
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet. , 01 November 2021
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.770134
This article is part of the Research Topic Machine Learning-Based Methods for RNA Data Analysis, Volume II View all 15 articles
 Kun Zheng1†
Kun Zheng1† Shuo Yao1†
Shuo Yao1† Wei Yao1
Wei Yao1 Qianxia Li1
Qianxia Li1 Yali Wang1
Yali Wang1 Lili Zhang1
Lili Zhang1 Xiuqiong Chen1
Xiuqiong Chen1 Huihua Xiong1
Huihua Xiong1 Xianglin Yuan1
Xianglin Yuan1 Yihua Wang2,3
Yihua Wang2,3 Yanmei Zou1*
Yanmei Zou1* Hua Xiong1*
Hua Xiong1*Background: Although ribosomal protein S6 kinases, 90 kDa, polypeptide 3 (RSK2, RPS6KA3) has been reported to play an important role in cancer cell proliferation, invasion, and migration, including breast cancer, its clinical implication in primary breast cancer patients is not well understood, and there were not many studies to explore the relationship between RSK2 and breast cancer on a clinical level.
Methods: A systematic series matrix file search uploaded from January 1, 2008 to November 31, 2017 was undertaken using ArrayExpress and Gene Expression Omnibus (GEO) databases. Search filters were breast cancer, RNA assay, and array assay. Files eligible for inclusion met the following criteria: a) sample capacity is over 100, b) tumor sample comes from unselected patient’s primary breast tumor tissue, and c) expression of RSK2 and any clinical parameters of patients were available from the files. We use median as the cutoff value to assess the association between the expression of RSK2 and the clinical indexes of breast cancer patients.
Finding: The meta-analysis identified 13 series matrix files from GEO database involving 3,122 samples that come from patients’ primary breast cancer tissue or normal tissue. The expression of RSK2 in tumor tissues is lower than that in normal tissues [odds ratio (OR), 0.54; 95% credible interval (CI), 0.44–0.67; Cochran’s Q test p = 0.14; I2 = 41.7%]. Patients with a high expression of RSK2 showed more favorable overall survival [hazard ratio (HR), 0.71; 95% CI, 0.49–0.94; Cochran’s Q test p = 0.95; I2 = 0.0%] and less potential of distant metastasis (OR, 0.59; 95% CI, 0.41–0.87; Cochran’s Q test p = 0.88; I2 = 0.0%) and lymph node infiltration (OR, 0.81; 95% CI, 0.65–0.998; Cochran’s Q test p = 0.09; I2 = 42.8%). Besides, the expression of RSK2 in luminal breast cancer is lower than Cochran’s Q test p = 0.06; I2 = 63.5%). RSK2 overexpression corresponded with higher histological grade (OR, 1.329; 95% CI, 1.03–1.721; Cochran’s Q test p = 0.69; I2 = 0.0%). RSK2 expression is also associated with estrogen receptor (ER) and age.
Conclusion: The meta-analysis provides evidence that RSK2 is a potential biomarker in breast cancer patients. The expression of RSK2 is distinctive in different intrinsic subtypes of breast cancer, indicating that it may play an important role in specific breast cancer. Further study is needed to uncover the mechanism of RSK2 in breast cancer.
Systematic Review Registration: (website), identifier (registration number).
Breast cancer is the most frequent cancer among women. Data from the World Health Organization shows that breast cancer impacts over 1.5 million women each year and also causes the greatest number of cancer-related deaths among women. In 2015, 570,000 women died from breast cancer—that is, approximately 15% of all cancer deaths among women. Although there has been a breakthrough in the treatment and prevention of breast cancer in the past few years, leading to the 5-years relative survival rate rising to 90%, the majority of breast cancer patients with distant metastasis succumb to cancer progression within 5 years (Siegel et al., 2018). Also, breast cancer is more than one single disease. Several molecular subtypes of breast cancer have been classified depending on their molecular characteristics (Perou et al., 2000), and each individual subtype corresponds to a different underlying biology, survival rate, and response to therapy (Prat et al., 2015; Nielsen et al., 2017). Therefore, the identification of biomarkers to screen high-risk patients, predict breast prognostic outcomes, and provide new therapeutic targets for specific breast cancer is urgently needed.
RSK2, ribosomal protein S6 kinase, 90 kDa, polypeptide 3, belongs to RSK serine/threonine kinase family and is a downstream of the mitogen-activated protein kinase (MAPK) pathway (Zhao et al., 2016). RSK is unique among serine–threonine kinases in that it contains two functional kinase domains: an N-terminal kinase that phosphorylates the substrates of RSK and a C-terminal kinase involved in the activation mechanism of RSK (Frödin and Gammeltoft, 1999). RSK isoforms are activated by virtually all extracellular signaling molecules including growth factors, peptide hormones, neurotransmitters, and environmental stresses (Arul and Cho, 2013). It has been demonstrated that RSK2 plays an important role in cancer cell proliferation, invasion, and migration, including breast cancer (Yoo et al., 2019; Guo and Kong, 2021).
In previous studies, it has been reported that RSK2 expression is different between breast cancer tissue and normal breast tissue and varies among different subtypes or histological grades of breast cancer. Some studies suggested that RSK2 overexpression is correlated with basal-like breast cancer and higher histological grade, and RSK2 mRNA is associated with poor survival in breast cancer patients who had not received chemotherapy (Stratford et al., 2012; Zhao et al., 2016). A protein downstream of RSK2 named Y-box binding protein-1(YB-1) was reported to transform human mammary epithelial cells in the development of basal-like breast cancer (Davies et al., 2014). Another study indicated that RSK2 activation status positively correlates with patient response to anti-estrogen hormonal therapies and is required for estrogen receptor+ (ER+) breast cancer tumorigenesis (Clark et al., 2001). Several drug trials illustrated that by suppressing RSK2 expression, the metastasis of human epidermal growth factor receptor 2+ (HER2+) breast cancer was repressed (Mao et al., 2016), the ability of migration and invasion of lung cancer cell was inhibited (Lee et al., 2015), and the carcinogenesis of ultraviolet radiation-induced skin cancer was prevented (Yao et al., 2014). These prompt us to investigate whether RSK2 might be a potential biomarker that can act as a promising biomarker of breast cancer or a novel therapy target for a specific subtype of breast cancer. However, RSK2 expression is rarely associated with clinical practice, which drives us to investigate the association between RSK2 expression and clinical parameters and prognosis of breast cancer patients.
In the case of very limited clinical studies of RSK2, we performed a meta-analysis using public electronic databases ArrayExpress (Parkinson et al., 2007) and Gene Expression Omnibus (GEO) (Clough and Barrett, 2016) to summarize and evaluate the clinical significance of RSK2 in breast cancer patients and in order to explore the possibility of RSK2 expression as a predictive marker of clinicopathological parameters and prognosis in primary breast cancer, so as to screen high-risk patients or to provide new targets and directions for the treatment of breast cancer patients with specific molecular subtypes.
We conducted a search of a series matrix files in the electronic database ArrayExpress (ArrayExpress, 2017) uploaded from January 1, 2008 to November 1, 2017 using the search filter “breast cancer,” “Homo sapiens,” “RNA assay,” “array assay,” and “all assay.” We also conducted a search of a series matrix files in the GEO database (NCBI, 2017) uploaded from January 1, 2008 to November 1, 2017 using the search filter “breast cancer,” “Homo sapiens,” “series,” and “expression profiling by array.” A total of 207 and 227 expressions by array dataset were listed in the ArrayExpress and GEO databases, respectively. Relevant literatures were found in GEO database using the GSE ID.
This meta-analysis collects data from primary breast cancer patient’s tumor tissue to assess the relationship of RSK2 expression and the clinical parameters of breast cancer patients, such as clinicopathological features and prognostic factors. Inclusion criteria are as follows: a) patients in the study have not been selected, or the selection had no effect on clinical indicators given the RSK2 expression might be different among different subtypes of breast cancer and different clinical status of patients; b) the sample size in each file is greater than 100 and comes from the same study; c) sample comes from breast cancer patients’ primary tumor tissue or normal tissue; d) the expression of RSK2 was efficient and available from the series matrix file; and e) any of a patient’s clinicopathological features and prognostic factors can be extracted from the file. When samples in a file are duplicated with samples in another file, we selected the larger or qualified one. The exclusion criteria are as follows: a) samples were gathered from different studies; b) the original study could not be found; c) the sample size in the document is inconsistent with the number of patients in the study, and the replicated samples could not be found in the file; and d) the file was ineligible after invalid samples were removed. Files were filtered by two researchers separately; the disagreement was resolved through discussion. The eligible files are listed on Table 1.
For observational studies, the Newcastle-Ottawa Quality Assessment Scale (NOS) was employed for assessing the quality of these studies. All data was abstracted by using a standardized data collection form, with information recorded as follows: first author’s name, publication year, country of origin, number of cases and controls, detection method, GSE ID, and platform of detection. Each sample’s RSK2 expression and corresponding clinicopathological features and prognostic factors were extracted from the series matrix file, including age, tissue, ER status, progesterone receptor (PR) status, HER2 status, lymph node infiltration, histological grade, TNM stage, tumor size, tumor type, metastasis, intrinsic subtype (by PAM50), overall survival time (OS), disease-free survival time (DFS), relapse-free survival time (RFS), and relevant status of the patient. Samples with incomplete information or data described above were removed.
Data extraction is conducted by two researchers, respectively, and disagreements were resolved by discussion.
Statistical analysis was conducted by the guidelines proposed by the Meta-Analysis of Observational Studies group (Stroup et al., 2000). Median was used as the cutoff value to determine the level of RSK2 expression because there was no suitable cutoff value to help us distinguish the expression status of RSK2. Odds ratio (OR) was employed for evaluating the association between RSK2 expression and clinicopathological features. Hazard ratio (HR) and 95% credible interval (CI) were appraised to assess the association between RSK2 expression and prognostic indicators, including OS, DFS, and RFS by using IBM SPSS Statistics 24. Heterogeneity of the OR and HR was calculated by using the Cochran’s Q and I2 test. A random-effect model was applied when p < 0.1 or I2 > 50%. When heterogeneity was absent, a fixed-effect model was employed. Begg’s rank correlation method and Egger’s weighted regression methods were employed to assess publication bias. STATA software package (version 12.0) was used to calculate pooled ORs, HRs, and corresponding 95% CI; all p values were two tailed.
Given the limited prognostic information of specific breast cancer patients in those GSE files, we used the Kaplan–Meier Plotter (Hou et al., 2017; Kaplan-Meier plotter), an online database including gene expression data and clinical data, to assess the prognostic value of RSK2 in breast cancer. The patient samples were divided into two cohorts according to the median expression of the gene (high vs. low expression).
The flow diagram for the recognition of eligible studies is presented in Figure 1. There were 207 and 227 GSE files identified from the ArrayExpress database and GEO database, respectively. After duplicates were removed, RSK2 expression, abstract, and full text were checked, and 13 GSE files from 13 independent studies involving 3,122 patients were identified by our search strategy. The features of the 13 studies are listed in Table 1. Ineligible samples and data were removed, such as samples from cell, blood, or distant metastasis. When sample was detected multiple times, the one with the highest RSK2 expression was selected. Histological grades Ⅰ and Ⅱ were grouped as low-grade disease, and Ⅲ was grouped as high-grade disease. Clinical stages Ⅰ and Ⅱ were grouped as early-stage disease, and III and IV were grouped as late-stage disease. Tumors larger than 2 cm were grouped as large tumors, and the rest were grouped as small tumors. Patients were divided into high-age group and low-age group, with 55 years old as the cutoff value. Clinical stage of GSE20685 was not available, which we estimated depending on the T, N, and M stage shown in the GSE file using the NCCN guidelines of breast cancer (version 2. 2011). Data in GSE39004 were only used for comparing RSK2 expression between normal tissues and tumor tissues, because its tumor sample size is not large enough.
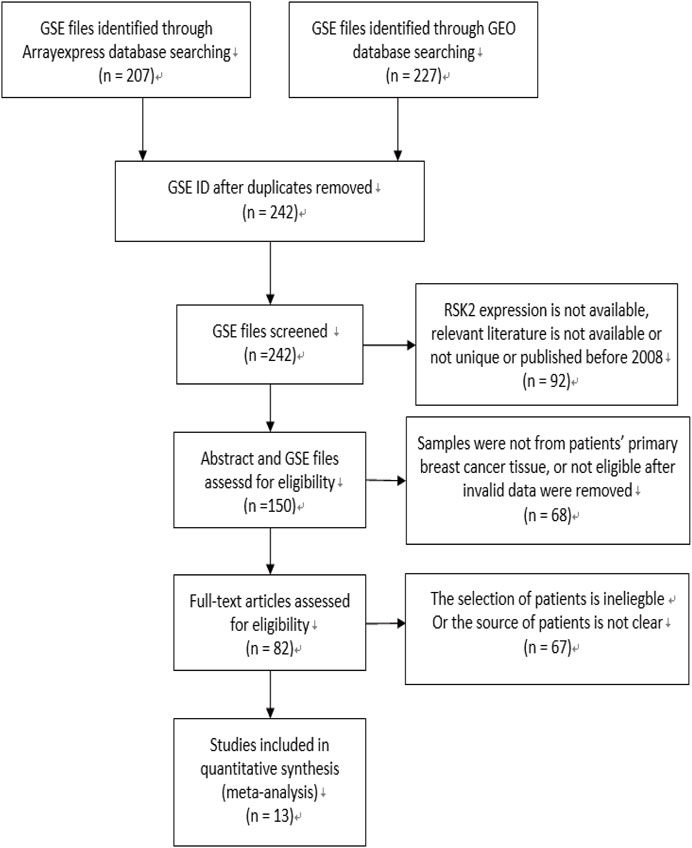
FIGURE 1. Flow diagram of literature selection. The search and selection process for the expression of RSK2 of breast cancer patients and the number of eligible studies.
Our meta-analysis demonstrated that RSK2 expression in breast cancer tissue was lower than that in normal tissue (pooled OR = 0.54, 95% CI: 0.44–0.67, Cochran’s Q test p = 0.14, I2 = 41.7%) (Figure 2A). There was no statistically significant difference between ductal carcinoma and lobular carcinoma (pooled OR = 0.75, 95% CI: 0.35–1.60, Cochran’s Q test p = 0.104, I2 = 51.3%) (Figure 2B). The relationship of RSK2 expression and molecular subtype was analyzed in our meta-analysis. The expression of RSK2 was obviously different between the luminal subtype and basal subtype of breast cancer (pooled OR = 0.25, 95% CI: 0.08–0.80, Cochran’s Q test p = 0.06, I2 = 63.5%) (Figure 2C), and no distinctive RSK2 expression was found between luminal A and luminal B subtypes of breast cancer (pooled OR = 0.73, 95% CI: 0.26–2.01, Cochran’s Q test p = 0.05, I2 = 66.5%) (Figure 2D).
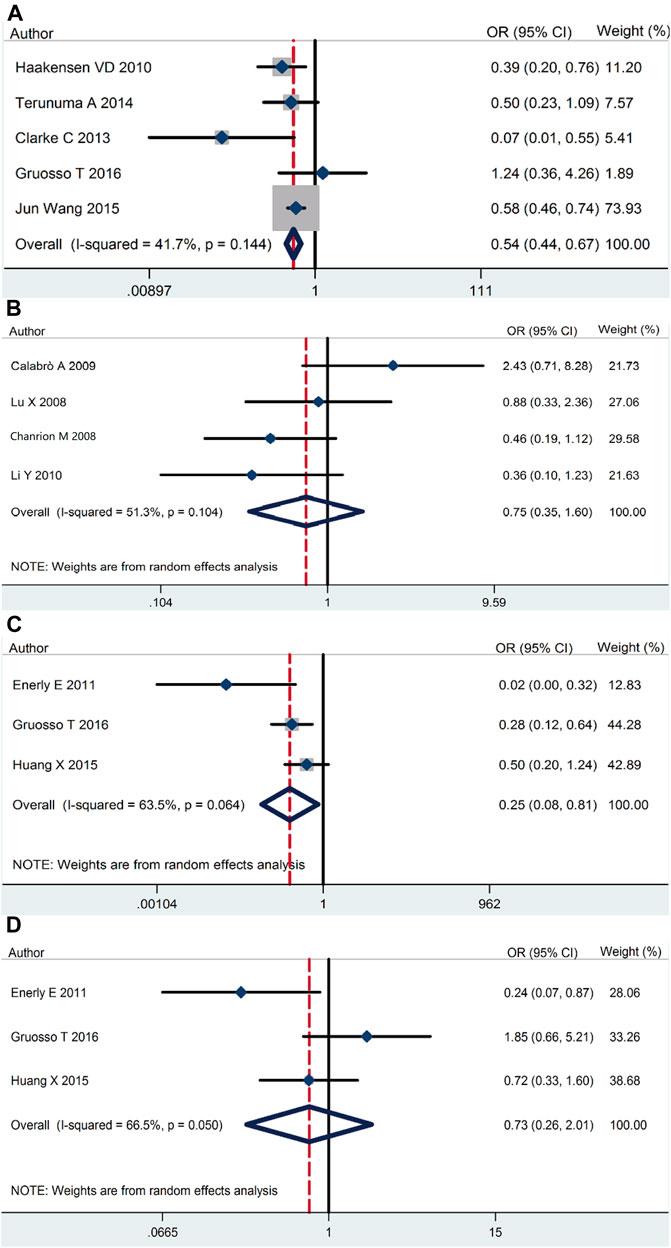
FIGURE 2. RSK2 expression among different types of breast tumor tissues as well as normal breast tissues. (A) RSK2 expression in breast cancer tissue compared with normal tissue. (B) The association between RSK2 expression and the ductal breast cancer relative to the lobular breast cancer. (C) RSK2 expression in luminal breast cancer compared with basal-like breast cancer. (D) RSK2 expression in luminal A breast cancer compared with luminal B breast cancer.
On the basic data obtained, we analyzed the relationship between RSK2 expression and the three main breast cancer biomarkers. RSK2 expression was inversely correlated with ER expression (pooled OR = 0.38, 95% CI: 0.25–0.58, Cochran’s Q test p = 0.009, I2 = 67.7%) (Figure 3A). There was no statistically significant relationship between PR expression (pooled OR = 0.83, 95% CI: 0.57–1.22, Cochran’s Q test p = 0.36, I2 = 1.3%) (Figure 3B) and HER2 expression (pooled OR = 0.91, 95% CI: 0.66–1.25, Cochran’s Q test p = 0.14, I2 = 39.2%) (Figure 3C).
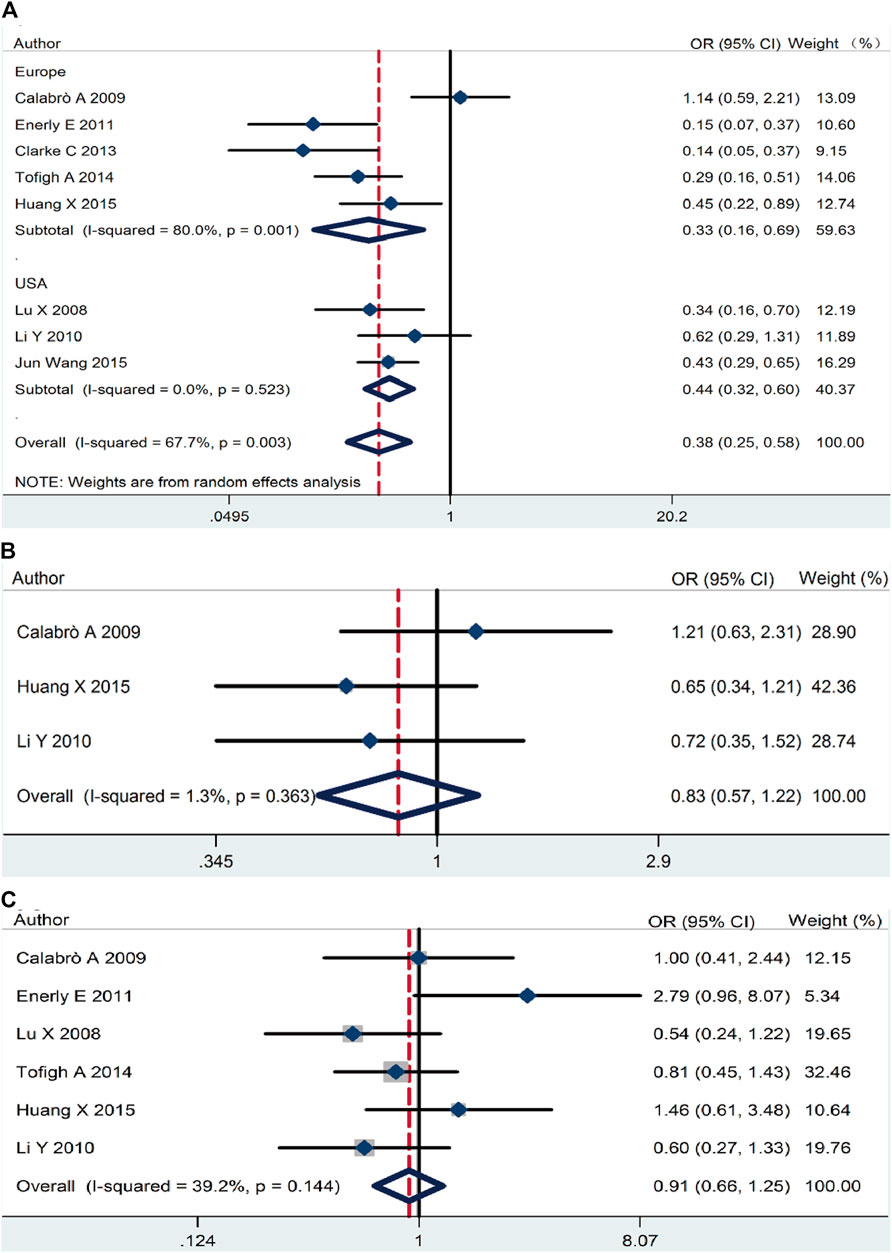
FIGURE 3. The association between RSK2 expression and the three main breast cancer biomarkers. (A) The association between RSK2 expression and estrogen receptor (ER) status. (B) The association between RSK2 expression and progesterone receptor (PR) status. (C) The association between RSK2 expression and HER2 status.
The association of RSK2 expression with breast cancer clinicopathological features was also analyzed. High RSK2 expression reduced the possibility of distant metastasis (pooled OR = 0.59, 95% CI: 0.41–0.87, Cochran’s Q test p = 0.88, I2 = 0.0%) (Figure 4A) and lymph node metastasis (pooled OR = 0.81, 95% CI: 0.65–0.998, Cochran’s Q test p = 0.09, I2 = 42.8%) (Figure 4B). The overexpression of RSK2 positively correlated with histological grade (pooled OR = 1.33, 95% CI: 1.03–1.72, Cochran’s Q test p = 0.69, I2 = 0.0%) (Figure 4C). However, RSK2 expression has no statistically significant relationship with other biological characters of breast cancer, including tumor size (pooled OR = 0.995, 95% CI: 0.77–1.28, Cochran’s Q test p = 0.43, I2 = 0.0%) (Figure 4D) and clinical stage (pooled OR = 1.09, 95% CI: 0.71–1.67, Cochran’s Q test p = 0.87, I2 = 0.0%) (Figure 4E).
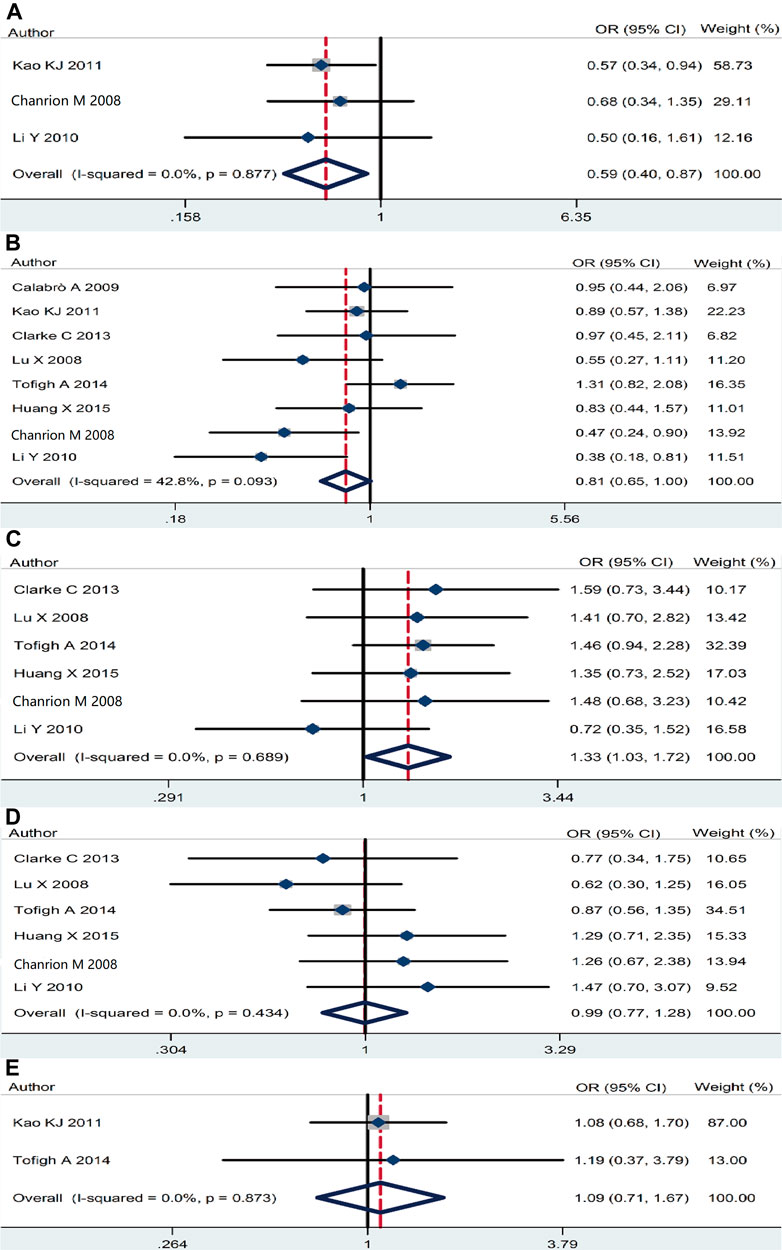
FIGURE 4. The association between RSK2 expression and other clinicopathological characters. (A) The association between RSK2 expression and distant metastasis. (B) The association between RSK2 expression and lymph node infiltration. (C) The association between RSK2 expression and histological grade. (D) The association between RSK2 expression and tumor size. (E) The association between RSK2 expression and clinical stage of breast cancer.
We evaluated the association between RSK2 expression level and survival outcome of breast cancer patients. The results indicate that RSK2 overexpression was statistically associated with the OS rate of breast cancer patients (pooled HR = 0.71, 95% CI: 0.48–0.94, Cochran’s Q test p = 0.95, I2 = 0.0%) (Figure 5A), while there was no significant relationship between RSK2 expression and DFS (pooled HR = 0.96, 95% CI: 0.63–1.29, Cochran’s Q test p = 0.94, I2 = 0.0%) (Figure 5B). Only one GSE file (GSE42568) involving 104 patients has the data of RFS, and there was no statistical significance between them (HR = 0.62, 95% CI: 0.32–1.22, log rank test p = 0.21) (Figure 5C). We did not find a significant difference in survival outcome between basal-like breast cancer and luminal breast cancer for the limited sample capacity and accessible data.

FIGURE 5. The association between RSK2 expression and survival outcome of breast cancer patients. (A) The association between RSK2 expression and breast cancer overall survival (OS). (B) The association between RSK2 expression and breast cancer disease-free survival (DFS). (C) The association between RSK2 expression and age of breast cancer patients.
We also recorded the corresponding age of every sample and grouped them into low-age group and high-age group with 55 years old as the cutoff value, which was randomly selected. Interestingly, we found that RSK2 expression is lower in the high-age group (pooled OR = 0.73, 95% CI: 0.60–0.89, Cochran’s Q test p = 0.45, I2 = 0.0%) (Figure 5D). In order to investigate whether RSK2 expression decreased with age, we calculated the relationship between RSK2 expression and age involving 508 normal tissue samples in GSE93601, and no statistically significant correlation was found (OR = 0.95, 95% CI: 0.66–1.36). The status of p53 was available in GSE19783, which involves 110 patients, indicating there was a positive relationship between RSK2 and P53 mutation (OR = 4.09, 95% CI: 1.74–9.22, Cochran’s Q test p = 0.0007).
The online database Kaplan–Meier was employed to evaluate the impact of RSK2 expression on the prognostic outcome in different molecular subtypes of breast cancer, indicating that elevated RSK2 expression predicts a favorable OS (luminal A breast cancer: HR = 1.04, 95% CI: 0.74–1.48, log rank p = 0.81; luminal B breast cancer: HR = 0.67, 95% CI: 0.46–0.97, log rank p = 0.034; basal-like breast cancer: HR = 0.48, 95% CI: 0.28–0.82, log rank p = 0.006; HER2+ breast cancer: HR = 0.7, 95% CI: 0.37–1.35, log rank p = 0.29) (Figure 6) and distant metastasis-free survival (DMFS) (luminal A breast cancer: HR = 0.96, 95% CI: 0.72–1.28, log rank p = 0.78; luminal B breast cancer: HR = 0.63, 95% CI: 0.44–0.9, log rank p = 0.009; basal-like breast cancer: HR = 0.54, 95% CI: 0.32–0.92, log rank p = 0.021; HER2+ breast cancer: HR = 1, 95% CI: 0.54–1.87, log rank p = 0.99) (Figure 7) in basal-like and luminal B breast cancer, but not in luminal A and HER2+ breast cancer. The overexpression of RSK2 predicts a favorable prognostic value of RFS (Figure 8) in all those subtypes of breast cancer (luminal A breast cancer: HR = 0.78, 95% CI: 0.65–0.92, log rank p = 0.004; luminal B breast cancer: HR = 0.68, 95% CI: 0.56–0.82, log rank p < 0.001; basal-like breast cancer: HR = 0.67, 95% CI: 0.52–0.87, log rank p = 0.002; HER2+ breast cancer: HR = 0.51, 95% CI: 0.35–0.76, log rank p < 0.001).
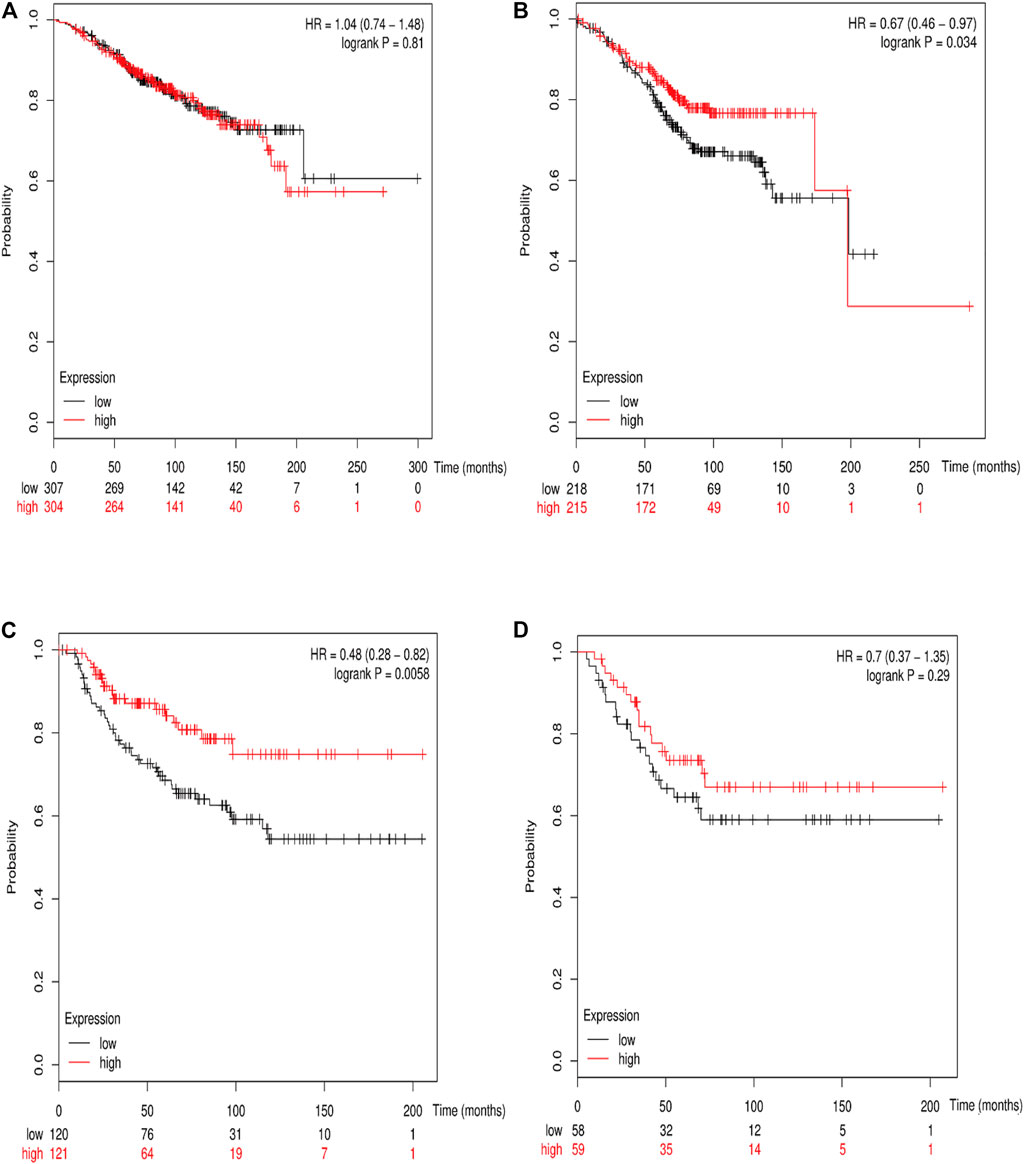
FIGURE 6. The association between RSK2 expression and OS in different molecular subtypes of breast cancer. (A) The association between RSK2 expression and OS in luminal A breast cancer. (B) The association between RSK2 expression and OS in luminal B breast cancer. (C) The association between RSK2 expression and OS in basal-like breast cancer. (D) The association between RSK2 expression and OS in HER2+ breast cancer.
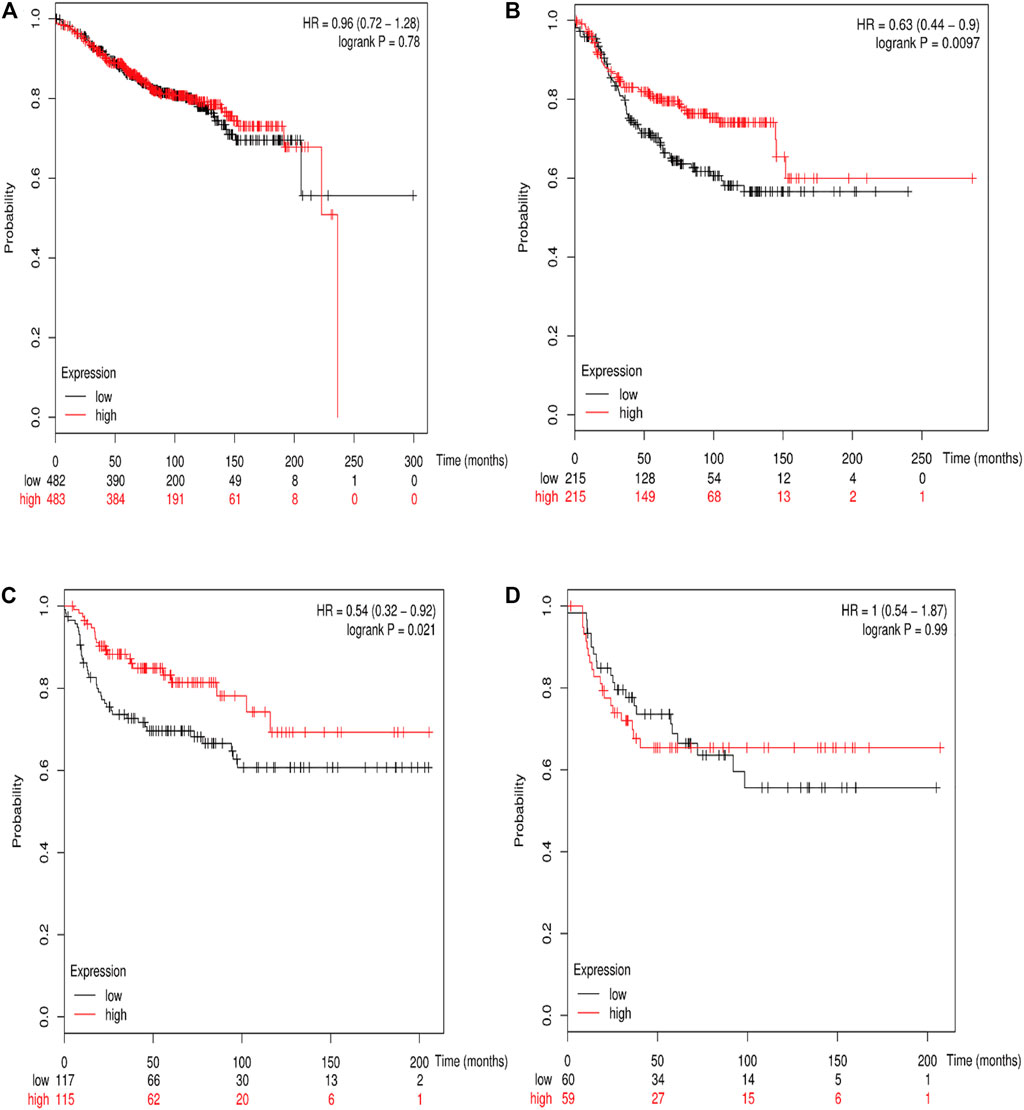
FIGURE 7. The association between RSK2 expression and distant metastasis-free survival (DMFS) in different molecular subtypes of breast cancer. (A) The association between RSK2 expression and DMFS in luminal A breast cancer. (B) The association between RSK2 expression and DMFS in luminal B breast cancer. (C) The association between RSK2 expression and DMFS in basal-like breast cancer. (D) The association between RSK2 expression and DMFS in HER2+ breast cancer.
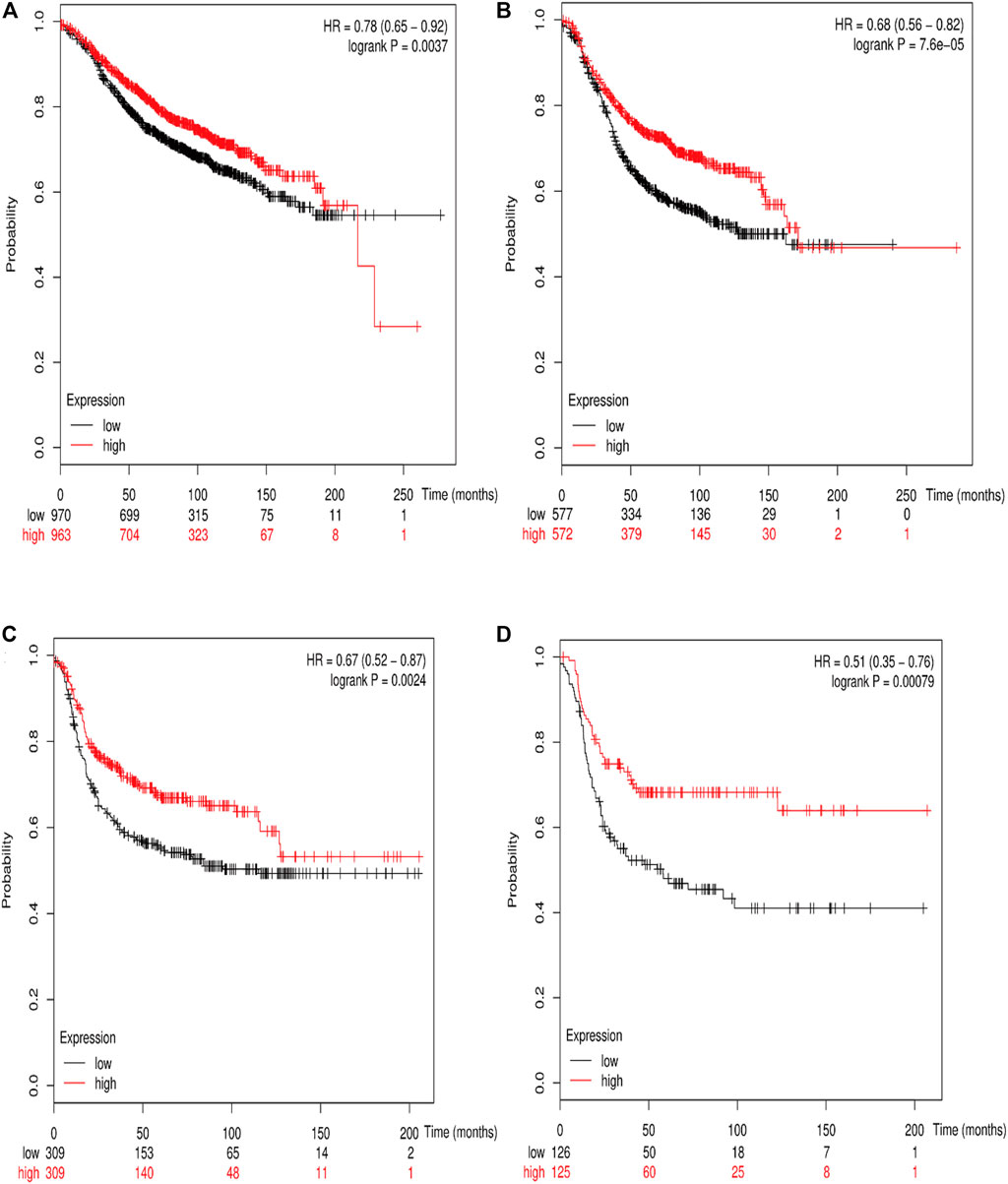
FIGURE 8. The association between RSK2 expression and relapse-free survival (RFS) in different molecular subtypes of breast cancer. (A) The association between RSK2 expression and RFS in luminal A breast cancer. (B) The association between RSK2 expression and RFS in luminal B breast cancer. (C) The association between RSK2 expression and RFS in basal-like breast cancer. (D) The association between RSK2 expression and RFS in HER2+ breast cancer.
The sample capacity is very large in GSE93601, which may have a great impact on the statistical results. We reanalyzed the data after removing the data from GSE93061 and got the same result as the previous one.
Publication bias statistics were obtained using the Begg’s test and Egger’s test, which did not indicate any significant publication bias (Table 2).
RSK2 is an X-linked dominant gene and acts as a modulator of craniofacial development, and the mutation of RSK2 was responsible for Coffin–Lowry syndrome (Laugel-Haushalter et al., 2014). It is generally believed that RSK2 plays an important role in the tumorigenesis, migration, invasion, cell proliferation, and response to stress (Sulzmaier and Ramos, 2013; Laugel-Haushalter et al., 2014; Alesi et al., 2016). Precisely measuring the prognostic value of RSK2 may help to guide individual therapies for breast cancer patients. Our meta-analysis takes advantage of a public electronic database to evaluate the association between the abundance of RSK2 mRNA and the clinical parameters of breast cancer patients for the first time. Although it is not possible to draw conclusions about causality, these findings suggest that RSK2 is a potential biomarker in breast cancer, especially in a specific subtype of breast cancer, and might provide new perspective of the interaction between RSK2 and breast cancer.
From our research, RSK2 expression was overexpressed in basal-like breast cancer and higher histological grade breast cancer and negatively correlated with estrogen receptor. These results corresponded with previous studies that suggest that RSK2 expression is highest in basal-like breast cancer and those with the highest histological grade (Stratford et al., 2012; Zhao et al., 2016). A protein downstream of RSK2, namely YB-1, transforms human mammary epithelial cells through chromatin remodeling leading to the development of basal-like breast cancer (Davies et al., 2014). Inactivating YB-1 can depress tumor-initiating cells of basal-like breast cancer. A study suggested that ER-α physically interacts with RKS2, resulting in the accumulation of RSK2 in nuclear sequestration, and RSK2 can promote neoplastic transformation and facilitate metastatic tumor growth of ER+ breast cancer (Ludwik et al., 2018), but there was no explanation for RSK2 expression negatively correlating with ER status. However, there is no obvious statistical significance between RSK2 expression and progesterone receptor on the basic data. Luminal A breast cancer is an ER-positive breast cancer with a lower histological grade, while luminal B breast cancer is an ER-positive breast cancer with a higher histological grade. Although RSK2 is overexpressed in higher histological grade breast tumor, there is no obvious distinction between luminal A and luminal B breast cancer from our data. It may be partly due to the limited sample size of luminal A and luminal B breast cancer, and the distinction of RSK2 expression was not large enough between them.
There were some unexpected results based on our data. It is generally believed that the expression of RSK2 in cancer tissue is higher than that in normal tissue, and reducing the expression of RSK2 can prevent tumorigenesis, tumor cell growth, and ability of migration and invasion (Lee et al., 2013; Yao et al., 2014; Mao et al., 2016; Zhao et al., 2016). However, based on our data, the contrary result was obtained. Furthermore, no evidence indicates that the immunohistochemistry outcome of RSK2 was different from that of mRNA microarray assay. The overexpression of RSK2 alone or RSK2 combined with other biomarkers indicates a poor prognostic outcome was reported. For example, it has been shown that targeting RSK2 with specific inhibitors or small interfering RNAs remarkably inhibits the growth and renewal of tumor-initiating cells in triple-negative breast cancer (TNBC) and that RSK2 promotes migration through the ERK/MEK pathway (Stratford et al., 2012). In addition, Czaplinska et al. found that fibroblast growth factor receptor 2 (FGFR2) can form an indirect complex with RSK2, which may be involved in the progression of breast cancer and lead to poor prognosis in breast cancer patients (Czaplinska et al., 2016).
Based on the data collected from microarray, the OS of breast cancer patients is higher in RSK2 high-expression patients than that in RSK2 low-expression patients, and with the increase of RSK2 expression, the potential of distant metastasis and lymph node infiltration decreased. Moreover, it is strange that the expression of RSK2 is highest in basal-like breast cancer (TNBC), which is defined by the absence of the three main breast cancer biomarkers—i.e., a lack of expression of ER and PR and a lack of amplification or overexpression of HER2—and cooperates with poor prognosis and high risk of distant metastasis (Carey et al., 2010), but the OS, the potential of distant metastasis, and the lymph node metastases were more favorable in the RSK2 high-expression group in the basic data from microarray. No relevant study was available to help us understand the mechanism under the paradoxical phenomenon. We hypothesize that RSK2 plays a different role in different subtypes of breast cancer. We did not find a significant difference in survival outcomes between basal-like breast cancer and luminal breast cancer for the limited sample capacity and accessible data. In reference to the result from the online Kaplan–Meier Plotter, the overexpression of RSK2 predicts more favorable prognostic value of RFS in all subtypes of breast cancer. As for the OS and DMFS, only basal-like and luminal B breast cancer patients were able to benefit from RSK2 overexpression.
We found that RSK2 expression is negatively correlated with the age of breast cancer patients for the first time, but there is no such relationship in normal tissue. There is also no statistically significant difference in RSK2 expression between the early-stage group and late-stage group of breast cancer patients. It was reported that RSK2 is sequestered in stress granules, which can aid cell survival in response to environmental stress by acting as sites of translational repression, and facilitates stress granule assembly to repress translation and to enhance cell survival (Eisinger-Mathason et al., 2008). The body’s response to stress decreases with age and may provide a possible explanation for the phenomenon.
Heterogeneity tests are an essential part of a meta-analysis. In this study, minor heterogeneities were observed with respect to OS, DFS, tumor size, clinical stage, and histological grade; however, there were substantial heterogeneities with respect to ER status, HER2 status, lymph node infiltration, and different subtypes of breast cancer. This unbalanced phenomenon could partly result from the detection method and accuracy of ER status, PR status, and HER2 status being different from each other, and the data completeness obtained from GSE files was not identical. Three GSE files have efficient PR status, and the heterogeneity was not obvious among them, while the other three GSE files have identified the molecular subtype of breast cancer, which shows a significant heterogeneity. Patients from different areas may respond to the heterogeneities. There were no heterogeneities in United States patients, while the main heterogeneity of ER status and HER2 status originates from different European countries when we conducted a subgroup analysis. Another significant heterogeneity was likely due to the detection platform. Publication bias is worth considering in a meta-analysis. In this study, there was no significant publication bias based on the Egger’s and Begg’s test.
There are still some limitations in this meta-analysis. First of all, the relevant studies and complete available data were limited, and the available clinical parameter is not homogenous among those matrix files. Secondly, the detection platform, method, and accuracy of hormone receptors are different among these studies. Thirdly, the therapy level and method are distinctive, and we cannot eliminate their effect. Lastly, we cannot ignore the publication bias. Some data are still unavailable.
For further verification, we could download the mRNA expression data and corresponding clinical information of breast cancer patients from other databases as a validation cohort to verify the relationship between the RSK2 expression level and the clinicopathological features as well as the prognosis of patients. In terms of experimental validation, future researchers could modify the expression of RSK2 in different breast cancer cell lines and then perform various in vitro and mice xenografts in vivo trials to observe the effects of altering RSK2 expression on the proliferation, apoptosis, cell cycle, metastasis, and invasion capabilities of breast cancer cells. Furthermore, the prognostic significance of RSK2 could also be verified by measuring the protein expression level of RSK2 by flow cytometry, western blotting, and immunohistochemistry staining on tumor and paracancerous normal tissues of breast cancer patients in conjunction with clinical information analysis. The strategies above could help to confirm the reliability of our meta-analysis findings based on RSK2 mRNA expression and prognosis.
In conclusion, our meta-analysis was the first study that used microarray assay to research the association between RSK2 expression and clinicopathological features and prognostic factors of primary breast cancer patients. Although some results corresponded with previous studies and some results were opposite to previous studies, both of them indicated RSK2 is a promising biomarker of breast cancer. This study provides a new research direction and area of RSK2, while more experimental studies and elaborate research are needed to uncover the sealed mechanism of RSK2 in breast cancer.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
YZ and HX contributed to conception and design of the study. KZ, SY, WY, and QL organized the database. YW and LZ performed the statistical analysis. KZ and SY wrote the first draft of the manuscript. XC, HX, XY, and YW wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (grant number 81772827).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer (FZ) declared a shared parent affiliation, with several of the authors (KZ, SY, WY, QL, YW, LZ, XC, HX, XY, YZ, and HX), to the handling editor at the time of the review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CI, credible interval; DFS, disease-free survival time; DMFS, distant metastasis-free survival; ER, estrogen receptor; FGFR2, fibroblast growth factor receptor 2; GEO, gene expression omnibus; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; MAPK, mitogen-activated protein kinase; NOS, newcastle-ottawa quality assessment scale; OR, odds ratio; OS, overall survival; PR, progesterone receptor; RFS, relapse-free survival time; RSK2/RPS6KA3, ribosomal protein S6 kinases, 90 kDa, polypeptide 3; TNBC, triple-negative breast cancer; YB-1, Y-box binding protein-1.
Alesi, G. N., Jin, L., Li, D., Magliocca, K. R., Kang, Y., Chen, Z. G., et al. (2016). RSK2 Signals through Stathmin to Promote Microtubule Dynamics and Tumor Metastasis. Oncogene 35 (41), 5412–5421. doi:10.1038/onc.2016.79
ArrayExpress (2017). ArrayExpress. Available at: https://www.ebi.ac.uk/arrayexpress/ArrayExpress (Accessed November 1, 2017).
Arul, N., and Cho, Y.-Y. (2013). A Rising Cancer Prevention Target of RSK2 in Human Skin Cancer. Front. Oncol. 3, 201. doi:10.3389/fonc.2013.00201
Calabrò, A., Beissbarth, T., Kuner, R., Stojanov, M., Benner, A., Asslaber, M., et al. (2009). Effects of Infiltrating Lymphocytes and Estrogen Receptor on Gene Expression and Prognosis in Breast Cancer. Breast Cancer Res. Treat. 116 (1), 69–77. doi:10.1007/s10549-008-0105-3
Carey, L., Winer, E., Viale, G., Cameron, D., and Gianni, L. (2010). Triple-negative Breast Cancer: Disease Entity or Title of Convenience. Nat. Rev. Clin. Oncol. 7 (12), 683–692. doi:10.1038/nrclinonc.2010.154
Chanrion, M., Negre, V., Fontaine, H., Salvetat, N., Bibeau, F., Grogan, G. M., et al. (2008). A Gene Expression Signature that Can Predict the Recurrence of Tamoxifen-Treated Primary Breast Cancer. Clin. Cancer Res. 14 (6), 1744–1752. doi:10.1158/1078-0432.CCR-07-1833
Clark, D. E., Poteet-Smith, C. E., Smith, J. A., and Lannigan, D. A. (2001). Rsk2 Allosterically Activates Estrogen Receptor Alpha by Docking to the Hormone-Binding Domain. EMBO J. 20 (13), 3484–3494. doi:10.1093/emboj/20.13.3484
Clarke, C., Madden, S. F., Doolan, P., Aherne, S. T., Joyce, H., O’Driscoll, L., et al. (2013). Correlating Transcriptional Networks to Breast Cancer Survival: a Large-Scale Coexpression Analysis. Carcinogenesis 34 (10), 2300–2308. doi:10.1093/carcin/bgt208
Clough, E., and Barrett, T. (2016). The Gene Expression Omnibus Database. Methods Mol. Biol. 1418, 93–110. doi:10.1007/978-1-4939-3578-9_5
Czaplinska, D., Mieczkowski, K., Supernat, A., Skladanowski, A. C., Kordek, R., Biernat, W., et al. (2016). Interactions between FGFR2 and RSK2-Implications for Breast Cancer Prognosis. Tumor Biol. 37 (10), 13721–13731. doi:10.1007/s13277-016-5266-9
Davies, A. H., Reipas, K. M., Pambid, M. R., Berns, R., Stratford, A. L., Fotovati, A., et al. (2014). YB-1 Transforms Human Mammary Epithelial Cells through Chromatin Remodeling Leading to the Development of Basal-like Breast Cancer. Stem Cells 32 (6), 1437–1450. doi:10.1002/stem.1707
Eisinger-Mathason, T. S. K., Andrade, J., Groehler, A. L., Clark, D. E., Muratore-Schroeder, T. L., Pasic, L., et al. (2008). Codependent Functions of RSK2 and the Apoptosis-Promoting Factor TIA-1 in Stress Granule Assembly and Cell Survival. Mol. Cel. 31 (5), 722–736. doi:10.1016/j.molcel.2008.06.025
Enerly, E., Steinfeld, I., Kleivi, K., Leivonen, S.-K., Aure, M. R., Russnes, H. G., et al. (2011). miRNA-mRNA Integrated Analysis Reveals Roles for miRNAs in Primary Breast Tumors. PLoS One 6 (2), e16915. doi:10.1371/journal.pone.0016915
Frödin, M., and Gammeltoft, S. (1999). Role and Regulation of 90 kDa Ribosomal S6 Kinase (RSK) in Signal Transduction. Mol. Cel Endocrinol. 151 (1-2), 65–77. doi:10.1016/s0303-7207(99)00061-1
Gruosso, T., Mieulet, V., Cardon, M., Bourachot, B., Kieffer, Y., Devun, F., et al. (2016). Chronic Oxidative Stress Promotes H2 AX Protein Degradation and Enhances Chemosensitivity in Breast Cancer Patients. EMBO Mol. Med. 8 (5), 527–549. doi:10.15252/emmm.201505891
Guo, Z.-F., and Kong, F.-L. (2021). Akt Regulates RSK2 to Alter Phosphorylation Level of H2A.X in Breast Cancer. Oncol. Lett. 21 (3), 187. doi:10.3892/ol.2021.12448
Haakensen, V. D., Biong, M., Lingjærde, O. C., Holmen, M. M., Frantzen, J. O., Chen, Y., et al. (2010). Expression Levels of Uridine 5'-Diphospho-Glucuronosyltransferase Genes in Breast Tissue from Healthy Women Are Associated with Mammographic Density. Breast Cancer Res. 12 (4), R65. doi:10.1186/bcr2632
Hou, G.-X., Liu, P., Yang, J., and Wen, S. (2017). Mining Expression and Prognosis of Topoisomerase Isoforms in Non-small-cell Lung Cancer by Using Oncomine and Kaplan-Meier Plotter. PLoS One 12 (3), e0174515. doi:10.1371/journal.pone.0174515
Huang, X., Dugo, M., Callari, M., Sandri, M., De Cecco, L., Valeri, B., et al. (2015). Molecular Portrait of Breast Cancer in China Reveals Comprehensive Transcriptomic Likeness to Caucasian Breast Cancer and Low Prevalence of Luminal A Subtype. Cancer Med. 4 (7), 1016–1030. doi:10.1002/cam4.442
Kao, K.-J., Chang, K.-M., Hsu, H.-C., and Huang, A. T. (2011). Correlation of Microarray-Based Breast Cancer Molecular Subtypes and Clinical Outcomes: Implications for Treatment Optimization. BMC Cancer 11, 143. doi:10.1186/1471-2407-11-143
Kaplan-Meier plotter Kaplan-meier Plotter. Available at: http://kmplot.com/private/ (Accessed November 1, 2017).
Laugel-Haushalter, V., Paschaki, M., Marangoni, P., Pilgram, C., Langer, A., Kuntz, T., et al. (2014). RSK2 Is a Modulator of Craniofacial Development. PLoS One 9 (1), e84343. doi:10.1371/journal.pone.0084343
Lee, C.-J., Lee, M.-H., Yoo, S.-M., Choi, K.-I., Song, J.-H., Jang, J.-H., et al. (2015). Magnolin Inhibits Cell Migration and Invasion by Targeting the ERKs/RSK2 Signaling Pathway. BMC Cancer 15, 576. doi:10.1186/s12885-015-1580-7
Lee, M.-H., Huang, Z., Kim, D. J., Kim, S.-H., Kim, M. O., Lee, S.-Y., et al. (2013). Direct Targeting of MEK1/2 and RSK2 by Silybin Induces Cell-Cycle Arrest and Inhibits Melanoma Cell Growth. Cancer Prev. Res. 6 (5), 455–465. doi:10.1158/1940-6207.CAPR-12-0425
Li, Y., Zou, L., Li, Q., Haibe-Kains, B., Tian, R., Li, Y., et al. (2010). Amplification of LAPTM4B and YWHAZ Contributes to Chemotherapy Resistance and Recurrence of Breast Cancer. Nat. Med. 16 (2), 214–218. doi:10.1038/nm.2090
Lu, X., Lu, X., Wang, Z. C., Iglehart, J. D., Zhang, X., and Richardson, A. L. (2008). Predicting Features of Breast Cancer with Gene Expression Patterns. Breast Cancer Res. Treat. 108 (2), 191–201. doi:10.1007/s10549-007-9596-6
Ludwik, K. A., McDonald, O. G., Brenin, D. R., and Lannigan, D. A. (2018). ERα-Mediated Nuclear Sequestration of RSK2 Is Required for ER+ Breast Cancer Tumorigenesis. Cancer Res. 78 (8), 2014–2025. doi:10.1158/0008-5472.CAN-17-2063
Mao, L., Summers, W., Xiang, S., Yuan, L., Dauchy, R. T., Reynolds, A., et al. (2016). Melatonin Represses Metastasis in Her2-Postive Human Breast Cancer Cells by Suppressing RSK2 Expression. Mol. Cancer Res. 14 (11), 1159–1169. doi:10.1158/1541-7786.MCR-16-0158
NCBI (2017). Gene Expression Omnibus. Available at: https://www.ncbi.nlm.nih.gov/gds (Accessed November 1, 2017).
Nielsen, T. O., Jensen, M.-B., Burugu, S., Gao, D., Jørgensen, C. L. T., Balslev, E., et al. (2017). High-Risk Premenopausal Luminal A Breast Cancer Patients Derive No Benefit from Adjuvant Cyclophosphamide-Based Chemotherapy: Results from the DBCG77B Clinical Trial. Clin. Cancer Res. 23 (4), 946–953. doi:10.1158/1078-0432.CCR-16-1278
Parkinson, H., Kapushesky, M., Shojatalab, M., Abeygunawardena, N., Coulson, R., Farne, A., et al. (2007). ArrayExpress--a Public Database of Microarray Experiments and Gene Expression Profiles. Nucleic Acids Res. 35 (Database issue), D747–D750. doi:10.1093/nar/gkl995
Perou, C. M., Sørlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., et al. (2000). Molecular Portraits of Human Breast Tumours. Nature 406 (6797), 747–752. doi:10.1038/35021093
Prat, A., Fan, C., Fernández, A., Hoadley, K. A., Martinello, R., Vidal, M., et al. (2015). Response and Survival of Breast Cancer Intrinsic Subtypes Following Multi-Agent Neoadjuvant Chemotherapy. BMC Med. 13, 303. doi:10.1186/s12916-015-0540-z
Siegel, R. L., Miller, K. D., and Jemal, A. (2018). Cancer Statistics, 2018. CA: A Cancer J. Clinic. 68 (1), 7–30. doi:10.3322/caac.21442
Stratford, A. L., Reipas, K., Hu, K., Fotovati, A., Brough, R., Frankum, J., et al. (2012). Targeting P90 Ribosomal S6 Kinase Eliminates Tumor-Initiating Cells by Inactivating Y-Box Binding Protein-1 in Triple-Negative Breast Cancers. Stem Cells 30 (7), 1338–1348. doi:10.1002/stem.1128
Stroup, D. F., Berlin, J. A., Morton, S. C., Olkin, I., Williamson, G. D., Rennie, D., et al. (2000). Meta-analysis of Observational Studies in EpidemiologyA Proposal for Reporting. JAMA 283 (15), 2008–2012. doi:10.1001/jama.283.15.2008
Sulzmaier, F. J., and Ramos, J. W. (2013). RSK Isoforms in Cancer Cell Invasion and Metastasis. Cancer Res. 73 (20), 6099–6105. doi:10.1158/0008-5472.CAN-13-1087
Terunuma, A., Putluri, N., Mishra, P., Mathé, E. A., Dorsey, T. H., Yi, M., et al. (2014). MYC-driven Accumulation of 2-hydroxyglutarate Is Associated with Breast Cancer Prognosis. J. Clin. Invest. 124 (1), 398–412. doi:10.1172/JCI71180
Tofigh, A., Suderman, M., Paquet, E. R., Livingstone, J., Bertos, N., Saleh, S. M., et al. (2014). The Prognostic Ease and Difficulty of Invasive Breast Carcinoma. Cel Rep. 9 (1), 129–142. doi:10.1016/j.celrep.2014.08.073
Wang, J., Zhang, X., Beck, A. H., Collins, L. C., Chen, W. Y., Tamimi, R. M., et al. (2015). Alcohol Consumption and Risk of Breast Cancer by Tumor Receptor Expression. Horm. Canc 6 (5-6), 237–246. doi:10.1007/s12672-015-0235-0
Yao, K., Chen, H., Liu, K., Langfald, A., Yang, G., Zhang, Y., et al. (2014). Kaempferol Targets RSK2 and MSK1 to Suppress UV Radiation-Induced Skin Cancer. Cancer Prev. Res. 7 (9), 958–967. doi:10.1158/1940-6207.CAPR-14-0126
Yoo, S.-M., Lee, C.-J., An, H.-J., Lee, J. Y., Lee, H. S., Kang, H. C., et al. (2019). RSK2-Mediated ELK3 Activation Enhances Cell Transformation and Breast Cancer Cell Growth by Regulation of C-Fos Promoter Activity. Int. J. Mol. Sci. 20 (8), 1994. doi:10.3390/ijms20081994
Keywords: ribosomal protein S6 kinase, 90kDa, polypeptide 3 (RSK2), breast cancer, molecular subtype, microarray, prognostic value, biomarkers
Citation: Zheng K, Yao S, Yao W, Li Q, Wang Y, Zhang L, Chen X, Xiong H, Yuan X, Wang Y, Zou Y and Xiong H (2021) Association Between RSK2 and Clinical Indexes of Primary Breast Cancer: A Meta-Analysis Based on mRNA Microarray Data. Front. Genet. 12:770134. doi: 10.3389/fgene.2021.770134
Received: 03 September 2021; Accepted: 30 September 2021;
Published: 01 November 2021.
Edited by:
Jialiang Yang, Geneis (Beijing) Co. Ltd., ChinaReviewed by:
Feng Zhu, Affiliated Hospital of Guilin Medical University, ChinaCopyright © 2021 Zheng, Yao, Yao, Li, Wang, Zhang, Chen, Xiong, Yuan, Wang, Zou and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanmei Zou, d2h0anp5bUB0amgudGptdS5lZHUuY24=; Hua Xiong, Y25oeGlvbmdAdGpoLnRqbXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.