
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 30 September 2022
Sec. Human and Medical Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1017302
This article is part of the Research Topic Genetic Factors in Male Infertility View all 8 articles
 Xuefeng Xie1*†
Xuefeng Xie1*† Mazhar Khan1*†
Mazhar Khan1*† Muhammad Zubair1
Muhammad Zubair1 Abbas Khan2
Abbas Khan2 Ranjha Khan1
Ranjha Khan1 Jianteng Zhou1
Jianteng Zhou1 Yuanwei Zhang1
Yuanwei Zhang1 Muzafar Said3
Muzafar Said3 Sher Ali Khan3
Sher Ali Khan3 Qamar Zaman1
Qamar Zaman1 Ghulam Murtaza1
Ghulam Murtaza1 Muzamil Ahmad Khan4
Muzamil Ahmad Khan4 Wei Liu1
Wei Liu1 Xiaoning Hou1
Xiaoning Hou1 Huan Zhang1
Huan Zhang1 Bo Xu1
Bo Xu1 Xiaohua Jiang1
Xiaohua Jiang1 Shun Bai1*
Shun Bai1* Qinghua Shi1*
Qinghua Shi1*Non-obstructive azoospermia (NOA) is a severe factor of male infertility; it affects approximately 1% of the global male population and accounts for 40% of male infertility cases. However, the majority of NOA cases remain idiopathic. This is the first study using whole-exome sequencing (WES) to identify a novel missense mutation in the DND1 gene (c.212A>C, p. E71A) from a Pakistani family, that includes three males with NOA. This mutation is predicted to cause DND1 protein misfolding and weaken the DND1 interaction with NANOS2, a significant regulator in primordial germ cell development. Our study identified a DND1 pathogenic mutation in NOA patients and highlighted its critical role in male fertility in humans.
Approximately 15% of couples worldwide experience infertility, while male infertility-associated factors are prevalent in half of these couples (Inhorn and Patrizio, 2015; Krausz and Riera-Escamilla, 2018). Azoospermia, the absence of sperm in the ejaculate, affects 10–20% of men with infertility and is classified as either obstructive azoospermia (OA) or non-obstructive azoospermia (NOA) (Peña et al., 2020). NOA is characterized by the absence or reduction of germ cells and is mainly caused by genetic defects, testicular tumors, and chemotherapy (Kasak and Laan, 2021). Mutations in testis-enriched genes, including mitosis genes (DAZL, NANOS1) and meiosis genes (DMC1, STAG3, C14ORF39, and MEIOB) have been implicated in NOA (Teng et al., 2002; Kusz-Zamelczyk et al., 2013; He et al., 2018; Gershoni et al., 2019; van der Bijl et al., 2019; Fan et al., 2021). However, genetic factors underlying the majority of NOA cases require further investigation.
The dead end 1 (DND1) gene encodes a protein that binds RNA and plays an essential role in gene regulation in biological activities, including embryonic development, cell differentiation, cell cycle, cell growth and apoptosis, and its dysfunction can result in a variety of diseases (Das et al., 2020). In mice, DND1 plays a crucial role in maintaining the population of primordial germ cells and suppressing tumors derived from germ cells (Noguchi and Noguchi, 1985; Sakurai et al., 1995; Youngren et al., 2005; Cook et al., 2009; Northrup et al., 2012). Previous studies have shown that DND1 is essential for the binding of target RNAs by NANOS2, a major regulatory factor in spermatogenesis (Suzuki et al., 2016; Yamaji et al., 2017; Zealy et al., 2017). Mutation in Dnd1 disrupted the interaction between DND1 and its partners, resulting in abnormal differentiation of male germ cells and male infertility in mice (Li et al., 2018). However, the function of the DND1 protein in human male fertility remains unknown.
Here, for the first time, we identified a homozygous missense mutation in DND1 (c.212A>C, p. E71A) from three males suffering from NOA in a Pakistani family. In vitro experiments showed that the mutation caused a remarkable decrease in DND1 protein levels and affected its interaction with NANOS2. These data provide functional evidence that pathogenic variants in DND1 cause male infertility in humans.
We recruited three males with primary infertility from a large family (Figure 1A). Each participant completed a detailed questionnaire regarding his reproductive history. According to clinical investigations, all of the affected individuals displayed normal erection and ejaculation, male external genitalia and secondary sexual characteristics. Semen parameters including sperm concentration, motility, and morphology were measured according to the World Health Organization (WHO) criteria for human semen analysis (Boitrelle et al., 2021). Blood samples were collected from all the available family members for genomic DNA extraction. This study was approved by the Institutional Ethical Committee of the University of Science and Technology of China (USTCEC202000003). Each family member provided written consent prior to the collection of biological samples.
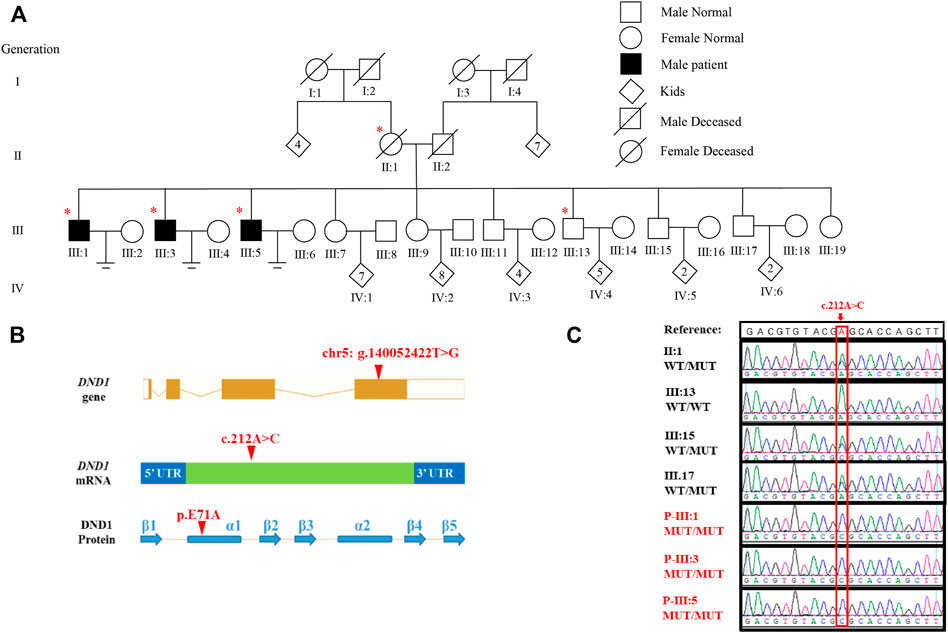
FIGURE 1. Identification of DND1 E71A Mutation in three patients with azoospermia. (A) Pedigree of the affected family. Circles and squares denote females and males, respectively. Deceased family members are denoted by slash symbols. Individuals marked with asterisks, undergone whole exome sequencing. (B) Location of the identified variant in DND1 at both cDNA and protein levels. The mutation site was located in α-subunit of RBD1. (C) Chromatograms of confirming DND1 mutation site by Sanger sequencing. WT, wild-type allele. MT, mutant allele. A, adenine; C, cytosine; G, guanine; T, thymine.
Total gDNA from peripheral blood of the patients and their family members was obtained using the QIAamp DNA Blood Mini Kit (QIAGEN) according to the manufacturer’s instructions. Exons were enriched via the Agilent SureSelect Human All Exon V5 Kit. The enriched DNA was sequenced on the Illumina HiSeq 2000 platform (Illumina, San Diego, CA). Sequencing reads (.qseq format) were aligned to the reference human genome (hg19) using Burrows-Wheeler Aligner (BWA) software with default parameters. SAMtool (http://samtools.sourceforge.net/) was used to convert the SAM files from each sample into BAM files. The PCR duplicates were removed with the Picard tool (http://picard.sourceforge.net/). Files were further processed with the Genome Analysis Toolkit (GATK) available from the Broad Institute (http://www.broadinstitute.org/gatk/), followed by realignment through indel realigner and variants (single-nucleotide variants (SNVs) and indels) calling using GATK’s Unified Genotyper (Zubair et al., 2022). PCR amplification and Sanger sequencing were performed to confirm the candidate pathogenic variants from whole-exome sequencing (WES). All primer sequences used for PCR and sequencing are shown in Supplementary Table S3.
The evolutionary conservation of altered amino acid residues was performed with MEGA (Tamura et al., 2013). The effect of pathogenic missense variants on protein structure was predicted by the SWISS-MODEL online tool.
The coding sequences of wild-type (WT) and mutant (MT) DND1 were fused to EGFP. Human embryonic kidney 293T cells (HEK293T) obtained from ATCC (CRL-1573) were cultured in DMEM (VivaCell, C3113-0500) supplemented with 10% fetal bovine serum (FBS, Gibco, 16,000-044) and penicillin-streptomycin (Gibco, 15,140-122). Plasmids harboring WT or MT eGFP-DND1 were transfected into HEK293T cells using Lipofectamine 3000 according to the manufacturer’s instructions (Invitrogen, L3000015). At 36 h post-transfection, plasmid expression was examined under a Nikon ECLIPSE 80i microscope. At 44 h post-transfection, the cells were harvested for Western blotting analysis.
The HEK293T cells were lysed in 4X Bolt LDS Sample Buffer (Invitrogen, B0008) with NuPAGE Antioxidant (Invitrogen, NP0005). The lysates were denatured for 10 min and then were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transferring the proteins to 0.45 μm pore size immobilon-P membranes (Millipore, IPVH00010). Membranes were blocked in TBST buffer (50 mM Tris, pH 7.4, 150 mm NaCl and 0.1% Tween-20) containing 5% nonfat milk for 1 h and incubated with primary antibodies (GFP, Abmart, M20004L; Myc, Abmart, M20002; mCherry, Abcam, AB125096; mCherry, Abmart, AB167453; β-Actin, Abcam, ab8227) at 4°C overnight. Following incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (HRP Donkey anti-rabbit IgG, BioLegend, Inc., 406,401; HRP Goat anti-mouse IgG, BioLegend, Inc., 405,306) for 1 h, the membranes were developed with chemiluminescence substrate by ImageQuant LAS 4000 imaging system (GE Healthcare).
The HEK293T cells were lysed in lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 5 mm EDTA and 0.1% NP-40) supplemented with PMSF, Protease Inhibitor Cocktail and MG132, followed by rotation at 4°C for 20 min. After centrifugation, 50 μL of the supernatant was taken out as the input. Beads (Pierce™ anti c-Myc beads, Thermo scientific, 88,842; Protein A/G Magnetic beads, bimake, B23202) were washed with pre-cooled lysis buffer for three times, then the beads were incubated with supernatant at 4°C for 2–3 h. After washing with lysis buffer for 6 times, the beads were boiled in 2×SDS sample buffer for 10 min. The samples were either analyzed by Western blotting immediately as described above or stored at −80°C.
In this study, we investigated a Pakistani family, having three infertile brothers suffering from NOA and male infertility (Figure 1A). The parents (II:1 and II:2) had three daughters and seven sons. The unmarried sister (III:19, 26 years old) had normal menstrual cycles and the two married sisters had six and five children, respectively. Among the seven brothers, four had children, whereas III:1 (43 years old), III:3 (36 years old) and III:5 (34 years old) were infertile despite being married for 22, 17, and 18 years, respectively.
The patients had no history of drinking, smoking, exposure to toxic chemicals, and any other diseases. The clinical information of each patient is summarized in Table 1. Routine semen analyses revealed that no spermatozoa were detected in their ejaculates, although the semen volume was normal. Ultrasound examination revealed normal vas deferens and small testes in all patients (right: 3.5*1.7*2.8 cm3 and left: 3.6*2.0*2.6 cm3 for III:1; right: 3.8*2.3*2.6 cm3 and left: 4.0*2.3*2.6 cm3 for III:3; right: 3.8*2.3*2.6 cm3 and left: 4.0*2.4*2.7 cm3 for III:5). Additionally, all enrolled patients had normal sex hormone levels and a 46, XY karyotype. Together, azoospermia in those patients is caused by spermatogenic failure rather than obstruction (Esteves, 2015).
To determine the genetic cause of azoospermia in these patients, WES was conducted in the three affected individuals (III:1, III:3, and III:5), their mother (II:1) and the healthy brother (III:13) (Figure 1A) using gDNA extracted from peripheral blood lymphocytes. Analysis of WES data identified three candidate pathogenic variants: PCDHGA8 (c. 862C>T, p. Q288X), ARHGAP26 (c. 2137G>A, p. V713I) and DND1 (c.212A>C, p. E71A), of which only DND1 is a highly testis-expressed gene (Supplementary Table S1) and its homozygous deficiency cause male infertility (Seelow et al., 2009; Zhang et al., 2013; Li et al., 2018; Niimi et al., 2019). Details regarding the exclusion of the two genes can be found in Supplementary Table S2. The autosomal recessive inheritance pattern of the missense variant of DND1 in this family was confirmed by Sanger sequencing (Figure 1C). In addition, the mutation was not present in several general population frequency databases, such as the 1000 Genomes Project and Genome Aggregation Database (gnomAD) (Supplementary Table S1).
DND1 gene is composed of four coding exons and produces 353 amino acids containing two consecutive RNA-binding domains (RBDs) and one double-strand RBD (dsRBD) with five β-subunits and two α-subunits. The variant p. E71A is located in the α-subunit of RBD1 (Figure 1B). Multiple sequence alignment of the DND1 protein showed the amino acid site of variant (p.E71A) is highly conserved across the eutherian species (Figure 2A). We further performed an in silico analysis to predict the structural changes in the DND1 protein (Figure 2B). As shown in Figure 2B (lower panel), disruption of the α-subunit was found in the DND1 mutated protein.
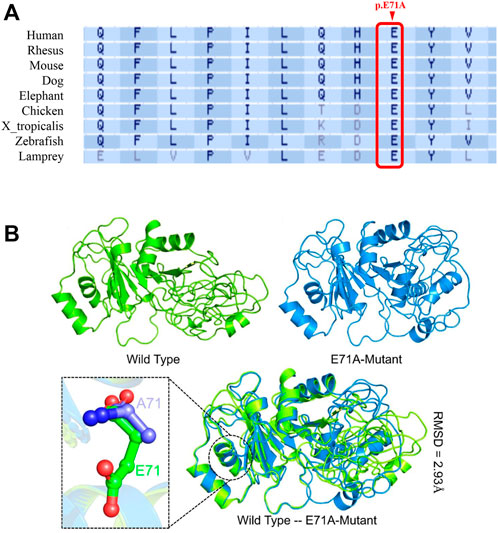
FIGURE 2. DND1 E71 is highly conserved, and the E71A mutation is likely to be pathogenic. (A) Multiple protein sequence alignment shows that the affected amino acid across species was highly conserved. Arrowheads, the mutation site. (B) The predicted structure of the DND1 protein. The wild type structure is shown in green while the E71A mutant is given in marine blue. The mutated residue is also shown as stick in the lower panel. As predicted, the E71 is exposed to the surface of the α-subunit and E71A the mutant may reduce protein stability.
To investigate the functional consequences of the DND1 missense variant, the wildtype (WT) and mutant (MT) DND1 protein with N-terminal EGFP-tags were expressed transiently in HEK293T cells. We found that the E71A mutant exhibited reduced DND1 signal intensity in both the cytoplasm and the nucleus (Figure 3A) and our results are consistent with previous findings, where by using the single guide RNA (sgRNA) and enhanced third generation base editing system (4B2N1) in oocytes and by generating mutant mice targeting Dnd1 gene with base mutation completely depleted PGCs and affected the protein-protein interaction of DND1 (Li et al., 2018). Moreover, the decrease in MT DND1 protein expression was confirmed by immunoblotting (Figure 3B).
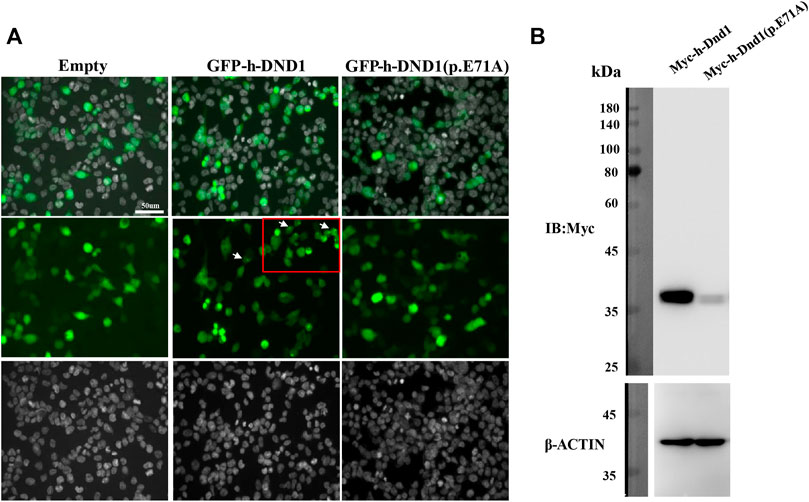
FIGURE 3. E71A mutation affected DND1 subcellular localization and protein expression. (A) Heterologous expression of human DND1 protein. GFP-fused wild-type and mutated DND1 (E71A) plasmids were transfected into in HEK293T cells. Green, GFP fluorescence. Cell nuclei were stained with DAPI (gray). Scale bar: 50 µm. (B) The expression levels of wild-type and mutant DND1 protein were detected by immunoblotting with an anti-GFP antibody. ACTIN served as a loading control.
Previous studies have shown that DND1 cooperatively functions with NANOS2 in male germ cell development. To test the effects of the missense DND1 variant on the interaction between NANOS2 and DND1, we conducted a co-IP assay in HEK293T cells transfected with Myc-tagged WT DND1, MT DND1 and mCherry-tagged NANOS2. In line with previous studies, Myc-tagged WT DND1 co-precipitated with mCherry-tagged NANOS2. However, the mutated DND1 (E71A) drastically reduced the interaction with NANOS2 (Figure 4A). We further examined whether mutation of DND1 interacts with CNOT1(1-551-aa), a component of the CCR4-NOT (CNOT) deadenylase complex that directly interacts with NANOS2. Similar to other mutants (E59K, V60M, P76L, G82R), DND1 with the E71A mutation almost did not disrupt the interaction with CNOT1 (Figure 4B) (Li et al., 2018). These findings indicated that the E71A mutation decreases protein stability and protein-protein interactions.
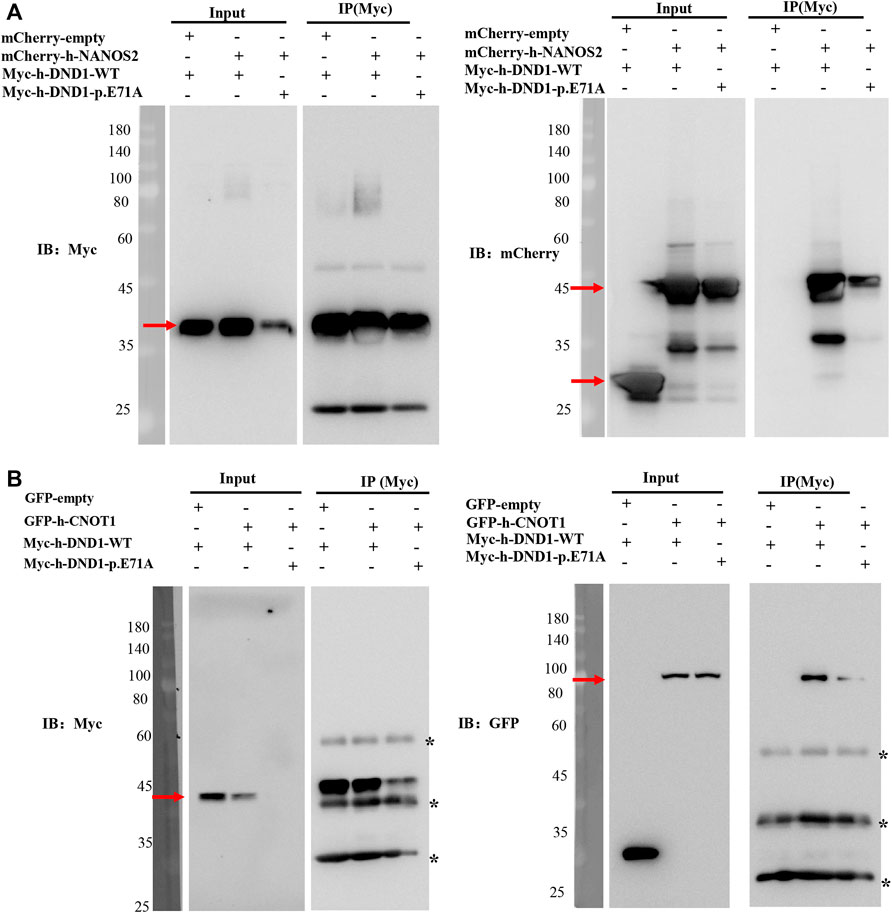
FIGURE 4. DND1 E71A Mutation disrupted its interactions with NANOS2. (A) HEK293T cells expressing the wild-type or mutant Myc-tagged DND1 protein and mCherry-tagged NANOS2 protein were immunoprecipitated with anti-Myc and immunoblotted with Myc and mCherry antibodies. (B) HEK293T cells expressing the wild-type or mutant Myc-tagged DND1 protein and GFP-tagged CNOT1(1-551) protein were immunoprecipitated with anti-Myc and immunoblotted with Myc and GFP antibodies.
The pathology of NOA is characteristic of testicular failure and aberrant spermatogenesis, including Sertoli cell-only syndrome (SCOS) and maturation arrest. A good determination of germ cell fate during the early development of testes is essential to maintain normal spermatogenesis. In the present study, we first reported that missense mutation in DND1, an RNA-binding protein that plays an essential role in primordial germ cells (PGCs) survival, caused NOA in patients with infertility from a Pakistani family. In silico analysis showed that the high conservation of the mutated amino acid caused structural changes in the DND1 protein, resulting in disruption of its interaction with other protein partners. Our study confirmed the indispensable role of DND1 in early germ cell development and male fertility.
DND1 deficiency results in male infertility and testis tumors due to the disruption of normal spermatogenesis. Loss of germ cells was observed in zebrafish, frogs and rodents (Weidinger et al., 2003; Youngren et al., 2005; Horvay et al., 2006). Dnd1 knockout mice displayed early loss of PGCs via the BAX-mediated apoptosis pathway (Cook et al., 2009). Likewise, conditional deletion of Dnd1 causes testis weight decrease and male sterility by loss of both undifferentiated and differentiated spermatogonia (Niimi et al., 2019). A recent study highlighted that PGCs were completely depleted in mouse embryos carrying Dnd1 missense mutations located in the RBD1 domain (E59K, V60M, P76L, and G82R) (Li et al., 2018). In the current study, we identified a novel DND1 variant (E71A) causing azoospermia, similar to previous observations that P76L mutation resulted in the absence of germ cells in the small testes in mice. Based on studies from several vertebrates, we speculate that there is an association between DND1 missense mutation and NOA in humans, although the characteristics of germ cells in seminiferous tubules were not observed due to a lack of testicular tissues.
Dnd1 was initially identified as a germ plasm component that functions in PGC migration and survival in zebrafish (Weidinger et al., 2003). Further study confirmed that DND1 was specifically expressed among PGCs to pre-meiotic germ cells in mice (Yamaji et al., 2017). DND1 transcripts were also enriched in spermatogonia and spermatids in humans (Karlsson et al., 2021), which may suggest a similar role for DND1 in germ cells from humans and mice. More importantly, the localization of DND1 was found in nucleus and germ cell granules or P-bodies in both zebrafish and mice, indicating that DND1 is involved in controlling RNA export and stability. Loss of C-terminal RBD regions alter the subcellular localization of DND1, which suggests that RBDs play an essential role in nucleocytoplasmic shuttling (Kedde et al., 2007). In addition, Li et al. reported that four critical loss-of-function mutations located in RBD1 caused DND1 protein misfolding and instability (Li et al., 2018). Similar to previous studies, our study showed that the new DND1 mutation in E71A disrupted the structure of RBD1 and affected either protein expression or subcellular localization, expanding RBD-dependent subcellular localization of DND1 in germ cells.
DND1 is an RBP protein that regulates gene expression by recruiting CCR4-NOT and NANOS2 proteins to target mRNAs in mice (Suzuki et al., 2016; Yamaji et al., 2017; Raisch et al., 2019). Mutating conserved amino acids in RBDs disrupted DND1 protein-protein interactions in vitro and in vivo (Slanchev et al., 2009; Li et al., 2018; Ruthig et al., 2019). Analyses of the three RBD structures and functions showed that RBD1 acts as the main binding platform required for direct interaction with NANOS2, while RBD2 cooperates with RBD1 (Hirano et al., 2022). In this study, we found that DND1 with the E71A mutation decreased the interaction with the NANOS2 protein, confirming that the RBD1 mutation could affect the interactions.
Overall, this study is the first to identify a missense mutation in DND1 that disrupts protein folding, decreases protein stability and subcellular localization and affects the protein-protein interactions, leading to NOA in humans. Our findings indicate the essential role of DND1 in maintaining germ cell development and the importance of genetic screening for patients with NOA.
The data presented in the study are deposited in the National Genomics Data Center (NGDC) repository, accession number PRJCA011527.
The studies involving human participants were reviewed and approved by the “ethical review board” of the University of Science and Technology of China and the National Bioethics Committee of Pakistan. The patients/participants provided their written informed consent to participate in this study.
XX and MK performed Experimental work. MK and MZ wrote original draft. MS and MK recruited the patients, MS preformed ultrasound and clinical investigation, SK, QZ, GM, RK, and MZ performed semen analysis and collected patient samples, AK performed in silico analysis. ZH, YZ, and JZ, performed WES sequencing, ZH, YZ, JZ, XJ, BX, and XH, performed Sanger sequencing and WES analyses. LW performed the homozygosity mapping. SB and QS reviewed and edited the final manuscript. QS has done the project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the China’s National Foundation for Natural Sciences (81971333, 31871514, 81971446, and 82071709), the China’s National Key Research and Development Program of China (2019YFA0802600 and 2021YFC2700202) and CAS-TWAS President’s PhD Fellowship Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1017302/full#supplementary-material
Boitrelle, F., Shah, R., Saleh, R., Henkel, R., Kandil, H., Chung, E., et al. (2021). The sixth edition of the WHO manual for human semen analysis: A critical review and SWOT analysis. Life 11, 1368. doi:10.3390/life11121368
Cook, M. S., Coveney, D., Batchvarov, I., Nadeau, J. H., and Capel, B. (2009). BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1Ter/Ter mice. Dev. Biol. 328, 377–383. doi:10.1016/j.ydbio.2009.01.041
Das, S., Mohamed, I. N., Teoh, S. L., Thevaraj, T., Ku Ahmad Nasir, K. N., Zawawi, A., et al. (2020). Micro-RNA and the features of metabolic syndrome: A narrative review. Mini Rev. Med. Chem. 20, 626–635. doi:10.2174/1389557520666200122124445
Esteves, S. C. (2015). Clinical management of infertile men with nonobstructive azoospermia. Asian J. Androl. 17, 459–470. doi:10.4103/1008-682X.148719
Fan, S., Jiao, Y., Khan, R., Jiang, X., Javed, A. R., Ali, A., et al. (2021). Homozygous mutations in C14orf39/SIX6OS1 cause non-obstructive azoospermia and premature ovarian insufficiency in humans. Am. J. Hum. Genet. 108, 324–336. doi:10.1016/j.ajhg.2021.01.010
Gershoni, M., Hauser, R., Barda, S., Lehavi, O., Arama, E., Pietrokovski, S., et al. (2019). A new MEIOB mutation is a recurrent cause for azoospermia and testicular meiotic arrest. Hum. Reprod. 34, 666–671. doi:10.1093/humrep/dez016
He, W. B., Tu, C. F., Liu, Q., Meng, L. L., Yuan, S. M., Luo, A. X., et al. (2018). DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J. Med. Genet. 55, 198–204. doi:10.1136/jmedgenet-2017-104992
Hirano, T., Wright, D., Suzuki, A., and Saga, Y. (2022). A cooperative mechanism of target RNA selection via germ-cell-specific RNA-binding proteins NANOS2 and DND1. Cell Rep. 39, 110894. doi:10.1016/j.celrep.2022.110894
Horvay, K., Claußen, M., Katzer, M., Landgrebe, J., and Pieler, T. (2006). Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 291, 1–11. doi:10.1016/j.ydbio.2005.06.013
Inhorn, M. C., and Patrizio, P. (2015). Infertility around the globe: New thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update 21, 411–426. doi:10.1093/humupd/dmv016
Karlsson, M., Zhang, C., Mear, L., Zhong, W., Digre, A., Katona, B., et al. (2021). A single-cell type transcriptomics map of human tissues. Sci. Adv. 7, eabh2169. doi:10.1126/sciadv.abh2169
Kasak, L., and Laan, M. (2021). Monogenic causes of non-obstructive azoospermia: Challenges, established knowledge, limitations and perspectives. Hum. Genet. 140, 135–154. doi:10.1007/s00439-020-02112-y
Kedde, M., Strasser, M. J., Boldajipour, B., Oude Vrielink, J. A., Slanchev, K., Le Sage, C., et al. (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273–1286. doi:10.1016/j.cell.2007.11.034
Krausz, C., and Riera-Escamilla, A. (2018). Genetics of male infertility. Nat. Rev. Urol. 15, 369–384. doi:10.1038/s41585-018-0003-3
Kusz-Zamelczyk, K., Sajek, M., Spik, A., Glazar, R., Jedrzejczak, P., Latos-Bielenska, A., et al. (2013). Mutations of NANOS1, a human homologue of the Drosophila morphogen, are associated with a lack of germ cells in testes or severe oligo-astheno-teratozoospermia. J. Med. Genet. 50, 187–193. doi:10.1136/jmedgenet-2012-101230
Li, Q., Li, Y., Yang, S., Huang, S., Yan, M., Ding, Y., et al. (2018). CRISPR–Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat. Cell Biol. 20, 1315–1325. doi:10.1038/s41556-018-0202-4
Niimi, Y., Imai, A., Nishimura, H., Yui, K., Kikuchi, A., Koike, H., et al. (2019). Essential role of mouse Dead end1 in the maintenance of spermatogonia. Dev. Biol. 445, 103–112. doi:10.1016/j.ydbio.2018.11.003
Noguchi, T., and Noguchi, M. (1985). A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J. Natl. Cancer Inst. 75, 385–392.
Northrup, E., Zschemisch, N.-H., Eisenblätter, R., Glage, S., Wedekind, D., Cuppen, E., et al. (2012). The ter mutation in the rat Dnd1 gene initiates gonadal teratomas and infertility in both genders. PLoS One 7, e38001. doi:10.1371/journal.pone.0038001
Peña, V. N., Kohn, T. P., and Herati, A. S. (2020). Genetic mutations contributing to non-obstructive azoospermia. Best. Pract. Res. Clin. Endocrinol. Metab. 34, 101479. doi:10.1016/j.beem.2020.101479
Raisch, T., Chang, C.-T., Levdansky, Y., Muthukumar, S., Raunser, S., and Valkov, E. (2019). Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nat. Commun. 10, 3173. doi:10.1038/s41467-019-11094-z
Ruthig, V. A., Friedersdorf, M. B., Garness, J. A., Munger, S. C., Bunce, C., Keene, J. D., et al. (2019). The RNA-binding protein DND1 acts sequentially as a negative regulator of pluripotency and a positive regulator of epigenetic modifiers required for germ cell reprogramming. Development 146, dev175950. doi:10.1242/dev.175950
Sakurai, T., Iguchi, T., Moriwaki, K., and Noguchi, M. (1995). The ter mutation first causes primordial germ cell deficiency in ter/ter mouse embryos at 8 days of gestation. Dev. Growth Differ. 37, 293–302. doi:10.1046/j.1440-169x.1995.t01-2-00007.x
Seelow, D., Schuelke, M., Hildebrandt, F., and Nürnberg, P. (2009). HomozygosityMapper—An interactive approach to homozygosity mapping. Nucleic Acids Res. 37, W593–W599. doi:10.1093/nar/gkp369
Slanchev, K., Stebler, J., Goudarzi, M., Cojocaru, V., Weidinger, G., and Raz, E. (2009). Control of Dead end localization and activity--implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 126, 270–277. doi:10.1016/j.mod.2008.10.006
Suzuki, A., Niimi, Y., Shinmyozu, K., Zhou, Z., Kiso, M., and Saga, Y. (2016). Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep. 17, 37–46. doi:10.15252/embr.201540828
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi:10.1093/molbev/mst197
Teng, Y. N., Lin, Y. M., Lin, Y. H., Tsao, S. Y., Hsu, C. C., Lin, S. J., et al. (2002). Association of a single-nucleotide polymorphism of the deleted-in-azoospermia-like gene with susceptibility to spermatogenic failure. J. Clin. Endocrinol. Metab. 87, 5258–5264. doi:10.1210/jc.2002-020016
Van Der Bijl, N., Röpke, A., Biswas, U., Wöste, M., Jessberger, R., Kliesch, S., et al. (2019). Mutations in the stromal antigen 3 (STAG3) gene cause male infertility due to meiotic arrest. Hum. Reprod. 34, 2112–2119. doi:10.1093/humrep/dez204
Weidinger, G., Stebler, J., Slanchev, K., Dumstrei, K., Wise, C., Lovell-Badge, R., et al. (2003). Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 13, 1429–1434. doi:10.1016/s0960-9822(03)00537-2
Yamaji, M., Jishage, M., Meyer, C., Suryawanshi, H., Der, E., Yamaji, M., et al. (2017). DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature 543, 568–572. doi:10.1038/nature21690
Youngren, K. K., Coveney, D., Peng, X., Bhattacharya, C., Schmidt, L. S., Nickerson, M. L., et al. (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435, 360–364. doi:10.1038/nature03595
Zealy, R. W., Wrenn, S. P., Davila, S., Min, K. W., and Yoon, J. H. (2017). microRNA-binding proteins: specificity and function. WIREs. RNA 8, e1414. doi:10.1002/wrna.1414
Zhang, Y., Zhong, L., Xu, B., Yang, Y., Ban, R., Zhu, J., et al. (2013). SpermatogenesisOnline 1.0: A resource for spermatogenesis based on manual literature curation and genome-wide data mining. Nucleic Acids Res. 41, D1055–D1062. doi:10.1093/nar/gks1186
Keywords: DND1, male infertility, NOA, gene mutation, homozygous missense mutation
Citation: Xie X, Khan M, Zubair M, Khan A, Khan R, Zhou J, Zhang Y, Said M, Khan SA, Zaman Q, Murtaza G, Khan MA, Liu W, Hou X, Zhang H, Xu B, Jiang X, Bai S and Shi Q (2022) A homozygous missense variant in DND1 causes non-obstructive azoospermia in humans. Front. Genet. 13:1017302. doi: 10.3389/fgene.2022.1017302
Received: 11 August 2022; Accepted: 13 September 2022;
Published: 30 September 2022.
Edited by:
Shenmin Yang, Suzhou Municipal Hospital, ChinaReviewed by:
Aline Fernanda De Souza, University of Guelph, CanadaCopyright © 2022 Xie, Khan, Zubair, Khan, Khan, Zhou, Zhang, Said, Khan, Zaman, Murtaza, Khan, Liu, Hou, Zhang, Xu, Jiang, Bai and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xei Xuefeng, eGlleHVlZmVuZ2xhQGZveG1haWwuY29t; Mazhar Khan, bWF6aGFyc3dAbWFpbC51c3RjLmVkdS5jbg==; Shun Bai, c2h1bmJhaUB1c3RjLmVkdS5jbg==; Qinghua Shi, cXNoaUB1c3RjLmVkdS5jbg==
†These authors have contributed equally to this work and shared first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.