
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 03 January 2022
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.771810
Background: Emerging research suggests that long non-coding RNAs (lncRNAs) play an important role in a variety of developmental or physiological processes of hepatocellular carcinoma (HCC). Various differentially expressed lncRNAs have been identified in HCC. Thus, a deeper analysis of recent research concerning lncRNA and HCC development could provide scientists with a valuable reference for future studies.
Methods: Related publications were retrieved from the Web of Science Core Collection database. CiteSpace version 5.6.R4 was employed to conduct bibliometric analysis. Several network maps were constructed to evaluate the collaborations between different countries, institutions, authors, journals, and keywords.
Results: A total of 2,667 records were initially found from the year of 2010–2020. The annual related publications output had increased dramatically during these years. Although China was the most prolific country in terms of research publication, the United States played a leading role in collaborative network. The Nanjing Medical University was the most productive institute in the field of lncRNAs in HCC development. Gang Chen was the most prolific researcher, while Yang F was the most frequently co-cited author. Oncotarget, Cell, and Oncogene were the most highly co-cited journals. The most recent burst keywords were interaction, database, and pathway.
Conclusion: This study provides a comprehensive overview for the field of lncRNAs in HCC development based on bibliometric and visualized methods. The results would provide a reference for scholars focusing on this field.
Liver cancer is expected to be the sixth most commonly diagnosed cancer, and it ranks as the fourth leading cause of cancer death. With the absence of early diagnosis and limited treatment methods, Bray et al. estimates the global incidence of liver cancer to be 841,000 new cases and 782,000 deaths in 2018, with greater prevalence in Northern and Western Africa and Eastern and South-Eastern Asia (Bray et al., 2018). Hepatocellular carcinoma (HCC) is the most prevalent form of primary liver cancer, which accounts for >90% of primary liver cancer cases (Llovet et al., 2021). The primary risk factors for HCC include chronic viral hepatitis infection (hepatitis B or C), aflatoxin B1 intake, obesity, and excessive alcohol consumption (Chhonker et al., 2021; Choi et al., 2021; Guan et al., 2021; Hwang et al., 2021; Lockart et al., 2021). These factors may be responsible for cancer-related mutations, DNA damage, and epigenetic alterations, resulting in inactivated oncogenes or inactivated tumor suppressor genes, ultimately leading to HCC development (Rovida et al., 2015; Ally et al., 2017; Dang et al., 2017). Current treatment strategies for HCC include specific kinase inhibitors, anti-hepatitis vaccine, and liver resection and transplantation (Dageforde et al., 2021; Pattyn et al., 2021; Sbenati et al., 2021; Yamamoto et al., 2021). These therapeutic approaches in combination with biomarker screening reduce HCC-related death. However, its treatment efficiency is far from satisfactory. Thus, novel treatment approaches with higher efficacy for HCC are an urgently needed.
Earlier studies related to HCC primarily concentrated on the coding genes in identified regions with their pivotal roles in many central biological processes (Ladero et al., 1996; Thorgeirsson et al., 2006; Park et al., 2016). However, only 2% of human genome transcripts comprise protein coding sequences, and nearly 98% of sequences have no coding for proteins; the transcripts that cannot be ultimately translated into proteins have been characterized as non-coding RNAs (ncRNAs) (Rosenbloom et al., 2012; Beermann et al., 2016). There is a growing body of evidences that support that these evolutionarily conserved ncRNAs, particularly in the long non-coding RNAs (lncRNAs), which are more than 200 nucleotides in length, play a regulatory role in various developmental or physiological processes of HCC (Huang et al., 2020; Qin et al., 2020; Zuo et al., 2020). The number of transcribed lncRNAs continues to grow rapidly (Xie et al., 2021). Several studies have proved that these lncRNAs are differentially expressed in a variety of tissues and HCC cells (Hongfeng et al., 2020; Topel et al., 2020; Gamaev et al., 2021). Currently, the function of a small fraction of lncRNAs in HCC has been clearly defined, such as DNA binding (Zhou et al., 2020), associating with proteins (Gou et al., 2018), RNA interaction (Zhou and Xia, 2020), and producing small peptides (Wu et al., 2020). Intriguingly, some HCC-related lncRNAs have been founded in bodily fluids, making them attractive alternative biomarkers for HCC (Burenina et al., 2021). Studies have shown that these lncRNAs serve as potential cancer biomarkers and therapeutic targets for treating HCC (Klingenberg et al., 2017; Ghafouri-Fard et al., 2020; Matboli et al., 2020; Sheng et al., 2021).
Recently, there has been increased interest in bibliometric studies. For example, Li et al. explored the research hotspots of external beam radiotherapy in prostate cancer (Li R. et al., 2021); the hotspots of the role of anesthesia on tumor prognosis was performed by Luo et al. (2021); Martynov et al. conducted a bibliometric study to find the hotspot trend related to neuroblastoma research (Martynov et al., 2020). However, few studies are available for the research hotspots of lncRNAs in HCC development by using bibliometric methods. Bibliometric analysis regarding the application of lncRNAs in HCC research can provide more detailed insights into how lncRNAs play a critical role in HCC development. In recent years, bibliometric approach has been used as the method for quantitative and qualitative analysis (Li Q. et al., 2021). Researchers may have a better understanding of research trends and research hotspots in particular areas from bibliometric analysis (Carollo et al., 2021; Kilicaslan et al., 2021). Thus, we preferentially used CiteSpace to explore the frontiers of the roles of lncRNAs in HCC development. The results would provide scientists with a valuable reference for future studies.
All relevant publications were retrieved from the Web of Science Core Collection (WoSCC) database with the following search strategies: TS = (Long non coding RNA OR LncRNA OR Lnc RNA OR Long noncoding RNA) AND TS = (Hepatocellular carcinoma OR Hepatocarcinoma OR HCC patient OR HCC cancer OR Hepatic carcinoma) AND Document types = (Article OR Review) AND Language = English, with a limited time frame from 2010 to 2020, index = Science Citation Index Expanded (SCI-EXPANDED). The data extraction was completed for all included studies within the day on August 20, 2021. In total, 2,667 publications were finally included. The data were retrieved by one author (ZL) and double checked by another author (NT). We then cleaned the data, such as merging “TAIWAN” into “China” in country cooperative analysis, unifying “long noncoding rna” and “long non-coding rna” as “LncRNA” in keyword co-occurrence analysis, and so on.
The CiteSpace version 5.6.R4 was used to conduct bibliometric analyses (Chen, 2006). Some valuable parameters were included, such as publication number, total number of citations, impact factor (IF), citation burst, and centrality. Productivity was calculated for individuals, countries, and institutions on the basis of the total number of publications. The total number of citations was used to evaluate the international impact on both authors and articles (Chen et al., 2020). IF was used for assessing the international impact of journals, and it was obtained from the 2020 Journal Citation Reports (Pacey, 2020). Burst keywords referred to keywords which were cited frequently over a period of time, and they were considered indicators of research frontiers (Zhu et al., 2020). Centrality was a critical parameter for measuring the importance of nodes in a network. The nodes with a higher centrality (≥0.1) in the network were highlighted using purple rings and were usually regarded as high influence in a network (Liang et al., 2017).
The retrieved data were used for bibliometric analysis, and analyses were performed with the software CiteSpace version 5.6.R4, such as cooperative analysis, document co-citation analysis, keyword co-occurrence, timeline view of keywords, and citation burst analysis of keywords. The top 50 most cited articles in each time splicing with 1 year were selected to create a network. In order to highlight the key network, minimum spanning tree was used to explore the co-author citation. Minimum duration was set to 1 in the burst analysis of keywords.
A total of 2,667 records were initially found based on the search criteria. Figure 1 shows the distribution of publications related to the field of lncRNAs in HCC development from year 2010 to year 2020. The literature showed an overall rising trend in the total number of scientific publications from 2 in 2010 to 647 in 2020. Remarkably, the annual output of papers was nearly doubled compared to the previous year after 2014. It indicated that the annual related publications output had increased dramatically during year 2014 to year 2020.
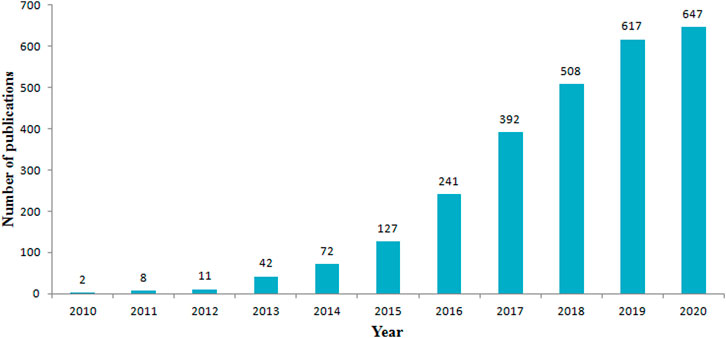
FIGURE 1. The number of annual publications on the roles of lncRNAs in HCC development research from 2010 to 2020.
There were 52 different countries which were identified in the network between year 2010 and year 2020. The country with at least 13 publications (T ≥ 13) are shown in Figure 2A. We computed the centrality for each node in order to identify strongly influential nodes in a network; the node with purple circle was considered more influential in the network. The centrality value for each country is shown in Table 1. The United States had an advantage in highest centrality (0.60), followed by China (0.39) and Germany (0.12). The top 10 countries which produced scientific publications concerning lncRNAs and HCC development are also presented in Table 1; the results indicated that China was the first leading country regarding the amount of publications (2,292), followed by the United States (231), Italy (39), and Germany (38). Among all the countries, China and the United States played a leading role in collaborative networks.
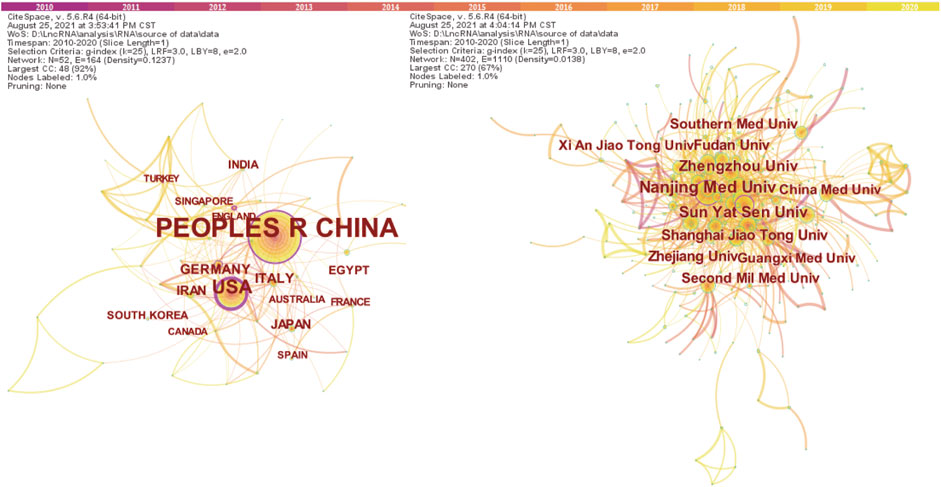
FIGURE 2. The co-occurrence map of countries (A) (T ≥ 13) and institutions (B) (T ≥ 62) in the field of lncRNAs in HCC development research.
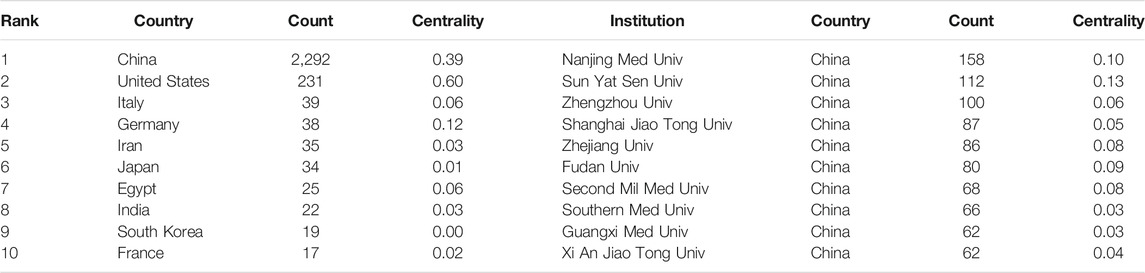
TABLE 1. The top 10 countries and institutions involved in the roles of lncRNAs in HCC development research.
Among the major international institutions, all of the top 10 contributing institutions were from China (Table 1), including the Nanjing Medical University (n = 158), Sun Yat Sen University (n = 112), and Zhengzhou University (n = 100). Institutions with centrality ≥0.1 included Nanjing Medical University and Sun Yat Sen University, suggesting that these institutions had an important role in this field. As Figure 3B (T ≥ 62) shows, the three aforementioned institutions were in a unique position in a collaborations network.
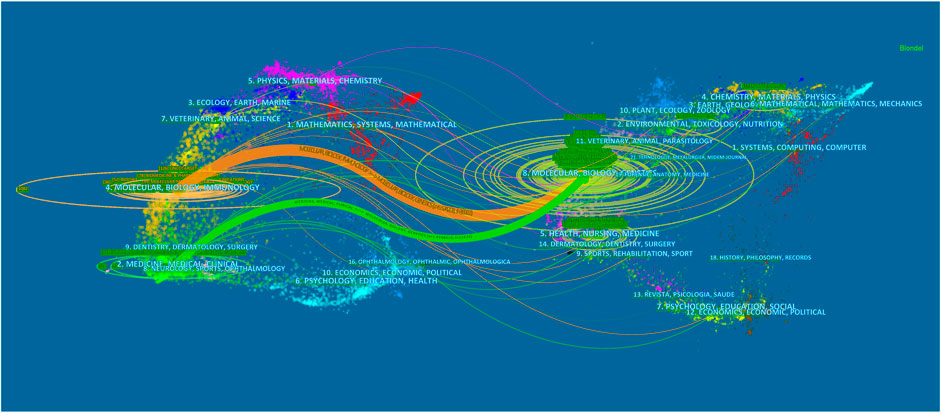
FIGURE 3. The dual-map overlay of journals related to lncRNA in HCC research. Notes: The citing journals are on the left, the cited journals are on the right, and the colored path represents citation relationship.
The dual-map overlay of journals stands for the topic distribution of academic journals (Chen and Leydesdorff, 2014) (Figure 3). The citing journals are located on the left, while the cited journals are on the right, and the colored paths indicate the citation relationships. There were two citation paths. The first orange path was for papers published in molecular/biology/genetics journals, which were mainly cited by the studies published in molecular/biology/immunology journals; the next green path was for papers published in molecular/biology/immunology journals, which were mainly cited by the studies published in medicine/medical/clinical area.
The top 10 co-cited journals were identified with highly frequent citations (Table 2). Oncotarget, one of the top 10 journals, had the most co-citations (1,751 citations, IF = none), followed by Cell (1,570 citations, IF = 41.582), Oncogene (1,460 citations, IF = 9.867), and Cancer Research (1,442 citations, IF = 12.701). Among these top 10 journals, it was noteworthy that the top six journals came from the United States, and eight of them were at the Q1 JCR division.
Figure 4 shows the authors who are distributed in the cluster with the largest size. Cooperation relationships were indicated by directed edges among nodes. When the relationship between the two authors was stronger, the line could be also thicker. Table 3 shows the top 10 prolific researchers. The number of published papers varied from 11 to 23 for different authors. Gang Chen published the largest amount of articles (23 publications), followed by Wei Wang (18 publications), and Yan Li (18 publications). Remarkably, only one of the top 10 prolific authors was included in the top 10 co-cited authors. The authors with at least 300 co-citations (T ≥ 300) were used to make the network (Figure 5). This type of knowledge map could clearly show the co-cited authors with high frequency. According to Figure 5, Yang F and Wang Y had the largest circles.
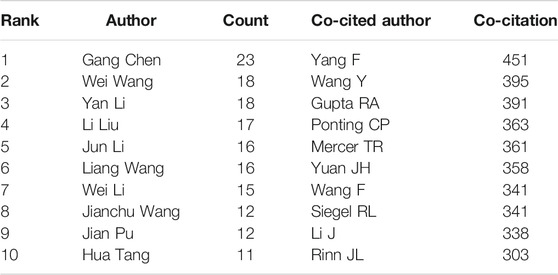
TABLE 3. The top 10 authors and co-cited authors that published articles on the roles of lncRNAs in HCC development research.
In total, 780 research keywords were found in the field of lncRNAs in HCC development; only the keywords with a frequency value larger than 260 are displayed in Figure 6. The top 20 cited keywords are shown in Table 4, including hepatocellular carcinoma, lncRNA, expression, and so on. These words were classified into 11 large clusters (Figure 7): “exosome,” “invasion,” “chemoresistance,” “glycolysis,” “liver fibrosis,” “egfr,” “heih,” “pvt1,” “dna methylation,” “wnt,” “inflammation,” and “laryngeal squamous cell carcinoma.” This timeline view (Figure 7) visualized the most important keywords in a specific field and showed the emergence, popularity, and decline of the research topic. Burst keywords were regarded as an indicator for the frontiers of the specific field during a period of time. The CiteSpace version 5.6.R4 was used to explore the keywords with the strongest citation bursts (Supplementary Figure 1), and 86 keywords were detected. By combining the timeline view with the keyword burst map, we found the evolutionary path of research hotspots. At the earlier exploration stage (2010–2013), some rising terms were identified with identification, promoter, chromatin, expression, enhancer, etc. However, at the rapid development stage (2014–2018), some new terms with association to biological functions and diseases emerged, such as DNA methylation, induction, suppression, gene regulation, oncogene, liver cancer, tumor initiating cell, ovarian cancer, therapeutic target, etc. At the stable-growth stage (2019–2020), some emerging topics were observed, such as interact, wnt, starbase, and myc.
Some papers which had been cited with a high frequency are listed in Table 5. “Yuan JH, DOI 10.1016/j.ccr.2014.03.010,” “Torre LA, DOI 10.3322/caac.21262,” and “Gupta RA, DOI 10.1038/nature08975” were the top three cited papers with the highest frequency. The total numbers of citations of the three aforementioned papers were 339, 321, and 258, respectively. These three cited papers were the fundamental cornerstone for this field.
To the best of our knowledge, this was the first study to explore the mapping intellectual structure for lncRNAs and HCC development by using CiteSpace. The number of published papers related to the field of lncRNAs in HCC development increased sharply in recent years. We found the knowledge base for the field of lncRNAs and HCC development by combining co-citation analysis with co-word analysis.
Regarding the top 10 prolific countries, four were developing countries, and six were developed countries. In terms of absolute publication numbers, China was the leading country, followed by the United States; this indicated that China had made significant progress in this area. However, China had a lower centrality value compared to the United States; it suggested that although China was the most prolific country in producing research publications, the United States played a leading role in collaborative network in this research field. There were extensive collaborations between western countries in the collaboration network. Transatlantic collaborations were the strongest among the United States, China, Germany, and Italy. The top 10 institutions were all from China. Among them, the papers published by Nanjing Medical University were cited with the highest frequency. For example, a study showed that Lnc-EGFR could suppress cytotoxic T lymphocyte (CTL) activity, stimulates Treg differentiation, and promotes HCC development through an EGFR-dependent signaling pathway (Jiang et al., 2017). Moreover, Zhuo et al. suggested that MEG3 serves as a prognostic biomarker for HCC (Zhuo et al., 2016). In addition to that, a previous study suggested that lncRNA ANRIL, as a cell growth modulator, may act as a novel potential therapeutic target for HCC (Huang et al., 2015).
In terms of the top 10 co-cited journals, six of the journals were from the United States. It was noteworthy that none of the top 10 co-cited journals were from China. However, Oncotarget was dropped by Science Citation Index Expanded. This demonstrated that these papers published in the journal need to be further improved. Moreover, impact factor ranged from 3.24 to 508.702. Additionally, this field had been more widely studied, and it covered more scientific disciplines in recent years, such as oncology (Lai et al., 2019; Lim et al., 2019; Wei et al., 2019), cell biology (Li et al., 2019; Song et al., 2019), and biochemistry molecular biology (Du et al., 2019; Guo et al., 2019). In the dual map, papers published in the molecular/biology/genetics journals were mostly cited by the studies published in molecular/biology/immunology and medicine/medical/clinical journals.
As for the top 10 prolific authors, each of them contributed to at least 11 publications. However, only one of the top 10 prolific authors was included in the top 10 co-cited authors, indicating that the authors should consider improving the quality of their articles in future. Co-cited authors include Yang F, who suggested that lncRNA-HEIH served as an oncogenic lncRNA that promotes tumor progression in HCC (Yang et al., 2011); Gupta RA, who indicated that lncRNAs played a positive role in regulating the cancer epigenome and could be the important targets in the diagnosis and therapy of HCC (Gupta et al., 2010); Torre LA, who reported that a large proportion of HCC cases and deaths could be effectively avoided by averting with appropriate preventive measures, such as vaccination, the use of early detection tests, and tobacco control (Torre et al., 2015); and Ponting CP, who explored the evolution of lncRNAs contributions to epigenetic gene regulation and transcriptional regulation (Ponting et al., 2009). Even though these authors were not prolific authors, they had already made significant contributions in this field.
Burst keywords were considered to discover emerging trends or research frontiers during a period of time (Chen, 2006). In this study, CiteSpace 5.6.R4 was used to capture the strongest citation bursts. At exploration stage (2010–2013), since the lncRNA HOTAIR had been proven to promote breast cancer metastasis by participating in chromatin remodeling (Gupta et al., 2010), researchers paid a lot of rising attention on lncRNAs. More and more functions of lncRNAs had been found. For example, lncRNAs could influence the development or progression of cancer (Ling et al., 2018; Chen and Xia, 2019). Subsequently, both the primary and secondary structures of lncRNAs had been discovered by enzymatic probing (Ilik et al., 2013) or chemical probing (Novikova et al., 2013; Smola et al., 2016). Researchers went one step further to divide lncRNAs into intronic, intergenic, sense, antisense, and bidirectional according to their genomic location relative to protein coding genes (Batagov et al., 2013).
At the rapid development stage (2014–2018), the mechanism, function, and application of lncRNA had been further studied. Based on the different mechanisms of lncRNAs acting on cellular processes, they are divided into five categories, such as signal (Pandey et al., 2008; Wang and Chang, 2011), decoy (Hung et al., 2011), guide (Wang and Chang, 2011), scaffold (Wang and Chang, 2011), and enhancer (Goyal et al., 2018; Liao et al., 2018). Moreover, some existing studies have suggested that the various functions of lncRNAs serve as critical roles in diverse biological processes, including epigenetic modification (Bhan and Mandal, 2014), transcriptional regulation (Wei et al., 2019), post-transcriptional regulation (Cesana et al., 2011), translational regulation (Yoon et al., 2012), and post-translational regulation (Wang et al., 2014). In addition, there had been more and more applications of lncRNAs discovered in cancer research. Initially, lncRNAs were reported to be associated with breast cancer (Shore et al., 2012). Some subsequent research were also reported in HCC (Shi et al., 2019), ovarian cancer (Oncul et al., 2020), and colorectal cancer (Huang et al., 2017). These lncRNAs could be used to work as novel prognostic biomarkers and intervention strategies for HCC.
At the stable-growth stage (2019–2020), with the rapid development of network public databases, there were many lncRNAs databases established, such as LNCipedia, LNCBook, TargetScan, Starbase (ENCORI), etc. Some of them were used to explore interactions (RNA–RNA interactions, RNA–DNA interactions, and RNA–Protein interactions) (Li et al., 2014) and pathway analyses (Qian et al., 2020), which were considered as a research frontier in the field of lncRNAs and HCC development. Furthermore, The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases could allow researchers to obtain raw data for free. These two databases were combined with some predictive or prognostic databases to perform bioinformatics analysis, such as UALCAN (Chandrashekar et al., 2017) and Kaplan–Meier Plotter (Hou et al., 2017), etc.
This was the first study to address the roles of lncRNAs in HCC development research with CiteSpace. However, some study limitations should not be overlooked. First, this study did not include all relevant literatures. Even though majority of the research papers in the field of lncRNAs and HCC were included in the WoSCC database, some other databases such as Pubmed and Scopus may provide a broader scope. Second, there were discrepancies between the bibliometric analysis results and the actual research conditions due to a lower citation count from the recently published studies. Third, although all the data extraction was included within 1 day to avoid bias, the database is still expanding due to the daily updates of databases. Despite this, the vast majority of studies were included in this study. Therefore, the conclusion may not be impacted with a small amount of recent papers updated.
This analysis urges other researchers to discover the research hotspots and frontiers in the field of lncRNAs and HCC development from year 2010 to year 2020. A total of 2,667 records on this topic were initially found worldwide. The annual related publications output had increased dramatically over this time. Although China was the most prolific country in terms of research publication, the United States played a leading role in collaborative network. The Nanjing Medical University was the most productive institute in this field. Gang Chen was the most prolific researcher, while Yang F was the most frequently co-cited author. Oncotarget, Cell, and Oncogene were the most highly co-cited journals. The most recent burst keywords were interaction, database, and pathway.
The datasets presented in this study can be found in online repositories https://www.webofscience.com/wos/woscc/basic-search. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
ZL performed the bibliometric analysis and wrote and revised the manuscript. XJ, NT, and YG revised the manuscript. LK conceived and designed the study, critically revised the manuscript, and gave final approval. All authors read and approved the final draft of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are grateful to all subjects who participated in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.771810/full#supplementary-material
Ally, A., Balasundaram, M., Carlsen, R., Chuah, E., Clarke, A., Dhalla, N., et al. (2017). Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 169, 1327–1341. doi:10.1016/j.cell.2017.05.046
Batagov, A. O., Yarmishyn, A. A., Jenjaroenpun, P., Tan, J. Z., Nishida, Y., and Kurochkin, I. V. (2013). Role of Genomic Architecture in the Expression Dynamics of Long Noncoding RNAs during Differentiation of Human Neuroblastoma Cells. BMC Syst. Biol. 7, S11. doi:10.1186/1752-0509-7-S3-S11
Beermann, J., Piccoli, M.-T., Viereck, J., and Thum, T. (2016). Non-Coding Rnas in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 96, 1297–1325. doi:10.1152/physrev.00041.2015
Bhan, A., and Mandal, S. S. (2014). Long Noncoding RNAs: Emerging Stars in Gene Regulation, Epigenetics and Human Disease. Chemmedchem 9, 1932–1956. doi:10.1002/cmdc.201300534
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clinicians 68, 394–424. doi:10.3322/caac.21492
Burenina, O. Y., Lazarevich, N. L., Kustova, I. F., Shavochkina, D. A., Moroz, E. A., Kudashkin, N. E., et al. (2021). Panel of Potential lncRNA Biomarkers Can Distinguish Various Types of Liver Malignant and Benign Tumors. J. Cancer Res. Clin. Oncol. 147, 49–59. doi:10.1007/s00432-020-03378-5
Carollo, A., Balagtas, J. P. M., Neoh, M. J., and Esposito, G. (2021). A Scientometric Approach to Review the Role of the Medial Preoptic Area (MPOA) in Parental Behavior. Brain Sci. 11, 1. doi:10.3390/brainsci11030393
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 147, 947. doi:10.1016/j.cell.2011.10.031
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Chen, C. (2006). CiteSpace II: Detecting and Visualizing Emerging Trends and Transient Patterns in Scientific Literature. J. Am. Soc. Inf. Sci. 57, 359–377. doi:10.1002/asi.20317
Chen, C., and Leydesdorff, L. (2014). Patterns of Connections and Movements in Dual-Map Overlays: A New Method of Publication Portfolio Analysis. J. Assn Inf. Sci. Tec 65, 334–351. doi:10.1002/asi.22968
Chen, S., and Xia, X. (2019). Long Noncoding RNA NEAT1 Suppresses Sorafenib Sensitivity of Hepatocellular Carcinoma Cells via Regulating miR‐335-c‐Met. J. Cell Physiol 234, 14999–15009. doi:10.1002/jcp.27567
Chen, S., Zhang, Y., Dai, W., Qi, S., Tian, W., Gu, X., et al. (2020). Publication Trends and Hot Spots in Postoperative Cognitive Dysfunction Research: A 20-year Bibliometric Analysis. J. Clin. Anesth. 67, 110012. doi:10.1016/j.jclinane.2020.110012
Chhonker, S. K., Rawat, D., and Koiri, R. K. (2021). Protective and Therapeutic Effects of Sildenafil and Tadalafil on Aflatoxin B1-Induced Hepatocellular Carcinoma. Mol. Cell Biochem 476, 1195–1209. doi:10.1007/s11010-020-03982-6
Choi, J., Jo, C., and Lim, Y. S. (2021). Tenofovir versus Entecavir on Recurrence of Hepatitis B Virus-Related Hepatocellular Carcinoma after Surgical Resection. Hepatology 73, 661–673. doi:10.1002/hep.31289
Dageforde, L. A., Vachharajani, N., Tabrizian, P., Agopian, V., Halazun, K., Maynard, E., et al. (2021). Multi-Center Analysis of Liver Transplantation for Combined Hepatocellular Carcinoma-Cholangiocarcinoma Liver Tumors. J. Am. Coll. Surgeons 232, 361–371. doi:10.1016/j.jamcollsurg.2020.11.017
Dang, H., Takai, A., Forgues, M., Pomyen, Y., Mou, H., Xue, W., et al. (2017). Oncogenic Activation of the RNA Binding Protein NELFE and MYC Signaling in Hepatocellular Carcinoma. Cancer Cell 32, 101–114. doi:10.1016/j.ccell.2017.06.002
Du, J., Chen, M., Liu, J., Hu, P., Guan, H., and Jiao, X. (2019). LncRNA F11‐AS1 Suppresses Liver Hepatocellular Carcinoma Progression by Competitively Binding with miR‐3146 to Regulate PTEN Expression. J. Cel Biochem 120, 18457–18464. doi:10.1002/jcb.29163
Gamaev, L., Mizrahi, L., Friehmann, T., Rosenberg, N., Pappo, O., Olam, D., et al. (2021). The Pro-oncogenic Effect of the lncRNA H19 in the Development of Chronic Inflammation-Mediated Hepatocellular Carcinoma. Oncogene 40, 127–139. doi:10.1038/s41388-020-01513-7
Ghafouri-Fard, S., Dashti, S., and Taheri, M. (2020). PCAT1: An Oncogenic lncRNA in Diverse Cancers and a Putative Therapeutic Target. Exp. Mol. Pathol. 114, 104429. doi:10.1016/j.yexmp.2020.104429
Gou, Q., Gao, L., Nie, X., Pu, W., Zhu, J., Wang, Y., et al. (2018). Long Noncoding RNA AB074169 Inhibits Cell Proliferation via Modulation of KHSRP-Mediated CDKN1a Expression in Papillary Thyroid Carcinoma. Cancer Res. 78, 4163–4174. doi:10.1158/0008-5472.can-17-3766
Goyal, N., Kesharwani, D., and Datta, M. (2018). Lnc-ing Non-coding RNAs with Metabolism and Diabetes: Roles of lncRNAs. Cell. Mol. Life Sci. 75, 1827–1837. doi:10.1007/s00018-018-2760-9
Guan, X., Xing, F., and Li, Y. (2021). Alcohol Consumption Increases the Incidence of Hepatocellular Carcinoma in Patients with Hepatitis B Cirrhosis but Not in Patients with Hepatitis C Cirrhosis. Eur. J. Gastroenterol. Hepatol. 33, 1218–1221. doi:10.1097/meg.0000000000001837
Guo, J.-C., Yang, Y.-J., Zheng, J.-F., Zhang, J.-Q., Guo, M., Yang, X., et al. (2019). Silencing of Long Noncoding RNA HOXA11-AS Inhibits the Wnt Signaling Pathway via the Upregulation of HOXA11 and Thereby Inhibits the Proliferation, Invasion, and Self-Renewal of Hepatocellular Carcinoma Stem Cells. Exp. Mol. Med. 51, 1–20. doi:10.1038/s12276-019-0328-x
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long Non-coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature 464, 1071–1076. doi:10.1038/nature08975
Hongfeng, Z., Andong, J., Liwen, S., Mingping, B., Xiaowei, Y., Mingyong, L., et al. (2020). lncRNA RMRP Knockdown Suppress Hepatocellular Carcinoma Biological Activities via Regulation miRNA‐206/TACR1. J. Cel Biochem 121, 1690–1702. doi:10.1002/jcb.29404
Hou, G. X., Liu, P., Yang, J., and Wen, S. (2017). Mining Expression and Prognosis of Topoisomerase Isoforms in Non-small-cell Lung Cancer by Using Oncomine and Kaplan-Meier Plotter. Plos One 12, e0174515. doi:10.1371/journal.pone.0174515
Huang, J.-Z., Chen, M., Chen, D., Gao, X.-C., Zhu, S., Huang, H., et al. (2017). A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell 68, 171–184. doi:10.1016/j.molcel.2017.09.015
Huang, M.-d., Chen, W.-m., Qi, F.-z., Xia, R., Sun, M., Xu, T.-p., et al. (2015). Long Non-coding RNA ANRIL Is Upregulated in Hepatocellular Carcinoma and Regulates Cell Proliferation by Epigenetic Silencing of KLF2. J. Hematol. Oncol. 8, 57. doi:10.1186/s13045-015-0153-1
Huang, Z., Zhou, J. K., Peng, Y., He, W., and Huang, C. (2020). The Role of Long Noncoding RNAs in Hepatocellular Carcinoma. Mol. Cancer 19, 77. doi:10.1186/s12943-020-01188-4
Hung, T., Wang, Y., Lin, M. F., Koegel, A. K., Kotake, Y., Grant, G. D., et al. (2011). Extensive and Coordinated Transcription of Noncoding RNAs within Cell-Cycle Promoters. Nat. Genet. 43, 621–629. doi:10.1038/ng.848
Hwang, S., Park, Y. M., Han, K. D., Yun, J. S., Ko, S. H., Ahn, Y. B., et al. (2021). Associations of General Obesity and central Obesity with the Risk of Hepatocellular Carcinoma in a Korean Population: A National Population‐based Cohort Study. Int. J. Cancer 148, 1144–1154. doi:10.1002/ijc.33305
Ilik, I. A., Quinn, J. J., Georgiev, P., Tavares-Cadete, F., Maticzka, D., Toscano, S., et al. (2013). Tandem Stem-Loops in roX RNAs Act Together to Mediate X Chromosome Dosage Compensation in Drosophila. Mol. Cell 51, 156–173. doi:10.1016/j.molcel.2013.07.001
Jiang, R., Tang, J., Chen, Y., Deng, L., Ji, J., Xie, Y., et al. (2017). The Long Noncoding RNA Lnc-EGFR Stimulates T-Regulatory Cells Differentiation Thus Promoting Hepatocellular Carcinoma Immune Evasion. Nat. Commun. 8, 15129. doi:10.1038/ncomms15129
Kiliçaslan, Ö. F., Nabi, V., Yardibi, F., Tokgöz, M. A., and Köse, Ö. (2021). Research Tendency in Lumbar Spinal Stenosis over the Past Decade: A Bibliometric Analysis. World Neurosurg. 149, e71–e84. doi:10.1016/j.wneu.2021.02.086
Klingenberg, M., Matsuda, A., Diederichs, S., and Patel, T. (2017). Non-coding RNA in Hepatocellular Carcinoma: Mechanisms, Biomarkers and Therapeutic Targets. J. Hepatol. 67, 603–618. doi:10.1016/j.jhep.2017.04.009
Ladero, J. M., Agundez, J. A., Rodriguez-Lescure, A., Diaz-Rubio, M., and Benitez, J. (1996). RsaI Polymorphism at the Cytochrome P4502E1 Locus and Risk of Hepatocellular Carcinoma. Gut 39, 330–333. doi:10.1136/gut.39.2.330
Lai, Y., Feng, B., Abudoureyimu, M., Zhi, Y., Zhou, H., Wang, T., et al. (2019). Non-coding RNAs: Emerging Regulators of Sorafenib Resistance in Hepatocellular Carcinoma. Front. Oncol. 9, 1156. doi:10.3389/fonc.2019.01156
Li, J.-H., Liu, S., Zhou, H., Qu, L.-H., and Yang, J.-H. (2014). starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and Protein-RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucl. Acids Res. 42, D92–D97. doi:10.1093/nar/gkt1248
Li, Q., Dai, W., Chen, X., Su, D., Yu, W., and Gu, X. (2021a). Publication Trends and Hot Spots in Chronic Postsurgical Pain (CPSP) Research: A 10-Year Bibliometric Analysis. Jpr 14, 2239–2247. doi:10.2147/jpr.s300744
Li, R., Liu, X., Yang, B., and Qiu, J. (2021b). External Beam Radiotherapy for Prostate Cancer: What Are the Current Research Trends and Hotspots? Cancer Med. 10, 772–782. doi:10.1002/cam4.3700
Li, Y., Ma, B., Yin, Z., Liu, P., Liu, J., Li, J., et al. (2019). Competing Endogenous RNA Network and Prognostic Nomograms for Hepatocellular Carcinoma Patients Who Underwent R0 Resection. J. Cell Physiol 234, 20342–20353. doi:10.1002/jcp.28634
Liang, Y.-D., Li, Y., Zhao, J., Wang, X.-Y., Zhu, H.-Z., and Chen, X.-H. (2017). Study of Acupuncture for Low Back Pain in Recent 20 years: a Bibliometric Analysis via CiteSpace. Jpr 10, 951–964. doi:10.2147/jpr.s132808
Liao, L.-M., Zhang, F.-H., Yao, G.-J., Ai, S.-F., Zheng, M., and Huang, L. (2018). Role of Long Noncoding RNA 799 in the Metastasis of Cervical Cancer through Upregulation of TBL1XR1 Expression. Mol. Ther. - Nucleic Acids 13, 580–589. doi:10.1016/j.omtn.2018.10.007
Lim, L. J., Wong, S. Y. S., Huang, F., Lim, S., Chong, S. S., Ooi, L. L., et al. (2019). Roles and Regulation of Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Res. 79, 5131–5139. doi:10.1158/0008-5472.can-19-0255
Ling, Z.-a., Xiong, D.-d., Meng, R.-m., Cen, J.-M., Zhao, N., Chen, G., et al. (2018). LncRNA NEAT1 Promotes Deterioration of Hepatocellular Carcinoma Based on In Vitro Experiments, Data Mining, and RT-qPCR Analysis. Cell Physiol Biochem 48, 540–555. doi:10.1159/000491811
Llovet, J. M., Kelley, R. K., Villanueva, A., Singal, A. G., Pikarsky, E., Roayaie, S., et al. (2021). Hepatocellular Carcinoma. Nat. Rev. Dis. Primers 7, 6. doi:10.1038/s41572-020-00240-3
Lockart, I., Hajarizadeh, B., Alavi, M., Davison, S., Prakoso, E., Levy, M. T., et al. (2021). Hepatitis C Virus Cure before Hepatocellular Carcinoma Diagnosis Is Associated with Improved Survival. J. Viral Hepat. 28, 710–718. doi:10.1111/jvh.13475
Luo, J., Shi, Y., Wang, X., Zhang, R., Chen, S., Yu, W., et al. (2021). A 20-Year Research Trend Analysis of the Influence of Anesthesia on Tumor Prognosis Using Bibliometric Methods. Front. Oncol. 11, 683232. doi:10.3389/fonc.2021.683232
Martynov, I., Klima-Frysch, J., and Schoenberger, J. (2020). A Scientometric Analysis of Neuroblastoma Research. BMC Cancer 20, 486. doi:10.1186/s12885-020-06974-3
Matboli, M., Labib, M. E., Nasser, H. E.-T., El-Tawdi, A. H. F., Habib, E. K., and Ali-Labib, R. (2020). Exosomal miR-1298 and lncRNA-RP11-583F2.2 Expression in Hepatocellular Carcinoma. Cg 21, 46–55. doi:10.2174/1389202920666191210111849
Novikova, I. V., Dharap, A., Hennelly, S. P., and Sanbonmatsu, K. Y. (2013). 3S: Shotgun Secondary Structure Determination of Long Non-coding RNAs. Methods 63, 170–177. doi:10.1016/j.ymeth.2013.07.030
Oncul, S., Amero, P., Rodriguez-Aguayo, C., Calin, G. A., Sood, A. K., and Lopez-Berestein, G. (2020). Long Non-coding RNAs in Ovarian Cancer: Expression Profile and Functional Spectrum. Rna Biol. 17, 1523–1534. doi:10.1080/15476286.2019.1702283
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-specific Transcriptional Silencing through Chromatin-Level Regulation. Mol. Cell 32, 232–246. doi:10.1016/j.molcel.2008.08.022
Park, Y.-Y., Sohn, B. H., Johnson, R. L., Kang, M.-H., Kim, S. B., Shim, J.-J., et al. (2016). Yes-associated Protein 1 and Transcriptional Coactivator with PDZ-Binding Motif Activate the Mammalian Target of Rapamycin Complex 1 Pathway by Regulating Amino Acid Transporters in Hepatocellular Carcinoma. Hepatology 63, 159–172. doi:10.1002/hep.28223
Pattyn, J., Hendrickx, G., Vorsters, A., and Van Damme, P. (2021). Hepatitis B Vaccines. J. Infect. Dis. 224, S343–S351. doi:10.1093/infdis/jiaa668
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and Functions of Long Noncoding RNAs. Cell 136, 629–641. doi:10.1016/j.cell.2009.02.006
Qian, G., Jin, X., and Zhang, L. (2020). LncRNA FENDRR Upregulation Promotes Hepatic Carcinoma Cells Apoptosis by Targeting miR-362-5p via NPR3 and P38-MAPK Pathway. Cancer Biother. Radiopharm. 35, 629–639. doi:10.1089/cbr.2019.3468
Qin, G., Tu, X., Li, H., Cao, P., Chen, X., Song, J., et al. (2020). Long Noncoding RNA p53‐Stabilizing and Activating RNA Promotes P53 Signaling by Inhibiting Heterogeneous Nuclear Ribonucleoprotein K deSUMOylation and Suppresses Hepatocellular Carcinoma. Hepatology 71, 112–129. doi:10.1002/hep.30793
Rosenbloom, K. R., Dreszer, T. R., Long, J. C., Malladi, V. S., Sloan, C. A., Raney, B. J., et al. (2012). ENCODE Whole-Genome Data in the UCSC Genome Browser: Update 2012. Nucleic Acids Res. 40, D912–D917. doi:10.1093/nar/gkr1012
Rovida, E., Di Maira, G., Tusa, I., Cannito, S., Paternostro, C., Navari, N., et al. (2015). The Mitogen-Activated Protein Kinase ERK5 Regulates the Development and Growth of Hepatocellular Carcinoma. Gut 64, 1454–1465. doi:10.1136/gutjnl-2014-306761
Sbenati, R. M., Zaraei, S.-O., El-Gamal, M. I., Anbar, H. S., Tarazi, H., Zoghbor, M. M., et al. (2021). Design, Synthesis, Biological Evaluation, and Modeling Studies of Novel Conformationally-Restricted Analogues of Sorafenib as Selective Kinase-Inhibitory Antiproliferative Agents against Hepatocellular Carcinoma Cells. Eur. J. Med. Chem. 210, 113081. doi:10.1016/j.ejmech.2020.113081
Sheng, J.-Q., Wang, M.-R., Fang, D., Liu, L., Huang, W.-J., Tian, D.-A., et al. (2021). LncRNA NBR2 Inhibits Tumorigenesis by Regulating Autophagy in Hepatocellular Carcinoma. Biomed. Pharmacother. 133, 111023. doi:10.1016/j.biopha.2020.111023
Shi, H., Xu, Y., Yi, X., Fang, D., and Hou, X. (2019). Current Research Progress on Long Noncoding RNAs Associated with Hepatocellular Carcinoma. Anal. Cell Pathol. 2019, 1–8. doi:10.1155/2019/1534607
Shore, A. N., Herschkowitz, J. I., and Rosen, J. M. (2012). Noncoding RNAs Involved in Mammary Gland Development and Tumorigenesis: There's a Long Way to Go. J. Mammary Gland Biol. Neoplasia 17, 43–58. doi:10.1007/s10911-012-9247-3
Smola, M. J., Christy, T. W., Inoue, K., Nicholson, C. O., Friedersdorf, M., Keene, J. D., et al. (2016). SHAPE Reveals Transcript-wide Interactions, Complex Structural Domains, and Protein Interactions across the Xist lncRNA in Living Cells. Proc. Natl. Acad. Sci. USA 113, 10322–10327. doi:10.1073/pnas.1600008113
Song, M., Zhong, A., Yang, J., He, J., Cheng, S., Zeng, J., et al. (2019). Large-scale Analyses Identify a Cluster of Novel Long Noncoding RNAs as Potential Competitive Endogenous RNAs in Progression of Hepatocellular Carcinoma. Aging 11, 10422–10453. doi:10.18632/aging.102468
Thorgeirsson, S. S., Lee, J.-S., and Grisham, J. W. (2006). Functional Genomics of Hepatocellular Carcinoma. Hepatology 43, S145–S150. doi:10.1002/hep.21063
Topel, H., Bagirsakci, E., Comez, D., Bagci, G., Cakan-Akdogan, G., and Atabey, N. (2020). lncRNA HOTAIR Overexpression Induced Downregulation of C-Met Signaling Promotes Hybrid Epithelial/mesenchymal Phenotype in Hepatocellular Carcinoma Cells. Cell Commun Signal 18, 110. doi:10.1186/s12964-020-00602-0
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global Cancer Statistics, 2012. CA: A Cancer J. Clinicians 65, 87–108. doi:10.3322/caac.21262
Wang, K. C., and Chang, H. Y. (2011). Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 43, 904–914. doi:10.1016/j.molcel.2011.08.018
Wang, P., Xue, Y., Han, Y., Lin, L., Wu, C., Xu, S., et al. (2014). The STAT3-Binding Long Noncoding RNA Lnc-DC Controls Human Dendritic Cell Differentiation. Science 344, 310–313. doi:10.1126/science.1251456
Wei, L., Wang, X., Lv, L., Liu, J., Xing, H., Song, Y., et al. (2019). The Emerging Role of microRNAs and Long Noncoding RNAs in Drug Resistance of Hepatocellular Carcinoma. Mol. Cancer 18, 147. doi:10.1186/s12943-019-1086-z
Wu, P., Mo, Y., Peng, M., Tang, T., Zhong, Y., Deng, X., et al. (2020). Emerging Role of Tumor-Related Functional Peptides Encoded by lncRNA and circRNA. Mol. Cancer 19, 22. doi:10.1186/s12943-020-1147-3
Xie, C., Li, S.-Y., Fang, J.-H., Zhu, Y., and Yang, J.-E. (2021). Functional Long Non-coding RNAs in Hepatocellular Carcinoma. Cancer Lett. 500, 281–291. doi:10.1016/j.canlet.2020.10.042
Yamamoto, M., Kobayashi, T., Hashimoto, M., Kuroda, S., Kawaoka, T., Aikata, H., et al. (2021). Significance of Liver Resection for Intermediate Stage Hepatocellular Carcinoma According to Subclassification. BMC Cancer 21, 668. doi:10.1186/s12885-021-08421-3
Yang, F., Zhang, L., Huo, X.-s., Yuan, J.-h., Xu, D., Yuan, S.-x., et al. (2011). Long Noncoding RNA High Expression in Hepatocellular Carcinoma Facilitates Tumor Growth through Enhancer of Zeste Homolog 2 in Humans. Hepatology 54, 1679–1689. doi:10.1002/hep.24563
Yoon, J.-H., Abdelmohsen, K., Srikantan, S., Yang, X., Martindale, J. L., De, S., et al. (2012). LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cell 47, 648–655. doi:10.1016/j.molcel.2012.06.027
Zhou, Y., Huan, L., Wu, Y., Bao, C., Chen, B., Wang, L., et al. (2020). LncRNA ID2-AS1 Suppresses Tumor Metastasis by Activating the HDAC8/ID2 Pathway in Hepatocellular Carcinoma. Cancer Lett. 469, 399–409. doi:10.1016/j.canlet.2019.11.007
Zhou, Z., and Xia, N. (2020). LncRNA DCST1-AS1 Sponges miR-107 to Upregulate CDK6 in Cervical Squamous Cell Carcinoma. Cmar 12, 7921–7928. doi:10.2147/cmar.s251582
Zhu, X., Kong, Q., Niu, X., Chen, L., and Ge, C. (2020). Mapping Intellectual Structure and Research Performance for the Nanoparticles in Pancreatic Cancer Field. Ijn 15, 5503–5516. doi:10.2147/ijn.s253599
Zhuo, H., Tang, J., Lin, Z., Jiang, R., Zhang, X., Ji, J., et al. (2016). The Aberrant Expression of MEG3 Regulated by UHRF1 Predicts the Prognosis of Hepatocellular Carcinoma. Mol. Carcinog. 55, 209–219. doi:10.1002/mc.22270
Keywords: lncRNA, hepatocellular carcinoma, bibliometric, citespace, database
Citation: Lin Z, Ji X, Tian N, Gan Y and Ke L (2022) Mapping Intellectual Structure for the Long Non-Coding RNA in Hepatocellular Carcinoma Development Research. Front. Genet. 12:771810. doi: 10.3389/fgene.2021.771810
Received: 09 September 2021; Accepted: 22 November 2021;
Published: 03 January 2022.
Edited by:
Chi-Ming Wong, Hong Kong Polytechnic University, Hong Kong SAR, ChinaReviewed by:
Peter Kokol, University of Maribor, SloveniaCopyright © 2022 Lin, Ji, Tian, Gan and Ke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Ke, a2VsaTEyMjFAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.