
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 11 January 2022
Sec. Neurogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.796812
Objective: The vulnerability of atherosclerotic plaques is among the leading cause of ischemic stroke. High wall shear stress (WSS) promotes the instability of atherosclerotic plaques by directly imparting mechanical stimuli, but the specific mechanisms remain unclear. We speculate that modulation of mechanosensitive genes may play a vital role in accelerating the development of plaques. The purpose of this study was to find mechanosensitive genes in vascular endothelial cells (ECs) through combining microarray data with bioinformatics technology and further explore the underlying dynamics–related mechanisms that cause the progression and destabilization of atherosclerotic plaques.
Methods: Microarray data sets for human vascular ECs under high and normal WSS were retrieved from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) were identified through the R language. The performance of enrichment analysis and protein–protein interaction (PPI) network presented the biological function and signaling pathways of the DEGs. Hub genes were identified based on the PPI network and validated by GEO data sets. Predicted transcription factor (TF) genes and miRNAs interaction with potential mechanosensitive genes were identified by NetworkAnalyst.
Results: A total of 260 DEGs, 121 upregulated and 139 downregulated genes, were screened between high and normal WSS from GSE23289. A total of 10 hub genes and four cluster modules were filtered out based on the PPI network. The enrichment analysis showed that the biological functions of the hub genes were mainly involved in responses to unfolded protein and topologically incorrect protein, and t to endoplasmic reticulum stress. The significant pathways associated with the hub genes were those of protein processing in the endoplasmic reticulum, antigen processing, and presentation. Three out of the 10 hub genes, namely, activated transcription factor 3 (ATF3), heat shock protein family A (Hsp70) member 6 (HSPA6), and dual specificity phosphatase 1 (DUSP1, also known as CL100, HVH1, MKP-1, PTPN10), were verified in GSE13712. The expression of DUSP1 was higher in the senescent cell under high WSS than that of the young cell. The TF–miRNA–mechanosensitive gene coregulatory network was constructed.
Conclusion: In this work, we identified three hub genes, ATF3, HSPA6, and DUSP1, as the potential mechanosensitive genes in the human blood vessels. DUSP1 was confirmed to be associated with the senescence of vascular ECs. Therefore, these three mechanosensitive genes may have emerged as potential novel targets for the prediction and prevention of ischemic stroke. Furthermore, the TF–miRNA–mechanosensitive genes coregulatory network reveals an underlying regulatory mechanism and the pathways to control disease progression.
Atherosclerosis (AS), the underlying cause of ischemic stroke, is the most common cause of morbidity, mortality, and disability in the world, which has become a major global health problem due to the lack of effective methods for screening at an early stage and limited therapeutic strategies. This disease imposes serious economic burden and health toll on the individuals, families, and health care systems indirectly (Hoshino et al., 2018; Geng et al., 2019). Atherosclerosis is a chronic inflammatory disorder of the arterial wall (Kimura et al., 2018). The aggressive control of the several risk factors for atherosclerosis, including smoking, hypertension, diabetes, and hyperlipidemia, or any other form of stroke is still nearly 14.9–24% (Derdeyn et al., 2014). It has been therefore suggested that various additional factors and pathogenic mechanisms may be involved, which urges physicians and biomedical scientists to find more effective approaches to diagnose, prevent, and treat this deadly disease.
The vascular endothelial cells (ECs) line the inner layer of the blood vessels, constituting a barrier that separates the blood from the vessel wall under normal physiological conditions (Pulous et al., 2019). Endothelial dysfunction increases endothelial permeability and stimulates a complex series of reactions such as lipid accumulation, inflammatory responses, neovascularization, unstable plaque progression, and oxidative stress to promote the development and progression of atherosclerosis (Zhang et al., 2015a; Badimon et al., 2017; Wiese et al., 2019). Numerous researches have shown that atherosclerotic lesions preferentially develop in areas of arterial turns or bifurcation where hemodynamic wall shear stress (WSS) is low, oscillatory, or multidirectional and complex, also known as disturbed flow (Malek et al., 1999; Ramírez et al., 2019). The pattern of complex blood flow and WSS in the vessels are thought to be associated with endothelial dysfunction, which may result in changes in endothelial cellular function by directly affecting WSS or regulating the expression of genes of ECs in the different regions of the arterial wall (Casey et al., 2012; Mack et al., 2017). Recent molecular and cellular studies have begun to elucidate that low shear stress acting directly on vascular ECs leads to the formation of atherosclerosis and the development of thin fibrous caps. It promotes the deposition of particulate matter on the arterial wall and the accumulation of plaque, resulting from persistent changes of inflammation, oxidative stress, and proliferation in ECs (Feldman et al., 2002; Chatzizisis et al., 2007; Milkiewicz et al., 2008; Hsu et al., 2019). Conversely, high WSS was not only more commonly associated with a positive remodeling but also induced inflammation and increased oxidative stress by directly acting on the plaques, making the atherosclerotic plaques destabilized and causing embolism (Cicha et al., 2011; Costopoulos et al., 2019; Leng et al., 2019). The vascular endothelial function is regulated by WSS through some shear-sensitive expressions of the genes, such as KLF2/KLF4, NRF2, HIF-1α, NF-κB, etc. (Spruell and Baker, 2013; Wang et al., 2016; Niu et al., 2019). However, the molecular mechanisms involved in this process remain largely unknown. The discovery of novel mechanosensitive genes and their potential functional mechanisms in ECs have great implications in the individualized early diagnosis and precision treatment of ischemic stroke.
Currently, genomic sequencing and microarray technology have been widely applied by numerous laboratories around the world in various biological problems and disease mechanisms to identify new biomarkers. Through bioinformatics analysis, it is possible to obtain tremendous gene expression information and explore disease-associated genes and their biological functions, processes, as well as signaling pathways. Furthermore, bioinformatics analysis plays an important role in molecular genetics and also provides a theoretical basis for “precision medicine.”
In this study, we aimed to perform the differentially expressed profiles associated with the mechanosensitive genes in the human umbilical vein ECs (HUVECs) exposed to normal and high shear stress, followed by gene function and pathway enrichment analyses. After constructing a protein–protein interaction (PPI) network, we identified hub genes related to high WSS in this network and further verified them using another Gene Expression Omnibus (GEO) data set. Then, a TF–miRNA coregulatory network with validated hub genes was established (Figure 1). Finally, this study explored the valuable information for hemodynamics related to hub genes and presented a corresponding discussion for hub genes as a new potentially predicted and protected target for patients with ischemic stroke.
The GEO database is a gene expression database, mainly containing a variety of microarray data and some sequencing data, created and maintained by The National Center for Biotechnology Information (Zhao et al., 2020). The GEO database was used to obtain microarray data of ECs cultured under different WSS for the analysis of ECs-expressed specific mechanosensitive genes involved in generating mechanical force on the vascular wall in our study. The filtering criteria are as follows: first, these data sets we selected for analysis were the gene expression data of HUVECs cultured under normal and relatively high WSS; second, each data set did possess both the above two samples; third, the type of data sets was RNA expression profile by arrays. Finally, two data sets, GSE23289 and GSE13712, were selected as the test and validation sets, respectively. The microarray expression profiling data set GSE23289, generated by White SJ et al., was download from the GEO data sets (https://www.ncbi.nlm.nih.gov/geo/), which was produced on the Illumina GPL6104 platform (Illumina HumanRef-8 v2.0 Expression BeadChips). A total of eight samples were contained in this experiment, consisting of two groups (four subjects per group) with the HUVECs cultured for 24 h on gelatin-coated slides under normal and high WSS (15 dynes/cm2 and 75 dynes/cm2, respectively) on the ECs. The other microarray data set GSE13712 was annotated in the GPL570 platform [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array], which included 12 samples; a comparison of the young and senescent HUVECs under normal and high WSS is given in Table 1.
We downloaded original files of the microarray data sets, GSE23289 and GSE13712, from the GEO database with the GEOquery R package (Davis and Meltzer, 2007). The probes matching multiple genes were removed, as where the same genes corresponding to multiple probes, and the maximum median intensity probe value was taken. After filtering, the data were shown by boxplots to reflect quantile normalization, and principal component analysis (PCA) was used to demonstrate clustering of samples in different groups (Figures 2A–D). The R package “limma” (bioconductor) was performed to analyze the differentially expressed genes (DEGs) between the normal and high WSS groups (Smyth et al., 2005). The DEGs were screening out according to the following two cutoff criteria: adjusted p-value <0.05 and absolute log2 [fold change (FC)] >1 (Rougeot et al., 2019; Gao et al., 2020). We removed the target DEGs that had no annotated gene names. These significant DEGs were visualized in volcano plots and heat maps, which were generated in R v4.0.1 with the ggplot2 package (Gu et al., 2016).
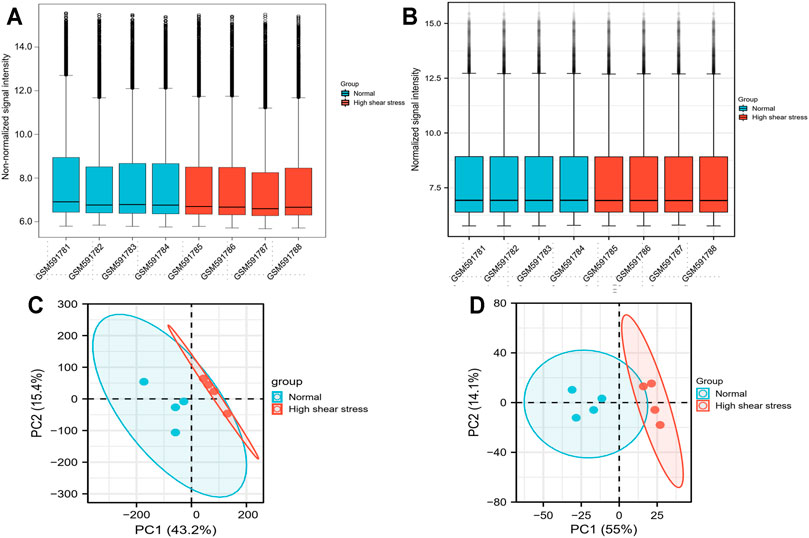
FIGURE 2. Data processing. (A) Boxplot for GSE23289 before quantile normalization. (B) Boxplot for GSE23289 after quantile normalization. (C) The PCA for GSE23289 before quantile normalization. (D) The PCA for GSE23289 after quantile normalization.
To get a better understanding of the biological functions of genes, we used the R package clusterProfiler and GOplot to perform gene ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the DEGs (Yu et al., 2012). GO annotations were gained from the Bioconductor R package org.Hs.eg.db for humans. The GO database consists of a set of terms that describe the properties of genes and gene products. These terms were divided into three main categories: biological process (BP), cellular component, and molecular function (MF). KEGG pathway analysis was used to assign DEGs to specific pathways for constructing networks of molecular interactions, reactions, and their relationships. The DEGs' functional enrichment and their pathway analyses were visualized by the R package ggplot2. The adjusted p-value <0.1 and q-value <0.2 (Benjamini–Hochberg method) was set as a default cutoff threshold.
A PPI network consisting of the significant DEGs were constructed by the online tool (https://string-db.org/)—with an interaction score of >0.4 (default parameters) (Hoyer et al., 2019; Li et al., 2020a; Gao et al., 2020)—the Search Tool for the Retrieval of Interacting Genes (STRING), which is a database to predict the predicted PPIs (Szklarczyk et al., 2018). The Cytoscape software (V3.8.2) was used to create and visualize the PPI network (Shannon et al., 2003). In this PPI network, each node stands for a target gene, and the lines between the nodes represents the associated interactions. The major gene clusters in the PPI network were identified by the plug-in Minimal Common Oncology Data Elements (mCODE) cluster in Cytoscape with the settings of degree cutoff = 2, node score cutoff = 0.2, k-core = 4, and max. depth = 100. The interface cytoHubba was used to explore important nodes in the biological networks. It provided many topological analysis methods; the Maximal Clique Centrality (MCC) algorithm was considered to be the most effective method to find the hub nodes. It had a better performance on the precision of predicting essential proteins from complex interaction networks (Chin et al., 2014; Li et al., 2020b). In our study, we detected the hub genes in this PPI network according to the threshold criteria of MCC calculated by a plug-in of cytoHubba in Cytoscape software.
The GO and KEGG pathway enrichment analyses were performed for hub genes using the R package Clusterprofiler and GOplot. We defined the adjusted p-value of <0.05 and q-value <0.2 (Benjamini–Hochberg method) as the cutoff criteria. The bubble graph for gene functions and the network diagram representing the gene interaction pathways were drawn by the R package ggplot2. Furthermore, the sensitivity and specificity of these hub genes in predictions for EC suffering high shear stress were evaluated by the receiver operator characteristic (ROC) curves and the area under the ROC curve. The pROC R package was used for ROC curve analysis, while the ggplot2 R package was used for data visualization.
Additional publicly available data set GSE13712 was used to further validate the specific expression patterns of such hub genes. Then two-sample t-test was used to compare the differential expression values of the hub genes between ECs lying in the normal and high WSS groups. Finally, a post-analysis was conducted between the young and old EC subgroups under high shear stress to identify the expression of hub genes. The hub gene expression levels between the two subgroups were performed using a t-test, and p-values <0.05 were regarded as statistically significant. The violin plots comparing the expression levels of the selected hub genes in the different groups were depicted by the R package ggplot2. Statistical significance was indicated as follows: *(p < 0.05), **(p < 0.01), and ns (p > 0.05, no significance).
We obtained TF–miRNA co-regulated interaction data from the RegNetwork repository, which contributes to predicting miRNAs and TFs that regulate the hub genes of interest at the transcriptional and post-transcriptional levels (Liu et al., 2015). The NetworkAnalyst platform (https://www.networkanalyst.ca/) was used to identify and visualize the TF genes–miRNAs interactions with selected hub genes using a cutoff degree >1.
The data set GSE23289, which included eight samples divided into two groups (HUVECs were cultured under the normal and the high WSS, respectively), identified and analyzed the DEGs using the R package (limma). As a result, using the cutoff criteria (adjusted p-value <0.05 and |log2FC|>1), compared with the normal WSS groups, a total of 260 DEGs (121 upregulated and 139 downregulated genes) were identified in the high WSS groups, which provided the underlying clues to study atherogenesis and atherosclerotic progression (Supplementary Table 1). These significantly DEGs in subsequence analysis were visualized via volcano plot and heat map. These DEGs with distinct changes (adjusted p-value <0.05 and |log2FC| >2) were marked by their symbols on the volcano plot diagram. Among all DEGs, the top 20 highly expressed genes and the top 20 lowly expressed genes, respectively, were selected to visualize in the heat map (Figures 3A,B).
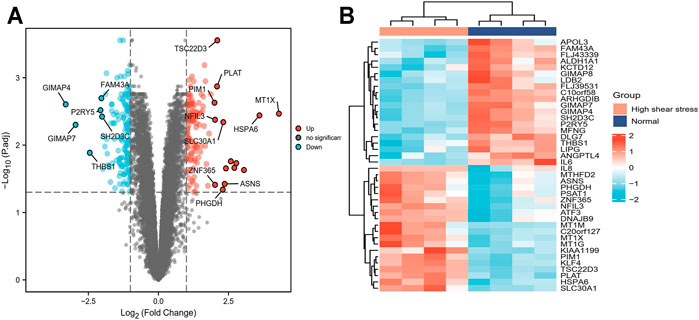
FIGURE 3. Screening of DEGs between normal and high WSS groups in GSE23289. (A) The volcano plot of DEGs, in which the red dots represent upregulated genes, blue dots represent downregulated genes, and grey dots represent genes with no significant difference. The y-axis indicates the −log10 (adjusted p-value), and the x-axis indicates the log2FC. FC is the fold change. (B) The cluster heat map of DEGs with each row corresponding to a gene and each column corresponding to a sample. The color scale ranges from light blue, showing relatively low gene expression levels, and passes through white to red, representing relatively high gene expression levels.
The GO and KEGG functional pathways enrichment analyses of 260 DEGs were performed at the threshold of adjusted p-value <0.1 and q-value <0.2 using the R package clusterProfiler. Overall, we found that 241 GO-BPs, 16 GO-MFs, and 16 KEGG pathways were highly enriched for these DEGs (Supplementary Table 2). The results of the GO terms showed that regulation of myeloid cell differentiation (GO:0045637), myeloid cell differentiation (GO:0030099), and response to unfolded protein (GO:0006986) were significantly enriched processes in the BP category. The apparently enriched entries for the MF category were mitogen-activated protein kinase (MAPK) tyrosine/serine/threonine phosphatase activity (GO:0017017), MAPK phosphatase activity (GO:0033549), and misfolded protein binding (GO:0051787). Additionally, the KEGG pathways enrichment analyses indicated that DEGs were enriched in the MAPK signaling pathway (hsa04010), protein processing in the endoplasmic reticulum (hsa04141), and longevity regulating pathway–multiple species (hsa04213). Finally, we displayed the top 15 GO terms and top 10 KEGG pathways enriched results (Figures 4A,B).
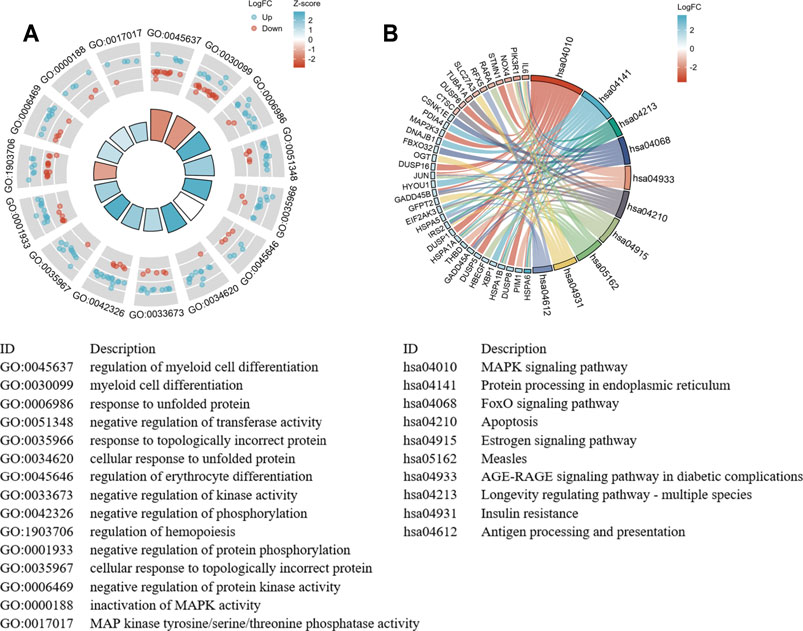
FIGURE 4. The significant enrichment GO terms and KEGG pathways of the DEGs. (A) GO functional enrichment Circos plots of DEGs. Each column of the inner circle corresponds to a GO term. The height represents the adjusted p-value. The higher the column, the smaller the adjusted p-value. The filled color of the column represents the z-score value corresponding to the GO terms. (B) Signaling pathways enrichment chord diagram of DEGs. The left half represents the DEGs, the right half shows the pathways, and the size of the color block represents the corresponding counts. The chord lines between the left and right parts represent links between DEGs and specialized signaling pathways.
With the STRING online tool and the Cytoscape software, we established a PPI network related to the above 260 DEGs. The PPI network included 167 nodes and 334 edges, as shown in Figure 5A. Using the mCODE plug-in, four significant cluster modules were obtained from the PPI network based on the screening criteria. Module 1 had the highest cluster score of 4.667, which included a total of 21 edges, 10 nodes, and a seed gene named KLHL21 (Figure 5B). The Module 2 subnetwork had a cluster score of 4.5, which consisted of nine edges, five nodes, and a seed gene named DNAJB9 (Figure 5C). Module 3 had a cluster score of 4, which included a total of six edges, four nodes, and a seed gene named MT1M (Figure 5D). The Module 4 subnetwork had the cluster score of 3.882, which consisted of 31 edges, 17 nodes, and a seed gene named ZFP36 (Figure 5E). Next, we filtered the top 10 hub genes ranked by the MCC algorithm [HSPA5, JUN, IL6, HSPA1A, activated transcription factor 3 (ATF3), XBP1, HSPA1B, GRPEL2, heat shock protein family A (Hsp70) member 6 (HSPA6), and dual specificity phosphatase 1 (DUSP1, also known as CL100, HVH1, MKP-1, PTPN10)], using cytoHubba plug-in (Figure 6; Table 2). These hub genes may be potentially mechanosensitive genes that have crucial roles in sensing the force stretch stimulation of ECs directly. Finally, these hub genes' functional enrichment analyses were performed by the R clusterProfiler package, and it was found that they were dramatically enriched in GO terms such as cellular response to unfolded protein, COP9 signalosome, heat shock protein binding, and KEGG signaling pathways such as protein processing in the endoplasmic reticulum and the MAPK signaling pathway. According to the criteria, all summary results of meaningful functional annotation analyses from the hub genes were shown in Figure 7; Supplementary Table 3.
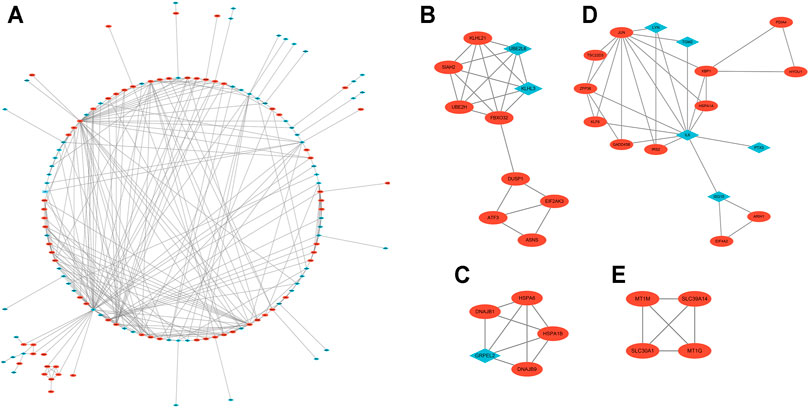
FIGURE 5. The PPI network and two cluster modules were extracted by mCODE. (A) The PPI network of DEGs in GSE23289, which comprised 167 nodes and 334 edges. Each node represents a protein, while each edge between the two proteins shows a protein–protein correlation. The up- or downregulated genes are highlighted as red circles and blue diamonds, respectively. (B) Cluster mCODE 1 consists of 10 nodes and 21 edges. mCODE score = 4.667. KLHL21 is the seed gene in this subnetwork. (C) Cluster mCODE 2 includes five nodes and nine edges. mCODE score = 4.5. DNAJB9 is the seed gene in this subnetwork. (D) Cluster mCODE 3 includes four nodes and six edges, MT1M is the seed gene, score = 4. (E) Cluster mCODE 4 includes 17 nodes and 31 edges, ZFP36 is the seed gene, and score = 3.882).
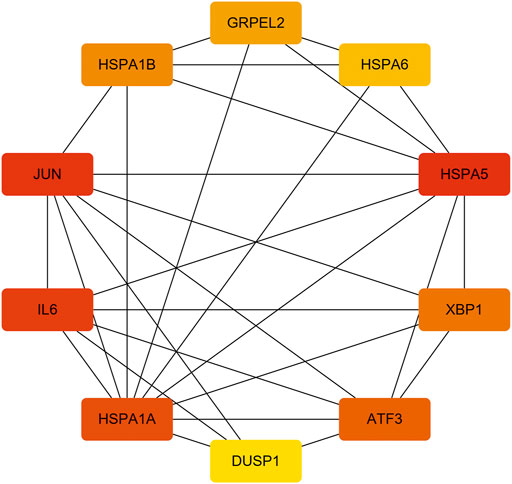
FIGURE 6. The 10 hub genes interaction network. Each node indicates a gene, and each edge between the two nodes indicates a possible interrelationship of those two genes. The color gradient of the node reflects the MCC score, and the darker the color, the higher the score.
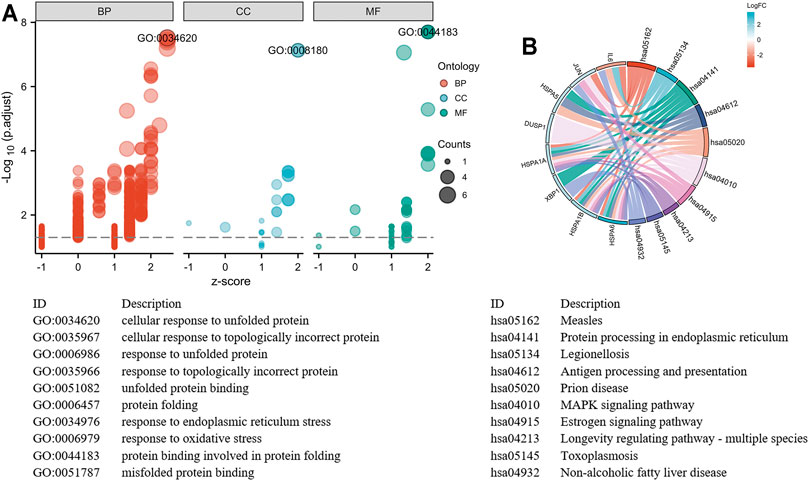
FIGURE 7. Top 10 hub genes functional enrichment analysis. (A) The GO functional enrichment bubble plot of hub genes. The abscissa represents the z-score, and the ordinate represents the adjusted p-value. The size of the bubble displayed is corresponding to the counts enriched in the GO terms, and the bubble's color correlated with the enriched GO categories. (B) The signaling pathways enrichment chord diagram of hub genes.
Due to the removal of probes matching multiple genes, only eight genes were retained, and the HSPA1A/HSPA1B genes were excluded in GSE13712. The expression profiles of these eight hub genes were validated by the GSE13712 data set. The ggplot2 package of R software was performed to draw violin plots and conducted a t-test for the comparison of the two groups. Compared with the normal group, the expression levels of the hub genes, ATF3 and DUSP1, in the relatively high WSS group were significantly increased (p < 0.01). In addition, the expression levels of the another two hub genes, HSPA6 and GRPEL2, were also strongly upregulated in high WSS (p < 0.05) (Figure 8A). To investigate whether aging can affect mechanosensitive genes, the expression of the abovementioned hub genes was statistically analyzed according to the different degrees of aging HUVECs. We observed that the expression levels of DUSP1, GRPEL2, and JUN genes in the senescent HUVECs group were significantly higher than those in the young HUVECs group under high WSS (p < 0.05) (Figure 8B). However, the expression level of GRPLE2 in the GSE13712 was inconsistent with that in the GSE23289 data set. In the GSE23289 data set, GRPLE2 was downregulated in the high WSS group, while in the GSE13712 data set, the gene was upregulated. The possible reasons for the inconsistency of results among the two data sets could be the different chip platforms for analyzing the data and the small sample size of these GEO data sets. Ultimately, we found that DUSP1 may be an important aging-related mechanosensitive gene because it is not only significantly upregulated in the high WSS group but also more significantly upregulated in the senescent HUVECs than in the young HUVECs.
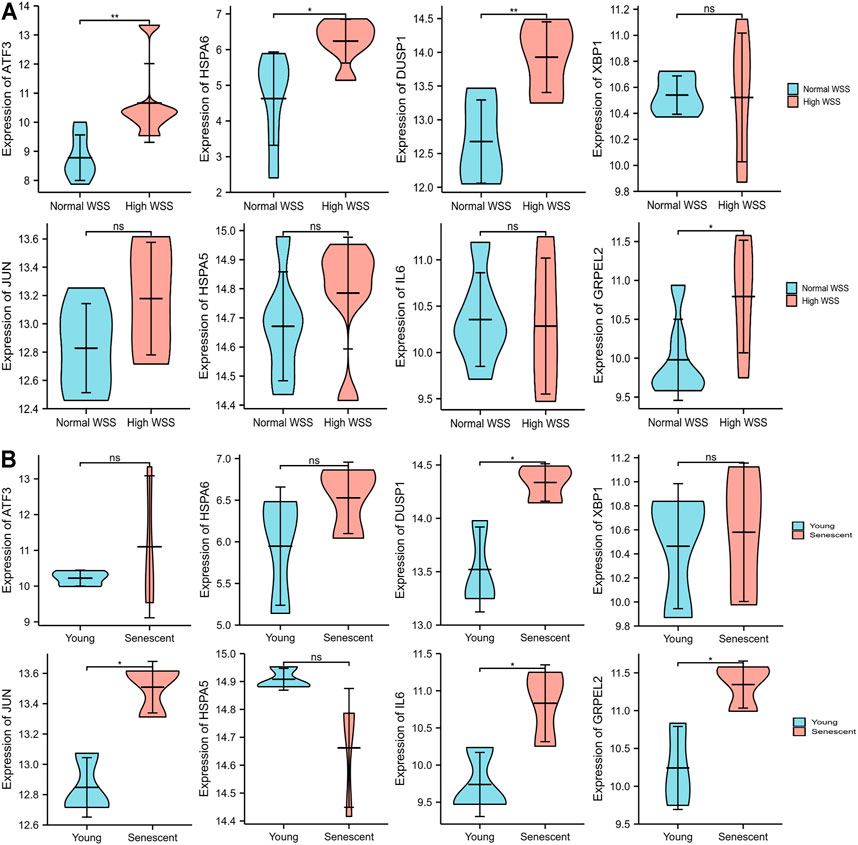
FIGURE 8. The expression level validation of hub genes. (A) A comparison of the expressed level of the hub genes between normal and high WSS groups in the GSE13712 data set. Compared with the normal group, hub genes (ATF3, DUSP1, HSPA6, and GRPLE2) are highly expressed in the high WSS group with significance. (B) The comparison of the expressed levels of the hub genes between the young and senescent HUVEC groups under high WSS in the GSE13712 data set. Compared with the young HUVECs group, DUSP1, GRPEL2, and JUN were highly expressed in the senescent HUVECs group, while others have no significance. *p < 0.05, **p < 0.01, ns: p > 0.05, no significance.
We used the pROC package in R software to analyze the expression profiles of the above specifically expressed hub genes in the normal group, high shear stress group, and young HUVECs and senescent HUVECs groups under high shear stress and drew the ROC curves by ggplot2. The area under the curve (AUC) is an indicator that combines sensitivity and specificity, which can describe the inherent validity of a diagnostic test. Compared with the normal group, these five specifically expressed hub genes (ATF3, HSPA6, DUSP1, GRPLE2, and JUN) have higher diagnostic value in the high shear stress group (0.7<AUC<1.0). Among them, ATF3 has the strongest diagnostic ability with an AUC of 0.944, followed by HSPA6 and DUSP1, which have the same AUC value of 0.917, and the AUC of GRPLE2 and JUN were 0.861 and 0.778, respectively. While in the senescent HUVECs group under high WSS, DUSP1 was found to have the highest diagnostic value (AUC = 1.0), which was consistent with the previous hub genes expression verification results between the young and senescent HUVEC groups (Figure 9). Therefore, after combining the above analysis of the eight hub genes expression levels with the ROC results, we concluded that ATF3, HSPA6, and DUSP1 might be the most potential mechanosensitive genes, being upregulated in high shear stress with statistical significance, which could have an important role in sensing the mechanical tension stimulus of the blood vessel wall directly. Moreover, under high shear stress, DUSP1 was expressed at a higher level in senescent HUVECs than in young HUVECs. Therefore, we speculated that DUSP1 might be a mechanosensitive gene related to the degree of aging based on our current samples data.
NetworkAnalyst online platform is used to generate TF–miRNA co-regulatory network. The interaction between TFs and miRNAs and the selected hub genes was delivered via the TF–miRNA co-regulatory network analysis. This interaction may be the underlying mechanism for the regulation of hub genes expression. The TF–miRNA co-regulatory network consisted of 14 nodes and 23 edges. A total of four miRNAs and seven TF genes interacted with the validated hub genes. Figure 10 shows the TF–miRNA co-regulatory network.
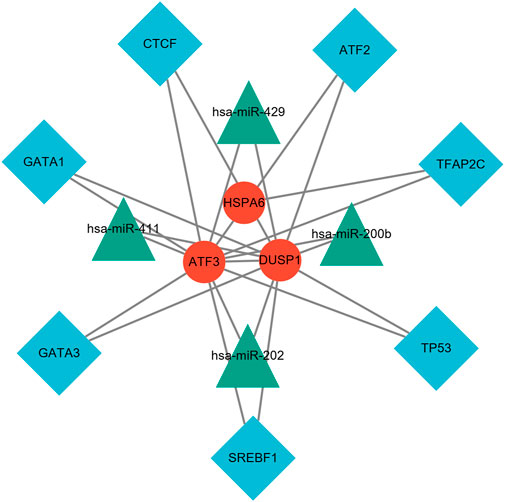
FIGURE 10. TF–miRNA coregulatory network. The network consisted of 14 nodes and 23 edges, including seven TF genes and four miRNAs, and three validated hub genes. The red round nodes are validated hub genes, the blue diamond nodes are TF genes, and the green triangle nodes are miRNA genes.
Stroke is the main cause of long-term disability and the second most common cause of death in the world with an increasing burden on global health (sD and Collaborators, 2016; MortalityD (2016). Cau, 2016; Feigin et al., 2017). Ischemic stroke, which accounts for nearly 80% of all types of stroke, is mainly caused by an in situ blockage of a blood flow, resulting from a narrowing large artery in the brain circulation, or emboli from a ruptured atherosclerotic plaque blocking downstream branches (Hademenos and Massoud, 1997). Previous studies have shown that there is a significant correlation between WSS and stroke events, and WSS is considered to be the main risk factor for atherosclerotic disease and ischemic stroke (Cicha et al., 2011; Leng et al., 2019). WSS is the frictional resistance exerted by the blood flow that acts on and parallel to the surface of the endothelium. Low WSS may promote the deposition of particles on the arterial wall and the accumulation of plaque, while high WSS not only stimulates adaptive outward remodeling but also destabilizes atherosclerotic plaques in narrowed blood vessels by directly acting on plaques, inducing inflammation, enhancing oxidative stress, and subsequently causing embolization of distal arteries (Dolan et al., 2013; Chen et al., 2019; Goudot et al., 2020). The ability of blood vessels to react to changes in hemodynamic stimuli is mediated primarily by ECs, which form the internal layer of blood vessel walls. Since WSS directly acts on vascular ECs, it is speculated that mechanical tension stimulates mechanics-sensitive genes on the endothelium, induces EC injury and dysfunction, and triggers a host of biological responses noted above. Another study illustrates that correcting hemodynamic factors plays a protective role in the blood vessels (Dhawan et al., 2010). Therefore, it is essential to study the mechanism of WSS-induced ischemic events and to identify new potential molecular targets for disease diagnosis and treatment. Bioinformatics technology has been widely used to discuss the specificity of gene expression during the occurrence and development of diseases. Moreover, it is a reliable way to explore validated diagnosis and treatment targets of disease.
At present, there is little research on the correlation between WSS and mechanics-sensitive genes expressed on vascular ECs and its mechanism of cerebrovascular health promotion and stroke prevention. In our study, using bioinformatics methods, we analyzed the expression profiles of different genes on vascular ECs under high and normal WSS, and a total of 260 DEGs were observed with 121 upregulated and 139 downregulated genes. Many, mainly the biological functions, of these DEGs were screened out, including cellular response to unfolded protein and topologically incorrect protein, response to endoplasmic reticulum stress, oxidative stress, heat shock protein binding, etc. The KEGG signaling pathways that we enriched included protein processing in the endoplasmic reticulum, antigen processing and presentation, the MAPK signaling pathway, and the signaling pathways associated with infection and inflammatory disease. After the 10 hub genes filtered by the PPI network were validated using another GEO data set, we determined eight hub genes (HSPA5, JUN, IL6, ATF3, XBP1, GRPEL2, HSPA6, and DUSP1). The ROC curve analysis shows that ATF3, HSPA6, DUSP1, GRPLE2, and JUN have significant diagnostic values in the high WSS group (0.7 < AUC < 1.0). Combined with these eight hub genes' expressed levels in high and normal WSS, ATF3, HSPA6, and DUSP1 were significantly different and upregulated in high WSS (p < 0.05). Subsequent analysis of the above genes in the subgroups revealed that in the high WSS group, compared with young cells, the expression level of DUSP1 was statistically significant in the senescent cells (AUC = 1, p = 0.032). Therefore, we hypothesized that ATF3, HSPA6, and DUSP1 may be mechanics-sensitive genes expressed on vascular ECs under high WSS based on our current samples. And among them, DUSP1 may be positively correlated with the degree of EC senescence. In addition, we constructed a TF–miRNA co-regulatory network, which provides the interaction between miRNA and TF with ATF3, HSPA6, and DUSP1. This interaction may be the reason for the regulation of mechanics-sensitive genes expression.
ATF3, an immediate, early transcription factor gene belonging to the activated protein 1 (AP-1) family, plays an important role in inflammation and infectious diseases, endoplasmic reticulum stress, mitochondrial oxidative stress, endothelial dysfunction, and prevention of atherosclerosis (Cai et al., 2000; Nawa et al., 2002; Aung et al., 2013; Rohini et al., 2018). Teasdale et al. (2017) found that elevated laminar flow in blood vessels can induce the expression of ATF3 and reduce the expression of inflammatory genes independent of nuclear factor-κB activation. They also identify that ATF3 is an important protective factor against vascular endothelial dysfunction, and regulating the expression of ATF3 may be a new way to modulate the expression of pro-inflammatory genes. It is suggested that ATF3 may play a pivotal role in the mechanism of vascular endothelial dysfunction and atherosclerotic plaque rupture induced by high WSS. Consistent with our study, we found that ATF3 was upregulated in HUVECs cultured under high WSS. In addition, the ROC curve showed that ATF3 has a high diagnostic value for high WSS (AUC = 0.944). Therefore, we considered ATF3 as a gene that is very sensitive to high WSS.
HSPA6 (also called Hsp70B'), is an inducible member of the HSPA (Hsp70) multigene family. Compared with the extensively studied HSP70 family genes, it has received little attention because it is the only gene expressed in the human genome but not in rodents such as mice and rats (Noonan et al., 2007; Deane and Brown, 2017). Previous studies have shown that HSP70 can be expressed in atherosclerotic plaques, and the high expression of HSP70 is positively correlated with the severity of atherosclerotic vascular lesions (Konstantinova et al., 2019). In addition, HSP70 has been shown to have crucial functions in inflammation/immune-related regulation and vascular endothelial dysfunction and to be linked to a series of inflammatory diseases such as atherosclerosis, arthritis, neurodegenerative diseases, as well as cancer (Thériault et al., 2005; Henderson and Pockley, 2010; Costa-Beber et al., 2020). A few studies referred to HSPA6. McCullagh et al. (2016) found that overexpression of HSPA6 can inhibit the proliferation of smooth muscle cells, which suggested that HSPA6 may play a crucial role in targeting therapy for vascular repair. Further studies have shown that HSPA6 is preferentially expressed in macrophages and is considered to have pro-inflammatory properties. It synergizes with circulating immune complexes to promote the progression of atherosclerotic processes, which could promise a new therapeutic target for regulating vulnerable atherosclerotic plaques (Hammad et al., 2009; Smith et al., 2010). At present, there is no research on the relationship between HSAP6 and WSS. However, in our study, HSP70 family genes, including HSPA1A, HSPA1B, HSPA5, and HSPA6, were significantly upregulated under high WSS in vascular ECs. Among them, HSPA6 served as the hub gene. It demonstrated that HSP70, especially the less studied HSPA6, maybe a mechanosensitive gene, which plays a key role in the progression of atherosclerotic disease and the ruptured atherosclerotic plaques.
DUSP1 belongs to a family of inducible nuclear dual-specific phosphatases that can dephosphorylate tyrosine and threonine/serine residues. Integrated analyses of ischemic stroke data sets from many previous studies have shown that DUSP1 is overexpressed in both males and females, which suggested that DUSP1 may be a diagnostic biomarker for ischemic stroke (Li et al., 2016; Zhang et al., 2019; Feng et al., 2021). Currently, the mechanism by which DUSP1 regulates atherosclerosis progression is still controversial. ZAKKAR M and HOPPSTADTER J et al. thought that the increased expression of DUSP1 is anti-atherogenic via inhibiting EC pro-inflammatory signaling pathways, such as MAPK and NF-κB (Zakkar et al., 2016; Hoppstädter and Ammit, 2019). Shen et al. (2010) considered that the deletion of DUSP1 leads to a reduction of atherosclerotic lesion formation, which is accompanied by decreases of multiple pro-inflammatory cytokines, including TNF and IL-1. Similar views have shown that DUSP1 is overexpressed in atherosclerotic lesion, correlated with hyperlipidemia, and reduced DUSP1 can inhibit the development of atherosclerosis (Reddy et al., 2004). In addition, a study by Lee et al. (2016) indicated that DUSP1 involves in the regulatory mechanism of cellular senescence via immunomodulatory properties. In our present study, unlike the abovementioned ATF3 and HSPA6, we identified that DUSP1 was not only highly expressed on vascular ECs under high WSS but also highly expressed in the senescent cells subgroup than in young cells (p = 0.0315 AUC = 1.000), indicating that DUSP1 may be a mechanosensitive gene related to high WSS and correlated with the senescence of vascular ECs.
Furthermore, the target TF genes and miRNAs were predicted for ATF3, HSPA6, and DUSP1, and a TF–miRNA–mechanosensitive genes coregulatory network was constructed by the NetworkAnalyst. This co-regulatory network revealed the regulatory mechanism of mechanosensitive genes at the transcriptome level. Among the most interactive TFs, the higher degree of TFAP2C was 2, and the higher betweenness was 1692. TFAP2C and ATF3 play multiple roles in shaping the sensitivity of ferroptosis through either transcription-independent or transcription-dependent mechanisms (Dai et al., 2020). Pharmacological components based on TFAP2C can be constructed to inhibit ferroptosis and cell death caused by excitotoxicity or endoplasmic reticulum stress and coordinate the repair of neurons after stroke (Alim et al., 2019). Evidence for changes in miRNA expression in the co-regulatory network has been established in various studies. For instance, has-miR-429 regulates the inflammatory response by targeting inhibition of DUSP1 expression (Qi and Wang, 2020). van der Kwast et al. (2020) demonstrated that hsa-miRNA-411, regarded as vasoactive miRNA, was abundantly expressed in the vasculature for chronic ischemic diseases. The TF genes are a reactor that regulates gene expression and complete regulation by combining with target genes or miRNA. On the other hand, miRNAs induce gene silencing and downregulate gene expression by binding to target genes (Zhang et al., 2015b).
Several limitations of this study should be considered. First, our study did not truly combine transcriptomic microarray data, since we use only one data set to discover mechanosensitive genes and another one as a validation data set. The sample size was not large enough, limited by the publicly available microarray data sets, which require further samples to be sent of our own for transcriptomic high-throughput sequencing or microarray data to expand the sample size. Second, recently, we did not illustrate the real co-relations between high shear stress, processes of atherogenesis, and atherosclerotic progression due to the lack of animal models in a different stage of atherosclerosis. Additional in-depth and more rigorous experimental studies will be designed to focus on the role of high shear stress in the processes of atherogenesis and atherosclerotic progression. Third, the work was solely based on bioinformatic strategies. Although we validated the potential three mechanosensitive genes in a public microarray data set, an in-depth validation of the hub genes would greatly enhance the amount of information in this article. Specifically, a mechanistic study based on the proposed TF–miRNA genes coregulatory network could better demonstrate the atherosclerotic disease progressions. But unfortunately, the experiment was halted due to the recurrent episodes of severe acute respiratory coronavirus 2 (SARS-CoV-2).
In this work, we identified three hub genes, namely, ATF3, HSPA6, and DUSP1, as potential mechanosensitive genes under high WSS in blood vessels. Among them, DUSP1 was confirmed to be associated with the senescence of vascular ECs that experienced high WSS. Therefore, these three mechanosensitive genes may have emerged as potential novel targets for the prediction and prevention of ischemic stroke. In addition, a TF–miRNA–mechanosensitive genes coregulatory network was constructed, which revealed an underlying regulatory mechanism and the pathways to control disease progression.
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LS and KZ conceived and designed the research protocol, drafted the manuscript, and analyzed data. HL, JY, and SH critically revised the manuscript and diagrams. FY and DH participated in work guidance and supervised the project. All authors contributed to the article and approved the submitted version.
This project was supported by the National Natural Science Foundation of China project (No. 81771258) and Shanghai Municipal Health Commission Research project (No. 20204Y0072).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to appreciate the support from Professor Yuzhen Zhang in Translational Medicine Center, Tongji University–affiliated Shanghai East Hospital. We also thank all contributors for providing highly qualified data to public databases.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.796812/full#supplementary-material
Alim, I., Caulfield, J. T., Chen, Y., Swarup, V., Geschwind, D. H., Ivanova, E., et al. (2019). Selenium Drives a Transcriptional Adaptive Program to Block Ferroptosis and Treat Stroke. Cell 177 (5), 1262–1279. doi:10.1016/j.cell.2019.03.032
Aung, H. H., Lame, M. W., Gohil, K., An, C.-I., Wilson, D. W., and Rutledge, J. C. (2013). Induction of ATF3 Gene Network by Triglyceride-Rich Lipoprotein Lipolysis Products Increases Vascular Apoptosis and Inflammation. Atvb 33 (9), 2088–2096. doi:10.1161/ATVBAHA.113.301375
Badimon, L., Suades, R., Arderiu, G., Peña, E., Chiva-Blanch, G., and Padró, T. (2017). Microvesicles in Atherosclerosis and Angiogenesis: From Bench to Bedside and Reverse. Front. Cardiovasc. Med. 4, 77. doi:10.3389/fcvm.2017.00077
Cai, Y., Zhang, C., Nawa, T., Aso, T., Tanaka, M., Oshiro, S., et al. (2000). Homocysteine-responsive ATF3 Gene Expression in Human Vascular Endothelial Cells: Activation of C-Jun NH2-terminal Kinase and Promoter Response Element. Blood 96 (6), 2140–2148. doi:10.1182/blood.v96.6.2140.h8002140_2140_2148
Casey, D. P., Padilla, J., and Joyner, M. J. (2012). α-Adrenergic Vasoconstriction Contributes to the Age-Related Increase in Conduit Artery Retrograde and Oscillatory Shear. Hypertension 60 (4), 1016–1022. doi:10.1161/HYPERTENSIONAHA.112.200618
Chatzizisis, Y. S., Coskun, A. U., Jonas, M., Edelman, E. R., Feldman, C. L., and Stone, P. H. (2007). Role of Endothelial Shear Stress in the Natural History of Coronary Atherosclerosis and Vascular Remodeling. J. Am. Coll. Cardiol. 49 (25), 2379–2393. doi:10.1016/j.jacc.2007.02.059
Chen, Z., Qin, H., Liu, J., Wu, B., Cheng, Z., Jiang, Y., et al. (2019). Characteristics of Wall Shear Stress and Pressure of Intracranial Atherosclerosis Analyzed by a Computational Fluid Dynamics Model: A Pilot Study. Front. Neurol. 10, 1372. doi:10.3389/fneur.2019.01372
Chin, C.-H., Chen, S.-H., Wu, H.-H., Ho, C.-W., Ko, M.-T., and Lin, C.-Y. (2014). cytoHubba: Identifying Hub Objects and Sub-networks from Complex Interactome. BMC Syst. Biol. 8 (Suppl. 4), S11. doi:10.1186/1752-0509-8-s4-s11
Cicha, I., Wörner, A., Urschel, K., Beronov, K., Goppelt-Struebe, M., Verhoeven, E., et al. (2011). Carotid Plaque Vulnerability. Stroke 42 (12), 3502–3510. doi:10.1161/STROKEAHA.111.627265
Costa-Beber, L. C., Hirsch, G. E., Heck, T. G., and Ludwig, M. S. (2020). Chaperone Duality: the Role of Extracellular and Intracellular HSP70 as a Biomarker of Endothelial Dysfunction in the Development of Atherosclerosis. Arch. Physiol. Biochem., 1–8. doi:10.1080/13813455.2020.1745850
Costopoulos, C., Timmins, L. H., Huang, Y., Hung, O. Y., Molony, D. S., Brown, A. J., et al. (2019). Impact of Combined Plaque Structural Stress and wall Shear Stress on Coronary Plaque Progression, Regression, and Changes in Composition. Eur. Heart J. 40 (18), 1411–1422. doi:10.1093/eurheartj/ehz132
Dai, C., Chen, X., Li, J., Comish, P., Kang, R., and Tang, D. (2020). Transcription Factors in Ferroptotic Cell Death. Cancer Gene Ther. 27 (9), 645–656. doi:10.1038/s41417-020-0170-2
DALYs GBD, Collaborators H (2016). Global, Regional, and National Disability-Adjusted Life-Years (DALYs) for 315 Diseases and Injuries and Healthy Life Expectancy (HALE), 1990-2015: a Systematic Analysis for the Global Burden of Disease Study 2015. Lancet 388 (10053), 1603–1658. doi:10.1016/S0140-6736(16)31460-X
Davis, S., and Meltzer, P. S. (2007). GEOquery: a Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23 (14), 1846–1847. doi:10.1093/bioinformatics/btm254
Deane, C. A. S., and Brown, I. R. (2017). Differential Targeting of Hsp70 Heat Shock Proteins HSPA6 and HSPA1A with Components of a Protein Disaggregation/Refolding Machine in Differentiated Human Neuronal Cells Following Thermal Stress. Front. Neurosci. 11, 227. doi:10.3389/fnins.2017.00227
Derdeyn, C. P., Chimowitz, M. I., Lynn, M. J., Fiorella, D., Turan, T. N., Janis, L. S., et al. (2014). Aggressive Medical Treatment with or without Stenting in High-Risk Patients with Intracranial Artery Stenosis (SAMMPRIS): the Final Results of a Randomised Trial. The Lancet 383 (9914), 333–341. doi:10.1016/S0140-6736(13)62038-3
Dhawan, S. S., Avati Nanjundappa, R. P., Branch, J. R., Taylor, W. R., Quyyumi, A. A., Jo, H., et al. (2010). Shear Stress and Plaque Development. Expert Rev. Cardiovasc. Ther. 8 (4), 545–556. doi:10.1586/erc.10.28
Dolan, J. M., Kolega, J., and Meng, H. (2013). High wall Shear Stress and Spatial Gradients in Vascular Pathology: a Review. Ann. Biomed. Eng. 41 (7), 1411–1427. doi:10.1007/s10439-012-0695-0
Feigin, V. L., Norrving, B., and Mensah, G. A. (2017). Global Burden of Stroke. Circ. Res. 120 (3), 439–448. doi:10.1161/CIRCRESAHA.116.308413
Feldman, C. L., Ilegbusi, O. J., Hu, Z., Nesto, R., Waxman, S., and Stone, P. H. (2002). Determination of In Vivo Velocity and Endothelial Shear Stress Patterns with Phasic Flow in Human Coronary Arteries: a Methodology to Predict Progression of Coronary Atherosclerosis. Am. Heart J. 143 (6), 931–939. doi:10.1067/mhj.2002.123118
Feng, B., Meng, X., Zhou, H., Chen, L., Zou, C., Liang, L., et al. (2021). Identification of Dysregulated Mechanisms and Potential Biomarkers in Ischemic Stroke Onset. Ijgm Vol. 14, 4731–4744. doi:10.2147/ijgm.S327594
Gao, S., Ding, B., and Lou, W. (2020). microRNA-Dependent Modulation of Genes Contributes to ESR1's Effect on ERα Positive Breast Cancer. Front. Oncol. 10, 753. doi:10.3389/fonc.2020.00753
Geng, S., Zhang, Y., Lee, C., and Li, L. (2019). Novel Reprogramming of Neutrophils Modulates Inflammation Resolution during Atherosclerosis. Sci. Adv. 5 (2), eaav2309. doi:10.1126/sciadv.aav2309
Goudot, G., Khider, L., Pedreira, O., Poree, J., Julia, P., Alsac, J.-M., et al. (2020). Innovative Multiparametric Characterization of Carotid Plaque Vulnerability by Ultrasound. Front. Physiol. 11, 157. doi:10.3389/fphys.2020.00157
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 32 (18), 2847–2849. doi:10.1093/bioinformatics/btw313
Hademenos, G. J., and Massoud, T. F. (1997). Biophysical Mechanisms of Stroke. Stroke 28 (10), 2067–2077. doi:10.1161/01.str.28.10.2067
Hammad, S. M., Twal, W. O., Barth, J. L., Smith, K. J., Saad, A. F., Virella, G., et al. (2009). Oxidized LDL Immune Complexes and Oxidized LDL Differentially Affect the Expression of Genes Involved with Inflammation and Survival in Human U937 Monocytic Cells. Atherosclerosis 202 (2), 394–404. doi:10.1016/j.atherosclerosis.2008.05.032
Henderson, B., and Pockley, A. G. (2010). Molecular Chaperones and Protein-Folding Catalysts as Intercellular Signaling Regulators in Immunity and Inflammation. J. Leukoc. Biol. 88 (3), 445–462. doi:10.1189/jlb.1209779
Hoppstädter, J., and Ammit, A. J. (2019). Role of Dual-Specificity Phosphatase 1 in Glucocorticoid-Driven Anti-inflammatory Responses. Front. Immunol. 10, 1446. doi:10.3389/fimmu.2019.01446
Hoshino, T., Sissani, L., Labreuche, J., Ducrocq, G., Lavallée, P. C., Meseguer, E., et al. (2018). Prevalence of Systemic Atherosclerosis Burdens and Overlapping Stroke Etiologies and Their Associations with Long-Term Vascular Prognosis in Stroke with Intracranial Atherosclerotic Disease. JAMA Neurol. 75 (2), 203–211. doi:10.1001/jamaneurol.2017.3960
Hoyer, F. F., Naxerova, K., Schloss, M. J., Hulsmans, M., Nair, A. V., Dutta, P., et al. (2019). Tissue-Specific Macrophage Responses to Remote Injury Impact the Outcome of Subsequent Local Immune Challenge. Immunity 51 (5), 899–914. doi:10.1016/j.immuni.2019.10.010
Hsu, P.-L., Chen, J.-S., Wang, C.-Y., Wu, H.-L., and Mo, F.-E. (2019). Shear-Induced CCN1 Promotes Atheroprone Endothelial Phenotypes and Atherosclerosis. Circulation 139 (25), 2877–2891. doi:10.1161/circulationaha.118.033895
Kimura, T., Kobiyama, K., Winkels, H., Tse, K., Miller, J., Vassallo, M., et al. (2018). Regulatory CD4 + T Cells Recognize Major Histocompatibility Complex Class II Molecule-Restricted Peptide Epitopes of Apolipoprotein B. Circulation 138 (11), 1130–1143. doi:10.1161/CIRCULATIONAHA.117.031420
Konstantinova, E. V., Chipigina, N. S., Shurdumova, M. H., Kovalenko, E. I., and Sapozhnikov, A. M. (2019). Heat Shock Protein 70 kDa as a Target for Diagnostics and Therapy of Cardiovascular and Cerebrovascular Diseases. Cpd 25 (6), 710–714. doi:10.2174/1381612825666190329123924
Lee, J. Y., Yu, K.-R., Kim, H.-S., Kang, I., Kim, J.-J., Lee, B.-C., et al. (2016). BMI1 Inhibits Senescence and Enhances the Immunomodulatory Properties of Human Mesenchymal Stem Cells via the Direct Suppression of MKP-1/DUSP1. Aging 8 (8), 1670–1689. doi:10.18632/aging.101000
Leng, X., Lan, L., Ip, H. L., Abrigo, J., Scalzo, F., Liu, H., et al. (2019). Hemodynamics and Stroke Risk in Intracranial Atherosclerotic Disease. Ann. Neurol. 85 (5), 752–764. doi:10.1002/ana.25456
Li, C. Y., Cai, J.-H., Tsai, J. J. P., and Wang, C. C. N. (2020). Identification of Hub Genes Associated with Development of Head and Neck Squamous Cell Carcinoma by Integrated Bioinformatics Analysis. Front. Oncol. 10, 681. doi:10.3389/fonc.2020.00681
Li, W.-X., Dai, S.-X., Wang, Q., Guo, Y.-C., Hong, Y., Zheng, J.-J., et al. (2016). Integrated Analysis of Ischemic Stroke Datasets Revealed Sex and Age Difference in Anti-stroke Targets. PeerJ 4, e2470. doi:10.7717/peerj.2470
Li, Z., Ding, B., Xu, J., Mao, K., Zhang, P., and Xue, Q. (2020). Relevance of STK11 Mutations Regarding Immune Cell Infiltration, Drug Sensitivity, and Cellular Processes in Lung Adenocarcinoma. Front. Oncol. 10, 580027. doi:10.3389/fonc.2020.580027
Liu, Z.-P., Wu, C., Miao, H., and Wu, H. (2015). RegNetwork: an Integrated Database of Transcriptional and post-transcriptional Regulatory Networks in Human and Mouse. Database 2015, bav095. Database : the journal of biological databases and curation. doi:10.1093/database/bav095
Mack, J. J., Mosqueiro, T. S., Archer, B. J., Jones, W. M., Sunshine, H., Faas, G. C., et al. (2017). NOTCH1 Is a Mechanosensor in Adult Arteries. Nat. Commun. 8 (1), 1620. doi:10.1038/s41467-017-01741-8
Malek, A. M., Alper, S. L., and Izumo, S. (1999). Hemodynamic Shear Stress and its Role in Atherosclerosis. Jama 282 (21), 2035–2042. doi:10.1001/jama.282.21.2035
McCullagh, K. J. A., Cooney, R., and O'Brien, T. (2016). Endothelial Nitric Oxide Synthase Induces Heat Shock Protein HSPA6 (HSP70B′) in Human Arterial Smooth Muscle Cells. Nitric Oxide 52, 41–48. doi:10.1016/j.niox.2015.11.002
Milkiewicz, M., Uchida, C., Gee, E., Fudalewski, T., and Haas, T. L. (2008). Shear Stress-Induced Ets-1 Modulates Protease Inhibitor Expression in Microvascular Endothelial Cells. J. Cel. Physiol. 217 (2), 502–510. doi:10.1002/jcp.21526
Mortality GBD (2016). Causes of Death C. Global, Regional, and National Life Expectancy, All-Cause Mortality, and Cause-specific Mortality for 249 Causes of Death, 1980-2015. Lancet 388 (10053), 1459–1544. a systematic analysis for the Global Burden of Disease Study 2015. doi:10.1016/S0140-6736(16)31012-1
Nawa, T., Nawa, M. T., Adachi, M. T., Uchimura, I., Shimokawa, R., Fujisawa, K., et al. (2002). Expression of Transcriptional Repressor ATF3/LRF1 in Human Atherosclerosis: Colocalization and Possible Involvement in Cell Death of Vascular Endothelial Cells. Atherosclerosis 161 (2), 281–291. doi:10.1016/s0021-9150(01)00639-6
Niu, N., Xu, S., Xu, Y., Little, P. J., and Jin, Z.-G. (2019). Targeting Mechanosensitive Transcription Factors in Atherosclerosis. Trends Pharmacol. Sci. 40 (4), 253–266. doi:10.1016/j.tips.2019.02.004
Noonan, E. J., Place, R. F., Giardina, C., and Hightower, L. E. (2007). Hsp70B′ Regulation and Function. Cell Stress Chaper 12 (4), 393–402. doi:10.1379/csc-278e.1
Pulous, F. E., Grimsley-Myers, C. M., Kansal, S., Kowalczyk, A. P., and Petrich, B. G. (2019). Talin-Dependent Integrin Activation Regulates VE-Cadherin Localization and Endothelial Cell Barrier Function. Circ. Res. 124 (6), 891–903. doi:10.1161/CIRCRESAHA.118.314560
Qi, R., and Wang, X. (2020). Inhibition of miR-429 Improves Neurological Recovery of Traumatic Brain Injury Mice and Attenuates Microglial Neuroinflammation. Int. Immunopharmacology 79, 106091. doi:10.1016/j.intimp.2019.106091
Ramírez, C. M., Zhang, X., Bandyopadhyay, C., Rotllan, N., Sugiyama, M. G., Aryal, B., et al. (2019). Caveolin-1 Regulates Atherogenesis by Attenuating Low-Density Lipoprotein Transcytosis and Vascular Inflammation Independently of Endothelial Nitric Oxide Synthase Activation. Circulation 140 (3), 225–239. doi:10.1161/CIRCULATIONAHA.118.038571
Reddy, S. T., Nguyen, J. T., Grijalva, V., Hough, G., Hama, S., Navab, M., et al. (2004). Potential Role for Mitogen-Activated Protein Kinase Phosphatase-1 in the Development of Atherosclerotic Lesions in Mouse Models. Atvb 24 (9), 1676–1681. doi:10.1161/01.ATV.0000138342.94314.64
Rohini, M., Haritha Menon, A., and Selvamurugan, N. (2018). Role of Activating Transcription Factor 3 and its Interacting Proteins under Physiological and Pathological Conditions. Int. J. Biol. Macromolecules 120 (Pt A), 310–317. doi:10.1016/j.ijbiomac.2018.08.107
Rougeot, J., Torraca, V., Zakrzewska, A., Kanwal, Z., Jansen, H. J., Sommer, F., et al. (2019). RNAseq Profiling of Leukocyte Populations in Zebrafish Larvae Reveals a Cxcl11 Chemokine Gene as a Marker of Macrophage Polarization during Mycobacterial Infection. Front. Immunol. 10, 832. doi:10.3389/fimmu.2019.00832
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shen, J., Chandrasekharan, U. M., Ashraf, M. Z., Long, E., Morton, R. E., Liu, Y., et al. (2010). Lack of Mitogen-Activated Protein Kinase Phosphatase-1 Protects ApoE-Null Mice against Atherosclerosis. Circ. Res. 106 (5), 902–910. doi:10.1161/CIRCRESAHA.109.198069
Smith, K. J., Twal, W. O., Soodavar, F., Virella, G., Lopes-Virella, M. F., and Hammad, S. M. (2010). Heat Shock Protein 70B′ (HSP70B′) Expression and Release in Response to Human Oxidized Low Density Lipoprotein Immune Complexes in Macrophages. J. Biol. Chem. 285 (21), 15985–15993. doi:10.1074/jbc.M110.113605
Smyth, G. K. (2005). “Limma: Linear Models for Microarray Data,” in Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Editors R. Gentleman, V. J. Carey, W. Huber, R. A. Irizarry, and S. Dudoit (New York, NY: Springer New York), 397–420.
Spruell, C., and Baker, A. B. (2013). Analysis of a High-Throughput Cone-And-Plate Apparatus for the Application of Defined Spatiotemporal Flow to Cultured Cells. Biotechnol. Bioeng. 110 (6), 1782–1793. doi:10.1002/bit.24823
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2018). STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-wide Experimental Datasets. Nucleic Acids Res. 47 (D1), D607–D13. doi:10.1093/nar/gky1131.%J.Nucleic.Acids.Research
Teasdale, J. E., Hazell, G. G. J., Peachey, A. M. G., Sala-Newby, G. B., Hindmarch, C. C. T., McKay, T. R., et al. (2017). Cigarette Smoke Extract Profoundly Suppresses TNFα-Mediated Proinflammatory Gene Expression through Upregulation of ATF3 in Human Coronary Artery Endothelial Cells. Sci. Rep. 7, 39945. doi:10.1038/srep39945
Thériault, J. R., Mambula, S. S., Sawamura, T., Stevenson, M. A., and Calderwood, S. K. (2005). Extracellular HSP70 Binding to Surface Receptors Present on Antigen Presenting Cells and Endothelial/epithelial Cells. FEBS Lett. 579 (9), 1951–1960. doi:10.1016/j.febslet.2005.02.046
van der Kwast, R. V. C. T., Woudenberg, T., Quax, P. H. A., and Nossent, A. Y. (2020). MicroRNA-411 and its 5′-IsomiR Have Distinct Targets and Functions and Are Differentially Regulated in the Vasculature under Ischemia. Mol. Ther. 28 (1), 157–170. doi:10.1016/j.ymthe.2019.10.002
Wang, L., Luo, J.-Y., Li, B., Tian, X. Y., Chen, L.-J., Huang, Y., et al. (2016). Integrin-YAP/TAZ-JNK cascade Mediates Atheroprotective Effect of Unidirectional Shear Flow. Nature 540 (7634), 579–582. doi:10.1038/nature20602
Wiese, C. B., Zhong, J., Xu, Z.-Q., Zhang, Y., Ramirez Solano, M. A., Zhu, W., et al. (2019). Dual Inhibition of Endothelial miR-92a-3p and miR-489-3p Reduces Renal Injury-Associated Atherosclerosis. Atherosclerosis 282, 121–131. doi:10.1016/j.atherosclerosis.2019.01.023
Yu, G., Wang, L-G., Han, Y., and He, Q-Y., clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. (2012) 16(5):284–287. doi:10.1089/omi.2011.0118
Zakkar, M., D Angelini, G., and Emanueli, C. (2016). Regulation of Vascular Endothelium Inflammatory Signalling by Shear Stress. Cvp 14 (2), 181–186. doi:10.2174/1570161114666151202205139
Zhang, H.-M., Kuang, S., Xiong, X., Gao, T., Liu, C., and Guo, A.-Y. (2015). Transcription Factor and microRNA Co-regulatory Loops: Important Regulatory Motifs in Biological Processes and Diseases. Brief. Bioinform. 16 (1), 45–58. doi:10.1093/bib/bbt085
Zhang, Q., Chen, W., Chen, S., Li, S., Wei, D., and He, W. (2019). Identification of Key Genes and Upstream Regulators in Ischemic Stroke. Brain Behav. 9 (7), e01319. doi:10.1002/brb3.1319
Zhang, Y., Sun, M., Han, Y., Zhai, K., Tang, Y., Qin, X., et al. (2015). The Saponin DT-13 Attenuates Tumor Necrosis Factor-α-Induced Vascular Inflammation Associated with Src/NF-Кb/MAPK Pathway Modulation. Int. J. Biol. Sci. 11 (8), 970–981. doi:10.7150/ijbs.11635
Keywords: mechanosensitive genes, wall shear stress, vascular endothelial cells, atherosclerotic plaques, ischemic stroke
Citation: Shen L, Zhou K, Liu H, Yang J, Huang S, Yu F and Huang D (2022) Prediction of Mechanosensitive Genes in Vascular Endothelial Cells Under High Wall Shear Stress. Front. Genet. 12:796812. doi: 10.3389/fgene.2021.796812
Received: 18 October 2021; Accepted: 13 December 2021;
Published: 11 January 2022.
Edited by:
Rossen Donev, MicroPharm Ltd., United KingdomReviewed by:
Rosa Suades, Sant Pau Institute for Biomedical Research, SpainCopyright © 2022 Shen, Zhou, Liu, Yang, Huang, Yu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Yu, cmVhbG1pbGFuQHNpbmEuY29t; Dongya Huang, ZG9uZ3lhaHVhbmc3N0Bob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.