
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 04 January 2022
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.816075
This article is part of the Research TopicThe Origination of Genetic Novelties: New Genes, New Regulations, and New Cell TypesView all 8 articles
The domesticated silkworm, Bombyx mori, is an economically important insect that synthesizes large amounts of silk proteins in its silk gland to make cocoons. In recent years, germline transformation strategies advanced the bioengineering of the silk gland as an ideal bioreactor for mass production of recombinant proteins. However, the yield of exogenous proteins varied largely due to the random insertion and gene drift caused by canonical transposon-based transformation, calling for site-specific and stable expression systems. In the current study, we established a targeted in-fusion expression system by using the transcription activator-like effector nuclease (TALEN)-mediated targeted insertion to target genomic locus of sericin, one of the major silk proteins. We successfully generated chimeric Sericin1-EGFP (Ser-2A-EGFP) transformant, producing up to 3.1% (w/w) of EGFP protein in the cocoon shell. With this strategy, we further expressed the medically important human epidermal growth factor (hEGF) and the protein yield in both middle silk glands, and cocoon shells reached to more than 15-fold higher than the canonical piggyBac-based transgenesis. This natural Sericin1 expression system provides a new strategy for producing recombinant proteins by using the silkworm silk gland as the bioreactor.
The lepidopteran model insect Bombyx mori is an important economic insect and possesses a highly specialized larval tissue, the silk gland, to synthesize and secret massive silk proteins in a few days of the late final larval instar. On average, each silkworm eats 20 g of mulberry leaves and produces about 0.5 g of pure silk protein, holding the great promise to be a cost-effective system for mass production of recombinant proteins (Ma et al., 2014). This efficient protein production capacity has been described as an important model for tissue-specific gene regulation and exogenous protein synthesis (Tomita et al., 2003; Takasu et al., 2016).
Silkworm silk proteins, which are the major components of cocoon shells, contain the insoluble fibroin protein and hydrophilic sericin protein (Iizuka et al., 2008). Fibroin protein is synthesized in the posterior silk gland (PSG), assembled in the lumen of the middle silk gland (MSG) with sericin, and then secreted into the anterior silk gland (ASG) to spin and form cocoon shells. Fibroin proteins account for 70%–80% of the total silk proteins, being composed by the heavy chain protein (FibH), light chain protein (FibL), and fibrohexamerin protein (Fhx) in a molar ratio of 6:6:1 (Inoue et al., 2005). In recent years, the transposon-based, transgenic silk gland expression system has been established to express recombinant proteins with the FibL promoter and its 5′-flanking sequences. The exogenous proteins were secreted into the lumen of PSG together with fibroin proteins and reached up to 0.84% of the whole cocoon shell weight (Tomita et al., 2003). Subsequently, another transgene-based expression system using the FibH promoter was established and promoted the recombinant protein amount to 15% (w/w) per cocoon shell (Tomita et al., 2007; Zhao et al., 2010). Most recently, a targeted expression of ampullate spidroin-1 gene with FibH gene replacement was developed successfully, with an unprecedented yield up to 35.2% wt/wt of cocoon shells (Xu et al., 2018). These cases suggested that fibroin genes can achieve high recombinant protein yield, however, extraction and purification of the proteins were complicated since the fibroin protein was insoluble and the exogenous proteins are tightly combined with silk fibers.
Sericin proteins weigh to nearly 20% of the total cocoon shell weight, while the extraction and purification processes from a cocoon shell are more practical since they are soluble. Sericin proteins are synthesized in the MSG and mainly consist of Sericin1 (Serl), Sericin2 (Ser2), and Sericin 3 (Ser3) proteins, in which Ser1 has the dominant expression (Takasu et al., 2007). They function as the glue protein and majorly coat and cement fibroin filaments to form the silk fibers (Xu et al., 2014; Dong et al., 2016). The transposon-based transgenic sericin expression system ectopically expressed exogenous protein with Ser1 promoter has been established and a series of modifications on the regulatory elements both at promoter and 3′ untranslated region (UTR) were performed to increase the transcriptional and translational level of exogenous proteins (Tomita et al., 2007; Iizuka et al., 2008; Wang et al., 2013; Wang, 2013). Additionally, exogenous protein yield could be increased under the mutant genetic background, which is deficient in fibroin secretion (Inoue et al., 2005). Altogether, these evidences suggested that transgenic production of exogenous proteins was largely depending on the regulatory elements, inspiring us to establish an in situ sericin expression system with the original regulatory sequences.
Transposon, especially the PiggyBac-mediated transgenesis advent the genomic era in the silkworm, however, targeted genome editing was still challengeable till the site-specific nuclease was engineered successfully (Maeder et al., 2008; Edgell, 2009; Boch, 2011; Mulepati et al., 2014). Along with the quick development and adaption of the site-specific nucleases, homing endonucleases, including ZFN (zinc-finger nuclease), TALEN (transcription activator-like effector nuclease), and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/RNA-guided Cas9 nucleases) have been wildly applied into a large range of host organisms and cells (Zhang et al., 2014). All these nucleases created double-stranded breaks (DSBs)at the targeted genomic DNA, which trigger and utilize the endogenous DSB repair machinery especially for homologous-directed repair to introduce designed modifications or insertions. Up to now, only TALEN-mediated targeting insertion was achieved in the silkworm successfully, which may be attributed to different DSB repair pathways that were used by these engineered nucleases (Wang et al., 2013; Xu et al., 2018; Zhang et al., 2018). Here we report establishment of an in situ Ser1 in-fusion expression system in B. mori by using the TALEN-mediated targeted insertion. High production of exogenous proteins of enhanced green fluorescent protein (EGFP) and the medically important human epidermal growth factor (hEGF) were successfully detected in both MSGs and cocoon shells. Compared with the transposon-based random insertion, our strategy promoted the hEGF production up to 15-fold. In conclusion, the current study established a natural Sericin1 bioreactor system in B. mori, showing great potential for mass production of recombinant proteins.
The multivoltine, nondiapausing silkworm strain, Nistari, was used for genetic transformation. Larvae were reared on fresh mulberry leaves under the standard condition at 25°C (Tan et al., 2013). Mammalian HEK293T cell was maintained in DMEM (Gibco) medium supplemented with 10% fetal bovine serum at 37°C under 5% CO2.
Pairs of TALENs were designed and constructed by ViewSolid Biotech using Golden-Gate assembly and ligated into the VK006-06 vector, under the control of T7 in vitro transcriptional promoter. The activity of the TALENs was examined using an SSA assay in HEK293T cell line (Wang et al., 2013). One TALEN targeting sites located around the stop codons (C) of the Sericin1 gene was chosen with the sequence listed as follows: 5′-TAAGAATATCGGTGTTTaatacaactaaacacgaCTTGGAGTATTCCTTGTA-3′, with the capital letters as the TALEN recognition sites and lowercase letters as the spacers. The targeting sites were verified by amplification from the genomic DNA to exclude the single nucleotide polymorphisms. A homology-directed recombination (HDR) donor plasmid was constructed based on the pGEM-T vector. For facilitating the HR recombination, the HR5-IE1-DsRed-SV40 cassette was cut from pXL-IE1-DsRed silkworm transgenic plasmid using BamHI single restriction enzyme, and subcloned into the pGEM-T easy vector (Promega) to generate pGEM-Red plain plasmid. A 1,000-bp 3′-HR arm amplified from the genomic DNA at the right flanking of the TALEN site was inserted into the pGEM-Red plasmid at the SpeI restriction enzyme site using in-fusion ClonExpress™ II One Step Cloning Kit (RA, Vazyme Biotech Co. Ltd.). The 1,000-bp left HR arm was amplified from the left of the TALEN site, which was then fused with the EGFP or hEGF expressing cassette. To achieve the sericin in-fusion expression, EGFP- or hEGF-coding sequences was inserted into the downstream of the right homologous arm using 2A self-cleavage sequence (GAGGGCAGAGGAAGTCTTCTAACATGCGGTGACGTGGAGGAGAATCCCGGCCCT) at the SacII restriction enzyme site. To optimize the exogenous proteins expression, the Sericin1 polyA (PA) sequence was cloned and ligated into the downstream of EGFP or hEGF sequence. In order to facilitate the integration, two TALENs targeting sequences were added to each side of the donors, to linearize the circular donor plasmids.
TALEN-expressing vector under the control of the T7 in vitro expression promoter was linearized using the NotI restriction enzyme, purified with phenol:chloroform:isoamyl alcohol (25:24:1), and sent for in vitro mRNA synthesis using the mMessage mMachine T7 Ultra Kit (Life Technologies). Mixture of the TALEN mRNA (250 ng/μl) and donor plasmid (300 ng/μl) was injected into silkworm embryos at the preblastoderm stage (Li et al., 2018). The injected eggs were incubated at 25°C for 10–12 days until hatched and reared on fresh mulberry leaves. G0 moths were crossed with wild-type (WT) animals and positive G1 embryos were screened under red fluorescence.
Total RNA extraction, first strand cDNA synthesis, and quantification of mRNA total RNA was extracted from the middle silk gland of WT, Ser-2A-EGFP, Ser-2A-hEGF, and Ser-T-hEGF animals during the whole fifth instar larvae. One microgram of the purified RNA was used for the cDNA synthesis using the ReverAid First Strand cDNA Synthesis Kit (Vazyme Biotech Co. Ltd.). The relative transcriptional levels of silkworm Sericin1, EGFP, and hEGF were examined by quantitative real-time PCR (qRT-PCR) using SYBR Green Real-time PCR Master Mix (TOYOBO) with the following primers sets, BmSer1RTF: 5′- GGCGAGCTCTACCATCTACG -3′ and BmSer1RTR: 5′- TCAGATTTGCTGCGTTTGTC-3, EGFPRTF: 5′- GGTGAACTTCAAGATCCGCC-3′ and EGFPRTR:5′- CTTGTACAGCTCGTCCATGC-3′, and hEGFRTF: 5′- TGTCCTCTCTCACATGACGG-3′ and hEGFRTR: 5′- ATGATGGCGTAATTCCCACC-3’. The primer set that amplified a 136-bp fragment of B. mori ribosomal protein 49 (Bmrp49) was used as the internal control (Li et al., 2018). Three independent biological replicates were used for all the qRT-PCR.
The G1 DsRed2-positive larvae were used for genomic DNA extraction. Insertion sites were confirmed using 5′- and 3′-end junction PCR using the primers as follows: DsRed5′F: 5′-CAGAAGTCATCGTTCAGGCG-3′ and DsRed5′R: 5′-TCCCACAACGAGGACTACAC-3′ for 5′-junction PCR, DsRed3′F: 5′-CAGTTCGGTTATGAGCCGTG-3′ and DsRed3′R: 5′-ATCACCCAGACGAAGAGCAA-3′ for 3′-junction PCR. Amplification products were cloned into the pJET1.0 cloning plasmid and sent for sequencing.
Silk proteins were extracted from the MSG of the wandering stage (W) larvae using the phosphate saline buffer (PBS) and silkworm cocoon shells were cut into small pieces for extraction using 8 M urea at 4°C overnight. The crude protein was quantified using BCA kit (Thermo) and sent for 10% SDS-PAGE. Separated proteins were treated with Coomassie brilliant blue (CBB) staining or transferred into the nitrocellulose membrane (GE Healthcare).
Silkworm middle silk glands extracted from the WT or Ser-2A-EGFP animals were prefixed with Qurnah’s fixative. A 5-μm cross section was cut with a Leica RM2235 microtome and sent for staining according to our previous publication (Li et al., 2018). The sections were incubated with an anti-EGFP (1:2,000, ABclonal) primary antibody for 48 h and then washed for three times with PBS, followed by treatment with an FITC-conjugated goat-anti-rabbit secondary antibody (1:100, YEASEN). The nuclei were stained with Hoechst (1:1,000, Beyotime) for 10 min. Samples were analyzed with a fluorescence microscope (Olympus, BX53).
All data were analyzed using GraphPad Prism (version 5.01) with two-way ANOVA and the Dunnett’s tests. The error bars are the means ± S.E.M. A p-value < 0.05 was used to determine significance in all cases.
In the current study, we targeted the Ser1 gene in B. mori to generate a Ser1-EGFP in-fusion expression transformant, as the proof-of-principle of our idea about the in situ expression of exogenous proteins in the silkworm silk gland. We used one pair of TALENs targeting sequence around the stop codons of Ser1 to generate an in-fusion gene expression (Figure 1A). The donor template carried 1,000-bp length left and right homologous fragments which matched exactly to the sequences flanking the TALEN target, as well as the 2A self-cleavage peptide followed by the EGFP coding sequence and Ser1 ployA sequences in between (Ser-2A-EGFP, Figure 1B). In theory, this scheme would express the target protein in the same manner with Ser1 since the common native regulatory elements were used. Given this idea, we designed three pairs of TALENs targeting the C-terminal of the Ser1 gene, and selected the one with the highest cutting efficiency (31.5-fold to the control, Figure 1D), which was determined by an in vitro SSA assay to perform subsequent experiments (Figures 1C, D).
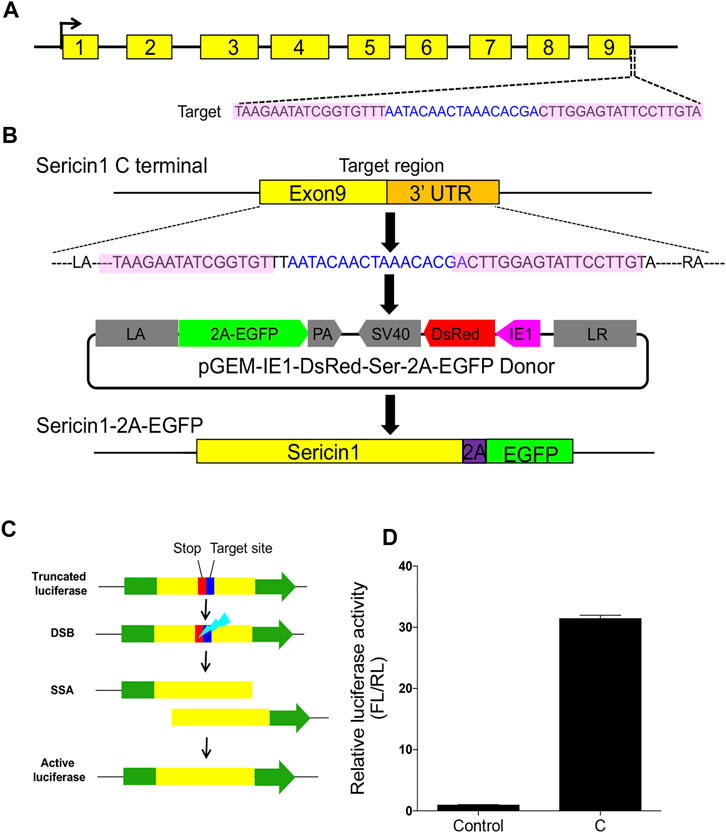
FIGURE 1. Designing and construction of TALENs. (A) Structure of silkworm Sericin1 gene and the targeting sites of TALENs. The black arrows indicate the start codons. The black line is the genome DNA sequence and brown boxes represent Sericin1 exons. Sequences of TALENs are also listed, with pink shaded letters indicating the TALEN recognition sites and blue letters are the spacers. (B) Scheme of 2A self-cleavage peptide (purple box)-mediated sericin-EGFP in-fusion expression on Sericin1 loci (Ser–2A–EGFP). (C) Schema of SSA assay for TALEN cutting efficiency. Green boxes are firefly luciferase coding sequences, yellow boxes indicate the repeat fragment flanking the target site. The stop codon and target site are presented as red and blue boxes, respectively. (D) Luciferase activity of the TALENs used for integration. The bars stand for mean ± S.E.M (n = 3).
In vitro synthesized TALENs mRNA and HR donors were coinjected into 640 silkworm preblastoderm eggs in each group to generate Ser–2A–EGFP transgenic line. In the G1 animals, five independent fluorescence-positive silkworm broods were obtained, achieving a 10.6% homologous recombination efficiency (Table 1). Genotype of the transformed animals were examined by through 5′- and 3′-junction PCR followed by Sanger sequencing using three animals from each G1 transformed silkworm broods. The results indicated that the integration events were precise and seamless (Figure 2A).

TABLE 1. Transformation efficiency of transcription activator-like effector nuclease (TALEN)-mediated transformation. Note. A total of 640 preblastoderm stage embryos were injected for each strain of animals.
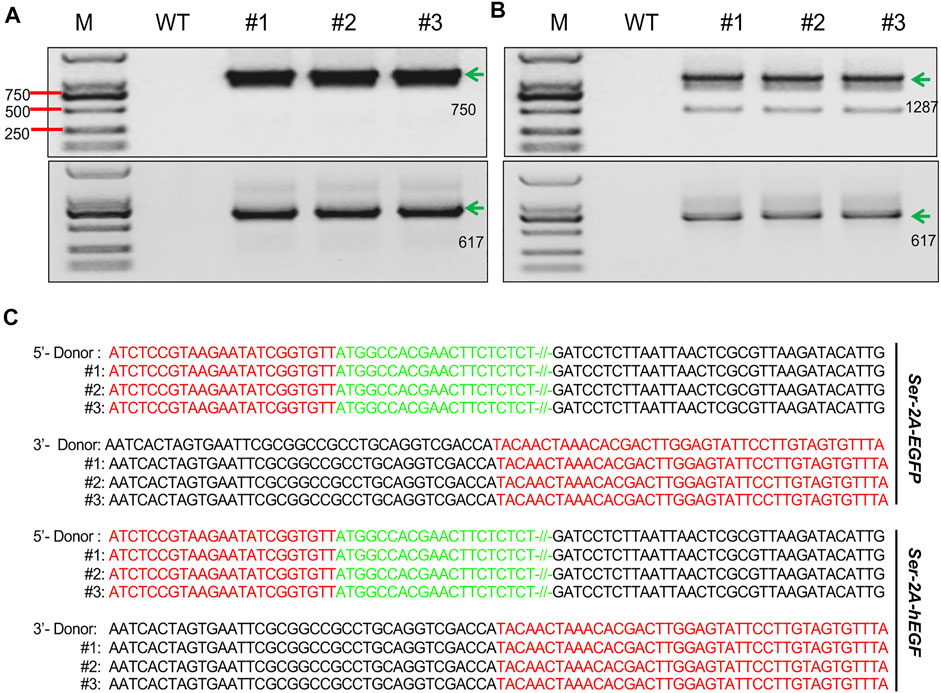
FIGURE 2. Genotyping of the transformed animals. (A, B) Junction PCR amplicons of the right and left flanking fragments in the SKI–2A–EGFP (A), and Ser–2A–hEGF (B) transformed silkworms. Three individual silkworms were used for detection. The green arrows indicate the correct amplicons, and their lengths are listed. M, marker; WT, wild type. (C) Sanger sequencing results for amplicons from (B) and (C). Red letters stand for the sequences of homologous arms, green letters are the partial sequences of EGFP or hEGF, and black letters are the sequences of donor backbones.
In order to make sure that the EGFP insertion did not affect native Ser1 gene expression, we first examined the transcription of Ser1 by using qRT-PCR. In the heterozygous animals, Ser1 presented relative low expression at the early stages of the final instar larvae, increased dramatically at the third day of larvae, and reached the peak at the wandering stage, being consistent with the WT animals (Figure 3A, Li et al., 2014). We observed that the expression level of Ser1 in the Ser–2A–EGFP animals was significantly decreased at L5D3 and L5D4, being comparable with the wild type at the late larval stages (Figure 3A). This result indicated that the integration of EGFP coding sequence had some degree of impact to the native Ser1 transcription (Figure 3A). We also detected EGFP proteins in the MSG of the Ser–2A–EGFP animals by Western blotting (Figure 3B). Observation of the bright EGFP fluorescence further confirmed the significant production of EGFP in Ser–2A–EGFP silkworm MSGs specifically (Figure 3C).
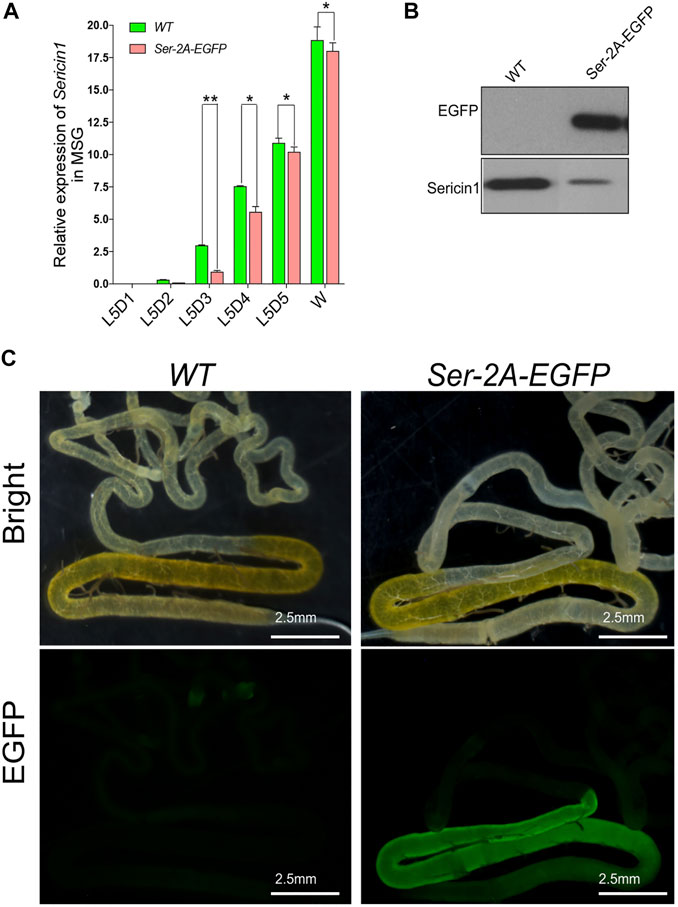
FIGURE 3. High production of enhanced green fluorescent protein (EGFP) protein by 2A mediated Sericin–EGFP in-fusion expression. (A) Relative transcriptional level of Sericin1 in WT and Ser–2A–EGFP silkworms. qRT-PCR was used to quantify the transcriptional level of Sericin1 and EGFP in (A) and (B). The silkworm ribosome protein 49 (Bmrp49) ortholog was used as the internal reference. Three individual replicates were used for qRT-PCR, and the error bars represent the mean ± S.E.M. (B) Western blotting of EGFP and Sericin1 protein in the middle silk gland (MSG) of wild-type (WT) and Ser–2A–EGFP silkworms at the wandering stage. (C) Fluorescence observation of the silk gland in the MSG of WT and Ser–2A–EGFP silkworms. Silk glands were extracted from the day 3 of the fifth instar larvae. Error bars stand for 2.5 mm.
In the Ser–2A–EGFP silkworms, we assumed EGFP protein colocalized with endogenous Ser1 in the surface of the middle silk gland, since they shared the same regulatory elements. In fact, EGFP proteins were exclusively detected in the cell layer of MSGs by using anti-EGFP primary antibody at day 3 of the final larval instar, which coated the inner fibers constructed majorly with fibroin proteins (Figure 4). Furthermore, a cytoplasmic distribution of EGFP was detected by staining the nucleus with Hoechst. These results confirmed the exogenous EGFP protein expressed in the MSG efficiently and specifically.
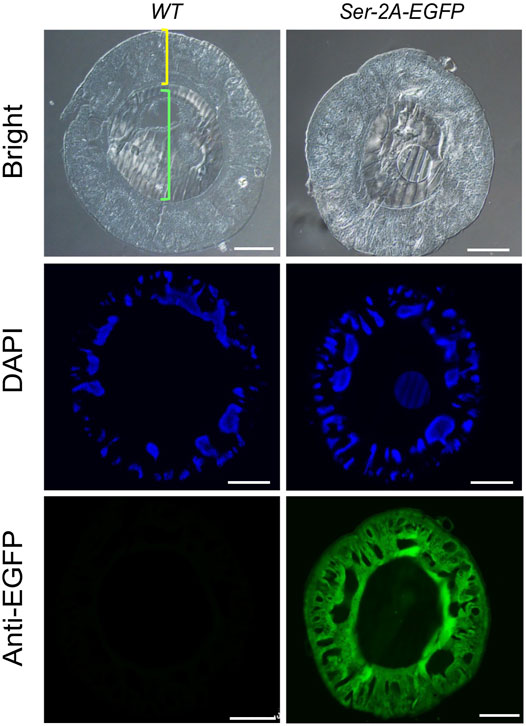
FIGURE 4. Immunohistochemistry of EGFP in the MSG of Ser–2A–EGFP animals. EGFP protein distributes in the sericin layer of the MSGs as indicated by the anti-EGFP primary antibody. The yellow bracket indicates the cellular layer and green bracket indicates the lumen of MSGs. MSGs were dissected from the WT or Ser–2A–EGFP animals at the third day of the fifth instar larvae (L5D3). Hoechst was used to stain the nucleus. Scale bars stand for 100 μm.
Being secreted together with Sericin1 protein, the EGFP protein was detected in the cocoon shells (Figure 5A). Nevertheless, the chimeric cocoon shells of Ser–2A–EGFP heterozygous animals were thinner and softer than the WT as we observed (Figure 5A). We measured the average weight of Ser–2A–EGFP silkworm cocoon shell, and found that it was decreased to 90.7% (0.107 ± 0.008 g) and 81.4% (0.092 ± 0.010 g) of the WT females and males, respectively (Table 2).
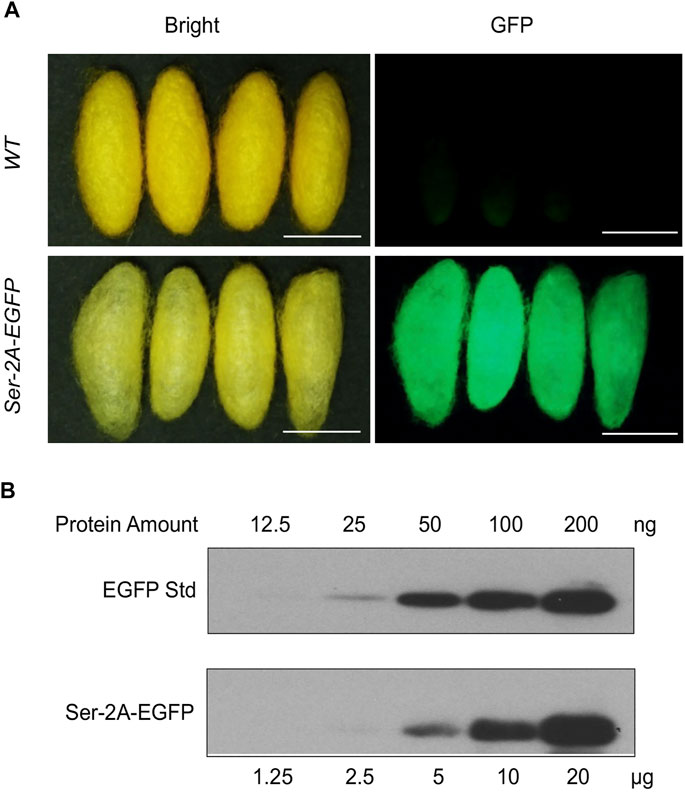
FIGURE 5. Quantification of EGFP protein in Ser–2A–EGFP cocoon shells. (A) Fluorescence observation of Ser–2A–EGFP cocoon shells. The upper row is the WT cocoon shells, and the lower row is the Ser–2A–EGFP cocoon shells. Both bright and GFP field are shown. Scale bars stand for 1 cm. (B) Quantification of the EGFP protein in the Ser–2A–EGFP cocoon shells. Proteins were diluted in a twofold series dilution manner and sent for analysis. The maximum amount for crude silk proteins and EGFP standard were 120 μg and 200 ng, respectively.
To quantify the production of EGFP protein in the cocoon shells of Ser–2A–EGFP animals, we extracted the crude proteins from the cocoon shells by using 8 M urea at 4°C overnight and performed Western blotting analysis. The experiment was conducted using both the extracted crude proteins and EGFP standard protein with a twofold series dilution (Figure 5B). In the heterozygous animals, the yield of EGFP protein reached to 1.05% (w/w) of the cocoon shell weight, which was higher than the existing transgenic-mediated production as reported previously (Figure 5B).
Since silkworm sericin protein is hydrophilic and does not cause allergic reactions, it has been widely used as a new medical material, including in wound healing (Aramwit et al., 2012). Here, we applied our sericin in-fusion expression system to produce the hEGF protein, the substrate of EGFR signaling which involves in re-epithelialization of epidermal wound healing and keratinocyte stem cell proliferation, in silkworm cocoons (Nanba et al., 2013). The hEGF gene was 183 bp and encoded a 7.2 KD short peptide. Here, we injected 640 WT eggs with the mixture of TALEN mRNA and donor plasmid, and four fluorescence-positive silkworm broods were obtained from a total of 65 G1 broods, the transformation efficiency reached 6.2% (Table 1). At the same time, transposon-based, transgenic silkworm expressing the hEGF driven by Ser1 promoter (Ser–T–hEGF) was also constructed as the control to compare protein yields with the Ser–2A–hEGF animals.
We first examined the expression level of Sericin1 gene, and no significant difference was observed when comparing both transformed lines to the WT silkworms (Figure 6A). However, the transcriptional level of hEGF was increased to 18-fold of that in Ser-T-hEGF animals at the W stage (Figure 6B), suggesting the native Ser1 regulatory elements or the local genomic DNA context were also important for target gene expression. Significant increase on the hEGF protein expression was detected in both the MSG and cocoon of Ser–2A–hEGF when compared with the Ser–T–hEGF animals, respectively (Figures 6C, D). Altogether, we successfully adapted the Ser1 in-fusion system to express the therapeutically important factor hEGF, which is promising for being used as a new biomedical substrate.
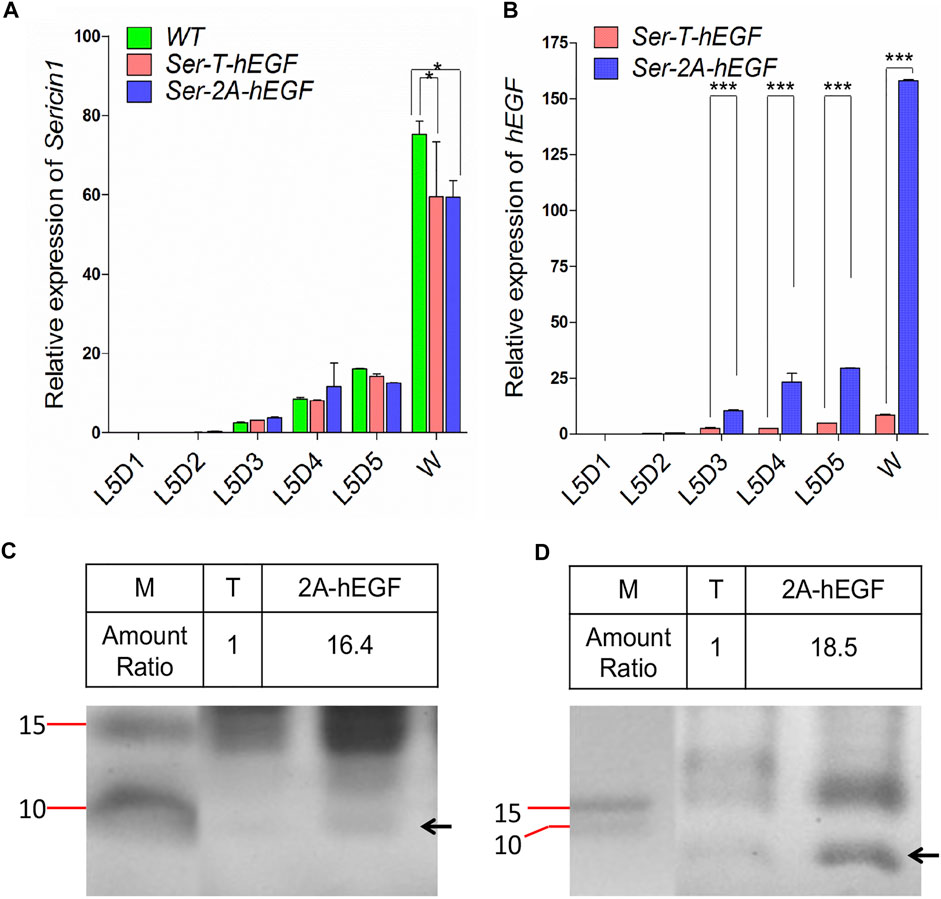
FIGURE 6. Expression of human epidermal growth factor (hEGF) protein in the Ser–2A–hEGF animals. (A) Comparison of hEGF transcriptional levels between Ser–T–hEGF and Ser–2A–hEGF animals. (B) Detection of Sericin1 in WT, Ser–T–hEGF, and Ser–2A–hEGF silkworms. qRT-PCR was used to quantify the transcriptional levels of Sericin1 and hEGF in A and B. The silkworm ribosome protein 49 (Bmrp49) ortholog was used as the internal reference gene. Three individual replicates were used for qRT-PCR, and the error bars represent the mean ± S.E.M. (C, D) Coomassie brilliant blue staining for the proteins extracted from the MSG (D) and cocoon (D) of Ser–T–hEGF and Ser–2A–hEGF silkworms. The black arrows indicate hEGF protein. M, protein marker; T, Ser–T–hEGF; 2A-hEGF, Ser–2A–hEGF. Size of protein marker bands are listed.
In the current study, we successfully established a Ser1 in-fusion expression system by using the TALEN-mediated, targeted gene integration in B. mori. This strategy was then used to express recombinant proteins specifically in the middle silk gland driven by the natural Ser1 regulatory elements.
The silkworm silk gland has been developed as an efficient bioreactor for a long period based on transposon-based transgenesis (Royer et al., 2005; Mori et al., 2014). Two systems, fibroin and sericin, have been developed for recombinant protein expression (Zhao et al., 2010; Wang et al., 2015; Xu et al., 2018). However, transposon-based transgenic production of the exogenous proteins was limited for several reasons. First, piggyBac-mediated transgene introduces the exogenous fragments into the undefined genome locus. Therefore, the genetic background and production of recombinant protein varied largely between different transgenic lines (Tomita et al., 2007; Wang et al., 2013). In addition, transposon-mediated transgene is instable and often causes gene drift. Besides, integrated fragments introduced by transgenesis are out of strict control and ubiquitously expressed, causing toxicity for the host and leading to high mortality (Tomita et al., 2007).
Here we performed a 2A-mediated Ser1 in-fusion expression by creating double-stranded breaks at the C-terminal of the Ser1 genomic loci, and integrated either EGFP or hEGF at the downstream seamlessly. High production of EGFP was detected in Ser–2A–EGFP animals in both the middle silk glands and cocoon shells. It was attributed to this system that uses the whole Ser1 promoter, which may optimize the promoter activity. Another benefit is that the in-fusion expression system did not disrupt the expression of the natural Ser1 gene and has less effect on that according to our results, loading few fitness costs on the host animals compared with the transgene strategy. However, we still observed that the overexpression of EGFP with this strategy produced a thinner cocoon shell (Figure 5A), which had the similar phenotype with the fibroin-deficient line Nd-sD, implying that the native Fibroin gene expression may be affected, which mechanism needs further exploration. In regard to the comparison between Ser–2A–EGFP and Ser–2A–hEGF lines, we also observed some degree of effect on the Ser1 expression with the smaller peptide (hEGF) integration (Figure 6A), we assumed that was caused by the toxicity by the high expression of hEGF in the MSG. It also should be noticed that the protein size-dependent effect on the fitness cost, since larger proteins being fused with sericin, the higher the possibility for deformed configuration on Sericin1 protein itself. Overall, this in-fusion expression strategy holds the great potential for recombinant protein expression in the silk gland.
In addition, the 2A-mediated in-fusion expression for hEGF increased to more than 15-fold than that in the transgenesis animals (Ser–T–hEGF, Figure 5A). In Ser–2A–hEGF animals, the completely native promoter and other upstream regulatory elements of Ser1 gene were subjected, rather than only the seed sequence of the promoter was used in the Ser–T–hEGF animals, which excludes the possibility that some potential enhancers existed in the upstream of the Sericin1 promoter. We also cloned a 404-bp 3′ UTR sequences of Ser1 and inserted it into the downstream of EGFP or hEGF, further mimicking the native Ser1 expression. Furthermore, we used the Thosea asigna virus-derived T2A self-cleavage peptides to achieve the in-fusion expression (Diao and White, 2012). T2A peptide forces the ribosome skip between the glycine and proline amino acids, without the peptide bond during translation, therefore the native Ser1 and linked EGFP or hEGF are transcribed together with Sericin1 but translated independently.
Actually, the Ser–2A system has wide usage on both genetics and biochemistry other than the application for single protein expression. One example is expressing tandem linkage of more than one copy of hEGF or other proteins in MSG, which may further increase the expression efficiency. In addition, 2A peptide can be used to link multiple protein-coding sequences tandemly and to be controlled by a single promoter, simplifying the construction aim to expression multiple factors. In addition to in-fusion with sericin, the exogenous proteins or sequences can be inserted into any site desired including the exon, intron, and even the untranslated regions, reducing the side effect on the targeted gene itself, and it also can be used for protein tagging or manipulation. In conclusion, the current work established a natural Ser1 expression system, providing us a new genetic strategy for the mass production of exogenous proteins and further promote the silk gland to be an excellent bioreactor system.
The original contributions presented in the study are included in the article/Supplementary Material, Further inquiries can be directed to the corresponding author.
ZL and AT designed the research and wrote the manuscript. ZL, LY, QZ, and YY performed the experiments and analyzed the data. AT revised the manuscript. All authors have approved the manuscript for publication.
This work was supported by the National Science Foundation of China (31925007 and U1738110).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aramwit, P., Siritientong, T., and Srichana, T. (2012). Potential Applications of Silk Sericin, a Natural Protein from Textile Industry By-Products. Waste Manag. Res. 30, 217–224. doi:10.1177/0734242x11404733
Diao, F., and White, B. H. (2012). A Novel Approach for Directing Transgene Expression in Drosophila: T2A-Gal4 In-Frame Fusion. Genetics 190, 1139–1144. doi:10.1534/genetics.111.136291
Dong, Z.-Q., Chen, T.-T., Zhang, J., Hu, N., Cao, M.-Y., Dong, F.-F., et al. (2016). Establishment of a Highly Efficient Virus-Inducible CRISPR/Cas9 System in Insect Cells. Antiviral Res. 130, 50–57. doi:10.1016/j.antiviral.2016.03.009
Edgell, D. R. (2009). Selfish DNA: Homing Endonucleases Find a home. Curr. Biol. 19, R115–R117. doi:10.1016/j.cub.2008.12.019
Iizuka, M., Tomita, M., Shimizu, K., Kikuchi, Y., and Yoshizato, K. (2008). Translational Enhancement of Recombinant Protein Synthesis in Transgenic Silkworms by a 5′-Untranslated Region of Polyhedrin Gene of Bombyx M Nucleopolyhedrovirus. J. Biosci. Bioeng. 105, 595–603. doi:10.1263/jbb.105.595
Inoue, S., Kanda, T., Imamura, M., Quan, G., Kojima, K., Tanaka, H., et al. (2005). A Fibroin Secretion-Deficient Silkworm Mutant, Provides an Efficient System for Producing Recombinant Proteins. Insect Biochem. Mol. Biol. 35, 51–59. doi:10.1016/j.ibmb.2004.10.002
Li, Z., You, L., Yan, D., James, A. A., Huang, Y., and Tan, A. (2018). Bombyx Mori Histone Methyltransferase BmAsh2 Is Essential for Silkworm piRNA-Mediated Sex Determination. Plos Genet. 14, e1007245. doi:10.1371/journal.pgen.1007245
Ma, S., Shi, R., Wang, X., Liu, Y., Chang, J., Gao, J., et al. (2014). Genome Editing of BmFib-H Gene Provides an Empty Bombyx Mori Silk Gland for a Highly Efficient Bioreactor. Sci. Rep. 4, 1–7. doi:10.1038/srep06867
Maeder, M. L., Thibodeau-Beganny, S., Osiak, A., Wright, D. A., Anthony, R. M., Eichtinger, M., et al. (2008). Rapid "Open-Source" Engineering of Customized Zinc-Finger Nucleases for Highly Efficient Gene Modification. Mol. Cel 31, 294–301. doi:10.1016/j.molcel.2008.06.016
Tatematsu, K.-I., Uchino, K., Sezutsu, H., and Tamura, T. (2014). Effect of ATG Initiation Codon Context Motifs on the Efficiency of Translation of mRNA Derived from Exogenous Genes in the Transgenic Silkworm, Bombyx mori. SpringerPlus 3, 1–12. doi:10.1186/2193-1801-3-136
Mulepati, S., Héroux, A., and Bailey, S. (2014). Crystal Structure of a CRISPR RNA-Guided Surveillance Complex Bound to a ssDNA Target. Science 345, 1479–1484. doi:10.1126/science.1256996
Nanba, D., Toki, F., Barrandon, Y., and Higashiyama, S. (2013). Recent Advances in the Epidermal Growth Factor Receptor/ligand System Biology on Skin Homeostasis and Keratinocyte Stem Cell Regulation. J. Dermatol. Sci. 72, 81–86. doi:10.1016/j.jdermsci.2013.05.009
Royer, C., Jalabert, A., Rocha, M. D., Grenier, A.-M., Mauchamp, B., Couble, P., et al. (2005). Biosynthesis and Cocoon-Export of a Recombinant Globular Protein in Transgenic Silkworms. Transgenic Res. 14, 463–472. doi:10.1007/s11248-005-4351-4
Takasu, Y., Yamada, H., Tamura, T., Sezutsu, H., Mita, K., and Tsubouchi, K. (2007). Identification and Characterization of a Novel Sericin Gene Expressed in the Anterior Middle Silk Gland of the Silkworm Bombyx Mori. Insect Biochem. Mol. Biol. 37, 1234–1240. doi:10.1016/j.ibmb.2007.07.009
Takasu, Y., Kobayashi, I., Tamura, T., Uchino, K., Sezutsu, H., and Zurovec, M. (2016). Precise Genome Editing in the Silkworm Bombyx M Using TALENs and Ds- and ssDNA Donors - A Practical Approach. Insect Biochem. Mol. Biol. 78, 29–38. doi:10.1016/j.ibmb.2016.08.006
Tan, A., Fu, G., Jin, L., Guo, Q., Li, Z., Niu, B., et al. (2013). Transgene-Based, Female-Specific Lethality System for Genetic Sexing of the Silkworm, Bombyx Mori. Proc. Natl. Acad. Sci. 110, 6766–6770. doi:10.1073/pnas.1221700110
Tomita, M., Munetsuna, H., Sato, T., Adachi, T., Hino, R., Hayashi, M., et al. (2003). Transgenic Silkworms Produce Recombinant Human Type III Procollagen in Cocoons. Nat. Biotechnol. 21, 52–56. doi:10.1038/nbt771
Tomita, M., Hino, R., Ogawa, S., Iizuka, M., Adachi, T., Shimizu, K., et al. (2007). A Germline Transgenic Silkworm that Secretes Recombinant Proteins in the Sericin Layer of Cocoon. Transgenic Res. 16, 449–465. doi:10.1007/s11248-007-9087-x
Wang, Y., Li, Z., Xu, J., Zeng, B., Ling, L., You, L., et al. (2013). The CRISPR/Cas System Mediates Efficient Genome Engineering in Bombyx Mori. Cell Res 23, 1414–1416. doi:10.1038/cr.2013.146
Wang, F., Wang, R., Wang, Y., Zhao, P., and Xia, Q. (2015). Large-Scale Production of Bioactive Recombinant Human Acidic Fibroblast Growth Factor in Transgenic Silkworm Cocoons. Sci. Rep. 5, 16323. doi:10.1038/srep16323
Xu, H., Yuan, L., Wang, F., Wang, Y., Wang, R., Song, C., et al. (2014). Overexpression of Recombinant Infectious Bursal Disease Virus (IBDV) Capsid Protein VP2 in the Middle Silk Gland of Transgenic Silkworm. Transgenic Res. 23, 809–816. doi:10.1007/s11248-014-9827-7
Xu, J., Dong, Q., Yu, Y., Niu, B., Ji, D., Li, M., et al. (2018). Mass Spider Silk Production through Targeted Gene Replacement in Bombyx Mori. Proc. Natl. Acad. Sci. USA 115, 8757–8762. doi:10.1073/pnas.1806805115
Zhang, F., Wen, Y., and Guo, X. (2014). CRISPR/Cas9 for Genome Editing: Progress, Implications and Challenges. Hum. Mol. Genet. 23, R40–R46. doi:10.1093/hmg/ddu125
Zhang, Z., Niu, B., Ji, D., Li, M., Li, K., James, A. A., et al. (2018). Silkworm Genetic Sexing through W Chromosome-Linked, Targeted Gene Integration. Proc. Natl. Acad. Sci. USA 115, 8752–8756. doi:10.1073/pnas.1810945115
Keywords: transcription activator-like effector nuclease (TALEN), silk gland, bioreactor, sericin, Bombyx mori
Citation: Li Z, You L, Zhang Q, Yu Y and Tan A (2022) A Targeted In-Fusion Expression System for Recombinant Protein Production in Bombyx mori. Front. Genet. 12:816075. doi: 10.3389/fgene.2021.816075
Received: 16 November 2021; Accepted: 30 November 2021;
Published: 04 January 2022.
Edited by:
Wei Zhang, Peking University, ChinaReviewed by:
Sanyuan Ma, Southwest University, ChinaCopyright © 2022 Li, You, Zhang, Yu and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anjiang Tan, YXRhbkBqdXN0LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.