- 1Department of Chemistry, Shia Postgraduate College, Lucknow, India
- 2Department of Zoology, Shia Postgraduate College, Lucknow, India
- 3Department of Medical Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, United States
- 4Department of Anatomy, University of Ghana Medical School, College of Health Sciences, University of Ghana, Accra, Ghana
- 5Department of Molecular Medicine, University of South Florida, Tampa, FL, United States
- 6Division of Hematology and Oncology, Department of Medicine, Case Western Reserve University, Cleveland, OH, United States
- 7Case Comprehensive Cancer Center, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 8Department of Pharmaceutical Sciences, Nova Southeastern University, Fort Lauderdale, FL, United States
Epigenetic regulation involves reversible changes in histones and DNA modifications that can be inherited without any changes in the DNA sequence. Dysregulation of normal epigenetic processes can lead to aberrant gene expression as observed in many diseases, notably cancer. Recent insights into the mechanisms of DNA methylation, histone modifications, and non-coding RNAs involved in altered gene expression profiles of tumor cells have caused a paradigm shift in the diagnostic and therapeutic approaches towards cancer. There has been a surge in search for compounds that could modulate the altered epigenetic landscape of tumor cells, and to exploit their therapeutic potential against cancers. Flavonoids are naturally occurring phenol compounds which are abundantly found among phytochemicals and have potentials to modulate epigenetic processes. Knowledge of the precise flavonoid-mediated epigenetic alterations is needed for the development of epigenetics drugs and combinatorial therapeutic approaches against cancers. This review is aimed to comprehensively explore the epigenetic modulations of flavonoids and their anti-tumor activities.
Introduction
Epigenetics can be defined as the study of heritable changes in gene expression without involving any changes in the DNA sequence (Weinhold, 2006). Epigenetic changes are established during early differentiation of cells and stably inherited through multiple cell divisions resulting in distinct cellular phenotypes without any changes in the underlying DNA sequence (Cheedipudi et al., 2014). The process of epigenetic regulation of gene expression involves chromatin remodeling mediated by events like DNA methylation, histone modifications and effects of non-coding RNAs as shown in Figure 1. These epigenetic changes are essential for normal development of cells, but their deregulation can lead to certain disease states including cancer (Egger et al., 2004).
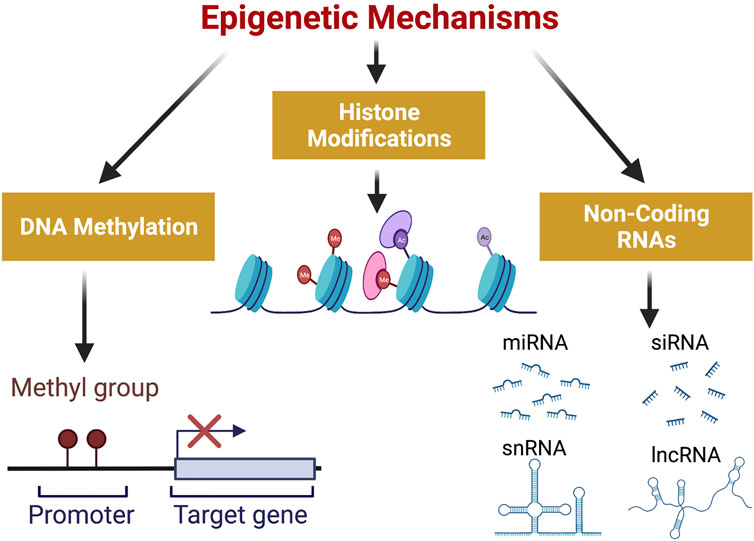
FIGURE 1. Epigenetic Mechanisms involved in gene expression: DNA methylation, Histone Modifications and Non-Coding RNAs constitute the three mechanisms involved in epigenetic regulation of gene expression. Methylation of DNA occurs at gene promoters and hypermethylation represses gene expression. Histone modifications include acetylation and methylation of histones which can either activate or repress gene expression. Non-coding RNAs which consist of short chain non-coding RNAs ( <200 nt) and long non-coding RNAs ( >200 nt) also play a significant role in regulation of gene expression. (Created with BioRender.com). Ac, acetylated histone; Me, methylated histone; miRNA, micro RNA; siRNA, small interfering RNA; snRNA, small nuclear RNA; lncRNA, long non-coding RNA.
There is a consensus among biologists that cancer can be considered as a genetic inevitability. DNA mutations accumulate in the cells as we age. The mutations can be either spontaneous due to errors of DNA replication or may be caused by physical or chemical mutagens. Hence, 90–95% of cancers involve mutations due to environmental or lifestyle factors and in the remaining 10% of cases, the genetic cause of cancer is hereditary (Anand et al., 2008). Interestingly, many of the gene defects in cancers are not due to changes in sequences but are a result of epigenetic changes that affect the expression profile of these genes (Taby and Issa, 2010; Cavalli and Heard, 2019). Recent advances in the field of epigenetics have highlighted global epigenetic abnormalities in cancer cells which can occur during the early stages of tumor development. Due to their reversible nature and early occurrence in the process of malignant transformation of normal cells, epigenetic modifications can serve as novel drug targets for the treatment of cancer (Dawson and Kouzarides, 2012; Bennett and Licht, 2018).
Even though, in 2020, the global estimate of new cancer cases was 19.3 million which resulted in almost 10 million deaths, cancer related deaths have been declining since 1991 leading to a decrease of 31% in 2018 (Siegel et al., 2021). This improvement in patient’s survival rate is a result of advancement in cancer treatments. Cancer treatments usually involve chemotherapeutic agents, radiotherapy, and immunotherapy, and these treatments have shown a lot of promise (Yu et al., 2019). Epigenetic modulators (e.g., histone deacetylases inhibitors) have been used to treat aberrant epigenetic modifications in cancer along with some novel approaches such as chimeric antigen receptor-engineered T cells (CAR-T cells) which have proven very efficient in treating malignancies of B-cells (Wang et al., 2020b). However, most of the cancer therapies including immunotherapy are associated with numerous side effects (Lacouture and Sibaud, 2018).
Thus, considering the adverse effects of radiation and chemotherapeutic agents on patients, there is an increased quest for alternative therapies without extreme side effects. Phytochemicals fit best in this criterion and are hence explored for their anticancer properties. Anticarcinogenic factors in plant-based foods are known to inhibit cancer by a variety of mechanisms ranging from antioxidant effects to immunomodulatory properties. Interestingly, it has been reported that phytochemicals can modulate epigenetic processes through DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) (Shukla et al., 2014). Flavonoids which constitute an important class of phytochemicals are implicated in modulation of gene expression patterns underlying cancer (Busch et al., 2015). In this review we first discuss the role of epigenetic aberrations in cancer and then present an overview of the usefulness of flavonoids as epigenetic modulators having chemotherapeutic and chemo-preventive properties.
Cancer Epigenetics
In eukaryotic cells, DNA is wrapped around a core of histone proteins to form nucleosomes which may wrap over themselves to adopt a condensed state temporarily or permanently. The degree to which a gene is expressed in each cell is controlled by a range of gene regulatory mechanisms, most of which interfere with chromatin condensation. The condensation of chromatin involves a compact packing of nucleosomes which renders the genes hetero-chromatinzed and hence inactive. On the contrary, decondensation of chromatin opens the nucleosomes and increased expression of genes. The transformation of normal cells into cancer cells involves epigenetic alterations and in most cases is preceded by genetic mutations (Brower, 2011). Amongst the key mechanisms involved in epigenetic regulation are DNA modifications (e.g., methylation), histone modifications (e.g., deacetylation, phosphorylation, and ubiquitination, etc.), nucleosome positioning, and non-coding RNAs (e.g., miRNA, siRNA, lncRNA, etc.) (Figure 1). These mechanisms have a significant impact on cellular homeostasis (Zhang et al., 2020).
DNA Methylation
The methylation or hydroxymethylation of DNA is an important epigenetic mechanism of gene regulation in cells (Gao and Das, 2014) and DNA methylation patterns contribute in establishing epigenetic memory (Bird, 2002). The methylation of DNA is usually negatively correlated with gene expression, but it also depends on the location of methylated bases in relation to coding regions of the genes being regulated (Blake et al., 2020). Cytosine methylation most commonly occurs at CpG sites which are widely distributed through the genome (Jones, 2012). The specific regions of CpG sites are designated as CpG islands when they have a length of more than 200 bp, a GC content of more than 55% and a >60% observed-to-expected CpG ratio (Takai and Jones, 2002). CpG islands are particularly abundant at promoter region of the gene, and are present in around 70% of human promoters (Deaton and Bird, 2011). When CpG sites are present in the promoter or enhancer regions of genes their methylation represses gene expression whereas it induces transcriptional activity if the CpG sites are present in the coding regions of genes (Greenberg and Bourc’his, 2019). The pattern of CpG methylation in cancer cells is different to that of normal cells (Esteller, 2007). In normal cells, whereas the CpG islands preceding promoters are unmethylated allowing active transcription while other CpG sites in the genome remain methylated. In cancer cells, the CpG dinucleotides have up to 50% less methylation than normal cells and CpG islands are also generally hypomethylated (Herman and Baylin, 2003). In general, promoters of tumor suppressor genes become hypermethylated thereby inhibiting their expression while those of oncogenes get hypomethylated and activated in cancer. Genes involved in DNA repair, cell cycle, migration and apoptosis are dysregulated due to aberrant DNA methylation in cancer cells (Cheung et al., 2009). This process of de novo DNA methylation is carried out by DNMTs and include three isoenzymes: DNMT1, DNMT3a and DNMT3b.
Histone Modifications
Histones are positively charged proteins that play a role in condensing and packaging the DNA into chromatin inside the nucleus. Open chromatin structure (euchromatin) is associated with transcriptional activation and closed chromatin structure (heterochromatin) is associated with repression of transcription. Histone modifications especially acetylation, phosphorylation, and methylation regulate structural changes in the chromatin influencing gene expression (Karlic et al., 2010; Bannister and Kouzarides, 2011).
Histone acetylation is defined as the addition of an acetyl group to lysine residues present at the histone tail by histone acetyl transferases (HATs). This modification weakens the DNA-histone interaction eventually leading to decondensation of chromatin and increased gene expression. On the contrary, HDACs constitute an important class of enzymes and are responsible for deacetylation of e-amino groups of lysine residues leading to condensation of chromatin and decreased gene expression. (Milazzo et al., 2020). The pattern of histone acetylation has been found to be remarkably different between normal and cancerous cells (Di Cerbo and Schneider, 2013). Histone H4 exhibits a decrease in monoacetylation of Lys20 and trimethylation of Lys16 in malignant cells (Fraga et al., 2005). Besides, decreased acetylation of histones H3 and H4 has also been observed in cancer progression (Audia and Campbell, 2016).
Histone methylation is another epigenetic modification that plays a role in regulating gene expression in cancer. These modifications are catalyzed by Histone Methyltransferases (HMTs) and Histone Demethylases (HDMs) of the specific amino acids in histones. Cancer specific gene transcription profiles are often related to the regulation of histone methylation (Varier and Timmers, 2011). For instance, there is a cancer-associated decrease in trimethylation on Lysine 4 of histone H3 (H3K4me3) along with a simultaneous increase of monomethylation on Lysine 9 of histone H3 that affects gene expression (Richon et al., 2000).
Phosphorylation is another histone posttranslational modification mediated by cell-cycle related kinases (Rossetto et al., 2014). The phosphorylation of serine at the C-terminus of a DNA double-strand break marker H2A histone family member X (H2AX), eventually contributes to genomic instability leading to cancer (Bonner et al., 2008).
Non-Coding RNAs
The next important epigenetic mechanism controlling cell function involves non-coding RNAs which are being shown to regulate gene expression to a great extent (Kurokawa et al., 2014; Statello et al., 2020). Small non-coding RNAs composed of 18–25 nucleotides are called MicroRNAs (miRNAs) which usually interacts with the 3’ region of the mRNA affecting mRNA stability and translation. One miRNA can regulate the expression of several genes or multiple miRNAs can affect the expression of the same gene (Mohr and Mott, 2015). Transcriptional activity of up to 60% of mammalian protein coding genes has been found to be controlled by miRNAs (Friedman et al., 2009). Several miRNAs have been associated with the regulation of oncogenes and tumor suppressor genes thereby having a role in cancer (Peng and Croce, 2016). The most common cancer associated miRNAs (onco-miRNAs) that are promising candidates for cancer treatment are let-7, miR-15, and miR-16 (Esquela-Kerscher and Slack, 2006). Also, miR-125b1 has been shown to act as a tumor suppressor as its decrease is associated with ovarian and prostate cancers (Soto-Reyes et al., 2012). The roles of micro RNAs in cancer progression are contributing to a great bulk of emerging knowledge about cancer (Ali Syeda et al., 2020). Events like genetic alterations, promoter hypermethylation, or other epigenetic modifications regulate the expression of miRNAs and promotes cellular transformation and cancer progression (Baer et al., 2013; Liu et al., 2013; Cammaerts et al., 2015).
Long non-coding RNAs (lncRNAs) are polyadenylated RNAs with a length of more than 200 nucleotides that can bind to DNA, RNA, and proteins. Epigenetic modulation is the most common method of lncRNAs based regulation of gene expression and is often associated with gene repression. Studies have reported that lncRNAs can function as oncogenes or tumor suppressors through a wide range of activities, including interaction with Polycomb Repressive Compex (PRC) regulating transcript stability, processing and translation; interaction with miRNAs, gene enhancers and repressors; and interaction with transcription factors affecting transcript production and transport (Marín-Béjar et al., 2013; Vance and Ponting, 2014; Marchese et al., 2017; Zhang et al., 2019b; Hou et al., 2019). LncRNAs like MALAT1 and HOTAIR which are associated with metastasis are over expressed in cancer and MEG3 and PTENP1 which inhibit cell proliferation and migration are downregulated in cancer (Huarte, 2015).
Flavonoids as Epigenetic Modulators
Flavonoids belong to an important class of natural low-molecular-weight polyphenolic compounds having basic benzo-
Despite the lifesaving role of chemotherapeutic agents in treating cancer, a disadvantage of these drugs is their potential cytotoxic effects on normal cells. Thus, there is a need for better substitutes without undesirable side-effects. In this regard, flavonoids show promising results as many of the anticarcinogenic flavonoids have relatively less toxicity towards normal cells (Galati and O'Brien, 2004); however, more in-depth toxicity studies are needed to evaluate their safety and side-effects. It is also documented that on normal cells they may act as pro-oxidants that generate free radicals, mutagens and act as inhibitors of key enzymes involved in hormone metabolism when consumed at higher doses (Skibola and Smith, 2000). Nonetheless, flavonoids having epigenetic modifying potential can be an attractive choice for potential cancer therapies, including combinatorial therapy (Figure 2; Table 1).
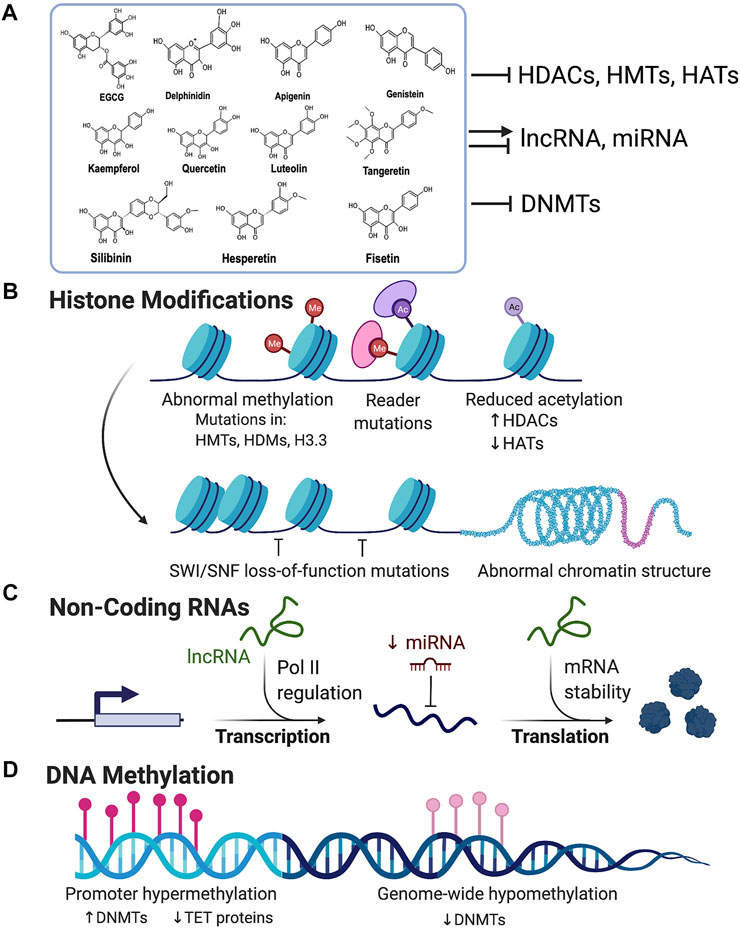
FIGURE 2. Flavonoids as epigenetic modulators in cancer: (A) Flavonoids are polyhydroxy compounds found in various plants and generally consist of two phenyl rings and a heterocyclic ring. Flavonoids are reported to exhibit inhibitory activity for HDACs, HMTs, HATs, and DNMTs. They can also either inhibit or activate miRNA and lncRNA. (B) Illustration showing various mechanisms of histone modifications associated with carcinogenesis. Flavonoids can block the aberrant expression of HMTs, HATs, and HDACs, and activate tumor suppressor genes and block the expression of oncogenes. (C) Flavonoids can either activate or repress non-coding RNAs which regulates chromatin structure and aberrant gene expression in cancer. (D) Promoter hypermethylation and genome-wide hypomethylation are associated with cancer. Flavonoids can inhibit DNA Methyl Transferases (DNMTs) and thus prevent hypermethylation of gene promoters like tumor suppressor genes thereby reactivating their expression. (Created with BioRender.com).
Cancer Prevention and Therapy by Epigenetically Active Flavonoids
Flavan-3-Ols/ Flavanols/ Catechins
Epigallocatechin gallate (EGCG) is a powerful polyphenolic, chemo-preventive compound isolated from green tea belongs to the catechin class of flavonoids (Singh et al., 2011). The other components of green tea consist of epigallocatechin, epicatechin-3-gallate and epicatechin. There are numerous in vitro, in vivo, and clinical studies that showed potential anti-proliferative, anti-angiogenic, pro-apoptotic and anti-invasive properties of EGCG (Singh et al., 2011). There are significant literatures demonstrating the impact of green tea components in involved in epigenetic modulations in cancer (Henning et al., 2013; Giudice et al., 2018; Jiang et al., 2021). Fang et al., first time revealed that EGCG act as inhibitor of DNA hypermethylation of CpG islands by acting on DNMT (Fang et al., 2003). Another study has shown the profound effects of EGCG on human prostate cancer cells (Paschka et al., 1998). Green tea causes an accumulation of acetylated histone H3 in total cellular chromatin resulting in epigenetically reactivation of p21/waf1 and Bax in prostate cancer cells that leads to cell cycle arrest and apoptosis (Thakur et al., 2012).
Lee et al., examined the regulation of androgen receptor acetylation by measuring histone acetyltransferase activity in androgen-dependent prostate cancer cells treated with green tea catechins (epicatechin, epigallocatechin and epigallocatechin-3-gallate). These catechins induce prostate cancer cell death, suppressed agonist-dependent androgen receptor (AR) activation and AR-regulated gene transcription (Lee et al., 2012). Another report showed that the combinatorial exposures of clofarabine and EGCG or genistein synergistically inhibited the growth of breast cancer cells (MCF7 and MDA-MB-231 cells) and induces apoptosis followed by RARB hypomethylation with consequent manifold increase in RARB, CDKN1A, and PTEN transcript levels. This combination promotes apoptosis and reactivates DNA methylation-silenced tumor suppressor genes in human breast cancer cells with unusual invasive prospective (Lubecka et al., 2018).
EGCG alters the expression of various tumor-suppressor genes by inhibiting DNA methyltransferases and histone deacetylases in human cervical cancer HeLa cells (Khan et al., 2015). Moreover, time-dependent exposure to EGCG resulted in reactivation of well-known tumor-suppressor genes (TSGs) in these cells due to marked transformations in the methylation of the promoter area of these genes (Khan et al., 2015). Recently Ciesielski et al., 2020 studied the impact of EGCG on the histone posttranslational modifications machinery along with chromatin remodeling in human endothelial cells (HMEC-1 and HUVECs origin). Results showed that EGCG increases methylation of both active (H3K4me3) and repressive (H3K9me3) chromatin marks and histone acetylation (H3K9/14ac, H3ac). These results indicated the broad epigenetic potential of EGCG concerning expression and action of epigenome modulators including HDAC5, and HDAC7, CREBP, KMT2A, p300 and LSD1 (Ciesielski et al., 2020).
Another study reported the anticancer mechanism of EGCG via synchronized transcriptional modification of numerous molecular targets through different signaling pathways in Hela cells (Kedhari Sundaram et al., 2020). In this study, transcriptional modulation of several epigenetic modifiers including histone modifiers and DNA methyltransferases (DNMT1, DNMT3A, DNMT3B, AURKA, AURKB, AURKC, PRMT6, PRMT7, KDM4A, KDM5C, HDAC5, HDAC6, HDAC7, HDAC11 and UBE2B) were observed. Downregulation of key signaling moieties of PI3K, Wnt and MAPK pathways, metastasis regulators, cell cycle regulators and pro-inflammatory moieties including CCNB1, CCNB2, TERT, PIK3C2B, PIK3CA, IL6, MMP2, MMP7 and MAPK8 were also detected (Kedhari Sundaram et al., 2020).
Kang et al., demonstrated that EGCG may hamper efficiently IR-induced injury to mouse normal hepatic cells (AML-12), and improve noticeably the radio-sensitivity of mouse hepatoma cells H22 to 60Coγ. They also revealed that EGCG played the key task of radio-sensitization on H22 cells because it activates the miR-34a/Sirt1/p53 signaling pathway (Kang et al., 2019). Deb et al., reported that treatment of human prostate cancer cell lines (DUPro and LNCaP) with green tea polyphenols (GTPs) and their major constituent, epigallocatechin-3-gallate (EGCG) induced TIMP3 expression by epigenetic mechanisms (Deb et al., 2019). Furthermore, a clinical study on patients undergoing prostatectomy consuming EGCG showed an increase in plasma TIMP3 levels (Deb et al., 2019).
Dietary flavonoids have potential to modulate non-coding RNAs, including miRNAs in cancer (Ahmed et al., 2020; Yang et al., 2020; Singh et al., 2021). In a recent in vivo study by Kang et al., revealed that oral administration of EGCG suppresses miR483–3p induced metastasis of hepatocellular carcinoma (Kang et al., 2021). EGCG modulate non-coding RNAs and inhibit tumor growth by targeting LINC00511/miR-29b/KDM2A axis in gastric cancer (Zhao et al., 2020). EGCG-capped gold nanoparticle significantly increased expression of tumor suppressor let-7a and miR-34a miRNAs (Mostafa et al., 2020).
Flavonols
Flavonols (3-hydroxylavones) are most prevalent flavonoids in food. Quercetin, myricetin, kaempferol and fisetin are the most common plant flavonols found in many vegetables and fruits, e.g., onions, apples, strawberries etc. The defensive impacts of quercetin on human health are facilitated by multidimensional, pleiotropic action still from an epigenetic perspective (Russo and Ungaro, 2019).
Quercetin modulates the expression of various chromatin modifiers and declines the activity of HDACs, DNMTs and HMTs in a dose-dependent manner in human cervical cancer (HeLa) cells (Kedhari Sundaram et al., 2019). It also downregulated global DNA methylation concentrations in a dose- and time-dependent manner and tested tumor suppressor genes showed steep dose-dependent decrease in promoter methylation with the restoration of their expression. Quercetin along with BET inhibitors promoted apoptosis and decreases the cell proliferation and sphere-forming ability by pancreatic cancer cells. It was also evidenced that quercetin also mediates some anti-tumor effects with the help of hnRNPA1 which is a nuclear protein well-known to monitor mRNA export and mRNA translation of anti-apoptotic proteins (Pham et al., 2019). Quercetin also induced let-7c which decreased pancreatic tumor growth by posttranscriptional activation of Numbl and indirect inhibition of Notch (Nwaeburu et al., 2016); Zheng et al. (2014) reported that nanoliposomal quercetin combined with butyrate modulated aberrant epigenetic alteration in Eca9706 cells via epigenetic-NF-κB signaling. In this study reverse expressions of global DNMT1, HDAC1, NF-κB p65 and Cyclin D1 were down-regulated, although expressions of p16INK4α and caspase-3 were up-regulated. Furthermore, quercetin modulate miR-1-3p/TAGLN2 (Wang et al., 2021b), miR-197/IGFBP5 (Hu et al., 2020), miR-16–5p/WEE1 (Wang et al., 2020a), miR-22/WNT1/β-catenin (Zhang et al., 2019a), miR-16/HOXA10 (Zhao et al., 2019), TP53/miR-15/miR-16 (Ahmed Youness et al., 2018) axes as well as miR15a/16 (Ramos et al., 2021), miR-200b-3p (Nwaeburu et al., 2017), miR-145 (Zhou et al., 2015), and miR-146a (Tao et al., 2015) in various cancers.
Kaempferol (3,4′,5,7-tetrahydroxyflavone) is a potential HDAC inhibitor and an anti-cancer agent against many types of cancers (Imran et al., 2019). Berger et al., reported first time that kaempferol has a distinctive epigenetic activity by inhibition of HDACs (Berger et al., 2013). The in-silico docking analysis fits kaempferol into binding pocket of HDAC2, 4, 7 or 8 and in vitro profiling of all conserved human HDACs of class I, II and IV demonstrated that it inhibited all tested HDACs. Furthermore, kaempferol stimulates hyperacetylation of histone H3 in HepG2 and Hep3B (hepatoma cancer cell lines) as well as on HCT-116 (colon cancer cell line) (Berger et al., 2013). Kaempferol induces autophagic cell death via IRE1-JNK-CHOP signaling and inhibiting HDAC/G9a axis in gastric cancer cells (Kim et al., 2018). In lung cancer A549 cell, kaempferol induces miR-340 expression (Han et al., 2018) which is known to induce apoptosis and inhibit cell proliferation in NSCLC (Fernandez et al., 2014).
Flavonol, fisetin is also a powerful anticancer agent, used to inhibit different stages of cancer cells, induce apoptosis, inhibit cell growth, prevent cell cycle progression, cause PARP cleavage, and modulate the expressions of Bcl-2 family proteins in various cancer cell lines (Imran et al., 2020). It also suppresses the activation of the ROS/ PKCα/ p38 MAPK and ERK1/2 signaling pathways, down-regulates the level of the oncoprotein securin and lowers the NF-κB activation (Pal et al., 2016). Recently, Ding et al., revealed that fisetin inhibits proliferation of pancreatic adenocarcinoma by inducing DNA damage via RFXAP/KDM4A-dependent histone H3K36 demethylation (Ding et al., 2020).
Flavones
Flavones are a group of flavonoids that contain the backbone of 2-phenylchromen-4-one (2-phenyl-1-benzopyran-4-one) having diverse pharmacological properties and are commonly found in herbs such as celery, parsley and in almost all edible cereal species. Apigenin, luteolin, tangeretin, chrysin, Tricin, baicalein, rhoifolin and 6-hydroxyflavone are some common flavones (Barreca et al., 2020).
Pandey et al. (2012) reported first time that apigenin inhibits class I HDACs, particularly HDAC1 and HDAC3, alters chromatin to induce growth arrest and apoptosis in human prostate cancer cells. Apigenin inhibited MDA-MB-231 breast cancer cell proliferation and tumor growth by induction of G2/M arrest and histone H3 acetylation-mediated p21 expression (Tseng et al., 2017). Apigenin enhances miR-16 (Chen et al., 2016) and miRNA215–5p (Cheng et al., 2021) expression to inhibits glioma and colon cancer growth respectively, as well as chemo-sensitize doxorubicin-resistant liver cancer cells by targeting miR-520b/ATG7 axis (Gao et al., 2018).
Recently Wu et al. (2021) discovered that luteolin inhibited the proliferation and metastasis of androgen receptor-positive triple-negative breast cancer cell by epigenetic regulation of MMP9 expression through a reduction in the levels of AKT/mTOR-inducing H3K56Ac and H3K27Ac. Earlier it was also revealed that luteolin suppresses the metastasis of triple-negative breast cancer, downregulates the ß-catenin expression for reversing epithelial-to-mesenchymal transition (Lin et al., 2017). In colorectal cancer cells Luteolin induces apoptosis by the downregulations of UHRF1, calpain, and DNMT1 expressions. This research further suggests that calpain might be involved in the epigenetic code inheritance by regulating the epigenetic integrator UHRF1(Krifa et al., 2014). In human prostate cancer (PC-3) cells, Luteolin and gefitinib regulate cell cycle pathway genes (CCNA2, CCNE2, CDC25A, CDKN1B, and PLK-1) through a mutual mechanism involving EGFR-associated tyrosine kinase (Markaverich et al., 2010). Authors suggested that these phytochemicals likely modulate the epigenetic control of gene expression as previously shown by their group that luteolin interacts with type II binding sites on histone H4 (Shoulars et al., 2010). Recently, Farooqi et al. and Mishan et al. have systematically reviewed the potent ability of luteolin to modulate miRNA expression in various cancers (Farooqi et al., 2020; Mishan et al., 2021). A derivative of tangeretin prevents the progress of human prostate cancer cells by epigenetically restoring p21 gene expression and inhibits cancer stem-like cell proliferation (Wei et al., 2019).
Baicalein (5,6,7-trihydroxyflavone) suppresses cancer cell proliferation, cell cycle arrest and induces apoptosis in human prostate, breast, T24 bladder and myeloma cancer cells (Gao et al., 2016). Lai et al., studied the epigenetic role of baicalin hydrate in nasopharyngeal carcinoma (NPC) and identified that it inhibits NPC cell growth both in vivo and in vitro. Furthermore, instead of DNA methylation, baicalin hydrate increased of m6A RNA methylation and promoted Suv39H1 gene splicing. (Lai et al., 2018). It is also documented that baicalin improves the developmental proficiency of in vitro cultured mouse embryos through reticence of cellular apoptosis and HSP70 expression and enhancement of DNA methylation (Qi et al., 2016). Several recent studies have shown that baicalein modulate the expression of miR-183 (Lei et al., 2021), miR-139–3p and miR-196b-5p (Ma et al., 2021), and miR-25 (Örenlil Yaylagül and Ülger, 2020) in various cancers.
Flavanones
Flavanones are aromatic, colourless ketones mainly present in citrus fruits (Barreca et al., 2017). Hesperetin, isosakuranetin, naringin, naringenin, isosakuranetin and eriodictyol and their particular glycosides are some main flavanones (Khan et al., 2013). Hesperetin is a common citrus flavanone that endorses DOT1L degradation and decreases histone H3K79 methylation to prevent gastric cancer metastasis, showing its epigenetic effect (Wang et al., 2021a). Natural flavonolignan, silibinin is the most effective phytochemical of silymarin. It is active both alone and in combination with other chemotherapeutic and epigenetic agents, substantially inhibit the growth of different cancer cells. It synergizes with DNA methyltransferase and histone deacetylase inhibitors in upregulating e-cadherin expression, also inhibits the invasion and migration of human non-small cell lung cancer cells. These results are highly substantial since failure of E-cadherin and metastatic dispersed of the illness via epithelial-to-mesenchymal transition is associated with poor prognosis and high mortalities in this type of cancer cells (Mateen et al., 2013). In human prostate cancer (DU145 and PC3) cells, silibinin reduces gene expression levels of EZH2 accompanied by an increase in H3K27me3 levels (Anestopoulos et al., 2016). Such responses were dependent on decreased expression levels of phosphorylated EZH2 (ser21) and phosphorylated Akt (ser473). Moreover, it also exerted other epigenetic impacts involving, decrease histone deacetylases 1–2 (HDACs1-2) expression levels while it increases total DNA methyltransferase (DNMT) activity, proving that it induces epigenetic alterations in human prostate cancer cells. (Anestopoulos et al., 2016). Hossainzadeh et al. reported that silibinin encapsulated in polymersome nanoparticles supress the expression of oncogenic miRNAs miR-125b and miR-182 (Hossainzadeh et al., 2019).
Naringenin (4,5,7 trihydroxyflavanone) is an aglycone form of naringin found in citrus fruits. When it combined with suberoylanilide hydroxamic acid (HDAC inhibitor), synergistically improved transamidation activity and suberoylanilide hydroxamic acid induced cytotoxicity in neuroblastoma cells which showed no cytotoxicity on normal non-malignant cells (Ling et al., 2012). This suggest that naringenin possesses effective histone deacetylase inhibitory activity; however, more comprehensive studies are needed understand its epigenetic potential.
Isoflavones
Isoflavones are naturally occurring isoflavonoids, mainly found in legumes, soy beans, and soy foods. They have several potent pharmacological activities like anti-inflammatory, antioxidant, antimicrobial, and anticancer (Panche et al., 2016). It is also well evidential that they exert estrogenic and/or antiestrogenic impacts. Isoflavones are considered as chemoprotective in nature and used in various type of alternative therapies for a broad range of hormonal ailments including several types of cancers, osteoporosis, menopausal problems and cardiovascular diseases (Taku et al., 2011; Vitale et al., 2012; Qin et al., 2013; Sathyapalan et al., 2018; Nakai et al., 2020). There are conflicting reports that isoflavones disrupt endocrine function; however, it appears the most common harmful effect is mild and appears at the gastrointestinal level (Křížová et al., 2019; Gómez-Zorita et al., 2020). Some common examples of isoflavones are Daidzein, Genistein, Genistin and Glycitein.
Among all isoflavones, genistein is the most potent and biologically active phytochemical, demonstrating different in vivo and in vitro anticancer and anti-proliferative effects on numerous types of human cancers (Spagnuolo et al., 2015). Prostate cell lines (DU-145 and PC-3) when treated with soy phytoestrogens, genistein and daidzein, cause decrease in DNA methylation at EPHB2, BRCA1 and GSTP1 promoters (Adjakly et al., 2011). Karsli-Ceppioglu et al., reported that phytoestrogens (genistein and daidzein) modulate genome-wide DNA methylation status in prostate cancer. They found that methylation profiles of 58 genes have been modified by genistein and daidzein treatments in prostate cancer DU-145 and LNCaP cell lines (Karsli-Ceppioglu et al., 2015). Notably, the methylation frequencies of the hTERT, MAD1L1, KDM4B and TRAF7 genes were remarkably altered by genistein treatment. Genistein regulates miRNAs expression in pan cancer (Javed et al., 2021). In head and neck cancer, genistein activate miR-34a/RTCB axis that results in ROS-associated apoptosis, decrease in stemness properties, and inhibition of EMT (Hsieh et al., 2020). Recently, Imai-Sumida et al., reported that genistein suppress kidney cancer by repressing HOTAIR/chromatin remodeling pathways (Imai-Sumida et al., 2020). Conversely, Allred et al., reported that dietary genistin as well as genistein can stimulates estrogen-dependent breast cancer cell growth in vivo (Allred et al., 2001). These conflicting reports compel a need for in-depth mechanistic studies to understand the biological effects of these dietary compounds, including their epigenetic modifying potentials, cytotoxicity, and anticancer properties.
Anthocyanidins
Anthocyanidins are the sugar-free counterparts of anthocyanins and are highly important water-soluble pigments found in plants. Among all anthocyanidins, delphinidin is the most potent and abundant flavonoid found in pigmented fruits (especially blueberry) and vegetables (Khoo et al., 2017).
Kuo et al., reported that delphinidin epigenetically re-activates Nrf2-ARE pathway and prevents neoplastic transformation of mouse skin JB6 P+ cells (Kuo et al., 2019). The Nrf2-ARE pathway activation was associated with demethylation of 15 CpG sites in the mouse Nrf2 promoter region between nucleotide -1,226 and -863 from the transcription start site. The decreased CpG methylation proportion in the Nrf2 promoter region was consistent with detected declines in the protein expression of DNA methyltransferases 1 (DNMT1), DNMT3a, and class I/II histone deacetylases (HDACs) (Kuo et al., 2019). Jeong et al., identified the epigenetic modulators that mediate the apoptotic effect of delphinidin (major anthocyanidin compound) in human prostate cancer cells (Jeong et al., 2016). They treated these cancer cells with delphinidin and observed an increase in caspase-3, -7, and -8 activity along with an increased histone deacetylase activity. Amongst all class I HDACs, the activity of HDAC3 was exclusively prevented by delphinidin. Moreover, the apoptosis induced by delphinidin was reliant on caspase-mediated cleavage of HDAC3, resulting in the stabilization and acetylation of p53. They also observed that this anthocyanidin effectively upregulated pro-apoptotic genes that are definitely regulated by p53 and also downregulated numerous anti-apoptotic genes (Jeong et al., 2016). Delphinidin targets HOTAIR/miR-34a axis to suppress the breast carcinogenesis (Han et al., 2019) and inhibits colorectal cancer metastasis by inducing miR-204–3p expression (Huang et al., 2019).
Conclusion
The existing literatures on cancer pathobiology has established that genetics and epigenetics play a central role in cancer initiation and progression. The reversible nature of epigenetic changes can be exploit for a better therapeutic intervention. Epigenetic modifiers such as DNMTs, HATs, HMTs, HDACs, and others can be modulated or inhibited by naturally occurring substances such as phytochemicals (Figure 2). In this review, several flavonoids (e.g., flavones, flavanones, flavonols) were described along with their epigenetic modulation, therapeutic, and chemo-preventive potentials in cancer (Table 1). Various plant-based drugs, including vinca alkaloids (e.g., vinblastine and vincristine), taxanes (e.g., paclitaxel and docetaxel), camptothecins, and others are routinely used for cancer treatment. There are numerous phytochemicals, including flavonoids with potent anticancer activity are in various preclinical and clinical trial stages. However, there are some limitations, including their low water solubility, poor bioavailability, rapid uptake by normal cells, poor therapeutic index and adverse effects on liver. Developing novel strategies such as nanocarriers can overcome these drawbacks.
The emerging role of phytochemicals in cancer therapy and prevention is enormous. Their potential to rectify epigenetic alterations of cancer cells add substantial armaments in fight against cancer. Combinatorial therapies using plant-based epigenetic modifiers with existing chemo and targeted therapies can help manage the disease better and reduce the side effects. However, an extensive research is needed into identification and characterization of anticarcinogenic phytochemicals and their respective mechanisms of actions. Given the fast pace of technological developments, this seems to be a promising pursuit in our battle against cancer.
Author Contributions
NF and SSRB wrote the initial manuscript draft; AB, NK-KK, and KH helped finalizing the figures, provided inputs in epigenetic components of the manuscript; and AA and RAA conceptualized, supervised, and finalized the draft.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, A., Hall, J. A., Patterson, W. L., Ho, E., Hsu, A., Al-Mulla, F., et al. (2016). Sulforaphane Modulates Telomerase Activity via Epigenetic Regulation in Prostate Cancer Cell Lines. Biochem. Cel Biol. 94, 71–81. doi:10.1139/bcb-2015-0038
Abbas, A., Patterson, W., and Georgel, P. T. (2013). The Epigenetic Potentials of Dietary Polyphenols in Prostate Cancer Management. Biochem. Cel Biol. 91, 361–368. doi:10.1139/bcb-2012-0044
Adjakly, M., Bosviel, R., Rabiau, N., Boiteux, J.-P., Bignon, Y.-J., Guy, L., et al. (2011). DNA Methylation and Soy Phytoestrogens: Quantitative Study in DU-145 and PC-3 Human Prostate Cancer Cell Lines. Epigenomics 3, 795–803. doi:10.2217/epi.11.103
Ahmed, F., Ijaz, B., Ahmad, Z., Farooq, N., Sarwar, M. B., and Husnain, T. (2020). Modification of miRNA Expression through Plant Extracts and Compounds against Breast Cancer: Mechanism and Translational Significance. Phytomedicine 68, 153168. doi:10.1016/j.phymed.2020.153168
Ahmed Youness, R., Amr Assal, R., Mohamed Ezzat, S., Zakaria Gad, M., and Abdel Motaal, A. (2018). A Methoxylated Quercetin Glycoside Harnesses HCC Tumor Progression in a TP53/miR-15/miR-16 Dependent Manner. Nat. Product. Res. 34, 1475–1480. doi:10.1080/14786419.2018.1509326
Ali Syeda, Z., Langden, S. S. S., Munkhzul, C., Lee, M., and Song, S. J. (2020). Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 21, 1723. doi:10.3390/ijms21051723
Allred, C. D., Ju, Y. H., Allred, K. F., Chang, J., and Helferich, W. G. (2001). Dietary Genistin Stimulates Growth of Estrogen-dependent Breast Cancer Tumors Similar to that Observed with Genistein. Carcinogenesis 22, 1667–1673. doi:10.1093/carcin/22.10.1667
Alvarez, M. C., Maso, V., Torello, C. O., Ferro, K. P., and Saad, S. T. O. (2018). The Polyphenol Quercetin Induces Cell Death in Leukemia by Targeting Epigenetic Regulators of Pro-apoptotic Genes. Clin. Epigenet 10, 139. doi:10.1186/s13148-018-0563-3
Anand, P., Kunnumakara, A. B., Sundaram, C., Harikumar, K. B., Tharakan, S. T., Lai, O. S., et al. (2008). Cancer Is a Preventable Disease that Requires Major Lifestyle Changes. Pharm. Res. 25, 2097–2116. doi:10.1007/s11095-008-9661-9
Anestopoulos, I., Sfakianos, A. P., Franco, R., Chlichlia, K., Panayiotidis, M. I., Kroll, D. J., et al. (2016). A Novel Role of Silibinin as a Putative Epigenetic Modulator in Human Prostate Carcinoma. Molecules 22. doi:10.3390/molecules22010062
Audia, J. E., and Campbell, R. M. (2016). Histone Modifications and Cancer. Cold Spring Harb Perspect. Biol. 8, a019521. doi:10.1101/cshperspect.a019521
Baer, C., Claus, R., and Plass, C. (2013). Genome-Wide Epigenetic Regulation of miRNAs in Cancer. Cancer Res. 73, 473–477. doi:10.1158/0008-5472.can-12-3731
Bannister, A. J., and Kouzarides, T. (2011). Regulation of Chromatin by Histone Modifications. Cell Res 21, 381–395. doi:10.1038/cr.2011.22
Barreca, D., Gattuso, G., Bellocco, E., Calderaro, A., Trombetta, D., Smeriglio, A., et al. (2017). Flavanones: Citrus Phytochemical with Health-Promoting Properties. BioFactors 43, 495–506. doi:10.1002/biof.1363
Barreca, D., Mandalari, G., Calderaro, A., Smeriglio, A., Trombetta, D., Felice, M. R., et al. (2020). Citrus Flavones: An Update on Sources, Biological Functions, and Health Promoting Properties. Plants (Basel) 9. doi:10.3390/plants9030288
Bennett, R. L., and Licht, J. D. (2018). Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol. 58, 187–207. doi:10.1146/annurev-pharmtox-010716-105106
Berger, A., Venturelli, S., Kallnischkies, M., Böcker, A., Busch, C., Weiland, T., et al. (2013). Kaempferol, a New Nutrition-Derived Pan-Inhibitor of Human Histone Deacetylases. J. Nutr. Biochem. 24, 977–985. doi:10.1016/j.jnutbio.2012.07.001
Bird, A. (2002). DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 16, 6–21. doi:10.1101/gad.947102
Blake, L. E., Roux, J., Hernando-Herraez, I., Banovich, N. E., Perez, R. G., Hsiao, C. J., et al. (2020). A Comparison of Gene Expression and DNA Methylation Patterns across Tissues and Species. Genome Res. 30, 250–262. doi:10.1101/gr.254904.119
Bonner, W. M., Redon, C. E., Dickey, J. S., Nakamura, A. J., Sedelnikova, O. A., Solier, S., et al. (2008). γH2AX and Cancer. Nat. Rev. Cancer 8, 957–967. doi:10.1038/nrc2523
Borutinskaitė, V., Virkšaitė, A., Gudelytė, G., and Navakauskienė, R. (2018). Green tea Polyphenol EGCG Causes Anti-cancerous Epigenetic Modulations in Acute Promyelocytic Leukemia Cells. Leuk. Lymphoma 59, 469–478. doi:10.1080/10428194.2017.1339881
Brower, V. (2011). Epigenetics: Unravelling the Cancer Code. Nature 471, S12–S13. doi:10.1038/471s12a
Busch, C., Burkard, M., Leischner, C., Lauer, U. M., Frank, J., and Venturelli, S. (2015). Epigenetic Activities of Flavonoids in the Prevention and Treatment of Cancer. Clin. Epigenet 7, 64. doi:10.1186/s13148-015-0095-z
Cammaerts, S., Strazisar, M., De Rijk, P., and Del Favero, J. (2015). Genetic Variants in microRNA Genes: Impact on microRNA Expression, Function, and Disease. Front. Genet. 6, 186. doi:10.3389/fgene.2015.00186
Carlos-Reyes, Á., López-González, J. S., Meneses-Flores, M., Gallardo-Rincón, D., Ruíz-García, E., Marchat, L. A., et al. (2019). Dietary Compounds as Epigenetic Modulating Agents in Cancer. Front. Genet. 10, 79. doi:10.3389/fgene.2019.00079
Cavalli, G., and Heard, E. (2019). Advances in Epigenetics Link Genetics to the Environment and Disease. Nature 571, 489–499. doi:10.1038/s41586-019-1411-0
Cheedipudi, S., Genolet, O., and Dobreva, G. (2014). Epigenetic Inheritance of Cell Fates during Embryonic Development. Front. Genet. 5, 19. doi:10.3389/fgene.2014.00019
Chen, X.-J., Wu, M.-Y., Li, D.-H., and You, J. (2016). Apigenin Inhibits Glioma Cell Growth through Promoting microRNA-16 and Suppression of BCL-2 and Nuclear Factor-Κb/mmp-9. Mol. Med. Rep. 14, 2352–2358. doi:10.3892/mmr.2016.5460
Cheng, Y., Han, X., Mo, F., Zeng, H., Zhao, Y., Wang, H., et al. (2021). Apigenin Inhibits the Growth of Colorectal Cancer through Down-Regulation of E2F1/3 by miRNA-215-5p. Phytomedicine 89, 153603. doi:10.1016/j.phymed.2021.153603
Cheung, H.-H., Lee, T.-L., Rennert, O. M., and Chan, W.-Y. (2009). DNA Methylation of Cancer Genome. Birth Defect Res. C 87, 335–350. doi:10.1002/bdrc.20163
Ciesielski, O., Biesiekierska, M., and Balcerczyk, A. (2020). Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules 25, 2326. doi:10.3390/molecules25102326
Dawson, M. A., and Kouzarides, T. (2012). Cancer Epigenetics: From Mechanism to Therapy. Cell 150, 12–27. doi:10.1016/j.cell.2012.06.013
Deaton, A. M., and Bird, A. (2011). CpG Islands and the Regulation of Transcription. Genes Dev. 25, 1010–1022. doi:10.1101/gad.2037511
Deb, G., Shankar, E., Thakur, V. S., Ponsky, L. E., Bodner, D. R., Fu, P., et al. (2019). Green tea-induced Epigenetic Reactivation of Tissue Inhibitor of Matrix Metalloproteinase-3 Suppresses Prostate Cancer Progression through Histone-Modifying Enzymes. Mol. Carcinog 58, 1194–1207. doi:10.1002/mc.23003
Di Cerbo, V., and Schneider, R. (2013). Cancers with Wrong HATs: the Impact of Acetylation. Brief. Funct. Genomics 12, 231–243. doi:10.1093/bfgp/els065
Ding, G., Xu, X., Li, D., Chen, Y., Wang, W., Ping, D., et al. (2020). Fisetin Inhibits Proliferation of Pancreatic Adenocarcinoma by Inducing DNA Damage via RFXAP/KDM4A-dependent Histone H3K36 Demethylation. Cell Death Dis 11, 893. doi:10.1038/s41419-020-03019-2
Egger, G., Liang, G., Aparicio, A., and Jones, P. A. (2004). Epigenetics in Human Disease and Prospects for Epigenetic Therapy. Nature 429, 457–463. doi:10.1038/nature02625
Esquela-Kerscher, A., and Slack, F. J. (2006). Oncomirs - microRNAs with a Role in Cancer. Nat. Rev. Cancer 6, 259–269. doi:10.1038/nrc1840
Esteller, M. (2007). Cancer Epigenomics: DNA Methylomes and Histone-Modification Maps. Nat. Rev. Genet. 8, 286–298. doi:10.1038/nrg2005
Fang, M. Z., Wang, Y., Ai, N., Hou, Z., Sun, Y., Lu, H., et al. (2003). Tea Polyphenol (-)-Epigallocatechin-3-Gallate Inhibits DNA Methyltransferase and Reactivates Methylation-Silenced Genes in Cancer Cell Lines. Cancer Res. 63, 7563–7570.
Fang, M. Z., Chen, D., Sun, Y., Jin, Z., Christman, J. K., and Yang, C. S. (2005). Reversal of Hypermethylation and Reactivation ofp16INK4a, RARβ, andMGMTGenes by Genistein and Other Isoflavones from Soy. Clin. Cancer Res. 11, 7033–7041. doi:10.1158/1078-0432.ccr-05-0406
Farooqi, A. A., Butt, G., El-Zahaby, S. A., Attar, R., Sabitaliyevich, U. Y., Jovic, J. J., et al. (2020). Luteolin Mediated Targeting of Protein Network and microRNAs in Different Cancers: Focus on JAK-STAT, NOTCH, mTOR and TRAIL-Mediated Signaling Pathways. Pharmacol. Res. 160, 105188. doi:10.1016/j.phrs.2020.105188
Fernandez, S., Risolino, M., Mandia, N., Talotta, F., Soini, Y., Incoronato, M., et al. (2014). miR-340 Inhibits Tumor Cell Proliferation and Induces Apoptosis by Targeting Multiple Negative Regulators of P27 in Non-small Cell Lung Cancer. Oncogene 34, 3240–3250. doi:10.1038/onc.2014.267
Fraga, M. F., Ballestar, E., Villar-Garea, A., Boix-Chornet, M., Espada, J., Schotta, G., et al. (2005). Loss of Acetylation at Lys16 and Trimethylation at Lys20 of Histone H4 Is a Common Hallmark of Human Cancer. Nat. Genet. 37, 391–400. doi:10.1038/ng1531
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res. 19, 92–105. doi:10.1101/gr.082701.108
Galati, G., and O'brien, P. J. (2004). Potential Toxicity of Flavonoids and Other Dietary Phenolics: Significance for Their Chemopreventive and Anticancer Properties. Free Radic. Biol. Med. 37, 287–303. doi:10.1016/j.freeradbiomed.2004.04.034
Gao, A.-M., Zhang, X.-Y., Hu, J.-N., and Ke, Z.-P. (2018). Apigenin Sensitizes Hepatocellular Carcinoma Cells to Doxorubic through Regulating miR-520b/ATG7 axis. Chemico-Biological Interactions 280, 45–50. doi:10.1016/j.cbi.2017.11.020
Gao, F., and Das, S. K. (2014). Epigenetic Regulations through DNA Methylation and Hydroxymethylation: Clues for Early Pregnancy in Decidualization. Biomol. Concepts 5, 95–107. doi:10.1515/bmc-2013-0036
Gao, Y., Snyder, S. A., Smith, J. N., and Chen, Y. C. (2016). Anticancer Properties of Baicalein: a Review. Med. Chem. Res. 25, 1515–1523. doi:10.1007/s00044-016-1607-x
Giudice, A., Montella, M., Boccellino, M., Crispo, A., D'arena, G., Bimonte, S., et al. (2018). Epigenetic Changes Induced by Green Tea Catechins a Re Associated with Prostate Cancer. Curr. Mol. Med. 17, 405–420. doi:10.2174/1566524018666171219101937
Gómez-Zorita, S., González-Arceo, M., Fernández-Quintela, A., Eseberri, I., Trepiana, J., and Portillo, M. P. (2020). Scientific Evidence Supporting the Beneficial Effects of Isoflavones on Human Health. Nutrients 12, 3853. doi:10.3390/nu12123853
Greenberg, M. V. C., and Bourc’his, D. (2019). The Diverse Roles of DNA Methylation in Mammalian Development and Disease. Nat. Rev. Mol. Cel Biol 20, 590–607. doi:10.1038/s41580-019-0159-6
Han, B., Peng, X., Cheng, D., Zhu, Y., Du, J., Li, J., et al. (2019). Delphinidin Suppresses Breast Carcinogenesis through the HOTAIR/micro RNA ‐34a axis. Cancer Sci. 110, 3089–3097. doi:10.1111/cas.14133
Han, X., Liu, C.-F., Gao, N., Zhao, J., and Xu, J. (2018). Kaempferol Suppresses Proliferation but Increases Apoptosis and Autophagy by Up-Regulating microRNA-340 in Human Lung Cancer Cells. Biomed. Pharmacother. 108, 809–816. doi:10.1016/j.biopha.2018.09.087
Henning, S. M., Wang, P., Carpenter, C. L., and Heber, D. (2013). Epigenetic Effects of green tea Polyphenols in Cancer. Epigenomics 5, 729–741. doi:10.2217/epi.13.57
Herman, J. G., and Baylin, S. B. (2003). Gene Silencing in Cancer in Association with Promoter Hypermethylation. N. Engl. J. Med. 349, 2042–2054. doi:10.1056/nejmra023075
Hossainzadeh, S., Ranji, N., Naderi Sohi, A., and Najafi, F. (2019). Silibinin Encapsulation in Polymersome: A Promising Anticancer Nanoparticle for Inducing Apoptosis and Decreasing the Expression Level of miR‐125b/miR‐182 in Human Breast Cancer Cells. J. Cel Physiol 234, 22285–22298. doi:10.1002/jcp.28795
Hou, Y., Zhang, R., and Sun, X. (2019). Enhancer LncRNAs Influence Chromatin Interactions in Different Ways. Front. Genet. 10, 936. doi:10.3389/fgene.2019.00936
Hsieh, P. L., Liao, Y. W., Hsieh, C. W., Chen, P. N., and Yu, C. C. (2020). Soy Isoflavone Genistein Impedes Cancer Stemness and Mesenchymal Transition in Head and Neck Cancer through Activating miR-34a/RTCB Axis. Nutrients 12, 1924. doi:10.3390/nu12071924
Hu, S. A., Cheng, J., Zhao, W. H., and Zhao, H. Y. (2020). Quercetin Induces Apoptosis in Meningioma Cells through the miR-197/IGFBP5 cascade. Environ. Toxicol. Pharmacol. 80, 103439. doi:10.1016/j.etap.2020.103439
Huang, C. C., Hung, C. H., Hung, T. W., Lin, Y. C., Wang, C. J., and Kao, S. H. (2019). Dietary Delphinidin Inhibits Human Colorectal Cancer Metastasis Associating with Upregulation of miR-204-3p and Suppression of the Integrin/FAK axis. Sci. Rep. 9, 18954. doi:10.1038/s41598-019-55505-z
Huarte, M. (2015). The Emerging Role of lncRNAs in Cancer. Nat. Med. 21, 1253–1261. doi:10.1038/nm.3981
Imai-Sumida, M., Dasgupta, P., Kulkarni, P., Shiina, M., Hashimoto, Y., Shahryari, V., et al. (2020). Genistein Represses HOTAIR/Chromatin Remodeling Pathways to Suppress Kidney Cancer. Cell Physiol Biochem 54, 53–70. doi:10.33594/000000205
Imran, M., Salehi, B., Sharifi-Rad, J., Aslam Gondal, T., Saeed, F., Imran, A., et al. (2019). Kaempferol: A Key Emphasis to its Anticancer Potential. Molecules 24. doi:10.3390/molecules24122277
Imran, M., Saeed, F., Gilani, S. A., Shariati, M. A., Imran, A., Afzaal, M., et al. (2020). Fisetin: An Anticancer Perspective. Food Sci. Nutr. 9, 3–16. doi:10.1002/fsn3.1872
Izzo, S., Naponelli, V., and Bettuzzi, S. (2020). Flavonoids as Epigenetic Modulators for Prostate Cancer Prevention. Nutrients 12, 1010. doi:10.3390/nu12041010
Javed, Z., Khan, K., Herrera-Bravo, J., Naeem, S., Iqbal, M. J., Sadia, H., et al. (2021). Genistein as a Regulator of Signaling Pathways and microRNAs in Different Types of Cancers. Cancer Cel Int 21, 388. doi:10.1186/s12935-021-02091-8
Jeong, M.-H., Ko, H., Jeon, H., Sung, G.-J., Park, S.-Y., Jun, W. J., et al. (2016). Delphinidin Induces Apoptosis via Cleaved HDAC3-Mediated P53 Acetylation and Oligomerization in Prostate Cancer Cells. Oncotarget 7, 56767–56780. doi:10.18632/oncotarget.10790
Jiang, W., Xia, T., Liu, C., Li, J., Zhang, W., and Sun, C. (2021). Remodeling the Epigenetic Landscape of Cancer-Application Potential of Flavonoids in the Prevention and Treatment of Cancer. Front. Oncol. 11, 705903. doi:10.3389/fonc.2021.705903
Jin, H., Chen, J. X., Wang, H., Lu, G., Liu, A., Li, G., et al. (2015). NNK-induced DNA Methyltransferase 1 in Lung Tumorigenesis in A/J Mice and Inhibitory Effects of (−)-Epigallocatechin-3-Gallate. Nutr. Cancer 67, 167–176. doi:10.1080/01635581.2015.976314
Jones, P. A. (2012). Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi:10.1038/nrg3230
Jucá, M. M., Cysne Filho, F. M. S., De Almeida, J. C., Mesquita, D. D. S., Barriga, J. R. D. M., Dias, K. C. F., et al. (2018). Flavonoids: Biological Activities and Therapeutic Potential. Nat. Product. Res. 34, 692–705. doi:10.1080/14786419.2018.1493588
Kang, Q., Zhang, X., Cao, N., Chen, C., Yi, J., Hao, L., et al. (2019). EGCG Enhances Cancer Cells Sensitivity under 60Coγ Radiation Based on miR-34a/Sirt1/p53. Food Chem. Toxicol. 133, 110807. doi:10.1016/j.fct.2019.110807
Kang, Q., Tong, Y., Gowd, V., Wang, M., Chen, F., and Cheng, K.-W. (2021). Oral Administration of EGCG Solution Equivalent to Daily Achievable Dosages of Regular tea Drinkers Effectively Suppresses miR483-3p Induced Metastasis of Hepatocellular Carcinoma Cells in Mice. Food Funct. 12, 3381–3392. doi:10.1039/d1fo00664a
Karlic, R., Chung, H.-R., Lasserre, J., Vlahovicek, K., and Vingron, M. (2010). Histone Modification Levels Are Predictive for Gene Expression. Proc. Natl. Acad. Sci. 107, 2926–2931. doi:10.1073/pnas.0909344107
Karsli-Ceppioglu, S., Ngollo, M., Adjakly, M., Dagdemir, A., Judes, G., Lebert, A., et al. (2015). Genome-wide DNA Methylation Modified by Soy Phytoestrogens: Role for Epigenetic Therapeutics in Prostate Cancer? OMICS: A J. Integr. Biol. 19, 209–219. doi:10.1089/omi.2014.0142
Kedhari Sundaram, M., Haque, S., Somvanshi, P., Bhardwaj, T., and Hussain, A. (2020). Epigallocatechin Gallate Inhibits HeLa Cells by Modulation of Epigenetics and Signaling Pathways. 3 Biotech. 10, 484. doi:10.1007/s13205-020-02473-1
Kedhari Sundaram, M., Hussain, A., Haque, S., Raina, R., and Afroze, N. (2019). Quercetin Modifies 5′CpG Promoter Methylation and Reactivates Various Tumor Suppressor Genes by Modulating Epigenetic marks in Human Cervical Cancer Cells. J. Cel Biochem 120, 18357–18369. doi:10.1002/jcb.29147
Khan, H., Belwal, T., Efferth, T., Farooqi, A. A., Sanches-Silva, A., Vacca, R. A., et al. (2021). Targeting Epigenetics in Cancer: Therapeutic Potential of Flavonoids. Crit. Rev. Food Sci. Nutr. 61, 1616–1639. doi:10.1080/10408398.2020.1763910
Khan, M. A., Hussain, A., Sundaram, M. K., Alalami, U., Gunasekera, D., Ramesh, L., et al. (2015). (−)-Epigallocatechin-3-gallate Reverses the Expression of Various Tumor-Suppressor Genes by Inhibiting DNA Methyltransferases and Histone Deacetylases in Human Cervical Cancer Cells. Oncol. Rep. 33, 1976–1984. doi:10.3892/or.2015.3802
Khan, M., Zill, E. H., and Dangles, O. (2013). A Comprehensive Review on Flavanones, the Major Citrus Polyphenols. J. Food Compost. Anal. 33, 85-104. doi:10.1016/j.jfca.2013.11.004
Khoo, H. E., Azlan, A., Tang, S. T., and Lim, S. M. (2017). Anthocyanidins and Anthocyanins: Colored Pigments as Food, Pharmaceutical Ingredients, and the Potential Health Benefits. Food Nutr. Res. 61, 1361779. doi:10.1080/16546628.2017.1361779
Kim, T. W., Lee, S. Y., Kim, M., Cheon, C., and Ko, S.-G. (2018). Kaempferol Induces Autophagic Cell Death via IRE1-JNK-CHOP Pathway and Inhibition of G9a in Gastric Cancer Cells. Cel Death Dis 9, 875. doi:10.1038/s41419-018-0930-1
Krifa, M., Leloup, L., Ghedira, K., Mousli, M., and Chekir-Ghedira, L. (2014). Luteolin Induces Apoptosis in BE Colorectal Cancer Cells by Downregulating Calpain, UHRF1, and DNMT1 Expressions. Nutr. Cancer 66, 1220–1227. doi:10.1080/01635581.2014.951729
Křížová, L., Dadáková, K., Kašparovská, J., and Kašparovský, T. (2019). Isoflavones. Molecules 24, 1076. doi:10.3390/molecules24061076
Kumar, S., and Pandey, A. K. (2013). Chemistry and Biological Activities of Flavonoids: an Overview. Sci.Wor.J. 2013, 162750. doi:10.1155/2013/162750
Kuo, H.-C. D., Wu, R., Li, S., Yang, A. Y., and Kong, A.-N. (2019). Anthocyanin Delphinidin Prevents Neoplastic Transformation of Mouse Skin JB6 P+ Cells: Epigenetic Re-activation of Nrf2-ARE Pathway. Aaps j 21, 83. doi:10.1208/s12248-019-0355-5
Kurokawa, R., Rosenfeld, M. G., and Glass, C. K. (2014). Transcriptional Regulation through Noncoding RNAs and Epigenetic Modifications. RNA Biol. 6, 233–236. doi:10.4161/rna.6.3.8329
Lacouture, M., and Sibaud, V. (2018). Toxic Side Effects of Targeted Therapies and Immunotherapies Affecting the Skin, Oral Mucosa, Hair, and Nails. Am. J. Clin. Dermatol. 19, 31–39. doi:10.1007/s40257-018-0384-3
Lai, W., Jia, J., Yan, B., Jiang, Y., Shi, Y., Chen, L., et al. (2018). Baicalin Hydrate Inhibits Cancer Progression in Nasopharyngeal Carcinoma by Affecting Genome Instability and Splicing. Oncotarget 9, 901–914. doi:10.18632/oncotarget.22868
Lee, Y. H., Kwak, J., Choi, H. K., Choi, K. C., Kim, S., Lee, J., et al. (2012). EGCG Suppresses Prostate Cancer Cell Growth Modulating Acetylation of Androgen Receptor by Anti-histone Acetyltransferase Activity. Int. J. Mol. Med. 30, 69–74. doi:10.3892/ijmm.2012.966
Lei, H., Shi, J., Teng, Y., Song, C., Zou, L., Ye, F., et al. (2021). Baicalein Modulates the Radiosensitivity of Cervical Cancer Cells In Vitro via miR-183 and the JAK2/STAT3 Signaling Pathway. Adv. Clin. Exp. Med. 30, 727–736. doi:10.17219/acem/135478
Lin, D., Kuang, G., Wan, J., Zhang, X., Li, H., Gong, X., et al. (2017). Luteolin Suppresses the Metastasis of Triple-Negative Breast Cancer by Reversing Epithelial-To-Mesenchymal Transition via Downregulation of β-catenin Expression. Oncol. Rep. 37, 895–902. doi:10.3892/or.2016.5311
Ling, D., Marshall, G. M., Liu, P. Y., Xu, N., Nelson, C. A., Iismaa, S. E., et al. (2012). Enhancing the Anticancer Effect of the Histone Deacetylase Inhibitor by Activating Transglutaminase. Eur. J. Cancer 48, 3278–3287. doi:10.1016/j.ejca.2012.02.067
Liu, X., Chen, X., Yu, X., Tao, Y., Bode, A. M., Dong, Z., et al. (2013). Regulation of microRNAs by Epigenetics and Their Interplay Involved in Cancer. J. Exp. Clin. Cancer Res. 32, 96. doi:10.1186/1756-9966-32-96
Lu, L., Wang, Y., Ou, R., Feng, Q., Ji, L., Zheng, H., et al. (2018). DACT2 Epigenetic Stimulator Exerts Dual Efficacy for Colorectal Cancer Prevention and Treatment. Pharmacol. Res. 129, 318–328. doi:10.1016/j.phrs.2017.11.032
Lubecka, K., Kaufman-Szymczyk, A., Cebula-Obrzut, B., Smolewski, P., Szemraj, J., and Fabianowska-Majewska, K. (2018). Novel Clofarabine-Based Combinations with Polyphenols Epigenetically Reactivate Retinoic Acid Receptor Beta, Inhibit Cell Growth, and Induce Apoptosis of Breast Cancer Cells. Int. J. Mol. Sci. 19, 3970. doi:10.3390/ijms19123970
Ma, D., Chen, S., Wang, H., Wei, J., Wu, H., Gao, H., et al. (2021). Baicalein Induces Apoptosis of Pancreatic Cancer Cells by Regulating the Expression of miR-139-3p and miR-196b-5p. Front. Oncol. 11, 653061. doi:10.3389/fonc.2021.653061
Majid, S., Dar, A. A., Shahryari, V., Hirata, H., Ahmad, A., Saini, S., et al. (2010). Genistein Reverses Hypermethylation and Induces Active Histone Modifications in Tumor Suppressor Gene B-Cell Translocation Gene 3 in Prostate Cancer. Cancer 116, 66–76. doi:10.1002/cncr.24662
Marchese, F. P., Raimondi, I., and Huarte, M. (2017). The Multidimensional Mechanisms of Long Noncoding RNA Function. Genome Biol. 18, 206. doi:10.1186/s13059-017-1348-2
Marín-Béjar, O., Marchese, F. P., Athie, A., Sánchez, Y., González, J., Segura, V., et al. (2013). Pint lincRNA Connects the P53 Pathway with Epigenetic Silencing by the Polycomb Repressive Complex 2. Genome Biol. 14, R104. doi:10.1186/gb-2013-14-9-r104
Markaverich, B. M., Vijjeswarapu, M., Shoulars, K., and Rodriguez, M. (2010). Luteolin and Gefitinib Regulation of EGF Signaling Pathway and Cell Cycle Pathway Genes in PC-3 Human Prostate Cancer Cells. J. Steroid Biochem. Mol. Biol. 122, 219–231. doi:10.1016/j.jsbmb.2010.06.006
Mateen, S., Raina, K., Agarwal, C., Chan, D., and Agarwal, R. (2013). Silibinin Synergizes with Histone Deacetylase and DNA Methyltransferase Inhibitors in Upregulating E-Cadherin Expression Together with Inhibition of Migration and Invasion of Human Non-small Cell Lung Cancer Cells. J. Pharmacol. Exp. Ther. 345, 206–214. doi:10.1124/jpet.113.203471
Mayr, C., Wagner, A., Neureiter, D., Pichler, M., Jakab, M., Illig, R., et al. (2015). The green tea Catechin Epigallocatechin Gallate Induces Cell Cycle Arrest and Shows Potential Synergism with Cisplatin in Biliary Tract Cancer Cells. BMC Complement. Altern. Med. 15, 194. doi:10.1186/s12906-015-0721-5
Milazzo, G., Mercatelli, D., Di Muzio, G., Triboli, L., De Rosa, P., Perini, G., et al. (2020). Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 11, 556. doi:10.3390/genes11050556
Miron, A., Aprotosoaie, A. C., Trifan, A., and Xiao, J. (2017). Flavonoids as Modulators of Metabolic Enzymes and Drug Transporters. Ann. N.Y. Acad. Sci. 1398, 152–167. doi:10.1111/nyas.13384
Mishan, M. A., Khazeei Tabari, M. A., Mahrooz, A., and Bagheri, A. (2021). Role of microRNAs in the Anticancer Effects of the Flavonoid Luteolin: a Systematic Review. Eur. J. Cancer Prev. 30, 413–421. doi:10.1097/cej.0000000000000645
Mohr, A., and Mott, J. (2015). Overview of MicroRNA Biology. Semin. Liver Dis. 35, 003–011. doi:10.1055/s-0034-1397344
Mostafa, S. M., Gamal-Eldeen, A. M., Maksoud, N. A. E., and Fahmi, A. A. (2020). Epigallocatechin Gallate-Capped Gold Nanoparticles Enhanced the Tumor Suppressors Let-7a and miR-34a in Hepatocellular Carcinoma Cells. Acad. Bras Cienc 92, e20200574. doi:10.1590/0001-3765202020200574
Nakai, S., Fujita, M., and Kamei, Y. (2020). Health Promotion Effects of Soy Isoflavones. J. Nutr. Sci. Vitaminol 66, 502–507. doi:10.3177/jnsv.66.502
Nwaeburu, C. C., Abukiwan, A., Zhao, Z., and Herr, I. (2017). Quercetin-induced miR-200b-3p Regulates the Mode of Self-Renewing Divisions in Pancreatic Cancer. Mol. Cancer 16, 23. doi:10.1186/s12943-017-0589-8
Nwaeburu, C. C., Bauer, N., Zhao, Z., Abukiwan, A., Gladkich, J., Benner, A., et al. (2016). Up-regulation of microRNA Let-7c by Quercetin Inhibits Pancreatic Cancer Progression by Activation of Numbl. Oncotarget 7, 58367–58380. doi:10.18632/oncotarget.11122
Örenlil Yaylagül, E., and Ülger, C. (2020). The Effect of Baicalein on Wnt/β-Catenin Pathway and miR-25 Expression in Saos-2 Osteosarcoma Cell Line. Turkish J. Med. Sci. 50, 1168–1179. doi:10.3906/sag-2001-161
Oya, Y., Mondal, A., Rawangkan, A., Umsumarng, S., Iida, K., Watanabe, T., et al. (2017). Down-regulation of Histone Deacetylase 4, −5 and −6 as a Mechanism of Synergistic Enhancement of Apoptosis in Human Lung Cancer Cells Treated with the Combination of a Synthetic Retinoid, Am80 and green tea Catechin. J. Nutr. Biochem. 42, 7–16. doi:10.1016/j.jnutbio.2016.12.015
Pal, H. C., Pearlman, R. L., and Afaq, F. (2016). Fisetin and its Role in Chronic Diseases. Adv. Exp. Med. Biol. 928, 213–244. doi:10.1007/978-3-319-41334-1_10
Panche, A. N., Diwan, A. D., and Chandra, S. R. (2016). Flavonoids: an Overview. J. Nutr. Sci. 5, e47. doi:10.1017/jns.2016.41
Pandey, M., Shukla, S., and Gupta, S. (2010). Promoter Demethylation and Chromatin Remodeling by green tea Polyphenols Leads to Re-expression of GSTP1 in Human Prostate Cancer Cells. Int. J. Cancer 126, 2520–2533. doi:10.1002/ijc.24988
Pandey, M., Kaur, P., Shukla, S., Abbas, A., Fu, P., and Gupta, S. (2012). Plant Flavone Apigenin Inhibits HDAC and Remodels Chromatin to Induce Growth Arrest and Apoptosis in Human Prostate Cancer Cells: In Vitro and In Vivo Study. Mol. Carcinog. 51, 952–962. doi:10.1002/mc.20866
Paschka, A. G., Butler, R., and Young, C. Y.-F. (1998). Induction of Apoptosis in Prostate Cancer Cell Lines by the green tea Component, (−)-Epigallocatechin-3-Gallate. Cancer Lett. 130, 1–7. doi:10.1016/s0304-3835(98)00084-6
Peng, Y., and Croce, C. M. (2016). The Role of MicroRNAs in Human Cancer. Signal. Transduct Target. Ther. 1, 15004. doi:10.1038/sigtrans.2015.4
Pham, T. N. D., Stempel, S., Shields, M. A., Spaulding, C., Kumar, K., Bentrem, D. J., et al. (2019). Quercetin Enhances the Anti-tumor Effects of BET Inhibitors by Suppressing hnRNPA1. Int. J. Mol. Sci. 20, 4293. doi:10.3390/ijms20174293
Pietta, P.-G. (2000). Flavonoids as Antioxidants. J. Nat. Prod. 63, 1035–1042. doi:10.1021/np9904509
Qi, X., Li, H., Cong, X., Wang, X., Jiang, Z., Cao, R., et al. (2016). Baicalin Increases Developmental Competence of Mouse Embryos In Vitro by Inhibiting Cellular Apoptosis and Modulating HSP70 and DNMT Expression. J. Reprod. Dev. 62, 561–569. doi:10.1262/jrd.2016-047
Qin, Y., Niu, K., Zeng, Y., Liu, P., Yi, L., Zhang, T., et al. (2013). Isoflavones for Hypercholesterolaemia in Adults. Cochrane Database Syst. Rev. 6, CD009518. doi:10.1002/14651858.cd009518.pub2
Ramos, Y. a. L., Souza, O. F., Novo, M. C. T., Guimarães, C. F. C., and Popi, A. F. (2021). Quercetin Shortened Survival of Radio-Resistant B-1 Cells In Vitro and In Vivo by Restoring miR15a/16 Expression. Oncotarget 12, 355–365. doi:10.18632/oncotarget.27883
Richon, V. M., Sandhoff, T. W., Rifkind, R. A., and Marks, P. A. (2000). Histone Deacetylase Inhibitor Selectively Induces p21WAF1 Expression and Gene-Associated Histone Acetylation. Proc. Natl. Acad. Sci. 97, 10014–10019. doi:10.1073/pnas.180316197
Rossetto, D., Avvakumov, N., and Côté, J. (2014). Histone Phosphorylation. Epigenetics 7, 1098–1108. doi:10.4161/epi.21975
Russo, G. L., and Ungaro, P. (2019). “Epigenetic Mechanisms of Quercetin and Other Flavonoids in Cancer Therapy and Prevention,” in Epigenetics of Cancer Prevention,( Amsterdam: Elsevier Inc)187–202. doi:10.1016/b978-0-12-812494-9.00009-3
Sathyapalan, T., Aye, M., Rigby, A. S., Thatcher, N. J., Dargham, S. R., Kilpatrick, E. S., et al. (2018). Soy Isoflavones Improve Cardiovascular Disease Risk Markers in Women during the Early Menopause. Nutr. Metab. Cardiovasc. Dis. 28, 691–697. doi:10.1016/j.numecd.2018.03.007
Sheng, J., Shi, W., Guo, H., Long, W., Wang, Y., Qi, J., et al. (2019). The Inhibitory Effect of (-)-Epigallocatechin-3-Gallate on Breast Cancer Progression via Reducing SCUBE2 Methylation and DNMT Activity. Molecules 24, 2899. doi:10.3390/molecules24162899
Shoulars, K., Rodriguez, M. A., Thompson, T., and Markaverich, B. M. (2010). Regulation of Cell Cycle and RNA Transcription Genes Identified by Microarray Analysis of PC-3 Human Prostate Cancer Cells Treated with Luteolin. J. Steroid Biochem. Mol. Biol. 118, 41–50. doi:10.1016/j.jsbmb.2009.09.016
Shukla, S., Meeran, S. M., and Katiyar, S. K. (2014). Epigenetic Regulation by Selected Dietary Phytochemicals in Cancer Chemoprevention. Cancer Lett. 355, 9–17. doi:10.1016/j.canlet.2014.09.017
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer Statistics, 2021. CA A. Cancer J. Clin. 71, 7–33. doi:10.3322/caac.21654
Singh, B. N., Shankar, S., and Srivastava, R. K. (2011). Green tea Catechin, Epigallocatechin-3-Gallate (EGCG): Mechanisms, Perspectives and Clinical Applications. Biochem. Pharmacol. 82, 1807–1821. doi:10.1016/j.bcp.2011.07.093
Singh, S., Raza, W., Parveen, S., Meena, A., and Luqman, S. (2021). Flavonoid Display Ability to Target microRNAs in Cancer Pathogenesis. Biochem. Pharmacol. 189, 114409. doi:10.1016/j.bcp.2021.114409
Skibola, C. F., and Smith, M. T. (2000). Potential Health Impacts of Excessive Flavonoid Intake. Free Radic. Biol. Med. 29, 375–383. doi:10.1016/s0891-5849(00)00304-x
Soto-Reyes, E., González-Barrios, R., Cisneros-Soberanis, F., Herrera-Goepfert, R., Pérez, V., Cantú, D., et al. (2012). Disruption of CTCF at the miR-125b1 Locus in Gynecological Cancers. BMC Cancer 12, 40. doi:10.1186/1471-2407-12-40
Spagnuolo, C., Russo, G. L., Orhan, I. E., Habtemariam, S., Daglia, M., Sureda, A., et al. (2015). Genistein and Cancer: Current Status, Challenges, and Future Directions. Adv. Nutr. 6, 408–419. doi:10.3945/an.114.008052
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2020). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cel Biol 22, 96–118. doi:10.1038/s41580-020-00315-9
Taby, R., and Issa, J.-P. J. (2010). Cancer Epigenetics. CA: A Cancer J. Clinicians 60, 376–392. doi:10.3322/caac.20085
Takai, D., and Jones, P. A. (2002). Comprehensive Analysis of CpG Islands in Human Chromosomes 21 and 22. Pnas 99, 3740–3745. doi:10.1073/pnas.052410099
Taku, K., Melby, M. K., Nishi, N., Omori, T., and Kurzer, M. S. (2011). Soy Isoflavones for Osteoporosis: An Evidence-Based Approach. Maturitas 70, 333–338. doi:10.1016/j.maturitas.2011.09.001
Tao, S.-F., He, H.-F., and Chen, Q. (2015). Quercetin Inhibits Proliferation and Invasion Acts by Up-Regulating miR-146a in Human Breast Cancer Cells. Mol. Cel Biochem 402, 93–100. doi:10.1007/s11010-014-2317-7
Thakur, V. S., Gupta, K., and Gupta, S. (2012). Green tea Polyphenols Increase P53 Transcriptional Activity and Acetylation by Suppressing Class I Histone Deacetylases. Int. J. Oncol. 41, 353–361. doi:10.3892/ijo.2012.1449
Tseng, T.-H., Chien, M.-H., Lin, W.-L., Wen, Y.-C., Chow, J.-M., Chen, C.-K., et al. (2017). Inhibition of MDA-MB-231 Breast Cancer Cell Proliferation and Tumor Growth by Apigenin through Induction of G2/M Arrest and Histone H3 Acetylation-Mediated p21WAF1/CIP1expression. Environ. Toxicol. 32, 434–444. doi:10.1002/tox.22247
Vance, K. W., and Ponting, C. P. (2014). Transcriptional Regulatory Functions of Nuclear Long Noncoding RNAs. Trends Genet. 30, 348–355. doi:10.1016/j.tig.2014.06.001
Varier, R. A., and Timmers, H. T. M. (2011). Histone Lysine Methylation and Demethylation Pathways in Cancer. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1815, 75–89. doi:10.1016/j.bbcan.2010.10.002
Vitale, D. C., Piazza, C., Melilli, B., Drago, F., and Salomone, S. (2012). Isoflavones: Estrogenic Activity, Biological Effect and Bioavailability. Eur. J. Drug Metab. Pharmacokinet. 38, 15–25. doi:10.1007/s13318-012-0112-y
Wang, Q., Chen, Y., Lu, H., Wang, H., Feng, H., Xu, J., et al. (2020a). Quercetin Radiosensitizes Non‐small Cell Lung Cancer Cells through the Regulation of miR‐16‐5p/WEE1 axis. IUBMB Life 72, 1012–1022. doi:10.1002/iub.2242
Wang, S.-w., Sheng, H., Zheng, F., and Zhang, F. (2021a). Hesperetin Promotes DOT1L Degradation and Reduces Histone H3K79 Methylation to Inhibit Gastric Cancer Metastasis. Phytomedicine 84, 153499. doi:10.1016/j.phymed.2021.153499
Wang, X., Waschke, B. C., Woolaver, R. A., Chen, S. M. Y., Chen, Z., and Wang, J. H. (2020b). HDAC Inhibitors Overcome Immunotherapy Resistance in B-Cell Lymphoma. Protein Cell 11, 472–482. doi:10.1007/s13238-020-00694-x
Wang, Y., Chen, X., Li, J., and Xia, C. (2021b). Quercetin Antagonizes Esophagus Cancer by Modulating miR-1-3p/TAGLN2 Pathway-dependent Growth and Metastasis. Nutr. Cancer, 1–10. doi:10.1080/01635581.2021.1972125
Wang, Z., and Chen, H. (2010). Genistein Increases Gene Expression by Demethylation of WNT5a Promoter in colon Cancer Cell Line SW1116. Anticancer Res. 30, 4537–4545.
Wei, G.-J., Chao, Y.-H., Tung, Y.-C., Wu, T.-Y., and Su, Z.-Y. (2019). A Tangeretin Derivative Inhibits the Growth of Human Prostate Cancer LNCaP Cells by Epigenetically Restoring P21 Gene Expression and Inhibiting Cancer Stem-like Cell Proliferation. Aaps j 21, 86. doi:10.1208/s12248-019-0345-7
Weinhold, B. (2006). Epigenetics: the Science of Change. Environ. Health Perspect. 114, A160–A167. doi:10.1289/ehp.114-a160
Woo, H.-H., Jeong, B. R., and Hawes, M. C. (2005). Flavonoids: from Cell Cycle Regulation to Biotechnology. Biotechnol. Lett. 27, 365–374. doi:10.1007/s10529-005-1521-7
Wu, H.-T., Lin, J., Liu, Y.-E., Chen, H.-F., Hsu, K.-W., Lin, S.-H., et al. (2021). Luteolin Suppresses Androgen Receptor-Positive Triple-Negative Breast Cancer Cell Proliferation and Metastasis by Epigenetic Regulation of MMP9 Expression via the AKT/mTOR Signaling Pathway. Phytomedicine 81, 153437. doi:10.1016/j.phymed.2020.153437
Yang, L., Zhang, W., Chopra, S., Kaur, D., Wang, H., Li, M., et al. (2020). The Epigenetic Modification of Epigallocatechin Gallate (EGCG) on Cancer. Cdt 21, 1099–1104. doi:10.2174/1389450121666200504080112
Yu, W.-D., Sun, G., Li, J., Xu, J., and Wang, X. (2019). Mechanisms and Therapeutic Potentials of Cancer Immunotherapy in Combination with Radiotherapy And/or Chemotherapy. Cancer Lett. 452, 66–70. doi:10.1016/j.canlet.2019.02.048
Zhang, C., Hao, Y., Sun, Y., and Liu, P. (2019a). Quercetin Suppresses the Tumorigenesis of Oral Squamous Cell Carcinoma by Regulating microRNA-22/wnt1/β-Catenin axis. J. Pharmacol. Sci. 140, 128–136. doi:10.1016/j.jphs.2019.03.005
Zhang, L., Lu, Q., and Chang, C. (2020). Epigenetics in Health and Disease. Adv. Exp. Med. Biol. 1253, 3–55. doi:10.1007/978-981-15-3449-2_1
Zhang, X., Wang, W., Zhu, W., Dong, J., Cheng, Y., Yin, Z., et al. (2019b). Mechanisms and Functions of Long Non-coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 20, 5573. doi:10.3390/ijms20225573
Zhang, Y., Wang, X., Han, L., Zhou, Y., and Sun, S. (2015). Green tea Polyphenol EGCG Reverse Cisplatin Resistance of A549/DDP Cell Line through Candidate Genes Demethylation. Biomed. Pharmacother. 69, 285–290. doi:10.1016/j.biopha.2014.12.016
Zhao, J., Fang, Z., Zha, Z., Sun, Q., Wang, H., Sun, M., et al. (2019). Quercetin Inhibits Cell Viability, Migration and Invasion by Regulating miR-16/HOXA10 axis in Oral Cancer. Eur. J. Pharmacol. 847, 11–18. doi:10.1016/j.ejphar.2019.01.006
Zhao, Y., Chen, X., Jiang, J., Wan, X., Wang, Y., and Xu, P. (2020). Epigallocatechin Gallate Reverses Gastric Cancer by Regulating the Long Noncoding RNA LINC00511/miR-29b/KDM2A axis. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. doi:10.1016/j.bbadis.2020.165856
Zheng, N.-G., Wang, J.-L., Yang, S.-L., and Wu, J.-L. (2014). Aberrant Epigenetic Alteration in Eca9706 Cells Modulated by Nanoliposomal Quercetin Combined with Butyrate Mediated via Epigenetic-NF-Κb Signaling. Asian Pac. J. Cancer Prev. 15, 4539–4543. doi:10.7314/apjcp.2014.15.11.4539
Zhou, J., Gong, J., Ding, C., and Chen, G. (2015). Quercetin Induces the Apoptosis of Human Ovarian Carcinoma Cells by Upregulating the Expression of microRNA-145. Mol. Med. Rep. 12, 3127–3131. doi:10.3892/mmr.2015.3679
Glossary
ABCG2 ATP-binding cassette G2
AKT Ak strain transforming
AMPK 5' adenosine monophosphate-activated protein kinase
APAF1 Apoptotic protease activating factor 1
AR Androgen receptor
ARE Antioxidant responsive element
ATG5 /7 Autophagy related 5 /7
AURKA /B /C Aurora kinase A /B /C
BAX BCL2 associated X
BCL-2 B-cell lymphoma 2
BCL2L11 Bcl-2-like protein 11
BDC Bile duct cancer
BET Bromodomain and extraterminal
BNIP3 BCL2 and adenovirus E1B 19-kDa-interacting protein 3
BNIP3L BCL2 and adenovirus E1B 19-kDa-interacting protein 3-like
BRCA1 /2 Breast cancer gene 1 /2
BTG3 B-cell translocation gene 3
C/EBPα CCAAT enhancer-binding protein alpha
CAF Chromatin assembly factor
CCNA2 /B1 /B2 / D1 /E1 /E2 Cyclin A2 /B1 /B2 /D1 /E1 /E2
CD Cluster of differentiation
CDC25A Cell division cycle 25 A
CDH1 /13 Cadherin 1 /13
CDK1 Cyclin-dependent kinase 1
CDKN1A /1B Cyclin dependent kinase inhibitor 1A /1B
CHOP C/EBP homologous protein
CREB cAMP-response element binding protein
DACT2 Disheveled-associated antagonist of β-catenin homolog 2
DAPK1 Death-associated protein kinase 1
DOT1L DOT1-like histone H3K79 methyltransferase
DR5 Death receptor 5
E2F1 E2F transcription factor 1
EED Embryonic ectoderm development
EGFR Epidermal growth factor receptor
EMT epithelial-mesenchymal transition
EPHB2 EPH receptor B2
ERK Extracellular signal-regulated protein kinase
EZH2 Enhancer of zeste homolog 2
GAS1 Growth arrest specific 1
GBC Gall bladder carcinoma
GSTP1 Glutathione S-transferase pi 1
hMLH1 Human mutL homolog 1
HMOX1 Heme oxygenase 1
hnRNPA1 Heterogeneous nuclear ribonucleoprotein A1
HOTAIR HOX transcript antisense RNA
HOXA10 Homeobox A10
HSP70 Heat shock proteins 70 kda;
hTERT Human telomerase reverse transcriptase
ICAM1 Intercellular adhesion molecule 1
IGFBP5 Insulin like growth factor binding protein 5
IL Interleukin
IRE1 Inositol-requiring enzyme1
JNK c-Jun N-terminal kinase
KDM4A /4B /5C Lysine (K)-specific demethylase 4A /4B /5C
KMT2A Lysine (K)-specific methyltransferase 2A
Let -7 Lethal-7
LSD1 Lysine-specific demethylase 1
m6A N⁶-methyl-adenosine
MAD1L1 MAD1 mitotic arrest deficient 1-like 1
MALAT1 Metastasis associated lung adenocarcinoma transcript 1
MAPK Mitogen-activated protein kinase
MEG3 Maternally expressed gene 3
MGMT O6-methylguanine-DNA methyltransferase
MMP Matrix metallopeptidase
mTOR Mammalian target of rapamycin
NF-κB Nuclear factor kappa B
NQO1 NAD(P)H quinone dehydrogenase 1
NRF2 Nuclear factor erythroid 2-related factor 2
NSCLC Non-small cell lung cancer
NUMBL NUMB Like Endocytic Adaptor Protein
p70S6K Ribosomal protein S6 kinase beta-1
PARP Poly (ADP-ribose) polymerase
PI3K Phosphoinositide 3-kinases
PIK3C2B Phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta
PIK3CA Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
PKCα Protein kinase C alpha
PLK-1 Polo-like kinase 1
PRC Polycomb repressive complex
PRDM2 PR domain zinc finger protein 2
PRMT6 /7 Protein arginine N-methyltransferase 6 /7
PTEN Phosphatase and tensin homolog
PTENP1 PTEN pseudogene 1
RARB Retinoic acid receptor beta
RFXAP Regulatory factor X associated protein
ROS Reactive oxygen species
RTCB RNA 2',3'-cyclic phosphate and 5'-OH ligase
RUNX3 Runt-related transcription factor 3
S100P S100 calcium binding protein P
SCUBE2 Signal peptide-CUB-EGF domain-containing protein 2
SOD1 Superoxide dismutase 1
SUV39H1 Suppressor of variegation 3-9 homolog 1
SUZ12 Suppressor of zeste homolog 12
TAGLN2 Transgelin 2
TIMP3 Tissue inhibitor of metalloproteinase 3
TRAF7 TNF receptor associated factor 7
UBE2B Ubiquitin conjugating enzyme E2 B
UHRF1 Ubiquitin like with PHD and ring finger domains 1
ULK1 Unc-51 like autophagy activating kinase
WEE1 WEE1 G2 Checkpoint Kinase
WISP2 WNT1-inducible-signaling pathway protein 2
WNT Wingless/Integrated
WNT5a Wnt family member 5A
γ-H2AX Gamma H2A histone family member X
Keywords: cancer, flavonoids, epigenetics, DNA methylation, histone modifications, non-coding RNAs
Citation: Fatima N, Baqri SSR, Bhattacharya A, Koney NK-, Husain K, Abbas A and Ansari RA (2021) Role of Flavonoids as Epigenetic Modulators in Cancer Prevention and Therapy. Front. Genet. 12:758733. doi: 10.3389/fgene.2021.758733
Received: 14 August 2021; Accepted: 26 October 2021;
Published: 09 November 2021.
Edited by:
Kai Tang, Purdue University, United StatesReviewed by:
Nabab Khan, University of North Dakota, United StatesYao Xu, Wuhan University of Science and Technology, China
Copyright © 2021 Fatima, Baqri, Bhattacharya, Koney, Husain, Abbas and Ansari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rais A. Ansari, cmE1NTdAbm92YS5lZHU=
 Nishat Fatima
Nishat Fatima Syed Shabihe Raza Baqri
Syed Shabihe Raza Baqri Atrayee Bhattacharya
Atrayee Bhattacharya Nii Koney-Kwaku Koney
Nii Koney-Kwaku Koney Kazim Husain
Kazim Husain Ata Abbas
Ata Abbas Rais A. Ansari
Rais A. Ansari