
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 06 December 2021
Sec. Human and Medical Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.784215
Dermatomyositis (DM), an inflammatory disorder, is often associated with interstitial lung disease (ILD). However, the underlying mechanism remains unclear. Our study performed RNA sequencing (RNA-seq) and integrative bioinformatics analysis of differentially expressed genes (DEGs) in patients with dermatomyositis-associated interstitial lung disease (DM-ILD) and healthy controls. A total of 2,018 DEGs were identified between DM-ILD and healthy blood samples. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that DEGs were mainly involved in immune- and inflammatory-related biological processes and pathways. Disease ontology (DO) enrichment analysis identified 35 candidate key genes involved in both skin and lung diseases. Meanwhile, a total of 886 differentially expressed alternative splicing (AS) events were found between DM-ILD and healthy blood samples. After overlapping DEGs with differential AS genes, the plasminogen activator and urokinase receptor (PLAUR) involved in immune-related biological processes and complement and coagulation cascades was screened and identified as the most important gene associated with DM-ILD. The protein–protein interaction (PPI) network revealed that PLAUR had interactions with multiple candidate key genes. Moreover, we observed that there were significantly more neutrophils and less naive B cells in DM-ILD samples than in healthy samples. And the expression of PLAUR was significantly positively correlated with the abundance of neutrophils. Significant higher abundance of PLAUR in DM-ILD patients than healthy controls was validated by RT-qPCR. In conclusion, we identified PLAUR as an important player in regulating DM-ILD by neutrophil-associated immune response. These findings enrich our understanding, which may benefit DM-ILD patients.
Dermatomyositis (DM) is an autoimmune disease characterized by the inflammation of skeletal muscles and skin lesions. DM is the most common subtype of idiopathic inflammatory myopathies (Lilleker et al., 2018), with interstitial lung disease (ILD) accounting for most of the mortality (Vuillard et al., 2018; Hervier and Uzunhan, 2019). However, patients with ILD may have minimal, absent, or late-onset myositis or skin lesions (Friedman et al., 1996; Tillie-Leblond et al., 2008). The lack of correlation between myositis and pulmonary symptoms often leads to delayed diagnosis and a compromised therapeutic response. In addition, MHC polymorphisms (Miller et al., 2013), epigenetic modification, and miRNA activity (Gao et al., 2018; Gao et al., 2019), which are important regulators of gene expression, have been reported to play a role in DM pathogenesis. For these reasons, the identification of key molecules involved in DM-ILD is highly required for improving the clinical outcome, and the role of their transcriptional regulations in the etiology of DM has great significance for investigation.
Alternative splicing (AS) is the process by which different splice sites in precursor messenger RNA (mRNA) are selected to generate multiple mRNA isoforms. It is an essential and ubiquitous mechanism for regulating gene expression. AS events increase the diversity of self-antigens in specific tissues and/or different developmental stages and create an additional challenge for the immune system to avoid self-reactivity (Nilsen and Graveley, 2010). AS is also closely related to immune function and autoimmune disease. Activated T cells expressed a γ-chain mRNA splice isoform as an active immunoregulatory molecule, which promotes inflammatory T-cell immune responses in vivo (Hong et al., 2014). Exons in autoimmune disease and type 1 diabetes candidate genes were more likely to be differentially expressed and alternatively spliced than the expressed exons of other genes (Newman et al., 2017). However, few studies on AS events in DM-ILD have been performed.
Therefore, the present study was designed to explore the role of AS in the pathogenesis of DM-ILD and identify AS-associated key genes involved in DM-ILD via RNA-seq.
Peripheral blood was obtained from 16 DM-ILD patients and 16 healthy controls. There was no significant difference in the general characteristics between the disease group and the control group (Supplementary Table S1). There was no significant difference in age and clinical data in the DM-ILD patients (Table 1, Table 2). All DM-ILD patients were treated with glucocorticoids and immunosuppressive agents, including prednisone acetate, methylprednisolone, methotrexate, cyclophosphamide, tacrolimus, and cyclosporin A.
Whole blood samples were collected from six DM-ILD patients (ID: LR20L26D X196, LR20L26DX197, LR20L26DX198, LR20L26DX199, LR20L26DX200, and LR20L26DX201) and six healthy controls (ID: LR20L26DX202, LR20L26DX203, LR20L26DX204, LR20L26DX205, LR20L26DX206, and LR20L26DX207) for RNA- seq. Total RNA was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. The RNA amount and purity of each sample were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, United States). The RNA integrity was assessed by Bioanalyzer 2100 (Agilent, CA, United States), with a RIN number >7.0, and confirmed by electrophoresis with denaturing agarose gel. Poly(A) RNA is purified from 1 μg total RNA using Dynabeads Oligo (dT)25-61005 (Thermo Fisher, CA, United States) using two rounds of purification. Then the poly(A) RNA was fragmented into small pieces using the Magnesium RNA Fragmentation Module (NEB, Cat. e6150, United States) under 94°C for 5–7 min. Then the cleaved RNA fragments were reverse-transcribed to create the cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, Cat. 1896649, United States), which were next used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I (NEB, Cat. m0209, United States), RNase H (NEB, cat. m0297, United States), and dUTP Solution (Thermo Fisher, Cat. R0133, United States). The A-base is then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contains a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Single- or dual-index adapters are ligated to the fragments, and size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme (NEB, Cat. m0280, United States) treatment of the U-labeled second-stranded DNA, the ligated products are amplified with PCR by the following conditions: initial denaturation at 95°C for 3 min; eight cycles of denaturation at 98 C for 15 s, annealing at 60°C for 15 s, extension at 72°C for 30 s; and then final extension at 72°C for 5 min. The average insert size for the final cDNA library was 300 ± 50 bp. At last, we performed the 2 × 150 bp paired-end sequencing (PE150) on an Illumina NovaSeq™ 6000 (LC-Bio Technology CO., Ltd. Hangzhou, China) following the vendor’s recommended protocol. The quality of sequencing data was evaluated by FastQC (version 0.11.9). The raw data were filtered by Trimmomatic (version 0.39). Then clean data were mapped to the reference genome of Homo sapiens GRCh38 using HISAT2 (version 2.2.1) with default parameters. Gene counts were calculated by FeatureCount (version 2.0.0). DEGs were identified by R package DESeq2 (version 1.26.0) with | log2 (fold change) | ≥1 and p-value < 0.05. AS events were analyzed by RMATS (version 3.1.1), and differentially expressed AS events were screened with FDR ≤0.05 and |IncLevelDifference| ≥ 0.1 (Shen et al., 2012) (Shen et al., 2014). Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and disease ontology (DO) enrichment analysis were analyzed using R package clusterProfiler. The quantitative analysis of interactive sets between different types of AS was displayed in UpSet plot. The detail of differentially expressed genes (DEGs) and differentially expressed AS events in the chromosome was visualized in the Circos plot.
After DO analysis, DEGs that were enriched into both skin and lung diseases were selected and identified as candidate key genes. Then by overlapping candidate key genes with parent genes of differentially expressed AS, key genes associated with DM-ILD were identified. Key genes and candidate key genes were input into the STRING database to analyze the interactions, and the PPI network was visualized by Cytoscape software (version 3.6.1).
CIBERSORT was applied to assess the infiltration of 22 immune cells in different subgroups, including naive B cells, memory B cells, plasma cells, CD8 T cells, naive CD4 T cells, resting memory CD4 T cells, activated memory CD4 T cells, follicular helper T cells, regulatory T cells (Tregs), gamma delta T cells, resting NK cells, activated NK cells, monocytes, M0 macrophages, M1 macrophages, M2 macrophages, resting dendritic cells, activated dendritic cells, resting mast cells, activated mast cells, eosinophils, and neutrophils. Then the t test was used to compare the difference of immune infiltration between six DM-ILD patients and six healthy controls. The correlations between key genes and differentially distributed immune cells were calculated.
Total RNA of ten DM-ILD patients and ten control subjects were isolated from peripheral blood using TRIzol reagent (15596018, Life Technologies, American). And RNA was reverse-transcribed to cDNA via the SureScript-First Strand cDNA Synthesis Kit (QP057, GeneCopoeia, Guangzhou, China). The expression of PLAUR was detected by Bio-Rad CFX96 (American) using BlazeTaq™ SYBR® Green qPCR Mix 2.0 (QP033, GeneCopoeia, Guangzhou, China). PCR amplification cycle conditions were 95°C 30 s, 95°C 10 s, 60°C 20 s, and 72°C 30 s for 40 cycles. GAPDH was used as the internal standardized reference. The relative expression level was calculated using the 2−ΔΔCt method. The specific primers were as follows: PLAUR, 5′-CGAGGTTGT GTGTGGGTTA-3’ (forward) and 5′-GGCACTGTTCTTCAGGGCT-3’ (reverse); GAPDH, 5′- CGCTGAGTACGTCGTGGAGTC-3′ (forward) and 5′- GCTGATGAT CTTGAGGCTGTTGTC-3′ (reverse).
All data were analyzed by R (version 4.0.0). Student’s t test was used to compare differences between healthy controls and DM-ILD. Pearson’s correlations between the expressions of key genes and abundance of differentially infiltrated immune cells were calculated. P-values less than or equal to 0.05 were considered statistically significant.
The quality scores, sequence content, and duplication levels of raw sequencing data were evaluated by FastQC and are shown in Supplementary Material S1. To obtain high-quality data for following analysis, low-quality reads were discarded, and the reads contaminated with adaptor sequences were trimmed using Trimmomatic. Thereafter, quality control of clean data was performed by FastQC and is shown in Supplementary Material S2. Statistical results of raw data and clean data are shown in Supplementary Material S3, proving that clean data can be used for following mapping. Then the clean data were mapped to the GRCh38 reference genome, and we found that the percentage of mapped reads was over 79% (Supplementary Material S4), indicating that the clean data meet the needs for following analyses. Next, gene counts were evaluated by FeatureCount (Supplementary Material S5), and the expressions of genes in each sample were calculated. Principal component analysis by R package ggplot2 showed that samples within the same group had good replication (Supplementary Figure S1). A total of 2,018 DEGs were found, including 1,084 upregulated and 934 downregulated genes in DM-ILD patients relative to healthy ones (Figure 1A). The expressions of top 50 upregulated and top 50 downregulated genes in each sample were displayed in the heatmap (Figure 1B).
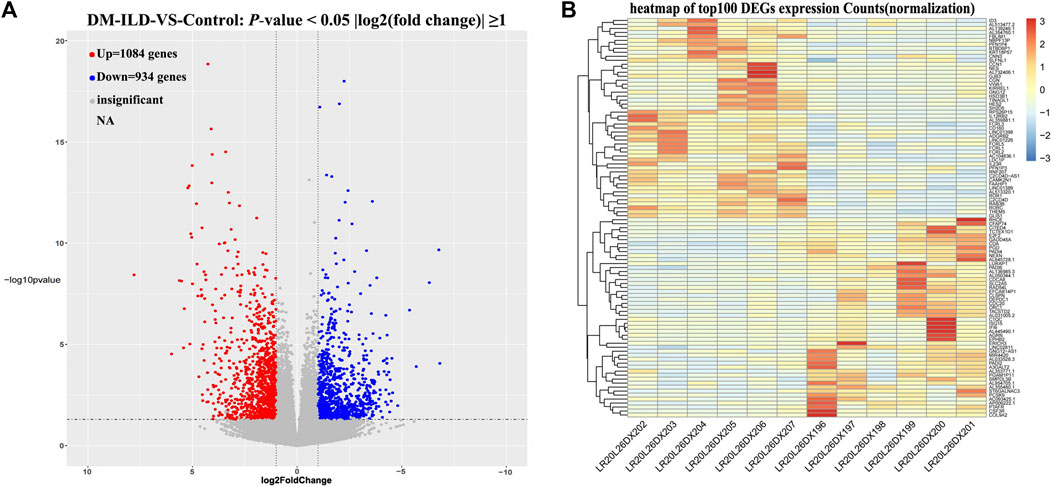
FIGURE 1. Differentially expressed mRNAs between DM-ILD and control samples. (A) 1,084 upregulated and 934 downregulated genes in DM-ILD patients relative to healthy ones were identified using a cutoff as “two-fold expression difference and p-value < 0.05.” (B) Expressions of top 50 upregulated and top 50 downregulated genes in each sample were displayed in the heatmap.
Thereafter, we investigated the biological function of DEGs in DM-ILD. We found that DEGs were mainly enriched into immune and inflammation biological processes, such as neutrophil activation, neutrophil-mediated immunity, neutrophil degranulation, neutrophil activation involved in immune response, leukocyte migration, humoral immune response, leukocyte chemotaxis and response to the molecule of bacterial origin (Figure 2A), and KEGG pathways, such as cytokine–cytokine receptor interaction, ECM-receptor interaction, IL-17 signaling pathway, NF-kappa B signaling pathway, PI3K-Akt signaling pathway, NOD-like receptor signaling pathway, complement and coagulation cascades, and JAK-STAT signaling pathway (Figure 2B). All of these results indicated that DEGs involved in DM-ILD had a strong relationship with immunity and inflammation. To explore the connection of DEGs and diseases, we further performed DO analysis. We found that DEGs were associated with multiple diseases, such as lung disease, skin disease, arteriosclerotic cardiovascular disease, bacterial infectious disease, obstructive lung disease, and female reproductive system disease (Figure 2C).
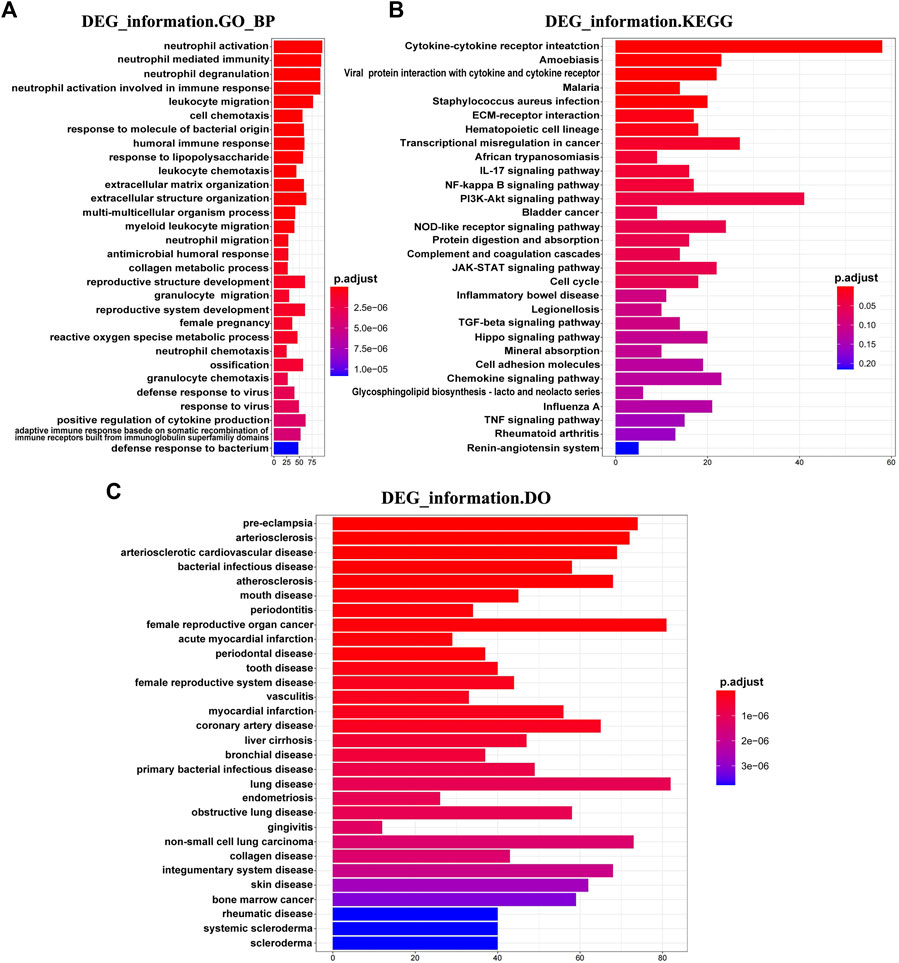
FIGURE 2. Functional enrichment of DEGs in DM-ILD. (A) DEGs were mainly enriched into immune and inflammation biological processes in GO analysis. (B) KEGG pathways indicated that DEGs involved in DM-ILD had a relationship with immune and inflammation. (C) DEGs were associated with multiple diseases in DO enrichment analysis.
By analyzing the transcriptome data, five AS forms, exon skipping (SE), alternative 5 and 3′ splice sites (A5SS and A3SS), mutually exclusive exons (MXE), and intron retention (RI) were identified in DM-ILD patients and healthy ones. After comparison, we obtained 866 differentially expressed AS events from 697 genes, including 97 A3SS in 92 genes, 53 A5SS in 53 genes, 97 MXE in 79 genes, 44 RI in 44 genes, and 575 SE in 494 genes (Figure 3A), indicating that one gene had one or more AS events. Therefore, we used UpSet plot to visualize the intersecting sets of each AS form. As shown in Figure 3B, most genes (91.25%) had only one AS type and 61 genes had two to four types of AS events, including a combination of MXE and SE, A3SS and SE, A5SS and SE, RI and A3SS, MXE and A3SS, RI and A5SS, A5SS and MXE, RI and SE, RI, A5SS, A3SS and SE, MXE, and A3SS and SE. The volcano plots shown in Figure 3C showed the upregulated and downregulated AS events of each AS form. Specifically, there were 54 upregulated and 43 downregulated A3SS, 23 upregulated and 30 downregulated A5SS, 308 upregulated and 267 downregulated SE, 32 upregulated and 12 downregulated RI, and 37 upregulated and 60 downregulated MXE events. Top 50 upregulated and top 50 downregulated AS events in each AS type were displayed in the heatmaps (Figure 3D).
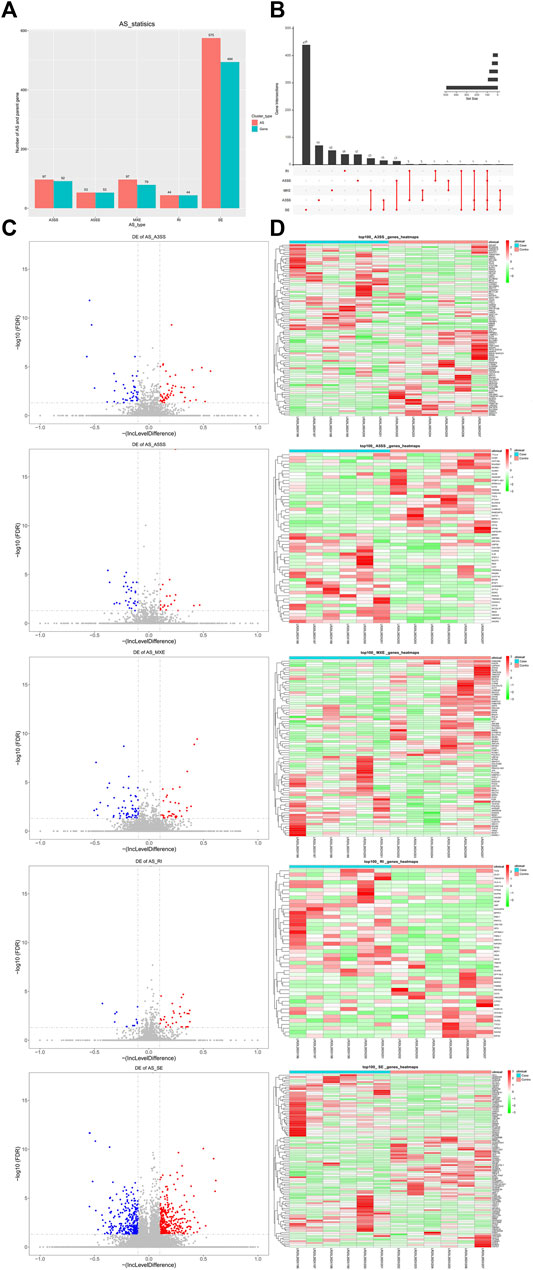
FIGURE 3. Differentially expressed AS events in between DM-ILD and control samples. (A) 866 differentially expressed AS events from 697 genes, including 97 A3SS in 92 genes, 53 A5SS in 53 genes, 97 MXE in 79 genes, 44 RI in 44 genes, and 575 SE in 494 genes were obtained by RMATS and screened with FDR ≤0.05 and |IncLevelDifference| ≥ 0.1. (B) Intersecting sets of each AS form. (C) Volcano plots showed the upregulated and downregulated AS events of each AS form. (D)Top 50 upregulated and top 50 downregulated AS events in each AS type were displayed in the heatmaps.
To screen key genes associated with DM-ILD, we extracted 35 common genes from lung disease and skin disease in DO analysis (Figure 4A), namely, BDKRB2, CAMP, COL1A1, F2RL1, F3, MS4A2, CXCR3, ICAM1, IGFBP3, IGHE, IL1A, IL1B, IL1RN, IL6, CXCL8, IL9R, IL10, IL18, CXCL10, KRT18, LAMA5, LAMC2, LEP, MET, CXCL9, MMP1, MMP2, MMP9, PLAUR, PPARG, RNASE2, RNASE3, SOD2, TLR4, and IL1RL1. Using “Dermatomyositis” or “Pulmonary interstitial fibrosis” or “Inflammation” or “Immunization” as key words on the VarElect database, we found that all of these 35 genes were associated with inflammation and immunization. Thus, these 35 genes were identified as candidate key genes. To further explore the role of AS, we overlapped 35 genes with parent genes of differentially expressed AS events and identified PLAUR as the key gene in AS-dependent DM-ILD (Figure 4B). We found that PLAUR had upregulated SE within chr19:43665316–43665459 in DM-ILD (Figure 4C). According to GO and KEGG pathway enrichment analyses, PLAUR genes were found to participate in complement and coagulation cascades, neutrophil-associated immune response, and coagulation-related biological responses, such as neutrophil activation, neutrophil-mediated immunity, neutrophil degranulation, neutrophil activation involved in immune response, blood coagulation, and negative regulation of coagulation. Next, we constructed the PPI network and found that PLAUR had interactions with multiple candidate key genes, including CXCL8, F3, ICAM1, IL1B, IL6, MET, MMP1, MMP2, and MMP9 (Figure 4D). Moreover, we generated a Circos plot to give a full view of DEGs, differentially expressed AS events, and the detail of PLAUR (Figure 5). By RNA-seq, we found that the abundance of PLAUR was higher in DM-ILD samples than the control ones (Figure 6A). Consistent with our sequencing results, RT-qPCR showed that the expression of PLAUR was significantly elevated in the DM-ILD samples (Figure 6B).
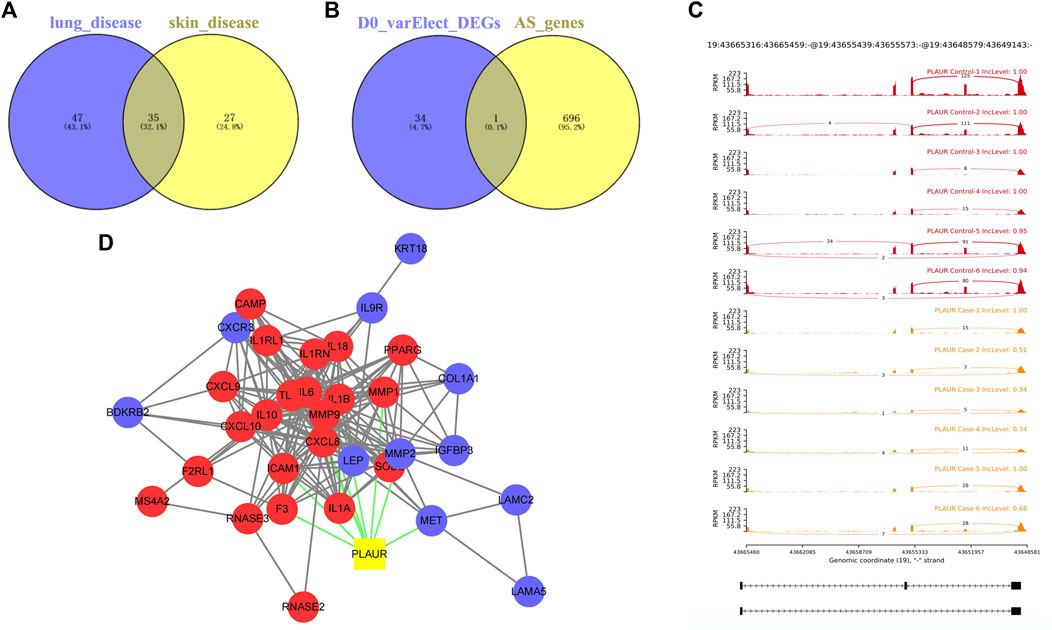
FIGURE 4. Key genes associated with DM-ILD. (A) 35 common genes from lung disease and skin disease in DO analysis. (B) PLAUR as the key gene in AS-dependent DM-ILD was identified by overlapping 35 genes with the parent genes of differentially expressed AS events. (C) PLAUR had upregulated SE within chr19:43665316–43665459 in DM-ILD. (D) PPI network showed that PLAUR had interactions with multiple candidate key genes.
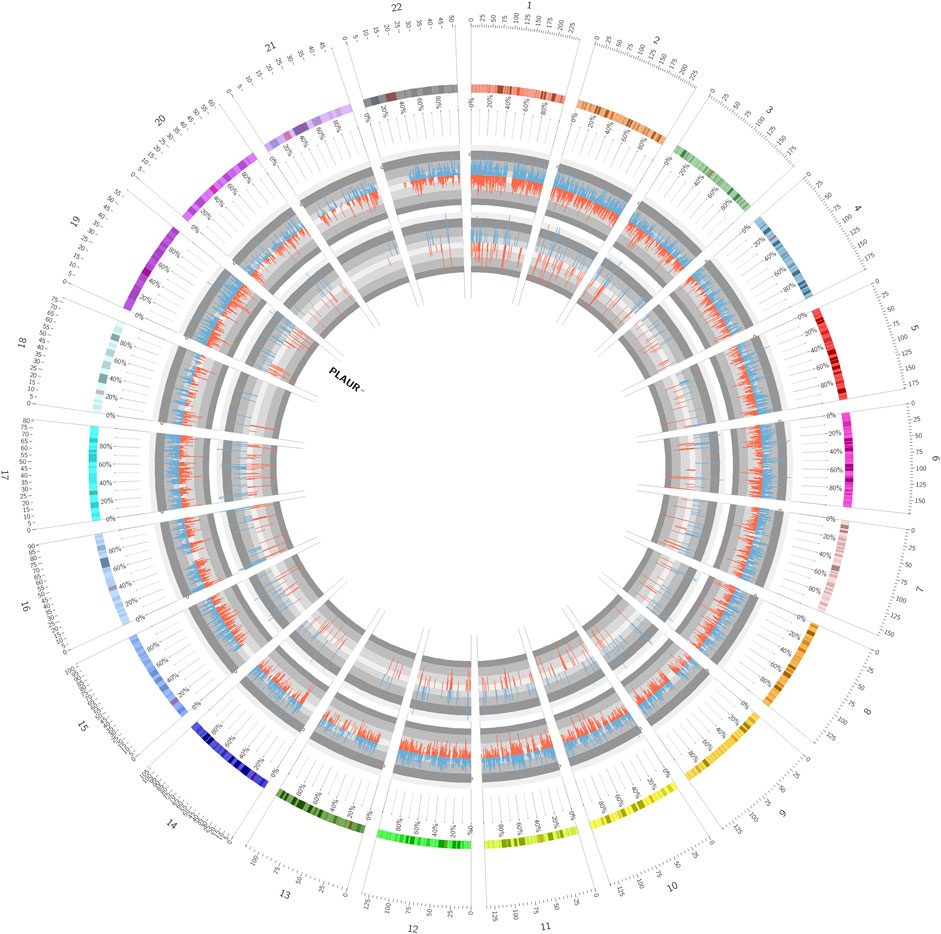
FIGURE 5. Circos plot showed a full view of DEGs, differentially expressed AS events, and the detail of PLAUR.
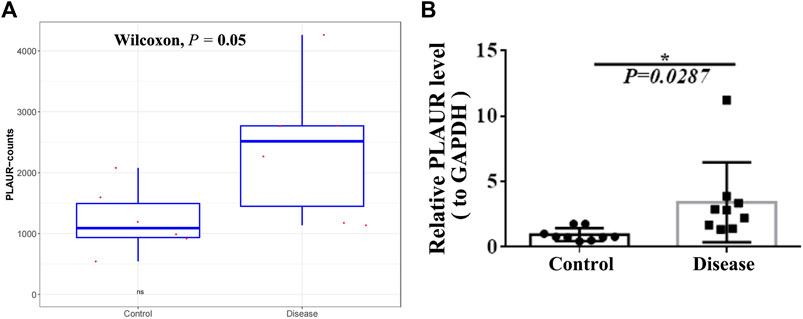
FIGURE 6. Expression of PLAUR in DM-ILD and control samples. (A) Abundance of PLAUR was higher in DM-ILD samples than control ones in RNA-sequencing. (B) Expression of PLAUR was significant elevated in the DM-ILD samples by RT-qPCR.
DM-ILD was associated with the immune cells. Given the role of immune cells in the immune system, we evaluated the abundance of 22 immune cell types in each sample from DM-ILD patients and healthy ones (Figure 7A). After removing activated memory CD4 T cells, follicular helper T cells, gamma delta T cells, M0 macrophages, M1 macrophages, resting mast cells, and eosinophils with no or few infiltrations in both DM-ILD patients and health ones, the abundance of the remaining 15 immune cell types was compared. We observed that there were significantly more neutrophils and less naive B cells in DM-ILD patients (Figure 7B and Supplementary Figure S2). Given that PLAUR was involved in immune-related biological processes and pathways, we calculated the correlations between PLAUR and neutrophils or naive B cells. Notably, a strong positive correlation (cor = 0.93) was observed between the expression of PLAUR and the abundance of neutrophils (Figure 7C).
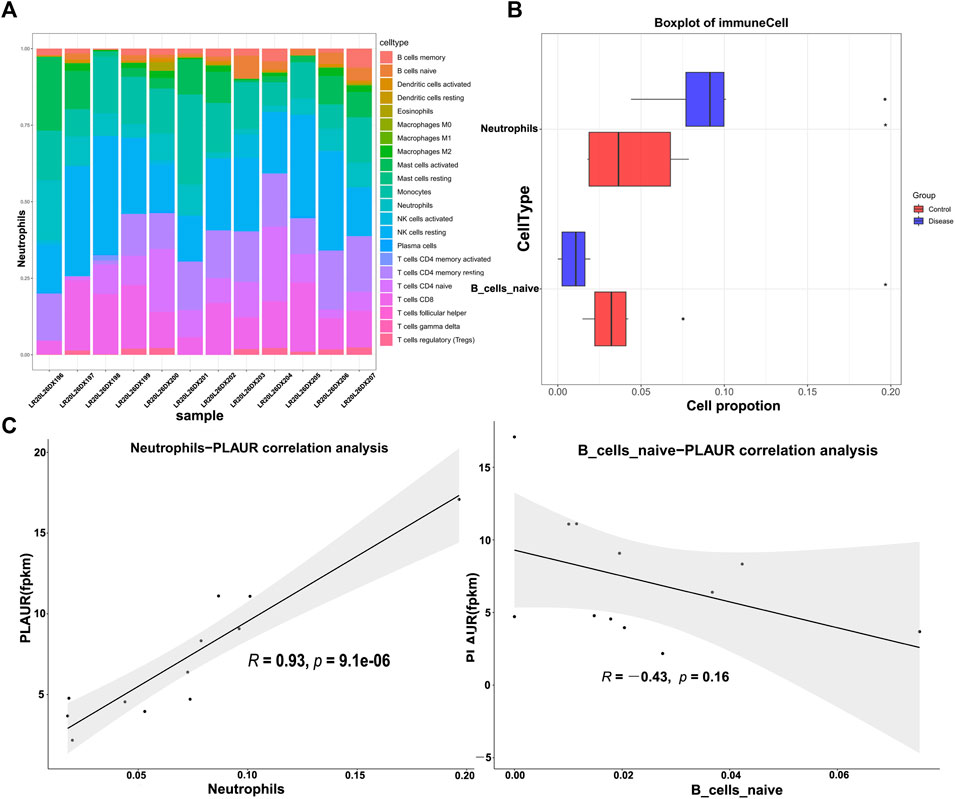
FIGURE 7. Immune infiltration analysis. (A) Abundance of 22 immune cell types in each sample of DM-ILD patients and healthy ones. (B) More neutrophils and less naive B cells were detected in DM-ILD patients. (C) Correlations between the expression of PLAUR and differentially infiltrated immune cells (neutrophils and naive B cells) were displayed in the plot.
Pulmonary involvement is a common complication of DM, causing a significant impact on patient morbidity and mortality. However, only a small number of gene markers in DM-ILD have been explored. In this study, we examined the blood samples in DM-ILD and healthy groups to identify novel candidate biomarkers and biological pathways involved in the etiology of DM.
We identified 2,018 DEGs between DM-ILD patients and healthy individuals. These DEGs were mainly enriched in the immune and inflammation biological processes and pathways. Among them, neutrophil-related biological processes were the ones at the top, with neutrophil activation ranking the first, followed by neutrophil-mediated immunity, neutrophil degranulation, and neutrophil activation involved in immune response. In DM-ILD, a number of different neutrophil disturbances have been described. For instance, the aberrant neutrophil extracellular trap (NET) formation may be involved in the pathogenesis of ILD in DM patients (Peng et al., 2018), and proteinase 3, one type of neutrophil serine proteinases, is lower in patients of DM without ILD (Gao et al., 2018). We also confirmed previous observations related to the pathogenesis of DM and DM-ILD, including the presence of the cytokine–cytokine receptor interaction (Zhu et al., 2017; Chen et al., 2018), IL−17 signaling pathway (Nasr et al., 2012). Additionally, we observed that complement and coagulation cascade pathway may play a critical role in DM-ILD. This pathway has been reported in immune diseases, such as multiple sclerosis (Li et al., 2021), Ig A nephropathy, and Henoch–Schönlein purpura nephritis (Fang et al., 2021). Complement activation has been demonstrated in previous reports (Roy et al., 2020; Tripathi et al., 2021) to be associated with type I interferon, which was significantly activated in DM and DM-ILD (Zhang et al., 2019a; de Jesus et al., 2020). These reports combined with our findings suggest direct and/or indirect links between the two pathways implicated in DM-ILD pathogenesis.
Considering that ILD is one of the severe complications of DM, we focused on 35 genes which were enriched in skin diseases and lung diseases in DO analysis. Consistent with our previous results of functional enrichment, these 35 genes were related to inflammation and immunization. Thus, these 35 genes, namely, BDKRB2, CAMP, COL1A1, F2RL1, F3, MS4A2, CXCR3, ICAM1, IGFBP3, IGHE, IL1A, IL1B, IL1RN, IL6, CXCL8, IL9R, IL10, IL18, CXCL10, KRT18, LAMA5, LAMC2, LEP, MET, CXCL9, MMP1, MMP2, MMP9, PLAUR, PPARG, RNASE2, RNASE3, SOD2, TLR4, and IL1RL1 were identified as candidate key genes. Many studies have characterized that IL-6, IL8, IL-18, and IL-10 were significantly elevated in DM-ILD and associated with disease activity and prognosis (Gono et al., 2010; Gono et al., 2014; Chen et al., 2018). The important role of CXCL9 and CXCL10 in the pathophysiology of DM-ILD has also been reported. The serum levels of CXCL9 and CXCL10 were higher in anti-Jo-1 antibody-positive DM-ILD patients than in those with idiopathic pulmonary fibrosis, which distinguished this disease entity from idiopathic pulmonary fibrosis and anti-signal recognition particle antibody-positive myositis (Richards et al., 2009). Oda et al. reported that serum levels of CXCL10 of DM-ILD patients at 2 weeks after treatment initiation were significantly higher in the death group, and the change ratio of serum levels of CXCL10 and CXCL11 was higher in the death group, suggesting that serum levels of CXCL10 may be possible biomarkers of disease activity and prognosis in DM-ILD (Oda et al., 2017). However, the role of other genes such as PLAUR in DM-ILD has not been reported. PLAUR overexpression in DM-ILD patients than healthy control samples was validated by RT-qPCR in our study. The mechanism how the genes participate in the etiology and development of DM-ILD requires further investigation.
AS events have been studied in many diseases, such as cancer (Kahles et al., 2018), autoimmune disease (Aravilli et al., 2021), and diabetes (Newman et al., 2017). However, the mechanisms of AS in DM-ILD remained unexplored. Here, we present a systemic analysis of AS in DM-ILD and found 866 differentially expressed AS events, including SE, A3SS, A5SS, MXE, and RI. Moreover, one gene had one or more AS events. To further explore the role of AS, we overlapped 35 genes with the parent genes of differentially expressed AS events and identified PLAUR as the key gene in AS-dependent DM-ILD. Zhang et al. reported that PLAUR had indirect connections with miR-155 and miR-146a, which are the top-ranked miRNAs responding to cystic fibrosis plasma (Zhang et al., 2019b). PLAUR, IL4, COL3A1, CXCL8, MMP2, LOX, MMP14, COL1A2, and PLOD2 were also found to be slightly upregulated in alveolar fibroelastosis and in organizing pneumonia but downregulated in non-specific interstitial pneumonia and usual interstitial pneumonia (Jonigk et al., 2019). Increased uPAR expression, as seen in the bronchial epithelium of patients with asthma, leads to attenuated wound repair which may contribute to the development and progression of airway remodeling in asthma (Stewart et al., 2012). In the current study, PLAUR had interactions with multiple candidate key genes and was found to participate in complement and coagulation cascades, neutrophil-associated immune response, and coagulation-related biological responses. Although the exact function of DM-ILD remains unclear, PLAUR has been shown to be associated with autoimmune disease (Göbel et al., 2016; Kobayashi et al., 2021) and relates to pulmonary dysfunction. Thus, PLAUR may be involved in the pathogenesis of DM-ILD through immune-mediated mechanism.
Based on the aforementioned results, we hypothesized that immune cell distribution of the disease group and the control group might be different. We found more neutrophils and less naive B cells in DM-ILD patients via CIBERSORT analysis. A strong positive correlation was observed between the expression of PLAUR and the abundance of neutrophils. The important role of neutrophils in DM-ILD was confirmed again, suggesting that PLAUR may regulate DM-ILD through neutrophil-related immune response. There are some limitations in our study. We identified PLAUR between DM-ILD patients and normal control blood samples. Further studies will be needed to explore the expression of PLAUR in DM-ILD patients and DM patients without ILD complications and in other sample types such as pulmonary tissue. Moreover, the detailed function of PLAUR in DM needs to be studied in future in vivo and in vitro experiments.
In conclusion, BDKRB2, CAMP, COL1A1, F2RL1, F3, MS4A2, CXCR3, ICAM1, IGFBP3, IGHE, IL1A, IL1B, IL1RN, IL6, CXCL8, IL9R, IL10, IL18, CXCL10, KRT18, LAMA5, LAMC2, LEP, MET, CXCL9, MMP1, MMP2, MMP9, PLAUR, PPARG, RNASE2, RNASE3, SOD2, TLR4, and IL1RL1, which were associated with inflammation and immunization, were identified as key genes in DM-ILD. Moreover, the present study is the first to systematically analyze AS events in patients with DM-ILD and find the association of PLAUR expression with DM-ILD. Furthermore, we preliminarily explore the possible mechanisms such as neutrophil-related immune response in DM-ILD. These findings enrich our understanding of DM-ILD, which may benefit DM-ILD patients in future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by The Second Affliated Hospital of Kunming Medical University (Protocol Number: PJ-2021-101). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
XL and JC contributed to conceptualization and methodology. RZ helped with data curation and funding acquisition. MX and CL assisted with formal analysis. JC contributed to investigation and writing of the original draft. All authors contributed to the article and approved the submitted version.
This research was funded by the Yunnan Fundamental Research projects (Grant Number 2019FE001-230 (JC)) for experimental materials. We also received support from the National Natural Science Foundation of China (Grant Number 82060305 (RZ)).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.784215/full#supplementary-material
Aravilli, R. K., Vikram, S. L., and Kohila, V. (2021). The Functional Impact of Alternative Splicing and Single Nucleotide Polymorphisms in Rheumatoid Arthritis. Curr Pharm Biotechnol. 22 (8), 1014–1029. doi:10.2174/1389201021666201001142416
Chen, M., Quan, C., Diao, L., Xue, F., Xue, K., Wang, B., et al. (2018). Measurement of Cytokines and Chemokines and Association With Clinical Severity of Dermatomyositis and Clinically Amyopathic Dermatomyositis. Br. J. Dermatol. 179 (6), 1334–1341. doi:10.1111/bjd.17079
de Jesus, A. A., Hou, Y., Brooks, S., Malle, L., Biancotto, A., Huang, Y., et al. (2020). Distinct Interferon Signatures and Cytokine Patterns Define Additional Systemic Autoinflammatory Diseases. J. Clin. Invest. 130 (4), 1669–1682. doi:10.1172/jci129301
Fang, X., Lu, M., Xia, Z., Gao, C., Cao, Y., Wang, R., et al. (2021). Use of Liquid Chromatography-Tandem Mass Spectrometry to Perform Urinary Proteomic Analysis of Children With IgA Nephropathy and Henoch-Schönlein Purpura Nephritis. J. Proteomics. 230, 103979. doi:10.1016/j.jprot.2020.103979
Friedman, A. W., Targoff, I. N., and Arnett, F. C. (1996). Interstitial Lung Disease With Autoantibodies Againstaminoacyl-tRNA Synthetases in the Absence of Clinically Apparent Myositis. Semin. Arthritis Rheum. 26 (1), 459–467. doi:10.1016/s0049-0172(96)80026-6
Gao, S., Zhang, H., Zuo, X., Xiao, Y., Liu, D., Zhu, H., et al. (2019). Integrated Comparison of the miRNAome and mRNAome in Muscles of Dermatomyositis and Polymyositis Reveals Common and Specific miRNA-mRNAs. Epigenomics. 11 (1), 23–33. doi:10.2217/epi-2018-0064
Gao, S., Zuo, X., Liu, D., Xiao, Y., Zhu, H., Zhang, H., et al. (2018). The Roles of Neutrophil Serine Proteinases in Idiopathic Inflammatory Myopathies. Arthritis Res. Ther. 20 (1), 134. doi:10.1186/s13075-018-1632-x
Göbel, K., Pankratz, S., Asaridou, C.-M., Herrmann, A. M., Bittner, S., Merker, M., et al. (2016). Blood Coagulation Factor XII Drives Adaptive Immunity During Neuroinflammation via CD87-Mediated Modulation of Dendritic Cells. Nat. Commun. 7, 11626. doi:10.1038/ncomms11626
Gono, T., Kaneko, H., Kawaguchi, Y., Hanaoka, M., Kataoka, S., Kuwana, M., et al. (2014). Cytokine Profiles in Polymyositis and Dermatomyositis Complicated by Rapidly Progressive or Chronic Interstitial Lung Disease. Rheumatology (Oxford). 53 (12), 2196–2203. doi:10.1093/rheumatology/keu258
Gono, T., Kawaguchi, Y., Sugiura, T., Ichida, H., Takagi, K., Katsumata, Y., et al. (2010). Interleukin-18 Is a Key Mediator in Dermatomyositis: Potential Contribution to Development of Interstitial Lung Disease. Rheumatology. 49 (10), 1878–1881. doi:10.1093/rheumatology/keq196
Hervier, B., and Uzunhan, Y. (2019). Inflammatory Myopathy-Related Interstitial Lung Disease: From Pathophysiology to Treatment. Front. Med. (Lausanne). 6, 326. doi:10.3389/fmed.2019.00326
Hong, C., Luckey, M. A., Ligons, D. L., Waickman, A. T., Park, J.-Y., Kim, G. Y., et al. (2014). Activated T Cells Secrete an Alternatively Spliced Form of Common γ-Chain that Inhibits Cytokine Signaling and Exacerbates Inflammation. Immunity. 40 (6), 910–923. doi:10.1016/j.immuni.2014.04.020
Jonigk, D., Stark, H., Braubach, P., Neubert, L., Shin, H. o., Izykowski, N., et al. (2019). Morphological and Molecular Motifs of Fibrosing Pulmonary Injury Patterns. J. Pathol. Clin. Res. 5 (4), 256–271. doi:10.1002/cjp2.141
Kahles, A., Lehmann, K. V., Toussaint, N. C., Hüser, M., Stark, S. G., Sachsenberg, T., et al. (2018). Comprehensive Analysis of Alternative Splicing Across Tumors From 8,705 Patients. Cancer Cell. 34 (2), 211–e6. e216. doi:10.1016/j.ccell.2018.07.001
Kobayashi, S., Nagafuchi, Y., Okubo, M., Sugimori, Y., Shirai, H., Hatano, H., et al. (2021). Integrated Bulk and Single-Cell RNA-Sequencing Identified Disease-Relevant Monocytes and a Gene Network Module Underlying Systemic Sclerosis. J. Autoimmun. 116, 102547. doi:10.1016/j.jaut.2020.102547
Li, Z., Liu, Y., Jia, A., Cui, Y., and Feng, J. (2021). Cerebrospinal Fluid Cells Immune Landscape in Multiple Sclerosis. J. Transl Med. 19 (1), 125. doi:10.1186/s12967-021-02804-7
Lilleker, J. B., Vencovsky, J., Wang, G., Wedderburn, L. R., Diederichsen, L. P., Schmidt, J., et al. (2018). The EuroMyositis Registry: an International Collaborative Tool to Facilitate Myositis Research. Ann. Rheum. Dis. 77 (1), 30–39. doi:10.1136/annrheumdis-2017-211868
Miller, F. W., Cooper, R. G., Vencovský, J., Rider, L. G., Danko, K., Wedderburn, L. R., et al. (2013). Genome-Wide Association Study of Dermatomyositis Reveals Genetic Overlap With Other Autoimmune Disorders. Arthritis Rheum. 65 (12), 3239–3247. doi:10.1002/art.38137
Nasr, R., Reed, A. M., and Peterson, E. J. (2012). Update. Curr. Opin. Rheumatol. 24 (6), 609–615. doi:10.1097/bor.0b013e3283585731
Newman, J. R. B., Conesa, A., Mika, M., New, F. N., Onengut-Gumuscu, S., Atkinson, M. A., et al. (2017). Disease-Specific Biases in Alternative Splicing and Tissue-Specific Dysregulation Revealed by Multitissue Profiling of Lymphocyte Gene Expression in Type 1 Diabetes. Genome Res. 27 (11), 1807–1815. doi:10.1101/gr.217984.116
Nilsen, T. W., and Graveley, B. R. (2010). Expansion of the Eukaryotic Proteome by Alternative Splicing. Nature. 463 (7280), 457–463. doi:10.1038/nature08909
Oda, K., Kotani, T., Takeuchi, T., Ishida, T., Shoda, T., Isoda, K., et al. (2017). Chemokine Profiles of Interstitial Pneumonia in Patients With Dermatomyositis: a Case Control Study. Sci. Rep. 7 (1), 1635. doi:10.1038/s41598-017-01685-5
Peng, Y., Zhang, S., Zhao, Y., Liu, Y., and Yan, B. (2018). Neutrophil Extracellular Traps May Contribute to Interstitial Lung Disease Associated with Anti-MDA5 Autoantibody Positive Dermatomyositis. Clin. Rheumatol. 37 (1), 107–115. doi:10.1007/s10067-017-3799-y
Richards, T. J., Eggebeen, A., Gibson, K., Yousem, S., Fuhrman, C., Gochuico, B. R., et al. (2009). Characterization and Peripheral Blood Biomarker Assessment of Anti-jo-1 Antibody-Positive Interstitial Lung Disease. Arthritis Rheum. 60 (7), 2183–2192. doi:10.1002/art.24631
Roy, E. R., Wang, B., Wan, Y.-w., Chiu, G., Cole, A., Yin, Z., et al. (2020). Type I Interferon Response Drives Neuroinflammation and Synapse Loss in Alzheimer Disease. J. Clin. Invest. 130 (4), 1912–1930. doi:10.1172/jci133737
Shen, S., Park, J. W., Huang, J., Dittmar, K. A., Lu, Z.-x., Zhou, Q., et al. (2012). MATS: a Bayesian Framework for Flexible Detection of Differential Alternative Splicing From RNA-Seq Data. Nucleic Acids Res. 40 (8), e61. doi:10.1093/nar/gkr1291
Shen, S., Park, J. W., Lu, Z.-x., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014). rMATS: Robust and Flexible Detection of Differential Alternative Splicing from Replicate RNA-Seq Data. Proc. Natl. Acad. Sci. USA. 111 (51), E5593–E5601. doi:10.1073/pnas.1419161111
Stewart, C. E., Nijmeh, H. S., Brightling, C. E., and Sayers, I. (2012). UPAR Regulates Bronchial Epithelial Repair In Vitro and Is Elevated in Asthmatic Epithelium. Thorax. 67 (6), 477–487. doi:10.1136/thoraxjnl-2011-200508
Tillie-Leblond, I., Wislez, M., Valeyre, D., Crestani, B., Rabbat, A., Israel-Biet, D., et al. (2008). Interstitial Lung Disease and Anti-jo-1 Antibodies: Difference between Acute and Gradual Onset. Thorax. 63 (1), 53–59. doi:10.1136/thx.2006.069237
Tripathi, A., Whitehead, C., Surrao, K., Pillai, A., Madeshiya, A., Li, Y., et al. (2021). Type 1 Interferon Mediates Chronic Stress-Induced Neuroinflammation and Behavioral Deficits via Compl Ement Component 3-Dependent Pathway. Mol. Psychiatry. 26 (7), 3043–3059. doi:10.1038/s41380-021-01065-6
Vuillard, C., Pineton de Chambrun, M., de Prost, N., Guérin, C., Schmidt, M., Dargent, A., et al. (2018). Clinical Features and Outcome of Patients With Acute Respiratory Failure Revealing Anti-synthetase or Anti-MDA-5 Dermato-Pulmonary Syndrome: a French Multicenter Retrospective Study. Ann. Intensive Care. 8 (1), 87. doi:10.1186/s13613-018-0433-3
Zhang, S. H., Zhao, Y., Xie, Q. B., Jiang, Y., Wu, Y. K., and Yan, B. (2019a). Aberrant Activation of the Type I Interferon System May Contribute to the Pathogenesis of Anti‐Melanoma Differentiation‐Associated Gene 5 Dermatomyositis. Br. J. Dermatol. 180 (5), 1090–1098. doi:10.1111/bjd.16917
Zhang, X., Pan, A., Jia, S., Ideozu, J. E., Woods, K., Murkowski, K., et al. (2019b). Cystic Fibrosis Plasma Blunts the Immune Response to Bacterial Infection. Am. J. Respir. Cel Mol Biol. 61 (3), 301–311. doi:10.1165/rcmb.2018-0114OC
Keywords: dermatomyositis, interstitial lung disease, RNA-sequencing, alternative splicing, plasminogen activator, urokinase receptor
Citation: Chen J, Zhang R, Xie M, Luan C and Li X (2021) Transcriptome Sequencing Identifies PLAUR as an Important Player in Patients With Dermatomyositis-Associated Interstitial Lung Disease. Front. Genet. 12:784215. doi: 10.3389/fgene.2021.784215
Received: 27 September 2021; Accepted: 03 November 2021;
Published: 06 December 2021.
Edited by:
Ramcés Falfán-Valencia, Instituto Nacional de Enfermedades Respiratorias (INER), MexicoReviewed by:
James A Poulter, University of Leeds, United KingdomCopyright © 2021 Chen, Zhang, Xie, Luan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolan Li, cHJvbGl4bEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.