- 1Tobacco Research Institute of Technology Centre, China Tobacco Hunan Industrial Corporation, Changsha, China
- 2State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
- 3MOA Key Laboratory of Plant Nutrition and Fertilization in Low-Middle Reaches of the Yangtze River, Nanjing Agricultural University, Nanjing, China
- 4Biological Science Research Center, Southwest University, Chongqing, China
The gibberellic acid stimulated Arabidopsis (GASA) gene family is critical for plant growth, development, and stress response. GASA gene family has been studied in various plant species, however, the GASA gene family in tobacco (Nicotiana tabacum) have not been characterized in detail. In this study, we identified 18 GASA genes in the tobacco genome, which were distributed to 13 chromosomes. All the proteins contained a conserved GASA domain and highly specific 12-cysteine residues at the C-terminus. Phylogenetic analysis divided the NtGASA genes into three well-conserved subfamilies. Synteny analysis suggested that tandem and segmental duplications played an important role in the expansion of the NtGASA gene family. Cis-elements analysis showed that NtGASA genes might influence different phytohormone and stress responses. Tissue expression analysis revealed that NtGASA genes displayed unique or distinct expression patterns in different tissues, suggesting their potential roles in plant growth and development. We also found that the expression of NtGASA genes were mostly regulated by abscisic and gibberellic acid, signifying their roles in the two phytohormone signaling pathways. Overall, these findings improve our understanding of NtGASA genes and provided useful information for further studies on their molecular functions.
Introduction
The gibberellic acid stimulated Arabidopsis (GASA) gene family is widespread in monocotyledonous and dicotyledonous plant species (Nahirñak et al., 2012). It encodes a class of cysteine-rich peptides characterized by a signaling amino acid region at the N-terminus and a conserved domain with 12 cysteines at the C-terminus (Silverstein et al., 2007). Previous studies indicated that peptides with a mutated or missing GASA domain are non-functional (Sun et al., 2013).
The GAST1 gene, which was first identified in tomato and characterized as a gibberellic acid (GA)-deficient (gib1) mutant gene (Shi et al., 1992). Subsequently, many GASA homologs were identified in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), wheat (Triticum aestivum), grapevine (Vitis vinifera L.), and tomato (Solanum lycopersicum) (Taylor and Scheuring, 1994; Aubert et al., 1998; Furukawa et al., 2006; Zhang et al., 2017; Ahmad et al., 2020). GASA gene family play important roles in plant growth and development. In Arabidopsis, AtGASA4 is involved in light signaling and promotes floral development, whereas overexpression of AtGASA5 delays flowering by downregulating the expression of LFY and FT and upregulating the expression of FLC (Zhang et al., 2009). In petunia, GASA are involved in floral transition and shoot elongation (Ben-Nissan et al., 2004).
Most GASA genes are involved in GA signaling pathways. In soybean (Glycine max), GmGASA32 is upregulated by GA and interacts with GmCDC25 to control plant height (Chen et al., 2021). In Gerbera corolla, GEG, a GASA family member, is stimulated by the exogenous application of GA3 and regulates cell expansion (Kotilainen et al., 1999). In strawberry (Fragaria×ananassa), FaGAST genes are upregulated by the exogenous application of GA and affect fruit ripening (de la Fuente et al., 2006). Besides, the expression of GASA genes is increased by other phytohormones such as brassinosteroid (BR), salicylic acid (SA), abscisic acid (ABA), naphthalene acetic acid (NAA), and indole-3-acetic acid (IAA) (Mutasa-Göttgens and Hedden, 2009; Lee et al., 2015; Qu et al., 2016; Boonpa et al., 2018). In rice, OsGSR1, a GASA family member, influences the BR signaling networks by interacting with the BR synthetase DIM/DWF1 (Wang et al., 2009). In Arabidopsis, AtGASA2, AtGASA5, and AtGASA14 are involved in ABA signaling and affect flower induction. AtGASA6 is an integrator of GA, ABA, and glucose signaling and controls seed germination and cell elongation (Zhang and Wang, 2008; Zhong et al., 2015). In apple (Malus domestica), the expression of MdGASA are upregulated by GA and ABA applications during the flowering stage (Fan et al., 2017).
GASA gene family also involved in plant response to abiotic and biotic stresses. In Arabidopsis, overexpression of AtGASA4 suppresses the accumulation of reactive oxygen species (ROS) and nitric oxide in wounded leaves (Rubinovich and Weiss, 2010). In transgenic Arabidopsis plants, overexpression of GASA4 from common beech (Fagus sylvatica) improves tolerance to salt, ROS, and heat stress (Alonso-Ramírez et al., 2009), overexpression of GsGASA1 from soybean inhibits root growth in low temperatures and upregulates the expression of RGL2 and RGL3 (Li et al., 2011). In tomato, Snakin-1 and Snakin-2, two GASA-like genes, are active in vitro against various bacteria (i.e., Clavibacter michiganensis subsp. Sepedonicus) and fungi (i.e., Fusarium solani and Botrytis cinerea) by regulating the redox levels (Almasia et al., 2008; Balaji and Smart, 2012). In rubber (Hevea brasiliensis), HbGASA genes are upregulated upon inoculation with Colletotrichum gloeosporioides and are involved in innate immunity by regulating ROS accumulation (An et al., 2018). Therefore, GASA gene family is involved in numerous physiological and biological processes, displaying complex and diverse functions.
Tobacco (Nicotiana tabacum L.) is widely cultivated and has been used as a model plant for biological research. GASA genes are important in plant growth and development, however, the tobacco GASA gene family were not characterized previously. In this study, we identified GASA gene family in the tobacco genome with bioinformatics methods, and characterized their gene structure, phylogenetic relationships, protein motifs, chromosomal locations, syntenic regions, cis-acting elements, and expression patterns in different tissues. Our findings provide useful clues for further studies of GASA gene family in tobacco.
Materials and Methods
Plant Materials and Growth Conditions
The cultivar K326 was used to analyze the expression of GASA genes in tobacco. Seeds were germinated in a nursery tray, seedlings were grown in a greenhouse with a cycle of 14 h light at 28°C/10 h dark at 25°C and relative humidity at 50–60%. Different tissues (root, stem, leaf, axillary bud, and flower) were collected at the flowering stage to analyze NtGASA expression. For phytohormone treatments, 3-week-old seedlings were transferred to plates containing 10 μM ABA, 10 μM GA, 10 μM IAA, 10 μM SA, 50 μM methyl-jasmonate (MeJA), or 1% (v/v) dimethyl sulfoxide (control) and incubated for 5 h under the same photoperiod, temperature, and humidity conditions (Kretzschmar et al., 2012; Zhang et al., 2018).
Genome-Wide Identification of NtGASA Genes
For NtGASA identification, 15 GASA sequences were obtained from the Arabidopsis database (TAIR; http://www.arabidopsis.org) and used as queries for BLAST search against the Solanaceae Genomics Network (https://solgenomics.net/). Subsequently, the Hidden Markov Model-based profile of the GASA domain PFAM 02704 was used to verify the presence of the complete GASA domain in NtGASA sequences. The non-redundant putative NtGASA sequences with a conserved GASA domain were used for further bioinformatics (phylogenetic relationships, chromosomal locations, Cis-regulatory elements, etc) and expression analysis.
Physicochemical Properties, Phylogenetic Relationships, Gene Structure, and Conserved Motifs Analysis
The isoelectric point, number of amino acids, and molecular weight of NtGASA were predicted using the ExPASy tool (http://web.expasy.org/protparam/). The sequences of GASA from Arabidopsis (AtGASA), rice (OsGASA), grapevine (Vitis vinifera; VvGASA), and tobacco (NtGASA) were used to construct a phylogenetic tree using MEGA 7.0 with the neighbor-joining (NJ) method and a bootstrap test of 1,000 replicates (Supplementary Table S1) (Tamura et al., 2007). The exon/intron structure of each NtGASA genes was illustrated using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn). The conserved motifs of NtGASA proteins were analyzed using MEME 5.1.1 (http://meme-suite. org/tools/meme) (Bailey et al., 2006).
Chromosomal Locations and Gene Duplications Analysis
To obtain the chromosomal locations of NtGASA genes, the DNA sequence of each gene was mapped using MG2C 2.0 (http://mg2c.iask.in/mg2c_v2.0/). Segmental and tandem duplicated gene pairs within the tobacco genome, as well as collinear gene pairs among the Arabidopsis, rice, grapevine, and tobacco genomes, were identified using MCScanX (Wang et al., 2012). The collinearity map was constructed using Circos (Krzywinski et al., 2009). The synonymous and non-synonymous substitution rates (Ks and Ka, respectively) were calculated using KaKs_Calculator 2.0 (Wang et al., 2010).
Expression Analysis of NtGASA Genes
Plant samples were collected from root, flower, leaf, stem and axillary bud of tobacco at flowering stage, total RNA was isolated from frozen samples using Trizol reagent (TaKaRa, Kusatsu, Shiga, Japan), and cDNA synthesis was performed using the M-MLV reverse transcriptase Kit (Thermo Fisher Scientific, Waltham, MA, United States), according to the manufacturer’s instructions. Quantitative reverse-transcription (qRT)-PCR was carried out using the Bio-Rad CFX96 real-time system (Bio-Rad, Hercules, CA, United States) with SYBR Green Master Mix (Bio-Rad). The NtGADPH gene was used as the internal control for data normalization, and the relative expression levels of selected genes were calculated using the 2−ΔΔCt method (Schmittgen and Livak, 2008). The primers used for qRT-PCR are listed in Supplementary Table S3.
Prediction and Classification of Cis-Regulatory Elements
The 3 kb DNA sequence upstream of the start codon of NtGASA genes was examined for the presence of cis-regulatory elements. Cis-regulatory elements in the promoters of each NtGASA gene were analyzed using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and classified according to their regulatory functions.
Results
Physicochemical Properties and Localization of NtGASA
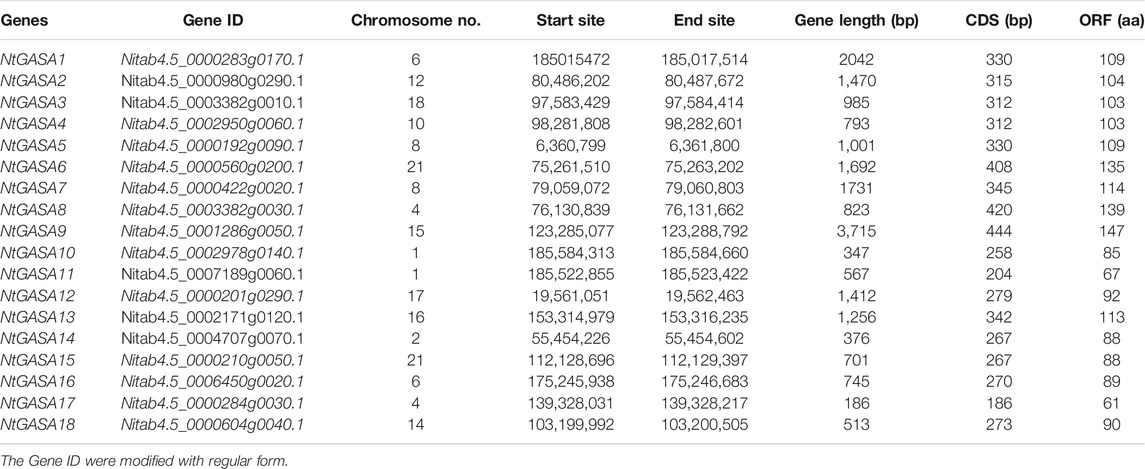
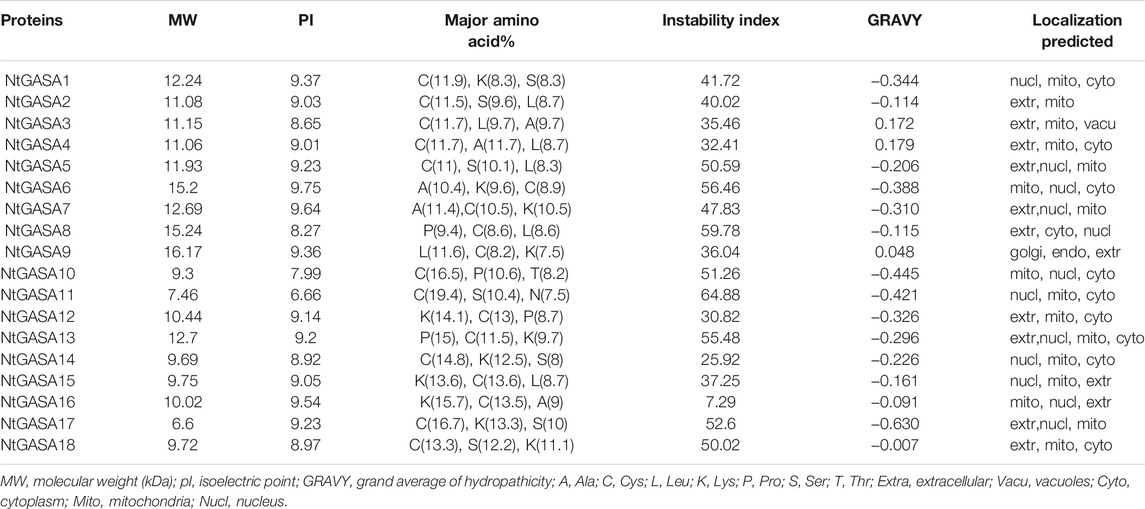
To identify the GASA genes in tobacco, we used 15 AtGASA sequences as queries for BLAST search, and identified 18 putative NtGASA based on amino acid similarities. As shown in Table 1, the total and coding sequence lengths of NtGASA genes were 186 to3,715 bp and 186 to 444 bp, respectively. The deduced NtGASA proteins varied from 61 to 147 amino acids with a molecular weight of 6.6–16.17 kDa, and the isoelectric point ranged from 6.66 to 9.75. Apart from these, the instability index for most of the proteins (77.8%) were more than 35. According to the Grand average of Hydropathicity (GRAVY), the NtGASA proteins were hydrophilic except for NtGASA3, NtGASA4, and NtGASA9. The amino acid content of NtGASA was conserved, cysteine, lysine, and leucine were predominant amino residues. Most NtGASA proteins were localized in the extracellular membrane, chloroplasts, and mitochondria. Detailed information about NtGASA physicochemical characteristics is presented in Table 2.
Phylogenetic Analysis of GASA Genes From Tobacco, Rice, Grape, and Arabidopsis
To characterize the phylogenetic relationships among GASA genes from Arabidopsis, rice, grapevine and tobacco, an unrooted NJ tree was constructed aligning 15 AtGASA, 10 OsGASA, 14 VvGASA, and 18 NtGASA. According to the phylogenetic tree, GASA genes could be classified into three subfamilies: subfamily I included six AtGASA genes (AtGASA1/2/3/9/11/13), three OsGASA genes (OsGASA3/5/7), six VvGASA genes (VvGASA1/5/8/10/11/12), and nine NtGASA genes (NtGASA1–9). Subfamily II included five AtGASA genes (AtGASA4/5/6/12/15), four OsGASA genes (OsGASA4/6/9/10), five VvGASA genes (VvGASA4/6/7/13/14), and four NtGASA genes (NtGASA10–13). Subfamily III included four AtGASA genes (AtGASA7/8/10/14), three OsGASA genes (OsGASA1/2/8), three VvGASA genes (VvGASA2/3/9), and five NtGASA genes (NtGASA14–18) (Figure 1). Therefore, subfamily I had more GASA members from Arabidopsis, grapevine, and tobacco, whereas subfamily II had more GASA members from rice.
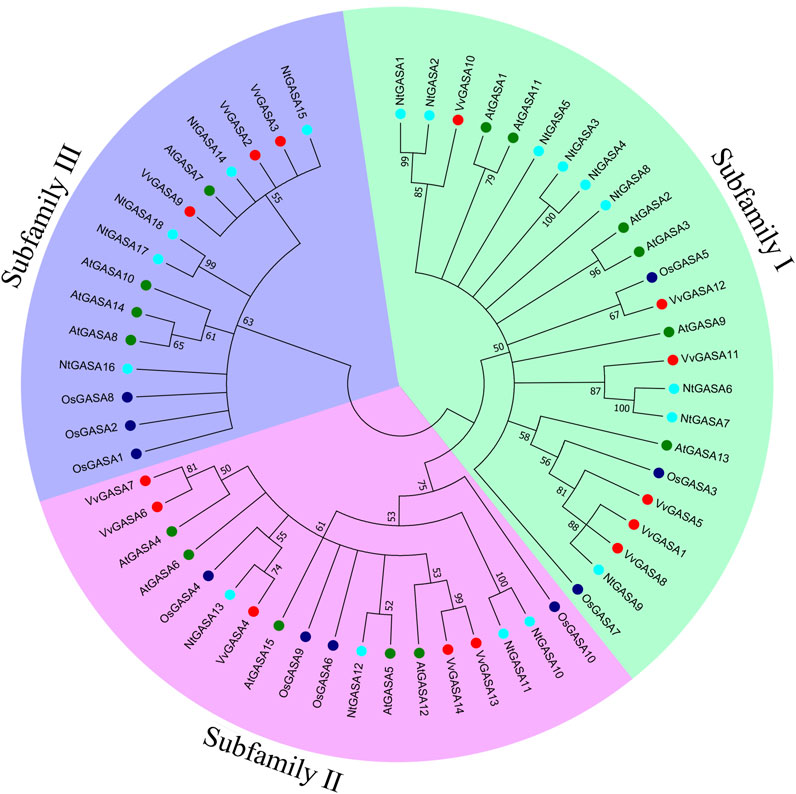
FIGURE 1. Phylogenetic analysis of GASA proteins from Arabidopsis, rice, grapevine and tobacco. A total of 15 GASA proteins from Arabidopsis, 10 GASA proteins from rice, 14 GASA proteins from grapevine, and 18 NtGASA proteins from tobacco were used to generate the unrooted neighbor-joining (NJ) tree with 1,000 bootstrap replicates. The GASA proteins are classified into three subfamilies (marked as I, II, III), and distinguished by different colors: AtGASA labeled in green, OsGASA labeled in blue, VvGASA labeled in red, and NtGASA labeled in cyan.
Chromosomal Distributions and Synteny Analysis of NtGASA Genes
The localizations of the NtGASA genes in the chromosomes of tobacco were further determined. Using a simplified physical map, we found that the 18 NtGASA genes were unevenly distributed in 11 chromosomes in the tobacco genome. Chromosome (Chr.) 1, 4, 6, 8, and 21 contained two copies each, whereas Chr. 2, 10, 12, 14, 15, 16, 17, and 18 contained one copy each (Figure 2).
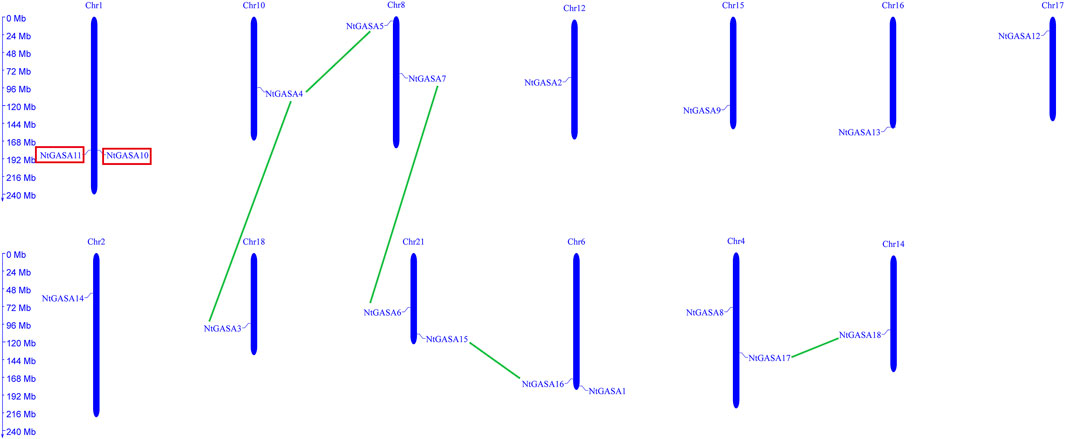
FIGURE 2. Chromosomal distributions and gene duplication of NtGASA genes. Chromosome size is indicated by its relative length. Segmental duplicated NtGASA genes are connected by green colored lines, and red box shows two tandem duplicated gene pairs.
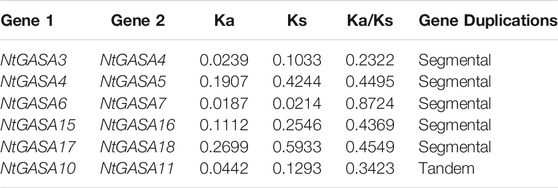
Tandem and segmental duplicates play an important role in the expansion of gene families. Two genes (NtGASA10 and NtGASA11) were tandemly duplicated on Chr.1. In addition, five pairs (NtGASA3/NtGASA4, NtGASA4/NtGASA5, NtGASA6/NtGASA7, NtGASA15/NtGASA16, and NtGASA17/NtGASA18) were segmental duplicated (Figure 2). All tandem and segmental duplicates had Ka/Ks values less than 1 (Table 3), indicating that the six gene pairs evolved under the influence of purifying selection.
We constructed a collinearity plot of the tobacco, rice, grapevine and Arabidopsis GASA gene families to further explore the evolutionary relationships among GASA genes from different species. A total of 2, 4, and 19 collinear gene pairs were identified between tobacco and rice, tobacco and Arabidopsis, and tobacco and grapevine, respectively. Most collinear relationships were many-to-one matches, such as (NtGASA3, NtGASA4, NtGASA5)/AtGASA1, (NtGASA16, NtGASA18)/OsGASA2, (NtGASA2, NtGASA3, NtGASA4, NtGASA5)/VvGASA10 and (NtGASA6, NtGASA7)/VvGASA11. There were also one-to-many matches, such as NtGASA9/(VvGASA1, VvGASA5, VvGASA8), NtGASA15(VvGASA2, VvGASA3, VvGASA9),NtGASA16/(VvGASA2, VvGASA3, VvGASA9) and NtGASA18/(VvGASA2, VvGASA3). The one-to-one matches were NtGASA16/AtGASA7,NtGASA13/VvGASA4 and NtGASA17/VvGASA3 (Figure 3, Supplementary Table S2). These results indicate that GASA genes were relatively conserved between different species and might originate from the same ancestor.
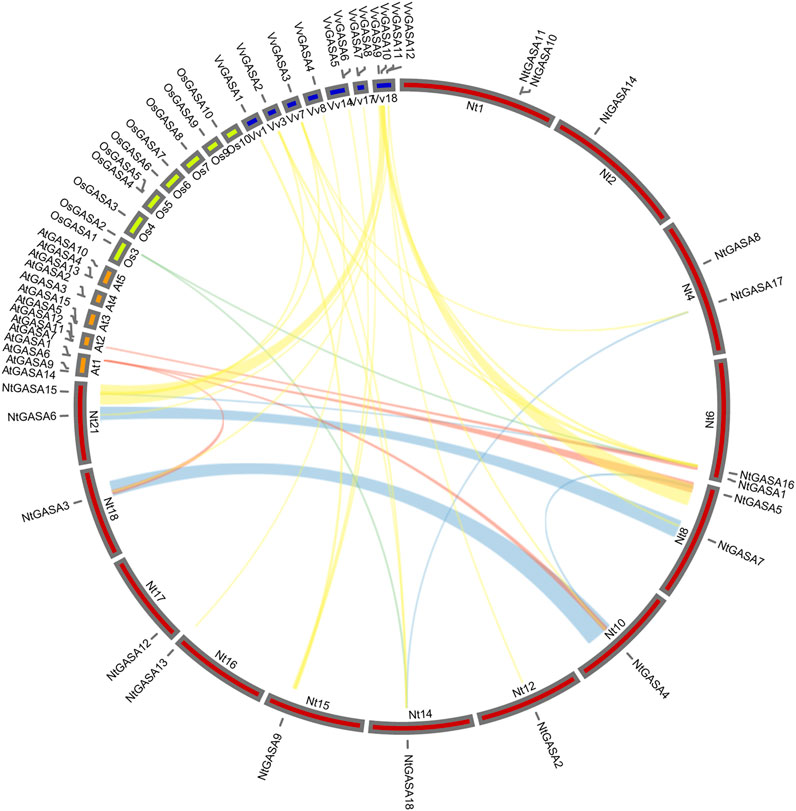
FIGURE 3. Collinear analysis of GASA genes between tobacco (Nt), grapevine (Vv), rice (Os), and Arabidopsis (At). Yellow, red and cyan lines represent the collinear gene pairs between tobacco and grapevine, tobacco and Arabidopsis, tobacco and rice chromosomes, respectively. Blue lines indicate the segmental duplicated NtGASA genes.
Analysis of Conserved Motifs and Gene Structure
To further explore the phylogenetic relationships among NtGASA genes, an unrooted tree was constructed between NtGASA genes. In concordance with the phylogenetic tree including the tobacco, Arabidopsis, grapevine, and rice GASA genes, this analysis also supported the classification of NtGASA genes into three subfamilies (Figure 4A). The number of conserved motifs in NtGASA proteins varied from three to 6 (Figure 4B). The highly conserved motifs 1, 2, and three were detected in all 18 NtGASA proteins, whereas motif five was only found in NtGASA6 and NtGASA7, motif eight were only found in NtGASA9 and NtGASA10. The diversity of motifs in different subfamilies suggests that NtGASA functions have tended to diversify during evolution.
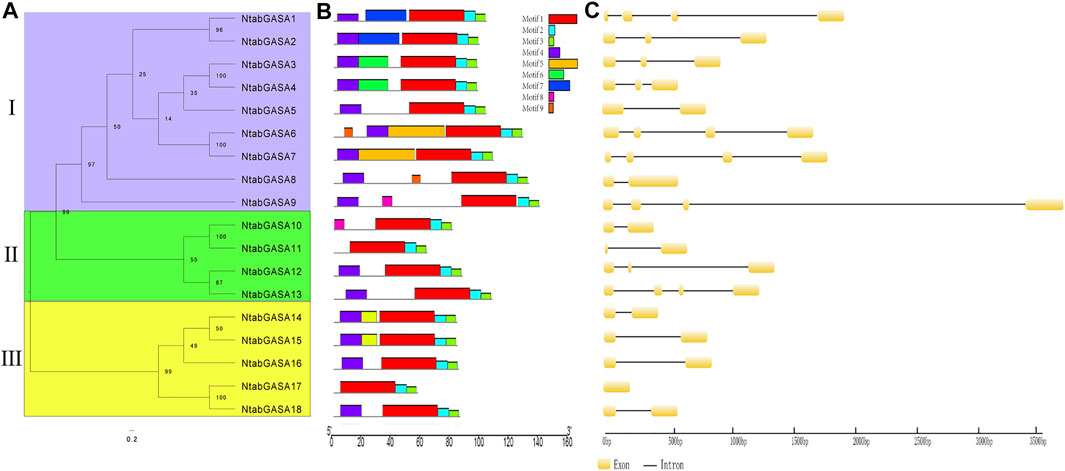
FIGURE 4. Phylogenetic relationship, conserved motifs of NtGASA proteins and exon–intron structures of NtGASA genes. (A) An unrooted phylogenetic tree constructed based on NtGASA protein sequences, different colors indicate different subgroups. (B) Conserved motifs in the NtGASA proteins. The conserved motifs were identified using MEME with complete protein sequences. Different motifs were displayed by various colors. (C) Exon–intron distribution of NtGASA genes.
Structural analysis revealed that the length, arrangement, and position of introns in NtGASA genes were relatively less conserved. For instance, subfamilies I and II contained one to three introns and subfamily III contained one intron, except for NtGASA8 that had only one exon and no intron (Figure 4C). Intron gain and loss is a frequent phenomenon during evolution and can increase the complexity of gene structures.
In previous findings, putative GASA protein possesses highly conserved C-terminal domain that containing 12 conserved cysteines (Aubert et al., 1998). Amino-acid sequence comparison of AtGASA, OsGASA, VvGASA, and NtGASA revealed that all putative NtGASA proteins shared a conserved GASA domain, except for NtGASA17, in which GASA domains were mutated by the insertion of several amino acids (Figure 5).

FIGURE 5. Alignment of the GASA domain from AtGASA, OsGASA, VvGASA, and NtGASA proteins, red asterisk represented their conserved cysteines.
Tissue-Specific Expression Profiling of NtGASA Genes
The spatio-temporal expression analysis of genes can provide information about gene function. We performed qRT-PCR for expression profiling of the NtGASA genes in the root, flower, leaf, stem, and axillary bud. The expression profiling showed that most NtGASA genes had diverse expression patterns in different tissues. NtGASA3, NtGASA11, NtGASA17, and NtGASA18 were expressed relatively ubiquitously. Whereas many NtGASA genes showed high expression in specific tissues, such as NtGASA9 had the highest expression levels in the stem, NtGASA7 in the leaf, NtGASA16 in the axillary bud, and NtGASA2, NtGASA5, NtGASA6, NtGASA10, NtGASA13, NtGASA14, and NtGASA15 in the flower. Notably, NtGASA12 had the lowest expression levels in the stem. In general, most NtGASA genes were highly expressed in reproductive organs (i.e., flower) compared with vegetative parts (i.e., leaf and stem) (Figure 6).
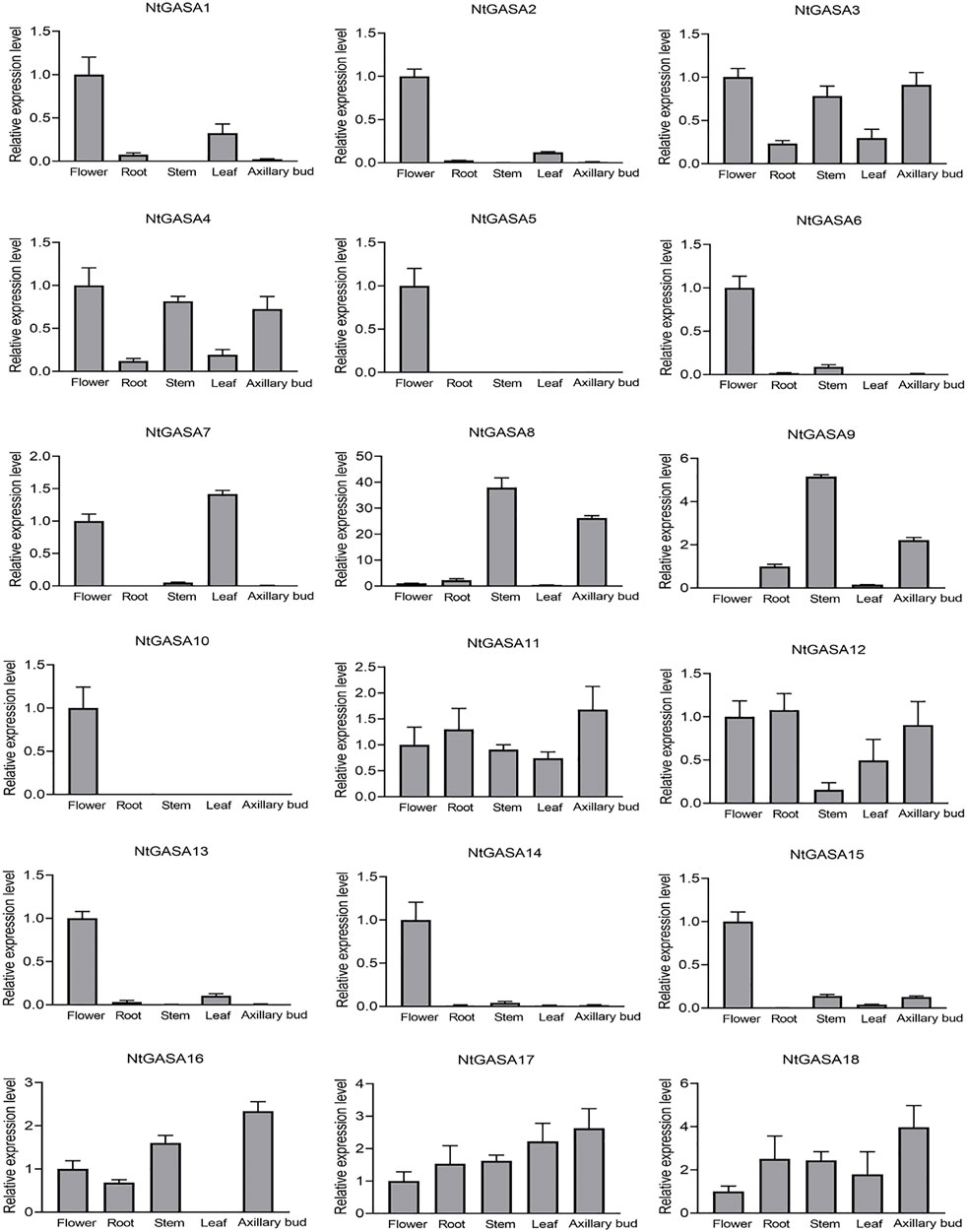
FIGURE 6. Expression patterns of NtGASA genes in different tissues and organs. Transcript levels in flowers were set to one, in NtGASA9, transcript levels in root was set to one. Each value represents the mean ± standard error of three biological replicates.
Analysis of Cis-Elements in the Promoters of NtGASA Genes
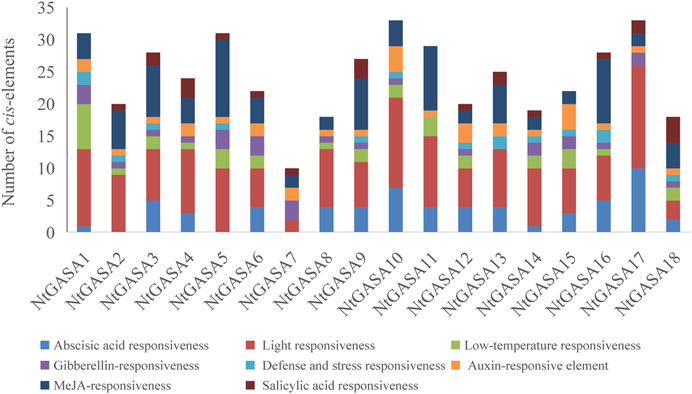
The study of cis-elements could provide clues about regulatory pathways of gene expression, then we analyzed the 3,000-bp upstream promoter sequences of NtGASA genes. The largest number of cis-elements observed across the NtGASA genes was associated with light-responsiveness. In addition, cis-elements involved in phytohormone (i.e., ABA, GA, IAA, SA, and MeJA) and stress (i.e., low temperature) responses were also identified in the promoter sequences of NtGASA genes (Figure 7). The diversity in response elements indicated the regulatory roles of NtGASA genes in various physiological and biological processes.
Expression Profiling of NtGASA Genes Under Various Phytohormone Treatments
The results of the cis-element analysis indicated that NtGASA genes might be related to many plant hormone responses. To elucidate the expression pattern of NtGASA genes and their possible roles in phytohormone signaling pathway, the transcript levels of all NtGASA genes under ABA, GA, IAA, MeJA, SA treatment were investigated. The expression profiling of NtGASA under different phytohormone treatments showed diverse patterns compared with the control. For instance, ABA significantly upregulated the expression of NtGASA1/2/3/4/8/9/13/14, but inhibited the expression of NtGASA5/10/17/18. Most of the NtGASA genes were highly expressed by GA treatment, except for NtGASA6/7/10/16/17/18. After IAA treatment, the expression of NtGASA1/3/4/8/9/14/15 were significantly upregulated, NtGASA11 was downregulated. The expression of NtGASA1/2/3/4/8/9/11/12 were significantly upregulated by SA treantment, NtGASA6/7/10/15/16 were downregulated. Moreover, after MeJA treantment, the expressions of NtGASA3/4/8/9/11/12/13/16 were significantly upregulated, and the expression of NtGASA16 was only upregulated by MeJA treatment, NtGASA1/2/5/6/7/15 were downregulated. Interestingly, the expression of NtGASA17 and NtGASA18 were downregulated by all phytohormones (Figure 8). These findings indicated that different NtGASA genes might play distinctive roles in response to various phytohormone signals.
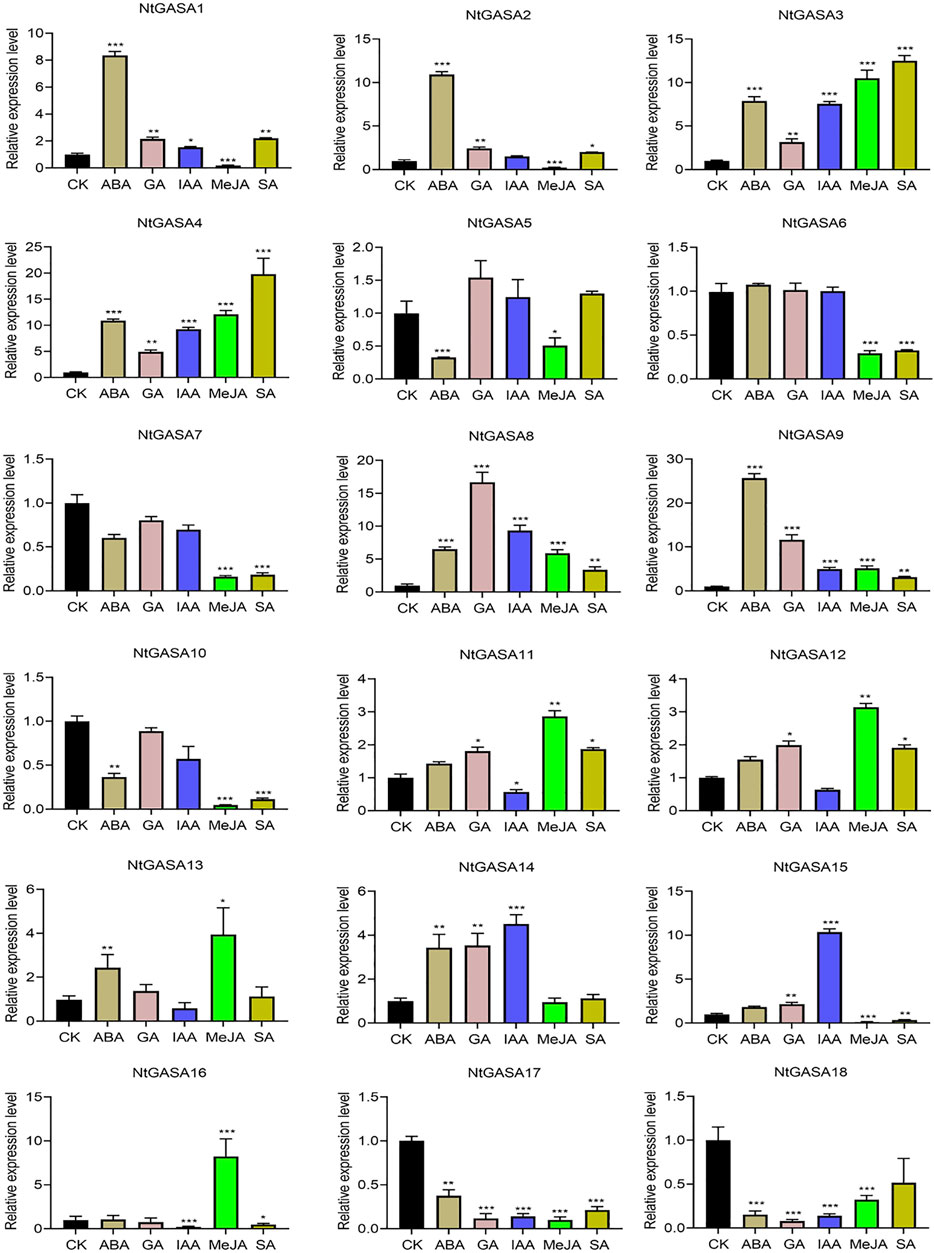
FIGURE 8. Expression patterns of NtGASA genes under various phytohormone treatments. 3-week-old seedlings treated with 10 μM ABA, 10 μM GA, 10 μM IAA, 10 μM SA, and 50 μM MeJA were collected for expression analysis, seedlings treated with 1% (v/v) DMSO served as controls. Each value represents the mean ± standard error of three biological replicates. Asterisks denote significant differences between the hormone treatment and control sample. (Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
GASA influence various biological processes and signal transduction pathways, and then playing critical roles in plant growth and development (Choi et al., 2017). Due to complexities in functional mechanisms, different members of the GASA gene family have identical or diverse functions during the vegetative and reproductive stages. In Arabidopsis, AtGASA5 is activated by ABA during seed dormancy, whereas AtGASA4 is expressed during germination (Zhang et al., 2009). In strawberry, FaGAST1 and FaGAST2 have distinct expression patterns and belong to different subfamilies, but they are both involved in similar physiological functions and synergistically affect the fruit cell size (Moyano-Cañete et al., 2013). The GASA gene family is found in many plant species, but little is known about the corresponding genes in tobacco. Here, we conducted a comprehensive genome-wide identification and expression profiling study of GASA gene family in tobacco.
We identified 18 NtGASA genes in the tobacco genome, more than those previously found in Arabidopsis, rice, grapevine, potato, and soybean (Roxrud et al., 2007; Nahirñak et al., 2016; Ahmad et al., 2019; Muhammad et al., 2019; Ahmad et al., 2020). Based on phylogenetic analyses, the identified NtGASA genes were divided into three subfamilies, of which subfamily I contained the highest number of genes (Figure 1). Physicochemical analysis showed that all the identified NtGASA had low molecular weight and were alkaline, except for NtGASA11 (Table 2), consistently with previously reported results in Arabidopsis, grapevine, and apple (Herzog et al., 1995; Berrocal-Lobo et al., 2002). In addition, cysteine was the predominant amino acid among NtGASA proteins, probably due to the highly conserved 12-cysteine residue at the C-terminus (Table 2; Figure 5).
We also found that motif 1, 2, and three were highly conserved and present in all 18 NtGASA proteins, whereas motif five and eight were only present in NtGASA6/7 and NtGASA9/10, respectively (Figure 4B). Variation in conserved motifs suggested that NtGASA functions were diversified during evolution. Indeed, NtGASA gene structure analysis revealed that the number of introns was varied from 0 to 3 (Figure 4C), indicating that a gain and loss of introns occurred over time, which may be caused by chromosomal rearrangements (Xu et al., 2012; Guo et al., 2013).
Tandem or segmental duplication, as well as whole-genome duplication, markedly affect the evolution of gene families (Vision Todd et al., 2000; Paterson et al., 2010). Our results showed that the presence of both tandem and segmental duplications contributed to the evolutionary process of NtGASA genes. We identified one pair of tandem duplicated NtGASA genes and five pairs of segmental duplicated NtGASA genes throughout the genome (Table 3), these results corroborates the previous findings that segmental duplications occur more frequently than tandem duplications (Zhang et al., 2020). The collinear analysis of GASA genes from Arabidopsis, rice, grapevine, and tobacco showed that the existence of more collinear gene pairs between grapevine and tobacco (Figure 3), suggesting a closer evolutionary distance between the two plant species.
We further analyzed the expression profiles of NtGASA genes in different tissues and found a large variety of expression patterns. Several genes (i.e., NtGASA11 and NtGASA17) showed ubiquitous expression, whereas most NtGASA genes were upregulated only in specific tissues (i.e., NtGASA9 in the stem; NtGASA7 in the leaf; NtGASA16 in the axillary bud; and NtGASA2/5/6/10/13/14/15 in the flower) (Figure 6). Previous studies indicated that GASA genes contribute to the regulation of flower induction in various species such as Petunia hybrida, Gerbera hybrida, rice, and cotton (Ben-Nissan et al., 2004; Peng et al., 2010; Muhammad et al., 2019; Qiao et al., 2021). Here, 13 NtGASA genes showed high expression in the flower, suggesting that they might play important roles in floral development.
The promoter region of a gene is related to its function, and thus, the analysis of cis-elements assists in its functional characterization (Lescot et al., 2002). Our results showed that NtGASA genes contained various regulation elements on their promoters, such as cis-acting regulatory elements essential for light, phytohormone, and stress responses (Figure 7), suggesting their involvement in multiple signaling pathways. GASA transcripts are responsive to phytohormones and share common phytohormone-related cis-elements. In the present study, we found that all NtGASA genes were regulated by multiple phytohormones, especially ABA and GA, except for NtGASA16, that was only induced by MeJA. Besides, NtGASA17 and NtGASA18 were downregulated by all applied phytohormones (ABA, GA, IAA, SA, or MeJA), indicating that unidentified cis-elements might regulate their expression (Figure 8). The complex expression patterns of NtGASA genes under phytohormone applications highlighted their potential integral roles in various physiological processes.
Conclusion
To our knowledge, this is the first report on the identification and characterization of GASA genes in tobacco. We identified 18 NtGASA genes and analyzed their physicochemical characteristics, phylogenetic relationships, gene structure, conserved motifs, chromosomal locations, synteny, and cis-elements in the promoters, which showed a clear evolutionary history for this family in tobacco. We also studied the expression patterns of NtGASA genes in various tissues and under different phytohormone applications. Overall, our results provided insights into the role of NtGASA genes in several physiological and biological pathways and laid a solid foundation for further exploring the underlying molecular and biochemical mechanisms.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
ZL and JG conceived and designed the study. GW, SW, and KC conducted the bioinformatics analysis. WP, YW, and QX assisted in data collection. ZL and XF wrote the paper. All authors read and approved the manuscript.
Funding
This work was supported by the Key funding of CNTC (No. 110202101003(JY-03) and No. 110201801030(JY-07)) and CTCCC (No. B20202NY1337). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
ZL, JG, SW, KC, WP, and YW were employed by the company China Tobacco Hunan Industrial Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This study received funding from CNTC (No.110202101003(JY-03) and No. 110201801030(JY-07)) and CTCCC (No.B20202NY1337). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. All authors declare no other competing interests.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.768942/full#supplementary-material
References
Ahmad, B., Yao, J., Zhang, S., Li, X., Zhang, X., Yadav, V., et al. (2020). Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development. Ijms 21, 1088. doi:10.3390/ijms21031088
Ahmad, M. Z., Sana, A., Jamil, A., Nasir, J. A., Ahmed, S., Hameed, M. U., et al. (2019). A Genome-wide Approach to the Comprehensive Analysis of GASA Gene Family in Glycine max. Plant Mol. Biol. 100, 607–620. doi:10.1007/s11103-019-00883-1
Almasia, N. I., Bazzini, A. A., Hopp, H. E., and Vazquez‐rovere, C. (2008). Overexpression of Snakin‐1 Gene Enhances Resistance to Rhizoctonia solani and Erwinia Carotovora in Transgenic Potato Plants. Mol. Plant Pathol. 9, 329–338. doi:10.1111/j.1364-3703.2008.00469.x
Alonso-Ramírez, A., Rodríguez, D., Reyes, D., Jiménez, J. A., Nicolás, G., López-Climent, M., et al. (2009). Evidence for a Role of Gibberellins in Salicylic Acid-Modulated Early Plant Responses to Abiotic Stress in Arabidopsis Seeds. Plant Physiol. 150, 1335–1344. doi:10.1104/pp.109.139352
An, B., Wang, Q., Zhang, X., Zhang, B., Luo, H., and He, C. (2018). Comprehensive Transcriptional and Functional Analyses of HbGASA Genes Reveal Their Roles in Fungal Pathogen Resistance in Hevea Brasiliensis. Tree Genet. Genomes 14, 41. doi:10.1007/s11295-018-1256-y
Aubert, D., Chevillard, M., Dorne, A.-M., Arlaud, G., and Herzog, M. (1998). Expression Patterns of GASA Genes in Arabidopsis thaliana: the GASA4 Gene Is Up-Regulated by Gibberellins in Meristematic Regions. Plant Mol. Biol. 36, 871–883. doi:10.1023/A:1005938624418
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: Discovering and Analyzing DNA and Protein Sequence Motifs. Nucleic Acids Res. 34, W369–W373. doi:10.1093/nar/gkl198
Balaji, V., and Smart, C. D. (2012). Over-expression of Snakin-2 and Extensin-like Protein Genes Restricts Pathogen Invasiveness and Enhances Tolerance to Clavibacter Michiganensis Subsp. Michiganensis in Transgenic Tomato (Solanum lycopersicum). Transgenic Res. 21, 23–37. doi:10.1007/s11248-011-9506-x
Ben-Nissan, G., Lee, J.-Y., Borohov, A., and Weiss, D. (2004). GIP, a Petunia Hybrida GA-induced Cysteine-Rich Protein: a Possible Role in Shoot Elongation and Transition to Flowering. Plant J. 37, 229–238. doi:10.1046/j.1365-313X.2003.01950.x
Berrocal-Lobo, M., Segura, A., Moreno, M., López, G., Garcı́a-Olmedo, F., and Molina, A. (2002). Snakin-2, an Antimicrobial Peptide from Potato Whose Gene Is Locally Induced by Wounding and Responds to Pathogen Infection. Plant Physiol. 128, 951–961. doi:10.1104/pp.010685
Boonpa, K., Tantong, S., Weerawanich, K., Panpetch, P., Pringsulaka, O., Yingchutrakul, Y., et al. (2018). Heterologous Expression and Antimicrobial Activity of OsGASR3 from rice (Oryza Sativa L.). J. Plant Physiol. 224-225, 95–102. doi:10.1016/j.jplph.2018.03.013
Chen, K., Liu, W., Li, X., and Li, H. (2021). Overexpression of GmGASA32 Promoted Soybean Height by Interacting with GmCDC25. Plant Signaling Behav. 16, 1855017. doi:10.1080/15592324.2020.1855017
Choi, H., Bae, e.-k., Choi, Y.-I., Yoon, S.-K., and Lee, H. (2017). Characterization of Gibberellic Acid-Stimulated Arabidopsis (GASA) Gene to Drought Stress Response in Poplar (Populus alba × P. Glandulosa). J. Plant Biotechnol. 44, 61–68. doi:10.5010/JPB.2017.44.1.061
de la Fuente, J. I., Amaya, I., Castillejo, C., Sánchez-Sevilla, J. F., Quesada, M. A., Botella, M. A., et al. (2006). The Strawberry Gene FaGAST Affects Plant Growth through Inhibition of Cell Elongation. J. Exp. Bot. 57, 2401–2411. doi:10.1093/jxb/erj213
Fan, S., Zhang, D., Zhang, L., Gao, C., Xin, M., Tahir, M. M., et al. (2017). Comprehensive Analysis of GASA Family Members in the Malus Domestica Genome: Identification, Characterization, and Their Expressions in Response to Apple Flower Induction. BMC genomics 18, 827. doi:10.1186/s12864-017-4213-5
Furukawa, T., Sakaguchi, N., and Shimada, H. (2006). Two OsGASR Genes, rice GAST Homologue Genes that Are Abundant in Proliferating Tissues, Show Different Expression Patterns in Developing Panicles. Genes Genet. Syst. 81, 171–180. doi:10.1266/ggs.81.171
Guo, R., Xu, X., Carole, B., Li, X., Gao, M., Zheng, Y., et al. (2013). Genome-wide Identification, Evolutionary and Expression Analysis of the Aspartic Protease Gene Superfamily in Grape. BMC genomics 14, 554. doi:10.1186/1471-2164-14-554
Herzog, M., Dorne, A.-M., and Grellet, F. o. (1995). GASA, a Gibberellin-Regulated Gene Family from Arabidopsis thaliana Related to the Tomato GAST1 Gene. Plant Mol. Biol. 27, 743–752. doi:10.1007/BF00020227
Kotilainen, M., Helariutta, Y., Mehto, M., Pollanen, E., Albert, V. A., Elomaa, P., et al. (1999). GEG Participates in the Regulation of Cell and Organ Shape during corolla and Carpel Development in Gerbera Hybrida. The Plant cell 11, 1093–1104. doi:10.2307/3870801
Kretzschmar, T., Kohlen, W., Sasse, J., Borghi, L., Schlegel, M., Bachelier, J. B., et al. (2012). A Petunia ABC Protein Controls Strigolactone-dependent Symbiotic Signalling and Branching. Nature 483, 341–344. doi:10.1038/nature10873
Krzywinski, M., Schein, J., Birol, İ., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 19, 1639–1645. doi:10.1101/gr.092759.109
Lee, S.-C., Han, S.-K., and Kim, S.-R. (2015). Salt- and ABA-Inducible OsGASR1 Is Involved in Salt Tolerance. J. Plant Biol. 58, 96–101. doi:10.1007/s12374-014-0497-z
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Li, K.-L., Bai, X., Li, Y., Cai, H., Ji, W., Tang, L.-L., et al. (2011). GsGASA1 Mediated Root Growth Inhibition in Response to Chronic Cold Stress Is Marked by the Accumulation of DELLAs. J. Plant Physiol. 168, 2153–2160. doi:10.1016/j.jplph.2011.07.006
Moyano-Cañete, E., Bellido, M. L., García-Caparrós, N., Medina-Puche, L., Amil-Ruiz, F., González-Reyes, J. A., et al. (2013). FaGAST2, a Strawberry Ripening-Related Gene, Acts Together with FaGAST1 to Determine Cell Size of the Fruit Receptacle. Plant Cel Physiol. 54, 218–236. doi:10.1093/pcp/pcs167
Muhammad, I., Li, W.-Q., Jing, X.-Q., Zhou, M.-R., Shalmani, A., Ali, M., et al. (2019). A Systematic In Silico Prediction of Gibberellic Acid Stimulated GASA Family Members: A Novel Small Peptide Contributes to floral Architecture and Transcriptomic Changes Induced by External Stimuli in rice. J. Plant Physiol. 234-235, 117–132. doi:10.1016/j.jplph.2019.02.005
Mutasa-Gottgens, E., and Hedden, P. (2009). Gibberellin as a Factor in floral Regulatory Networks. J. Exp. Bot. 60, 1979–1989. doi:10.1093/jxb/erp040
Nahirñak, V., Almasia, N. I., Hopp, H. E., and Vazquez-Rovere, C. (2012). Snakin/GASA Proteins. Plant Signaling Behav. 7, 1004–1008. doi:10.4161/psb.20813
Nahirñak, V., Rivarola, M., Gonzalez de Urreta, M., Paniego, N., Hopp, H. E., Almasia, N. I., et al. (2016). Genome-wide Analysis of the Snakin/GASA Gene Family in Solanum tuberosum Cv. Kennebec. Am. J. Potato Res. 93, 172–188. doi:10.1007/s12230-016-9494-8
Paterson, A. H., Freeling, M., Tang, H., and Wang, X. (2010). Insights from the Comparison of Plant Genome Sequences. Annu. Rev. Plant Biol. 61, 349–372. doi:10.1146/annurev-arplant-042809-112235
Peng, J., Lai, L., and Wang, X. (2010). Temporal and Spatial Expression Analysis of PRGL in Gerbera Hybrida. Mol. Biol. Rep. 37, 3311–3317. doi:10.1007/s11033-009-9917-4
Qiao, K., Ma, C., Lv, J., Zhang, C., Ma, Q., and Fan, S. (2021). Identification, Characterization, and Expression Profiles of the GASA Genes in Cotton. J. Cotton Res. 4. doi:10.1186/s42397-021-00081-9
Qu, J., Kang, S. G., Hah, C., and Jang, J.-C. (2016). Molecular and Cellular Characterization of GA-Stimulated Transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 246, 1–10. doi:10.1016/j.plantsci.2016.01.009
Roxrud, I., Lid, S. E., Fletcher, J. C., Schmidt, E. D. L., and Opsahl-Sorteberg, H.-G. (2007). GASA4, One of the 14-Member Arabidopsis GASA Family of Small Polypeptides, Regulates Flowering and Seed Development. Plant Cel Physiol. 48, 471–483. doi:10.1093/pcp/pcm016
Rubinovich, L., and Weiss, D. (2010). The Arabidopsis Cysteine-Rich Protein GASA4 Promotes GA Responses and Exhibits Redox Activity in Bacteria and in Planta. Plant J. 64, 1018–1027. doi:10.1111/j.1365-313X.2010.04390.x
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Shi, L., Gast, R., Gopalraj, M., and Olszewski, N. (1992). Characterization of a Shoot-specific, GA3- and ABA-Regulated Gene from Tomato. Plant J. 2, 153–159. doi:10.1046/j.1365-313x.1992.t01-39-00999.x
Silverstein, K. A. T., Moskal, W. A., Wu, H. C., Underwood, B. A., Graham, M. A., Town, C. D., et al. (2007). Small Cysteine-Rich Peptides Resembling Antimicrobial Peptides Have Been Under-predicted in Plants. Plant J. 51, 262–280. doi:10.1111/j.1365-313X.2007.03136.x
Sun, S., Wang, H., Yu, H., Zhong, C., Zhang, X., Peng, J., et al. (2013). GASA14 Regulates Leaf Expansion and Abiotic Stress Resistance by Modulating Reactive Oxygen Species Accumulation. J. Exp. Bot. 64, 1637–1647. doi:10.1093/jxb/ert021
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi:10.1093/molbev/msm092
Taylor, B. H., and Scheuring, C. F. (1994). A Molecular Marker for Lateral Root Initiation: The RSI-1 Gene of Tomato (Lycopersicon esculentum Mill) Is Activated in Early Lateral Root Primordia. Mol. Gen. Genet. 243, 148–157. doi:10.1007/BF00280311
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The Origins of Genomic Duplications in Arabidopsis. Science 290, 2114–2117. doi:10.1126/science.290.5499.2114
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genomics, Proteomics & Bioinformatics 8, 77–80. doi:10.1016/S1672-0229(10)60008-3
Wang, L., Wang, Z., Xu, Y., Joo, S.-H., Kim, S.-K., Xue, Z., et al. (2009). OsGSR1is Involved in Crosstalk between Gibberellins and Brassinosteroids in rice. Plant J. 57, 498–510. doi:10.1111/j.1365-313X.2008.03707.x
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40, e49. doi:10.1093/nar/gkr1293
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of Duplicate Genes in Exon-Intron Structure. Proc. Natl. Acad. Sci. 109, 1187–1192. doi:10.1073/pnas.1109047109
Zhang, J., Li, Z., Jin, J., Xie, X., Zhang, H., Chen, Q., et al. (2018). Genome-wide Identification and Analysis of the Growth-Regulating Factor Family in Tobacco ( Nicotiana Tabacum ). Gene 639, 117–127. doi:10.1016/j.gene.2017.09.070
Zhang, L., Geng, X., Zhang, H., Zhou, C., Zhao, A., Wang, F., et al. (2017). Isolation and Characterization of Heat-Responsive Gene TaGASR1 from Wheat (Triticum aestivum L.). J. Plant Biol. 60, 57–65. doi:10.1007/s12374-016-0484-7
Zhang, S., and Wang, X. (2008). Expression Pattern of GASA, Downstream Genes of DELLA, in Arabidopsis. Sci. Bull. 53, 3839–3846. doi:10.1007/s11434-008-0525-9
Zhang, S., Yang, C., Peng, J., Sun, S., and Wang, X. (2009). GASA5, a Regulator of Flowering Time and Stem Growth in Arabidopsis thaliana. Plant Mol. Biol. 69, 745–759. doi:10.1007/s11103-009-9452-7
Zhang, Z., Chen, J., Liang, C., Liu, F., Hou, X., and Zou, X. (2020). Genome-Wide Identification and Characterization of the bHLH Transcription Factor Family in Pepper (Capsicum Annuum L.). Front. Genet. 11, 570156. doi:10.3389/fgene.2020.570156
Keywords: GASA, Nicotiana tabacum, expression analysis, phylogenetic analysis, cis-elements
Citation: Li Z, Gao J, Wang G, Wang S, Chen K, Pu W, Wang Y, Xia Q and Fan X (2022) Genome-Wide Identification and Characterization of GASA Gene Family in Nicotiana tabacum. Front. Genet. 12:768942. doi: 10.3389/fgene.2021.768942
Received: 01 September 2021; Accepted: 29 December 2021;
Published: 01 February 2022.
Edited by:
Zefeng Yang, Yangzhou University, ChinaReviewed by:
Haiyang Jiang, Anhui Agricultural University, ChinaMuhammad Zulfiqar Ahmad, Gomal University, Pakistan
Copyright © 2022 Li, Gao, Wang, Wang, Chen, Pu, Wang, Xia and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingyou Xia, eGlhcXlAc3d1LmVkdS5jbg==; Xiaorong Fan, eGlhb3JvbmdmYW5AbmphdS5lZHUuY24=
 Zhaowu Li
Zhaowu Li Junping Gao1
Junping Gao1 Xiaorong Fan
Xiaorong Fan