
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 20 January 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.821281
This article is part of the Research Topic Legume Breeding in Transition: Innovation and Outlook View all 28 articles
 Yasir Sharif1,2
Yasir Sharif1,2 Hua Chen2,3
Hua Chen2,3 Ye Deng1,2
Ye Deng1,2 Niaz Ali2,3
Niaz Ali2,3 Shahid Ali Khan2,3
Shahid Ali Khan2,3 Chong Zhang2,3
Chong Zhang2,3 Wenping Xie1,2
Wenping Xie1,2 Kun Chen1,2
Kun Chen1,2 Tiecheng Cai2,3
Tiecheng Cai2,3 Qiang Yang2,3
Qiang Yang2,3 Yuhui Zhuang2
Yuhui Zhuang2 Ali Raza2,3
Ali Raza2,3 Weijian Zhuang1,2,3*
Weijian Zhuang1,2,3*Peanut (Arachis hypogaea L.) is an important oil and food legume crop grown in tropical and subtropical areas of the world. As a geocarpic crop, it is affected by many soil-borne diseases and pathogens. The pericarp, an inedible part of the seed, acts as the first layer of defense against biotic and abiotic stresses. Pericarp promoters could drive the defense-related genes specific expression in pericarp for the defense application. Here, we identified a pericarp-abundant promoter (AhGLP17-1P) through microarray and transcriptome analysis. Besides the core promoter elements, several other important cis-elements were identified using online promoter analysis tools. Semiquantitative and qRT-PCR analyses validated that the AhGLP17-1 gene was specifically expressed only in the pericarp, and no expression was detected in leaves, stem, roots, flowers, gynophore/peg, testa, and embryo in peanut. Transgenic Arabidopsis plants showed strong GUS expression in siliques, while GUS staining was almost absent in remaining tissues, including roots, seedlings, leaf, stem, flowers, cotyledons, embryo, and seed coat confirmed its peanut expressions. Quantitative expression of the GUS gene also supported the GUS staining results. The results strongly suggest that this promoter can drive foreign genes’ expression in a pericarp-abundant manner. This is the first study on the functional characterization of the pericarp-abundant promoters in peanut. The results could provide practical significance to improve the resistance of peanut, and other crops for seed protection uses.
Peanut (Arachis hypogaea L.), also known as groundnut, is an important legume crop (Fabaceae family) that is mainly cultivated for its edible seeds. Botanically peanut is a legume, but it is frequently eaten as a “nut”, and its nutritional value is comparable to other nuts (Settaluri et al., 2012). It is a major nutrition source for humanity, providing edible oil and proteins (Khan et al., 2020) after soybean and rapeseeds. Peanut is full of nutrients, including carbohydrates, vitamins, lipids, minerals, etc. (Akhtar et al., 2014). Peanut is widely cultivated globally in tropical and subtropical regions, producing 48.75 million tons of shells annually. China is the leading producer with 17.52 million tons of shells and shares 36% of the Worlds’ total production, followed by India (14%), Nigeria, Sudan, and the USA (FAOSTAT, 2019).
Like other crops, the peanut is affected by several bacterial, fungal, and viral pathogens (Ali et al., 2020). To manage these pathogens, nature has provided peanuts with several defense mechanisms. As peanut seeds are of primary importance, they have evolved to form non-edible outer coverings (pericarp and testa) to protect the seeds from insects, pathogens, and physical damage and maintain seed viability from generation to generation (Souza and Marcos-Filho, 2001). The shape, color, thickness, etc., of the seed coat, may differ among species during the evolutionary process and due to different growing environments. Biologically active chemicals in seed coats provide the right solution for infections, especially seed coat-based pathogen resistance attributed to hydrophobic molecules like lignin and tannins (Dardick and Callahan, 2014). Seed inner pericarp tissues also accumulate different antifungal and antibacterial metabolic by-products and flavonoids (Dixon and Paiva, 1995). Plenty of work is available on the genetic basis of seed coat development in the model dicot plant, i.e., Arabidopsis, and many genes have been worked out for their functions in seed coat development (Windsor et al., 2000; Haughn and Chaudhury, 2005; Dardick and Callahan, 2014). Similarly, detailed knowledge about the genes involved in seed coat development in peanut can help manipulate specific genes to get resistance against specific biotic or abiotic stresses. Paik-Ro and his team have reported that the cDNA of PSC33I is specifically expressed in peanut seed tissues without showing any expression in other tissues (Paik-Ro et al., 2002).
The physical appearance of any plant and its response to the surrounding environment is mainly controlled by a cascade of genes. Expression of any gene depends upon the binding of specific transcription factors (TFs) or proteins with unique upstream elements (cis-elements) and regulation at the transcription site. Promoters are the non-coding regulatory DNA sequences present upstream of a genes’ coding region and contain specific cis-elements that regulate the spatio-temporal expression of a gene. A promoter has multiple binding sequences for TFs and RNA polymerase for activation and regulation of functions of the downstream gene (Ong and Corces, 2011; Gupta et al., 2012; Hernandez-Garcia and Finer, 2014). Promoters are important tools for molecular research to study the functions of a gene (Xu et al., 2016). Promoters can be divided into constitutive, spatio-temporal/tissue-specific, inducible, and synthetic promoters based on regulating the gene functions. Constitutive promoters drive the expression of a gene in all tissues, such as maize ubiquitin promoter (Christensen et al., 1992) and cauliflower mosaic virus promoter (CaMV35S) (Odell et al., 1985), are widely used for functional gene studies in plants.
Similarly, Figwort mosaic virus sub-genomic transcript promoter FMV Sgt is another example of constitutive promoters (Bhattacharyya et al., 2002). The constitutive expression of an exogenous gene can cause adverse effects on a plants’ normal growth and functioning as it causes an extra burden on plant metabolism for expressing a gene in tissues where it is not required, and sometimes it results in undesired phenotypes (Nakashima et al., 2007). Tissue-specific and inducible promoters are more important as they can drive a genes’ expression in a tissue-specific manner or under specific stress; hence, avoiding the adverse effects of constitutive expression. The selection of a suitable promoter is the key step for developing transgenic plants since it influences the cell, tissue, or stage specificity and determines the expression level of the transgene (Koyama et al., 2005). Therefore, comprehensive knowledge of promoter activity is a prerequisite for transgenic breeding.
In this paper, we studied the promoter of a peanut pericarp abundant expression gene belonging to the Germin-like protein (GLPs) family. Germin-like proteins were first identified in germinating wheat seedlings as germination specific markers during the 1980s (Lane et al., 1993). These proteins belong to a group of water-soluble glycoproteins with diverse functions. GLPs are ubiquitously found in gymnosperms, and angiosperms (Lu et al., 2010) and are broadly involved in plants’ defense against biotic and abiotic stresses (Godfrey et al., 2007). Germin-like proteins are a group of “Cupin superfamily” containing the Cupin domain (PF00190). Structurally these proteins are composed of the β-sheet barrel (jellyroll beta-barrel structure), and the protein’s C-terminus contains a metal ion binding site (Agarwal et al., 2009). GLPs contain two conserved motifs known as “germin-box”. These motifs G(x)5HxH(x)3,4E(x)6G and G(x)5PxG(x)2H(x)3N are packed in a classic jellyroll beta-barrel structural domain (Yamahara et al., 1999). Thus far, there is no report available which investigated its specific expression and characterized its upstream promoter in a plant pericarp.
Here, this study identified and characterized a peanut GLP gene, “Germin-like protein subfamily 1 member 7 (AhGLP17-1)," showing abundant expression in seed pericarp. This gene was selected based on the transcriptome and microarray expression data, and its pericarp abundant expression was verified by semiquantitative and qRT-PCR. Further, the cis-regulatory elements of the promoter were analyzed, and their expression was characterized in transgenic Arabidopsis plants.
Seeds of peanut cultivars Minhua-6 (M-6), and Xinhuixiaoli (XHXL), and Arabidopsis thaliana ecotype Columbia-0 (Col-0) were obtained from the Institute of Oil Crops, Fujian Agriculture and Forestry University, Fuzhou, China. Peanut plants (M-6) were grown in research fields at Yangzhong, Sanming county of Fujian province during the summer season (April-August, 2018), and samples from different tissues, including leaves, stem, flower, peg, pericarp, testa, embryo, and roots were collected at different growing stages for semiquantitative and qRT-PCR-based confirmation of AhGLP17-1 gene. For isolation of promoter, peanut plants (XHXL) were grown in the greenhouse in small pots (10 cm diameter) filled with compost, and leaf samples were collected from one-week-old plants. Arabidopsis plants were germinated on MS medium (Murashige and Skoog, 1962), transferred into 8-cm diameter plastic pots containing compost after 2 weeks, and further grown in the greenhouse, where 25°C temperature and 16/8 h photoperiod for day/night cycle was maintained for Arabidopsis seedlings. All samples were washed with 75% ethanol and sterilized water, packed in previously labeled plastic bags, and immediately frozen in liquid nitrogen and stored at −80°C for further use.
A pericarp abundant gene with Peanut Genome Resource (PGR) ID AH06G08990.1 was identified by the microarray and transcriptome expression data, which is available at the PGR database http://peanutgr.fafu.edu.cn/ (accessed on 20th April 2018) (Zhuang et al., 2019). This gene belongs to the germin-like protein family viz. “germin-like protein subfamily 1 member 7”. Due to the polyploid nature of the peanut genome, peanut contains nine different copies of germin-like protein subfamily 1 member 7, and we named this gene AhGLP17-1. The protein 3D structure of AhGLP17-1 was constructed using 3D Ligand Site https://www.wass-michaelislab.org/3dlig/index.html (accessed on 15th June 2018) (Wass et al., 2010), physio-chemical properties were predicted by Expasy server https://web.expasy.org/protparam/ (accessed on 20th June 2018) (Gasteiger et al., 2005), and subcellular localization was predicted by CELLO V2.5 http://cello.life.nctu.edu.tw/ (accessed on 22nd June 2018) (Yu et al., 2006). Gene functional annotation “gene ontology” information was retrieved from the PGR database (http://peanutgr.fafu.edu.cn/). Semiquantitative and qRT-PCR analysis were used to confirm its expression in different tissues.
A 2296 bp region upstream of the start codon of the AhGLP17-1 gene was selected for promoter analysis. The promoter was named AhGLP17-1P. Cis-regulatory elements of promoter region were predicted by PLACE https://www.dna.affrc.go.jp/PLACE/?action=newplace (accessed on 30th June 2018) (Higo et al., 1998) and PlantCARE databases http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 30th June 2018) (Rombauts et al., 1999).
RNA from different tissues (leaves, stem, flowers, peg, pericarp, testa, embryo, and roots) of peanut cultivar Minhua-6 was extracted by the Cetyl Trimethyl Ammonium Bromide (CTAB) (200 mM Tris HCL, 20 mM EDTA, 2% CTAB, 1.4 M NaCl, and 2% PVP-40, pH = 8.0, 0.2% β-Mercapto Ethanol added before use) method with some modification (Chen et al., 2016). First-strand cDNA was synthesized using 1 µg RNA with the PrimeScript 1st strand cDNA Synthesis Kit (Cat# 6110A) (Takara, Dalian, China) according to manufacturer guidelines. Semiquantitative PCR analysis was performed to check the expression of the AhGLP17-1 gene in different tissues using the peanut Actin gene and gene-specific primers. The semiquantitative PCR results were viewed on 1% agarose gel. For qRT-PCR analysis, three different cDNA preparations for each tissue were used. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed by MonAmp™ ChemoHS qPCR Mix (Cat# 160431) (Monad Biotech, Wuhan, China). The PCR reaction mixture contained 10 µL MonAmp Master Mix, 1 µL forward and reverse primer (diluted at 10 mM concentration), and 1 µL cDNA template. The peanut Actin gene (Chi et al., 2012) was used as an internal control to normalize the cDNA. The qRT-PCR reaction was performed using Applied Biosystems 7,500 real-time PCR system (ThermoFisher Scientific, USA). The cycling program was as follows: 94°C (1 min), 60°C (1 min), and 72°C (1 min) for 40 cycles. Primers used for semiquantitative and qRT-PCR analysis are given in (Supplementary Table S1).
Genomic DNA from peanut (XHXL variety) leaves was extracted using the CTAB (1 M Tris-HCl, 0.5 M Na-EDTA, 2% CTAB, 1.4 M NaCl, 3% PVP-40, and 0.2% β-Mercapto Ethanol added before use) method (Li et al., 2013) with some modifications. Promoter-specific primers (Supplementary Table S1) covering 2,296 bp upstream region before start codon were used to clone AhGLP17-1P. The promoter region was amplified with PrimeSTAR® Max DNA polymerase (Cat# Ro45B) (Takara, Dalian, China), according to manufacturers’ guidelines. The PCR amplified product was visualized on 2% agarose gel and purified by TIANGEN Universal DNA Purification Kit (Cat# DP103-03) (Tiangen Biotech Beijing, China). Purified PCR products were sub-cloned into pMD19T vector (Cat# 3271) (Takara, Dalian, China) and sequenced by Beijing Genomics Institute (BGI, Shenzhen, China).
The sequence-verified PCR clones were used to construct the expression vector using Two-step Gateway cloning. First, the promoter was amplified by primers containing universal overlapping sequences for gateway vectors (Supplementary Table S1) using the TA-cloning product as template and purified through TIANGEN Universal DNA Purification Kit (Cat# DP214-03) (Tiangen Biotech Beijing, China). The purified PCR products were ligated by Gateway BP reaction using BP Clonase enzyme (Cat# 11789020) (Invitrogen, ThermoFisher Scientific USA) into entry vector pDONR-207 between attP sites. Vector pDONR-207 containing AhGLP17-1P was transferred to E. coli (DH5α), and positive clones were selected for sequence confirmation. After sequence confirmation, AhGLP17-1P was ligated into destination vector pMDC164 by Gateway LR reaction using LR Clonase enzyme (Cat# 11791020) (Invitrogen, ThermoFisher Scientific USA). Expression vector pMDC164 contains the hygromycin resistance gene for positive plants’ selection and the GUS reporter gene. The vector was named pMDC164-AhGLP17-1P.
Expression vector pMDC164-AhGLP17-1P was transferred into Agrobacterium tumefaciens strain GV3101, and positive colonies were selected on yeast extract beef (YEB) selection medium plates supplemented with 50 μg ml−1 kanamycin. Positive A. tumefaciens cells harboring the expression vector were grown overnight at 28°C, 220 rpm to get logarithmic growth phase (OD600 = 1.0-1.5) in liquid YEB medium supplemented with kanamycin 50 μg ml−1, and 75 μg ml−1 rifampicin. Bacterial cells were harvested by centrifugation at 4,000 rpm for 10 min and resuspended in 5% sucrose solution containing 0.02% Silwet L-77 and 100 μg ml−1 acetosyringone was also added to achieve higher transformation efficiency (Sheikholeslam and Weeks, 1987). Mature Arabidopsis plants were used for transformation by floral dip method (Clough and Bent, 1998) by dipping the unopened flowers into the prepared solution for 10–15 s, and then placed under dark for 24 h. Siliques and opened flowers were removed before the transformation, and the floral dipping was repeated after 5 days. After that, plants were grown under optimum growth conditions until seeds were ready to harvest, and finally, the T0 seeds were harvested.
To identify positively transformed plants, T0 transgenic seeds were screened on MS medium containing 50 μg ml−1 hygromycin. First, seeds were surface sterilized with 75% ethanol for 2 min and then treated with 10% H2O2 for 2 min, followed by 4–5 times washing with distilled water. Then seeds were spread over MS medium containing plates supplemented with Hygromycin antibiotic. Eight randomly selected hygromycin-resistant plants were verified by PCR amplification with promoter-specific forward and GUS gene specific reverse primers (Supplementary Table S1). The selected transgenic plants were covered with plastic sheets to avoid the chances of cross-pollination, and T1 transgenic seeds were obtained. Transgenic seeds were grown to further generations to get homozygous T3 lines.
For the GUS staining assay (Jefferson et al., 1987), tissue samples were incubated in 2 mM 5-Bromo-4-chloro-3-indolyl β-D-glucuronide (X-Gluc) solution prepared in 0.1% Triton X-100, 50 mM sodium phosphate buffer, 10 mM EDTA, 2 mM potassium ferricyanide, and 2 mM potassium ferrocyanide. Plant samples were incubated in the above-prepared staining solution at 37°C for 12 h. After staining, samples were washed with 50, 75, and 100% ethanol for 5 min separately. Finally, samples were decolorized by incubating in 75% ethanol at 37°C until all green color was removed, while ethanol was changed every 4 hours. Samples were photographed with the digital camera and Olympus microscope Model BX3-CBH with attached Olympus DP80 digital camera (Olympus, Tokyo, Japan). Quantitative expression of the GUS gene in different tissues of transgenic Arabidopsis plants was analyzed by qRT-PCR with GUS gene-specific primers (Supplementary Table S1).
For pericarp sampling, young siliques were carefully opened by sharp needles and forceps, and seeds were removed. Siliques without seeds were stored for RNA extraction. Similarly, seeds were used to examine testa, cotyledon, and embryo expression. It was impossible to separate the testa from cotyledons and get enough samples for RNA extraction from Arabidopsis seeds, so whole seeds were used to extract the RNA. RNA from different tissues of transgenic Arabidopsis was extracted using TriQuick Reagent (Cat# R1100) (Solarbio, Beijing, China), following the manufacturers’ instructions.
The seeds of transgenic Arabidopsis plants were ruptured and incubated in the GUS staining solution to confirm whether GUS staining is present in testa, cotyledons, and embryo or not. After overnight incubation in GUS solution, seeds were further processed for cryostat sectioning in Leica CM1950 Cryostat Microtome (Leica Biosystems, Germany). The cryostat microtome was turned on for 5 hours before use, and the temperature was set at −20°C. Specimen discs, brushes, and forceps were put inside the cooling chamber. The freezing compound was applied on specimen discs, and seeds were gently placed on specimen discs containing the freezing compound. Specimen discs containing the seeds were kept at −20°C for 30 min. After that, 50 µm sections were made and placed on glass slides. Later, images of sectioned specimens were taken by Olympus IX73 microscope with attached Olympus DP80 digital camera (Olympus, Tokyo, Japan).
We searched the Peanut Genome Resource database (http://peanutgr.fafu.edu.cn/) (Zhuang et al., 2019) for the candidate gene with high transcriptome and microarray expression in pericarp and with no or very low expression in other tissues for cloning of promoter. A member of the peanut germin-like protein family (AhGLP) named germin-like protein subfamily 1 member 7 (AhGLP17-1) with the PGR gene ID AH06G08990 and mRNA ID AH06G08990.1 was found to be specifically showing high expression in pericarp as compared to other tissues (Figure 1A). Although microarray expression was found in root tissues and gynophore/peg (Supplementary Table S2), some transcriptome expression was also present in roots and peg (Supplementary Table S3). Still, the tendency of expression of the AhGLP17-1 gene was higher in the pericarp (Figure 1A). This gene is present on the 6th chromosome of sub-genome A at 12182490-12204462 position on the negative strand and has a CDS length of 666 base pairs and 21973 base pairs genomic length. The protein, CDS, and promoter sequences of the AhGLP17-1 gene are given in Supplementary file S1. It consists of three exons of almost the same size and two introns, one of which is 21170 bp and the second intron is 134 bp long (Figure 1B). In silico subcellular localization showed that AhGLP17-1 is localized in extracellular spaces and plasma membrane. Protein comprises 222 amino acid residues with a molecular weight of 24.49 KDa and a theoretical isoelectric point of 9.36. Further, it contains the Cupin_1 domain (PF00190) at 87-187 aa position (Figure 1C). The protein 3D structure prediction showed that AhGLP17-1 is composed of the β-sheet barrel (jellyroll beta-barrel structure) with the ligands (Figure 1D). GO functional annotation revealed that the AhGLP17-1 gene participates in three categories, including biological process in oxalate metabolic process (GO:0033609), molecular functions in nutrient reservoir activity (GO:0045735), oxalate decarboxylase activity (GO:0046564), manganese ion binding (GO:0030145), and cellular components apoplast (GO:0048046), cell wall (GO:0005618), and extracellular region (GO:0005576) (Supplementary Table S4). Other related information (orthologues in other plant species) is given in Supplementary Table S5.
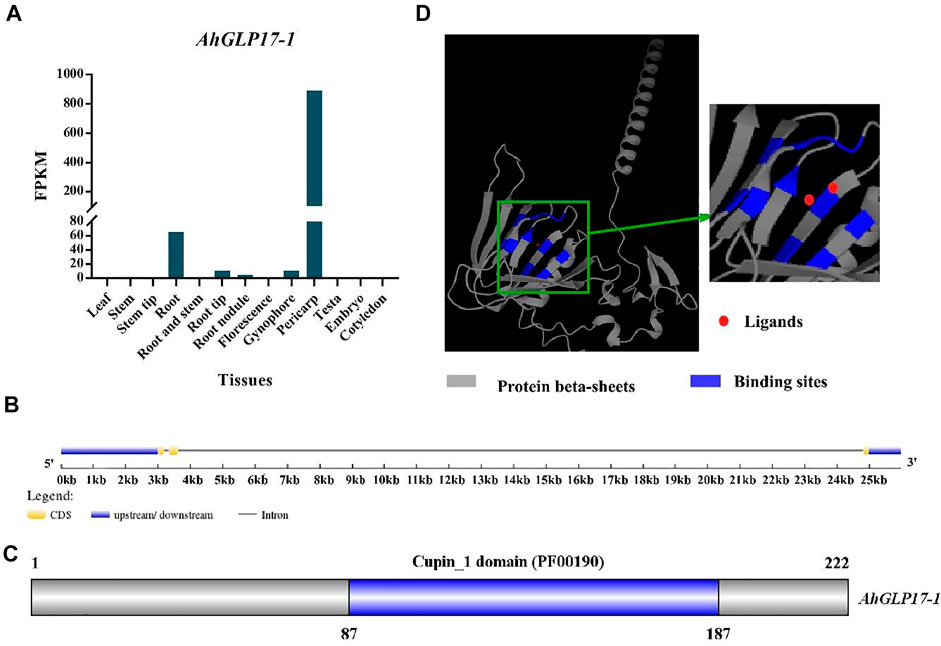
FIGURE 1. Expression and characterization of AhGLP17-1 gene. (A) Transcriptome expression (FPKM values) of the AhGLP17-1 gene in different tissues of peanut (average values of the pericarp, testa, and embryo transcriptome expression are used). (B) Gene structure of AhGLP17-1. (C) The position of cupin_1 domain. (D)The 3D protein structure of AhGLP17-1. Where grey color shows the protein β-sheets, blue color shows binding sites, and red dots show ligands.
Pericarp-abundant expression of AhGLP17-1 among different tissues was confirmed by semiquantitative PCR and qRT-PCR. The peanut Actin gene was used as an internal control for both semiquantitative and qRT-PCR analysis. The peanut actin gene showed a bright band in RNA samples of all tissues, while the AhGLP17-1 gene showed a bright band in the pericarp samples, but no expression was detected in all other tissues (Figure 2A). Results of semiquantitative PCR showed that AhGLP17-1 was preferentially expressed in the pericarp. On the other hand, the qRT-PCR results clearly showed that a high level of transcripts of AhGLP17-1 was present in the pericarp with minute expression in all other tissues (Figure 2B). Both semiquantitative PCR and qRT-PCR results showed that the AhGLP17-1 gene was specifically expressed in the pericarp. It showed very minute expression in all other tissues.
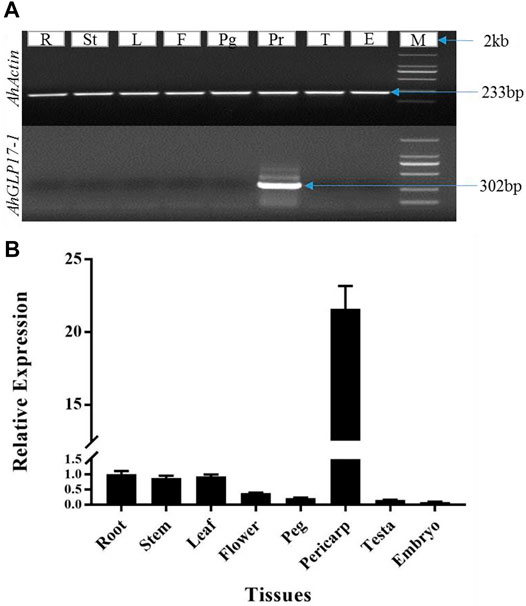
FIGURE 2. Expression analysis of AhGLP17-1 gene expression in different tissues. (A) Semiquantitaive PCR-based expression analysis, (B) qRT-PCR-based expression analysis of AhGLP17-1 gene expression. Both semiquantitative and qRT-PCR results are consistent with transcriptome and microarray expression data (as shown in Figure 1A). L = leaf, St = stem, Fl = flower, Em = embryo, Ts = testa, peg = peg/gynophore, peri = pericarp, and R = root. Root expression was used as a control to analyze the data.
A 2296 bp upstream sequence of AhGLP17P-1 contained the basic promoter elements, including the TATA box, the key element for precise transcription initiation (Grace et al., 2004), and the CAAT box required for tissue-specific activity (Shirsat et al., 1989). Many other important regulatory elements, including light-responsive elements (ATCT-motif, Box 4, G-Box, GA-motif, GATA-motif, GT1-motif, and Gap-box); hormones-responsive elements including salicylic acid (TCA-element), gibberellin (TATC-box), ethylene (ERE), and abscisic acid (ABRE) were also predicted in the AhGLP17-1P. Moreover, defense-related elements (TC-rich repeats and MYB binding sites), wound responsive element (WUN-motif), and anaerobic induction responsive element (ARE) were also found inside the promoter region. Further information on cis-regulatory elements and their position in AhGLP17-1P is presented in Figure 3.

FIGURE 3. Sequence analysis of AhGLP17-1P promoter. Presence of cis-elements in promoter sequences predicted by the PlantCARE database.
Analysis of cis-elements by the PLACE database revealed the presence of a number of important elements, including seed-specific elements (RY-element) and transcription factor binding sites. The details of transcription factor binding sites and other elements are given in Supplementary Table S6. The presence of these regulatory elements strongly suggests that this promoter can be suitable substitute for a genes’ native promoter. Except for these already reported cis-elements, some unknown elements were also found in the promoter region of the AhGLP17-1 gene (Figure 3).
A 2296bp region for AhGLP17-1P was PCR amplified (Figure 4A) from the DNA template of a high yielding and fungal pathogens resistant peanut variety XHXL (Khan et al., 2020) by promoter-specific primers (Supplementary Table S1). After confirmation of sequence, the amplified promoter fragment was again amplified with gateway primers containing gateway adapter sequences and then ligated into the attP sites of entry vector pDONR207 by Gateway BP-cloning reaction (Figure 4B). The sequence was confirmed again after BP-cloning, and the promoter fragment was ligated into the attR sites of expression vector pMDC164 by Gateway LR-cloning reaction. The resulting expression vector pMDC164-AhGLP17-1P (Figure 4C) was transformed to Agrobacterium tumefaciens competent cells by heat shock method. Positive Agrobacterium colonies harboring the expression vector were selected on selection medium (YEB plates containing kanamycin and rifampicin antibiotics). Positive agrobacterium colonies were used to transform the Arabidopsis plants through the floral dip method, and hygromycin resistant T0 transgenic plants were screened on MS plates containing 50 μg ml−1 hygromycin antibiotic. Non-transformed plants turned yellow on hygromycin selection medium, while positively transformed plants were dark green and healthy, and these plants were transplanted into plastic pots containing compost. These hygromycin-resistant plants were also verified by PCR amplification.
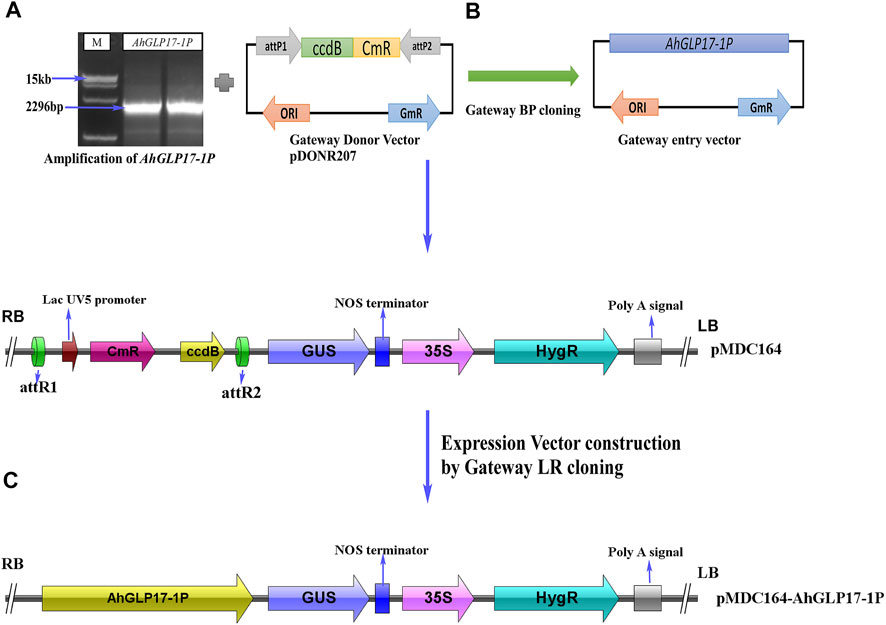
FIGURE 4. Construction of vectors using the backbone of pMDC164 vector by Gateway cloning. (A) Amplification of AhGLP17-1P promoter, (B) construction of Gateway entry vector by Gateway BP-cloning, (C) construction of Gateway expression vector using the backbone of binary vector pMDC164.
Hygromycin resistant positive transgenic plants were confirmed by PCR amplification using DNA as the template with promoter-specific forward and GUS gene specific reverse primers. While Arabidopsis Col-0 plants were used as a negative control, and Gateway LR constructs were used as a positive control for PCR confirmation. Eight hygromycin-resistant plants were confirmed by PCR amplification (Figure 5A). Seeds of eight positively transformed plants were sown to get the T1 generation. In T1 generation again, eight plants were selected based on hygromycin resistance and PCR confirmation. Eight selected plants of the T1 generation were covered to avoid cross-pollination, and in this way, homozygous T3 generation was obtained. Histochemical GUS expression was checked in different tissues at different growth stages in the T3 generation.
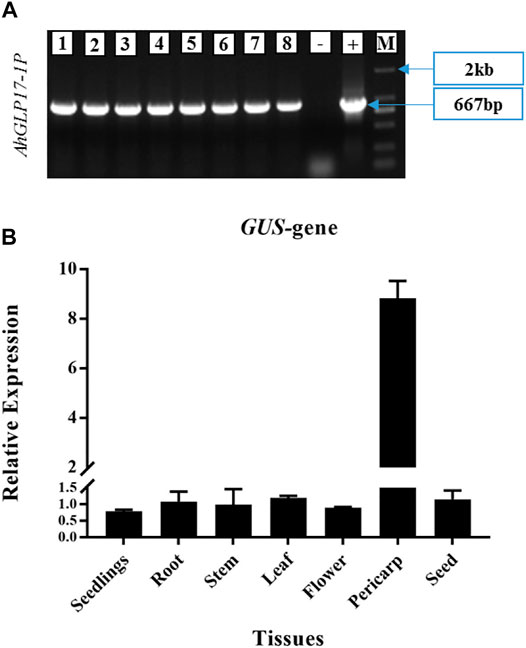
FIGURE 5. (A) Confirmation of T0 transgenic Arabidopsis plants transformed with AhGLP17-1P (667 bp fragment). Eight hygromycin-resistant plants verified by PCR amplification with promoter-specific forward and GUS gene specific reverse primer. Arabidopsis Col-0 was used as –ve control, and Gateway LR constructs were used as + ve control for PCR verification. M shows 2 kb marker, (B) Quantitative expression of GUS gene driven by AhGLP17-1P in different tissues of transgenic Arabidopsis plants. Root expression was used as control to analyse the data.
The quantitative expression of the GUS gene was analyzed in different tissues of transgenic plants by qRT-PCR (Figure 5B). qRT-PCR results showed a relatively higher transcript level of the GUS gene in the pericarp of transgenic plants and a very low transcript level in other tissues (Figure 5B). GUS staining was not detected in all vegetative tissues and young seedlings (Figure 6). Among reproductive organs, a moderate level of GUS staining was present in siliques, and mild staining was also present in flowers in some cases; staining was not present in seeds. To confirm that GUS staining is not present in seed coat/testa, cotyledons, and embryo, the ruptured seeds incubated in GUS staining solution were further processed for cryostat sectioning. Staining was not found in any of the seed tissues (Figure 7). Non-transformed Arabidopsis Col-0 plants were used as a control to compare the GUS staining. Staining results showed dark blue color only in the pericarp (outer covering of siliques). In all other tissues, staining was not present except a minute staining in flowers in rare cases. These results clearly showed that AhGLP17-1P is abundantly expressed in the pericarp and almost no expression in other tissues. Overall, the results strongly suggest that this promoter is a suitable candidate to guide the expression of a gene in a pericarp-specific manner.

FIGURE 6. GUS staining of different vegetative tissues of Arabidopsis transgenic plants. AhGLP17-1P plants showed no staining in any vegetative tissue (seedlings, roots, leaf, and stem). Different vegetative tissues of wild-type (Col-0) plants were also used for GUS staining to compare the results.
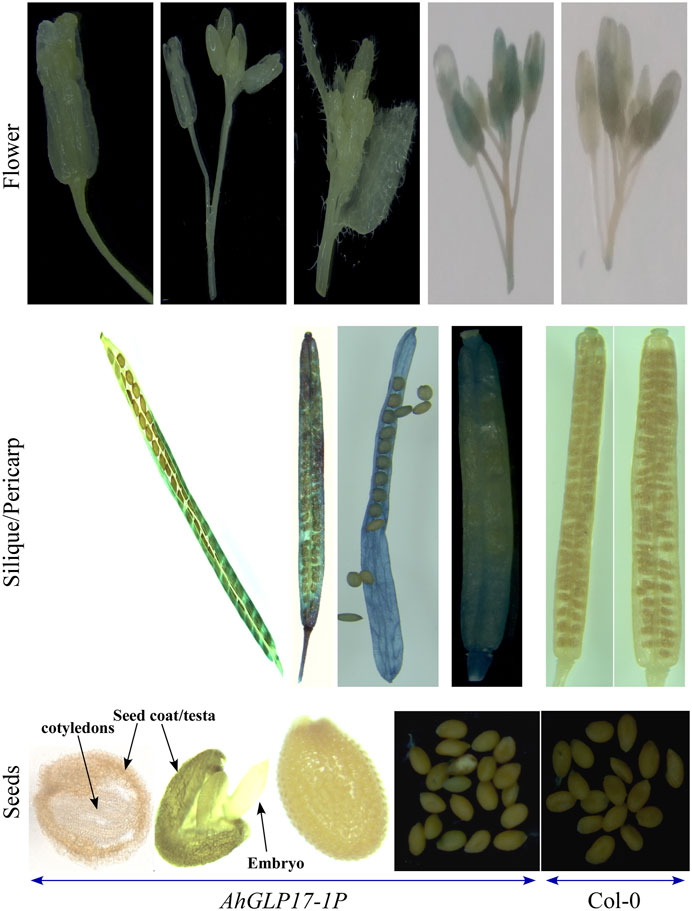
FIGURE 7. GUS staining of different reproductive tissues/organs of transgenic Arabidopsis plants. AhGLP17-1P plants showed no staining in flowers (a minute staining in some cases). Seed outer covering (pericarp) showed good staining. While staining was not present in any seed tissue (testa, cotyledons, and embryo).
Constitutive expression of a gene in transgenic plants results in an additional metabolic burden on the plant system, and constitutive promoters can produce undesired phenotypes (Yuan et al., 2019b) and reduced production. Plants need to direct the valuable resources to the target areas for survival and smooth growth under normal and stressed conditions (Divya et al., 2019). Therefore, tissue-specific or stress-inducible promoters are ideal for altering the plants’ genetic architecture to perform better according to a researchers’ desired ideotype. Previous crop biofortification programs that resulted in present-day purple embryo maize (Liu et al., 2018), purple endosperm rice (Zhu et al., 2017), golden rice (Paine et al., 2005), and Brassica juncea for fish oil docosahexaenoic acid (DHA) production (Wu et al., 2005) were carried out to introduce new metabolic pathways in endosperm and seeds of these crop species by employing endosperm and seed-specific promoters. For example, a strong endosperm-specific rice glutenin GluT01 promoter (Glu) was used to drive a novel rice phytoene synthase (psy) gene and Erwinia uredovora crtI gene fused with pea Rubisco small subunit plastid peptide to produce the high amount of β-carotene in Golden Rice (Paine et al., 2005). As peanut is an important oil and protein providing crop and primary source of nutrition in many Asian and African countries, it is prone to many biotic and abiotic stresses (Zhang et al., 2017). Changing its genetic makeup is key for its better performance under stressed conditions. The pericarp is a non-edible part of peanut seeds and serves as the first layer of defense against pathogens. Using pericarp-specific promoters to drive resistance-related genes is ideal for improving its fighting ability against soil and seed-borne pathogens and diseases. Although, in recent years, some studies have reported the identification and functional characterization of seed-specific promoters and genes in peanut (Yuan et al., 2019a; Yuan et al., 2019b; Tang et al., 2021), but no detail is viable for pericarp-specific promoters.
Therefore, the current study was based on identifying and functionally characterizing the promoter of a gene with unique expression in peanut pericarp and no/minimum expression in all other tissues. We identified a pericarp-specific gene (germin-like protein subfamily 1 member 7) by available microarray and transcriptome expression data. Germin-like proteins GLPs are a group of well-known proteins ubiquitously found in the plant kingdom. GLPs are “cupin superfamily” domain-containing proteins, composed of β-sheet barrel structure and metal ion binding site at their C-terminus (Dunwell et al., 2008; Agarwal et al., 2009). GLPs actively participate in plant defense against various fungal, bacterial, and viral pathogens (Godfrey et al., 2007; Knecht et al., 2010; Guevara-Olvera et al., 2012). We checked the expression specificity of the AhGLP17-1 gene in different tissues by semiquantitative and qRT-PCR using a widely grown peanut cultivar, “Minhua-6”. Further, we cloned the promoter region of the AhGLP17-1 gene from a high-yielding and disease-resistant cultivar, “Xinhuxiaoli”. We used two different peanut varieties as Minhua-6 is a largely cultivated variety, and our microarray expression is based on this cultivar. Its samples were easily available for RNA extraction and expression verification. At the same time, Xinhuixiaoli cultivar (disease-resistant; fungal and bacterial diseases), was used for cloning of promoter based on its possible future use to develop transgenic peanut for disease resistance. Online databases for promoter analysis predicted many cis-acting elements, including core promoter element as TATA Box and proximal control elements as CAAT Box, GC Box (Muthusamy et al., 2017), and many other light-responsive, hormones responsive, growth and regulation responsive, and stress-responsive elements. The number and types of cis-regulatory elements are important determinants of promoter strength and specificity (Stålberg et al., 1993; Tang et al., 2015). RY-repeat elements, known for seed-specific expression (Fujiwara and Beachy, 1994), were also present in the promoter region of the AhGLP17-1 gene. One copy of (CA)n element “CNAACAC” was also found in the promoter region. (CA)n is known to be involved in the embryo and endosperm-specific transcription (Ellerström et al., 1996). But, to date, not a single element has been reported to be involved in the pericarp-specific expression. From in silico analysis of AhGLP17-1P, some unnamed elements were found that include Unnamed_1 (GAATTTAATTAA), Unnamed_2 (AACCTAACCT), Unnamed_4 (CTCC), and Unnamed_6 (taTAAATATct). There is a possibility that some of these elements or some other element have role in pericarp-specific activity.
Peanut is a suitable crop for genomic studies, but the main bottleneck is difficulties in peanut transformation, so Arabidopsis becomes an ideal alternative for functional studies of genes and promoters (Grace et al., 2004; Zavallo et al., 2010; Sunkara et al., 2014). Here, we verified the pericarp-abundant behavior of a germin-like protein family gene (AhGLP17-1) of peanut. Peanut is affected by several biotic and abiotic stresses, and the pericarp is the primary defense organ that protects peanut seeds from stresses and harsh conditions. Hence, altering the composition of this organ can result in new peanut cultivars with enhanced defense capabilities. If stress-related genes under the control of pericarp-specific promoters are successfully transformed into peanuts, it will be a milestone achievement in peanut breeding.
Pod rot is a complex disease associated with several Pythium species, deteriorates young pods and seeds (Yu et al., 2019). Aspergillus flavus is a serious threat to food safety which causes crop yield and quality deterioration by producing aflatoxins (Deng et al., 2018). Similarly, gray mold disease of peanut caused by Botrytis cinerea (Alam et al., 2019), web blight disease of groundnut caused by Rhizoctonia solani (Ganesan and Sekar, 2011), and a huge seed-borne fungal microflora attack peanut seeds and deteriorates yield and quality. Their first target is the pericarp or pod; after that, these pathogens invade edible seeds. Changing the genetic makeup of peanut pods is an ideal solution to avoid the damages of these pathogens. Chitinases, stilbene/resveratrol synthase, glucanases are well-known genes showing resistance to bacterial and fungal diseases (Medeiros et al., 2018; Vestergaard and Ingmer, 2019; Loc et al., 2020; Ueki et al., 2020). These genes can be derived by pericarp abundant/specific promoter to show high expression in pericarp tissues. In this way, the defensive ability of this tissue can be enhanced to protect the edible seeds.
Similarly, there are several other seed-borne diseases and pathogens that attack the growing peanut kernels. The pericarp is a potential barrier against these diseases and pathogens (Cobos et al., 2018). Transformation of these genes under the control of pod-specific promoters into high-yielding varieties can improve their disease resistance. Our results showed that the AhGLP17-1 promoter showed expression specificity in pericarp tissues. Although there are some variations in expression patterns like in peanut, this gene also showed some expression in roots and no expression in flowers. But in transgenic Arabidopsis plants, the GUS gene under AhGLP17-1P showed some staining in flowers in some cases and no staining in roots. These variations are possibly attributed to diverse species. We are fully convinced that the AhGLP17-1P promoter investigated in this study could potentially drive the resistance-related genes in pericarp specific manner and alter the peanut pericarp genetic architecture to protect the edible seed from biotic stresses and environmental stresses.
In this study, we cloned and functionally characterized a novel pericarp-specific promoter (AhGLP17-1P) of peanut for the first time. This specifically expressed gene was cloned based on the microarray and transcriptome expression data. Both semiquantitative and qRT-PCR confirmed its pericarp-specific and abundant expressions. The GUS staining and qPCR analysis of the GUS gene under AhGLP17-1P in different vegetative and reproductive tissues/organs of transgenic Arabidopsis plants clearly showed its expression in pericarp tissues and no expression in all other tissues including, roots, seedlings, stem, leaf, seeds, except minute expression in flowers in some cases. Our studied promoter can potentially improve disease/pathogen resistance in transgenic peanuts and other agronomically important crops by employing resistance-related genes.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
YS, HC, and WZ conceived the idea. YS analyzed the data and wrote the manuscript. YD, NA, SK, CZ, WX, KC, TC, QY, YZ, and AR helped in literature search, data analysis, and provided technical guidance. HC, CZ, and WZ supervised the work and edited the final version. All authors have read and approved the final version of the manuscript.
This work was supported by grants from the National Science Foundation (NSF) of China (U1705233, 31601337, and 32072103), the Science and Technology Foundation of Fujian Province of China (2017N0006, 2018N0004, and 2021N5007), and Fujian Agriculture and Forestry University, Fuzhou, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
YS wants to say special thanks to Fujian Agriculture and Forestry University, and Chinese Government Scholarship Council, China, for providing Ph.D. scholarship and research environment. Further, we are also thankful to all lab/institute members for their support and guidance in conducting this research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.821281/full#supplementary-material
Agarwal, G., Rajavel, M., Gopal, B., and Srinivasan, N. (2009). Structure-based Phylogeny as a Diagnostic for Functional Characterization of Proteins with a Cupin Fold. PLoS ONE 4, e5736. doi:10.1371/journal.pone.0005736
Akhtar, S., Khalid, N., Ahmed, I., Shahzad, A., and Suleria, H. A. R. (2014). Physicochemical Characteristics, Functional Properties, and Nutritional Benefits of Peanut Oil: a Review. Crit. Rev. Food Sci. Nutr. 54, 1562–1575. doi:10.1080/10408398.2011.644353
Alam, A., Gupta, S., and Sharma, V. J. M. (2019). Biochemical Screening of Ten Peanut Cultivars for Tolerance to Botrytis Cinerea, 15.
Ali, N., Chen, H., Zhang, C., Khan, S. A., Gandeka, M., Xie, D., et al. (2020). Ectopic Expression of AhGLK1b (GOLDEN2-like Transcription Factor) in Arabidopsis Confers Dual Resistance to Fungal and Bacterial Pathogens. Genes 11, 343. doi:10.3390/genes11030343
Bhattacharyya, S., Dey, N., and Maiti, I. B. (2002). Analysis of Cis-Sequence of Subgenomic Transcript Promoter from the Figwort Mosaic Virus and Comparison of Promoter Activity with the Cauliflower Mosaic Virus Promoters in Monocot and Dicot Cells. Virus. Res. 90, 47–62. doi:10.1016/s0166-0934(02)00146-5
Chen, H., Zhang, C., Cai, T. C., Deng, Y., Zhou, S., Zheng, Y., et al. (2016). Identification of Low Ca2+stress-Induced Embryo Apoptosis Response Genes inArachis Hypogaeaby SSH-Associated Library Lift (SSHaLL). Plant Biotechnol. J. 14, 682–698. doi:10.1111/pbi.12415
Chi, X., Hu, R., Yang, Q., Zhang, X., Pan, L., Chen, N., et al. (2012). Validation of Reference Genes for Gene Expression Studies in Peanut by Quantitative Real-Time RT-PCR. Mol. Genet. Genomics 287, 167–176. doi:10.1007/s00438-011-0665-5
Christensen, A. H., Sharrock, R. A., and Quail, P. H. (1992). Maize Polyubiquitin Genes: Structure, thermal Perturbation of Expression and Transcript Splicing, and Promoter Activity Following Transfer to Protoplasts by Electroporation. Plant Mol. Biol. 18, 675–689. doi:10.1007/bf00020010
Clough, S. J., and Bent, A. F. (1998). Floral Dip: a Simplified Method forAgrobacterium-Mediated Transformation ofArabidopsis Thaliana. Plant J. 16, 735–743. doi:10.1046/j.1365-313x.1998.00343.x
Cobos, C. J., Tengey, T. K., Balasubramanian, V. K., Williams, L. D., Sudini, H. K., Varshney, R. K., et al. (2018). Employing Peanut Seed Coat Cell Wall Mediated Resistance against Aspergillus flavus Infection and Aflatoxin Contamination.
Dardick, C., and Callahan, A. M. (2014). Evolution of the Fruit Endocarp: Molecular Mechanisms Underlying Adaptations in Seed protection and Dispersal Strategies. Front. Plant Sci. 5, 284. doi:10.3389/fpls.2014.00284
Deng, Y., Chen, H., Zhang, C., Cai, T., Zhang, B., Zhou, S., et al. (2018). Evolution and Characterisation of the AhRAF4 NB-ARC Gene Family Induced by Aspergillus flavus Inoculation and Abiotic Stresses in Peanut. Plant Biol. J. 20, 737–750. doi:10.1111/plb.12726
Divya, K., Kavi Kishor, P. B., Bhatnagar-Mathur, P., Singam, P., Sharma, K. K., Vadez, V., et al. (2019). Isolation and Functional Characterization of Three Abiotic Stress-Inducible (Apx, Dhn and Hsc70) Promoters from Pearl Millet (Pennisetum Glaucum L.). Mol. Biol. Rep. 46, 6039–6052. doi:10.1007/s11033-019-05039-4
Dixon, R. A., and Paiva, N. L. (1995). Stress-induced Phenylpropanoid Metabolism. The plant cell 7, 1085. doi:10.2307/3870059
Dunwell, J. M., Gibbings, J. G., Mahmood, T., and Saqlan Naqvi, S. M. (2008). Germin and Germin-like Proteins: Evolution, Structure, and Function. Crit. Rev. Plant Sci. 27, 342–375. doi:10.1080/07352680802333938
Ellerström, M., Stålberg, K., Ezcurra, I., and Rask, L. J. P. m. b. (1996). Functional Dissection of a Napin Gene Promoter: Identification of Promoter Elements Required for Embryo and Endosperm-specific Transcription. 32, 1019–1027.
Fujiwara, T., and Beachy, R. N. (1994). Tissue-specific and Temporal Regulation of a ?-conglycinin Gene: Roles of the RY Repeat and Other Cis-Acting Elements. Plant Mol. Biol. 24, 261–272. doi:10.1007/bf00020166
Ganesan, S., and Sekar, R. J. P. I. T. M. W. (2011). Screening of Biocontrol Agents against Rhizoctonia solani Causing Web Blight Disease of Groundnut. Arachis hypogaea L., 127–140.
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S. e., Wilkins, M. R., Appel, R. D., et al. (2005). Protein Identification and Analysis Tools on the ExPASy Server, 571–607. doi:10.1385/1-59259-890-0:571Protein Identification and Analysis Tools on the ExPASy Server
Godfrey, D., Able, A. J., and Dry, I. B. (2007). Induction of a grapevine Germin-like Protein (VvGLP3) Gene Is Closely Linked to the Site of Erysiphe Necator Infection: a Possible Role in Defense. Mpmi 20, 1112–1125. doi:10.1094/mpmi-20-9-1112
Grace, M. L., Chandrasekharan, M. B., Hall, T. C., and Crowe, A. J. (2004). Sequence and Spacing of TATA Box Elements Are Critical for Accurate Initiation from the β-Phaseolin Promoter. J. Biol. Chem. 279, 8102–8110. doi:10.1074/jbc.m309376200
Guevara-Olvera, L., Ruíz-Nito, M. L., Rangel-Cano, R. M., Torres-Pacheco, I., Rivera-Bustamante, R. F., Muñoz-Sánchez, C. I., et al. (2012). Expression of a Germin-like Protein Gene (CchGLP) from a Geminivirus-Resistant Pepper (Capsicum Chinense Jacq.) Enhances Tolerance to Geminivirus Infection in Transgenic Tobacco. Physiol. Mol. Plant Pathol. 78, 45–50. doi:10.1016/j.pmpp.2012.01.005
Gupta, N. C., Jain, P. K., Bhat, S. R., and Srinivasan, R. (2012). Upstream Sequence of Fatty Acyl-CoA Reductase (FAR6) of Arabidopsis thaliana Drives Wound-Inducible and Stem-specific Expression. Plant Cel Rep 31, 839–850. doi:10.1007/s00299-011-1205-9
Haughn, G., and Chaudhury, A. (2005). Genetic Analysis of Seed Coat Development in Arabidopsis. Trends Plant Science 10, 472–477. doi:10.1016/j.tplants.2005.08.005
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and Validation of Promoters and Cis-Acting Regulatory Elements. Plant Sci. 217-218, 109–119. doi:10.1016/j.plantsci.2013.12.007
Higo, K., Ugawa, Y., Iwamoto, M., and Higo, H. (1998). PLACE: a Database of Plant Cis-Acting Regulatory DNA Elements. Nucleic Acids Res. 26, 358–359. doi:10.1093/nar/26.1.358
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS Fusions: Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher Plants. EMBO J. 6, 3901–3907. doi:10.1002/j.1460-2075.1987.tb02730.x
Khan, S. A., Chen, H., Deng, Y., Chen, Y., Zhang, C., Cai, T., et al. (2020). High-density SNP Map Facilitates fine Mapping of QTLs and Candidate Genes Discovery for Aspergillus flavus Resistance in Peanut (Arachis hypogaea). Theor. Appl. Genet. 133, 2239–2257. doi:10.1007/s00122-020-03594-0
Knecht, K., Seyffarth, M., Desel, C., Thurau, T., Sherameti, I., Lou, B., et al. (2010). Expression of BvGLP-1 Encoding a Germin-like Protein from Sugar Beet in Arabidopsis thaliana Leads to Resistance against Phytopathogenic Fungi. Mpmi 23, 446–457. doi:10.1094/mpmi-23-4-0446
Koyama, T., Ono, T., Shimizu, M., Jinbo, T., Mizuno, R., Tomita, K., et al. (2005). Promoter of Arabidopsis thaliana Phosphate Transporter Gene Drives Root-specific Expression of Transgene in rice. J. Biosci. Bioeng. 99, 38–42. doi:10.1263/jbb.99.38
Lane, B. G., Dunwell, J. M., Ray, J. A., Schmitt, M. R., and Cuming, A. C. (1993). Germin, a Protein Marker of Early Plant Development, Is an Oxalate Oxidase. J. Biol. Chem. 268, 12239–12242. doi:10.1016/s0021-9258(18)31377-2
Li, J., Wang, S., Yu, J., Wang, L., and Zhou, S. J. C. B. o. B. (2013). A Modified CTAB Protocol for Plant DNA Extraction. 48, 72.
Liu, X., Yang, W., Mu, B., Li, S., Li, Y., Zhou, X., et al. (2018). Engineering of 'Purple Embryo Maize' with a Multigene Expression System Derived from a Bidirectional Promoter and Self-Cleaving 2A Peptides. Plant Biotechnol. J. 16, 1107–1109. doi:10.1111/pbi.12883
Loc, N. H., Huy, N. D., Quang, H. T., Lan, T. T., and Thu Ha, T. T. (2020). Characterisation and Antifungal Activity of Extracellular Chitinase from a Biocontrol Fungus, Trichoderma Asperellum PQ34. Mycology 11, 38–48. doi:10.1080/21501203.2019.1703839
Lu, M., Han, Y. P., Gao, J. G., Wang, X. J., and Li, W. B. (2010). Identification and Analysis of the Germin-like Gene Family in Soybean. BMC Genomics 11, 620–715. doi:10.1186/1471-2164-11-620
Medeiros, S. C. d., Monteiro-Júnior, J. E., Sales, G. W. P., Grangeiro, T. B., and Nogueira, N. A. P. (2018). Chitinases as Antibacterial Proteins: A Systematic Review.
Murashige, T., and Skoog, F. (1962). A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant 15, 473–497. doi:10.1111/j.1399-3054.1962.tb08052.x
Muthusamy, S., Sivalingam, P., Sridhar, J., Singh, D., Haldhar, S., and Kaushal, P. (2017). Biotic Stress Inducible Promoters in Crop Plants-A Review. Jae 04, 14–24. doi:10.53911/jae.2017.4202
Nakashima, K., Tran, L.-S. P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional Analysis of a NAC-type Transcription Factor OsNAC6 Involved in Abiotic and Biotic Stress-Responsive Gene Expression in rice. Plant J. 51, 617–630. doi:10.1111/j.1365-313x.2007.03168.x
Odell, J. T., Nagy, F., and Chua, N.-H. (1985). Identification of DNA Sequences Required for Activity of the Cauliflower Mosaic Virus 35S Promoter. Nature 313, 810–812. doi:10.1038/313810a0
Ong, C.-T., and Corces, V. G. (2011). Enhancer Function: New Insights into the Regulation of Tissue-specific Gene Expression. Nat. Rev. Genet. 12, 283–293. doi:10.1038/nrg2957
Paik-Ro, O. G., Seib, J. C., and Smith, R. L. (2002). Seed-specific, Developmentally Regulated Genes of Peanut. Theor. Appl. Genet. 104, 236–240. doi:10.1007/s001220100763
Paine, J. A., Shipton, C. A., Chaggar, S., Howells, R. M., Kennedy, M. J., Vernon, G., et al. (2005). Improving the Nutritional Value of Golden Rice through Increased Pro-vitamin A Content. Nat. Biotechnol. 23, 482–487. doi:10.1038/nbt1082
Rombauts, S., Dehais, P., Van Montagu, M., and Rouze, P. (1999). PlantCARE, a Plant Cis-Acting Regulatory Element Database. Nucleic Acids Res. 27, 295–296. doi:10.1093/nar/27.1.295
Settaluri, V., Kandala, C., Puppala, N., and Sundaram, J. (2012). Peanuts and Their Nutritional Aspects—A Review.
Sheikholeslam, S. N., and Weeks, D. P. (1987). Acetosyringone Promotes High Efficiency Transformation of Arabidopsis thaliana Explants by Agrobacterium Tumefaciens. Plant Mol. Biol. 8, 291–298. doi:10.1007/bf00021308
Shirsat, A., Wilford, N., Croy, R., and Boulter, D. (1989). Sequences Responsible for the Tissue Specific Promoter Activity of a Pea Legumin Gene in Tobacco. Mol. Gen. Genet. 215, 326–331. doi:10.1007/bf00339737
Souza, F. H. D. D., and Marcos-Filho, J. (2001). The Seed Coat as a Modulator of Seed-Environment Relationships in Fabaceae. Rev. Bras. Bot. 24, 365–375. doi:10.1590/s0100-84042001000400002
Stålberg, K., Ellerström, M., Josefsson, L.-G., and Rask, L. (1993). Deletion Analysis of a 2S Seed Storage Protein Promoter of Brassica Napus in Transgenic Tobacco. Plant Mol. Biol. 23, 671–683.
Sunkara, S., Bhatnagar-Mathur, P., and Sharma, K. K. (2014). Isolation and Functional Characterization of a Novel Seed-specific Promoter Region from Peanut. Appl. Biochem. Biotechnol. 172, 325–339. doi:10.1007/s12010-013-0482-x
Tang, G., Xu, P., Li, P., Zhu, J., Chen, G., Shan, L., et al. (2021). Cloning and Functional Characterization of Seed-specific LEC1A Promoter from Peanut (Arachis hypogaea L.). PLoS ONE 16, e0242949. doi:10.1371/journal.pone.0242949
Tang, G., Xu, P., Liu, W., Liu, Z., and Shan, L. (2015). Cloning and Characterization of 5′ Flanking Regulatory Sequences of AhLEC1B Gene from Arachis Hypogaea L. PloS one 10, e0139213. doi:10.1371/journal.pone.0139213
Ueki, A., Takehara, T., Ishioka, G., Kaku, N., Ueki, K., and biotechnology, (2020). β-1,3-Glucanase Production as an Anti-fungal Enzyme by Phylogenetically Different Strains of the Genus Clostridium Isolated from Anoxic Soil that Underwent Biological Disinfestation. Appl. Microbiol. Biotechnol. 104, 5563–5578. doi:10.1007/s00253-020-10626-8
Vestergaard, M., and Ingmer, H. (2019). Antibacterial and Antifungal Properties of Resveratrol. Int. J. Antimicrob. Agents 53, 716–723. doi:10.1016/j.ijantimicag.2019.02.015
Wass, M. N., Kelley, L. A., and Sternberg, M. J. E. (2010). 3DLigandSite: Predicting Ligand-Binding Sites Using Similar Structures. 38, W469–W473.doi:10.1093/nar/gkq406
Windsor, J. B., Symonds, V. V., Mendenhall, J., and Lloyd, A. M. (2000). Arabidopsis Seed Coat Development: Morphological Differentiation of the Outer Integument. Plant J. 22, 483–493. doi:10.1046/j.1365-313x.2000.00756.x
Wu, G., Truksa, M., Datla, N., Vrinten, P., Bauer, J., Zank, T., et al. (2005). Stepwise Engineering to Produce High Yields of Very Long-Chain Polyunsaturated Fatty Acids in Plants. Nat. Biotechnol. 23, 1013–1017. doi:10.1038/nbt1107
Xu, N., Wang, R., Zhao, L., Zhang, C., Li, Z., Lei, Z., et al. (2016). The Arabidopsis NRG2 Protein Mediates Nitrate Signaling and Interacts with and Regulates Key Nitrate Regulators. Plant Cell 28, 485–504. doi:10.1105/tpc.15.00567
Yamahara, T., Shiono, T., Suzuki, T., Tanaka, K., Takio, S., Sato, K., et al. (1999). Isolation of a Germin-like Protein with Manganese Superoxide Dismutase Activity from Cells of a Moss, Barbula Unguiculata. J. Biol. Chem. 274, 33274–33278. doi:10.1074/jbc.274.47.33274
Yu, C.-S., Chen, Y.-C., Lu, C.-H., and Hwang, J.-K. (2006). FunctionPrediction of Protein Subcellular Localization. Proteins 64, 643–651. doi:10.1002/prot.21018
Yu, J., Xu, M., Liang, C., Zhang, X., Guo, Z., Wu, J., et al. (2019). First Report of Pythium Myriotylum Associated with Peanut Pod Rot in China. Plant Dis. 103, 1794. doi:10.1094/pdis-02-19-0321-pdn
Yuan, C., Li, C., Yan, C., Zhao, X., Wang, J., Sun, Q., et al. (2019a). Isolation and Characterization of a Novel Seed-specific Promoter from Peanut (Arachis hypogaea L.). Mol. Biol. Rep. 46, 3183–3191. doi:10.1007/s11033-019-04775-x
Yuan, C., Sun, Q., and Kong, Y. (2019b). Genome-wide Mining Seed-specific Candidate Genes from Peanut for Promoter Cloning. PLoS ONE 14, e0214025. doi:10.1371/journal.pone.0214025
Zavallo, D., Lopez Bilbao, M., Hopp, H. E., and Heinz, R. (2010). Isolation and Functional Characterization of Two Novel Seed-specific Promoters from sunflower (Helianthus Annuus L.). Plant Cel Rep 29, 239–248. doi:10.1007/s00299-010-0816-x
Zhang, C., Chen, H., Cai, T., Deng, Y., Zhuang, R., Zhang, N., et al. (2017). Overexpression of a Novel Peanut NBS-LRR geneAhRRS5enhances Disease Resistance toRalstonia Solanacearumin Tobacco. Plant Biotechnol. J. 15, 39–55. doi:10.1111/pbi.12589
Zhu, Q., Yu, S., Zeng, D., Liu, H., Wang, H., Yang, Z., et al. (2017). Development of "Purple Endosperm Rice" by Engineering Anthocyanin Biosynthesis in the Endosperm with a High-Efficiency Transgene Stacking System. Mol. Plant 10, 918–929. doi:10.1016/j.molp.2017.05.008
Keywords: cis-elements, GUS staining, pathogens, tissue-specific expression, transgenic arabidopsis
Citation: Sharif Y, Chen H, Deng Y, Ali N, Khan SA, Zhang C, Xie W, Chen K, Cai T, Yang Q, Zhuang Y, Raza A and Zhuang W (2022) Cloning and Functional Characterization of a Pericarp Abundant Expression Promoter (AhGLP17-1P) From Peanut (Arachis hypogaea L.). Front. Genet. 12:821281. doi: 10.3389/fgene.2021.821281
Received: 24 November 2021; Accepted: 31 December 2021;
Published: 20 January 2022.
Edited by:
Aamir Raina, Aligarh Muslim University, IndiaReviewed by:
Farhat Abbas, South China Agricultural University, ChinaCopyright © 2022 Sharif, Chen, Deng, Ali, Khan, Zhang, Xie, Chen, Cai, Yang, Zhuang, Raza and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijian Zhuang, d2VpamlhbnpAZmFmdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.