- 1Fetal Care Center, Department of Obstetrics and Gynecology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
- 2National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, China
- 3Department of Obstetrics and Gynecology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
- 4Fetal Medicine Center, Department of Obstetrics and Gynecology, First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
Background: The etiology of preeclampsia (PE) remains unclear. With the utilization of metabolomics, dysregulated production of several metabolic components in human plasma, such as lipids, amino acids, androgens and estrogens, was found to be important in the pathogenesis of PE. Transcriptomics adds more in-depth information, and the integration of transcriptomics and metabolomics may yield further insight into PE pathogenesis than either one alone.
Objectives: We investigated the placental metabolomics and transcriptomics of PE patients to identify affected metabolic pathways and potential biological targets for exploring the disease pathogenesis.
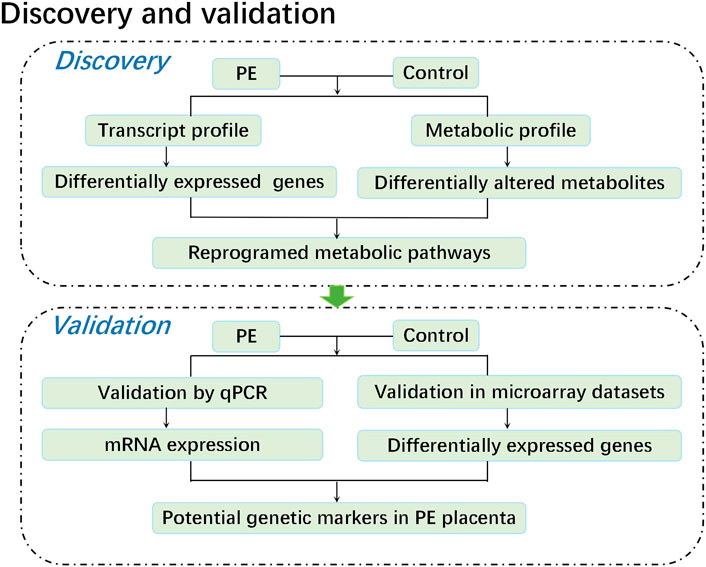
Methods: Integrated transcriptomics and metabolomics were used to analyze five paired human placentas from patients with severe PE and normal pregnancies. This was followed by further validation of our findings in a publicly available dataset of 173 PE vs. 157 control placentas. In addition, weighted gene coexpression network construction was performed to assess the correlation between genetic alterations and diseases.
Results: We identified 66 and 41 differentially altered metabolites in negative and positive ion modes, respectively, in the PE group compared to the control group, and found 2,560 differentially expressed genes. Several pathways were aberrantly altered in the PE placenta at both the metabolic and transcriptional levels, including steroid hormone biosynthesis, the cAMP signaling pathway, neuroactive ligand–receptor interactions, taste transduction and prion diseases. Additionally, we found 11 differential metabolites and 11 differentially expressed genes involved in the steroid hormone biosynthesis pathway, indicating impaired metabolism of steroid hormones in the PE placenta. Furthermore, we found that CYP11A1, HSD3B2, and HSD17B6 are highly correlated with diseases.
Conclusion: Our findings provide a profile of the dysregulated steroid hormone biosynthesis in PE placenta, we observed a dysregulated cortisol-to-cortisone ratio, testosterone accumulation, decreased testosterone downstream metabolites, impaired production of estrone and estriol, and aberrant hydroxylation and methylation of estradiol. Disorders of placental steroid hormone metabolism might be a consequence or a compensatory change in pathological placentation in PE, which underscores the need to investigate the physiology of steroid hormone metabolites in the etiology of PE.
1 Background
Preeclampsia (PE) is a heterogeneous gestational disorder characterized by new-onset maternal hypertension and proteinuria after 20 weeks of gestation, and PE affects approximately 3%–7% of all pregnancies worldwide (Kelly et al., 2017). It can affect virtually every organ system, leading to maternal and fetal morbidity and mortality (Berg et al., 2009; Duley, 2009; Kuklina et al., 2009; Bansil et al., 2010). Abnormal trophoblastic invasion and insufficient remodeling of spiral arteries impair placental perfusion, leading to hypoxia and necrosis, which provoke a cascade of events in the systematic inflammatory response and oxidative stress, eventually resulting in systematic endothelial activation and vasospasm (Rana et al., 2019). Although several theories have been proposed regarding the pathogenesis of preeclampsia, there is no consensus on its etiology or on its early identification and prophylaxis. In recent years, omics approaches have been utilized to understand how it develops and whether we can predict it or treat it, and this has become the focus of numerous studies (Nobakht, 2018; Benny et al., 2020).
Transcriptional profiling or transcriptomics is being increasingly utilized to discover key molecular changes underlying the etiology and early identification of PE (Liu et al., 2018; Tong et al., 2018; Maeda et al., 2019). Metabolic profiling or metabolomics provides data-rich information on metabolic alterations that reflect genetic and epigenetic factors influencing cellular physiology (Nobakht, 2018; Benny et al., 2020). The integration of “-omic” studies may provide a more in-depth understanding of PE pathogenesis (Liu et al., 2013; Leavey et al., 2018; Benny et al., 2020). A metabolomic study showed that several metabolites, such as lipids, amino acids and their derivatives, are promising in the early identification of preeclampsia patients (Nobakht, 2018); moreover, combined approaches of transcriptomics and metabolomics have elucidated altered lipid and amino acid metabolism and dysregulation of the immune response in preeclampsia (Kelly et al., 2017; Shin et al., 2018). In addition, metabolic studies have demonstrated that the mechanism of imbalanced androgen and estrogen production is of great value in the pathogenesis of the disease (Jobe et al., 2013; Shao et al., 2017; Shin et al., 2018; Cantonwine et al., 2019; Shao et al., 2019) and is likely to be associated with alterations in angiogenesis and vascular function (Jobe et al., 2013). Integration of transcriptomics and metabolomics may yield further insight into PE pathogenesis than either approach alone.
In this study, we performed a metabolomic study of five paired human placental samples comprising PE placentas and normal pregnancy placentas by liquid chromatography–mass spectrometry (LC–MS), aiming to identify key metabolite alterations unique to human placentas from PE patients. We then performed transcriptome sequencing on these samples to identify genes with significant changes in expression. Through KEGG enrichment analysis, we integrated metabolome and transcriptome data to identify significantly perturbed pathways at both the metabolic and transcriptional levels, revealing potential causes that may aid in understanding the pathogenesis of PE. Finally, these corresponding genetic alterations were further validated by both qPCR data from the same specimens and mRNA expression profile microarray datasets from published studies. Weighted gene coexpression network analysis (WGCNA) was conducted on public mRNA expression profile microarray datasets to assess the correlation between genetic alterations and diseases, providing an effective biological target for exploring the pathogenesis of PE (Figure 1).
2 Methods
2.1 Study participants
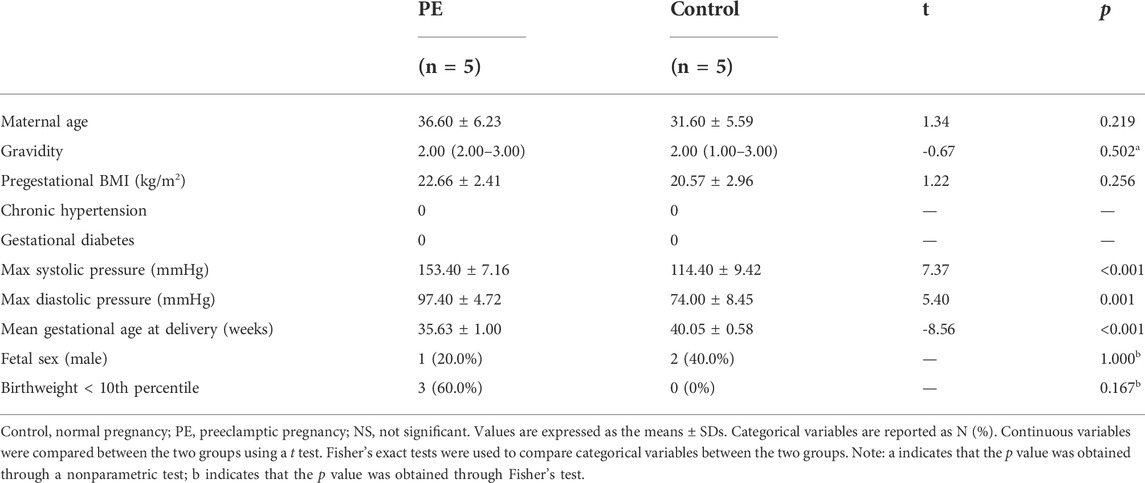
Women aged 20–40 years with singleton pregnancies undergoing selective cesarean section at Guangzhou Women and Children’s Medical Center from January 1 to 31 March 2019, were recruited. The inclusion criteria were as follows: The PE group was included based on the diagnostic criteria described in Gestational Hypertension and Preeclampsia: (ACOG Practice Bulletin 2019). The control group included pregnant women with no preexisting medical diseases or antenatal complications. The exclusion criteria were as follows: pregnancy conceived by assisted reproductive technology; lupus or antiphospholipid antibody syndrome; or placental abruption, chorioamnionitis or meconium-stained fluids. Obtaining samples from qualified controls for early-onset severe PE cases is particularly difficult, since spontaneous late abortion or preterm birth usually result from various pregnancy complications, which are very likely to affect the metabolic state of the placenta. To match the gestational age as closely as possible, we finally included only five severe PE cases (near-term deliveries) and five qualified controls (term deliveries). The included PE cases presented with severe features were given intravenous magnesium sulfate, oral labetalol, and delivered without delay. Preeclampsia was defined as new-onset BP > 140/90 mmHg plus proteinuria (0.3 g/24 h) after 20 weeks in previously normotensive women. Those who met one or more of the following indicators were considered severe cases: 1) systolic blood pressure of 160 mm Hg or more or diastolic blood pressure of 110 mm Hg or more on two occasions at least 4 h apart (unless antihypertensive therapy was initiated before this time), 2) thrombocytopenia (platelet count less than 100 × 109/L), 3) impaired liver function that was not accounted for by alternative diagnoses and as indicated by abnormally elevated blood concentrations of liver enzymes, 4) renal insufficiency (serum creatinine concentration more than 1.1 mg/dl or a doubling of the serum creatinine concentration in the absence of other renal disease), 5) pulmonary edema, 6) new-onset headache unresponsive to medication and not accounted for by alternative diagnoses, and 7) visual disturbances (scotomata, amaurosis fugax, blurred vision, diplopia, homonymous hemianopsia) (Wagner et al., 2011). The institutional review board at the Guangzhou Women and Children’s Medical Center reviewed and approved the study protocol. Written informed consent were obtained from all of the study participants. The clinical and demographic characteristics of the participants included in the study are given in Table 1.
2.2 Placental tissue biopsies
After the delivery of the infant, placental tissues were obtained from the maternal central side of the placenta by biopsy forceps at a maximum depth of 0.5 cm. The tissues did not contain the decidual or chorionic plate but had the basal plate and intervillous space and/or placental parenchyma. The specimens were carefully cleaned of visible blood clots before being placed in a cryopreservation tube, quickly frozen in liquid nitrogen, and then transferred to −80°C within 20 min.
2.3 Untargeted metabolomic profiling
2.3.1 Metabolite extraction from tissues
One hundred milligrams of tissue was ground with liquid nitrogen, and the homogenate was resuspended in prechilled 80% methanol and 0.1% formic acid by vortexing. The samples were incubated on ice for 5 min and then centrifuged at 15,000 rpm and 4°C for 5 min. A portion of the supernatant was diluted to a final concentration containing 60% methanol by LC–MS grade water. The samples were subsequently transferred to a fresh Eppendorf tube with a 0.22-μm filter and then centrifuged at 15,000 g and 4°C for 10 min. Finally, the filtrate was injected into the LC–MS/MS system for analysis.
2.3.2 UHPLC–MS/MS analysis
LC–MS/MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Samples were injected onto a Hyperil Gold column (100 × 2.1 mm, 1.9 μm) using a 16-min linear gradient at a flow rate of 0.2 ml/min. The eluents for the positive polarity mode were eluent A (0.1% FA in water) and eluent B (methanol). The eluents for the negative polarity mode were eluent A (5 mM ammonium acetate, pH 9.0) and eluent B (methanol). The solvent gradient was set as follows: 2% B, 1.5 min; 2%–100% B, 12.0 min; 100% B, 14.0 min; 100%–2% B, 14.1 min; and 2% B, 16 min. A Q Exactive HF-X mass spectrometer was operated in positive/negative polarity mode with a spray voltage of 3.2 kV, capillary temperature of 320°C, sheath gas flow rate of 35 arb and aux gas flow rate of 10 arb.
2.3.3 Data analysis
The raw data files generated by UHPLC–MS/MS were processed using Compound Discoverer 3.0 (CD 3.0, Thermo Fisher) to perform peak alignment, peak picking, and quantitation for each metabolite. The main parameters were set as follows: retention time tolerance, 0.1 min; actual mass tolerance, 5 ppm; signal intensity tolerance, 30%; signal/noise ratio, three; and minimum intensity, 100,000. Thereafter, peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks and fragment ions. Then, peaks were matched with the mzCloud (https://www.mzcloud.org/) and ChemSpider (http://www.chemspider.com/) databases to obtain accurate qualitative and relative quantitative results. PCA and PLS-DA analysis use R language ropls package and ggbiplot package. The value of Variable Importance in the Projection (VIP) from the first principal component of the PLS-DA model was used, combined with the p value of the t-test to find differentially expressed metabolites, the threshold was set to VIP > 1.0, and the fold difference FC > 1.5 or FC < 0.667 and p value < 0.05. KEGG Pathway was used as the unit and applied the hypergeometric test to find out the Pathways that are significantly enriched in the differential metabolites compared with all the identified metabolite backgrounds.
2.4 Transcriptomic profiling
2.4.1 Sample RNA extraction
Five paired placental samples of pregnant women were treated by the following steps. First, total RNA was extracted as previously described (Lian et al., 2016); second, agarose gel electrophoresis was performed to analyze the extent of RNA degradation and contamination; third, Nanodrop was used to assess RNA purity (OD260/280 ratio); fourth, we used Qubit to accurately quantify the RNA concentration; and finally, the Agilent 2,100 Bioanalyzer was used to accurately detect RNA integrity.
2.4.2 Library construction and sequencing
A Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) (Epicenter, Madison, WI, United States) was used to remove rRNA from total RNA. The library was constructed using an Illumina TruSeq RNA Sample Prep Kit v2 (Illumina Inc., San Diego, CA). Briefly, RNA was sheared into 200–300-bp fragments. In addition, first- and second-strand cDNA was synthesized. Then, short cDNA fragments were purified, end repaired and tailed with a single A (adenine) addition. Adapters were ligated to the A-tailed cDNA fragments. These cDNA fragments were enriched by 12 PCR cycles. Purified libraries were quantified using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, United States) and validated using an Agilent 2,100 Bioanalyzer (Agilent, Beijing, China). The reads that passed the Illumina quality filter were kept for sequence analysis.
2.4.3 Data analysis.
High-quality reads were mapped to the human genome by using SOAP aligner/SOAP2 (Li et al., 2009) with five maximum alignment errors. The mRNA abundance was normalized using TPM (Transcripts per million). Differential gene expression analysis was performed by edgeR (edgeR: a Bioconductor package for differential expression analysis of digital gene expression data). Genes with p values less than 0.05 and fold changes > 1.5 or < 0.667 were considered differentially expressed genes (DEGs). Gene list enrichment analysis used Enrichr online tools (https://maayanlab.cloud/Enrichr/).
2.4.4 Public dataset acquisition
The mRNA expression profile microarray dataset was from Gene Expression Omnibus (GEO), NCBI’s publicly available genomics database, and the data number is GSE75010. Next, we performed differential analysis (adjusted p value < 0.05, and fold changes > 1.5 or < 0.667) by comparing PE placentas to normal pregnancy placentas in the R computing environment using the limma package. Weighted gene coexpression network construction was performed by the WGCNApackage in R as previously described (Zhang and Horvath, 2005; Langfelder and Horvath, 2008). The expression of the genes for all samples were inputted to construct the weighted gene coexpression network.
2.5 RT-qPCR
The qRT-PCR was performed with SYBR Premix Ex Taq (Takara, Dalian, China) in an iQ5 thermal cycler (Biorad, Hercules, CA) according to the manufacturer’s instructions. The cycling conditions were as follows: 94°C for 5 min, followed by 35 cycles at 94°C for 1 min, at 55°C for 1 min and 72°C for 1 min with a final step of 72°C for 5 min. Melting curves were read from 60 to 95°C in steps of 1°C. Normalized expression levels of the target gene transcripts were calculated relative to GAPDH. Then t-test was used to RT-qPCR statistical analysis. The primer information is listed in Supplementary Table S9.
3 Results
3.1 Demographic characteristics and infant outcomes
Five PE cases and five maternal age-matched controls were included in the study. The maternal demographic characteristics and infant outcomes of the PE cases and controls are presented in Table 1. Fetal birth weight and gestational age at delivery were lower in preeclamptic pregnancies than in normotensive pregnancies.
3.2 Metabolomic and transcriptional analysis revealed the major pathways that are altered in PE
Ten placental specimens from PE and normotensive controls were analyzed by liquid chromatography–mass spectrometry (LC–MS/MS) using a Vanquish UHPLC system coupled with an Orbitrap Q Exactive HF-X mass spectrometer. Principal component analysis (PCA) and partial least squares discrimination analysis (PLS-DA) were carried out, as shown in Supplementary Figure S2. A total of 980 metabolites were identified, and the relative changes in metabolites were compared between the two groups. Fold changes and p values are presented. This approach yielded 66 differentially altered metabolites in negative ion mode (variable importance in projection, VIP > 1, p < 0.05, fold change, FC > 1.5 or FC < 0.667), among which 49 were downregulated and the remaining metabolites were upregulated. In positive ion mode, there were 41 differentially altered metabolites, of which 15 were downregulated and the rest were upregulated (Supplementary Table S1).
To reveal the enzymes involved, transcriptional profiling of the same specimens was conducted to discover DEGs. Of the 2,560 significantly DEGs (p < 0.05, FC ≥ 1.5) (Supplementary Table S2), 627 were upregulated, and the remaining genes were downregulated. Using KEGG pathway significant enrichment analysis, five pathways were identified to be significantly altered at both the metabolomic and mRNA expression levels in the PE group (Figure 2A, Supplementary Tables S3,S4), including the metabolism of steroid hormone biosynthesis, cAMP signaling pathway, neuroactive ligand–receptor interaction, taste transduction and prion diseases. Figure 2B demonstrates the altered pathways based on metabolic profiling, among which we identified steroid hormone biosynthesis as the metabolic pathway of greatest interest, as it exhibited lower p values and the most enriched differential metabolites. There were 11 differential metabolites and 11 DEGs involved in the steroid hormone biosynthesis pathway (Figures 2C,D).
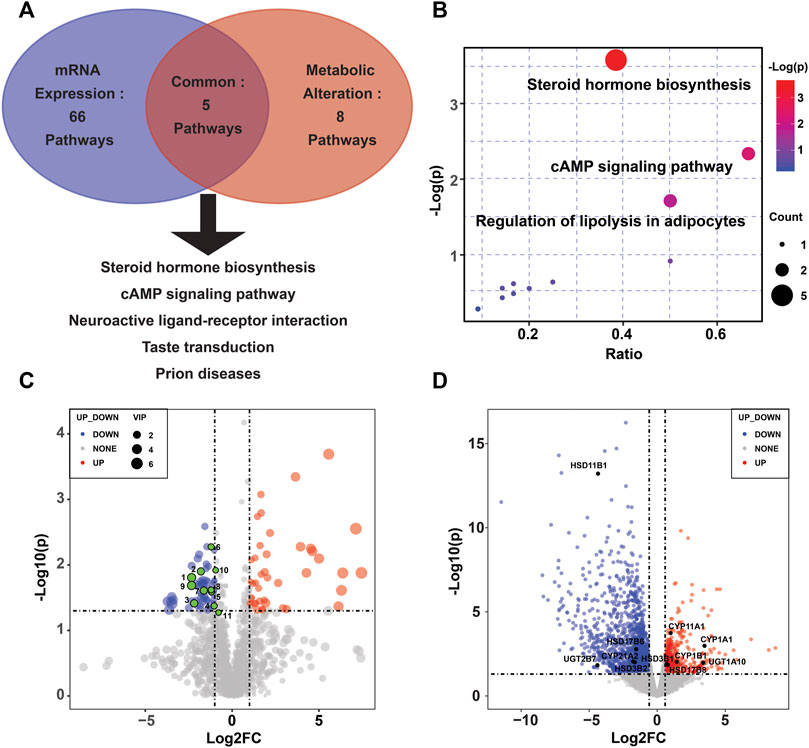
FIGURE 2. Altered pathways in the PE placenta. (A) Metabolic and transcriptional analysis identification of five pathways that were significantly altered in PE placentas. (B) Overview of the pathway analysis based on metabolite alterations. (C) Volcano plots of the differential metabolites in the steroid hormone biosynthesis pathway. (D) Volcano plots of the differentially expressed genes in the steroid hormone biosynthesis pathway.
3.3 Alterations in metabolite and mRNA expression in the steroid hormone biosynthesis pathway in the PE placenta
Steroid hormones are all derived from cholesterol and are metabolized into glucocorticoids, mineralocorticoids, and sex hormones mainly through the cytochrome P450 (CYP) family and the hydroxysteroid dehydrogenase (HSD) family (Beaudoin et al., 1997). We found alterations in glucocorticoids and sex hormones at the metabolic and transcriptomic levels in the present study, and the relative levels of metabolites of interest and gene mRNAs are highlighted in Supplementary Tables S2,S3.
Regarding glucocorticoid metabolism, PE placentas exhibited a significant reduction in 17α-hydroxypregnenolone, corticosterone, tetrahydrocortisol, and tetrahydrocortisone compared to control placentas. A lower placental cortisol/cortisone ratio was observed in the PE group, but the difference was not significant (Table 2). Furthermore, we examined the corresponding genes involved in glucocorticoid biosynthesis. Compared with the controls, upregulated CYP11A1 and HSD3B1 and decreased HSD3B2, HSD11B1, and CYP21A2 were observed in the PE group. CYP11A1, a key enzyme initiating the catalysis of cholesterol, was significantly upregulated in PE. HSD3B, which has two isoforms (HSD3B1 and HSD3B2), is a rate-limiting enzyme involved in the conversion of 17α-hydroxypregnenolone to 17α-hydroxyprogesterone. HSD3B1 was significantly upregulated, while HSD3B2 was downregulated. CYP21A2 plays a key role in the transformation of progesterone to corticosterone and exhibited a significant reduction. HSD11B1 and HSD11B2 are vital to proper cortisol metabolism. HSD11B2 catalyzes the metabolization of cortisol to cortisone and protects the fetus from exposure to excessive maternal cortisol during pregnancy, whereas HSD11B1 performs the reverse function of HSD11B2 and reactivates cortisol by converting cortisone to cortisol (Berkane et al., 2018). We observed significantly decreased HSD11B1, nonsignificantly elevated HSD11B2, and a lower cortisol/cortisone ratio in PE cases. These findings are summarized in Figure 3A.
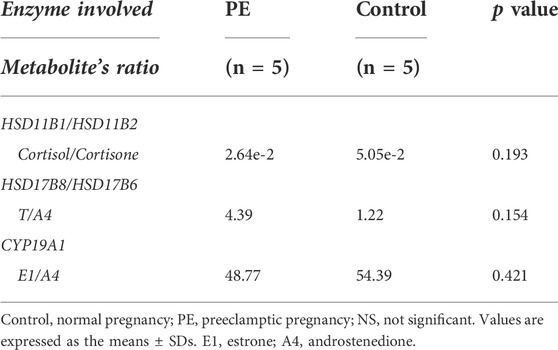
TABLE 2. Activity of the main placental enzymes involved in steroidogenesis in the PE and control groups.
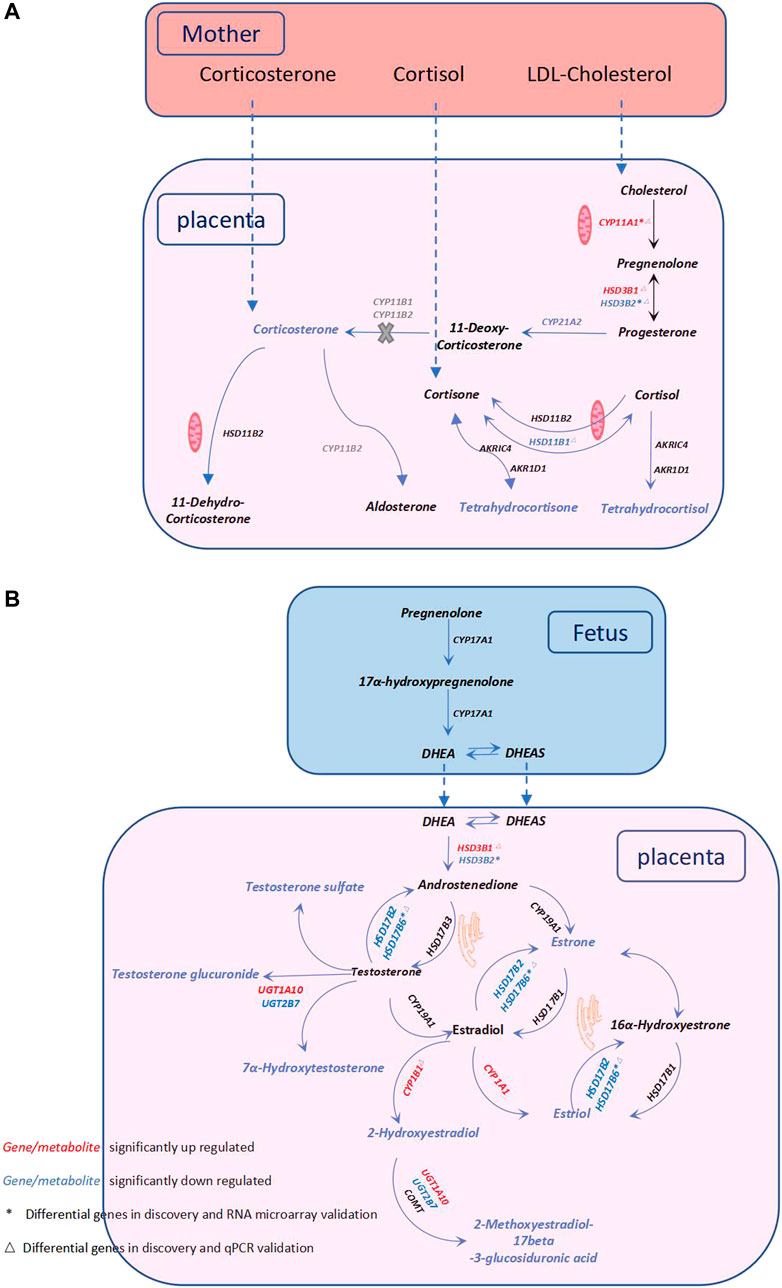
FIGURE 3. Metabolic pathway alterations in PE—Steroid hormone biosynthesis. (A) Description of glucocorticoid metabolism. (B) Description of sex hormone metabolism. Steroid hormones are synthesized from cholesterol mainly through the catalysis of the cytochrome P450 (CYP) family and the hydroxysteroid dehydrogenase (HSD) family. The catalysis of cholesterol is initiated by CYP11A1, and cholesterol is subsequently metabolized into glucocorticoids, mineralocorticoids, and sex hormones. The dysregulation of glucocorticoid metabolism and sex hormones in the PE placenta is illustrated in (A) and (B), respectively. Red and blue fonts indicate significant increases and decreases, respectively. * Differential genes in the discovery and RNA microarray validation. △ Differential genes in the discovery and qPCR validation. The genes in grey represent they are lack in the placenta, and the X in grey indicates that this reaction is blocked in the placenta.
With regard to sex hormone biosynthesis, PE exhibited no significant increase in testosterone in the PE placenta and a significant decrease in testosterone metabolites such as testosterone sulfate, testosterone glucuronide, and 7α-hydroxytestosterone. In addition, we found significantly decreased estradiol metabolites, including estrone (E1) (VIP = 0.919), estriol (E3) (VIP = 0.986), 2-hydroxyestradiol and 2-methoxyestradiol-17beta-3-glucosiduronic acid. Genes of this pathway were enriched in the PE placenta. HSD17B6 (an isoform of HSD17B) was significantly downregulated, and the corresponding testosterone (T)/androstenedione (A4) ratio was found to be increased (Table 2). CYP1A1 and CYP1B1 were significantly increased in PE; however, the corresponding downstream metabolites E3 and 2-hydroxyestradiol were significantly decreased. CYP19A1 in the PE group was not significantly altered, nor was the corresponding E1/A4 ratio (Table 2). UGT1A10 was found to be increased; conversely, UGT2B7 was found to be decreased, while the glucuronide forms of testosterone and 2-hydroxyestradiol were significantly decreased in PE. All alterations at both the metabolic and transcriptional levels in this pathway are illustrated in Figure 3B.
3.4 Validation of the metabolomic and transcriptomic alterations in PE
To validate our findings, qPCR was conducted on the same specimens, and we revealed consistent alterations between the transcriptional analysis and qPCR-quantified mRNA expression levels (Supplementary Figure S1). Furthermore, we validated the transcriptional findings based on the mRNA expression profile microarray datasets of 330 human placenta samples from eight previously published studies (Leavey et al., 2016), including 173 preeclamptic and 157 control placentas. A total of 2,650 DEGs were found in our RNA-seq dataset (p < 0.05, FC ≥ 1.5) (Supplementary Table S2), while 4,665 were found in the mRNA expression profile microarray dataset (FDR<0.05) (Supplementary Table S5), among which 767 were identified to be altered in both datasets (Figure 4A). The comparisons of the enriched genes in the steroid hormone biosynthesis pathway between these two datasets are shown in Supplementary Table S8. Furthermore, three genes (CYP11A1, HSD3B2, and HSD17B6) involved in the steroid hormone biosynthesis pathway were consistently altered in the RNA-seq data, publicly available mRNA expression profile microarray datasets, and qPCR validation (Figure 4B). In addition, we conducted WGCNA. Five modules were constructed. The red and blue colors showed strong positive and negative correlations, respectively. HSD17B6 and HSD3B2 were strongly negatively correlated with diseases, and CYP11A1 presented a strong positive correlation. These genes were located in three modules, namely, turquoise, yellow, and blue. These three modules were highly correlated with diseases (Figures 4C,D, Supplementary Table S6).
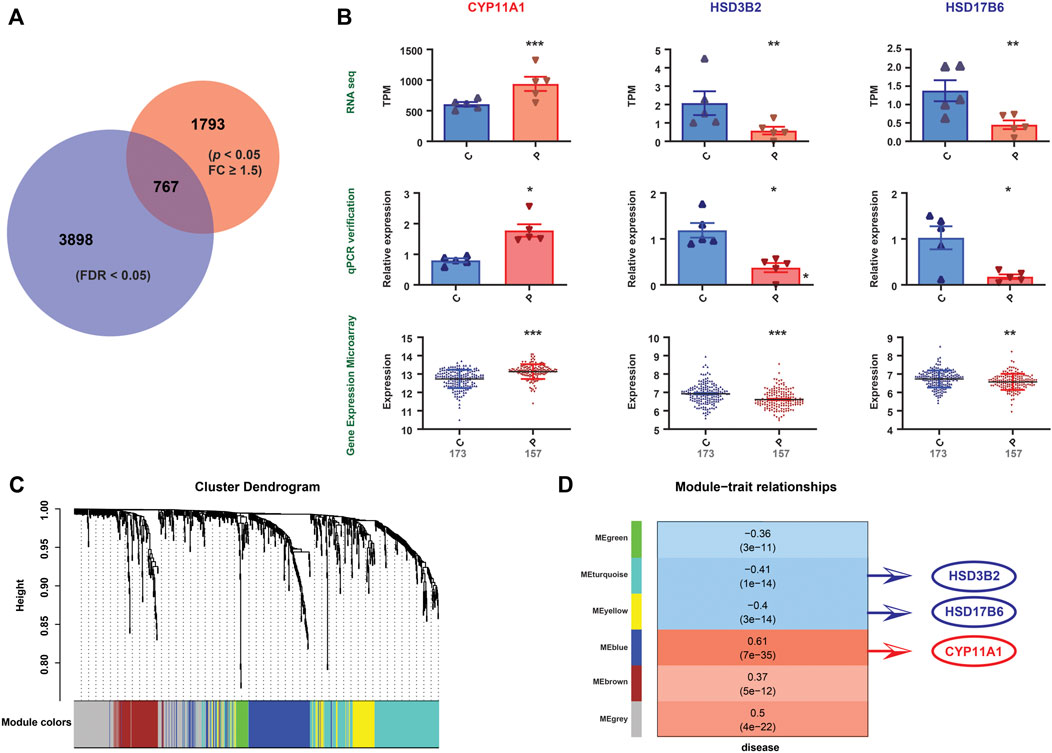
FIGURE 4. Validation of the gene expression patterns. (A) Differential gene expression between the PE and control groups from RNA-seq data (indicated by the red circle). Differential gene expression between the PE and control groups from mRNA expression profile microarray datasets (indicated by the blue circle). The Venn diagram represents the intersection between the two sets (767 genes). (B) Among the 767 genes, three differentially expressed genes (CYP11A1, HSD3B2, and HSD17B6), which correlate with steroid hormone biosynthesis, were altered uniformly in the RNA-seq data, qPCR validation and mRNA expression profile microarray datasets. Significant differences, *: p <0.05, **: p <0.01, ***: p <0.001. (C) The figure demonstrates the differentially expressed gene cluster dendrogram analysis in the preeclampsia vs. control groups. The five colors, namely, green, turquoise, yellow, blue, and brown, represent the modules. The dynamic tree cut method was implemented for analysis. The degree of coexpression between the genes assigned by the same module was relatively high. (D) The figure represents the correlation between mRNA module eigengenes and phenotypic traits. Each row represents the module eigengene or ME (ME is the correlation matrix of the module and sample, labeled by color), and each column represents a trait. Each square contains Pearson’s correlation coefficient between the MEs and traits and their associated p values. The red and blue colors show a strong positive and negative correlation, respectively. The HSD17B6, HSD3B2 and CYP11A1 genes are located in three modules, that is, turquoise, yellow, and blue, respectively. These three modules are highly correlated with diseases.
4 Discussion
In the present study, we performed a comprehensive analysis based on metabolic and transcriptomic data and found a metabolic reprogramming in the PE placenta. The multi-omics analysis in this study found five pathways that were significantly altered in the PE placenta compared to the control, and steroid hormone biosynthesis was identified as the most significantly enriched metabolic pathway and was further investigated. CYP11A1 encodes the P450scc enzyme that catalyzes the first and rate-limiting step of steroid biosynthesis (Hu et al., 2004), converting cholesterol to pregnenolone, which is further metabolized into three major steroid hormone classes: mineralocorticoids, glucocorticoids, and sex steroids. The main corresponding enzymes involved in this metabolic process include HSD3B, CYP17, CYP21, CYP11B, HSD11B, HSD17B, and CYP19 (Shih et al., 2011). To date, the pathogenesis of PE is poorly understood, and defective placentation is thought to be causal. Dysregulation of sex hormone metabolism plays a critical role in PE placental dysplasia by affecting trophoblast differentiation and uterine-placenta angiogenesis (Morales et al., 1995; Maliqueo et al., 2016; Berkane et al., 2017; Berkane et al., 2018). Our findings provide a profile of the metabolic and genetic changes in the PE placenta, as well as the alterations in steroid hormone biosynthesis at both the metabolic and transcriptional levels in the PE placenta.
4.1 Initiation of steroid hormone biosynthesis—conversion from cholesterol to progesterone
Steroid hormones are initially synthesized from cholesterol, which is essentially taken up from the maternal circulation. In the first step, the conversion of cholesterol to pregnenolone was catalyzed by cytochrome P450scc (CYP11A1) in mitochondria. Subsequently, pregnenolone is converted to progesterone by HSD3B (HSD3B1 and HSD3B2) (Hu et al., 2004; Shih et al., 2011).
In the present study, we found that CYP11A1 expression is upregulated in the PE placenta compared to that of the control group. We validated this finding and demonstrated that CYP11A1 presents a strong positive correlation with PE (Figure 4B). One study reported overexpression of CYP11A1 in preeclamptic maternal serum with cotemporally increased serum pregnenolone and progesterone concentrations (Moon et al., 2014). In the placenta, CYP11A1 is located in syncytial trophoblasts (Hill et al., 2014). Upregulated CYP11A1 enhances trophoblast autophagy and inhibits trophoblastic invasion, which is associated with the etiology of PE (Pan et al., 2017). Yu Chien et al. established a CYP11A1 transgenic mouse model and found that overexpression of CYP11A1 induces the loss of decidual cell polarity and results in anomalous placentation (Chien et al., 2013). Placentation defects are leading causes of PE, and CYP11A1 might play a critical role in placental dysplasia via oxidative stress and hypoxia.
HSD3B is a key rate-limiting enzyme in steroid biosynthesis pathways. It is expressed as two tissue-specific isoforms (HSD3B1 and HSD3B2) (Thomas et al., 2005). HSD3B1 is almost exclusively expressed in the placenta and peripheral tissues, while HSD3B2 is predominantly expressed in the adrenal gland, ovary, and testis (Simard et al., 1996). HSD3B2 is a bifunctional enzyme that plays a key role in the conversion of pregnenolone to progesterone and DHEA to A4. In our study, we found significantly upregulated HSD3B1 and downregulated HSD3B2 in PE but confirmed only the alteration in HSD3B2 in the validation course, no difference in HSD3B1 expression between PE and the controls was found in the validation sets (Leavey et al., 2016), finding that HSD3B2 was highly negatively correlated with disease in WGCNA (Figure 4D). The reduction in HSD3B2 might play a role in the dysregulated synthesis of progesterone in the PE placenta. However, the low placental expression of HSD3B2 and the expression difference of HSD3B1 between RNA-seq and validation microarray data sets bring concerns, thus confirming HSD3B1, HSD3B2 and their downstream proteins at protein level may provide more valuable information.
4.2 Metabolic profile of glucocorticoid biosynthesis in the preeclamptic placenta
Transplacental transfer of biologically active glucocorticoids is unidirectionally catalyzed by HSD11B2, which converts corticosterone to 11-dehydrocorticosterone and cortisol to cortisone. HSD11B1 catalyzes the reverse reaction to HSD11B2, and it reactivates cortisol by converting cortisone to cortisol (Causevic and Mohaupt, 2007; Jayasuriya et al., 2019). These conversions inactivate cortisol and corticosterone, thereby protecting the fetus from exposure to high levels of active glucocorticoids. This exposure may lead to gestational hypertension (Siiteri and MacDonald, 1966; Staud et al., 2006; Abbassi-Ghanavati et al., 2009; Aufdenblatten et al., 2009; Jayasuriya et al., 2019). In PE, enhanced systematic activity of maternal HSD11B2 or reduced HSD11B1 activity were demonstrated, which is expected to result in a decreased maternal serum cortisol-to-cortisone ratio (Jayasuriya et al., 2019). However, other studies came to the opposite conclusion, showing reduced placental HSD11B2 expression in PE and paradoxically indicated that reduced HSD11B2 activity would be expected to result in increased maternal cortisol-to-cortisone ratios (McCalla et al., 1998; Schoof et al., 2001; Hu et al., 2015). In the present study, we demonstrated significantly reduced HSD11B1 and nonsignificantly increased HSD11B2 at the transcriptional level and a decreased cortisol-to-cortisone ratio in the PE placenta. This might be a placenta-specific compensatory response to increased fetal exposure to systematic cortisol, resulting from the increased transplacental transfer of cortisol in preeclampsia patients, but this hypothesis requires further confirming.
4.3 Metabolic profile of androgen and estrogen biosynthesis in the preeclamptic placenta
The conversion from pregnenolone to dehydroepiandrosterone (DHEA), dehydroepiandrosterone sulfate (DHEAS), and 16OH-DHEAS was accomplished in the fetal or maternal adrenal gland and liver. The human placenta lacks CYP17A1 (Christensen, 1974), but it does express HSD17B, CYP19A1, and HSD3B (Mason et al., 1993; Takeyama et al., 1998). Therefore, the placenta can synthesize A4 from fetal DHEAS and 16OH-DHEAS and undertake the subsequent synthesis of both testosterone and estradiol (Morel et al., 2016).
High circulating testosterone and aberrant estrogen synthesis in PE were observed in previous studies (Zhorzholadze et al., 2006; Shao et al., 2017; Kumar et al., 2018; Keya et al., 2019; Shao et al., 2019; Lan et al., 2020). Augmented placental androgen synthesis and decreased placental aromatase expression in PE were observed in recent studies (Hertig et al., 2010); however, the role of placenta-derived androgen and estrogen in preeclampsia remains elusive. We observed the T to A4 ratios in the PE and control groups were 4.39 and 1.22, respectively (p = 0.154), whereas the production of testosterone derivatives, such as testosterone sulfate, testosterone glucuronide, and 7α-hydroxytestosterone, were significantly decreased. Moreover, the production of metabolites derived from estradiol (E2), including E1, E3, 16α-hydroxyestrone, 2-hydroxyestradiol and 2-methoxyestradiol-17beta-3-glucosiduronic acid were likely to be impaired. These findings may partially explain the accumulation of testosterone and the impaired production of estrogens in PE placenta.
HSD3B were involved in the conversion of DHEA to A4 (Berkane et al., 2017). In the present study, increased HSD3B1 and decreased HSD3B2 in PE were demonstrated. The effect of HSD3B on androgen metabolism in the PE placenta is controversial. Hogg, K demonstrated significant hypomethylation of HSD3B1 in the placenta of early-onset preeclampsia (Hogg et al., 2013). Shimodaira, M showed that polymorphisms of maternal HSD3B1 and HSD3B2 were not associated with preeclampsia (Shimodaira et al., 2012). Therefore, the protein expression of HSD3B1, HSD3B2 in PE placenta, and the role of HSD3B in abnormal androgen synthesis in the placenta of PE remains intriguing unsolved mysteries.
HSD17B plays a role in the conversion of A4 to T and E1 to E2, it has multiple isoforms (HSD17B1 to HSD17B8). Shao, X found that HSD17B3 is markedly increased in PE placenta and that it is the key enzyme that converts A4 to T (Shao et al., 2017). However, other researchers claimed that the activity of HSD17B3 in the preeclamptic placenta was compromised (Shin et al., 2018; Shao et al., 2019). In our study, a 0.43-fold elevation in HSD17B3 expression was found in PE placentas (p = 0.64). In the validation datasets of 330 human placental samples, placental HSD17B3 increased 0.03-fold in the PE group (p = 0.63). These findings indicated that HSD17B3 did not change significantly at the transcriptional level in the PE placenta; however, the status of its posttranscriptional modification, protein expression, and enzyme activity in PE remains to be elucidated. Conversion of E1 to E2 in the placenta depends on the presence of HSD17B1 (Husen et al., 2003), and it has been reported to be an independent risk factor for predicting PE (Ishibashi et al., 2012; Ohkuchi et al., 2012). The transcriptional level of HSD17B1 was not altered significantly in our study; however, it demonstrated a 0.51-fold reduction in the PE group in the validation dataset analysis (p = 2.52E-21). Decreased placental HSD17B1 expression might partly explain the reduction in E2 in PE patients. In addition, HSD17B2 and HSD17B6 were noticeably decreased in PE placentas in the present study (p = 0.179 and p = 0.002, respectively). These two genes were observed to decrease markedly in the PE group in the validation datasets (p = 4.41E-10 and p = 0.007, respectively). HSD17B2 and HSD17B6 perform the reverse reactions of isoforms 1 and 3, converting E2 into E1 and A4 into T, respectively (Saloniemi et al., 2012). Together with other studies, we highlight a possible correlation between the reduction in HSD17B2 and HSD17B6 and the elevated T level in PE. Further studies are warranted to validate these findings and to reveal the underlying mechanism.
CYP19A1 converts A4 into E1 and T into E2. In several studies, the protein and mRNA levels of placental aromatase were significantly decreased in PE placentas compared to normotensive pregnancies. Accordingly, the ratios of the corresponding metabolites, that is, the circulating concentrations of 17-β- E2/T and E1/A4, were significantly decreased in PE patients (Hertig et al., 2010; Perez-Sepulveda et al., 2015; Zhang et al., 2016; Berkane et al., 2018). However, our observations demonstrated that placental CYP19A1 was not altered significantly at the transcriptional level (p = 0.42), which was confirmed in the analysis of validation datasets of 330 human placental samples (p = 0.60). E1/A4 was reduced in the PE group (48.77 vs. 54.93, p = 0.42) in our research, and similarly, it was found to decrease significantly in another study (Berkane et al., 2018), suggesting a deficiency of placental aromatase in preeclampsia, which might not be at the transcriptional level but rather at the level of posttranscriptional modification and regulation.
4.4 Possible connections between the pathogenesis of PE and the aberrant placental synthesis and metabolism of steroid hormones
Increased trophoblast progesterone in PE may act by a paracrine mechanism to inhibit prostacyclin synthesis in the placental vasculature, leading to compromised vasodilatation (Walsh and Coulter, 1989). Testosterone has been shown to be associated with the inhibition of angiogenesis at the fetal-maternal interface and increases uterine vascular constriction (Baker et al., 1978; Blesson et al., 2015; Maliqueo et al., 2016). However, estrogens and their metabolites act as vasodilators, leading to an increase in uterine blood flow, promoting early placental angiogenesis and regulating the remodeling of the uterine spiral artery by the invasion of placental trophoblast cells (Jobe et al., 2013; Lechuga et al., 2015; Shao et al., 2017). Inactivation of cortisol and corticosterone may protect the fetus from exposure to high levels of active glucocorticoids in gestational hypertension (Siiteri and MacDonald, 1966; Staud et al., 2006; Abbassi-Ghanavati et al., 2009; Aufdenblatten et al., 2009; Jayasuriya et al., 2019). Thus, we propose a working hypothesis that disrupted steroid hormone production in preeclampsia may lead to insufficient vascular dilation, enhanced oxidative stress or hypoxia, compromised angiogenesis, insufficient trophoblast invasion and uterine spiral artery remodeling, thereby triggering some of the pathophysiology of preeclampsia (Figure 5). These metabolic disorders might be consequent or compensatory changes in pathological placentation in PE. The above alterations in the PE placenta may lead to corresponding changes in maternal plasma and underscore the need for prospective longitudinal studies covering the first, second, and third trimesters to further assess and validate whether these metabolites and genes involved in steroid hormone biosynthesis can be useful biomarkers in the early detection of PE and to understand the pathophysiology of the disease.
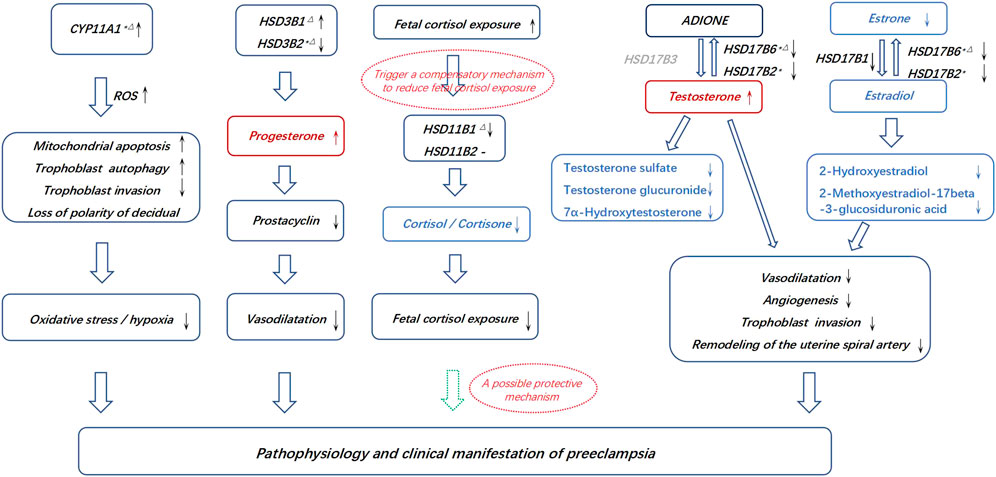
FIGURE 5. Possible connections between the pathogenesis of PE and the aberrant placental synthesis and metabolism of steroid hormones. * Differential genes in the discovery and RNA microarray validation. △Differential genes in the discovery and qPCR validation. The dotted hollow arrow in green represents a potential protective effect. The metabolites in red represents an accumulation, and the metabolites in blue represents aberrantly affected biosynthesis. The gene in grey represents that it is lack in placenta.
4.5 Limitations
The sample size of this research was limited. The validation microarray datasets showed mild difference in the expressions of CYP11A1, HSD3B2, and HSD17B6 between PE and control groups, compared to mRNA seq. The possible reasons might be that, these data were generated from seven independent researches, participants were from different races, the gestational age at delivery varied widely, but the three genes showed the same trend of change in different researches. Furthermore, we failed to match each case with controls of the same gestational age. The reason for not including gestational age-matched preterm delivery patients into the control group is that they are often complicated by chorioamnionitis, placental abruption, fetal growth restriction with fetal distress, maternal-fetal hemorrhage, etc., which were likely to affect the metabolic state of the placenta. Another concern is that the observed transcriptional differences might not lead to corresponding protein or functional difference. Further investigations should aim to identify these genes and their downstream proteins at the protein level and explore their functions.
5 Conclusion
In summary, the perturbed metabolism in PE placenta affected a diverse array of intracellular pathways, including steroid hormone biosynthesis, the cAMP signaling pathway, neuroactive ligand–receptor interaction, taste transduction and prion diseases. Steroid hormone biosynthesis was the most significantly enriched metabolic pathway. A dysregulated cortisol-to-cortisone ratio, accumulation of testosterone, impaired estrogen synthesis, and aberrant hydroxylation and methylation of estradiol were observed in PE placentas. DEGs identified in this metabolic process could be the cornerstone of the dysregulated steroid hormone biosynthesis in severe PE placentas. However, the disorders of placental steroid hormone metabolism found in the present study may also be a consequence or a compensatory change in the pathological placentation of PE. Prospective longitudinal studies covering the first, second, and third trimesters are needed to further assess and validate these conclusions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/gsa-human/s/0m207aH0, PRJCA0098 67.
Ethics statement
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of the Guangzhou Women and Children’s Medical Center. The patients/participants provided their written informed consent to participate in this study.
Author contributions
Conception and design of the work, XH, data analysis; XL, funding acquisition; YF and KG, sample acquisition; KG, interpretation of data; GZ and XH, visualization; YF and XL, writing—original draft; YF and XL, writing—review and editing.
Funding
This study was funded by the Guangzhou Municipal Science and Technology Bureau. (Funding number: 202102010318) and the National Natural Science Foundation of China (Funding number: 81801466).
Acknowledgments
We are indebted to the patients and staff of the Guangzhou Women and Children’s Medical Center who participated in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1010657/full#supplementary-material
Abbreviations
A4, Androstenedione; BMI, Body Mass Index; CYP, Cytochrome P450; DHEAS, Dehydroepiandrosterone Sulfate; DEGs, Differentially Expressed Genes; DHEA, Dehydroepiandrosterone; E1, Estrone; E2, Estradiol; E3, Estriol; FC, Fold Change; GEO, Gene Expression Omnibus; HSD, Hydroxysteroid Dehydrogenase; LC–MS, Liquid chromatography–mass spectrometry; NS, Not Significant; PCA, Principal Component Analysis; PLS-DA, Partial Least Squares Discrimination Analysis; PE, Preeclampsia; SGA, Small for Gestational Age; T, Testosterone; VIP, Variable Importance in the Projection; WGCNA, Weighted Gene Coexpression Network Analysis.
References
Abbassi-Ghanavati, M., Greer, L. G., and Cunningham, F. G. (2009). Pregnancy and laboratory studies: A reference table for clinicians. Obstet. Gynecol. 114 (6), 1326–1331. doi:10.1097/AOG.0b013e3181c2bde8
ACOG Practice Bulletin No (2019). ACOG Practice Bulletin No. 202: Gestational hypertension and preeclampsia. Obstet. Gynecol. 133 (1), 1–e25. doi:10.1097/aog.0000000000003018
Aufdenblatten, M., Baumann, M., Raio, L., Dick, B., Frey, B. M., Schneider, H., et al. (2009). Prematurity is related to high placental cortisol in preeclampsia. Pediatr. Res. 65 (2), 198–202. doi:10.1203/PDR.0b013e31818d6c24
Baker, P. J., Ramey, E. R., and Ramwell, P. W. (1978). Androgen-mediated sex differences of cardiovascular responses in rats. Am. J. Physiol. 235 (2), H242–H246. doi:10.1152/ajpheart.1978.235.2.H242
Bansil, P., Kuklina, E. V., Meikle, S. F., Posner, S. F., Kourtis, A. P., Ellington, S. R., et al. (2010). Maternal and fetal outcomes among women with depression. J. Womens Health 19 (2), 329–334. doi:10.1089/jwh.2009.1387
Beaudoin, C., Bonenfant, M., and Tremblay, Y. (1997). Regulation of cytochrome P450 cholesterol side-chain cleavage, 3 beta-hydroxysteroid dehydrogenase/delta 5-delta 4 isomerase type 1 and estradiol-17 beta-hydroxysteroid dehydrogenase mRNA levels by calcium in human choriocarcinoma JEG-3 cells. Mol. Cell. Endocrinol. 133 (1), 63–71. doi:10.1016/s0303-7207(97)00143-3
Benny, P. A., Alakwaa, F. M., Schlueter, R. J., Lassiter, C. B., and Garmire, L. X. (2020). A review of omics approaches to study preeclampsia. Placenta 92, 17–27. doi:10.1016/j.placenta.2020.01.008
Berg, C. J., MacKay, A. P., Qin, C., and Callaghan, W. M. (2009). Overview of maternal morbidity during hospitalization for labor and delivery in the United States: 1993-1997 and 2001-2005. Obstet. Gynecol. 113 (5), 1075–1081. doi:10.1097/AOG.0b013e3181a09fc0
Berkane, N., Liere, P., Lefevre, G., Alfaidy, N., Nahed, R. A., Vincent, J., et al. (2018). Abnormal steroidogenesis and aromatase activity in preeclampsia. Placenta 69, 40–49. doi:10.1016/j.placenta.2018.07.004
Berkane, N., Liere, P., Oudinet, J. P., Hertig, A., Lefèvre, G., Pluchino, N., et al. (2017). From pregnancy to preeclampsia: A key role for estrogens. Endocr. Rev. 38 (2), 123–144. doi:10.1210/er.2016-1065
Blesson, C. S., Chinnathambi, V., Hankins, G. D., Yallampalli, C., and Sathishkumar, K. (2015). Prenatal testosterone exposure induces hypertension in adult females via androgen receptor-dependent protein kinase Cδ-mediated mechanism. Hypertension 65 (3), 683–690. doi:10.1161/hypertensionaha.114.04521
Cantonwine, D. E., McElrath, T. F., Trabert, B., Xu, X., Sampson, J., Roberts, J. M., et al. (2019). Estrogen metabolism pathways in preeclampsia and normal pregnancy. Steroids 144, 8–14. doi:10.1016/j.steroids.2019.01.005
Causevic, M., and Mohaupt, M. (2007). 11beta-Hydroxysteroid dehydrogenase type 2 in pregnancy and preeclampsia. Mol. Asp. Med. 28 (2), 220–226. doi:10.1016/j.mam.2007.04.003
Chien, Y., Cheng, W. C., Wu, M. R., Jiang, S. T., Shen, C. K., and Chung, B. C. (2013). Misregulated progesterone secretion and impaired pregnancy in CYP11A1 transgenic mice. Biol. Reprod. 89 (4), 91. doi:10.1095/biolreprod.113.110833
Christensen, A. (1974). Hormone and enzyme assays in pregnancy. I. Studies on the placental and the tissue cystine-aminopeptidase activity in peripheral plasma from non-pregnant and pregnant women, and in plasma from the umbilical cord. Acta Endocrinol. (Copenh). 76 (1), 189–200. doi:10.1530/acta.0.0760189
Duley, L. (2009). The global impact of pre-eclampsia and eclampsia. Semin. Perinatol. 33 (3), 130–137. doi:10.1053/j.semperi.2009.02.010
Hertig, A., Liere, P., Chabbert-Buffet, N., Fort, J., Pianos, A., Eychenne, B., et al. (2010). Steroid profiling in preeclamptic women: Evidence for aromatase deficiency. Am. J. Obstet. Gynecol. 203 (5), 477.e471–e9. doi:10.1016/j.ajog.2010.06.011
Hill, M., Pašková, A., Kančeva, R., Velíková, M., Kubátová, J., Kancheva, L., et al. (2014). Steroid profiling in pregnancy: A focus on the human fetus. J. Steroid Biochem. Mol. Biol. 139, 201–222. doi:10.1016/j.jsbmb.2013.03.008
Hogg, K., Blair, J. D., McFadden, D. E., von Dadelszen, P., and Robinson, W. P. (2013). Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PLoS One 8 (5), e62969. doi:10.1371/journal.pone.0062969
Hu, M. C., Hsu, H. J., Guo, I. C., and Chung, B. C. (2004). Function of CYP11A1 in animal models. Mol. Cell. Endocrinol. 215 (1-2), 95–100. doi:10.1016/j.mce.2003.11.024
Hu, W., Wang, H., and Huang, H. (2015). Analysis of gene expression and preliminary study of methylation about 11β-HSD2 gene in placentas of Chinese pre-eclampsia patients of Han ethnicity. J. Obstet. Gynaecol. Res. 41 (3), 343–349. doi:10.1111/jog.12555
Husen, B., Adamski, J., Brüns, A., Deluca, D., Fuhrmann, K., Möller, G., et al. (2003). Characterization of 17beta-hydroxysteroid dehydrogenase type 7 in reproductive tissues of the marmoset monkey. Biol. Reprod. 68 (6), 2092–2099. doi:10.1095/biolreprod.102.012476
Ishibashi, O., Ohkuchi, A., Ali, M. M., Kurashina, R., Luo, S. S., Ishikawa, T., et al. (2012). Hydroxysteroid (17-β) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: A novel marker for predicting preeclampsia. Hypertension 59 (2), 265–273. doi:10.1161/hypertensionaha.111.180232
Jayasuriya, N. A., Hughes, A. E., Sovio, U., Cook, E., Charnock-Jones, D. S., and Smith, G. C. S. (2019). A lower maternal cortisol-to-cortisone ratio precedes clinical diagnosis of preterm and term preeclampsia by many weeks. J. Clin. Endocrinol. Metab. 104 (6), 2355–2366. doi:10.1210/jc.2018-02312
Jobe, S. O., Tyler, C. T., and Magness, R. R. (2013). Aberrant synthesis, metabolism, and plasma accumulation of circulating estrogens and estrogen metabolites in preeclampsia implications for vascular dysfunction. Hypertension 61 (2), 480–487. doi:10.1161/hypertensionaha.111.201624
Kelly, R. S., Croteau-Chonka, D. C., Dahlin, A., Mirzakhani, H., Wu, A. C., Wan, E. S., et al. (2017). Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics. 13 (1), 7. doi:10.1007/s11306-016-1149-8
Keya, S. L., Khanam, N. N., Chowdhury, A. A., Ripon, R., Tasnim, T., and Sharmin, A. (2019). Relationship between free testosterone and preeclampsia. Mymensingh Med. J. 28 (3), 574–581.
Kuklina, E. V., Ayala, C., and Callaghan, W. M. (2009). Hypertensive disorders and severe obstetric morbidity in the United States. Obstet. Gynecol. 113 (6), 1299–1306. doi:10.1097/AOG.0b013e3181a45b25
Kumar, S., Gordon, G. H., Abbott, D. H., and Mishra, J. S. (2018). Androgens in maternal vascular and placental function: Implications for preeclampsia pathogenesis. Reproduction 156 (5), R155–r167. doi:10.1530/rep-18-0278
Lan, K. C., Lai, Y. J., Cheng, H. H., Tsai, N. C., Su, Y. T., Tsai, C. C., et al. (2020). Levels of sex steroid hormones and their receptors in women with preeclampsia. Reprod. Biol. Endocrinol. 18 (1), 12. doi:10.1186/s12958-020-0569-5
Langfelder, P., and Horvath, S. (2008). Wgcna: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559. doi:10.1186/1471-2105-9-559
Leavey, K., Benton, S. J., Grynspan, D., Kingdom, J. C., Bainbridge, S. A., and Cox, B. J. (2016). Unsupervised placental gene expression profiling identifies clinically relevant subclasses of human preeclampsia. Hypertension 68 (1), 137–147. doi:10.1161/hypertensionaha.116.07293
Leavey, K., Wilson, S. L., Bainbridge, S. A., Robinson, W. P., and Cox, B. J. (2018). Epigenetic regulation of placental gene expression in transcriptional subtypes of preeclampsia. Clin. Epigenetics 10, 28. doi:10.1186/s13148-018-0463-6
Lechuga, T. J., Zhang, H. H., Sheibani, L., Karim, M., Jia, J., Magness, R. R., et al. (2015). Estrogen replacement therapy in ovariectomized nonpregnant ewes stimulates uterine artery hydrogen sulfide biosynthesis by selectively up-regulating cystathionine β-synthase expression. Endocrinology 156 (6), 2288–2298. doi:10.1210/en.2015-1086
Lian, X., Guo, J., Gu, W., Cui, Y., Zhong, J., Jin, J., et al. (2016). Genome-wide and experimental resolution of relative translation elongation speed at individual gene level in human cells. PLoS Genet. 12 (2), e1005901. doi:10.1371/journal.pgen.1005901
Li, R., Yu, C., Li, Y., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi:10.1093/bioinformatics/btp336
Liu, J. L., Zhang, W. Q., and Huang, M. Y. (2018). Transcriptional signature of the decidua in preeclampsia. Proc. Natl. Acad. Sci. U. S. A. 115 (24), E5434–e5436. doi:10.1073/pnas.1802234115
Liu, L. Y., Yang, T., Ji, J., Wen, Q., Morgan, A. A., Jin, B., et al. (2013). Integrating multiple 'omics' analyses identifies serological protein biomarkers for preeclampsia. BMC Med. 11, 236. doi:10.1186/1741-7015-11-236
Maeda, K. J., Showmaker, K. C., Johnson, A. C., Garrett, M. R., and Sasser, J. M. (2019). Spontaneous superimposed preeclampsia: Chronology and expression unveiled by temporal transcriptomic analysis. Physiol. Genomics 51 (8), 342–355. doi:10.1152/physiolgenomics.00020.2019
Maliqueo, M., Echiburú, B., and Crisosto, N. (2016). Sex steroids modulate uterine-placental vasculature: Implications for Obstetrics and neonatal outcomes. Front. Physiol. 7, 152. doi:10.3389/fphys.2016.00152
Mason, J. I., Ushijima, K., Doody, K. M., Nagai, K., Naville, D., Head, J. R., et al. (1993). Regulation of expression of the 3 beta-hydroxysteroid dehydrogenases of human placenta and fetal adrenal. J. Steroid Biochem. Mol. Biol. 47 (1-6), 151–159. doi:10.1016/0960-0760(93)90069-9
McCalla, C. O., Nacharaju, V. L., Muneyyirci-Delale, O., Glasgow, S., and Feldman, J. G. (1998). Placental 11 beta-hydroxysteroid dehydrogenase activity in normotensive and pre-eclamptic pregnancies. Steroids 63 (10), 511–515. doi:10.1016/s0039-128x(98)00056-7
Moon, J. Y., Moon, M. H., Kim, K. T., Jeong, D. H., Kim, Y. N., Chung, B. C., et al. (2014). Cytochrome P450-mediated metabolic alterations in preeclampsia evaluated by quantitative steroid signatures. J. Steroid Biochem. Mol. Biol. 139, 182–191. doi:10.1016/j.jsbmb.2013.02.014
Morales, D. E., McGowan, K. A., Grant, D. S., Maheshwari, S., Bhartiya, D., Cid, M. C., et al. (1995). Estrogen promotes angiogenic activity in human umbilical vein endothelial cells in vitro and in a murine model. Circulation 91 (3), 755–763. doi:10.1161/01.cir.91.3.755
Morel, Y., Roucher, F., Plotton, I., Goursaud, C., Tardy, V., and Mallet, D. (2016). Evolution of steroids during pregnancy: Maternal, placental and fetal synthesis. Ann. Endocrinol. 77 (2), 82–89. doi:10.1016/j.ando.2016.04.023
Nobakht, M. G. B. F. (2018). Application of metabolomics to preeclampsia diagnosis. Syst. Biol. Reprod. Med. 64 (5), 324–339. doi:10.1080/19396368.2018.1482968
Ohkuchi, A., Ishibashi, O., Hirashima, C., Takahashi, K., Matsubara, S., Takizawa, T., et al. (2012). Plasma level of hydroxysteroid (17-β) dehydrogenase 1 in the second trimester is an independent risk factor for predicting preeclampsia after adjusting for the effects of mean blood pressure, bilateral notching and plasma level of soluble fms-like tyrosine kinase 1/placental growth factor ratio. Hypertens. Res. 35 (12), 1152–1158. doi:10.1038/hr.2012.109
Pan, T., He, G., Chen, M., Bao, C., Chen, Y., Liu, G., et al. (2017). Abnormal CYP11A1 gene expression induces excessive autophagy, contributing to the pathogenesis of preeclampsia. Oncotarget 8 (52), 89824–89836. doi:10.18632/oncotarget.21158
Perez-Sepulveda, A., Monteiro, L. J., Dobierzewska, A., España-Perrot, P. P., Venegas-Araneda, P., Guzmán-Rojas, A. M., et al. (2015). Placental aromatase is deficient in placental ischemia and preeclampsia. PLoS One 10 (10), e0139682. doi:10.1371/journal.pone.0139682
Rana, S., Lemoine, E., Granger, J. P., and Karumanchi, S. A. (2019). Preeclampsia: Pathophysiology, challenges, and perspectives. Circ. Res. 124 (7), 1094–1112. doi:10.1161/circresaha.118.313276
Saloniemi, T., Jokela, H., Strauss, L., Pakarinen, P., and Poutanen, M. (2012). The diversity of sex steroid action: Novel functions of hydroxysteroid (17β) dehydrogenases as revealed by genetically modified mouse models. J. Endocrinol. 212 (1), 27–40. doi:10.1530/joe-11-0315
Schoof, E., Girstl, M., Frobenius, W., Kirschbaum, M., Dörr, H. G., Rascher, W., et al. (2001). Decreased gene expression of 11beta-hydroxysteroid dehydrogenase type 2 and 15-hydroxyprostaglandin dehydrogenase in human placenta of patients with preeclampsia. J. Clin. Endocrinol. Metab. 86 (3), 1313–1317. doi:10.1210/jcem.86.3.7311
Shao, X., Liu, Y., Liu, M., Wang, Y., Yan, L., Wang, H., et al. (2017). Testosterone represses estrogen signaling by upregulating miR-22: A mechanism for imbalanced steroid hormone production in preeclampsia. Hypertension 69 (4), 721–730. doi:10.1161/hypertensionaha.116.08468
Shao, X., Wang, Y., Liu, Y., Guo, X., Li, D., Huo, R., et al. (2019). Association of imbalanced sex hormone production with excessive procoagulation factor SerpinF2 in preeclampsia. J. Hypertens. 37 (1), 197–205. doi:10.1097/hjh.0000000000001862
Shih, M. C., Chiu, Y. N., Hu, M. C., Guo, I. C., and Chung, B. C. (2011). Regulation of steroid production: Analysis of CYP11A1 promoter. Mol. Cell. Endocrinol. 336 (1-2), 80–84. doi:10.1016/j.mce.2010.12.017
Shimodaira, M., Nakayama, T., Sato, I., Sato, N., Izawa, N., Mizutani, Y., et al. (2012). Estrogen synthesis genes CYP19A1, HSD3B1, and HSD3B2 in hypertensive disorders of pregnancy. Endocrine 42 (3), 700–707. doi:10.1007/s12020-012-9699-7
Shin, Y. Y., Jeong, J. S., Park, M. N., Lee, J. E., An, S. M., Cho, W. S., et al. (2018). Regulation of steroid hormones in the placenta and serum of women with preeclampsia. Mol. Med. Rep. 17 (2), 2681–2688. doi:10.3892/mmr.2017.8165
Siiteri, P. K., and MacDonald, P. C. (1966). Placental estrogen biosynthesis during human pregnancy. J. Clin. Endocrinol. Metab. 26 (7), 751–761. doi:10.1210/jcem-26-7-751
Simard, J., Durocher, F., Mébarki, F., Turgeon, C., Sanchez, R., Labrie, Y., et al. (1996). Molecular biology and genetics of the 3 beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. J. Endocrinol. 150, S189–S207.
Staud, F., Mazancová, K., Miksík, I., Pávek, P., Fendrich, Z., and Pácha, J. (2006). Corticosterone transfer and metabolism in the dually perfused rat placenta: Effect of 11beta-hydroxysteroid dehydrogenase type 2. Placenta 27 (2-3), 171–180. doi:10.1016/j.placenta.2005.01.001
Takeyama, J., Sasano, H., Suzuki, T., Iinuma, K., Nagura, H., and Andersson, S. (1998). 17Beta-hydroxysteroid dehydrogenase types 1 and 2 in human placenta: An immunohistochemical study with correlation to placental development. J. Clin. Endocrinol. Metab. 83 (10), 3710–3715. doi:10.1210/jcem.83.10.5212
Thomas, J. L., Boswell, E. L., Scaccia, L. A., Pletnev, V., and Umland, T. C. (2005). Identification of key amino acids responsible for the substantially higher affinities of human type 1 3beta-hydroxysteroid dehydrogenase/isomerase (3beta-HSD1) for substrates, coenzymes, and inhibitors relative to human 3beta-HSD2. J. Biol. Chem. 280 (22), 21321–21328. doi:10.1074/jbc.M501269200
Tong, J., Zhao, W., Lv, H., Li, W. P., Chen, Z. J., and Zhang, C. (2018). Transcriptomic profiling in human decidua of severe preeclampsia detected by RNA sequencing. J. Cell. Biochem. 119 (1), 607–615. doi:10.1002/jcb.26221
Wagner, S. J., Acquah, L. A., Lindell, E. P., Craici, I. M., Wingo, M. T., Rose, C. H., et al. (2011). Posterior reversible encephalopathy syndrome and eclampsia: Pressing the case for more aggressive blood pressure control. Mayo Clin. Proc. 86 (9), 851–856. doi:10.4065/mcp.2011.0090
Walsh, S. W., and Coulter, S. (1989). Increased placental progesterone may cause decreased placental prostacyclin production in preeclampsia. Am. J. Obstet. Gynecol. 161 (1), 1586–1592. doi:10.1016/0002-9378(89)90931-9
Zhang, B., and Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17. Article17. doi:10.2202/1544-6115.1128
Zhang, M., Muralimanoharan, S., Wortman, A. C., and Mendelson, C. R. (2016). Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc. Natl. Acad. Sci. U. S. A. 113 (45), E7069–e7076. doi:10.1073/pnas.1607849113
Keywords: preeclampsia, metabolomic, transcriptomic, placenta, metabolic pathways, biomarker
Citation: Feng Y, Lian X, Guo K, Zhang G and Huang X (2022) A comprehensive analysis of metabolomics and transcriptomics to reveal major metabolic pathways and potential biomarkers of human preeclampsia placenta. Front. Genet. 13:1010657. doi: 10.3389/fgene.2022.1010657
Received: 03 August 2022; Accepted: 14 September 2022;
Published: 03 October 2022.
Edited by:
Jihan Xia, University of Helsinki, FinlandReviewed by:
Huanan Shi, Baylor College of Medicine, United StatesGuangda Peng, Georgia State University, United States
Xiangxiang Hu, University of North Carolina at Chapel Hill, United States
Tong Wu, Indiana University, Purdue University Indianapolis, United States
Copyright © 2022 Feng, Lian, Guo, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuan Huang, SHVhbmd4MzVAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yan Feng
Yan Feng Xinlei Lian
Xinlei Lian Kaimin Guo
Kaimin Guo Guanglan Zhang
Guanglan Zhang Xuan Huang
Xuan Huang