- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
- 2Chengdu Institute of Biology, Chinese Academy of Science, Chengdu, China
- 3Key Laboratory of Crop Genetic Resources and Improvement, Ministry of Education, Sichuan Agricultural University, Chengdu, China
Two advanced wheat lines BAd7-209 and BAd23-1 without the functional gene GPC-B1 were obtained from a cross between common wheat cultivar Chuannong 16 (CN16) and wild emmer wheat accession D97 (D97). BAd7-209 showed superior quality parameters than those of BAd23-1 and CN16. We found that the components of glutenins and gliadins in BAd7-209 and BAd23-1 were similar, whereas BAd7-209 had higher amount of glutenins and gliadins than those of BAd23-1. RNA sequencing analysis on developing grains of BAd7-209 and BAd23-1 as well as their parents revealed 382 differentially expressed genes (DEGs) between the high–grain protein content (GPC) (D97 + BAd7-209) and the low-GPC (CN16 + BAd23-1) groups. DEGs were mainly associated with transcriptional regulation of the storage protein genes, protein processing in endoplasmic reticulum, and protein export pathways. The upregulated gluten genes and transcription factors (e.g., NAC, MYB, and bZIP) may contribute to the high GPC in BAd7-209. Our results provide insights into the potential regulation pathways underlying wheat grain protein accumulation and contribute to make use of wild emmer for wheat quality improvement.
Introduction
Wheat (Triticum aestivum L.) is one of the most important staple crops, which can be processed into a wide range of products such as bread, noodle, and biscuit. Grain protein content (GPC) is an important quality trait in wheat and determines the nutritional value and processing quality (Shewry et al., 1995, 2009). However, GPC and grain yield-related traits are usually negative correlated (Groos et al., 2003), which hampered their simultaneous improvement in conventional wheat breeding program. The characterization and transferring of gene(s)/quantitative trait loci (QTLs) from wheat wild relatives is an effective strategy in the development of elite varieties with high GPC and/or yield (Krugman et al., 2018; Liu et al., 2019; Xiang et al., 2019; Gong et al., 2021).
Wild emmer (T. turgidum ssp. dicoccoides, 2n = 4x = 28, AABB), the tetraploid ancestor of common wheat, provides a valuable reservoir of genetic variation for GPC (Uauy et al., 2006; Liu et al., 2019). A number of QTLs affecting GPC were reported in wild emmer wheat (Joppa et al., 1997; Gonzalez-Hernandez et al., 2004; Uauy et al., 2006; Fatiukha et al., 2019; Liu et al., 2019). To date, however, only Gpc-B1 on chromosome 6BS has been cloned. The introgression of Gpc-B1 in wheat breeding programs can significantly improve GPC while reduce grain-yield related traits in some wheat lines and environments (Uauy et al., 2006; Chen et al., 2017). Therefore, it is desirable to identify GPC-QTLs that are not negative correlated or less correlated with grain yield-related traits.
GPC was regulated by a plethora of genes and easily affected by environment (Liu et al., 2019). In forward genetics, identification of candidate genes related to GPC is a time-consuming and laborious process. RNA sequencing (RNA-seq) provides high-resolution methods for deciphering quality traits and quantifying expression levels of candidate genes on a genome-wide scale (Furtado et al., 2015; Rangan et al., 2017). Currently, RNA-seq has been utilized to study the differentially expressed genes (DEGs) and regulation networks that associated with wheat grain protein accumulation (Cantu et al., 2011; Gong et al., 2021).
Grain protein synthesis in cereal crops were determined by several pathways, mainly including transcriptional regulation of the storage protein genes (glutenin and gliadin) and protein processing in endoplasmic reticulum (ER) and Golgi apparatus. Transcription factors (TFs) belong to bZIP, Dof, MYB, and NAC families have been widely reported in transcriptional regulation of grain protein genes in rice, maize, and barley (Suzuki et al., 1998; Onodera et al., 2001; Diaz et al., 2002; Shewry and Halford, 2002; Zhang et al., 2019). Previous studies demonstrated that the protein processing in ER and Golgi apparatus had crucial role in grain protein synthesis in rice (Takemoto et al., 2002; Wang et al., 2016; Ren et al., 2020), whereas their function for GPC was less reported in wheat.
In our previous studies, the agronomically stable advanced wheat lines were developed from a cross between common wheat cultivar Chuannong 16 (CN16) and wild emmer accession D97 (D97) followed by successive selfing (Jiang et al., 2017; Wang et al., 2018; Liu et al., 2019). Some advanced lines with simultaneous improvement of GPC and thousand-kernel weight (TKW) were obtained. In the present study, two sister lines, BAd7-209 and BAd23-1, with desirable agronomic traits were obtained from CN16×D97. These two lines showed contrasting GPC, while both of them did not contain the functional Gpc-B1. A comparison of the transcriptomes of developing grains from BAd7-209, BAd23-1, CN16, and D97 revealed candidate genes and regulation pathways that may be contributed to wheat grain protein accumulation.
Materials and Methods
Plant Materials
Two sister lines BAd7-209 and BAd23-1 as well as their parents D97 and CN16 were kept at the Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China. CN16 and BAd23-1 were characterized as low-gluten wheat lines, whereas BAd7-209 and D97 were high-GPC lines (Table 1). Wheat plants were grown in the field with three biological replicates (10 rows each replicate) at the experimental field of the Sichuan Agricultural University over two wheat growing seasons (2016 and 2017) at Wenjiang (2016WJ and 2017WJ) and Chongzhou (2016CZ and 2017CZ). Individuals were planted 10-cm apart in a 2-m row with 30 cm between rows. A compound fertilizer [N: P: K (15: 15: 15)] was applied before sowing at a rate of 450 kg per hectare. Developing grains were sampled at 10, 14, 18, 22, 26, 30, 34, and 38 days after pollination (DAP) and snap-frozen in liquid nitrogen and then kept at −80°C for RNA-seq.
Characterization of Agronomic Traits and Karyotype
Agronomic traits of BAd7-209 and BAd23-1 and their parents were measured as listed in Table 1. The traits of plant height (PH), number of spikelets per spike (NSp), and spike number per plant (SN) were averaged by 10 plants. The weight of 300 randomly selected seeds (GB/T 5519-2008, 2008) was recorded to represent the 1,000-kernel weight (TKW) as described by Wang et al. (2018).
At least 30 root-tip cells of BAd7-209 and BAd23-1 and their parents D97 and CN16 were observed for detection the chromosome number using the methods as described by Zhang et al. (2007).
Single-Nucleotide Polymorphism (SNP) Genotyping
Genomic DNA from leaves was isolated using a plant genomic DNA kit (Tiangen Biotech, Beijing Co. Ltd. Beijing, China). Chip-based genotyping was conducted using the wheat 55 K SNP array (www.capit albio.com). The flanking sequence of each SNP was used to map onto the bread wheat reference sequence (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Assemblies/v1.0/), using BLASTN with E-value threshold of 10−10 and a maximum mismatch of one base. SNP markers showed homozygous genotype among the parents, and advanced wheat lines were used to estimate D97 introgressions. The ratios of same SNP to the total SNPs scored between D97 and two advanced lines were computed using a sliding window of 10 Mb and step length of 1 Mb as described by Hao et al. (2019). Graphical representations were constructed using the R package ggplot2 (Wickham 2016).
Quality Parameters Determination
Mature seeds were harvested for measurement of wheat quality parameters (Supplementary Table S1). Seeds of D97, CN16, BAd7-209, and BAd23-1 were conditioned to 14% moisture and milled using the Chopin CD1 AUTO (Renault, Boulogne-Billancourt, France) (Wang et al., 2018). Wet gluten content (WGC) was determined following the American Association of Cereal Chemists (AACC) method 38-12A. The sedimentation volume (SV) was measured following the AACC 44-15A (FOSS, Denmark). GPC (0.5 g) was recorded according to the AACC 39-10. Hydroscopic rate (HR) was measured following the AACC-54-21. All of the experiments were repeated three times.
Grain Protein Component Measurement
Albumins, globulins, glutenins, and gliadins were extracted from mature grains using the methods as described by Duan and Zhao, 2004 and Wan et al. (2000) with slight modifications. Protein content was tested using Kjeldahl method (KjeltecTM 8400). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the glutenins following the method of Hu et al. (2010) using the protein extraction buffer consisting of 62.5 mM Tris-HCl, pH 6.8, 10% (v/v) glycerol, 2% (w/v) SDS, 0.002% (w/v) bromophenol blue, and 1.5% (w/v) dithiothreitol (DTT).
RNA-Seq Analysis
Total RNA was extracted using a plant RNA extraction kit v1.5 (Biofit Biotechnologies, Chengdu). The RNA concentration and integrity were assessed using an Qubit®RNA assay kit on a Qubit® 2.0 Flurometer (Life Technologies, CA, United States) and an RNA Nano 6000 assay kit, respectively. RNAs from different grain developing stages were pooled in equi-molar concentrations (1 µg of RNA per sample) for cDNA library construction (three biological replicates per library). The sequencing libraries were completed using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, United States) and then sequenced using the Illumina Hiseq platform (Novogene Bioinformatics Technology Co. Ltd. Beijing, China).
The adaptors, reads with more than 10% N, and reads with phred quality scored Q < 20 from RNA-seq raw data were removed using Trimmomatic (Bolger et al., 2014). The generated clean reads from each library were assessed using the Q20, Q30, and GC contents and aligned to Chinese Spring reference genome (https://urgi.versailles.inra.fr/blast_iwgsc/) using Hisat2 (Kim et al., 2019). FPKM (Elowitz et al., 2002; Jiang and Wong, 2009) was estimated to represent the expression level. DESeq was used for differential expression analysis as described by Wang et al. (2010). Genes with adjusted FDR <0.001 and |log2FC| ≥ 2 found by EBSeq (Leng et al., 2013) were considered as DEGs. Short Time-series Expression Miner (STEM) software (v1.3.11) was used to explore expression patterns of DEGs as described by Ernst and Bar-Joseph (2006) with log2 standardization, p-value ≤ 0.05, and |log2FC|≥2. Functional annotation of the DEGs was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases (Kanehisa et al., 2004).
Prediction of Transcription Factors and Cis-elements
The putative plant TFs and cis-elements were predicted using iTAK software (Zheng et al., 2016) and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al., 1999).
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) was performed to validate the RNA-seq data. Gene-specific primers were designed using Primer3Plus software (http://www.primer3plus.com/) (Hung and Weng, 2016). Reactions were performed using the Bio-Rad CFX96 Real-Time PCR System (Bio-Rad, United States) in 10 µl of reaction volume containing 2 ng of cDNA, 5 µl of 1 × SYBR Premix ExTaq (TaKaRa), 0.5 µl (300 nM) of each primer, and ddH2O up to 10 µl (Wang et al., 2020). The 2−ΔΔCT method (Schmittgen 2002) was used to quantify the gene expression with endogenous GAPDH as the internal reference.
Statistical Analysis
Statistical analyses were conducted using SPSS version 22 software (SPSS Inc. Chicago, IL, United States). The morphological traits and quality parameters were compared by analysis of variance (ANOVA) complemented by Tukey’s test.
Results
Agronomic and Karyotype Characteristics
The NSp of BAd7-209 and BAd23-1 were less than that of CN16 (p < 0.05), while higher than D97 (p < 0.05). The BAd7-209 resembled the BAd23-1 with respect to spike and SN. We have observed significantly differences in PH and TKW among BAd7-209, BAd23-1, and CN16 (Figure 1; Table 1). The D97 had significantly less NSp and TKW, while significantly higher PH than that of advanced wheat lines (Table 2).
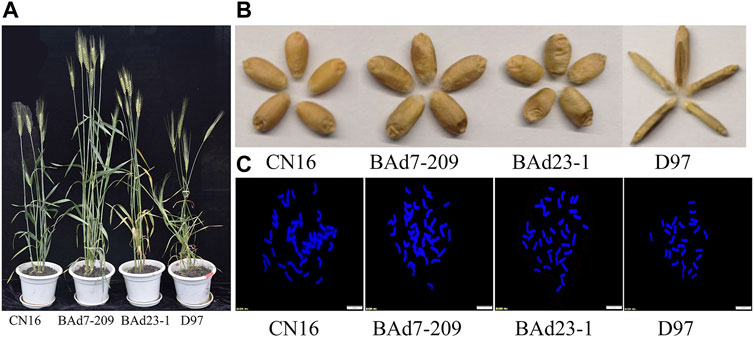
FIGURE 1. Agronomic traits and chromosome patterns of CN16, BAd7-209, BAd23-1, and D97. (A) Plants and (B) seeds of CN16, BAd7-209, BAd23-1, and D97. (C) The number of root-tip chromosomes; scale bar: 10 um.
The BAd7-209 had higher SV (41.3%), WGC (39.49%), HR (64.6%), and stabilization time (7.93%) than those of BAd23-1 and CN16. The GPC in D97 was highest (23.70%), followed by BAd7-209 (15.8%), BAd23-1 (13.8%), and CN16 (12.6%) (Table 2). The WGC, SV, HR, and GPC levels of BAd7-209 were higher than the strong-gluten wheat and those of BAd23-1 were between weak-gluten and medium-gluten wheat.
Cytological observations indicated that both BAd23-1 and BAd7-209 have 42 chromosomes (Figure 1C), which reached the genetic background of common wheat. SNP genotyping analyses revealed 150 and 108 potential D97 segments on chromosomes of BAd23-1 and BAd7-209. The number of CN16 segments on BAd7-209 and BAd23-1 chromosomes was 212 and 232 (Supplementary Figure S1).
Grain Protein Component and Glutenin Subunits
The total amount of glutenins, gliadins, albumins, and globulins in D97 was higher than those of CN16. There were no differences in albumin and globulin contents between BAd23-1 and BAd7-209. The glutenin and gliadin contents of BAd7-209 were significantly higher than those of BAd23-1 (Table 2).
SDS-PAGE analysis showed different high-molecular-glutenin subunits (HMW-GSs) in CN16 (1Ax1, 1Dx5, 1Dy10, and 1Bx+1By20) and D97 (1Ax2.2, 1Bx, 1By8.1, and 1Ay). The BAd7-209 possessed five HWM-GSs (1Ax1.2, 1Bx7 + 1By8, and 1Dx5 + 1Dy10), which were consistent with previous reports (Jiang et al., 2017; Xiang et al., 2019). The BAd23-1 had five HMW-GSs, including 1Bx7 and 1Dx5+1Dy10 that are identical to BAd7-209, and one different 1Ax subunit (1Ax1) and one 1By subunit (1By8.1) (Figure 2).
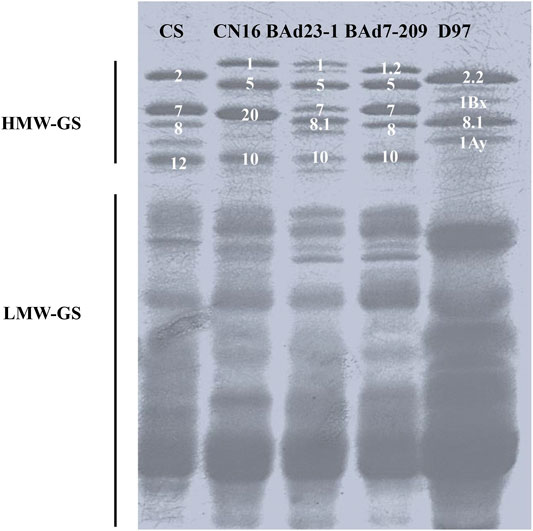
FIGURE 2. Separation of glutenin proteins from BAd7-209 and BAd23-1 and their parents by SDS-PAGE and Chinese Spring (CS) as the references.
The expression levels of 1Ax and 1By in high-GPC group (D97 + BAd7-209) were higher than those of low-GPC group (CN16 + BAd23-1). The composition of low-molecular-glutenin subunits (LMW-GSs) in CN16, BAd7-209, and BAd23-1 was similar. The wild emmer D97 showed less LWM-GS subunits probably due to the absence of the D genome. The expression levels of LMW-GSs in high-GPC group (D97 + BAd7-209) were higher than those of low-GPC group (CN16 + BAd23-1) (Figure 2).
Analysis of RNA-Seq Data
RNA-seq generated 189.93 million raw reads from developing grains of BAd7-209, BAd23-1, CN16, and D97 pools. A total of 94.96 million clean reads were retained and further mapped to the T. aestivum cDNA database (IWGSC1.0). The mapping ratios ranged from 79.42% to 84.33%, 86.92% to 88.12%, 85.76% to 89.33%, and 79.15% to 80.89% in BAd7-209, BAd23-1, CN16, and D97, respectively. The GC contents among replicates were almost identical, and the Q30 was over 90% in each library (Supplementary Table S2). These results demonstrate that the RNA-seq data were qualified for subsequent analysis.
Differential Expression Analysis
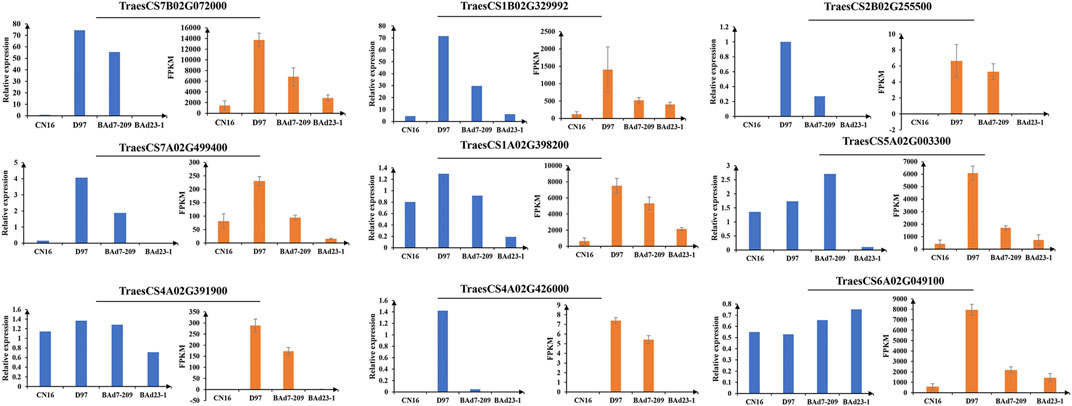
In total, 62,404 genes mapped to IWGSC1.0 genome were expressed (FPKM≥1) in D97, CN16, BAd7-209, and BAd23-1 libraries. A total of 24,700, 2,332, 21,555, and 2,459 DEGs (|log2FC|≥2) were found in D97 vs. CN16, BAd7-209 vs. CN16, D97 vs. BAd23-1, and BAd7-209 vs. BAd23-1, respectively (Figure 3A). The comparison between the high-GPC group (D97 + BAd7-209) and low-GPC group (CN16 + BAd23-1) revealed 382 DEGs (Figure 3B), of which 148 genes were common expressed, 34 genes only expressed in D97 and BAd7-209, whereas 56 genes only expressed in CN16 and BAd23-1. Nine DEGs encoding HWM-GS, alpha-gliadin, nodulin protein, trypsin inhibitor, purothionin, and pre-mRNA-splicing factor proteins were selected to perform qRT-PCR (Supplementary Table S3). The expression changes of seven DEGs were quite consistent with those obtained from the RNA-seq (Figure 4, Supplementary Table S3).
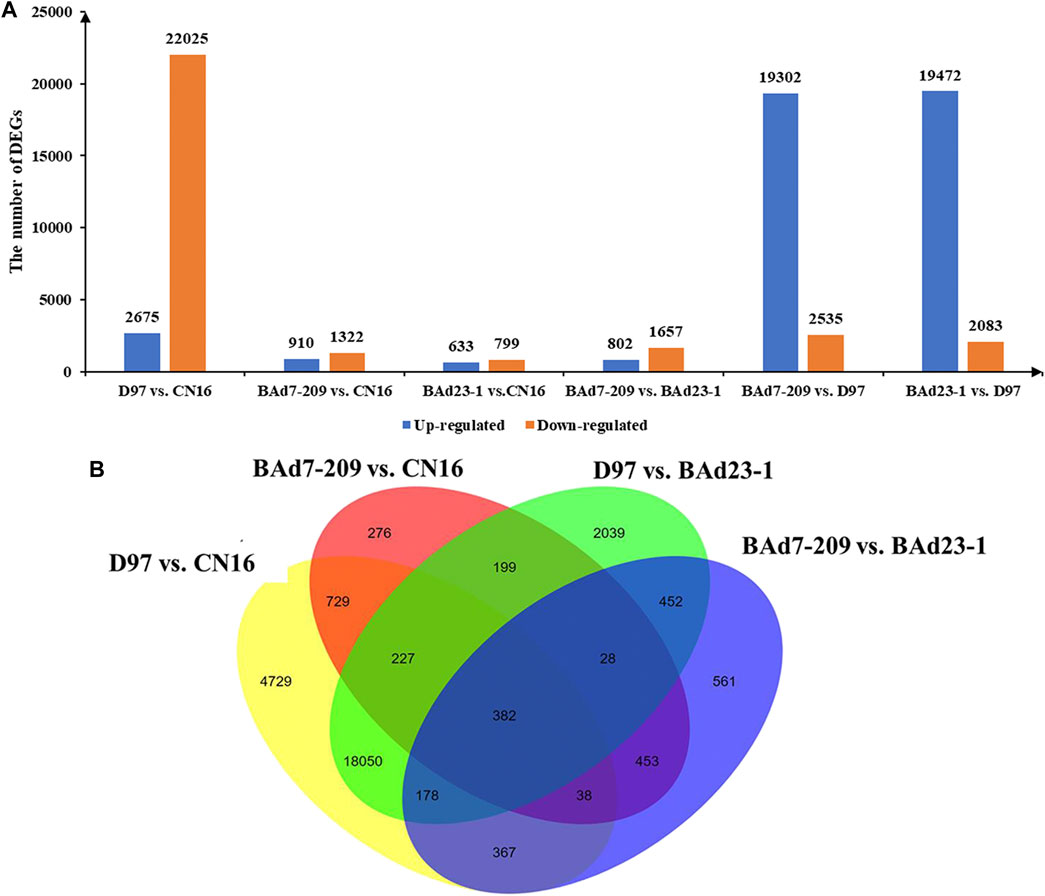
FIGURE 3. The histograms and Venn diagrams of DEGs in different comparisons. (A) Up- and downregulated DEGs in BAd7-209 and BAd23-1 and their parents. (B) The Venn diagram of DEGs in the high-GPC (D97 + BAd7-209) vs. low-GPC (CN16 + BAd23-1) groups.
All genes were further analyzed with the STEM software (v1.3.11) (Ernst and Bar-Joseph, 2006) to obtain the temporal expression patterns. Nineteen expression profiles were clustered, and seven profiles (9, 10, 0, 19, 4, 7, and 6) were regarded as significantly changed (p ≤ 0.05) (Figure 5A). The profile 19 had similar tendency to the GPC in advanced lines and the parents (Figure 5B), indicating the positive role in regulation of grain protein accumulation.
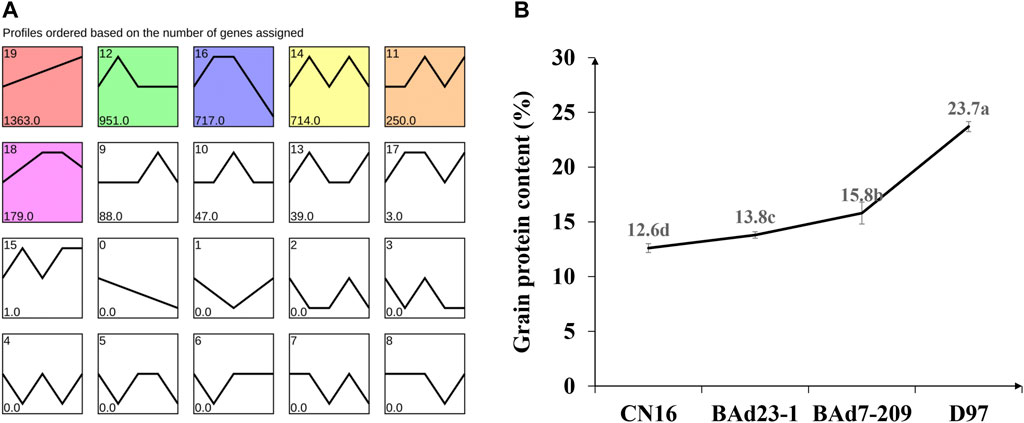
FIGURE 5. Temporal expression patterns of DEGs (A) and grain protein content in CN16, BAd23-1, BAd7-209, and D97 (B). Small letters a, b, c, and d indicate the significant difference at 0.05 level with Tukey’s two-way test.
The Functional Annotation of DEGs
A total of 459 DEGs in profile 19 were subjected to GO and KEGG analyses. On the basis of GO terms for those genes, three categories can be classified: biological process, cellular component, and molecular function. The GO term metabolic process (GO: 0008152, 172 DEGs), cellular process (GO: 0009987, 131 DEGs), and single-organism process (GO: 0044699, 93 DEGs) were highly enriched in biological process; those of cell (GO: 0005623, 137 DEGs), cell part (GO: 0044464, 136 DEGs), and organelle (GO: 0043226, 105 DEGs) were dominant in cellular component; and those of binding (GO: 0005488, 138 DEGs), catalytic activity (GO: 0003824, 145 DEGs), and nutrient reservoir activity (GO: 0045735, 19 DEGs) were primary in molecular function (Figure 6A, Supplementary Table S4).
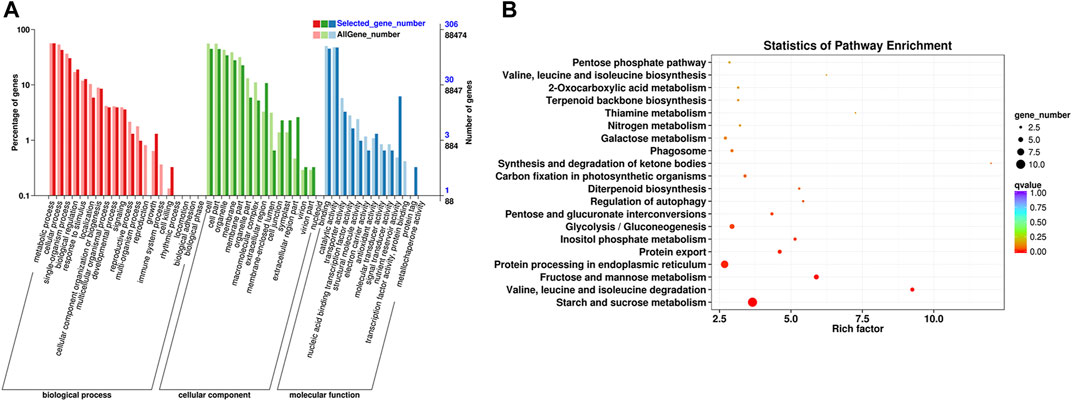
FIGURE 6. Functional annotation of the DEGs based on GO and KEGG pathway enrichment analyses. (A) GO classification of the DEGs. (B) KEGG classification of the DEGs.
KEGG pathway enrichment analysis showed that starch and sucrose metabolism (ko00500, 10 DEGs), protein processing in ER (ko04141, eight DEGs), fructose and mannose metabolism (ko00051, five DEGs), glycolysis/gluconeogenesis (ko00010, five DEGs), protein export (ko03060, four DEGs), inositol phosphate metabolism (ko00562, three DEGs), pentose and glucuronate interconversions (ko00040, three DEGs), valine, leucine, and isoleucine degradation (ko002800, one DEG) were significantly enriched pathways (Figure 6B, Supplementary Table S5). Eight DEGs in pathway of protein processing in ER are related to protein transport protein Sec61 (Sec61, 3 genes), heat shock protein (HSP, 3 genes), SKP1-like protein (SKP1, 1 gene), and E3 ubiquitin-protein ligase RMA3 (RMA3, 1 gene). Four genes in pathway of protein export are related to Sec61 (3 genes) and signal recognition particle 54 kDa protein 2-like (SRP, 1 gene).
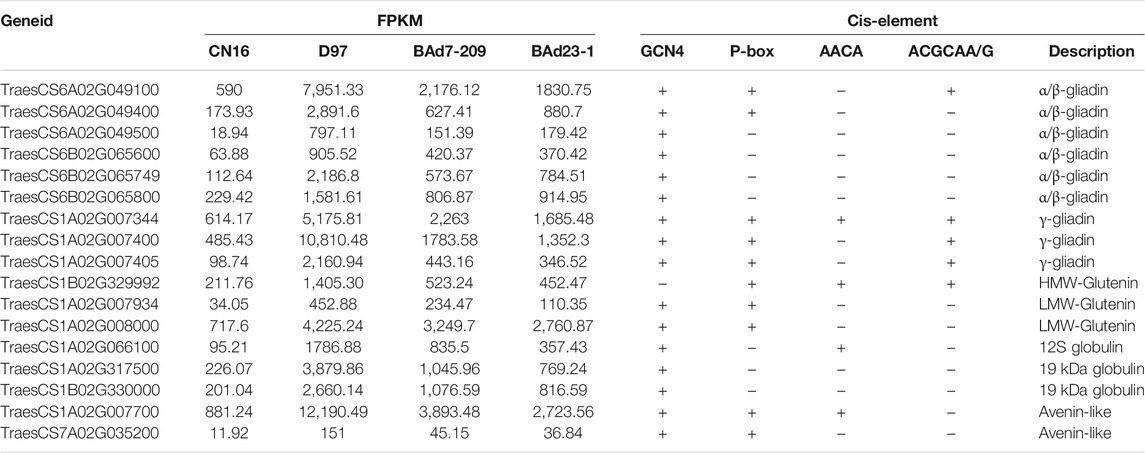
Characterization of DEGs Encoded Gluten and TFs
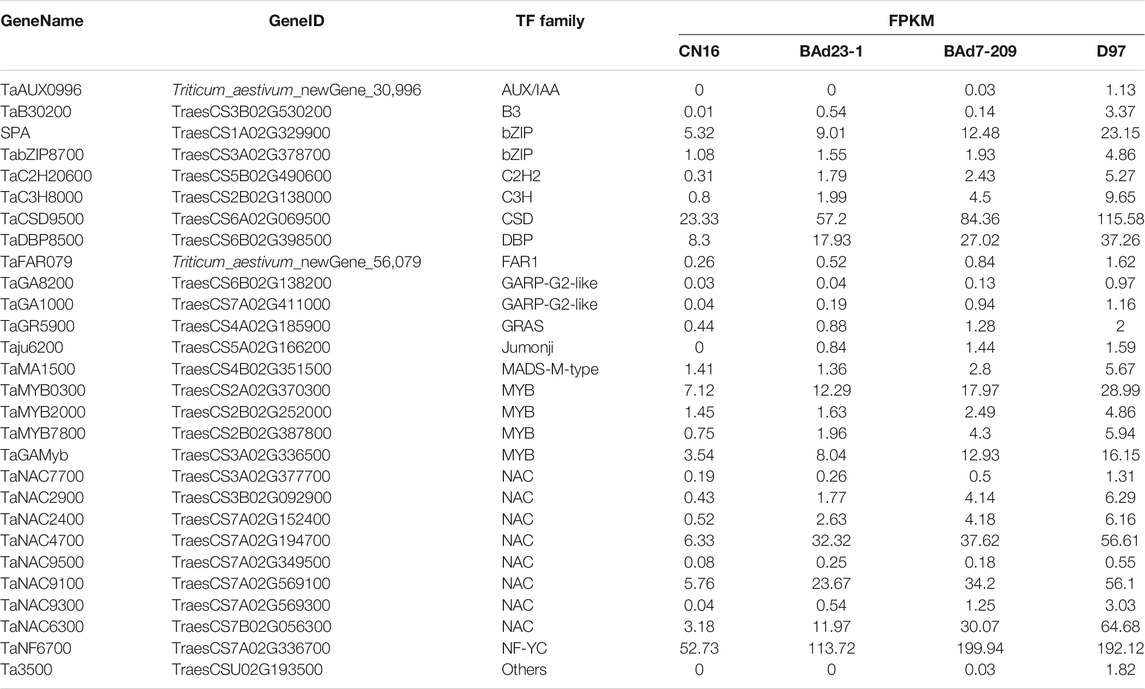
Twelve out of 459 genes in profile 19 were related to α/β-gliadins (six genes), γ-gliadins (three genes), LMW-GSs (two genes), and HWM-GSs (one gene) (Table 3). All these genes were upregulated in the high-GPC group compared to the low-GPC group. Twenty-eight genes belonging to 16 TF families, such as NAC, MYB, GARP-G2-like, bZIP, and C2H2, were identified (Table 4). To understand the expression patterns, the expression levels of TFs in different tissues sampled at different time points were retrieved from public available RNA-seq data of IWGSC (Thomas et al., 2014). We found that seven genes (e.g., TabZIP8700, TaC2H20600, and TaMYB0300) were expressed in different tissues at different time points. Ten genes (e.g., SPA, TaC3H8000, TaGAMyb, and TaNAC019) were only expressed in grains. Two genes (TaMYB7800 and TaNAC9500) were root specific. The others were not detected in all tissues (Supplementary Table S6).
Discussion
GPC is an important quality trait in common wheat. The wild emmer gene GPC-B1 positively impacts protein, Zn, and Fe in wheat grain (Uauy et al., 2006). In the current study, two sister wheat lines BAd7-209 and BAd23-1 derived from wild emmer showed contrasting GPC, while both of them did not contain the Gpc-B1. Processing quality parameters of BAd7-209 were significantly higher than those of BAd23-1. These two lines had genetic background of common wheat and introgression segments from wild emmer D97. Our results indicate the presence of other wild emmer gene(s) that contributed to high GPC in BAd7-209.
Previous reports revealed that the components and expression levels of glutenins and gliadins could affect GPC (Xi and Zheng, 2011; Jiang et al., 2017; Wang et al., 2018; Xiang et al., 2019; Shen et al., 2020). In this study, similar glutenins subunits were identified in BAd7-209 and BAd23-1, whereas BAd7-209 had significant higher amount of glutenins and gliadins than BAd23-1. Nineteen DEGs including gliadin (nine genes), glutenin (three genes), globulin (three genes), avenin-like (three genes), and serpin (two genes) were enriched in the GO term nutrient reservoir activity (GO: 0045735). These genes were upregulated in the high GPC (D97 + BAd7-209) vs. the low-GPC (CN16 + BAd23-1) groups. Previous reports revealed that overexpression gliadin and glutenin genes could increase GPC in wheat (Guo et al., 2015; Li et al., 2019) and rice (Cho et al., 2019). Serpin genes were positively associated with wheat grain development (Benbow et al., 2019) and seed germination (Dong et al., 2015). The results in this study together with previous reports (Jiang et al., 2017; Wang et al., 2018; Xiang et al., 2019; Gong et al., 2021) show that the GPC is closely associated with the expression levels of glutenin and gliadin genes in wheat.
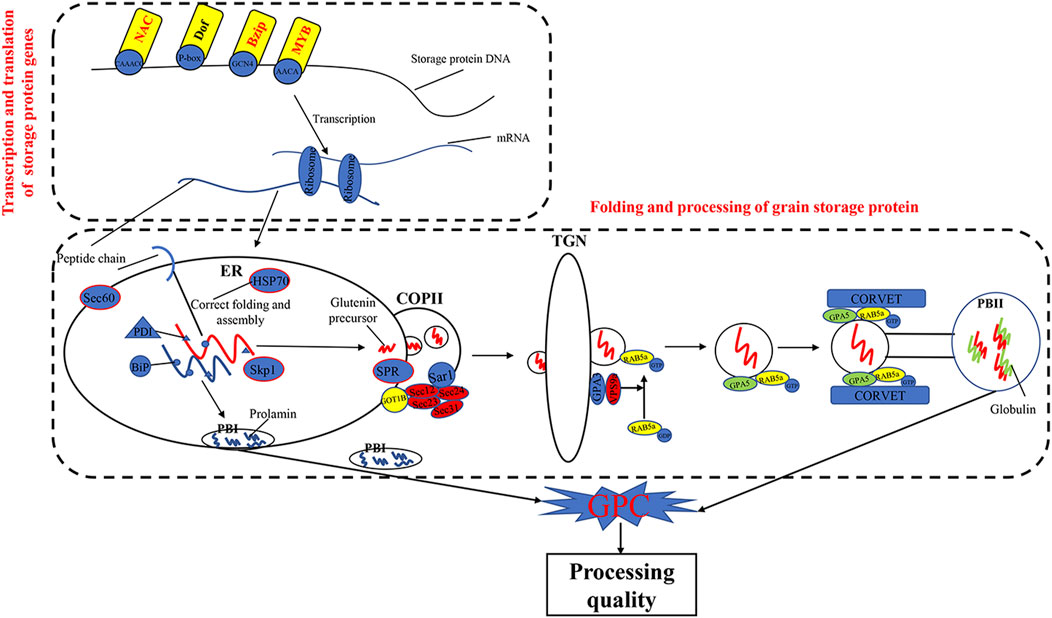
The KEGG pathways of processing in ER (ko04141) and protein export (ko03060) were previously reported to play important roles in the folding and maturation processing of grain storage protein in cereals (Crofts et al., 1998; Foresti et al., 2003; Strasser, 2018; Yu et al., 2018; Yu et al., 2020; Wang et al., 2016; Zhu et al., 2014) (Figure 7). In the present study, we found that DEGs, such as HSP, Sec61, SKP1, RMA3, and SRP, were significantly enriched in the two pathways (ko04141 and ko03060). Previous studies have shown that Hsp70 (Dupont et al., 1998) was a key chaperone for the processing and polymerization processes of grain storage proteins in ER. The Sec proteins (e.g., Sec13, Sec23, Sec24, and Sec31) were involved in transferring proteins from the ER to the Golgi apparatus (D'Arcangelo et al., 2013). SKP1 protein is a key member of the SCF (SKP–cullin–F-box protein) E3 ligase complex and mediates the regulation of plant ABA sensitivity (Hu et al., 2013; Rao et al., 2018). In wheat plants, ABA treatment could accelerate nitrogen remobilization from vegetative organs to grains (Saeed, 2012; Xie et al., 2004). In addition, the SRP proteins are essential for protein translocation across the ER (Walter and Blobel, 1982). These results demonstrated that the pathways of protein processing in ER and protein export and related DEGs are important in wheat grain protein accumulation.
TFs were involved in transcriptional regulation of gluten genes through binding their cis-elements in promoters (Albani et al., 1997; Xi and Zheng, 2011). For example, the bZIP TFs could bind GCN4 motifs of gliadin and glutenin genes in maize (O2) (Schmidt et al., 1992; Vicente-Carbajosa et al., 1998). The Dof TFs (PBF) could activate α-gliadin and Glu-1 genes in wheat (Zhu et al., 2018) by binding P-box elements. MYB TFs were specifically bound to the AACA motifs of gluten genes in rice (OsMYB5) (Suzuki et al., 1998), barley (HvGAMYB, HvMYBS2, and HvMCB1) (Gubler et al., 1995; Diaz et al., 2002), and wheat (TaGAMyb) (Guo et al., 2015). The TaNAC019 activated the expression of HMW-GS gene by binding to the [AT]NNNNNNNNNNN [ATC][CG]A [CA]GN [ACT]A motif in the promoter region (Gao et al., 2021). In the present study, we have identified 28 TFs, such as NAC, MYB, GARP-G2-like, bZIP, and C2H2, from the high-GPC and low-GPC groups. Some TFs (e.g., TaC3H8000, TaNAC2400, TaNAC4700, and TaNF6700) were specifically expressed in grains. In addition, we have identified either NAC (ACGCAA/G), MYB (AACA), bZIP (GCN4), or Dof (P-box) motifs in the promoters of gluten genes that were differentially expressed in the high-GPC vs. low-GPC groups. Taken together, our results indicate that some TFs are involved in regulation of gluten and gliadin genes that contributed to the GPC accumulation in BAd7-209 (Figure 7).
Conclusion
In the present study, we have characterized the agronomic, cytological, grain protein characteristics, and transcriptomic profile of two advanced wheat lines (BAd7-209 and BAd23) from a cross between CN16 and D97. We found that BAd7-209 and BAd23 had genetic background of common wheat and introgression segments from wild emmer D97. The two advanced lines had contrasting GPC, while both absence of the functional GPC-B1. BAd7-209 had superior processing quality parameters and higher amount of glutenins and gliadins than those of BAd23-1, while their glutenin and gliadin subunits were similar. RNA-seq revealed that the contrasting GPC in BAd7-209 and BAd23-1 may be closely associated with the expression levels of glutenin and gliadin genes which regulated by TFs. The protein processing in ER and protein export pathways and related DEGs are important in wheat grain protein accumulation.
Data Availability Statement
The original contributions presented in the study are publicly available in NCBI under accession number PRJNA777016.
Author Contributions
FG: formal analysis and writing—original draft preparation. TQ: investigation. YLu: investigation. JL: investigation. XZ: investigation. JH: investigation. YLi: investigation. YZ: supervision. DL: supervision. LH: writing and editing. BW: conceptualization, project administration, funding acquisition.
Funding
This work was financially supported by the Key Research and Development Program of Sichuan Province, China (2021YFYZ0002) and the International Cooperation Program of Science & Technology Department of Sichuan Province (2021YFH0110).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.804481/full#supplementary-material
References
Albani, D., Hammond-Kosack, M. C., Smith, C., Conlan, S., Colot, V., Holdsworth, M., et al. (1997). The Wheat Transcriptional Activator SPA: a Seed-specific bZIP Protein that Recognizes the GCN4-like Motif in the Bifactorial Endosperm Box of Prolamin Genes. Plant Cell 9, 171–184. doi:10.1105/tpc.9.2.171
Benbow, H. R., Jermiin, L. S., and Doohan, F. M. (2019). Serpins: Genome-Wide Characterisation and Expression Analysis of the Serine Protease Inhibitor Family In Triticum aestivum. G3. 9, 2709–2722. doi:10.1534/g3.119.400444
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Cantu, D., Pearce, S. P., Distelfeld, A., Christiansen, M. W., Uauy, C., Akhunov, E., et al. (2011). Effect of the Down-Regulation of the High Grain Protein Content (GPC) Genes on the Wheat Transcriptome during Monocarpic Senescence. BMC Genomics 12, 492. doi:10.1186/1471-2164-12-492
Chen, X.-y., Song, G.-q., Zhang, S.-j., Li, Y.-l., Gao, J., Shahidul, I., et al. (2017). The Allelic Distribution and Variation Analysis of the NAM-B1 Gene in Chinese Wheat Cultivars. J. Integr. Agric. 16, 1294–1303. doi:10.1016/S2095-3119(16)61459-4
Cho, K., Jo, Y. M., Lim, S. H., Kim, J. Y., Han, O., and Lee, J. Y. (2019). Overexpressing Wheat Low-Molecular-Weight Glutenin Subunits in Rice (Oryza Sativa L. Japonica Cv. Koami) Seeds. Biotech. 9, 49. 3. doi:10.1007/s13205-019-1579-x
Crofts, A. J., Leborgne-Castel, N., Pesca, M., Vitale, A., and Denecke, J. (1998). BiP and Calreticulin Form an Abundant Complex that Is Independent of Endoplasmic Reticulum Stress. The Plant Cell 10, 813–824. doi:10.1105/tpc.10.5.813
D'Arcangelo, J. G., Stahmer, K. R., and Miller, E. A. (2013). Vesicle-mediated export from the ER: COPII Coat Function and Regulation. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1833, 2464–2472. doi:10.1016/j.bbamcr.2013.02.003
Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Martinez, M., Isabel-La Moneda, I., Carbonero, P., et al. (2002). The GAMYB Protein from Barley Interacts with the DOF Transcription Factor BPBF and Activates Endosperm-specific Genes during Seed Development. Plant J. 29, 453–464. doi:10.1046/j.0960-7412.2001.01230.x
Dong, K., Zhen, S., Cheng, Z., Hui, C., Ge, P., and Yan, Y. (2015). Proteomic Analysis Reveals Key Proteins and Phosphoproteins Upon Seed Germination of Wheat (Triticum aestivum L.). Front. Plant Sci., 6, 1017. doi:10.3389/fpls.2015.01017
Duan, S. E., and Zhao, W. M. (2004). Rapid Separation and SDS-PAGE Analysis of Wheat Glutenin Subunits. J. Shaanxi Normal Univ. 32, 77–79.
Dupont, F. M., Hurkman, W. J., Tanaka, C. K., and Chan, R. (1998). BiP, HSP70, NDK and PDI in Wheat Endosperm. I. Accumulation of mRNA and Protein during Grain Development. Physiologia Plantarum 103, 70–79. doi:10.1034/j.1399-3054.1998.1030109.x
Elowitz, M. B., Levine, A. J., Siggia, E. D., and Swain, P. S. (2002). Stochastic Gene Expression in a Single Cell. Science 297, 1183–1186. doi:10.1126/science.1070919
Ernst, J., and Bar-Joseph, Z. (2006). STEM: A Tool for the Analysis of Short Time Series Gene Expression Data. BMC Bioinform. 7, 191. doi:10.1186/1471-2105-7-191
Fatiukha, A., Lupo, I., Lidzbarsky, G., Klymiuk, V., Korol, A. B., Pozniak, C., et al. (2019). Grain Protein Content QTLs Identified in a Durum × Wild Emmer Wheat Mapping Population Tested in Five Environments. Theor. Appl. Genet. 133, 119–131. doi:10.1101/601773
Foresti, O., Frigerio, L., Holkeri, H., de Virgilio, M., Vavassori, S., and Vitale, A. (2003). A Phaseolin Domain Involved Directly in Trimer Assembly Is a Determinant for Binding by the Chaperone BiP. The Plant Cell Online 15, 2464–2475. doi:10.2307/387197710.1105/tpc.013052
Furtado, A., Bundock, P. C., Banks, P. M., Fox, G., Yin, X., and Henry, R. J. (2015). A Novel Highly Differentially Expressed Gene in Wheat Endosperm Associated with Bread Quality. Sci. Rep. 5, 10446. doi:10.1038/srep10446
Gao, Y., An, K., Guo, W., Chen, Y., Zhang, R., Zhang, X., et al. (2021). The Endosperm-specific Transcription Factor TaNAC019 Regulates Glutenin and Starch Accumulation and its Elite Allele Improves Wheat Grain Quality. The Plant Cell 33, 603–622. doi:10.1093/plcell/koaa040
Gong, F., Huang, L., Qi, T., Tang, G., Liu, J., Xiang, L., et al. (2021). Comparative Analysis of Developing Grain Transcriptome Reveals Candidate Genes and Pathways Improving GPC in Wheat Lines Derived from Wild Emmer. J. Appl. Genet. 62, 17–25. doi:10.1007/s13353-020-00588-y
Gonzalez-Hernandez, J. L., Elias, E. M., and Kianian, S. F. (2004). Mapping Genes for Grain Protein Concentration and Grain Yield on Chromosome 5B of Triticum Turgidum (L.) Var. Dicoccoides. Euphytica 139, 217–225. doi:10.1007/s10681-004-3157-4
Groos, C., Robert, N., Bervas, E., and Charmet, G. (2003). Genetic Analysis of Grain Protein-Content, Grain Yield and Thousand-Kernel Weight in Bread Wheat. Theor. Appl. Genet. 106, 1032–1040. doi:10.1007/s00122-002-1111-1
Gubler, F., Kalla, R., Roberts, J. K., and Jacobsen, J. V. (1995). Gibberellin-regulated Expression of a Myb Gene in Barley Aleurone Cells: Evidence for Myb Transactivation of a High-pI Alpha-Amylase Gene Promoter. Plant Cell 7, 1879–1891. doi:10.2307/387019510.1105/tpc.7.11.1879
Guo, W., Yang, H., Liu, Y., Gao, Y., Ni, Z., Peng, H., et al. (2015). The Wheat Transcription Factor TaGAMyb Recruits Histone Acetyltransferase and Activates the Expression of a High-Molecular-Weight Glutenin Subunit Gene. Plant J. 84, 347–359. doi:10.1111/tpj.13003
Hao, M., Zhang, L., Zhao, L., Dai, S., Li, A., Yang, W., et al. (2019). A Breeding Strategy Targeting the Secondary Gene Pool of Bread Wheat: Introgression from a Synthetic Hexaploid Wheat. Theor. Appl. Genet. 132, 2285–2294. doi:10.1007/s00122-019-03354-9
Hu, X.-g., Wu, B.-h., Yan, Z.-h., Liu, D.-c., Wei, Y.-m., and Zheng, Y.-l. (2010). Characterization of a Novel 1Ay Gene and its Expression Protein in Triticum Urartu. Agric. Sci. China 9, 1543–1552. doi:10.1016/S1671-2927(09)60250-9
Hu, D. L., Chen, Q. Z., Zhang, C. J., Wang, Y., Zhang, B. J., and Tang, C. M. (2013). Identification of Cotton Skp1-Like Gene Ghskp1 and its Function in Seed Germination and Taproot Growth in Tobacco. Canadian J. Plant Sci. 93, 817–825. doi:10.1139/CJPS2012-312
Hung, J. H., and Weng, Z. (2016). Designing Polymerase Chain Reaction Primers Using Primer3Plus. Cold Spring Harbor Protoc. pdb, prot093096. doi:10.1101/pdb.prot093096
Jiang, H., and Wong, W. H. (2009). Statistical Inferences for Isoform Expression in RNA-Seq. Bioinformatics 25, 1026–1032. doi:10.1093/bioinformatics/btp113
Jiang, Z.-L., Wu, B.-H., Wang, Z.-Z., Hu, J.-L., Yuan, J., Chen, H.-L., et al. (2017). Enriching Novel Glu-Ax Alleles and Significantly Strengthening Gluten Properties of Common Wheat through Wide Hybridization with Wild Emmer. J. Cereal Sci. 76, 271–279. doi:10.1016/j.jcs.2017.04.018
Joppa, L. R., Du, C., Hart, G. E., and Hareland, G. A. (1997). Mapping Gene(s) for Grain Protein in Tetraploid Wheat ( Triticum Turgidum L.) Using a Population of Recombinant Inbred Chromosome Lines. Crop Sci. 37, 1586–1589. doi:10.2135/cropsci1997.0011183X003700050030x
Kanehisa, M., Goto, S., Kawashima, S., and Okuno, Y. (2004). The KEGG Resource for Deciphering the Genome. Nucleic Acids Res. 32, 277D–280D. doi:10.1093/nar/gkh063
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Krugman, T., Nevo, E., Beharav, A., Sela, H., and Fahima, T. (2018). The Institute of Evolution Wild Cereal Gene Bank at the University of Haifa. Isr. J. Plant Sci. 65, 129–146. doi:10.1163/22238980-00001065
Leng, N., Dawson, J. A., Thomson, J. A., Ruotti, V., Rissman, A. I., Smits, B. M. G., et al. (2013). EBSeq: an Empirical Bayes Hierarchical Model for Inference in RNA-Seq Experiments. Bioinformatics 29, 1035–1043. doi:10.1093/bioinformatics/btt087
Liu, J., Huang, L., Wang, C., Liu, Y., Yan, Z., Wang, Z., et al. (2019). Genome-wide Association Study Reveals Novel Genomic Regions Associated with High Grain Protein Content in Wheat Lines Derived from Wild Emmer Wheat. Front. Plant Sci. 10, 464. doi:10.3389/fpls.2019.00464
Onodera, Y., Suzuki, A., Wu, C.-Y., Washida, H., and Takaiwa, F. (2001). A Rice Functional Transcriptional Activator, RISBZ1, Responsible for Endosperm-Specific Expression of Storage Protein Genes Through GCN4 Motif. J. Biol. Chem. 276, 14139–14152. doi:10.1074/jbc.M007405200
Rangan, P., Furtado, A., and Henry, R. J. (2017). The Transcriptome of the Developing Grain: a Resource for Understanding Seed Development and the Molecular Control of the Functional and Nutritional Properties of Wheat. BMC Genomics 18, 766. doi:10.1186/s12864-017-4154-z
Rao, V., Petla, B. P., Verma, P., Salvi, P., and Majee, M. (2018). Arabidopsis Skp1-Like Protein13 (ASK13) Positively Regulates Seed Germination and Seedling Growth Under Abiotic Stress. J. Exp. Botany. 69, 3899–3915. doi:10.1093/jxb/ery191Saeedipour
Rombauts, S., Dehais, P., VanMontagu, M., and Rouze, P. (1999). Plantcare, a Plant Cis-Acting Regulatory Element Database. Nucleic Acids Res. 27, 295–296. doi:10.1093/nar/27.1.295
Saeed, S. (2012). Relationship of Endogenous Aba and Iaa to Accumulation of Grain Protein and Starch in two Winter Wheat Cultivars Under Post-Anthesis Water Deficit. J. Agricult. Sci. 4, 147–156. doi:10.5539/jas.v4n2p147
Schmidt, R. J., Ketudat, M., Aukerman, M. J., and Hoschek, G. (1992). Opaque-2 Is a Transcriptional Activator that Recognizes a Specific Target Site in 22-kD Zein Genes. Plant Cell 4, 689–700. doi:10.1105/tpc.4.6.689
Shen, L., Luo, G., Song, Y., Xu, J., Ji, J., Zhang, C., et al. (2021). A Novel NAC Family Transcription Factor SPR Suppresses Seed Storage Protein Synthesis in Wheat. Plant Biotechnol. J. 19, 992–1007. doi:10.1111/pbi.13524
Shewry, P. R., and Halford, N. G. (2002). Cereal Seed Storage Proteins: Structures, Properties and Role in Grain Utilization. J. Exp. Bot. 53, 947–958. doi:10.1093/jexbot/53.370.947
Shewry, P. R., Tatham, A. S., Barro, F., Barcelo, P., and Lazzeri, P. (1995). Biotechnology of Breadmaking: Unraveling and Manipulating the Multi-Protein Gluten Complex. Nat. Biotechnol. 13, 1185–1190. doi:10.1038/nbt1195-1185
Shewry, P. R., Underwood, C., Wan, Y., Lovegrove, A., Bhandari, D., Toole, G., et al. (2009). Storage Product Synthesis and Accumulation in Developing Grains of Wheat. J. Cereal Sci. 50, 106–112. doi:10.1016/j.jcs.2009.03.009
Strasser, R. (2018). Protein Quality Control in the Endoplasmic Reticulum of Plants. Annu. Rev. Plant Biol. 69, 147–172. doi:10.1146/annurev-arplant-042817-040331
Suzuki, A., Wu, C.-Y., Washida, H., and Takaiwa, F. (1998). Rice MYB Protein OSMYB5 Specifically Binds to the AACA Motif Conserved Among Promoters of Genes for Storage Protein Glutelin. Plant Cel Physiol. 39, 555–559. doi:10.1093/oxfordjournals.pcp.a029404
Takemoto, Y., Coughlan, S. J., Okita, T. W., Satoh, H., Ogawa, M., and Kumamaru, T. (2002). The rice Mutant Esp2 Greatly Accumulates the Glutelin Precursor and Deletes the Protein Disulfide Isomerase. Plant Physiol. 128, 1212–1222. doi:10.1104/pp.010624
Thomas, M., Lise, H., Manuel, S., and Matthias, P. (2014). A Chromosome-Based Draft Sequence of the Hexaploid Bread Wheat (Triticum aestivum) Genome. Science 345, 1251788. doi:10.1126/science.1251788
Uauy, C., Brevis, J. C., and Dubcovsky, J. (2006). The High Grain Protein Content Gene Gpc-B1 Accelerates Senescence and Has Pleiotropic Effects on Protein Content in Wheat. J. Exp. Bot. 57, 2785–2794. doi:10.1093/jxb/erl047
Vicente-Carbajosa, J., Onate, L., Lara, P., Diaz, I., and Carbonero, P. (1998). Barley BLZ1: a bZIP Transcriptional Activator that Interacts with Endosperm‐specific Gene Promoters. Plant J. 13, 629–640. doi:10.1111/j.1365-313X.1998.00068.x
Wan, Y., Liu, K., Wang, D., and Shewry, P. R. (2000). High-molecular-weight Glutenin Subunits in the Cylindropyrum and Vertebrata Section of the Aegilops Genus and Identification of Subunits Related to Those Encoded by the Dx Alleles of Common Wheat. Theor. Appl. Genet. 101, 879–884. doi:10.1007/s001220051556
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R Package for Identifying Differentially Expressed Genes from RNA-Seq Data. Bioinformatics 26, 136–138. doi:10.1093/bioinformatics/btp612
Wang, Y., Liu, F., Ren, Y., Wang, Y., Liu, X., Long, W., et al. (2016). GOLGI TRANSPORT 1B Regulates Protein export from the Endoplasmic Reticulum in rice Endosperm Cells. Plant Cell 28, 2850–2865. doi:10.1105/tpc.16.00717
Wang, Y. F., Huang, L., Luo, W., Jin, Y. R., Gong, F. Y., He, J. S., et al. (2020). Transcriptome Analysis Provides Insights into the Mechanisms Underlying Wheat Cultivar Shumai126 Responding to Stripe Rust. Gene. 768, 145290. doi:10.1016/j.gene.2020.145290
Wang, Z., Huang, L., Wu, B., Hu, J., Jiang, Z., Qi, P., et al. (2018). Characterization of an Integrated Active Glu-1ay Allele in Common Wheat from Wild Emmer and its Potential Role in Flour Improvement. Int. J. Mol. Sci. 19, 923. doi:10.3390/ijms19040923
Walter, , and Blobel, . (1982). Signal Recognition Particle Contains A 7S RNA Essential for Protein Translocation Across the Endoplasmic Reticulum. Nature. 299, 691–698. doi:10.1038/299691a0
Xi, D.-M., and Zheng, C.-C. (2011). Transcriptional Regulation of Seed Storage Protein Genes inArabidopsisand Cereals. Seed Sci. Res. 21, 247–254. doi:10.1017/S0960258511000237
Xiang, L., Huang, L., Gong, F., Liu, J., Wang, Y., Jin, Y., et al. (2019). Enriching LMW-GS Alleles and Strengthening Gluten Properties of Common Wheat through Wide Hybridization with Wild Emmer. 3 Biotech. 9, 355. doi:10.1007/s13205-019-1887-1
Xie, Z., Dong, J., Dai, T., Qi, J., and Cao, W. (2004). Effects of Exogenous Aba and Cytokinin on Leaf Photosynthesis and Grain Protein Accumulation in Wheat Ears Cultured In Vitro. Plant Growth Regul. 44, 25–32. doi:10.1007/s10725-004-1880-4
Yu, Z., Islam, S., She, M., Diepeveen, D., Zhang, Y., Tang, G., et al. (2018). Wheat Grain Protein Accumulation and Polymerization Mechanisms Driven by Nitrogen Fertilization. Plant J. 96, 1160–1177. doi:10.111/tpj.1409610.1111/tpj.14096
Zhang, L.-Q., Yen, Y., Zheng, Y.-L., and Liu, D.-C. (2007). Meiotic Restriction in Emmer Wheat Is Controlled by One or More Nuclear Genes that Continue to Function in Derived Lines. Sex. Plant Reprod. 20, 159–166. doi:10.1007/s00497-007-0052-x
Zhang, Z., Dong, J., Ji, C., Wu, Y., and Messing, J. (2019). NAC-type Transcription Factors Regulate Accumulation of Starch and Protein in maize Seeds. Proc. Natl. Acad. Sci. USA 116, 11223–11228. doi:10.1073/pnas.1904995116
Zheng, Y., Jiao, C., Sun, H., Rosli, H. G., Pombo, M. A., Zhang, P., et al. (2016). iTAK: a Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 9, 1667–1670. doi:10.1016/j.molp.2016.09.014
Zhu, J., Hao, P., Chen, G., Han, C., Li, X., Zeller, F. J., et al. (2014). Molecular Cloning, Phylogenetic Analysis, and Expression Profiling of Endoplasmic Reticulum Molecular Chaperone BiP Genes from Bread Wheat (Triticum aestivum L.). BMC Plant Biol. 14, 260. doi:10.1186/s12870-014-0260-0
Keywords: wild emmer wheat, GPC, transcriptome, processing quality, gluten
Citation: Gong F, Qi T, Zhang T, Lu Y, Liu J, Zhong X, He J, Li Y, Zheng Y, Liu D, Huang L and Wu B (2022) Comparison of the Agronomic, Cytological, Grain Protein Characteristics, as Well as Transcriptomic Profile of Two Wheat Lines Derived From Wild Emmer. Front. Genet. 12:804481. doi: 10.3389/fgene.2021.804481
Received: 29 October 2021; Accepted: 13 December 2021;
Published: 28 January 2022.
Edited by:
Suxu Tan, Michigan State University, United StatesReviewed by:
Xiaojun Nie, Northwest A and F University, ChinaSatinder Kaur, Punjab Agricultural University, India
Copyright © 2022 Gong, Qi, Zhang, Lu, Liu, Zhong, He, Li, Zheng, Liu, Huang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Huang, bGh1YW5nQHNpY2F1LmVkdS5jbg==; Bihua Wu, d3ViaWh1YTIwMTdAMTI2LmNvbQ==
 Fangyi Gong
Fangyi Gong Tiangang Qi1
Tiangang Qi1 Youliang Zheng
Youliang Zheng Lin Huang
Lin Huang