- 1Biosciences Eastern and Central Africa-International Livestock Research Institute Hub (BecA-ILRI Hub), Nairobi, Kenya
- 2Central Veterinary Research Laboratory, Khartoum, Sudan
- 3Faculty of Veterinary Medicine, University of Khartoum, Khartoum, Sudan
- 4Atbara Veterinary Research Laboratory, Atbara, Sudan
- 5Faculty of Veterinary Medicine, University of Al-Butana, Tamboul, Sudan
- 6School of Life Sciences, Centre for Genetics and Genomics, University of Nottingham, Nottingham, United Kingdom
- 7International Livestock Research Institute, Nairobi, Kenya
- 8Central Laboratory, Khartoum, Sudan
Theileria annulata, which causes tropical theileriosis, is a major impediment to improving cattle production in Sudan. Tropical theileriosis disease is prevalent in the north and central regions of Sudan. Outbreaks of the disease have been observed outside the known endemic areas, in east and west regions of the country, due to changes in tick vector distribution and animal movement. A live schizont attenuated vaccination based on tissue culture technology has been developed to control the disease. The parasite in the field as well as the vaccine strain need to be genotyped before the vaccinations are practiced, in order to be able to monitor any breakthrough or breakdown, if any, after the deployment of the vaccine in the field. Nine microsatellite markers were used to genotype 246 field samples positive for T. annulata DNA and the vaccine strain. North and central populations have a higher multiplicity of infection than east and west populations. The examination of principal components showed two sub-structures with a mix of all four populations in both clusters and the vaccine strain used being aligned with left-lower cluster. Only the north population was in linkage equilibrium, while the other populations were in linkage disequilibrium, and linkage equilibrium was found when all samples were regarded as single population. The genetic identity of the vaccine and field samples was 0.62 with the north population and 0.39 with west population. Overall, genetic investigations of four T. annulata populations in Sudan revealed substantial intermixing, with only two groups exhibiting regional origin independence. In the four geographically distant regions analyzed, there was a high level of genetic variation within each population. The findings show that the live schizont attenuated vaccine, Atbara strain may be acceptable for use in all Sudanese regions where tropical theileriosis occurs.
Introduction
Tropical theileriosis is a tick-borne disease, caused by Theileria annulata that continues to be a major concern for livestock in tropical countries affecting millions of animals, particularly crossbreed and exotic cattle, and resulting in significant economic loss and mortality (Dolan, 1989). Hyalomma anatolicum transmits T. annulata sporozoites causing a lymphoproliferative disease (Ghosh and Azhahianambi, 2007). The sporozoites develop into schizonts which reside inside the host’s lymphocyte and macrophages (Sager et al., 1998). The schizont stage is the only symptomatic stage among different parasite stages in the host, on which attenuated schizont vaccine was designed (Pipano and Shkap, 2006). Many countries, including, Israel (Pipano et al., 1981), Iran (Hashemi-Fesharki, 1988), Russia (Stepanova and Zablotskii, 1989), India (Beniwal et al., 1997), China (Zhang, 1997), Turkey (Sayin et al., 1997), Spain (Viseras et al., 1997), Morocco (Ouhelli et al., 1997), Tunisia (Darghouth, 2008), and India (Roy et al., 2019), have employed the attenuated schizont vaccines to control T. annulata infection.
The importance of genetic diversity research in providing information on protozoan parasites, such as epidemiology, control, evolution, virulence, antigenicity, infectivity, treatment sensitivity, and host preference, has been demonstrated (Weir, et al., 2011; Sivakumar et al., 2014). T. annulata from other endemic regions, such as China, Oman, Turkey, Tunisia, and Portugal have all been researched utilizing a multilocus genotyping technique for assessing genetic diversity, population structure, and transmission patterns (Weir et al., 2007, 2011; Al-Hamidhi et al., 2015; Gomes et al., 2016; Yin et al., 2018; Roy et al., 2021). T. annulata genetic populations studies were notable for its genetic variation, the availability of many genotypes per sample, and sub-structuring by geography (Weir et al., 2007, 2011; Al-Hamidhi et al., 2015; Gomes et al., 2016; Yin et al., 2018; Roy et al., 2019).
Microsatellite-based genotyping was utilized in this work to better understand genetic diversity, population structure, and geographical sub structuring of the T. annulata vaccine and parasite samples collected from four different regions in Sudan. The diversity of the markers used would reflect the genetic makeup of the samples as well as the vaccine genetic makeup. The recognition of the vaccine strain would be as fast and efficient if the genetic makeup of the vaccine is the same as the field strain. The findings offer the first glimpse of the T. annulata parasites population genetics and diversity in Sudan.
Materials and Methods
Cattle Blood Samples
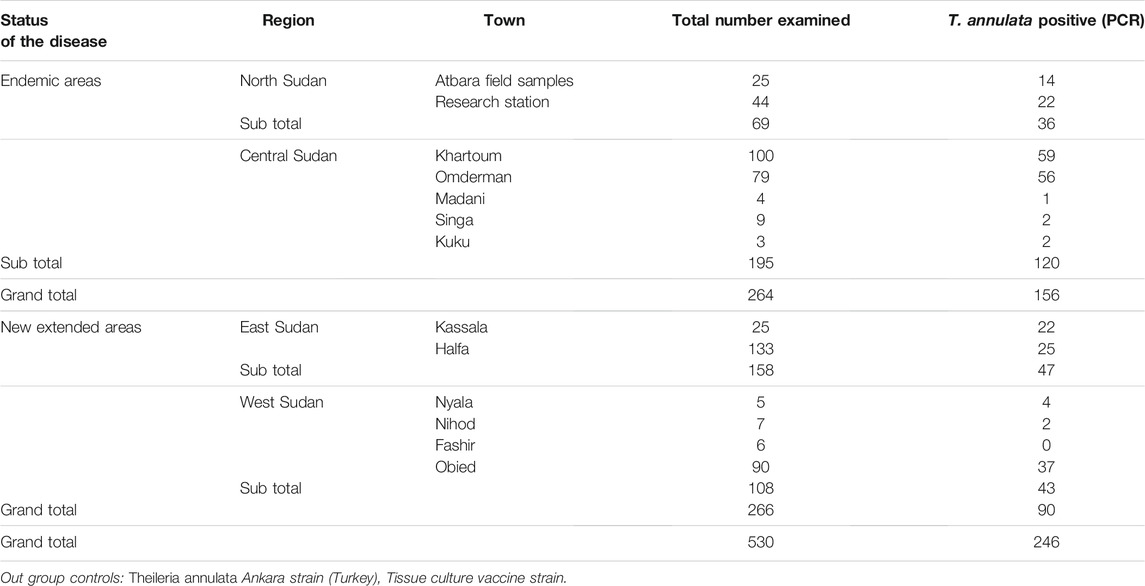
A total of 530 blood samples were collected from cattle in four Sudanese regions using FTA™ cards (Whatman Biosciences, United Kingdom). The four regions were north (n = 69), central (n = 195), east (n = 158), and west (n = 108). North and central regions were designated endemic region, while east and west were designated new extension regions. Information on the sampling locations, whether from endemic or new extension regions, total number examined and T. annulata positive samples by PCR are provided in Table 1.
Extraction of DNA and Small Subunit “SSU” rRNA PCR
Extraction of DNA from cattle blood samples and Atbara vaccine strain was carried out using the PureLink™ Genomic DNA Mini extraction kit (Invitrogen, Germany). For diagnosis of T. annulata, the primer used was SSU rRNA gene 989 5′AGTTTCTGACCTATCAG3′ and the reverse primer was 1,347 5′TGCACAGACCCCAGAG G 3′ giving an amplicon of 370 bp (Allsopp et al., 1993; Taha et al., 2013).
Microsatellite PCR Assay
In this study, the primers used were designed by Weir et al. (2007). For detection in capillary electrophoresis, the forward primer was labeled with standard labeling dyes at the 5′ end (Supplementary Table S1). The PCR amplification was carried out as described in Salih et al. (2018). For negative control, the nuclease free water was used, while DNA extracted from a schizont-infected lymphocyte culture derived from T. annulata Ankara strain was used as a positive control.
Capillary Electrophoresis and Genotyping
The ABI 3730 Genetic Analyzer (Applied Biosystems-USA) was used to analyze the PCR amplicons at the BecA-ILRI Hub, SegoliP sequencing unit, Nairobi, Kenya. For size fractionation, the Gene Scan 500 LIZ internal lane size standard (Applied Biosystems-USA) was employed. The Gene Mapper tool (Applied Biosystems-USA) was used to score the results, which allowed for the resolution of 1 base pair (bp) changes with many products from a single PCR reaction. The predominant allele was determined as the one with the biggest area under the curve, and amplicons with highest peak height were scored. Allelobin software (Idury and Cardon, 1997) was used to re-sized all Gene Mapper data based on consensus sequence repeats of each marker (Table 2). The inaugural form of file, designated multi locus genotype (MLG) consisted of genotypes created from only the predominant allele at each locus (Weir et al., 2007). The allelic profile dataset, on the other hand, contained genotypic profiles derived from all alleles observed at each locus (where minor peaks were greater than 33% the height of the predominant allele present). The MLG file was used to determine population genetic diversity and structure, while the allelic profile file was utilized to calculate the multiplicity of infection (MOI), as well as to rule out linkage disequilibrium as a null hypothesis.
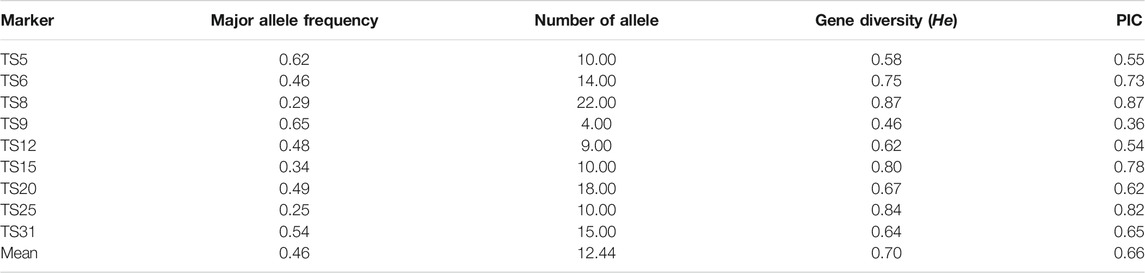
TABLE 2. Major allele frequency, gene diversity, number of alleles and polymorphic information content (PIC) of the nine microsatellite markers used in this study.
Analyses of Population Genetic
Arlequin v. 3.5 http://cmpg.unibe.ch/software/arlequin 35/ (Excoffier, and Lischer, 2010) was used to calculate the expected heterozygosity, as Theileria is haploid and heterozygosity cannot be observed directly. To investigate the genetic relationships between population, principal component analysis (PCA) was calculated in GenAlEx6.5 (Peakall and Smouse, 2006; 2012). Analysis of molecular variance (AMOVA) was performed using ARLEQUIN to test for hierarchical population structure. Nei’s genetic distance (D) (Nei, 1978) was calculate between each group of samples from different populations and the vaccine strain using the genetic data analysis tool (GDA) (http://lewis.eeb.uconn.edu/lewishome/gda.html).
The standardized index of association (ISA) between each group of samples was estimated using the LIAN 3.7 program, as well as, the degree of linkage disequilibrium (LD) within and between populations (Haubold and Hudson, 2000). After each population was studied separately, the samples were pooled and processed as a single dataset.
STRUCTURE 2.3.4 (http://pritchardlab.stanford.edu/structure.html) was used to investigate population structure employing Bayesian clustering analysis with sample sites as a basis and the admixture scenario with linked allele polymorphism (Pritchard et al., 2000; Evanno et al., 2005). Initial runs of one million steps were used to investigate the datasets (burn-in of 20%). For every value of K scale from one (considering all are T. annulata) to five (assuming all the five populations are genetically distinct), triplicates were performed. To identify which K produced the greatest representation of the data, STRUCTUREHARVESTER 0.6.1 (Earl and von Holdt, 2012) was employed. CLUMPP 1.1 (Jakobsson and Rosenberg, 2007) and DISTRUCT 1.1.2 (Rosenberg, 2004) were used to parse and format the data in order to assess the STRUCTURE output. CLUMPP 1.1 aligns cluster assignments across duplicate analyses, while DISTRUCT 1.1.2 assists with visual representation.
Multiplicity of Infection
MOI was considered as “existence of numerous genotypes per isolate” when more than one allele was detected at a locus and the smaller peaks were exceed 33% of the height of the predominant allele expressed (Weir et al., 2011; Salih et al., 2018). The mean number of alleles across all nine loci was determined for every sample, and this number was used to indicate the multiplicity of infection in that sample. The multiplicity of infection for each population was calculated by taking the overall mean for each sample’s index value.
Results
Verification of Positive Samples for T. annulata DNA
T. annulata DNA was tested in 530 cattle blood samples. The SSU rRNA PCR assay verified 246 (46.4%) samples positive for T. annulata DNA which were subjected to genotyping in addition to the vaccine strain (Table 1). Distribution of the positive samples were as follow, endemic regions n = 156 (North n = 36, Central n = 120) and new extension regions n = 90 (East n = 47, West n = 43) (Figure 1).
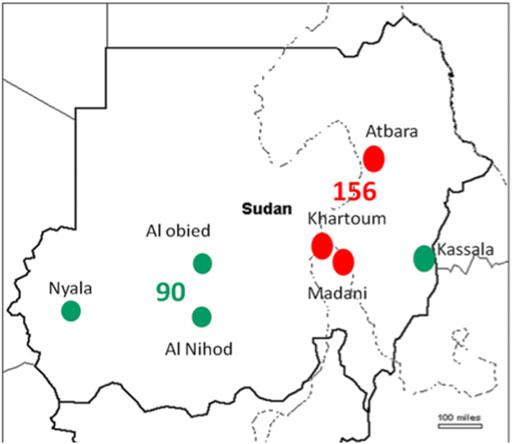
FIGURE 1. The regions of Sudan where samples were obtained are depicted on the map, the red dots indicates endemic region, while the green ones indicates new extended regions.
Satellite Marker Diversity
In all of the samples, each marker was highly polymorphic. The polymorphic information content (PIC) of marker TS8 had the highest (0.87), whereas TS9 had the lowest (0.36) (Table 2). This finding argued in favor that these markers could be effective in determining linkage disequilibrium analysis in T. annulata populations. The existence of more than one allele at one or more loci confirmed the presence of several genotypes in the samples. For each marker, the number of alleles identified varied from four in TS9 to 22 for TS8 with the mean of 12.44 per marker (Table 2). The dominant allele frequencies varied from 0.25 (TS25) to 0.65 (TS9), with an average of 0.46 (Table 2).
Population Diversity and Structure
Principal components analysis (PCA) revealed that there is no clustering according to geographical origin (Figure 2). Two sub-structures with a mix of all four populations in both clusters and the vaccine stain being aligned with left-lower cluster were demonstrated, indicating that the parasite populations are rather distinct, with considerable genetic mixing and gene flow between parasites in the four distinct geographical populations investigated.
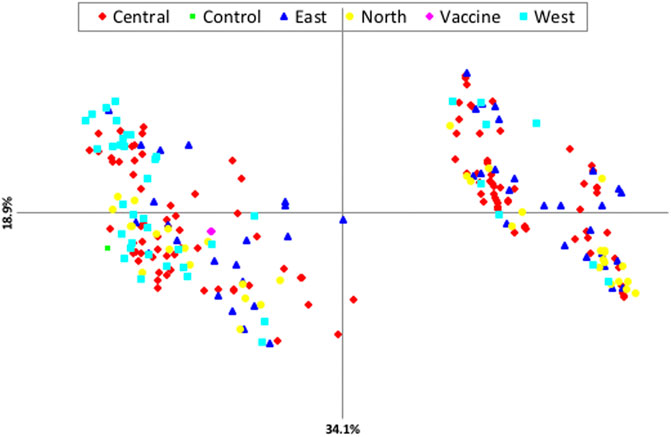
FIGURE 2. Principle component analysis (PCA) showing the genetic structure of T. annulata populations from the four regions of Sudan.
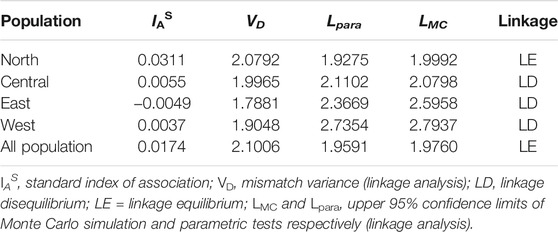
The allelic profile data set was examined to see if the T. annulata populations observed in Sudan were in linkage equilibrium or disequilibrium. When all the four sub-populations were analysed together (as a single population), the (ISA) was positive and greater than zero and the pairwise variance (VD) was more than the 95% critical value (L) suggesting that the merged populations are in linkage equilibrium (LE) (Table 3). The analysis was performed for each population individually to assess for geographic sub-structuring, and three of the populations central, east and west, were shown to be in linkage disequilibrium (LD) (Table 3). Only 11% of the genetic variation was explained by variations between populations, which account for a considerable portion of the genetic diversity (89%) detected within populations (Figure 3).
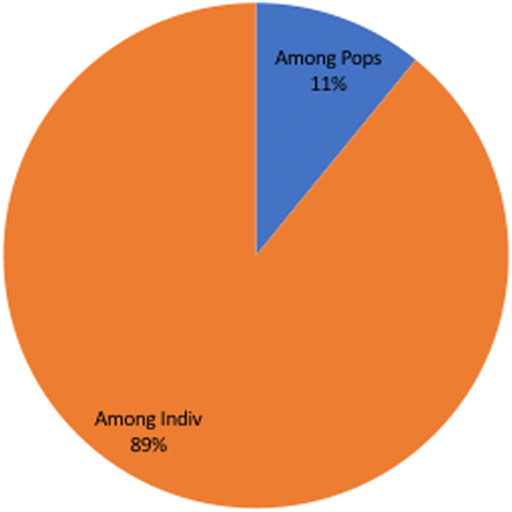
FIGURE 3. Analysis of Molecular Variation (AMOVA) showing only 11% of the genetic variation was explained by populations differences, despite the fact that population differences account for the majority of genetic variety (89%) observed within populations.
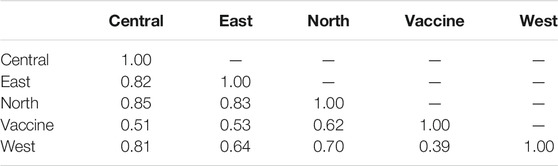
Estimating Nei’s genetic distance (D) between each of the four regionally sampled populations as well as between them and the vaccine strain, was used to evaluate genetic differentiation between the four populations (Table 4). The genetic differentiation between central and east populations (D = 0.82) was greater than that observed between the east and west populations (D = 0.64). The population with the lowest genetic distance from the vaccine genotype was west (D = 0.39), while the most genetically similar was north (D = 0.62) (Table 4).
Based on the Evanno et al. delta K technique, the STRUCTURE results imply that K = 3 is the optimal number of genetic groups to define the genotypes of Sudanese T. annulata populations as well as in T. annulata vaccine strain (Figure 4). The three clusters are designated as gene pool 1, 2 and 3 respectively. Gene pool 1 (purple colour) prevailed in central and east, while gene pool 2 (blue colour) were most prevalent in north and vaccine, and pool 3 (yellow colour) predominated in west (Figure 4). In the vaccine strain, gene pool 2 appears to be more common than gene pool 1.
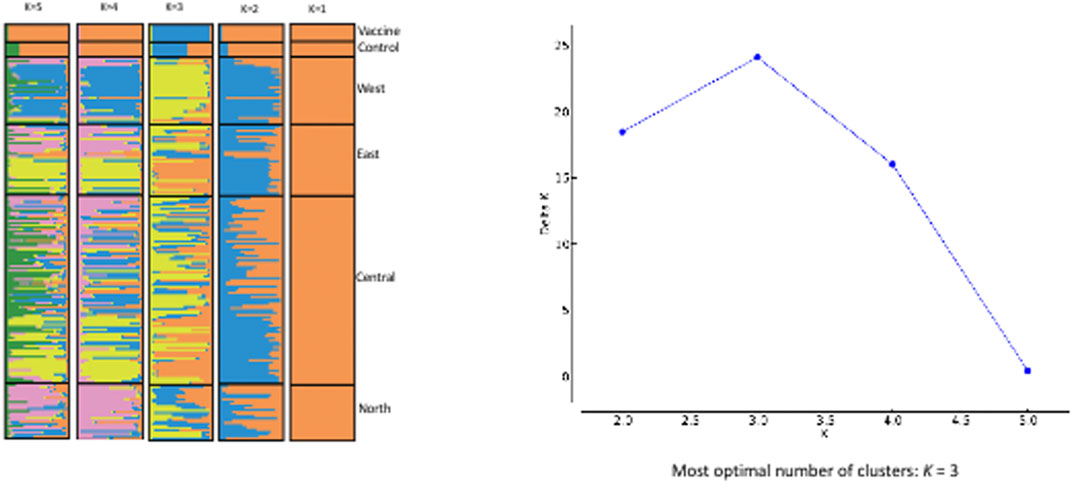
FIGURE 4. EVANNO Method Delta and STRUCTURE. The graph shows optimal number of clusters from the STRUCTURE analysis; STRUCTURE analysis from K = 2 to K = 5 with samples from the four regions of Sudan.
Multiplicity of Infection
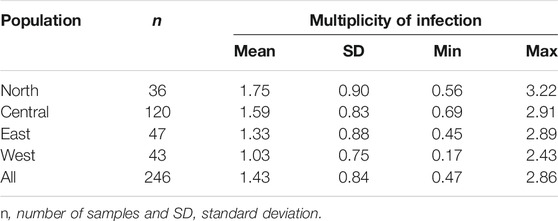
Multiple genotypes were observed in T. annulata populations from the four geographic regions, with multiple alleles being found at one or more loci. The mean number of alleles for the nine loci was determined for each sample, to obtain an index value that denoted multiplicity of infection. Table 5 summarizes the multiplicity of infection for each population and across all four populations analyzed. North and central populations had high mean of values of 1.75 and 1.59, respectively, while east and west had values of 1.33 and 1.03, respectively.
Discussion
Using microsatellite markers, this study investigated the diversity and population structure of T. annulata in Sudan. The study’s samples (n = 246) were obtained from four different geographical regions. North and central regions known to be endemic of T. annulata since the eighties, and the remaining two (west and east) witnessed the spreading of the disease in the nineties. In addition, the T. annulata vaccine from Sudan was also included in the study. In order to gain insight into the epidemiology of a parasite, ascertain sources of infection and modes of transmission, it is critical to assess population, genetic diversity and structure (Weir et al., 2007, 2011; Salih et al., 2018).
The genetic diversity and population structure of T. annulata found in Sudan were studied using a panel of nine microsatellite markers. The highest mean genetic diversity was observed in north, a finding which could be due to significant tick infestations in this region, where the disease has been established for long time (El Hussein, et al., 2012; Gharbi et al., 2020). The lower degree of T. annulata diversity detected in the parasite population from the west corresponded to the recent reported of tropical theileriosis (Mohammed-Ahmed et al., 2020). In other countries where tropical theileriosis is endemic, a comparable scale of genetic variation has been observed among T. annulata populations (Weir et al., 2011; Al-Hamidhi et al., 2015; Gomes et al., 2016; Yin et al., 2018; Roy et al., 2021).
The results revealed relatively slight geographical sub-structuring among the four populations of T. annulata in Sudan with no evidence of grouping based on geographical origin. The fact that resources (feeds and water) are collectively utilized under the nomadic cattle systems prominent in Sudan is essential to enhance genetic uniformity. This result is supported by PCA analysis as well as STRUCRURE results. AMOVA revealed a high percentage of crossing between various T. annulata samples as well as recombination within the parasite population. Individual samples, rather than groups derived from a specific geographic region, accounted for the majority of genetic variation. In the future, other aspects such as parasite challenge and quantifying the extent of tick infestation should be examined. PCA and AMOVA results figured out no evident link between population genetic structure and the geographical origin of the isolates investigated. However, PCA analysis revealed a close genetic link between the north and T. annulata vaccine genotypes, with the T. annulata vaccine and majority of north genotypes clustered together.
When the PCA and STRUCTURE data are combined, it can be expected that there are three potential populations of T. annulata in Sudan. It’s possible that gene pool 1 is introgressing into gene pool 2 or vice versa, with the two gene pools will eventually merging into one. This conclusion could be a result of cattle migration being unfettered across the country, due to the lack of trade barriers and policies restricting livestock movement (Oura et al., 2005; Roy et al., 2019). The mobility of parasite-infected/tick-infested cattle from one region to another assists in population homogenization.
The extent of linkage equilibrium between alleles at pairs of loci was evaluated, to see if the T. annulata populations in the four regions of Sudan constituted a single panmictic population with a high degree of genetic exchange. When the samples from the four regions were analyzed as a single population, an ISA value of 0.0174 was obtained as well as a VD value (2.1006) that was greater than L (1.9591), demonstrating LE. The presence of LE in the combined populations could be due to an epidemic population structure (Smith et al., 1993), or it could be due to occasional genetic exchange, resulting in a clonal population structure (Wier et al., 2011). Other factors that could contribute to the reported LE include inbreeding, recombination rate and the size of the regional parasite functional population (Charlesworth, 2009). More samples from Sudan are needed to clarify which characteristics are most essential, especially because a limited number of genetically identical parasites in the vertebrate host could result in substantial linkage disequilibrium (Anderson et al., 2000).
The highest level of multiplicity of infection (MOI) was identified in the north, with a highest value of 3.22, followed by east, with lowest and maximum values of 0.45 and 2.89, respectively showing a significant degree of variability in the dataset. In the midgut of the tick vector, multiple infections stimulate cross-mating and recombination among distinct parasite genotypes, as well as the formation of unique recombinant genotypes (Weir et al., 2001; Al-Hamidhi et al., 2015; Salih et al., 2018). The higher number of T. annulata genotypes in north could enable a high rate of cross-mating and recombination, resulting in increased genetic diversity in the bovine host (Conway et al., 1999). It could be also explained by the high tick load reported in north compared to the other regions (Salih et al., 2004).
In conclusion, the application of polymorphic microsatellite loci has offered preliminary insight into the population genetic diversity and structure of T. annulata population in Sudan. Extensive genetic intermixing between the four T. annulata populations studied was indicated, as well as minimal evidence of genetic differentiation and a high level of genetic diversity within each population. The findings show that the vaccine (Atbara strain) could be used in all areas where tropical theileriosis present.
T. annulata populations found in north African countries where tropical theileriosis is currently an economically important disease, should be examined and compared to see how genetically similar they are. Such data can assist veterinary control policy makers in determined if preventative measures, such as immunization, should be deployed at the national, regional or continental level.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by ILRI IACUC (The ILRI’s Institutional Animal Care and Use Committee). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
DS study design, conceptualization, data curation, data analysis, visualization, writing original draft, AA helped in sample collection, data curation MN Methodology, KT helped in sample collection, review manuscript, MM helped in sample collection, JM data analysis, review manuscript NM-K data analysis, AE conceptualization, supervision, manuscript review and editing, RB conceptualization, supervision, RS conceptualization, supervision, funding acquisition. All authors have read and approved the final version of the manuscript.
Funding
We gratefully acknowledge the financial support provided to the Biosciences eastern and central Africa Hub at the International Livestock Research Institute (BecA-ILRI Hub, Nairobi) by the Australian Agency for International Development (AusAID) through a partnership between Australia’s Commonwealth Scientific and Industrial Research Organization (CSIRO) and the BecA-ILRI Hub; and by the Syngenta Foundation for Sustainable Agriculture (SFSA); the Bill and Melinda Gates Foundation (BMGF); and the Swedish Ministry of Foreign Affairs through the Swedish International Development Agency (Sida), which made this work possible. DAS was a recipient of an Africa Biosciences Challenge Fund (ABCF) Fellowship. The work was also supported in part by a grant from the DFG (Germany-African Cooperation; “Molecular epidemiology network for promotion and support of delivery of live vaccines against T. parva and T. annulata infection in Eastern and Northern Africa” (SE862/2-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.742808/full#supplementary-material
Supplementary Table 1 | Primers information used.
References
Al-Hamidhi, S., H. Tageldin, M., Weir, W., Al-Fahdi, A., Johnson, E. H., Bobade, P., et al. (2015). Genetic Diversity and Population Structure of Theileria Annulata in Oman. PLoS One 10, e0139581. doi:10.1371/journal.pone.0139581
Allsopp, B. A., Baylis, H. A., Allsoppi, M. T. E. P., Cavalier-smith, T., Bishop, R. P., Carrington, D. M., et al. (1993). Discrimination between Six Species of Theileria Using Oligonucleotide Probes Which Detect Small Subunit Ribosomal RNA Sequences. Parasitology 107, 157–165. doi:10.1017/s0031182000067263
Anderson, T. J. C., Haubold, B., Williams, J. T., Estrada-Franco§, J. G., Richardson, L., Mollinedo, R., et al. (2000). Microsatellite Markers Reveal a Spectrum of Population Structures in the Malaria Parasite Plasmodium Falciparum. Mol. Biol. Evol. 17, 1467–1482. doi:10.1093/oxfordjournals.molbev.a026247
Beniwal, R. K., Nichani, A. K., Sharma, R. D., Rakha, N. K., Suri, D., and Sarup, S. (1997). Responses in Animals Vaccinated with theTheileria Annulata (Hisar) Cell Culture Vaccine. Trop. Anim. Health Prod. 29 (4Suppl. l), 109S–113S. doi:10.1007/BF02632947
Charlesworth, B. (2009). Effective Population Size and Patterns of Molecular Evolution and Variation. Nat. Rev. Genet. 10, 195–205. doi:10.1038/nrg2526
Conway, D. J., Roper, C., Oduola, A. M. J., Arnot, D. E., Kremsner, P. G., Grobusch, M. P., et al. (1999). High Recombination Rate in Natural Populations of Plasmodium Falciparum. Proc. Natl. Acad. Sci. 96, 4506–4511. doi:10.1073/pnas.96.8.4506
Darghouth, M. A. (2008). Review on the Experience with Live Attenuated Vaccines against Tropical Theileriosis in Tunisia: Considerations for the Present and Implications for the Future. Vaccine 26 (Suppl. 6), G4–G10. doi:10.1016/j.vaccine.2008.09.065
Dolan, T. T. (1989). Theileriasis : a Comprehensive Review. Rev. Sci. Tech. OIE 8 (1), 11–78. doi:10.20506/rst.8.1.398
Earl, D. A., and vonHoldt, B. M. (2012). STRUCTURE HARVESTER: a Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conservation Genet. Resour. 4, 359–361. doi:10.1007/s12686-011-9548-7
El Hussein, A. M., Hassan, S. M., and Salih, D. A. (2012). Current Situation of Tropical Theileriosis in the Sudan. Parasitol. Res. 111, 503–508. doi:10.1007/s00436-012-2951-5
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the Number of Clusters of Individuals Using the Software Structure: a Simulation Study. Mol. Ecol. 14, 2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi:10.1111/j.1755-0998.2010.02847.x
Gharbi, M., Darghouth, M. A., Elati, K., AL‐Hosary, A. A. T., Ayadi, O., Salih, D. A., et al. (2020). Current Status of Tropical Theileriosis in Northern Africa: A Review of Recent Epidemiological Investigations and Implications for Control. Transbound Emerg. Dis. 67 (Suppl. 1), 8–25. doi:10.1111/tbed.13312
Ghosh, S., and Azhahianambi, P. (2007). Laboratory Rearing of Theileria Annulata-free Hyalomma Anatolicum Anatolicum Ticks. Exp. Appl. Acarol. 43, 137–146. doi:10.1007/s10493-007-9100-3
Gomes, J., Salgueiro, P., Inácio, J., Amaro, A., Pinto, J., Tait, A., et al. (2016). Population Diversity of Theileria Annulata in Portugal. Infect. Genet. Evol. 42, 14–19. doi:10.1016/j.meegid.2016.04.023
Hashemi-Fesharki, R. (1988). Control of Theileria Annulata in Iran. Parasitol. Today 4 (2), 36–40. doi:10.1016/0169-4758(88)90062-2
Haubold, B., and Hudson, R. R. (2000). LIAN 3.0: Detecting Linkage Disequilibrium in Multilocus Data. Bioinformatics 16, 847–849. doi:10.1093/bioinformatics/16.9.847
Idury, R. M., and Cardon, L. R. (1997). A Simple Method for Automated Allele Binning in Microsatellite Markers. Genome Res. 7, 1104–1109. doi:10.1101/gr.7.11.1104
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a Cluster Matching and Permutation Program for Dealing with Label Switching and Multimodality in Analysis of Population Structure. Bioinformatics 23, 1801–1806. doi:10.1093/bioinformatics/btm233
Mohammed-Ahmed, G. M., Hassan, S. M., El Hussein, A. M., and Salih, D. A. (2018). Molecular, Serological and Parasitological Survey of Theileria Annulata in North Kordofan State, Sudan. Vet. Parasitol. Reg. Stud. Rep. 13, 24–29. doi:10.1016/j.vprsr.2018.03.006
Nei, M. (1978). Estimation of Average Heterozygosity and Genetic Distance from a Small Number of Individuals. Genetics 89, 583–590. doi:10.1093/genetics/89.3.583
Ouhelli, H., Kachani, M., Flach, E., Williamson, S., El Hasnaoui, M., and Spooner, R. (1997). Investigations on Vaccination against Theileriosis in Morocco. Trop. Anim. Health Prod. 29, 103S. doi:10.1007/bf02632945
Oura, C. A. L., Asiimwe, B. B., Weir, W., Lubega, G. W., and Tait, A. (2005). Population Genetic Analysis and Sub-structuring of Theileria Parva in Uganda. Mol. Biochem. Parasitol. 140, 229–239. doi:10.1016/j.molbiopara.2004.12.015
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research-Aan Update. Bioinformatics 28, 2537–2539. doi:10.1093/bioinformatics/bts460
Peakall, R., and Smouse, P. E. (2006). Genalex 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 6, 288–295. doi:10.1111/j.1471-8286.2005.01155.x
Pipano, E., Samish, M., Kriegel, Y., and Yeruham, I. (1981). Immunization of Friesian Cattle against Theileria Annulata by the Infection-Treatment Method. Br. Vet. J. 137 (4), 416–420. doi:10.1016/s0007-1935(17)31640-8
Pipano, E., and Shkap, V. (2006). Vaccination against Tropical Theileriosis. Ann. N. Y. Acad. Sci. 916, 484–500. doi:10.1111/j.1749-6632.2000.tb05328.x
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of Population Structure Using Multilocus Genotype Data. Genetics 155, 945–949.
Rosenberg, N. A. (2004). DISTRUCT: A Program for the Graphical Display of Population Structure. Mol. Ecol. Notes 4, 137–138.
Roy, S., Bhandari, V., Barman, M., Kumar, P., Bhanot, V., Arora, J. S., et al. (2021). Population Genetic Analysis of the Theileria Annulata Parasites Identified Limited Diversity and Multiplicity of Infection in the Vaccine from India. Front. Microbiol. 11, 579929. doi:10.3389/fmicb.2020.579929
Roy, S., Bhandari, V., Dandasena, D., Murthy, S., and Sharma, P. (2019). Genetic Profiling Reveals High Allelic Diversity, Heterozygosity and Antigenic Diversity in the Clinical Isolates of the Theileria Annulata from India. Front. Physiol. 10, 673. doi:10.3389/fphys.2019.00673
Sager, H., Bertoni, G., and Jungi, T. W. (1998). Differences between B Cell and Macrophage Transformation by the Bovine Parasite, Theileria Annulata: a Clonal Approach. J. Immunol. 1161 (1), 335–341.
Salih, D. A., Hassan, S. M., El Hussein, A. M., and Jongejan, F. (2004). Preliminary Survey of Ticks (Acari: Ixodidae) on Cattle in Northern Sudan. Onderstepoort J. Vet. Res. 71, 319–326. doi:10.4102/ojvr.v71i4.252
Salih, D. A., Mwacharo, J. M., Pelle, R., Njahira, M. N., Odongo, D. O., Mbole-Kariuki, M. N., et al. (2018). Genetic Diversity and Population Structure of Theileria Parva in South Sudan. Ticks Tick-borne Dis. 9, 806–813. doi:10.1016/j.ttbdis.2018.03.002
Sivakumar, T., Hayashida, K., Sugimoto, C., and Yokoyama, N. (2014). Evolution and Genetic Diversity of Theileria. Infect. Genet. Evol. 27, 250–263.
Smith, J. M., Smith, N. H., O'Rourke, M., and Spratt, B. G. (1993). How Clonal Are Bacteria? Proc. Natl. Acad. Sci. 90, 4384–4388. doi:10.1073/pnas.90.10.4384
Stepanova, N. I., Zablotskii, V. T., Çakmak, A., Inci, A., Yukari, B. A., Vatanserver, Z., et al. (1989). Bovine Theileriosis in the USSR. Rev. Sci. Tech. OIE 8 (1), 89–92. doi:10.20506/rst.8.1.396
Taha, K. M., Salih, D. A., Ali, A. M., Omer, R. A., and El Hussein, A. M. (2013). Naturally Occurring Infections of Cattle with Theileria Lestoquardi and Sheep with Theileria Annulata in the Sudan. Vet. Parasitol. 191, 143–145. doi:10.1016/j.vetpar.2012.08.003
Viseras, J., García-Fernández, P., and Adroher, F. J. (1997). Isolation and Establishment in In Vitro Culture of a Theileria Annulata - Infected Cell Line from Spain. Parasitol. Res. 83 (4), 394–396. doi:10.1007/s004360050270
Weir, W., Ben-Miled, L., Karagenç, T., Katzer, F., Darghouth, M., Shiels, B., et al. (2007). Genetic Exchange and Sub-structuring in Theileria Annulata Populations. Mol. Biochem. Parasitol. 154 (2), 170–180. doi:10.1016/j.molbiopara.2007.04.015
Weir, W., Karagenç, T., Gharbi, M., Simuunza, M., Aypak, S., Aysul, N., et al. (2011). Population Diversity and Multiplicity of Infection in Theileria Annulata. Int. J. Parasitol. 41, 193–203. doi:10.1016/j.ijpara.2010.08.004
Yin, F., Liu, Z., Liu, J., Liu, A., Salih, D. A., Li, Y., et al. (2018). Population Genetic Analysis of Theileria Annulata from Six Geographical Regions in China, Determined on the Basis of Micro- and Mini-Satellite Markers. Front. Genet. 9 (9), 50. doi:10.3389/fgene.2018.00050
Keywords: cattle, cell culture vaccine, Theileria annulata, genotyping, population genetics, sub-structure, Sudan
Citation: Salih DA, Ali AM, Njahira M, Taha KM, Mohammed MS, Mwacharo JM, Mbole-Kariuki N, El Hussein AM, Bishop R and Skilton R (2021) Population Genetic Analysis and Sub-Structuring of Theileria annulata in Sudan. Front. Genet. 12:742808. doi: 10.3389/fgene.2021.742808
Received: 16 July 2021; Accepted: 25 October 2021;
Published: 19 November 2021.
Edited by:
Moses Okpeku, University of KwaZulu-Natal, South AfricaReviewed by:
Liang Wang, Institut Pasteur of Shanghai (CAS), ChinaAleksandr G. Bulaev, Federal Center Research Fundamentals of Biotechnology (RAS), Russia
Copyright © 2021 Salih, Ali, Njahira, Taha, Mohammed, Mwacharo, Mbole-Kariuki, El Hussein, Bishop and Skilton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diaeldin A. Salih, ZGlhZWxkaW4yMDAwQGhvdG1haWwuY29t
†Present address: Moses Njahira, International Centre of Insect Physiology and Ecology (icipe), Nairobi, Kenya; Joram M. Mwacharo, International Centre for Agricultural Research in the Dry Areas (ICARDA), Addis Ababa, Ethiopia; Richard Bishop, Department of Veterinary Microbiology and Pathology, Washington State University, Pullman, WA, United States; Robert Skilton, International Centre of Insect Physiology and Ecology (icipe), Nairobi, Kenya
 Diaeldin A. Salih
Diaeldin A. Salih Awadia M. Ali3
Awadia M. Ali3 Joram M. Mwacharo
Joram M. Mwacharo Abdelrhim M. El Hussein
Abdelrhim M. El Hussein Richard Bishop
Richard Bishop