- Key Laboratory of Pig Molecular Quantitative Genetics of Anhui Academy of Agricultural Sciences, Anhui Provincial Key Laboratory of Livestock and Poultry Product Safety Engineering, Institute of Animal Husbandry and Veterinary Medicine, Anhui Academy of Agricultural Sciences, Hefei, China
The genetic resources among pigs in Anhui Province are diverse, but their value and potential have yet to be discovered. To illustrate the genetic diversity and population structure of the Anhui pigs population, we resequenced the genome of 150 pigs from six representative Anhui pigs populations and analyzed this data together with the sequencing data from 40 Asian wild boars and commercial pigs. Our results showed that Anhui pigs were divided into two distinct types based on ancestral descent: Wannan Spotted pig (WSP) and Wannan Black pig (WBP) origins from the same ancestor and the other four populations origins from another ancestor. We also identified several potential selective sweep regions associated with domestication characteristics among Anhui pigs, including reproduction-associated genes (CABS1, INSL6, MAP3K12, IGF1R, INSR, LIMK2, PATZ1, MAPK1), lipid- and meat-related genes (SNX19, MSTN, MC5R, PRKG1, CREBBP, ADCY9), and ear size genes (MSRB3 and SOX5). Therefore, these findings expand the catalogue and how these genetic differences among pigs and this newly generated data will be a valuable resource for future genetic studies and for improving genome-assisted breeding of pigs and other domesticated animals.
1 Introduction
Domestic pig (Sus scrofa) is an important livestock species, and has served as a source of stable food, organic fertilizer, industrial raw material, and medicine for humans since its domestication in the early Neolithic period. Since then, the domestic pig has promoted population growth and socioeconomic transformation from a hunter-gatherer society to sedentary agricultural settlements. Through domestication, natural and artificial selection has generated 48 pig breeds in China (China National Commission of Animal Genetic Resources. 2011). Among evolutionary biologists, there is substantial interest in elucidating how natural and artificial selection have shaped modern pig genomes and the how these genetic differences contribute to the complex phenotypes seen among pigs.
Whole-genome resequencing has the potential to resolve pig population structure and identify phenotype-associated functional genes that arose during domestication and breeding of different pig breeds. In recent years, whole-genome sequencing studies on germplasm characteristics among different pig breeds have revealed the evolutionary history of the pig population and elucidated the genetic basis of complex phenotypes, while also providing new focuses in the future breeding of Chinese indigenous pigs (Rubin et al., 2012; Li et al., 2013; Ai et al., 2015; Frantz et al., 2015). Several causative genes have been reported as responsible for phenotypic variation among Chinese indigenous pigs. For instance, HIFA has been identified as the gene responsible for hypoxic adaptation in Tibetan pigs (Li et al., 2013). Moreover, TGFB3 and DAB2IP have been shown to be important in regulating the number of ribs (Zhu et al., 2015). Additionally, MITF, EDNRB, and MC1R have been identified as the genes responsible for color variations among Chinese breeds (Fang et al., 2009; Wang et al., 2015; Zhao et al., 2018). However, most of the aforementioned studies focused only on a few phenotypes among a limited number of breeds or populations. Therefore, considering that China has such a large and a diverse number of pig breeds, more genes with germplasm characteristics still need to be identified.
The Anhui Province, located in the Yangtze River Delta, is one of the important birthplaces of Chinese civilization. Additionally, the province is home to a wide variety of indigenous pig populations, including the Dingyuan pig (DYP), Huoshou Black pig (HBP), Wannan Spotted pig (WSP), Wannan Black pig (WBP), Anqing Six-end-white pig (ASP), and Wei pig (WP). These pig breeds are representative autochthonous Chinese breeds with long histories of breeding in the country. Of mention is that Anhui indigenous pig populations are facing crises of population decline and loss of genetic characteristics. The reasons for this are complex, including long cycles of feeding, the introduction of modern commercial pigs, etc. however, insufficient protection is the most direct cause. To date, there have been no systematic studies (i.e., based on a large panel of Anhui indigenous pig populations) seeking to illustrate the genomic diversity and signatures of selection among Anhui indigenous pig populations. Therefore, a comprehensive study of the genetic diversity, phylogenetic relationships, population structure, demographic history, and selection signatures among the Anhui pig populations are critical. Furthermore, the Anhui pig populations represent unique genetic resources for understanding the role of functional diversity, which can be applied in other contexts as well. Moreover, characterizing genetic variations and identifying the associated phenotypes are vital for future breeding of indigenous pig populations.
For this purpose, we performed whole-genome sequencing of six representative pig populations in Anhui (WSP, n = 25; WBP, n = 25; ASP, n = 25; WP, n = 25; DYP, n = 25; HBP, n = 25). Using whole-genome sequencing data in conjunction with downloaded sequence data, we explored the genetic diversity, phylogenetic relationships, population structure, and demographic history of the six pig populations. In addition, we calculated the FST (fixation index) and log2 (θπ ratio) values to elucidate the signatures of selection and then characterized these candidates’ genes are present in the genome of Anhui pigs. Our findings provide valuable insights into the evolutionary history of Anhui pig populations and reveal several promising candidate genes for future indigenous pig breeding.
2 Materials and methods
2.1 Sample collection and genome sequencing
To survey the overall genetic diversity among autochthonous pig breeds in Anhui province, blood samples from 150 individuals within six indigenous Anhui pig populations were collected from pig conservation farms (Figure 1; Supplementary Table S1). These breeds included: ASP (N = 25, ♀15, ♂10), WSP (N = 25, ♀10, ♂15), WBP (N = 25, ♀11, ♂14), WP (N = 25, ♀13, ♂12), DYP (N = 25, ♀4, ♂21), HBP (N = 25, ♀10, ♂15). This study was conducted in accordance with and was approved by the Animal Care Committee of the Anhui Academy of Agricultural Sciences (Hefei, China; no. AAAS2020-04). For the blood samples, genomic DNA was extracted using the standard phenol–chloroform method (Sambrook and Russel 2001) and stored at 4°C to avoid freeze–thawing. The quality and concentration of the DNA were assessed using a 0.5% agarose gel (run for >8 h at 25 V) and a Nanodrop spectrophotometer (Thermo Fisher Scientific). DNA was then fragmented and treated in accordance with the Illumina DNA sample preparation protocol. Sequencing libraries were constructed and sequenced on the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, United States) using paired-end 150 bp reads through the Novogene service (Beijing, China).
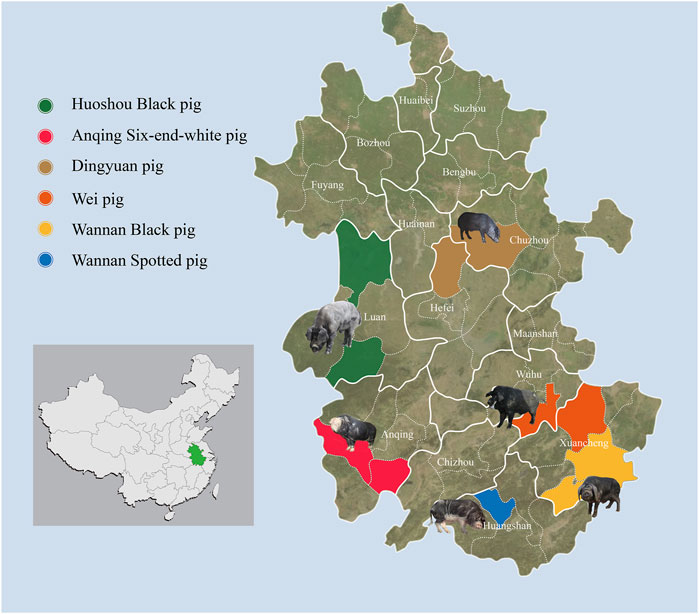
FIGURE 1. Geographic distribution and appearances of typical pigs. 150 Anhui indigenous pigs representing six populations were included.
For comparison, 40 pig resequenced data sets from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra/) were downloaded (Ai et al., 2015; Rubin et al., 2012; Zhao et al., 2018; Groenen et al., 2012) (Supplementary Table S2), including Asian wild boar, Duroc, Pietrain, Yorkshire, and Landrace.
2.2 Read mapping and variant calling
Before alignment, adapter and low-quality reads [e.g., reads with 10% unidentified nucleotides (N); reads with >10 nt aligned to the adapter; reads with >30% bases with Phred quality ≤25] were filtered using the NGSQCToolkit_v2.3.351. The remaining, high-quality reads were aligned with the pig reference genome (ftp://ftp.ensembl.org/pub/release-99/variation/gvf/sus_scrofa/) using BWA-0.7.12 software (https://sourceforge.net/projects/bio-bwa/) default parameters (Li and Durbin. 2009). For the mapping process, a BAM file index was built using SAMtools (https://github.com/samtools/samtools/releases/) (Li et al., 2009). The BAM file was sorted using the Picard-tools-1.105 software (https://github.com/broadinstitute/picard/releases). Duplicate read data were excluded. Next, we performed variant calling for all samples on a population-scale using SAMtools mpileup with the parameters “-m 2 -F 0.002 -d 1000” and then bcftools to view variants (Li and Durbin 2011). We further resolved the SNPs according to the following criteria: QD (Variant Confidence/Quality by Depth) < 5.0, MQ (RMS Mapping Quality) < 40.0, FS (Phred-scaled p value using Fisher’s exact test to detect strand bias) > 60.0, SOR (Strand Odd Ratio) > 3.0, MQRankSum (Z-score from Wilcoxon rank sum test of Alt vs. Ref read mapping qualities) < −10.0, ReadPosRankSum (Z-score from Wilcoxon rank sum test of Alt vs. Ref read position bias) < −8.0 and QUAL < 30. SNPs that were bi-allelic missed >40% of calls and had a MAF < 0.05 were removed and the remaining SNPs comprised the basic set. Variants were annotated using ANNOVAR (Wang et al., 2011). The downloaded resequencing data were analyzed using the process described above.
2.3 Genomic diversity and population genetic structure
Nucleotide diversity (θπ) was calculated based on the maximum likelihood estimates of the fold-site frequency spectrum using a sliding window approach (50-kb windows with 50-kb steps; Kim et al., 2011). To illustrate the population structure, principal component analysis was performed using the GCTA software (v.1.25; Yang et al., 2011). Additionally, phylogenetic trees were inferred through the neighbor-joining (NJ) method and implemented in TreeBest (http://treesoft.sourceforge.net/treebest.shtml) with 1000 bootstraps. The population structure was deduced using the ADMIXTURE software (https://dalexander.github.io/admixture/index.html) with a kinship (K) set from 2 to 6 (Alexander et al., 2009).
2.4 Pairwise sequential Markovian coalescence analysis
The demographic history of the six Anhui indigenous pig populations was examined using the hidden Markov model approach and implemented according to the pairwise sequential Markovian coalescence (PSMC) model (Li and Durbin 2011). To estimate the distribution time, we used the parameters of g = 5 and a rate of 2.5 × 10−8 mutations per generation.
2.5 Selective sweeps analysis
To identify genetic variants associated with adaptation of the Anhui pig populations, comparisons were performed between three populations: 1) the Anhui pig populations versus the Asian wild boar, and 2) the Anhui pig populations versus the commercial pig population. The θπ and FST were calculated using a 40-kb sliding window approach with a 20-kb step-size, in PopGenome (Pfeifer et al., 2014). The overlapping windows within the top 1% or 5% of the FST and θπ ratio (log2-transformed) distributions were considered putative selective regions (Li et al., 2013). Gene contents in “significant” genomic regions were retrieved from the Ensembl Genes 102 Database and analyzed in the BioMart software (http://asia.ensembl.org/biomart/martview/). To further explore the potential biological significance of the genes within these sweep regions, Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted to perform functional annotation for these genes using KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/). The terms and pathways exhibiting p-values < 0.05 were considered significant.
3 Results
3.1 Whole-genome sequencing and genetic variations
In total, 150 individuals from six pig populations indigenous to Anhui were selected for whole-genome resequencing. In total, 31 billion reads (or ∼4,678 Gb of sequences) were generated. Average data sets consisted of 31.2 Gb (10.85 X) per individual and 800.36, 750.16, 724.8, 797.18, 834.25, and 771.53 Gb for the HBP, ASP, WSP, DYP, WBP, and WP collective populations, respectively. We also downloaded data for 40 non-indigenous individuals (Asian wild boars, Duroc, Landrace, Yorkshire, and Pitelan), resulting in 5,732 Gb of data in total. Using BWA, reads were aligned to the pig reference genome with an average alignment rate of 98.49% ± 0.14% and an estimated error rate of 0.03% ± 0.005% (Supplementary Tables S3, S4). After stringent variant calling and filtering, approximately 23 million single nucleotide polymorphisms (SNPs) were identified. For all detected SNPs, the average transition/transversion ratio was 2.42, which is consistent with a previous study (Kerstens et al., 2009). Further annotation of these SNPs revealed that they were most abundant in intergenic regions (53.58%) and intronic regions (42.97%), followed by downstream (0.62%) and upstream (0.56%) regions, whereas only 0.7% were located in coding sequences. Most variations were located in non-coding sequences, indicating that during domestication and breeding, the non-coding sequences could potentially change protein function by regulating gene expression.
3.2 Genomic diversity and population genetic structure
Nucleotide diversity reflects the degree of polymorphism within a population (Nei and Li. 1979). On a genome-wide window scale of 50-kb, with steps of 50-kb, the foreign pig breeds displayed reduced nucleotide diversity compared with the indigenous Anhui populations (Figure 2A). This is likely the result of extensive artificial selection over generations. We performed PCA analysis for the experimental populations in GCTA (Figure 2B). This analysis was successful in separating breed clusters based on genotypic data, where the foreign breeds, ASP and WP, HBP and DYP, and WBP and WSP were clustered together. These findings are consistent with the established information on the Anhui populations. To elucidate the phylogenetic relationship among the Anhui pig breeds, unrooted phylogenetic trees were constructed, revealing genetically distinct clusters (Figure 2C). This was consistent with the results of the PCA analysis, revealing clustering into distinct genetic groups. We used the ADMIXTURE software to determine the degree of admixture when K is increased from 2 to 6, where K is the assumed number of ancestral populations. K = 2 was suggested as the most plausible number among our samples (Figure 2D), reflecting the divergence of the Anhui populations and commercial breeds within the pig population. These results show that the current pig populations (WP, DYP, ASP, and HBP) may have some of the same genetic information as foreign pigs, possibly owing to the introduction of modern commercial pigs. We also conducted an admixture analysis of the Anhui populations (Figure 2E), and the results showed that K = 2 was the probable number of genetically distinct groups within the Anhui populations. It was revealed that WSP and WBP originated from the same ancestor and the other four populations from another ancestor. At K = 3, WBP was separated from WSP, and the others still descended from the same ancestor. At K = 4, WBP was still distinct from WSP, WP was grouped with ASP, and DYP was grouped with HBP, in agreement with prior knowledge of Anhui pig populations.
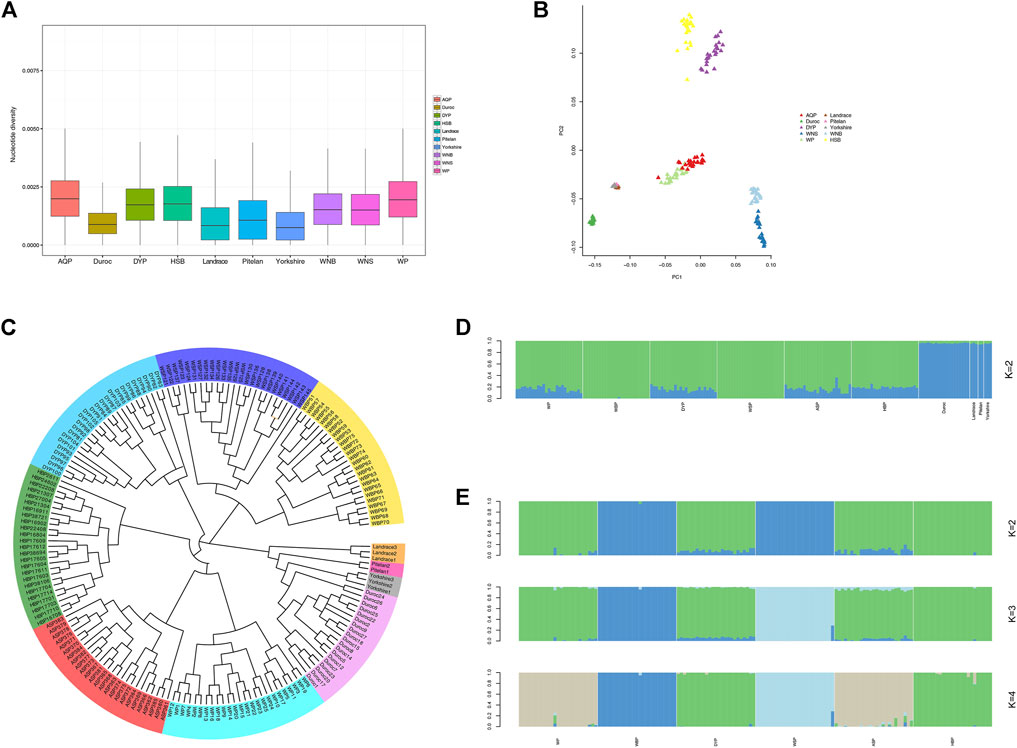
FIGURE 2. Population structure and relationship of Anhui in comparison to commercial pig. (A) Genome-wide distribution of nucleotide diversity among Anhui pig breeds and four commercial pig breeds with a 50-kb window. (B) Principal component analysis of Anhui pig breeds and four commercial pig breeds, PC1 against PC2. (C) Neighbor-joining tree of the relationships between the ten pig populations. The scale bar represents the identity-by-state (IBS) score between pairs of animals. (D) Proportion of ancestry for each individual assuming different number of ancestral population (K = 2). Colors in each vertical line represent the likelihood proportion of an animal genome assigned to a source population. (E) Proportion of ancestry for each individual in Anhui pig populations.
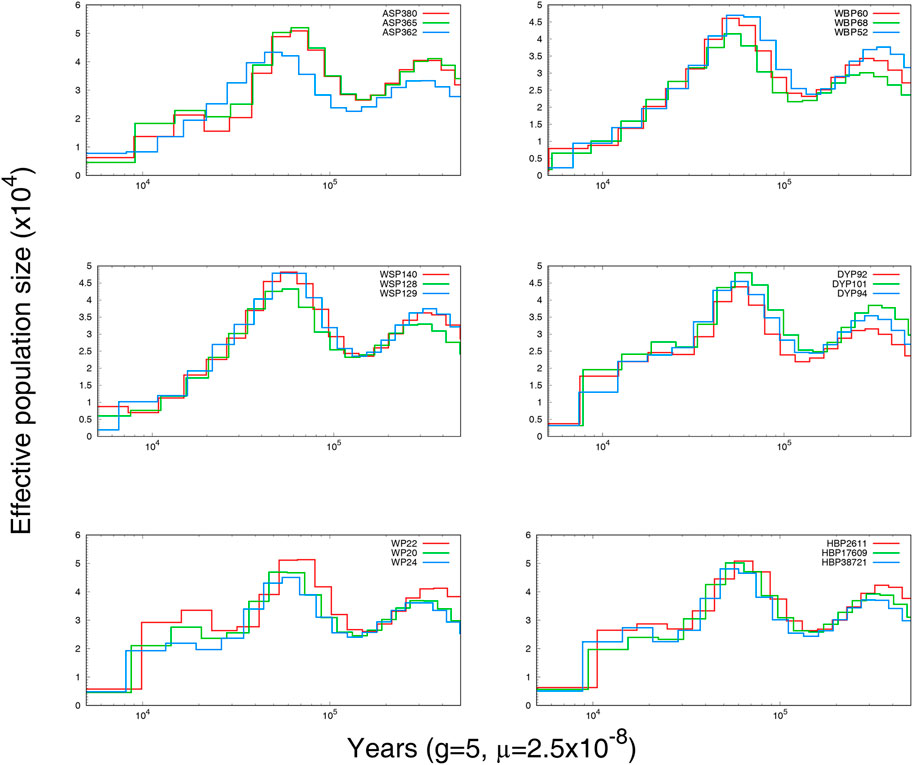
3.3 Demographic history
To explore the demographic histories of the six Anhui pig populations, PSMC was used to infer the historical population size. The variations in effective population size (Ne) over time are shown in Figure 3. Individuals from the same breed displayed similar historical fluctuations in effective population size. All breeds shared a collective population decline beginning at approximately 10,000 BP, likely a consequence of Neolithic domestication events (Zeder, 2008; Boitard et al., 2016).
3.3.1 The selective regions of Anhui pig populations compared to those of the Asian wild boar population
To identify the signatures of selection, we compared the genomes of Anhui pig populations to those of Asian wild boars. Specifically, we used both θπ- and FST-based cross approaches to investigate the signatures of selection. For this study, the regions within the top 1% were defined as the selected regions. The genome distribution of the two statistics were shown in Figures 4A,B. A total of 402 selective regions harboring 187 genes were identified in the top 1% of the FST and log2 (θπ, ratios) distributions (16.1 Mb of the genome, Figure 4C and Supplementary Tables S5, S6). All 187 genes were used for GO and KEGG analyses (Supplementary Tables S7, S8 and Supplementary Figures S1, S2). Functional annotation revealed that the selected genes may have effects on reproduction (CABS1, INSL6, MAP3K12, and MAPK1) and lipid- and meat-related processes (SNX19 and MSTN).
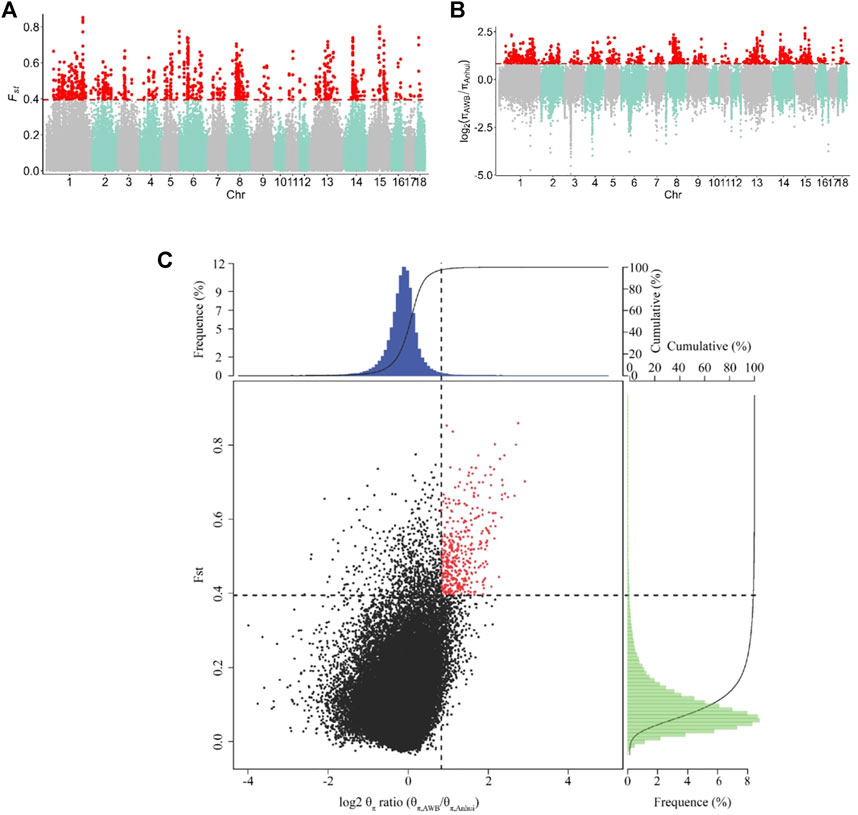
FIGURE 4. Identification of genomic regions with strong selective sweep signals in Anhui six pig population, which are calculated in a 40-kb sliding window approach with 20 kb step-size. (A) Distribution of FST values among autosome chromosomes. The red line represents the 0.01 level. (B) Distribution of log2 (θπ ratio) among autosomal chromosomes. The red line represents the 0.01 level. (C) The final selection regions based on two statistics. Points located to the right of the vertical dashed lines (corresponding to 1% right tails of the empirical log2 (θπ ratio) distribution, where log2 (θπ ratio) is 0.824992) and above the horizontal dashed line (1% above tail of the empirical FST distribution, where FST is 0.395220) were identified as selected regions for Anhui pig population (red points). The AWB refer to Asia wild boar, the Anhui refer to Anhui pig population.
3.3.2 The selective regions of Anhui pig populations compared to those of the commercial pig population
To capture potential genes that underwent divergent selection between the Anhui and commercial pig populations, we estimated the genome-wide values of log2 (θπ ratio) and FST based on the whole-genome resequencing data of 177 samples from ten pig populations. The genome distribution of the two statistics were shown in Figures 5A,B. Using the top 5% FST cutoff (FST ≥ 0.2527) and log2 (θπ ratio) cutoff [log2 (θπ ratio) ≥ 0.0011], we detected nine hundred and sixty-seven selective regions containing 331 candidate genes (38.7 Mb of the genome; Figure 5C and Supplementary Tables S9, S10). All 331 genes were used for GO and KEGG analyses (Supplementary Figures S3, S4; Supplementary Tables S11, S12). Functional enrichment analyses revealed that the selected genes may play an important role in reproduction (CABS1, MAPK1, IGF1R, INSR, LIMK2, and PATZ1), meat quality and fat deposition capacity (MC5R, MSTN, PRKG1, CREBBP, and ADCY9), and ear size (MSRB3 and SOX5).
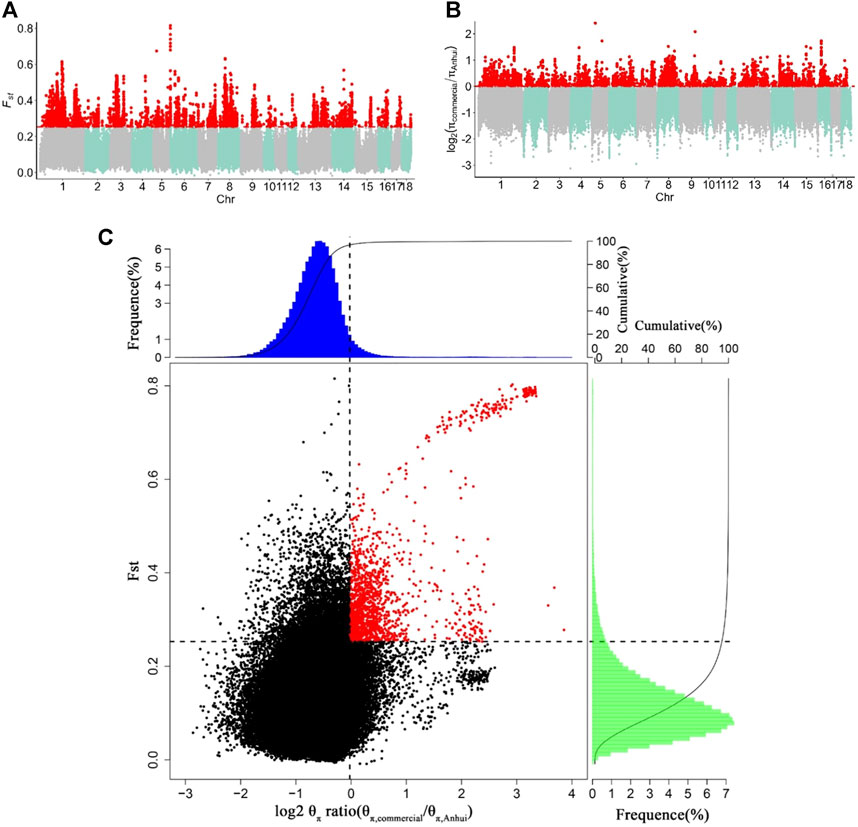
FIGURE 5. Identification of genomic regions with strong selective sweep signals between Anhui six pig population and four commercial pig population, which are calculated in a 40-kb sliding window approach with 20 kb step-size. (A) Distribution of FST values among autosome chromosomes. The red line represents the 0.05 level. (B) Distribution of log2 (θπ ratio) among autosomal chromosomes. The red line represents the 0.05 level. (C) The final selection regions based on two statistics. Points located to the right of the vertical dashed lines (corresponding to 5% right tails of the empirical log2 (θπ ratio) distribution, where log2 (θπ ratio) is 0.0.0011) and above the horizontal dashed line (5% above tail of the empirical FST distribution, where FST is 0.2527) were identified as selected regions for Anhui pig population (red points). The commercial refers to four commercial pigs, the Anhui refer to Anhui pig population.
4 Discussion
The pig, as one of the first domesticated animals, plays an important role in many aspects of human life. In recent years, the continued protection and utilization of the biological seed industry has attracted significant attention in China. The successively implemented “The third national census of livestock and poultry genetic resources” and “Precise identification of livestock and poultry germplasm resources” could help us to better understand the differences among pig breeds, and to protect and utilize the unique Chinese pig germplasm. To systematically analyze the genetic relationships, genetic diversity, population structure, and selection signatures resulting from natural and artificial selection in the Anhui pig populations, 150 unrelated samples were collected from six pig breeds along with sequencing data from 40 foreign pigs. In this study, the genetic variation in the Anhui pig populations was surveyed, using blood samples from pig conservation farms, from which DNA was isolated and sequenced in order to construct a genetic conservation system. Going forward, this information can serve to guide the protection and utilization of indigenous pig breeds in Anhui. Moreover, the genetic structure of this population has also been elucidated. The WSP and WBP descended from the same ancestor, and the other four breeds from another ancestor. We also found that the Anhui pig populations exhibited a highly similar pattern of historical fluctuation in effective population size at approximately 10,000 BP, which is likely the consequence of Neolithic domestication events. In addition, several genes have been found to be associated with important economic traits.
4.1 The selected genes of Anhui pig populations compared to those of the Asian wild boar population
Domestic pigs have higher fertility rates than those of wild boars. GO analysis revealed several genes related to reproduction, including those related to reproductive process (GO: 0022414, five genes), reproduction (GO: 0000003, six genes), and fertilization (GO: 0007283, two genes). The acrosome reactions decreased significantly after the porcine capacitated sperm were treated with anti-pCABS1 antiserum, suggesting that porcine expression of CABS1 plays an important role in acrosome reactions (Shawki et al., 2016). In this study, two variants of the CABS1 gene were identified (Supplementary Table S13; exon1: c; G1047T: p.S349S; exon1:c.C1069G: p.L357V). Notably, the “exon1:c.C1069G: p.L357V” variant results in a Leu to Val substitution, which may in turn alter the expression of CABS1. The CABS1 gene was identified as a selected gene in ASP in our previous study (Zhang et al., 2020), which suggests that CABS1 is commonly selected within the Anhui pig populations. Another gene involved in reproduction, insulin-like 6 (INSL6), is a member of the insulin superfamily that plays an important role in the progression of spermatogenesis. Moreover, a deficiency in INSL6 can result in varying levels of male infertility (Lok et al., 2000; Brailoiu et al., 2005; Ivell and Grutzner. 2009; Chan et al., 2011). In this study, a missense variant of INSL6 (Supplementary Table S14; exon2:c.C403G: p.Q135E) was found among the Anhui pig breeds. This variation promotes a Gln-to-Glu substitution, implying that variations may impact the progression of spermatogenesis. Mitogen-activated protein kinase 12 (MAP3K12) is involved in the MAPK signaling pathway and is specifically selected in a high fecundity goat lineage, indicating its role in reproduction (Lai et al., 2016). MAPK1 plays an important role in embryonic and placental development (Hatano et al., 2003). Furthermore, a signal adequate for promoting trophoblast proliferation and invasion can decrease below thresholds if MAPK1 protein expression is insufficient (Saba et al., 2003; Jeong et al., 2013). In a previous study, population high-density SNP array analysis of multiparous and uniparous sheep revealed that MAPK1 is a selected gene among Kazakhstan sheep (Wang et al., 2020), highlighting its role in reproduction. One synonymous SNV was also identified in MAPK1 (Supplementary Table S15; exon1:c.C144T: p.L48L).
Genes related to lipid and meat traits have also been identified. Sorting nexin 19 (SNX19) is primarily associated with meat quality and plays a vital role in the fat subnetwork (Kogelman et al., 2014). Interestingly, SNX19 was identified as a selected gene in a previous pig study (Guo et al., 2021). Five nonsynonymous variants were identified for SNX19 (Supplementary Table S16; exon11: c. A1037G: p.N346S, exon7:c.C487T: p.P163S, exon4: c. G94T: p.V32L, exon2:c.C1834T: p.L612F, exon1: c. G37C: p.A13P), and the variants with observable phenotypes were used to discriminate the functional site. We also identified myostatin (MSTN), a transcription factor belonging to the transforming growth factor beta (TGFβ) superfamily which plays an important role in muscle and fat development in pigs. Gene ontology annotation showed that among the Anhui pig populations, MSTN was enriched for growth (GO: 0040007), growth factor activity (GO: 0008083), and protein binding (GO: 0003824). Furthermore, a study by Deng et al. (2012) revealed that MSTN impacts the development of the longissimus muscle and the rectus superior muscle. Transgenic pigs with MSTN displayed decreased muscle growth and significant increases in intramuscular fat (Yang et al., 2009). Additionally, it has been shown that MSTN has the potential to regulate adipogenesis of mesenchymal stem cells during the determination and differentiation phases (Sun et al., 2016). MSTN inhibits adipogenesis in preadipocytes, (Hirai et al., 2007; Carrarelli et al., 2015), whereas it promotes adipogenesis in pluripotent stem cells (Feldman et al., 2006; Pantoja et al., 2008). In animals, MSTN-knockout typically reduces fat mass and resistance to diet-induced obesity (Guo et al., 2009; Gu et al., 2016).
4.2 The selected genes of Anhui pig populations compared to those of the commercial pig population
We identified several genes related to reproduction among the GO terms and KEGG pathways, with each playing an important role in spermatogenesis, ovarian steroidogenesis, the estrogen signaling pathway, the FoxO signaling pathway, or oocyte meiosis. Of these genes, calcium-binding protein spermatid associated 1(CABS1) and mitogen-activated protein kinase 1 (MAPK1) were those found in the selective regions of the Anhui pig population, but not in the selective regions of the Asian wild boar. The insulin-like growth factor 1 receptor (IGF1R) is widely expressed in mammals (including pigs, sheep, goats, and ducks) and is involved in growth, carcass traits, and reproductive performance (Liu et al., 1993; Terman, 2011; Hoopes et al., 2012; Kijas et al., 2012; Zhao et al., 2018; Jin et al., 2020; Grochowska et al., 2021; Jiang et al., 2021). In female mice, IGF1R was found to play a vital role in steroidogenesis, follicle survival, and fertility (Baumgarten et al., 2017), where it helped transfer nutrients to the fetus (Hellström et al., 2016). Zhou et al. (2021) found that IGF1R is also necessary for epithelial differentiation and normal uterine preparation for embryo implantation (Zhou et al., 2021). Given this background, IGF1R likely contributes to phenotypic differences in growth and reproduction among pigs. In this study, 450 variations were identified in the IGF1R gene (Supplementary Table S18), and only seven were located in exons, none of which resulted in amino acid changes. Conversely, three variations were found within the 3′UTR, and 30 within the 5′UTR that may regulate the function of the IGF1R. Insulin receptor (INSR), a tyrosine kinase receptor, affects development and growth (Hubbard. 2013; Bedinger and Adams. 2015). INSR has been validated as a polycystic ovary syndrome risk locus using a very large, well-designed case-control GWAS (Shi et al., 2012). Meanwhile, INSR has been confirmed to be necessary for optimal endometrial proliferation and implantation (Sekulovski et al., 2021). Furthermore, INSR plays a crucial role in controlling adipose tissue development and adipocyte survival (Boucher et al., 2012; Cignarelli et al., 2019). INSR may be responsible for fat accumulation in adipose tissues, which could induce angiopoietin-like 8, inhibit lipolysis, control postprandial fat storage in white adipose tissue, and direct that fatty acids be stored in the adipose tissue during the fed state (Ahbara et al., 2019). The testis-specific isoform of LIM kinase 2 (tLIMK2) plays a key role in the progression of spermatogenesis (Takahashi et al., 2002). The weight of LIMK2−/− mice sharply decreased by 20% compared to that of control mice. Furthermore, inhibition of LIMK1/2 activity results in failure of embryo cleavage and blastocyst formation (Duan et al., 2018). POZ/BTB and AT hook containing zinc finger 1(PATZ1), a zinc finger protein, can affect the basal activity of different promoters. PATZ1 plays a crucial role in embryonic development (Valentino et al., 2013) and normal male gametogenesis and impairment of PATZ1 result in disruption of testis cytoarchitecture and block spermatogenesis (Fedele et al., 2008).
According to previous research, indigenous Anhui pig breeds exhibit significant differences in meat quality traits and fat deposition capacity compared to commercial pig breeds (Xu et al., 2018; Hu et al., 2019; Li et al., 2021). In the present study, several genes were found to be associated with fat deposition and other carcass quality traits. The melanocortin five receptor gene (MC5R), a member of the G protein-coupled receptor superfamily, participates in lipid production, fatty acid oxidation of skeletal muscle, and lipolysis of dipocytes. MC5R is associated with the metabolism of skeletal muscle, fatty acids, and fat cells, as well as dysfunction of the exocrine gland under abnormal expression (Zhang et al., 2011). The polymorphisms of MC5R are associated with obesity in humans (Chagnon et al., 1997; Valli et al., 2008). Furthermore, MC5R deficiency accelerates lipolysis and reduces oil secretion (Chen et al., 1997). Whole genome analysis of 46 cattle from six representative Chinese breeds together with international breeds has shown that MC5R was under-selected (Mei et al., 2018). In the pig genome, MC5R was found close to the 97.625–98.725 Mb regions on swine chromosome 6 (SSC6). The regions of SSC6 are associated with lipid metabolism, back fat thickness, and intramuscular fat percentage (Switonski et al., 2013; Ma et al., 2018), and play a vital role in proinflammatory activity (Jun et al., 2010). Among the pigs in this study, three variations were found in the 3′UTR of the MC5R gene, and four variations were located in the exon (Supplementary Table S19). Two exonic variants (exon3:c.A952G:p. S318G; exon3:c.A580G:p.T194A) resulted in amino acid changes from Ser to Gly and Thr to Ala. The exon1: c. G92A created a premature stop codon in the MC5R amino acid sequence. Additionally, kinase cGMP-dependent 1 (PRKG1) has been shown to regulate adipocytes lipolysis and plays an important role in pig fatty acid composition (Revilla et al., 2017). Interestingly, PRKG1 exhibits differential expression between the high and low fatty acid composition groups in muscle according to RNA-Seq analysis (Puig et al., 2014). Moreover, PRKG1 knockout mice have decreased triglyceride stores in brown adipose tissue (Amieux and Mcknight, 2010). The taste and quality of cooked and cured meat are directly affected by the oxidative stability of the muscle, which is related to fatty acid (FA) composition (Serra et al., 1998; Wood et al., 2008). We also found two genes, CREB binding protein (CREBBP) and adenylate cyclase 9 (ADCY9), which are involved in cAMP signaling. cAMP signaling regulates energy homeostasis in multiple tissues by mediating the action of metabolism-controlling hormones such as glucagon and epinephrine (Ravnskjaer et al., 2016). A previous GWAS and post-GWAS study discovered that CREBBP and ADCY9 are located inside regions that are significantly associated with meat pH (Verardo et al., 2017). For ADCY9, there were 13 exonic variants, three of which were missense variants (Supplementary Table S20, exon8:c.A2594G:p.H865R; exon8:c.T2669C:p.M890T; exon11:c.G3943T:p.G1315C), results in amino acid changes that may impact meat quality. CREBBP regulates a plethora of metabolic target genes involved in glucose metabolism (Ravnskjaer et al., 2016), which can also potentially affect meat quality due to acidification. In this study, one missense variant was identified (Supplementary Table S21, exon22:c.A3907G:p. I1303V) in which the amino acid Ile was changed to Val.
Two selected genes were found to be associated with ear size phenotypic traits. The size and type of the ears are important conformational characteristics that distinguish pig breeds. Many indigenous Chinese pig breeds, such as the six Anhui pig populations, have unusually large floppy ears. In contrast, commercial breeds (Duroc, Landrace, Yorkshire, and Pitelan) have smaller, more erect ears. In this study, we identified two important candidate genes influenced by selection. The first is methionine sulfoxide reductase B3 (MSRB3), which has been shown to be associated with ear size in sheep, goats, dogs, and pigs (Webster et al., 2015; Zhang et al., 2015; Chen et al., 2018; Kumar et al., 2018; Zhao et al., 2020; Posbergh and Huson. 2021). In the present study, a variant in the 3′-UTR of MSRB3 was identified among the Anhui pig breeds (Supplementary Table S17; g. T29862412C), suggesting that it could be the source of their erect ear phenotype. SRY-box transcription factor 5 (SOX5) encodes a member of the SOX transcription factor family, and has been shown to play a role in chondrogenesis (Lefebvre et al., 1998; Smits et al., 2001). Additionally, it has been identified as a selected gene in Duroc pigs and Duroc × Korean native pigs, in which it plays a key role in ear morphology (Edea et al., 2017). Moreover, mutations in genes encoding homeobox transcription factors are often responsible for the defective development of the outer ear (Fekete, 1999). Positive selection of MSRB3 and SOX5 among the Anhui pig populations, but not among the commercial pig breeds, may be the genetic mechanism behind the ear size phenotype.
Although the present study has produced many interesting findings, it has its limitations. Firstly, as the phenotypic values were not collected, functional experimental assays are still needed to further validate the associations between the phenotypes and genotypes of the mentioned variants and to identify the targets involved in reproduction, lipid and meat quality, and ear size.
In this study, we generated a novel catalog of population genomic data on the Anhui pig populations using whole-genome resequencing. Population genomic analyses have further elucidated genomic variation, population structure, demographic history, and signatures of selection among Anhui pigs. Moreover, we discovered several potential signatures of selection associated with the domestication characteristics of the Anhui pig populations, with selected regions involved in reproduction, lipid and meat characteristics, and ear size. These findings substantially expand the catalogue of genetic variants among pigs, and the newly generated genome-wide data are a valuable resource for future genetic studies and those to improve genome-assisted breeding of pigs and other domestic animals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by the recommendations of the Animal Care Committee of Anhui Academy of Agricultural Sciences (Hefei, People’s Republic of China). The protocol was approved by the Animal Care Committee of Anhui Academy of Agricultural Sciences (No. AAAS2020-04).
Author contributions
Conceptualization: WZ and CW; Methodology: WZ and YJ; Investigation: WZ and XiL; Resources: XiL, YJ, SS, CX, and XuL; Data curation: WZ; Writing—original draft preparation: WZ; Funding acquisition: WZ, MZ, and CW.
Funding
This work was supported by the grants from Anhui Academy of Agricultural Sciences Key Laboratory Project (No. 2021YL023), Anhui Province Financial Fund for Modern Seed Industry Project, Anhui Province Natural Science Foundation Youth Fund Project (2108085QC135); the Special Fund for Anhui Agricultural Research System (AHCYJSTX-05-12, AHCYJSTX-05-23); the Anhui Provincial Key Laboratory of Livestock and Poultry Product Safety Engineering Young Talents Support Engineering Innovation Guidance Fund (XMT 2022-09).
Acknowledgments
We thank many people not listed as authors who provided help, encouragement and feedback.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1022261/full#supplementary-material
References
Ahbara, A., Bahbahani, H., Almathen, F., Al Abri, M., Agoub, M. O., Abeba, A., et al. (2019). Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Front. Genet. 9, 699. doi:10.3389/fgene.2018.00699
Ai, H., Fang, X., Yang, B., Huang, Z., Chen, H., Mao, L., et al. (2015). Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 47, 217–225. doi:10.1038/ng.3199
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi:10.1101/gr.094052.109
Amieux, P. S., and McKnight, G. S. (2010). Cyclic nucleotides converge on Brown adipose tissue differentiation. Sci. Signal. 3, pe2. doi:10.1126/scisignal.3104pe2
Baumgarten, S. C., Armouti, M., Ko, C., and Stocco, C. (2017). IGF1R expression in ovarian granulosa cells is essential for steroidogenesis, follicle survival, and fertility in female mice. Endocrinology 158, 2309–2318. doi:10.1210/en.2017-00146
Bedinger, D. H., and Adams, S. H. (2015). Metabolic, anabolic, and mitogenic insulin responses: A tissue-specific perspective for insulin receptor activators. Mol. Cell. Endocrinol. 415, 143–156. doi:10.1016/j.mce.2015.08.013
Boitard, S., Rodriguez, W., Jay, F., Mona, S., and Austerlitz, F. (2016). Inferring population size history from large samples of genome-wide molecular data-an approximate Bayesian computation approach. PLoS Genet. 12, e1005877. doi:10.1371/journal.pgen.1005877
Boucher, J., Mori, M. A., Lee, K. Y., Smyth, G., Liew, C. W., Macotela, Y., et al. (2012). Impaired thermogenesis and adipose tissue development in mice with fat-specific disruption of insulin and IGF-1 signalling. Nat. Commun. 3, 902. doi:10.1038/ncomms1905
Brailoiu, G. C., Dun, S. L., Yin, D., Yang, J., Chang, J. K., and Dun, N. J. (2005). Insulin-like 6 immunoreactivity in the mouse brain and testis. Brain Res. 1040, 187–190. doi:10.1016/j.brainres.2005.01.077
Carrarelli, P., Yen, C. F., Arcuri, F., Funghi, L., Tosti, C., Wang, T. H., et al. (2015). Myostatin, follistatin and activin type II receptors are highly expressed in adenomyosis. Fertil. Steril. 104, 744–752. doi:10.1016/j.fertnstert.2015.05.032
Chagnon, Y. C., Chen, W. J., Pérusse, L., Chagnon, M., Nadeau, A., Wilkison, W. O., et al. (1997). Linkage and association studies between the melanocortin receptors 4 and 5 genes and obesity-related phenotypes in the Québec Family Study. Mol. Med. 3, 663–673. doi:10.1007/bf03401705
Chan, L. J., Hossain, M. A., Samuel, C. S., Separovic, F., and Wade, J. D. (2011). The relaxin peptide family-structure, function and clinical applications. Protein Pept. Lett. 18, 220–229. doi:10.2174/092986611794578396
Chen, C., Liu, C., Xiong, X., Fang, S., Yang, H., Zhang, Z., et al. (2018). Copy number variation in the MSRB3 gene enlarges porcine ear size through a mechanism involving miR-584-5p. Genet. Sel. Evol. 50, 72. doi:10.1186/s12711-018-0442-6
Chen, W., Kelly, M. A., Opitz-Araya, X., Thomas, R. E., Low, M. J., and Cone, R. D. (1997). Exocrine gland dysfunction in MC5-R-deficient mice: Evidence for coordinated regulation of exocrine gland function by melanocortin peptides. Cell. 91, 789–798. doi:10.1016/s0092-8674(00)80467-5
China National Commission of Animal Genetic Resource (2011). Animal genetic resource in China. Pigs. Beijing, China: China Agriculture Press.
Cignarelli, A., Genchi, V. A., Perrini, S., Natalicchio, A., Laviola, L., and Giorgino, F. (2019). Insulin and insulin receptors in adipose tissue development. Int. J. Mol. Sci. 20, 759. doi:10.3390/ijms20030759
Deng, B., Wen, J., Ding, Y., Gao, Q., Huang, H., Ran, Z., et al. (2012). Functional analysis of pig myostatin gene promoter with some adipogenesis- and myogenesis-related factors. Mol. Cell. Biochem. 363, 291–299. doi:10.1007/s11010-011-1181-y
Duan, X., Zhang, H. L., Wu, L. L., Liu, M. Y., Pan, M. H., Ou, X. H., et al. (2018). Involvement of LIMK1/2 in actin assembly during mouse embryo development. Cell. Cycle 17, 1381–1389. doi:10.1080/15384101.2018.1482138
Edea, Z., Hong, J. K., Jung, J. H., Kim, D. W., Kim, Y. M., Kim, E. S., et al. (2017). Detecting selection signatures between Duroc and Duroc synthetic pig populations using high-density SNP chip. Anim. Genet. 48, 473–477. doi:10.1111/age.12559
Fang, M., Larson, G., Ribeiro, H. S., Li, N., and Andersson, L. (2009). Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5, e1000341. doi:10.1371/journal.pgen.1000341
Fedele, M., Franco, R., Salvatore, G., Paronetto, M. P., Barbagallo, F., Pero, R., et al. (2008). PATZ1 gene has a critical role in the spermatogenesis and testicular tumours. J. Pathol. 215, 39–47. doi:10.1002/path.2323
Fekete, D. M. (1999). Development of the vertebrate ear: Insights from knockouts and mutants. Trends Neurosci. 22, 263–269. doi:10.1016/s0166-2236(98)01366-6
Feldman, B. J., Streeper, R. S., Farese, R. V., and Yamamoto, K. R. (2006). Myostatin modulates adipogenesis to generate adipocytes with favorable metabolic effects. Proc. Natl. Acad. Sci. U. S. A. 103, 15675–15680. doi:10.1073/pnas.0607501103
Frantz, L. A., Schraiber, J. G., Madsen, O., Megens, H. J., Cagan, A., Bosse, M., et al. (2015). Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet. 47, 1141–1148. doi:10.1038/ng.3394
Grochowska, E., Lisiak, D., Akram, M. Z., Adeniyi, O. O., Lühken, G., and Borys, B. (2021). Association of a polymorphism in exon 3 of the IGF1R gene with growth, body size, slaughter and meat quality traits in Colored Polish Merino sheep. Meat Sci. 172, 108314. doi:10.1016/j.meatsci.2020.108314
Groenen, M. A., Archibald, A. L., Uenishi, H., Tuggle, C. K., Takeuchi, Y., Rothschild, M. F., et al. (2012). Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398. doi:10.1038/nature11622
Gu, H., Cao, Y., Qiu, B., Zhou, Z., Deng, R., Chen, Z., et al. (2016). Establishment and phenotypic analysis of an Mstn knockout rat. Biochem. Biophys. Res. Commun. 477, 115–122. doi:10.1016/j.bbrc.2016.06.030
Guo, L., Sun, H., Zhao, Q., Xu, Z., Zhang, Z., Liu, D., et al. (2021). Positive selection signatures in Anqing six-end-white pig population based on reduced-representation genome sequencing data. Anim. Genet. 52, 143–154. doi:10.1111/age.13034
Guo, T., Jou, W., Chanturiya, T., Portas, J., Gavrilova, O., and McPherron, A. C. (2009). Myostatin inhibition in muscle, but not adipose tissue, decreases fat mass and improves insulin sensitivity. PLoS One 4, e4937. doi:10.1371/journal.pone.0004937
Hatano, N., Mori, Y., Oh-hora, M., Kosugi, A., Fujikawa, T., Nakai, N., et al. (2003). Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes. cells. 8, 847–856. doi:10.1046/j.1365-2443.2003.00680.x
Hellström, A., Ley, D., Hansen-Pupp, I., Hallberg, B., Ramenghi, L. A., Löfqvist, C., et al. (2016). Role of insulinlike growth factor 1 in fetal development and in the early postnatal life of premature infants. Am. J. Perinatol. 33, 1067–1071. doi:10.1055/s-0036-1586109
Hirai, S., Matsumoto, H., Hino, N., Kawachi, H., Matsui, T., and Yano, H. (2007). Myostatin inhibits differentiation of bovine preadipocyte. Domest. Anim. Endocrinol. 32, 1–14. doi:10.1016/j.domaniend.2005.12.001
Hoopes, B. C., Rimbault, M., Liebers, D., Ostrander, E. A., and Sutter, N. B. (2012). The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm. Genome 23, 780–790. doi:10.1007/s00335-012-9417-z
Hu, H., Wu, C. D., Ding, Y. Y., Zhang, X. D., Yang, M., Wen, A. Y., et al. (2019). Comparative analysis of meat sensory quality, antioxidant status, growth hormone and orexin between Anqingliubai and Yorkshire pigs. J. Appl. Anim. Res. 47, 357–361. doi:10.1080/09712119.2019.1643729
Hubbard, S. R. (2013). The insulin receptor: Both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb. Perspect. Biol. 5 (3), a008946. doi:10.1101/cshperspect.a008946
Ivell, R., and Grutzner, F. (2009). Evolution and male fertility: Lessons from the insulin-like factor 6 gene (Insl6). Endocrinology 150, 3986–3990. doi:10.1210/en.2009-0691
Jeong, W., Kim, J., Bazer, F. W., and Song, G. (2013). Epidermal growth factor stimulates proliferation and migration of porcine trophectoderm cells through protooncogenic protein kinase 1 and extracellular-signal-regulated kinases 1/2 mitogen-activated protein kinase signal transduction cascades during early pregnancy. Mol. Cell. Endocrinol. 81, 302–311. doi:10.1016/j.mce.2013.08.024
Jiang, F., Lin, R., Xiao, C., Xie, T., Jiang, Y., Chen, J., et al. (2021). Analysis of whole-genome re-sequencing data of ducks reveals a diverse demographic history and extensive gene flow between Southeast/South Asian and Chinese populations. Genet. Sel. Evol. 53, 35. doi:10.1186/s12711-021-00627-0
Jin, M., Lu, J., Fei, X., Lu, Z., Quan, K., Liu, Y., et al. (2020). Genetic Signatures of Selection for Cashmere Traits in Chinese Goats, 10.Anim. (Basel)
Jun, D. J., Na, K. Y., Kim, W., Kwak, D., Kwon, E. J., Yoon, J. H., et al. (2010). Melanocortins induce interleukin 6 gene expression and secretion through melanocortin receptors 2 and 5 in 3T3-L1 adipocytes. J. Mol. Endocrinol. 44, 225–236. doi:10.1677/JME-09-0161
Kerstens, H. H., Kollers, S., Kommadath, A., Del Rosario, M., Dibbits, B., Kinders, S. M., et al. (2009). Mining for single nucleotide polymorphisms in pig genome sequence data. BMC genomics 10, 4. doi:10.1186/1471-2164-10-4
Kijas, J. W., Lenstra, J. A., Hayes, B., Boitard, S., Porto Neto, L. R., San Cristobal, M., et al. (2012). Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10, e1001258. doi:10.1371/journal.pbio.1001258
Kim, S. Y., Lohmueller, K. E., Albrechtsen, A., Li, Y., Korneliussen, T., Tian, G., et al. (2011). Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinforma. 12, 231. doi:10.1186/1471-2105-12-231
Kogelman, L. J., Cirera, S., Zhernakova, D. V., Fredholm, M., Franke, L., and Kadarmideen, H. N. (2014). Identification of co-expression gene networks, regulatory genes and pathways for obesity based on adipose tissue RNA sequencing in a porcine model. BMC Med. Genomics 7, 57. doi:10.1186/1755-8794-7-57
Kumar, C., Song, S., Dewani, P., Kumar, M., Parkash, O., Ma, Y., et al. (2018). Population structure, genetic diversity and selection signatures within seven indigenous Pakistani goat populations. Anim. Genet. 49, 592–604. doi:10.1111/age.12722
Kuo, I. Y., and Ehrlich, B. E. (2015). Signaling in muscle contraction. Cold Spring Harb. Perspect. Biol. 7, a006023. doi:10.1101/cshperspect.a006023
Lai, F. N., Zhai, H. L., Cheng, M., Ma, J. Y., Cheng, S. F., Ge, W., et al. (2016). Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (Capra hircus). Sci. Rep. 6, 38096. doi:10.1038/srep38096
Lefebvre, V., Li, P., and de Crombrugghe, B. (1998). A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 17, 5718–5733. doi:10.1093/emboj/17.19.5718
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi:10.1093/bioinformatics/btp324
Li, H., and Durbin, R. (2011). Inference of human population history from individual whole genome sequences. Nature 475, 493–496. doi:10.1038/nature10231
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi:10.1093/bioinformatics/btr509
Li, M., Tian, S., Jin, L., Zhou, G., Li, Y., Zhang, Y., et al. (2013). Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45, 1431–1438. doi:10.1038/ng.2811
Li, X. J., Liu, L. Q., Dong, H., Yang, J. J., Wang, W. W., Zhang, Q., et al. (2021). Comparative genome-wide methylation analysis of longissimus dorsi muscles in Yorkshire and Wannanhua pigs. Anim. Genet. 52, 78–89. doi:10.1111/age.13029
Liu, J. P., Baker, J., Perkins, A. S., Robertson, E. J., and Efstratiadis, A. (1993). Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell. 75, 59–72. doi:10.1016/s0092-8674(05)80084-4
Lok, S., Johnston, D. S., Conklin, D., Lofton-Day, C. E., Adams, R. L., Jelmberg, A. C., et al. (2000). Identification of INSL6, a new member of the insulin family that is expressed in the testis of the human and rat. Biol. Reprod. 62, 1593–1599. doi:10.1095/biolreprod62.6.1593
Ma, Y., Zhang, S., Zhang, K., Fang, C., Xie, S., Du, X., et al. (2018). Genomic Analysis to Identify Signatures of Artificial Selection and Loci Associated with Important Economic Traits in Duroc Pigs, 8. Bethesda, Md: G3, 3617–3625.
Mei, C., Wang, H., Liao, Q., Wang, L., Cheng, G., Wang, H., et al. (2018). Genetic architecture and selection of Chinese cattle revealed by whole genome resequencing. Mol. Biol. Evol. 35, 688–699. doi:10.1093/molbev/msx322
Nei, M., and Li, W-H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U. S. A. 76, 5269–5273. doi:10.1073/pnas.76.10.5269
Pantoja, C., Huff, J. T., and Yamamoto, K. R. (2008). Glucocorticoid signaling defines a novel commitment state during adipogenesis in vitro. Mol. Biol. Cell. 19, 4032–4041. doi:10.1091/mbc.e08-04-0420
Pfeifer, B., Wittelsbürger, U., Ramos-Onsins, S. E., and Lercher, M. J. (2014). PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936. doi:10.1093/molbev/msu136
Posbergh, C. J., and Huson, H. J. (2021). All sheeps and sizes: A genetic investigation of mature body size across sheep breeds reveals a polygenic nature. Anim. Genet. 52, 99–107. doi:10.1111/age.13016
Puig, A., Ramayo-Caldas, Y., Corominas, J., Estelle, J., Perez-Montarelo, D., Hudson, N. J., et al. (2014). Differences in muscle Transcriptome among pigs phenotypically extreme for fatty acid composition. PLoS One 9, e99720. doi:10.1371/journal.pone.0099720
Ravnskjaer, K., Madiraju, A., and Montminy, M. (2016). Role of the cAMP pathway in glucose and lipid metabolism. Handb. Exp. Pharmacol. 233, 29–49. doi:10.1007/164_2015_32
Revilla, M., Puig-Oliveras, A., Castelló, A., Crespo-Piazuelo, D., Paludo, E., Fernández, A. I., et al. (2017). A global analysis of CNVs in swine using whole genome sequence data and association analysis with fatty acid composition and growth traits. PLoS One 12, e0177014. doi:10.1371/journal.pone.0177014
Rubin, C. J., Megens, H. J., Martinez Barrio, A., Maqbool, K., Sayyab, S., Schwochow, D., et al. (2012). Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. U. S. A. 109, 19529–19536. doi:10.1073/pnas.1217149109
Saba, M. K., Vella, F. D., Vernay, B., Voisin, L., Chen, L., Labrecque, N., et al. (2003). An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 4, 964–968. doi:10.1038/sj.embor.embor939
Sambrook, J., and Russell, D. W. (2001). Molecular cloning: A laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press.
Sekulovski, N., Whorton, A. E., Shi, M., Hayashi, K., and MacLean, J. A. (2021). Insulin signaling is an essential regulator of endometrial proliferation and implantation in mice. FASEB J. 35, e21440. doi:10.1096/fj.202002448R
Serra, X., Gil, C., Pérez-Enciso, M., Oliver, M., Vázquez, J., Gispert, M., et al. (1998). A comparison of carcass, meat quality and histochemical characteristics of Iberian (Guadyerbas line) and Landrace pigs. Livest. Prod. Sci. 56, 215–223. doi:10.1016/s0301-6226(98)00151-1
Shawki, H. H., Kigoshi, T., Katoh, Y., Matsuda, M., Ugboma, C. M., Takahashi, S., et al. (2016). Identification, localization, and functional analysis of the homologues of mouse CABS1 protein in porcine testis. Exp. Anim. 65, 253–265. doi:10.1538/expanim.15-0104
Shi, Y., Zhao, H., Shi, Y., Cao, Y., Yang, D., Li, Z., et al. (2012). Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat. Genet. 44, 1020–1025. doi:10.1038/ng.2384
Smits, P., Li, P., Mandel, J., Zhang, Z., Deng, J. M., Behringer, R. R., et al. (2001). The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell. 1, 277–290. doi:10.1016/s1534-5807(01)00003-x
Sun, W. X., Dodson, M. V., Jiang, Z. H., Yu, S. G., Chu, W. W., and Chen, J. (2016). Myostatin inhibits porcine intramuscular preadipocyte differentiation in vitro. Domest. Anim. Endocrinol. 55, 25–31. doi:10.1016/j.domaniend.2015.10.005
Switonski, M., Mankowska, M., and Salamon, S. (2013). Family of melanocortin receptor (MCR) genes in mammals-mutations, polymorphisms and phenotypic effects. J. Appl. Genet. 54, 461–472. doi:10.1007/s13353-013-0163-z
Takahashi, H., Koshimizu, U., Miyazaki, J., and Nakamura, T. (2002). Impaired spermatogenic ability of testicular germ cells in mice deficient in the LIM-kinase 2 gene. Dev. Biol. 241, 259–272. doi:10.1006/dbio.2001.0512
Terman, A. (2011). The IGF1R gene: A new marker for reproductive performance traits in sows? Acta Agric. Scand. Sect. A - Animal Sci. 61, 67–71. doi:10.1080/09064702.2011.570780
Valentino, T., Palmieri, D., Vitiello, M., Simeone, A., Palma, G., Arra, C., et al. (2013). Embryonic defects and growth alteration in mice with homozygous disruption of the Patz1 gene. J. Cell. Physiol. 228, 646–653. doi:10.1002/jcp.24174
Valli, K., Suviolahti, E., Schalin-Jäntti, C., Ripatti, S., Silander, K., Oksanen, L., et al. (2008). Further evidence for the role of ENPP1 in obesity: Association with morbid obesity in Finns. Obes. (Silver Spring, Md.) 16, 2113–2119. doi:10.1038/oby.2008.313
Verardo, L. L., Sevón-Aimonen, M. L., Serenius, T., Hietakangas, V., and Uimari, P. (2017). Whole-genome association analysis of pork meat pH revealed three significant regions and several potential genes in Finnish Yorkshire pigs. BMC Genet. 18, 13. doi:10.1186/s12863-017-0482-x
Wang, C., Wang, H., Zhang, Y., Tang, Z., Li, K., and Liu, B. (2015). Genome-wide analysis reveals artificial selection on coat colour and reproductive traits in Chinese domestic pigs. Mol. Ecol. Resour. 15, 414–424. doi:10.1111/1755-0998.12311
Wang, K., Li, M., and Hakonarson, H. (2011). Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi:10.1093/nar/gkq603
Wang, Y., Niu, Z., Zeng, Z., Jiang, Y., Jiang, Y., Ding, Y., et al. (2020). Using high-density SNP array to reveal selection signatures related to prolificacy in Chinese and Kazakhstan sheep breeds. Animals. 10, 1633. doi:10.3390/ani10091633
Webster, M. T., Kamgari, N., Perloski, M., Hoeppner, M. P., Axelsson, E., Hedhammar, Å., et al. (2015). Linked genetic variants on chromosome 10 control ear morphology and body mass among dog breeds. BMC genomics 16, 474. doi:10.1186/s12864-015-1702-2
Wood, J. D., Enser, M., Fisher, A. V., Nute, G. R., Sheard, P. R., Richardson, R. I., et al. (2008). Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 78, 343–358. doi:10.1016/j.meatsci.2007.07.019
Xu, J., Wang, C., Jin, E., Gu, Y., Li, S., and Li, Q. (2018). Identification of differentially expressed genes in longissimus dorsi muscle between Wei and Yorkshire pigs using RNA sequencing. Genes. Genomics 40, 413–421. doi:10.1007/s13258-017-0643-3
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). Gcta: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi:10.1016/j.ajhg.2010.11.011
Yang, J., Schulman, L., Pursel, V. G., Solomon, M. B., Mitchell, A. D., Zhao, B., et al. (2009). Expression of porcine myostatin prodomain genomic sequence leads to a decrease in muscle growth, but significant intramuscular fat accretion in transgenic pigs. Transgenic Animal Res. Conf. VIII, 15.
Zeder, M. A. (2008). Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. U. S. A. 105, 11597–11604. doi:10.1073/pnas.0801317105
Zhang, L., Li, W. H., Anthonavage, M., Pappas, A., Rossetti, D., Cavender, D., et al. (2011). Melanocortin-5 receptor and sebogenesis. Eur. J. Pharmacol. 660, 202–206. doi:10.1016/j.ejphar.2010.10.100
Zhang, W., Yang, M., Zhou, M., Wang, Y., Wu, X., Zhang, X., et al. (2020). Identification of signatures of selection by whole-genome resequencing of a Chinese native pig. Front. Genet. 11, 566255. doi:10.3389/fgene.2020.566255
Zhang, Y., Liang, J., Zhang, L., Wang, L., Liu, X., Yan, H., et al. (2015). Porcine methionine sulfoxide reductase B3: Molecular cloning, tissue-specific expression profiles, and polymorphisms associated with ear size in Sus scrofa. J. Anim. Sci. Biotechnol. 6, 60. doi:10.1186/s40104-015-0060-x
Zhao, F., Deng, T., Shi, L., Wang, W., Zhang, Q., Du, L., et al. (2020). Genomic scan for selection signature reveals fat deposition in Chinese indigenous sheep with extreme tail types. Animals. 10, 773. doi:10.3390/ani10050773
Zhao, P., Yu, Y., Feng, W., Du, H., Yu, J., Kang, H., et al. (2018). Evidence of evolutionary history and selective sweeps in the genome of Meishan pig reveals its genetic and phenotypic characterization. GigaScience 7, giy058. doi:10.1093/gigascience/giy058
Zhou, C., Lv, M., Wang, P., Guo, C., Ni, Z., Bao, H., et al. (2021). Sequential activation of uterine epithelial IGF1R by stromal IGF1 and embryonic IGF2 directs normal uterine preparation for embryo implantation. J. Mol. Cell. Biol. 13, 646–661. doi:10.1093/jmcb/mjab034
Keywords: whole genome sequencing, SNP, signatures of selection, Anhui pig population, population structure
Citation: Zhang W, Li X, Jiang Y, Zhou M, Liu L, Su S, Xu C, Li X and Wang C (2022) Genetic architecture and selection of Anhui autochthonous pig population revealed by whole genome resequencing. Front. Genet. 13:1022261. doi: 10.3389/fgene.2022.1022261
Received: 18 August 2022; Accepted: 28 September 2022;
Published: 17 October 2022.
Edited by:
Muhammad Zahoor Khan, University of Agriculture, Dera Ismail Khan, PakistanReviewed by:
Serafino M. A. Augustino, University of Juba, South SudanXianyong Lan, Northwest A&F University, China
Copyright © 2022 Zhang, Li, Jiang, Zhou, Liu, Su, Xu, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chonglong Wang, YWh3Y2hsQDE2My5jb20=
 Wei Zhang
Wei Zhang Xiaojin Li
Xiaojin Li Chonglong Wang
Chonglong Wang