- 1Key Laboratory of Soybean Biology of Chinese Education Ministry, Northeast Agricultural University, Harbin, China
- 2State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, China
Lipoxygenases (LOXs; EC1.13.11.12) are a family of iron- or manganese-containing dioxygenases that catalyze the oxygenation of polyunsaturated fatty acids (PUFAs) and play important roles in plant growth, development, and stress response. In this study, a total of 36 LOX gene family members were identified and annotated in Zhonghuang 13, a soybean cultivar bred by Chinese scientists in 2001. Sanger sequencing of the GmLOX1-coding sequence and colorimetric assays for the GmLOX1 protein showed that Zhonghuang 13 possessed the GmLOX1 gene. These LOX genes are divided into three subfamilies: 9-LOX, type Ⅰ 13-LOX and type II 13-LOX. In the 13-LOX group, the number of GmLOX members was the highest. These GmLOX genes are unevenly distributed on chromosomes 3, 7, 8, 10, 11, 12, 13, 15, 16, 19, and 20. Most of the 13-LOX genes exist in the form of gene clusters, indicating that these genes may originate from tandem duplications. The analysis of duplicated gene pairs showed that GmLOX genes underwent purifying selective pressure during evolution. The gene structures and conserved functional domains of these genes are quite similar. Compared to the orthologous gene pairs of LOX genes between wild soybean (Glycine soja W05) and Zhonghuang 13, the sequences of most gene pairs are relatively conserved. Many cis-elements are present in the promoter region and are involved in stress response, growth and development, hormone response and light response. The tissue-specific gene expression of GmLOX genes was evaluated. Represented by GmLOX1, GmLOX2, and GmLOX3, which were expressed at extremely high levels in seeds, they showed the characteristics of specific expression. This study provides detailed information on soybean lipoxygenase gene family members in Zhonghuang 13, which lays a foundation for further research.
Introduction
Lipoxygenases (LOXs) are iron- or manganese-containing dioxygenases (Feussner and Wasternack, 2002). LOXs of animals, plants and prokaryotes contain Fe as the catalytic metal, whereas fungi express LOXs with Fe or Mn (Wennman et al., 2016). LOXs can oxidize polyunsaturated fatty acids containing one or more 1Z, 4Z-pentadiene units to hydroperoxides (Brash, 1999; Song et al., 2016). The resulting hydroperoxides are further metabolized into biologically active oxylipins, including jasmonic acid (JA) and green leaf volatiles (GLV) (Upadhyay et al., 2019). LOXs and their products accumulate transiently upon developmental or environmental stimuli (Viswanath et al., 2020). JA can be involved in defense responses against abiotic and biotic environmental stresses (Vogt et al., 2013). LOXs are also known to play a decisive role in the production of volatiles that influence the flavor and aroma of floral fruit, which are important to the agri-food industry (Upadhyay et al., 2020). For instance, hydroperoxides, LOX-catalyzed products in mature soybean seeds, are converted to volatile compounds, which are associated with an unpleasant beany taste (Wang et al., 2020). In addition, LOXs have been associated with some processes in a series of developmental stages, including the mobilization of storage lipids during germination and fruit ripening (Porta and Rocha-Sosa, 2002; Viswanath et al., 2020). LOXs contain two major domains, a lipoxygenase domain at the C-terminus and a polycystin-1, lipoxygenase, alpha-toxin (PLAT) domain at the N-terminus (Song et al., 2016). The plant lipoxygenase domain is involved in iron binding and contains approximately 38 amino acids with five conserved histidines (Steczko et al., 1992). The PLAT domain, which contains an eight-stranded antiparallel β-barrel, is found in a variety of membrane- or lipid-associated proteins (Bateman and Sandford, 1999). This domain can bind to procolipase, which mediates membrane associations (Tomchick et al., 2001). According to the substrate oxygenated at either carbon atom 9 (9-LOX) or carbon atom 13 (13-LOX) of the hydrocarbon backbone of the fatty acid (Feussner et al., 2001), plant LOXs can be classified into two types, 9-LOX and 13-LOX (Kolomiets et al., 2001). Furthermore, it can be divided into I-LOXs and II-LOXs according to the consistency of sequence structure and whether there is a chloroplast targeting peptide: I-LOXs have high consistency but do not contain the targeted peptide that II-LOXs contain (Brash, 1999). All LOXs belong to the same gene family, and they are widely distributed. To date, many LOX genes have been identified, such as those in Arabidopsis thaliana (Park and Polacco, 1989), Citrullus lanatus (Liu J et al., 2020), Fagopyrum tataricum (Ji et al., 2020), Arachis duranensis (Song et al., 2016), Malus pumila (Vogt et al., 2013), Spirodela polyrhiza (Upadhyay et al., 2020) and Populus L (Chen et al., 2015). Zhou et al. (2020) demonstrated that overexpression of CsLOX1 in cucumber may impair the production of endogenous JA. The function of CsLOX1 may involve plant growth and development, such as shoot growth, male fertility and fruit development. Vogt et al. (2013) found that MdLOX1a and MdLOX5e were candidate genes involved in the production of apple fruit aroma volatiles. Chen et al. (2015) found that the expression patterns of PtLOX genes in response to stress were different in poplar. This result reflected that these genes may be involved in related functions of specific types of stress.
Soybean (Glycine max [L.] Merr.), one of the most important leguminous plants, is widely cultivated all over the world (Wang et al., 2020). The utilization of soybeans relates to multiple fields of production and life. Soybean’s oil content is approximately 20%, protein is approximately 40%, and it is a crucial raw material for food processing, which provides energy and nutrition (Chaudhary et al., 2015). In addition, soybean meal is used in a variety of animal feed, such as that for fish and pigs (Dunsford et al., 1989; Tomas et al., 2005), because it is high in protein with a good amino acid balance. Furthermore, soybeans are also used in industrial and pharmaceutical applications as well as in the production of biodiesel (Chaudhary et al., 2015). Soybean production has a great impact on human life. Studies on the evolution of the soybean genome indicate that the soybean underwent two whole genome replication events and has an ancient tetraploid genome (Schmutz et al., 2010). The soybean genome size is approximately nine times that of A. thaliana (Haas et al., 2003). At present, genome information of several soybean varieties has been identified (Schmutz et al., 2010; Shen et al., 2018; Liu Y et al., 2020). Williams 82, a soybean cultivar domesticated in America, was the first soybean genome to be sequenced, and the emergence of this reference genome made it possible to explore functional genomics in soybean (Chan et al., 2012). As an undomesticated relative of cultivated soybean, the sequencing of the wild soybean (G. soja W05) genome was helpful to explore the diversity of lost genes in cultivated soybean and provided a novel gene source for soybean breeding and improvement (Li et al., 2014). For cultivated soybeans, there are wide genetic differences among different varieties. The soybean cultivar “Zhonghuang 13” possesses the widest planting range in China (Liu J et al., 2020). The genome of Zhonghuang 13 was sequenced, and the reference genome represents the characteristics of Asian soybean (Shen et al., 2018). Compared with other reported plant genomes, the genome assembly of Zhonghuang 13 is more complete and more continuous (Shen et al., 2019). The elucidation of genome sequencing in Zhonghuang 13 is of great significance for further exploration of the trait-related genes of Asian soybean. Multiple LOX isozymes distinct from seed isozymes have been found in many tissues of soybean (Park and Polacco, 1989). The expression patterns of LOXs are different during the different stages from seed germination to plant maturity and in different parts of the plant (Veldink et al., 1977). Considering the potential roles of LOX genes, it is necessary to study the genome-wide analysis of LOX genes in Zhonghuang 13.
In this study, we identified the members of the LOX gene family using hidden Markov model (HMM) searches against a reference soybean genome of a soybean cultivar, Zhonghuang 13. For genes that were missing annotations, such as GmLOX1, Sanger sequencing and colorimetric assays were utilized to detect their existence at the transcription level and translation level, respectively. A phylogenetic tree was constructed with four outgroup species. Next, we analyzed the gene structures, conserved motifs, chromosomal distributions, collinearity, and cis-regulatory elements. Subsequently, we explored the evolution of LOX genes by comparing the discrepancy in LOX genes between wild soybean (G. soja W05) and Zhonghuang 13. Furthermore, we evaluated the expression of GmLOX genes in different parts of soybean to explore their spatial expression. Our results provide information on the soybean LOX gene family and lay a foundation for further study on the specific function of LOX genes in soybeans.
Materials and methods
Plant materials and treatment
The Zhonghuang 13 seeds were planted in a round pot with a diameter of 15 cm in a greenhouse in May 2021. Soybean plants grew naturally in the soil under greenhouse conditions. When the plants were at the R6 stage (pod stage), the roots, stems, leaves, flowers, pods, and seeds of the soybean plants were taken as the research materials [flowers were obtained at the R2 stage (flowering period)], placed in liquid nitrogen and stored at −80°C. Each treatment consisted of three biological replicates.
Identification of LOX family members in Zhonghuang 13
The genome information of Zhonghuang 13 (Gmax_ZH13_v2.0) was downloaded from the Genome Warehouse in BIG Data Center under accession number GWHAAEV00000000.1 (Shen et al., 2019). To compare LOX genes between different species, we also downloaded the genome sequences of Arabidopsis thaliana (TAIR10), Populus trichocarpa (v4.1), Vitis vinifera (v2.1) and Oryza sativa (v7.0) from the Phytozome database (https://phytozome.jgi.doe.gov/). The genome data of Williams 82 (version G. max Wm82.a4.v1) were also downloaded from the Phytozome database to compare LOX genes in different soybean varieties. In addition, to acquire all the possible LOX genes, the HMM profiles of the lipoxygenase domain (PF00305) and the PLAT domain (PF01477) were downloaded from the Pfam database (http://pfam.xfam.org/). The protein sequences of LOX genes were searched against the HMMER program (v 3.3.2) (Finn et al., 2011). If both of these domains exist, the gene was recognized as an LOX gene. To ensure the accuracy of gene identification, the sequences were submitted to the Search Pfam site (http://pfam.xfam.org/search) for domain visualization and manual inspection to delete the unqualified genes. The isoelectric point (pI) and molecular weight (MW) of soybean LOXs were calculated by the Compute pI/Mw tool in ExPASy (https://web.expasy.org/compute_pi/). Protein subcellular localization prediction was performed by the WoLF PSORT website (https://wolfpsort.hgc.jp/).
The GmLOX1 gene is an important soybean lipoxygenase gene. Primers were designed for the full-length GmLOX1-encoding sequence. The RNA of Zhonghuang 13 seeds was extracted and reverse transcribed into cDNA as a template. A 2×Taq PCR MasterMix II kit (TIANGEN, KT211-02, Beijing, China) was used to amplify the target sequence, and the PCR product was examined by DNA agarose gel electrophoresis with a 5,000 bp DNA marker (Trans, BM111-01, Beijing, China). The PCR product that matched the expected length was sent to GENEWIZ for Sanger sequencing. The colorimetric assay method mentioned in previous studies (Wang et al., 2020) was used to identify the presence of GmLOX1 protein in Zhonghuang 13 seeds, with Williams 82 and Dongfudou-3 being the positive and negative controls, respectively. Dongfudou-3, bred by Northeast Agricultural University, is a soybean cultivar lacking the GmLOX1 protein.
Multiple sequence alignment and phylogenetic analysis
To investigate the protein sequences, multiple sequence alignment of the LOX protein sequences identified in five species was performed using the MAFFT program (version 7.490) (Katoh et al., 2002). A phylogenetic tree was created using the maximum likelihood (ML) method with 1000 ultrafast bootstrap replications by the IQ-TREE program (version 2.2.0) (Nguyen et al., 2015). The classification of LOX protein sequences depends on the characteristics of 9-LOX and 13-LOX. The phylogenetic tree was visualized using the Evolview (He et al., 2016) online site (https://www.evolgenius.info/evolview/).
Gene structure and conserved motif prediction
The intron and exon localization information of GmLOX genes was extracted from genome annotation files. The Multiple Expectation Maximization for Motif Elicitation (MEME) program (Timothy and Bailey, 1994) (https://meme-suite.org/meme/tools/meme) was employed to confirm conserved motifs with the following parameters: the site distribution site was set to zero or one occurrence per sequence (zoops), the number of motifs was set to 10, and the minimum and maximum widths were 6 and 50, respectively. The Gene Structure View (Advanced) program in TBTools software (Chen et al., 2020) was used to visualize gene structure and conserved motifs.
Chromosomal location and duplication analysis
The chromosomal location information of GmLOX genes was obtained from the soybean genome database. TBtools software was used to visualize the location of genes on chromosomes. Duplication analysis among the GmLOX genes was conducted with the MCScanX (Wang et al., 2012) tools in TBtools software. The parameters of BLAST (Camacho et al., 2009) were as follows: the E-value was set to 1e-5, the number of hits was set to 5, and the number of alignments was set to 5. When running the MCScanX program, the minimum block size was set to 5. KaKs_Calculator 3.0 (Zhang, 2022) was used to calculate the Ka/Ks values of homologous gene pairs.
Prediction of cis-regulatory elements in the GmLOX promoters
The 2,000 bp upstream sequences of the GmLOX gene coding sequences were obtained from genomic data of soybean and then uploaded to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) with default parameters to predict stress- and hormone-responsive cis-regulatory elements. The visualization of the regulatory elements was performed with the Evolview online tool.
Evolution of LOX genes between wild soybean (G. soja W05) and Zhonghuang 13
To analyze the evolution of LOX genes in wild soybean and cultivated soybean, genomic data of wild soybean (G. soja W05) were downloaded from DDBJ/ENA/GenBank under accession QZWG00000000 (Xie et al., 2019), and members of the LOX gene family of wild soybean were identified according to the above procedure (Song et al., 2016). Data from the identified duplicated blocks and orthologous gene pairs in two soybean varieties were obtained by the MCscanX tool in TBtools software. The One Step MCScanX function was used to process two genomic datasets, with the E-value set to 1e-5 and the number of BlastHits set to 5. In addition, the orthologous gene pairs of LOX genes were checked by BLAST. KaKs_Calculator 3.0 (Zhang, 2022) was used to calculate the Ka/Ks values of coding sequences and the Kn/Ks values of intro sequences and promoter sequences of the orthologous gene pairs of LOX genes. The MA method (Zhang B et al., 2006) was used for estimating Ka/Ks, and 0.618 was used as the neutral mutation rate for estimating Kn/Ks.
Expression analysis based on RNA-Seq data in different tissues
The samples were sent to Wuhan Frasergen Bioinformatics Co., Ltd. on dry ice for library preparation and sequencing using the BGISEQ-500 platform. After data quality control by FastQC (v 0.11.9), raw reads were cleaned by fastp (v 0.23.1) (Chen et al., 2018) and 10 bases with poor quality at the starting position of reads, bases with base quality values less than 20, reads with lengths less than 90 bases, and reads that more than 20% did not meet the quality control standards were removed. The clean reads were aligned to the genome using HISAT2 (v 2.2.1) (Kim et al., 2015). SAMtools (v. 1.12) (Danecek et al., 2021) was used to convert SAM files to BAM files. The Trimmed Mean of M Value (TMM) normalization method was used to calculate transcripts per million (TPM) values. The TPM value after log10 processing was used to reflect the gene expression. The heatmap was drawn with Omicshare online cloud tools.
RNA isolation and quantitative real-time PCR (qRT‒PCR)
The samples were pulverized in liquid nitrogen, and total RNA was extracted using a Generic Plant Total RNA Rapid Extraction kit (Bio Teke, RP3302, Wuxi, China) according to the manufacturer’s instructions. The concentration and purity of RNA were measured by an ultramicro nucleic acid analyzer (Nano400A). Genomic DNA was removed, and RNA (1 μg) was reverse transcribed into cDNA using the PrimeScript RT reagent Kit with gDNA Eraser (Takara, RR047A, Beijing, China). The Primer-BLAST tools in NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and the qPrimerDB online tools (https://biodb.swu.edu.cn/qprimerdb/) were used to select specific primers for 12 GmLOX genes and three internal reference genes. Detailed primer sequences are shown in Supplementary Table S1. Three technical replicate qRT-PCRs were performed with SYBR Green Realtime PCR Master Mix (Toyobo, QPK-201, Shanghai, China) in an AriaMx real-time PCR system G8830A for each biological replicate. The amplification conditions were as follows: pre-denaturation reaction at 95°C for 1 min, denaturation reaction at 95°C for 15 s, 45 cycles, extension reaction at 60°C for 1 min, with Rox as the reference dye. After the last polymerase chain reaction cycle, the melting curve was drawn under the conditions of 95°C for 15 s, 65°C for 1 min and 95°C for 15 s. The 2−ΔΔCt method (Livak and Schmittgen, 2001) was used to calculate the gene expression data. According to the previous literature (Hu et al., 2009; Gao et al., 2017), we screened three internal reference genes, β-actin (ACT), cyclophilin (CYP2) and elongation factor 1-beta (ELF1B), which are involved in different biological regulatory mechanisms. After selecting the stably expressed internal reference genes, the geometric mean expression levels of the selected reference genes were used to normalize the expression levels of our target genes.
Results
Identification of LOX family members in Zhonghuang 13
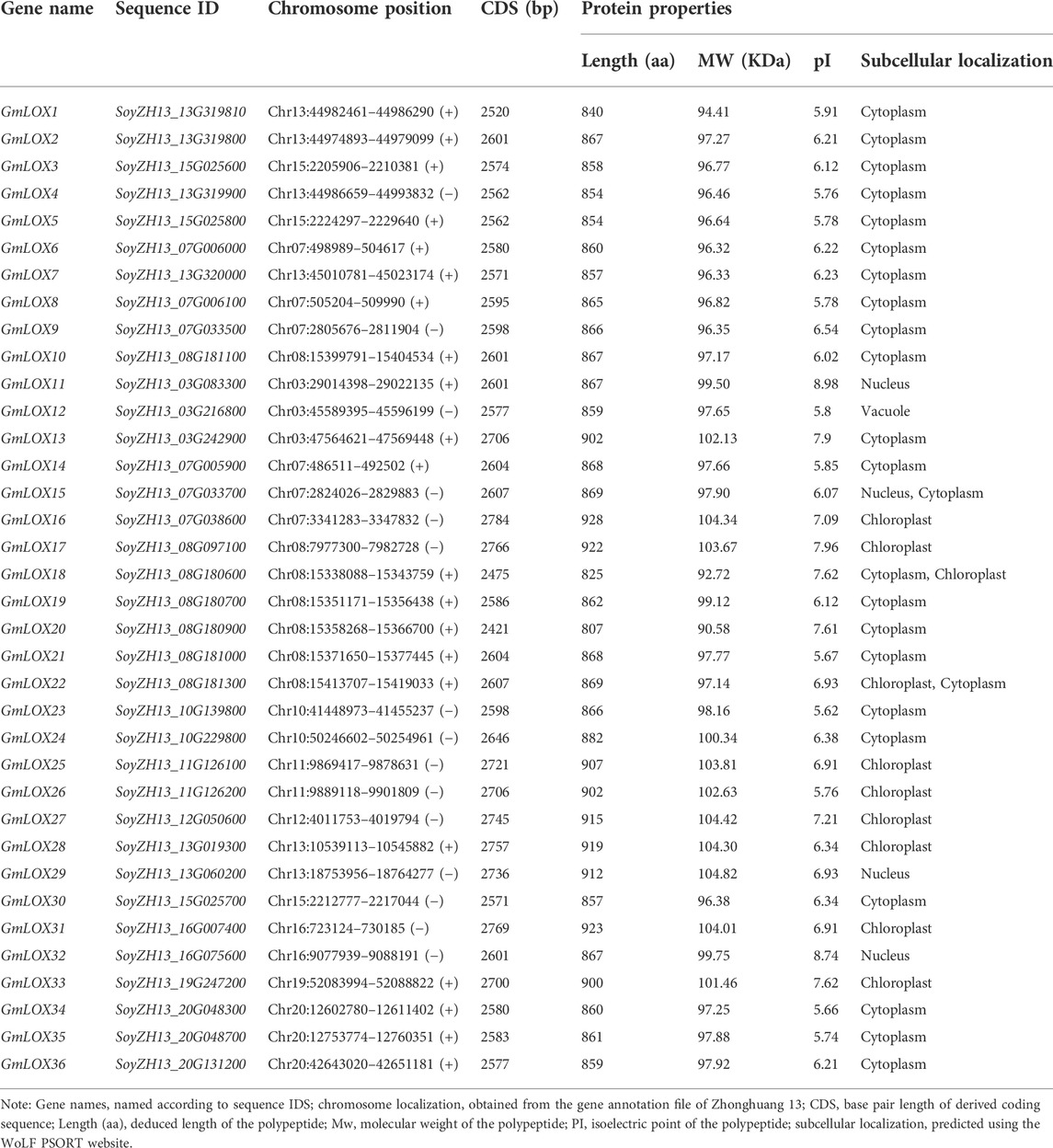
A total of 36 soybean LOX proteins were ultimately identified. All of the deduced proteins contained the lipoxygenase domain (PF00305) and the PLAT domain (PF01477) based on Pfam. According to the 10 LOX genes registered on the National Center for Biotechnology Information (NCBI) in soybeans, we found the same genes in Zhonghuang 13 through BLAST (Camacho et al., 2009). Other genes were named GmLOX11–GmLOX36 according to their locations on chromosomes. Detailed information on the GmLOX genes identified in Zhonghuang 13 (including chromosome position, CDS length, protein length, MW, pI, and subcellular localization) is displayed in Table 1. The GmLOX protein lengths ranged from 807 (GmLOX20) to 928 (GmLOX16) amino acids. The predicted molecular weight of the GmLOX proteins varied from 90.58 (GmLOX26) to 104.82 (GmLOX30) kDa, and the pI values ranged from 5.62 (GmLOX29) to 8.98 (GmLOX18). Most of the GmLOX proteins were predicted to be localized within the cytoplasm and chloroplast, and several were located within the nucleus and vacuole.
At present, there are three strategies for gene sequence annotation: de novo prediction, homology-based prediction and transcription-based prediction. The accuracy of these prediction methods is limited. It is necessary to manually determine the accuracy of GmLOX genes. We compared the LOX gene family members identified in Zhonghuang 13 with those identified in previous studies in Williams 82 (Song et al., 2016) and listed homologous gene pairs in Supplementary Table S2. There were three unpaired genes, SoyZH13_10G229800 (GmLOX24), Glyma.07G196800 and Glyma.20G054000. SoyZH13_10G229800 was retained because its domain and protein sequence structure met the requirements of LOX protein. A BLAST alignment (E-value < 1e-5) was performed on Williams 82 genome data using the coding sequence of SoyZH13_10G229800 as the query sequence. The results showed that a part of the alignment was on chromosome 10 and the other part was on scaffold_524 (identity >99%). It could be inferred that scaffold_524 should be attached to chromosome 10, and there may be an unannotated LOX gene on chromosome 10 in Williams 82. The corresponding regions of Glyma.07G196800 and Glyma.20G054000 in Zhonghuang 13 were also found by BLAST alignment. Glyma.07G196800 corresponded to SoyZH13_07G182200, which resulted in premature termination of protein translation due to a single base mutation. The corresponding region of Glyma.20G054000 was frame-shifted due to single base insertion. As a result, both regions lack the necessary structure for the LOX gene (verification information in Supplementary Figure S1). Based on comparison with LOX gene family members in Williams 82 and the support of RNA-seq data, we modified the LOX gene annotation information in Zhonghuang 13 (corrected the gene annotation information of GmLOX12, GmLOX2 and GmLOX35 and added the new gene GmLOX1). It should be noted that although these corrections refer to multiple model plants and experimental results, they cannot be guaranteed to be complete and accurate and need to be verified by further experiments in the future. The modified gene annotation file result has been uploaded to http://www.wangsui.net.cn/resource/database/public/plant/Glycine_max/ ZH13/Lox_gene_family/Gmax_ZH13_v2.0_corrected.v1.0.gff3.gz. The extraction results of the GmLOX genes coding sequences (CDSs), GmLOXs protein sequences and 2,000 bp of the nucleotide sequences upstream of the GmLOX translation initiation codon were in Supplementary Data S1–S3, respectively.
Modified GmLOX1 gene, Sanger sequencing and protein identification of GmLOX1
To further confirm the information of GmLOX1, Sanger sequencing and colorimetric assays were used to detect its existence at the transcription level and translation level, respectively. The length of the GmLOX1-encoding sequence is 2,520 bp. The result of electrophoresis in agarose gel was consistent with the expected fragment length (Figure 1A). The raw sequencing results are provided in Supplementary Data S4. The full sequence of GmLOX1 was sequenced. The initiation codon and the termination codon of sequencing are shown in Figure 1B. The results of the colorimetric assay showed that Zhonghuang 13 contained the GmLOX1 protein. As shown in Figure 1C, the negative control Dongfudou-3 is blue, and Zhonghuang 13 is colorless, the same as the positive control Williams 82. According to the sequencing results and the GmLOX1 gene sequences in different versions of Williams 82, the annotation of the GmLOX1 gene of Zhonghuang 13 was modified. The alignment result of GmLOX1 protein sequences between Zhonghuang 13 and different versions of Williams 82 is shown in Figure 2. The start position of GmLOX1 in Zhonghuang 13 is the same as that in Williams 82.a2.v1, and the end position is the same as that in Williams 82.a4.v1. A single base discrepancy (Wm82.a4.v1 one more base than Wm82.a2.v1) caused the difference in GmLOX1 protein sequences between the two versions of Williams 82.
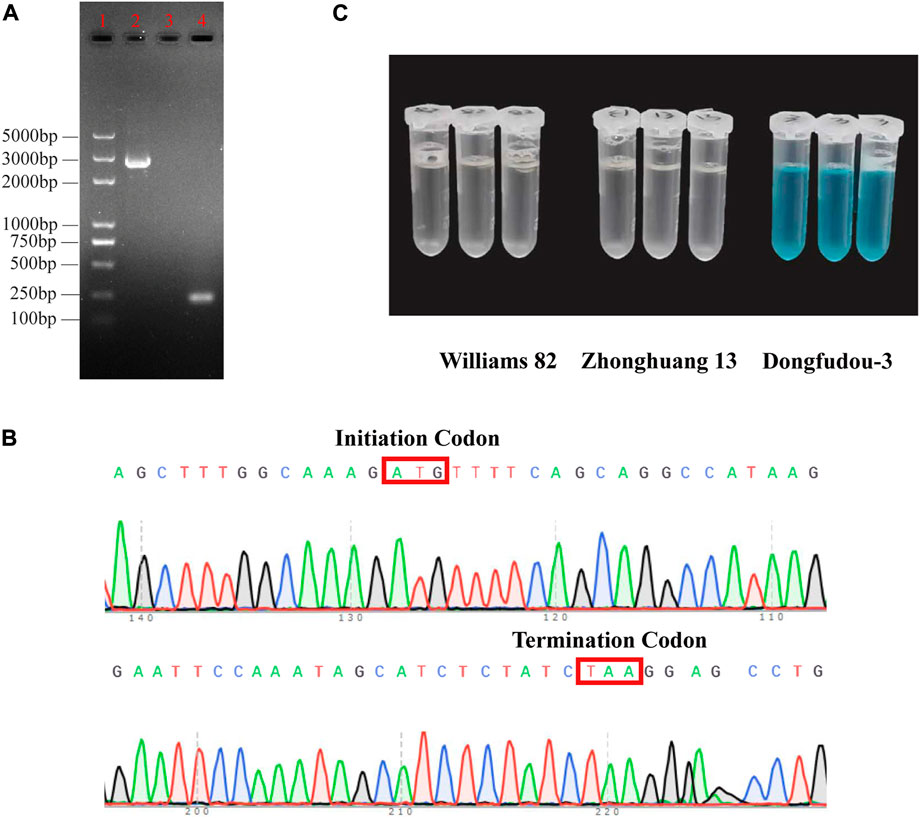
FIGURE 1. Identification results of the GmLOX1 gene (A) Gel image showing PCR products of GmLOX1 in Zhonghuang 13 seeds. (B) The initiation codon and termination codon in Sanger sequencing. (C) The detection results of the colorimetric assay method. Note: (A) 5,000 bp marker in lane 1, PCR products in lane 2, negative control in lane 3 and positive control in lane 4. The negative control used nuclease-free water as the cDNA template. The positive control used the primers for qRT‒PCR of GmLOX1. (C) The sample with GmLOX1 protein will be colourless; without GmLOX1 it will be blue.

FIGURE 2. Alignment result of the modified GmLOX1 protein sequence in Zhonghuang 13 with different versions of GmLOX1 of Williams 82 Note: SoyZH13_13G319810 is the sequence ID of GmLOX1 in Zhonghuang 13. Glyma.13G347600 is the sequence ID of GmLOX1 in Williams82, and there is one transcript in Wm82.a4.v1 and two transcripts in Wm82.a2.v1.
Multiple sequence alignment and phylogenetic analysis
To acquire more information about the characteristics and obtain the number of members in each LOX subfamily, we compared the LOX proteins in five plant species (A. thaliana, P. trichocarpa, V. vinifera, O. sativa and G. max). We used the same method to identify LOX proteins in these species, and the numbers of LOX gene family members in A. thaliana, P. trichocarpa, V. vinifera and O. sativa were 6, 18, 11, and 11, respectively. Gene names corresponding to sequence IDs are shown in Supplementary Table S3. Soybeans have the highest number of members of the LOX gene family in the five species.
To clarify the genetic relationships of these LOX genes and which subfamily they belong to, a total of 82 LOX proteins in these five species were subjected to multiple sequence alignment, and an ML phylogenetic tree was constructed among 6AtLOXs, 11OsLOXs, 18PtLOXs, 11VvLOXs, and 36GmLOXs (Figure 3). According to the Bayesian Information Criterion (BIC), the Q.plant + I + G4 model was chosen as the best-fit model. The LOX genes that were reported in the paper (Umate, 2011; Chen et al., 2015; Song et al., 2016) have already been classified into three subfamilies: the type Ⅰ 13-LOX subfamily, type II 13-LOX subfamily, and 9-LOX subfamily. Here, based on the clusters displayed on the phylogenetic tree, we were able to classify GmLOX into the above three subfamilies. The number of members of GmLOX among the type Ⅰ 13-LOX subfamily, type II 13-LOX subfamily, and 9-LOX subfamily was 20, 12, and 4, respectively (the red font gene names are shown in Figure 3). We found that in the type Ⅰ 13-LOX subfamily, there are four times as many GmLOX members as the LOX members of other species combined, while there are no AtLOX members. The GmLOX members occupy an entire branch (the purple highlight branch shown in Figure 3). In the 9-LOX subfamily, the number of GmLOX members is relatively rare compared to that in other species. There is also a small branch that includes all GmLOX members in the type II 13-LOX.
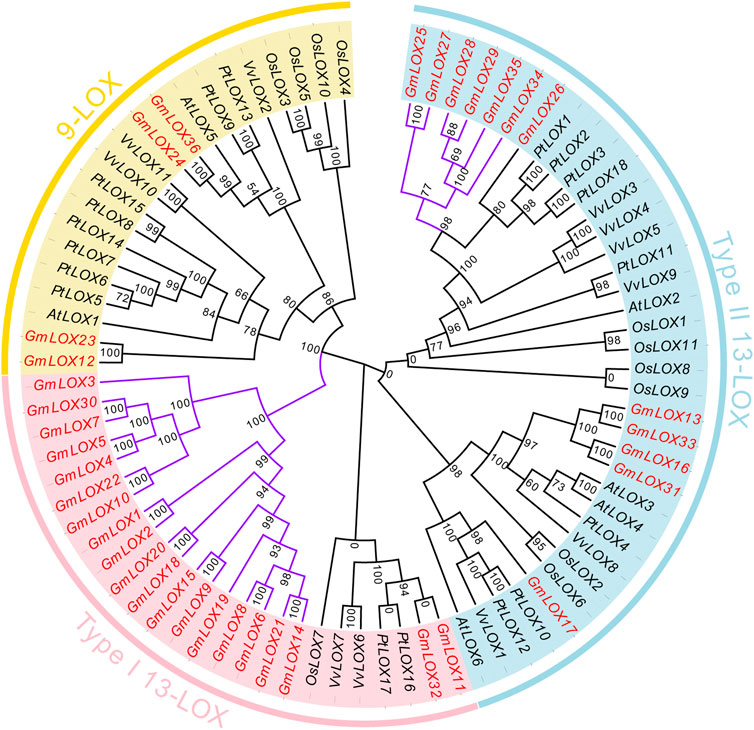
FIGURE 3. Phylogenetic analysis of GmLOX proteins in Zhonghuang 13 with other plant species Note: AtLOX: Arabidopsis thaliana. PtLOX: Populus trichocarpa. OsLOX: Oryza sativa. VvLOX: Vitis vinifera. Phylogenetic trees were constructed using the full-length amino acid sequences of LOX proteins from five species by the maximum likelihood (ML) method with 1000 ultrafast bootstrap replicates, and the ultrafast bootstrap support values are indicated on each node.
Gene structure and conserved motif prediction
A separate phylogenetic tree was constructed using the 36 GmLOX coding sequences, and the intron and exon locations of each gene sequence were extracted and compared (Figure 4A) to acquire further insight into the gene structure of the soybean lipoxygenase gene family in detail. Each gene contains at least seven exons and at most 13 exons, with nine exons being the most common among the genes. The genetic structure of the soybean lipoxygenase gene was displayed similarly by comparing the position distribution of the GmLOX gene coding regions.
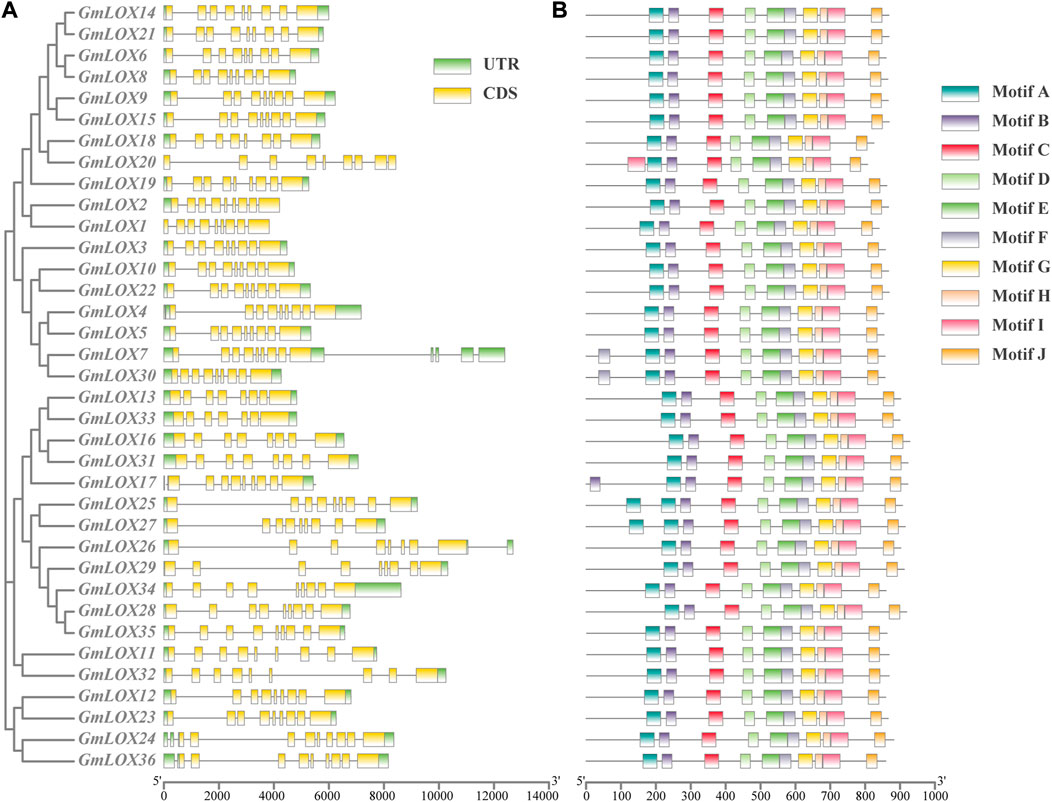
FIGURE 4. Gene structure analysis and conserved motif prediction of GmLOX genes in Zhonghuang 13 Note: (A) The result of gene structure analysis with a cluster analysis tree of polypeptide sequences by the ML method. CDS (coding sequence) regions are marked with yellow boxes, UTR (untranslated region) with green boxes and intron with black lines. (B) The motifs represented by different coloured boxes refer to the legend on the right. The scales at the bottom are used to measure the length of the sequence.
As shown in Figure 4B, 10 types of motifs were detected (Supplementary Figure S2). The motifs that were identified contained between 21 and 50 amino acids. All members shared 10 motifs, which indicates the high sequence similarity of GmLOXs, and all the motifs were arranged in the same order, which suggests that the motif locations of GmLOXs are conserved. For certain motifs, several genes possess more than one motif. GmLOX20 has an extra motif I, GmLOX17 has an extra motif B, GmLOX7 and GmLOX30 have an extra motif F, and GmLOX25 and GmLOX27 have an extra motif A.
Chromosomal location, duplication and synteny analysis
The distribution of genes across chromosomes indicated that the 36 GmLOX genes were located on chromosomes 3, 7, 8, 10, 11, 12, 13, 15, 16, 19 and 20, including 3 on Chr3, 7 on Chr7, 7 on Chr8, 2 on Chr11, 1 on Chr12, 6 on Chr13, 3 on Chr15, 2 on Chr16, 1 on Chr19 and 4 on Chr20 (Figure 5). In the figure, 9-LOX members are orange font, type Ⅰ 13-LOX is red and type II 13-LOX is blue. Some of the GmLOX genes on chromosomes 7, 8, 11, 13, 15, and 20 are quite close together. Tandem repeats based on positional relationships can be inferred.
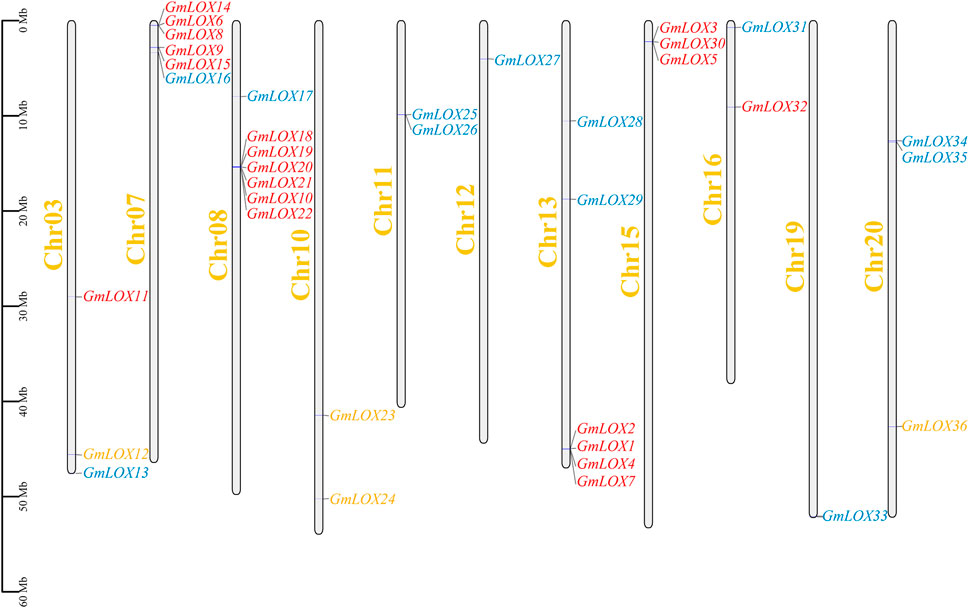
FIGURE 5. Chromosomal distribution of GmLOX family genes in Zhonghuang 13 Note: The scale is chromosome length (Mb). The grey bars represent chromosomes. Chromosome numbers are shown on the left side of the bar. Chr03-20, chromosomes in Zhonghuang 13. The GmLOX genes are marked on the right side of the chromosome.
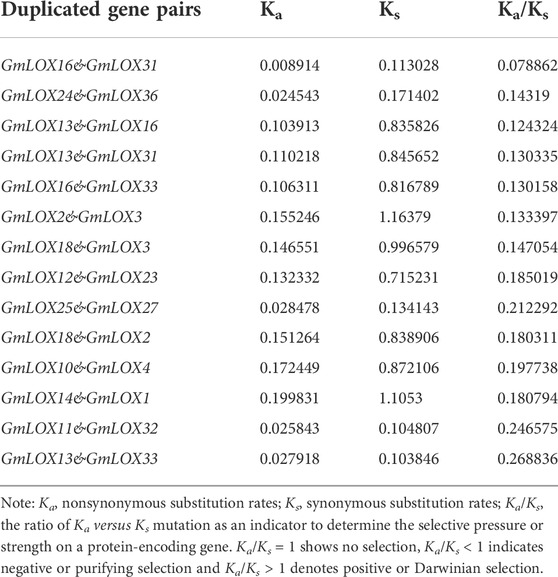
Soybean is an ancient tetraploid plant that underwent two genome-wide duplication events (Song et al., 2017). Approximately 75% of the genes in the soybean genome have multiple copies (Schmutz et al., 2010). Duplication analysis of GmLOX genes was performed, and a large number of genes were found to be collinear. There were 14 duplicate gene pairs identified among 36 GmLOX (Figure 6). This result suggested that the GmLOX gene family underwent extensive duplication during the evolution of soybean. Notably, GmLOX2 and GmLOX3 were a pair of duplicated genes. In parallel, we calculated the Ka/Ks of these duplicated gene pairs (Table 2). The GmLOX gene family was subjected to purifying selection during the evolution of soybean.
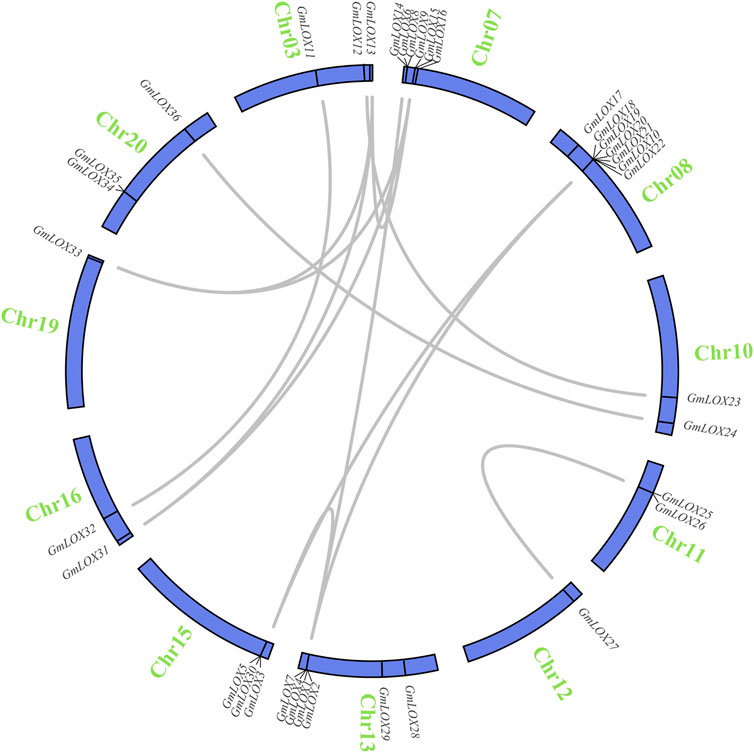
FIGURE 6. Circle plot of segmental duplicated gene pairs of GmLOX genes on chromosomes of Zhonghuang 13 Note: The blue circle bars represent chromosomes. Chromosome numbers are located outside the bar in green. The genes connected by the grey line are deplication gene pairs.
Prediction of cis-regulatory elements in the GmLOX promoters
To explore the regulatory mechanism of GmLOX genes, the cis-regulatory elements of promoter sequences were predicted. As shown in Figure 7, a total of 51 cis-acting elements were identified, including nine growth- and development-related cis-elements, 12 hormone-responsive cis-elements, five stress-responsive cis-elements and 25 light-responsive cis-elements. The hormonal response cis-elements include abscisic acid responsiveness, auxin responsiveness, gibberellin responsiveness, MeJA responsiveness, salicylic acid responsiveness and flavonoid biosynthetic gene regulation. Stress-responsive cis-elements included anaerobic induction, drought inducibility, low-temperature responsiveness and defense and stress responsiveness. A summary of the number of functional cis elements of each gene is given in Supplementary Table S4. These results suggest that the GmLOX gene family may play a role in growth and development and its response to light, hormones, and stress.
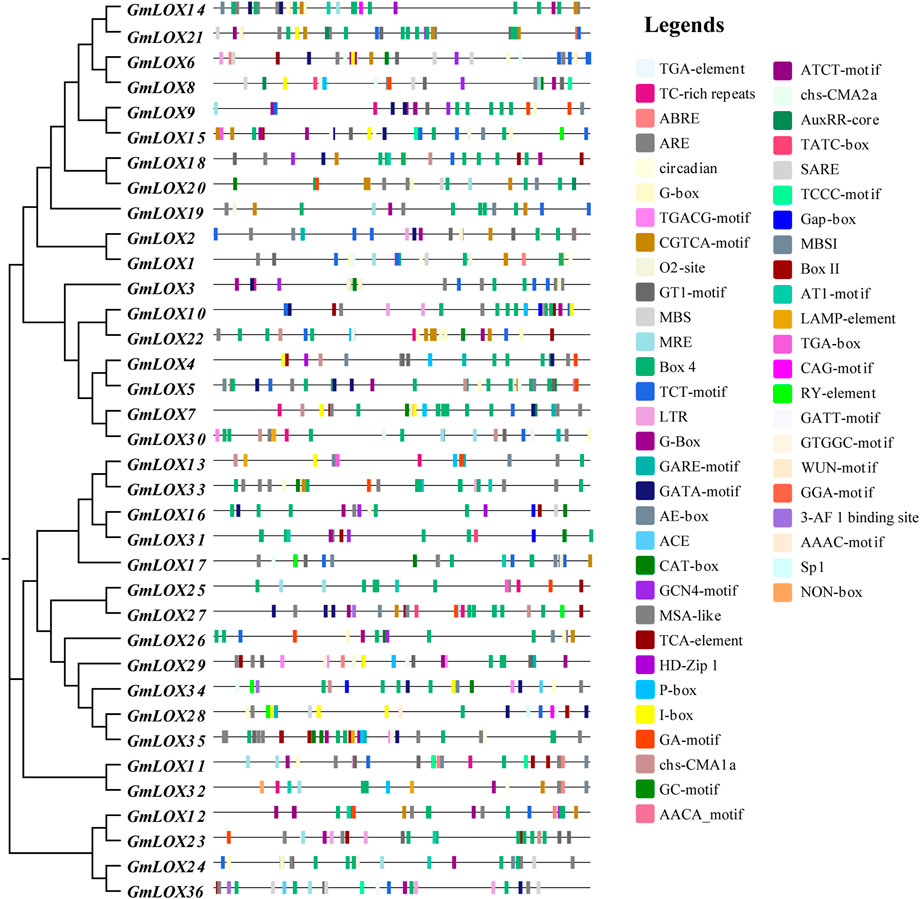
FIGURE 7. Analysis of cis-elements in the promoters of GmLOX genes Note: 2,000 bp upstream from the transcription initiation site was intercepted for analysis, and the sequence lengths in the figure all represent 2000. Response elements involve stress, growth and development, hormones, light response and so on.
Evolution of LOX genes between wild soybean (G. soja W05) and Zhonghuang 13
G. soja is the undomesticated ancestor of G. max, and soybean domestication is complicated (Kim et al., 2010), indicating that it is of great significance to explore the changes in the LOX gene during soybean domestication. The LOX gene family members of G. soja W05 were identified according to the method described above. Through BLAST alignment with LOX genes in Williams 82 and Zhonghuang 13, we found that two tandem genes were annotated into one gene in W05. We manually modified the LOX gene annotations in W05 that were identified as faulty. The evolution of LOX genes was observed by identifying the orthologous gene pairs of LOX genes between G. soja W05 and Zhonghuang 13 (Figure 8). The previous results (Xie et al., 2019) of chromosome sequence comparison between Zhonghuang 13 and W05 showed that translocation occurred on chromosomes 11 and 13 of soybean. The translocation segment contains a gene cluster of the LOX gene family, which is on chromosome 11 in W05 but on chromosome 13 in Zhonghuang 13. The results of calculating the Ka/Ks and Kn/Ks of the orthologous gene pairs are shown in Supplementary Table S5. The Ka/Ks values of most orthologous gene pairs were less than 1, while a few gene pairs had very high values. The Kn/Ks values of both the intron sequence and promoter sequence were very low, indicating that the noncoding sequences of the LOX gene between the two varieties were relatively conserved.
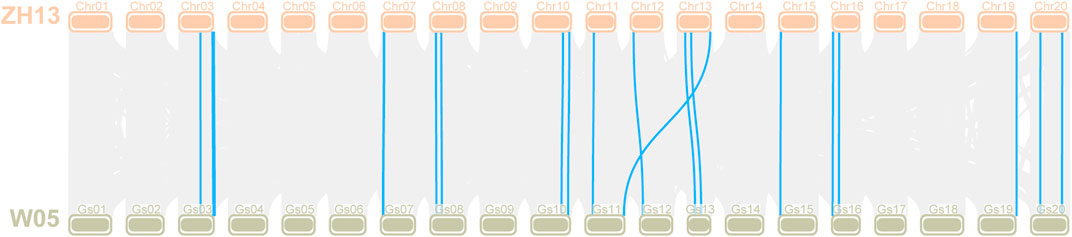
FIGURE 8. Collinearity analysis of the LOX gene between Zhonghuang 13 and G. soja W05 Note: The orange bars at the top marked with Chr01-20 represent chromosomes in Zhonghuang 13, and the green bars labelled Gs01-20 at the bottom represent the wild soybean (G. soja W05) chromosomes. Orthologous gene pairs of genes in the two varieties are linked by blue highlighted lines.
Expression analysis based on RNA-Seq data in different tissues of soybean
The expression pattern of each GmLOX is shown in Figure 9, and the genes were arranged based on hierarchical clustering analysis. The heatmap of RNA-Seq analysis reflects that many GmLOXs appeared at low transcript abundance levels. The expression levels of GmLOX1, GmLOX2 and GmLOX3 in seeds were particularly high, and GmLOX4 in soybean roots was extremely high. The RNA-Seq atlas data revealed that some GmLOX genes are expressed specifically in soybean tissues. GmLOX6, GmLOX7 and GmLOX9 are specifically expressed in flowers and pods, GmLOX21 and GmLOX14 are specifically expressed in soybean roots, and GmLOX29 is specifically expressed in stems. However, two genes (GmLOX20 and GmLOX22) had very low expression levels and could be considered unexpressed in our samples.
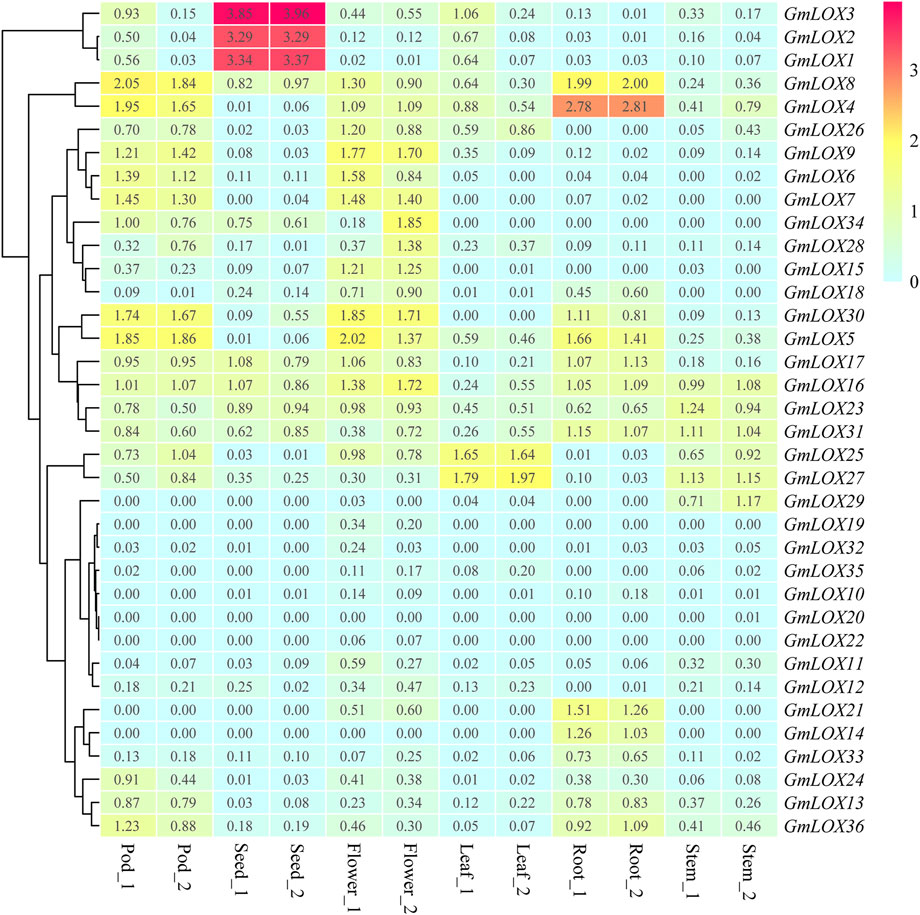
FIGURE 9. Heatmap of the expression patterns of GmLOX genes in different tissues based on transcriptome data Note: Root_1 and Root_2 are two biological duplications in roots (R2 > 0.85), and so on for other organizations. The log10 conversion value of expression values +1 (transcripts per million, TPM) was used for plotting. Light blue represents low expression, yellow represents medium expression, and red represents high expression.
RNA isolation and quantitative real-time PCR (qRT‒PCR)
By analyzing the qRT‒PCR data of the three reference genes, the expression levels of the ELF1b gene were significantly different in each tissue, while the expression levels of the CPY2 and ACT genes tended to be stable, so the geometric mean of the Ct values of these two genes was used to calculate the expression of 12 GmLOX genes. Due to the large difference in GmLOX gene expression, samples with appropriate expression levels for each gene were selected as calibration samples. The qRT‒PCR results showed that GmLOX had extremely significant tissue-specific expression, which was represented by GmLOX1, GmLOX2 and GmLOX3, and the expression of these three genes was super hyper-expressed in seeds (Figures 10A–C). The expression levels of GmLOX4 and GmLOX5 were low in seeds but high in other tissues to varying degrees (Figures 10D,E). The expression of GmLOX34 was only high in flowers (Figure 10F). The expression levels of GmLOX8, GmLOX9 and GmLOX30 were fairly low in seeds and leaves; GmLOX8 and GmLOX30 were expressed to different degrees in other tissues, while GmLOX9 had a strong specific expression in flowers and pods (Figures 10G–I). The expression patterns of GmLOX25 and GmLOX27 were also similar in that they were not expressed in roots and seeds (Figures 10J,K). The expression of GmLOX16 is high in flowers and is also found in other tissues (Figure 10L). The trend of the qRT‒PCR results was consistent with that of the transcriptome analysis.
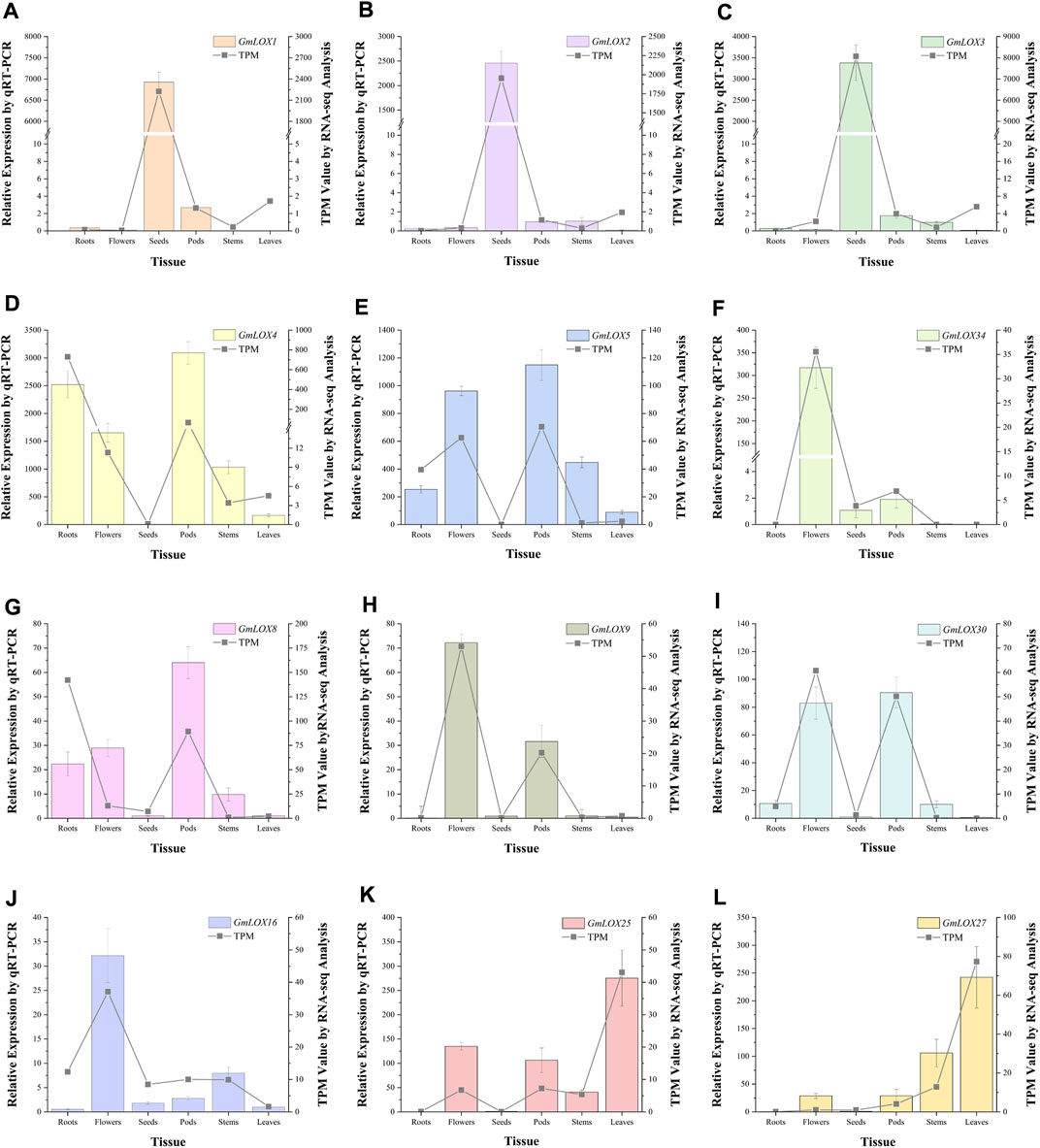
FIGURE 10. Twelve GmLOX genes were selected for qRT‒PCR analysis in different tissues Note: The left vertical axis represents the relative expression obtained by qRT‒PCR analysis, which is shown by the bar graphs in the figure. The standard deviation was calculated from the results of three independent experiments. The right vertical axis is the TPM value calculated by RNA-seq analysis, which is presented in the form of line graphs in the figure. Labels (A–L) show the qRT-PCR analysis results of GmLOX1, GmLOX2, GmLOX3, GmLOX4, GmLOX5, GmLOX34, GmLOX8, GmLOX9, GmLOX30, GMLOX16, GmLOX25, and GmLOX27 genes, respectively.
Discussion
In soybean, lipoxygenase protein is encoded by a multigene family (Liu Y et al., 2020). In this study, we identified lipoxygenase gene family members in Zhonghuang 13. Although Zhonghuang 13 is the most complete soybean genome at present, its gene structure annotation results are not very accurate for practical use. BUSCO scores assessed the genome assembly up to 99.7%, but the annotated protein-coding genes were only up to 96.3%. Therefore, we first calibrate the annotation of LOX genes manually. In the phylogenetic analysis, to confirm the accuracy of the LOX gene family members results in alien species, we verified the existing but unidentified results in the literature (Umate, 2011; Chen et al., 2015) and deleted the genes lacking the PLAT domain or lipoxygenase domain. The results confirmed that the numbers of LOX genes in O. sativa and P. trichocarpa were 11 and 18, respectively. Consistent with previous research (Liu et al., 2011; Shaban et al., 2018; Wang et al., 2019), the GmLOX genes were divided into three groups (type Ⅰ 13-LOX, type II 13-LOX, and 9-LOX) according to protein structure and similarity. The proteins of these subfamilies bind to substrates at different locations. The carbon atom nine of the hydrocarbon main chain of 9-LOX fatty acids binds to substrates and mainly participates in the fat oxidation reaction of linoleic acids (LA, 18:2). Carbon atom 13 of the hydrocarbon main chain of 13-LOX fatty acids binds to substrates and is mainly involved in the fat oxidation reaction of linolenic acids (LeA, 18:3) (Podolyan et al., 2010). Combined with the analysis of chromosome location, more than half of the genes are concentrated on chromosomes 7, 8, and 13 and exist as gene clusters. Almost all the genes of the gene clusters belong to the 13-LOX subfamily, indicating that specific amplification of the soybean LOX gene family occurred in the evolution of species. Tandem duplication occurs for a number of reasons. It is currently believed that tandem duplication is presumed to arise through unequal chromosomal crossing over (Leister, 2004), which is responsible for much of the tandem duplication gene number and allelic variation within species. Tandem duplicated genes are likely to be important for adaptive evolution to rapidly changing environments (HUANG et al., 2022). Some studies have shown that the formation of tandem duplication genes during evolution is related to immunity and stress (Rodgers-Melnick et al., 2012), which verifies the viewpoint that the LOX genes is related to stress (Rosahl, 1996). Relevant studies have found transposable element sequences in LOX genes of wheat (Garbus et al., 2009). In this study, the RepeaMasker program (Tempel, 2012) was used to analyze the repeat sequence in the Zhonghuang 13 genome and found that LTR (long-term relationship) sequences existed in more than 70% of the GmLOX genes, suggesting that transposition may be one of the reasons for the amplification of the GmLOX gene. In addition, pseudogenes may also be formed due to the loss of normal function caused by improper transposon insertion sites or sequence mutation. The spatiotemporal expression data of genes also reflect the hypothesis that many genes are not involved in transcriptional expression. In the comparison of LOX genes between Zhonghuang 13 and Williams 82, two LOX genes that have been eliminated by mutations supported the above view. Gene duplication plays an important role in the generation of new genes and sub-functionalization or new functionalization (Zhang et al., 2016a). There are a large number of duplicated gene pairs in the GmLOX gene family, and the duplicated genes have undergone purification selection pressure. In the conserved motifs, motif E is a specific sequence of the LOX family, which contains a sequence of 38 amino acid residues, His-(X)4-His-(X)4-His-(X)17-His-(X)8-His (Supplementary Figure S3). All GmLOX proteins contain this highly conserved structure (Zhang Z et al., 2006). In the prediction of GmLOX subcellular localization, these genes were mainly distributed in the cytoplasm and chloroplast. This result was consistent with that of subfamily clustering. Most of the Type II 13-LOX genes that had a chloroplast targeting peptide were located in chloroplasts, which may be related to the structure of the gene sequence. Relevant reviews indicate that Type II-LOX proteins are chloroplast proteins that carry N-terminal plastid transport peptides, while Type I-LOX without transport peptides usually have cytoplasmic localization (Zhang et al., 2014). It could be inferred that different lipoxygenases play a catalytic role in different positions.
In A. thaliana, AtLOX genes belong to only the 9-LOX and type II 13-LOX subfamilies (Bannenberg et al., 2009). In the 9-LOX subfamily, GmLOX12 and GmLOX23 are clustered with AtLOX1, and GmLOX24 and GmLOX36 are associated with AtLOX5. It has been reported that the products of 9-LOX genes in A. thaliana can protect plants from various biological and abiotic stresses (Vellosillo et al., 2007). The synthesis of JA compounds requires participation of the LOX gene (Ruan et al., 2019). A relevant article shows that AtLOX1 plays an important regulatory role in the synthesis of metabolites such as JA in response to pathogen attack (Chen et al., 2015), which implies that GmLOX12 and GmLOX23 may participate in the formation of JA. The response cis-regulatory elements associated with the regulation of methyl jasmonate (MeJA) were identified in the analysis of the GmLOX gene promoter region. Nevertheless, further research and confirmation are needed for the specific potential mechanism of this function. Some studies indicated that external application of MeJA had a certain effect on LOX gene expression. In a study of the LOX gene family in C. lanatus, the most significant increases were observed for ClLOX7 upon JA treatment, suggesting that ClLOX7 plays a primary role in JA signaling in leaves and roots (Liu J et al., 2020). Exogenous MeJA treatment was found to significantly enhance the transcript expression of most SpLOX genes belonging to the 13-LOX subfamily (Upadhyay et al., 2020).
Wild soybean is a close relative ancestor of cultivated soybean, and they both belong to Leguminosae, Papilionoideae, Glycine, and Soja (Zhang et al., 2016b). Cultivated soybean is the result of artificial selection according to people’s needs. In contrast, there are more diverse original genes in wild soybean. At present, many studies trace the evolutionary origin of genes by comparing the differences between wild soybean and cultivated soybean (Rehman et al., 2021). The LOX gene of wild soybean (G. soja W05) was also identified and compared. Notably, the translocation segment between chromosome 11 and chromosome 13 contained a LOX gene cluster including four LOX genes. Many studies have shown that noncoding sequences play an important regulatory role in biological evolution (Zhang, 2022). The Kn/Ks values of intron and promoter sequences are very low, indicating that the noncoding sequence of the GmLOX gene is highly conserved. For the coding sequence, the Ka/Ks results showed that most gene pairs were subject to purification selection pressure. However, there were several gene pairs for which Ka/Ks values were far greater than 1. We speculated that on the one hand, these genes may have experienced positive selection, or on the other hand, they may have been caused by population polymorphism. Additionally, during the evolution of species, there would be regional population differences. The species that evolved from the same region would have the highest similarity with the ancestors of the region, but there would be more SNP (single nucleotide polymorphism) loci with the ancestors of other regions. It could be speculated that these regions may come from different populations that have wide nucleic acid polymorphisms, resulting in Ka/Ks values of individual genes far greater than 1. We analyzed the location of introns and exons of LOX genes in W05. The result is very similar to that of Zhonghuang 13. Theoretically, in the recent evolution of eukaryotes, a high intron loss rate is a more common phenomenon (Roy and Penny, 2007). The number of introns of LOX genes in W05 and Zhonghuang 13 basically did not change. LOX genes are very conserved. In the long-term domestication and improvement of soybeans, only a small number of genetic resources were selected (Huihui et al., 2018). We speculate that the LOX gene family may not belong to strong artificial selection loci.
The cis-regulatory elements in the GmLOX promoters mainly include defense and stress, circadian control, development and tissue specificity, light and hormones. The number of light response elements was the largest, and relevant studies have investigated the relationship between the expression of the LOX gene and light. In the study of the expression pattern of the LOX gene in response to light in Fagopyrum tataricum, red and blue light induced the expression of FtLOX1 and FtLOX7, whereas FtLOX4 and FtLOX6 were inhibited by light. FtLOX5-7 expression levels were the highest under far-red light (Ji et al., 2020). Previous studies have also shown that LOX expression is strongly associated with the application of plant hormones such as MeJA, salicylic acid, abscisic acid, and ethylene. A study of the differential expression of the LOX gene family in ripening kiwifruit found that the expression levels of AdLOX2, AdLOX3 and AdLOX4 were negatively correlated with ethylene accumulation, and ethylene application promoted a decrease in the expression levels (Zhang B et al., 2006). In Taxus chinensis, TcLOX1 was upregulated by MeJA and abscisic acid, while TcLOX2 was upregulated by ABA (Li et al., 2012). The verification of the above functions of GmLOX genes needs to be further studied.
In qRT‒PCR analysis, some reference genes or housekeeping genes may sometimes respond considerably to changes in experimental conditions or tissue types (Gutierrez et al., 2008; Hu et al., 2009). Therefore, multiple internal reference genes were selected to reduce the influence of uncertainty. In our study, three reference genes were selected: GmACT, GmCYP2, and GmELF1B. The ACT gene is involved in the formation of cytoskeletal structural proteins. The CYP2 gene is involved in protein folding, and the ELF1B gene is involved in translation elongation. Previous studies have shown that unique expression patterns occur in some LOX genes, which can provide a favorable starting point for studying the mechanism and physiological function of LOX genes. For example, analysis of Oep2LOX1 expression levels in different olive tissues from Picual and Arbequina cultivars showed that Oep2LOX1 expression was higher in leaves but lower in other tissues (Padilla et al., 2012). These genes exist in different tissue sites, which may indicate differences in function. Transcriptional data analysis of different soybean tissues showed that GmLOX gene expression had significant tissue specificity. Represented by GmLOX1, GmLOX2 and GmLOX3, the expression levels in seeds were particularly high. Wang et al. (2020) obtained mutants without GmLOX1, GmLOX2 and GmLOX3 using a pooled CRISPR‒Cas9 system, confirming the role of these three LOXs in causing the beany flavor. Related articles have shown that LOXs stored in seeds can be used as nutritional storage proteins and participate in the process of seed maturation and seedling growth (Viswanath et al., 2020). It can be speculated that these three GmLOX genes with high expression in seeds may play a role in seed maturation and seedling growth. These findings provide important clues for further exploring the function of GmLOX genes in the future.
Conclusion
In this study, 36 GmLOX genes were identified in Zhonghuang 13. We modified the annotation information of four GmLOX genes in Zhonghuang 13 by combining transcriptome analysis data with LOX gene information from William 82. We verified the existence of GmLOX1 at both the gene and protein levels. Phylogenetic analysis showed that the LOX gene was mainly divided into three clusters: type I 13-LOX, type II 13-LOX, and 9-LOX. The type I 13-LOX gene of soybean accounted for a whole branch. Combined with gene localization and collinearity, it could be speculated that the type I 13-LOX subfamily of GmLOX genes may have replicated on a large scale. The GmLOX genes were subjected to negative selection pressure during evolution. The highly conserved characteristics of GmLOX genes were confirmed by both gene structure and conserved motifs. There are many cis-regulatory elements, including those related to stress response, growth and development, hormone response and light response, in the promoter region. The Ka/Ks analysis of coding sequences and Kn/Ks analysis of noncoding sequences (intron sequence and promoter sequence) of orthologous gene pairs between Zhonghuang 13 and G. soja W05 showed that the noncoding sequences of GmLOX genes were highly conserved, and the coding sequences of most GmLOX genes were subject to negative selection. The spatial expression of GmLOX genes was studied and the results showed that some genes had obvious tissue-specific expression. These findings lay a foundation for further exploration of the function and evolution of the LOX gene.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. RNA-Seq data were submitted to the NCBI Sequence Read Archive (SRA) under accession numbers SRR21075390 to SRR21075406 (17 accession numbers in total).
Author contributions
ShW and SuW conceived and designed the experiments and revised the manuscript. JZ and CN analyzed the data and wrote the manuscript. YJ provided experimental samples and XW participated in data analysis. All authors read and approve the final manuscript.
Funding
This work was financially supported by Hundred-Thousand and Million Project of Heilongjiang Province for Engineering and Technology Science’ Soybean Breeding Technology Innovation and New Cultivar Breeding (SC2019ZX16B0039), Young Talents Program of Northeast Agricultural University (19QC01) and Funding for the Opening Project of Key Laboratory of Soybean Biology of Chinese Education Ministry (SBKF01). The funding bodies had no role in the design of the study, collection, analysis, or interpretation of data or in the writing of the manuscript.
Acknowledgments
The authors gratefully acknowledge Guan-Zheng Qu at Northeast Forestry University for his help with the RNA-Seq data analysis. We sincerely thank laboratory peers for the support and assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1020554/full#supplementary-material
Supplementary Table S1 | Primers for 12 GmLOX genes and three internal reference genes.
Supplementary Table S2 | Homologous gene pairs between Zhonghuang 13 and Williams 82.
Supplementary Table S3 | Gene names corresponding to sequence IDs in four outgroup species.
Supplementary Table S4 | The summary of functional cis elements number of each GmLOX gene.
Supplementary Table S5 | Estimation of Ka/Ks and Kn/Ks of orthologous gene pairs between G. soja W05 and Zhonghuang13.
Supplementary Data S1 | GmLOX genes coding sequences.
Supplementary Data S2 | GmLOXs protein sequences.
Supplementary Data S3 | GmLOX genes promoter sequences -2,000 bp upstream of the GmLOX translation initiation codon.
Supplementary Data S4 | Raw sequencing results of GmLOX1 gene full-length sequence.
Supplementary Figure S1 | Two LOX genes terminated prematurely of protein translation due to mutation.
Supplementary Figure S2 | Sequence details of 10 motifs.
Supplementary Figure S3 | Conserved structure constituted by 38 amino acid residues in LOX gene.
References
Bailey, L., and Elkan, C. E. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bannenberg, G., Martínez, M., Hamberg, M., and Castresana, C. (2009). Diversity of the enzymatic activity in the lipoxygenase gene family of arabidopsis thaliana. Lipids 44, 85–95. doi:10.1007/s11745-008-3245-7
Bateman, A., and Sandford, R. (1999). The PLAT domain: A new piece in the PKD1 puzzle. Curr. Biol. 9, R588–R590. doi:10.1016/S0960-9822(99)80380-7
Brash, A. R. (1999). Lipoxygenases: Occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem. 274, 23679–23682. doi:10.1074/jbc.274.34.23679
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinforma. 10, 421. doi:10.1186/1471-2105-10-421
Chan, C., Qi, X., Li, M., Wong, F., and Lam, H. (2012). Recent developments of genomic research in soybean. J. Genet. Genomics 39, 317–324. doi:10.1016/j.jgg.2012.02.002
Chaudhary, J., Patil, G. B., Sonah, H., Deshmukh, R. K., Vuong, T. D., Valliyodan, B., et al. (2015). Expanding omics resources for improvement of soybean seed composition traits. Front. Plant Sci. 6, 1021. doi:10.3389/fpls.2015.01021
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi:10.1093/bioinformatics/bty560
Chen, Z., Chen, X., Yan, H., Li, W., Li, Y., Cai, R., et al. (2015). The lipoxygenase gene family in poplar: Identification, classification, and expression in response to MeJA treatment. Plos One 10, e0125526. doi:10.1371/journal.pone.0125526
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. GigaScience 10, giab008. doi:10.1093/gigascience/giab008
Dunsford, B. R., Knabe, D. A., and Haensly, W. E. (1989). Effect of dietary soybean meal on the microscopic anatomy of the small intestine in the early-weaned pig. J. Animal Sci. 67, 1855. doi:10.2527/jas1989.6771855x
Feussner, I., Kühn, H., and Wasternack, C. (2001). Lipoxygenase-dependent degradation of storage lipids. England: Elsevier.
Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. doi:10.1146/annurev.arplant.53.100301.135248
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Gao, M., Liu, Y., Ma, X., Shuai, Q., Gai, J., and Li, Y. (2017). Evaluation of reference genes for normalization of gene expression using quantitative RT-PCR under aluminum, cadmium, and heat stresses in soybean. PLoS One 12, e0168965. doi:10.1371/journal.pone.0168965
Garbus, I., Carrera, A. D., Dubcovsky, J., and Echenique, V. (2009). Physical mapping of durum wheat lipoxygenase genes. J. Cereal Sci. 50, 67–73. doi:10.1016/j.jcs.2009.02.008
Gutierrez, L., Mauriat, M., Gunin, S., Pelloux, J., Lefebvre, J. F., Louvet, R., et al. (2008). The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618. doi:10.1111/j.1467-7652.2008.00346.x
Haas, B. J., Delcher, A. L., Mount, S. M., Wortman, J. R., Smith, R. K., Hannick, L. I., et al. (2003). Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666. doi:10.1093/nar/gkg770
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi:10.1093/nar/gkw370
Hu, R., Fan, C., Li, H., Zhang, Q., and Fu, Y. F. (2009). Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 10, 93. doi:10.1186/1471-2199-10-93
Huang, Y., Zhang, L., Zhang, K., Chen, S., Hu, J., and Cheng, F. (2022). The impact of tandem duplication on gene evolution in Solanaceae species. J. Integr. Agric. 21, 1004–1014. doi:10.1016/S2095-3119(21)63698-5
Huihui, G., Wang, Y., and Li, W. (2018). Transcriptomic comparison reveals genetic variation potentially underlying seed developmental evolution of soybeans. J. Exp. Bot. 69 (21), 5089–5104. doi:10.1093/jxb/ery29
Ji, Q., Zhang, T., Zhang, D., Lv, S., and Tan, A. (2020). Genome-wide identification and expression analysis of lipoxygenase genes in Tartary buckwheat. Biotechnol. Biotechnol. Equip. 34, 273–286. doi:10.1080/13102818.2020.1738956
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066. doi:10.1093/nar/gkf436
Kim, D., Langmead, B., and Salzberg, S. L. (2015). Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Kim, M. Y., Lee, S., Van, K., Kim, T. H., Jeong, S. C., Choi, I.-Y., et al. (2010). Whole-genome sequencing and intensive analysis of the undomesticated soybean ( Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci. U.S.A. 107, 22032–22037. doi:10.1073/pnas.1009526107
Kolomiets, M. V., Hannapel, D. J., Chen, H., Tymeson, M., and Gladon, R. J. (2001). Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13, 613–626. doi:10.1105/tpc.13.3.613
Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122. doi:10.1016/j.tig.2004.01.007
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Li, S., Zhang, M., Fu, C., Xie, S., Zhang, Y., and Yu, L.-j. (2012). Molecular cloning and characterization of two 9-lipoxygenase genes from taxus chinensis. Plant Mol. Biol. Rep. 30, 1283–1290. doi:10.1007/s11105-012-0439-1
Li, Y. H., Zhou, G., Ma, J., Jiang, W., Jin, L. G., Zhang, Z., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052. doi:10.1038/nbt.2979
Liu, J., Zhou, Y., Li, J., Wang, F., and Yang, Y. (2020). Comprehensive genomic characterization and expression analysis of the lipoxygenase gene family in watermelon under hormonal treatments. Agriculture 10, 429. doi:10.3390/agriculture10100429
Liu, S. Q., Liu, X. H., and Jiang, L. W. (2011). Genome-wide identification, phylogeny and expression analysis of the lipoxygenase gene family in cucumber. Genet. Mol. Res. 10, 2613–2636. doi:10.4238/2011.October.25.9
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020). Pan-Genome of wild and cultivated soybeans. Cell 182, 162–176. doi:10.1016/j.cell.2020.05.023
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi:10.1093/molbev/msu300
Padilla, M. N., Hernández, M. L., Sanz, C., and Martínez-Rivas, J. M. (2012). Molecular cloning, functional characterization and transcriptional regulation of a 9-lipoxygenase gene from olive. Phytochemistry 74, 58–68. doi:10.1016/j.phytochem.2011.11.006
Park, T. K., and Polacco, J. C. (1989). Distinct lipoxygenase species appear in the hypocotyl/radicle of germinating soybean. Plant Physiol. 90, 285–290. doi:10.1104/pp.90.1.285
Podolyan, A., White, J., Jordan, B., and Winefield, C. (2010). Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct. Plant Biol. 37, 767. doi:10.1071/FP09271
Porta, H., and Rocha-Sosa, M. (2002). Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 130, 15–21. doi:10.1104/pp.010787
Rehman, S. U., Qanmber, G., Tahir, M., Irshad, A., Fiaz, S., Ahmad, F., et al. (2021). Characterization of Vascular plant One-Zinc finger (VOZ) in soybean (Glycine max and Glycine soja) and their expression analyses under drought condition. PLoS One 16, e0253836. doi:10.1371/journal.pone.0253836
Rodgers-Melnick, E., Mane, S. P., Dharmawardhana, P., Slavov, G. T., Crasta, O. R., Strauss, S. H., et al. (2012). Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22, 95–105. doi:10.1101/gr.125146.111
Rosahl, S. (1996). Lipoxygenases in plants -their role in development and stress response. Z Naturforsch C J. Biosci. 51, 123–138. doi:10.1515/znc-1996-3-401
Roy, S. W., and Penny, D. (2007). Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O. Sativa and a. Thaliana. Mol. Biol. Evol. 24, 171–181. doi:10.1093/molbev/msl159
Ruan, J., Zhou, Y., Zhou, M., Yan, J., Khurshid, M., Weng, W., et al. (2019). Jasmonic acid signaling pathway in plants. Ijms 20, 2479. doi:10.3390/ijms20102479
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi:10.1038/nature08670
Shaban, M., Ahmed, M. M., Sun, H., Ullah, A., and Zhu, L. (2018). Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genomics 19, 599. doi:10.1186/s12864-018-4985-2
Shen, Y., Du, H., Liu, Y., Ni, L., Wang, Z., Liang, C., et al. (2019). Update soybean Zhonghuang 13 genome to a golden reference. Sci. China Life Sci. 62, 1257–1260. doi:10.1007/s11427-019-9822-2
Shen, Y., Liu, J., Geng, H., Zhang, J., Liu, Y., Zhang, H., et al. (2018). De novo assembly of a Chinese soybean genome. Sci. China Life Sci.Life Sci. 61, 871–884. doi:10.1007/s11427-018-9360-0
Song, H., Wang, P., Li, C., Han, S., Lopez-Baltazar, J., Zhang, X., et al. (2016). Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 6, 35245. doi:10.1038/srep35245
Song, J., Li, Z., Liu, Z., Guo, Y., and Qiu, L. J. (2017). Next-Generation sequencing from Bulked-Segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front. Plant Sci. 8, 919. doi:10.3389/fpls.2017.00919
Steczko, J., Donoho, G. P., Clemens, J. C., Dixon, J. E., and Axelrod, B. (1992). Conserved histidine residues in soybean lipoxygenase: Functional consequences of their replacement. Biochemistry 31, 4053–4057. doi:10.1021/bi00131a022
Tempel, S. (2012). Using and understanding RepeatMasker. Methods Mol. Biol. 859, 29–51. doi:10.1007/978-1-61779-603-6_2
Tomas, A., De La Gandara, F., Garcia-gomez, A., Perez, L., and Jover, M. (2005). Utilization of soybean meal as an alternative protein source in the Mediterranean yellowtail, Seriola dumerili. Aquac. Nutr. 11, 333–340. doi:10.1111/j.1365-2095.2005.00365.x
Tomchick, D. R., Phan, P., Cymborowski, M., Minor, W., and Holman, T. R. (2001). Structural and functional characterization of Second-Coordination sphere mutants of soybean lipoxygenase-1. Biochemistry 40, 7509–7517. doi:10.1021/bi002893d
Umate, P. (2011). Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 6, 335–338. doi:10.4161/psb.6.3.13546
Upadhyay, R. K., Edelman, M., and Mattoo, A. K. (2020). Identification, phylogeny, and comparative expression of the lipoxygenase gene family of the aquatic duckweed, spirodela polyrhiza, during growth and in response to methyl jasmonate and salt. Ijms 21, 9527. doi:10.3390/ijms21249527
Upadhyay, R. K., Handa, A. K., and Mattoo, A. K. (2019). Transcript abundance patterns of 9- and 13-lipoxygenase subfamily gene members in response to abiotic stresses (heat, cold, drought or salt) in tomato (Solanum lycopersicum L.) highlights member-specific dynamics relevant to each stress. Genes 10, 683. doi:10.3390/genes10090683
Veldink, G. A., Vliegenthart, J. F. G., and Boldingh, J. (1977). Plant lipoxygenases. Prog. Chem. Fats Other Lipids 15, 131–166. doi:10.1016/0079-6832(77)90014-3
Vellosillo, T., Martínez, M., López, M. A., Vicente, J., Cascón, T., Dolan, L., et al. (2007). Oxylipins produced by the 9-lipoxygenase pathway inArabidopsisRegulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19, 831–846. doi:10.1105/tpc.106.046052
Viswanath, K. K., Varakumar, P., Pamuru, R. R., Basha, S. J., Mehta, S., and Rao, A. D. (2020). Plant lipoxygenases and their role in plant physiology. J. Plant Biol. 63, 83–95. doi:10.1007/s12374-020-09241-x
Vogt, J., Schiller, D., Ulrich, D., Schwab, W., and Dunemann, F. (2013). Identification of lipoxygenase (LOX) genes putatively involved in fruit flavour formation in apple (Malus × domestica). Tree Genet. Genomes 9, 1493–1511. doi:10.1007/s11295-013-0653-5
Wang, J., Hu, T., Wang, W., Hu, H., Wei, Q., Wei, X., et al. (2019). Bioinformatics analysis of the lipoxygenase gene family in radish (Raphanus sativus) and functional characterization in response to abiotic and biotic stresses. Ijms 20, 6095. doi:10.3390/ijms20236095
Wang, J., Kuang, H., Zhang, Z., Yang, Y., Yan, L., Zhang, M., et al. (2020). Generation of seed lipoxygenase-free soybean using CRISPR-Cas9. Crop J. 8, 432–439. doi:10.1016/j.cj.2019.08.008
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi:10.1093/nar/gkr1293
Wennman, A., Oliw, E. H., Karkehabadi, S., and Chen, Y. (2016). Crystal structure of manganese lipoxygenase of the rice blast fungus magnaporthe oryzae. J. Biol. Chem. 291, 8130–8139. doi:10.1074/jbc.M115.707380
Xie, M., Chung, C. Y., Li, M. W., Wong, F. L., Wang, X., Liu, A., et al. (2019). A reference-grade wild soybean genome. Nat. Commun. 10, 1216. doi:10.1038/s41467-019-09142-9
Zhang, B., Chen, K., Bowen, J., Allan, A., Espley, R., Karunairetnam, S., et al. (2006). Differential expression within the LOX gene family in ripening kiwifruit. J. Exp. Bot. 57, 3825–3836. doi:10.1093/jxb/erl151
Zhang, C., Jin, Y., Liu, J., Tang, Y., Cao, S., and Qi, H. (2014). The phylogeny and expression profiles of the lipoxygenase (LOX) family genes in the melon (Cucumis melo L.) genome. Sci. Hortic. 170, 94–102. doi:10.1016/j.scienta.2014.03.005
Zhang, J., Jia, H., Li, J., Li, Y., Lu, M., and Hu, J. (2016a). Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 6, 30050. doi:10.1038/srep30050
Zhang, J., Yang, D., Li, M., and Shi, L. (2016b). Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. PLoS One 11, e0159622. doi:10.1371/journal.pone.0159622
Zhang, Z. (2022). KaKs_calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genomics, Proteomics Bioinforma. 1, 1. doi:10.1016/j.gpb.2021.12.002
Zhang, Z., Li, J., Zhao, X., Wang, J., Wong, G. K., and Yu, J. (2006). KaKs_calculator: Calculating ka and ks through model selection and model averaging. Genomics, Proteomics Bioinforma. 4, 259–263. doi:10.1016/S1672-0229(07)60007-2
Keywords: soybean, Zhonghuang 13, lipoxygenase, genome-wide analysis, gene expression
Citation: Zhang J, Ng C, Jiang Y, Wang X, Wang S and Wang S (2022) Genome-wide identification and analysis of LOX genes in soybean cultivar “Zhonghuang 13”. Front. Genet. 13:1020554. doi: 10.3389/fgene.2022.1020554
Received: 16 August 2022; Accepted: 20 September 2022;
Published: 07 October 2022.
Edited by:
Vandana Jaiswal, Institute of Himalayan Bioresource Technology (CSIR), IndiaReviewed by:
Xiugui Chen, Cotton Research Institute (CAAS), ChinaWuwei Ye, Cotton Research Institute (CAAS), China
Copyright © 2022 Zhang, Ng, Jiang, Wang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaodong Wang, d3NkaGxqQG5lYXUuZWR1LmNu; Sui Wang, d2FuZ3N1aS53c0AxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Jing Zhang
Jing Zhang Cheungchuk Ng1†
Cheungchuk Ng1† Yan Jiang
Yan Jiang Sui Wang
Sui Wang