
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 08 November 2022
Sec. Genetics of Common and Rare Diseases
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1009589
This article is part of the Research Topic Genetics of Inflammatory and Immune Diseases View all 19 articles
 Chang Zhang1,2,3,4,5†
Chang Zhang1,2,3,4,5† Qin Qin1,2,3,4,5†
Qin Qin1,2,3,4,5† Yuanyuan Li1,2,3,4,5†
Yuanyuan Li1,2,3,4,5† Xiaodong Zheng1,2,3,4,5
Xiaodong Zheng1,2,3,4,5 Weiwei Chen1,2,3,4,5
Weiwei Chen1,2,3,4,5 Qi Zhen1,2,3,4,5
Qi Zhen1,2,3,4,5 Bao Li1,2,3,4,5
Bao Li1,2,3,4,5 Wenjun Wang1,2,3,4,5*
Wenjun Wang1,2,3,4,5* Liangdan Sun1,2,3,4,5*
Liangdan Sun1,2,3,4,5*Background: Psoriasis is a common immune-mediated hyperproliferative skin dysfunction with known genetic predisposition. Gene–gene interaction (e.g., between HLA-C and ERAP1) in the psoriasis context has been reported in various populations. As ERAP1 has been recognized as a psoriasis susceptibility gene and plays a critical role in antigen presentation, we performed this study to identify interactions between ERAP1 and other psoriasis susceptibility gene variants.
Methods: We validated psoriasis susceptibility gene variants in an independent cohort of 5,414 patients with psoriasis and 5,556 controls. Multifactor dimensionality reduction (MDR) analysis was performed to identify the interaction between variants significantly associated with psoriasis in the validation cohort and ERAP1 variants. We then conducted a meta-analysis of those variants with datasets from exome sequencing, target sequencing, and validation analyses and used MDR to identify the best gene–gene interaction model, including variants that were significant in the meta-analysis and ERAP1 variants.
Results: We found that 19 of the replicated variants were identified with p < 0.05 and detected six single-nucleotide polymorphisms of psoriasis susceptibility genes in the meta-analysis. MDR analysis revealed that the best predictive model was that between the rs27044 polymorphism of ERAP1 and the rs7590692 polymorphism of IFIH1 (cross-validation consistency = 9/10, test accuracy = 0.53, odds ratio = 1.32 (95% CI, 1.09–1.59), p < 0.01).
Conclusion: Our findings suggest that the interaction between ERAP1 and IFIH1 affects the development of psoriasis. This hypothesis needs to be tested in basic biological studies.
Psoriasis is a common immune-mediated hyperproliferative skin dysfunction with known genetic predisposition that affects almost 125 million people worldwide (Armstrong and Read, 2020). Psoriasis morbidity rates range from 0.09% to 5.1% (Michalek et al., 2017), with major regional differences. The condition results from a combination of interacting genetic, environmental, and immunological factors (Myers et al., 2005; Mahil et al., 2015; Greb et al., 2016). Although progress has been made in the interpretation of its heredity, the pathogenesis of psoriasis remains unclear. Early correlation studies established that the genetic contribution to psoriasis is complex and multifactorial.
Since 2005, genome-wide association studies (GWASs) have been established as the most effective approach to the identification of genetic variations related to complex diseases via the use of a complex single-nucleotide polymorphism (SNP) map of the human genome to identify differences in allele frequency between patients and controls (Kruglyak, 2008; Zhang, 2012). Although GWASs have led to the robust identification of numerous susceptibility loci, they capture small proportions of estimated disease heritability; this issue is referred to as the “missing heritability” problem (Manolio et al., 2009; Eichler et al., 2010). Most correlational research involves the detection of SNPs in isolation, without examination of the mutual effects of their interaction, leading intricate reciprocities in the biosystem to be overlooked. With the accumulation of large amounts of genome data, further gene–gene interaction and association studies are essential to explore the biological pathways and pathogeneses of common diseases (Cordell, 2009).
Most common diseases are induced by the nonlinear interaction of numerous genetic and environmental factors. Complex diseases are always generated by gene–environment and gene–gene interactions, the latter of which can enhance their danger (Evans et al., 2006; Cordell, 2009). Many methods can be used to test for gene–gene interaction, including those involving logistic regression models, neural networks, and multifactor dimensionality reduction (MDR) (Cordell, 2009). The use of a traditional (e.g., logistic regression) model to detect multiple interactions at the same time imposes a huge computational burden due to the exponential increase in the number of interaction terms (Chattopadhyay and Lu, 2019). Although neural networks are considered good nonlinear models, they are prone to overfitting (Ritchie et al., 2003). MDR, developed by Ritchie et al. (Ritchie et al., 2001) in 2001, is a non-parametric model-free method that can be applied directly in case-control and discordant sib-pair studies. It involves no a priori genetic model assumption, unlike other approaches (e.g., linear regression or generalized linear models); this enables the detection of any genetic interaction in relation to disease. With the division of samples into high- and low-risk groups, MDR enables the reduction of n-dimensional models to one-dimension and reduces type-I and-II errors. It is currently the most popular method for detection of gene–gene interaction, and it has been used to identify potential interaction loci in many diseases, including sporadic breast cancer (Ritchie et al., 2001), hypertension (Moore and Williams, 2002), type-2 diabetes mellitus (Cho et al., 2004), and the autoimmune diseases rheumatoid arthritis (Julià et al., 2007; Liu et al., 2011), systemic lupus erythematosus (Zhang et al., 2016), and ankylosing spondylitis (Evans et al., 2011).
Based on existing GWAS data (Zhang et al., 2009), our group has analyzed the interaction of various susceptibility genes (including HLA–IL12B, HLA–ICE (Zheng et al., 2011), HLA-C–ERAP1, and HLA-C–TRAF3IP2 (Yin et al., 2013) interactions) in the Han Chinese population. Such gene interactions, including those related to psoriasis, have been reported in different populations (Strange et al., 2010). ERAP1 has been reported as being related to psoriasis in multiple populations, and our group has genotyped and replicated its polymorphisms (Chen et al., 2022). In this study, we detected interactions of ERAP1 with other psoriasis susceptibility genes in the Han Chinese population.
This study was conducted with data generated in our previous research—an exome sequencing study conducted with 781 patients with psoriasis and 676 controls and a targeted sequencing study conducted with 9,946 psoriasis cases and 9,906 controls (Tang et al., 2014). We performed a case-control study with an independent sample cohort consisting of 5,414 psoriasis cases and 5,556 controls to validate the psoriasis susceptibility genes. All patients and controls were of Han Chinese origin and had attended the First Affiliated Hospital of Anhui Medical University. The controls were healthy volunteers without psoriasis, other autoimmune diseases, systemic disease, or a family history of psoriasis (in first-, second-, and third-degree relatives). Each patient was examined by medical professionals and diagnosed with psoriasis by two experienced dermatologists, with rigorous recording of clinical data to ensure their specificity and reliability. After obtaining informed consent, medical professionals collected peripheral blood samples from all patients and controls. This research was approved by the Ethics Committee of Anhui Medical University Committee and performed following Declaration of Helsinki guidelines.
Genomic DNA was extracted from the participants’ peripheral blood mononuclear cells. DNA concentrations and optical density ratios (A260/A280) were determined using a NanoDrop 1,000 spectrophotometer (Thermo Fisher Scientific, United States). The Sequenom MassArray system (Sequenom, United States) was used for SNP genotyping. We used 15 ng standardized genomic DNA per sample for subsequent genetic typing. We used multiplex polymerase chain reactions to amplify the genomic DNA and then performed site-specific monacyl elongation reactions to obtain products. The amplified products were purified with AgencourtAM SPRI XP microbeads (Beckman Coulter Life Sciences, United States). After desalination, the final products were transferred to a 384-element SpectrCHIP array (Applied Biosystems, United States). Alleles were detected with a matrix-assisted laser desorption ionization time-of-flight 70 mass spectrometer and analyzed using Massarray Typer 71 software (Sequenom, United States). In the independent cohort, validated SNPs were selected using the following terms: position within 500 kb of a psoriasis susceptibility gene tag, p < 1.0 × 10−4 in the exome sequencing and targeted sequencing studies, genotyping detection rate >90%, and Hardy–Weinberg test p > 1.0 × 10−4 in controls.
Based on our group’s previous study (Chen et al., 2022), ERAP1 variants were genotyped and replicated in the present cohort. The statistical analysis was performed with PLINK 1.07 and a significance level of p < 0.05. Gene–gene interactions were analyzed with open-source MDR software (ver. 2.0 Beta 2; available from http://sourceforge.net/projects/mdr). As the MDR software requires complete datasets with no missing value, we replaced missing values with numbers that did not represent genotypes; the second method used was k-nearest neighbor estimation to fill in the missing values (k was equal to the square root of the number of SNPs) (Schwender, 2012).
To expand the sample size and detect more gene–gene interactions, we conducted a meta-analysis of summary GWAS statistics from the exome sequencing (n = 1,457), targeted sequencing (n = 19,007), and validation (n = 10,970) cohorts. We performed the analysis with METAL (available from http://www.sph.umich.edu/csg/abecasis/Metal/), with the test statistics weighted by sample size. The genome-wide significance threshold was p < 5 × 10−8. We performed genomic correction of all results to control for potential inflation of the test statistics. Effector allele frequencies were tracked selectively across all files.
Of 39 SNPs selected for genotyping verification, 19 were significant (p < 0.05) in the independent cohort validation analysis. IFIH1 showed the strongest correlation with psoriasis (chr_163737871, p = 6.52 × 10−7, odds ratio (OR) = 0.81; chr2_163136771, p = 1.72 × 10−6, OR = 0.81; Table 1). Other validated psoriasis susceptibility genes were ERAP2 (5q15), IL18R1 (2q12.1), LTB (12p13.31), IL1RL1 (2q12.1), CARD14 (17q25.3), and SLC9A4 (2q12.1).
The MDR analysis was conducted with the 19 SNPs showing significance in the replicated cohort and six ERAP1 SNPs. The rs27044 polymorphism of ERAP1 was the best single-locus model (CVC = 10, test accuracy = 0.52, OR = 1.32 (95% CI, 1.07–1.63), p < 0.01). The best predictive model (i.e., that with maximum testing accuracy) was that between the rs27044 polymorphism of ERAP1 and the rs7590692 polymorphism of IFIH1 (CVC = 9/10, test accuracy = 0.53, OR = 1.32 (95% CI, 1.09–1.59), p < 0.01; Table 2). In this model, high-risk genotypes were TT × CC, TC × GG, TC × GC, TC × CC, CC × GC, and CC × CC, and low-risk genotypes were TT × GC, TT × GG, and CC × GG (Figure 1). Figures 2, 3 show measures of characterized epistatic of the models from the interaction analysis.
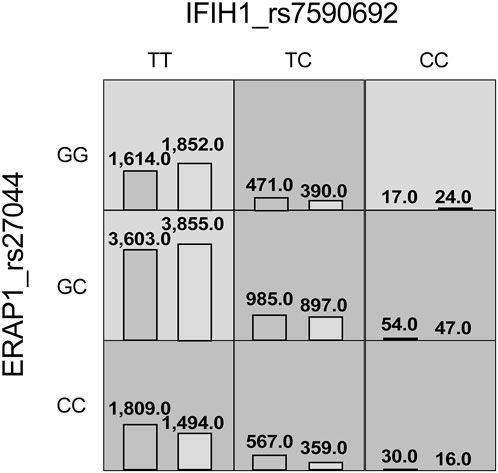
FIGURE 1. High- and low-risk genotypes in the best psoriasis interaction model. Dark-gray cells represent high risk (combinations exceeding the r ratio of the percentage of cases to controls ≥1.0) and light-gray cells represent low risk (combinations below the ratio threshold).
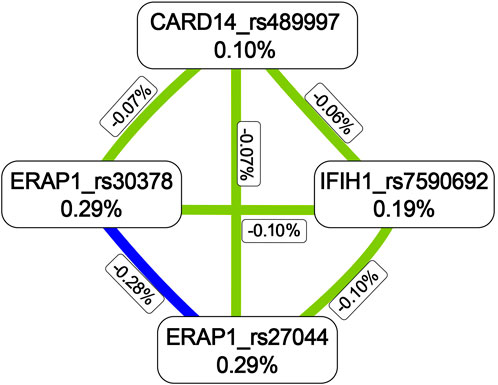
FIGURE 2. Circular graph of the best models. The numbers between two SNPs was information gain, which was used to detect and characterize epistatics of the interaction model and calculated by constructing two attributes using the MDR function. Red: positive information gain (synergistic or non-additive effect); blue: negative information gain (redundancy or correlation); yellow: no information gain (independence).

FIGURE 3. Dendrogram between polymorphisms in psoriasis susceptibility genes. Colors depict degrees of synergy, ranging from red (most information gain) to yellow–green and blue (most information redundancy).
In the meta-analysis, six of the 39 SNPs reached the genome-wide significance threshold (p < 10−8). These SNPs were in IFIH1 (rs12479043, p = 2.85 × 10−15; rs7590692, p = 1.57 × 10−14), CARD14 (rs4889997, p = 4.03 × 10−12), GJB2 (rs72474224, p = 8.22 × 10−11), NFKB1 (rs3817685, p = 4.24 × 10−10), and ERAP2 (rs2303208, p = 7.50 × 10−10; Table 3). In the MDR analysis of these SNPs and six SNPs of ERAP1, the best model was still that between the rs27044 polymorphism of ERAP1 and the rs7590692 polymorphism of IFIH1 (CVC = 9/10, test accuracy = 0.53, OR = 1.32 (95% CI, 1.09–1.59) p < 0.01).
With the deep development of psoriasis-related genetic analysis, our group has generated systemic genomic data on psoriasis through exome and targeted sequencing. In this study, we verified the psoriasis susceptibility genes IFIH1 (2q24.3), ERAP2 (5q15), IL18R1 (2q12.1), LTBR (12p13.31), IL1RL1 (2q12.1), CARD14 (17q25.3), and SLC9A4 (2q12.1). We found that the most significant interaction was between the rs27044 polymorphism of ERAP1 and the rs7590692 of IFIH1; this result was confirmed in the MDR analysis.
ERAP1 is an immune-related gene encoding multifunctional aminopeptidase that is induced by interferon-γ. It plays critical roles in antigen presentation and N-terminal peptide pruning to reach the best presenting size of the major histocompatibility complex I molecule. These molecules can present polypeptides containing 8–10 amino acids directly outside of cells, enhancing antigen presentation and T-cell activation in disease processes (Kochan et al., 2011). ERAP1 is more likely to be present in individuals carrying the HLA-C susceptibility allele, with the first and most important psoriasis susceptibility gene identified (Bowcock, 2005). A recent study revealed that ERAP1 causes psoriasis by affecting HLA-C production via melanocyte autoantigens (Arakawa et al., 2021). Thus, it may cause psoriasis lesions by interacting with HLA-C. ERAP1 has been related to psoriasis in Chinese (rs151823) (Sun et al., 2010) and European (rs27524) (Strange et al., 2010) populations. In a meta-analysis of nine case-control studies, the rs27044 polymorphism of ERAP1 was associated significantly with psoriasis (Wu and Zhao, 2021). This gene has also been associated with ankylosing spondylitis (Tsui et al., 2010), Behcet’s disease, multiple sclerosis, and inflammatory bowel disease (Reeves and James, 2018).
We replicated IFIH1 in our independent sample cohort with p = 6.52 × 10−7. IFIH1 encodes MDA5, an innate pattern recognition receptor. MDA5 is a cytoplasmic sensor of picornavirus nucleic acids that triggers a cascade of antiviral responses, including the induction of type I interferons and pro-inflammatory cytokines. Once it is bound to pathogen-like viral RNA, activated MDA5 interacts with mitochondrial antiviral-signaling proteins and then phosphorylated interferon regulatory factors (IRFs) 3 and 7. Dimers are formed by phosphorylated IRF-3 and -7 and move into cell nuclei to stimulate the secretion of type 1 interferon (IFN1) (Brisse and Ly, 2019). IFN1 mediates autoimmune responses to invaders such as viruses. MDA5 has been observed to significantly increase psoriatic plaques and keratinocytes, and its overexpression acts as a negative regulator for the differentiation of calcium-induced keratinocytes in psoriatic skin lesions (Hong et al., 2021). IFIH1 has been related to psoriasis in a European population (rs17716942) (Strange et al., 2010), and its rs35667974 and rs10930046 polymorphisms were found to have protective effects against psoriasis in a white North American population (Li et al., 2010). In addition, evidence supports the involvement of IFIH1 SNPs in inflammatory diseases such as type 1 diabetes (Nejentsev et al., 2009), inflammatory bowel disease (Cananzi et al., 2021), and dermatomyositis (Kurtzman and Vleugels, 2018).
Our MDR analysis revealed a significant association between ERAP1 and IFIH1, suggesting that the presence of the ERAP1 rs27044 polymorphism affects the expression of the IFIH1 rs7590692 polymorphism. In an in vitro experiment (Aldhamen et al., 2013), increased levels of proinflammatory cytokines such as tumor necrosis factor (TNF)-α were observed in ERAP1-knockout mice, suggesting that the lack of ERAP1 expression induces increased autoimmune activity. TNF-α, a cytokine commonly elevated in patients with psoriasis, promotes the transcription of MDA5 and RIG-I in keratinocytes (Kitamura et al., 2007; Racz et al., 2011). Taken together, these molecular biological findings suggest that ERAP1 might regulate the expression of IFIH1, but more basic biological studies are needed to determine how this gene interaction affects psoriasis.
This study has several limitations. The results provide evidence of correlations between gene–gene interactions and psoriasis but no information about the mechanisms of resulting effects, and the accuracy of the best model was not high. We will carry out further basic biological studies to verify this result. In addition, the study was conducted with data from a Han Chinese population; similar studies need to be conducted with data from other ethnic groups.
In summary, we screened for interactions between genes that affect psoriasis susceptibility and found that the most significant interaction was between the rs27044 polymorphism of ERAP1 and the rs7590692 polymorphism of IFIH1. Psoriasis is a multifactorial autoimmune disease with many genetic contributions. Although our comprehension of the genetic mechanisms underlying it has increased and the interaction of susceptibility genes has been identified based on SNPs, many questions remain. A better understanding of how these gene interactions affect the development of psoriasis, generated from basic biological studies, may provide answers to these questions.
The datasets presented in this study can be found in online repositories. The names of there pository/repositories and accession number(s) can be found below: Zenodo with accession 7260050—(https://doi.org/10.5281/zenodo.7260050).
The studies involving human participants were reviewed and approved by the Clinical Research Ethics Committee of Anhui Medical University. The patients/participants provided their written informed consent to participate in this study.
LS and WW conceived and designed the research. CZ wrote this manuscript. QQ and YL were responsible for selecting blood samples and extracting DNA. CZ, QQ, and YL contributed equally to this work. XZ, WC, BL, and QZ performed data analysis. All authors contributed to the study and approved the final manuscript.
This study was supported by grants from the Anhui Institute of Translation Medicine (ZHY 2020A005) and the University Synergy Innovation Program of Anhui Province (GXXT-2020-064).
We thank the patients and their families who contributed to this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
GWAS, genome-wide association studies; KNN, K-nearest neighbors; MAF, minor allele frequency; MDR, multifactor dimensionality reduction; PCR, polymerase chain reaction; SNP, single-nucleotide polymorphism; SNVs, single-nucleotide variations.
Aldhamen, Y. A., Seregin, S. S., Rastall, D. P. W., Aylsworth, C. F., Pepelyayeva, Y., and Busuito, C. J., (2013). Endoplasmic reticulum aminopeptidase-1 functions regulate key aspects of the innate immune response. PloS one 8 (7), e69539. doi:10.1371/journal.pone.0069539
Arakawa, A., Reeves, E., Vollmer, S., Arakawa, Y., He, M., Galinski, A., et al. (2021). ERAP1 controls the autoimmune response against melanocytes in psoriasis by generating the melanocyte autoantigen and regulating its amount for HLA-C*06:02 presentation. J. Immunol. 207 (9), 2235–2244. doi:10.4049/jimmunol.2100686
Armstrong, A. W., and Read, C. (2020). Pathophysiology, clinical presentation, and treatment of psoriasis: A review. JAMA 323 (19), 1945–1960. doi:10.1001/jama.2020.4006
Bowcock, A. M. (2005). The genetics of psoriasis and autoimmunity. Annu. Rev. Genomics Hum. Genet. 6, 93–122. doi:10.1146/annurev.genom.6.080604.162324
Brisse, M., and Ly, H. (2019). Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front. Immunol. 10, 1586. doi:10.3389/fimmu.2019.01586
Cananzi, M., Wohler, E., Marzollo, A., Colavito, D., You, J., and Jing, H., (2021). IFIH1 loss-of-function variants contribute to very early-onset inflammatory bowel disease. Hum. Genet. 140 (9), 1299–1312. doi:10.1007/s00439-021-02300-4
Chattopadhyay, A., and Lu, T. P. (2019). Gene-gene interaction: The curse of dimensionality. Ann. Transl. Med. 7 (24), 813. doi:10.21037/atm.2019.12.87
Chen, W., Yong, L., Ge, H., Xu, Q., Zhen, Q., and Li, B., (2022). Polymorphisms in ERAP1 gene are associated with psoriasis. Meta Gene 31, 100995. doi:10.1016/j.mgene.2021.100995
Cho, Y. M., Ritchie, M. D., Moore, J. H., Park, J. Y., Lee, K. U., and Shin, H. D., (2004). Multifactor-dimensionality reduction shows a two-locus interaction associated with Type 2 diabetes mellitus. Diabetologia 47 (3), 549–554. doi:10.1007/s00125-003-1321-3
Cordell, H. J. (2009). Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 10 (6), 392–404. doi:10.1038/nrg2579
Eichler, E. E., Flint, J., Gibson, G., Kong, A., Leal, S. M., and Moore, J. H., (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11 (6), 446–450. doi:10.1038/nrg2809
Evans, D. M., Marchini, J., Morris, A. P., and Cardon, L. R. (2006). Two-stage two-locus models in genome-wide association. PLoS Genet. 2 (9), e157. doi:10.1371/journal.pgen.0020157
Evans, D. M., Spencer, C. C. A., Pointon, J. J., Su, Z., Harvey, D., and Kochan, G., (2011). Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43 (8), 761–767. doi:10.1038/ng.873
Greb, J. E., Goldminz, A. M., Elder, J. T., Lebwohl, M. G., Gladman, D. D., and Wu, J. J., (2016). Nat. Rev. Dis. Prim. 2, 16082. doi:10.1038/nrdp.2016.82
Hong, D. K., Choi, M. R., Hwang, Y. L., Lee, J. K., Lee, Y., and Seo, Y. J., (2021). Potential role of cytosolic RNA sensor MDA5 as an inhibitor for keratinocyte differentiation in the pathogenesis of psoriasis. Ann. Dermatol. 33 (4), 339–344. doi:10.5021/ad.2021.33.4.339
Julià, A., Moore, J., Miquel, L., Alegre, C., Barcelo, P., and Ritchie, M., (2007). Identification of a two-loci epistatic interaction associated with susceptibility to rheumatoid arthritis through reverse engineering and multifactor dimensionality reduction. Genomics 90 (1), 6–13. doi:10.1016/j.ygeno.2007.03.011
Kitamura, H., Matsuzaki, Y., Kimura, K., Nakano, H., Imaizumi, T., and Satoh, K., (2007). Cytokine modulation of retinoic acid-inducible gene-I (RIG-I) expression in human epidermal keratinocytes. J. Dermatol. Sci. 45 (2), 127–134. doi:10.1016/j.jdermsci.2006.11.003
Kochan, G., Krojer, T., Harvey, D., Fischer, R., Chen, L., and Vollmar, M., (2011). Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. U. S. A. 108 (19), 7745–7750. doi:10.1073/pnas.1101262108
Kruglyak, L. (2008). The road to genome-wide association studies. Nat. Rev. Genet. 9 (4), 314–318. doi:10.1038/nrg2316
Kurtzman, D. J. B., and Vleugels, R. A. (2018). Anti-melanoma differentiation-associated gene 5 (MDA5) dermatomyositis: A concise review with an emphasis on distinctive clinical features. J. Am. Acad. Dermatol. 78 (4), 776–785. doi:10.1016/j.jaad.2017.12.010
Li, Y., Liao, W., Cargill, M., Chang, M., Matsunami, N., and Feng, B. J., (2010). Carriers of rare missense variants in IFIH1 are protected from psoriasis. J. Invest. Dermatol. 130 (12), 2768–2772. doi:10.1038/jid.2010.214
Liu, C., Ackerman, H. H., and Carulli, J. P. (2011). A genome-wide screen of gene-gene interactions for rheumatoid arthritis susceptibility. Hum. Genet. 129 (5), 473–485. doi:10.1007/s00439-010-0943-z
Mahil, S. K., Capon, F., and Barker, J. N. (2015). Genetics of psoriasis. Dermatol. Clin. 33 (1), 1–11. doi:10.1016/j.det.2014.09.001
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B., Hindorff, L. A., and Hunter, D. J., (2009). Finding the missing heritability of complex diseases. Nature 461 (7265), 747–753. doi:10.1038/nature08494
Michalek, I. M., Loring, B., and John, S. M. (2017). A systematic review of worldwide epidemiology of psoriasis. J. Eur. Acad. Dermatol. Venereol. 31 (2), 205–212. doi:10.1111/jdv.13854
Moore, J. H., and Williams, S. M. (2002). New strategies for identifying gene-gene interactions in hypertension. Ann. Med. 34 (2), 88–95. doi:10.1080/07853890252953473
Myers, A., Kay, L. J., Lynch, S. A., and Walker, D. J. (2005). Recurrence risk for psoriasis and psoriatic arthritis within sibships. Rheumatol. Oxf. 44(6): p. 773–776. doi:10.1093/rheumatology/keh589
Nejentsev, S., Walker, N., Riches, D., Egholm, M., and Todd, J. A. (2009). Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324 (5925), 387–389. doi:10.1126/science.1167728
Racz, E., Prens, E. P., KantM., , , FlorEncia, E., Jaspers, N. G., and Laman, J. D., (2011). Narrowband ultraviolet B inhibits innate cytosolic double-stranded RNA receptors in psoriatic skin and keratinocytes. Br. J. Dermatol. 164 (4), 838–847. doi:10.1111/j.1365-2133.2010.10169.x
Reeves, E., and James, E. (2018). The role of polymorphic ERAP1 in autoinflammatory disease. Biosci. Rep. 38 (4), BSR20171503. doi:10.1042/BSR20171503
Ritchie, M. D., Hahn, L. W., RoodiN., , , Bailey, L. R., Dupont, W. D., Parl, F. F., et al. (2001). Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 69 (1), 138–147. doi:10.1086/321276
Ritchie, M. D., White, B. C., Parker, J. S., Hahn, L. W., and Moore, J. H. (2003). Optimization of neural network architecture using genetic programming improves detection and modeling of gene-gene interactions in studies of human diseases. BMC Bioinforma. 4, 28. doi:10.1186/1471-2105-4-28
Schwender, H. (2012). Imputing missing genotypes with weighted k nearest neighbors. J. Toxicol. Environ. Health. A 75 (8-10), 438–446. doi:10.1080/15287394.2012.674910
Strange, A., Capon, F., Spencer, C. C. A., Knight, J., Weale, M. E., Allen, M. H., et al. (2010). A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42 (11), 985–990. doi:10.1038/ng.694
Sun, L.-D., Cheng, H., Wang, Z. X., Zhang, A. P., Wang, P. G., and Xu, J. H., (2010). Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat. Genet. 42 (11), 1005–1009. doi:10.1038/ng.690
Tang, H., Jin, X., Li, Y., Jiang, H., Tang, X., and Yang, X., (2014). A large-scale screen for coding variants predisposing to psoriasis. Nat. Genet. 46 (1), 45–50. doi:10.1038/ng.2827
Tsui, F. W. L., Haroon, N., Reveille, J. D., Rahman, P., Chiu, B., and Tsui, H. W., (2010). Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Ann. Rheum. Dis. 69 (4), 733–736. doi:10.1136/ard.2008.103804
Wu, X., and Zhao, Z. (2021). Associations between ERAP1 gene polymorphisms and psoriasis susceptibility: A meta-analysis of case-control studies. Biomed. Res. Int. 2021, 5515868. doi:10.1155/2021/5515868
Yin, X. Y., Zhang, R., Cheng, H., Pan, Q., Shen, C. B., and Fan, X., (2013). Gene-gene interactions between HLA-C, ERAP1, TNFAIP3 and TRAF3IP2 and the risk of psoriasis in the Chinese Han population. Br. J. Dermatol. 169 (4), 941–943. doi:10.1111/bjd.12442
Zhang, X. (2012). Genome-wide association study of skin complex diseases. J. Dermatol. Sci. 66 (2), 89–97. doi:10.1016/j.jdermsci.2012.02.017
Zhang, X. J., Huang, W., Yang, S., Sun, L. D., Zhang, F. Y., and Zhu, Q. X., (2009). Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41 (2), 205–210. doi:10.1038/ng.310
Zhang, Y., Yang, J., Zhang, J., Sun, L., Hirankarn, N., and Pan, H. F., (2016). Genome-wide search followed by replication reveals genetic interaction of CD80 and ALOX5AP associated with systemic lupus erythematosus in Asian populations. Ann. Rheum. Dis. 75 (5), 891–898. doi:10.1136/annrheumdis-2014-206367
Keywords: psoriasis, gene–gene interaction, multifactor dimensionality reduction, genome-wide association studies, IFIH1, ERAP1
Citation: Zhang C, Qin Q, Li Y, Zheng X, Chen W, Zhen Q, Li B, Wang W and Sun L (2022) Multifactor dimensionality reduction reveals the effect of interaction between ERAP1 and IFIH1 polymorphisms in psoriasis susceptibility genes. Front. Genet. 13:1009589. doi: 10.3389/fgene.2022.1009589
Received: 02 August 2022; Accepted: 18 October 2022;
Published: 08 November 2022.
Edited by:
Ahmed Rebai, Centre of Biotechnology of Sfax, TunisiaReviewed by:
Antonio Victor Campos Coelho, Albert Einstein Israelite Hospital, BrazilCopyright © 2022 Zhang, Qin, Li, Zheng, Chen, Zhen, Li, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjun Wang, d2FuZ3dqMDcyMEBzaW5hLmNvbQ==; Liangdan Sun, YWhtdXNsZEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.