- College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang, China
MADS-box gene, one of the largest transcription factor families in plants, is a class of transcription factors widely present in eukaryotes. It plays an important role in plant growth and development and participates in the growth and development of flowers and fruits. Sweet potato is the seventh most important food crop in the world. Its tuberous roots, stems, and leaves contain a large number of proteins, lipids, carotenoids, anthocyanins, conjugated phenolic acids, and minerals, which have high edible, forage, and medicinal value, and is also an important energy crop. At present, MADS-box genes in sweet potato have rarely been reported, and there has been no study on the genome-wide identification and classification of MADS-box genes in Ipomoea batatas. This study provided the first comprehensive analysis of sweet potato MADS-box genes. We identified 95 MADS-box genes, analyzed the structure and protein of sweet potato MADS-box genes, and categorized them based on phylogenetic analysis with Arabidopsis MADS-box proteins. Chromosomal localization indicated an unequal number of MADS-box genes in all 14 chromosomes except LG3, with more than 10 MADS-box genes located on chromosomes LG7, LG11, and LG15. The MADS domain and core motifs of the sweet potato MADS-box genes were identified by motif analysis. We identified 19 MADS-box genes with collinear relationships and analyzed duplication events. Cis-acting elements, such as light-responsive, auxin-responsive, drought-inducible, and MeJA-responsive elements, were found in the promoter region of the MADS-box genes in sweet potato, which further indicates the basis of MADS-box gene regulation in response to environmental changes and hormones. RNA-seq suggested that sweet potato MADS-box genes exhibit tissue-specific expression patterns, with 34 genes highly expressed in sweet potato flowers and fruits, and 19 genes highly expressed in the tuberous root, pencil root, or fibrous root. qRT-PCR again validated the expression levels of the 10 genes and found that IbMADS1, IbMADS18, IbMADS19, IbMADS79, and IbMADS90 were highly expressed in the tuberous root or fibrous root, and IbMADS18, IbMADS31, and IbMADS83 were highly expressed in the fruit. In this study, the molecular basis of MADS-box genes of sweet potato was analyzed from various angles. The effects of MADS-box genes on the growth and development of sweet potato were investigated, which may provide a certain theoretical basis for molecular breeding of sweet potato.
1 Introduction
MADS-box family gene is a kind of transcription factor (TF) widely existing in eukaryotes. MADS is the first letter abbreviation of Mini chromosome maintenance 1 (MCM1) gene in Saccharomyces cerevisiae (Passmore et al., 1988), AGAMOUS (AG) gene in Arabidopsis thaliana (Yanofsky et al., 1990), DEFICIENS (DEF) gene in Antirrhinum majus (Sommer et al., 1990), and serum response factor (SRF) gene in human serum (Norman et al., 1988). It is one of the largest TF families in plants and plays an important role in plant growth and development. Based on phylogenetic analysis, MADS-box genes in plants are categorized into type I and type II (Parenicova et al., 2003). Type I MADS-box gene, also known as M Type, usually has one to two exons, encoding proteins with highly conserved MADS domain. According to the differences in MADS domain, it is further divided into Mα, Mβ, and Mγ subfamilies (De Bodt et al., 2003). Type II gene, also known as MIKC type MADS-box gene, is a kind of plant-specific MADS-box gene with six to eight exons. The encoded protein contains four conserved domains: MADS-box (M domain), Intervening domain (I domain), Keratin-like domain (K domain), and C-terminal domain (Nam et al., 2005).
The completion of the whole genome sequences of many plant species has led to the identification and characterization of important gene families. MADS-box genes are integral to the ABCDE model of flowering regulation. Fan et al. identified three flower development class E genes, AGL2, AGL4, and AGL9 (Fan et al., 1997). Class E genes cooperate with several other classes to control calyx, petal, stamen, carpel, and ovule development (Theissen, 2001). AP3 homolog genes in Chrysanthemum play a role in the development of ray floret and disc floret in chrysanthemum (Won et al., 2021), and nitrate signaling-associated TFs such as AGL8, AGL21, and LBD29 are involved in CmANR1-modulated control of root development. In addition, CmANR1 also acts as a positive regulator to control shoot growth and development (Sun et al., 2021). Wang et al. found 15 tomato MADS-box genes involved in floral organ identification and five tomato MADS-box genes related to fruit development (Wang et al., 2019). AGL17 towards the elongation zone is expressed in lateral root cap epidermal cells; AGL12 and AGL21 are also expressed in the central cylinder of differentiated roots. Both are expressed in developing embryos of Arabidopsis (Burgeff et al., 2002).
Sweet potato [Ipomoea batatas (L.) Lam.], 2n = 6x = 90, is an annual herb, and its underground parts are round, oval, or spindle-shaped tuberous roots. The shape, skin color, and flesh color of the tuberous root vary with varieties or soil (Wu et al., 1998). Sweet potato is also the seventh important food crop in the world, with high yield and wide use. The tuberous roots, stems, and leaves contain a large number of proteins, lipids, carotenoids, anthocyanin, conjugated phenolic acids, and minerals, which have high edible, feed, and medicinal values, and is also essential energy crop (Wang et al., 2016). In 2002, Kim et al. cloned two MADS-box genes named IbMADS3 and IbMADS4 from sweet potato for the first time and found that these two MADS-box genes were significantly expressed in pencil roots and tuberous roots of sweet potato (Kim et al., 2002). In 2005, Kim et al. reported three MADS-box genes of IbMADS79, IbAGL17, and IbAGL20; and they found that IbMADS79 was only expressed in roots of sweet potato (Kim et al., 2005). Subsequently, Ku et al. (2008) cloned IbMADS1, indicating that IbMADS1 was involved in the initiation of tuberous root differentiation of sweet potato. Dong et al. (2018) cloned nine MADS-box TFs in 2018 and found that IbMBP2, IbMBP3, IbMBP4, and IbMBP9 showed high expression during storage roots development and were upregulated by storage roots development-related hormones. In 2020, Zhu et al. identified 37 MADS-box genes in Ipomoea trifida, the close wild ancestor of sweet potato. They found that MIKCC genes may be significant for regulating the floral organ development in I. trifida (Zhu et al., 2020). The MADS-box genes within the sweet potato genome have not been completely mined; the function and mechanism of action need to be verified; and the identification of more MADS-box genes is beneficial to elucidating the molecular mechanism of growth and development of sweet potato, providing a certain theoretical basis for molecular breeding.
2 Materials and Methods
2.1 Identification and Gene Structure Analysis of MADS-Box Family Members in Sweet Potato
The sweet potato genomic information was derived from I. batatas “Taizhong 6” genomic data provided by the Ipomoea genome hub (https://ipomoea-genome.org/). The MADS-box family HMM model files SRF-TF (PF00319) and K-box (PF01486) were downloaded from the Pfam database (https://pfam.xfam.org/). The whole genome MADS-box family genes of sweet potato were identified by HMMER software (Krejci et al., 2016), and the MADS-box genes were identified by the SMART website (http://smart.embl-heidel-berg.de/) again. The genes containing the MADS-box domain were named from IbMADS1 to IbMADS95, and the three genes matching the published sequences of IbMADS1 (Ku et al., 2008), IbMADS4 (Kim et al., 2002), and IbMADS79 (Kim et al., 2005) were still named as IbMADS1, IbMADS4, and IbMADS79, respectively. IbMBP1, IbMBP4, and IbMBP8 (Dong et al., 2018) were named as IbMADS47, IbMADS38, and IbMADS51, respectively, in this study. According to the sweet potato genome gff3 annotation information, GSDS online website was used (http://gsds.gao-lab.org/). The MADS-box gene structure of sweet potato was analyzed visually. TBtools (Chen et al., 2020) was used to plot the innovative prediction results visually. TBtools (Chen et al., 2020) was used for structure visualization. According to the annotation information of the sweet potato genome, the MADS-box genes structure of sweet potato was visualized using GSDS online website tool (http://gsds.gao-lab.org/).
2.2 MADS-Box Phylogenetic Tree Construction
The Arabidopsis MADS-box protein sequence was obtained from the Plant TFDB website (http://planttfdb.gao-lab.org/index.php) (Jin et al., 2017). I. trifida MADS-box proteins were obtained according to the research of Zhu et al. (Zhu et al., 2020). The MADS-box genes of sweet potato were divided into two categories, type I and type II, based on the phylogenetic analysis of the MADS gene with Arabidopsis. ClustalW was used to compare the two types of MADS-box proteins; a neighbor-joining (NJ) tree was constructed using bootstrap analysis with 1,000 replicates, the amino acid p-distance model, missing data treatment option set at partial deletion, and a site coverage cutoff of 60%; and MEGA-X (Kumar et al., 2018) was used to construct the NJ phylogenetic tree (Saitou and Nei, 1987). The parameters for constructing the evolutionary tree were as follows: Poisson model, uniform rates, Sam (homogeneous), and pairwise deletion. According to the classification method of Sheng et al. (2019), the MADS-box gene subfamily of sweet potato was categorized based on the MADS-box gene subfamily of Arabidopsis, using Evolview online tools (https://www.evolgenius.info/evolview/) to beautify the tree.
2.3 Chromosomal Localization of MADS-Box Genes
According to the annotation information of the sweet potato genome downloaded from the Ipomoea genome hub, the positions of 95 sweet potato MADS-box genes on chromosomes were obtained, and the chromosome mapping was performed using Map Chart (Voorrips, 2002) software. Origin 8.5 (https://www.originlab.com) was used to draw the distribution cake map of sweet potato MADS-box genes.
2.4 Conserved Motif Analysis of MADS-Box Proteins
The sweet potato MADS-box protein sequence was submitted to MEME online tool (http://meme-suite.org/) (Bailey et al., 2009) for the motif prediction. The discovery number of Motif was set to be 8, the width of motif was 6–50, and other default settings were used. The MADS-box motif domain of sweet potato was visualized by TBtools.
2.5 Cis-Acting Elements Analysis of MADS-Box Genes
According to the annotation information of the sweet potato genome and sweet potato genome sequence, referring to Sumiya (2021) and Zhang et al. (2021), the 2-kb upstream sequence of sweet potato MADS-box genes was submitted to PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) for cis-acting element prediction. The PlantCare analysis results were simplified, and TBtools was used for visualization.
2.6 Collinearity Analysis of MADS-Box Genes
According to the annotation information and the whole genome protein sequence of sweet potato, MCScanX (Wang et al., 2012) software was used to analyze the MADS-box collinearity of sweet potato, and Circos (Krzywinski et al., 2009) software was used for visualization.
2.7 Expression Analysis of MADS-Box Genes
The tuberous root (greater than 10 mm in diameter), pencil root (2–5 mm in diameter), fibrous root (diameter of less than 1.5 mm), flower (an incompletely opened bud), fruit (the peel is green and the fruit form full), and stem tip (1–2 rachises of the rolled leaf at the stem apex, excluding the rachis where the expanded leaf is located) of “Jishu 26” (I. batatas cv. Ji 26) cultivated in an open field for 100 days were selected. After rapid freezing in liquid nitrogen, with dry ice sent to Biomarker Technologies Company (Beijing, China, https://www.biocloud.net) for total RNA extraction, library construction, and transcriptome sequencing, raw data were uploaded to National Center for Biotechnology Information (NCBI) project PRJNA744414; the expression of sweet potato MADS-box genes was analyzed based on genome information and local blast; the FPKM value (Florea et al., 2013) was calculated to reflect the gene expression; and the calculation formula is as follows: FPKM = {cDNA Fragments\over{Mapped Fragments(Millions) * TranscriptLength(kb)}}. Pheatmap package and ggplot2 package (Ito and Murphy, 2013) of R Statistics (https://www.r-project.org) were used for data processing and visualization.
2.8 Verification of Gene Expression by qRT-PCR
The tuberous root is the main harvest organ of sweet potato. According to the expression profile of the MADS-box gene in different parts, several genes highly expressed in tuberous root were selected for qRT-PCR expression verification. The double-stranded cDNA of fibrous root (F.R, sampling details were the same as 1.7), tuberous root (T.R, sampling details were the same at 1.7), leaf (L, expanded leaf 1–2 at the apex of stem), stem (S, 1–2 rachises of the expanded leaf at the stem apex, excluding the rachis where the rolled leaf is located), stem tip (S.T, sampling details were the same at 1.7), fruit (F.T, sampling details were the same at 1.7), and other tissues cultivated in open field for 100 days “Jishu 26” were used as templates; β-actin was used as a reference gene, using the CFX96™ quantitative PCR instrument to perform the fluorescent quantitative PCR. The reaction program was as follows: predenaturation at 95°C for 3 min, 95°C for 5 s, 60°C for 30 s, 40 cycles; and 95°C for 15 s, 60°C for 6 s, 95°C for 15 s. The reaction system was as follows: 2× universal SYBR green fast qPCR mix (ABclonal Technology, Wuhan, China) for 5 μl, ddH2O for 3 μl, cDNA for 1 μl, forward primer for 0.5 μl, and reverse primer for 0.5 μl. Primers were designed using Primer premier 6 (Singh et al., 1998) based on the sequence of the amplified CDs of the MADS-box genes, and the primer sequences are listed in Supplementary Table S1. Three biological replicates and three technical replicates were set up, and relative gene expression was calculated by the 2−ΔΔCt method (Livak and Schmitten, 2001). Data processing and analysis were performed using SPSS software, and column graphs were drawn using origin 8.5.
3 Results and Analysis
3.1 Identification and Structural Analysis of the MADS-Box Family of Sweet Potato
A total of 95 MADS-box genes were identified from the whole genome of sweet potato and named according to IbMADS1–IbMADS95 (Supplementary Table S2). According to the gene structure, the sweet potato MADS-box genes were divided into two categories: type I and type II (Figure 1). The different types of MADS-box gene structures of sweet potato have apparent differentiation. Most type I genes are less than 2 kb in length and have one to two long fragment exons. On the other hand, the type II gene is usually over 3 kb and contains more than six exons, and most exonic fragments are short. Smart protein structure prediction found that the sweet potato MADS-box gene contains other domains in addition to the MADS domain. Most type I genes contained variable numbers of low-complexity region (LCR) structures and coiled-coil region (CCR) structures. The 21 type II genes contained a K-box domain; the K-box domain was commonly found to be associated with SRF-type TFs, also the signature domain of type II MADS-box genes. And the type II genes without a K-box domain usually belong to the MIKC* subfamily.

FIGURE 1. SMART identification and structure of sweet potato MADS-box gene. (A) Type I gene structure. (B) Type I protein structure prediction by SMART. (C) Type II gene structure. (D) Type II protein structure prediction by SMART.
3.2 Phylogenetic Analysis of MADS Proteins
The NJ phylogenetic tree of MADS-box proteins from I. batatas, I. trifida, and Arabidopsis were constructed by MEGA-X. Based on the subfamily classification of Arabidopsis MADS proteins, sweet potato type I MADS proteins were divided into three subfamilies: Mα, Mβ, and Mγ (Figure 2A). In comparison, type II MADS proteins were divided into 12 subfamilies (Figure 2B). The type I MADS proteins of sweet potato and Arabidopsis were cross-distributed in each subfamily. Among them, both Mα subfamily and Mβ subfamily had 27 proteins each, and Mγ subfamily was less abundant with 16 proteins, which was similar to the distribution of the Arabidopsis protein subfamily. However, the type I proteins of I. trifida were mainly concentrated in Mα subfamily, with only one gene each in the Mβ and Mγ subfamilies.
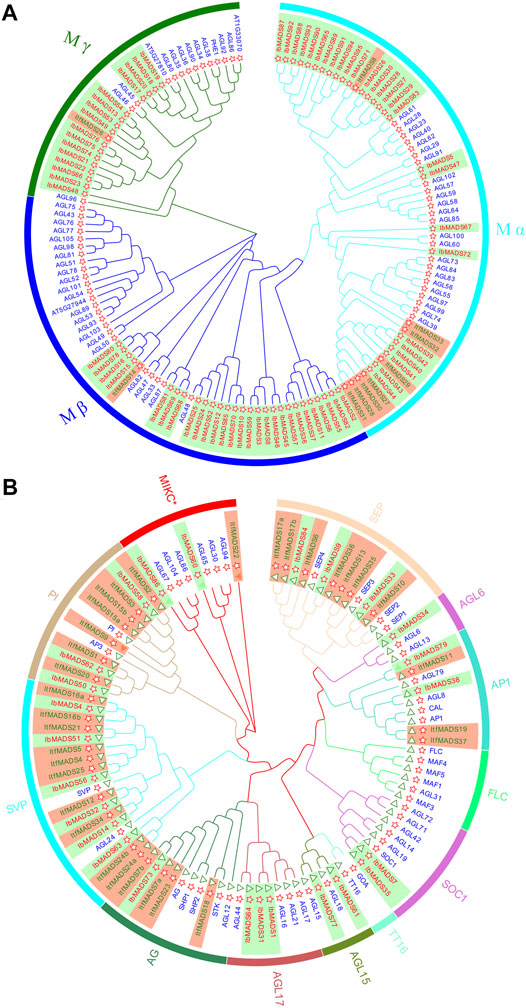
FIGURE 2. Phylogenetic tree of type I (A) and type II (B) MADS-box proteins in Ipomoea batatas, Ipomoea trifida, and Arabidopsis. Red ID indicates I. batatas gene, green ID indicates I. trifida gene, and blue ID indicates Arabidopsis gene. The red star indicates the sequence containing the SRF-box domain, and the green triangle indicates the sequence containing K-Box domain.
The type II gene of I. batatas is phylogenetically closer to that of I. trifida, as indicated by gene number and subfamily distribution. The type II gene of I. batatas did not appear in the FLC subfamily or TT16 subfamily, and other subfamilies were distributed, with one to six proteins varying in number. The proteins for I. trifida are mainly distributed in subfamilies such as SEP, PI, and SVP and show deletions in several subfamilies. Twenty-two type II proteins contain K-box region, while three sweet potato type II proteins have no K-box region, one of them belongs to the AP1 subfamily, and the other two belong to the MIKC* subfamily.
3.3 Chromosomal Localization of MADS-Box Genes
Chromosome localization of 95 MADS-box genes in sweet potato was performed (Figure 3). It was found that except LG3 chromosome, the distribution of MADS-box genes in sweet potato varied in quantity in the other 14 chromosomes. More than 10 MADS-box genes were distributed on LG7, LG8, LG11, and LG15 chromosomes. LG1, LG5, and LG12 chromosomes contain fewer MADS-box genes, only one to three. Mα subfamily is mainly distributed on chromosomes LG7, LG8, and LG15, with more than five genes. In contrast, the Mβ subfamily (blue) is more fragmented, with one to four genes distributed in all chromosomes except LG16 and LG12. Seven MADS-box genes on LG6 were Mγ subfamily, and the rest were sporadically distributed on LG4, LG9, LG10, LG12, and LG14. The MIKC subfamily genes are widely distributed, except for LG1, LG3, LG6, LG9, and LG12, which are more numerous on chromosome LG11, with seven genes belonging to the MIKC subfamily. Gene cluster exists for LG6, LG7, LG8, LG14, and LG15, especially in the interval 10413181 to 10505372 of LG15 containing nine MADS-box genes. Gene clusters were present on chromosomes LG6, LG7, LG8, LG14, and LG15. In particular, the interval 10413181 to 10505372 of LG15 contains nine MADS-box gene presence distributed centrally.
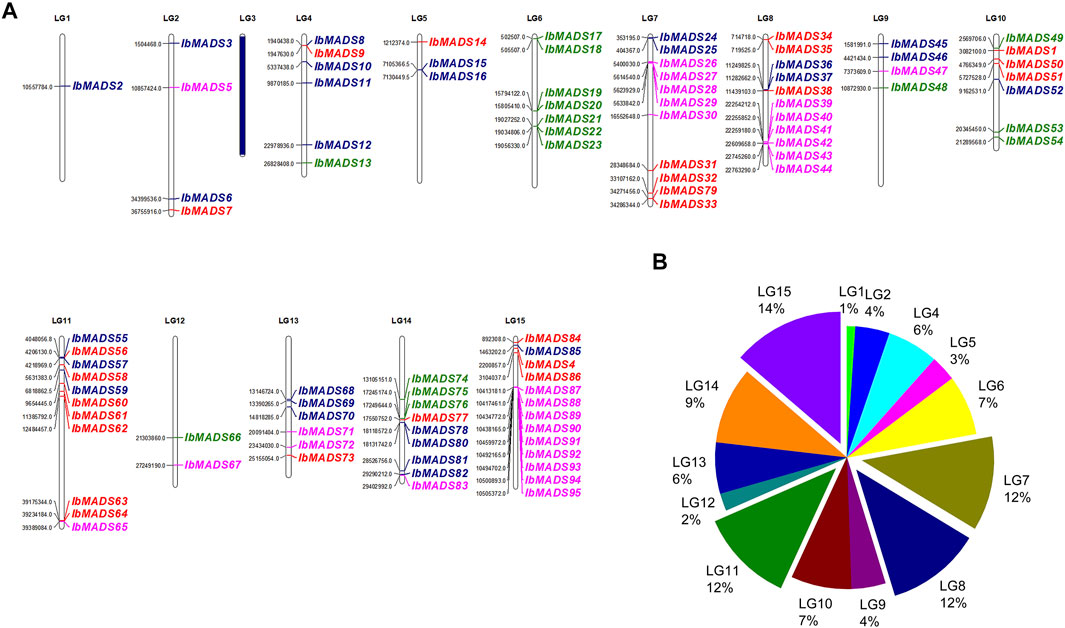
FIGURE 3. Chromosome location (A) and distribution (B) of MADS-box gene in sweet potato. The purple name represents Mα subfamily, the blue name represents Mβ subfamily, the green name represents Mγ subfamily, and the red name represents type II MADS-box genes. LG3 has no MADS-box gene distribution.
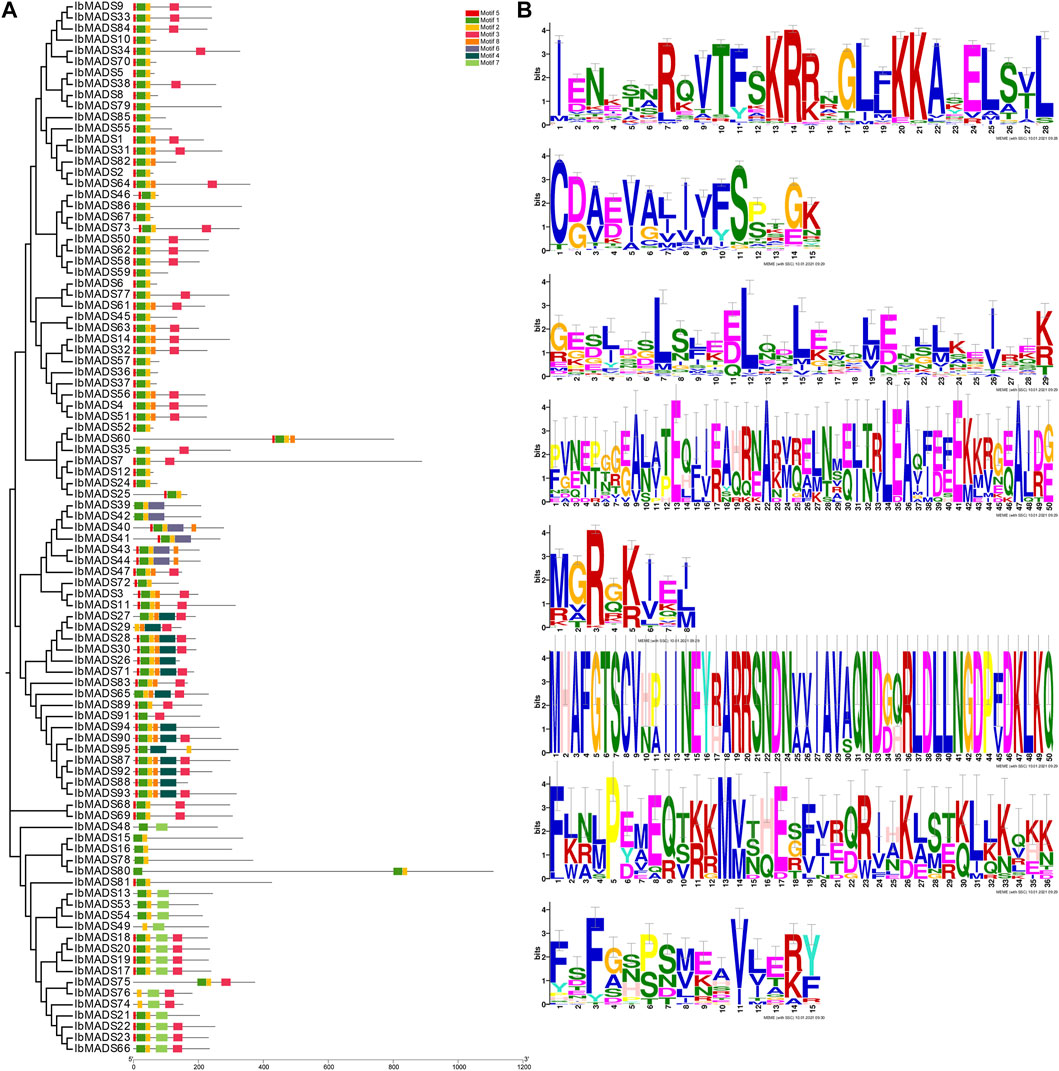
3.4 Analysis of MADS Proteins Conserve Motif
Conserve motif analysis of sweet potato MADS-protein sequence was performed by online MEME tool. There are some differences in the number of MADS-protein motif in sweet potato. Each protein contains two to six motifs (Figure 4). All genes have motif 1 domain encoding 28 amino acids, and more than 90 genes contain motif 2 and motif 5. The 51 amino acids encoded by these three motifs are the core motifs of MADS-box genes, which are related to the SRF-box region. In addition, the motif 3 domain also plays an important role, and more than 50 proteins contain highly conserved motif 3 domains encoding 29 amino acids, including 22 type II MADS proteins.
3.5 Analysis of Cis-Acting Elements of MADS-Box Gene Promoter Regions
The 2-kb upstream CDS sequences were extracted from the promoter regions of 95 MADS-box genes of sweet potato. The cis-acting elements in the promoter region of sweet potato MADS-box were predicted by PlantCARE online tool. All sweet potato MADS-box genes contain three to 12 light-responsive elements, including Box 4, G-box, GT1-motif, and TCT-motif. In addition, the promoter regions of sweet potato MADS-box genes are mainly more than 200 MeJA-responsive elements, including CGTCA-motif and TGACG-motif. And ABA-responsive elements (ABRE), auxin-responsive elements (TGA-element and AuxRR-core), and about 100 MYB response elements are involved in drought inducibility and light-responsiveness. In addition, there are a small number of regulatory elements such as circadian control elements, meristem expression elements, salicylic acid-responsive elements, low-temperature-responsive elements, gibberellin-responsive elements, defense, and stress-responsive elements. They indicated that MADS-box genes could respond to various environmental changes and coordinate the average growth and development of sweet potato (Figure 5).
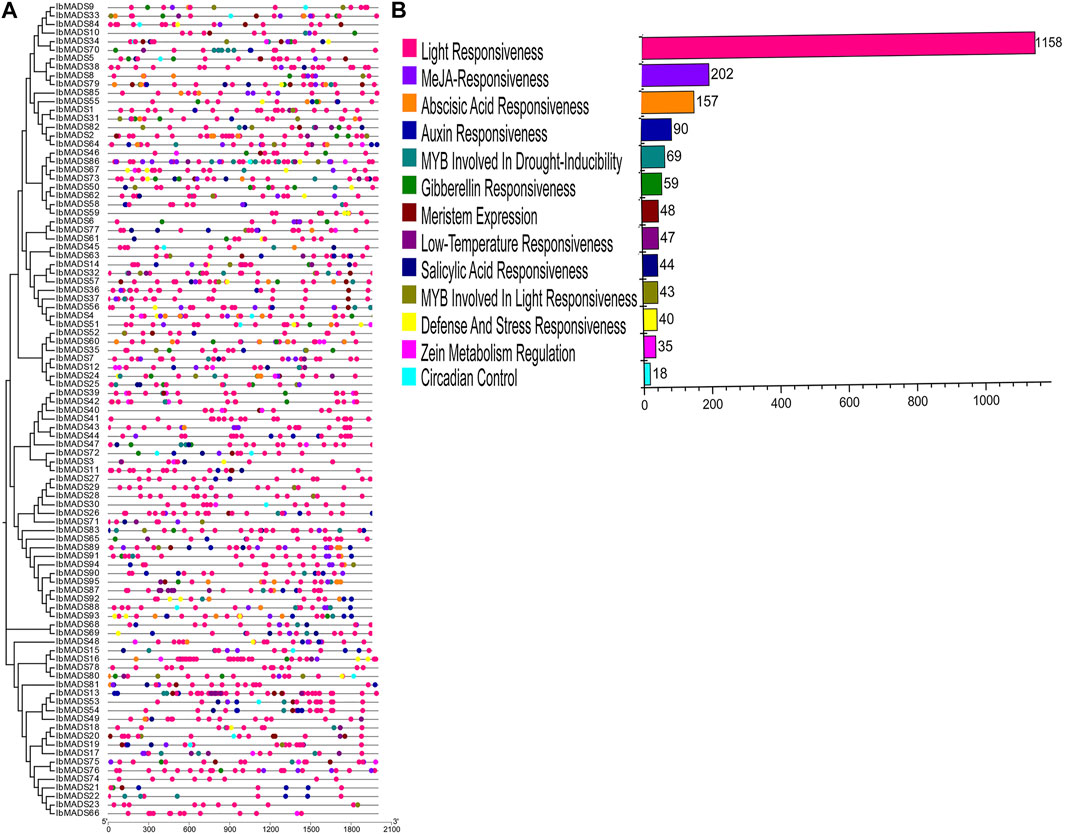
FIGURE 5. Cis-acting elements analysis of sweet potato MADS-box. (A) Cis-acting element distribution. (B) Cis-acting element numbers.
3.6 Collinearity Analysis of MADS-Box Gene in Sweet Potato
Based on the collinearity analysis by MCScanX, a large number of genes were found to be collinear relational (light pink and light green) between and within chromosomes in sweet potato. There are 19 sweet potato MADS-box genes with collinearity, of which 15 genes belonged to inter-chromosome segmental duplications and four genes belonged to tandem duplications. Among them, IbMADS4, IbMADS14, IbMADS32, and IbMADS56 genes share a reciprocal segmental duplication relationship, with IbMADS51 showing a segmental duplication relationship with IbMADS56 and IbMADS4 but not with IbMADS14 or IbMADS32. There are tandem duplications between IbMADS58 and IbMADS59, and between IbMADS68 and IbMADS69. And there are segmental duplications in the other 10 intergenic pairs. The most MADS-box genes with collinear relationships were on chromosome LG11 with five, and the remaining chromosomes had fewer than two or no MADS-box genes with collinear relationships (Figure 6).
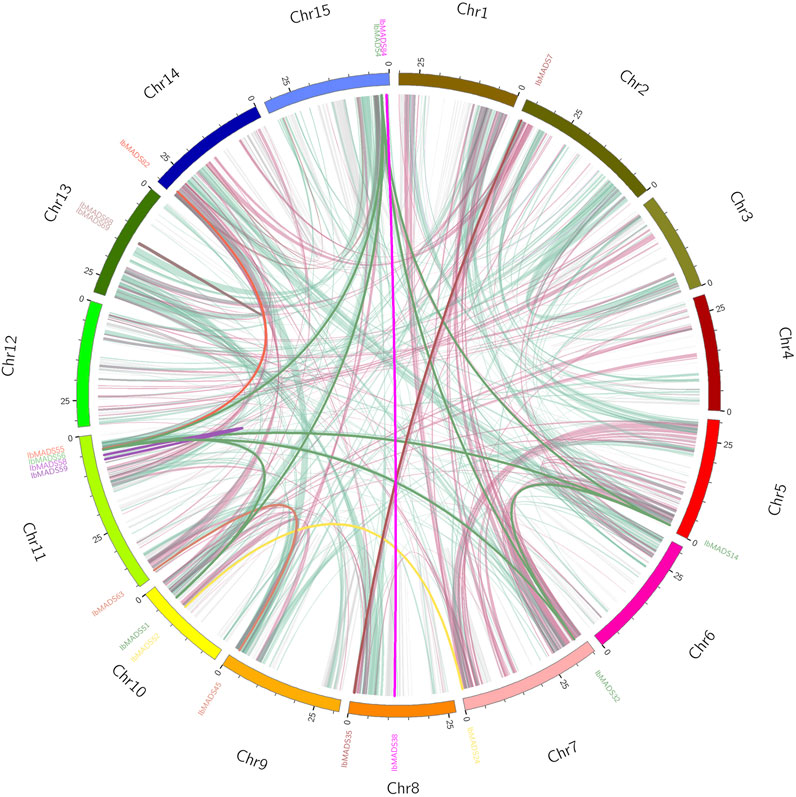
FIGURE 6. Collinearity analysis of MADS-box gene in sweet potato. The same color name indicates that the gene has a tandem repeat.
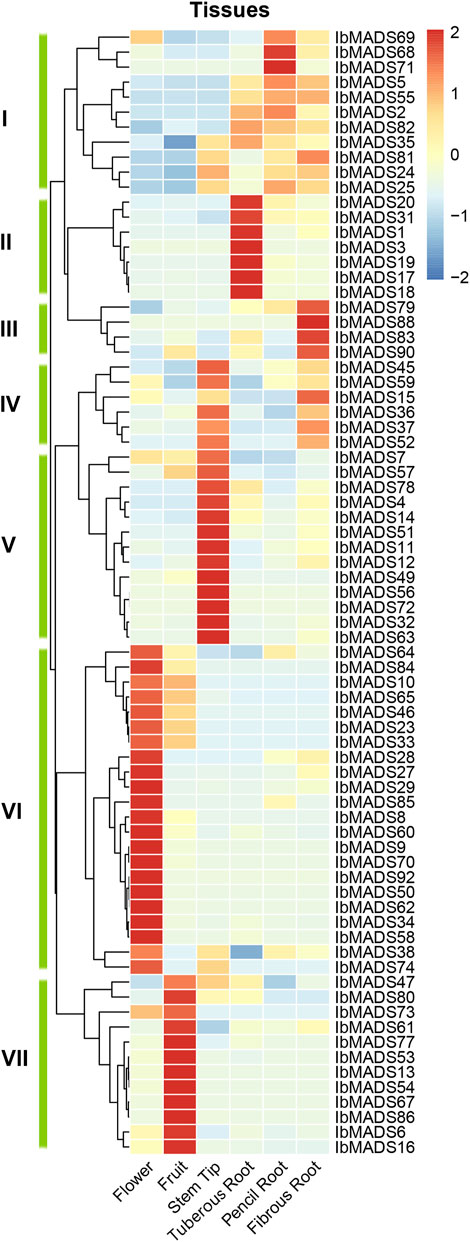
3.7 Expression Profiles of MADS-Box Genes in Sweet Potato
To investigate the expression patterns of MADS-box genes in sweet potato, the 75 MADS-box gene expression profiles were detected in six different tissues by transcriptome sequencing. The results showed that sweet potato MADS-box genes were expressed and differentiated in different tissues (Figure 7). Cluster analysis clustered the sweet potato MADS-box gene expression profiles into seven classes, with class I, II, and III genes mainly expressed in different types of roots. Among them, seven genes in class II were highly and specifically expressed in the tuberous root. The class IV genes were expressed in the stem tip and fibrous root. The class V genes were highly and specifically expressed in the stem tip. The class VI genes were predominantly expressed in flower, and a small proportion of these genes were also somewhat expressed in fruits. The class VII genes are highly and specifically expressed in fruit. Overall, 22 MADS-box genes were highly expressed in flowers, 19 were highly expressed in stem tips, 12 were highly expressed in fruits, and the remaining genes were differentially expressed in different types of roots.
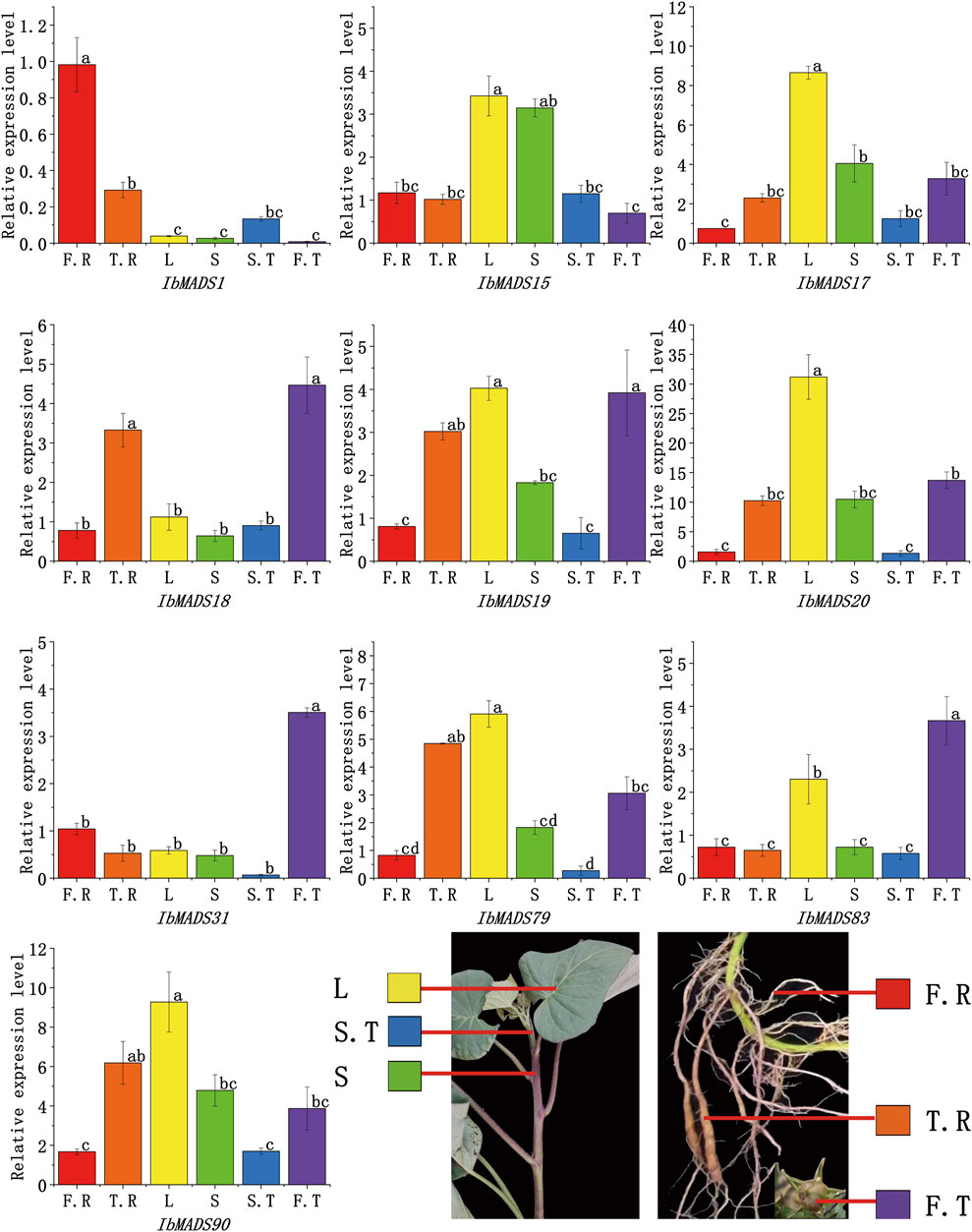
3.8 Expression Analysis of MADS-Box Genes by qRT-PCR
qRT-PCR was used to further validate the expression profiles of the 10 genes, which were more highly expressed in roots than other tissues, according to transcriptome sequencing of MADS-box genes in different tissues (Figure 8). Ten genes showed tissue-specific expression at different levels in all tissues; and IbMADS1, IbMADS18, IbMADS19, IbMADS79, and IbMADS90 were highly expressed in the fibrous root or tuberous root. Seven genes were more highly expressed in the leaf, only IbMADS15 was highly expressed in the stem, and the remaining genes were relatively poorly expressed in the stem. Ten genes were all expressed at lower levels in the stem tip. IbMADS18, IbMADS31, and IbMADS83 were significantly expressed in the fruit. Some genes were highly expressed only in a single tissue, and others were not significantly expressed, such as IbAMDS1, which was significantly expressed only in the fibrous root; IbMADS17 and IbMADS20, which were highly expressed only in the leaf; and IbMADS31 and IbMADS83, which were significantly expressed only in the fruit. The five remaining genes were more highly expressed in two to three tissues.
4 Discussion
MADS-box genes are widely present in eukaryotes. There are 108, 72, 168, and 160 MADS-box genes identified in Arabidopsis (Livak and Schmittgen, 2001), Oryza sativa (Ray et al., 2007), Nicotiana tabacum (Bai et al., 2019), and Brassica rapa (Duan et al., 2015). Interestingly, I. trifida, a diploid plant belonging to the genus Ipomoea, has 37 MADS-box genes (Zhu et al., 2020). In this study, 95 sweet potato MADS-box genes were identified, the number of which was approximately three times that of I. trifida. Phylogenetic analysis found that the MADS-box genes of I. batatas are not simply tripled by I. trifida. We discovered more type I MADS genes in I. batatas, and the distribution of type II genes was also more abundant than that of I. trifida. Nevertheless, I. batatas are missing only in the FLC subfamily and the TT16 subfamily. It is suggested that FLC genes are repressors of flowering which affect multiple pathways for controlling the flowering time (Michaels and Amasino, 1999; Scott et al., 2003). Loss of FLC subfamily genes may indicate that sweet potato vernalization is not regulated by its repression. Consistently, a previous study also showed that the emergence of FLC subfamily members is not detected in I. trifida (Zhu et al., 2020). TT16 encodes a TF involved in the transcriptional regulation of proanthocyanidin biosynthesis in the seed coat (Nesi et al., 2002). The absence of sweet potato TT16 subfamily genes suggests the potential involvement of other pathways for regulating proanthocyanidin biosynthesis in sweet potato.
Moreover, we again found differentiation in the MADS-box gene structure, which may be attributed to the presence of I-domain, K-domain, and C-domain. Type II genes typically exhibit a longer gene length and a greater number of exons, which is also one of the basis for classifying subtypes of MADS-box genes (Kaufmann et al., 2005). Consistent with the structure feature, the type I genes of sweet potato have one to two exons, and most of them range at 1 kb, while the type II genes are generally larger than 3 kb and have more than six exons. In addition, the MADS-box genes of sweet potato are distributed more dispersedly on the chromosome. Their numbers differ from those of chromosomes, which are similar to the chromosome distribution of Arabidopsis (Lamesch et al., 2012), Jatropha curcas (Tang et al., 2020), Cardamine hirsuta (Ghorbani et al., 2020), and I. trifida. The MADS-box proteins in sweet potato have a strong motif conservation among various subfamilies, with different numbers and motifs. The core motifs are motif 1, motif 2, and motif 5, which encode about 50 amino acids, and they are related to the SRF-box domain (Ng et al., 2001). Sweet potato is a hexaploid plant with both homologous and heterologous chromosomes, which has a complex genetic background (Yang et al., 2017). MCScanX analysis showed that a large number of genes had collinear relationships within the sweet potato genome, and the MADS-box genes also had a certain homology of evolutionary relationship. Among them, 19 MADS-box-genes had collinear relationships. In particular, reciprocal segmental duplications existed in IbMADS4, IbMADS14, IbMADS32, and IbMADS56. IbMAD51 shared a segmental duplication relationship with IbMADS4 and IbMADS56, while it did not have a segmental duplication relationship with IbMADS14 and IbMADS32. It is presumed that IbMADS4, IbMADS14, IbMADS32, and IbMADS56 were derived from a single duplication event, while IbMAD51 was derived from another separate duplication event, possibly a single segmental duplication event.
The MADS-box family is considered to be as a key family of genes responsible for regulating reproductive growth and vegetative organ development, which was initially characterized as homeotic genes in floral organs. Also, the importance of the MADS-box family in the morphogenesis and growth of other organs of plants, especially roots and fruits, has been highlighted (Riechmann et al., 1997; Zhao et al., 2019). Multiple types of cis-acting elements have been identified in the promoter region of the MADS-box gene from sweet potato, mainly including light-responsive elements, auxin-responsive elements, MeJA-responsive elements, abscisic acid-responsive elements, and drought-inducible elements. It is indicated that MADS-box genes in sweet potato respond to both hormones and environmental factors, and they are closely related to the normal growth and development of sweet potato. Expression levels of MADS-box genes can reveal the relevant physiological functions, Hasebe et al. (1998) found that most MADS-box genes in ferns are expressed in reproductive and vegetative organs, while most MADS-box genes in seed plants are only expressed in one organism, and only a few are expressed in reproductive and vegetative organisms. Transcriptome sequencing revealed that the majority of MADS-box genes in sweet potato exhibit the expression specialization. Thirty-four genes were specifically expressed in sweet potato flowers and fruits, while the remaining were mainly expressed in vegetative tissues like the root and stem tip. The expression profiles of 10 genes in different tissues of sweet potato were further verified by qRT-PCR. IbMADS1 was highly expressed in the fibrous root; and IbMADS79, IbMADS18, IbMADS19, and IbMADS90 were highly expressed in the tuberous root. It has been reported that IbMADS1 and IbMADS79 are associated with the root development of sweet potato. In particular, IbMADS1 is an important integrator at the initiation of tuberization (Kim et al., 2005; Ku et al., 2008), which is consistent with our finding. Cis-acting elements like the light-responsive elements, Zein metabolism-regulation elements, and auxin-responsive elements were found in the promoter region of IbMADS1; and light-responsive elements, meristem-express elements, MeJA-responsive elements, and salicylic acid-responsive element were found in the promoter region of IbMADS79. IbMADS1 and IbMADS79 may play an important role in the root development process and may participate in the formation of the tuberous root. The homologous genes IbMADS18 and IbMADS19 of FEM11 were highly expressed not only in the fruit but also in the tuberous root. And the homologous gene IbMADS90 of AGL62 was highly expressed in the tuberous root and leaf. FEM111 and AGL62 are associated with the flower and fruit development in Arabidopsis (Kang et al., 2008; Portereiko et al., 2006; Steffen et al., 2008). IbMADS18, IbMADS19, and IbMADS90 are involved in the tuberous root development of sweet potato and the fruit development to some extent. In addition, IbMADS31 and IbMADS83 are also specifically expressed in the fruit. AGL61, a homolog of IbMADS83, functions as a TF that controls the expression of downstream genes during central cell development and is involved in the seed endosperm development (Steffen et al., 2008). And AGL17, the homolog of IbMADS31, is a significant downstream target of CURLY LEAF in floral transition control (Shu et al., 2020). We found that cis-acting elements like the MeJA-responsive elements, auxin-responsive elements, abscisic acid-responsive elements, and gibberellin-responsive elements in the promoter region of IbMADS31 and IbMADS83 provided indirect evidences for the involvement of IbMADS31 and IbMADS83 in the flower and fruit development of sweet potato. IbMADS15, IbMADS17, IbMADS19, IbMADS20, IbMADS79, and IbMADS90 were highly expressed in the leaf or stem, which were all identified as type I genes. In addition, a large number of type I genes were detected mainly expressed in the stem tip by RNA-seq. It is suggested that type I genes were involved in the nutritional growth process of the aerial parts of sweet potato, to some extent.
Overall, the sweet potato MADS-box genes are involved in the growth and development of reproductive organs like the flower and fruit of sweet potato. They were vital in the process of nutritional growth and development such as the root, stem, and leaf of sweet potato.
5 Conclusion
In this study, the 95 MADS-box family genes were identified from the sweet potato genome by Hmmer, SMART, and other tools, and they were divided into type I and type II subgroups according to the gene structure. The phylogenetic relationship of MADS-box genes between sweet potato and Arabidopsis was analyzed based on the NJ phylogenetic tree. The type I genes of sweet potato were further divided into three subfamilies, Mα, Mβ, and Mγ, whereas the type II genes were divided into 12 subfamilies.
The distribution of the sweet potato MADS-box genes on chromosomes was analyzed by chromosomal localization, and the analysis of the motif identified the MADS-box conserved domain motifs. Collinearity between and within chromosomes of sweet potato was analyzed using MCScanX to identify 19 MADS-box genes with segmental duplications or tandem duplications. Analysis of the MADS-box promoter region of sweet potato revealed that the MADS-box genes contain light-responsive elements, auxin-responsive elements, MeJA-responsive elements, abscisic acid-responsive elements, drought induction elements, and other cis-acting elements that respond to multiple environmental changes and coordinate growth and development. Furthermore, we analyzed the expression patterns of MADS-box genes in various tissues of sweet potato by transcriptome sequencing and qRT-PCR, and we found that the sweet potato MADS-box genes are involved in the growth and development of reproductive organs such as the flower and fruit, but also the growth and development of nutritional organs such as the root, stem, and leaf of sweet potato.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZS conceived and designed the experiments. ZS and MH performed the experiments. ZZ and YC prepared the materials. ZS analyzed the data and wrote the paper. A-DH and HZ helped to revise the paper. All authors read and approved the final articles.
Funding
This research was funded by the NSFC-Guangdong Natural Science Foundation Joint Project (U1701234).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.750137/full#supplementary-material
References
Bai, G., Yang, D. H., Cao, P., Yao, H., Zhang, Y., Chen, X., et al. (2019). Genome-Wide Identification, Gene Structure and Expression Analysis of the MADS-Box Gene Family Indicate Their Function in the Development of Tobacco (Nicotiana Tabacum l). Int. J. Mol. Sci. 20 (20), 5043. doi:10.3390/ijms20205043
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37 (2), W202–W208. doi:10.1093/nar/gkp335
Burgeff, C., Liljegren, S. J., Tapia-López, R., Yanofsky, M. F., and Alvarez-Buylla, E. R. (2002). MADS-Box Gene Expression in Lateral Primordia, Meristems and Differentiated Tissues of Arabidopsis thaliana Roots. Planta 214 (3), 365–372. doi:10.1007/s004250100637
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
De Bodt, S., Raes, J., Florquin, K., Rombauts, S., Rouz, P., Theien, G., et al. (2003). Genomewide Structural Annotation and Evolutionary Analysis of the Type I MADS-Box Genes in Plants. J. Mol. Evol. 56 (5), 573–586. doi:10.1007/s00239-002-2426-x
Dong, T., Song, W., Tan, C., Zhou, Z., Yu, J., Han, R., et al. (2018). Molecular Characterization of Nine Sweet Potato (Ipomoea Batatas Lam.) MADS-Box Transcription Factors during Storage Root Development and Following Abiotic Stress. Plant Breed 137 (5), 790–804. doi:10.1111/pbr.12613
Duan, W., Song, X., Liu, T., Huang, Z., Ren, J., Hou, X., et al. (2015). Genome-wide Analysis of the MADS-Box Gene Family in Brassica Rapa (Chinese Cabbage). Mol. Genet. Genomics. 290 (1), 239–255. doi:10.1007/s00438-014-0912-7
Fan, H.-Y., Hu, Y., Tudor, M., and Ma, H. (1997). Specific Interactions between the K Domains of AG and AGLs, Members of the MADS Domain Family of DNA Binding Proteins. Plant J. 12 (5), 999–1010. doi:10.1046/j.1365-313X.1997.12050999.x
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of Exon Skipping Events Differentiate Among Splicing Patterns in Sixteen Human Tissues. F1000Res 2, 188. doi:10.12688/f1000research.2-188.v2
Ghorbani Marghashi, M., Bagheri, H., and Gholami, M. (2020). Genome-Wide Study of Flowering-Related MADS-Box Genes Family in Cardamine Hirsuta. 3 Biotech. 10 (12), 518. doi:10.1007/s13205-020-02521-w
Hasebe, M., Wen, C.-K., Kato, M., and Banks, J. A. (1998). Characterization of MADS Homeotic Genes in the Fern Ceratopteris Richardii. Proc. Natl. Acad. Sci. 95 (11), 6222–6227. doi:10.1073/pnas.95.11.6222
Ito, K., and Murphy, D. (2013). Application of Ggplot2 to Pharmacometric Graphics. CPT: Pharmacometrics Syst. Pharmacol. 2, 79. doi:10.1038/psp.2013.56
Jin, J., Tian, F., Yang, D.-C., Meng, Y.-Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 45 (D1), D1040–D1045. doi:10.1093/nar/gkw982
Kang, I.-H., Steffen, J. G., Portereiko, M. F., Lloyd, A., and Drews, G. N. (2008). The AGL62 MADS Domain Protein Regulates Cellularization during Endosperm Development inArabidopsis. Plant Cell 20 (3), 635–647. doi:10.1105/tpc.107.055137
Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-Type MADS-Domain Proteins: Structural Modularity, Protein Interactions and Network Evolution in Land Plants. Gene 347 (2), 183–198. doi:10.1016/j.gene.2004.12.014
Kim, S.-H., Hamada, T., Otani, M., and Shimada, T. (2005). Isolation and Characterization of MADS Box Genes Possibly Related to Root Development in Sweetpotato(Ipomoea Batatas L. Lam). J. Plant Biol. 48 (4), 387–393. doi:10.1007/BF03030580
Kim, S.-H., Mizuno, K., and Fujimura, T. (2002). Isolation of MADS-Box Genes from Sweet Potato (Ipomoea Batatas (L.) Lam.) Expressed Specifically in Vegetative Tissues. Plant Cel Physiol 43 (3), 314–322. doi:10.1093/pcp/pcf043
Krejci, A., Hupp, T. R., Lexa, M., Vojtesek, B., and Muller, P. (2016). Hammock: A Hidden Markov Model-Based Peptide Clustering Algorithm to Identify Protein-Interaction Consensus Motifs in Large Datasets. Bioinformatics 32 (1), btv522–16. doi:10.1093/bioinformatics/btv522
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 19 (9), 1639–1645. doi:10.1101/gr.092759.109
Ku, A. T., Huang, Y.-S., Wang, Y.-S., Ma, D., and Yeh, K.-W. (2008). IbMADS1 (Ipomoea Batatas MADS-Box 1 Gene) Is Involved in Tuberous Root Initiation in Sweet Potato (Ipomoea Batatas). Ann. Bot. 102 (1), 57–67. doi:10.1093/aob/mcn067
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): Improved Gene Annotation and New Tools. Nucleic Acids Res. 40 (Database issue), D1202–D1210. doi:10.1093/nar/gkr1090
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C Encodes a Novel MADS Domain Protein that Acts as a Repressor of Flowering. Plant Cell 11 (5), 949–956. doi:10.1105/tpc.11.5.949
Michaels, S. D., He, Y., Scortecci, K. C., and Amasino, R. M. (2003). Attenuation of FLOWERING LOCUS C Activity as a Mechanism for the Evolution of Summer-Annual Flowering Behavior in Arabidopsis. Proc. Natl. Acad. Sci. 100 (17), 10102–10107. doi:10.1073/pnas.1531467100
Nam, J., Kaufmann, K., Theissen, G., and Nei, M. (2005). A Simple Method for Predicting the Functional Differentiation of Duplicate Genes and its Application to MIKC-Type MADS-Box Genes. Nucleic Acids Res. 33 (2), e12. doi:10.1093/nar/gni003
Nesi, N., Debeaujon, I., Jond, C., Stewart, A. J., Jenkins, G. I., Caboche, M., et al. (2002). The Transparent Testa16 Locus Encodes the Arabidopsis Bsister Mads Domain Protein and Is Required for Proper Development and Pigmentation of the Seed Coat. Plant Cell 14 (10), 2463–2479. doi:10.1105/tpc.004127
Ng, M., and Yanofsky, M. F. (2001). Function and Evolution of the Plant MADS-Box Gene Family. Nat. Rev. Genet. 2 (3), 186–195. doi:10.1038/35056041
Norman, C., Runswick, M., Pollock, R., and Treisman, R. (1988). Isolation and Properties of cDNA Clones Encoding SRF, a Transcription Factor that Binds to the C-Fos Serum Response Element. Cell 55 (6), 989–1003. doi:10.1016/0092-8674(88)90244-9
Par̆enicová, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and Phylogenetic Analyses of the Complete MADS-Box Transcription Factor Family in Arabidopsis. Plant Cell 15 (7), 1538–1551. doi:10.1105/tpc.011544
Passmore, S., Maine, G. T., Elble, R., Christ, C., and Tye, B.-K. (1988). Saccharomyces cerevisiae Protein Involved in Plasmid Maintenance Is Necessary for Mating of MATα Cells. J. Mol. Biol. 204 (3), 593–606. doi:10.1016/0022-2836(88)90358-0
Portereiko, M. F., Lloyd, A., Steffen, J. G., Punwani, J. A., Otsuga, D., and Drews, G. N. (2006). AGL80Is Required for Central Cell and Endosperm Development inArabidopsis. Plant Cell 18 (8), 1862–1872. doi:10.1105/tpc.106.040824
Ray, S., Agarwal, P., Arora, R., Kapoor, S., and Tyagi, A. K. (2007). Expression Analysis of Calcium-Dependent Protein Kinase Gene Family during Reproductive Development and Abiotic Stress Conditions in rice (Oryza Sativa L. Ssp. Indica). Mol. Genet. Genomics. 278 (5), 493–505. doi:10.1007/s00438-007-0267-4
Riechmann, J. L., and Meyerowitz, E. M. (1997). MADS Domain Proteins in Plant Development. Biol. Chem. 378 (10), 1079–1101. doi:10.1515/bchm.1997.378.10.1079
Saitou, N., and Nei, M. (1987). The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 4 (4), 406–425. doi:10.1093/oxfordjournals.molbev.a040454
Sheng, X.-G., Zhao, Z.-Q., Wang, J.-S., Yu, H.-F., Shen, Y.-S., Zeng, X.-Y., et al. (2019). Genome Wide Analysis of MADS-Box Gene Family in Brassica oleracea Reveals Conservation and Variation in Flower Development. BMC Plant Biol. 19 (1), 106. doi:10.1186/s12870-019-1717-y
Shu, J., Chen, C., Kohalmi, S. E., and Cui, Y. (2020). Evidence that AGL17 Is a Significant Downstream Target of CLF in floral Transition Control. Plant Signaling Behav. 15 (7), 1766851. doi:10.1080/15592324.2020.1766851
Singh, V. K., Mangalam, A. K., Dwivedi, S., and Naik, S. (1998). Primer Premier: Program for Design of Degenerate Primers from a Protein Sequence. Biotechniques 24 (2), 318–319. doi:10.2144/98242pf02
Sommer, H., Beltrán, J. P., Huijser, P., Pape, H., Lönnig, W. E., Saedler, H., et al. (1990). Deficiens, a Homeotic Gene Involved in the Control of Flower Morphogenesis in Antirrhinum Majus: The Protein Shows Homology to Transcription Factors. EMBO J. 9 (3), 605–613. doi:10.1002/j.1460-2075.1990.tb08152.x
Steffen, J. G., Kang, I.-H., Portereiko, M. F., Lloyd, A., and Drews, G. N. (2008). AGL61 Interacts with AGL80 and Is Required for Central Cell Development in Arabidopsis. Plant Physiol. 148 (1), 259–268. doi:10.1104/pp.108.119404
Sumiya, N. (2021). Cis-Acting Elements Involved in the G2/M-Phase-Specific Transcription of the Cyclin B Gene in the Unicellular Alga Cyanidioschyzon Merolae. J. Plant Res. 134 (6), 1301–1310. doi:10.1007/s10265-021-01334-z
Sun, C.-H., Wang, J.-H., Gu, K.-D., Zhang, P., Zhang, X.-Y., Zheng, C.-S., et al. (2021). New Insights into the Role of MADS-Box Transcription Factor Gene CmANR1 on Root and Shoot Development in chrysanthemum (Chrysanthemum Morifolium). BMC Plant Biol. 21 (1), 79. doi:10.1186/s12870-021-02860-7
Tang, Y., Wang, J., Bao, X., Wu, Q., Yang, T., Li, H., et al. (2020). Genome-Wide Analysis of Jatropha Curcas MADS-Box Gene Family and Functional Characterization of the JcMADS40 Gene in Transgenic rice. BMC Genomics 21 (1), 325. doi:10.1186/s12864-020-6741-7
Theissen, G. (2001). Development of floral Organ Identity: Stories from the MADS House. Curr. Opin. Plant Biol. 4 (1), 75–85. doi:10.1016/s1369-5266(00)00139-4
Voorrips, R. E. (2002). MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 93 (1), 77–78. doi:10.1093/jhered/93.1.77
Wang, S., Nie, S., and Zhu, F. (2016). Chemical Constituents and Health Effects of Sweet Potato. Food Res. Int. 89 (Pt 1), 90–116. doi:10.1016/j.foodres.2016.08.032
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wang, Y., Zhang, J., Hu, Z., Guo, X., Tian, S., and Chen, G. (2019). Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum Lycopersicum. Int. J. Mol. Sci. 20 (12), 2961. doi:10.3390/ijms20122961
Won, S. Y., Jung, J.-A., and Kim, J. S. (2021). Genome-Wide Analysis of the MADS-Box Gene Family in Chrysanthemum. Comput. Biol. Chem. 90, 107424. doi:10.1016/j.compbiolchem.2020.107424
Yang, J., Moeinzadeh, M.-H., Kuhl, H., Helmuth, J., Xiao, P., Haas, S., et al. (2017). Haplotype-Resolved Sweet Potato Genome Traces Back its Hexaploidization History. Nat. Plants 3 (9), 696–703. doi:10.1038/s41477-017-0002-z
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The Protein Encoded by the Arabidopsis Homeotic Gene Agamous Resembles Transcription Factors. Nature 346 (6279), 35–39. doi:10.1038/346035a0
Zhang, S., Guo, Y., Zhang, Y., Guo, J., Li, K., Fu, W., et al. (2021). Genome-wide Identification, Characterization and Expression Profiles of the CCD Gene Family in Gossypium Species. 3 Biotech. 11 (5), 249. doi:10.1007/s13205-021-02805-9
Zhao, H.-B., Jia, H.-M., Wang, Y., Wang, G.-Y., Zhou, C.-C., Jia, H.-J., et al. (2019). Genome-Wide Identification and Analysis of the MADS-Box Gene Family and its Potential Role in Fruit Development and Ripening in Red Bayberry (Morella Rubra). Gene 717, 144045. doi:10.1016/j.gene.2019.144045
Keywords: sweet potato, MADS-box, MIKC type, bioinformatics analysis, growth development, expression analysis
Citation: Shao Z, He M, Zeng Z, Chen Y, Hanna A-D and Zhu H (2021) Genome-Wide Identification and Expression Analysis of the MADS-Box Gene Family in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Genet. 12:750137. doi: 10.3389/fgene.2021.750137
Received: 30 July 2021; Accepted: 05 October 2021;
Published: 17 November 2021.
Edited by:
Himabindu Kudapa, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaReviewed by:
Muhammad Ramzan Khan, Quaid-i-Azam University, PakistanJin Xiao, Nanjing Agricultural University, China
Copyright © 2021 Shao, He, Zeng, Chen, Hanna and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbo Zhu, dGR6aHVAMTI2LmNvbQ==
 Zhengwei Shao
Zhengwei Shao Minhong He
Minhong He