
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 21 January 2022
Sec. Toxicogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.820778
This article is part of the Research Topic Long-Term Toxicity and Epigenetic Effects of Environmental Exposures View all 8 articles
Spodoptera frugiperda is the world’s major agricultural pest and has the distinctive features of high fecundity, strong migratory capacity, and high resistance to most insecticides. At present, the control of S. frugiperda in China relies mainly on the spraying of chemical insecticides. MicroRNAs (miRNAs) are a class of small, single-stranded, non-coding RNAs and play crucial regulatory roles in various physiological processes, including the insecticide resistance in insects. However, little is known about the regulatory roles of miRNAs on the resistance of S. frugiperda to insecticides. In the present research, the miRNAs that were differentially expressed after cyantraniliprole, spinetoram, and emamectin benzoate treatment were analyzed by RNA-Seq. A total of 504 miRNAs were systematically identified from S. frugiperda, and 24, 22, and 31 miRNAs were differentially expressed after treatments of cyantraniliprole, spinetoram, and emamectin benzoate. GO and KEGG enrichment analyses were used to predict the function of differentially expressed target genes of miRNAs. Importantly, ten miRNAs were significantly differentially expressed among the treatments of three insecticides. miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p were significantly downregulated among the treatments of three insecticides by RT-qPCR. Furthermore, the overexpression of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p significantly increased the mortality of S. frugiperda to cyantraniliprole and emamectin benzoate. The mortality was significantly increased with spinetoram treatment after the overexpression of miR-13b-3p, miR-10485-5p, and miR-10483-5p. These results suggest that miRNAs, which are differentially expressed in response to insecticides, may play a key regulatory role in the insecticide tolerance in S. frugiperda.
MicroRNAs (miRNAs) are a class of small, single-stranded, non-coding RNAs of approximately 18–25 nucleotides length (Ambros, 2004; Bartel, 2004). miRNAs are derived from the stem-loop RNA precursors (pre-miRNAs) and are widely found in animals, plants, bacteria, and viruses (Lee et al., 2004). miRNAs have been shown to be involved in posttranscriptional regulation of mRNAs by forming silencing complexes through complementary forms of exact or incomplete nucleotide matches with target messenger RNAs (mRNAs) (Lee et al., 2004). Previous studies have shown that miRNAs account for 1–5% of all genes in animals and that 20–30% of genes are regulated by miRNAs (Stark et al., 2005). A single miRNA can regulate multiple target genes and is involved in a number of complex regulatory pathways (Tsai et al., 2009; Salvi et al., 2019).
In arthropods, a growing number of research studies have indicated that miRNAs play crucial roles in physiological and developmental pathways, such as metamorphosis, embryogenesis, molting, reproduction, immunity, and wing development (Etebari and Asgari, 2013; Lucas et al., 2015a; Lozano et al., 2015; Zhang et al., 2016; Ling et al., 2017; Song et al., 2018; Li et al., 2021a; Shang et al., 2020). Recently, miRNAs have been shown to be involved in the metabolism of plant toxin and the insecticide resistance in different insects. For example, miR-4133-3p regulates gossypol and tannic acid detoxification via targeting cytochrome P450 4CJ1 (CYP4CJ1) in Aphis gossypii (Ma et al., 2019). In Nilaparvata lugens, modulating the expressions of miRNAs novel-85 and novel-191 significantly altered the susceptibility of N. lugens to nitenpyram by targeting CYP6ER1 and carboxylesterase 1 (CarE1) (Mao et al., 2021). In addition, the microinjection of miR-7a or miR-8519 mimics in Plutella xylostella decreases the expression of the ryanodine receptor and significantly increases the susceptibility of P. xylostella to chlorantraniliprole (Li et al., 2015a).
Spodoptera frugiperda is native to North America, South America, and subtropical regions and feeds on a variety of crops including corn, soybeans, cotton, and rice (Gouin et al., 2017). Two biotypes, the “corn-strain” and “rice-strain”, have been identified for S. frugiperda (Hafeez et al., 2021). S. frugiperda has strong migratory capacity, first found in Central and Western Africa in 2016, moving into India in 2018, and invading China in January 2019 (Junior et al., 2012; Goergen et al., 2016; Early et al., 2018; Li et al., 2020). S. frugiperda migrates to a new location, and resistant alleles persist in resistant populations (Arias et al., 2019). S. frugiperda is now resistant to a variety of chemical insecticides, mainly including organophosphates, carbamates, pyrethroids, and other traditional insecticides (Yu and Jr, 2007; Carvalho et al., 2013). Recent studies have shown that S. frugiperda has also developed varying degrees of resistance to new chemical insecticides such as diamides, spinosyns, and benzoylureas (Nascimento et al., 2016; Bolzan et al., 2019; Lira et al., 2020).
A part of miRNAs has been identified in S. frugiperda, and their functional study mainly focuses on antiviral immune defense and adaptive evolution (Moné et al., 2018; Karamipour et al., 2019), but the systemic identification and functional analysis of miRNAs involved in insecticide resistance are still unknown. In this research, the expression profiles of miRNAs in S. frugiperda have been investigated based on high-throughput sequencing of the small RNA library. The differentially expressed miRNAs related to the tolerance of cyantraniliprole, spinetoram, and emamectin benzoate were systematically detected. More importantly, the effects of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p on the susceptibility of S. frugiperda to insecticides were verified.
The S. frugiperda population was collected from Tancheng (Shandong, China) in 2019 and maintained in the laboratory. S. frugiperda was provided by Prof. Xingyuan Men (Shandong Academy of Agricultural Sciences, Jinan, China) and reared in an illumination incubator at 26 ± 1°C with 60–70% humidity and 16 h light:8 h dark photoperiod. S. frugiperda was fed on artificial diet which was reported in the previous research (He et al., 2021).
Cyantraniliprole technical (98.00%) and spinetoram technical (85.80%) were purchased from Sigma-Aldrich (St. Louis, MO, United States). Emamectin benzoate technical (95.00%) was provided by Shandong Lukang Biopesticides Co. Ltd (Dezhou, China). Acetone (reagent grade) and Triton X-100 were purchased from Sigma-Aldrich (St. Louis, MO, United States).
Cyantraniliprole, spinetoram, and emamectin benzoate were dissolved in acetone and diluted to five concentrations of 0.04, 0.2, 1, 5, and 25 mg L−1 in the distilled water containing 0.05% Triton X-100. The distilled water, containing 0.05% Triton X-100 and 1% acetone, was used as the control. The toxicity bioassay was performed by the method of leaf-dipping (Fouad et al., 2016). Maize leaves were cut into small pieces of 20 mm long and immersed in each concentration of insecticides for 15s. Third-instar S. frugiperda was placed in the air-drying maize leaves. A total of twenty insects were treated in each concentration, and each concentration was repeated three times. All insects were reared at 25 ± 1°C with 50–70% humidity and 16 h light:8 h dark photoperiod. The mortality was checked after treatment in 24 h.
Third-instar S. frugiperda was treated with cyantraniliprole, spinetoram, and emamectin benzoate at doses of LC50 by the leaf-dipping method. After 24 h, the survival insects were collected, and the distilled water, containing 0.05% Triton X-100 and 1% acetone, was used as the control. Each treatment contained 20 insects and was repeated three times. S. frugiperda was frozen in liquid nitrogen for RNA extraction.
The total RNA was extracted by TRIzol reagent (Invitrogen, United States) following the manufacturer’s procedure. The total RNA quantity and purity were analyzed using a Bioanalyzer 2,100 (Agilent, United States) with RIN number >7.0. Approximately, 3 μg of total RNAs were used to prepare a small RNA library according to the protocol of TruSeq Small RNA Sample Prep Kits (Illumina, United States). The single-end sequencing (50 bp) was performed by an Illumina HiSeq 2,500 at the LC-BIO (Hangzhou, China) following the vendor’s recommended protocol. The adapter dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, and snoRNA), and repeats were removed by ACGT101-miR (LC Sciences, Houston, Texas, United States). In order to identify known miRNAs and novel 3p- and 5p-derived miRNAs, the unique sequences with a length of 18–26 nucleotides were mapped to specific species precursors in miRBase 22.0 by BLAST. The unmapped sequences were BLASTed the specific genomes, and the hairpin RNA structures containing sequences were predicated from the flank 80 nt sequences using RNAfold software (http://rna.tbi.univie.ac. at/cgi-bin/RNAfold.cgi). The differential expression of miRNAs was analyzed by ANOVA based on normalized deep-sequencing counts. The different expression analysis of miRNA was selected with log2 (fold change) > 1 or log2 (fold change) < −1 and with statistical significance (p < 0.05 or p < 0.01). The target genes of differentially expressed miRNAs were predicted by two computational target prediction algorithms (TargetScan 50 and miRanda 3.3a), and the 3-untranslated region (UTR) of the candidate target sequences was used to miRNA target prediction. The data predicted by both algorithms were combined, and the overlaps were calculated. The Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes database (KEGG) pathway of these miRNA targets were also annotated.
The relative miRNA expression was analyzed by the quantitative real-time PCR (RT-qPCR). PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Japan). It was used to synthesize the cDNA from 1 µg of total RNA. The quantitative PCR reaction was completed using an iQ™5 Multicolor Real-Time PCR detection system (Bio-Rad, United States) with the SYBR PrimeScript™ RT-PCR Kit (Takara, Japan). The U6 snRNA was used as an endogenous control to normalize the expression levels of miRNAs (Zhang et al., 2021a). The 2−∆∆Ct method was used to calculate the relative expression levels of miRNAs (Livak and Schmittegen, 2001). The RT-qPCR reaction included three independent technical and biological replications. The RT-qPCR primers are listed in Supplementary Table S1.
The tissues of the salivary gland, fat body, Malpighian tubule, and midgut were dissected from third-instar S. frugiperda. Before dissection, the third-instar S. frugiperda was rinsed with 75% ethanol and dissected in chilled 1 × phosphate-buffered saline (pH 7.4) under a stereomicroscope. Each sample contained fifty insects and repeated three times. The tissues collected were kept and analyzed by RT-qPCR.
The agomir-miR-278-5p, agomir-miR-13b-3p, agomir-miR-10485-5p, agomir-miR-10483-5p, and agomir-NC were synthesized in GenePharma (Shanghai, China). After anesthetized on ice, approximately 500 nl 20 μM agomir-NC, agomir-miR-278-5p, agomir-miR-13b-3p, agomir-miR-10485-5p, and agomir-miR-10483-5p were injected into the antesternum of third-instar S. frugiperda using a Nanoject II (Drummond, United States). The insects were treated with cyantraniliprole, spinetoram, and emamectin benzoate at the LC50 dose for 1 day after injection. All insects were reared at 25 ± 1°C with 50–70% humidity and 16 h light:8 h dark photoperiod. The mortality was checked after insecticide treatment in 24 h. Each treatment contained thirty insects and repeated three times.
SPSS 20.0 software (IBM Corporation, United States) was used to perform the statistical analyses. The Student’s t-test was used to compare the differences between treatments. Data were shown as mean ± standard error (SE). The p values <0.05 and 0.01 were considered statistically significant and very significant differences, respectively.
The toxicity of cyantraniliprole, spinetoram, and emamectin benzoate against the susceptible strain of S. frugiperda was tested. The LC50 values of cyantraniliprole, spinetoram, and emamectin benzoate were 0.304, 0.161, and 0.194 mg L−1, respectively (Table 1).
The sequencing data had been deposited in the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) with the accession number GSE189968. The twelve small RNA (sRNA) libraries derived from the control, cyantraniliprole-treated, spinetoram-treated, and emamectin benzoate-treated S. frugiperda. The total number of clean reads was 6,273,553, 11,856,426, 8,602,367, and 6,849,604 with unique reads of 1,252,300, 1,640,252, 1,173,199, and 1,478,489 from the cyantraniliprole treatment, spinetoram treatment, emamectin benzoate treatment, and the control libraries, respectively (Table 2). In the total reads, ribosomal RNAs (rRNAs) accounted for 64.66, 67.22, 68.61, and 72.79% among the non-miRNA sRNAs in the control, cyantraniliprole treatment, spinetoram treatment, and emamectin benzoate treatment libraries, respectively (Supplementary Figure S1). In the unique reads, rRNAs accounted for 69.63, 71.77, 70.17, and 70.72% in the control, cyantraniliprole treatment, spinetoram treatment, and emamectin benzoate treatment libraries, respectively (Supplementary Figure S1). The twelve sRNA libraries shared a similar length distribution pattern, with 22 nt being the most abundant (Supplementary Figure S2).
A total of 504 miRNAs in S. frugiperda were identified, including 379 conserved miRNAs and 125 novel miRNAs (Supplementary Table S2). The length distribution of miRNAs in S. frugiperda was ranged from 18 nt to 26 nt, with 22 nt miRNAs being the most abundant, accounting for 35.52% (Figure 1). miR-1, miR-6497-5p, and miR-276-3p were the three most abundant miRNAs, and the information of the most highly expressed ten miRNAs is listed in Supplementary Table S3. The conservative profile of miRNAs in S. frugiperda was explored, and 58 miRNA sequence families were identified from the miRBase database (Figure 2A). miR-1 was the family with the largest number of miRNAs (Figure 2A). Furthermore, the frequency of miRNA identified in S. frugiperda in other insect species was analyzed. Bombyx mori and Manduca sexta shared 61 and 56 of their conserved miRNAs with S. frugiperda, respectively (Figure 2B). The sequencing data would greatly expand the number of miRNAs in S. frugiperda.
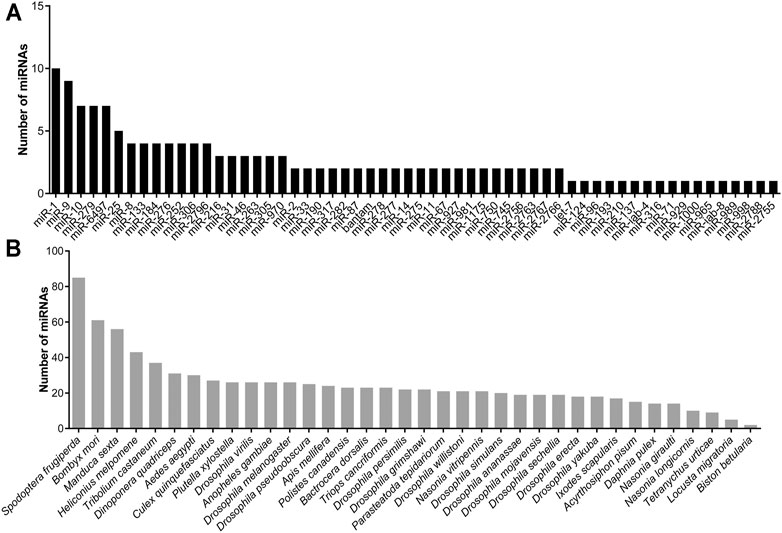
FIGURE 2. Characterization analysis of miRNAs in S. frugiperda. (A) Number of miRNAs in different family. (B) Number of miRNAs shared with other insect species.
After insecticide treatment, the differentially expressed miRNAs were determined by normalized read counts. Compared to control, 24 miRNAs were differentially expressed in cyantraniliprole treatment, 16 were significantly upregulated, and 8 were significantly downregulated (Figure 3A). A total of 22 miRNAs were significantly upregulated and 7 were significantly downregulated between spinetoram treatment and control (Figure 3B). In addition, 31 miRNAs were found to be differentially expressed in both emamectin benzoate treatment and control, 23 were upregulated, and 8 were downregulated (Figure 3C). These results indicated that miRNAs could be induced by insecticides in S. frugiperda.

FIGURE 3. Number of differentially expressed miRNAs after insecticide treatment. (A) Number of differentially expressed miRNAs after cyantraniliprole treatment. (B) Number of differentially expressed miRNAs after spinetoram treatment. (C) Number of differentially expressed miRNAs after emamectin benzoate treatment.
In order to understand the miRNA functions in insecticide resistance, the targets of differentially expressed miRNAs were predicted (Supplementary Table S4). To predict the functions of these target genes, the GO and KEGG enrichment analyses were performed. The target genes were assigned the terms of the biological process, cellular component, and molecular function, and the cytoplasm was the most enriched subgroup (Figure 4A). The top twenty significantly enriched KEGG pathways of the miRNA target genes are presented in Figure 5. The MAPK signaling pathway-fly, endocytosis, Wnt signaling pathway, autophagy-animal, and apoptosis-fly were the five most significantly enriched pathways (Figure 4B).
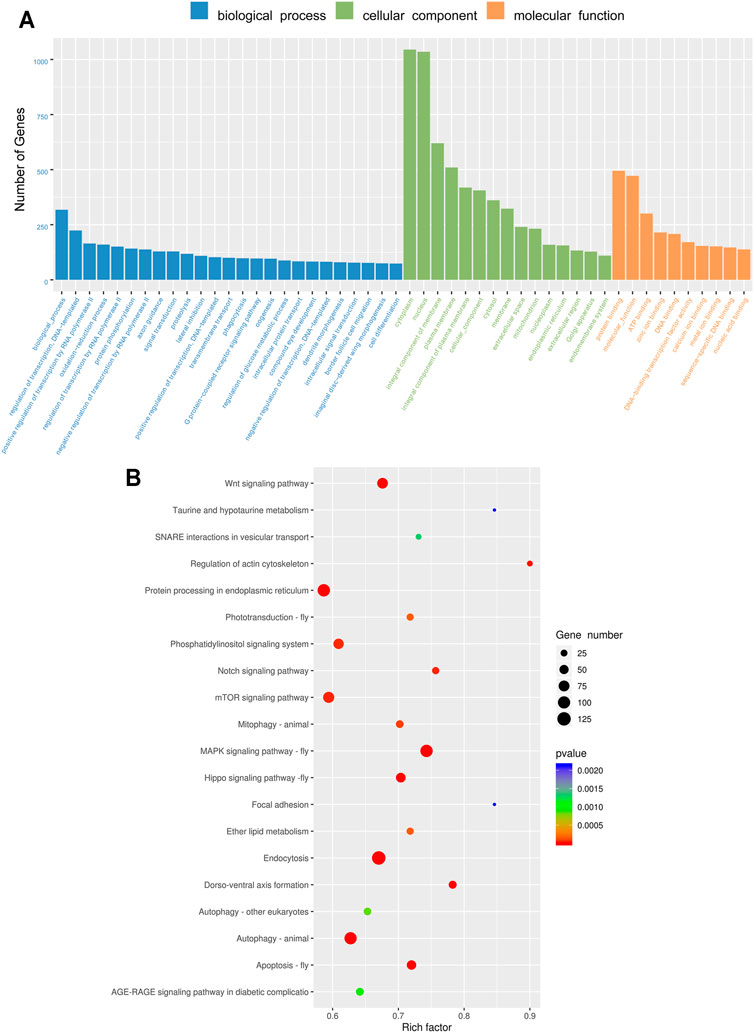
FIGURE 4. Gene Ontology (GO) term and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of the target genes. (A) Gene Ontology (GO) term of the target genes of differentially expressed miRNAs. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the target genes of differentially expressed miRNAs.
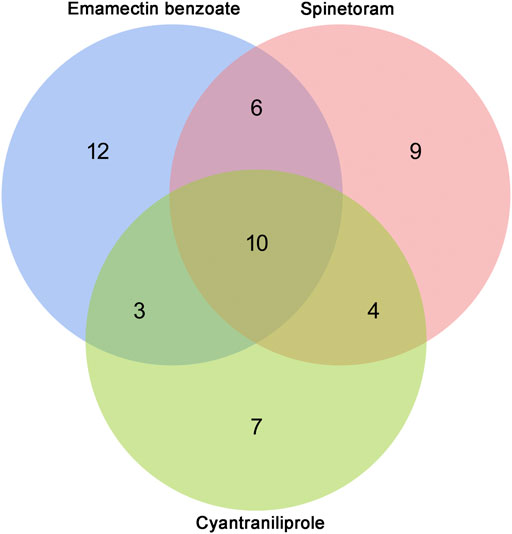
FIGURE 5. Venn diagram of differentially expressed miRNAs after cyantraniliprole, spinetoram, and emamectin benzoate treatment.
A total of ten miRNAs were significantly differentially expressed among the treatments of cyantraniliprole, spinetoram, and emamectin benzoate, including miR-278-5p, miR-13b-3p, miR-10485-5p, PC-5p-65875, miR-2766-3p, miR-10483-5p, miR-981, miR-2b-5p, miR-2a-5p, and PC-3p-2374 (Supplementary Table S5, Figure 5). The relative expression of ten differentially expressed miRNAs was validated by RT-qPCR. Compared to control, 4 miRNAs, including miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p, were significantly downregulated among the treatments of cyantraniliprole, spinetoram, and emamectin benzoate (Figures 6A–C,F). miR-981 was significantly upregulated among the treatments of cyantraniliprole, spinetoram, and emamectin benzoate (Figure 6G). miR-2766-3p was significantly upregulated between spinetoram and emamectin benzoate treatments (Figure 6E). miR-2b-5p was significantly downregulated and upregulated after cyantraniliprole and spinetoram treatments, respectively (Figure 6H). PC-3p-2374 was significantly downregulated after the spinetoram treatment (Figure 6J). The relative expression levels of PC-5p-65875 and miR-2a-5p showed no significant difference among the treatments of cyantraniliprole, spinetoram, and emamectin benzoate compared to control (Figures 6D–I).
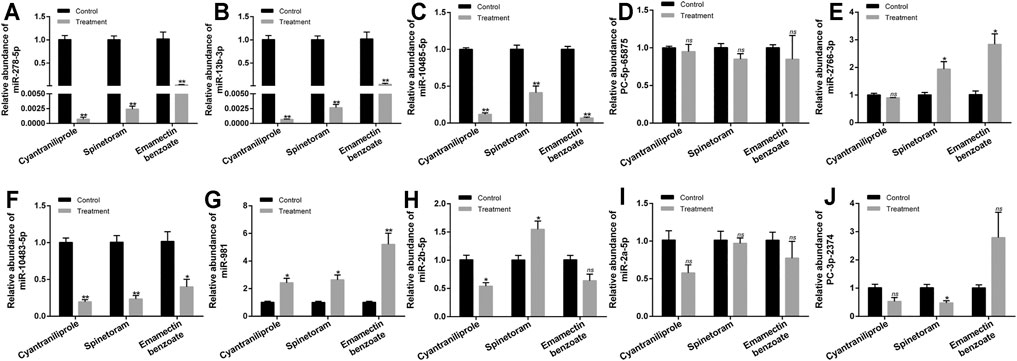
FIGURE 6. Relative expression of ten miRNAs after the treatments of cyantraniliprole, spinetoram, and emamectin benzoate. (A–J) Relative expressions of miR-278-5p, miR-13b-3p, miR-10485-5p, PC-5p-65875, miR-2766-3p, miR-10483-5p, miR-981, miR-2b-5p, miR-2a-5p, and PC-3p-2374 after the treatments of cyantraniliprole, spinetoram, and emamectin benzoate. The asterisk (*) and asterisks (**) indicate p < 0.05 and p < 0.01. ns represents no significant difference. Results are shown as the average±standard error (SE).
Furthermore, the expression profiles of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p were further investigated in different tissues of S. frugiperda. As shown in Figure 7A, miR-278-5p had the highest expression in the Malpighian tubule and the lowest expression in the midgut. The expression of miR-13b-3p had no significant difference in the tissues of the salivary gland, fat body, Malpighian tubule, and midgut (Figure 7B). Similar to miR-278-5p, miR-10485-5p had the highest expression in the Malpighian tubule and the lowest expression in the midgut (Figure 7C). The expression of miR-10483-5p had the highest expression in the salivary gland and the lowest expression in the midgut (Figure 7D).
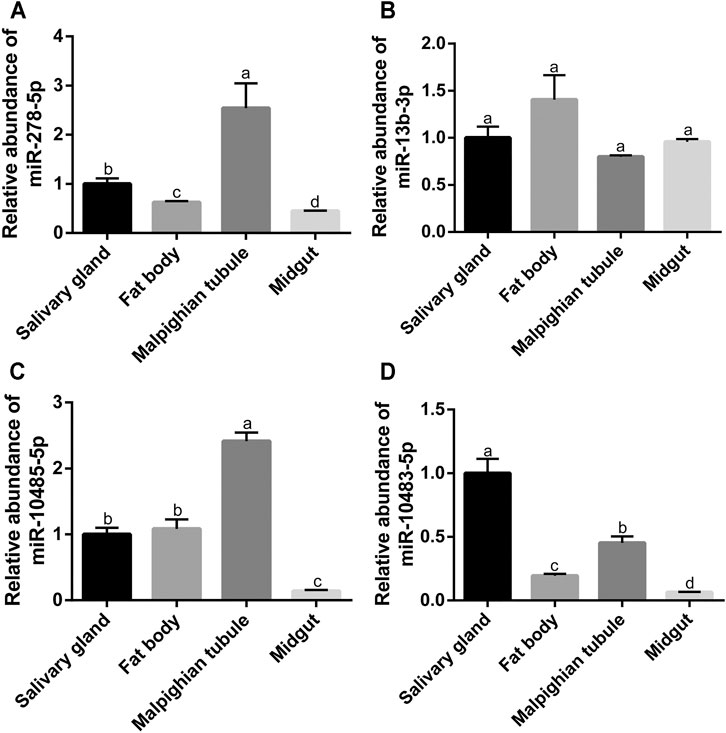
FIGURE 7. Expression of four miRNAs in different tissues of S. frugiperda. (A–D) Analysis of the miR-278-5p expression at the different tissues of the salivary gland, fat body, Malpighian tubule, and midgut. The letters a, b, c, d, e, and f represent significant differences (p < 0.05). Results are shown as the average±standard error (SE).
The predicted target genes of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p that might participate in the xenobiotic metabolism were further investigated. Some target genes have been verified to play vital functions in responses to xenobiotics, such as G-protein–coupled receptor, cytochrome P450 monooxygenases, ATP-binding cassette transporters, and UDP-glycosyltransferase (Supplementary Table S6).
The functions of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p in the tolerance of cyantraniliprole, spinetoram, and emamectin benzoate were studied by miRNA agomir injection. The relative abundance of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p was significantly elevated by injection of agomir-miR-278-5p, agomir-miR-13b-3p, agomir-miR-10485-5p, and agomir-miR-10483-5p (Supplementary Figure S3). The mortality was significantly increased in agomir-miR-278-5p, agomir-miR-13b-3p, agomir-miR-10485-5p, and agomir-miR-10483-5p injection, with LC50 doses of cyantraniliprole and emamectin benzoate treatment (Figures 8A–D). In addition, the mortality was significantly increased in agomir-miR-13b-3p, agomir-miR-10485-5p, and agomir-miR-10483-5p injection, with an LC50 dose of spinetoram treatment (Figures 8B–D). The mortality showed no significant difference in agomir-miR-278-5p injection, with an LC50 dose of spinetoram treatment (Figure 8A). These results indicated that miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p could regulate the tolerance of S. frugiperda to insecticides.
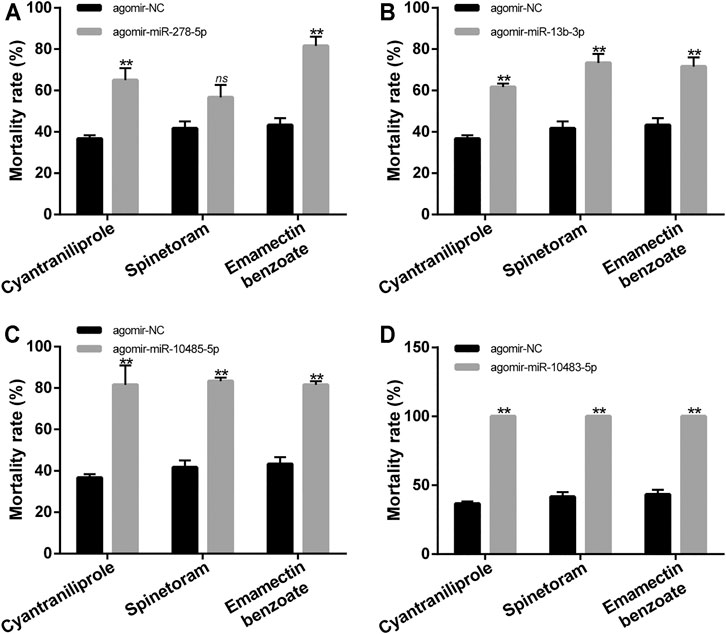
FIGURE 8. Mortality of S. frugiperda with LC50 doses of cyantraniliprole, spinetoram, and emamectin benzoate treatments after injection of miRNA agomir. (A) Mortality of S. frugiperda with LC50 doses of treatment of three insecticides after injection of agomir-miR-278-5p. (B) Mortality of S. frugiperda with LC50 doses of treatment of three insecticides after injection of agomir-miR-13b-3p. (C) Mortality of S. frugiperda with LC50 doses of treatment of three insecticides after injection of agomir-miR-10485-5p. (D) Mortality of S. frugiperda with LC50 doses of treatment of three insecticides after injection of agomir-miR-10483-5p. The asterisks (**) indicate p < 0.01. ns represents no significant difference. Results are shown as the average±standard error (SE).
With the advent of new computational methods and deep-sequencing technologies, an increasing number of miRNAs have been identified in several eukaryotes (Asgari, 2013). The functions of miRNAs are mainly focused on a variety of physiological processes in insects (Lucas et al., 2015b). Recently, major progress has been made in the miRNA regulatory roles in insecticide resistance by targeting detoxification genes in several insect species (Zhang et al., 2021b). It has been reported that a series of miRNAs participate in the antiviral immune defense in S. frugiperda (Karamipour et al., 2019). However, the profiles and functions of miRNAs in insecticide resistance and detoxification are still unknown in S. frugiperda.
Our present research systematically identified the miRNAs that might play regulatory roles in the tolerance of cyantraniliprole, spinetoram, and emamectin benzoate in S. frugiperda. A total of 379 conserved and 125 novel miRNAs existing in S. frugiperda were systematically identified in this study. A previous study has identified 215 and 239 miRNAs in the “corn-strain” and “rice-strain” S. frugiperda, respectively (Moné et al., 2018). Our sequencing data provide an important basis for further research about miRNAs in different physiological processes. The number of miRNAs identified in S. frugiperda was similar to that of other insects, such as Plutella xylostella (Zhu et al., 2017). The 22 nt length was the most abundant miRNAs in S. frugiperda which was similar to the previous reports in insects (Zhang et al., 2021c; Li et al., 2015b; Zhu et al., 2017; Mao et al., 2021; Zhang et al., 2021a). In addition, miR-1, miR-6497-5p, and miR-276-3p were the three most abundant miRNAs in S. frugiperda. miR-1 is a widely studied conserved miRNA, and a recent research showed that miR-1-3p was significantly downregulated in cyflumetofen-resistant strain of Tetranychus cinnabarinus and involved in the resistance to cyflumetofen via targeting the glutathione S-transferase gene (Zhang et al., 2018). These results indicated that the high abundance of miRNAs identified in S. frugiperda might also participate in the insecticide tolerance.
A total of 24, 22, and 31 miRNAs were differentially expressed after treatments of cyantraniliprole, spinetoram, and emamectin benzoate, respectively, suggesting that these miRNAs might play a regulatory role in the detoxification of insecticides in S. frugiperda. miRNAs play critical roles by regulating the transcript levels of target mRNA genes, thereby understanding the functions of target genes will be helpful to reveal the roles of miRNAs. In this study, the target genes of differentially expressed miRNAs were predicted by GO and KEGG enrichment analyses. Interestingly, the MAPK signaling pathway was the most significantly enriched pathway. Recent research has shown that cyantraniliprole could regulate the reproduction of Bactrocera dorsalis by affecting 20-hydroxyecdysone through the MAPK signaling pathway (Li et al., 2021b). These results indicated that miRNAs could affect the physiological and developmental pathways of S. frugiperda by the MAPK signaling pathway.
Furthermore, we found the relative expressions of miR-278-5p, miR-13b-3p, miR-10485-5p, PC-5p-65875, miR-2766-3p, miR-10483-5p, miR-981, miR-2b-5p, miR-2a-5p, and PC-3p-2374 were all significantly differentially expressed among the treatments of cyantraniliprole, spinetoram, and emamectin benzoate. The common differentially expressed miRNAs were likely to be involved in the tolerance of cyantraniliprole, spinetoram, and emamectin benzoate. However, as insects have complex insecticide-resistant mechanisms, these 10 miRNAs may also reveal common mechanisms of resistance to these three insecticides. Previous reports have shown that miR-2 and miR-13 could regulate the resistance to deltamethrin in Culex pipiens pallens (Guo et al., 2017). In addition, miR-278-3p could regulate the resistance to pyrethroid in C. pipiens pallens by targeting CYP6AG11 (Lei et al., 2015). In order to further explore the roles of these 10 miRNAs in the tolerance of insecticides, the relative expressions were detected in S. frugiperda after treatments of cyantraniliprole, spinetoram, and emamectin benzoate by RT-qPCR. miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p were all significantly downregulated among the treatments of these three insecticides. Through tissue expression profiling, we found miR-278-5p, miR-10485-5p, and miR-10483-5p that were lowly expressed in the midgut of S. frugiperda, suggesting that their target genes were highly expressed in the midgut. Some target genes of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p which previously reported relevant to insecticide resistance were discovered, mainly including P450s, ABC transporters, and UDP-glycosyltransferase (Lin et al., 2013; Perreault et al., 2013; Meng et al., 2020). However, a relationship between miRNAs and their target genes needs to be further researched. These results indicated that miRNAs might regulate the resistance to insecticides in S. frugiperda by targeting genes putatively participated in insecticide detoxication.
To further reveal the functions of miRNAs in the insecticide tolerance, we analyzed the mortality of S. frugiperda to cyantraniliprole, spinetoram, and emamectin benzoate after injection of miRNA agomir. The overexpression of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p significantly increased the mortality of S. frugiperda to cyantraniliprole and emamectin benzoate treatment. Furthermore, the mortality was significantly increased with spinetoram treatment after overexpression of miR-13b-3p, miR-10485-5p, and miR-10483-5p. These results indicated that miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p could regulate the tolerance of S. frugiperda to cyantraniliprole, spinetoram, and emamectin benzoate and indicated that these four miRNAs might reveal a common mechanism to insecticide resistance.
In summary, we identified the differentially expressed miRNAs in S. frugiperda after cyantraniliprole, spinetoram, and emamectin benzoate treatments. The effects of miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p on the tolerance of S. frugiperda to cyantraniliprole, spinetoram, and emamectin benzoate were analyzed. These results indicate that miRNAs play key roles in the resistance of S. frugiperda to insecticides.
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number GSE189968.
ZJ, ZM, and YY conceived and designed the study. YY and ZJ performed the experiments and analyzed the data with the help of ZY, WA, DA, and XC. ZJ wrote the first draft of the manuscript. ZJ, YY, ZM, and WK participated in manuscript drafting and modification.
This study was funded by the Technology Innovation Guidance Plan of Shandong Province, China (2020LYXZ030), National Natural Science Foundation of China (32102211), and Natural Science Foundation of Shandong Province, China (ZR2019BC055).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.820778/full#supplementary-material
Arias, O., Cordeiro, E., Corrêa, A. S., Domingues, F. A., Guidolin, A. S., and Omoto, C. (2019). Population Genetic Structure and Demographic History of Spodoptera Frugiperda (Lepidoptera: Noctuidae): Implications for Insect Resistance Management Programs. Pest Manag. Sci. 75, 2948–2957. doi:10.1002/ps.5407
Asgari, S. (2013). MicroRNA Functions in Insects. Insect Biochem. Mol. Biol. 43, 388–397. doi:10.1016/j.ibmb.2012.10.005
Bartel, D. P. (2004). MicroRNAs: Genomics, Biogenesis, Mechanism, and functionMicroRNAs. Cell 116, 281–297. doi:10.1016/s0092-8674(04)00045-5
Bolzan, A., Padovez, F. E., Nascimento, A. R., Kaiser, I. S., Lira, E. C., Amaral, F. S., et al. (2019). Selection and Characterization of the Inheritance of Resistance ofSpodoptera frugiperda(Lepidoptera: Noctuidae) to Chlorantraniliprole and Cross‐resistance to Other Diamide Insecticides. Pest Manag. Sci. 75, 2682–2689. doi:10.1002/ps.5376
Carvalho, R. A., Omoto, C., Field, L. M., Williamson, M. S., and Bass, C. (2013). Investigating the Molecular Mechanisms of Organophosphate and Pyrethroid Resistance in the Fall Armyworm Spodoptera Frugiperda. PLoS One 8, e62268. doi:10.1371/journal.pone.0062268
Early, R., González-Moreno, P., Murphy, S. T., and Day, R. (2018). Forecasting the Global Extent of Invasion of the Cereal Pest Spodoptera Frugiperda, the Fall Armyworm. Nb 40, 25–50. doi:10.3897/neobiota.40.28165
Etebari, K., and Asgari, S. (2013). Conserved microRNA miR-8 Blocks Activation of the Toll Pathway by Upregulating Serpin 27 Transcripts. RNA Biol. 10, 1356–1364. doi:10.4161/rna.25481
Fouad, E. A., Abou-Yousef, H. M., Abdallah, I. S., and Kandil, M. A. (2016). Resistance Monitoring and Enzyme Activity in Three Field Populations of Cowpea Aphid ( Aphis Craccivora ) from Egypt. Crop Prot. 81, 163–167. doi:10.1016/j.cropro.2015.12.015
Goergen, G., Kumar, P. L., Sankung, S. B., Togola, A., and Tamò, M. (2016). First Report of Outbreaks of the Fall Armyworm Spodoptera Frugiperda (J E Smith) (Lepidoptera, Noctuidae), a New Alien Invasive Pest in West and central Africa. PLoS One 11, e0165632. doi:10.1371/journal.pone.0165632
Gouin, A., Bretaudeau, A., Nam, K., Gimenez, S., Aury, J.-M., Duvic, B., et al. (2017). Two Genomes of Highly Polyphagous Lepidopteran Pests (Spodoptera Frugiperda, Noctuidae) with Different Host-Plant Ranges. Sci. Rep. 7, 11816. doi:10.1038/s41598-017-10461-4
Guo, Q., Huang, Y., Zou, F., Liu, B., Tian, M., Ye, W., et al. (2017). The Role of miR-2∼13∼71 Cluster in Resistance to Deltamethrin in Culex pipiens Pallens. Insect Biochem. Mol. Biol. 84, 15–22. doi:10.1016/j.ibmb.2017.03.006
Hafeez, M., Li, X. W., Zhang, J. M., Zhang, Z. J., Huang, J., Wang, L. K., et al. (2021). Role of Digestive Protease Enzymes and Related Genes in Host Plant Adaptation of a Polyphagous Pest, Spodoptera Frugiperda. Insect Sci. 28, 611–626. doi:10.1111/1744-7917.12906
He, L.-m., Wang, T.-l., Chen, Y.-c., Ge, S.-s., Wyckhuys, K. A. G., and Wu, K.-m. (2021). Larval Diet Affects Development and Reproduction of East Asian Strain of the Fall Armyworm, Spodoptera Frugiperda. J. Integr. Agric. 20, 736–744. doi:10.1016/S2095-3119(19)62879-0
Junior, I. L., Degrande, P. E., Melo, E. P. D., Bertoncello, T. F., and Suekane, R. (2012). Spodoptera Frugiperda (J.E. Smith, 1797) (Lepidoptera: Noctuidae) Infestation and its Natural Enemies on Corn in Dry and Irrigated Conditions. Revista Agrarian 5, 14–19.
Karamipour, N., Fathipour, Y., Talebi, A. A., Asgari, S., and Mehrabadi, M. (2019). The microRNA Pathway Is Involved in Spodoptera Frugiperda (Sf9) Cells Antiviral Immune Defense against Autographa Californica Multiple Nucleopolyhedrovirus Infection. Insect Biochem. Mol. Biol. 112, 103202. doi:10.1016/j.ibmb.2019.103202
Lee, Y. S., Nakahara, K., Pham, J. W., Kim, K., He, Z., Sontheimer, E. J., et al. (2004). Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA Silencing Pathways. Cell 117, 69–81. doi:10.1016/s0092-8674(04)00261-2
Lei, Z., Lv, Y., Wang, W., Guo, Q., Zou, F., Hu, S., et al. (2015). MiR-278-3p Regulates Pyrethroid Resistance in Culex pipiens Pallens. Parasitol. Res. 114, 699–706. doi:10.1007/s00436-014-4236-7
Li, J.-M., Zhou, Y.-R., Sun, Z.-T., Wang, X., Xie, L., and Chen, J.-P. (2015b). Identification and Profiling of Conserved and Novel microRNAs in Laodelphax Striatellus in Response to rice Black-Streaked dwarf Virus (RBSDV) Infection. Genomics Data 3, 63–69. doi:10.1016/j.gdata.2014.08.010
Li, J., Liu, J., Chi, B., Chen, P., and Liu, Y. (2021b). 20E and MAPK Signal Pathway Involved in the Effect of Reproduction Caused by Cyantraniliprole in Bactrocera Dorsalis Hendel (Diptera: Tephritidae). Pest Manag. Sci. 78, 63–72. doi:10.1002/ps.6607
Li X, X., Zhao, M. H., Tian, M. M., Zhao, J., Cai, W. L., and Hua, H. X. (2021a). An InR/Mir‐9a/NlUbx Regulatory cascade Regulates wing Diphenism in Brown Planthoppers. Insect Sci. 28, 1300–1313. doi:10.1111/1744-7917.12872
Li, X., Guo, L., Zhou, X., Gao, X., and Liang, P. (2015a). miRNAs Regulated Overexpression of Ryanodine Receptor Is Involved in Chlorantraniliprole Resistance in Plutella Xylostella (L.). Sci. Rep. 5, 14095. doi:10.1038/srep14095
Li, X. J., Wu, M. F., Ma, J., Gao, B. Y., WuChen, Q. L. A. D., Chen, A. D., et al. (2020). Prediction of Migratory Routes of the Invasive Fall Armyworm in Eastern China Using a Trajectory Analytical Approach. Pest Manag. Sci. 76, 454–463. doi:10.1002/ps.5530
Lin, Q., Jin, F., Hu, Z., Chen, H., Yin, F., Li, Z., et al. (2013). Transcriptome Analysis of Chlorantraniliprole Resistance Development in the Diamondback Moth Plutella Xylostella. PLoS One 8, e72314. doi:10.1371/journal.pone.0072314
Ling, L., Kokoza, V. A., Zhang, C., Aksoy, E., and Raikhel, A. S. (2017). MicroRNA-277 Targetsinsulin-like Peptides 7and8to Control Lipid Metabolism and Reproduction inAedes Aegyptimosquitoes. Proc. Natl. Acad. Sci. USA 114, E8017–E8024. doi:10.1073/pnas.1710970114
Lira, E. C., Bolzan, A., Nascimento, A. R., Amaral, F. S., Kanno, R. H., Kaiser, I. S., et al. (2020). Resistance of Spodoptera Frugiperda (Lepidoptera: Noctuidae) to Spinetoram: Inheritance and Cross‐resistance to Spinosad. Pest Manag. Sci. 76, 2674–2680. doi:10.1002/ps.5812
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lozano, J., Montañez, R., and Belles, X. (2015). MiR-2 Family Regulates Insect Metamorphosis by Controlling the Juvenile Hormone Signaling Pathway. Proc. Natl. Acad. Sci. USA 112, 3740–3745. doi:10.1073/pnas.1418522112
Lucas, K. J., Zhao, B., Liu, S., and Raikhel, A. S. (2015a). Regulation of Physiological Processes by microRNAs in Insects. Curr. Opin. Insect Sci. 11, 1–7. doi:10.1016/j.cois.2015.06.004
Lucas, K. J., Zhao, B., Roy, S., Gervaise, A. L., and Raikhel, A. S. (2015b). Mosquito-specific microRNA-1890 Targets the Juvenile Hormone-Regulated Serine Protease JHA15 in the Female Mosquito Gut. RNA Biol. 12, 1383–1390. doi:10.1080/15476286.2015.1101525
Ma, K., Li, F., Tang, Q., Liang, P., Liu, Y., Zhang, B., et al. (2019). CYP4CJ1-mediated Gossypol and Tannic Acid Tolerance in Aphis Gossypii Glover. Chemosphere 219, 961–970. doi:10.1016/j.chemosphere.2018.12.025
Mao, K., Jin, R., Ren, Z., Zhang, J., Li, Z., He, S., et al. (2021). miRNAs Targeting CYP6ER1 and CarE1 Are Involved in Nitenpyram Resistance in Nilaparvata Lugens. Insect Sci. doi:10.1111/1744-7917.12910
Meng, X., Yang, X., Wu, Z., Shen, Q., Miao, L., Zheng, Y., et al. (2020). Identification and Transcriptional Response of ATP ‐binding Cassette Transporters to Chlorantraniliprole in the rice Striped Stem Borer, Chilo Suppressalis. Pest Manag. Sci. 76, 3626–3635. doi:10.1002/ps.5897
Moné, Y., Nhim, S., Gimenez, S., Legeai, F., Seninet, I., Parrinello, H., et al. (2018). Characterization and Expression Profiling of microRNAs in Response to Plant Feeding in Two Host-Plant Strains of the Lepidopteran Pest Spodoptera Frugiperda. BMC Genomics 19, 804. doi:10.1186/s12864-018-5119-6
Nascimento, A. R. B. D., Farias, J. R., Bernardi, D., Horikoshi, R. J., and Omoto, C. (2016). Genetic Basis ofSpodoptera frugiperda(Lepidoptera: Noctuidae) Resistance to the Chitin Synthesis Inhibitor Lufenuron. Pest Manag. Sci. 72, 810–815. doi:10.1002/ps.4057
Perreault, M., Gauthier-Landry, L., Trottier, J., Verreault, M., Caron, P., Finel, M., et al. (2013). The Human UDP-Glucuronosyltransferase UGT2A1 and UGT2A2 Enzymes Are Highly Active in Bile Acid Glucuronidation. Drug Metab. Dispos 41, 1616–1620. doi:10.1124/dmd.113.052613
Salvi, V., Gianello, V., Tiberio, L., Sozzani, S., and Bosisio, D. (2019). Cytokine Targeting by miRNAs in Autoimmune Diseases. Front. Immunol. 10, 10–15. doi:10.3389/fimmu.2019.00015
Shang, F., Niu, J., Ding, B.-Y., Zhang, W., Wei, D.-D., Wei, D., et al. (2020). The miR-9b microRNA Mediates Dimorphism and Development of wing in Aphids. Proc. Natl. Acad. Sci. USA 117, 8404–8409. doi:10.1073/pnas.1919204117
Song, J., Li, W., Zhao, H., Gao, L., Fan, Y., and Zhou, S. (2018). MicroRNA Let-7 and miR-278 Regulate Insect Metamorphosis and Oogenesis via Targeting Juvenile Hormone Early Response Gene Krüppel-Homolog 1. Development 145, 170670. doi:10.1242/dev.170670
Stark, A., Brennecke, J., Bushati, N., Russell, R. B., and Cohen, S. M. (2005). Animal MicroRNAs Confer Robustness to Gene Expression and Have a Significant Impact on 3′UTR Evolution. Cell 123, 1133–1146. doi:10.1016/j.cell.2005.11.023
Tsai, W.-C., Hsu, P. W.-C., Lai, T.-C., Chau, G.-Y., Lin, C.-W., Chen, C.-M., et al. (2009). MicroRNA-122, a Tumor Suppressor microRNA that Regulates Intrahepatic Metastasis of Hepatocellular Carcinoma. Hepatology 49, 1571–1582. doi:10.1002/hep.22806
Yu, S. J., and McCord, E. (2007). Lack of Cross-Resistance to Indoxacarb in Insecticide-resistantSpodoptera Frugiperda (Lepidoptera: Noctuidae) andPlutella Xylostella (Lepidoptera: Yponomeutidae). Pest Manag. Sci. 63, 63–67. doi:10.1002/ps.1309
Zhang, B.-Z., Hu, G.-L., Lu, L.-Y., Hu, S.-F., Li, Y.-S., Su, X., et al. (2021c). Identification of Differentially Expressed microRNAs under Imidacloprid Exposure in Sitobion Miscanthi. Pestic. Biochem. Physiol. 177, 104885. doi:10.1016/j.pestbp.2021.104885
Zhang, J., Dong, Y., Wang, M., Wang, H., Yi, D., Zhou, Y., et al. (2021a). MicroRNA ‐315‐ 5p Promotes rice Black‐streaked dwarf Virus Infection by Targeting a Melatonin Receptor in the Small Brown Planthopper. Pest Manag. Sci. 77, 3561–3570. doi:10.1002/ps.6410
Zhang, Q., Dou, W., Taning, C. N. T., Smagghe, G., and Wang, J.-J. (2021b). Regulatory Roles of microRNAs in Insect Pests: Prospective Targets for Insect Pest Control. Curr. Opin. Biotechnol. 70, 158–166. doi:10.1016/j.copbio.2021.05.002
Zhang, Y., Feng, K., Hu, J., Shi, L., Wei, P., Xu, Z., et al. (2018). A microRNA-1 Gene, Tci-miR-1-3p , Is Involved in Cyflumetofen Resistance by Targeting a Glutathione S-Transferase Gene, TCGSTM4 , in Tetranychus Cinnabarinus. Insect Mol. Biol. 27, 352–364. doi:10.1111/imb.12375
Zhang, Y., Zhao, B., Roy, S., Saha, T. T., Kokoza, V. A., Li, M., et al. (2016). microRNA-309 Targets the Homeobox Gene SIX4 and Controls Ovarian Development in the Mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 113, E4828–E4836. doi:10.1073/pnas.1609792113
Keywords: Spodoptera frugiperda, insecticides, miRNA-13b-3p, miRNA-278-5p, miRNA-10483-5p, miRNA-10485-5p
Citation: Yang Y, Zhang Y, Wang A, Duan A, Xue C, Wang K, Zhao M and Zhang J (2022) Four MicroRNAs, miR-13b-3p, miR-278-5p, miR-10483-5p, and miR-10485-5p, Mediate Insecticide Tolerance in Spodoptera frugiperda. Front. Genet. 12:820778. doi: 10.3389/fgene.2021.820778
Received: 23 November 2021; Accepted: 30 December 2021;
Published: 21 January 2022.
Edited by:
Kangxu Wang, Nanjing University of Finance and Economics, ChinaReviewed by:
Kang He, Zhejiang University, ChinaCopyright © 2022 Yang, Zhang, Wang, Duan, Xue, Wang, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Zhang, emhhbmdqaWFuaHVhMTk4OTA0QDE2My5jb20=; Ming Zhao, c2NyY3pobUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.