
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 07 December 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1004467
This article is part of the Research TopicFunctional screening for cancer drug discovery: from experimental approaches to data integrationView all 10 articles
DDX56, a member of the RNA helicase family, is upregulated in colon adenocarcinoma, lung squamous cell carcinoma, and osteosarcoma. However, the relationships between DDX56 and other tumors are not clear, and the molecular mechanism of its action is not fully understood. Here, we explore the biological functions of DDX56 in 31 solid tumors and clarify that DDX56 can promote oncogenesis and progression in multiple tumor types based on multi-omics data. Bioinformatics analysis revealed that the cancer-promoting effects of DDX56 were achieved by facilitating tumor cell proliferation, inhibiting apoptosis, inducing drug resistance, and influencing immune cell infiltration. Furthermore, we found that copy number alterations and low DNA methylation of DDX56 were likely to be related to aberrantly high DDX56 expression. Our results suggest that DDX56 is a potential pan-cancer biomarker that could be used to predict survival and response to therapy, as well as a potential novel therapeutic target. We validated some of our results and illustrated their reliability using CRISPR Screens data. In conclusion, our results clarify the role of DDX56 in the occurrence and development of multiple cancers and provide insight into the molecular mechanisms involved in the process of pathogenesis, indicating a direction for future research on DDX56 in cancers.
Cancer poses a great threat to human health and is a leading cause of death, with more than 19.3 million people diagnosed with cancer and more than 10.0 million deaths each year (Sung et al., 2021). The identification of key molecular targets in various cancers has helped to enhance treatment effects and improve the prognosis of cancer patients. For instance, sorafenib, which can inhibit multiple tyrosine kinases including VEGFR1, VEGFR2, KIT, and PDGFR-α, is widely used in hepatocellular carcinoma (HCC) and renal cell cancer (Bedard et al., 2020). In breast cancer, individual treatment strategies targeting molecular subtypes have dramatically improved survival outcomes in 70–80% of patients (Loibl et al., 2021). These reports show that identification of critical molecules can lead to innovation in cancer therapeutic strategies, with massive application potential. Therefore, the exploration of new key molecules and underlying mechanisms is of great significance.
In addition to molecular targeted therapies, immunotherapy is a prospective treatment for multiple cancer types. The focus of immunotherapy has shifted from the tumor itself to the host’s immune system and tumor microenvironment, with the aim of mobilizing immune cells to discern and eventually eliminate cancer cells (Sharma et al., 2017). Immunotherapy based on immune checkpoint inhibitors has dramatically changed management strategies for various advanced cancers, including non-small-cell lung cancer, extensive small-cell lung cancer, HCC, and classical Hodgkin lymphoma) (Lee et al., 2022). Combination therapy with anti-CTLA4 and anti-PD-1 checkpoint inhibition is an effective option for advanced melanoma and unresectable malignant pleural mesothelioma (Baas et al., 2021; Carlino et al., 2021). Treatment targeting LAG3 has also shown good response (Andrews et al., 2017). However, not all patients can benefit from immunotherapy, and there is a lack of effective markers to predict response to immunotherapy. Therefore, it is urgent to screen more therapeutic targets and identify predictive biomarkers of immunotherapy.
DDX56 is a member of the DEAD box RNA helicase family that plays a key part in various RNA-related biological processes (Cordin et al., 2006; Linder and Jankowsky, 2011). Previous studies have shown that DDX56 can promote the occurrence and development of colon cancer, osteosarcoma, glioblastoma, and lung squamous cell carcinoma (Kouyama et al., 2019; Zhu et al., 2020; Pryszlak et al., 2021; Wu et al., 2021). However, the role of DDX56 in other tumors has not been reported. In addition, it has been reported that other members of DEAD box RNA helicase family can induce resistance of tumor cells to chemotherapeutic agents (Park et al., 2018; Mani et al., 2020). Whether DDX56 contributes to tumor progression or could be used as a biomarker remains to be determined. Here, based on bioinformatics analysis of multi-omics data, we illustrate that DDX56 is involved in the occurrence and development of multiple tumors. Further, we conduct co-expression and enrichment analyses of the biological functions of DDX56 in various solid cancers. We also investigate the potential associations between DDX56 expression and immune infiltration levels and immune-related markers. Finally, we explore the possible mechanisms of high DDX56 expression in tumor tissues. Our results demonstrate the role of DDX56 in oncogenesis in multiple tumors and its potential to serve as a therapeutic target and prognostic indicator.
We compared DDX56 RNA expression among different tissues using RNA sequencing (RNA-seq) datasets from The Cancer Genome Atlas (TCGA). RNA-seq data (TPM) and related clinical data were downloaded from UCSC Xena (http://xena.ucsc.edu/) (Goldman et al., 2020). The data corresponded to 31 solid tumor types: adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS), uveal melanoma (UVM). Using the “Gene” module of TISCH (http://tisch.comp-genomics.org/search-gene/), we determined the RNA expression of DDX56 in multiple cell types based on single-cell RNA-seq data (Sun et al., 2021). UALCAN carried out a comparative analysis of protein expression (http://ualcan.path.uab.edu/index.html) (Chandrashekar et al., 2017). Only tumors with matched normal tissue data were used for differential analysis. We defined clinical stages as follows: early stages, TNM stages I/II; advanced stages, TNM stages III/IV.
Univariate Cox regression was used to assess the prognostic significance of DDX56 across cancer types. Multivariate Cox regression was used to identify independent prognostic factors. The surv_cutpoint function (from R package “survminer”, https://github.com/kassambara/survminer) was used to determine the optimal cutoff values of DDX56 expression level. Only solid tumor data with complete clinical information were included in the survival analysis.
Genes co-expressed with DDX56 were screened by calculating the Spearman correlation coefficient between DDX56 and all other genes in each cancer. The Benjamini–Hochberg method was used to decrease the false discovery rate (adjusted p-value). To gain functional and mechanistic insights regarding DDX56, we performed a pre-ranked gene set enrichment analysis (GSEA) (Subramanian et al., 2005) based on Hallmarker’s gene sets, which were downloaded from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/) (Liberzon et al., 2015). The genes with significant correlation coefficients (adjusted p < 0.05) were sorted according to Spearman correlation coefficient and then used in GSEA.
The CRISPR pooled libraries consist of thousands of plasmids, each containing multiple gRNAs for each target gene (Shalem et al., 2014). In a CRISPR screening experiment, target cells are treated with the pooled library to create a population of mutant cells that are then screened for a phenotype of interest. Essential genes for specific phenotypic screens can be curated by comparing sgRNA abundance. Therefore, the database includes research information on whether a particular gene is essential for a certain phenotype in a particular tumor cell line (Joung et al., 2017). Compiled CRISPR screen data were obtained from the Biological General Repository for Interaction Datasets (BioGRID) (https://orcs.thebiogrid.org/) (Oughtred et al., 2019). Data from proliferation-based CRISPR screens in solid tumor cancer cell lines were selected. These datasets were used to verify the mitogenic activity of DDX56. Resistance-related CRISPR screening evidence was also retrieved from the BioGRID database. The methods were as previously described except that the studies chosen focused on the “response to chemicals” phenotype.
Half-maximal inhibitory concentration (IC50) data and associated RNA-seq data from multiple cancer cell lines were obtained from the CellMiner database (https://discover.nci.nih.gov/cellminer/) (Reinhold et al., 2012). We conducted Spearman correlation analysis between DDX56 expression and drug IC50 in order to investigate the relationship between DDX56 expression and drug sensitivity.
Proportions of TILs were estimated using the CIBERSORT function (https://cibersort.stanford.edu/). (Newman et al., 2015; Chen et al., 2018). Based on the Spearman correlation coefficient, we assessed the statistical correlation between DDX56 expression level and the proportion of TILs.
We evaluated the value of DDX56 in predicting immunotherapy response in an immune checkpoint blockade therapy cohort. The transcriptomic data and relevant clinical data from this cohort were obtained from dbGaP (phs000452) (Liu et al., 2019a). Univariate Cox regression was used to assess the prognostic significance of DDX56.
We conducted gene mutation analysis on cBioportal (https://www.cbioportal.org/) based on TCGA Pan-Cancer Atlas data (Cerami et al., 2012). The correlation of DDX56 RNA expression level with copy number variation (CNV) was evaluated by MEXPRESS (https://mexpress.be/) (Koch et al., 2019). The correlation between the expression data and the DNA methylation data was estimated by using MEXPRESS (https://mexpress.be/). Oncodb (http://oncodb.org/) was used to compare methylation profiles between tumor tissues and normal tissues (Koch et al., 2019; Tang et al., 2022). We used the Toolkit for CistromeDB (http://dbtoolkit.cistrome.org/) to predict which transcription factors had the greatest potential to enhance expression of DDX56 (Zheng et al., 2019).
We compared non-normally distributed continuous variables using Wilcoxon rank-sum test and compared categorical variables using chi-square test between two groups. Kaplan–Meier and log-rank tests were conducted for survival analysis. Unless otherwise stated, all data analysis was performed in R (version 4.1.0). Unless otherwise specified, p < 0.05 was considered to indicate statistical significance in all analyses. The tumors involved in each analysis are recorded in Supplementary Table S1.
We compared DDX56 RNA expression between different cancer tissues and matched normal tissues based on TCGA data. The pan-cancer analysis showed that the DDX56 RNA expression level was higher in 16 solid cancers than in their matched normal tissues (Wilcoxon test, p < 0.05) (Figure 1A), consistent with findings in lung squamous cell carcinoma (LUSC), osteosarcoma (OV), and colon cancer (COAD) (Zhu et al., 2020; Pryszlak et al., 2021; Wu et al., 2021). UALCAN was used to determine the protein expression of DDX56 in different types of cancer. According to significant unique analyses (provided by UALCAN, Student’s t-test), DDX56 protein was significantly over-expressed in nine tumor types (p < 0.05, Figure 1B). An effective tumor biomarker and drug target requires specific and high expression in tumor cells compared with other components of the tumor microenvironment. Therefore, we analyzed pan-cancer single-cell RNA-seq data using the TISCH database and observed that DDX56 was mainly expressed in tumor cells rather than immune cells, stromal cells, and others (Figure 1C), indicating the potential of DDX56 as a tumor biomarker and drug target.
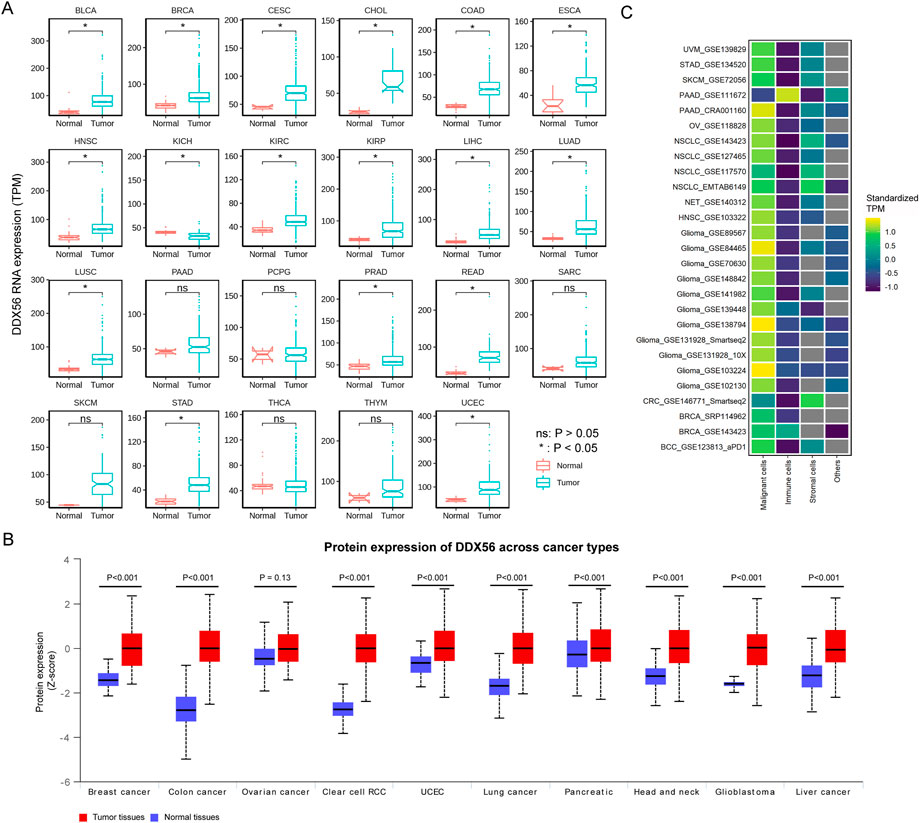
FIGURE 1. RNA and protein expression of DDX56 in various tumors. (A) Box plots showing RNA expression of DDX56 in different tissues (Wilcoxon test). P-values are marked. (B) Box plots showing protein expression levels of DDX56 in different tissues (T test). P-values are marked. (C) Heat map showing RNA expression in different cell types of various tumors. TPM values were standardized for each tumor. The colors indicate the TPM value after standardization. The X-axis indicates single-cell RNA-seq datasets. The Y-axis indicates the cell type. Abbreviations: BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; BCC, basal cell carcinoma; NET, Neuroendocrine tumor; CRC, colorectal cancer; RCC, renal cell carcinoma.
Next, we assessed the correlation between DDX56 expression and the clinical features of patients. Although few significant differences were observed between groups in age or gender (Supplementary Figure S1), the expression level of DDX56 was consistently higher in patients in the advanced stages than those in the early stages of diseases including ACC, BRCA, COAD, HNSC, KIRC, and LUAD (Wilcoxon test, p < 0.05, Figure 2A and Supplementary Figure S1). Further, we investigated the prognostic value of DDX56 at a pan-cancer level. The results showed that high DDX56 expression was correlated with worse outcomes in 11 cancer types (univariate Cox regression, p < 0.05, hazard ratio >1) (Figure 2B and Supplementary Figure S2). Then, we implemented multivariate Cox regression analysis based on DDX56 expression level and other clinical factors and found that DDX56 was an independent predictor for cancer prognosis in nine tumor types; moreover, worse clinical outcomes in patients with higher expression of DDX56 were observed in eight tumor types, comprising ACC, HNSC, KICH, KIRC, LIHC, LUAD, THCA, and UVM (Figure 2C). These results suggest that DDX56 high expression may be a significant predictor of prognosis and function as a promotor in multiple tumor types.
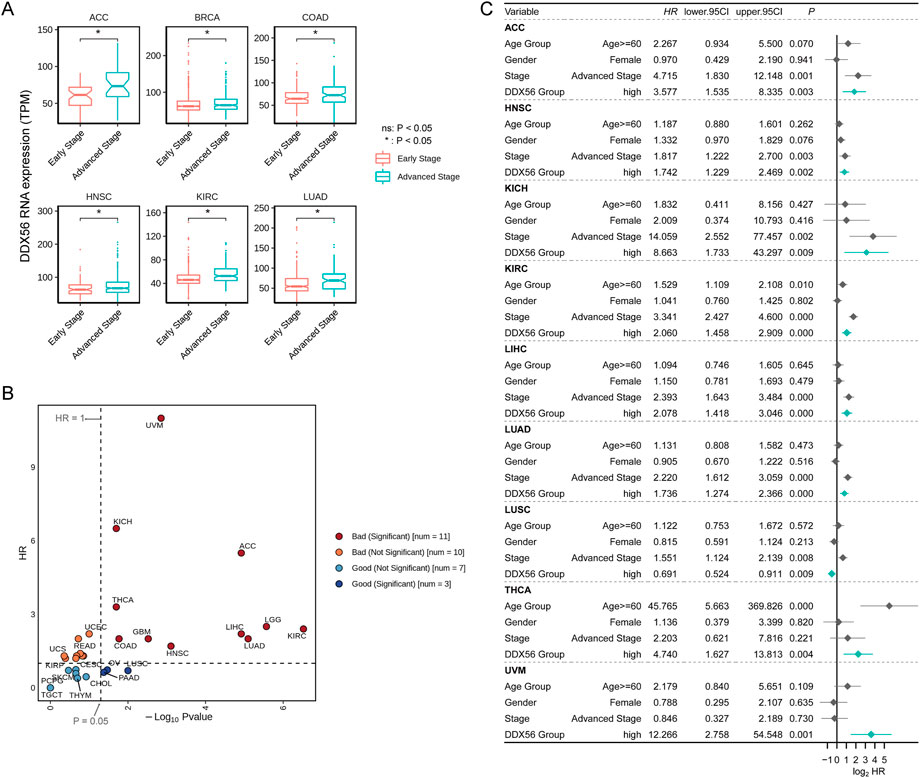
FIGURE 2. Prognostic value of DDX56 in various cancers. (A) Box plots showing RNA expression levels of DDX56 at different clinical stages. For each tumor, stages Ⅰ and Ⅱ were classified as early stage; stages Ⅲ and Ⅳ were classified as advanced stage. (B) Scatter plot showing outcomes in cancer patients with different DDX56 expression levels. Statistics were obtained by univariate Cox regression analysis. (C) Forest plot showing the results of multivariate Cox regression for multiple tumor types. Abbreviations: ACC, adrenocortical carcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
To explore the molecular mechanism of DDX56 in carcinogenesis, a network of genes co-expressed with DDX56 was first conducted in each tumor dataset, and then pathway enrichment analysis was performed using GSEA. The results revealed that DDX56 mainly participated in cell-proliferation-related signaling pathways, including G2/M checkpoint, MYC targets v1, MYC targets v2, and E2F targets (Figure 3A) (Liberzon et al., 2015). Concomitantly, we observed enrichment of tumor metabolism-related pathways including oxidative phosphorylation and unfolded protein response. In order to validate the possible pro-proliferation function of DDX56, we collected and analyzed CRISPR screens data from the BioGRID Open Respository of CRISPR Screens. CRISPR-based genetic screening is a powerful tool to identify the genes required for specific functions such as cell viability and chemical resistance. We found 512 CRISPR screens studies in 19 tumor types, which confirmed the effects of DDX56 on the proliferation of tumor cells at the pan-cancer level (Figure 3B and Supplementary Table S2).
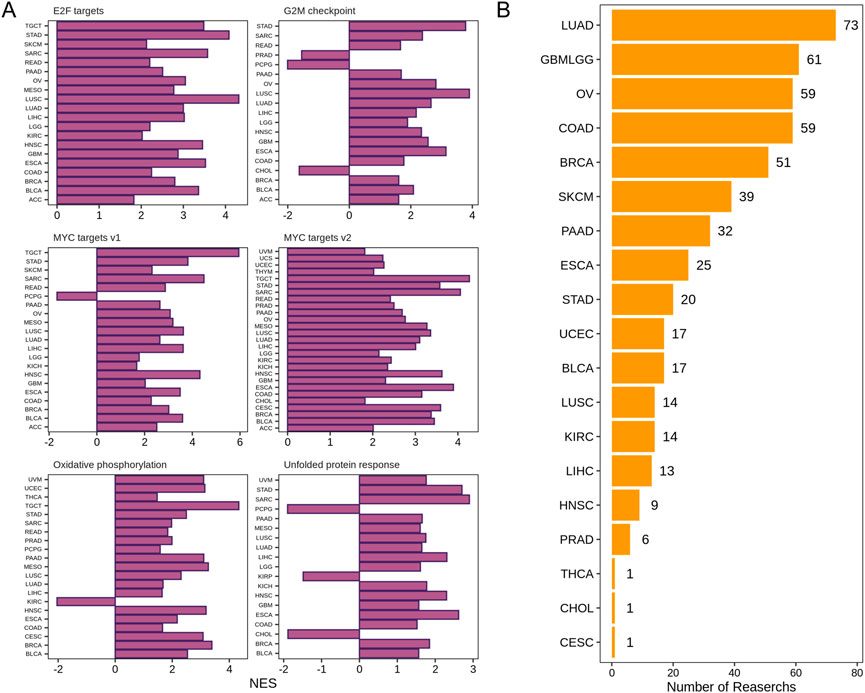
FIGURE 3. DDX56 positively modulates the proliferation of cancer cells. (A) GSEA enrichment results for DDX56 in multiple tumor types. Only pathways deemed to be significantly enriched based on GSEA (p < 0.05) are illustrated. (B) Multiple CRISPR screening studies verifying that DDX56 can positively regulate the proliferation of cancer cells. X-axis indicates the number of CRISPR screening studies supporting a pro-proliferative phenotypic role of this gene in tumor cell lines. Y-axis indicates the tumor type of the CRISPR screens studies. Abbreviations: ACC: adrenocortical carcinoma; BRCA: breast invasive carcinoma; COAD: colon adenocarcinoma; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MESO: mesothelioma; OV: ovarian serous cystadenocarcinoma; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; SARC: sarcoma; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THCA: thyroid carcinoma; THYM: thymoma; UCEC: uterine corpus endometrial carcinoma; UCS: uterine carcinosarcoma; UVM: uveal melanoma.
As the GSEA results had shown enrichment of unfolded protein response (Figure 3A), a chemoresistance-related pathway, (Bahar et al., 2019; Sisinni et al., 2019), we speculated that DDX56 was associated with chemotherapy resistance. To investigate whether DDX56 was relevant to drug resistance, we searched the CRISPR screens evidence in the BioGRID database. We found that DDX56 was an essential gene required for drug resistance in five different tumors (Figure 4A and Supplementary Table S3). Moreover, we obtained IC50 levels of all available drugs for tumor cell lines from the CellMiner database and calculated the Spearman correlation coefficient between the IC50 values and DDX56 RNA expression in matched tumor cell lines. Our results revealed that high expression of DDX56 was correlated with increased resistance to cladribine, entinostat, amuvatinib, triethylenemelamine, irofulven, IWR-1, JZL-195, XL-147, and Cpd-401 (p < 0.05, Figure 4B). On the other hand, high expression of DDX56 could lead to enhanced sensitivity to a geldanamycin analog and alectinib (p < 0.05, Figure 4B).
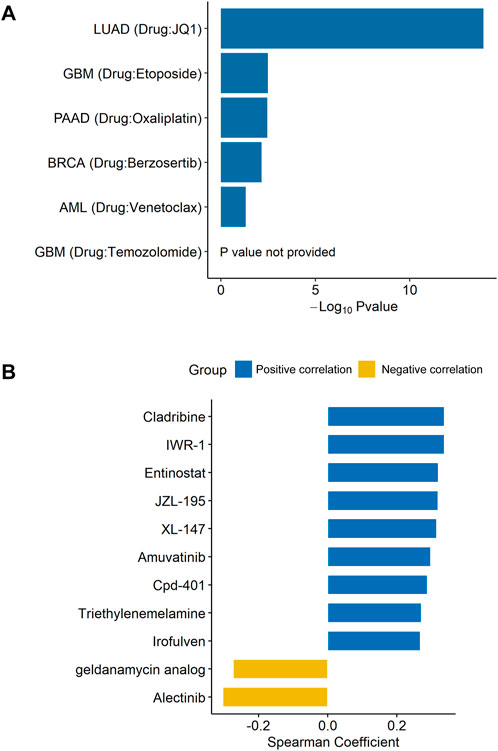
FIGURE 4. DDX56 promotes drug resistance in multiple tumor types. (A) Multiple CRISPR screening studies showing that DDX56 is associated with drug resistance. Y-axis indicates the tumor type and the associated drug. X-axis indicates the logarithm of the negative p-value of DDX56 in each CRISPR screening study. (B) Bar plot showing the Spearman coefficients for DDX56 RNA expression and drug IC50. Eleven statistically significant results were obtained from 450 drugs in the screen. Abbreviations: LUAD: lung adenocarcinoma; GBM: glioblastoma multiforme; PAAD: pancreatic adenocarcinoma; BRCA: breast invasive carcinoma; AML: acute myeloid leukemia.
The GSEA results suggested that DDX56 may have a critical role in inhibiting cell apoptosis and suppressing antitumor immunity (IL-2 STAT5 signaling, IL-6 JAK STAT3 signaling, and interferon gamma response, Figure 5A) (Liberzon et al., 2015). To further explore the potential relationships between DDX56 and immune cells, we examined the correlations between DDX56 and several immune cell markers including immune cells (PTPRC), T cells (CD3D, CD4, CD8A), B cells (CD19), and MHC class II molecules (Supplementary Figure S3). The results suggested that the expression of DDX56 was associated with immune infiltration in the tumor microenvironment. Next, we estimated the proportions of immune cells in each TCGA tumor type by using CIBERSORT. We observed that DDX56 expression was negatively correlated with immune infiltration levels of plasma cells, resting dendritic cells (DC), and CD4+ memory T cells (Figure 5B). Given that DDX56 participates in the immune infiltration process, it may have crucial biological functions in immunotherapy. Using transcriptome and clinical data from a recently published immunotherapy study (Liu et al., 2019b), we found that anti-PD-1-treated patients with high DDX56 expression had shorter progression-free survival than those with low DDX56 expression (Figure 5C).
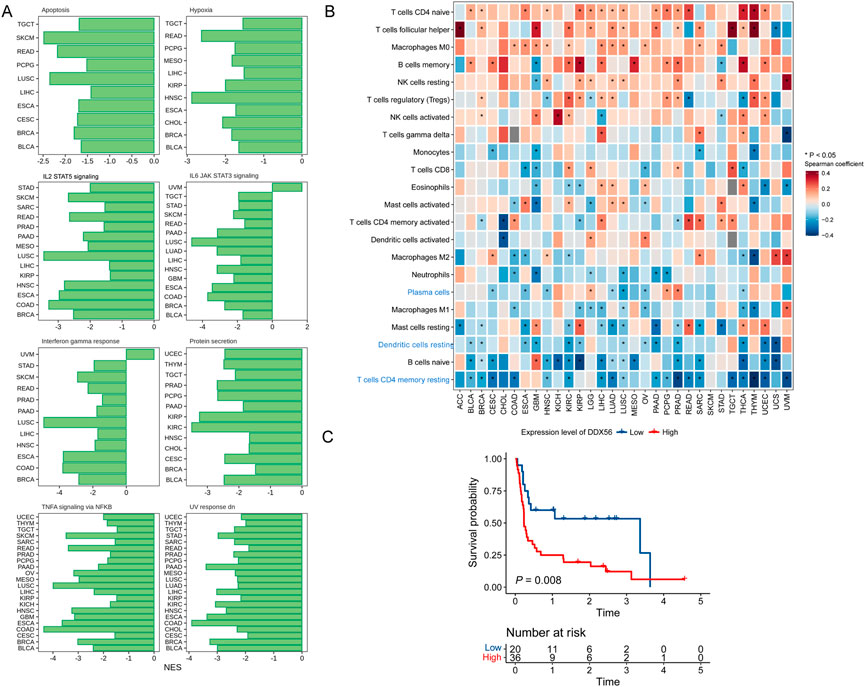
FIGURE 5. Relationships of DDX56 with tumor immune features. (A) GSEA enrichment results for DDX56 in multiple tumor types. Only pathways deemed significant enriched based on GSEA (p < 0.05) are illustrated. (B) Heat map showing Spearman coefficients for DDX56 RNA expression and proportions of tumor-infiltrating immune cells. (C) Kaplan–Meier plots showing worse clinical outcome in immunotherapy cohort patients with higher expression of DDX56. Univariate Cox regression was used to assess the statistics.
The occurrence and development of cancer is related to gene alterations. To determine the genomic characteristics of DDX56 in cancers, comparative analysis of DDX56 was performed using cBioPortal. We observed that the main alterations of DDX56 were amplifications and mutations in multiple tumors (Figure 6A). The most common mutation type was missense mutation, which could lead to alteration of protein structure and functions (Figure 6B). As CNV can lead to higher gene mRNA levels (Haraksingh and Snyder, 2013), we analyzed the correlation between RNA expression and CNV at the pan-cancer levels. The results showed that the CNV and RNA expression of DDX56 DNA in tumor tissues were significantly associated (Pearson coefficient >0.2, p < 0.05, Figure 6C). In addition, increased RNA transcription may result from low levels of DNA methylation (Bradner et al., 2017). We compared DDX56 methylation status between various tumors and normal tissues and observed that DDX56 methylation in five tumor types was lower compared with that in paired normal tissues (Wilcoxon test, p < 0.05) (Figure 6D). Furthermore, we estimated the correlation between expression and DNA methylation data using MEXPRESS. We observed that methylation of some CpG dinucleotides and CpG islands, including cg25257687 and cg01998345, was significantly negatively correlated with the RNA expression of DDX56 (Pearson correlation coefficient <0 and p < 0.05, Supplementary Figure S4). Our results suggested that lower methylation levels of DDX56 DNA may result in high expression levels of DDX56 RNA in some cancers.
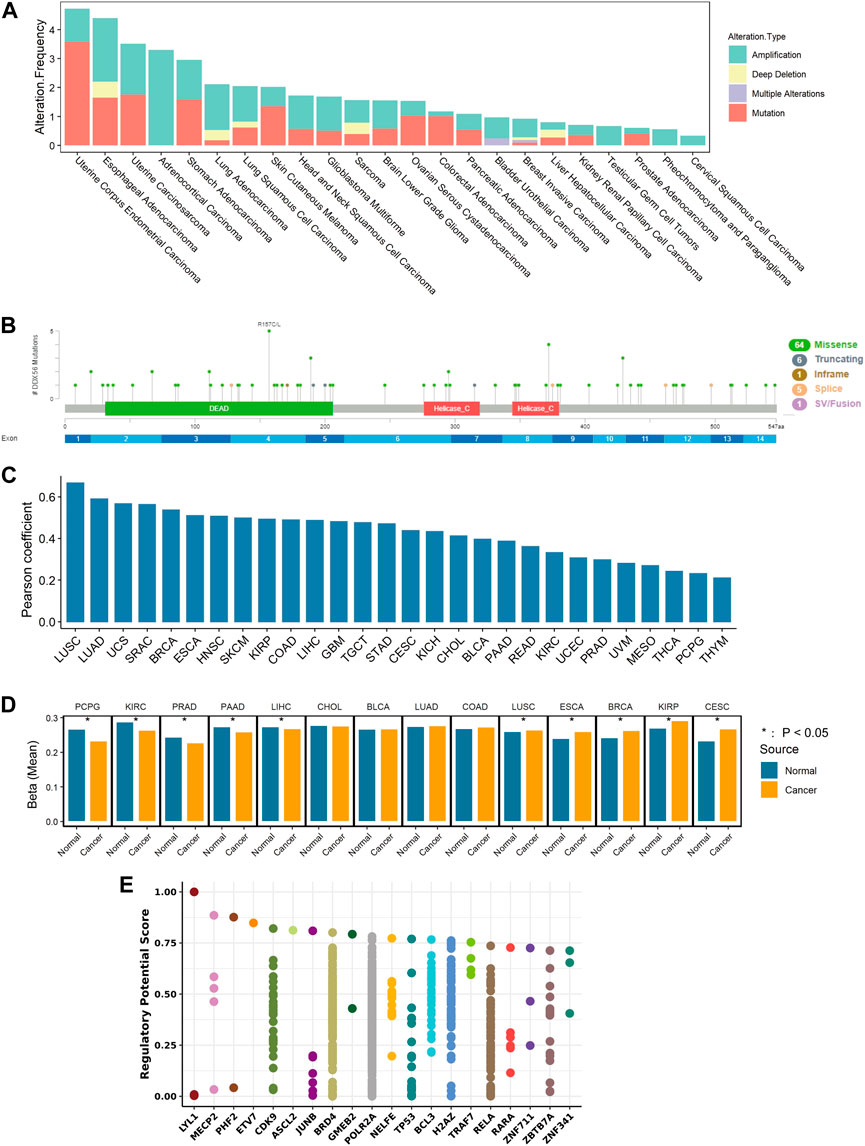
FIGURE 6. Mechanism of regulation of DDX56 expression. (A) Analysis of DDX56 alteration frequency in multiple cancer types, colored by mutation type. (B) Sites of different mutation types of DDX56. (C) Bar plot showing the Pearson correlation of CNV and DDX56 RNA expression in different tumor types. (D) Bar plot showing the DNA methylation status of DDX56 in tumors and normal tissues. Y-axis indicates the mean of the beta value. *p < 0.05. The statistics are from OncoDB. (E) Transcription factors potentially regulating DDX56 (results obtained from Toolkit). The plot illustrates the top 20 factors. Dots on a single axis line indicate the same factor. Abbreviations: ACC: adrenocortical carcinoma; BRCA: breast invasive carcinoma; COAD: colon adenocarcinoma; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MESO: mesothelioma; OV: ovarian serous cystadenocarcinoma; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; SARC: sarcoma; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THCA: thyroid carcinoma; THYM: thymoma; UCEC: uterine corpus endometrial carcinoma; UCS: uterine carcinosarcoma; UVM: uveal melanoma.
Finally, screening was performed to search candidate transcription factors possibly regulating DDX56 expression. According to our results, LYL1, MECP2, PHF2, ETV7, and CDK9 were the top five transcription factors with the potential to alter the expression level of DDX56 (Figure 6E). Furthermore, we estimated the co-expression relation of these top five transcription factors and DDX56 (Spearman correlation, Supplementary Figure S5). We compared the expression levels of the top five transcription factors in tumor tissues and normal tissues (Wilcoxon test, Supplementary Figure S6). For example, we observed that the expression of CDK9 was highly related to the expression of DDX56 in THCA (Spearman correlation coefficient 0.384, p < 0.05, Supplementary Figure S5 and Supplementary Table S4). Simultaneously, we observed that the expression levels of CDK9 and DDX56 were both high in THCA tumor tissue (Wilcoxon test, p < 0.05, Supplementary Figure S6 and Figure 1A). We thus consider that CDK9 might be responsible for the high expression of DDX56 in THCA.
The role of DDX56 in tumorigenesis and development has attracted increasing attention in recent years (Zhu et al., 2020; Pryszlak et al., 2021; Sung et al., 2021; Wu et al., 2021). Although previous studies have reported upregulation of DDX56 in several tumor types, the underlying mechanisms of its pro-oncogenic function remain indistinct. Our results revealed that RNA expression of DDX56 was indeed higher in 16 tumor types, which was confirmed at the protein level in nine tumor types. On this basis, we found that DDX56 may exert pro-oncogenic effects by enhancing proliferation and restraining apoptosis of tumor cells, affecting the infiltration of immune cells into the tumor microenvironment, and inducing tumor drug resistance. Our results suggest that DDX56 is involved in the occurrence and development of multiple cancers, as well as therapeutic response to chemotherapeutic agents. DDX56 could be an independent predictor of prognosis in a variety of tumor types and may also have value in prediction of immunotherapy efficacy.
Here, we report for the first time that the RNA and protein expression levels of DDX56 are significantly higher in tumor tissues than that in control normal tissues in various tumor types. Based on multi-omics data, we found that the high DDX56 expression in tumor tissues may be due to CNV and aberrant methylation. However, these changes were not present in all patients and may only partially explain the high DDX56 expression. Transcription factors are important components that regulate RNA transcription. We screened the potential transcription factors involved in regulating DDX56 using Toolkit. These transcription factors may be partially responsible for the abnormally high expression of DDX56, but more work needs to be done to reach such a conclusion.
We investigated the molecular mechanisms by which DDX56 promotes tumorigenesis and development through functional enrichment analysis. Our results suggested that DDX56 may promote the proliferation of tumor cells and inhibit tumor apoptosis. Zhu et al. showed that the knockdown of DDX56 could reduce the proliferation and promoted the apoptosis of osteosarcoma cells (Zhu et al., 2020). Kouyama et al. (2019) showed that the overexpression of DDX56 could enhance the proliferation of colon cancer cells. These results were consistent with our findings. Wu et al. (2021) further revealed that DDX56 could promote the proliferation of tumor cells through the WNT signaling pathway. Our results suggested that there may also be other molecular mechanisms involved, such as “E2F targets” and “MYC targets”, which have not previously been reported in DDX56-related studies. Therefore, we provide additional insights into the signaling pathways by which DDX56 promotes tumor proliferation.
Our functional enrichment results for DDX56 also suggested that DDX56 may be related to the infiltration of immune cells. We found that high DDX56 expression was closely related to low infiltration levels of DC, plasma cells, and CD4+ T cells. DC are the most powerful antigen-presenting cells and can initiate immune responses (Gardner and Ruffell, 2016; Wculek et al., 2020). As critical mediators in anti-tumor immunity, plasma cells are capable of producing antibodies and CD4+ T cells secrete diverse cytokines that enhance humoral and cellular immunity (Borst et al., 2018; Sharonov et al., 2020). All of them are essential ancillary components in anti-tumor immunity, and their absence is detrimental to the immune reaction to cancer. This may also contribute to the poor prognosis and poor efficacy of immunotherapy in patients with high DDX56 expression. Here, DDX56 was reported for the first time to be associated with infiltration of several immune cell types in the tumor microenvironment.
Combining all the data, we can conclude that DDX56 has a tumor-promoting function in most solid tumors. However, it may promote tumor progression in different ways in different cancer types. Therefore, we summarized the consistency of those aspects among different cancer types (Supplementary Table S3). For instance, there was no evidence that DDX56 could promote the proliferation of pheochromocytoma and paraganglioma (Figure 3A, NSE<0 or insignificant in all proliferation-related pathways). However, we observed that it was related to low level of apoptosis (Figure 5A, Supplementary Table S3) and could affect the infiltration of some immune cell types in pheochromocytoma and paraganglioma (Figure 5B, Supplementary Table S3).
Furthermore, we found that higher expression of DDX56 was associated with worse patient prognosis in multiple tumor types. High DDX56 expression was also found to related to lower efficacy of PD-1 antibody immunotherapy. All these results indicate that DDX56 could be used to predict not only prognosis but also the efficacy of immune checkpoint inhibitors. Our results also indicate that DDX56 may promote multiple anticancer drug resistance in tumor cells, which has not been previously reported.
We acknowledge several limitations of our study. Much more research needs to be done to determine whether DDX56 has the potential to predict prognosis and efficacy of chemotherapy drugs and immunotherapy. Although we has proposed potential pro-tumor molecular mechanisms involving DDX56 based on bioinformatics analysis, we did not perform biological experiments to validate these results. A large amount of CRISPR screens data verified our partial conjecture, giving some credibility to our results (Chow and Chen, 2018). However, the CRISPR screening evidence could not confirm the promoted proliferation ability of DDX56 and further functional experiments are needed to confirm the function of DDX56 and to explore the underlying mechanisms.
In summary, we have clarified the tumor-promoting role of DDX56 based on multi-omics data at a pan-cancer level and elucidated the possible molecular mechanisms involved. These results contribute to our understanding of the biological function of DDX56 in tumors and provide evidence and potential research directions for future studies on DDX56 as an oncogenic driver.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
ZR, YZ, and RP designed the study. ZR and YZ performed the data analyses. All authors contributed to the conception of the study and drafted the manuscript. All authors contributed significantly to writing the manuscript. ZR and YZ contributed equally to this work and share first authorship.
The project was supported by the Basic and Applied Basic Research Fund of Guangdong Province (2022A1515012387).
We are grateful to all the researchers who provided the data we used.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1004467/full#supplementary-material
Supplementary Figure S1 | Comparison of DDX56 expression in populations with different clinical characteristics.
Supplementary Figure S2 | Pan-cancer overall survival analysis of DDX56.
Supplementary Figure S3 | Pan-cancer correlation analysis of DDX56 RNA expression with several cellular marker genes and MHC class II molecules.
Supplementary Figure S4 | Correlation analysis of methylation levels of DDX56 DNA and expression levels of DDX56 RNA.
Supplementary Figure S5 | Pan-cancer correlation analysis of DDX56 RNA expression with top five transcription factors screened by Toolkit (http://dbtoolkit.cistrome.org/).
Supplementary Figure S6 | Bar plots showing median RNA expression levels of top five DDX56-related transcription factors in different tissues (Wilcoxon test). Significant: P<0.05; Insignificant: P>0.05 or P = 0.05.
Supplementary Table S1 | Tumors involved in each analysis.
Supplementary Table S2 | List of CRISPR screening studies showing that DDX56 is associated with the proliferation of cancer cells.
Supplementary Table S3 | List of CRISPR screening studies showing that DDX56 is associated with drug resistance.
Supplementary Table S4 | Results of pan-cancer correlation analysis of DDX56 RNA expression with top five transcription factors screened by Toolkit.
Supplementary Table S5 | Summary of the results.
Andrews, L. P., Marciscano, A. E., Drake, C. G., and Vignali, D. A. (2017). LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 276, 80–96. doi:10.1111/imr.12519
Baas, P., Scherpereel, A., Nowak, A. K., Fujimoto, N., Peters, S., Tsao, A. S., et al. (2021). First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): A multicentre, randomised, open-label, phase 3 trial. Lancet 397, 375–386. doi:10.1016/S0140-6736(20)32714-8
Bahar, E., Kim, J. Y., and Yoon, H. (2019). Chemotherapy resistance explained through endoplasmic reticulum stress-dependent signaling. Cancers (Basel) 11 (3), 338. doi:10.3390/cancers11030338
Bedard, P. L., Hyman, D. M., Davids, M. S., and Siu, L. L. (2020). Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 395, 1078–1088.
Borst, J., Ahrends, T., Bąbała, N., Melief, C. J. M., and Kastenmüller, W. (2018). CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 18, 635–647. doi:10.1038/s41577-018-0044-0
Bradner, J. E., Hnisz, D., and Young, R. A. (2017). Transcriptional addiction in cancer. Cell 168, 629–643. doi:10.1016/j.cell.2016.12.013
Carlino, M. S., Larkin, J., and Long, G. V. (2021). Immune checkpoint inhibitors in melanoma. Lancet 398, 1002–1014. doi:10.1016/S0140-6736(21)01206-X
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. doi:10.1158/2159-8290.CD-12-0095
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. a. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M., and Alizadeh, A. A. (2018). Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 1711, 243–259. doi:10.1007/978-1-4939-7493-1_12
Chow, R. D., and Chen, S. (2018). Cancer CRISPR screens in vivo. Trends Cancer 4, 349–358. doi:10.1016/j.trecan.2018.03.002
Cordin, O., Banroques, J., Tanner, N. K., and Linder, P. (2006). The DEAD-box protein family of RNA helicases. Gene 367, 17–37. doi:10.1016/j.gene.2005.10.019
Gardner, A., and Ruffell, B. (2016). Dendritic cells and cancer immunity. Trends Immunol. 37, 855–865. doi:10.1016/j.it.2016.09.006
Goldman, M. J., Craft, B., Hastie, M., Repečka, K., Mcdade, F., Kamath, A., et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678. doi:10.1038/s41587-020-0546-8
Haraksingh, R. R., and Snyder, M. P. (2013). Impacts of variation in the human genome on gene regulation. J. Mol. Biol. 425, 3970–3977. doi:10.1016/j.jmb.2013.07.015
Joung, J., Konermann, S., Gootenberg, J. S., Abudayyeh, O. O., Platt, R. J., Brigham, M. D., et al. (2017). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863. doi:10.1038/nprot.2017.016
Koch, A., Jeschke, J., Van criekinge, W., Van engeland, M., and De meyer, T. (2019). MEXPRESS update 2019. Nucleic Acids Res. 47, W561–W565. doi:10.1093/nar/gkz445
Kouyama, Y., Masuda, T., Fujii, A., Ogawa, Y., Sato, K., Tobo, T., et al. (2019). Oncogenic splicing abnormalities induced by DEAD-Box Helicase 56 amplification in colorectal cancer. Cancer Sci. 110, 3132–3144. doi:10.1111/cas.14163
Lee, J. B., Kim, H. R., and Ha, S. J. (2022). Immune checkpoint inhibitors in 10 Years: Contribution of basic research and clinical application in cancer immunotherapy. Immune Netw. 22, e2. doi:10.4110/in.2022.22.e2
Liberzon, A., Birger, C., Thorvaldsdóttir, H., Ghandi, M., Mesirov, J. P., and Tamayo, P. (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. doi:10.1016/j.cels.2015.12.004
Linder, P., and Jankowsky, E. (2011). From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516. doi:10.1038/nrm3154
Liu, D., Schilling, B., Liu, D., Sucker, A., Livingstone, E., Jerby-Arnon, L., et al. (2019a). Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927. doi:10.1038/s41591-019-0654-5
Liu, D., Schilling, B., Liu, D., Sucker, A., Livingstone, E., Jerby-Arnon, L., et al. (2019b). Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927. doi:10.1038/s41591-019-0654-5
Loibl, S., Poortmans, P., Morrow, M., Denkert, C., and Curigliano, G. (2021). Breast cancer. Lancet 397, 1750–1769. doi:10.1016/S0140-6736(20)32381-3
Mani, S. K. K., Yan, B., Cui, Z., Sun, J., Utturkar, S., Foca, A., et al. (2020). Restoration of RNA helicase DDX5 suppresses Hepatitis B virus (HBV) biosynthesis and Wnt signaling in HBV-related hepatocellular carcinoma. Theranostics 10, 10957–10972. doi:10.7150/thno.49629
Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., et al. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457. doi:10.1038/nmeth.3337
Oughtred, R., Stark, C., Breitkreutz, B. J., Rust, J., Boucher, L., Chang, C., et al. (2019). The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–d541. doi:10.1093/nar/gky1079
Park, S. Y., Kim, W. J., Byun, J. H., Lee, J. J., Jeoung, D., Park, S. T., et al. (2018). Role of DDX53 in taxol-resistance of cervix cancer cells in vitro. Biochem. Biophys. Res. Commun. 506, 641–647. doi:10.1016/j.bbrc.2018.10.145
Pryszlak, M., Wiggans, M., Chen, X., Jaramillo, J. E., Burns, S. E., Richards, L. M., et al. (2021). The DEAD-box helicase DDX56 is a conserved stemness regulator in normal and cancer stem cells. Cell Rep. 34, 108903. doi:10.1016/j.celrep.2021.108903
Reinhold, W. C., Sunshine, M., Liu, H., Varma, S., Kohn, K. W., Morris, J., et al. (2012). CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72, 3499–3511. doi:10.1158/0008-5472.CAN-12-1370
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelson, T., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. doi:10.1126/science.1247005
Sharma, P., Hu-Lieskovan, S., Wargo, J. A., and Ribas, A. (2017). Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723. doi:10.1016/j.cell.2017.01.017
Sharonov, G. V., Serebrovskaya, E. O., Yuzhakova, D. V., Britanova, O. V., and Chudakov, D. M. (2020). B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol. 20, 294–307. doi:10.1038/s41577-019-0257-x
Sisinni, L., Pietrafesa, M., Lepore, S., Maddalena, F., Condelli, V., Esposito, F., et al. (2019). Endoplasmic reticulum stress and unfolded protein response in breast cancer: The balance between apoptosis and autophagy and its role in drug resistance. Int. J. Mol. Sci. 20, 857. doi:10.3390/ijms20040857
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi:10.1073/pnas.0506580102
Sun, D., Wang, J., Han, Y., Dong, X., Ge, J., Zheng, R., et al. (2021). Tisch: A comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 49, D1420–d1430. doi:10.1093/nar/gkaa1020
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249.
Tang, G., Cho, M., and Wang, X. (2022). OncoDB: An interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 50, D1334–d1339. doi:10.1093/nar/gkab970
Wculek, S. K., Cueto, F. J., Mujal, A. M., Melero, I., Krummel, M. F., and Sancho, D. (2020). Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 20, 7–24. doi:10.1038/s41577-019-0210-z
Wu, Q., Luo, X., Terp, M. G., Li, Q., Li, Y., Shen, L., et al. (2021). DDX56 modulates post-transcriptional Wnt signaling through miRNAs and is associated with early recurrence in squamous cell lung carcinoma. Mol. Cancer 20, 108. doi:10.1186/s12943-021-01403-w
Zheng, R., Wan, C., Mei, S., Qin, Q., Wu, Q., Sun, H., et al. (2019). Cistrome data browser: Expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 47, D729–d735. doi:10.1093/nar/gky1094
Keywords: DDX56, pan-cancer, multi-omics data, bio-marker, survival
Citation: Ruan Z, Zhang Y, Quan Q, Jiang J, Wang Q, Zhang Y and Peng R (2022) Pan-cancer analysis identifies DDX56 as a prognostic biomarker associated with immune infiltration and drug sensitivity. Front. Genet. 13:1004467. doi: 10.3389/fgene.2022.1004467
Received: 27 July 2022; Accepted: 24 November 2022;
Published: 07 December 2022.
Edited by:
Hua Tan, National Human Genome Research Institute (NIH), United StatesReviewed by:
Avantika Lal, National Centre for Biological Sciences, IndiaCopyright © 2022 Ruan, Zhang, Quan, Jiang, Wang, Zhang and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roujun Peng, cGVuZ3JqQHN5c3VjYy5vcmcuY24=; Yujing Zhang, emhhbmd5akBzeXN1Y2Mub3JnLmNu
†These authors contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.