- 1Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China, Ministry of Education, International Ecological Foresty Research Center of Kunming, Horticulture and Landscape Architecture, Southwest Forestry University, Kunming, Yunnan, China
- 2Yunnan Institute of Tropical Crops, Xishuangbanna, Yunnan, China
- 3Key Laboratory of Hainan Province for Postharvest Physiology and Technology of Tropical Horticultural Products, South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, China
- 4Office of Academic Affairs, Yunnan University of Finance and Economics, Kunming, China
- 5French Associates Institute for Agriculture and Biotechnology of Dryland, The Jacob Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Be’er Sheva, Israel
- 6State Key Laboratory of Component-based Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 7Institute of Tropical and Subtropical Cash Crops, Yunnan Academy of Agricultural Sciences, Baoshan, China
Macadamia ternifolia is a dynamic oil-producing nut crop in the world. However, the nutshell is frequently considered as a low-quality material. Further, its metabolic profile is still uncharacterized. In order to explore the industrial significance of the nutshell, this study performed metabolic and transcriptomic analyses at various developmental stages of the nutshell. The qualitative and quantitative metabolic data analysis identified 596 metabolic substances including several species of phenolic acids, flavonoids, lipids, organic acids, amino acids and derivatives, nucleotides and derivatives, alkaloids, lignans, coumarins, terpenoids, tannins, and others. However, phenolic acids and flavonoids were predominant, and their abundance levels were significantly altered across various developmental stages of the nutshell. Comparative transcriptome analysis revealed that the expression patterns of phenolic acid and flavonoid pathway related genes were significantly changed during the nutshell growth. In particular, the expression of phenylalanine ammonia-lyase, C4H, 4CL, CHS, CHI, F3H, and FLS had dynamic differences at the various developmental stages of the nutshell. Our integrative metabolomic and transcriptomic analyses identified the key metabolic substances and their abundance levels. We further discussed the regulatory mechanism of phenolic and flavonoid biosynthesis in the nutshell of M. ternifolia. Our results provide new insights into the biological profiles of the nutshell of M. ternifolia and help to elucidate the molecular mechanisms of phenolic and flavonoid biosynthesis in the nutshell of M. ternifolia.
Introduction
Macadamia nuts belong to the Proteaceae family, native to Australia, and are considered one of the highest oil quality nuts in the world (Mast et al., 2008; Hu et al., 2019). It is commercially cultivated in China, Hawaii, South Africa, New Zealand, Kenya, and several South American countries (Rengel et al., 2015; Navarro and Rodrigues, 2016). There are nine species in the Macadamia genus but only four of them have a wider public acceptance. In particular, Macadamia integrifolia, and M. tetraphylla had a large scale commercial production than M. jansenii and M. ternifolia (Howlett et al., 2015). The fruit of Macadamia contains a delicate and delicious cream to white colored kernel which covers in overall a thick dark green colored nutshell (Mast et al., 2008). The large part of the fruit is nutshell that is commonly wasted, and sometime used as low-quality raw material (O’Hare et al., 2019). The edible Macadamia nut contains essential nutrients, vitamins, minerals, and monounsaturated fatty acids (Malvestiti et al., 2017). Oleic and palmitoleic acids are predominately present in higher amounts in Macadamia nuts oil compared to other nut crops (Kaijser et al., 2000; Hu et al., 2019). Because of the high nutritive characteristics, Macadamia products are thought to be effective in medical field against cardiovascular diseases like myocardial infarction, cancer, arteriosclerosis, and dyslipidemia (Garg et al., 2003; Wang and Hu, 2017). In addition, Macadamia is also used in cosmetic industry to produce various skincare and anti-aging creams (Rafia, 2013). Macadamia is largely cultivated in Australia and approximately 35,000 tons of the shell is produced alone in Australia (Fan et al., 2018). For the environmental protection, there is a need to change the high amount of macadamia nutshell into useful by products to mitigate industrial based waste management problems.
The nutshell is very important in dry fruits. It synthesizes and transports essential nutrients to the kernel during stressful growth periods. Moreover, it not only regulate the fruit shape and size but also prevents mechanical and chemical injuries (Francoz et al., 2018). The Macadamia nutshell has an exceptional cell wall thickness which is considered as an isotropic wood by some researchers (Wang and Mai, 1994). Some researches described that two or three different layers with lignified cells are present in the nutshell which increases the hardness of the wall (Hartung and Storey, 1939). Macadamia nutshell is widely used as a direct burning material. It is utilized as a compost to prevent weed growth and moisture loss (Conesa et al., 2000; Girdis et al., 2017). In addition, Macadamia nutshell is used to produce biomass pyrolysis through controlled high-temperature serial reactions (Poinern et al., 2011). Several pyrolysis-based biochar applications include steel production (Kumar et al., 2017), soil amendments (Wrobel-Tobiszewska et al., 2018), and de-nitrification mechanism (Bakly et al., 2019). Similarly, the nutshells of almond, hazelnut, walnut, and apricot are a cost-effective source of activated carbons for treatment of industrial effluents (Aygün et al., 2003). Interestingly, extracts from Macadamia nutshell have been widely utilized to develop fragrance, hair, and skincare products in recent years (Syed and Walele, 2006). However, research in Macadamia has been largely focused on the physical, biological, and health properties of the kernel oil (Akhtar et al., 2006; Garg et al., 2007). Limited information is available about the chemical composition of the active metabolic compounds of the nutshell. There is a need of increased knowledge to synthesize high-quality by-products from Macadamia nutshell.
In recent years, multi-omic analyses such as the integration of transcriptomic and metabolomic data have gained much importance to investigate the regulatory mechanism of metabolite substances in plants (Chen et al., 2014; Shen et al., 2021). In this regard, mass spectrometry has been broadly applied to analyze the composition and concentration of metabolic substances (Matsuda et al., 2009; Fang et al., 2019). Whereas, transcriptome sequencing provides effective means to unveil gene to metabolite networks in oilseed crops (Zhao et al., 2018; Wu et al., 2020). Previous high-throughput research already characterized the biological mechanism of different metabolites such as fatty acids, sugars, amino acids, phenolic acids, and flavonoids biosynthesis in various oilseed crops (Bourgis et al., 2011; Alagna et al., 2012; Zheng et al., 2017; Rehman et al., 2018; Zhang et al., 2019). The key genes that regulate the biosynthesis and degradation of key metabolites have been uncharacterized in the nutshell of M. ternifolia. To accomplish this research gap, our research group performed a comprehensive metabolome and transcriptome analyses at various developmental stages of the nutshell. Our study identified the metabolic substances, characterized their abundance, and established biological profiles of key metabolites in M. ternifolia nutshell. These results will be important to improve the existing knowledge allied with qualitative and quantitative metabolic substances of the nutshell. Furthermore, it provides an important theoretical reference for future research which aims to investigate the molecular mechanism of polyphenol biosynthesis and a better valorization of the nutshell of M. ternifolia.
Results
Metabolite Composition and Concentration in M. Ternifolia Nutshell
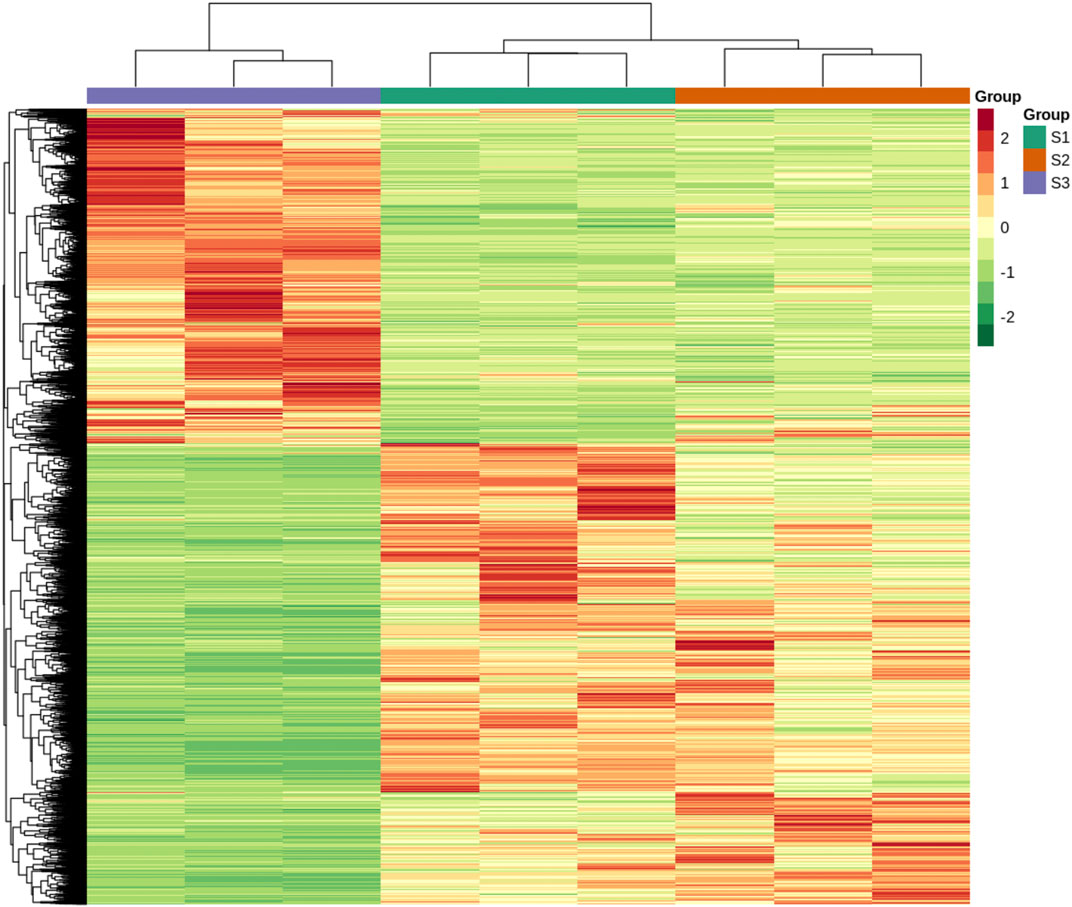
To explore the metabolic profiles of M. ternifolia nutshell, the nutshell samples were selected at the stages of young fruit (S1), medium fruit (S2), and mature fruit (S3). A total of 596 metabolites were identified which were categorized into 12 different classes. The major metabolites were phenolic acids, flavonoids, and lipids followed by other classes. For example, there were 114 phenolic acids, 109 flavonoids, 89 lipids, 72 others, 57 organic acids, 51 amino acids and derivatives, 37 nucleotides and derivatives, 24 alkaloids, 20 lignans, and coumarins, 16 terpenoids, 6 tannins, and 1 quinone (Figure 1A). All metabolites had distinct abundance at various development stages of a nutshell (Figure 1B). Specifically, many metabolites exhibited a higher abundance at the mature fruit stage as compared to early stages. The metabolites associated with different types of benzoic acid, coumaroylquinic acid, and chlorogenic acid were key phenolic acids (Supplementary Table S1). Among flavonoids, the different forms of methoxyflavone, glucuronideflavone, glucosideflavone, and rutinosideflavone had most abundance. The metabolites in the lipid class were mainly decenoic acid, palmitic acid, linolenic acid, and linolenoyl glycerol. Cluster analysis arranged the 275 metabolites into six major classes. Each class was based on the metabolic abundance during the different stages of nutshell development (Figure 2A). Classes 1 and 3 contained 45 and 9 metabolites, respectively, with a higher abundance at the S2 stage of the nutshell. Class 5 had 51 metabolites that showed the highest abundance at the S1 stage, whereas classes 2, 4, and 6 contained 45, 56, and 57 metabolites, respectively. All metabolites of these classes exhibited the highest abundance during the S3 stage. These results indicated that phenolic acids and flavonoids are predominant metabolites in the nutshell. In addition, metabolite composition and concentration changed during the developmental stages of nutshell in M. ternifolia.
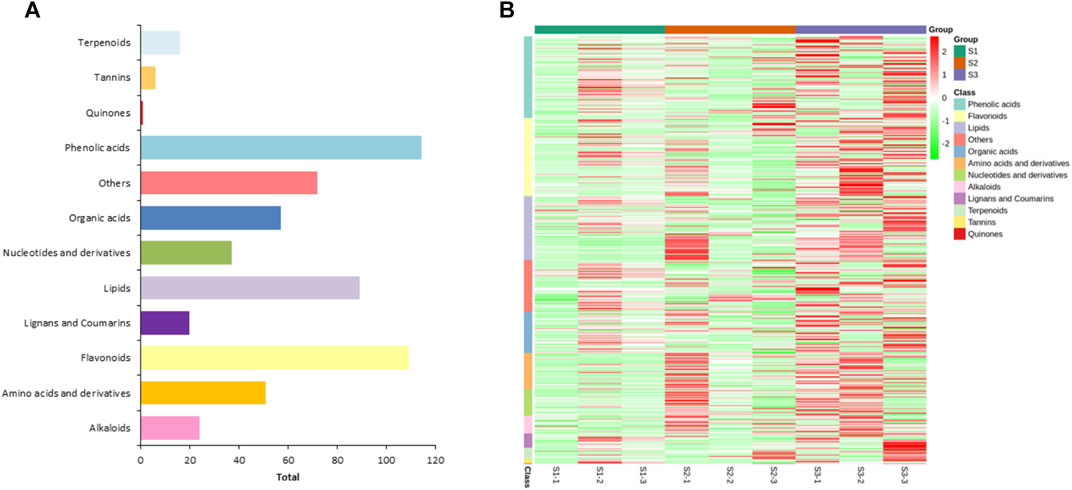
FIGURE 1. Classification and abundance of the metabolites at three developmental stages of M. ternifolia nutshell. (A) The classification results of different metabolites (B) The abundance level of different metabolites.
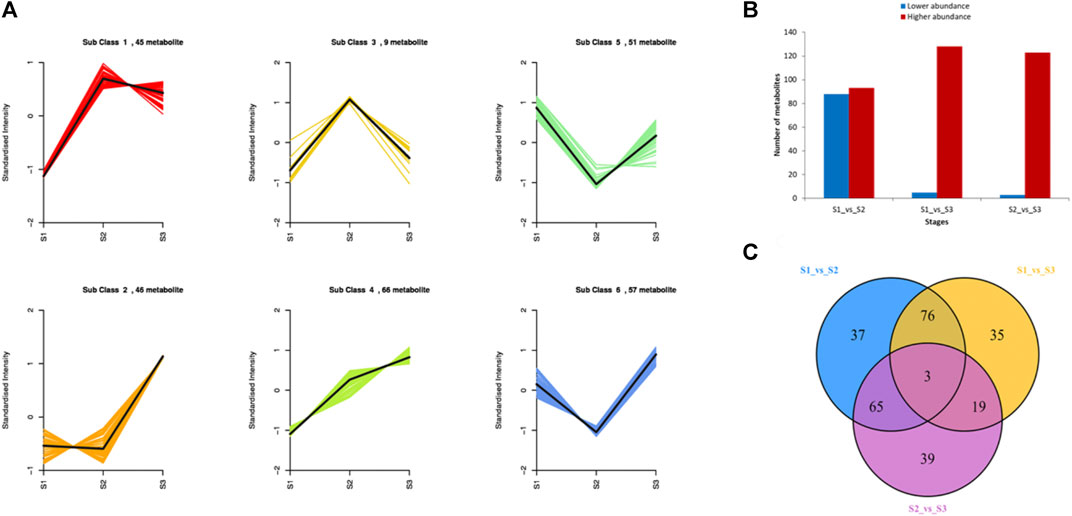
FIGURE 2. The K-means clustering and statistics of differential metabolites at three developmental stages of M. ternifolia nutshell (A) The clustering results of metabolites (B) Total number of up regulated and down regulated metabolites (C) The distribution of differential metabolites at various development stages.
Differential Metabolites Analysis in M. Ternifolia Nutshell
The abundance of each metabolite was compared among growth stages to identify differential metabolites. There were 181 differentially accumulated metabolites in the comparison of S1 and S2, 88 of which showed a lower abundance and 93 showed a higher abundance at S1 than S2 (Figure 2B). In the comparison of S1 and S3, 133 metabolites showed a significantly different accumulation. Of which, only 5 metabolites showed a decreased abundance and 128 metabolites showed a higher abundance at S1 as compared to S3. A total of 126 metabolites had a significant difference between the comparison S2 and S3. Relative to S3, only 3 metabolites showed lower abundance and 123 metabolites showed a higher abundance at the S2 stage. Venn diagram revealed that a major portion of the metabolites was overlapped among the different stages, whereas few metabolites were stage-specific (Figure 2C). For example, only 37, 35, and 39 metabolites were specific to S1vsS2, S1vsS3, and S2vsS3, respectively. As expected, metabolites related to phenolic acids, flavonoids, lipids, and amino acid derivatives had dynamic variations among the different stages. The metabolites belonging to these classes exhibited significantly higher abundance at the late growth stage than early growth stages (Supplementary Figure S1). Overall, M. ternifolia displays significant metabolic differences at the different stages of nutshell growth. In particular, different species of phenolic acids and flavonoids were found with dynamic changes.
Identification of Key Metabolites in M. Ternifolia Nutshell
The results of metabolic composition and profiling revealed many metabolites with differential abundance in nutshell. To identify putative key metabolites which are critical components of the nutshell, we further selected those metabolites which showed significant abundance throughout various development stages of the nutshell. In this way, a total of 67 highly differential abundant metabolites were identified (Figure 3). A total of 29 metabolites belong to phenolic acids and 38 to flavonoids. It was observed that vanillic acid 4-O-glucoside, 3-O-p-coumaroylquinic acid, syringaldehyde 1-O-gentisoyl-D-glucoside, 3,4,5 trimethoxyphenyl-1-O-glucoside, chlorogenic acid (3-O-caffeoylquinic acid), cryptochlorogenic acid, vnilloylmalic acid, and protocatechuic acid-4-O-glucoside were major the phenolic components of the nutshell (Figure 3A). However, vanillic acid 4-O-glucoside, chlorogenic acid (3-O-caffeoylquinic acid), cryptochlorogenic acid, 3-O-p-coumaroylquinic acid, and syringaldehyde were detected in a very high abundance. While benzyl-β-gentiobioside, sinapoyl malate, and grevirobstol-B had the lowest abundance among all identified phenolic acids. Quercetin-linked glucoside related flavonoids were the major components of the nutshell, which exhibited dynamic differences among different developmental stages (Figure 3B). However, catechin, epicatechin, mearnsetin-3-O-glucoside, mearnsetin-3-O-glucuronide, and quercetin-3-O-galactoside showed the highest abundance among all identified flavonoids. In contrast, ayanin, eupatorin (5,3-dihydroxy-6,7,4-trimethoxyflavone), 5,4 dihydroxy-3,6,7,3-tetramethoxyflavone, and 6,7,8-tetrahydroxy-5-methoxyflavone had lower abundances at all stages of nutshell growth in this study. These results suggest that different classes of phenolic acids and flavonoids linked with glucosides are the primary components of nutshell in M. ternifolia.
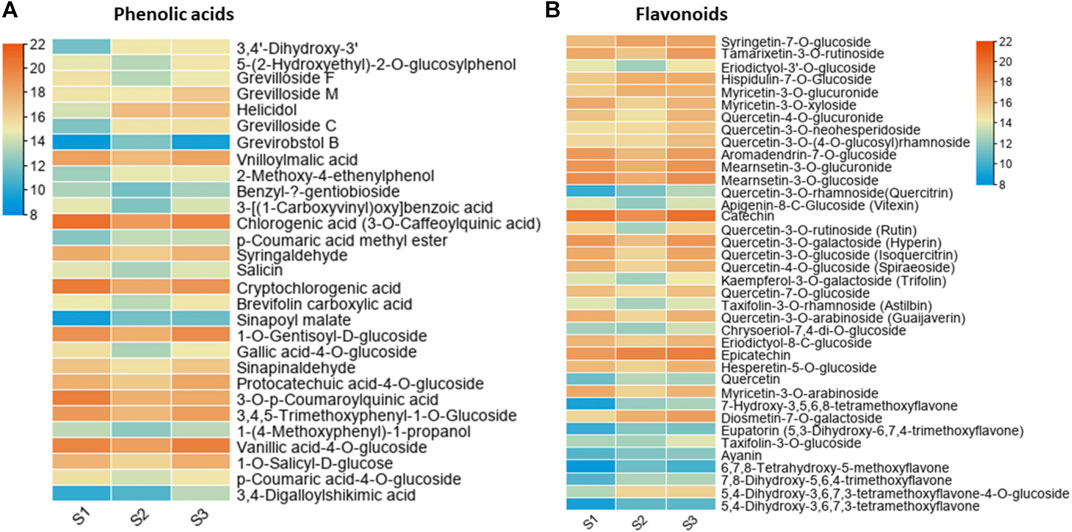
FIGURE 3. Heatmaps of key metabolite substances at three developmental stages of M. ternifolia nutshell (A) Phenolic acids (B) Flavonoids.
Ribonucleic Acid Sequencing and Unigenes Functional Annotations
The nutshell samples including S1, S2, and S3 were RNA-sequenced to explore the gene expression changes associated with regulation of different metabolites in M. ternifolia nutshell. According to the results of the RNA-sequencing, on average 45,282,444, 53,082,622, and 50,630,658 raw reads were acquired for S1, S2, and S3, respectively (Table 1). More than 7 GB of clean data were generated after removing low-quality reads for each sample. The Q20 score was greater than 98% and Q30 was 94% in our study, whereas the guanine-cytosine (GC) content was almost 45% for all samples. A total of 20,8119 unigenes were assembled with Trinity software (Trinity release v2.4.0, United States) from clean reads with a mean length of 1,256, N50 length of 1,883 bp, and N90 length of 569 bp (Supplementary Table S2). These unigenes had a length distribution range from 200 to >2,000 bp. A total of 16,565 unigenes had a length range from 200 to 300 bp, 21,579 unigenes had a length range from 300 to 400 bp. However, most unigenes had a length range >2,000 bp (Supplementary Figure S2A). The blast results revealed that the majority of unigenes had functional annotation within NR and Trembl databases followed by GO, Pfam, KEGG, SwissProt, and KOG databases (Supplementary Figure S2B).
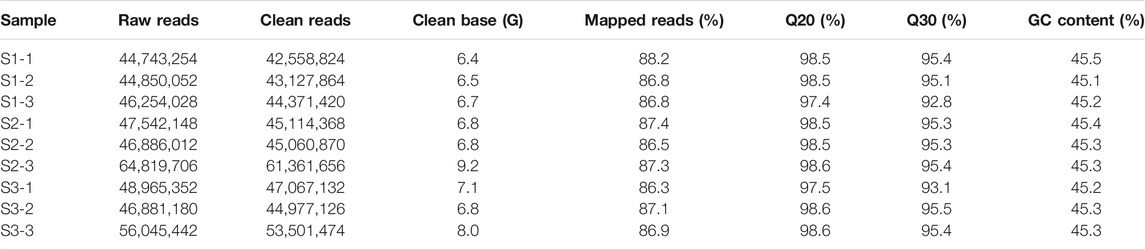
TABLE 1. Summary of transcriptome analysis for three developmental stages of M. ternifolia nutshell.
Differential Gene Expression and Functional Enrichment Analysis
The expression level of each unigene was measured in the form of FPKM. There was a strong correlation among the biological replicates. In contrast, S3 samples had a weak correlation with different repeats of S1 and S2 stages (Figure 4A). The correlation coefficient among different repeats was almost greater than 0.95 for each stage, while S1 and S2 repeats had a correlation coefficient of 0.25 with the S3 stage. The gene expression-based clustering analysis showed distinct gene expression profiles across S1, S2, and S3. Cluster analysis divided the expression profiles of unigenes into six major classes (Figure 4B). Classes 1 and 2 contained 6,959 and 2,143 unigenes, respectively, characterized by a high expression at the S3 stage. Classes 3, 4, 5, and 6 contained 2,845, 1,824, 4,420, and 3,600 unigenes, respectively. All unigenes of these classes exhibited a lower expression during the S3 stage than the other two stages of nutshell growth. In addition, unigenes belonging to classes 3 and 5 exhibited a higher expression at the S1 stage compared to others, whereas classes 4 and 6 contained unigenes with a higher expression at the S2 stage.
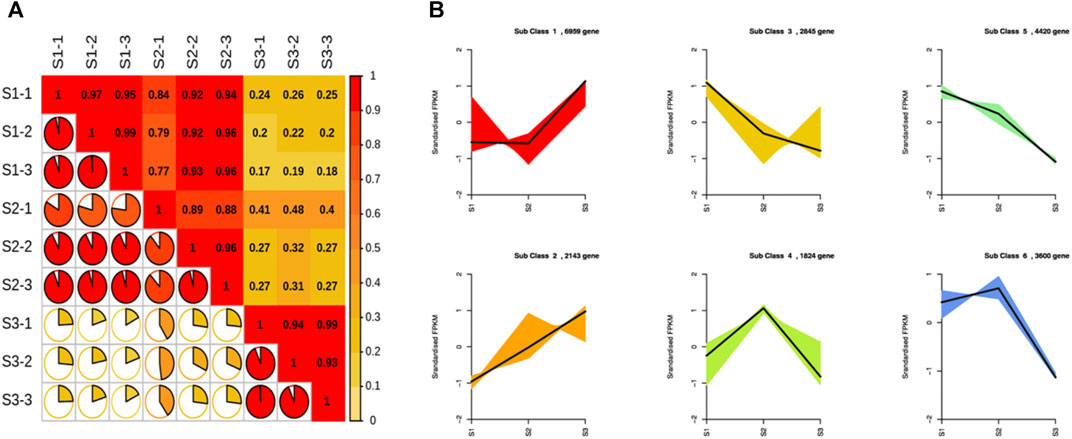
FIGURE 4. Correlation analysis and K-means clustering of unigenes at three developmental stages of M. ternifolia nutshell (A) Correlation analysis (B) K-means clustering of unigene.
To investigate the gene expression differences between the three developmental stages, we made pairwise comparison of the transcript profiles in S1, S2, and S3. The DEGs were considered significant if they had log2 Fold Change ≥1 or ≤ -1 and the false discovery rate <0.05. Comparative analysis showed a total number of 1,810 differentially expressed genes (DEGs) in S1vsS2 (Figure 5A). In S1vsS3, there were 18,214 DEGs in total, which stated much higher expression differences among these stages. Comparative analysis in S2vsS3 revealed a total of 14,831 DEGs. The Venn diagram distribution revealed that a total of 21,791 DEGs existed across the three development stages (Figure 5B). There were only 673 overlapped DEGs between the three stages. However, the comparison of S1vsS3 and S2vsS3 had 10,935 overlapped DEGs. All DEGs expression heatmap stated an undulating expression trend at different developmental stages. But, most DEGs were found with higher expressions level at S1 and S3 (Figure 6). Functional characterization showed that metabolic and biosynthesis of secondary metabolites-related pathways had more significance in comparisons of S1vsS2, S1vsS3, and S2vsS3 (Supplementary Figure S3). However, S1vsS3 and S2vsS3 had almost more than 2,000 metabolic DEGs, whereas the metabolic DEGs between S1 and S2 were only 300. Theoretically, we deduce that a large number of metabolic DEGs with undulating expression profiles are associated with the differences in metabolite abundances/composition observed at the three developmental stages of nutshell in M. ternifolia.
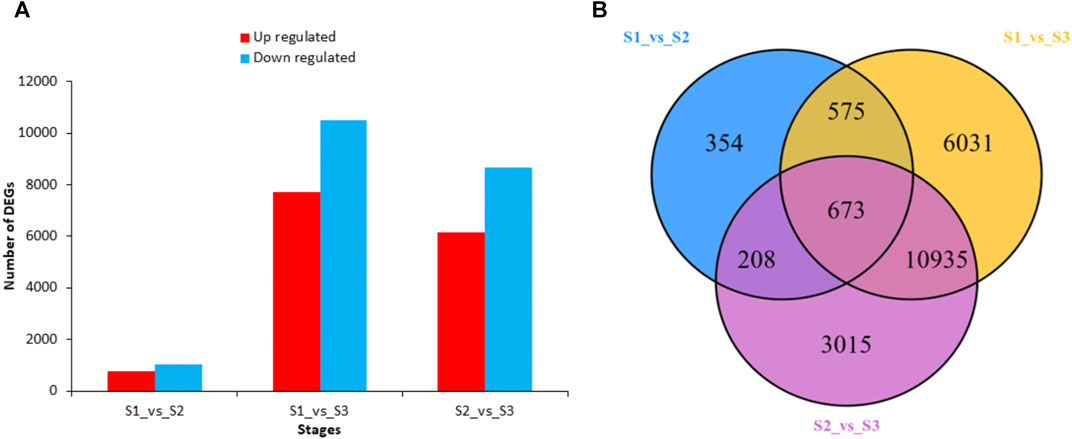
FIGURE 5. The total number of up-regulated and down-regulated DEGs and their distribution at three developmental stages of M. ternifolia nutshell (A) Total number of up-regulated and down regulated DEGs (B) The distribution of DEGs among the developmental stages.
Identification of Key Genes Involved in Phenolic Acids and Flavonoids Related Metabolism
Our metabolomics results showed that phenolic acid is the major component of nutshell in M. ternifolia. Therefore, genes with significant expression differences across stages and involved in the regulation of phenolic acids metabolism were excavated. The phenylalanine is essential to produce phenolics and flavonoids metabolites via different enzymes including phenylalanine ammonia-lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumaroyl coenzyme A ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone -3-hydroxylase (F3H), and flavonol synthase (FLS) (Figure 7A). In our study, 73 genes annotated with phenolic and flavonoids metabolites exhibited significant expression differences at different developmental stages of the nutshell (Figures 7B,C). Specifically, the expression of 14 PAL unigenes (unigene 13,908-PAL1, unigene 45269-PAL, unigene 47,506-PAL2, unigene 57,087-PAL2, unigene 66,217-PAL1, unigene 74899-PAL, unigene 79549-PAL, unigene 82143-PAL, unigene 84,042-PAL2, unigene 84,043-PAL2, unigene 84362-PAL, unigene 84,515-PAL1, unigene 86387-PAL, and unigene 92394-PAL) had a significant lower expression at S2 and S3 (Figure 7B). Similarly, unigene 71,603-C4H and unigene 71,608-C4H showed a lower expression at late stages of nutshell growth. A total of 15 unigenes were identified to encode 4CL, including three for 4CL1, three for 4CL2, five for 4CL5, three for 4CL9, and one for 4CL16. The expression profiles of all these 4CL genes were peaked during the early stages (Figure 7B).
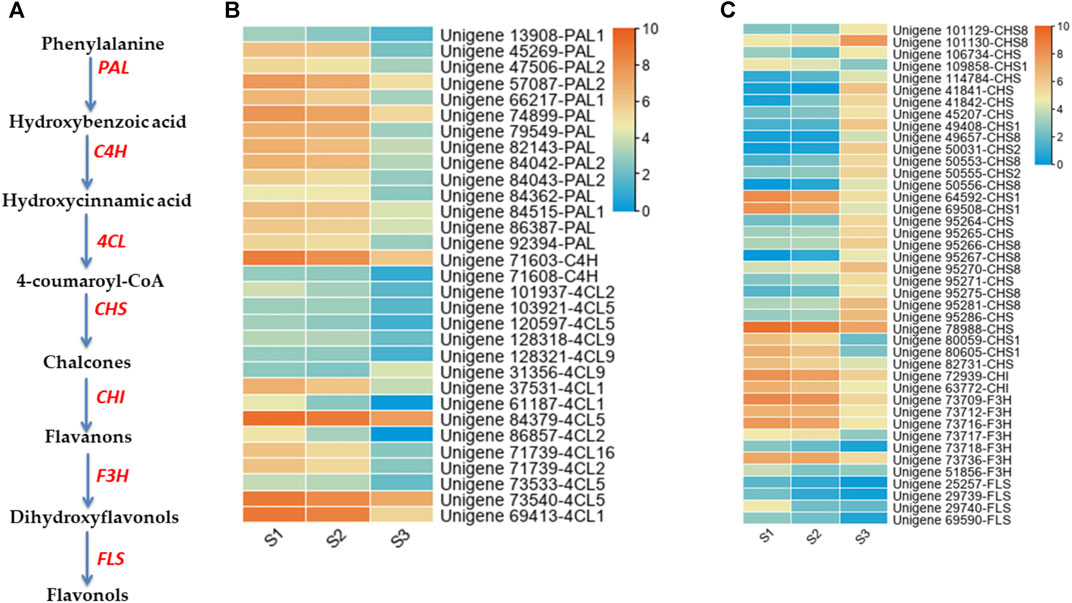
FIGURE 7. Expression profiles of phenolic acid and flavonoid pathway genes distribution at three developmental stages of M. ternifolia nutshell (A) Simplest diagram of phenolic acid and flavonoid biosynthesis pathway (B) Phenolic acid metabolism related DEGs (C) Flavonoids pathway related DEGs.
Interestingly, there were 11 CHS unigenes with significant differential expression patterns across the three stages of nutshell growth. In particular, unigene 95286-CHS had a strong expression at the late stage (Figure 7C). In contrast, unigene 78988-CHS showed much high expression during early stages. Among other identified CHS encoding unigenes, there were 6 for CHS1, two for CHS2, and ten for CHS8. The expression trends of two genes denoted as unigene 72939-CHI and unigene 63772-CHI exhibited up regulated expression at the early stage compared to the late stage. The F3H and FLS enzymes encoding genes are vital to synthesize different kinds of flavonoids. The expression of seven F3H genes including unigene 73,709-F3H, unigene 73,712-F3H, unigene 73,716-F3H, unigene 73,717-F3H, unigene 73,718-F3H, unigene 73,736-F3H, and unigene 51,856-F3H showed peak expression at S1. Similarly, four FLS enzymes encoding genes such as unigene 25257-FLS, unigene 29739-FLS, unigene 29740-FLS, and unigene 69590-FLS had remarkably low expression levels at late stages of nutshell growth (Figure 7C). The dynamic expression changes in phenolic acid and flavonoid pathway genes indicate their critical role in the regulation of metabolites in the nutshell of M. ternifolia.
qRT-Polymerase Chain Reaction Validation of Key Genes Related to Polyphenol Compounds
Based on the metabolic and transcriptome analysis, 11 genes (unigene 57,087-PAL2, unigene 74899-PAL, unigene 71,603-C4H, unigene 71,608-C4H, unigene 84,379-4CL5, unigene 73,540-4CL5, unigene 64,592-CHS1, unigene 78988-CHS, unigene 73,709-F3H, unigene 29740-FLS, and unigene 69590-FLS) involved in polyphenol compounds metabolism were selected to perform a qRT-PCR analysis. All genes showed a dynamic expression difference at the three developmental stages of the nutshell (Figure 8). These results speculated that these genes most likely play important role in metabolites accumulation in the nutshell of M. ternifolia. Furthermore, these findings validate our transcriptome data analysis.
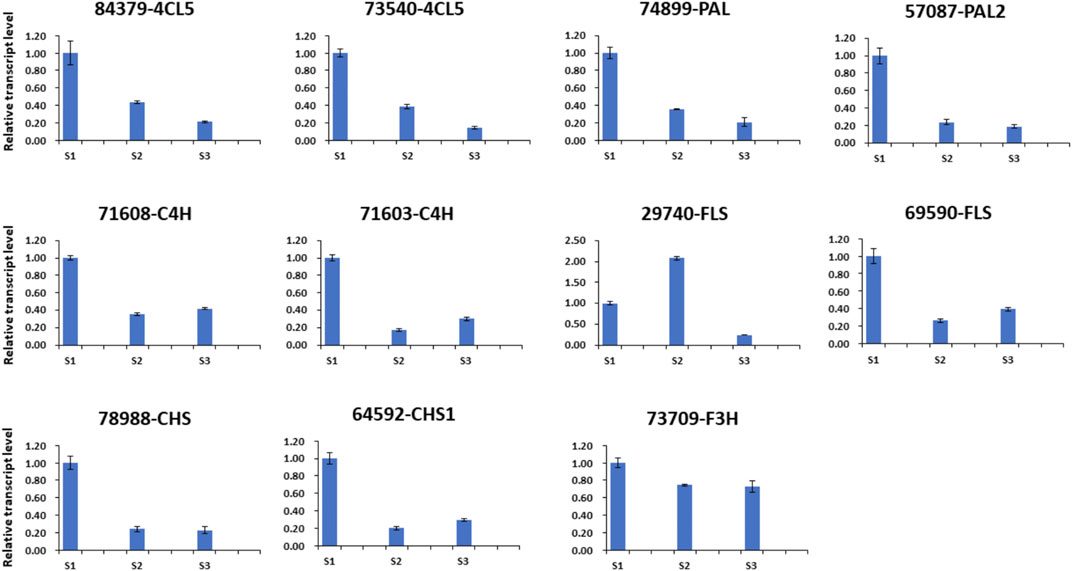
FIGURE 8. The qRT-PCR of polyphenol compounds related genes at three developmental stages of nutshell.
Discussion
Polyphenol Compounds Profiles in the Nutshell of M. Ternifolia
In Macadamia nuts, the kernel is covered in a hard nutshell and considered as a premium product. It has been utilized to produce healthy diet for humans. However, the nutshell is still not well valorized in Macadamia nuts. Our study showed that M. ternifolia had a diversity of metabolic compounds in the nutshell with polyphenols especially phenolic acids and flavonoids being the major metabolites present in the nutshell. In addition, both the composition and concentration of these metabolites significantly changed at the three developmental stages of the nutshell. These functional metabolites may have useful applications in medical and ornamental industries. In plants, phenolic acids and flavonoids are low-molecular-weight pervasive groups (Ghasemzadeh and Ghasemzadeh, 2011; Rehman et al., 2018). Phenolic compounds are carbon-based secondary metabolites, which act as powerful antioxidants, antiviral, and anti-allergic agents (Yagi et al., 2013; Rouphael et al., 2017). They can absorb free radical oxygen, which subsequently reduced aging, heart attack, and cancer diseases (Hollman, 2001). Additionally, environmental factors such as light, temperature, nutrient availability, and salinity affected the abundance of phenolic compounds in plants (Marin et al., 2015; Sgherri et al., 2017; Henry-Kirk et al., 2018). It has been stated that the biosynthesis of different types of phenolic compounds play a vital role against pathogen and insect defense (Beckman, 2000). In this regard, increased information about phenolic profile can be important to mitigate anthracnose husk rot disease, which is a major threat to Macadamia nut production worldwide (Campbell, 2015). In M. ternifolia, glucosides and chlorogenic acid derived phenolic acids had a dynamic abundance in nutshell. A previous research has clarified that chlorogenic acid is characterized by antioxidant and anti-inflammatory properties (Sasaki et al., 2010). Therefore, it serves as a precursor in therapeutics, cosmetics, and food industries. In addition, phenolic acids are useful to enhance the organoleptic characters of food items, reduce skin damage, and avoid itching (Croft, 1998; Tan, 2000). Flavonoids in plants belong to polyphenols, synthesized by aromatic rings, and classified into different kinds of flavones, flavanones, chalcones, isoflavones, flavonols, deoxy-flavonoids, and anthocyanins (Panche et al., 2016). They have been widely applied in food, pharmaceutical, medicinal, and cosmetic industries. Almost every group of flavonoids has potential health benefits through antioxidant defense mechanisms (Nijveldt et al., 2001). Interestingly, flavonoids were major component in the nutshell of M. ternifolia. In particular, quercetin linked glucoside, catechin, epicatechin, mearnsetin-3-O-glucoside, mearnsetin-3-O-glucuronide, and quercetin-3-O-galactoside had the highest abundance among all the identified flavonoids. The stone fruits such as peach, sweet cherry, apricot, plums, and almond skin have a diversity of polyphenols. However, catechin, epicatechin, chlorogenic acid derivatives are the most common among detected polyphenols in these fruits (Garrido et al., 2008; Campbell and Padilla-Zakour, 2013; Pinu, 2016; Koprivica et al., 2018). Previous research revealed several health-promoting benefits of polyphenols obtained from plants (Lara et al., 2020). It is noteworthy that polyphenols by-products obtained from almond skin is used to reduce viral, inflammatory, allergic, mutagenic, and carcinogenic disorders (Siriwardhana and Shahidi, 2002). Improved knowledge about bioactive compounds present in the nutshell of M. ternifolia could be useful to produce several kinds of antioxidant materials, which can be utilized in the food industry and could also be used as functional ingredients in pharmaceutical and cosmetic industries.
Regulatory Mechanism of Polyphenols in the Nutshell of M. Ternifolia
The phenolic compounds are mainly synthesized from the shikimate/phenylpropanoid pathway in plants (Tzin and Galili, 2010; Vogt, 2010). In this pathway, phenylalanine act as a precursor of different kinds of phenolic acids and flavonoids (Weisshaar and Jenkins, 1998). Nearly, all genes involved in the phenylpropanoid pathway has been investigated in Arabidopsis, poplar, and rice (Hamberger et al., 2007). In brief, phenylalanine in the first step generates derivatives of hydroxybenzoic acid by the action of the PAL gene family. Later, serial reactions regulated by C4H and 4CL gene families synthesized various forms of gallic acids, cinnamic acids, caffeic acids, ferulic, acids, sinapic acids, and chlorogenic acids. In the nutshell of M. ternifolia, a dynamic expression of 14 PAL, 2 C4H, and 15 4CL genes may regulate the composition of different phenolic acids (Ragg et al., 1981; Chen et al., 2007; Chang et al., 2008). Additionally, the expression of PAL, C4H, and 4CL genes is altered significantly in response to growth conditions, abiotic and biotic stresses (Lee et al., 1995; Bellés et al., 2008; Huang et al., 2010). Flavonoids are synthesized from branches of the phenylpropanoid pathway (Dixon and Steele, 1999). CHS genes catalyzed the first committed step to generate chalcone, which is a primary precursor of all flavonoids. In the next step, chalcones are isomerized into flavanones by the action of CHI genes. The gene family F3H generates dihydroflavanols from flavanones, which finally form flavonols by the action of FLS genes. Several genes allied with CHS, CHI, and FLS showed significantly altered expression in developing nutshell of M. ternifolia. These results predict that flavonoids pathway genes expression and the abundance of flavonoids compounds are most likely interlinked with each other. A recent study in Brassica juncea revealed a strong correlation between flavonoids pathway genes expression and the abundance of flavonoids compounds (Shen et al., 2021). Flavonoids pathway genes are well understood in Arabidopsis (Saito et al., 2013), brassica (Liu et al., 2013), and pear (Feng et al., 2010). Transgenic studies reported that the phenylpropanoid pathway is comprised of several complex chemical reactions. The increased or decreased expression of a specific gene may not lead to a clear phenotype, which is probably due to the synthesis of the certain compound via an alternative pathway (Sewalt et al., 1997). However, a strong correlation between metabolite and transcript profiling of specific compounds is critical to identify a single gene by following advance transgenic quantifications (Rohde et al., 2004). Because the genetic transformation protocol of M. ternifolia is not yet well established, it is not possible at this stage to study the functions of some candidate genes identified in this study. Nonetheless, our integrative metabolome and transcriptome analysis are effective to identify key potential genes responsible for polyphenols biosynthesis in the nutshell of M. ternifolia.
Materials and Methods
Plant Materials and Analysis of Metabolic Substances
Plant materials used in this study were taken from M. ternifolia. The nutshell samples were collected in three biological replicates at the young fruit (S1), medium fruit (S2), and mature fruit (S3) stages. The harvested samples were placed immediately in liquid nitrogen and stored in an ultra-low temperature refrigerator at −80°C for metabolomics and transcriptomics analysis. In order to explore the differences in metabolites in different samples, 1 g biological sample was ground into powder. Then 100 mg of powder was dissolved in 1.2 ml 70% aqueous methanol. Later on, samples were vortex for 30 min and saved into the refrigerator at 4°C overnight. The samples were centrifuged at 12000 rpm for 10 min. The upper clear liquid was collected in a sample bottle, filtered with a micro-porous membrane (0.22 um pore size), and stored for UPLC-MS/MS analysis. The ultra-performance liquid chromatography (UPLC) (Shimadzu Nexera X2) and tandem mass spectrometry (MS/MS) (Applied Biosystems 4500 QTRAP) were used in this study to perform metabolic analysis. The instrument running parameters and data acquisition methods were followed as previously described (Yin et al., 2019). Based on the local metabolic databases e.g., MzCloud, Massbank, Metlin, HMDB, and KEGG, qualitative and quantitative analysis of the metabolites for each sample was carried out. The identification of significantly differential metabolites was achieved with variable importance in projection (VIP) value combined with fold change (FC). The metabolites with FC ≤ 0.5 or ≥0.5 and VIP ≥1 were generally considered to be significant among the control group and the experimental group. The metabolites that differed more than 2 times or less than 0.5 were considered significant high or low differences between the control and experimental groups.
Ribonucleic Acid Sequencing and Downstream Analysis
High quality total RNA were extracted with the CTAB method (Gambino et al., 2008) from samples of young (S1), medium (S2), and mature fruit (S3) nutshell of M. ternifolia. The Agarose gel electrophoresis was used to analyze the integrity of RNA and the presence of contaminations. RNA concentration was measured with Qubit 2.0 fluorescence meter. Moreover, RNA purity and integrity were assessed with Nanodrop spectrophotometer (Thermo Fisher Scientific, United States) and Bioanalyzer Agilent 2,100 (Agilent Technologies, United States, respectively. The 9 cDNA libraries were prepared with NEBNext®mRNA Library Prep Master Mix Set by following the manufacturer’s standard protocol. Then, the Illumina HiSeq2000 platform was used to perform de novo pair-end RNA sequencing by Novogene Bioinformatic Technology Co. Ltd. (Tianjin, China). After sequencing, the original sequenced data was filtered to remove low quality reads and to obtain high-quality standard reads. The M. ternifolia is without a reference genome, therefore high quality clean reads were assembled with Trinity software (Trinity release v2.4.0, United States) to obtain a reference sequence for succeeding analysis (Grabherr et al., 2011). The functional annotations for assembled transcripts were retrieved from publicly available protein databases including the NCBI Non-redundant nucleotide and protein database (Nr), EuKaryotic Orthologous Group (KOG), Kyoto Encyclopedia of Genes and Genomes database (KEGG), Gene Ontology (GO) Swiss-Prot, Trembl, and Pfam with Basic Local Alignment Search Tool (BLAST). The expression levels of each unigene were calculated as FPKM. The DESeq tool was used to identify DEGs among the control group and the experimental group (Anders and Huber, 2010). The identification of significant DEGs between different samples was achieved using log2 (fold change) > 1 or < − 1 and p-value < 0.05 as selection criteria. The FPKM of genes is first centralized and standardized and then Kmeans clustering is performed with Mfuzz software (Kumar and Futschik, 2007). The TBtools version 1.055 was utilized to make desired heatmaps (Chen et al., 2020). The pathway enrichment analysis for DEGs was retrieved as detailed previously by Kanehisa et al. (2007).
Gene Expression Analyses With qRT-Polymerase Chain Reaction
The TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, China) kit was utilized to synthesize the first standard cDNA for qRT-PCR. The specific primers for each gene were prepared with Oligo 7 and listed in Supplementary Table S3. For the qRT-PCR reaction mixture, QIAGEN SYBR Green PCR Kit was used with already standardized volume of each component. The GAPDH was used as the reference gene and the relative expression of each gene was acquired with the 2−ΔΔCt method (Livak and Schmittgen, 2001). The qRT-PCR was conducted with three biological replicates and three technical replicates.
Conclusion
In this study, 596 metabolites were identified at three developmental stages of the nutshell in M. ternifolia. The qualitative and quantitative metabolic data analysis revealed that phenolic acids and flavonoids were the predominant components in the nutshell. Importantly, their abundance was significantly changed during the nutshell growth periods. These results are vital for the synthesis of different by-products from the nutshell. Integration of metabolome and transcriptome analyses identified significant expression changes in phenolic acid and flavonoid pathway genes. Our results provide novel insights into the biosynthesis of polyphenols in the nutshell of M. ternifolia. However, functional research is essential to understand the potential genetic mechanism of phenolic acids and flavonoids metabolism in the nutshell of M. ternifolia.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Conceptualization, JH and FH; methodology, LT, and CZ; software, LT, XT, CZ, and ZX; validation, JH, AR, JA, and FH; formal analysis, XT, LT, CZ, ZX, and JH; investigation, AR, JA, and CZ; resources, XT, FH, and RS; data curation, CZ, ZX, and LT; writing—original draft preparation, RS, JA, JH, and FH; writing—review and editing, RS and FH; visualization, FH; supervision, RS, XT, and JH; project administration, JA, AR, and ZX; funding acquisition, JH, and FH; All authors have read and approved final version of the manuscript.
Funding
This work was supported by National Key R&D Program of China (2021YFD1000202), China Agriculture Research System of MOF & MARA (CARS-21-05B), Major Science and Technology Project of Yunnan (202102AE090042, 202002AA10007, 2019ZG00901), Major Science and Technology Project of Kunming (2021JH002), High-end Foreign Experts Program (G2021039002, 202105AQ130011, 2019013),Yunnan Provincial Financial Forestry Science and Technology Promotion Demonstration Special Project in 2020 (2020) TS09.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.809986/full#supplementary-material
References
Akhtar, N., Ahmad, M., Madni, A., and Bakhsh, S. (2006). Evaluation of Basic Properties of Macadamia Nut Oil. Gomal Univ. J. Res. 22, 21–27.
Alagna, F., Mariotti, R., Panara, F., Caporali, S., Urbani, S., Veneziani, G., et al. (2012). Olive Phenolic Compounds: Metabolic and Transcriptional Profiling during Fruit Development. BMC Plant Biol. 12, 162. doi:10.1186/1471-2229-12-162
Anders, S., and Huber, W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol., 11, R106. doi:10.1038/npre.2010.4282.1
Aygün, A., Yenisoy-Karakaş, S., and Duman, I. (2003). Production of Granular Activated Carbon from Fruit Stones and Nutshells and Evaluation of Their Physical, Chemical and Adsorption Properties. Microporous Mesoporous Mater. 66, 189–195. doi:10.1016/j.micromeso.2003.08.028
Bakly, S., Al-Juboori, R. A., and Bowtell, L. (2019). Macadamia Nutshell Biochar for Nitrate Removal: Effect of Biochar Preparation and Process Parameters. J. Carbon. Res. 5, 47. doi:10.3390/c5030047
Beckman, C. H. (2000). Phenolic-storing Cells: Keys to Programmed Cell Death and Periderm Formation in Wilt Disease Resistance and in General Defence Responses in Plants? Physiol. Mol. Plant Pathol. 57, 101–110. doi:10.1006/pmpp.2000.0287
Bellés, J. M., López-Gresa, M. P., Fayos, J., Pallás, V., Rodrigo, I., and Conejero, V. (2008). Induction of Cinnamate 4-hydroxylase and Phenylpropanoids in Virus-Infected Cucumber and Melon Plants. Plant Sci. 174, 524–533. doi:10.1016/j.plantsci.2008.02.008
Bourgis, F., Kilaru, A., Cao, X., Ngando-Ebongue, G.-F., Drira, N., Ohlrogge, J. B., et al. (2011). Comparative Transcriptome and Metabolite Analysis of Oil palm and Date palm Mesocarp that Differ Dramatically in Carbon Partitioning. Proc. Natl. Acad. Sci. 108, 12527–12532. doi:10.1073/pnas.1106502108
Campbell, O. E., and Padilla-Zakour, O. I. (2013). Phenolic and Carotenoid Composition of Canned Peaches (Prunus Persica) and Apricots (Prunus Armeniaca) as Affected by Variety and Peeling. Food Res. Int. 54, 448–455. doi:10.1016/j.foodres.2013.07.016
Chang, A., Lim, M.-H., Lee, S.-W., Robb, E. J., and Nazar, R. N. (2008). Tomato Phenylalanine Ammonia-Lyase Gene Family, Highly Redundant but Strongly Underutilized. J. Biol. Chem. 283, 33591–33601. doi:10.1074/jbc.m804428200
Chen, A.-H., Chai, Y.-R., Li, J.-N., and Chen, L. (2007). Molecular Cloning of Two Genes Encoding Cinnamate 4-hydroxylase (C4H) from Oilseed Rape (Brassica Napus). BMB Rep. 40, 247–260. doi:10.5483/bmbrep.2007.40.2.247
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, W., Gao, Y., Xie, W., Gong, L., Lu, K., Wang, W., et al. (2014). Genome-wide Association Analyses Provide Genetic and Biochemical Insights into Natural Variation in rice Metabolism. Nat. Genet. 46, 714–721. doi:10.1038/ng.3007
Conesa, J. A., Sakurai, M., and Antal, M. J. (2000). Synthesis of a High-Yield Activated Carbon by Oxygen Gasification of Macadamia Nut Shell Charcoal in Hot, Liquid Water. Carbon 38, 839–848. doi:10.1016/s0008-6223(99)00182-7
Croft, K. D. (1998). The Chemistry and Biological Effects of Flavonoids and Phenolic Acidsa. Ann. NY Acad Sci 854, 435–442. doi:10.1111/j.1749-6632.1998.tb09922.x
Dixon, R. A., and Steele, C. L. (1999). Flavonoids and Isoflavonoids - a Gold Mine for Metabolic Engineering. Trends Plant Science 4, 394–400. doi:10.1016/s1360-1385(99)01471-5
Fan, F., Yang, Z., Li, H., Shi, Z., and Kan, H. (2018). Preparation and Properties of Hydrochars from Macadamia Nut Shell via Hydrothermal Carbonization. R. Soc. Open Sci. 5, 181126. doi:10.1098/rsos.181126
Fang, C., Fernie, A. R., and Luo, J. (2019). Exploring the Diversity of Plant Metabolism. Trends Plant Sci. 24, 83–98. doi:10.1016/j.tplants.2018.09.006
Feng, S., Wang, Y., Yang, S., Xu, Y., and Chen, X. (2010). Anthocyanin Biosynthesis in Pears Is Regulated by a R2R3-MYB Transcription Factor PyMYB10. Planta 232, 245–255. doi:10.1007/s00425-010-1170-5
Francoz, E., Lepiniec, L., and North, H. M. (2018). Seed coats as an Alternative Molecular Factory: Thinking outside the Box. Plant Reprod. 31, 327–342. doi:10.1007/s00497-018-0345-2
Gambino, G., Perrone, I., and Gribaudo, I. (2008). A Rapid and Effective Method for RNA Extraction from Different Tissues of grapevine and Other Woody Plants. Phytochem. Anal. 19, 520–525. doi:10.1002/pca.1078
Garg, M. L., Blake, R. J., Wills, R. B. H., and Clayton, E. H. (2007). Macadamia Nut Consumption Modulates Favourably Risk Factors for Coronary Artery Disease in Hypercholesterolemic Subjects. Lipids 42, 583–587. doi:10.1007/s11745-007-3042-8
Garg, M. L., Blake, R. J., and Wills, R. B. H. (2003). Macadamia Nut Consumption Lowers Plasma Total and LDL Cholesterol Levels in Hypercholesterolemic Men. J. Nutr. 133, 1060–1063. doi:10.1093/jn/133.4.1060
Garrido, I., Monagas, M., Gómez-Cordovés, C., and Bartolomé, B. (2008). Polyphenols and Antioxidant Properties of almond Skins: Influence of Industrial Processing. J. Food Sci. 73, C106–C115. doi:10.1111/j.1750-3841.2007.00637.x
Ghasemzadeh, A., and Ghasemzadeh, N. (2011). Flavonoids and Phenolic Acids: Role and Biochemical Activity in Plants and Human. J. Med. plants Res. 5, 6697–6703. doi:10.5897/jmpr11.1404
Girdis, J., Gaudion, L., Proust, G., Löschke, S., and Dong, A. (2017). Rethinking Timber: Investigation into the Use of Waste Macadamia Nut Shells for Additive Manufacturing. Jom 69, 575–579. doi:10.1007/s11837-016-2213-6
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 29, 644–652. doi:10.1038/nbt.1883
Hamberger, B., Ellis, M., Friedmann, M., De Azevedo Souza, C., Barbazuk, B., and Douglas, C. J. (2007). Genome-wide Analyses of Phenylpropanoid-Related Genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza Sativa: the Populus Lignin Toolbox and Conservation and Diversification of Angiosperm Gene familiesThis Article Is One of a Selection of Papers Published in the Special Issue on Poplar Research in Canada. Can. J. Bot. 85, 1182–1201. doi:10.1139/b07-098
Hartung, M., and Storey, W. (1939). The Development of the Fruit of Macadamia. J. Agric. Res. 59, 397.
Henry-Kirk, R. A., Plunkett, B., Hall, M., Mcghie, T., Allan, A. C., Wargent, J. J., et al. (2018). Solar UV Light Regulates Flavonoid Metabolism in Apple (Malus X Domestica). Plant Cel Environ 41, 675–688. doi:10.1111/pce.13125
Hollman, P. C. H. (2001). Evidence for Health Benefits of Plant Phenols: Local or Systemic Effects? J. Sci. Food Agric. 81, 842–852. doi:10.1002/jsfa.900
Howlett, B. G., Nelson, W. R., Pattemore, D. E., and Gee, M. (2015). Pollination of Macadamia: Review and Opportunities for Improving Yields. Scientia Horticulturae 197, 411–419. doi:10.1016/j.scienta.2015.09.057
Hu, W., Fitzgerald, M., Topp, B., Alam, M., and O'hare, T. J. (2019). A Review of Biological Functions, Health Benefits, and Possible De Novo Biosynthetic Pathway of Palmitoleic Acid in Macadamia Nuts. J. Funct. Foods 62, 103520. doi:10.1016/j.jff.2019.103520
Huang, J., Gu, M., Lai, Z., Fan, B., Shi, K., Zhou, Y.-H., et al. (2010). Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress. Plant Physiol. 153, 1526–1538. doi:10.1104/pp.110.157370
Kaijser, A., Dutta, P., and Savage, G. (2000). Oxidative Stability and Lipid Composition of Macadamia Nuts Grown in New Zealand. Food Chem. 71, 67–70. doi:10.1016/s0308-8146(00)00132-1
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2007). KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 36, D480–D484. doi:10.1093/nar/gkm882
Koprivica, M. R., Trifković, J. Đ., Dramićanin, A. M., Gašić, U. M., Akšić, M. M. F., and Milojković-Opsenica, D. M. (2018). Determination of the Phenolic Profile of Peach (Prunus Persica L.) Kernels Using UHPLC-LTQ OrbiTrap MS/MS Technique. Eur. Food Res. Technol. 244, 2051–2064. doi:10.1007/s00217-018-3116-2
Kumar, L., and Futschik, M. E. (2007). Mfuzz: a Software Package for Soft Clustering of Microarray Data. Bioinformation 2, 5–7. doi:10.6026/97320630002005
Kumar, U., Maroufi, S., Rajarao, R., Mayyas, M., Mansuri, I., Joshi, R. K., et al. (2017). Cleaner Production of Iron by Using Waste Macadamia Biomass as a Carbon Resource. J. Clean. Prod. 158, 218–224. doi:10.1016/j.jclepro.2017.04.115
Lara, M. V., Bonghi, C., Famiani, F., Vizzotto, G., Walker, R. P., and Drincovich, M. F. (2020). Stone Fruit as Biofactories of Phytochemicals with Potential Roles in Human Nutrition and Health. Front. Plant Sci. 11, 1323. doi:10.3389/fpls.2020.562252
Lee, D., Ellard, M., Wanner, L. A., Davis, K. R., and Douglas, C. J. (1995). The Arabidopsis thaliana 4-coumarate:CoA Ligase (4CL) Gene: Stress and Developmentally Regulated Expression and Nucleotide Sequence of its cDNA. Plant Mol. Biol. 28, 871–884. doi:10.1007/bf00042072
Liu, X., Lu, Y., Yuan, Y., Liu, S., Guan, C., Chen, S., et al. (2013). De Novo transcriptome of Brassica Juncea Seed Coat and Identification of Genes for the Biosynthesis of Flavonoids. PLoS One 8, e71110. doi:10.1371/journal.pone.0071110
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. methods 25, 402–408. doi:10.1006/meth.2001.1262
Malvestiti, R., Borges, L. d. S., Weimann, E., Junior, E. P. d. S., Levada‐Pires, A. C., Dermargos, A., et al. (2017). The Effect of Macadamia Oil Intake on Muscular Inflammation and Oxidative Profile Kinetics after Exhaustive Exercise. Eur. J. Lipid Sci. Technol. 119, 1600382. doi:10.1002/ejlt.201600382
Marin, A., Ferreres, F., Barberá, G. G., and Gil, M. I. (2015). Weather Variability Influences Color and Phenolic Content of Pigmented Baby Leaf Lettuces throughout the Season. J. Agric. Food Chem. 63, 1673–1681. doi:10.1021/acs.jafc.5b00120
Mast, A. R., Willis, C. L., Jones, E. H., Downs, K. M., and Weston, P. H. (2008). A Smaller Macadamia from a More Vagile Tribe: Inference of Phylogenetic Relationships, Divergence Times, and Diaspore Evolution in Macadamia and Relatives (Tribe Macadamieae; Proteaceae). Am. J. Bot. 95, 843–870. doi:10.3732/ajb.0700006
Matsuda, F., Yonekura-Sakakibara, K., Niida, R., Kuromori, T., Shinozaki, K., and Saito, K. (2009). MS/MS Spectral Tag-Based Annotation of Non-targeted Profile of Plant Secondary Metabolites. Plant J. 57, 555–577. doi:10.1111/j.1365-313x.2008.03705.x
Navarro, S. L. B., and Rodrigues, C. E. C. (2016). Macadamia Oil Extraction Methods and Uses for the Defatted Meal Byproduct. Trends Food Sci. Tech. 54, 148–154. doi:10.1016/j.tifs.2016.04.001
Nijveldt, R. J., Van Nood, E., Van Hoorn, D. E., Boelens, P. G., Van Norren, K., and Van Leeuwen, P. A. (2001). Flavonoids: a Review of Probable Mechanisms of Action and Potential Applications. Am. J. Clin. Nutr. 74, 418–425. doi:10.1093/ajcn/74.4.418
O’hare, T. J., Trieu, H. H., Topp, B., Russell, D., Pun, S., Torrisi, C., et al. (2019). Assessing Fatty Acid Profiles of Macadamia Nuts. HortScience 54, 633–637. doi:10.21273/hortsci13418-18
Panche, A. N., Diwan, A. D., and Chandra, S. R. (2016). Flavonoids: an Overview. J. Nutr. Sci. 5, e47. doi:10.1017/jns.2016.41
Pinu, F. R. (2016). Early Detection of Food Pathogens and Food Spoilage Microorganisms: Application of Metabolomics. Trends Food Sci. Tech. 54, 213–215. doi:10.1016/j.tifs.2016.05.018
Poinern, G. E. J., Senanayake, G., Shah, N., Thi-Le, X. N., Parkinson, G. M., and Fawcett, D. (2011). Adsorption of the Aurocyanide, Complex on Granular Activated Carbons Derived from Macadamia Nut Shells - A Preliminary Study. Minerals Eng. 24, 1694–1702. doi:10.1016/j.mineng.2011.09.011
Ragg, H., Kuhn, D. N., and Hahlbrock, K. (1981). Coordinated Regulation of 4-coumarate:CoA Ligase and Phenylalanine Ammonia-Lyase mRNAs in Cultured Plant Cells. J. Biol. Chem. 256, 10061–10065. doi:10.1016/s0021-9258(19)68741-7
Rehman, R. N. U., You, Y., Ali, S., Wang, Y., Zhang, L., Li, P., et al. (2018). Phenolic Compounds as Biochemical Markers of Senescence in Woody Ornamental Flowers of Malus Crabapple. Hortic. Environ. Biotechnol. 59, 1–10. doi:10.1007/s13580-018-0001-7
Rengel, A., Pérez, E., Piombo, G., Ricci, J., Servent, A., Tapia, M. S., et al. (2015). Lipid Profile and Antioxidant Activity of Macadamia Nuts (Macadamia Integrifolia) Cultivated in Venezuela. Nat. Sci. 07, 535. doi:10.4236/ns.2015.712054
Rohde, A., Morreel, K., Ralph, J., Goeminne, G., Hostyn, V., De Rycke, R., et al. (2004). Molecular Phenotyping of the Pal1 and Pal2 Mutants of Arabidopsis thaliana Reveals Far-Reaching Consequences on Phenylpropanoid, Amino Acid, and Carbohydrate Metabolism. Plant Cell 16, 2749–2771. doi:10.1105/tpc.104.023705
Rouphael, Y., Colla, G., Graziani, G., Ritieni, A., Cardarelli, M., and De Pascale, S. (2017). Phenolic Composition, Antioxidant Activity and mineral Profile in Two Seed-Propagated Artichoke Cultivars as Affected by Microbial Inoculants and Planting Time. Food Chem. 234, 10–19. doi:10.1016/j.foodchem.2017.04.175
Saito, K., Yonekura-Sakakibara, K., Nakabayashi, R., Higashi, Y., Yamazaki, M., Tohge, T., et al. (2013). The Flavonoid Biosynthetic Pathway in Arabidopsis: Structural and Genetic Diversity. Plant Physiol. Biochem. 72, 21–34. doi:10.1016/j.plaphy.2013.02.001
Sasaki, K., Alamed, J., Weiss, J., Villeneuve, P., López Giraldo, L. J., Lecomte, J., et al. (2010). Relationship between the Physical Properties of Chlorogenic Acid Esters and Their Ability to Inhibit Lipid Oxidation in Oil-In-Water Emulsions. Food Chem. 118, 830–835. doi:10.1016/j.foodchem.2009.05.070
Sewalt, V., Ni, W., Blount, J. W., Jung, H. G., Masoud, S. A., Howles, P. A., et al. (1997). Reduced Lignin Content and Altered Lignin Composition in Transgenic Tobacco Down-Regulated in Expression of L-Phenylalanine Ammonia-Lyase or Cinnamate 4-hydroxylase. Plant Physiol. 115, 41–50. doi:10.1104/pp.115.1.41
Sgherri, C., Pérez-López, U., Micaelli, F., Miranda-Apodaca, J., Mena-Petite, A., Muñoz-Rueda, A., et al. (2017). Elevated CO 2 and Salinity Are Responsible for Phenolics-Enrichment in Two Differently Pigmented Lettuces. Plant Physiol. Biochem. 115, 269–278. doi:10.1016/j.plaphy.2017.04.006
Shen, S., Tang, Y., Zhang, C., Yin, N., Mao, Y., Sun, F., et al. (2021). Metabolite Profiling and Transcriptome Analysis Provide Insight into Seed Coat Color in Brassica Juncea. Ijms 22, 7215. doi:10.3390/ijms22137215
Siriwardhana, S. S. K. W., and Shahidi, F. (2002). Antiradical Activity of Extracts of almond and its By-Products. J. Amer Oil Chem. Soc. 79, 903–908. doi:10.1007/s11746-002-0577-4
Syed, S., and Walele, I. (2006). Macadamia Lipid Based Surfactants and Derivatives and Process for Preparing Same. Google Patents.
Tan, S. C. (2000). “Determinants of Eating Quality in Fruit and Vegetables,” in Proceedings of the Nutrition Society of Australia, Freemantle, Australia, December 3–6, 2000 (Nutrition Society of Australia), 183–190.
Tzin, V., and Galili, G. (2010). New Insights into the Shikimate and Aromatic Amino Acids Biosynthesis Pathways in Plants. Mol. Plant 3, 956–972. doi:10.1093/mp/ssq048
Wang, C.-H., and Mai, Y.-W. (1994). Deformation and Fracture of Macadamia Nuts. Int. J. Fracture 69, 67–85.
Wang, D. D., and Hu, F. B. (2017). Dietary Fat and Risk of Cardiovascular Disease: Recent Controversies and Advances. Annu. Rev. Nutr. 37, 423–446. doi:10.1146/annurev-nutr-071816-064614
Weisshaar, B., and Jenkins, G. I. (1998). Phenylpropanoid Biosynthesis and its Regulation. Curr. Opin. Plant Biol. 1, 251–257. doi:10.1016/s1369-5266(98)80113-1
Wrobel-Tobiszewska, A., Boersma, M., Sargison, J., Adams, P., Singh, B., Franks, S., et al. (2018). Nutrient Changes in Potting Mix and Eucalyptus Nitens Leaf Tissue under Macadamia Biochar Amendments. J. For. Res. 29, 383–393. doi:10.1007/s11676-017-0437-0
Wu, Q., Cao, Y., Chen, C., Gao, Z., Yu, F., and Guy, R. D. (2020). Transcriptome Analysis of Metabolic Pathways Associated with Oil Accumulation in Developing Seed Kernels of Styrax Tonkinensis, a Woody Biodiesel Species. BMC Plant Biol. 20, 121. doi:10.1186/s12870-020-2327-4
Yagi, H., Tan, J., and Tuan, R. S. (2013). Polyphenols Suppress Hydrogen Peroxide-Induced Oxidative Stress in Human Bone-Marrow Derived Mesenchymal Stem Cells. J. Cel. Biochem. 114, 1163–1173. doi:10.1002/jcb.24459
Yin, N.-W., Wang, S.-X., Jia, L.-D., Zhu, M.-C., Yang, J., Zhou, B.-J., et al. (2019). Identification and Characterization of Major Constituents in Different-Colored Rapeseed Petals by UPLC-HESI-MS/MS. J. Agric. Food Chem. 67, 11053–11065. doi:10.1021/acs.jafc.9b05046
Zhang, C., Yao, X., Ren, H., Wang, K., and Chang, J. (2019). Isolation and Characterization of Three Chalcone Synthase Genes in Pecan (Carya Illinoinensis). Biomolecules 9, 236. doi:10.3390/biom9060236
Zhao, S., Zhang, X., Su, Y., Chen, Y., Liu, Y., Sun, M., et al. (2018). Transcriptome Analysis Reveals Dynamic Fat Accumulation in the walnut Kernel. Int. J. genomics 2018, 1. doi:10.1155/2018/8931651
Zheng, L., Shockey, J., Guo, F., Shi, L., Li, X., Shan, L., et al. (2017). Discovery of a New Mechanism for Regulation of Plant Triacylglycerol Metabolism: the Peanut Diacylglycerol Acyltransferase-1 Gene Family Transcriptome Is Highly Enriched in Alternative Splicing Variants. J. Plant Physiol. 219, 62–70. doi:10.1016/j.jplph.2017.09.009
Keywords: macadamia, nutshell, metabolic, transcript, phenolic, flavonoid
Citation: Shi R, Tao L, Tu X, Zhang C, Xiong Z, Rami Horowitz A, Asher JB, He J and Hu F (2022) Metabolite Profiling and Transcriptome Analyses Provide Insight Into Phenolic and Flavonoid Biosynthesis in the Nutshell of Macadamia Ternifolia. Front. Genet. 12:809986. doi: 10.3389/fgene.2021.809986
Received: 05 November 2021; Accepted: 29 December 2021;
Published: 21 February 2022.
Edited by:
Deepmala Sehgal, International Maize and Wheat Improvement Center, MexicoReviewed by:
Kalenahalli Yogendra, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaFuyou Fu, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2022 Shi, Tao, Tu, Zhang, Xiong, Rami Horowitz, Asher, He and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunsheng Zhang, Y3N6aGFuZ0B5bnVmZS5lZHUuY24=; Jun He, aGVqdW42NzNAdGp1dGNtLmVkdS5jbg==; Faguang Hu, aGZnMjYzMkAxMjYuY29t
†These authors have contributed equally to this work
 Rui Shi
Rui Shi Liang Tao2†
Liang Tao2† Xinghao Tu
Xinghao Tu Chunsheng Zhang
Chunsheng Zhang Jun He
Jun He Faguang Hu
Faguang Hu