- 1Department of Laboratory Medicine, The Third Xiangya Hospital, Central South University, Changsha, China
- 2Department of Laboratory Medicine, Xiangya School of Medicine, Central South University, Changsha, China
- 3School of Basic Medical Science, Institute of Reproductive and Stem Cell Engineering, Central South University, Changsha, China
- 4Reproductive and Genetic Hospital of CITIC-Xiangya, Changsha, China
Background: Structural chromosome abnormality (SCA) is an important cause of human diseases, including recurrent pregnancy loss (RPL). DNA double-strand breaks (DSBs) repair-related genes play critical roles in SCA. The present study aims to investigate the potential contribution of DSBs repair-related gene polymorphisms to SCA.
Methods: Fifty-four affected RPL individuals with SCA, 88 affected RPL individuals without SCA, and 84 controls were analyzed. Targeted whole-exome sequencing (WES) was used for screening single nucleotide polymorphisms in six DSBs repair-related genes (EP300, XRCC6, LIG4, XRCC4, PRKDC, and DCLRE1C), and validation was performed by Sanger sequencing. Finally, we detected the frequency of radiation-induced chromosome translocations in no SCA samples with significant polymorphisms by fluorescence in situ hybridization (FISH).
Results: A total of 35 polymorphisms have been identified and confirmed. Frequencies of EP300 rs20551, XRCC6 rs132788, and LIG4 rs1805388 were significantly different between SCA RPL and no SCA RPL (p = 0.030, 0.031, and 0.040 respectively). Frequencies of those three gene polymorphisms between SCA RPL and controls also were significantly different (p = 0.017, 0.028, and 0.029 respectively). Moreover, the frequency of the G allele at rs20551 locus, the T allele at rs132788 locus and the A allele at rs1805388 locus was significantly higher in SCA RPL than no SCA RPL (OR = 3.227, p = 0.005; OR = 1.978, p = 0.008 and OR = 1.769, p = 0.036 respectively) and controls (OR = 7.130, p = 0.000; OR = 2.157, p = 0.004; OR = 2.397, p = 0.003 respectively). Additionally, the frequency of radiation-induced translocation in no SCA samples with rs20551, rs132788 or rs1805388 was significantly higher compared with the wild type samples (p = 0.015, 0.012, and 0.007 respectively).
Conclusion: Our results suggest that rs20551, rs132788, and rs1805388 might be associated with the risk of SCA. Larger scales of genetic variations studies and functional experiments are necessary to further confirm these findings.
Introduction
Structural chromosome abnormality (SCA) is an important cause of human diseases including recurrent pregnancy loss (RPL) (Rai and Regan, 2006). In approximately 2–5% of couples with RPL, one partner (more often the woman) will have a genetically balanced SCA (RCOOG, 2011).
Types of SCA include translocation, inversion, deletion, Tandem duplication, ring chromosome, etc. (Morin et al., 2017; Menghi et al., 2018; Panday et al., 2021). The most common SCA in women with RPL is translocation (usually 60% reciprocal and 40% Robertsonian approximately), and the segregation during meiosis can result in gametes with duplication or deficiency of chromosome segments (Prosée et al., 2020). Chromosome inversion is also associated with a higher risk of RPL, and the risk of RPL is affected by the size and genetic content of the rearranged chromosomal segments (Nagirnaja et al., 2014; Page and Silver, 2016).
The biogenesis of SCA is remarkably poorly understood. Generally, the formation of SCA is considered a multistep process, and the initial event is the concomitant occurrence of DNA double-strand breaks (DSB) in multiple chromosomal locations (Nambiar and Raghavan, 2011). It is generally agreed that DSBs repair, especially non-homologous end joining (NHEJ) repair, plays an important role in the formation of SCA (Chang et al., 2017).
The human EP300, XRCC6, LIG4, XRCC4, PRKDC, and DCLRE1C were identified as playing critical roles in NHEJ repair (Tropberger et al., 2013; Wang et al., 2013; Ochi et al., 2015; Manickavinayaham et al., 2019). EP300 encodes the E1A binding protein p300 which functions as histone acetyltransferase and regulates transcription via chromatin remodeling (Tropberger et al., 2013). XRCC6 locates on chromosome 22q13, coding the X-ray repair cross-complementing protein 6 (also named Ku70), which can be readily participated in repairing a DSB (Zhao et al., 2020). Moreover, DNA LIG4 is also essential for DSBs repair (Grawunder et al., 1998). The protein encoded by XRCC4 functions together with DNA LIG4 and the DNA-dependent protein kinase in the repair of DSBs (Zolner et al., 2011), and polymorphisms within these genes have been shown contributing to cancers and other disorders caused by genomic instability (Singh et al., 2018; Garcia et al., 2019). PRKDC encodes the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), is a candidate regulator of DSBs repair (Bunting and Nussenzweig, 2013). Additionally, DCLRE1C encodes Artemis, as one co-chaperone of DNA-PKcs, could bind to Ku70-Ku80-DNA complex and processes the DSBs (Bunting and Nussenzweig, 2013). We hypothesize that polymorphisms within those six DSBs repair related genes might contribute to the formation of SCA.
In the present study, we investigated the potential contribution of EP300, XRCC6, LIG4, XRCC4, PRKDC, and DCLRE1C gene polymorphisms to structural chromosome abnormalities (SCA) based on recurrent pregnancy loss. We used targeted WES in a relatively small exploratory sample at the first stage, and then confirmed by Sanger sequencing in a lager cohort including all exploratory sample and confirmatory sample. Finally, we also detected the frequency of radiation-induced chromosome translocations in no SCA samples with significant polymorphisms by fluorescence in situ hybridization (FISH).
Materials and Methods
Ethics Approval Statement
The study was approved by the Ethics Committee of the Third Xiangya Hospital (Quick 19159). Informed consent was obtained from all subjects involved in the study.
Study Subjects
The 142 affected individuals, all were RPL (54 with SCA and 88 without SCA), had no history of endocrine, metabolic, autoimmune, or other systemic disorders, thrombophilia, or uterine anatomic abnormalities. We recruited the RPL in strict accordance with the Practice Committee of the American Society for Reproductive Medicine (Practice Committee of the American Society for Reproductive Medicine, 2020). The controls included 84 age-matched fertile women in pregnancy and had no history of complicated pregnancies, miscarriages, still births, small for gestational age fetuses, preeclampsia, ectopic pregnancy, preterm delivery or any other pregnancy complication. Chromosomal abnormalities were excluded in the control by karyotype results. The demographic and clinical characteristics also were collected.
The flowchart for the study design was shown in Figure 1. We first used targeted WES to identify significant SNPs in relatively small exploratory samples (n = 75) at the first stage, and then confirmed by Sanger sequencing in a larger cohort (n = 226) including all exploratory samples (n = 75) and confirmatory samples (n = 151). Finally, to further confirm the association of significant SNPs with SCA, we detected the frequency of radiation-induced (2Gy X-ray) chromosome translocations in normal karyotype RPL peripheral blood lymphocytes (PBLs) with significant gene polymorphisms by FISH.
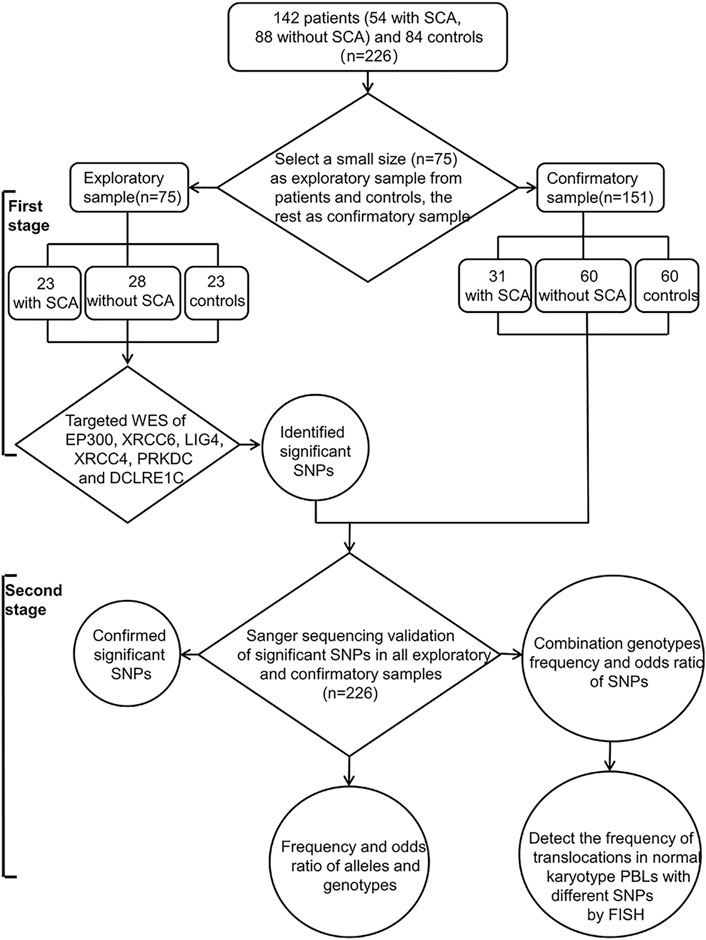
FIGURE 1. Flowchart of this study design. There are two stages in our study: The first stage, to identify the significant SNPs by targeted WES in a relatively small size exploratory sample (n = 75); Second, validation in a larger sample size (n = 226, exploratory and confirmatory sample) using Sanger sequencing, and then detect the frequency of radiation-induced translocations in normal karyotype PBLs with different genotype by FISH. SCA: Structural chromosome abnormalities; WES: Whole-exome sequencing; SNPs: Single nucleotide polymorphisms; PBLs: Peripheral blood lymphocytes; FISH: fluorescent in situ hybridization.
Peripheral Blood Karyotype Analysis
A standard 72-h lymphocyte culture of peripheral blood (2–5 ml) from each patient was performed to produce Metaphases for karyotyping. G banding was performed by a trypsin pretreatment of chromosomes followed by Giemsa staining. Chromosomes’ analysis was done using MetaSystems Ikaros (ZEISS, Germany) and karyotypes were reported according to International System for Human Cytogenetic Nomenclature (Simons et al., 2013). Karyotype analysis was performed using at least 20 Metaphases for each sample. The number was expanded to 100 metaphases in case of suspected mosaicism.
Screening Single Nucleotide Polymorphisms by Targeted Whole-Exome Sequencing
We first detected 75 samples (23 with SCA, 28 without SCA and 24 controls) by targeted whole-exome sequencing (WES). Genotyping of SNPs was performed with the WES-based targeted sequence analysis and Sanger sequencing. The library was constructed with the kit (Vazyme VAHTS UniveRPLl Plus DNA Library Prep Kit for Illumina, United States) by the standard procedure according to the manufacturer’s instructions. xGen® Lockdown® Probes (Nanodigmbio, United States, Sequences presented in Supplementary Table S1) were used to capture the target genes. Sequencing was carried out in NovaSeq 6000 (Illumina). FastQC was used to filter the raw data. The sequenced reads were aligned to the human reference genome 19 (HG19) using BWA MEN, and PCR duplicates were marked with PICARD. Variants were called by GATK HaplotypeCaller with default parameters, and retained considering DP (reads depth) ≥8, MQ (Mapping Quality) ≥20. Variation annotation was done in ANNOVAR software, variants with VAF≤0.01 [1000 genome project (2015) and ExAC Project] in the coding region and splicing site were filtered out, and the VAF >0.01 were kept.
SNPs Validation (Sanger Sequencing)
All significant SNPs detected were verified by Sanger sequencing (ABI 3730XL, United States). SNPs were reported according to Human Genome Variation Society nomenclature (Dunnen and Antonarakis, 2000). The sequences for PCR primers are listed in Supplementary Table S2.
Detection of the Translocations by Fluorescence in situ Hybridization (FISH)
FISH was used to detect the radiation-induced chromosome translocations in peripheral blood lymphocytes (PBLs) from normal karyotype RPL after 2Gy X-rays as previously described (Nakano et al., 2001). Metaphases were harvested after co-cultured with colchicine for 2 h. Chromosomes 1 and 4 were painted green by in situ hybridization with composite probes labeled with SYBR green (Cytocell, United Kingdom), chromosomes 2 were painted red by in situ hybridization with composite probes labeled with Rhodamine B (Cytocell, United Kingdom). The observed frequency of translocations (Fp) detected by FISH represents the frequency between painted chromosomes 1, 2, and 4 and the remaining counterstained chromosomes. To compare Fp with the values for translocations detected by the conventional method that detects aberrations involving the entire chromosome set, it is necessary to estimate the genome-equivalent frequency of translocations (FG). Thus, since the fraction of the total genomic DNA content represented by painted chromosomes 1, 2, and 4 to the total genome is 0.228 for males and 0.224 for females, Fp was multiplied by 2.771 for males and 2.806 for females to estimate FG; the basic method used is essentially that described by Pearce (Pearce et al., 2012). 400 metaphase splitting images were observed for each sample by three observers. The experiments were repeated three times.
Statistical Analysis
All statistical analyses were performed using SSPS 25.0 (IBM Corp, Chicago). Prism 7.0 software (GraphPad, United States) was utilized to draw figures. Qualitative data were expressed as n (%) and analyzed using Pearson chi-square (χ2) test (n ≥ 40 and expected frequencies ≥5). Measurement data were expressed as‾x (SD). Differences in genotype and allele frequencies (n < 40 or expected frequencies <5) between affected individuals and controls, as well as deviations from Hardy-Weinberg equilibrium were determined using Fisher exact test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to evaluate the contribution of EP300, XRCC6, LIG4, XRCC4, PRKDC, and DCLRE1C gene polymorphisms to SCA. p < 0.05 was considered to be statistically significant.
Results
Demographic and Clinical Characteristics of Subjects
The demographic and clinical characteristics of the affected individuals and controls are summarized in Table 1. Overall, the mean age, education level, body mass index, smoking, alcohol use, menarche age, menstrual cycle, age of pregnancy, TSH, PRL, Ureaplasma urealyticum, Mycoplasma hominus, Toxoplasma gondii, rubella, cytomegalovirus, herpes virus, LA, ß2-GPI, and aCL were similar between affected individuals and controls. No significant differences were identified (p > 0.05) (Table 1).
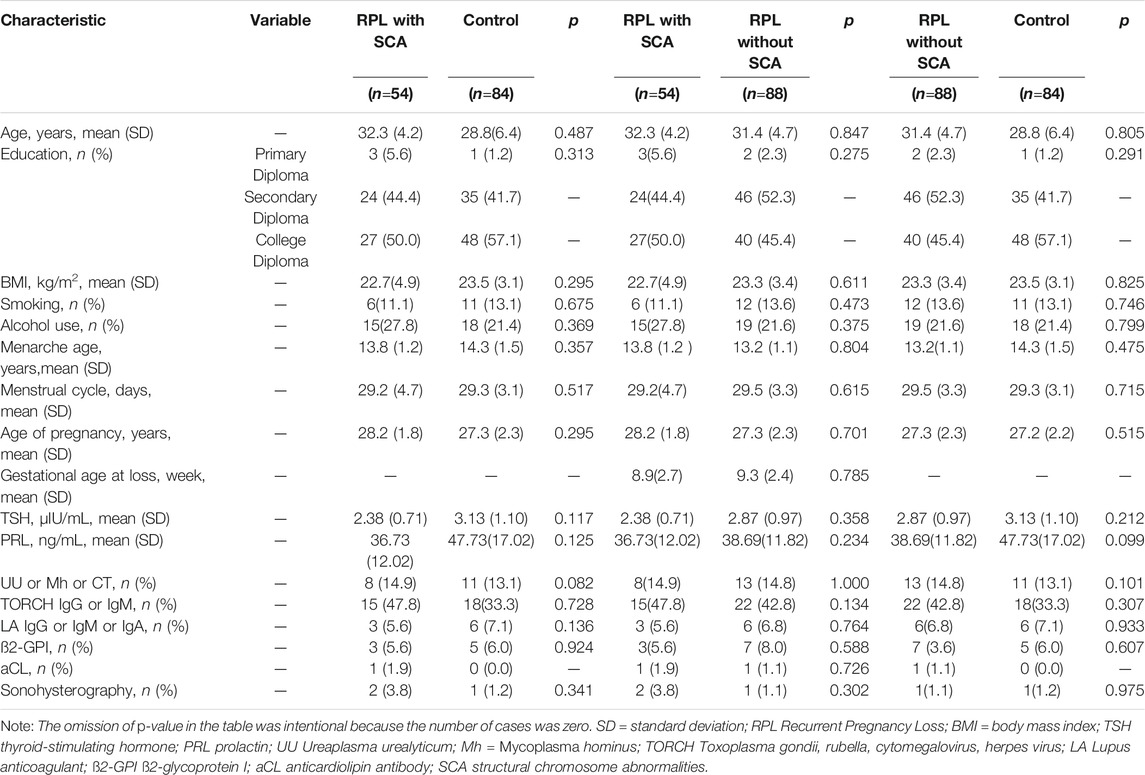
TABLE 1. Demographic and clinical characteristics of the affected RPL individuals and the controls. SD = standard deviation.
Distribution of Structural Chromosome Abnormalities (SCA)
There were 54 RPL with SCA: 32 carried a balanced reciprocal translocation, among them, chromosome 2 were involved in the translocation most frequently (n = 7) followed by chromosomes 1, 4, 11, and 12 (n = 5, each), chromosomes 3, 5, and 9 (n = 2, each), chromosomes 6, 17, and 19 were not involved; 8 carriers of inversions were observed, 7 were inversions of chromosome 9 and 1 inversion of chromosome 8; 14 carried a balanced Robertsonian translocation (Supplementary Table S3).
Results of Sequencing
A total of 35 polymorphisms had been identified in our samples (Table 2), nine within EP300, two within XRCC6, four within LIG4, three within XRCC4, ten within PRKDC and seven within DCLRE1C by WES. In EP300 polymorphisms, three were non-synonymous variants, six were synonymous variants. All XRCC6 polymorphisms identified were synonymous variants, while all LIG4 polymorphisms were non-synonymous variants and only one non-synonymous variant was identified in XRCC4. Additionally, most polymorphisms in PRKDC and DCLRE1C were non-synonymous variants (Table 2). There was no missing data. The alleles and genotype frequencies of all the polymorphism loci in control were consistent with the Hardy-Weinberg equilibrium (p > 0.05, data not shown).
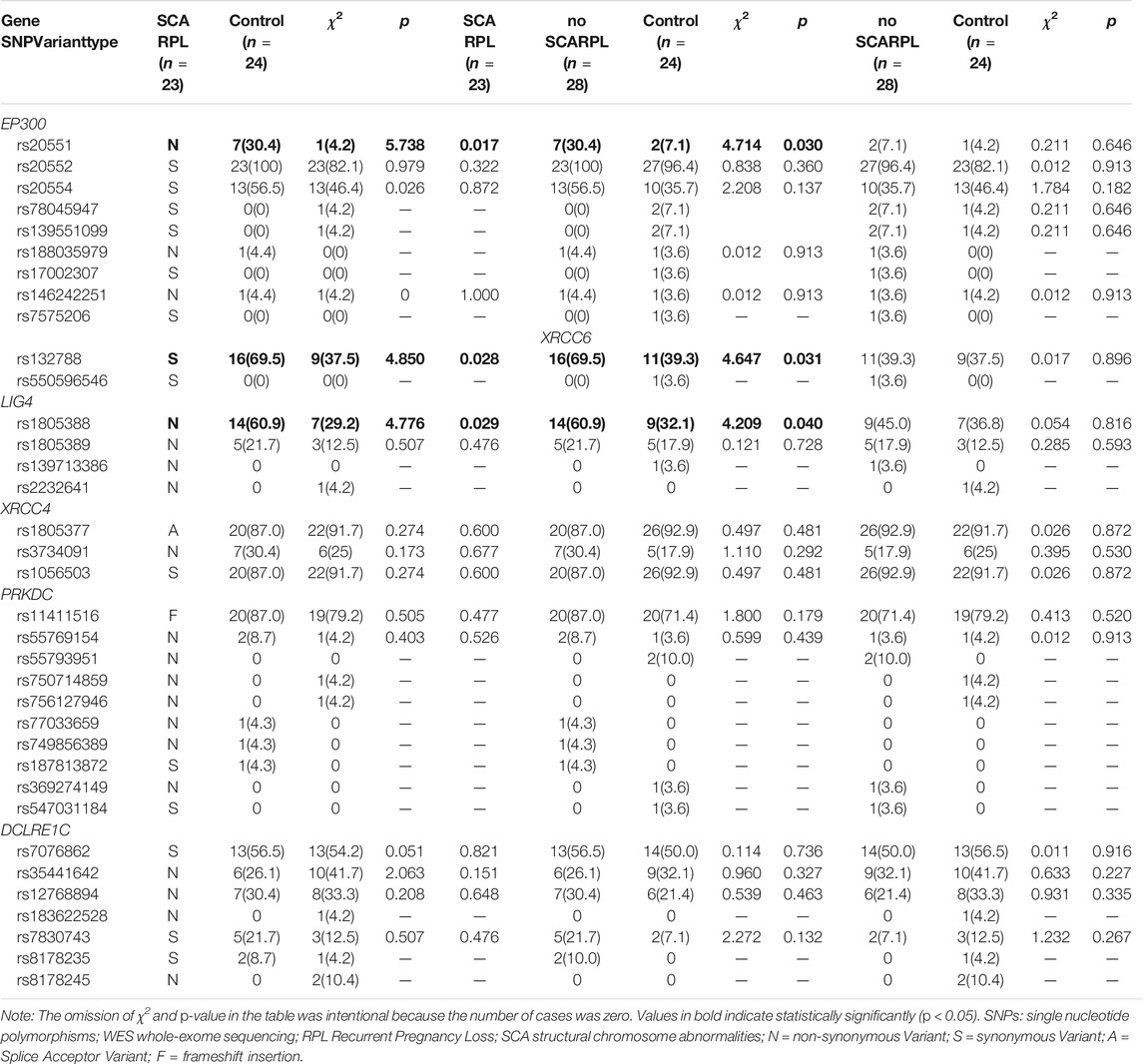
TABLE 2. Frequency of SNPs found in exploratory sample among affected RPL individuals and controls identified by the WES, n (%).
Frequencies of EP300 rs20551, XRCC6 rs132788, and LIG4 rs1805388 were statistically significantly different between RPL with SCA and RPL without SCA group (p = 0.030, 0.031, 0.040 respectively). Frequencies of those three gene polymorphisms between RPL with SCA group and controls were also shown significantly different (p = 0.017, 0.028, and 0.029 respectively). All rs20551 were heterozygous, while rs132788 and rs1805388 consisted of heterozygotes and homozygotes, and verified by Sanger sequencing (Figure 2), the concordance rate was 100%. The frequency of the G allele at rs20551 locus, the T allele at rs132788 locus and the A allele at rs1805388 locus in SCA RPL was statistically significantly higher than the no SCA RPL (OR = 3.227, p = 0.005; OR = 1.978, p = 0.008; OR = 1.769, p = 0.036 respectively) and the control group (OR = 7.130, p = 0.000; OR = 2.157, p = 0.004; OR = 2.397, p = 0.003 respectively) (Table 3), indicating that these three significant polymorphisms could be risk factors of SCA.
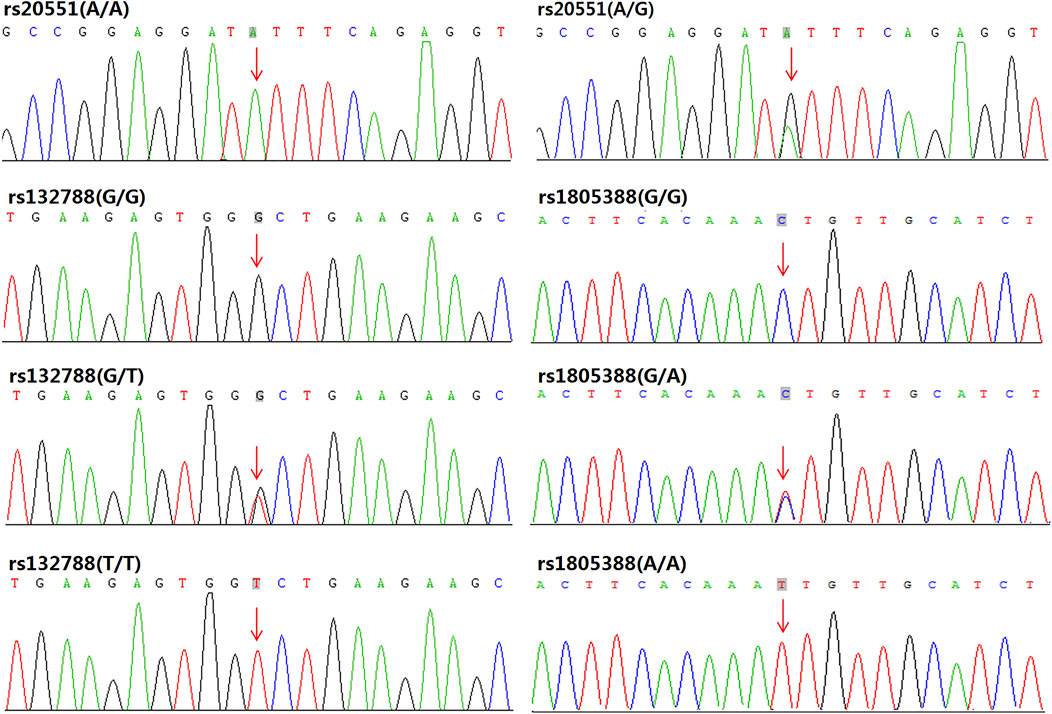
FIGURE 2. Sanger sequencing validation results for EP300 rs20551, XRCC6 rs132788, and LIG4 rs1805388: In our samples, all the rs20551 loci were heterozygous of A/G (except for the wild type A/A), while rs132788 consisted of heterozygotes (G/T) and homozygotes (T/T), and rs1805388 consisted of heterozygotes (G/A) and homozygotes (A/A), the consistency with results from WES was 100%.
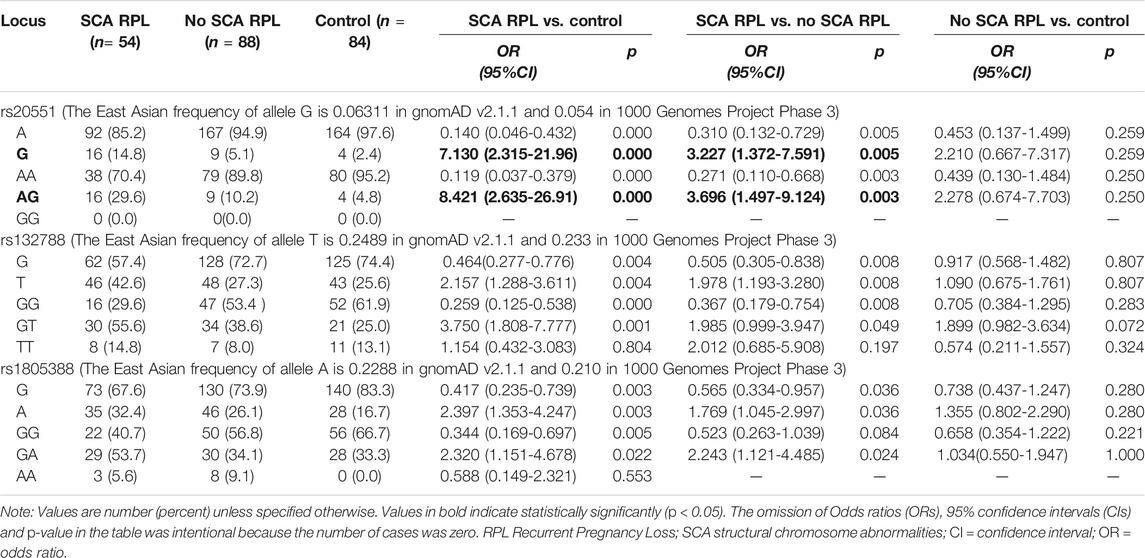
TABLE 3. Alleles and genotypes frequency of rs20551, rs132788, and rs1805388 in all RPL involved exploratory and confirmatory sample (integrated).
Genotype frequencies of the rs20551, rs132788, and rs1805388 also were analyzed, as shown in Table 3, the rs20551 (AG), rs132788 (GT) and rs1805388 (GA) odds ratios were significantly greater in SCA RPL vs. no SCA RPL (Table 3, OR 3.696, 95% CI, 1.497–9.124, p = 0.003; OR 1.985, 95% CI, 0.999–3.947, p = 0.049 and OR 2.243, CI, 1.121–4.485, p = 0.024 respectively). However, no significant differences were observed in the frequencies of any combination of genotypes between affected RPL individuals and controls (p > 0.05, Table 4).
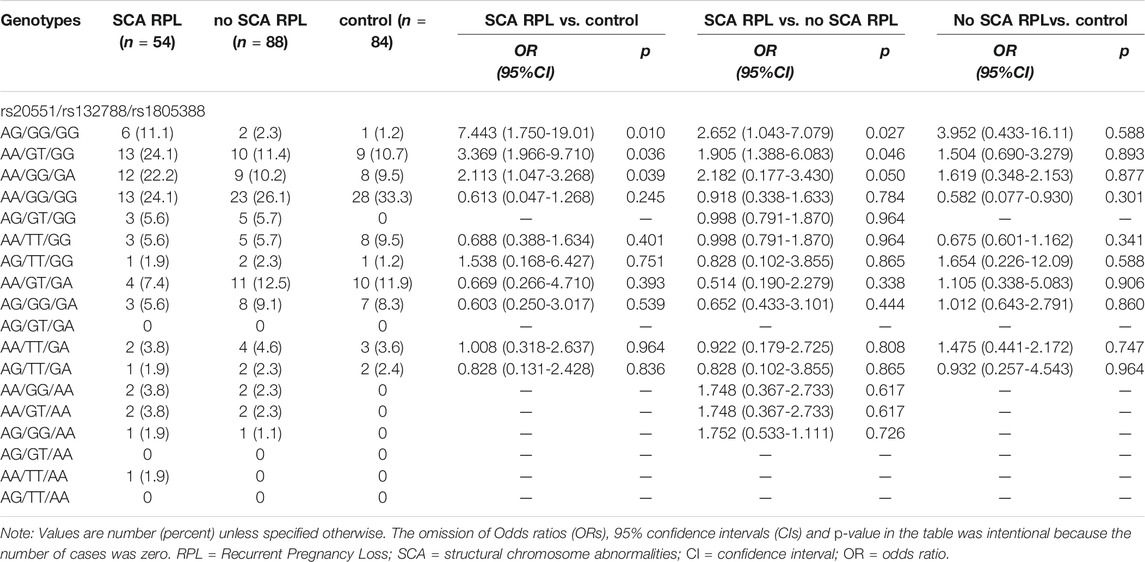
TABLE 4. Combination genotypes frequency of rs20551, rs132788, and rs1805388 in all RPL involved exploratory and confirmatory sample (integrated).
Frequencies of Translocations in No SCA Samples With Different Genotypes
To further confirm the association of significant SNPs (rs20551/rs132788/rs1805388) with SCA, FISH was used to detect the radiation-induced chromosome translocations (the most common SCA) in different genotype peripheral blood lymphocytes (PBLs) from no SCA RPL after 2Gy X-rays. The result demonstrates the frequencies of radiation-induced chromosome translocations in AG/GG/GG, AA/GT/GG and AA/GG/GA PBLs were significantly higher than that in AA/GG/GG (wild type) PBLs (Figure 3, p = 0.015, p = 0.012, p = 0.007 respectively).
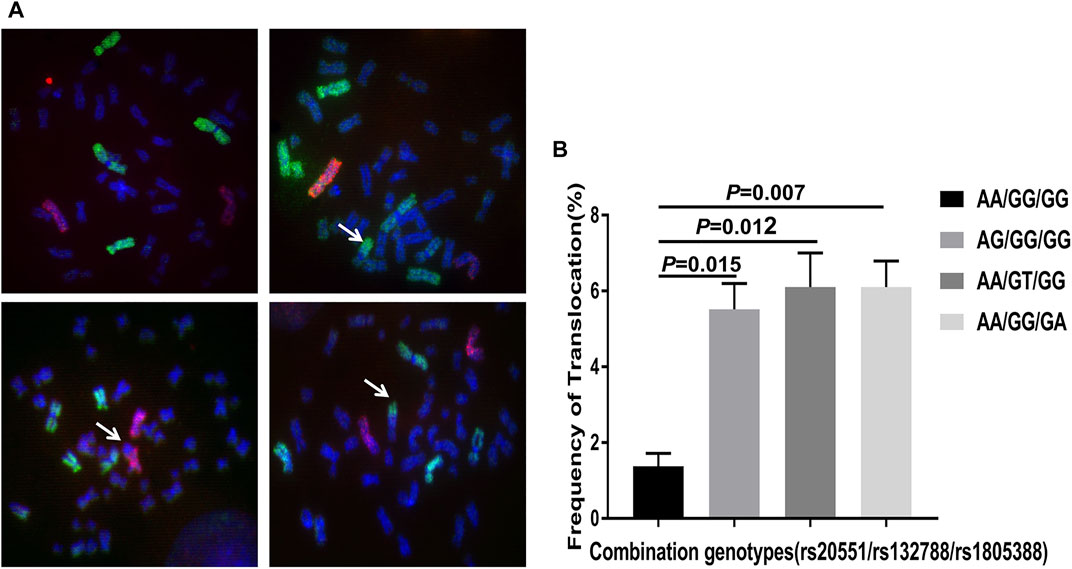
FIGURE 3. Fluorescence in situ hybridization (FISH) using chromosome 1 probe (green), chromosome 2 probe (red) and chromosome 4 probe (green) (Cytocell, Cambridge, United Kingdom). (A) Representative images of photomicrographs showing FISH painted human chromosome 1, 4 (green) and chromosome 2 (red) in metaphase lymphocytes. No translocation in the upper left image, the upper right showing translocation involving chromosome 1 (arrow), the bottom left showing translocation involving chromosome 2 (arrow), the bottom right showing translocation involving chromosome 4 (arrow). (B) Analysis of the frequency of radiation-induced chromosome translocations in no SCA patients with different genotypes. The statistical analysis chart showing the frequencies of radiation-induced translocations in patients with AG/GG/GG (rs20551), AA/GT/GG (rs132788) and AA/GG/GA (rs1805388) genotype were significantly higher than that in AA/GG/GG (wild type) (p = 0.015, p = 0.012, p = 0.007 respectively). The experiments were repeated three times. FISH: fluorescent in situ hybridization; SCA: structural chromosome abnormality.
Discussion
In the present study, the potential association of EP300, XRCC6, LIG4, XRCC4, PRKDC, and DCLRE1C genes polymorphisms with structural chromosome abnormality (SCA) has been investigated by targeted whole-exome sequencing for the first time. EP300 rs20551, XRCC6 rs132788, and LIG4 rs1805388 frequencies were statistically significantly different between RPL with SCA and RPL without SCA. Moreover, no SCA peripheral blood lymphocytes (PBLs) with rs20551, rs132788, or rs1805388 locus were more prone to translocation after radiation. These findings provide evidence that DNA repair related genes polymorphisms could be an important contributor to the risk of SCA.
From few studies on the association of gene polymorphisms with SCA, one found a significant decrease in the distribution of T allele in MTHFR 677C > T polymorphisms among patients with chromosomal abnormalities (Sinthuwiwat et al., 2012). The rs231775 and rs3087243 of CTLA4, as well as rs2232365 and rs2232368 of Foxp3, all appeared to have chromosomal abnormalities (Fan et al., 2018). Before the present study, no gene polymorphism within EP300, XRCC6 and LIG4 genes was reported associated with SCA.
EP300 functions as histone acetyltransferase that regulates transcription via chromatin remodeling (Lundblad et al., 1995), plays a critical role in SCA. Histone acetyltransferase modification is considered to be an important factor in the formation of chromosomal translocation (Burgess, 2015). The acetylation of histone enrolls chromatin remodeling complexes to the nearby double-strand breaks (DSBs) sites, promoting the process of DNA damage repair (DDR) (Lee et al., 2010). It is known that DDR is considered to be the initiating molecular event in the formation of chromosome translocation (Nambiar and Raghavan, 2011). The rs20551 is a non-synonymous single nucleotide variant in EP300 locates on chromosome 22, with the change of c.2989A > G, resulting in the substitution of valine for isoleucine at codon 997 close to the Bromodomain (Li et al., 2017). It is known that the Bromodomain is a protein domain that recognizes acetylated lysine residues, and the recognition could be affected when some changes occur nearby. In our study, the frequency of G allele in rs20551 was significantly higher in SCA group than no SCA group, indicating that G allele in rs20551 could be a risk factor to SCA. The tentative explanation is that the acetylation of EP300 may be affected when the EP300 rs20551 is present, and the normal DNA repair pathways EP300 involved may also be affected as a consequence.
XRCC6 encodes the Ku 70 protein, which is crucial to repairing DSBs in identifying broken ends of DNA. In the process of DNA damage repair (DDR), Ku heterodimer composed of Ku 70 and Ku 80 binds to the broken DNA as the first molecule (Chanut et al., 2016), and a recruitment platform for subsequent repair enzymes is established (Williams et al., 2014). The basic steps of DDR have been biochemically defined to require DSBs detection by the Ku heterodimer, which functions in combination with XRCC4 and XLF (Williams et al., 2014). The rs132788 is a synonymous variant in XRCC6 with the change of c.1629G > T. Although the encoded amino acids not be changed (Gly > Gly), rate of protein synthesis could be influenced as the codon changes (Koutmou et al., 2015). A review and meta-analysis on risk factors for breast cancer showed that rs132788 (G > T) might be protective (Zhou et al., 2012), while another study suggested that the rs132788 polymorphism may be a susceptibility factor for radiation-induced oral mucositis in Chinese nasopharyngeal carcinoma patients (Ren et al., 2014). In our study, the frequency of the T allele in the XRCC6 rs132788 locus was significantly higher in the SCA affected individuals, clearly suggesting that rs132788 could be a susceptibility factor to SCA, filling in the gap of clinical significance reported in ClinVar database.
DNA LIG4 is essential for V(D)J recombination and DNA double-strand breaks (DSBs) repair through non-homologous end joining (NHEJ) (Grawunder et al., 1998; Zhao et al., 2020). Defects in LIG4 could lead to pronounced radio-sensitivity and confer a predisposition to leukemia (Riballo et al., 1999). Rs1805388 in LIG4 was also reported associated with increased radio-resistance (Mumbrekar et al., 2016). One study claimed the rs1805388 gene polymorphism is not a risk factor of cancer (Xie et al., 2014), while another study reported rs1805388 was associated with an increased glioma risk among smokers (Zhao et al., 2013). Additionally, LIG4 rs1805388 was also associated with susceptibility to male infertility (Ji et al., 2013). Our results showed the rs1805388 was strongly associated with SCA.
Although the SCA cases we used were derived from recurrent pregnancy loss (RPL), the three significant polymorphisms we found were not associated with RPL. When the no SCA.
RPL was compared to normal control, no significant polymorphism was found. The evidence is more robust that rs20551, rs132788 and rs1805388 are associated with the risk of SCA rather than RPL.
As one of the most important types of SCA, translocation is often assumed to form because of the joining of DSBs that arise at different sites on non-homologous chromosomes (Bunting and Nussenzweig, 2013). One study suggests that Ku70 can increase DSB rejoining and translocation levels in LIG4-deficient G1-arrested progenitor B cells (Liang et al., 2021). Translocations were also increased in a reporter system in mouse embryonic stem cells when XRCC4–XLF was inactivated (Simsek and Jasin, 2010). Our results also show that polymorphisms within EP300, XRCC6 (Ku70), and LIG4 might affect the risk of translocation.
Despite sufficient powerful mastery and analysis, one of the limitations of our study might be the relatively small sample size, which does not allow definite conclusion, especially for the analysis of the interaction between combined genotypes. Another limitation is only six genes in RPL women have been analyzed. Future studies of the other SCA cases are needed. Nevertheless, this study has several strengths including the use of human peripheral blood samples for analysis, case-control and inclusion of typical clinical affected individuals with SCA. Significantly higher frequencies of EP300 rs20551 (A/G), XRCC6 rs132788 (G/T) and LIG4 rs1805388 (G/A) were found in SCA group.
In conclusion, our study improved the understanding of genetic polymorphisms within the EP300, XRCC6, LIG4, XRCC4, PRKDC, and DCLRE1C genes with structure chromosomal abnormalities (SCA). EP300 rs20551, XRCC6 rs132788 and LIG4 rs1805388 might be associated with the risk of SCA. This all could be useful in guiding future research into molecular mechanisms of SCA and uncovering the partial pathogenesis of human diseases caused by SCA. Moreover, these significant polymorphisms might also be valuable diagnostic markers and potential therapy targets for the affected RPL individuals with SCA.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics’ Committee of the Third Xiangya Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
ZC and KX conceived and designed the study. LG and YH collected the data, managed the database, and analyzed the data. WZ and JL contributed to the interpretation of the data. YL and CZ provided peripheral blood karyotype technology and information. DC performed the experiments. ZC and KX drafted and revised the manuscript. All authors have approved the final version of the manuscript to be published.
Funding
This study was supported by the National Natural Science Foundation of China (81471499 to KX), the Natural Science Foundation of Hunan province (2019JJ40347 to KX) and the Fundamental Research Funds for the Central Universities of Central South University (2018zzts243 to ZC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to all the participants in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.787718/full#supplementary-material
References
Bunting, S. F., and Nussenzweig, A. (2013). End-joining, Translocations and Cancer. Nat. Rev. Cancer 13 (7), 443–454. doi:10.1038/nrc3537
Burgess, D. J. (2015). Chromatin: Histone Influences on Chromosomal Translocations. Nat. Rev. Genet. 16 (8), 438–439. doi:10.1038/nrg3979
Chang, H. H. Y., Pannunzio, N. R., Adachi, N., and Lieber, M. R. (2017). Non-homologous DNA End Joining and Alternative Pathways to Double-Strand Break Repair. Nat. Rev. Mol. Cell Biol 18 (8), 495–506. doi:10.1038/nrm.2017.48
Chanut, P., Britton, S., Coates, J., Jackson, S. P., and Calsou, P. (2016). Coordinated Nuclease Activities Counteract Ku at Single-Ended DNA Double-Strand Breaks. Nat. Commun. 7, 12889. doi:10.1038/ncomms12889
Dunnen, J. T. d., and Antonarakis, S. E. (2000). Mutation Nomenclature Extensions and Suggestions to Describe Complex Mutations: a Discussion. Hum. Mutat. 15 (1), 7–12. doi:10.1002/(sici)1098-1004(200001)15:1<7:aid-humu4>3.0.co;2-n
Fan, Q. e., Zhang, J., Cui, Y., Wang, C., Xie, Y., Wang, Q., et al. (2018). The Synergic Effects of CTLA-4/Foxp3-Related Genotypes and Chromosomal Aberrations on the Risk of Recurrent Spontaneous Abortion Among a Chinese Han Population. J. Hum. Genet. 63 (5), 579–587. doi:10.1038/s10038-018-0414-2
Garcia, J. A., Kalacas, N. A., Sy Ortin, T., Ramos, M. C., and Albano, P. M. (2019). XRCC4 c.1394G>T Single Nucleotide Polymorphisms and Breast Cancer Risk Among Filipinos. Asian Pac. J. Cancer Prev. 20 (4), 1097–1101. doi:10.31557/apjcp.2019.20.4.1097
Grawunder, U., Zimmer, D., Fugmann, S., Schwarz, K., and Lieber, M. R. (1998). DNA Ligase IV Is Essential for V(D)J Recombination and DNA Double-Strand Break Repair in Human Precursor Lymphocytes. Mol. cell 2 (4), 477–484. doi:10.1016/s1097-2765(00)80147-1
Ji, G., Yan, L., Liu, W., Huang, C., Gu, A., and Wang, X. (2013). Polymorphisms in Double-Strand Breaks Repair Genes Are Associated with Impaired Fertility in Chinese Population. Reproduction 145 (5), 463–470. doi:10.1530/rep-12-0370
Koutmou, K. S., Radhakrishnan, A., and Green, R. (2015). Synthesis at the Speed of Codons. Trends Biochemical Sciences 40 (12), 717–718. doi:10.1016/j.tibs.2015.10.005
Lee, H.-S., Park, J.-H., Kim, S.-J., Kwon, S.-J., and Kwon, J. (2010). A Cooperative Activation Loop Among SWI/SNF, γ-H2AX and H3 Acetylation for DNA Double-Strand Break Repair. Embo J. 29 (8), 1434–1445. doi:10.1038/emboj.2010.27
Li, J., Ding, N., Wang, X., Mi, L., Ping, L., Jin, X., et al. (2017). EP300 Single Nucleotide Polymorphism Rs20551 Correlates with Prolonged Overall Survival in Diffuse Large B Cell Lymphoma Patients Treated with R-CHOP. Cancer Cell Int 17, 70. doi:10.1186/s12935-017-0439-1
Liang, Z., Kumar, V., Le Bouteiller, M., Zurita, J., Kenrick, J., Lin, S. G., et al. (2021). Ku70 Suppresses Alternative End Joining in G1-Arrested Progenitor B Cells. Proc. Natl. Acad. Sci. USA 118 (21), e2103630118. doi:10.1073/pnas.2103630118
Lundblad, J. R., Kwok, R. P. S., Laurance, M. E., Harter, M. L., and Goodman, R. H. (1995). Adenoviral ElA-Associated Protein P300 as a Functional Homologue of the Transcriptional Co-activator CBP. Nature 374 (6517), 85–88. doi:10.1038/374085a0
Manickavinayaham, S., Vélez-Cruz, R., Biswas, A. K., Bedford, E., Klein, B. J., Kutateladze, T. G., et al. (2019). E2F1 Acetylation Directs p300/CBP-Mediated Histone Acetylation at DNA Double-Strand Breaks to Facilitate Repair. Nat. Commun. 10 (1), 4951. doi:10.1038/s41467-019-12861-8
Menghi, F., Barthel, F. P., Yadav, V., Tang, M., Ji, B., Tang, Z., et al. (2018). The Tandem Duplicator Phenotype Is a Prevalent Genome-wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer cell 34 (2), 197–210. doi:10.1016/j.ccell.2018.06.008
Morin, S. J., Eccles, J., Iturriaga, A., and Zimmerman, R. S. (2017). Translocations, Inversions and Other Chromosome Rearrangements. Fertil. sterility 107 (1), 19–26. doi:10.1016/j.fertnstert.2016.10.013
Mumbrekar, K. D., Goutham, H. V., Vadhiraja, B. M., and Bola Sadashiva, S. R. (2016). Polymorphisms in Double Strand Break Repair Related Genes Influence Radiosensitivity Phenotype in Lymphocytes from Healthy Individuals. DNA repair 40, 27–34. doi:10.1016/j.dnarep.2016.02.006
Nagirnaja, L., Palta, P., Kasak, L., Rull, K., Christiansen, O. B., Nielsen, H. S., et al. (2014). Structural Genomic Variation as Risk Factor for Idiopathic Recurrent Miscarriage. Hum. Mutat. 35 (8), 972–982. doi:10.1002/humu.22589
Nakano, M., Kodama, Y., Ohtaki, K., Itoh, M., Delongchamp, R., Awa, A. A., et al. (2001). Detection of Stable Chromosome Aberrations by FISH in A-Bomb Survivors: Comparison with Previous Solid Giemsa Staining Data on the Same 230 Individuals. Int. J. Radiat. Biol. 77 (9), 971–977. doi:10.1080/09553000110050065
Nambiar, M., and Raghavan, S. C. (2011). How Does DNA Break during Chromosomal Translocations? Nucleic Acids Res. 39 (14), 5813–5825. doi:10.1093/nar/gkr223
Ochi, T., Blackford, A. N., Coates, J., Jhujh, S., Mehmood, S., Tamura, N., et al. (2015). PAXX, a Paralog of XRCC4 and XLF, Interacts with Ku to Promote DNA Double-Strand Break Repair. Science 347 (6218), 185–188. doi:10.1126/science.1261971
Page, J. M., and Silver, R. M. (2016). Genetic Causes of Recurrent Pregnancy Loss. Clin. Obstet. Gynecol. 59 (3), 498–508. doi:10.1097/grf.0000000000000217
Panday, A., Willis, N. A., Elango, R., Menghi, F., Duffey, E. E., Liu, E. T., et al. (2021). FANCM Regulates Repair Pathway Choice at Stalled Replication forks. Mol. cell 81 (11), 2428–2444. doi:10.1016/j.molcel.2021.03.044
Pearce, M. S., Salotti, J. A., Little, M. P., McHugh, K., Lee, C., Kim, K. P., et al. (2012). Radiation Exposure from CT Scans in Childhood and Subsequent Risk of Leukaemia and Brain Tumours: a Retrospective Cohort Study. The Lancet 380 (9840), 499–505. doi:10.1016/s0140-6736(12)60815-0
Practice Committee of the American Society for Reproductive Medicine Definitions of Infertility and Recurrent Pregnancy Loss: a Committee Opinion. Fertil. Steril (2020) 113(3):533–535. doi:10.1016/j.fertnstert.2019.11.025
Prosée, R. F., Wenda, J. M., and Steiner, F. A. (2020). Adaptations for Centromere Function in Meiosis. Essays Biochem. 64 (2), 193–203. doi:10.1042/ebc20190076
Rai, R., and Regan, L. (2006). Recurrent Miscarriage. The Lancet 368 (9535), 601–611. doi:10.1016/s0140-6736(06)69204-0
RCOOG (2011). The Investigation and Treatment of Couples with Recurrent First Trimester and Second Trimester Miscarriage. London: RCOG.
Ren, J.-H., Dai, X.-F., Yan, G.-L., Jin, M., Liu, C.-W., Yang, K.-Y., et al. (2014). Acute Oral Mucositis in Nasopharyngeal Carcinoma Patients Treated with Radiotherapy: Association with Genetic Polymorphism in DNA DSB Repair Genes. Int. J. Radiat. Biol. 90 (3), 256–261. doi:10.3109/09553002.2014.873558
Riballo, E., Critchlow, S. E., Teo, S.-H., Doherty, A. J., Priestley, A., Broughton, B., et al. (1999). Identification of a Defect in DNA Ligase IV in a Radiosensitive Leukaemia Patient. Curr. Biol. 9 (13), 699–S2. doi:10.1016/s0960-9822(99)80311-x
Simons, A., Shaffer, L. G., and Hastings, R. J. (2013). Cytogenetic Nomenclature: Changes in the ISCN 2013 Compared to the 2009 Edition. Cytogenet. Genome Res. 141 (1), 1–6. doi:10.1159/000353118
Simsek, D., and Jasin, M. (2010). Alternative End-Joining Is Suppressed by the Canonical NHEJ Component Xrcc4-Ligase IV during Chromosomal Translocation Formation. Nat. Struct. Mol. Biol. 17 (4), 410–416. doi:10.1038/nsmb.1773
Singh, P. K., Mistry, K. N., Chiramana, H., Rank, D. N., and Joshi, C. G. (2018). Exploring the Deleterious SNPs in XRCC4 Gene Using Computational Approach and Studying Their Association with Breast Cancer in the Population of West India. Gene 655, 13–19. doi:10.1016/j.gene.2018.02.040
Sinthuwiwat, T., Poowasanpetch, P., Wongngamrungroj, A., Soonklang, K., Promso, S., Auewarakul, C., et al. (2012). Association of MTHFR Polymorphisms and Chromosomal Abnormalities in Leukemia. Dis. markers 32 (2), 115–121. doi:10.1155/2012/292507
Tropberger, P., Pott, S., Keller, C., Kamieniarz-Gdula, K., Caron, M., Richter, F., et al. (2013). Regulation of Transcription through Acetylation of H3K122 on the Lateral Surface of the Histone Octamer. Cell 152 (4), 859–872. doi:10.1016/j.cell.2013.01.032
Wang, Z., Lin, H., Hua, F., and Hu, Z.-w. (2013). Repairing DNA Damage by XRCC6/KU70 Reverses TLR4-Deficiency-Worsened HCC Development via Restoring Senescence and Autophagic Flux. Autophagy 9 (6), 925–927. doi:10.4161/auto.24229
Williams, G. J., Hammel, M., Radhakrishnan, S. K., Ramsden, D., Lees-Miller, S. P., and Tainer, J. A. (2014). Structural Insights into NHEJ: Building up an Integrated Picture of the Dynamic DSB Repair Super Complex, One Component and Interaction at a Time. DNA repair 17, 110–120. doi:10.1016/j.dnarep.2014.02.009
Xie, S., Shan, X.-F., Shang, K., Xu, H., He, J., and Cai, Z.-G. (2014). Relevance of LIG4 Gene Polymorphisms with Cancer Susceptibility: Evidence from a Meta-Analysis. Sci. Rep. 4, 6630. doi:10.1038/srep06630
Zhao, B., Rothenberg, E., Ramsden, D. A., and Lieber, M. R. (2020). The Molecular Basis and Disease Relevance of Non-homologous DNA End Joining. Nat. Rev. Mol. Cell Biol 21 (12), 765–781. doi:10.1038/s41580-020-00297-8
Zhao, P., Zou, P., Zhao, L., Yan, W., Kang, C., Jiang, T., et al. (2013). Genetic Polymorphisms of DNA Double-Strand Break Repair Pathway Genes and Glioma Susceptibility. BMC cancer 13, 234. doi:10.1186/1471-2407-13-234
Zhou, L.-P., Luan, H., Dong, X.-H., Jin, G.-J., Man, D.-L., and Shang, H. (2012). Association between XRCC5, 6 and 7 Gene Polymorphisms and the Risk of Breast Cancer: a HuGE Review and Meta-Analysis. Asian Pac. J. Cancer Prev. 13 (8), 3637–3643. doi:10.7314/apjcp.2012.13.8.3637
Zolner, A. E., Abdou, I., Ye, R., Mani, R. S., Fanta, M., Yu, Y., et al. (2011). Phosphorylation of Polynucleotide Kinase/Phosphatase by DNA-dependent Protein Kinase and Ataxia-Telangiectasia Mutated Regulates its Association with Sites of DNA Damage. Nucleic Acids Res. 39 (21), 9224–9237. doi:10.1093/nar/gkr647
Keywords: structural chromosome abnormalities, gene polymorphisms, DNA double-strand breaks, non-homologous end joining, EP300, whole-exome sequencing, recurrent pregnancy loss
Citation: Cheng Z, Cheng D, Li J, Guo L, Zhang W, Zhang C, Liu Y, Huang Y and Xu K (2021) Polymorphisms Within DNA Double-Strand Breaks Repair-Related Genes Contribute to Structural Chromosome Abnormality in Recurrent Pregnancy Loss. Front. Genet. 12:787718. doi: 10.3389/fgene.2021.787718
Received: 01 October 2021; Accepted: 06 December 2021;
Published: 23 December 2021.
Edited by:
Fan Jin, Zhejiang University, ChinaReviewed by:
Arvind Panday, Harvard Medical School, United StatesZhang Zhihua, Fudan University, China
Najmeh Ahangari, Mashhad University of Medical Sciences, Iran
Copyright © 2021 Cheng, Cheng, Li, Guo, Zhang, Zhang, Liu, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keqian Xu, eHVrZXFpYW5AY3N1LmVkdS5jbg==
 Zhenbo Cheng
Zhenbo Cheng Dehua Cheng
Dehua Cheng Jiancheng Li1,2
Jiancheng Li1,2