- Genome Integrity, Immunity and Cancer Unit, Institut Pasteur, Université de Paris, INSERM U1223, Equipe Labellisée Ligue Contre Le Cancer, Paris, France
DNA double-strand breaks (DSBs) are highly toxic lesions that can be mended via several DNA repair pathways. Multiple factors can influence the choice and the restrictiveness of repair towards a given pathway in order to warrant the maintenance of genome integrity. During V(D)J recombination, RAG-induced DSBs are (almost) exclusively repaired by the non-homologous end-joining (NHEJ) pathway for the benefit of antigen receptor gene diversity. Here, we review the various parameters that constrain repair of RAG-generated DSBs to NHEJ, including the peculiarity of DNA DSB ends generated by the RAG nuclease, the establishment and maintenance of a post-cleavage synaptic complex, and the protection of DNA ends against resection and (micro)homology-directed repair. In this physiological context, we highlight that certain DSBs have limited DNA repair pathway choice options.
Introductory Remarks
The integrity of a cell’s genome is continuously threatened by exogenous or endogenous factors generating DNA damage of various nature, which can impact a single nucleotide or result in lesions of the DNA backbone. Independently of the type or circumstances leading to the DNA damage, it must robustly be sensed, signaled, and repaired, ideally resulting in no or minimal alterations to the genetic code and recovery of an intact genome. Mammalian cells are equipped with several molecular tool kits warranting efficient repair of damaged DNA, where the nature of the DNA lesion largely dictates the selected repair apparatus. Nevertheless, multiple DNA repair pathways exist for a single type of damage such as the case for the mending of DNA double-strand breaks (DSBs). DNA DSBs are often considered as the most deleterious form of DNA damage for a cell, resulting in the physical separation of DNA molecules. Failure to accurately repair DSBs can lead to cell death or to DNA structural changes (i.e., loss of genetic material, sequence alterations or joining of the wrong couple of DNA ends generating chromosomal translocations) potentially triggering carcinogenesis or onset of pathologies, including neurodegenerative diseases or immunodeficiencies (Mitelman et al., 2007; Jackson and Bartek, 2009; Ciccia and Elledge, 2010; Goldstein and Kastan, 2015). Maybe recklessly, chromosomal breakage has been co-opted by the immune system as an integral part of B- and T-cell development when V(D)J recombination–a programmed DNA rearrangement process–generates a vast array of antigen receptor molecules. V(D)J recombination is initiated when the lymphoid-restricted recombination-activating genes RAG1 and RAG2 are expressed and form a site-specific endonuclease (the RAG nuclease or RAG recombinase) that induces DSBs within T cell receptor (TCR, α/δ, β, γ) and Ig (h, κ, λ) gene loci. Despite the existence of multiple DSB repair pathways, including the canonical non-homologous end-joining (NHEJ) and homologous recombination (HR) pathways as well as additional (micro)homology-directed sub-pathways, RAG-initiated DSBs are “almost” exclusively repaired by NHEJ. In the following review, we address various parameters which restrict DNA DSB repair pathway choice in lymphocytes undergoing V(D)J recombination and discuss how NHEJ-mediated repair impacts on successful antigen receptor gene assembly or association to immunodeficiencies and lymphoid cancers.
V(D)J Recombination and Double-Strand Break Repair
V(D)J recombination is a somatic antigen receptor gene rearrangement process occurring in developing B and T cells, involving rearrangement of V (variable), D (diversity) and J (joining) gene segments located within the Ig or TCR locus (Figure 1A) (reviewed in (Gellert, 2002; Litman et al., 2010; Schatz and Ji, 2011; Roth, 2014; Lescale and Deriano, 2016)). This locus-specific reaction is initiated by the RAG nuclease which introduces two DSBs at recombination signal sequences (RSSs) flanking selected V, D and J segments. RAG-DSBs pose a threat to overall genome stability and thus the activity of the RAG recombinase is tightly controlled (Roth, 2003; Roth, 2014; Lescale and Deriano, 2016).
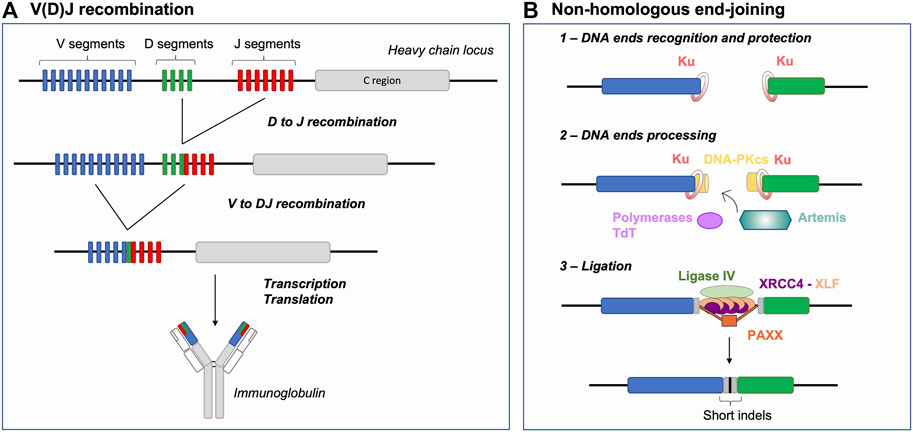
FIGURE 1. V(D)J recombination and NHEJ Basics: Generating antigen receptor diversity. (A) V(D)J recombination at the immunoglobulin heavy chain locus (depicted as an example) consists in a sequential 2-step rearrangement of V, D and J segments. This combinatorial process generates the diversity of antigen receptors. (B) After RAG cleavage, the NHEJ repair pathway is initiated by binding of the Ku70/80 heterodimer (Ku) to DNA ends. Ku together with DNA-PKcs form the DNA-PK holoenzyme. RAG DNA ends are then processed by the endonuclease Artemis and polymerases (e.g., Pol µ), specifically the terminal deoxynucleotidyl transferase (TdT), resulting in increased junctional diversity (in gray). This additional diversity is generated, prior to joining, in two forms: 1) P- palindromic sequences, produced through the endonuclease action of Artemis at RAG-induced hairpin-sealed ends and 2) N-nucleotide sequences, the addition of non-templated nucleotides by TdT. Finally, the ligation complex composed of Ligase IV, XRCC4 and XLF joins the processed ends. Joining of DNA ends via NHEJ further participates to generating indels, moreover favoring junctional diversity. NHEJ: non-homologous end-joining, indels: insertions or deletions.
Lymphocytes, as any other cell types, possess several DSB repair pathways including HR and NHEJ, which are considered the main DNA DSB repair pathways. HR is based on the capacity of the cellular machinery to find and access an intact template (sister chromatid or chromosome homolog) used to mediate error-free repair of the break. Initiation of HR involves the identification of broken DNA end(s), a 5′-3′ nucleolytic digestion process generating 3′ single-stranded DNA (ssDNA) (end resection) permitting homology search and DNA synthesis (Elbakry and Löbrich, 2021). NHEJ is thought to be a rapid and efficient way of repairing DSBs, as it involves the identification and (quasi) direct ligation of the two DNA ends without search for (extended) homology (Figure 1B) (reviewed in (Chang et al., 2017)). Briefly, upon detection of a DSB, the Ku70/80 heterodimer (Ku) is loaded onto DNA ends and acts as a scaffold for recruitment of additional NHEJ factors (Gottlieb and Jackson, 1993; Nick McElhinny et al., 2000; Walker et al., 2001; Fell and Schild-Poulter, 2015; Ochi et al., 2015). Ku recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the DNA-PK holoenzyme that phosphorylates multiple substrates, promoting synapsis of DNA ends and facilitating the recruitment of end processing and ligation enzymes (Chen X. et al., 2021; Zha et al., 2021). The ligation complex, composed of Ligase IV-XRCC4-XLF, joins the ends together (Ahnesorg et al., 2006; Buck et al., 2006). PAXX, a paralog of XRCC4 and XLF, also contributes to end-joining during NHEJ (Ochi et al., 2015; Lescale et al., 2016b) notably by promoting accumulation of Ku at DSBs (Liu et al., 2017). This repair pathway, as opposed to HR, is sometimes defined as error-prone because it can generate small insertions and deletions (indels) (Figure 1B) (Stinson et al., 2020). DNA 5′ end resection is a major determining factor for the NHEJ to HR choice in cells, as mentioned above HR involves the formation of extensive 3′ ssDNA. The chromatin-bound protein 53BP1, together with downstream effectors, counteracts DNA end resection and thus act as a pro-NHEJ regulator upon DSB injury (Mirman and de Lange, 2020). Alternative end-joining (alt-EJ) and single-strand annealing (SSA) are yet other DSB repair pathways relying on intermediate length of DNA end resection and bias towards usage of (micro)homologies (Symington, 2016). These pathways are intrinsically unfaithful as they generate deletions between microhomology tracts and as alt-EJ is associated to genomic instability, notably chromosomal translocations (Corneo et al., 2007; Soulas-Sprauel et al., 2007; Yan et al., 2007). Alt-EJ is thought to be particularly active in cells deficient for HR or NHEJ (Corneo et al., 2007; Soulas-Sprauel et al., 2007; Yan et al., 2007; Wyatt et al., 2016), although some studies indicate that this pathway is also utilized in DNA repair proficient cells (Lee et al., 2004; Corneo et al., 2007; Coussens et al., 2013; Deriano and Roth, 2013).
During V(D)J recombination, antigen receptor gene diversity is achieved by 1) unique combinations of V, D and J coding segments, so called combinatorial diversity and 2) the imprecision of the DSB repair reaction at segment joints–driven by NHEJ and the action of the terminal deoxynucleotidyl transferase (TdT), termed junctional diversity (Figure 1B) (Gilfillan et al., 1993; Komori et al., 1993; Ramsden, 2011). NHEJ thus offers the ideal repair pathway to permit Ig and TCR gene diversification in early lymphocytes as opposed to resection- and homology-based repair pathways that would ultimately restore germline sequences or generate genetic instability in the cell. In the following sections, we address different parameters that limit DSB repair pathway choice to NHEJ during V(D)J recombination, including 1) the nature of DSB end structures, 2) the establishment of DSB end synapsis and 3) the impediment of DNA end resection and (micro)homology-driven repair.
DNA End Structures–Meant to Be Repaired by Non-Homologous End-Joining
Broken DNA ends often cannot be directly reattached and require processing prior to mending, thus the nature of the broken ends acts as an important factor influencing repair pathway choice (Chang et al., 2017). During V(D)J recombination, the RAG complex promotes the assembly of a pre-synaptic complex that includes a 12 and a 23 RSS prior to conducting its nuclease activity (Schatz and Ji, 2011). DNA cleavage occurs in two steps and relies on RAG1, RAG2, a divalent metal ion, and the ubiquitous bending factors HMGB1 or HMGB2. RAG introduces a nick between each RSS and its flanking coding sequence, generating a free 3′-OH group which then attacks the opposite strand by transesterification. This cleavage reaction results in four broken DNA ends with specific structures: two hairpin-sealed coding ends (CE) at gene segments and two blunt signal ends (SE) at RSSs (Figure 2A). Upon cleavage, RAG-induced DNA breaks activate Ataxia telangiectasia mutated (ATM), an important mediator of the DNA DSB response (Helmink and Sleckman, 2012). Activated ATM phosphorylates numerous proteins that promote the G1/S checkpoint and participate in DNA end protection (see below), favoring NHEJ. Ku has a strong affinity for hairpin sealed, blunt or short overhang DNA ends (Falzon et al., 1993; Downs and Jackson, 2004), directing RAG-DSBs towards NHEJ repair. Ku not only binds avidly broken ends but also serves as a scaffold for the recruitment of DNA-PKcs, forming the DNA-PK holoenzyme, and downstream NHEJ factors that permit the processing and ligation of RAG-induced DSB ends. This second attribute of Ku is particularly important for CEs as blunt SEs can be directly ligated by XRCC4-Ligase IV to form DNA circles (i.e., in the case of deletional recombination). Indeed, CEs necessitate the action of the endonuclease Artemis to open the hairpin structure (Figure 2A). Proper Artemis endonuclease activity requires DNA-PK and leads to the formation of protruding 3′ ends with an -OH group (Ma et al., 2002). This latter DNA end topology favors repair by NHEJ, as XRCC4-Ligase IV necessitate a -OH at both DNA ends for ligation. Additionally, the XRCC4-Ligase IV complex can stimulate the removal of few nucleotides-long overhangs–generated by Artemis–prior to ligation (Gerodimos et al., 2017). Notably, Ku also promotes the recruitment of TdT–the third lymphoid-specific protein in addition to RAG1 and RAG2 – that adds nucleotides at Artemis-opened CEs preceding the ligation by XRCC4-Ligase IV. This links NHEJ to the generation of junctional diversity at coding joins (Gilfillan et al., 1993; Komori et al., 1993; Purugganan et al., 2001; Ramsden, 2011), increasing the genetic diversity of V(D)J rearrangement outcomes. Therefore, the topology of RAG-induced DSB ends significantly biases repair towards NHEJ, by generating an NHEJ prone environment.
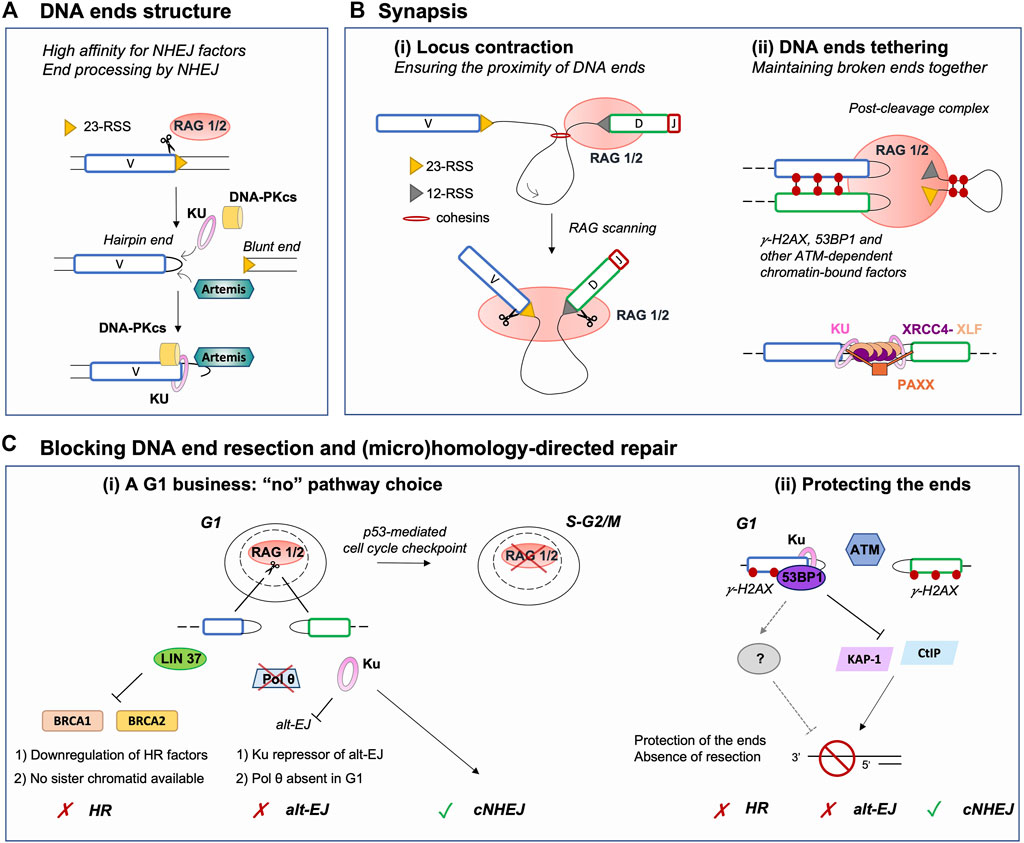
FIGURE 2. Major parameters restricting DNA repair pathway choice to NHEJ during V(D)J recombination. (A) RAG-induced breakage generates a covalently sealed hairpin end (coding end) and a blunt end (signal end). This facilitates the loading of Ku, which acts as a scaffold for other NHEJ factors, as it has a high affinity for blunt or hairpin sealed ends. In addition, hairpin sealed ends require to be opened by another NHEJ factor Artemis, which renders ends compatible for ligation. Thus, this DNA end topology contributes to the establishment of a NHEJ-prone environment. (B) (i) Upon binding an RSS, RAG scans the adjacent chromatin by a loop extrusion mechanism. Breakage is induced only upon reaching a compatible RSS, ensuring the induction of DSBs in close proximity despite the large size of the immunoglobulin locus. (ii) Following DSB induction, RAG remains bound to DNA ends in a post-cleavage complex (PCC). The PCC together with NHEJ and ATM-dependent chromatin-bound DNA factors (e.g., phosphorylated H2AX and 53BP1) favor DNA ends tethering and stabilization. This likely prevents the search for distant partner DNA ends and channels broken DSB ends to NHEJ for safe repair. (C) (i) V(D)J recombination is a G1-restricted process, as RAG is degraded upon entry in the S phase. In G1, HR cannot operate as pre-replicative cells do not harbor a sister chromatid, used as a template for repair. In addition, several factors required for HR are transcriptionally repressed in G0/G1. Similarly, Pol θ, an important factor for alt-EJ, is poorly expressed in G1 consequently limiting the use of this repair pathway. Furthermore, alt-EJ is blocked by Ku upon binding DNA ends, yet again promoting processing and repair by NHEJ. (ii) Chromatin DSB-response factors γH2AX, 53BP1 and possibly additional downstream effectors contribute to the protection of RAG-DSB ends by blocking the activity of nucleases such as CtIP or acting via transcriptional repressors such as KAP-1. This protection prevents DNA end resection, an essential intermediate step for (micro)homology-directed repair (e.g., alt-EJ, HR, etc.), hence promoting NHEJ. NHEJ: non-homologous end-joining, RSS: Recombination Signal Sequence, Alt-EJ: alternative end-joining, HR: homologous recombination.
Synapsis - Keeping DNA Ends Together for Safe Repair by Non-Homologous End-Joining
Maintaining broken DNA ends in close proximity is a major parameter that influences pathway choice, notably because NHEJ requires the physical proximity of both DNA ends, while it is dispensable for certain HR reactions such as break-induced replication (Pham et al., 2021). Synapsis of DSB ends during V(D)J recombination is quite challenging as it involves the sequestration of four DSB ends (i.e., two CEs and two SEs). Additionally, V(D)J recombination implicates gene segments that can be situated at considerable distances from one another; for instance the murine germline immunoglobulin heavy chain (IgH) locus spans approximately 2.75 Mbp of chromosome 12 (Lucas et al., 2015). For efficient V(D)J recombination, 1) V, D and J gene segments must be brought in vicinity of each other prior to cleavage and 2) DNA ends, specifically CEs which require processing, must be kept together for ligation.
Contraction and spatial reorganization of antigen receptor loci during V(D)J recombination rely largely on the formation of chromatin loops through a cohesin-dependent extrusion process (Bossen et al., 2012; Ebert et al., 2015; Ba et al., 2020; Hill et al., 2020; Dai et al., 2021; Davidson and Peters, 2021), as well as transcription and subnuclear relocation (Rogers et al., 2021). Remarkably, this mechanism poises the loci for recombination independently of RAG, but also endows RSS-bound RAG with the ability to scan chromatin for a partner RSS, providing directionality and spatial restriction to RAG activity within the chromatin loop domain (Figure 2Bi) (Lin et al., 2018; Zhang et al., 2019; Ba et al., 2020; Hill et al., 2020; Dai et al., 2021). Additional chromatin-bound factors such as 53BP1 contribute to bringing V(D)J segments close-by, as depletion of this latter factor results in a reduction of distal V to DJ segments joins (Difilippantonio et al., 2008). Induction of two DSBs in the vicinity of one another likely contributes to favoring rapid repair of RAG-induced DNA breaks by NHEJ, without the need to search for partner DNA ends.
After cleavage, the RAG proteins stay associated with the DNA ends in a post-cleavage complex (PCC) (Figure 2Bii). Mutations resulting in RAG-PCC destabilization were shown to increase repair of RAG-mediated DSBs via HR and alt-EJ (Lee et al., 2004; Corneo et al., 2007; Coussens et al., 2013; Deriano and Roth, 2013), pathways considered as unconventional for V(D)J recombination. These observations suggested that the RAG-PCC might contribute to shepherding DNA ends to the NHEJ machinery for repair, thus protecting them from error prone end-joining pathways and aberrant recombination events (Roth, 2003; Lee et al., 2004; Deriano and Roth, 2013). Indeed, a RAG2 mutant - possessing deletion of C-terminal residues 352–527 (core RAG2) - destabilizes the RAG-PCC and is associated with an increased rate of aberrant recombination outcomes in vitro and to inter-chromosomal translocations involving the V(D)J loci in vivo (Sekiguchi et al., 2001; Talukder et al., 2004; Corneo et al., 2007; Curry and Schlissel, 2008; Deriano et al., 2011; Coussens et al., 2013). Additionally, core RAG2/p53-deficient mice present increased genomic instability and accelerated lymphomagenesis via alt-EJ, generating tumors bearing a complex landscape of chromosomal rearrangements (Deriano et al., 2011; Mijušković et al., 2012; Mijušković et al., 2015). Strikingly, the lymphomas and translocations observed in the latter animals resemble those of ATM-deficient mice, suggesting that a similar DNA end destabilization mechanism might underlie genomic instability and lymphomagenesis in both mouse models (Deriano et al., 2011). Consistent with this, ATM - beyond its role in activating checkpoints - is important for the stability of RAG-PCCs in vivo (Bredemeyer et al., 2006). Upon DSB damage, ATM phosphorylates chromatin- and DNA-associated proteins, including the histone variant H2AX (forming γH2AX), 53BP1, MDC1 and factors of the MRN complex (MRE11, RAD50, and NBS1) that assemble on both sides of DNA breaks forming so-called nuclear DNA repair foci. The stabilization function of ATM depends on its kinase activity. Thus, formation of ATM-dependent DNA repair foci has been proposed to tether DNA ends for proper joining via NHEJ (Figure 2Bii). In ATM-deficient cells undergoing V(D)J recombination, the fraction of CEs which evade the PCC are occasionally joined aberrantly, forming chromosomal deletions, inversions, and translocations (Bredemeyer et al., 2006; Helmink and Sleckman, 2012). Altogether, these results indicate that RAG2 (by extension the RAG-PCC) and ATM share mechanistic properties during V(D)J recombination, via the stabilization of broken DNA-ends consequently avert the use of alternative repair pathways.
Additional insights into the mechanisms responsible for the stabilization of RAG-cleaved DNA ends come from the analysis of animal models double-deficient for XLF and ATM or core RAG2. XLF and XRCC4 are two distantly related members of the same protein family and share structural similarity (Callebaut et al., 2006; Andres et al., 2007; Li Y. et al., 2008). Together, they form long filaments, thought to help DNA end tethering and synapsis during repair (Figure 2Bii) (Tsai et al., 2007; Riballo et al., 2009; Hammel et al., 2011; Ropars et al., 2011; Reid et al., 2015; Chen S. et al., 2021). In contrast to other NHEJ-deficient mice, XLF-deficient mice are not markedly immune-deficient and early lymphoid cells from these animals perform nearly normal V(D)J recombination. These observations suggest that other factors or pathways compensate for XLF function during V(D)J recombination (Li G. et al., 2008). In fact, cells deficient for both XLF and ATM-dependent DSB response (e.g., XLF and ATM, 53BP1, or H2AX double mutants) display severe block in lymphocyte development and a significant defect in the repair of RAG-mediated DSBs. This reveals functional redundancy between XLF and ATM-DSB response factors during V(D)J recombination (Li G. et al., 2008; Zha et al., 2011; Liu et al., 2012; Oksenych et al., 2012; Oksenych et al., 2013; Vera et al., 2013; Kumar et al., 2014). Similarly, core RAG2/XLF double deficiency leads to a profound lymphopenia associated with a severe defect in joining of RAG-cleaved DNA ends (Lescale et al., 2016a). These findings are consistent with a two-tier model in which the RAG proteins, together with the ATM chromatin DSB-response, collaborate with NHEJ factors to promote functional V(D)J recombination and emphasize the importance of DNA end tethering for proper repair.
Blocking DNA End Resection and (micro)Homology-Driven Double-Strand Break Repair
A G1-Phase Business
RAG-induced V(D)J recombination is limited to the G1 phase of the cell cycle, which offers an additional level of restriction to NHEJ-driven repair. This is due to the specific destruction of RAG2 during the G1-to-S transition that is triggered by phosphorylation of the T490 residue (Li et al., 1996). Additionally, RAG-induced DSBs trigger an ATM/p53-dependent DSB response that promotes G1/S cell cycle arrest and eventually cell death (Figure 2Ci). Finally, RAG-DSBs activate a specific checkpoint that opposes the pre-B cell receptor proliferative signals and prevent cells from entering into S phase before resolving the damage (Bredemeyer et al., 2008; Bednarski and Sleckman, 2012; Bednarski et al., 2016).
As the ideal template for HR is the sister chromatid, HR is restricted to the S and G2 phases of the cell cycle and cannot fully operate in G1-phase cells. Alt-EJ (and SSA) do not possess this constraint, thus could potentially serve as alternatives to NHEJ for repair of DSBs in non-dividing cells. Using high-throughput sequencing techniques, it was recently shown that end joining of RAG-induced DSBs is virtually null in G0/G1-arrested progenitor (pro-) B cells deficient for XRCC4 (Yu et al., 2020). Within the same setting, Cas9-induced DSBs are also poorly repaired, suggesting that additional factors, other than RAG, limit the access of broken DNA ends to alt-EJ pathways in G0/G1-phase cells. Similarly, DSBs generated by RAG, Cas9 or zinc finger endonucleases in G0/G1-arrested pro-B cells remain unjoined in the absence of Ligase IV (Liang et al., 2021). However, Ku70-deficient or Ku70/Ligase IV-deficient G0/G1-arrested pro-B cells perform quite robust end-joining, albeit at lower levels than wild type cells, indicating that Ku acts as a strong repressor of alt-EJ in G0/G1-phase cells (Figure 2Ci) (Frock et al., 2021; Liang et al., 2021). Cells might also not be fully equipped to perform resection- and homology-dependent repair in G0/G1. For instance, LIN37, a component of the DREAM transcriptional repressor, inhibits resection and HR in G0/G1-blocked pro-B cells by repressing the expression of HR proteins such as BRCA1, BRCA2, PALB2 and RAD51 (Chen B. R. et al., 2021). Similarly, DNA polymerase theta (Pol θ, encoded by Polq in mice), implicated in alt-EJ (Ramsden et al., 2021), is not expressed in G0/G1-arrested pro-B cells (Figure 2Ci) (Yu et al., 2020).
Nevertheless, analysis of mice harboring combined deficiency in p53 and in NHEJ (i.e., Ku, XRCC4 or Ligase IV) irremediably develop aggressive pro-B lymphomas displaying RAG-dependent translocations and amplifications between Igh and c-Myc by alt-EJ (Nussenzweig and Nussenzweig, 2010; Gostissa et al., 2011; Ramsden and Nussenzweig, 2021). It was suggested that p53 deficiency enables cells to move inappropriately into S phase and acquire DSBs that initiate chromosomal translocations and amplifications (Paulson et al., 1998; Zhu et al., 2002). In fact, an in vitro study using XRCC4/p53-deficient pro-B cell lines shows that the transition from G0/G1-phase to S-G2/M-phases of the cell cycle enables alt-EJ repair, promoting massive genetic instability in the form of chromosomal deletions and translocations (Yu et al., 2020). It is tempting to speculate that unrepaired G1-DNA breaks progressing to S-G2/M get lost in the cellular space with unprotected DNA ends being subjected to repetitive nuclease attacks until (micro)homology-driven alt-EJ stabilizes them in cis or trans. In that regard, although multiple homology-directed sub-pathways would theoretically be able to process these lost DNA ends (Elbakry and Löbrich, 2021), the repair of G0/G1-DSBs in S-G2/M would strictly depend on Pol θ. Indeed, in XRCC4/Pol θ/p53-triple deficient pro-B cells, DSBs induced in G1 accumulate in the form of chromosomal breaks resulting in lethality at the next mitosis (Yu et al., 2020). Whether Pol θ contributes to the development of pro-B cell lymphomas, carrying Igh/c-Myc translocations, in NHEJ/p53-deficient animals remains to be addressed.
DNA End Protection
The extent of resection is actively regulated by the protection of DNA ends, which limits the access of nucleases to the break sites. In addition to the above-mentioned parameters (i.e., DNA end structures and synapsis), several DSB response chromatin-bound factors localize at RAG-DSBs and are thought to protect DNA ends against resection, including γH2AX and 53BP1 (Difilippantonio et al., 2008; Helmink et al., 2011; Zha et al., 2011; Dorsett et al., 2014; Lescale et al., 2016a; Chen B. R. et al., 2021). In G1, yH2AX prevents CtIP-mediated nucleolytic resection (Helmink et al., 2011). Similarly, KAP-1, a transcriptional repressor modulating chromatin structure, was shown to promote resection in G1 lymphocytes in the absence of yH2AX and 53BP1 (Tubbs et al., 2014). Moreover, depletion of 53BP1 in Ligase IV-deficient G0/G1-blocked pro-B cells results in increased levels of resection at irradiation-induced DSB ends, demonstrating that 53BP1 is crucial for DNA end protection in this cell-cycle phase (Figure 2Cii) (Chen B. R. et al., 2021). The Shieldin complex, composed of SHLD1, SHLD2, SHLD3 and MAD2L2/REV7, acts downstream of 53BP1-RIF1 to antagonize DNA end resection and favor NHEJ over HR (Greenberg, 2018; Setiaputra and Durocher, 2019; Mirman and de Lange, 2020; de Krijger et al., 2021). It acts in a paradoxical manner as it requires to bind >50 nt-long ssDNA ends in order to hinder DNA end resection (Dev et al., 2018; Findlay et al., 2018; Gao et al., 2018; Noordermeer et al., 2018). Mechanistically, it is thought to directly inhibit resection by physically blocking access of nucleases to the free ssDNA-dsDNA ends. Additionally, this complex promotes the recruitment and coordination of additional factors leading to the processing of ssDNA-dsDNA intermediates prior to NHEJ repair such as ASTE1, which cleaves the protruding ssDNA, and the CST (CTC1–STN1–TEN1)–DNA polymerase-α–primase complex, to fill in the residual ssDNA (Mirman et al., 2018; Zhao et al., 2021). Although the Shieldin complex counteracts HR in BRCA1-deficient cells and is important for NHEJ-driven repair during class switch recombination or in the fusion of unprotected telomeres, it seems dispensable for V(D)J recombination (Dev et al., 2018; Ghezraoui et al., 2018; Mirman et al., 2018; Ling et al., 2020). Indeed, SHLD2 or REV7 deficiencies in mice do not significantly alter lymphocyte development and V(D)J recombination (Ghezraoui et al., 2018; Ling et al., 2020). It is to note that in wild-type cells, the processing of RAG-induced DNA ends does not generate >50 nt-long ssDNA intermediates, thus potentially explaining why Shieldin-mediated protection prior to joining seems dispensable during V(D)J recombination. However, whether the Shieldin complex plays a role in protecting RAG-generated DNA ends against resection in the context of crippled NHEJ remains to be investigated. In XLF-deficient mice (impaired NHEJ), 53BP1 plays an essential role in counteracting resection at RAG-DSB ends, promoting V(D)J recombination and lymphocyte differentiation (Liu et al., 2012; Oksenych et al., 2012). Nonetheless, it is unclear if this DNA end protection is mediated through 53BP1 downstream effectors (e.g., Shieldin complex) or via intrinsic properties of 53BP1 (Figure 2Cii). Notably, Ku also antagonizes DNA end resection through at least two distinct mechanisms 1) by blocking the access of nucleases to DSB ends (Zahid et al., 2021) and 2) by recruiting TdT which promotes template-independent and -dependent synthesis prior to ligation (Loc’h and Delarue, 2018). Taken together, in the context of V(D)J recombination the downregulation of the DSB end resection machinery and the protection of DNA ends by chromatin-bound factors and Ku seem to act as forefront anti-resection barriers, promoting repair via NHEJ but not HR or alt-EJ (Figure 2C).
Conclusion and Perspectives
In this review, we present V(D)J recombination as a relevant biological setting to investigate factors influencing DSB repair pathway choice, specifically those constraining the repair of DSBs to NHEJ. This repair pathway is essential for V(D)J combinatorial rearrangement as well as for the generation of diversity at V(D)J junctions, two pre-requisites for antigen receptor gene diversification and the establishment of a primary immune repertoire. During V(D)J recombination, several factors prime for repair via NHEJ, including the spatial organization of the genomic loci subjected to these rearrangement events. Additionally, the RAG nuclease, the type of generated DSBs, the G1 phase-specific environment and the dedicated DSB response predispose (arguably dictate) repair through NHEJ. Albeit numerous the studies which shed light onto the mechanisms through which DSBs generated during V(D)J recombination are biased towards repair by NHEJ, several questions remain unanswered. For instance, while the RAG-PCC plays a role in favoring repair via NHEJ, possibly by stabilizing DNA ends, it remains unclear if the RAG proteins directly interact with certain NHEJ factors and whether such interaction(s) would contribute to NHEJ pathway choice. We also discussed the importance of blocking DNA end resection during V(D)J recombination through the action of specific chromatin DSB response factors, most notably 53BP1. Whether this end protection only relies on the capacity of the chromatin-bound factors to maintain a stable PCC or whether it also requires specific downstream effectors to act at the DSB ends is unclear. In that regard, it is interesting to note that the mode of action of the Shieldin complex on DSB ends generated by AID during IgH class switch recombination is somewhat reminiscent to that of Ku during V(D)J recombination. Both Ku and the Shieldin complex have the capacity to physically obstruct resection at DSB ends and to actively recruit factors implicated in DNA end modifications (i.e., action of Ku and TdT/Artemis versus Shieldin-complex and ASTE1/CST-DNA polymerase α). In Ku-deficient G0/G1-arrested pro-B cells (and to a much lesser extent in XRCC4- or Ligase IV-deficient cells), V(D)J joints harbor rather short resection tracks (typically less than 100 nucleotides) (Yu et al., 2020; Liang et al., 2021). Could RAG-DSB ends benefit from Shieldin complex protection against resection in such circumstances? Additionally, the nature of alt-EJ and the factors implicated in alt-EJ in G0/G1-phase cells as opposed to Pol θ-mediated alt-EJ in S-G2/M remain to be established. The role of these sub-pathways in the onset of pro-B cell lymphomas in NHEJ/p53-deficient animals also remains to be investigated. Finally, antigen receptor loci relocate from the nuclear periphery to permissive euchromatin in the nuclear interior before V(D)J recombination (Rogers et al., 2021). This subnuclear relocation likely provides specific local chromatin environments that might influence downstream DSB repair events (Mitrentsi et al., 2020). Recent studies have also highlighted the importance of 3D genome (re)organization and dynamics in DSB repair, for instance through the establishment of γH2AX/53BP1 DSB response foci (Arnould et al., 2021) or the restriction of homology search during HR (Piazza et al., 2021), two crucial chromatin events influencing DSB repair outcome and pathway choice. How such chromosome dynamics contributes to (lack of) DNA DSB pathway choice and overall genome integrity maintenance during V(D)J recombination remains a question for future studies.
Author Contributions
AL, TM and LD wrote the manuscript and approved the submitted version.
Funding
The Deriano laboratory is funded by Institut Pasteur, Institut National de la Santé et de la Recherche Médicale (INSERM), Wordwide Cancer Research (grant # 19–0333), Ligue Nationale Contre le Cancer (Equipe labellis ée 2019) and Institut National du Cancer (INCa, grant # PLBIO19-122). AL received funding from Université de Paris, Sorbonne Paris Cité.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahnesorg, P., Smith, P., and Jackson, S. P. (2006). XLF Interacts with the XRCC4-DNA Ligase IV Complex to Promote DNA Nonhomologous End-Joining. Cell 124, 301–313. doi:10.1016/j.cell.2005.12.031
Andres, S. N., Modesti, M., Tsai, C. J., Chu, G., and Junop, M. S. (2007). Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining. Mol. Cel 28, 1093–1101. doi:10.1016/j.molcel.2007.10.024
Arnould, C., Rocher, V., Finoux, A.-L., Clouaire, T., Li, K., Zhou, F., et al. (2021). Loop Extrusion as a Mechanism for Formation of DNA Damage Repair Foci. Nature 590, 660–665. doi:10.1038/s41586-021-03193-z
Ba, Z., Lou, J., Ye, A. Y., Dai, H.-Q., Dring, E. W., Lin, S. G., et al. (2020). CTCF Orchestrates Long-Range Cohesin-Driven V(D)J Recombinational Scanning. Nature 586, 305–310. doi:10.1038/s41586-020-2578-0
Bednarski, J. J., Pandey, R., Schulte, E., White, L. S., Chen, B.-R., Sandoval, G. J., et al. (2016). RAG-Mediated DNA Double-Strand Breaks Activate a Cell Type-Specific Checkpoint to Inhibit Pre-B Cell Receptor Signals. J. Exp. Med. 213, 209–223. doi:10.1084/jem.20151048
Bednarski, J. J., and Sleckman, B. P. (2012). Lymphocyte Development: Integration of DNA Damage Response Signaling. Adv. Immunol. 116, 175–204. doi:10.1016/B978-0-12-394300-2.00006-5
Bossen, C., Mansson, R., and Murre, C. (2012). Chromatin Topology and the Regulation of Antigen Receptor Assembly. Annu. Rev. Immunol. 30, 337–356. doi:10.1146/annurev-immunol-020711-075003
Bredemeyer, A. L., Helmink, B. A., Innes, C. L., Calderon, B., McGinnis, L. M., Mahowald, G. K., et al. (2008). DNA Double-Strand Breaks Activate a Multi-Functional Genetic Program in Developing Lymphocytes. Nature 456, 819–823. doi:10.1038/nature07392
Bredemeyer, A. L., Sharma, G. G., Huang, C.-Y., Helmink, B. A., Walker, L. M., Khor, K. C., et al. (2006). ATM Stabilizes DNA Double-Strand-Break Complexes during V(D)J Recombination. Nature 442, 466–470. doi:10.1038/nature04866
Buck, D., Malivert, L., de Chasseval, R., Barraud, A., Fondanèche, M.-C., Sanal, O., et al. (2006). Cernunnos, a Novel Nonhomologous End-Joining Factor, Is Mutated in Human Immunodeficiency with Microcephaly. Cell 124, 287–299. doi:10.1016/j.cell.2005.12.030
Callebaut, I., Malivert, L., Fischer, A., Mornon, J.-P., Revy, P., and de Villartay, J.-P. (2006). Cernunnos Interacts with the XRCC4·DNA-Ligase IV Complex and Is Homologous to the Yeast Nonhomologous End-Joining Factor Nej1. J. Biol. Chem. 281, 13857–13860. doi:10.1074/jbc.C500473200
Chang, H. H. Y., Pannunzio, N. R., Adachi, N., and Lieber, M. R. (2017). Non-homologous DNA End Joining and Alternative Pathways to Double-Strand Break Repair. Nat. Rev. Mol. Cel Biol. 18, 495–506. doi:10.1038/nrm.2017.48
Chen, B.-R., Wang, Y., Tubbs, A., Zong, D., Fowler, F. C., Zolnerowich, N., et al. (2021). LIN37-DREAM Prevents DNA End Resection and Homologous Recombination at DNA Double-Strand Breaks in Quiescent Cells. Elife 10, e68466. doi:10.7554/eLife.68466
Chen, S., Lee, L., Naila, T., Fishbain, S., Wang, A., Tomkinson, A. E., et al. (2021). Structural Basis of Long-Range to Short-Range Synaptic Transition in NHEJ. Nature 593, 294–298. doi:10.1038/s41586-021-03458-7
Chen, X., Xu, X., Chen, Y., Cheung, J. C., Wang, H., Jiang, J., et al. (2021). Structure of an Activated DNA-PK and its Implications for NHEJ. Mol. Cel. 81, 801–810.e3. doi:10.1016/j.molcel.2020.12.015
Ciccia, A., and Elledge, S. J. (2010). The DNA Damage Response: Making it Safe to Play with Knives. Mol. Cel 40, 179–204. doi:10.1016/j.molcel.2010.09.019
Corneo, B., Wendland, R. L., Deriano, L., Cui, X., Klein, I. A., Wong, S.-Y., et al. (2007). Rag Mutations Reveal Robust Alternative End Joining. Nature 449, 483–486. doi:10.1038/nature06168
Coussens, M. A., Wendland, R. L., Deriano, L., Lindsay, C. R., Arnal, S. M., and Roth, D. B. (2013). RAG2's Acidic Hinge Restricts Repair-Pathway Choice and Promotes Genomic Stability. Cel Rep. 4, 870–878. doi:10.1016/j.celrep.2013.07.041
Curry, J. D., and Schlissel, M. S. (2008). RAG2's Non-Core Domain Contributes to the Ordered Regulation of V(D)J Recombination. Nucleic Acids Res. 36, 5750–5762. doi:10.1093/nar/gkn553
Dai, H.-Q., Hu, H., Lou, J., Ye, A. Y., Ba, Z., Zhang, X., et al. (2021). Loop Extrusion Mediates Physiological Igh Locus Contraction for RAG Scanning. Nature 590, 338–343. doi:10.1038/s41586-020-03121-7
Davidson, I. F., and Peters, J.-M. (2021). Genome Folding through Loop Extrusion by SMC Complexes. Nat. Rev. Mol. Cel Biol. 22, 445–464. doi:10.1038/s41580-021-00349-7
de Krijger, I., Boersma, V., and Jacobs, J. J. L. (2021). REV7: Jack of many Trades. Trends Cel Biol. 31, 686–701. doi:10.1016/j.tcb.2021.04.002
Deriano, L., Chaumeil, J., Coussens, M., Multani, A., Chou, Y., Alekseyenko, A. V., et al. (2011). The RAG2 C Terminus Suppresses Genomic Instability and Lymphomagenesis. Nature 471, 119–123. doi:10.1038/nature09755
Deriano, L., and Roth, D. B. (2013). Modernizing the Nonhomologous End-Joining Repertoire: Alternative and Classical NHEJ Share the Stage. Annu. Rev. Genet. 47, 433–455. doi:10.1146/annurev-genet-110711-155540
Dev, H., Chiang, T.-W. W., Lescale, C., de Krijger, I., Martin, A. G., Pilger, D., et al. (2018). Shieldin Complex Promotes DNA End-Joining and Counters Homologous Recombination in BRCA1-Null Cells. Nat. Cel Biol. 20, 954–965. doi:10.1038/s41556-018-0140-1
Difilippantonio, S., Gapud, E., Wong, N., Huang, C.-Y., Mahowald, G., Chen, H. T., et al. (2008). 53BP1 Facilitates Long-Range DNA End-Joining during V(D)J Recombination. Nature 456, 529–533. doi:10.1038/nature07476
Dorsett, Y., Zhou, Y., Tubbs, A. T., Chen, B.-R., Purman, C., Lee, B.-S., et al. (2014). HCoDES Reveals Chromosomal DNA End Structures with Single-Nucleotide Resolution. Mol. Cel. 56, 808–818. doi:10.1016/j.molcel.2014.10.024
Downs, J. A., and Jackson, S. P. (2004). A Means to a DNA End: The Many Roles of Ku. Nat. Rev. Mol. Cel Biol. 5, 367–378. doi:10.1038/nrm1367
Ebert, A., Hill, L., and Busslinger, M. (2015). Spatial Regulation of V-(D)J Recombination at Antigen Receptor Loci. Adv. Immunol. 128, 93–121. doi:10.1016/bs.ai.2015.07.006
Elbakry, A., and Löbrich, M. (2021). Homologous Recombination Subpathways: A Tangle to Resolve. Front. Genet. 12, 723847. doi:10.3389/fgene.2021.723847
Falzon, M., Fewell, J. W., and Kuff, E. L. (1993). EBP-80, a Transcription Factor Closely Resembling the Human Autoantigen Ku, Recognizes Single- to Double-Strand Transitions in DNA. J. Biol. Chem. 268, 10546–10552. doi:10.1016/s0021-9258(18)82233-5
Fell, V. L., and Schild-Poulter, C. (2015). The Ku Heterodimer: Function in DNA Repair and Beyond. Mutat. Research/Reviews Mutat. Res. 763, 15–29. doi:10.1016/j.mrrev.2014.06.002
Findlay, S., Heath, J., Luo, V. M., Malina, A., Morin, T., Coulombe, Y., et al. (2018). SHLD 2/FAM 35A Co‐Operates with REV 7 to Coordinate DNA Double‐Strand Break Repair Pathway Choice. EMBO J. 37, e100158. doi:10.15252/embj.2018100158
Frock, R. L., Sadeghi, C., Meng, J., and Wang, J. L. (2021). DNA End Joining: G0-Ing to the Core. Biomolecules 11, 1487. doi:10.3390/biom11101487
Gao, S., Feng, S., Ning, S., Liu, J., Zhao, H., Xu, Y., et al. (2018). An OB-Fold Complex Controls the Repair Pathways for DNA Double-Strand Breaks. Nat. Commun. 9, 3925. doi:10.1038/s41467-018-06407-7
Gellert, M. (2002). V(D)J Recombination: RAG Proteins, Repair Factors, and Regulation. Annu. Rev. Biochem. 71, 101–132. doi:10.1146/annurev.biochem.71.090501.150203
Gerodimos, C. A., Chang, H. H. Y., Watanabe, G., and Lieber, M. R. (2017). Effects of DNA End Configuration on XRCC4-DNA Ligase IV and its Stimulation of Artemis Activity. J. Biol. Chem. 292, 13914–13924. doi:10.1074/jbc.M117.798850
Ghezraoui, H., Oliveira, C., Becker, J. R., Bilham, K., Moralli, D., Anzilotti, C., et al. (2018). 53BP1 Cooperation with the REV7-Shieldin Complex Underpins DNA Structure-Specific NHEJ. Nature 560, 122–127. doi:10.6019/PXD00965010.1038/s41586-018-0362-1
Gilfillan, S., Dierich, A., Lemeur, M., Benoist, C., and Mathis, D. (1993). Mice Lacking TdT: Mature Animals with an Immature Lymphocyte Repertoire. Science 261, 1175–1178. doi:10.1126/science.8356452
Goldstein, M., and Kastan, M. B. (2015). The DNA Damage Response: Implications for Tumor Responses to Radiation and Chemotherapy. Annu. Rev. Med. 66, 129–143. doi:10.1146/annurev-med-081313-121208
Gostissa, M., Alt, F. W., and Chiarle, R. (2011). Mechanisms that Promote and Suppress Chromosomal Translocations in Lymphocytes. Annu. Rev. Immunol. 29, 319–350. doi:10.1146/annurev-immunol-031210-101329
Gottlieb, T. M., and Jackson, S. P. (1993). The DNA-Dependent Protein Kinase: Requirement for DNA Ends and Association with Ku Antigen. Cell 72, 131–142. doi:10.1016/0092-8674(93)90057-w
Greenberg, R. A. (2018). Assembling a Protective Shield. Nat. Cel Biol. 20, 862–863. doi:10.1038/s41556-018-0152-x
Hammel, M., Rey, M., Yu, Y., Mani, R. S., Classen, S., Liu, M., et al. (2011). XRCC4 Protein Interactions with XRCC4-Like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair. J. Biol. Chem. 286, 32638–32650. doi:10.1074/jbc.M111.272641
Helmink, B. A., and Sleckman, B. P. (2012). The Response to and Repair of RAG-Mediated DNA Double-Strand Breaks. Annu. Rev. Immunol. 30, 175–202. doi:10.1146/annurev-immunol-030409-101320
Helmink, B. A., Tubbs, A. T., Dorsett, Y., Bednarski, J. J., Walker, L. M., Feng, Z., et al. (2011). H2AX Prevents CtIP-Mediated DNA End Resection and Aberrant Repair in G1-Phase Lymphocytes. Nature 469, 245–249. doi:10.1038/nature09585
Hill, L., Ebert, A., Jaritz, M., Wutz, G., Nagasaka, K., Tagoh, H., et al. (2020). Wapl Repression by Pax5 Promotes V Gene Recombination by Igh Loop Extrusion. Nature 584, 142–147. doi:10.1038/s41586-020-2454-y
Jackson, S. P., and Bartek, J. (2009). The DNA-Damage Response in Human Biology and Disease. Nature 461, 1071–1078. doi:10.1038/nature08467
Komori, T., Okada, A., Stewart, V., and Alt, F. W. (1993). Lack of N Regions in Antigen Receptor Variable Region Genes of TdT-Deficient Lymphocytes. Science 261, 1171–1175. doi:10.1126/science.8356451
Kumar, V., Alt, F. W., and Oksenych, V. (2014). Functional Overlaps between XLF and the ATM-Dependent DNA Double Strand Break Response. DNA Repair 16, 11–22. doi:10.1016/j.dnarep.2014.01.010
Lee, G. S., Neiditch, M. B., Salus, S. S., and Roth, D. B. (2004). RAG Proteins shepherd Double-Strand Breaks to a Specific Pathway, Suppressing Error-Prone Repair, but RAG Nicking Initiates Homologous Recombination. Cell 117, 171–184. doi:10.1016/s0092-8674(04)00301-0
Lescale, C., Abramowski, V., Bedora-Faure, M., Murigneux, V., Vera, G., Roth, D. B., et al. (2016a). RAG2 and XLF/Cernunnos Interplay Reveals a Novel Role for the RAG Complex in DNA Repair. Nat. Commun. 7, 10529. doi:10.1038/ncomms10529
Lescale, C., and Deriano, L. (2016). V(D)J Recombination: Orchestrating Diversity Without Damage. Encyclopedia of Cell Biology 3, 550–566.
Lescale, C., Lenden Hasse, H., Blackford, A. N., Balmus, G., Bianchi, J. J., Yu, W., et al. (2016b). Specific Roles of XRCC4 Paralogs PAXX and XLF during V(D)J Recombination. Cel Rep. 16, 2967–2979. doi:10.1016/j.celrep.2016.08.069
Li, G., Alt, F. W., Cheng, H.-L., Brush, J. W., Goff, P. H., Murphy, M. M., et al. (2008). Lymphocyte-Specific Compensation for XLF/cernunnos End-Joining Functions in V(D)J Recombination. Mol. Cel. 31, 631–640. doi:10.1016/j.molcel.2008.07.017
Li, Y., Chirgadze, D. Y., Bolanos-Garcia, V. M., Sibanda, B. L., Davies, O. R., Ahnesorg, P., et al. (2008). Crystal Structure of Human XLF/Cernunnos Reveals Unexpected Differences from XRCC4 with Implications for NHEJ. EMBO J. 27, 290–300. doi:10.1038/sj.emboj.7601942
Li, Z., Dordai, D. I., Lee, J., and Desiderio, S. (1996). A Conserved Degradation Signal Regulates RAG-2 Accumulation during Cell Division and Links V(D)J Recombination to the Cell Cycle. Immunity 5, 575–589. doi:10.1016/S1074-7613(00)80272-1
Liang, Z., Kumar, V., Le Bouteiller, M., Zurita, J., Kenrick, J., Lin, S. G., et al. (2021). Ku70 Suppresses Alternative End Joining in G1-Arrested Progenitor B Cells. Proc. Natl. Acad. Sci. USA. 118, e2103630118. doi:10.1073/pnas.2103630118
Lin, S. G., Ba, Z., Alt, F. W., and Zhang, Y. (2018). RAG Chromatin Scanning during V(D)J Recombination and Chromatin Loop Extrusion Are Related Processes. Adv. Immunol. 139, 93–135. doi:10.1016/bs.ai.2018.07.001
Ling, A. K., Munro, M., Chaudhary, N., Li, C., Berru, M., Wu, B., et al. (2020). SHLD 2 Promotes Class Switch Recombination by Preventing Inactivating Deletions within the Igh Locus. EMBO Rep. 21, e49823. doi:10.15252/embr.201949823
Litman, G. W., Rast, J. P., and Fugmann, S. D. (2010). The Origins of Vertebrate Adaptive Immunity. Nat. Rev. Immunol. 10, 543–553. doi:10.1038/nri2807
Liu, X., Jiang, W., Dubois, R. L., Yamamoto, K., Wolner, Z., and Zha, S. (2012). Overlapping Functions between XLF Repair Protein and 53BP1 DNA Damage Response Factor in End Joining and Lymphocyte Development. Proc. Natl. Acad. Sci. 109, 3903–3908. doi:10.1073/pnas.1120160109
Liu, X., Shao, Z., Jiang, W., Lee, B. J., and Zha, S. (2017). PAXX Promotes KU Accumulation at DNA Breaks and Is Essential for End-Joining in XLF-Deficient Mice. Nat. Commun. 8, 13816. doi:10.1038/ncomms13816
Loc’h, J., and Delarue, M. (2018). Terminal Deoxynucleotidyltransferase: the story of an Untemplated DNA Polymerase Capable of DNA Bridging and Templated Synthesis across Strands. Curr. Opin. Struct. Biol. 53, 22–31. doi:10.1016/j.sbi.2018.03.019
Lucas, J. S., Murre, C., Feeney, A. J., and Riblet, R. (2015). The Structure and Regulation of the Immunoglobulin Loci. Mol. Biol. B Cell (Second Edition) 2015, 1–11. doi:10.1016/B978-0-12-397933-9.00001-1
Ma, Y., Pannicke, U., Schwarz, K., and Lieber, M. R. (2002). Hairpin Opening and Overhang Processing by an Artemis/DNA-Dependent Protein Kinase Complex in Nonhomologous End Joining and V(D)J Recombination. Cell 108, 781–794. doi:10.1016/S0092-8674(02)00671-2
Mijušković, M., Brown, S. M., Tang, Z., Lindsay, C. R., Efstathiadis, E., Deriano, L., et al. (2012). A Streamlined Method for Detecting Structural Variants in Cancer Genomes by Short Read Paired-End Sequencing. PLoS One 7, e48314. doi:10.1371/journal.pone.0048314
Mijušković, M., Chou, Y.-F., Gigi, V., Lindsay, C. R., Shestova, O., Lewis, S. M., et al. (2015). Off-Target V(D)J Recombination Drives Lymphomagenesis and Is Escalated by Loss of the Rag2 C Terminus. Cel Rep. 12, 1842–1852. doi:10.1016/j.celrep.2015.08.034
Mirman, Z., and de Lange, T. (2020). 53BP1: A DSB Escort. Genes Dev. 34, 7–23. doi:10.1101/gad.333237.119
Mirman, Z., Lottersberger, F., Takai, H., Kibe, T., Gong, Y., Takai, K., et al. (2018). 53BP1-RIF1-Shieldin Counteracts DSB Resection through CST- and Polα-Dependent Fill-In. Nature 560, 112–116. doi:10.1038/s41586-018-0324-7
Mitelman, F., Johansson, B., and Mertens, F. (2007). The Impact of Translocations and Gene Fusions on Cancer Causation. Nat. Rev. Cancer 7, 233–245. doi:10.1038/nrc2091
Mitrentsi, I., Yilmaz, D., and Soutoglou, E. (2020). How to Maintain the Genome in Nuclear Space. Curr. Opin. Cel Biol. 64, 58–66. doi:10.1016/j.ceb.2020.02.014
Nick McElhinny, S. A., Snowden, C. M., McCarville, J., and Ramsden, D. A. (2000). Ku Recruits the XRCC4-Ligase IV Complex to DNA Ends. Mol. Cel. Biol. 20, 2996–3003. doi:10.1128/MCB.20.9.2996-3003.2000
Noordermeer, S. M., Adam, S., Setiaputra, D., Barazas, M., Pettitt, S. J., Ling, A. K., et al. (2018). The Shieldin Complex Mediates 53BP1-Dependent DNA Repair. Nature 560, 117–121. doi:10.1038/s41586-018-0340-7
Nussenzweig, A., and Nussenzweig, M. C. (2010). Origin of Chromosomal Translocations in Lymphoid Cancer. Cell 141, 27–38. doi:10.1016/j.cell.2010.03.016
Ochi, T., Blackford, A. N., Coates, J., Jhujh, S., Mehmood, S., Tamura, N., et al. (2015). PAXX, a Paralog of XRCC4 and XLF, Interacts with Ku to Promote DNA Double-Strand Break Repair. Science 347, 185–188. doi:10.1126/science.1261971
Oksenych, V., Alt, F. W., Kumar, V., Schwer, B., Wesemann, D. R., Hansen, E., et al. (2012). Functional Redundancy between Repair Factor XLF and Damage Response Mediator 53BP1 in V(D)J Recombination and DNA Repair. Proc. Natl. Acad. Sci. 109, 2455–2460. doi:10.1073/pnas.1121458109
Oksenych, V., Kumar, V., Liu, X., Guo, C., Schwer, B., Zha, S., et al. (2013). Functional Redundancy between the XLF and DNA-PKcs DNA Repair Factors in V(D)J Recombination and Nonhomologous DNA End Joining. Proc. Natl. Acad. Sci. 110, 2234–2239. doi:10.1073/pnas.1222573110
Paulson, T. G., Almasan, A., Brody, L. L., and Wahl, G. M. (1998). Gene Amplification in a P53-Deficient Cell Line Requires Cell Cycle Progression under Conditions that Generate DNA Breakage. Mol. Cel. Biol. 18, 3089–3100. doi:10.1128/MCB.18.5.3089
Pham, N., Yan, Z., Yu, Y., Faria Afreen, M., Malkova, A., Haber, J. E., et al. (2021). Mechanisms Restraining Break‐Induced Replication at Two‐Ended DNA Double‐strand Breaks. EMBO J. 40, e104847. doi:10.15252/embj.2020104847
Piazza, A., Bordelet, H., Dumont, A., Thierry, A., Savocco, J., Girard, F., et al. (2021). Cohesin Regulates Homology Search during Recombinational DNA Repair. Nat. Cel Biol. 23, 1176–1186. doi:10.1038/s41556-021-00783-x
Purugganan, M. M., Shah, S., Kearney, J. F., and Roth, D. B. (2001). Ku80 Is Required for Addition of N Nucleotides to V(D)J Recombination Junctions by Terminal Deoxynucleotidyl Transferase. Nucleic Acids Res. 29, 1638–1646. doi:10.1093/nar/29.7.1638
Ramsden, D. A., Carvajal-Garcia, J., and Gupta, G. P. (2021). Mechanism, Cellular Functions and Cancer Roles of Polymerase-Theta-Mediated DNA End Joining. Nat. Rev. Mol. Cel Biol. Online ahead of print. doi:10.1038/s41580-021-00405-2
Ramsden, D. A., and Nussenzweig, A. (2021). Mechanisms Driving Chromosomal Translocations: Lost in Time and Space. Oncogene 40, 4263–4270. doi:10.1038/s41388-021-01856-9
Ramsden, D. A. (2011). Polymerases in Nonhomologous End Joining: Building a Bridge over Broken Chromosomes. Antioxid. Redox Signaling 14, 2509–2519. doi:10.1089/ars.2010.3429
Reid, D. A., Keegan, S., Leo-Macias, A., Watanabe, G., Strande, N. T., Chang, H. H., et al. (2015). Organization and Dynamics of the Nonhomologous End-Joining Machinery during DNA Double-Strand Break Repair. Proc. Natl. Acad. Sci. USA 112, E2575–E2584. doi:10.1073/pnas.1420115112
Riballo, E., Woodbine, L., Stiff, T., Walker, S. A., Goodarzi, A. A., and Jeggo, P. A. (2009). XLF-Cernunnos Promotes DNA Ligase IV-XRCC4 Re-Adenylation Following Ligation. Nucleic Acids Res. 37, 482–492. doi:10.1093/nar/gkn957
Rogers, C. H., Mielczarek, O., and Corcoran, A. E. (2021). Dynamic 3D Locus Organization and its Drivers Underpin Immunoglobulin Recombination. Front. Immunol. 11, 1–9. doi:10.3389/fimmu.2020.633705
Ropars, V., Drevet, P., Legrand, P., Baconnais, S., Amram, J., Faure, G., et al. (2011). Structural Characterization of Filaments Formed by Human Xrcc4-Cernunnos/XLF Complex Involved in Nonhomologous DNA End-Joining. Proc. Natl. Acad. Sci. 108, 12663–12668. doi:10.1073/pnas.1100758108
Roth, D. B. (2003). Restraining the V(D)J Recombinase. Nat. Rev. Immunol. 3, 656–666. doi:10.1038/nri1152
Roth, D. B. (2014). V(D)J Recombination: Mechanism, Errors, and Fidelity. Microbiol. Spectr. 2, 1–19. doi:10.1128/microbiolspec.MDNA3-0041-2014
Schatz, D. G., and Ji, Y. (2011). Recombination Centres and the Orchestration of V(D)J Recombination. Nat. Rev. Immunol. 11, 251–263. doi:10.1038/nri2941
Sekiguchi, J., Whitlow, S., and Alt, F. W. (2001). Increased Accumulation of Hybrid V(D)J Joins in Cells Expressing Truncated versus Full-Length RAGs. Mol. Cel 8, 1383–1390. doi:10.1016/s1097-2765(01)00423-3
Setiaputra, D., and Durocher, D. (2019). Shieldin - the Protector of DNA Ends. EMBO Rep. 20, e47560. doi:10.15252/embr.201847560
Soulas-Sprauel, P., Le Guyader, G., Rivera-Munoz, P., Abramowski, V., Olivier-Martin, C., Goujet-Zalc, C., et al. (2007). Role for DNA Repair Factor XRCC4 in Immunoglobulin Class Switch Recombination. J. Exp. Med. 204, 1717–1727. doi:10.1084/jem.20070255
Stinson, B. M., Moreno, A. T., Walter, J. C., and Loparo, J. J. (2020). A Mechanism to Minimize Errors during Non-Homologous End Joining. Mol. Cel. 77, 1080–1091.e8. doi:10.1016/j.molcel.2019.11.018
Symington, L. S. (2016). Mechanism and Regulation of DNA End Resection in Eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51, 195–212. doi:10.3109/10409238.2016.1172552
Talukder, S. R., Dudley, D. D., Alt, F. W., Takahama, Y., and Akamatsu, Y. (2004). Increased Frequency of Aberrant V(D)J Recombination Products in Core RAG-Expressing Mice. Nucleic Acids Res. 32, 4539–4549. doi:10.1093/nar/gkh778
Tsai, C. J., Kim, S. A., and Chu, G. (2007). Cernunnos/XLF Promotes the Ligation of Mismatched and Noncohesive DNA Ends. Proc. Natl. Acad. Sci. 104, 7851–7856. doi:10.1073/pnas.0702620104
Tubbs, A. T., Dorsett, Y., Chan, E., Helmink, B., Lee, B.-S., Hung, P., et al. (2014). KAP-1 Promotes Resection of Broken DNA Ends Not Protected by γ-H2AX and 53BP1 in G 1 -Phase Lymphocytes. Mol. Cel. Biol. 34, 2811–2821. doi:10.1128/mcb.00441-14
Vera, G., Rivera-Munoz, P., Abramowski, V., Malivert, L., Lim, A., Bole-Feysot, C., et al. (2013). Cernunnos Deficiency Reduces Thymocyte Life Span and Alters the T Cell Repertoire in Mice and Humans. Mol. Cel. Biol. 33, 701–711. doi:10.1128/mcb.01057-12
Walker, J. R., Corpina, R. A., and Goldberg, J. (2001). Structure of the Ku Heterodimer Bound to DNA and its Implications for Double-Strand Break Repair. Nature 412, 607–614. doi:10.1038/35088000
Wyatt, D. W., Feng, W., Conlin, M. P., Yousefzadeh, M. J., Roberts, S. A., Mieczkowski, P., et al. (2016). Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks. Mol. Cel. 63, 662–673. doi:10.1016/j.molcel.2016.06.020
Yan, C. T., Boboila, C., Souza, E. K., Franco, S., Hickernell, T. R., Murphy, M., et al. (2007). IgH Class Switching and Translocations Use a Robust Non-Classical End-Joining Pathway. Nature 449, 478–482. doi:10.1038/nature06020
Yu, W., Lescale, C., Babin, L., Bedora-Faure, M., Lenden-Hasse, H., Baron, L., et al. (2020). Repair of G1 Induced DNA Double-Strand Breaks in S-G2/M by Alternative NHEJ. Nat. Commun. 11, 5239. doi:10.1038/s41467-020-19060-w
Zahid, S., Seif El Dahan, M., Iehl, F., Fernandez-Varela, P., Le Du, M.-H., Ropars, V., et al. (2021). The Multifaceted Roles of Ku70/80. Int. J. Mol. Sci. 22, 4134. doi:10.3390/ijms22084134
Zha, S., Guo, C., Boboila, C., Oksenych, V., Cheng, H.-L., Zhang, Y., et al. (2011). ATM Damage Response and XLF Repair Factor Are Functionally Redundant in Joining DNA Breaks. Nature 469, 250–254. doi:10.1038/nature09604
Zha, S., Shao, Z., and Zhu, Y. (2021). The Plié by DNA-PK: Dancing on DNA. Mol. Cel 81, 644–646. doi:10.1016/j.molcel.2021.01.025
Zhang, Y., Zhang, X., Ba, Z., Liang, Z., Dring, E. W., Hu, H., et al. (2019). The Fundamental Role of Chromatin Loop Extrusion in Physiological V(D)J Recombination. Nature 573, 600–604. doi:10.1038/s41586-019-1547-y
Zhao, F., Kim, W., Gao, H., Liu, C., Zhang, Y., Chen, Y., et al. (2021). ASTE1 Promotes Shieldin-Complex-Mediated DNA Repair by Attenuating End Resection. Nat. Cel Biol. 23, 894–904. doi:10.1038/s41556-021-00723-9
Keywords: DNA double-strand break, V(D)J recombination, non-homologous end-joining, homology-directed repair, DNA end resection, DNA double-strand break repair pathway choice
Citation: Libri A, Marton T and Deriano L (2022) The (Lack of) DNA Double-Strand Break Repair Pathway Choice During V(D)J Recombination. Front. Genet. 12:823943. doi: 10.3389/fgene.2021.823943
Received: 28 November 2021; Accepted: 13 December 2021;
Published: 05 January 2022.
Edited by:
Jenny Kaur Singh, Institut Curie, FranceReviewed by:
Magdalena Rother, Leiden University Medical Center, NetherlandsRichard L. Frock, Stanford University, United States
Copyright © 2022 Libri, Marton and Deriano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ludovic Deriano, bHVkb3ZpYy5kZXJpYW5vQHBhc3RldXIuZnI=
†These authors have contributed equally to this work
 Alice Libri†
Alice Libri† Timea Marton
Timea Marton Ludovic Deriano
Ludovic Deriano