
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 15 November 2021
Sec. Genetics of Common and Rare Diseases
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.781272
This article is part of the Research Topic Advancing Our Understanding of the Genetic and Functional Basis of Skeletal Dysplasia View all 15 articles
Osteoclasts are the sole bone resorbing cells, which undertake opposing roles to osteoblasts to affect skeletal mass and structure. However, unraveling the comprehensive molecular mechanisms behind osteoclast differentiation is necessitated to overcome limitations and scarcity of available data, particularly in relation with the emerging roles of long non-coding RNAs (LncRNAs) in gene expression. In this study, we performed comprehensive and progressive analyses of the dynamic transcriptomes of murine osteoclasts, generated in vitro. We compared the total RNA-based transcriptomes of murine bone marrow derived cells with differentiated osteoclasts, while focusing on potentially novel genes and LncRNAs, to uncover critical genes and their associated pathways, which are differentially regulated during osteoclast differentiation. We found 4,214 differentially regulated genes during osteoclast differentiation, which included various types of LncRNAs. Among the upregulated protein coding genes not previously associated with osteoclast are Pheta1, Hagh, Gfpt1 and Nol4, while downregulated genes included Plau, Ltf, Sell and Zfp831. Notably, we report Nol4 as a novel gene related to osteoclast activity since Nol4 knockout mice Nol4em1(International Mouse Phenotyping Consortium)J exhibit increased bone mineral density. Moreover, the differentially expressed LncRNAs included antisense and long intergenic non-coding RNAs, among others. Overall, immune-related and metabolism-related genes were downregulated, while anatomical morphogenesis and remodeling-related genes were upregulated in early-differentiated osteoclasts with sustained downregulation of immune-related genes in mature osteoclasts. The gene signatures and the comprehensive transcriptome of osteoclast differentiation provided herein can serve as an invaluable resource for deciphering gene dysregulation in osteoclast-related pathologic conditions.
The balance between osteoclast and osteoblast activity can dictate pathogenesis of bone diseases. Osteoclasts are the exclusive bone resorbing cells involved in bone remodeling and resorption, and perform opposing roles to osteoblasts to affect skeletal mass and structure (Teitelbaum and Ross, 2003). Augmented osteoclast activity can lead to bone loss in osteoporosis, inflammatory arthritis and tumor invasion in bone, while osteopetrosis is characterized by increased bone mass and results from attenuation in osteoclast function/recruitment or arrested osteoclastogenesis (Teitelbaum, 2007). Abnormalities in osteoclasts are considered the primary cause of many bone diseases including osteoporosis, the most common bone disorder, and Paget disease of the bone (PDB) in which accelerated osteoclastic bone resorption leads to osteolytic or osteosclerotic bone lesions (Shaker, 2009).
Osteoclasts are multi-nucleated cells belonging to myeloid lineage, and generated via the fusion of monocytes/macrophage precursor cells (Roodman, 1999). Importantly, signaling via tumor necrosis factor (TNF)-family cytokine, receptor activator of nuclear factor (NF)-kappaB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF) are identified as the primary pathways associated with osteoclast differentiation (Asagiri and Takayanagi, 2007). Osteoclast precursors express receptor activator of nuclear factor kappa B (RANK), while its ligand (RANKL) is expressed on osteoblasts/stromal cell precursors, and inhibited by the decoy receptor osteoprotegerin (OPG) (Yasuda et al., 1998). Moreover, various genes including TNF receptor-associated factor (TRAF) 6, NFKB1, FOS, nuclear factor of activated T cells 1 (NFATC1), and dendritic cell-specific transmembrane protein (DC-STAMP) have been associated with osteoclastogenesis (Asagiri and Takayanagi, 2007), while the tartrate-resistant acid phosphatase (TRAcP) and cathepsin K (CTSK), released by osteoclasts during resorption are identified as specific osteoclast markers (Boyle et al., 2003; Kirstein et al., 2006). Osteoclast functionality in bone resorption is dependent on αvβ3 integrin-mediated induction, binding and polarization, while deficiencies in acidified bone resorptive components disrupt regulated bone remodeling, leading to osteopetrosis. Amplified stimulation of osteoclastogenesis primarily via NFKB signaling can lead to increased osteolysis in osteoporosis (Novack and Teitelbaum, 2008).
Genome-wide screening and the in vitro induction of osteoclastogenesis using osteoclast precursor cells and soluble mediators has enabled the identification of molecular mechanisms governing osteoclast differentiation (Asagiri and Takayanagi, 2007). Sequencing techniques and microarray analyses have also contributed to disclosing important genes related to osteoclast differentiation (Cappellen et al., 2002; Day et al., 2004; Nomiyama et al., 2005; Garlet et al., 2008; Coudert et al., 2014; Kim and Lee, 2014; Purdue et al., 2014; Madel et al., 2020). In addition, recent reports have highlighted the roles of long noncoding RNAs (LncRNAs), which have emerged as vital regulators of gene expression, epigenetics and protein translation (Fatica and Bozzoni, 2014), in osteoclast differentiation (Fei et al., 2020; Liu et al., 2020; Yang et al., 2021). However, available data have primarily focused on differences between fully differentiated/generated osteoclasts compared to precursors or focused predominantly on targeted sets of genes, previously linked with osteoclast differentiation/activity. Therefore, a comprehensive insight into the differentially regulated genes and their associated pathways during osteoclast differentiation is warranted.
In this study, we performed sequential and comprehensive transcriptomic profiling of murine osteoclasts generated in vitro by comparing the total RNA transcriptomes of bone marrow derived precursor cells with differentiated osteoclasts over different time points. Importantly, we focused on novel genes and LncRNAs, which have not been previously associated with osteoclast differentiation. These progressive analyses provide important insights into the differentially regulated genes and their associated pathways during osteoclast differentiation. Identification of novel gene signatures and LncRNAs associated with osteoclastogenesis can serve as molecular biomarkers for osteoclast differentiation or explored for therapeutic benefits, while the comprehensive transcriptome of osteoclast differentiation provided herein can serve as an invaluable resource for deciphering gene dysregulation in diseases related to osteoclast differentiation/activity.
C57 black 6 (CBL/6) mice, with access to adequate nutrition (pelleted RM1; SDS diets, Essex, United Kingdom) and hydration, in a standard animal research facility were utilized in this study. All protocols were performed as per guidelines of the United Kingdom Animals Act of 1986 (Scientific Procedures). Bone marrow (BM) was flushed from long bones of 3-4-month-old CBL/6 mice and cultured in specialized media designed to induce osteoclast differentiation, as described below. All experiments were performed adhering to applicable guidelines and regulations.
Osteoclast cultures were maintained as described previously (Alonso et al., 2021). Briefly, BM cells from CBL/6 mice were cultured in complete growth media (α-MEM supplemented with 10% fetal calf serum, penicillin/streptomycin and glutamine) and in the presence of soluble M-CSF (Prospec Technology, United Kingdom) at 100 ng/ml to generate bone marrow derived macrophages (BMDM). Non-adherent cells were washed after 48 h, while adherent cells were removed using cell dissociation buffer (Gibco, United Kingdom) and re-cultured in parallel in 6-well plates at a density of 3 × 105 cells per well and in 96-well plates at 1 × 104 cells per well in the presence of M-CSF (25 ng/ml) and soluble RANKL (R&D Systems, United Kingdom) at 100 ng/ml. Cultures in 96-well plates were used to monitor osteoclast differentiation and mature osteoclasts were detected by staining for tartrate-resistant acid phosphatase (TRAcP). Cells with more than three nuclei and positive for TRAcP staining were considered osteoclasts. In addition, the resorptive activity of generated osteoclasts was determined using Osteo Assay plates (Corning, New York, United States), as per manufacturer’s guidelines. Quantification of resorption areas was carried out using ImageJ software (National Institutes of Health, Maryland, United States). Unpaired t-test was used to investigate statistical significance between two groups, while one-way Anova test was performed to determine statistical significance between more than two groups (GraphPad Prism, version 9.0; GraphPad Software, California, United States). A p value of <0.05 was considered statistically significant.
Cultures in 6-well plates were used to collect RNA for gene expression profiling. Cells were collected at the following time points for gene expression analysis: day 0 (BMDM), day 3 (osteoclast precursor) and day 4 (osteoclast) for subsequent investigations.
RNA was isolated from BMDM cells (Day 0) and differentiated osteoclasts from different time points (Day 3 and Day 4) using GenElute Mammalian Total RNA Kit (Sigma-Aldrich, Missouri, United States) by following manufacturer’s protocol. Integrity and purity of RNA was assessed using Bioanalyzer 2100 (Agilent Technologies, California, United States). A total of nine RNA samples were obtained (representing three biological replicates at Day 0, Day 3 and Day 4).
cDNA libraries were generated from isolated total RNA samples using TruSeq Stranded Total RNA kit with Ribo-Zero Globin (Illumina, California, United States) by following manufacturer’s protocol. Quality-passed libraries were sequenced on NovaSeq6000 system (Illumina) using 100 bp paired-end protocol.
Data were analyzed and illustrated using multiple bioinformatics software under default settings unless otherwise stated. Reads were quality-trimmed using Cutadapt1 (version 1.9. dev2) and those with low quality and short reads (<35 bp) were trimmed along with Illumina TruSeq RNA kit adapters. Reads were aligned to the reference genome (Mus Musculus GRmc38) using STAR2 (version 2.5.2b) specifying paired-end reads. Reads were assigned to features of type ‘exon’ in the input annotation grouped by gene_id in the reference genome using featureCounts3 (version 1.5.1). The raw counts table was filtered to remove genes consisting predominantly of near-zero counts, filtering on counts per million (CPM) to avoid artefacts due to library depth. Overall, three biological replicate datasets were generated for each time point (Day 0, Day 3, and Day 4). Abundance data were successively subjected to differential gene expression analyses. Z-scores were calculated from CPM values as described previously (Malone et al., 2011) and heatmaps generated using GraphPad Prism software (GraphPad Software).
Differential gene expression analyses and gene ontology (GO) clustering analyses were performed using Integrated Differential Expression and Pathway (iDEP.92, South Dakota State University, United States) online tool. Raw CPM values were uploaded and computed (min. CPM = 1) to identify and generate various illustrations for gene clustering and differentially expressed genes. Differential expression analysis was performed using the DEseq2 method (Love et al., 2014). PCA and Volcano plots were generated with Log2-fold change (FC) > 2 and false discovery rate (FDR) cutoff <0.05. K-Means clustering were used for performing gene enrichment analyses using GO biological processes pathway database (Ashburner et al., 2000).
An overview of the study design to decipher the transcriptomic changes during murine osteoclast differentiation in vitro is depicted in Figure 1A. The morphological changes in osteoclast differentiation showed the progressive and statistically significant differences in the number of TRAcP+ osteoclasts (≥3 nuclei) formed during differentiation of BMDM cells to osteoclast (Figures 1B,C). Osteoclast precursors exhibit a pre-fusion state whereby the cells cluster together to form multi-nucleated mature osteoclasts (Day 4). In addition, the functional activity of generated osteoclasts was assessed by Osteo Assay (resorptive activity assay), which showed statistically significant, distinct and large resorption areas with osteoclasts compared to osteoclast precursors and undifferentiated BMDM cells (Figures 1D,E). Comprehensive investigations were performed on the differentially regulated genes disclosed during osteoclast differentiation, using stringent criteria and cutoffs as described above.
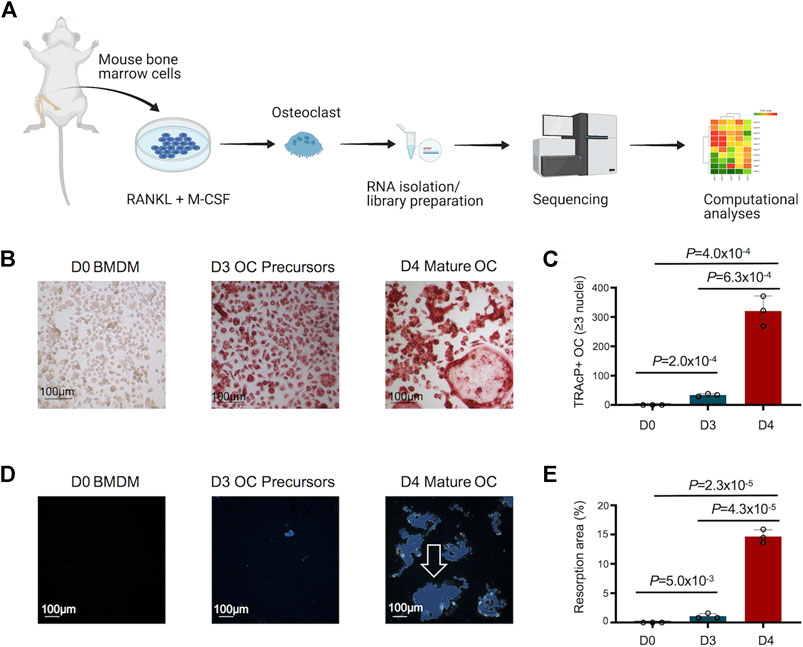
FIGURE 1. Study design, and morphological and functional changes during osteoclast (OC) differentiation. (A) Bone marrow cells flushed from long bones of CBL/6 mice were cultured in the presence of soluble M-CSF and RANKL to induce OC differentiation. Libraries were generated from bone marrow derived macrophages (BMDM) (Day 0) and differentiated osteoclasts at different time points (Days 3 and 4) to perform comprehensive and progressive analyses of the dynamic transcriptomes of murine osteoclasts by RNA-Seq. Various bioinformatics tools were utilized for data analyses/visualization. Schematic representation of the study design is shown. (B) The generated OC were detected by staining for tartrate-resistant acid phosphatase (TRAcP). Cells positive for TRAcP staining and with more than three nuclei were identified as OC representative images of TRAcP stained cells show Bone marrow derived macrophages (BMDM) at day 0 (D0), OC precursors at D3 and mature OC at D4 of the culture. (C) Scatter/bar-hybrid plot shows the numbers with mean ± standard deviation (SD) of TRAcP+ OC in D0, D3 and D4 comparisons. (D) The bone resorption activity of OC was investigated using Osteo Assay. Representative images show cultures from BMDM cells at D0, OC precursors at D3 and mature OC at D4 of the culture. Resorption areas are indicated by white arrow. (E) Scatter/bar-hybrid plot shows the differences in percentage resorption area (mean ± SD) in D0, D3 and D4 cultures.
We generated comprehensive datasets for the transcriptomes of undifferentiated BMDM cells (Day 0) and differentiated osteoclasts on Days 3 and 4 (Supplementary Tables S1-3). One replicate from day 3 failed the quality control measures and was removed from the analysis. PCA analyses showed close proximity of biological replicates for each time point: PC1 showed 87% variance and PC2 showed 7% variance (Figure 2A). Hierarchical gene clustering during osteoclast differentiation showed distinct gene clusters (Figure 2B). Overall, 4,214 differentially regulated genes (2,251 downregulated and 1,963 upregulated) were identified, which showed some overlap between the different timepoint comparisons, primarily between Day 3 versus Day 0 and Day 4 versus Day 0 comparisons (Figure 2C).
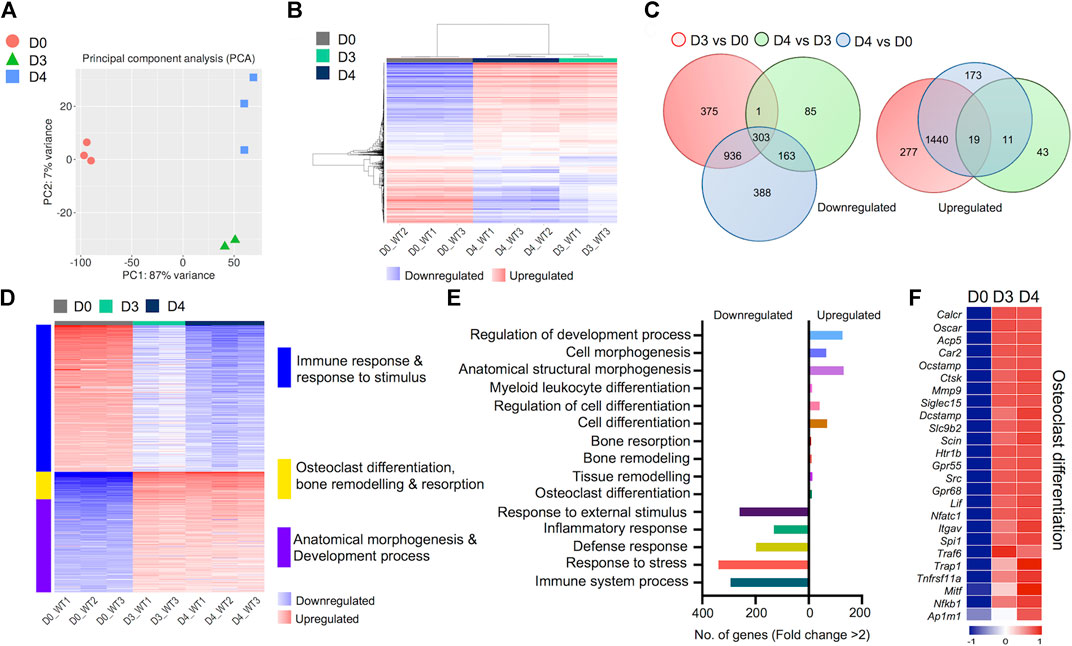
FIGURE 2. Transcriptomic profiling of osteoclast differentiation. The transcriptomes of bone marrow derived macrophages (BMDM; D0) were compared with osteoclast precursors (D3) and fully differentiated osteoclasts (D4) by performing differential gene expression analyses. (A) PCA plot shows variability in gene expression among biological replicates between D0, D3 and D4. (B) Heatmap shows hierarchical clustering of gene expression in cells from D0, D3 and D4. (C) Venn diagram depicts the total numbers of overlapping differentially regulated (downregulated and upregulated) genes between D0, D3 and D4. (D) Heatmap shows gene enrichment and associated pathways in D0, D3 and D4. (E) Bar plot shows the numbers of genes corresponding to significantly up/downregulated functional pathways from gene ontology enrichment analyses. (F) Heat map shows the gene expression (Z-scores) of osteoclast-related gene panel for confirmation of osteoclast generation.
Importantly, K-Means clustering analyses revealed the corresponding pathways of gene enrichment observed during osteoclast differentiation (Figure 2D). Our data showed that genes related to immune and inflammatory response, and response to stimulus were downregulated during osteoclast differentiation. Concurrently, genes related to anatomical morphogenesis and developmental process were upregulated in differentiated osteoclasts. Moreover, upregulation of genes related to osteoclast differentiation, bone remodeling and resorption was observed during osteoclast differentiation. Further analyses revealed that the number of upregulated genes related to osteoclast differentiation (n = 12), tissue remodeling (n = 14), bone remodeling (n = 11) and bone resorption (n = 9) was lower compared to the number of downregulated genes associated with immune response (n = 294), response to stress (n = 339), defense response (n = 198), inflammatory response (n = 131) and response to external stimulus (n = 260) (Figure 2E).
Of note, to confirm the in vitro generation of osteoclasts, we investigated the expression levels of critical genes known to be expressed in osteoclasts (Teitelbaum and Ross, 2003; Asagiri and Takayanagi, 2007). These selective genes included Oscar, Ocstamp, Acp5 (TRAcP), Ctsk, Dcstamp, Nfatc1, Traf6, Trap1 and Nfkb1, among others. We found that all genes in our selected panel of osteoclast-related genes were upregulated following induction of osteoclast differentiation (Figure 2F). These confirmatory data provided additional evidence for successful osteoclast generation.
An important aspect of this study was to uncover gene expression profiles during the course of osteoclast differentiation. In this pursuit, we first compared the transcriptomes of early-differentiated osteoclasts (Day 3) with BMDM cells (Day 0; Figure 3). Our data revealed 3,351 differentially regulated genes, of which 1,736 genes were upregulated while 1,615 genes were downregulated in early-differentiated osteoclasts (Figure 3A). Importantly, GO biological process enrichment analyses revealed distinct gene clusters between the transcriptomes of Day 3 versus Day 0 analyses (Figure 3B). Upregulated genes predominantly corresponded to metabolic processes, whereas downregulated genes primarily corresponded to immune and stimulus response. Generation of precursor metabolites and energy- (n = 102), small molecule metabolic process- (n = 256), phosphate containing compound metabolic process- (n = 365) and nucleotide metabolic process-related genes (n = 114) comprised upregulated genes. In contrast, response to stress- (n = 475), regulation of response to stimulus- (n = 467), immune system process- (n = 342) and inflammatory response-related genes (n = 139) comprised downregulated genes during early osteoclast differentiation (Figure 3C).
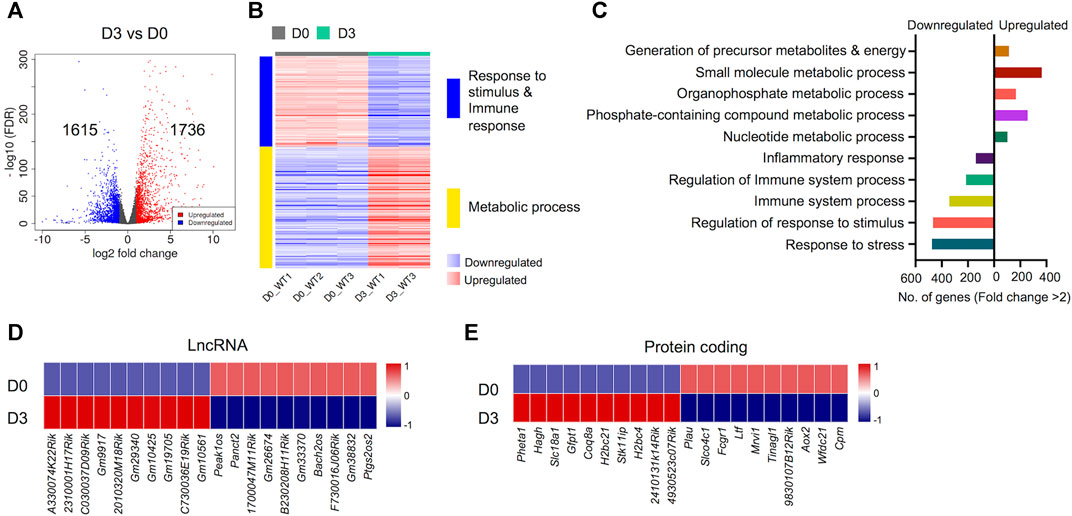
FIGURE 3. Differentially regulated genes in early osteoclast differentiation. The transcriptomes of osteoclast precursors (D3) were compared with bone marrow derived macrophages (BMDM) (D0). (A) Volcano plot represents significantly upregulated (red), downregulated (blue) or genes with unchanged expression levels (gray) between D3 and D0 comparison. (B) Heatmap shows gene enrichment and associated pathways in D3 versus D0. (C) Bar plot shows the numbers of genes corresponding to significantly up/downregulated functional pathways from gene ontology enrichment analyses. Heat maps show the gene expression (Z-scores) of top differentially-regulated long non-coding RNAs (LncRNA) (D) and protein coding genes (E) in D3 versus D0 comparison (Log2-FC ≥ 2).
Apart from protein coding genes, the importance of LncRNAs in the regulation of osteoclast differentiation has been recently described (Fei et al., 2020; Liu et al., 2020; Yang et al., 2021). We compiled a list of differentially regulated LncRNAs during osteoclast differentiation (Supplementary Figure S4). These differentially regulated LncRNAs comprised of different subtypes as listed in Table 1. We found that 35 potentially novel LncRNAs were upregulated of which 20 LncRNAs showed Log2-FC ≥ 2, while 37 LncRNAs were downregulated with 15 LncRNAs showing Log2-FC ≥ 2 during early osteoclast differentiation. The top 20 differentially expressed LncRNAs during early osteoclast differentiation are presented in Figure 3D. These data reflect that these LncRNA genes may have significant roles in osteoclastogenesis and warrant further scrutiny.
In addition, we also identified the top upregulated protein coding genes, which have not been previously reported to be associated with osteoclast differentiation. The top 20 differentially expressed genes based on significance are presented in Figure 3E while, the complete list of potentially novel differentially expressed genes during osteoclast differentiation is provided in Supplementary Table S5. Additionally, the top 10 upregulated and 10 downregulated genes during osteoclast differentiation, based on FC and significance (p value), irrespective of novelty, are presented in Supplementary Figure S1.
Next, we compared the transcriptomes of early-differentiated osteoclasts (Day 3) with fully differentiated osteoclasts (Day 4). Interestingly, we found that the transcriptomes of committed osteoclasts did not show immense differences with fully differentiated osteoclasts. Overall, only 641 genes showed differential regulation between Day 4 and Day 3 comparison, of which 73 genes were upregulated while 552 genes were downregulated (Figure 4A). Importantly, these differentially regulated genes showed further enrichment in downregulation of immune response-related genes, whereas cell cycle and DNA replication-related processes were upregulated (Figure 4B). The downregulated genes corresponding to immune response-related processes were associated with process such as cytokine production (n = 68), leukocyte activation (n = 73), inflammatory (n = 68) and immune response (n = 112). The potentially novel genes not previously reported in association with osteoclast differentiation/activity are shown in Figure 4D, while the top differentially regulated genes based on FC and significance, irrespective of novelty are presented in Supplementary Figure S1.
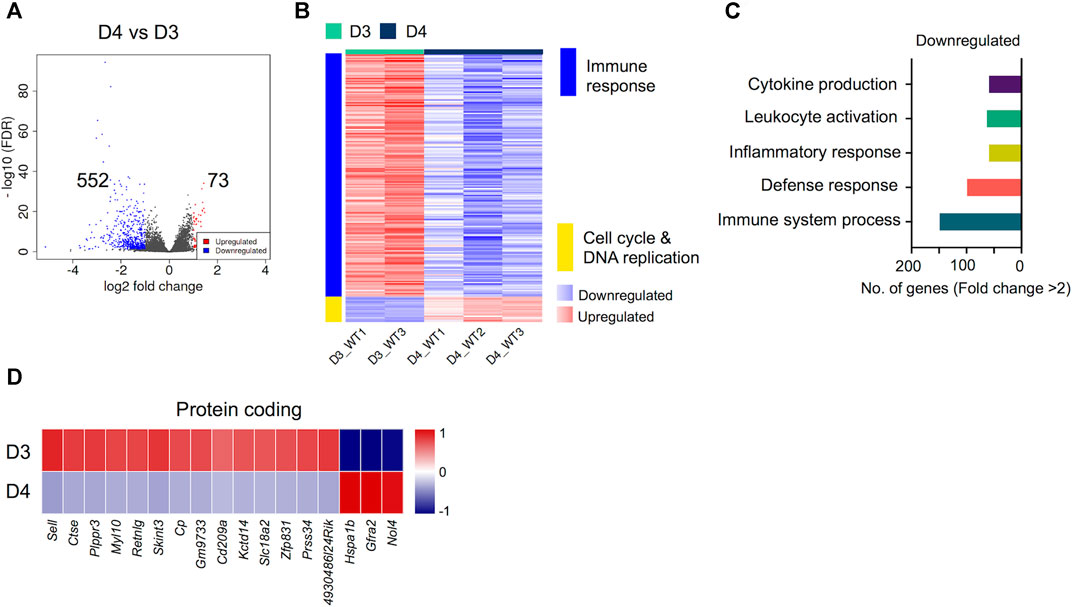
FIGURE 4. Differentially regulated genes in late osteoclast differentiation. The transcriptomes of fully differentiated osteoclasts (D4) were compared with osteoclast precursors (D3). (A) Volcano plot represents significantly upregulated (red), downregulated (blue) or genes with unchanged expression levels (gray) between D4 and D3 comparison. (B) Heatmap shows gene enrichment and associated pathways in D4 versus D3. (C) Bar plot shows the numbers of genes corresponding to significantly up/downregulated functional pathways from gene ontology enrichment analyses. (D) Heat maps show the gene expression (Z-scores) of downregulated and upregulated protein coding genes in D4 versus D3 comparison.
The upregulation of selected potentially novel protein coding genes in mature osteoclasts prompted us to explore their prospective roles in osteoclast functionality. Since these genes have not been previously linked with osteoclast generation or activity, we investigated their associated phenotypes in the International Mouse Phenotyping Consortium (IMPC) database (Dickinson et al., 2016; International Mouse Phenotyping Consortium, 2021). Interestingly, we found that Selectin L (Sell), Myosin Light Chain 10 (Myl10), Resistin-like gamma (Retnlg) and Ceruloplasmin (Cp) Signal Regulatory Protein Delta (Sirpd/Gm9733), Zinc Finger Protein 831 (Zfp831) and Nuclear Protein 4 (Nol4) were previously investigated in relation with bone-related phenotypes. However, only Nol4 showed statistically significant associations with bone mineral density (BMD) (Figure 5A), while Zfp831 showed significant associations with tibia length (Figure 5B) in data from IMPC. Performing a body composition (DEXA lean/fat) phenotypic assay on 4,448 mice (male and female) showed that homozygous Nol4 knockout mic (Nol4em1(IMPC)J, n = 15) exhibited a significant increase in BMD compared to wild type mice (Figure 5A). While, performing phenotypic assays on Zfp831 knockout mice (Zfp831tm1b(KOMP)Wtsi) homozygote mutant mice (n = 10) compared to controls (n = 295) showed a significant reduction in bone (tibia) length (Figure 5B). The downregulation of Zfp831 in mature osteoclasts in our dataset therefore indicated its potential role in bone development/growth.
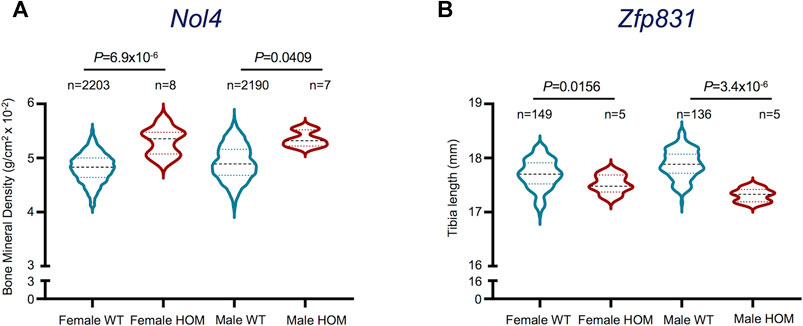
FIGURE 5. Bone-related phenotypes of Nol4 and Zfp831 knockout mice in the International Mouse Phenotyping (IMPC) datasets. (A) Violin plot shows the differences in Bone Mineral Density (excluding skull) in female (n = 8), male (n = 7) homozygote (HOM) mutants for the Nol4em1(IMPC)J allele compared to female (n = 2,203), male (n = 2,190) wild type (WT) controls. (B) Violin plot shows the differences in Tibia length in female (n = 5), male (n = 5) HOM mutants for Zfp831tm1b(KOMP)Wtsi allele compared to female (n = 149) and male controls (n = 136). The interquartile range, minimum and maximum data points, and individual points representing outliers of each dataset are presented. p values represent the statistical significance between each comparison [Data taken from the International Mouse Phenotyping Consortium (IMPC) (Dickinson et al., 2016; International Mouse Phenotyping Consortium, 2021)].
The molecular mechanisms behind osteoclast differentiation have been extensively explored. For instance, Capellen et al., used microarray analyses to decipher changes in gene expression during osteoclast differentiation and showed synergy between MCSF and RANKL-induced gene expression (Cappellen et al., 2002). More recently, several potential genetic regulators of osteoclast differentiation have been identified. Nishikawa et al., reported the epigenetic control of osteoclast differentiation via DNA (cytosine-5)-methyltransferase 3A (Dnmt3a) (Nishikawa et al., 2015) and Laha et al., reported regulation of osteoclastogenesis by KLF2 (kruppel-like factor 2 [lung]) via reduction in autophagic cells (Laha et al., 2019). In addition, low-density lipoprotein receptor-related protein1 (LRP1), and COMMD1 were reported as critical regulators of osteoclastogenesis, osteoclast activity and bone mass (Murata et al., 2017; Bartelt et al., 2018), while leucine-rich repeat-containing G-protein-coupled receptor 4 (LGR4) has been identified as another putative receptor for RANKL (Luo et al., 2016).
While these studies revealed important osteoclast-related targets, a comprehensive list of genes provided in our study is of paramount significance in understanding the dynamic changes in gene expression during osteoclast differentiation. Irrefutably, the preexistent available literature related to osteoclast identification and functionality enabled us to confirm the generation of osteoclasts in vitro. However, our data revealed the dynamic changes in osteoclast transcriptome during osteoclastogenesis, showing diminishing of genes related to immune processes with concurrent upregulation of genes associated with anatomical morphogenesis and development. Moreover, these genes encoded for important mediators of cell differentiation, structural morphogenesis and bone and tissue remodeling.
The potentially novel genes related to osteoclast differentiation disclosed herein include PH Domain Containing Endocytic Trafficking Adaptor 1 (Pheta1), Hydroxyacylglutathione hydrolase (Hagh), Solute Carrier Family 18 Member A1 (Slc18a1), Glutamine-Fructose-6-Phosphate transaminase 1 (Gfpt1), Coenzyme Q8A (Coq8a), Solute Carrier Family 14 Member 2 (Slc14a2), Whirlin (Whrn) and C-Type Lectin Domain Family four Member A (Clec4a4), among others. The proteins encoded by these genes are primarily associated with biochemical or signaling pathways at the cellular level and defects in expression may lead to certain disorders. For instance, anomalies in PHETA1/2 have been associated with abnormal bone formation resulting in craniofacial defects (Ates et al., 2020), HAGH has been associated with skin, bone and joint infections in Yaws disease (Cheng et al., 2018) and mutations in GFPT1 have been associated with muscle weakness in congenital myasthenic syndrome (Helman et al., 2019). In addition, histone-related genes H2B Clustered Histone 21 (H2bc21) and H2bc4 were also significantly upregulated during osteoclast differentiation. Epigenetic regulation of bone development and remodeling has been extensively reported (Yi et al., 2019). The upregulation of histone-related genes in osteoclast differentiation indicates chromatin remodeling as a critical step in osteoclast generation.
One of the pivotal novel findings of this study is the upregulation and confirmatory evidence of the potential role of Nol4 in mature osteoclasts, which has not been reported previously. Investigating bone-related phenotypes of some of the potentially novel genes we recorded showed that selective genes including Pheta1, Hagh, Plasminogen Activator urokinase (Plau), Sell, Retnlg and Cp are related to bone phenotypes in the IMPC database (Dickinson et al., 2016; International Mouse Phenotyping Consortium, 2021). However, only Nol4 and Zfp831 showed statistically significant associations with BMD or tibia length, respectively. Importantly, the knockout of Nol4 led to increase in BMD compared to wild type mice, thereby providing evidence for its role in bone biology. Our differential gene analyses showed upregulation of Nol4 expression in differentiated osteoclasts, which supports their contribution in affecting BMD in bone resorption and presents Nol4 as a key gene related to osteoclast functionality. Of note, downregulation of Zfp831 in mature osteoclasts in our data and association of Zfp831 knockout with reduced bone (tibia) length in IMPC datasets suggest its potential roles in osteoclast-mediated effects on bone growth and remodeling. However, further investigations are necessitated to fully explore the effects of these genes on bone biology. The protein encoded by Nol4 is associated with RNA binding and has been identified as a cancer/testis antigen in humans, recently presented as a candidate target in small cell lung cancer (Kim et al., 2021). However, the role of Nol4 in bone-related pathologies remains to be fully explored. Similarly, the protein encoded by Zfb831 is associated with nucleic acid binding and was downregulated in tumor-infiltrating cytotoxic T lymphocytes (CTLs) compared to CTLs from spleen in a murine transplantable tumor model (Waugh et al., 2016), but its effects in bone-related diseases are not explored.
Accumulating evidences have highlighted the roles of LncRNAs in controlling gene expression to affect cell differentiation and development (Fatica and Bozzoni, 2014). Importantly, in relation with osteoclasts. Liu et al., reported dysregulation in 1,117 LncRNAs in human osteoclasts differentiated from CD14+ monocytes in vitro (Liu et al., 2020). Fei et al., also reported 204 differentially expressed LncRNAs in male osteoporosis (Fei et al., 2020), while Yang et al., identified 46 differentially expressed LncRNAs between osteoarthritis and osteolysis following total hip arthroplasty, and reported potential roles of specific LncRNA-mRNA pairs in regulating CD8A, CD8B and osteoclastogenesis in these patients (Yang et al., 2021). Moreover, Bu et al., reported the role of LncRNA TSIX in promoting osteoblast apoptosis in particle induced osteolysis (PIO), evident from decreased BMD following implantation, via modulation of the microRNA miR-30a-5p (Bu et al., 2018). These reports rationalize the significance of LncRNAs in osteoclast generation and activity. We performed comprehensive total RNA-based analyses to disclose the differentially regulated LncRNAs during osteoclast differentiation and exclusively reported LncRNA not previously reported or associated with osteoclasts. Importantly, we have highlighted the top 20 novel LncRNAs upregulated during murine osteoclast generation.
Osteoporosis is characterized by attenuated bone strength due to deterioration in bone microarchitecture and reduction in bone mass (Sozen et al., 2017). While various risk factors have been identified for predisposition to osteoporosis, endocrine diseases have also been liked with osteoporosis (Sozen et al., 2017). For instance, in diabetes-associated osteoporosis altered bone metabolism also leads to changes in BMD and has been linked with high osteoclast activity (Kemink et al., 2000; Reni et al., 2016). Importantly, insulin is identified as an essential mediator in osteoclast energy metabolism. Kim et al., investigated the effects on gene expression in insulin-induced osteoclast differentiation and reported that insulin conducts similar roles as RANKL in osteoclast activity (Kim and Lee, 2014). These reports reflect the significance of energy and metabolism in osteoclast activity, which leads to osteoporosis. We found that numerous genes encoding important metabolism/energy-related mediators were significantly increased during early osteoclast generation.
Osteoclast progenitors are essentially immune cells, due to their origins from monocytes/macrophage precursors and may present as innate immune cells within the bone (Wu et al., 2008). Jacquin et al., showed that osteoclast progenitor populations in murine BM comprise CD45R− CD11b−/low populations (Jacquin et al., 2006), while Jacome-Galarza et al., reported that B220−CD3−CD11b−/lowCD115+CD117high mouse BM cells possess high osteoclastic potential (Jacome-Galarza et al., 2013). Our data showed that osteoclast differentiation deviates their associations with other immunomodulatory cells, evident from downregulation of genes related to immune/inflammatory response and response to stress/stimulus in differentiated osteoclasts. Moreover, we found that the differences between the transcriptomes of fully-committed/differentiated and early-differentiated osteoclasts were predominantly limited to sustained downregulation of genes related to immune cell characteristics. These differentially expressed genes primarily encoded for cytokine production, leukocyte activation and immune response-related functions. Importantly, upregulation of genes related to cell cycle and DNA replication in fully differentiated osteoclasts exhibited the cellular expansion of committed osteoclasts. Of note, induction of bone resorption in inflammatory disorders and immune-related disorders results from the plethora of secreted cytokines, which induce osteoclastogenesis (Adamopoulos, 2018).
Overall, this study disclosed genes and their associated pathways during osteoclast differentiation which can be explored further in successive studies. Importantly, we highlighted the potentially novel genes and LncRNAs in relation to osteoclast differentiation and activity. However, functional studies are necessitated to disclose the roles of specific targets. The comprehensive list of differentially regulated genes provided herein can serve as an expedient tool in osteoclast-related research.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA, Accession no: PRJNA769960 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA769960/).
The animal study was reviewed and approved by Animal Welfare and the Ethical Review Body, University of Edinburgh.
ST: Data curation, Formal analysis, Visualization, Writing–original draft. SW: Data curation, Investigation, Methodology, Writing–review and editing. OA: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing.
This work was supported by a grant from the Medical Research Council United Kingdom (Grant No. G0800933/87,390) and by the European Research Council (ERC) consolidator grant awarded to Prof. Omar ME Albagha (Grant No. 311723-GENEPAD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.781272/full#supplementary-material
Adamopoulos, I. E. (2018). Inflammation in Bone Physiology and Pathology. Curr. Opin. Rheumatol. 30 (1), 59–64. doi:10.1097/bor.0000000000000449
Alonso, N., Wani, S., Rose, L., Van't Hof, R. J., Ralston, S. H., and Albagha, O. M. E. (2021). Insertion Mutation in Tnfrsf11a Causes a Paget's Disease-like Phenotype in Heterozygous Mice and Osteopetrosis in Homozygous Mice. J. Bone Miner Res. 36, 1376. doi:10.1002/jbmr.4288
Asagiri, M., and Takayanagi, H. (2007). The Molecular Understanding of Osteoclast Differentiation. Bone 40 (2), 251–264. doi:10.1016/j.bone.2006.09.023
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 25 (1), 25–29. doi:10.1038/75556
Ates, K. M., Wang, T., Moreland, T., Veeranan-Karmegam, R., Ma, M., Jeter, C., et al. (2020). Deficiency in the Endocytic Adaptor Proteins PHETA1/2 Impairs Renal and Craniofacial Development. Dis. Model. Mech. 13 (5), dmm041913. doi:10.1242/dmm.041913
Bartelt, A., Behler-Janbeck, F., Beil, F. T., Koehne, T., Müller, B., Schmidt, T., et al. (2018). Lrp1 in Osteoblasts Controls Osteoclast Activity and Protects against Osteoporosis by Limiting PDGF-RANKL Signaling. Bone Res. 6, 4. doi:10.1038/s41413-017-0006-3
Boyle, W. J., Simonet, W. S., and Lacey, D. L. (2003). Osteoclast Differentiation and Activation. Nature 423 (6937), 337–342. doi:10.1038/nature01658
Bu, Y., Zheng, D., Wang, L., and Liu, J. (2018). LncRNA TSIX Promotes Osteoblast Apoptosis in Particle-Induced Osteolysis by Down-Regulating miR-30a-5p. Connect. Tissue Res. 59 (6), 534–541. doi:10.1080/03008207.2017.1413362
Cappellen, D., Luong-Nguyen, N.-H., Bongiovanni, S., Grenet, O., Wanke, C., and Šuša, M. (2002). Transcriptional Program of Mouse Osteoclast Differentiation Governed by the Macrophage Colony-stimulating Factor and the Ligand for the Receptor Activator of NFκB. J. Biol. Chem. 277 (24), 21971–21982. doi:10.1074/jbc.m200434200
Cheng, Z., VanPelt, J., Bergstrom, A., Bethel, C., Katko, A., Miller, C., et al. (2018). A Noncanonical Metal Center Drives the Activity of the Sediminispirochaeta Smaragdinae Metallo-β-Lactamase SPS-1. Biochemistry 57 (35), 5218–5229. doi:10.1021/acs.biochem.8b00728
Coudert, A. E., Del Fattore, A., Baulard, C., Olaso, R., Schiltz, C., Collet, C., et al. (2014). Differentially Expressed Genes in Autosomal Dominant Osteopetrosis Type II Osteoclasts Reveal Known and Novel Pathways for Osteoclast Biology. Lab. Invest. 94 (3), 275–285. doi:10.1038/labinvest.2013.140
Day, C. J., Kim, M. S., Stephens, S. b. R. J., Simcock, W. E., Aitken, C. J., Nicholson, G. C., et al. (2004). Gene Array Identification of Osteoclast Genes: Differential Inhibition of Osteoclastogenesis by Cyclosporin A and Granulocyte Macrophage colony Stimulating Factor. J. Cel. Biochem. 91 (2), 303–315. doi:10.1002/jcb.10780
Dickinson, M. E., Flenniken, A. M., Flenniken, A. M., Ji, X., Teboul, L., Wong, M. D., et al. (2016). High-throughput Discovery of Novel Developmental Phenotypes. Nature 537 (7621), 508–514. doi:10.1038/nature19356
Fatica, A., and Bozzoni, I. (2014). Long Non-coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 15 (1), 7–21. doi:10.1038/nrg3606
Fei, Q., Li, X., Lin, J., Yu, L., and Yang, Y. (2020). Identification of Aberrantly Expressed Long Non-coding RNAs and Nearby Targeted Genes in Male Osteoporosis. Cia 15, 1779–1792. doi:10.2147/cia.s271689
Garlet, T. P., Coelho, U., Repeke, C. E., Silva, J. S., Cunha, F. d. Q., and Garlet, G. P. (2008). Differential Expression of Osteoblast and Osteoclast Chemmoatractants in Compression and Tension Sides during Orthodontic Movement. Cytokine 42 (3), 330–335. doi:10.1016/j.cyto.2008.03.003
Helman, G., Sharma, S., Crawford, J., Patra, B., Jain, P., Bent, S. J., et al. (2019). Leukoencephalopathy Due to Variants in GFPT1-Associated Congenital Myasthenic Syndrome. Neurology 92 (6), e587–e593. doi:10.1212/wnl.0000000000006886
International Mouse Phenotyping Consortium (2021). IMPC Data 2021. Available at: www.mousephenotype.org (Accessed August 18, 2021).
Jacome-Galarza, C. E., Lee, S.-K., Lorenzo, J. A., and Aguila, H. L. (2013). Identification, Characterization, and Isolation of a Common Progenitor for Osteoclasts, Macrophages, and Dendritic Cells from Murine Bone Marrow and Periphery. J. Bone Miner Res. 28 (5), 1203–1213. doi:10.1002/jbmr.1822
Jacquin, C., Gran, D. E., Lee, S. K., Lorenzo, J. A., and Aguila, H. L. (2006). Identification of Multiple Osteoclast Precursor Populations in Murine Bone Marrow. J. Bone Miner Res. 21 (1), 67–77. doi:10.1359/JBMR.051007
Kemink, S. A. G., Hermus, A. R. M. M., Swinkels, L. M. J. W., Lutterman, J. A., and Smals, A. G. H. (2000). Osteopenia in Insulin-dependent Diabetes Mellitus; Prevalence and Aspects of Pathophysiology. J. Endocrinol. Invest. 23 (5), 295–303. doi:10.1007/bf03343726
Kim, H. S., and Lee, N. K. (2014). Gene Expression Profiling in Osteoclast Precursors by Insulin Using Microarray Analysis. Mol. Cell 37 (11), 827–832. doi:10.14348/molcells.2014.0223
Kim, Y.-R., Kim, K.-U., Lee, J.-H., Kim, D.-W., Chung, J.-H., Kim, Y.-D., et al. (2021). Cancer Testis Antigen, NOL4, Is an Immunogenic Antigen Specifically Expressed in Small-Cell Lung Cancer. Curr. Oncol. 28 (3), 1927–1937. doi:10.3390/curroncol28030179
Kirstein, B., Chambers, T. J., and Fuller, K. (2006). Secretion of Tartrate-Resistant Acid Phosphatase by Osteoclasts Correlates with Resorptive Behavior. J. Cel. Biochem. 98 (5), 1085–1094. doi:10.1002/jcb.20835
Laha, D., Deb, M., and Das, H. (2019). KLF2 (Kruppel-like Factor 2 [lung]) Regulates Osteoclastogenesis by Modulating Autophagy. Autophagy 15 (12), 2063–2075. doi:10.1080/15548627.2019.1596491
Liu, W., Li, Z., Cai, Z., Xie, Z., Li, J., Li, M., et al. (2020). LncRNA‐mRNA Expression Profiles and Functional Networks in Osteoclast Differentiation. J. Cel Mol. Med. 24 (17), 9786–9797. doi:10.1111/jcmm.15560
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Luo, J., Yang, Z., Ma, Y., Yue, Z., Lin, H., Qu, G., et al. (2016). LGR4 Is a Receptor for RANKL and Negatively Regulates Osteoclast Differentiation and Bone Resorption. Nat. Med. 22 (5), 539–546. doi:10.1038/nm.4076
Madel, M. B., Ibáñez, L., Ciucci, T., Halper, J., Rouleau, M., Boutin, A., et al. (2020). Dissecting the Phenotypic and Functional Heterogeneity of Mouse Inflammatory Osteoclasts by the Expression of Cx3cr1. Elife 9, e54493. doi:10.7554/eLife.54493
Malone, B. M., Tan, F., Bridges, S. M., and Peng, Z. (2011). Comparison of Four ChIP-Seq Analytical Algorithms Using rice Endosperm H3K27 Trimethylation Profiling Data. PLoS One 6 (9), e25260. doi:10.1371/journal.pone.0025260
Murata, K., Fang, C., Terao, C., Giannopoulou, E. G., Lee, Y. J., Lee, M. J., et al. (2017). Hypoxia-Sensitive COMMD1 Integrates Signaling and Cellular Metabolism in Human Macrophages and Suppresses Osteoclastogenesis. Immunity 47 (1), 66–79. doi:10.1016/j.immuni.2017.06.018
Nishikawa, K., Iwamoto, Y., Kobayashi, Y., Katsuoka, F., Kawaguchi, S.-i., Tsujita, T., et al. (2015). DNA Methyltransferase 3a Regulates Osteoclast Differentiation by Coupling to an S-Adenosylmethionine-Producing Metabolic Pathway. Nat. Med. 21 (3), 281–287. doi:10.1038/nm.3774
Nomiyama, H., Egami, K., Wada, N., Tou, K., Horiuchi, M., Matsusaki, H., et al. (2005). Short Communication: Identification of Genes Differentially Expressed in Osteoclast-like Cells. J. Interferon Cytokine Res. 25 (4), 227–231. doi:10.1089/jir.2005.25.227
Novack, D. V., and Teitelbaum, S. L. (2008). The Osteoclast: Friend or Foe? Annu. Rev. Pathol. Mech. Dis. 3, 457–484. doi:10.1146/annurev.pathmechdis.3.121806.151431
Purdue, P. E., Crotti, T. N., Shen, Z., Swantek, J., Li, J., Hill, J., et al. (2014). Comprehensive Profiling Analysis of Actively Resorbing Osteoclasts Identifies Critical Signaling Pathways Regulated by Bone Substrate. Sci. Rep. 4, 7595. doi:10.1038/srep07595
Reni, C., Mangialardi, G., Meloni, M., and Madeddu, P. (2016). Diabetes Stimulates Osteoclastogenesis by Acidosis-Induced Activation of Transient Receptor Potential Cation Channels. Sci. Rep. 6, 30639. doi:10.1038/srep30639
Roodman, G. D. (1999). Cell Biology of the Osteoclast. Exp. Hematol. 27 (8), 1229–1241. doi:10.1016/s0301-472x(99)00061-2
Shaker, J. L. (2009). Paget's Disease of Bone: A Review of Epidemiology, Pathophysiology and Management. Ther. Adv. Musculoskelet. 1 (2), 107–125. doi:10.1177/1759720x09351779
Sozen, T., Ozisik, L., and Calik Basaran, N. (2017). An Overview and Management of Osteoporosis. Eur. J. Rheumatol. 4 (1), 46–56. doi:10.5152/eurjrheum.2016.048
Teitelbaum, S. L. (2007). Osteoclasts: what Do They Do and How Do They Do it? Am. J. Pathol. 170 (2), 427–435. doi:10.2353/ajpath.2007.060834
Teitelbaum, S. L., and Ross, F. P. (2003). Genetic Regulation of Osteoclast Development and Function. Nat. Rev. Genet. 4 (8), 638–649. doi:10.1038/nrg1122
Waugh, K. A., Leach, S. M., Moore, B. L., Bruno, T. C., Buhrman, J. D., and Slansky, J. E. (2016). Molecular Profile of Tumor-specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model. J.I. 197 (4), 1477–1488. doi:10.4049/jimmunol.1600589
Wu, Y., Humphrey, M. B., and Nakamura, M. C. (2008). Osteoclasts-the Innate Immune Cells of the Bone. Autoimmunity 41 (3), 183–194. doi:10.1080/08916930701693180
Yang, G., Tang, K., Qiao, L., Li, Y., and Sun, S. (2021). Identification of Critical Genes and lncRNAs in Osteolysis after Total Hip Arthroplasty and Osteoarthritis by RNA Sequencing. Biomed. Res. Int. 2021, 6681925. doi:10.1155/2021/6681925
Yasuda, H., Shima, N., Nakagawa, N., Yamaguchi, K., Kinosaki, M., Mochizuki, S.-i., et al. (1998). Osteoclast Differentiation Factor Is a Ligand for Osteoprotegerin/osteoclastogenesis-Inhibitory Factor and Is Identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. 95 (7), 3597–3602. doi:10.1073/pnas.95.7.3597
Keywords: osteoclast, osteopororosis, bone resorption, RANK ligand (RANKL), differentiation
Citation: Toor SM, Wani S and Albagha OME (2021) Comprehensive Transcriptomic Profiling of Murine Osteoclast Differentiation Reveals Novel Differentially Expressed Genes and LncRNAs. Front. Genet. 12:781272. doi: 10.3389/fgene.2021.781272
Received: 22 September 2021; Accepted: 22 October 2021;
Published: 15 November 2021.
Edited by:
Long Guo, RIKEN Center for Integrative Medical Sciences, JapanReviewed by:
Vincent Everts, VU University Amsterdam, NetherlandsCopyright © 2021 Toor, Wani and Albagha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Omar M. E. Albagha, b2FsYmFnaGFAaGJrdS5lZHUucWE=
†These authors contributed equally to this study
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.