
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 10 January 2022
Sec. Statistical Genetics and Methodology
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.779455
This article is part of the Research Topic Data Mining and Statistical Methods for Knowledge Discovery in Diseases Based on Multimodal Omics View all 15 articles
Aim: This study aimed to accurately identification of potential miRNAs for gastric cancer (GC) diagnosis at the early stages of the disease.
Methods: We used GSE106817 data with 2,566 miRNAs to train the machine learning models. We used the Boruta machine learning variable selection approach to identify the strong miRNAs associated with GC in the training sample. We then validated the prediction models in the independent sample GSE113486 data. Finally, an ontological analysis was done on identified miRNAs to eliciting the relevant relationships.
Results: Of those 2,874 patients in the training the model, there were 115 (4%) patients with GC. Boruta identified 30 miRNAs as potential biomarkers for GC diagnosis and hsa-miR-1343-3p was at the highest ranking. All of the machine learning algorithms showed that using hsa-miR-1343-3p as a biomarker, GC can be predicted with very high precision (AUC; 100%, sensitivity; 100%, specificity; 100% ROC; 100%, Kappa; 100) using with the cut-off point of 8.2 for hsa-miR-1343-3p. Also, ontological analysis of 30 identified miRNAs approved their strong relationship with cancer associated genes and molecular events.
Conclusion: The hsa-miR-1343-3p could be introduced as a valuable target for studies on the GC diagnosis using reliable biomarkers.
Gastric cancer (GC) is a significant global health issue due to being the fifth leading cancer worldwide as well as the third cancer-related death leading cause, which leads to nearly 8,00,000 deaths annually (Bray, 2018). Morbidity and mortality due to GC have reduced in recent years, though the rate of 5-year survival is still fairly low (Howlader, 2014). A significant prognostic factor is the stage of cancer at the diagnosis time. The 5-year survival of GC patients is below 30% if the disease is diagnosed at the advanced stages (Hundahl et al., 2000), while the 5-year survival of patients ranges between 70 and 90% if diagnosed at the early stages (Choi, 2015). Thus, GC will remain among the toughest challenges for physicians and researchers for so long since GC is not symptomatic until the advanced stages; this is why effective screening approaches for the early detection of GC are mandatory to overcome GC mortalities (Penon et al., 2014). Presently, gastroscopy is yet the standard test to diagnose GC (Veitch et al., 2015). Nonetheless, this screening approach is invasive and costly. Furthermore, minimally invasive or non-invasive markers, including carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) have been commonly used clinically, though these markers are neither specific nor sensitive enough for GC early diagnosis (Carpelan-Holmström et al., 2002). Due to non-specific symptoms and the absence of an early diagnosis, a great number of patients with GC are diagnosed at the advanced stages (Hundahl et al., 2000; Hartgrink et al., 2009). Thus, cost-effective and non-invasive biomarkers are immediately required for the early diagnosis of GC.
Recent genome analysis revealed several biomarkers which are related to RNA, DNA, exosome, et cetera. A class of endogenous non-coding RNAs is MicroRNAs (miRNAs) (nearly 22 nt) which module the expression of the gene after transcription through degradation or translation blockage of target mRNAs (Bartel, 2004; Caldas and Brenton, 2005). It is well-known that cancer cells may release miRNAs via exosomes to enhance proliferation and migration (Li, 2018; Yoshimura, 2018; Zeng, 2018). The exosomal miRNAs released into biofluids, including serum, plasma, tear, urine, and gastric juice, may escape being degraded by RNases (Gilad, 2008). Moreover, miRNAs have been suggested as potential biomarkers which may be used to diagnose several types of cancers, including testicular germ cell tumors (using miRNA-371a-3p: specificity 94.0% and sensitivity 90.1%) (Dieckmann, 2019), bladder cancer (using 7-miRNA panel: specificity 87% and sensitivity 95%) (Usuba, 2019a), and hepatocellular carcinoma (using miR-424: specificity 87.13% and sensitivity 95.12%) (Lin, 2015), and lung cancer (Aftabi, 2021). Moreover, several studies reported that numerous miRNAs might be potentially used as biomarkers for GC diagnosis (Zhou, 2010; Cui, 2013; Su, 2014). Nonetheless, most of the miRNA biomarkers are not developed using comprehensive data mining according to miRNA profiling, and even they lack proper external efficacy validation (Link and Kupcinskas, 2018; Wei, 2019). Instead, recently, Artificial intelligence Technology (AT) usage in the field of microarray Data has attracted more attention. The disadvantage of the conventional statistical models, including logistic regression, was that they excluded the possible interaction terms and highly correlated variables; thus, they might lose a part of useful information, which might decrease their accuracy, specifically in the case of high dimensional miRNA data analysis (Alpaydin, 2020). Furthermore, the traditional models are not able to capture variables’ non-linear associations (James et al., 2013; Gilani et al., 2017; Gilani et al., 2019). Instead, Machine Learning (ML) is able to deal with non-linear structures as well as detecting all the possible interactions which may exist between predictors (Gilani et al., 2018; Wiemken and Kelley, 2020).
Machine learning has several algorithms of which the decision trees (DT), random forests (RF), extreme gradient boosted trees (XGBT), and artificial neural networks (ANN) that have been frequently applied in medicine (Cleophas and Zwinderman, 2015; Deo, 2015), particularly in prediction of cancer (DeGregory, 2018; Fakhari et al., 2019). Random forest is a tree-based classification algorithm, and as the name indicates, the algorithm creates a forest with a huge number of trees. It is an ensemble algorithm that combines multiple algorithms. The random forest creates a set of decision trees from a random sample of the training set. It repeats the process with multiple random samples and makes a final decision based on majority voting (Zhou, 2012). Briefly, gradient boosted trees combine multiple classification trees into an additively weighted classifier. Boosting refers to the method where sequentially ascertained trees were trained, meaning each observation was weighted by its error obtained by minimizing the appropriate loss of function in the previous iteration. In this way, boosting is a gradient descent algorithm (Christensen and Bastien, 2016) and forces the classifier to focus on aspects of the data that are difficult to learn (Hastie et al., 2009).
Artificial neural networks have been broadly used in medical studies (Darsey et al., 2015; DeGregory, 2018; Shahid et al., 2019). Such models perform satisfactorily, especially for classification problems with complex and non-linear associations between variables (Hastie et al., 2009). Briefly, artificial neural networks are based on a collection of artificial neurons, which receive and process inputs (predictors), transmit them to other artificial neurons, and produce an output (Zhou, 2012).
Considering the important role of GC early diagnosis in patient’s survival rate and the lack of published article on identifying potential miRNAs for GC prediction at an early stage by AT, the present study aims to identify the potential miRNA for predicting GC by AT in the datasets of Gene Expression Omnibus (GEO) specifically with the stat of the art machine learning models. Traditional statistical models such as linear models previously has been used in looking for GC biomarkers and identified miRNAs with the potential prediction power (Yao, 2020), however, they have not implemented advanced methods such as machine learning and new variable selection approaches such as Synthetic Minority Oversampling Technique (SMOTE). In the present study, for the first time, we aimed to use those new techniques for identification of GC related miRNAs with a reliable cut-of and highest possible accurecy in the external validation.
For training sample, we used GSE106817 dataset that is available at https://www.ncbi.nlm.nih.gov/geo/. The dataset consist of the data of 2,566 miRNAs obtained from 2,759 non-cancer controls, and 115 GC cases (4%). In the original study the serum samples of cancer cases and non-cancer controls have been analyzed by microarray for miRNA expression profiles (Yokoi, 2018). For test sample we used GSE113486 dataset, which includes data of miRNA expression profiles from the serum samples of 40 GC cases (28.6%) and 100 normal controls (71.4%) (Usuba, 2019b). All the datasets were serum miRNA profiles based on the same microarray platform, 3D-Gene Huma miRNA V21_1.0.0 (39). The study was approved by the NCCH Institutional Review Board (2015-376, 2016-29) and the Research Ethics Committee of Medical Corporation Shintokai Yokohama Minoru Clinic (6019-18-3772). Written informed consent was obtained from each participant (42). This study was approved by the Ethics Committee of Tabriz University of Medical Sciences (No: IR. TBZMED.REC.1400.006).
We used the Boruta machine learning algorithm to select the most critical miRNAs related to GC in the training sample that produce the highest prediction accuracy. In short, Boruta selects the variables that have a high impact on the prediction accuracy by providing the “variable importance” (Kursa and Rudnicki, 2018). We used SMOTE random oversampling to balance the outcome in the GSE106817 data. We then used five-fold cross-validation to find the optimal hyper parameters on DT, RF, LR, XGBT, and ANN to choose the best approaches in the balanced sample using the most important variables selected by Boruta. Once the prediction models were developed, we applied them on the test sample GSE113486 to verify the accuracy of developed prediction approach. We looked for an algorithm that may generate a higher predictive power among the 5 ML algorithms in terms of the yielded areas under the ROC curves (AUCs). Sensitivity, specificity, positive predictive value, negative predictive value, misclassification rate, and Kappa were assessed. The guidelines of developing transparent multivariable prediction models was followed for these analysis (Moons, 2015).
GeneCodis is a web-based tool for the ontological analysis of lists of genes, proteins, and regulatory elements like miRNAs, transcription factors, and CpGs. It can be used to determine biological annotations or combinations of annotations that are significantly associated to a list of genes under study with respect to a reference list. As well as single annotations, this tool allows users to simultaneously evaluate annotations from different sources, for example GO Biological Process and KEGG. To this end, and before computing p-values, it uses the apriori algorithm to extract sets of annotations that frequently co-occur in the analyzed list of genes (Garcia-Moreno, 2021). We used GeneCodis 4 (https://genecodis.genyo.es/) for ontological analysis of the identified miRNAs list.
Of those 2,874 patients included in this study, there were 115 (4%) patients with gastric cancer. This analysis consists of 2,566 miRNAs.
Of those 2,566 miRNA in GSE106817 data, the Boruta algorithm initially selected 108 miRNA using Gini Index measurement (results are not shown here). The processing time was 17.24 minuts There were 77 tentative variables at the first stage. After fixing the tentative features, Boruta identified 156 miRNA for the analysis (results are not shown here). The process took 99 iterations convergence. It was observed that hsa-miR-1343-3p had the highest importance for prediction accurecy (minimum importance; 6.47, median importance; 11.44, mean importance; 10.81; maximum importance; 13.63) among all identified miRNAs. The hsa-miR-1290 and hsa-miR-5100 had the second and third highest importance, with mean importance of 8.69 and 8.66, respectively (Table 1).
The balanced training data using SMOTE random oversampling technique had 1,376 cancer cases and 1,498 non-cancer controls. We trained DT, RF, LR, XGBT, and ANN perdition models with the selected miRNAs in the balanced training data.
The external validation data GSE113486 had 40 (28.6%) gastric cancer and 100 (71.4%) non-cancer (controls). hsa-miR-1343-3 produced the highest prediction accuracy for GC prediction (Table 1). For the hsa-miR-1343-3, all of the accuracy measures including AUC, sensitivity and specificity, positive predictive value, negative predictive value, Kappa were 100%. According to the decision trees, the cut-off point for this miRNA was 8.2 (Figure 1). Further, hsa-miR-8073 and hsa-miR-1228-5p produced 100% AUC but other accuracy measures were not 100%. On the other hsa-miR-1185-1-3p had the lowest AUC which has the least contribution to the prediction of GC.
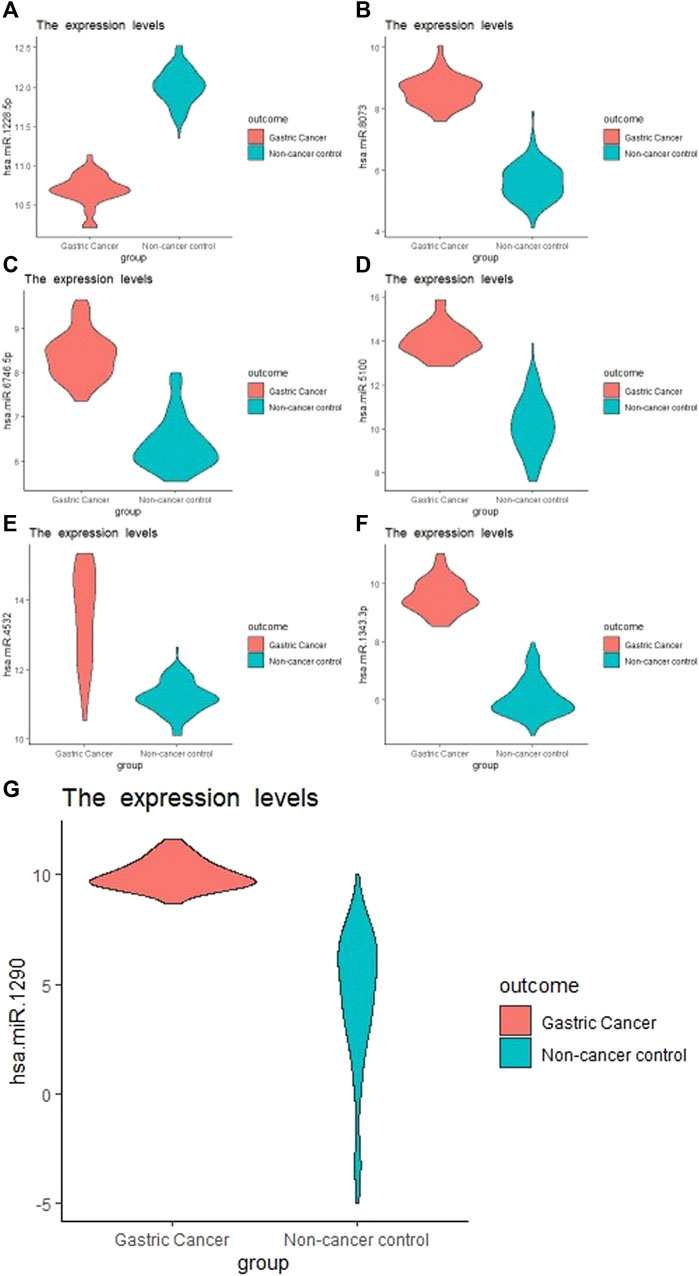
FIGURE 1. Boxplot of the selected miRNA from Boruta Algorithm. (A), hsa-miR-1228-5p; (B), hsa-miR-8073; (C), hsa-miR-6746-5p; (D), hsa-miR-5100; (E), hsa-miR-4532; (F): hsa-miR-1343-3p; (G), hsa-miR-1290.
Among several models discussed in the study, the XGBT algorithm had better prediction accuracy overall (Table S1-S4). However, for hsa-miR-1343-3 all models had consistently 100% accuracy which indicates that this miRNA may strongly predict GC. For some miRNA such as hsa-miR-422a XGBT algorithm could predict GC with higher accuracy than the logistic regression and decision trees. Figure 2 shows the correlation of the important miRNAs. It can be observed that most of the identified miRNAs except hsa-miR-422a, hsa-miR-1185-1-3p, hsa-miR-1185-2-3p, hsa-miR-6087, and hsa-miR-1199-5p are highly correlated. Consequently, clustering of correlated those miRNAs is helpful for the identification of cancerous and non-cancerous patients. Finally, Heatmap plot indicates the result of the hierarchical clustering analysis of the 30 selected miRNAs, which represents that identified miRNAs can easily distinguish GC cases and controls in test sample obtained from GSE113486 dataset (Figure 3).
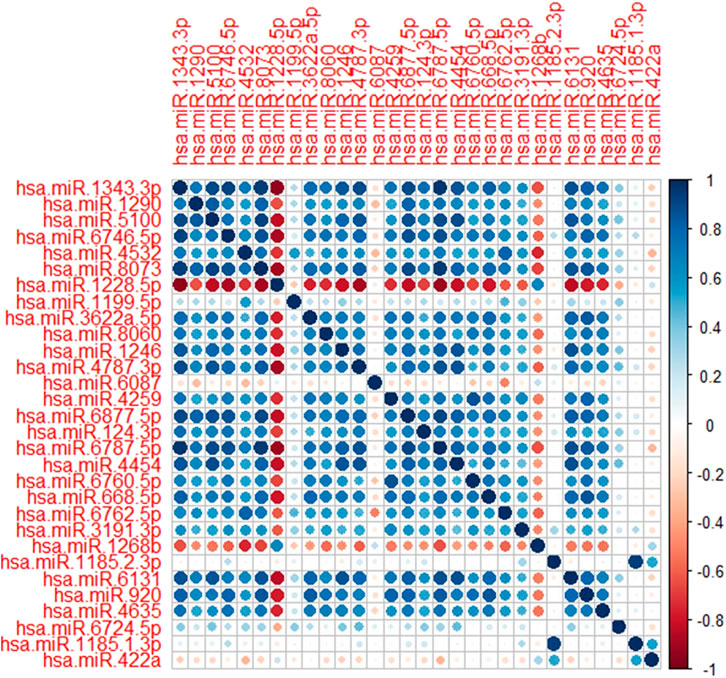
FIGURE 2. Correlation plot of the selected miRNAs. Dark blue and dark red shows the strength of the correlations between miRNAs.
Regulatory, functional, and perturbation analysis by GeneCodis 4 showed that 30 identified miRNAs (Table 1) are related strongly to the cancer-associated genes and molecular events (Figure 4). Visualizations generated for 10 top terms of associations with Transcription Factors (Figure 4A), co-annotation of HMDD v3 (the Human microRNA disease database), MNDR (Mammalian ncRNA-Disease Repository), and TAM2 (The tool for annotations of human miRNAs) databases (Figure 4B), GO (Gene Ontology and GO Annotations) Biological Process (Figure 4C), GO Molecular Function (Figure 4D), co-annotation of KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathways, Panther (Protein ANalysis THrough Evolutionary Relationships) Pathways, and WikiPathways (Figure 4E), and co-annotation of HPO (The Human Phenotype Ontology) and OMIM (Online Mendelian Inheritance in Man) databases (Figure 4F).
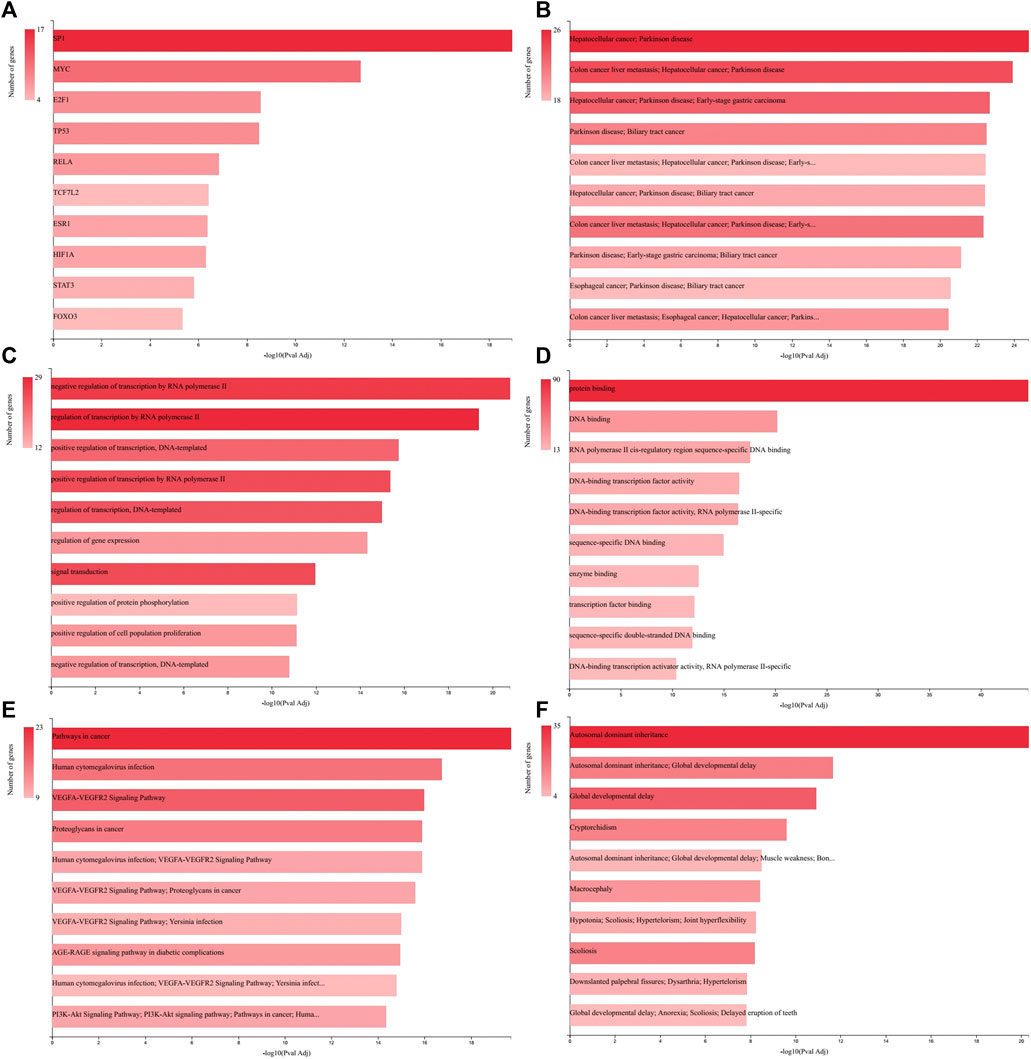
FIGURE 4. GeneCodis Ontological analysis. Visualizations generated for 10 top terms of related categories with our identified miRNAs list are presented here for Transcription Factors (A), Co-annotation of miRNAs-based analysis using HMDD v3, MNDR, and TAM2 (B), GO Biological Process (C), GO Molecular Function (D), Co-annotation of KEGG Pathways, Panther Pathways, and WikiPathways databases (E), and Co-annotation of HPO and OMIM databases (F).
Using artificial intelligence technology, we identified hsa-miR-1343-3 as a very strong nominate for biomarker analysis of GC diagnosis. The value of hsa-miR-1343-3 higher than 8.2 indicates that it could be a strong predictor for GC (100% of AUC, 100% of Sensitivity and Sepecificity). We also found three other miRNAs (hsa-miR-8073 and hsa-miR-1228-5p) with a great contribution to the GC prediction. A medical expert can use these findings for the early detection of GC instead of using costly and time-consuming tools such as colonoscopy Yao et al. (Yao, 2020).
This study had several strengths compared to the previous studies. Compared to Shi et al. that identified the miR-1246 as the potential biomarker of GC that generated the AUC of 83%, our study identified the hsa-miR-1343-3p using the Boruta algorithm that led to a significant increase in the AUC (Shi and Zhang, 2019). The study of Yao et al., selected three miRNAs that produced similar precision to our study that using only single miRNA that may have economical merits. Further, their study used a limited sample size (70 gastric cancer patients and 374 non-cancer controls) in the training set that may lead to an inferior model. The current study used very advanced variable selection methods and the state of the art machine learning approaches that produced consistent results. Another merit of the study is introducing a simple cut-off point of 8.2 using decision trees that may has very practical value in GC classification.
Figure 4A depicted that among the transcription factors related to the genes associated with the identified miRNAs list (Table 1) the SP1, MYC, and E2F1 have higher priorities. SP1 protein expression is up regulated in GC tissues compared with normal tissues and is positively associated with depth of invasion and TNM stage of GC (Shi and Zhang, 2019). MYC is an oncogene responsible for excessive cell growth in cancer, enabling transcriptional activation of genes involved in cell cycle regulation, metabolism, and apoptosis, and is usually overexpressed in GC (Maués, 2018). E2F1 is a member of the E2F family that functions in cell cycle progression and apoptosis induction in response to DNA damage. Deregulated E2F1 acts as a driving force in GC progression and promotes tumor invasion and metastasis independently from its other cellular activities (Yan, 2014).
As depicted in Figure 4B gastrointestinal cancers including hepatocellular cancer, colon cancer, biliary tract cancer, and especially early-stage GC are among the most related diseases to the analyzed miRNAs list. From biological process and molecular function perspectives as showed in Figures 4C,D, the regulation of transcription and gene expression, and protein and DNA binding are the most targeted aspects, which are the general aspects of molecular biology of GC (Cervantes et al., 2007; Vauhkonen et al., 2006; Tan and Yeoh, 2015). Co-annotation of three pathway databases (Figure 4E) has shown that the miRNAs list is general in relation with pathways in cancer, VEGFA-VEGFR2 signaling pathway and PI3K-Akt signaling pathway. The increased expression of VEGFA in the tubular glands and VEGFR2 in the endothelium of GC samples mainly in the T2, T3 and T4 stages of tumor progression has been reported previously (Tamma, 2018). Also, it is showed that the PI3K/AKT/mTOR pathway is activated in GC with overexpression in tumor tissue, which is correlated with the depth of tumor infiltration and the presence of lymph node metastases (Tapia, 2014). Surprisingly, relation with Human cytomegalovirus infection, which was identified in our pathway analysis, has been reported to be associated with the development of GC (Jin, 2014) and GC lymphatic metastasis (Zhang, 2017).
The analysis of human phenotype and Mendelian inheritance ontologies identified Autosomal dominant inheritance and Global developmental delay among the most related phenomena with our miRNAs list. It is reported that gastric adenocarcinoma and proximal polyposis of the stomach is an autosomal dominant syndrome (Worthley, 2012). Also, some common variants have been described for GC and developmental delay (Hansford, 2015; Zhang, 2020).
In our study, we have shown theoreticaly that ther is a strong relationship between hsa-miR-1343-3p and GC. Hsa-miR-1343-3p has been indicated as a tumor suppressor for many types of cancer. It has been suggested that miR-1343-3p, which regulates the oncogenic effect of TEA domain transcription factors is associated with GC (Zhou, 2017). The correlation between hsa-miR-1343-3p and lung adenocarcinoma was evaluated and its expression was found to be low in patients with vascular invasion (Kim, 2017). Yuan et al. demonstrated that hsa-miR-1343-3p is consistently down-regulated in colon, prostate, and pancreatic cancers. Also, hsa-miR-1343-3p has been proposed as a biomarker to distinguish pancreato-biliary malignancy from non-malignant diseases. The major genes targeted by miR-1343-3p have been identified (Yuan, 2016). In this context, these target genes and their interaction with GC should also be investigated. The hsa-miR-1343-3p targets including SHISA7, TGFBR1, DLGAP3, SPRED1, ATXN7L3, and PLXDC2 genes are listed at MIRDB (http://mirdb.org/). Among them transforming growth factor beta-1 (TGFβ1) play an important role in carcinogenesis upon binding its receptor (TGFBR1). It acts as a tumor suppressor by inhibiting cellular proliferation or by promoting cellular differentiation and apoptosis. However, it turns to be a tumor promoter by stimulating angiogenesis and cell motility, suppressing the immune response, and increasing progressive invasion and metastasis (Yuan, 2016). Other reports have also revealed that hsa-miR-1343-3p reduces the expression of transforming growth factor-β (TGF-β) receptor-1, which induces angiogenesis through vascular endothelial growth factor (VEGF)-mediated apoptosis. Therefore, hsa-miR-1343-3p may also play an anti-angiogenic role (Ferrari et al., 2009; Stolzenburg et al., 2016; Kim, 2017). He et al. determined that TGFBR1 genes’ two polymorphisms (rs334348, rs10512263) were associated with the risk of GC (He, 2018). In another study, Zhang et al. have shown that silencing of TGFBR1 inhibited cell proliferation, migration, invasion, and EMT in GC cells (Zhang, 2019).
Discs large associated proteins (DLGAPs) family has been implicated in psychological and neurological diseases. However, few studies have explored the association between the expression of DLGAPs and different types of cancer. Liu et al. has suggested that the significant overexpression of DLGAP4 in GC may be a promising potential prognostic marker for GC (Liu et al., 2018). Aslo, Liu et al. have determined decreased expression of SPRED1 in GC tissues (Liu et al., 2020).
However, there were certain limitations in our study. We had relatively small sample size in GC group. Other limitations were the pathological information such as the tumor stage, age or other factors which were not available in our datasets. Nonetheless, the prediction accuracy of our model has high enough (100% AUC) for clinical use. Further, we were unable to do the survival analysis to further validate the markers identified in this paper based on public available data (Howlader, 2014).
Using several state of the art machine learning methods and Boruta algorithm, we identified several miRNAs that can predict GC. Specifically, hsa-miR-1343-3p, which identified by cut-off point of 8.2 may be nominated as a highly reliable biomarker for, GC diagnosis after meticulous empirical tests.
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/.
NG and RB conducted this study and performed statistical analysis and machine learning models. YA and TE provided ontological analysis. The paper was reviewed by EF and MS before final submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the Associate Editor and three anonymous expert reviewers who significantly helped to improve the presentation of the paper.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.779455/full#supplementary-material
Aftabi, Y. (2021). Long Non‐coding RNAs as Potential Biomarkers in the Prognosis and Diagnosis of Lung Cancer: A Review and Target Analysis. 73, 307–327. doi:10.1002/iub.2430
Bartel, D. P. (2004). MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. cell 116, 281–297. doi:10.1016/s0092-8674(04)00045-5
Bray, F. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a Cancer J. clinicians 68, 394–424. doi:10.3322/caac.21492
Caldas, C., and Brenton, J. D. (2005). Sizing up miRNAs as Cancer Genes. Nat. Med. 11, 712–714. doi:10.1038/nm0705-712
Carpelan-Holmström, M., Louhimo, J., Stenman, U.-H., Alfthan, H., and Haglund, C. C. E. A. (2002). CA 19-9 and CA 72-4 Improve the Diagnostic Accuracy in Gastrointestinal Cancers. Anticancer Res. 22, 2311–2316.
Cervantes, A., Braun, E. R., Fidalgo, A. P., González, I. C. J. C., and Oncology, T. (2007). Mol. Biol. gastric Cancer 9, 208–215. doi:10.1007/s12094-007-0041-4
Choi, I. J. (2015). Long-term Outcome Comparison of Endoscopic Resection and Surgery in Early Gastric Cancer Meeting the Absolute Indication for Endoscopic Resection. Gastrointest. Endosc. 81, 333–341. doi:10.1016/j.gie.2014.07.047
Christensen, J., and Bastien, C. (2016). Introduction to General Optimization Principles and Methods. Nonlinear Optimization Vehicle Saf. Structures, 107–168. doi:10.1016/b978-0-12-417297-5.00003-1
Cleophas, T. J., and Zwinderman, A. H. (2015). Machine Learning in Medicine-A Complete Overview. Springer.
Cui, L. (2013). Gastric Juice MicroRNAs as Potential Biomarkers for the Screening of Gastric Cancer. Cancer 119, 1618–1626. doi:10.1002/cncr.27903
Darsey, J. A., Griffin, W. O., Joginipelli, S., and Melapu, V. K. (2015). Artificial Neural Networks 269-283. Springer.
DeGregory, K. (2018). A Review of Machine Learning in Obesity. Obes. Rev. 19, 668–685. doi:10.1111/obr.12667
Deo, R. C. (2015). Machine Learning in Medicine. Circulation 132, 1920–1930. doi:10.1161/circulationaha.115.001593
Dieckmann, K.-P. (2019). Serum Levels of microRNA-371a-3p (M371 Test) as a New Biomarker of Testicular Germ Cell Tumors: Results of a Prospective Multicentric Study. J. Clin. Oncol. 37, 1412. doi:10.1200/jco.18.01480
Fakhari, A., Gharepapagh, E., Dabiri, S., and Gilani, N. (2019). Correlation of Cancer Antigen 15-3 (CA15-3) Serum Level and Bony Metastases in Breast Cancer Patients. Med. J. Islamic Republic Iran 33, 142. doi:10.47176/mjiri.33.142
Ferrari, G., Cook, B. D., Terushkin, V., Pintucci, G., and Mignatti, P. (2009). Transforming Growth Factor‐beta 1 (TGF‐β1) Induces Angiogenesis through Vascular Endothelial Growth Factor (VEGF)‐mediated Apoptosis. J. Cell. Physiol. 219, 449–458. doi:10.1002/jcp.21706
Garcia-Moreno, A. (2021). GeneCodis 4: Expanding the Modular Enrichment Analysis to Regulatory Elements.
Gilad, S. (2008). Serum microRNAs Are Promising Novel Biomarkers. PloS one 3, e3148. doi:10.1371/journal.pone.0003148
Gilani, N., Esmaeili, A., and Haghshenas, R. (2018). The Effect of Eight Weeks Concurrent Training and Supplementation of L_Arginine on Plasma Level of 8-hydroxydeoxyguanosine (8-OHdG), Malondialdehyde and Total Antioxidant Capacity in Elderly Men (Multivariate Longitudinal Modeling).
Gilani, N., Haghshenas, R., and Esmaeili, M. (2019). Application of Multivariate Longitudinal Models in SIRT6, FBS, and BMI Analysis of the Elderly. The Aging Male 22, 260–265. doi:10.1080/13685538.2018.1477933
Gilani, N., Kazemnejad, A., Zayeri, F., Asghari, J. M., and Izadi, A. F. S. (2017). Predicting Outcomes in Traumatic Brain Injury Using the glasgow Coma Scale: A Joint Modeling of Longitudinal Measurements and Time to Event. Iran Red Crescent Med J 19 (2), e29663.
Hansford, S. (2015). Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and beyond. 1, 23–32.
Hartgrink, H. H., Jansen, E. P., van Grieken, N. C., and van de Velde, C. J. (2009). Gastric Cancer. Lancet (London, England) 374, 477–490. doi:10.1016/s0140-6736(09)60617-6
Hastie, T., Tibshirani, R., and Friedman, J. (2009). The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Switzerland AG: Springer Science & Business Media.
He, B. (2018). Polymorphisms of TGFBR1, TLR4 Are Associated with Prognosis of Gastric Cancer in a Chinese Population. Cancer Cel. Int. 18, 1–10. doi:10.1186/s12935-018-0682-0
Howlader, N. (2014). SEER Cancer Statistics Review, 1975–2011, 19. Bethesda, MD: National Cancer Institute.
Hundahl, S. A., Phillips, J. L., and Menck, H. R. (2000). The National Cancer Data Base Report on Poor Survival of US Gastric Carcinoma Patients Treated with Gastrectomy: American Joint Committee on Cancer Staging, Proximal Disease, and the “Different Disease” Hypothesis. Cancer 88, 921–932. doi:10.1002/(sici)1097-0142(20000215)88:4<921:aid-cncr24>3.0.co;2-s
James, G., Witten, D., Hastie, T., and Tibshirani, R. (2013). An Introduction to Statistical Learning, Vol. 112. Springer.
Jin, J. (2014). Latent Infection of Human Cytomegalovirus Is Associated with the Development of Gastric Cancer, 8, 898–904. doi:10.3892/ol.2014.2148
Kim, H. (2017). MicroRNA Expression Profiles and Clinicopathological Implications in Lung Adenocarcinoma According to EGFR, KRAS, and ALK Status. Oncotarget 8, 8484. doi:10.18632/oncotarget.14298
Kursa, M. B., and Rudnicki, W. R. (2018). Wrapper Algorithm for All Relevant Feature Selection. Boruta: CRAN-Package.
Li, Z. (2018). Tumor-secreted Exosomal miR-222 Promotes Tumor Progression via Regulating P27 Expression and Re-localization in Pancreatic Cancer. Cell Physiol. Biochem. 51, 610–629. doi:10.1159/000495281
Lin, X.-J. (2015). A Serum microRNA Classifier for Early Detection of Hepatocellular Carcinoma: a Multicentre, Retrospective, Longitudinal Biomarker Identification Study with a Nested Case-Control Study. Lancet Oncol. 16, 804–815. doi:10.1016/s1470-2045(15)00048-0
Link, A., and Kupcinskas, J. (2018). MicroRNAs as Non-invasive Diagnostic Biomarkers for Gastric Cancer: Current Insights and Future Perspectives. World J. Gastroenterol. 24, 3313. doi:10.3748/wjg.v24.i30.3313
Liu, J., Liu, Z., Zhang, X., Gong, T., and Yao, D. (2018). Examination of the Expression and Prognostic Significance of DLGAPs in Gastric Cancer Using the TCGA Database and Bioinformatic Analysis. Mol. Med. Rep. 18, 5621–5629. doi:10.3892/mmr.2018.9574
Liu, W., Fang, S., and Zuo, G. (2020). A Study on the Expression of SPRED1 and PBRM1 (Baf180) and Their Clinical Significances in Patients with Gastric Cancer, 66. Clinical Laboratory. doi:10.7754/clin.lab.2020.200312
Maués, J. H. d. S. (2018). Gastric Cancer Cell Lines Have Different MYC-Regulated Expression Patterns but Share a Common Core of Altered Genes.
Moons, K. G. (2015). Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and Elaboration. Ann. Intern. Med. 162, W1–W73. doi:10.7326/m14-0698
Penon, D., Cito, L., and Giordano, A. (2014). Novel Findings about Management of Gastric Cancer: a Summary from 10th IGCC. World J. Gastroenterol. WJG 20, 8986. doi:10.3748/wjg.v20.i27.8986
Shahid, N., Rappon, T., and Berta, W. (2019). Applications of Artificial Neural Networks in Health Care Organizational Decision-Making: A Scoping Review. PloS one 14, e0212356. doi:10.1371/journal.pone.0212356
Shi, S., and Zhang, Z. G. J. O. l. (2019). Role of Sp1 Expression in Gastric Cancer: A Meta-Analysis and Bioinformatics Analysis. 18, 4126–4135. doi:10.3892/ol.2019.10775
Stolzenburg, L. R., Wachtel, S., Dang, H., and Harris, A. (2016). miR-1343 Attenuates Pathways of Fibrosis by Targeting the TGF-β Receptors. Biochem. J. 473, 245–256. doi:10.1042/bj20150821
Su, Z.-X. (2014). Diagnostic and Prognostic Value of Circulating miR-18a in the Plasma of Patients with Gastric Cancer. Tumor Biol. 35, 12119–12125. doi:10.1007/s13277-014-2516-6
Tamma, R. (2018). VEGFA and VEGFR2 RNAscope Determination in Gastric Cancer, 49, 429–435. doi:10.1007/s10735-018-9777-0
Tan, P., and Yeoh, K.-G. J. G. (2015). Genet. Mol. pathogenesis gastric adenocarcinoma 149, 1153–1162. doi:10.1053/j.gastro.2015.05.059
Tapia, O. (2014). The PI3K/AKT/mTOR Pathway Is Activated in Gastric Cancer with Potential Prognostic and Predictive Significance. 465, 25–33. doi:10.1007/s00428-014-1588-4
Usuba, W. (2019). Circulating miRNA Panels for Specific and Early Detection in Bladder Cancer. Cancer Sci. 110, 408. doi:10.1111/cas.13856
Usuba, W. (2019). Circulating miRNA Panels for Specific and Early Detection in Bladder Cancer. 110, 408–419. doi:10.1111/cas.13856
Vauhkonen, M., Vauhkonen, H., Sipponen, P. J. B. P., and Gastroenterology, R. C. (2006). Pathology and Molecular Biology of Gastric Cancer, 20, 651–674. doi:10.1016/j.bpg.2006.03.016
Veitch, A. M., Uedo, N., Yao, K., and East, J. E. (2015). Optimizing Early Upper Gastrointestinal Cancer Detection at Endoscopy. Nat. Rev. Gastroenterol. Hepatol. 12, 660. doi:10.1038/nrgastro.2015.128
Wei, H. (2019). The Diagnostic Value of Circulating microRNAs as a Biomarker for Gastric Cancer: A Meta-Analysis. Oncol. Rep. 41, 87–102. doi:10.3892/or.2018.6782
Wiemken, T. L., and Kelley, R. R. (2020). Machine Learning in Epidemiology and Health Outcomes Research. Annu. Rev. Public Health 41, 21–36. doi:10.1146/annurev-publhealth-040119-094437
Worthley, D. (2012). Gastric Adenocarcinoma and Proximal Polyposis of the Stomach (GAPPS): a New Autosomal Dominant Syndrome. 61, 774–779. doi:10.1136/gutjnl-2011-300348
Yan, L.-H. (2014). Overexpression of E2F1 in Human Gastric Carcinoma Is Involved in Anti-cancer Drug Resistance. 14, 1–10. doi:10.1186/1471-2407-14-904
Yao, Y. (2020). Identification of Serum Circulating MicroRNAs as Novel Diagnostic Biomarkers of Gastric Cancer. Front. Genet. 11, 515. doi:10.3389/fgene.2020.591515
Yokoi, A. (2018). Integrated Extracellular microRNA Profiling for Ovarian Cancer Screening. Nat. Commun. 9, 1–10. doi:10.1038/s41467-018-06434-4
Yoshimura, A. (2018). Exosomal miR-99a-5p Is Elevated in Sera of Ovarian Cancer Patients and Promotes Cancer Cell Invasion by Increasing Fibronectin and Vitronectin Expression in Neighboring Peritoneal Mesothelial Cells. BMC cancer 18, 1–13. doi:10.1186/s12885-018-4974-5
Yuan, T. (2016). Plasma Extracellular RNA Profiles in Healthy and Cancer Patients. Scientific Rep. 6, 1–11. doi:10.1038/srep19413
Zeng, Z. (2018). Cancer-derived Exosomal miR-25-3p Promotes Pre-metastatic Niche Formation by Inducing Vascular Permeability and Angiogenesis. Nat. Commun. 9, 1–14. doi:10.1038/s41467-018-07810-w
Zhang, C. (2020). YARS as an Oncogenic Protein that Promotes Gastric Cancer Progression through Activating PI3K-Akt Signaling. 146, 329–342. doi:10.1007/s00432-019-03115-7
Zhang, L. (2019). Circular RNA CircCACTIN Promotes Gastric Cancer Progression by Sponging MiR-331-3p and Regulating TGFBR1 Expression. Int. J. Biol. Sci. 15, 1091. doi:10.7150/ijbs.31533
Zhang, L. (2017). Human Cytomegalovirus Detection in Gastric Cancer and its Possible Association with Lymphatic Metastasis. 88, 62–68. doi:10.1016/j.diagmicrobio.2017.02.001
Zhou, H. (2010). Detection of Circulating Tumor Cells in Peripheral Blood from Patients with Gastric Cancer Using microRNA as a Marker. J. Mol. Med. 88, 709–717. doi:10.1007/s00109-010-0617-2
Zhou, Y. (2017). TEAD1/4 Exerts Oncogenic Role and Is Negatively Regulated by miR-4269 in Gastric Tumorigenesis. Oncogene 36, 6518–6530. doi:10.1038/onc.2017.257
Keywords: miRNA, machine learning, boruta algorithm, gastric cancer, hsa-miR-1343-3p, AUC, GSE106817, GSE113486
Citation: Gilani N, Arabi Belaghi R, Aftabi Y, Faramarzi E, Edgünlü T and Somi MH (2022) Identifying Potential miRNA Biomarkers for Gastric Cancer Diagnosis Using Machine Learning Variable Selection Approach. Front. Genet. 12:779455. doi: 10.3389/fgene.2021.779455
Received: 18 September 2021; Accepted: 22 November 2021;
Published: 10 January 2022.
Edited by:
Tao Wang, Northwestern Polytechnical University, ChinaReviewed by:
Xiawei Li, Zhejiang University, ChinaCopyright © 2022 Gilani, Arabi Belaghi, Aftabi, Faramarzi, Edgünlü and Somi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neda Gilani, Z2lsYW5pbkB0YnptZWQuYWMuaXI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.