Zhang, Y., Yao, J.-L., Feng, H., Jiang, J., Fan, X., Jia, Y.-F., et al. (2019). Identification of the defense-related gene VdWRKY53 from the wild grapevine Vitis davidii using RNA sequencing and ectopic expression analysis in Arabidopsis. Hereditas 156, 14. doi:10.1186/s41065-019-0089-5
Zhao, J., Davis, L. C., and Verpoorte, R. (2005). Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 23, 283–333. doi:10.1016/j.biotechadv.2005.01.003
Zhou, J.-M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant. Microbe. Interact. 13, 191–202. doi:10.1094/MPMI.2000.13.2.191
Keywords: Salicylic acid, PR1, white rot, Cis-elements, transcriptomics, grapevine
Citation: Rahman FU, Khan IA, Aslam A, Liu R, Sun L, Wu Y, Aslam MM, Khan AU, Li P, Jiang J, Fan X, Liu C and Zhang Y (2022) Transcriptome analysis reveals pathogenesis-related gene 1 pathway against salicylic acid treatment in grapevine (Vitis vinifera L). Front. Genet. 13:1033288. doi: 10.3389/fgene.2022.1033288
Received: 31 August 2022; Accepted: 30 September 2022;
Published: 20 October 2022.
Edited by:
Ali Raza, Fujian Agriculture and Forestry University, ChinaReviewed by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanMuhammad Uzair, National Institute for Genomics and Advanced Biotechnology (NIGAB), Pakistan
Sunny Ahmar, University of Silesia in Katowice, Poland
Copyright © 2022 Rahman, Khan, Aslam, Liu, Sun, Wu, Aslam, Khan, Li, Jiang, Fan, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chonghuai Liu, bGl1Y2hvbmdodWFpQGNhYXMuY24=; Ying Zhang, emhhbmd5aW5nMDVAY2Fhcy5jbg==
 Faiz Ur Rahman
Faiz Ur Rahman Irshad Ahmad Khan
Irshad Ahmad Khan Ali Aslam
Ali Aslam Ruitao Liu
Ruitao Liu Lei Sun
Lei Sun Yandi Wu
Yandi Wu Muhammad Muzammal Aslam1
Muhammad Muzammal Aslam1 Peng Li
Peng Li Chonghuai Liu
Chonghuai Liu Ying Zhang
Ying Zhang