- 1Department of Biosciences, Jamia Millia Islamia, New Delhi, India
- 2Department of Laboratory Medicine, Faculty of Applied Medical Sciences, Albaha University, Albaha, Saudi Arabia
- 3School of Biosciences, Apeejay Stya University, Gurugram, India
- 4Department of Medical Laboratories, College of Applied Medical Sciences, Qassim University, Buraydah, Saudi Arabia
- 5Department of Surgical Oncology, All India Institute of Medical Sciences, New Delhi, India
Background: FOXP3 gene, known to be a potential tumor suppressor, has been identified to interact with HER2 in mammary cancer. Moreover, the high expression of FOXP3 serves as a good predictor of the survival of patients in breast cancer, prostate cancer, and gastric cancer. The expression and epigenetic alterations were evaluated in female breast cancer patients.
Material and Methods: Expression studies at the mRNA level and protein level were conducted in 140 breast cancer cases by real-time PCR and immunohistochemistry, respectively. Epigenetic studies were also conducted by analyzing the methylation status at the promoter region of the gene using MS-PCR.
Results: FOXP3 mRNA expression and protein expression were downregulated in breast cancer patients. The absence of FOXP3 protein expression is significantly associated with promoter methylation, where 70 methylated cases exhibited protein loss (70/95, 73.6%). Statistically, we also found a significant correlation between FOXP3 protein expression and TNM stage, promoter methylation, and histological grade. The methylated FOXP3 cases that did not express protein were also significantly associated with positive lymph node metastasis and HER-2 status.
Conclusion: The expression profile of FOXP3 may serve as a prognostic factor. In short, FOXP3 may stand in the most crucial list of biomarkers for breast cancer, bringing compelling results in terms of treatment and management of the disease.
Introduction
It is a well-known fact that females around the world are mostly affected by breast cancer (1.7 million cases, 11.9%); however, it positions fifth as the cause of death (6.4%) because of the comparatively conducive prognosis. Whereas breast cancer accounted for the most prominent cause of mortality in females in undeveloped and developing regions of the world (Ferlay et al., 2015), the development of biomarkers and their clinical use in therapy for prediction and expected response hold a remarkable potential.
FOXP3(Forkhead box P3), located on Xp11.23, is a member of the Forkhead/winged-helix family of transcription factors which is responsible for X-linked autoimmune diseases in mice as well as humans (Wildin et al., 2001; Brunkow et al., 2001; Schubert et al., 2001). Transcription factor FOXP3 regulates the development and function of Treg cells, and Treg cells are known to regulate homeostasis (Chen et al., 2015) and immunosuppression (Müller et al., 2010) and also recognized as the most peculiar marker for Treg (Hori et al., 2003). Before the advanced research on FOXP3 expression, it was thought to be expressed only in hematopoietic cells but now seems to be present in human tumors, particularly tumors of the breast (Merlo et al., 2009). In mammary cancers, FOXP3 is found to regulate HER-2 and SKP2 by repressing their expression, and importantly these genes are linked to a poor prognosis in the cases with breast carcinoma (Martin et al., 2010). The downregulation and many functional somatic mutations in the FOXP3 gene were usually found in human breast cancer samples. These mutations may also account for the overall down-regulation at the protein level (Karanikas et al., 2008).
FOXP3 halts the transcription of HER-2 by attaching to the promoter region of the ERBB2 gene (Redpath et al., 2011; Zuo et al., 2007), and it is a well-known fact that HER-2 is a potent marker in terms of prediction and effective therapy (Presson et al., 2011; Weigelt and Reis-Filho, 2010). The study has also pointed out the high expression of FOXP3 as a good predictor of the survival of a patient in prostate cancer, breast cancer, gastric cancer, and bladder cancer (Li et al., 2007; Wang et al., 2009; Winerdal et al., 2011; He et al., 2013; Fiori Lopes et al., 2014; Hao et al., 2014; Ma et al., 2014). Previous studies also reported that many SNPs in the FOXP3 gene had been associated with breast cancer (Jiang and Ruan, 2014).
FOXP3 is also linked to p21 and LATS2, where it is involved in transcriptional control (Li et al., 2011). Due to these captivating characteristics of FOXP3, the present work examines the correlation of FOXP3 protein expression with the clinicopathological variables to strengthen its role as a putative biomarker for breast carcinoma in the Indian population.
Methodology
Ethical Statement
The University Ethical Committee of Jamia Millia Islamia (JMI), New Delhi, and the Ethical Committee for Human Study of AIIMS (All India Institute of Medical Sciences), New Delhi, have officially approved the study. The experimental work had been undertaken with written consent obtained from each subject, and the study complies with the rules and standards set by the Ethics Code of the Medical Association of the world, which have been noted as per the Declaration of Helsinki as published in British Medical Journal (1964).
Sample Collection
A total of 140 participants were included in the present case–control study. Cancer tissue from the breast and non-cancerous adjacent tissue were both obtained from the surgical oncology department of the collaborating institute (AIIMS). The samples were collected in three vials containing RNALater, phosphate-buffered saline (PBS), and formalin, respectively, for further processing.
The classification of breast cancer stages was done under the TNM staging system, and the histological grading of tumors was classified based on the Nottingham grade system. The exclusion criteria in the current study included familial cancer, any previous type of cancer, other metastasized cancer that has spread from different organs, and chemotherapy and radiotherapy exposure. In addition, included were various clinicopathological variables such as tumor distinctiveness [age, tumor size, metastasis at the lymph node level, TNM staging, grade of tumor, molecular subtype of tumor, hormonal receptor status (ER, PR, and Her2neu), and reproductive history (menopausal status parity)].
Quantitative- PCR
RNALater (Qiagen) was used to store excised tissues from normal and breast cancer patients, and then RNA was extracted by using the TRIzol method as per the manual. cDNA was synthesized using a Thermo Fisher verso kit from the extracted RNA. The qPCR is processed using Roche LightCycler® 96 machine with SYBR Green I Master mix reagent (Roche) with the help of FOXP3 primers (Fwd- 5′-TCCCAGAGTTCCTCCACAAC-3′ and Rev-5′ATTGAGTGTCCGCTGCTTCT-3′) that give an amplified 122-bp product. The internal control used was GAPDH gene which was also amplified in the same PCR reactions. The program used for the amplification cycles was as follows: preheating at 95°C for 1 min, 30 cycles of denaturation at 95°C for 20 s, annealing at 58°C for 15 s, extension at 72°C for 15 s, and further elongation at 72°C for 7 min. The experiments were repeated thrice.
The relative quantification of expression was calculated as the calibrator normalized ratio using LightCycler 96 (Roche) Software 1.5. The formula used, RQ = 2–∆∆Ct, was according to MIQE.
Genomic DNA Extraction
The phenol-chloroform extraction method was used to isolate high-molecular-weight total gDNA from both tumor and normal tissues stored in PBS. The genomic DNA isolated was quantified on a Nanodrop spectrophotometer, and its quality was also assessed using an A260/280 ratio. It was further visualized on the 1% agarose gel stained with ethidium bromide under a UV transilluminator.
MS-PCR for Epigenetic Analysis
Isolated gDNA from the tissues were given bisulfite treatment using Zymo research EZ DNA Methylation-Gold™ Kit per the instructions. The treated gDNA was amplified using two sets of methylated and unmethylated primers for the FOXP3 promoter. MethPrimer tool was used to design the set of primers for methylation and unmethylation. (Li and Dahiya, 2002). The methylated primer pairs for the promoter region of the FOXP3 gene were: forward 5′- TGTAGGGGGTGTAGAATTTTTTTC-3′ and reverse 5′- AAACTAAATTCCCAAAAACCTCG-3′ and for the unmethylated were forward 5′- GTAGGGGGTGTAGAATTTTTTTTGT-3 and reverse 5′- TAAAACTAAATTCCCAAAAACCTCA-3′. The positive controls used in the experiment were commercially available Completely methylated and unmethylated human genomic DNA.
MS-PCR was performed under the following cycles: First denaturation at 95°C for 7 min, denaturation at 95°C for 30s 52.5°C annealing for both types of primers for 30 s, 72°C for 30 s and final elongation at 72°C for 7 min which was amplified for 35 cycles. Then, 2% agarose gel stained with EtBr was used to visualize and analyze the PCR product, which was finally photographed using the Bio-Rad Gel Documentation system. All experiments were conducted in triplicates.
Immunohistochemistry
The tissue biopsies of the breast carcinoma and adjoining non-cancerous tissue were conserved in formalin-fixed blocks. The blocks were then sectioned and engraved on slides coated with poly-L-lys and exposed to deparaffinization by dipping in different concentrations of xylene and further rehydrated with grades of ethanol. By using 0.3% H2O2 for 30 min, the endogenous peroxidase activity was blocked, and sodium citrate buffer (pH 9.0) was used for Ag retrieval.
The sections were treated for 30 min with TENG-T (10 mM Tris; 5 mM ethylenediaminetetraacetic acid, 0.15 mol/L NaCl, 0.25% gelatin, and 0.05% (v/v) Tween20 (pH 8.0) to block the samples. Bovine serum albumin was used to limit unspecific binding to the protein. The slides were treated with the primary antibody (mAbCam#ab22510 FOXP3 1:50) and incubated overnight at 4°C. The slides were then treated for 30 min with a biotinylated anti-mouse secondary antibody along with streptavidin–horseradish peroxidase conjugate. The DAB chromogen was finally used as a substrate to give a brown-colored precipitate. The slides were also treated with hematoxylin as a counterstain for better contrast.
Histopathologists interpreted the slides after immunohistochemistry; the slides were photographed under a light microscope at ×400 magnification. The pathologist further graded the expression on the number scale 0–4, with 0 as no expression and 4 as highest expression; the slides with >50% protein staining were considered in the highest scale.
Statistics
SPSS version 22.0 for Windows was used for all the statistical correlations between the outcomes and the clinicopathological parameters. All data were expressed as mean ± standard error. Fisher’s exact test was used to obtain p-values between mRNA levels, methylation status, and protein expression with the clinicopathological parameters. Non-parametric Wilcoxon signed-rank test is used to estimate FOXP3/GAPDH mRNA expression levels significantly in both cancer and normal tissue samples. The p-values >0.05 were considered as significant.
Results
mRNA Expression of the FOXP3 Gene Is Downregulated in the Cases of Breast Cancer
FOXP3 mRNA expression revealed downregulation in 63.5% (89/140) of cases (Figure 1A), out of which nearly 86.5% (77/89) of the cases registered in the study were linked to histological grade type 1 and type 2. As per the fold change analysis, 89 cases out of 140 samples seemed to be downregulated (5.09-fold), while the expression pattern of FOXP3 at the level of mRNA, when normalized accordingly with the internal control GAPDH in tumor and non-tumor tissues, was 2.07 ± 0.7 (mean ± standard error) and 2.51 ± 0.4 (mean ± standard error) (Figure 1B), (p-value of <0.0001; Table 1) respectively. However, there was no significant association observed between FOXP3 mRNA level and various clinical variables.

FIGURE 1. (A) Heat map plot showing the mRNA relative expression of the FOXP3 gene (fold change) in breast cancer patients. (B) Graphical representation of the relative expression of FOXP3/GAPDH mRNA in breast tumor and adjoining non-tumorous tissues.
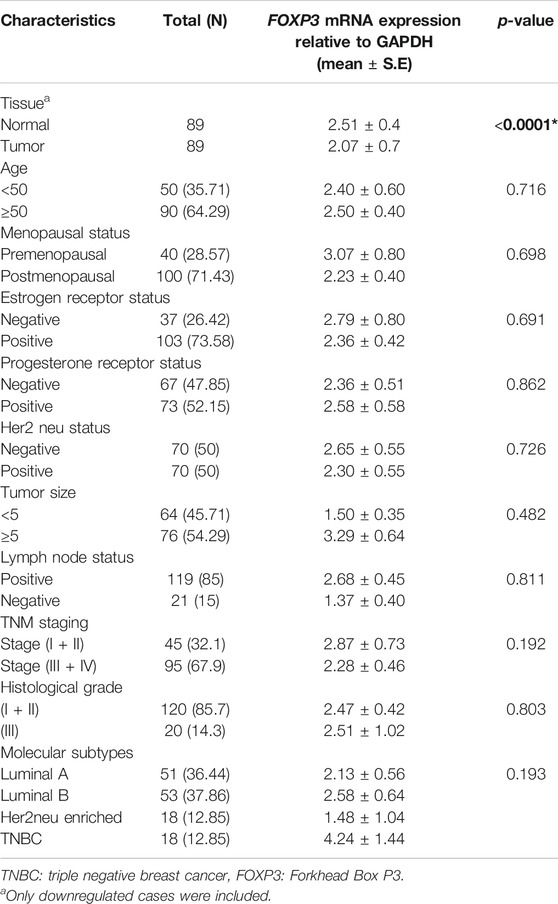
TABLE 1. Correlation study of FOXP3 mRNA expression with clinicopathological parameters in North Indian breast cancer patients.
Expression of FOXP3 Protein is Either Lost or Low in Breast Cancer
The expression of FOXP3 at the protein level was found to be either low or absent in 95 cases of the total 140 samples involved in the study (67.85%) (Figure 2), while in the other 45 cases, the expression pattern was either in the high or moderate range as interpreted by a histopathologist on the basis expression scale (45/140, 32.14%). The protein expression pattern was in relation to the mRNA expression. The FOXP3 protein, as visualized by immunohistochemistry, was mainly located in the nuclear region.
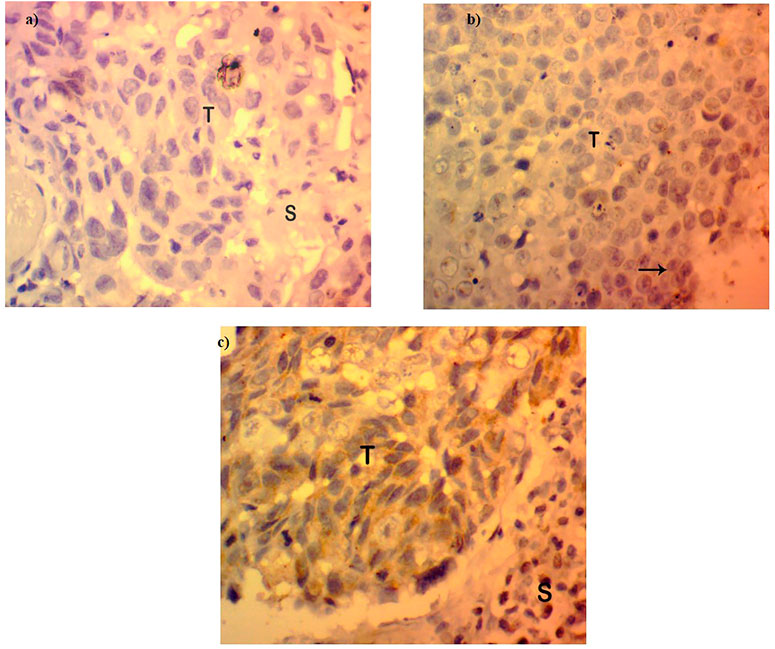
FIGURE 2. Protein expression in FOXP3 by immunohistochemistry. (A) Absence or loss of FOXP3 expression in Breast cancer tissues. (B) FOXP3 low expression in breast tumor tissues. (C) FOXP3 moderate/high expression in breast tumor tissues.
FOXP3 Protein Expression and Its Significant Correlation With Clinicopathological Parameters
As revealed by immunohistochemistry staining, the majority of FOXP3 proteins in the samples were found to be significantly downregulated. Moreover, when we tried to statistically associate the protein expression with the clinical parameters of the patients, we observed a significant correlation between TNM staging and FOXP3 protein expression (p < 0.035) (Table 2). However, with other parameters, no significant association was obtained statistically (Table 2), though most cases with tumor grades 1 and 2 seem to have protein loss (82/95, 86.3%).
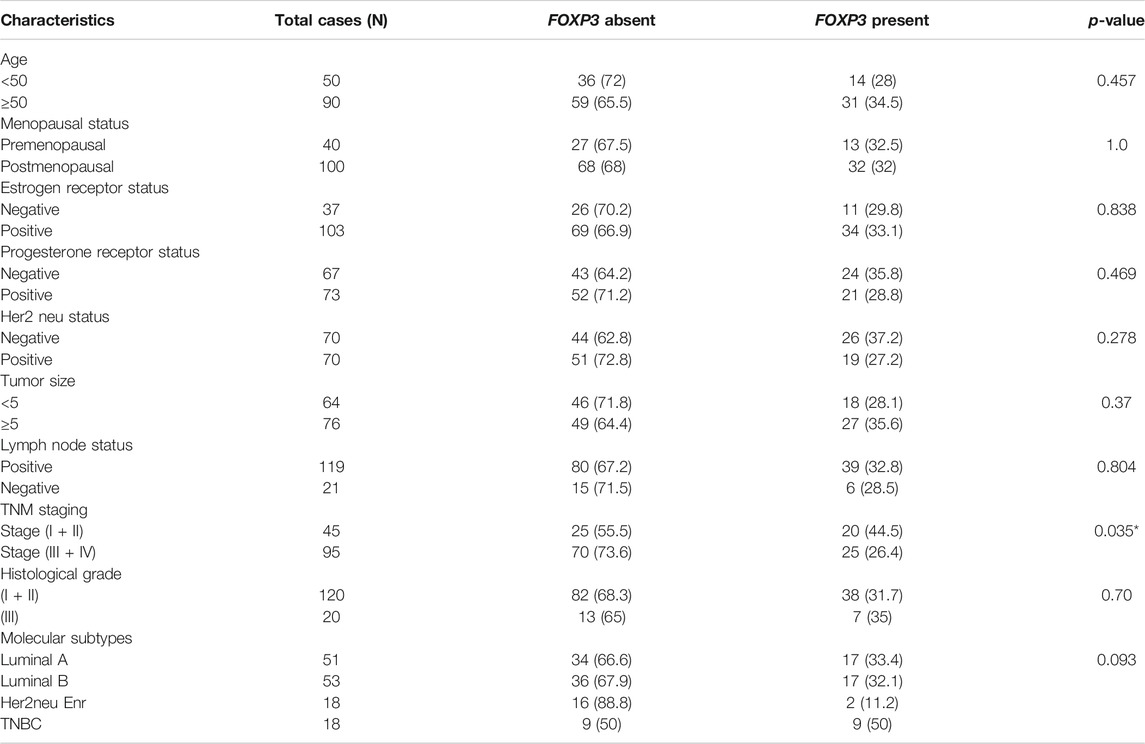
TABLE 2. Statistical analysis of the protein expression pattern of FOXP3 with the clinicopathological variables of patients with breast cancer.
FOXP3 Promoter Methylation and Its Association With Clinical Variables
Methylation at various CpG present in the upstream promoter area of FOXP3 gene was observed in 73 cases (73/140, 52.14%) (Figure 3) and, once linked with clinical parameters, revealed a significant association with the Nottingham histological grades 1 and 2 type tumors of breast cancer patients (0.031). Though no significant associations were seen with other parameters included in the study (Table 3), we did note a significantly higher number of methylated cases in metastatic lymph node (59/73, 80.8%), estrogen receptor-positive (52/73, 71.2%), and menopausal female patients (51/73, 69.8%) (Table 3).
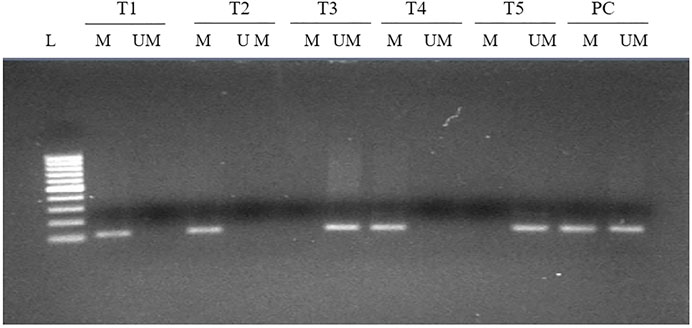
FIGURE 3. MS-PCR analysis of the promoter region of FOXP3 in breast carcinoma. Promoter methylation was evaluated by two types of primers to amplify either unmethylated gDNA (UM) or methylated gDNA (M). L, 100-bp DNA ladder; T, tumor tissue; PC, positive control for completely methylated and unmethylated DNA.

TABLE 3. Statistical association study of methylated FOXP3 gene with clinicopathological parameters of patients having breast carcinoma.
Convincing Association of FOXP3 Protein Expression With FOXP3 Promoter Methylation in the Cases of Breast Carcinoma
The correlation study of methylated FOXP3 gene and its respective protein expression displayed the significant link, in which out of 95 cases with protein loss, 70 cases possessed methylation at the promoter region (70/95, 73.68%) (Table 4), whereas 25 cases were completely unmethylated (25/95, 26.3%) (Table 4). To add more, in 67 unmethylated samples (67/140, 47.8%), noticeably 62.6% (42/67) cases showed the presence of protein. Therefore, a potential statistical relation was observed between the FOXP3 protein expression and its promoter methylation (p = 0.0001) (Table A4).
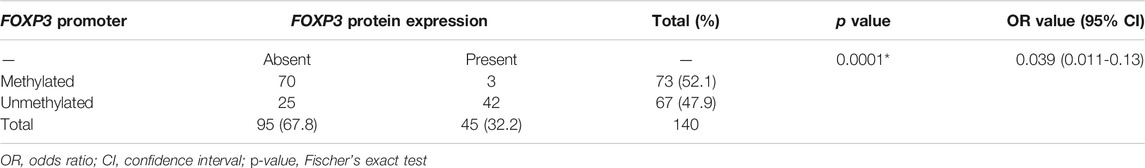
TABLE 4. Correlation study between FOXP3 protein expression and its promoter methylation in patients with breast cancer.
Association of Methylated FOXP3 Gene Exhibiting Loss of Protein With Numerous Clinicopathological Variables
The methylated promoter region of FOXP3 cases demonstrating either the absence or presence of protein exhibited a statistically significant relation with Her 2 neu receptor (p = 0.004) and metastatic lymph node tumors (p = 0.01) (Table 5). Moreover, 95.7% of methylated cases (67/70) with lymph node metastasis have protein loss. (Table 5). Additionally, the cases having protein loss exhibiting either a methylated or unmethylated FOXP3 promoter region shows a convincing association with positive Her-2 receptor (p = 0.03) and tumors of grades 1 and 2 (p = 0.01) (Table 5). Furthermore, it is seen that there is a strong statistical relation between FOXP3 protein loss and the promoter methylation with the various clinicopathological parameters (Table 6), where most of the features were associated in a highly significant manner (p-value < 0.05).
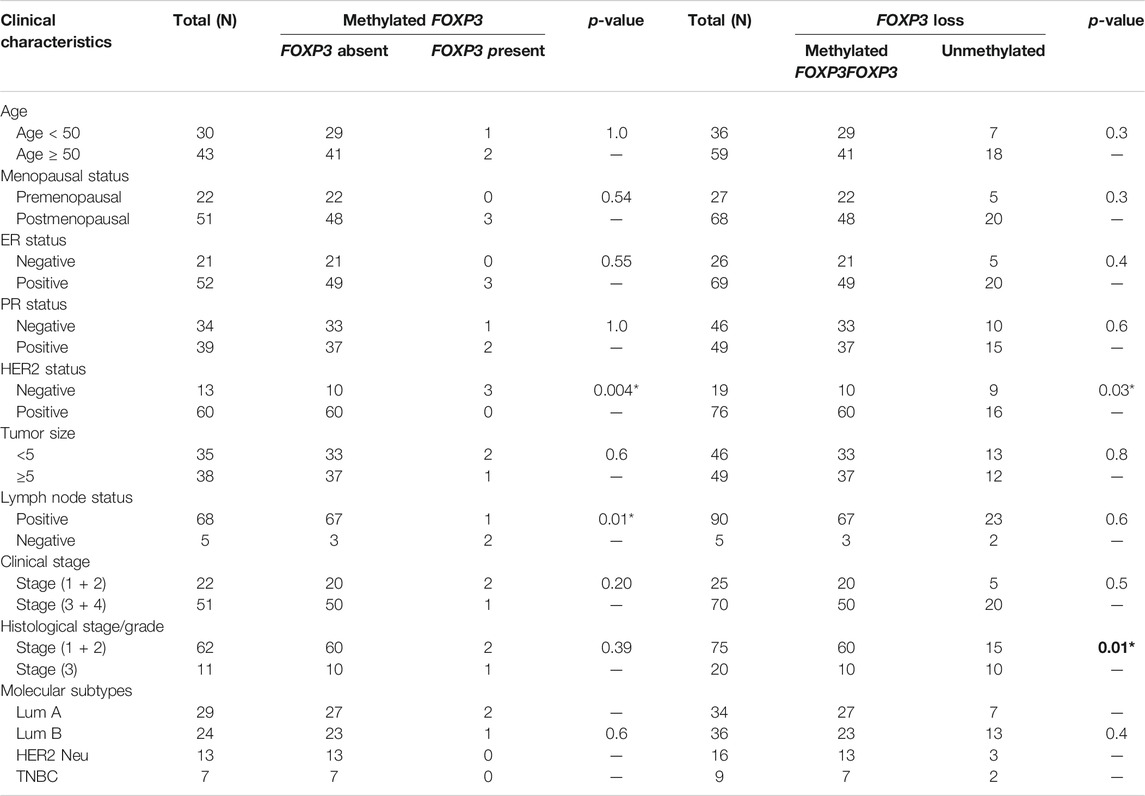
TAble 5. Significant association of promoter methylation and its protein expression in patients with methylated FOXP3 promoter or FOXP3 Protein expression loss with various clinicopathological features of breast carcinoma.
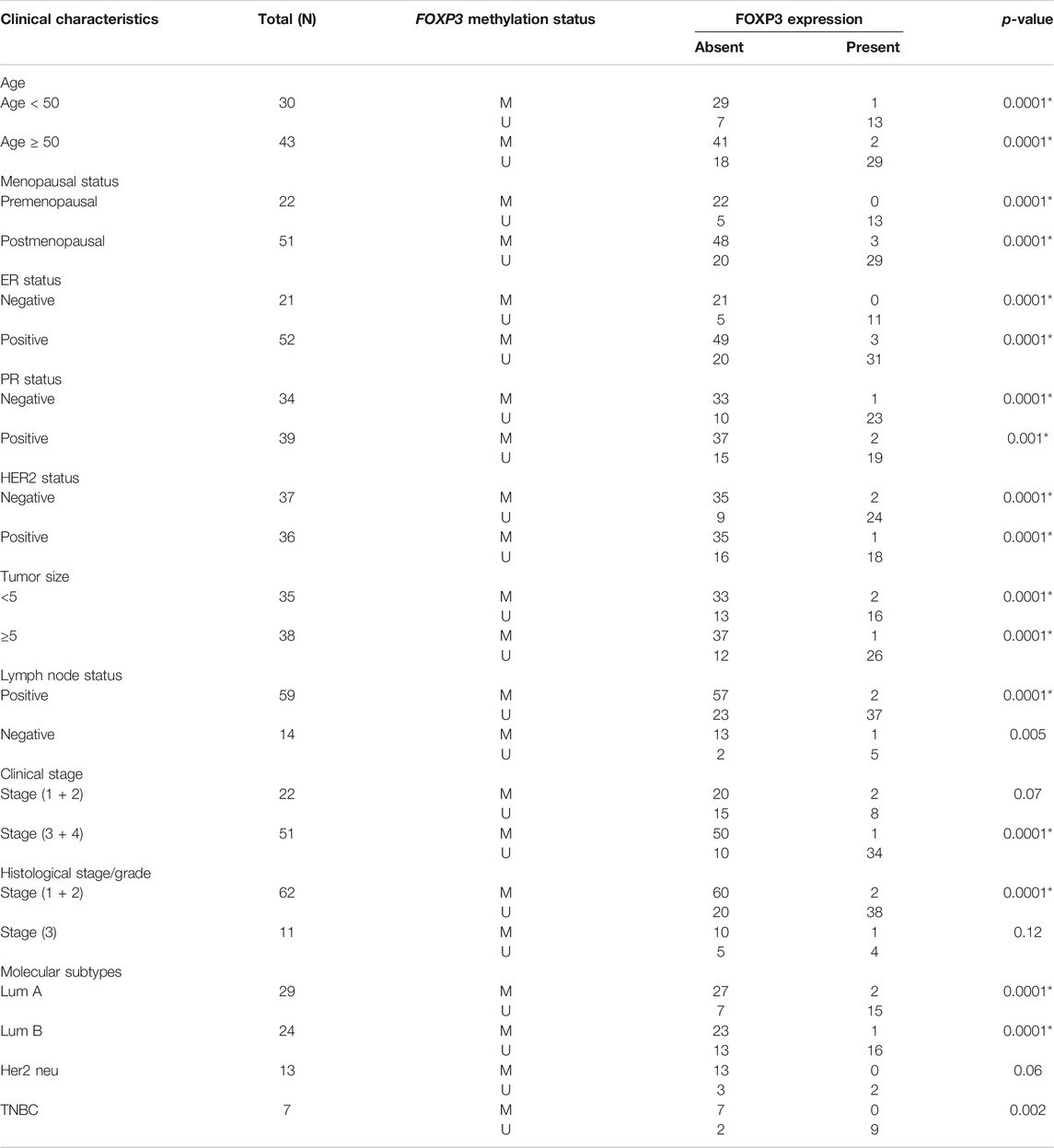
TABLE 6. Association study between methylated FOXP3 and FOXP3 protein expression in stratification with clinicopathological features.
Discussion
FOXP3 expression is identified in tumors of the breast, prostate, lung, gastric, and thyroid (Liu et al., 2015; Yang et al., 2017; Ma et al., 2014; the Chu et al., 2015) suggesting its crucial role in the biology of cancer. The previous study demonstrated the inverse correlation between breast cancer angiogenesis and nuclear FOXP3 expression. Adding to the same observation, the significant downregulation of FOXP3 also resulted in the reduced survival in breast cancer (Li et al., 2018) FOXP3 has been reported to modulate the expression of various genes involved in the process of carcinogenesis to exert its suppressing role in tumor development (Szylberg et al., 2016). At the same time, we cannot forget that significant studies have suggested the positive association between FOXP3 expression and better survival in patients and the tumor-suppressive role of the FOXP3 gene in breast cancer. Therefore, the present work investigated the FOXP3 expression pattern and its correlation with various clinicopathological variables to strengthen its prognostic value and tumor-suppressive property. An earlier study demonstrated a quantitative method to assess the methylation status of FOXP3 to understand the role of Treg cells in immunomodulation (Wieczorek et al., 2009). In our study transcription factor, FOXP3 promoter methylation and expression were studied and analyzed in breast cancer patients of the northern region of India using methylation-specific PCR, real-time PCR, and immunohistochemistry to assess its role as a potential biomarker. The study correlated the findings with the clinicopathological variables (age, histological grade, ER status, HER2 status, etc.) of the procured cases.
In our study, at the mRNA level, nearly 63.5% (89/140) of cases were found to be downregulated (5.09 fold), and interestingly 86.5% (77/89) were linked with the histological grades I and II, suggesting the possible role of FOXP3 in the early development of the disease. The study is supported by the previous studies on FOXP3 expression at the transcript level (Zhang and Sun, 2010; Hinz et al., 2007). Furthermore, apart from breast cancer, one of the studies pointed out that the upregulation of FOXP3 in gastric cancer cells put a brake on GC cell growth in both in vivo and in vitro studies (Hao et al., 2014), unraveling the crucial role of FOXP3 expression in different carcinomas.
The protein expression profile exhibited low or no expression in nearly 69% (95/140) of breast cancer cases, followed by either moderate or high expression in 32% (45/140) of the cases. The expression was either cytoplasmic or nuclear, which was demonstrated in different types of cancer (Merlo et al., 2009; Karanikas et al., 2008; Zhang and Sun, 2010; Hinz et al., 2007; Ladoire et al., 2011; Tao et al., 2012). We did find a significant association between FOXP3 protein expression and the TNM stage (p-value, 0.035). Interestingly almost 74% (95/140) of the cases of stage (III and IV) harbored protein loss. Because of the above mentioned condition, it has been observed earlier that, in the most aggressive cancer of epithelial tissues, FOXP3 may help in the suppression of cancer as these aggressive cancer tissues harbored very low or no expression of FOXP3 at the transcript and protein levels (Wang et al., 2009; Jiang and Ruan, 2014; Li et al., 2011).
The promoter methylation of FOXP3 was observed in 52% of the cases (73/140) and significantly associated with Nottingham histological grades 1 and 2 (p-value 0.031). The finding depicts a strong association between promoter hyper-methylation and mRNA expression in deactivating or down-regulating the possible role of FOXP3 in the suppression of breast cancer (Esteller, 2007; Li et al., 2007).
While analyzing an association between protein loss and hypermethylated promoter cases, we found a compelling association as out of 95 protein loss cases, 70 cases possessed methylation at the promoter region (70/95, 73.68%), whereas in a total of 67 unmethylated cases, 62.6% (42/67) exhibited the presence of protein. More intriguing results came out while analyzing methylation and protein loss with each other, as 95.7% methylated cases (67/70) with lymph node metastasis displayed protein loss. The findings are strongly supported by previous studies that have shown the association to be significantly lower in tumor grade and lymph node involvement in the breast tumor cells with positive FOXP3 expression (Ladoire et al., 2012). Thus, the loss in FOXP3 due to epigenetic change like methylation may serve as a potential biomarker in cases with lymph node metastasis.
In summary, our data provide some intriguing findings of FOXP3 expression and its association with different clinicopathological parameters. However, the present study on a smaller sample size may weaken the statistical power. Therefore, further investigation on different sets of the population with a larger sample size is required to establish FOXP3 as a potential cancer biomarker for diagnostic and prognostics purposes.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
This study was in agreement with the ethical standards of the host institute Jamia Millia Islamia University and AIIMS, New Delhi, India, and also as per the guidelines of the Helsinki Declaration in 1964 and its amendments. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Sa and SH took charge of the experimental design and execution. The experiments were executed by Sa. The reagents, materials, and analysis tools were contributed by Sa, MN, MK, and SD. Manuscript preparation was carried out by Sa and co-authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
UGC-MANF for Higher Studies (India).
References
Brunkow, M. E., Jeffery, E. W., Hjerrild, K. A., Paeper, B., Clark, L. B., Yasayko, S.-A., et al. (2001). Disruption of a New Forkhead/winged-helix Protein, Scurfin, Results in the Fatal Lymphoproliferative Disorder of the Scurfy Mouse. Nat. Genet. 27 (1), 68–73. doi:10.1038/83784
Chen, Y., Chen, C., Zhang, Z., Liu, C.-C., Johnson, M. E., Espinoza, C. A., et al. (2015). DNA Binding by FOXP3 Domain-Swapped Dimer Suggests Mechanisms of Long-Range Chromosomal Interactions. Nucleic Acids Res. 43 (2), 1268–1282. doi:10.1093/nar/gku1373
Chu, R., Liu, S. Y. W., Vlantis, A. C., van Hasselt, C. A., Ng, E. K. W., Fan, M. D., et al. (2015). Inhibition of Foxp3 in Cancer Cells Induces Apoptosis of Thyroid Cancer Cells. Mol. Cell. Endocrinol. 399, 228–234. doi:10.1016/j.mce.2014.10.006
Esteller, M. (2007). Epigenetic Gene Silencing in Cancer: the DNA Hypermethylome. Hum. Mol. Genet. 16 (R1), R50–R59. doi:10.1093/hmg/ddm018
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., et al. (2015). Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 136 (5), E359–E386. doi:10.1002/ijc.29210
Fiori Lopes, L., Losi Guembarovski, R., Guembarovski, A. L., Okuyama Kishima, M., Campos, C. Z., Oda, J. M. M., et al. (2014). FOXP3 Transcription Factor: a Candidate Marker for Susceptibility and Prognosis in Triple Negative Breast Cancer. Biomed. Research International 2014, 1–7. doi:10.1155/2014/341654
Hao, Q., Zhang, C., Gao, Y., Wang, S., Li, J., Li, M., et al. (2014). FOXP3 Inhibits NF-Κb Activity and Hence COX2 Expression in Gastric Cancer Cells. Cell Signal. 26 (3), 564–569. doi:10.1016/j.cellsig.2013.11.030
He, Y.-Q., Bo, Q., Yong, W., Qiu, Z.-X., Li, Y.-L., and Li, W.-M. (2013). FoxP3 Genetic Variants and Risk of Non-small Cell Lung Cancer in the Chinese Han Population. Gene 531 (2), 422–425. doi:10.1016/j.gene.2013.08.066
Hinz, S., Pagerols-Raluy, L., Oberg, H.-H., Ammerpohl, O., Grüssel, S., Sipos, B., et al. (2007). Foxp3 Expression in Pancreatic Carcinoma Cells as a Novel Mechanism of Immune Evasion in Cancer. Cancer Res. 67 (17), 8344–8350. doi:10.1158/0008-5472.CAN-06-3304
Hori, S., Nomura, T., and Sakaguchi, S. (2003). Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 299 (5609), 1057–1061. doi:10.1126/science.1079490
Jiang, L.-L., and Ruan, L.-W. (2014). Association between FOXP3 Promoter Polymorphisms and Cancer Risk: A Meta-Analysis. Oncol. Lett. 8 (6), 2795–2799. doi:10.3892/ol.2014.2585
Karanikas, V., Speletas, M., Zamanakou, M., Kalala, F., Loules, G., Kerenidi, T., et al. (2008). Foxp3 Expression in Human Cancer Cells. J. Transl Med. 6 (1), 1–8. doi:10.1186/1479-5876-6-19
Ladoire, S., Arnould, L., Mignot, G., Coudert, B., Rébé, C., Chalmin, F., et al. (2011). Presence of Foxp3 Expression in Tumor Cells Predicts Better Survival in HER2-Overexpressing Breast Cancer Patients Treated with Neoadjuvant Chemotherapy. Breast Cancer Res. Treat. 125 (1), 65–72. doi:10.1007/s10549-010-0831-1
Ladoire, S., Mignot, G., Dalban, C., Chevriaux, A., Arnould, L., Rébé, C., et al. (2012). FOXP3 Expression in Cancer Cells and Anthracyclines Efficacy in Patients with Primary Breast Cancer Treated with Adjuvant Chemotherapy in the Phase III UNICANCER-PACS 01 Trial. Ann. Oncol. 23 (10), 2552–2561. doi:10.1093/annonc/mds028
Li, B., Samanta, A., Song, X., Iacono, K. T., Bembas, K., Tao, R., et al. (2007). FOXP3 Interactions with Histone Acetyltransferase and Class II Histone Deacetylases Are Required for Repression. Proc. Natl. Acad. Sci. 104 (11), 4571–4576. doi:10.1073/pnas.0700298104
Li, L.-C., and Dahiya, R. (2002). MethPrimer: Designing Primers for Methylation PCRs. Bioinformatics 18 (11), 1427–1431. doi:10.1093/bioinformatics/18.11.1427
Li, W., Wang, L., Katoh, H., Liu, R., Zheng, P., and Liu, Y. (2011). Identification of a Tumor Suppressor Relay between the FOXP3 and the Hippo Pathways in Breast and Prostate Cancers. Cancer Res. 71 (6), 2162–2171. doi:10.1158/0008-5472.CAN-10-3268
Li, X., Gao, Y., Li, J., Zhang, K., Han, J., Li, W., et al. (2018). FOXP3 Inhibits Angiogenesis by Downregulating VEGF in Breast Cancer. Cell Death Dis 9 (7), 1–12. doi:10.1038/s41419-018-0790-8
Liu, R., Liu, C., Chen, D., Yang, W.-H., Liu, X., Liu, C.-G., et al. (2015). FOXP3 Controls an miR-146/nf-Κb Negative Feedback Loop that Inhibits Apoptosis in Breast Cancer Cells. Cancer Res. 75 (8), 1703–1713. doi:10.1158/0008-5472.CAN-14-2108
Ma, G.-F., Miao, Q., Liu, Y.-M., Gao, H., Lian, J.-J., Wang, Y.-N., et al. (2014). High FoxP3 Expression in Tumour Cells Predicts Better Survival in Gastric Cancer and its Role in Tumour Microenvironment. Br. J. Cancer 110 (6), 1552–1560. doi:10.1038/bjc.2014.47
Martin, F., Ladoire, S., Mignot, G., Apetoh, L., and Ghiringhelli, F. (2010). Human FOXP3 and Cancer. Oncogene 29, 4121–4129. doi:10.1038/onc.2010.174
Merlo, A., Casalini, P., Carcangiu, M. L., Malventano, C., Triulzi, T., Mènard, S., et al. (2009). FOXP3 Expression and Overall Survival in Breast Cancer. Jco 27 (11), 1746–1752. doi:10.1200/JCO.2008.17.9036
Müller, S., Poehnert, D., Müller, J. A., Scheumann, G. W. F., Koch, M., and Lück, R. (2010). Regulatory T Cells in Peripheral Blood, Lymph Node, and Thyroid Tissue in Patients with Medullary Thyroid Carcinoma. World J. Surg. 34 (7), 1481–1487. doi:10.1007/s00268-010-0484-6
Presson, A. P., Yoon, N. K., Bagryanova, L., Mah, V., Alavi, M., Maresh, E. L., et al. (2011). Protein Expression Based Multimarker Analysis of Breast Cancer Samples. BMC cancer 11 (1), 1–14. doi:10.1186/1471-2407-11-230
Redpath, M., Xu, B., van Kempen, L. C., and Spatz, A. (2011). The Dual Role of the X-Linked FoxP3 Gene in Human Cancers. Mol. Oncol. 5 (2), 156–163. doi:10.1016/j.molonc.2011.03.001
Schubert, L. A., Jeffery, E., Zhang, Y., Ramsdell, F., and Ziegler, S. F. (2001). Scurfin (FOXP3) Acts as a Repressor of Transcription and Regulates T Cell Activation. J. Biol. Chem. 276 (40), 37672–37679. doi:10.1074/jbc.M104521200
Szylberg, Ł., Karbownik, D., and Marszałek, A. (2016). The Role of FOXP3 in Human Cancers. Anticancer Res. 36 (8), 3789–3794.
Tao, H., Mimura, Y., Aoe, K., Kobayashi, S., Yamamoto, H., Matsuda, E., et al. (2012). Prognostic Potential of FOXP3 Expression in Non-small Cell Lung Cancer Cells Combined with Tumor-Infiltrating Regulatory T Cells. Lung cancer 75 (1), 95–101. doi:10.1016/j.lungcan.2011.06.002
Wang, L., Liu, R., Li, W., Chen, C., Katoh, H., Chen, G.-Y., et al. (2009). Somatic Single Hits Inactivate the X-Linked Tumor Suppressor FOXP3 in the Prostate. Cancer cell 16 (4), 336–346. doi:10.1016/j.ccr.2009.08.016
Weigelt, B., and Reis-Filho, J. S. (2010). Molecular Profiling Currently Offers No More Than Tumour Morphology and Basic Immunohistochemistry. Breast Cancer Res. 12 (4), 1–4. doi:10.1186/bcr2734
Wieczorek, G., Asemissen, A., Model, F., Turbachova, I., Floess, S., Liebenberg, V., et al. (2009). Quantitative DNA Methylation Analysis of FOXP3 as a New Method for Counting Regulatory T Cells in Peripheral Blood and Solid Tissue. Cancer Res. 69 (2), 599–608. PMID: 19147574. doi:10.1158/0008-5472.CAN-08-2361
Wildin, R. S., Ramsdell, F., Peake, J., Faravelli, F., Casanova, J.-L., Buist, N., et al. (2001). X-linked Neonatal Diabetes Mellitus, Enteropathy and Endocrinopathy Syndrome Is the Human Equivalent of Mouse Scurfy. Nat. Genet. 27 (1), 18–20. doi:10.1038/83707
Winerdal, M. E., Marits, P., Winerdal, M., Hasan, M., Rosenblatt, R., Tolf, A., et al. (2011). FOXP3 and Survival in Urinary Bladder Cancer. BJU Int. 108 (10), 1672–1678. doi:10.1111/j.1464-410x.2010.10020.x
Yang, S., Liu, Y., Li, M.-Y., Ng, C. S. H., Yang, S.-l., Wang, S., et al. (2017). FOXP3 Promotes Tumor Growth and Metastasis by Activating Wnt/β-Catenin Signaling Pathway and EMT in Non-small Cell Lung Cancer. Mol. Cancer 16 (1), 1–12. doi:10.1186/s12943-017-0700-1
Zhang, H.-Y., and Sun, H. (2010). Up-regulation of Foxp3 Inhibits Cell Proliferation, Migration and Invasion in Epithelial Ovarian Cancer. Cancer Lett. 287 (1), 91–97. doi:10.1016/j.canlet.2009.06.001
Keywords: PCR-real time, methylation specific PCR (MS PCR), immunohistochemistry (IHC), biomarkers, mammary cancer, gene experession
Citation: Sadaf , Akhter N, Alharbi RA, Sindi AAA, Najm MZ, Alhumaydhi FA, Khan MA, Deo S and Husain SA (2021) Epigenetic Alteration and its Association With Downregulated FOXP3 Gene in Indian Breast Cancer Patients. Front. Genet. 12:781400. doi: 10.3389/fgene.2021.781400
Received: 22 September 2021; Accepted: 26 October 2021;
Published: 29 November 2021.
Edited by:
Edivaldo Herculano Correa De Oliveira, Evandro Chagas Institute, BrazilReviewed by:
Jogendra Pawar, Purdue University, United StatesSaif Ahmad, Barrow Neurological Institute (BNI), United States
Copyright © 2021 Sadaf, Akhter, Alharbi, Sindi, Najm, Alhumaydhi, Khan, Deo and Husain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Syed Akhtar Husain, c2h1c2FpbkBqbWkuYWMuaW4=
 Sadaf
Sadaf Naseem Akhter1
Naseem Akhter1 Mohammad Zeeshan Najm
Mohammad Zeeshan Najm Fahad A. Alhumaydhi
Fahad A. Alhumaydhi Mohammad Aasif Khan
Mohammad Aasif Khan Syed Akhtar Husain
Syed Akhtar Husain