- 1Department of Biomedical Sciences, University of North Dakota School of Medicine, Grand Forks, ND, United States
- 2Skolkovo Institute of Science and Technology, Moscow, Russia
Transcription is a step in gene expression that defines the identity of cells and its dysregulation is associated with diseases. With advancing technologies revealing molecular underpinnings of the cell with ever-higher precision, our ability to view the transcriptomes may have surpassed our knowledge of the principles behind their organization. The human RNA polymerase II (Pol II) machinery comprises thousands of components that, in conjunction with epigenetic and other mechanisms, drive specialized programs of development, differentiation, and responses to the environment. Parts of these programs are repurposed in oncogenic transformation. Targeting of cancers is commonly done by inhibiting general or broadly acting components of the cellular machinery. The critical unanswered question is how globally acting or general factors exert cell type specific effects on transcription. One solution, which is discussed here, may be among the events that take place at genes during early Pol II transcription elongation. This essay turns the spotlight on the well-known phenomenon of promoter-proximal Pol II pausing as a step that separates signals that establish pausing genome-wide from those that release the paused Pol II into the gene. Concepts generated in this rapidly developing field will enhance our understanding of basic principles behind transcriptome organization and hopefully translate into better therapies at the bedside.
Introduction
Treatments of cancers should ideally be tailored to their specific molecular signatures. The central place and complexity of transcription regulation in normal and cancer cells offer tantalizing opportunities for precise targeting (Bushweller, 2019; Van Hoeck et al., 2019; Malone et al., 2020). Many anticancer drugs in use and in development today target the transcriptional machinery or epigenetic regulators (Villicaña et al., 2014; Mohammad et al., 2019; Park and Han, 2019; Laham-Karam et al., 2020). However, rather than specific transcription factors, many drugs work against factors with broad-spectrum functionality including epigenome modifiers and components of the basal transcriptional machinery (Bywater et al., 2013). This targeting strategy draws from long-standing observations that perturbation of general or globally acting factors often results in distinct cell type specific effects for reasons that remain poorly understood (Ptashne, 2013). Because many targets of anti-cancer therapies have broad or essential roles in normal cells, their use remains heavily based on empirical findings. Targeting specific factors such as transcription factors causative of certain cancers is becoming feasible (Sievers et al., 2018; Duffy and Crown, 2021), but is limited by an uncertainty in how these and other factors may function in different cellular contexts.
Transcription is the first step in expression of genes and genomes. The combined activity of some 20,000 human genes results in genome-wide RNA transcriptome patterns that reflect the biology and define the identity of every cell. Despite the flood of technologies describing the transcriptomes with increasing precision, our understanding of gene regulation remains fundamentally based on the knowledge gained from studies of individual genes. A long-standing gene-centric paradigm describes regulation by sequence-specific transcription factors that serve as repressors and activators, in contrast to factors broadly involved in the process of transcription that are considered basal or general (Nikolov and Burley, 1997; Juven-Gershon and Kadonaga, 2010). This paradigm does not explain network-level events especially the molecular rules governing interactions among thousands of genes in different cell types.
Two features of Pol II transcription are of note. On the one hand, the Pol II machinery is constantly modulated by a host of activating and repressing inputs that connect transcription to the environment within and outside the cell. These second to minute-scale events underlie rapid responses to stimuli and have generated the bulk of our understanding of transcription regulation in cellular responses to environmental triggers such as heat shock, hormones such as estrogen, or innate immune responses (Adelman et al., 2009; Hah et al., 2011; Mahat et al., 2016). In addition, metazoan cells have a special ability to form distinct stable steady states and undergo regulated transitions between them. These transitions take place on longer timescales, lasting hours to years, and involve predefined programs that are commonly visualized through the concept of the epigenetic landscape (Waddington, 1957). These transitions underlie development and differentiation of normal cells and involve transcriptional and epigenetic control mechanisms and factors that can be ectopically activated in cancers (Rousseaux et al., 2013).
Targeting strategies should benefit from better understanding of principles that govern the transcriptomes. This problem can be conceptually narrowed down to defining the molecular interactions that link individual genes within transcriptional networks. Given the overall conservation of the RNA polymerase II machinery (Hampsey, 1998), mechanisms that drive this quantum leap in complexity in higher organisms presumably do not involve too many additional players and instead must rely on repurposing of existing components. In this essay, we discuss some of the challenges in targeting the transcriptional machinery and suggest a potential avenue for improving the precision of broad-stroke interventions.
Pervasive Uncertainty in Targeting Cellular Components
In this section we describe some of the challenges in targeting the transcriptional machinery. These arise not only from unintended effects of drugs, which can be improved by identifying better targets and better drug design, but also from the inherent uncertainty of transcription regulation in different cellular contexts.
Targeting the Transcription Machinery: Knocking on the Black Box
Transcription is the ultimate target of numerous anticancer drugs that act on Pol II or the epigenetic machinery. Cancer targeting aims to either kill or reprogram cells into more benign states (Gong et al., 2019). To gain selectivity over normal cells, targeting strategies exploit distinct properties of cancer cells. One property is addiction to transcription (Bradner et al., 2017), which increases the demands of cancer cells for Pol II activity and makes them more sensitive to its inhibition. Inhibitors of general transcription factors such as TFIIH and P-TEFb have been used (Villicaña et al., 2014; Wang et al., 2015; Sava et al., 2020). Known oncogene transcription factors such as c-Myc, KRAS, etc, are tempting targets because of their key roles in cancer initiation and progression (Hallin et al., 2020; Madden et al., 2021). However, some of these factors have been considered undruggable or difficult to target for various reasons including their critical roles in normal cells and/or difficulty to specifically target interactors as compared to enzymes (Lazo and Sharlow, 2016; Zhang et al., 2018). Recent studies suggest that these challenges will be at least to some extent overcome (Duffy and Crown, 2021; Trkulja et al., 2021; Wang et al., 2021). However, the highly changeable nature of cancers that can outselect therapies will always remain a formidable caveat.
A second property of cancer cells is broad dysregulation of the transcriptional machinery (Bywater et al., 2013; Lee and Young, 2013), which alters the requirements of cancer cells for its components and leads to unusual sensitivity to inhibition of certain factors. Several classes of epigenetic drugs are in development or already on the market, with the more common including Histone Deacetylase (HDAC) Inhibitors, Histone Acetyltransferase (HAT) inhibitors, Bromodomain Inhibitors, DNA methylation inhibitors, etc. These and others are described in detail in reviews elsewhere, for example, in (Heerboth et al., 2014; Mohammad et al., 2019; Nepali and Liou, 2021). Broad-stroke targeting has been rather successful in generating drugs, placing a burden on better understanding of when to target distinct components.
A third property of cancer cells is altered expression of genes outside of those involved in transcription. Differentially expressed genes are often marked as sources for therapeutic targets. Identifying therapeutic targets is perhaps the most common justification for basic studies over the years. Increasing our understanding of how different types of cancers work has indeed resulted in identification of targets including surface and nuclear receptors, kinases, etc (Kannaiyan and Mahadevan, 2018; Zhao et al., 2019; Skidmore et al., 2020). While revealing molecular mechanisms of various processes and perhaps holding the keys to successful therapies in a long run, translation of these findings into therapies takes years with no guaranteed success. The ability to tailor a drug to a living cancer patient therefore remains limited (Krzyszczyk et al., 2018).
Ambiguous Roles of Transcription Factors
Identifying causative factors for precise targeting of cancers is an attractive goal that has met serious challenges (Vishnoi et al., 2020). Cancers with well-known etiology such as fusion protein driven pediatric cancers remain difficult to target (Uren and Toretsky, 2005; Brien et al., 2019). This uncertainty is only amplified in adult cancers (Dawson et al., 2011; Fung et al., 2015; Shan et al., 2017; Winters and Bernt, 2017). One major reason behind this uncertainty is that the functions of individual factors can be dramatically altered across cell types and different individuals. This property may be inherent to transcription factors themselves and might not always be controllable.
DNA-binding transcription factors are commonly labeled as either activators or repressors (Wolffe et al., 1997). It is becoming increasingly clear, however, that most if not all transcription factors can, and likely do, function both as repressors and activators. There are several possible reasons for this duality. First, a factor itself may play different roles at the same loci. A number of transcription factors involved in Drosophila development and stimulus responses in human cells show default repression of target genes unless activated, usually by a co-factor, thereby appearing both as repressors and activators (Barolo and Posakony, 2002). In E. coli bacteriophage T4, the transcription factor gp33 causes default repression of late promoters, but becomes their potent co-activator in the presence of its specific co-factors (Kolesky et al., 2002; Nechaev and Geiduschek, 2006). Drosophila Hunchback and Dorsal (Pan and Courey, 1992; Dubnicoff et al., 1997; Bauer et al., 2010) serve as repressors or activators depending on co-factors on different promoters to drive highly coordinated embryo development (Staller et al., 2015). These well studied examples show that switching between a repressor and an activator in principle does not require complex changes.
Dual functions of transcription factors may arise through other mechanisms. One involves distinct activities for different isoforms of the same gene. Given a large number of known gene isoforms and frequent creation of new gene isoforms in cancers (Belluti et al., 2020), alternative splicing may be a significant contributor to the ambiguity of transcription factor designation, at least at the level of a gene (Walker et al., 1996). The same factors may also have different roles because they function in distinct complexes. The Polycomb Repressive Complex 2, PRC2, introduces the H3K27 histone mark with essential roles in development, cell differentiation, and cancer (Aranda et al., 2015; Schuettengruber et al., 2017; Healy et al., 2019). Ezh2 is the catalytic component of PRC2 responsible for introducing the mark. Ezh2 has been also shown to activate transcription in a separate role that does not involve its catalytic activity and is independent of PRC2 complex (Kim et al., 2018). Ezh2 role as an activator involves the binding at a promoter of a target (AR) gene as a DNA-binding transcription factor.
While the Drosophila factors have been well known to have dual roles, as more studies become available in human systems, even long-studied factors “acquire” opposing functions (Ip, 1995). For example, Snail is a conserved member of a family of E-box motif binding transcription factors that is involved in development though its role in the epithelial-mesenchymal transition with close relevance to cancer metastasis (Alberga et al., 1991; Carver et al., 2001; Peinado et al., 2004). Snail was initially considered to be exclusively a transcriptional repressor that binds to target gene promoters such as E-Cadherin. However, Snail was later shown to also activate genes in Drosophila during mesoderm development (Wu et al., 2017). Conversion of Snail from a repressor to an activator was shown to involve acetylation of Snail by the CREB-binding protein (CBP) (Hsu et al., 2014). Post-translational modifications can convert transcriptional repressors to activators (Mosley et al., 2003; Zhang et al., 2012), suggesting that this mechanism may be used in cancers as well. The duality of transcription factor roles as activators and repressors, therefore, is likely to be their inherent property rather than an exception.
Another major mechanism that can contribute to dual roles of transcription factors has to do not with their direct function, but with compensatory changes in the rest of the cell. On a short time scale, such as during rapid responses to stimuli, these changes may be driven by redistribution of cellular machinery components (Bregman et al., 1995). Such effects are considered secondary or nonspecific and are not well understood, but are pervasive and may be just as important as direct roles of transcription factors. For example, it is common for experimental perturbation of a factor by assays such as RNA-interference to cause both activation and repression of gene cohorts regardless of its actual mechanism of action. One possible exception is the transcriptional amplifier c-Myc that supports unbiased amplification of gene activity from all promoters genome-wide (Lin et al., 2012; Nie et al., 2012). Which genes are indirectly activated or repressed through secondary interactions should depend on the cellular context of individual cancers. The apparent dual roles of P53 tumor suppressor may fall into a similar category. The transcription factor p53 is widely considered to be an activator. Several studies, however, have proposed p53 as a direct repressor of genes (Banerjee et al., 2009; Allen et al., 2014). Its repressor role is controversial and has been suggested to be indirect (Fischer et al., 2014).
The ambiguity of functional designations for transcription factors extends to their phenotypic classification as oncogenes versus tumor suppressors (Shen et al., 2018; Datta et al., 2020), which may or may not be connected to their molecular mechanisms of action. The duality of transcriptional effects as well as cancer targeting outcomes persists through all levels of transcription factor function. These opposing functions are inherent to transcription factors and might not be separable even by specific targeting.
Epigenetic Marks – A Knot Around Transcription
Epigenetic marks are known to be frequently altered in cancers (Jones and Baylin, 2007; Baylin and Jones, 2011; Bennett and Licht, 2018), making potentially reversible epigenetic reprogramming an attractive targeting strategy (Jin and Kim, 2017). There are caveats, however. First, known epigenetic marks have a broad scope, either covering large regions of the genome or distributed across multiple punctate regions, limiting the specificity of direct targeting. Second, many known histone marks are closely tied to transcription, either associated with repressed or active states of the nearby genes or regulatory elements such as enhancers (Henikoff and Shilatifard, 2011; Li et al., 2012; Kang et al., 2020) through mechanisms that remain to be fully understood. For example, histone H3 Lysine 4 trimethylation (H3K4Me3) preferentially marks active promoters, whereas monomethylation (H3K4Me1) mark appears to prefer regions outside of promoters including active and poised enhancers (Heintzman et al., 2007), and might have to do with transcriptional memory (Saeed et al., 2014; Bae and Lesch, 2020). Even with these well studied marks there are overlaps between distinct elements such as promoters and enhancers and the rules behind their deposition merit further studies (Pekowska et al., 2011; Scruggs et al., 2015; Soares et al., 2017). Histone H3K27 acetylation is commonly used to profile open genomic regions including active enhancers (Creyghton et al., 2010), but its functional roles remain not fully clear (Zhang et al., 2020). Acetylation patterns of various histones may be a good predictor for various types of regulatory elements (Rajagopal et al., 2014). Because genes and other regulatory regions are hotspots for multiple epigenetic marks with at least partial redundancy (Takeshima et al., 2009; Benveniste et al., 2014; Ahsendorf et al., 2017), targeting individual marks inevitably affects other marks and possibly the entire transcriptome.
The histone code hypothesis (Strahl and Allis, 2000) implies that the patterns of covalent histone modifications on a gene should reflect its dynamic regulatory state, likely contributing to widespread interest in epigenetics. Indeed, some histone modifications can be uncoupled from transcription activity. The conserved Polycomb Group (PcG) and Tritorax group of genes (trxG) complexes play crucial roles in development and introduce, respectively, repressive and activating histone modifications. The so-called bivalent genes that simultaneously harbor repressive and activating marks are poised for fate commitment in development and differentiation (Bernstein et al., 2006; Vastenhouw and Schier, 2012). The histone H3K36 modifications in the gene body regions may regulate alternative splicing (Kim et al., 2011). The histone H3K9 methylation, a repressive mark associated with heterochromatin, may be involved in arranging chromatin domains at the nuclear periphery (Towbin et al., 2012; Bian et al., 2020), protection against mechanical damage of the nucleus (Nava et al., 2020), remodeling of chromatin domains during differentiation (Wang et al., 2018; Burton et al., 2020) and maintenance of cell identity (Nicetto and Zaret, 2019). Many known histone modifications do not have clearly assigned functions yet, and will likely generate new findings (Tan et al., 2011).
When considering the dynamics of chromatin modifications, it is important to distinguish differences across loci within a cell type from differences at the same locus across cell types. Different genes in the same cell type clearly show distinct epigenomic patterns (Ernst and Kellis, 2010). However, across cell types, genome-wide patterns for histone modifications that we examined (ENCODE et al., 2020), at least in bulk experiments, appear to be similar for the same loci (Figure 1 and not shown). This is mirrored in Pol II distribution as well (Figure 1), consistent with an earlier study, for example, (Day et al., 2016). Even examining a study that highlighted differences between epigenomes, most of the epigenomic features on a given gene are quantitatively similar (Yen and Kellis, 2015). Differences in epigenome patters between cell types may therefore be in relatively subtle shifts in balance among different marks (Gopi and Kidder, 2021), for example, in the breadth of regions harboring the marks (Benayoun et al., 2014), possibly representing distinct overall states of the nucleus or its compartments (Zhao et al., 2007), or distinct signaling pathways (Yen and Kellis, 2015). Comparing epigenetic marks remains difficult at this level due to quantitative limitations of omics technologies. Future studies employing emerging technologies and better integration of datasets should reveal new insights into the tightly interconnected workings of epigenetic marks. For example, recent work using a mouse model identified epigenetic reprogramming as an essential step for the initiation of pancreatic cancer, wherein cells carrying certain oncogenic mutations require an environmental insult that causes epigenetic changes and triggers cancer cell fate (Alonso-Curbelo et al., 2021). To the best of our knowledge, this study is among the first to directly demonstrate a role for an epigenetic “hit” to trigger carcinogenesis. Finding the reasons behind why some cells are more sensitive to epigenetic reprogramming by drugs or environment, and identifying the weak points for their reprogramming, is an exciting direction to explore.
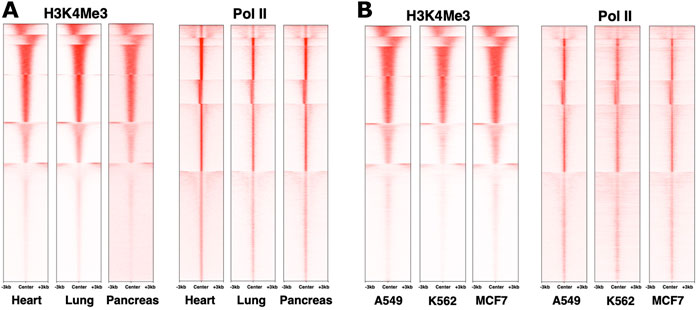
FIGURE 1. Similarity in genome-wide distributions of Pol II and histone marks across distinct systems. ChIP-sequencing datasets from the ENCODE database (Davis et al., 2018) for human heart, lung and pancreas normal tissues (A) and A549, MCF7 and K562 cancer cell lines (B) were used to plot density heatmaps around peak regions ± 3 kb from a peak center using computeMatrix reference-point (version 3.3.0) and plotHeatmap (version 3.3.0) with k-means clustering (k = 6). The numbers of peaks are 22,944 for Pol II and 28,287 for H3K4Me3. The datasets used were ENCSR901SIL, ENCSR701FGA, ENCSR876DCP, ENCSR336YRS, ENCSR033NHF, ENCSR610EFT, ENCSR000DMZ, ENCSR388QZF, ENCSR000DMT, ENCSR203XPU, ENCSR668LDD, ENCSR985MIB, respectively.
Multiple Sides of the Cellular Context
Cancers readily repurpose mechanisms that normally govern cell state transitions in differentiation, development, and responses to stimuli. Targeting of cancers borrows some of the overall concepts from the stem cells. Despite major advances in the stem cell field, reprogramming of stem cells rarely if ever approaches one hundred percent and can generate heterogeneous populations (Paik et al., 2018; Terryn et al., 2018). This heterogeneity, which is expected to be even higher in cancer cells, is likely to affect reprogramming and/or response to drugs. A recent study compared the effect of c-Myc on distinct fates of murine pre-B cells: transdifferentiating into macrophages and their reprogramming into iPSCs (Francesconi et al., 2019). Cells with high Myc activity reprogrammed to iPSCs more efficiently than transdifferentiated into macrophages, whereas cells with low Myc, in contrast, transdifferentiated readily, but failed to reprogram. That the levels of one factor can dramatically influence cell fate decisions highlights the importance and complexity of the cellular context.
Despite sharing certain features such as immortality and tendency for dedifferentiation, even recently transformed cells, without high if any mutation load, can readily deviate from normal cellular programs. A recent study showed that overexpression of the RAS oncogene in wild type mouse embryonic fibroblasts (MEFs) enhanced their dedifferentiation, but transformation of the same cells by deleting p53 or Arf tumor suppressors precluded it (Ferreirós et al., 2019). Similar interactions and context dependency were observed for histone marks as well (Nagaraja et al., 2019; Vidal et al., 2020). These data indicate that cancer cells diverge into distinct epigenetic programs, and likely change their cellular context, at the early stages of transformation. Thus, relatively small changes can lead to different responses to the same signal. These and other studies highlight the uncertainty that the cellular context can affect the apparent mechanism of transcription factor action as well as functional outcomes of its targeting.
A Common Framework of Transcriptome Organization
In this section we discuss a genome-wide view of transcription noting overall similarities in gene expression and highlighting potential mechanisms that might be pertinent for understanding transcriptome regulation.
The Transcriptome Rests on DNA
mRNA profiling has been widely used to identify genes with differential expression. Genome-wide gene expression patterns have been proposed as a basis to classify cancers (Perou et al., 2000). RNA-sequencing is arguably the easiest omics tool from the user’s point of view today, and some thousands of mRNA datasets are publicly available. Comparing RNA profiles of publicly available datasets of human tissues, a majority of transcripts are classified as present in all or in most tissues (Uhlén et al., 2016), indicating that, at least in bulk experiments, most genes show similar patterns of expression across distinct cell types. Despite substantial differences, cancer cells and primary tumors show unexpectedly high correlation of RNA signal even among distant cancer types (for example, (Yu et al., 2019)). Comparison of RNA profiles showed that the group of genes that are differentially expressed is rather similar between distant cell lines (Crow et al., 2019). Analysis of gene expression across several species showed that variation in expression among different genes exceeds that of the same gene in different conditions (Zrimec et al., 2020). Accordingly, genes that account for differential transcriptome profiles are relatively few and seem to fall into categories related to stress responses and immune function regardless of the cell type (Crow et al., 2019). A small number of genes may be sufficient to classify cancer subtypes as, for example, is done for breast cancers (Parker et al., 2009). RNA-sequencing in cancers is just as, if not more frequently used for the detection of exonic mutations (PCAWG et al., 2020). These observations point to overall similarity of gene expression states across the genome, that is, imply a fundamentally common transcriptome structure in human cells.
One way to breach the boundaries of transcription control is through changing gene copy numbers. Local and chromosome-wide changes of DNA copy number is a frequent occurrence and is one of the hallmarks of cancer (Hanahan and Weinberg, 2011). Changes in copy number enable cancer cells to alter expression of genes without any other regulatory inputs (Shao et al., 2019). This may reflect a fundamental property of mammalian transcriptomes wherein changes in gene copy numbers are not by default compensated (Disteche, 2016) and can alter the entire transcriptome. Indeed, genetic haploinsufficiency is associated with many diseases (Han et al., 2018). Dosage compensation is best known in X-chromosome inactivation (Brockdorff and Turner, 2015). Forced reactivation of the inactive X-chromosome copy by knocking down Xist levels in mice results in an increase of total X-linked gene expression (Yang et al., 2016) that can lead to cancer (Yildirim et al., 2013). Interestingly, autosomal polysomy in human cells can be compensated at the level of protein, but not at the level of RNA (Stingele et al., 2012). Sensitivity to DNA copy number raises an intriguing possibility that the transcriptome may be fundamentally structured by the process of transcription rather than its products.
Pol II Pausing – A Common Step in Complex Organisms
Widespread accumulation of Pol II signal at promoter regions of genes has been well documented (Kim et al., 2005). Rather than preinitiation complexes, this signal comes largely from elongating Pol II that began RNA synthesis, but paused within the first ∼50 nucleotides (Pugh and Venters, 2016) (Figure 2). Because of its proximity to gene transcription start sites, the prevalence of Pol II pausing across the genome became clear only as technologies attained sufficient resolution (Muse et al., 2007; Zeitlinger et al., 2007; Nechaev et al., 2010). Perhaps because Pol II pausing was originally described on inducible genes such as MYC and heat shock HSP70 (O'Brien and Lis, 1991; Spencer and Groudine, 1990; Krumm et al., 1995), it was long believed to be a specialized mechanism that prepares highly inducible genes for activation. However, based on analyses of short RNA transcripts, Pol II pausing signatures are not confined to inducible genes, but are present on all genes, and likely accompany all transcription, whether initiating at or outside of promoters, including divergent transcription, intergenic transcription, enhancers, etc (Scheidegger et al., 2019). This makes pausing unlikely to be a mechanism that universally prepares genes for activation, although it likely contributes to it (see below) (Muse et al., 2007). Many details of Pol II pausing such as its relationship with the burst mode of transcription (Corrigan and Chubb, 2014; Fukaya et al., 2016; Tunnacliffe and Chubb, 2020) and its dynamics remain to be established. Whether Pol II pausing can be bypassed for individual transcription events or not, at least a certain percentage of Pol II complexes appear to undergo pausing at every start site of transcription (Scheidegger et al., 2019). The so-called “nonpaused” genes still show the same small RNA pausing signatures in terms of their size distributions, but are either less active or have a lower pausing index (Muse et al., 2007; Zeitlinger et al., 2007) (or higher traveling ratio (Rahl et al., 2010)) compared to other genes (Nechaev et al., 2010; Scheidegger et al., 2019). Examination of genome-wide datasets shows that different genes within a dataset show higher or lower pausing index, but these signatures, just like some histone marks, are overall stable across cell lines (Figure 1). These observations reinforce a notion on fundamental conservation of the human transcriptome and also raise a question about roles of pausing in regulation.
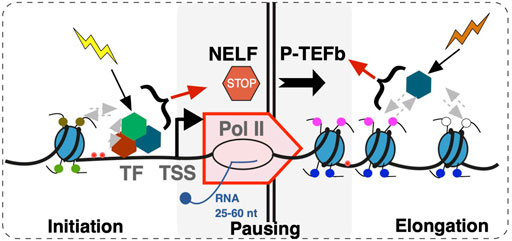
FIGURE 2. A scheme of events during early transcription elongation. Early transcription elongation within 25–60 nucleotides (nt) from the Transcription Start Site (TSS) is a step integrating upstream and downstream regulatory inputs. Upstream inputs from multiple factors including transcription factors (TF) and epigenetic marks result in transcription initiation at the promoter and culminate in NELF-dependent pausing. DSIF complex is not included in the model. Pause release into transcription elongation is dependent on P-TEFb. Uncoupling of transcription initiation and elongation at the site of pausing through mechanisms such as promoter proximal termination contributes to transcriptome organization.
In considering potential biological roles of promoter-proximal Pol II pausing, it might be of significance that Pol II activity at this site is controlled by at least two distinct groups of factors: those that establish pausing and, on the other hand, those that release the Pol II from the paused state (Figure 2). The Negative Elongation Factor (NELF) is a five-subunit complex (Yamaguchi et al., 1999) that in conjunction with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF) (Yamaguchi et al., 2001) is sufficient to cause Pol II pausing in vitro (Renner et al., 2001; Wu et al., 2003). NELF is absent from yeast and C. elegans, indicating that NELF-dependent pausing is a function of higher organisms. Budding yeast S. cerevisiae do show Pol II accumulation at promoter regions at least at some genes (Radonjic et al., 2005). High-resolution nascent RNA analysis shows major differences between budding and fission (S. pombe) yeast, with the latter showing pausing signatures at a significant proportion of genes (Booth et al., 2016). However, fission yeast show clear differences from Drosophila and mammals in terms of the +1 nucleosome positioning presumably due to absence of NELF. These observations indicate that Pol II pausing is more prevalent and may be more tightly regulated in higher organisms.
Targeting of individual NELF subunit alters the levels of its other subunits (Sun et al., 2008), indicating that NELF components function as a complex. NELF is downregulated in breast cancers (Sun et al., 2008), but is potentially oncogenic in other cancers including prostate (Yun et al., 2018), liver (Dang et al., 2017; El Zeneini et al., 2017) and pancreas (Han et al., 2019). These observations indicate that the functional outcome of NELF perturbation is defined by the cellular context. The Positive Transcription Elongation factor B (P-TEFb) (Marshall and Price, 1995) is a two-component complex consisting of a cyclin T1, which can be substituted with cyclin T2 or possibly cyclin K, and a cyclin-dependent kinase 9 (CDK9) (Peng et al., 1998; Peterlin and Price, 2006; Kohoutek, 2009). P-TEFb releases the paused complex into productive elongation by phosphorylation of several proteins including DSIF subunit Spt5 at several sites (Wada et al., 1998; Kim and Sharp, 2001; Yamada et al., 2006; Parua et al., 2020), NELF (Fujinaga et al., 2004; Lu et al., 2016) and Pol II C-terminal domain at Ser-2 residues (Marshall et al., 1996). P-TEFb appears to be more conserved than NELF. Yeast may have more than one kinase (Hsin and Manley, 2012), although mammalian homologs of these kinases such as CDK12 appear to phosphorylate Pol II Ser-2 during elongation downstream of pause release by P-TEFb (Bartkowiak et al., 2010; Tellier et al., 2020). Neither NELF nor P-TEFb are essential for Pol II enzymatic function in vitro, but do appear to be essential for proper transcription in vivo (Sun et al., 2011; Aoi et al., 2020; Fujinaga, 2020). P-TEFb is a hub for regulatory inputs for multiple factors including c-MYC and NF-kB, and possibly many others (Zhou et al., 2012; Mahat et al., 2016; Aoi et al., 2020). The requirement for distinct essential factors individually controlling the on and off rates of a Pol II complex that is already committed to elongation is notable.
Regulation by Globally Acting Factors
Here we discuss how transcription can be regulated by essential factors at the site of promoter-proximal Pol II pausing.
Encoding the Change in the Static Genome
One genome must specify the entire structure and dynamics of the transcriptomes for in every cell, but the rules of how it does so remain obscure. It is evident, however, that promoters, including core promoter basal sequence elements such as the TATA box, initiator motif, Downstream Promoter Element, etc, do not merely serve as a passive platform for the binding of regulatory factors, but actively shape the outcomes of regulatory inputs (Butler and Kadonaga, 2002). Flexibility of promoters is fundamental and starts with bacteria: in E. coli, no single promoter, including for the highest expressed rRNA genes, contains a full consensus sequence of basal elements (Bervoets and Charlier, 2019). Instead, promoters are pre-wired to require additional inputs for highest-level transcription and thus to be inherently controllable. In eukaryotes, properties of promoters depend on the presence of distinct basal sequence elements (Butler and Kadonaga, 2002). Mechanisms for such selectivity may include preference for distinct cohorts of general transcription factors, localization within the nucleus, or interaction with enhancers (Juven-Gershon and Kadonaga, 2010; Zabidi et al., 2015; Russo et al., 2018). Differences in core promoter properties should lead to differences in their responses to the same signal, thereby creating patterns that can be complex especially with multiple signals. In this regard, about ∼350 human genes were recently noted to contain 5′-untranslated regions (UTRs) that show conservation across vertebrates, especially among specific categories of homeobox genes, kinases and genes involved in neurogenesis (Zuccotti et al., 2020).
Apart from differences between promoters, another question is whether and how the genome enables the same promoters to assume distinct regulatory states. In Drosophila, transcription start sites of regulatory genes contain sequences that favor both Pol II pausing and nucleosome binding at their transcription start sites (Gilchrist et al., 2010), which result in, respectively, active and repressed gene states. Housekeeping gene promoters, in contrast, do not contain these marks. By favoring mutually exclusive marks leading to distinct regulatory states, some promoters are intrinsically primed to be regulated. How these dynamic states are encoded in human promoters, which are highly enriched in CpG sequences, remains to be determined.
Transcriptional Responses Expose the Transcriptome
Global requirement for NELF- and P-TEFb raises a question about their functional relationship at the site of Pol II pausing. It has been proposed that activation of genes should proceed through pause “release,” that is, relatively increased P-TEFb activity (Boehm et al., 2003; Hah et al., 2011; Liu et al., 2015; Mahat et al., 2016). Sustained transcription activation must also recruit additional Pol II, leaving a question as to how Pol II recruitment and pause release are related. A consistently rigid connection between pausing establishment and release would not be conducive for regulation. Evidence suggests that promoter Pol II recruitment and pause release can indeed be uncoupled. Chemical inhibition of P-TEFb - dependent Pol II pause release by flavopiridol causes accumulation of Pol II at promoters of active genes (Rahl et al., 2010; Henriques et al., 2013; Jonkers et al., 2014). Work by the Lis lab on heat shock response demonstrated that genes repressed by heat shock can accumulate Pol II signal at promoters (Mahat et al., 2016). Global Pol II accumulation at promoters has also been shown during acute oxidative stress response (Nilson et al., 2017). By demonstrating that Pol II pausing takes place even when P-TEFb activity is perturbed, these studies show that pausing establishment and release can be functionally uncoupled, and that this connection can be regulatory. Conversely, changes in Pol II recruitment without affecting pause release are possible as well. First, gene activation can take place through increased Pol II recruitment to promoters without changes in pause release (Samarakkody et al., 2015). Second, the Shilatifard lab showed that pause release can take place without additional Pol II recruitment when Pol II is being cleared from promoters prior to mitosis (Liang et al., 2015). Overall, these and other observations suggest that processes upstream of pausing, such as Pol II recruitment to promoters and formation of preinitiation complexes, can be functionally decoupled from pause release. Furthermore, the function of Pol II pausing as a limiting step for transcription may be more significant not at the steady state, but during rapid responses to stimuli (Gressel et al., 2019). Observing cells during rapid responses to stimuli outside of the steady state may help reveal their regulatory architecture (Danko et al., 2013; Vihervaara et al., 2017).
A study from the Young lab showed that overexpression of c-Myc causes uniform amplification of all genes (Lin et al., 2012; Nie et al., 2012). Since c-Myc was shown to function through recruitment of P-TEFb to promoters (Rahl et al., 2010), this amplification is likely due to uniformly increased genome-wide pause release. These findings are important because they imply that the transcriptome is fundamentally modular. Throttling up of pause release siphons Pol II traffic through all available transcription start sites without altering the upstream steps such as promoter architecture. These observations reveal pausing as a central step that can globally separate regulatory inputs at distinct points of the transcription cycle. In this view, rather than integrating regulatory signals, Pol II pausing separates them.
Regulation of the Transcriptome at the Level of Pol II Pausing
Either NELF or P-TEFb activities may become limiting at certain circumstances such as during rapid responses to stimuli. This raises a question of how these factors are redistributed across the genome when limited amounts or activity are available. This question is broadly related to an earlier concept of enhancer insufficiency (Barolo and Posakony, 2002) wherein restriction of the activator availability limits transcription from certain genes such as those with low activity promoters, thereby reducing noise. Simple simulation of a closed cell containing promoters with randomly distributed strengths shows that restricting a factor that is essential for transcription at a post-recruitment step, such as NELF, is sufficient to generate pools of genes that appear as activated and repressed (Figure 3). Addition of a second post-recruitment step such as that controlled by P-TEFb should likewise prioritize pause release among Pol II complexes that had reached the previous step. Prioritization of essential factors at multiple steps of the transcription cycle may lead to nonlinear effects including cooperativity and stabilization of transcription output. Prioritization of essential factors, we suggest, is a key principle organizing the transcriptome.
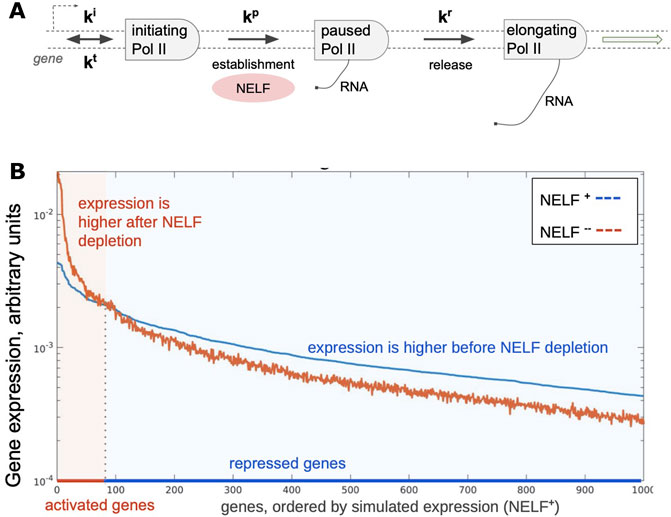
FIGURE 3. Simulated changes in gene expression upon depletion of a pausing factor. (A) A scheme of steps in Pol II pausing. Initiation (ki), pausing (kp), and release (kr) constants were randomly distributed for 1000 simulated genes (with cauchy and two gaussian probability distribution functions, respectively); termination constant (kt) was the same for all simulated genes. (B) Results of simulation showing steady-state distribution of gene expression (light colored arrow) levels with two different amounts of total available NELF in the system, which results in activation and repression of genes.
Step-wise prioritization of essential factors retrospectively accounts for known features of mammalian transcriptomes. Some transcription factors such as HSF1 and NF-kB have been shown to act on pause release by directly or indirectly recruiting P-TEFb (Zhou et al., 2012; Mahat et al., 2016; Aoi et al., 2020). P-TEFb function can be either global across the genome or involve specific groups of genes (Luo et al., 2012). Secondly, the model is agnostic to the exact promoter activity patterns and can work in all cells wherein promoter activity defines the network structure while pausing factors stabilize it. Notably, this regulation should be highly sensitive to gene copy numbers but does not require lateral interactions between gene products. This model is in principle similar to the concept of phase separation (Hnisz et al., 2017), except that the phases here are defined not spatially, but through availability of a factor that may be governed by diffusion or over-representation in different nuclear compartments. In this view, transcription of every gene must involve transitions between phases. Multiple inputs upstream and downstream of pausing including epigenetic and other broad-stroke mechanisms may form additional layers of regulation or converge on the two major limiting steps at the site of pausing. A caveat to our view is that a complex makeup of the cell is essentially reduced to a binary readout, which is certainly an oversimplification. However, the concept of binary readouts distinguishing complex systems such as cancers is gaining experimental ground (Pearson et al., 2021) and may form a paradigm for understanding and targeting these complex diseases. Transcriptional superenhancers (Hnisz et al., 2013; Hnisz et al., 2015) as well as the recently reported partitioning of small molecule drugs at distinct nuclear loci (Klein et al., 2020) must affect the specificity of globally acting factors and outcomes of their targeting through principles that remain to be explored. We note that our model is readily compatible with transcriptional bursting (Rodriguez and Larson, 2020) because bursting creates local and transient demand for transcriptional machinery components. Lastly, despite the prevalence of pause release during responses to stimuli, Pol II recruitment to promoters may still be the step limiting for overall transcription.
Discussion
Substantial advances are made in targeting cancers based on transcription and epigenetic signatures (Malone et al., 2020). However, progress remains complicated by the inherent uncertainty in the roles of transcription machinery components and critical yet poorly understood contribution of the cellular context. Pausing regulators including NELF and P-TEFb have been proposed (Yun et al., 2018) or used as therapeutic targets including P-TEFb (CDK9) small molecule inhibitors in clinical trials (Morales and Giordano, 2016; Cassandri et al., 2020). However, the main value of pausing factors may be not in serving as therapy targets, but in providing a conceptual platform to better understand targeting of other components. Dividing the myriad factors directly or indirectly affecting transcription into those that act upstream or downstream of Pol II pausing (Figure 2) highlights the balance between these steps as an important readout of cancers that can help expose their vulnerabilities.
The process of Pol II pausing is much more granular at the molecular level than described above (Elrod et al., 2019) and includes multiple additional steps such as premature transcription termination likely involving multiple mechanisms (London et al., 1991; Brannan et al., 2012; Huang et al., 2020; Rimel et al., 2020). These and other steps should affect the balance between Pol II functions upstream and downstream of pausing. The model of transcriptome regulation by stepwise restriction of globally acting factors proposed here is agnostic to the actual nature of its components and does not need to be limited to Pol II pausing. Additional transcription elongation checkpoints such as those controlled by CDK12 (Tellier et al., 2020) may define new steps where a balance would need to be considered. Despite the ongoing quest for therapeutics targeting specific factors, cancer treatments largely rely on broad-stroke interventions and are expected to do so for the foreseeable future. Studies that integrate experiments with refined models should reveal new insights into cancer transcriptomes to improve the treatments targeting the cellular machinery via globally acting or general factors.
Author Contributions
DP - analyzed data, prepared figures and wrote the manuscript; MV - analyzed data and prepared figures; LK - analyzed data; SN, conceived the manuscript and wrote the manuscript.
Funding
Funding for this work was provided by the National Science Foundation CAREER award 1750379 to SN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Motoki Takaku and Archana Dhasarathy for critical reading of the manuscript as well as Eda Yildirim and Al Courey for helpful suggestions.
References
Adelman, K., Kennedy, M. A., Nechaev, S., Gilchrist, D. A., Muse, G. W., Chinenov, Y., et al. (2009). Immediate Mediators of the Inflammatory Response Are Poised for Gene Activation Through RNA Polymerase II Stalling. Proc. Natl. Acad. Sci. U S A. 106 (43), 18207–18212. doi:10.1073/pnas.0910177106
Ahsendorf, T., Müller, F. J., Topkar, V., Gunawardena, J., and Eils, R. (2017). Transcription Factors, Coregulators, and Epigenetic Marks Are Linearly Correlated and Highly Redundant. PLoS One 12, e0186324. doi:10.1371/journal.pone.0186324
Alberga, A., Boulay, J. L., Kempe, E., Dennefeld, C., and Haenlin, M. (1991). The Snail Gene Required for Mesoderm Formation in Drosophila Is Expressed Dynamically in Derivatives of All Three Germ Layers. Development 111 (4), 983–992. doi:10.1242/dev.111.4.983
Allen, M. A., Andrysik, Z., Dengler, V. L., Mellert, H. S., Guarnieri, A., Freeman, J. A., et al. (2014). Global Analysis of P53-Regulated Transcription Identifies its Direct Targets and Unexpected Regulatory Mechanisms. Elife 3, e02200. doi:10.7554/eLife.02200
Alonso-Curbelo, D., Ho, Y. J., Burdziak, C., Maag, J. L. V., Morris, J. P., Chandwani, R., et al. (2021). A Gene-Environment-Induced Epigenetic Program Initiates Tumorigenesis. Nature 590 (7847), 642–648. doi:10.1038/s41586-020-03147-x
Aoi, Y., Smith, E. R., Shah, A. P., Rendleman, E. J., Marshall, S. A., Woodfin, A. R., et al. (2020). NELF Regulates A Promoter-Proximal Step Distinct from RNA Pol II Pause-Release. Mol. Cel. 78 (2), 261–274.e5. doi:10.1016/j.molcel.2020.02.014
Aranda, S., Mas, G., and Di Croce, L. (2015). Regulation of Gene Transcription by Polycomb Proteins. Sci. Adv. 1 (11), e1500737. doi:10.1126/sciadv.1500737
Bae, S., and Lesch, B. J. (2020). H3K4me1 Distribution Predicts Transcription State and Poising at Promoters. Front Cel. Dev. Biol. 8, 289. doi:10.3389/fcell.2020.00289
Banerjee, T., Nath, S., and Roychoudhury, S. (2009). DNA Damage Induced P53 Downregulates Cdc20 by Direct Binding to its Promoter Causing Chromatin Remodeling. Nucleic Acids Res. 37 (8), 2688–2698. doi:10.1093/nar/gkp110
Barolo, S., and Posakony, J. W. (2002). Three Habits of Highly Effective Signaling Pathways: Principles of Transcriptional Control by Developmental Cell Signaling. Genes Dev. 16 (10), 1167–1181. doi:10.1101/gad.976502
Bartkowiak, B., Liu, P., Phatnani, H. P., Fuda, N. J., Cooper, J. J., Price, D. H., et al. (2010). CDK12 Is a Transcription Elongation-Associated CTD Kinase, the Metazoan Ortholog of Yeast Ctk1. Genes Dev. 24 (20), 2303–2316. doi:10.1101/gad.1968210
Bauer, D. C., Buske, F. A., and Bailey, T. L. (2010). Dual-functioning Transcription Factors in the Developmental Gene Network of Drosophila melanogaster. BMC Bioinformatics 11, 366. doi:10.1186/1471-2105-11-366
Baylin, S. B., and Jones, P. A. (2011). A Decade of Exploring the Cancer Epigenome - Biological and Translational Implications. Nat. Rev. Cancer 11 (10), 726–734. doi:10.1038/nrc3130
Belluti, S., Rigillo, G., and Imbriano, C. (2020). Transcription Factors in Cancer: When Alternative Splicing Determines Opposite Cell Fates. Cells 9 (3), E760. doi:10.3390/cells9030760
Benayoun, B. A., Pollina, E. A., Ucar, D., Mahmoudi, S., Karra, K., Wong, E. D., et al. (2014). H3K4me3 Breadth Is Linked to Cell Identity and Transcriptional Consistency. Cell 158 (3), 673–688. doi:10.1016/j.cell.2014.06.027
Bennett, R. L., and Licht, J. D. (2018). Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol. 58, 187–207. doi:10.1146/annurev-pharmtox-010716-105106
Benveniste, D., Sonntag, H. J., Sanguinetti, G., and Sproul, D. (2014). Transcription Factor Binding Predicts Histone Modifications in Human Cell Lines. Proc. Natl. Acad. Sci. U S A. 111 (37), 13367–13372. doi:10.1073/pnas.1412081111
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 125 (2), 315–326. doi:10.1016/j.cell.2006.02.041
Bervoets, I., and Charlier, D. (2019). Diversity, Versatility and Complexity of Bacterial Gene Regulation Mechanisms: Opportunities and Drawbacks for Applications in Synthetic Biology. FEMS Microbiol. Rev. 43 (3), 304–339. doi:10.1093/femsre/fuz001
Bian, Q., Anderson, E. C., Yang, Q., and Meyer, B. J. (2020). Histone H3K9 Methylation Promotes Formation of Genome Compartments in Caenorhabditis elegans via Chromosome Compaction and Perinuclear Anchoring. Proc. Natl. Acad. Sci. U S A. 117 (21), 11459–11470. doi:10.1073/pnas.2002068117
Boehm, A. K., Saunders, A., Werner, J., and Lis, J. T. (2003). Transcription Factor and Polymerase Recruitment, Modification, and Movement on Dhsp70 In Vivo in the Minutes Following Heat Shock. Mol. Cel Biol. 23 (21), 7628–7637. doi:10.1128/mcb.23.21.7628-7637.2003
Booth, G. T., Wang, I. X., Cheung, V. G., and Lis, J. T. (2016). Divergence of a Conserved Elongation Factor and Transcription Regulation in Budding and Fission Yeast. Genome Res. 26 (6), 799–811. doi:10.1101/gr.204578.116
Bradner, J. E., Hnisz, D., and Young, R. A. (2017). Transcriptional Addiction in Cancer. Cell 168 (4), 629–643. doi:10.1016/j.cell.2016.12.013
Brannan, K., Kim, H., Erickson, B., Glover-Cutter, K., Kim, S., Fong, N., et al. (2012). mRNA Decapping Factors and the Exonuclease Xrn2 Function in Widespread Premature Termination of RNA Polymerase II Transcription. Mol. Cel 46 (3), 311–324. doi:10.1016/j.molcel.2012.03.006
Bregman, D. B., Du, L., van der Zee, S., and Warren, S. L. (1995). Transcription-dependent Redistribution of the Large Subunit of RNA Polymerase II to Discrete Nuclear Domains. J. Cel Biol. 129 (2), 287–298. doi:10.1083/jcb.129.2.287
Brien, G. L., Stegmaier, K., and Armstrong, S. A. (2019). Targeting Chromatin Complexes in Fusion Protein-Driven Malignancies. Nat. Rev. Cancer 19 (5), 255–269. doi:10.1038/s41568-019-0132-x
Brockdorff, N., and Turner, B. M. (2015). Dosage Compensation in Mammals. Cold Spring Harb Perspect. Biol. 7 (3), a019406. doi:10.1101/cshperspect.a019406
Burton, A., Brochard, V., Galan, C., Ruiz-Morales, E. R., Rovira, Q., Rodriguez-Terrones, D., et al. (2020). Heterochromatin Establishment During Early Mammalian Development Is Regulated by Pericentromeric RNA and Characterized by Non-repressive H3K9me3. Nat. Cel Biol. 22 (7), 767–778. doi:10.1038/s41556-020-0536-6
Bushweller, J. H. (2019). Targeting Transcription Factors in Cancer - From Undruggable to Reality. Nat. Rev. Cancer 19 (11), 611–624. doi:10.1038/s41568-019-0196-7
Butler, J. E., and Kadonaga, J. T. (2002). The RNA Polymerase II Core Promoter: A Key Component in the Regulation of Gene Expression. Genes Dev. 16 (20), 2583–2592. doi:10.1101/gad.1026202
Bywater, M. J., Pearson, R. B., McArthur, G. A., and Hannan, R. D. (2013). Dysregulation of the Basal RNA Polymerase Transcription Apparatus in Cancer. Nat. Rev. Cancer 13 (5), 299–314. doi:10.1038/nrc3496
Carver, E. A., Jiang, R., Lan, Y., Oram, K. F., and Gridley, T. (2001). The Mouse Snail Gene Encodes a Key Regulator of the Epithelial-Mesenchymal Transition. Mol. Cel Biol. 21 (23), 8184–8188. doi:10.1128/mcb.21.23.8184-8188.2001
Cassandri, M., Fioravanti, R., Pomella, S., Valente, S., Rotili, D., Del Baldo, G., et al. (2020). CDK9 as a Valuable Target in Cancer: From Natural Compounds Inhibitors to Current Treatment in Pediatric Soft Tissue Sarcomas. Front. Pharmacol. 11, 1230. doi:10.3389/fphar.2020.01230
Corrigan, A. M., and Chubb, J. R. (2014). Regulation of Transcriptional Bursting by a Naturally Oscillating Signal. Curr. Biol. 24 (2), 205–211. doi:10.1016/j.cub.2013.12.011
Creyghton, M. P., Cheng, A. W., Welstead, G. G., Kooistra, T., Carey, B. W., Steine, E. J., et al. (2010). Histone H3K27ac Separates Active from Poised Enhancers and Predicts Developmental State. Proc. Natl. Acad. Sci. U S A. 107 (50), 21931–21936. doi:10.1073/pnas.1016071107
Crow, M., Lim, N., Ballouz, S., Pavlidis, P., and Gillis, J. (2019). Predictability of Human Differential Gene Expression. Proc. Natl. Acad. Sci. U S A. 116 (13), 6491–6500. doi:10.1073/pnas.1802973116
Dang, H., Takai, A., Forgues, M., Pomyen, Y., Mou, H., Xue, W., et al. (2017). Oncogenic Activation of the RNA Binding Protein NELFE and MYC Signaling in Hepatocellular Carcinoma. Cancer Cell 32 (1), 101–114.e8. doi:10.1016/j.ccell.2017.06.002
Danko, C. G., Hah, N., Luo, X., Martins, A. L., Core, L., Lis, J. T., et al. (2013). Signaling Pathways Differentially Affect RNA Polymerase II Initiation, Pausing, and Elongation Rate in Cells. Mol. Cel. 50 (2), 212–222. doi:10.1016/j.molcel.2013.02.015
Datta, N., Chakraborty, S., Basu, M., and Ghosh, M. K. (2020). Tumor Suppressors Having Oncogenic Functions: The Double Agents. Cells 10 (1), E46. doi:10.3390/cells10010046
Davis, C. A., Hitz, B. C., Sloan, C. A., Chan, E. T., Davidson, J. M., Gabdank, I., et al. (2018). The Encyclopedia of DNA Elements (ENCODE): Data portal Update. Nucleic Acids Res. 46 (D1), D794–D801. doi:10.1093/nar/gkx1081
Dawson, M. A., Prinjha, R. K., Dittmann, A., Giotopoulos, G., Bantscheff, M., Chan, W. I., et al. (2011). Inhibition of BET Recruitment to Chromatin as an Effective Treatment for MLL-Fusion Leukaemia. Nature 478 (7370), 529–533. doi:10.1038/nature10509
Day, D. S., Zhang, B., Stevens, S. M., Ferrari, F., Larschan, E. N., Park, P. J., et al. (2016). Comprehensive Analysis of Promoter-Proximal RNA Polymerase II Pausing Across Mammalian Cell Types. Genome Biol. 17 (1), 120. doi:10.1186/s13059-016-0984-2
Disteche, C. M. (2016). Dosage Compensation of the Sex Chromosomes and Autosomes. Semin. Cel Dev Biol. 56, 9–18. doi:10.1016/j.semcdb.2016.04.013
Dubnicoff, T., Valentine, S. A., Chen, G., Shi, T., Lengyel, J. A., Paroush, Z., et al. (1997). Conversion of Dorsal from an Activator to a Repressor by the Global Corepressor Groucho. Genes Dev. 11 (22), 2952–2957. doi:10.1101/gad.11.22.2952
Duffy, M. J., and Crown, J. (2021). Drugging “Undruggable” Genes for Cancer Treatment: Are We Making Progress. Int. J. Cancer 148 (1), 8–17. doi:10.1002/ijc.33197
El Zeneini, E., Kamel, S., El-Meteini, M., and Amleh, A. (2017). Knockdown of COBRA1 Decreases the Proliferation and Migration of Hepatocellular Carcinoma Cells. Oncol. Rep. 37 (3), 1896–1906. doi:10.3892/or.2017.5390
Elrod, N. D., Henriques, T., Huang, K. L., Tatomer, D. C., Wilusz, J. E., Wagner, E. J., et al. (2019). The Integrator Complex Attenuates Promoter-Proximal Transcription at Protein-Coding Genes. Mol. Cel. 76 (5), 738–752.e7. doi:10.1016/j.molcel.2019.10.034
Encode, P. C., Moore, J. E., Purcaro, M. J., Pratt, H. E., Epstein, C. B., Shoresh, N., et al. (2020). Expanded Encyclopaedias of DNA Elements in the Human and Mouse Genomes. Nature 583 (7818), 699–710. doi:10.1038/s41586-020-2493-4
Ernst, J., and Kellis, M. (2010). Discovery and Characterization of Chromatin States for Systematic Annotation of the Human Genome. Nat. Biotechnol. 28 (8), 817–825. doi:10.1038/nbt.1662
Ferreirós, A., Pedrosa, P., Da Silva-Álvarez, S., Triana-Martínez, F., Vilas, J. M., Picallos-Rabina, P., et al. (2019). Context-Dependent Impact of RAS Oncogene Expression on Cellular Reprogramming to Pluripotency. Stem Cel Rep. 12 (5), 1099–1112. doi:10.1016/j.stemcr.2019.04.006
Fischer, M., Steiner, L., and Engeland, K. (2014). The Transcription Factor P53: Not a Repressor, Solely an Activator. Cell Cycle 13 (19), 3037–3058. doi:10.4161/15384101.2014.949083
Francesconi, M., Di Stefano, B., Berenguer, C., de Andrés-Aguayo, L., Plana-Carmona, M., Mendez-Lago, M., et al. (2019). Single Cell RNA-Seq Identifies the Origins of Heterogeneity in Efficient Cell Transdifferentiation and Reprogramming. Elife 8, e41627. doi:10.7554/eLife.41627
Fujinaga, K. (2020). P-TEFb as A Promising Therapeutic Target. Molecules 25 (4), E838. doi:10.3390/molecules25040838
Fujinaga, K., Irwin, D., Huang, Y., Taube, R., Kurosu, T., and Peterlin, B. M. (2004). Dynamics of Human Immunodeficiency Virus Transcription: P-TEFb Phosphorylates RD and Dissociates Negative Effectors from the Transactivation Response Element. Mol. Cel Biol. 24 (2), 787–795. doi:10.1128/mcb.24.2.787-795.2004
Fukaya, T., Lim, B., and Levine, M. (2016). Enhancer Control of Transcriptional Bursting. Cell 166 (2), 358–368. doi:10.1016/j.cell.2016.05.025
Fung, J. J., Kosaka, A., Shan, X., Danet-Desnoyers, G., Gormally, M., Owen, K., et al. (2015). Registered Report: Inhibition of BET Recruitment to Chromatin as an Effective Treatment for MLL-Fusion Leukemia. Elife 4, e08997. doi:10.7554/eLife.08997
Gilchrist, D. A., Dos Santos, G., Fargo, D. C., Xie, B., Gao, Y., Li, L., et al. (2010). Pausing of RNA Polymerase II Disrupts DNA-Specified Nucleosome Organization to Enable Precise Gene Regulation. Cell 143 (4), 540–551. doi:10.1016/j.cell.2010.10.004
Gong, L., Yan, Q., Zhang, Y., Fang, X., Liu, B., and Guan, X. (2019). Cancer Cell Reprogramming: A Promising Therapy Converting Malignancy to Benignity. Cancer Commun. (Lond) 39 (1), 48. doi:10.1186/s40880-019-0393-5
Gopi, L. K., and Kidder, B. L. (2021). Integrative Pan Cancer Analysis Reveals Epigenomic Variation in Cancer Type and Cell Specific Chromatin Domains. Nat. Commun. 12 (1), 1419. doi:10.1038/s41467-021-21707-1
Gressel, S., Schwalb, B., and Cramer, P. (2019). The Pause-Initiation Limit Restricts Transcription Activation in Human Cells. Nat. Commun. 10 (1), 3603. doi:10.1038/s41467-019-11536-8
Hah, N., Danko, C. G., Core, L., Waterfall, J. J., Siepel, A., Lis, J. T., et al. (2011). A Rapid, Extensive, and Transient Transcriptional Response to Estrogen Signaling in Breast Cancer Cells. Cell 145 (4), 622–634. doi:10.1016/j.cell.2011.03.042
Hallin, J., Engstrom, L. D., Hargis, L., Calinisan, A., Aranda, R., Briere, D. M., et al. (2020). The KRASG12C Inhibitor MRTX849 Provides Insight Toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 10 (1), 54–71. doi:10.1158/2159-8290.CD-19-1167
Hampsey, M. (1998). Molecular Genetics of the RNA Polymerase II General Transcriptional Machinery. Microbiol. Mol. Biol. Rev. 62 (2), 465–503. doi:10.1128/MMBR.62.2.465-503.1998
Han, L., Zan, Y., Huang, C., and Zhang, S. (2019). NELFE Promoted Pancreatic Cancer Metastasis and the Epithelial-To-Mesenchymal Transition by Decreasing the Stabilization of NDRG2 mRNA. Int. J. Oncol. 55 (6), 1313–1323. doi:10.3892/ijo.2019.4890
Han, X., Chen, S., Flynn, E., Wu, S., Wintner, D., and Shen, Y. (2018). Distinct Epigenomic Patterns Are Associated with Haploinsufficiency and Predict Risk Genes of Developmental Disorders. Nat. Commun. 9 (1), 2138. doi:10.1038/s41467-018-04552-7
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of Cancer: The Next Generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Healy, E., Mucha, M., Glancy, E., Fitzpatrick, D. J., Conway, E., Neikes, H. K., et al. (2019). PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol. Cel 76 (3), 437–452.e6. doi:10.1016/j.molcel.2019.08.012
Heerboth, S., Lapinska, K., Snyder, N., Leary, M., Rollinson, S., and Sarkar, S. (2014). Use of Epigenetic Drugs in Disease: An Overview. Genet. Epigenet 6, 9–19. doi:10.4137/GEG.S12270
Heintzman, N. D., Stuart, R. K., Hon, G., Fu, Y., Ching, C. W., Hawkins, R. D., et al. (2007). Distinct and Predictive Chromatin Signatures of Transcriptional Promoters and Enhancers in the Human Genome. Nat. Genet. 39 (3), 311–318. doi:10.1038/ng1966
Henikoff, S., and Shilatifard, A. (2011). Histone Modification: Cause or Cog. Trends Genet. 27 (10), 389–396. doi:10.1016/j.tig.2011.06.006
Henriques, T., Gilchrist, D. A., Nechaev, S., Bern, M., Muse, G. W., Burkholder, A., et al. (2013). Stable Pausing by RNA Polymerase II Provides an Opportunity to Target and Integrate Regulatory Signals. Mol. Cel 52 (4), 517–528. doi:10.1016/j.molcel.2013.10.001
Hnisz, D., Abraham, B. J., Lee, T. I., Lau, A., Saint-André, V., Sigova, A. A., et al. (2013). Super-enhancers in the Control of Cell Identity and Disease. Cell 155 (4), 934–947. doi:10.1016/j.cell.2013.09.053
Hnisz, D., Schuijers, J., Lin, C. Y., Weintraub, A. S., Abraham, B. J., Lee, T. I., et al. (2015). Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-enhancers. Mol. Cel 58 (2), 362–370. doi:10.1016/j.molcel.2015.02.014
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K., and Sharp, P. A. (2017). A Phase Separation Model for Transcriptional Control. Cell 169 (1), 13–23. doi:10.1016/j.cell.2017.02.007
Hsin, J. P., and Manley, J. L. (2012). The RNA Polymerase II CTD Coordinates Transcription and RNA Processing. Genes Dev. 26 (19), 2119–2137. doi:10.1101/gad.200303.112
Hsu, D. S., Wang, H. J., Tai, S. K., Chou, C. H., Hsieh, C. H., Chiu, P. H., et al. (2014). Acetylation of Snail Modulates the Cytokinome of Cancer Cells to Enhance the Recruitment of Macrophages. Cancer Cell 26 (4), 534–548. doi:10.1016/j.ccell.2014.09.002
Huang, K. L., Jee, D., Stein, C. B., Elrod, N. D., Henriques, T., Mascibroda, L. G., et al. (2020). Integrator Recruits Protein Phosphatase 2A to Prevent Pause Release and Facilitate Transcription Termination. Mol. Cel. 80 (2), 345–358.e9. doi:10.1016/j.molcel.2020.08.016
Ip, Y. T. (1995). Transcriptional Regulation. Converting an Activator into a Repressor. Curr. Biol. 5 (1), 1–3. doi:10.1016/s0960-9822(95)00001-7
Jin, X., and Kim, H. (2017). Cancer Stem Cells and Differentiation Therapy. Tumour Biol. 39 (10), 1010428317729933. doi:10.1177/1010428317729933
Jones, P. A., and Baylin, S. B. (2007). The Epigenomics of Cancer. Cell 128 (4), 683–692. doi:10.1016/j.cell.2007.01.029
Jonkers, I., Kwak, H., and Lis, J. T. (2014). Genome-wide Dynamics of Pol II Elongation and its Interplay with Promoter Proximal Pausing, Chromatin, and Exons. Elife 3, e02407. doi:10.7554/eLife.02407
Juven-Gershon, T., and Kadonaga, J. T. (2010). Regulation of Gene Expression via the Core Promoter and the Basal Transcriptional Machinery. Dev. Biol. 339 (2), 225–229. doi:10.1016/j.ydbio.2009.08.009
Kang, H., Shokhirev, M. N., Xu, Z., Chandran, S., Dixon, J. R., and Hetzer, M. W. (2020). Dynamic Regulation of Histone Modifications and Long-Range Chromosomal Interactions During Postmitotic Transcriptional Reactivation. Genes Dev. 34 (13-14), 913–930. doi:10.1101/gad.335794.119
Kannaiyan, R., and Mahadevan, D. (2018). A Comprehensive Review of Protein Kinase Inhibitors for Cancer Therapy. Expert Rev. Anticancer Ther. 18 (12), 1249–1270. doi:10.1080/14737140.2018.1527688
Kim, J. B., and Sharp, P. A. (2001). Positive Transcription Elongation Factor B Phosphorylates hSPT5 and RNA Polymerase II Carboxyl-Terminal Domain Independently of Cyclin-dependent Kinase-Activating Kinase. J. Biol. Chem. 276 (15), 12317–12323. doi:10.1074/jbc.m010908200
Kim, J., Lee, Y., Lu, X., Song, B., Fong, K. W., Cao, Q., et al. (2018). Polycomb- and Methylation-independent Roles of EZH2 as a Transcription Activator. Cell Rep. 25 (10), 2808–2820.e4. doi:10.1016/j.celrep.2018.11.035
Kim, S., Kim, H., Fong, N., Erickson, B., and Bentley, D. L. (2011). Pre-mRNA Splicing Is a Determinant of Histone H3K36 Methylation. Proc. Natl. Acad. Sci. U S A. 108 (33), 13564–13569. doi:10.1073/pnas.1109475108
Kim, T. H., Barrera, L. O., Zheng, M., Qu, C., Singer, M. A., Richmond, T. A., et al. (2005). A High-Resolution Map of Active Promoters in the Human Genome. Nature 436 (7052), 876–880. doi:10.1038/nature03877
Klein, I. A., Boija, A., Afeyan, L. K., Hawken, S. W., Fan, M., Dall’Agnese, A., et al. (2020). Partitioning of Cancer Therapeutics in Nuclear Condensates. Science 368 (6497), 1386–1392. doi:10.1126/science.aaz4427
Kolesky, S. E., Ouhammouch, M., and Geiduschek, E. P. (2002). The Mechanism of Transcriptional Activation by the Topologically DNA-Linked Sliding Clamp of Bacteriophage T4. J. Mol. Biol. 321 (5), 767–784. doi:10.1016/s0022-2836(02)00732-5
Krumm, A., Hickey, L. B., and Groudine, M. (1995). Promoter-proximal Pausing of RNA Polymerase II Defines a General Rate-Limiting Step After Transcription Initiation. Genes Dev. 9 (5), 559–572. doi:10.1101/gad.9.5.559
Krzyszczyk, P., Acevedo, A., Davidoff, E. J., Timmins, L. M., Marrero-Berrios, I., Patel, M., et al. (2018). The Growing Role of Precision and Personalized Medicine for Cancer Treatment. Technology (Singap World Sci) 6 (3-4), 79–100. doi:10.1142/S2339547818300020
Laham-Karam, N., Pinto, G. P., Poso, A., and Kokkonen, P. (2020). Transcription and Translation Inhibitors in Cancer Treatment. Front. Chem. 8, 276. doi:10.3389/fchem.2020.00276
Lazo, J. S., and Sharlow, E. R. (2016). Drugging Undruggable Molecular Cancer Targets. Annu. Rev. Pharmacol. Toxicol. 56, 23–40. doi:10.1146/annurev-pharmtox-010715-103440
Lee, T. I., and Young, R. A. (2013). Transcriptional Regulation and its Misregulation in Disease. Cell 152 (6), 1237–1251. doi:10.1016/j.cell.2013.02.014
Li, G., Ruan, X., Auerbach, R. K., Sandhu, K. S., Zheng, M., Wang, P., et al. (2012). Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell 148 (1-2), 84–98. doi:10.1016/j.cell.2011.12.014
Liang, K., Woodfin, A. R., Slaughter, B. D., Unruh, J. R., Box, A. C., Rickels, R. A., et al. (2015). Mitotic Transcriptional Activation: Clearance of Actively Engaged Pol II via Transcriptional Elongation Control in Mitosis. Mol. Cel. 60 (3), 435–445. doi:10.1016/j.molcel.2015.09.021
Lin, C. Y., Loven, J., Rahl, P. B., Paranal, R. M., Burge, C. B., Bradner, J. E., et al. (2012). Transcriptional Amplification in Tumor Cells with Elevated C-Myc. Cell 151 (1), 56–67. doi:10.1016/j.cell.2012.08.026
Liu, X., Kraus, W. L., and Bai, X. (2015). Ready, Pause, Go: Regulation of RNA Polymerase II Pausing and Release by Cellular Signaling Pathways. Trends Biochem. Sci. 40 (9), 516–525. doi:10.1016/j.tibs.2015.07.003
London, L., Keene, R. G., and Landick, R. (1991). Analysis of Premature Termination in C-Myc During Transcription by RNA Polymerase II in a HeLa Nuclear Extract. Mol. Cel Biol. 11 (9), 4599–4615. doi:10.1128/mcb.11.9.4599-4615.1991
Lu, X., Zhu, X., Li, Y., Liu, M., Yu, B., Wang, Y., et al. (2016). Multiple P-TEFbs Cooperatively Regulate the Release of Promoter-Proximally Paused RNA Polymerase II. Nucleic Acids Res. 44, 6853–6867. doi:10.1093/nar/gkw571
Luo, Z., Lin, C., Guest, E., Garrett, A. S., Mohaghegh, N., Swanson, S., et al. (2012). The Super Elongation Complex Family of RNA Polymerase II Elongation Factors: Gene Target Specificity and Transcriptional Output. Mol. Cel Biol. 32 (13), 2608–2617. doi:10.1128/MCB.00182-12
Madden, S. K., de Araujo, A. D., Gerhardt, M., Fairlie, D. P., and Mason, J. M. (2021). Taking the Myc Out of Cancer: Toward Therapeutic Strategies to Directly Inhibit C-Myc. Mol. Cancer 20 (1), 3. doi:10.1186/s12943-020-01291-6
Mahat, D. B., Salamanca, H. H., Duarte, F. M., Danko, C. G., and Lis, J. T. (2016). Mammalian Heat Shock Response and Mechanisms Underlying its Genome-wide Transcriptional Regulation. Mol. Cel. 62 (1), 63–78. doi:10.1016/j.molcel.2016.02.025
Malone, E. R., Oliva, M., Sabatini, P. J. B., Stockley, T. L., and Siu, L. L. (2020). Molecular Profiling for Precision Cancer Therapies. Genome Med. 12 (1), 8. doi:10.1186/s13073-019-0703-1
Marshall, N. F., Peng, J., Xie, Z., and Price, D. H. (1996). Control of RNA Polymerase II Elongation Potential by A Novel Carboxyl-Terminal Domain Kinase. J. Biol. Chem. 271 (43), 27176–27183. doi:10.1074/jbc.271.43.27176
Marshall, N. F., and Price, D. H. (1995). Purification of P-TEFb, A Transcription Factor Required for the Transition into Productive Elongation. J. Biol. Chem. 270 (21), 12335–12338. doi:10.1074/jbc.270.21.12335
Mohammad, H. P., Barbash, O., and Creasy, C. L. (2019). Targeting Epigenetic Modifications in Cancer Therapy: Erasing the Roadmap to Cancer. Nat. Med. 25 (3), 403–418. doi:10.1038/s41591-019-0376-8
Morales, F., and Giordano, A. (2016). Overview of CDK9 as a Target in Cancer Research. Cell Cycle 15 (4), 519–527. doi:10.1080/15384101.2016.1138186
Mosley, A. L., Lakshmanan, J., Aryal, B. K., and Ozcan, S. (2003). Glucose-mediated Phosphorylation Converts the Transcription Factor Rgt1 from a Repressor to an Activator. J. Biol. Chem. 278 (12), 10322–10327. doi:10.1074/jbc.M212802200
Muse, G. W., Gilchrist, D. A., Nechaev, S., Shah, R., Parker, J. S., Grissom, S. F., et al. (2007). RNA Polymerase Is Poised for Activation Across the Genome. Nat. Genet. 39 (12), 1507–1511. doi:10.1038/ng.2007.21
Nagaraja, S., Quezada, M. A., Gillespie, S. M., Arzt, M., Lennon, J. J., Woo, P. J., et al. (2019). Histone Variant and Cell Context Determine H3K27M Reprogramming of the Enhancer Landscape and Oncogenic State. Mol. Cel. 76 (6), 965–980.e12. doi:10.1016/j.molcel.2019.08.030
Nava, M. M., Miroshnikova, Y. A., Biggs, L. C., Whitefield, D. B., Metge, F., Boucas, J., et al. (2020). Heterochromatin-Driven Nuclear Softening Protects the Genome Against Mechanical Stress-Induced Damage. Cell 181 (4), 800–817.e22. doi:10.1016/j.cell.2020.03.052
Nechaev, S., Fargo, D. C., dos Santos, G., Liu, L., Gao, Y., and Adelman, K. (2010). Global Analysis of Short RNAs Reveals Widespread Promoter-Proximal Stalling and Arrest of Pol II in Drosophila. Science 327 (5963), 335–338. doi:10.1126/science.1181421
Nechaev, S., and Geiduschek, E. P. (2006). The Role of an Upstream Promoter Interaction in Initiation of Bacterial Transcription. EMBO J. 25 (8), 1700–1709. doi:10.1038/sj.emboj.7601069
Nepali, K., and Liou, J. P. (2021). Recent Developments in Epigenetic Cancer Therapeutics: Clinical Advancement and Emerging Trends. J. Biomed. Sci. 28 (1), 27. doi:10.1186/s12929-021-00721-x
Nicetto, D., and Zaret, K. S. (2019). Role of H3K9me3 Heterochromatin in Cell Identity Establishment and Maintenance. Curr. Opin. Genet. Dev. 55, 1–10. doi:10.1016/j.gde.2019.04.013
Nie, Z., Hu, G., Wei, G., Cui, K., Yamane, A., Resch, W., et al. (2012). c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell 151 (1), 68–79. doi:10.1016/j.cell.2012.08.033
Nikolov, D. B., and Burley, S. K. (1997). RNA Polymerase II Transcription Initiation: A Structural View. Proc. Natl. Acad. Sci. U S A. 94 (1), 15–22. doi:10.1073/pnas.94.1.15
Nilson, K. A., Lawson, C. K., Mullen, N. J., Ball, C. B., Spector, B. M., Meier, J. L., et al. (2017). Oxidative Stress Rapidly Stabilizes Promoter-Proximal Paused Pol II Across the Human Genome. Nucleic Acids Res. 45 (19), 11088–11105. doi:10.1093/nar/gkx724
O’Brien, T., and Lis, J. T. (1991). RNA Polymerase II Pauses at the 5’ End of the Transcriptionally Induced Drosophila Hsp70 Gene. Mol. Cel Biol. 11 (10), 5285–5290. doi:10.1128/mcb.11.10.5285
Paik, D. T., Tian, L., Lee, J., Sayed, N., Chen, I. Y., Rhee, S., et al. (2018). Large-Scale Single-Cell RNA-Seq Reveals Molecular Signatures of Heterogeneous Populations of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Circ. Res. 123 (4), 443–450. doi:10.1161/CIRCRESAHA.118.312913
Pan, D., and Courey, A. J. (1992). The Same Dorsal Binding Site Mediates Both Activation and Repression in a Context-dependent Manner. EMBO J. 11 (5), 1837–1842. doi:10.1002/j.1460-2075.1992.tb05235.x
Park, J. W., and Han, J. W. (2019). Targeting Epigenetics for Cancer Therapy. Arch. Pharm. Res. 42 (2), 159–170. doi:10.1007/s12272-019-01126-z
Parker, J. S., Mullins, M., Cheang, M. C., Leung, S., Voduc, D., Vickery, T., et al. (2009). Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 27 (8), 1160–1167. doi:10.1200/JCO.2008.18.1370
Parua, P. K., Kalan, S., Benjamin, B., Sansó, M., and Fisher, R. P. (2020). Distinct Cdk9-Phosphatase Switches Act at the Beginning and End of Elongation by RNA Polymerase II. Nat. Commun. 11 (1), 4338. doi:10.1038/s41467-020-18173-6
Pcawg, T. C. G., Calabrese, C., Davidson, N. R., Demircioğlu, D., Fonseca, N. A., He, Y., et al. (2020). Genomic Basis for RNA Alterations in Cancer. Nature 578 (7793), 129–136. doi:10.1038/s41586-020-1970-0
Pearson, J. D., Huang, K., Pacal, M., McCurdy, S. R., Lu, S., Aubry, A., et al. (2021). Binary Pan-Cancer Classes with Distinct Vulnerabilities Defined by Pro- or Anti-cancer YAP/TEAD Activity. Cancer Cell 39 (8), 1115–1134.e12. doi:10.1016/j.ccell.2021.06.016
Peinado, H., Ballestar, E., Esteller, M., and Cano, A. (2004). Snail Mediates E-Cadherin Repression by the Recruitment of the Sin3A/histone Deacetylase 1 (HDAC1)/HDAC2 Complex. Mol. Cel Biol. 24 (1), 306–319. doi:10.1128/mcb.24.1.306-319.2004
Pekowska, A., Benoukraf, T., Zacarias-Cabeza, J., Belhocine, M., Koch, F., Holota, H., et al. (2011). H3K4 Tri-methylation Provides an Epigenetic Signature of Active Enhancers. EMBO J. 30 (20), 4198–4210. doi:10.1038/emboj.2011.295
Peng, J., Zhu, Y., Milton, J. T., and Price, D. H. (1998). Identification of Multiple Cyclin Subunits of Human P-TEFb. Genes Dev. 12 (5), 755–762. doi:10.1101/gad.12.5.755
Perou, C. M., Sørlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., et al. (2000). Molecular Portraits of Human Breast Tumours. Nature 406 (6797), 747–752. doi:10.1038/35021093
Peterlin, B. M., and Price, D. H. (2006). Controlling the Elongation Phase of Transcription with P-TEFb. Mol. Cel. 23 (3), 297–305. doi:10.1016/j.molcel.2006.06.014
Ptashne, M. (2013). Epigenetics: Core Misconcept. Proc. Natl. Acad. Sci. U S A. 110 (18), 7101–7103. doi:10.1073/pnas.1305399110
Pugh, B. F., and Venters, B. J. (2016). Genomic Organization of Human Transcription Initiation Complexes. PLoS One 11 (2), e0149339. doi:10.1371/journal.pone.0149339
Radonjic, M., Andrau, J. C., Lijnzaad, P., Kemmeren, P., Kockelkorn, T. T., van Leenen, D., et al. (2005). Genome-wide Analyses Reveal RNA Polymerase II Located Upstream of Genes Poised for Rapid Response upon S. cerevisiae Stationary Phase Exit. Mol. Cel. 18 (2), 171–183. doi:10.1016/j.molcel.2005.03.010
Rahl, P. B., Lin, C. Y., Seila, A. C., Flynn, R. A., McCuine, S., Burge, C. B., et al. (2010). c-Myc Regulates Transcriptional Pause Release. Cell 141 (3), 432–445. doi:10.1016/j.cell.2010.03.030
Rajagopal, N., Ernst, J., Ray, P., Wu, J., Zhang, M., Kellis, M., et al. (2014). Distinct and Predictive Histone Lysine Acetylation Patterns at Promoters, Enhancers, and Gene Bodies. G3 (Bethesda) 4 (11), 2051–2063. doi:10.1534/g3.114.013565
Renner, D. B., Yamaguchi, Y., Wada, T., Handa, H., and Price, D. H. (2001). A Highly Purified RNA Polymerase II Elongation Control System. J. Biol. Chem. 276 (45), 42601–42609. doi:10.1074/jbc.m104967200
Rimel, J. K., Poss, Z. C., Erickson, B., Maas, Z. L., Ebmeier, C. C., Johnson, J. L., et al. (2020). Selective Inhibition of CDK7 Reveals High-Confidence Targets and New Models for TFIIH Function in Transcription. Genes Dev. 34 (21-22), 1452–1473. doi:10.1101/gad.341545.120
Rodriguez, J., and Larson, D. R. (2020). Transcription in Living Cells: Molecular Mechanisms of Bursting. Annu. Rev. Biochem. 89, 189–212. doi:10.1146/annurev-biochem-011520-105250
Rousseaux, S., Debernardi, A., Jacquiau, B., Vitte, A. L., Vesin, A., Nagy-Mignotte, H., et al. (2013). Ectopic Activation of Germline and Placental Genes Identifies Aggressive Metastasis-Prone Lung Cancers. Sci. Transl Med. 5 (186), 186ra66. doi:10.1126/scitranslmed.3005723
Russo, M., Natoli, G., and Ghisletti, S. (2018). Housekeeping and Tissue-specific Cis-Regulatory Elements: Recipes for Specificity and Recipes for Activity. Transcription 9 (3), 177–181. doi:10.1080/21541264.2017.1378158
Saeed, S., Quintin, J., Kerstens, H. H., Rao, N. A., Aghajanirefah, A., Matarese, F., et al. (2014). Epigenetic Programming of Monocyte-To-Macrophage Differentiation and Trained Innate Immunity. Science 345 (6204), 1251086. doi:10.1126/science.1251086
Samarakkody, A., Abbas, A., Scheidegger, A., Warns, J., Nnoli, O., Jokinen, B., et al. (2015). RNA Polymerase II Pausing Can Be Retained or Acquired During Activation of Genes Involved in the Epithelial to Mesenchymal Transition. Nucleic Acids Res. 43 (8), 3938–3949. doi:10.1093/nar/gkv263
Sava, G. P., Fan, H., Coombes, R. C., Buluwela, L., and Ali, S. (2020). CDK7 Inhibitors as Anticancer Drugs. Cancer Metastasis Rev. 39 (3), 805–823. doi:10.1007/s10555-020-09885-8
Scheidegger, A., Dunn, C. J., Samarakkody, A., Koney, N. K., Perley, D., Saha, R. N., et al. (2019). Genome-wide RNA Pol II Initiation and Pausing in Neural Progenitors of the Rat. BMC Genomics 20 (1), 477. doi:10.1186/s12864-019-5829-4
Schuettengruber, B., Bourbon, H. M., Di Croce, L., and Cavalli, G. (2017). Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171 (1), 34–57. doi:10.1016/j.cell.2017.08.002
Scruggs, B. S., Gilchrist, D. A., Nechaev, S., Muse, G. W., Burkholder, A., Fargo, D. C., et al. (2015). Bidirectional Transcription Arises from Two Distinct Hubs of Transcription Factor Binding and Active Chromatin. Mol. Cel. 58 (6), 1101–1112. doi:10.1016/j.molcel.2015.04.006
Shan, X., Fung, J. J., Kosaka, A., Danet-Desnoyers, G., and Reproducibility, P. C. B. (2017). Replication Study: Inhibition of BET Recruitment to Chromatin as an Effective Treatment for MLL-Fusion Leukaemia. Elife 6, e25306. doi:10.7554/eLife.25306
Shao, X., Lv, N., Liao, J., Long, J., Xue, R., Ai, N., et al. (2019). Copy Number Variation Is Highly Correlated with Differential Gene Expression: A Pan-Cancer Study. BMC Med. Genet. 20 (1), 175. doi:10.1186/s12881-019-0909-5
Shen, L., Shi, Q., and Wang, W. (2018). Double Agents: Genes with Both Oncogenic and Tumor-Suppressor Functions. Oncogenesis 7 (3), 25. doi:10.1038/s41389-018-0034-x
Sievers, Q. L., Petzold, G., Bunker, R. D., Renneville, A., Słabicki, M., Liddicoat, B. J., et al. (2018). Defining the Human C2H2 Zinc Finger Degrome Targeted by Thalidomide Analogs Through CRBN. Science 362 (6414), eaat0572. doi:10.1126/science.aat0572
Skidmore, L., Sakamuri, S., Knudsen, N. A., Hewet, A. G., Milutinovic, S., Barkho, W., et al. (2020). ARX788, A Site-specific Anti-HER2 Antibody-Drug Conjugate, Demonstrates Potent and Selective Activity in HER2-Low and T-DM1-Resistant Breast and Gastric Cancers. Mol. Cancer Ther. 19 (9), 1833–1843. doi:10.1158/1535-7163.MCT-19-1004
Soares, L. M., He, P. C., Chun, Y., Suh, H., Kim, T., and Buratowski, S. (2017). Determinants of Histone H3K4 Methylation Patterns. Mol. Cel. 68 (4), 773–785.e6. doi:10.1016/j.molcel.2017.10.013
Spencer, C. A., and Groudine, M. (1990). Transcription Elongation and Eukaryotic Gene Regulation. Oncogene 5 (6), 777–785.
Staller, M. V., Vincent, B. J., Bragdon, M. D., Lydiard-Martin, T., Wunderlich, Z., Estrada, J., et al. (2015). Shadow Enhancers Enable Hunchback Bifunctionality in the Drosophila Embryo. Proc. Natl. Acad. Sci. U S A. 112 (3), 785–790. doi:10.1073/pnas.1413877112
Stingele, S., Stoehr, G., Peplowska, K., Cox, J., Mann, M., and Storchova, Z. (2012). Global Analysis of Genome, Transcriptome and Proteome Reveals the Response to Aneuploidy in Human Cells. Mol. Syst. Biol. 8, 608. doi:10.1038/msb.2012.40
Strahl, B. D., and Allis, C. D. (2000). The Language of Covalent Histone Modifications. Nature 403 (6765), 41–45. doi:10.1038/47412
Sun, J., Pan, H., Lei, C., Yuan, B., Nair, S. J., April, C., et al. (2011). Genetic and Genomic Analyses of RNA Polymerase II-Pausing Factor in Regulation of Mammalian Transcription and Cell Growth. J. Biol. Chem. 286 (42), 36248–36257. doi:10.1074/jbc.M111.269167
Sun, J., Watkins, G., Blair, A. L., Moskaluk, C., Ghosh, S., Jiang, W. G., et al. (2008). Deregulation of Cofactor of BRCA1 Expression in Breast Cancer Cells. J. Cel Biochem 103 (6), 1798–1807. doi:10.1002/jcb.21568
Takeshima, H., Yamashita, S., Shimazu, T., Niwa, T., and Ushijima, T. (2009). The Presence of RNA Polymerase II, Active or Stalled, Predicts Epigenetic Fate of Promoter CpG Islands. Genome Res. 19 (11), 1974–1982. doi:10.1101/gr.093310.109
Tan, M., Luo, H., Lee, S., Jin, F., Yang, J. S., Montellier, E., et al. (2011). Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell 146 (6), 1016–1028. doi:10.1016/j.cell.2011.08.008
Tellier, M., Zaborowska, J., Caizzi, L., Mohammad, E., Velychko, T., Schwalb, B., et al. (2020). CDK12 Globally Stimulates RNA Polymerase II Transcription Elongation and Carboxyl-Terminal Domain Phosphorylation. Nucleic Acids Res. 48 (14), 7712–7727. doi:10.1093/nar/gkaa514
Terryn, J., Tricot, T., Gajjar, M., and Verfaillie, C. (2018). Recent Advances in Lineage Differentiation from Stem Cells: Hurdles and Opportunities. F1000Res 7, 220. doi:10.12688/f1000research.12596.1
Towbin, B. D., González-Aguilera, C., Sack, R., Gaidatzis, D., Kalck, V., Meister, P., et al. (2012). Step-wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery. Cell 150 (5), 934–947. doi:10.1016/j.cell.2012.06.051
Trkulja, C. L., Jungholm, O., Davidson, M., Jardemark, K., Marcus, M. M., Hägglund, J., et al. (2021). Rational Antibody Design for Undruggable Targets Using Kinetically Controlled Biomolecular Probes. Sci. Adv. 7 (16), eabe6397. doi:10.1126/sciadv.abe6397
Tunnacliffe, E., and Chubb, J. R. (2020). What Is a Transcriptional Burst. Trends Genet. 36 (4), 288–297. doi:10.1016/j.tig.2020.01.003
Uhlén, M., Hallström, B. M., Lindskog, C., Mardinoglu, A., Pontén, F., and Nielsen, J. (2016). Transcriptomics Resources of Human Tissues and Organs. Mol. Syst. Biol. 12 (4), 862. doi:10.15252/msb.20155865
Uren, A., and Toretsky, J. A. (2005). Pediatric Malignancies Provide Unique Cancer Therapy Targets. Curr. Opin. Pediatr. 17 (1), 14–19. doi:10.1097/01.mop.0000147904.84978.ae
Van Hoeck, A., Tjoonk, N. H., van Boxtel, R., and Cuppen, E. (2019). Portrait of a Cancer: Mutational Signature Analyses for Cancer Diagnostics. BMC Cancer 19 (1), 457. doi:10.1186/s12885-019-5677-2
Vastenhouw, N. L., and Schier, A. F. (2012). Bivalent Histone Modifications in Early Embryogenesis. Curr. Opin. Cel Biol. 24 (3), 374–386. doi:10.1016/j.ceb.2012.03.009
Vidal, S. E., Polyzos, A., Chatterjee, K., Ee, L. S., Swanzey, E., Morales-Valencia, J., et al. (2020). Context-Dependent Requirement of Euchromatic Histone Methyltransferase Activity During Reprogramming to Pluripotency. Stem Cel Rep. 15 (6), 1233–1245. doi:10.1016/j.stemcr.2020.08.011
Vihervaara, A., Mahat, D. B., Guertin, M. J., Chu, T., Danko, C. G., Lis, J. T., et al. (2017). Transcriptional Response to Stress Is Pre-wired by Promoter and Enhancer Architecture. Nat. Commun. 8 (1), 255. doi:10.1038/s41467-017-00151-0
Villicaña, C., Cruz, G., and Zurita, M. (2014). The Basal Transcription Machinery as a Target for Cancer Therapy. Cancer Cel Int 14 (1), 18. doi:10.1186/1475-2867-14-18
Vishnoi, K., Viswakarma, N., Rana, A., and Rana, B. (2020). Transcription Factors in Cancer Development and Therapy. Cancers (Basel) 12 (8), E2296. doi:10.3390/cancers12082296
Wada, T., Takagi, T., Yamaguchi, Y., Ferdous, A., Imai, T., Hirose, S., et al. (1998). DSIF, A Novel Transcription Elongation Factor that Regulates RNA Polymerase II Processivity, Is Composed of Human Spt4 and Spt5 Homologs. Genes Dev. 12 (3), 343–356. doi:10.1101/gad.12.3.343
Waddington, C. H. (1957). The Strategy of the Genes A Discussion of Some Aspects of Theoretical Biology with an Appendix by H Kacser. London: George Allen & Unwin, Ltd., 262.
Walker, W. H., Girardet, C., and Habener, J. F. (1996). Alternative Exon Splicing Controls a Translational Switch from Activator to Repressor Isoforms of Transcription Factor CREB During Spermatogenesis. J. Biol. Chem. 271 (33), 20145–21050. doi:10.1074/jbc.271.33.20219
Wang, C., Liu, X., Gao, Y., Yang, L., Li, C., Liu, W., et al. (2018). Reprogramming of H3K9me3-dependent Heterochromatin During Mammalian Embryo Development. Nat. Cel Biol. 20 (5), 620–631. doi:10.1038/s41556-018-0093-4
Wang, C., Zhang, J., Yin, J., Gan, Y., Xu, S., Gu, Y., et al. (2021). Alternative Approaches to Target Myc for Cancer Treatment. Signal. Transduct Target. Ther. 6 (1), 117. doi:10.1038/s41392-021-00500-y
Wang, Y., Zhang, T., Kwiatkowski, N., Abraham, B. J., Lee, T. I., Xie, S., et al. (2015). CDK7-dependent Transcriptional Addiction in Triple-Negative Breast Cancer. Cell 163 (1), 174–186. doi:10.1016/j.cell.2015.08.063
Winters, A. C., and Bernt, K. M. (2017). MLL-rearranged Leukemias-An Update on Science and Clinical Approaches. Front. Pediatr. 5, 4. doi:10.3389/fped.2017.00004
Wolffe, A. P., Wong, J., and Pruss, D. (1997). Activators and Repressors: Making Use of Chromatin to Regulate Transcription. Genes Cells 2 (5), 291–302. doi:10.1046/j.1365-2443.1997.1260323.x
Wu, C. H., Yamaguchi, Y., Benjamin, L. R., Horvat-Gordon, M., Washinsky, J., Enerly, E., et al. (2003). NELF and DSIF Cause Promoter Proximal Pausing on the Hsp70 Promoter in Drosophila. Genes Dev. 17 (11), 1402–1414. doi:10.1101/gad.1091403
Wu, W. S., You, R. I., Cheng, C. C., Lee, M. C., Lin, T. Y., and Hu, C. T. (2017). Snail Collaborates with EGR-1 and SP-1 to Directly Activate Transcription of MMP 9 and ZEB1. Sci. Rep. 7 (1), 17753. doi:10.1038/s41598-017-18101-7
Yamada, T., Yamaguchi, Y., Inukai, N., Okamoto, S., Mura, T., and Handa, H. (2006). P-TEFb-mediated Phosphorylation of hSpt5 C-Terminal Repeats Is Critical for Processive Transcription Elongation. Mol. Cel. 21 (2), 227–237. doi:10.1016/j.molcel.2005.11.024
Yamaguchi, Y., Filipovska, J., Yano, K., Furuya, A., Inukai, N., Narita, T., et al. (2001). Stimulation of RNA Polymerase II Elongation by Hepatitis Delta Antigen. Science 293 (5527), 124–127. doi:10.1126/science.1057925
Yamaguchi, Y., Takagi, T., Wada, T., Yano, K., Furuya, A., Sugimoto, S., et al. (1999). NELF, A Multisubunit Complex Containing RD, Cooperates with DSIF to Repress RNA Polymerase II Elongation. Cell 97 (1), 41–51. doi:10.1016/s0092-8674(00)80713-8
Yang, L., Kirby, J. E., Sunwoo, H., and Lee, J. T. (2016). Female Mice Lacking Xist RNA Show Partial Dosage Compensation and Survive to Term. Genes Dev. 30 (15), 1747–1760. doi:10.1101/gad.281162.116
Yen, A., and Kellis, M. (2015). Systematic Chromatin State Comparison of Epigenomes Associated with Diverse Properties Including Sex and Tissue Type. Nat. Commun. 6, 7973. doi:10.1038/ncomms8973
Yildirim, E., Kirby, J. E., Brown, D. E., Mercier, F. E., Sadreyev, R. I., Scadden, D. T., et al. (2013). Xist RNA Is a Potent Suppressor of Hematologic Cancer in Mice. Cell 152 (4), 727–742. doi:10.1016/j.cell.2013.01.034
Yu, K., Chen, B., Aran, D., Charalel, J., Yau, C., Wolf, D. M., et al. (2019). Comprehensive Transcriptomic Analysis of Cell Lines as Models of Primary Tumors Across 22 Tumor Types. Nat. Commun. 10 (1), 3574. doi:10.1038/s41467-019-11415-2
Yun, H., Bedolla, R., Horning, A., Li, R., Chiang, H. C., Huang, T. H., et al. (2018). BRCA1 Interacting Protein COBRA1 Facilitates Adaptation to Castrate-Resistant Growth Conditions. Int. J. Mol. Sci. 19 (7), E2104. doi:10.3390/ijms19072104
Zabidi, M. A., Arnold, C. D., Schernhuber, K., Pagani, M., Rath, M., Frank, O., et al. (2015). Enhancer-core-promoter Specificity Separates Developmental and Housekeeping Gene Regulation. Nature 518 (7540), 556–559. doi:10.1038/nature13994
Zeitlinger, J., Stark, A., Kellis, M., Hong, J. W., Nechaev, S., Adelman, K., et al. (2007). RNA Polymerase Stalling at Developmental Control Genes in the Drosophila melanogaster Embryo. Nat. Genet. 39 (12), 1512–1516. doi:10.1038/ng.2007.26
Zhang, G., Andersen, J., and Gerona-Navarro, G. (2018). Peptidomimetics Targeting Protein-Protein Interactions for Therapeutic Development. Protein Pept. Lett. 25 (12), 1076–1089. doi:10.2174/0929866525666181101100842
Zhang, L. J., Vogel, W. K., Liu, X., Topark-Ngarm, A., Arbogast, B. L., Maier, C. S., et al. (2012). Coordinated Regulation of Transcription Factor Bcl11b Activity in Thymocytes by the Mitogen-Activated Protein Kinase (MAPK) Pathways and Protein Sumoylation. J. Biol. Chem. 287 (32), 26971–26988. doi:10.1074/jbc.M112.344176
Zhang, T., Zhang, Z., Dong, Q., Xiong, J., and Zhu, B. (2020). Histone H3K27 Acetylation Is Dispensable for Enhancer Activity in Mouse Embryonic Stem Cells. Genome Biol. 21 (1), 45. doi:10.1186/s13059-020-01957-w
Zhao, L., Zhou, S., and Gustafsson, J. Å. (2019). Nuclear Receptors: Recent Drug Discovery for Cancer Therapies. Endocr. Rev. 40 (5), 1207–1249. doi:10.1210/er.2018-00222
Zhao, X. D., Han, X., Chew, J. L., Liu, J., Chiu, K. P., Choo, A., et al. (2007). Whole-genome Mapping of Histone H3 Lys4 and 27 Trimethylations Reveals Distinct Genomic Compartments in Human Embryonic Stem Cells. Cell Stem Cell 1 (3), 286–298. doi:10.1016/j.stem.2007.08.004
Zhou, Q., Li, T., and Price, D. H. (2012). RNA Polymerase II Elongation Control. Annu. Rev. Biochem. 81, 119–143. doi:10.1146/annurev-biochem-052610-095910
Zrimec, J., Börlin, C. S., Buric, F., Muhammad, A. S., Chen, R., Siewers, V., et al. (2020). Deep Learning Suggests that Gene Expression Is Encoded in All Parts of a Co-evolving Interacting Gene Regulatory Structure. Nat. Commun. 11 (1), 6141. doi:10.1038/s41467-020-19921-4
Keywords: transcriptome regulation, epigenetics, transcription elongation, RNA pol II pausing, NELF model
Citation: Parrello D, Vlasenok M, Kranz L and Nechaev S (2021) Targeting the Transcriptome Through Globally Acting Components. Front. Genet. 12:749850. doi: 10.3389/fgene.2021.749850
Received: 30 July 2021; Accepted: 02 September 2021;
Published: 16 September 2021.
Edited by:
Rais Ahmad Ansari, Nova Southeastern University, United StatesReviewed by:
Rong Li, George Washington University, United StatesPabitra Parua, Albert Einstein College of Medicine, United States
Copyright © 2021 Parrello, Vlasenok, Kranz and Nechaev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergei Nechaev, c2VyZ2VpLm5lY2hhZXZAVU5ELmVkdQ==
 Damien Parrello1
Damien Parrello1 Sergei Nechaev
Sergei Nechaev