- 1Department of Biotechnology, University of Agricultural Sciences, Dharwad, India
- 2Department of Food Science and Nutrition, University of Agricultural Sciences, Dharwad, India
- 3Department of Microbiology, Cornell University, Ithaca, NY, United States
- 4President, International Phytomedomics and Nutriomics Consortium (ipnc.info), Daejeon, South Korea
- 5New Mexico State University-Agricultural Science Center at Clovis, New Mexico, NM, United States
Functional foods are natural products of plants that have health benefits beyond necessary nutrition. Functional foods are abundant in fruits, vegetables, spices, beverages and some are found in cereals, millets, pulses and oilseeds. Efforts to identify functional foods in our diet and their beneficial aspects are limited to few crops. Advances in sequencing and availability of different omics technologies have given opportunity to utilize these tools to enhance the functional components of the foods, thus ensuring the nutritional security. Integrated omics approaches including genomics, transcriptomics, proteomics, metabolomics coupled with artificial intelligence and machine learning approaches can be used to improve the crops. This review provides insights into omics studies that are carried out to find the active components and crop improvement by enhancing the functional compounds in different plants including cereals, millets, pulses, oilseeds, fruits, vegetables, spices, beverages and medicinal plants. There is a need to characterize functional foods that are being used in traditional medicines, as well as utilization of this knowledge to improve the staple foods in order to tackle malnutrition and hunger more effectively.
Introduction
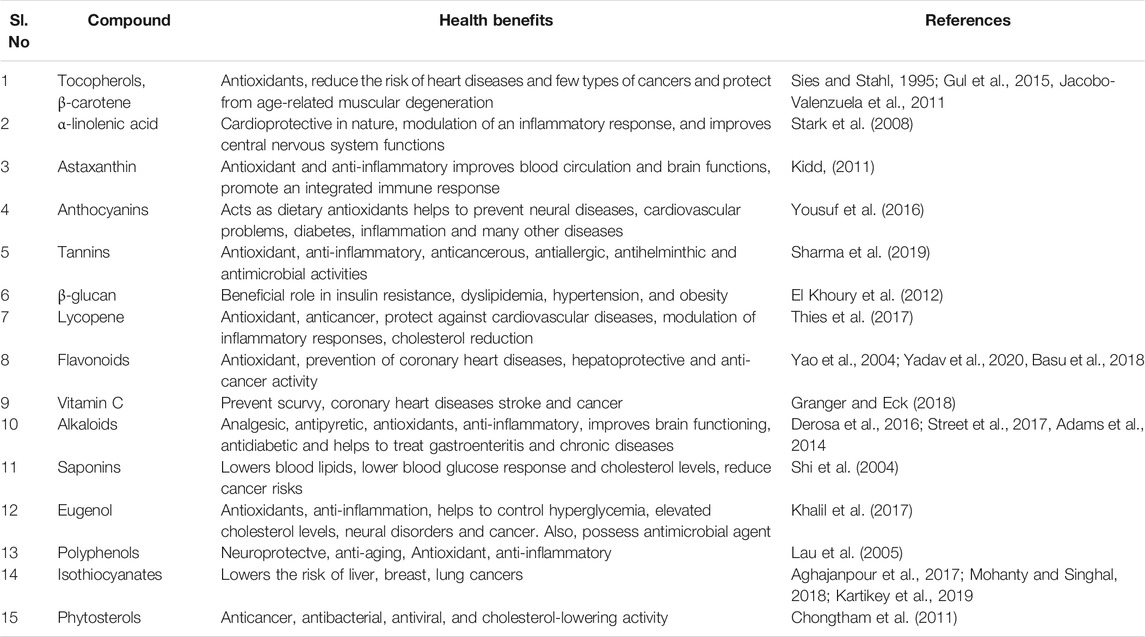
To address global food and nutritional security, there is a need to increase the agricultural production and nutritive value of food. Assured access to nutritionally adequate and safe food is essential for attaining the nutritional security. With urbanization and changing food habits, “smart foods with higher nutrition per bite” is the need. The awareness of utilization of these foods for prevention and treatment of certain diseases prompted the researchers to discover active compounds that render health benefits. Foods that have an additional physiological benefits besides providing basic nutritional needs were first referred to as “functional foods” in Japan in the mid-1980s. Broadly, functional foods can be categorized according to the active components that have health benefits. Based on their origin, they can be classified as naturally derived products (plant or animal sources) or synthetic products (synbiotics, nutraceuticals) (Mohanty and Singhal, 2018). The functional products from plant origin include phytochemicals such as polyphenolic compounds, alkaloids, flavonoids, carotenoids, saponins, allyl sulfides, catechins, nutraceuticals, etc. (Table 1; Figure 1). There are clear evidences from epidemiological studies and clinical trials that a plant-based diet can reduce the risk of chronic diseases and disorders such as cancer (Velmurugan et al., 2005; Aghajanpour et al., 2017; Sayeed et al., 2017), diabetes (Hannan et al., 2007; Ballali and Lanciai, 2012; Alkhatib et al., 2017), obesity (Hill and Peters, 2002; Riccardi et al., 2005; Baboota et al., 2013), cardiovascular ailments (Alissa and Ferns, 2012; Hamid and Abd Hamid, 2019) and other effects on human health (Lobo et al., 2010). Most of the functional foods with scientific supporting evidence are the native/familiar foods that were used in traditional medicine for generations (Hasler, 1998; Fokunang et al., 2011; Abbott, 2014).
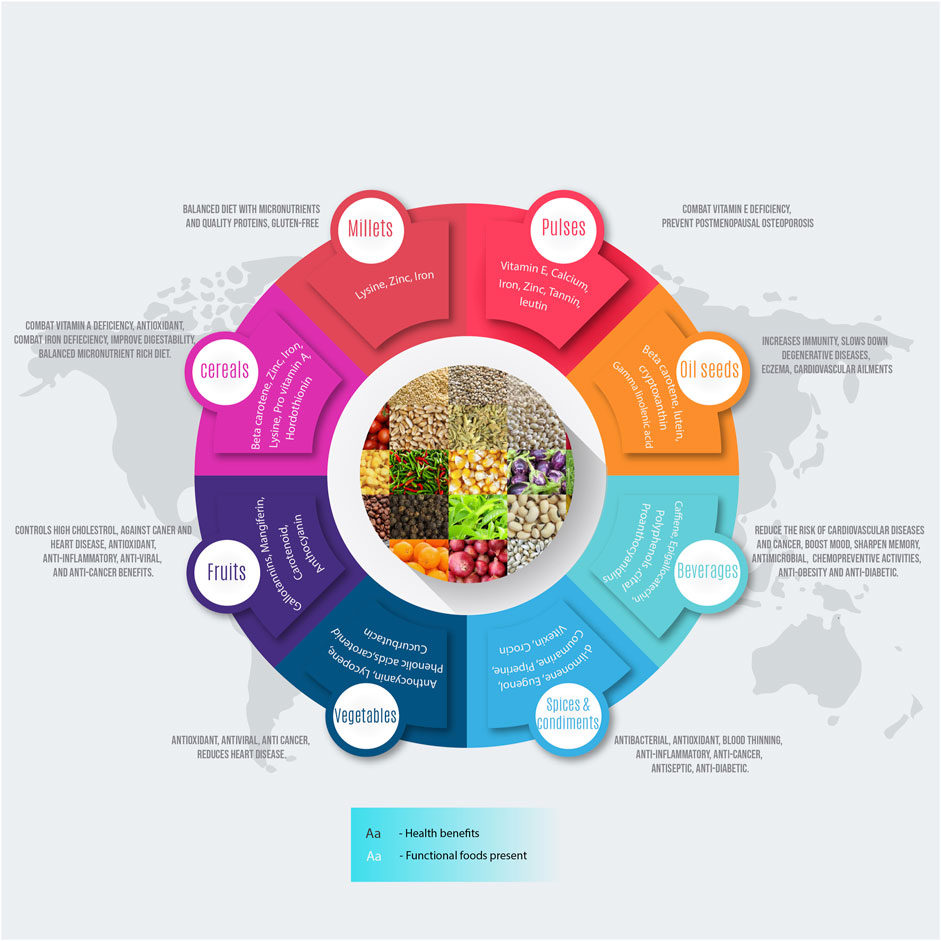
FIGURE 1. An infographics showing the functional components of the food and their potential health benefits to human beings.
The scientific advances and next generation sequencing technologies available in recent years have impacted significantly on crop breeding and food science (Varshney et al., 2009; Kato et al., 2011). There is a lot of scope to utilize these technologies to understand the functional compounds that have health benefits and to improve the crops with respect to its nutritional status along with productivity related traits. Utilization of different omics technologies in research related to food and nutrition with the objective of improving the human health and well-being is referred as foodomics (Capozzi and Bordoni, 2013). An integrated use of omics technologies approaches to increase the nutrient potential of any crop, further applications in food processing and formulations can influence the nutritional security to greater extent (Bagchi et al., 2015; Tian et al., 2016).
The omics discipline comprises of four major broad areas like genomics, transcriptomics, proteomics and metabolomics. Integrated use of the omics technologies provides a holistic approach to study the systems biology (Pazhamala et al., 2021). Genomics includes the sequencing of whole genomes, assembly and annotation of the sequences, study of the genes, identification and development of molecular markers and quantitative trait loci (QTLs) for target traits, genomics assisted breeding, genomic selection, etc. (Varshney et al., 2005; Kole et al., 2015; McGuire et al., 2020). Transcriptomics deals with the dynamic expression of gene products in specific tissue at particular stage. The study of differential expression is quantified by using different molecular biology tools such as RNA sequencing, microarrays, Serial analysis of Gene Expression (SAGE), qRT-PCR, etc. While microarray, SAGE and qRT-PCR technologies determine the abundance of defined transcripts, the RNA-sequencing utilizes the advantage of high-throughput sequencing to identify the novel transcripts (Lowe et al., 2017). Proteomics can be effectively used to study protein structure, function, and interaction with other proteins or ligands such as bioactive compounds. Advanced techniques like Matrix-assisted laser desorption/ionization Time of flight (MALDI-TOF) and Liquid chromatography coupled to mass spectrometry (LC-MS) are able to detect expression of specific proteins. Metabolomics identifies and quantifies specific metabolites present in a sample. Metabolomics can be beneficial for quantification of biologically active compounds, food fingerprinting, and food profiling. Techniques like Gas chromatography coupled to mass spectrometry (GC-MS), Liquid chromatography coupled to mass spectrometry (LC-MS), Inductive couple plasma (ICP), nuclear magnetic resonance (NMR), Near infrared spectrometry (NIR) have been used for characterization of metabolites (Prakash et al., 2018; Kumar et al., 2019). Besides these omics approaches, genome editing tools like RNAi, CRISPR/Cas9, TALENs, ZFNs can be utilized to improve the crop plants. Use of computational and bioinformatics tools is indispensable while using all the above mentioned technologies. Advances in data science with applications of artificial intelligence and machine learning has enabled deep learning of the data for better understanding of the biological processes and crop prediction modelling in genomic selections (Figure 2). In this review, we discuss about the utilization of omics technologies in determining and enhancing the active food compounds in major crop plants including cereals, millets, pulses, oil seeds, fruits, vegetables, spices and medicinal plants.
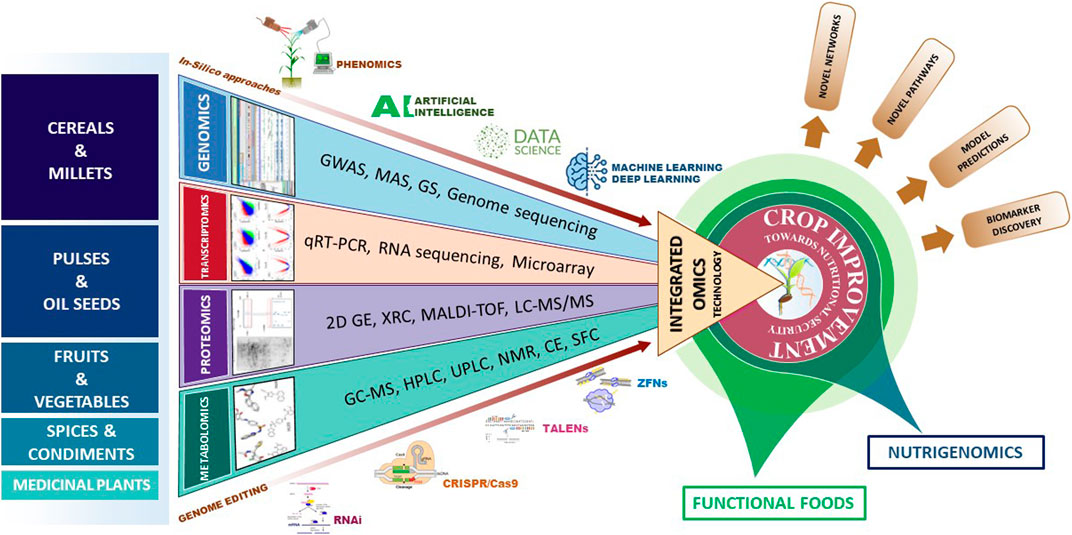
FIGURE 2. Integrated omics approaches for enhancing functional foods. In this figure, the crop categories specified are orderly arranged in descending manner with respect to the utilization of omics technologies to improve functional foods. Abbreviations: GWAS, Genome-Wide Association Studies; MAS, Molecular Assisted Selection; GS, Genomic Selection; qRT-PCR, Quantitative Real Time polymerase Chain Reaction; 2D GE, 2-Dimensional Gel electrophoresis; XRC, X-Ray Crystallography; MALDI-TOF, Matrix-Assisted Laser Desorption/Ionization-Time Of Flight; LC-MS/MS, Liquid Chromatography—Mass Spectrophotometry; GC-MS, Gas Chromatography—Mass Spectrophotometry, HPLC: High Performance Liquid Chromatography; UPLC, Ultra Performance Liquid Chromatography; NMR, Nuclear Magnetic Resonance; CE, Capillary Electrophoresis and SFC, Supercritical Fluid Chromatography; RNAi, RNA interference; TALENs, Transcription Activator-Like Effector Nucleases; ZFNs, Zinc Finger Nucleases.
Cereals
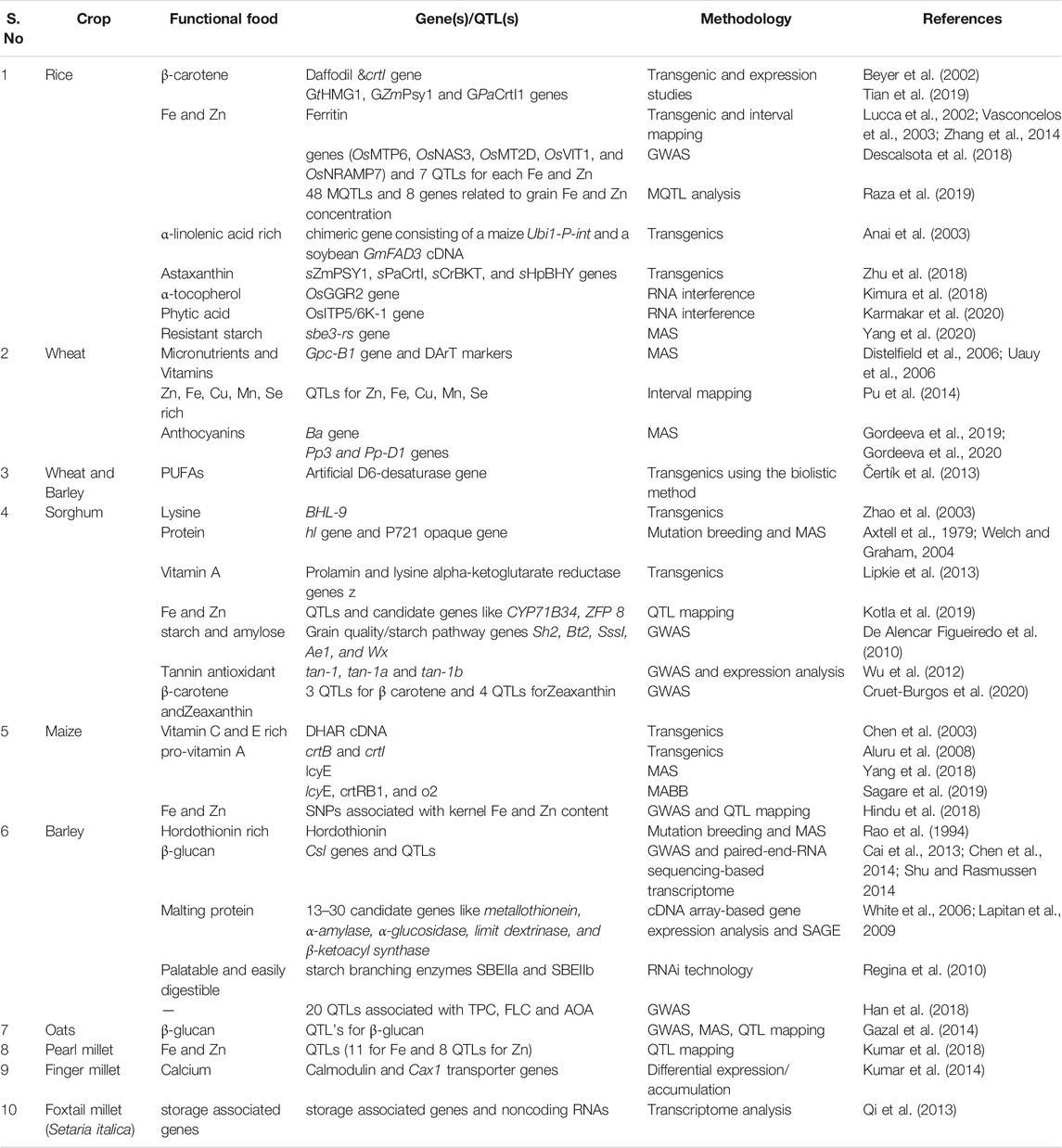
Cereals are the major part of our daily diet and source of carbohydrates but lack an adequate amount of nutrition in terms of vitamins, and essential amino acids (Munck, 1972). Hence there is a need to improve the quality and nutritional parameters of cereals. Recent advances in genomics and genetic engineering are useful in targeted improvements especially by improving the quality and nutritional value in crop plants (Sedeek et al., 2019). Several omics technologies have been used to improve rice, wheat, barley especially for disease resistance and improving the yield of the crops (Zenda et al., 2021). However, there are only a few reports related to deciphering the functional compounds in cereal crops using modern biotechnological tools (Table 2). For instance, rice is improved with higher carotenoid content leading to increased Vitamin A (Dubock, 2019) and biofortified with micronutrients like Fe and Zn (Welch and Graham, 2004; Trijatmiko et al., 2016). A genetic engineering approach was successfully used to develop “Golden Rice” with significant levels of β-carotene that will help to combat vitamin A deficiency. Ye et al. (2000) and Shao et al. (2011) reported the marker loci/QTLs underlying the naturally occurring variations of grain color and nutritional quality traits in 416 rice germplasm accessions, including 361 white rice, 50 red rice, and six black rice across 41 marker loci. These markers could be further used for marker-assisted breeding to improve rice for nutritional qualities. The efforts were also made to dissect the nutrient traits especially Fe, Zn and anthocyanin content using genome-wide association studies on diversity panel consisting of 156 accessions of colored rice (Descalsota-Empleo et al., 2019). QTLs for functional components like phenolic content, flavonoid content and antioxidant capacity were identified using 127 double haploid lines developed through anther culture (Jin et al., 2009). Genome editing tools like CRISPR-Cas9 have also been utilized to enhance the amylose content (Sun Y. et al., 2017b). There are also efforts to develop fragrant rice by knocking out of betaine aldehyde dehydrogenase (BADH2) gene using TALEN technology (Shan et al., 2015).
Similar to rice, there are many reports to utilize the omics approaches in wheat to improve the nutrition and functional components. To list a few, an enriched wheat with high vitamin A content was developed by transforming the two bacterial carotenoid biosynthetic genes CrtB and CrtI into wheat cultivar Bobwhite (Wang et al., 2014). In addition to this, candidate genes involved in carotenoid biosynthesis and catabolism have been elucidated using GWAS studies in wheat (Colasuonno et al., 2017). Genomic regions for the color, carotenoids, and polyphenol oxidase activity of flour in wheat have been studied using linkage-based QTL analysis (Zhao et al., 2013). Hussain et al. (2017) reported QTLs for several nutrients, including Zn, Fe, Mn, Cu, Ca, Mg, etc. under saline conditions. In wheat, candidate genes for enhancing the grain Zn content have been identified by GWAS using high-density genotyping arrays on 369 wheat genotypes (Alomari et al., 2018). Genetic improvement in grain quality and micronutrients has been instrumental in quality breeding for wheat (Distelfeld et al., 2006; Balyan et al., 2013; Pu et al., 2014). Apart from this, there is also a need to utilize genomics approaches to decrease heavy metal (for example, Cadmium) uptake (Knox et al., 2009) and improve digestibility with reduced flatulence (Sharma et al., 2002). Genetically modified maize and wheat have showed increased accumulation of folate (Vitamin B9) levels (Liang et al., 2019).
Many economically backward countries rely on crops such as sorghum and maize as their staple food. In maize, biofortification with micronutrients like Zn and Fe (Zhao, 2007), enhanced β-carotene (Muthusamy et al., 2014), and amino acids like Lysine (Mertz et al., 1964; Shetti et al., 2020) have been carried out to ensure the nutritional security. To dissect the genomic regions for various metabolites in maize (Zea mays), a metabolome-based GWAS was carried out (Zhou et al., 2019). An integrated omics-based mapping to unravel flavonoid biosynthesis was also attempted in maize (Jin et al., 2017). There have been extensive efforts to breed for quality protein maize (QPM) with nearly as twice lysine and tryptophan content than the usual (Gibbon and Larkins, 2005). In addition to this, a transgenic approach has been used to increase protein by reducing zein content (Huang et al., 2006), and increasing Provitamin A content (Aluru et al., 2008). CRISPR-Cas9 and TALEN approaches have been used in maize to reduce phytic acid content, a food inhibitor that chelates micronutrients and prevents their bioavailability for mono gastric animals, including humans (Liang et al., 2014).
The reports related to the use of omics approaches to enhance functional compounds in barley (Hordeum vulgare) and oats (Avena sativa) are limited as compared to major cereals. In barley, the β-glucan content greatly improves the malting properties and its presence has been found to increase palatability (Chen et al., 2014). A gene expression study using SAGE analysis identified six proteins associated with the malting property (White et al., 2006). Other functional compounds like total polyphenols, flavonoids, and antioxidant properties were studied in 67 cultivated and 156 Tibetan wild barley accessions using GWAS (Han et al., 2018). In oats, a GWAS study was conducted in a global germplasm collection to identify molecular markers associated with β-glucan content (Newell et al., 2012; Gazal et al., 2014).
Sorghum and Millets
Sorghum and millets are small-grained cereals and seed grasses that are traditional staple foods in African and Asian countries. In recent times, sorghum and millets are being utilized as an alternative to major cereals because of their higher nutritional, mineral, dietary fiber content along with climate-resilient nature. Besides, they are gluten-free and play a pivotal role in preventing and curing several lifestyle health issues like diabetes (Anitha et al., 2021). The genetic and genomic resources have been developed in some of the small millets (Vetriventhan et al., 2020) and efforts to utilize the genomic tools to improve the nutrient components are underway (Table 2). The nutraceutical property is mainly based on the kernel color in these crops. In this regard, QTL analysis of endosperm color and carotenoid (provitamin A) content in sorghum grains utilized in breeding high provitamin sorghum crop (Fernandez et al., 2008). Another effort using GWAS analysis with 404,628 SNP markers identified novel marker-trait association for polyphenols in a global diversity panel of 381 sorghum accessions (Rhodes et al., 2014).
There are limited efforts to utilize genomics tools in all other millets for improving the nutritional properties. The biofortification of millets seemed to be a good option for improving the nutritionally rich millets (Vinoth and Ravindhran, 2017). The QTLs controlling the content of micronutrients like Zn and Fe were identified in pearl millet (Kumar et al., 2018; Govindraj et al., 2019). Finger millet (Eleusine coracana) has been studied at various stages of growth and development using transcriptomics and was found to have high absorption and accumulation of calcium during grain development (Mirza et al., 2014). Glucosinolates in millets were found to reduce carcinogen-DNA interaction resulting in detoxification. Similarly, the isoflavones (phytoestrogens), genistein, and daidzein were found to reduce the incidence of many cancers, coronary heart diseases, and osteoporosis (Bandyopadhyay et al., 2017). The bioavailability of the micronutrients present in millets needs to be elucidated and utilized in crop improvement.
Pulses and Oilseeds
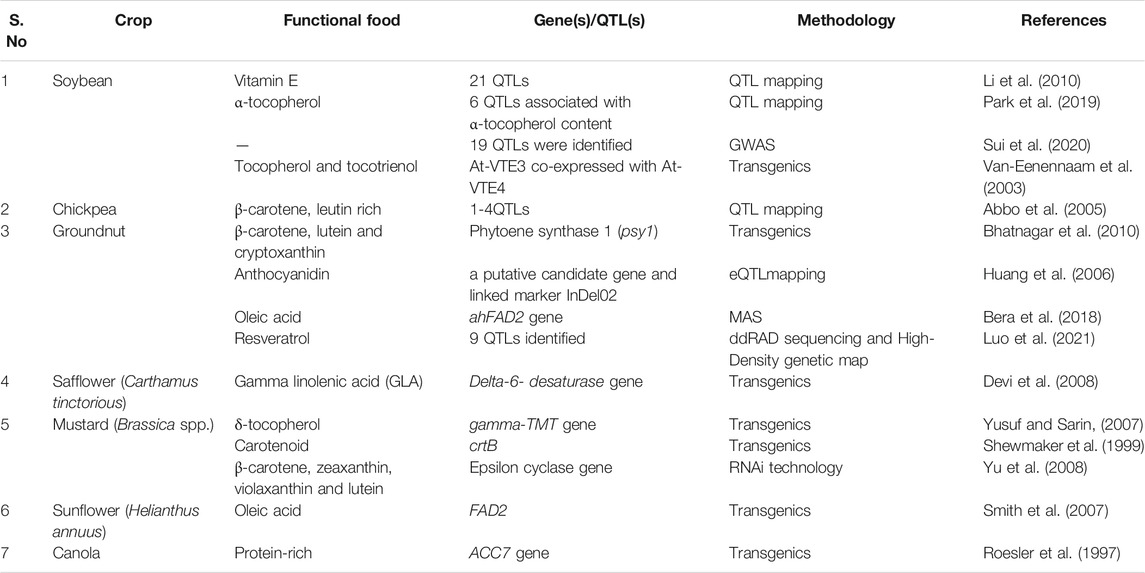
Pulses are a rich source of protein, with low fat, high fiber content and low glycemic index. Soluble fiber helps to decrease blood cholesterol levels and control blood sugar levels, and insoluble fiber helps with digestion. The biotechnological application for nutritional improvement mainly concentrates on enriching micronutrients and vitamins in pulses (Table 3). Pulses are known for the functional component saponins and several health benefits associated with them (Singh et al., 2017). Although pulses have been studied for several biotic and abiotic stresses at the molecular level, there are very few reports related to the genetic dissection of antioxidant activity and nutrition-related traits. Biofortification of pulses with Fe and Zn in lentils, chickpeas and field pea (Pisum sativum) has been carried out to address global malnutrition and micronutrient deficiencies (Thavarajah and Gupta, 2014). In chickpea, the GWAS study conducted in 94 diverse chickpea genotypes showed eight SNPs associated with Fe and Zn content in the seeds (Diapari et al., 2014). Similarly, a GWAS study in lentils identified two tightly linked SNP markers for Fe and Zn content (Khazaei et al., 2017).
Omega-3 fatty acids are considered to be essential for brain development, which is mainly available through oil seed crops in human diet. The areas of crop improvement in terms of nutrition in oilseeds rely on improving oil quality, resveratrol content and improved shelf life (Pandey et al., 2014; Qi et al., 2014; Shasidhar et al., 2017; Luo et al., 2021). Profiling of nutraceutical properties of 60 groundnut cultivars differentiating in kernel colors has been carried out and marker-trait association studies have been carried out (Nayak et al., 2020). The expression of phytoene synthase showed 50-fold increased levels of carotenoids in rapeseed using genetic engineering (Shewmaker et al., 1999). An increased expression of zeaxanthin, violaxanthin and lutein by targeting the downregulation of the epsilon cyclase gene using RNAi technology) has been reported in mustard (Yu et al., 2008). The molecular mapping and QTL analysis of flavonoid genes was also elucidated in rapeseed (Qu et al., 2016), soybean (Li et al., 2016), and groundnut (Mondal et al., 2015). Efforts are being carried out to use advanced biotechnological applications to improve oilseeds nutritionally for further crop improvement (Table 3).
Fruits
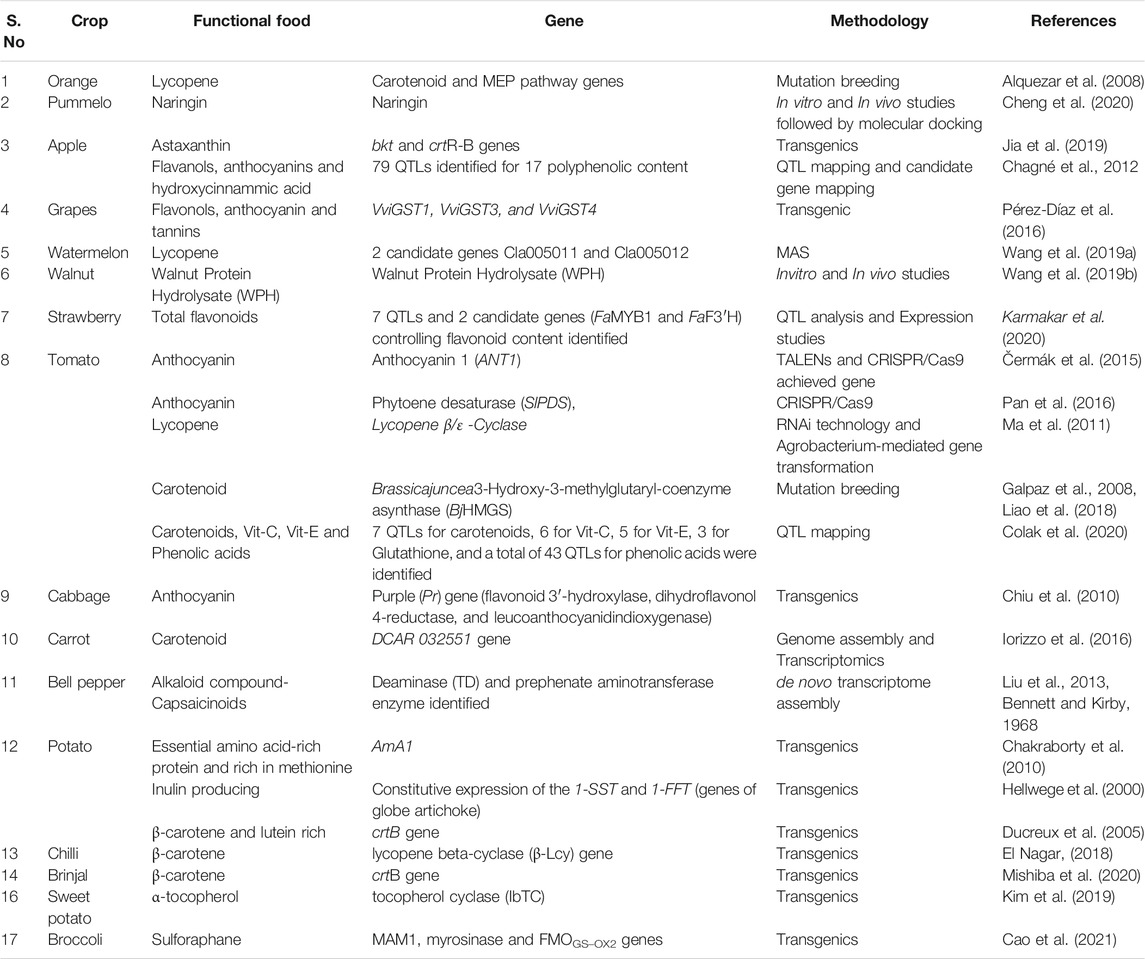
Fruits are promoted as functional foods as they are a rich source of several antioxidants, polyphenols, minerals, soluble fibers, vitamins especially C, A and E. They primarily consist of flavonoids including flavonols, flavones, isoflavones, flavanones and anthocyanins, and non-flavonoid polyphenolics including phenolic acids, lignans and stilbenes (Joy et al., 2018). Though fruits are the major source of functional foods, systematic experimental reports on the utilization of omics technologies to improve functional components are limited to a few fruit crops (Table 4).
In citrus, GWAS studies were conducted on 787 different citrus fruits using 1,841 SNP markers, and marker-trait associations were studied on fruit quality traits, including acid %, taste, and aroma (Minamikawa et al., 2017). Specific locus amplified fragment (SLAF) sequencing was performed over C. reticulata × P. trifoliata F1 pseudo testcross population and have constructed a high density integrated genetic map with 3,817 markers. This study has identified 17 significant QTLs of which three colocalized genomic regions were observed for multiple carotenoid constituents (Zheng et al., 2018). In another study, a navel orange (Citrus sinensis L. Osbeck) mutant (“Cara Cara”) was developed with bright red pulp with presence of lycopene (Alquezar et al., 2008). The expression analysis of genes involved in the carotenoid pathway using HPLC, northern hybridization, and RT-PCR indicated the increased accumulation of lycopene content in the mutant compared to navel orange. To elucidate the basis of lycopene accumulation in Cara Cara, the carotenoid profile and expression of three isoprenoids and nine carotenoid genes in flavedo and pulp of Cara Cara and Navel fruits throughout development and maturation were studied. The results indicated the accumulation of lycopene along with phytoene and phytofluene from early developmental stages in pulp as well as peel (Alquezar et al., 2008). Lemons are known for several functional components, including phenolics, vitamins, minerals, dietary fiber, essential oils and carotenoids (González-Molina et al., 2010). In the case of Sicilian blood oranges, retrotransposons were shown to induce seed-specific accumulation of anthocyanins during cold stress (Butelli et al., 2012). Edmunds et al. (2012) reported that the anti-inflammatory property of kiwifruit extract is due to the changes in the expression level of genes involved in the immune signaling pathway and metabolic processes using microarray technique.
The king of fruits “mango” (Mangifera indica) is a rich source of various polyphenolic compounds and is found in all the parts of the plant including pulp, peel, seed, bark, leaf, and flower. Mango polyphenols, especially mangiferin, acts as an antioxidant and has several health benefits (Masibo and He, 2008). The transcriptomics and proteomics studies in mango have predicted the involvement of genes involved in the anthocyanin biosynthesis pathway during the fruit development stage of mango (Wu et al., 2014). There is little effort towards the use of biotechnological approaches to improve the functional components of mango.
Red grapes are significant sources of anthocyanins, the main compounds responsible for the color of red grapes and wine (Mazza and Francis, 1995). Metabolite profiling of bioactive components of grapes especially flavonols, anthocyanins, and tannins indicated the presence of several bioactive compounds. The quercetin and kaempferol content was found to be greater in white grapes than red ones, but the red grapes were reservoirs of other bioactive components such as myricetin, laricitrin, syringetin and isorhamnetin (Mattivi et al., 2006). Resveratrol, an antioxidant that is known to lower blood pressure, and act as a chemopreventive with antiaging benefits are present in grapes. These flavonoids not only provide health benefits to humans but also help plants to fight against several biotic and abiotic stresses. For instance, transformation of bHLH transcription factor gene, VvbHLH1 from grapes into Arabidopsis, resulted in an increased accumulation of flavonoids and enhanced salt and drought tolerance (Wang et al., 2016).
In Japanese plum (Prunus salicina), the molecular marker associated with transcription factors found in the flavonoid pathway was used to study population diversity (González et al., 2016). Date palm (Phoenix dactylifera) fruits are composed of minerals (Se, Cu, K, and Mg), vitamins (C, A, B6, B9, B2, B3) besides being a good source of total phenolics and natural antioxidants (such as anthocyanins, ferulic acid). Phenolic compounds and selenium present in date fruit impart antioxidant activity (Guizani, 2013). Similarly, transcriptome sequencing in Indian gooseberry (Phyllanthus emblica) revealed the genes involved in flavonoid and vitamin C biosynthesis (Kumar et al., 2016). In many fruits, biotechnological approaches, including “omics” studies and use of molecular markers for trait mapping to improve bioactive components are very limited.
Vegetables
Among vegetables, most of the genomics studies have been carried out in tomatoes as this crop is considered to be one of the model plants in genetic transformation and other genomics studies. The most critical functional component present in the tomato is carotenoids, especially lycopene and anthocyanins. To obtain lycopene-rich tomatoes, the genes encoding lycopene β/ε-cyclase, responsible for the conversion of lycopene to carotenoid, were silenced using RNAi technology. Significant increases in lycopene content were observed in transgenic plants (Ma et al., 2011). A mutation breeding approach was also used to increase the carotenoid content of tomatoes by 30%. Abscisic acid-deficient mutants in tomatoes have been shown to increase the lycopene content (Galpaz et al., 2008). Further, vegetables rich in anthocyanins were developed by overexpression of specific genes of the carotenoid biosynthesis pathway that induced a purple color, especially in tomato and cauliflower (Brassica oleracea var. botrytis) (Gonzali et al., 2009; Chiu et al., 2010).
Genome-editing technologies, especially CRISPR-Cas9, has potential use in horticultural crops (Karkute et al., 2017). Recently, this technique was used to edit five genes that are involved in the carotenoid pathway to increase lycopene content by inhibiting the conversion from lycopene to β- and α-carotene in tomatoes that increased lycopene content by five-folds (Li et al., 2018). In another study, intense purple-colored tomato plants were obtained by overexpressing an Anthocyanin mutant 1 (ANT1) gene that encodes for Myb transcription factors using TALENs and CRISPR/Cas9 approaches (Čermák et al., 2015). Furthermore, phytoene desaturase (S1PDS), an essential enzyme in carotenoid biosynthesis, and phytochrome interaction factor PIF 4 (S1PIF4) were targeted using gRNAs with the stable transformed CRISPR/Cas9 system (Pan et al., 2016). Most of the flavonoids in tomatoes are present in the peel of the fruit. Hence, a holistic approach of pathway engineering to increase the content of novel flavonoids especially stilbenes in the flesh of the tomato fruit was reported (Schijlen et al., 2006).
In carrot (Daucus carota subsp. Carota), a candidate gene, DCAR_032551 that is responsible for carotenoid accumulation in carrot taproot and is co-expressed with several isoprenoid biosynthetic genes was identified from genome assembly and transcriptomic studies (Iorizzo et al., 2016). A candidate gene-based association study was carried out in carrots using 109 SNPs in 17 candidates/carotenoid biosynthesis genes over 380 diverse carrot cultivars, indicated the association of carotenoid content with the root color (Jourdan et al., 2015). A terpene synthase gene family of carrot was studied using QTL analysis and candidate gene-based association on a panel of carrot diversity set of 85 cultivars. GBS approach was used to genotype the panel with >168,000 SNPs (Keilwagen et al., 2017). Similarly, in bell pepper, several putative candidate genes are involved in the biosynthesis of capsaicinoids, such as Dihydroxyacid dehydratase (DHAD), Thr deaminase (TD) and Prephenate aminotransferase (PAT) were predicted from de novo transcriptome assembly (Liu et al., 2013).
Besides, several transcriptomics studies related to functional foods are available in crops such as lettuce (Lactuca sativa) (Zhang et al., 2017). In general, there is much scope to use genomics approaches to understand the molecular mechanisms and to increase the functional components in fruits and vegetables as evident by reports (Table 4).
Spices and Condiments
In spices and condiments, several studies have been carried out to profile metabolites, especially flavonoids, tannins, and alkaloids (Lee and Shibamoto, 2001; Shahidi and Ambigaipalan, 2015). In cinnamon (Cinnamomum verum), DART-QToF-MS method was utilized to discriminate true cinnamon from other species (Avula et al., 2015). There are limited reports on trait mapping in the case of spices (Table 5).
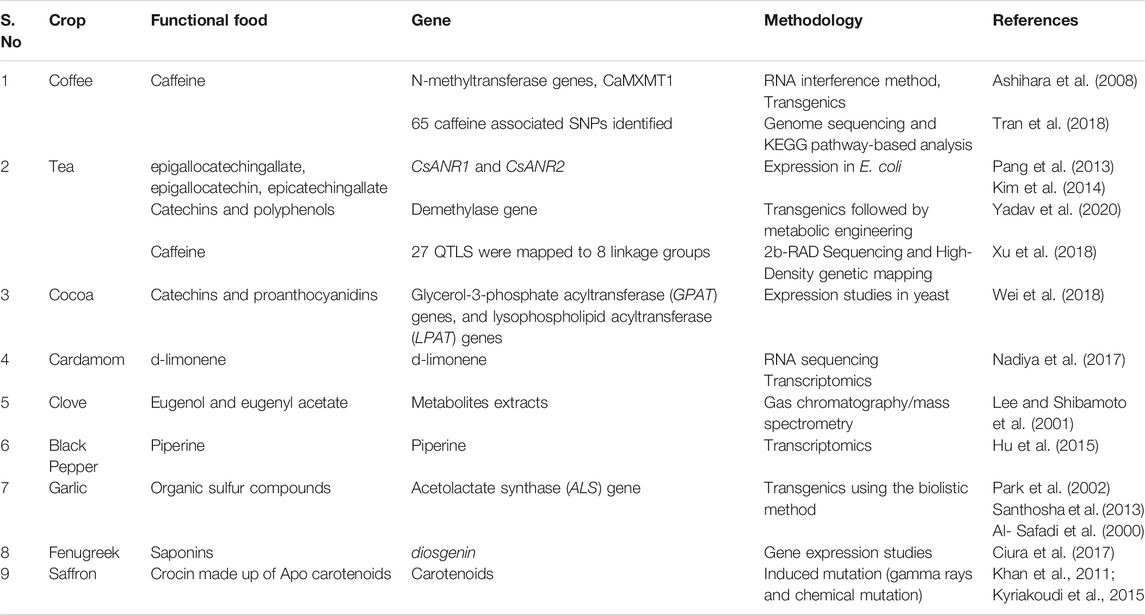
TABLE 5. Study of functional foods in beverages, spices and condiments using biotechnological approaches.
Most of the research in spices is related to the discovery of functional components. For instance, garlic has organic sulfur compounds as primary functional foods that have medicinal properties to reduce common cold, blood pressure and harmful cholesterol levels (Martin-Lagos et al., 1995). The functional food in turmeric is referred as curcumin, which acts as a acid neutralizer, blood purifier, tonic and antiseptic. The biological properties of curcumin were explored by using protein expression studies (Fang et al., 2011). The functional component of cardamom (Elettaria cardamomum) is d-limonene with antibacterial, anti-inflammatory, analgesic, and antispasmodic activities (Nadiya et al., 2017). Similarly, eugenol and eugenyl acetate, the functional components of clove (Syzygium aromaticum) are natural oxidants (Lee and Shibamoto, 2001). Coumarin, a functional component of cinnamon at lower doses has blood-thinning, anti-fungicidal and anti-tumor activities (Kawatra and Rajagopalan, 2015). Piperine from black pepper (Piper nigrum) has antioxidant, anti-inflammatory, and anticancer properties (Gorgani et al., 2017). Cumin (Cuminum cyminum) has cuminaldehyde that enhances appetite, taste perception, digestion, vision, strength, and lactation. It is also used to treat diseases such as fever, loss of appetite, diarrhea, vomiting, abdominal distension, edema and puerperal disorders (Sowbhagya, 2013). Ginger (Zingiber officinale) has gingerols, shagols, and paradols with antioxidant, antimicrobial, and anti-inflammatory potential (Butt and Sultan, 2011). Nutmeg (Myristica fragrans) has tannin, flavonoid, and terpenoid which are natural antioxidants (Assa et al., 2014). Coriander (Coriandrum sativum) has carotenoids, polyphenols and essential oils, which provides vitamin A and vitamin C (Laribi et al., 2015). Fenugreek (Trigonella foenum-graecum) has quercetin, kaempferol and vitexin derivatives which are anti-diabetic and anti-nociceptive properties (Ghosh et al., 2015). Saffron (Crocus sativus) has crocins, picrocrocin, and safranal, which is antispasmodic, eupeptic, gingival sedative, carminative, diaphoretic activities (Melnyk et al., 2010). Using mutation breeding in saffron has increased yields (Khan et al., 2011) that in turn increases the overall bioactive components per plant.
Transgenic research is still an emerging area in spices and condiments. In garlic and turmeric (Curcuma longa), genetic engineering approaches were utilized for developing herbicide tolerant plants (Park et al., 2002; Shirgurkar et al., 2006). Although there are some reports on the transcriptome of black pepper fruits (Hu et al., 2015), ginseng (Panax ginseng) (Rai et al., 2016), and cardamom (Nadiya et al., 2017) to study global transcriptome, there are no reports related to functional components in most of the spices. There is tremendous potential to use genomics approaches including trait mapping, transcriptomics, whole-genome studies and allele mining in case of spices to demonstrate and increase the functional components.
Beverages
Beverage crops produce potable beverages other than water. Major beverage crops include Coffee (Coffea spp.), Tea (Camelia sinensis), Cocoa (Theobroma cacao), and Lemongrass (Cymbopogon citratus). Coffee has caffeine as the primary phenolic compound and is known to reduce the risk of stroke and cancer. Caffeine in higher doses is harmful as it may lead to insomnia, nervousness, restlessness, irritability, an upset stomach, a fast heartbeat, and even muscle tremors. As a result, there are efforts to improve decaffeinated coffee plants using RNAi technology (Ashihara et al., 2008). Tea has catechins and epicatechin as primary functional foods, and they are known to possess chemopreventive activities against prostate and ovarian cancers, anti-obesity and anti-diabetic effects. Efforts are underway to elucidate the proanthocyanidin pathway, also to reduce caffeine content (Pang et al., 2013). Lemongrass has citral as its primary functional food which has antimicrobial and medicinal properties. Little research has been performed on this crop.
QTLs for flavonoid-related traits in a tea were identified using a high-density genetic map (Xu et al., 2018). Several transcriptomics studies have been carried out in tea to elucidate genes involved in polyphenol synthesis, Catechin biosynthesis and other regulatory networks (Mamati et al., 2006; Wu et al., 2016; Sun P. et al., 2017). To knock down the expression of the genes involved in caffeine biosynthesis, RNAi was used to repress the expression of the gene encoding theobromine synthase (CaMXMT1) that reduced the caffeine content in the transgenic coffee plants up to 70% (Ogita et al., 2003). In the case of tea, the functional characterization of the proanthocyanidin pathway and potential applications in metabolic engineering was elucidated (Pang et al., 2013). Cocoa rich in catechins and proanthocyanidins has a promising effect on lowering blood pressure, boosting moods, and sharpening memory. Metabolic engineering of yeast for cocoa butter production was attempted by cloning the genes involved in triglycerol synthesis viz., glycerol-3-phosphate acyltransferase (GPAT), lysophospholipid acyltransferase (LPAT) from cocoa into yeast (Wei et al., 2018). Efforts are being made to develop lemongrass varieties such as Jor Lab L-8 with higher amounts of essential oil and herbage production (Mohan et al., 2016). The biotechnological applications have not been effectively utilized to increase the functional components in beverages and there are few reports related to this (Table 5).
Medicinal Plants
Medicinal plants are called so because of their antibiotic, antidiabetic, antihyperglycemic, and antihyperlipidemic properties. Most medicinal plants are not consumed as staple foods, but as preventive medicines for several diseases ranging from the common cold to complex diseases like cancer. Herbal genomics has high potential to explore, though there are few efforts related to molecular breeding and genetic engineering in the medicinal crops (Chakraborty, 2018). However, metabolite profiling of some medicinal plants has been studied. In the case of a famous Ayurvedic crop Haritaki (Terminalia chebula), a component of Triphala (an ayurvedic composition), the metabolite profiling of polyphenols and evaluation of the decoction as a chemopreventive agent was studied (Pellati et al., 2013). Similarly, metabolite profiling was examined in a highly traded South African medicinal plant commonly known as pain brush lily (Scadoxus puniceus) and the bioactive compounds were isolated (Naidoo et al., 2018). Efforts for profiling polyphenols, alkaloids and other bioactive compounds are being carried out in other Asian medicinal plants (Gibon et al., 2006; Vega-Gálvez et al., 2011; Gantait et al., 2014; Hao and Xiao, 2015; Saito, 2018). For instance, in the case of Candyleaf (Stevia rebaudiana), the water extracts from leaf and calli were shown to have antioxidant activity and contain bioactive compounds including folic acid, vitamin C, catechin, quercetin and pyrogallol. Higher reactive oxygen species (ROS) scavenging activities were found in leaf extracts (Kim et al., 2011). Transcriptomics studies have also been carried out in some of the important medicinal plants including Ashwagandha (Withania somnifera) to understand the secondary metabolites which have therapeutic utilization (Tripathi et al., 2020).
Recent advances in metabolite and pathway engineering and their utilization in medicinal plant research have positively contributed to herbal genomics research. Most of the molecular studies in medicinal plants involved either discovery of the genes/enzymes/pathways related to secondary metabolites or increasing the production of the secondary metabolites using elicitors, hairy root cultures or metabolite engineering approaches.
Future Prospects
Current approaches in crop sciences using integrated omics platform aims at providing a nutritionally rich, diverse balanced diet to the society. Several leading edge technologies in understanding and manipulating different segments of scientific research areas viz. genomics, proteomics, metabolomics etc. has enabled the researchers to enhance contents of key nutrients in crop plants. Not just nutrition, but reducing the unflavorful compounds (phytic acid, acrylamide-forming amino acids, etc.) in food crops has allowed people to consume a wide range of food crops. Bio fortification has potential to solve nutrition deficiencies and in this view several food crops viz. rice, maize, wheat, etc. have been biofortified to have enhanced amounts of Fe, Zn, etc. (Ye et al., 2000; Gil-Humanes et al., 2014; Mugode et al., 2014; Trijatmiko et al., 2016). Crop improvement with new advancements in field phenomics, employing applications of machine learning (Niazian and Niedbala., 2020), nanotechnology and artificial intelligence (Ben Ayed and Hanana, 2021; Zhang et al., 2021), biosensors like lidar (Jin et al., 2021) followed by statistical analysis using data science (Tong and Nikoloski, 2021) approaches will enable researchers to precisely assess traits for plant breeding and development (Deery and Jones, 2021).
In order to ensure the nutrional security, along with enhancing the nutritional value, we need to work on reduced food-wastes that has a significant economic, environmental and social impact (FAO, 2019). Several initiatives in estimating food waste and prevention has been proposed (Moraes et al., 2021), however, devising methodologies in estimating and reducing food wastage is still a paradox (Richards et al., 2021). This can be featured as an opportunity to overcome malnutrition in addition to food waste reduction and stabilize bio-economy with sustainable processing of food waste into bio-based products (Sharma et al., 2021). The innovative technologies for extraction and microencapsulation of bioactives using novel technologies in metabolomics can be utilized in enhancing plant based functional foods (Pattnaik et al., 2021).
The research in nutrition and omics technologies in food science with epidemiological techniques should be classically established (Palou et al., 2004). In the future, the advances in foodomics and nutrigenomics can enable to achieve nutritional security in most of the crops. Utilization of omics technologies to identify the functional components in less explored crops like fruits, vegetables, spices and medicinal plants is essential to improve the functional components. There is a need to integrate multi-omics technologies in functional food research to elucidate and enhance the nutrition components in plants. Nutrigenomics can provide insights into the interaction of functional foods in human health and would provide allusion towards scientifically personalized diet.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SNN along with BA, SSM, and RP wrote the first draft. BSS, PB, KH, CK, and NP revised the manuscript. All the authors have read and approved the manuscript.
Funding
USDA-NIFA-Hatch funds provided to New Mexico Agricultural Experiment Station, New Mexico State University. SNN, BA and BSS acknowledge DBT-GoI for funds and fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbo, S., Molina, C., Jungmann, R., Grusak, M. A., Berkovitch, Z., Reifen, R., et al. (2005). Quantitative Trait Loci Governing Carotenoid Concentration and Weight in Seeds of Chickpea (Cicer Arietinum L.). Theor. Appl. Genet. 111 (2), 185–195. doi:10.1007/s00122-005-1930-y
Abbott, R. (2014). Documenting Traditional Medical Knowledge. Ryan Abbott, Documenting Traditional Medical Knowledge. Geneva, Switzerland: World Intellectual Property Organization.
Adams, G. G., Imran, S., Wang, S., Mohammad, A., Kok, M. S., Gray, D. A., et al. (2014). The Hypoglycemic Effect of Pumpkin Seeds, Trigonelline (TRG), Nicotinic Acid (NA), and D-Chiro-Inositol (DCI) in Controlling Glycemic Levels in Diabetes Mellitus. Crit. Rev. Food Sci. Nutr. 54 (10), 1322–1329. doi:10.1080/10408398.2011.635816
Aghajanpour, M., Nazer, M. R., Obeidavi, Z., Akbari, M., Ezati, P., and Kor, N. M. (2017). Functional Foods and Their Role in Cancer Prevention and Health Promotion: a Comprehensive Review. Am. J. Cancer Res. 7 (4), 740–769.
Al-Safadi, B., Mir, A., and Arabi, M. (2000). Improvement of Garlic (Allium Sativum L.) Resistance to white Rot and Storability Using Gamma Irradiation Induced Mutations. J. Genet. Breed. 54 (3), 175–182.
Alissa, E. M., and Ferns, G. A. (2012). Functional Foods and Nutraceuticals in the Primary Prevention of Cardiovascular Diseases. J. Nutr. Metab. doi:10.1155/2012/569486
Alkhatib, A., Tsang, C., Tiss, A., Bahorun, T., Arefanian, H., Barake, R., et al. (2017). Functional Foods and Lifestyle Approaches for Diabetes Prevention and Management. Nutrients 9 (12), 1310. doi:10.3390/nu9121310
Alomari, D. Z., Eggert, K., von Wirén, N., Alqudah, A. M., Polley, A., Plieske, J., et al. (2018). Identifying Candidate Genes for Enhancing Grain Zn Concentration in Wheat. Front. Plant Sci. 9, 1313. doi:10.3389/fpls.2018.01313
Alquezar, B., Rodrigo, M. J., and Zacarías, L. (2008). Regulation of Carotenoid Biosynthesis during Fruit Maturation in the Red-Fleshed orange Mutant Cara Cara. Phytochemistry 69 (10), 1997–2007. doi:10.1016/j.phytochem.2008.04.020
Aluru, M., Xu, Y., Guo, R., Wang, Z., Li, S., White, W., et al. (2008). Generation of Transgenic maize with Enhanced Provitamin A Content. J. Exp. Bot. 59 (13), 3551–3562. doi:10.1093/jxb/ern212
Anai, T., Koga, M., Tanaka, H., Kinoshita, T., Rahman, S. M., and Takagi, Y. (2003). Improvement of rice (Oryza Sativa L.) Seed Oil Quality through Introduction of a Soybean Microsomal omega-3 Fatty Acid Desaturase Gene. Plant Cel Rep. 21 (10), 988–992. doi:10.1007/s00299-003-0609-6
Anitha, S., Kane-Potaka, J., Tsusaka, T. W., Botha, R., Rajendran, A., Givens, D. I., et al. (2021). A Systematic Review and Meta-Analysis of the Potential of Millets and Sorghum for Managing and Preventing Diabetes Mellitus. Front. Nutr. 386.
Ashihara, H., Sano, H., and Crozier, A. (2008). Caffeine and Related Purine Alkaloids: Biosynthesis, Catabolism, Function and Genetic Engineering. Phytochemistry 69 (4), 841–856. doi:10.1016/j.phytochem.2007.10.029
Assa, J. R., Widjanarko, S. B., Kusnadi, J., and Berhimpon, S. (2014). Antioxidant Potential of Flesh, Seed and Mace of Nutmeg (Myristica Fragrans Houtt). Int. J.ChemTech Res. 6 (4), 2460–2468.
Avula, B., Smillie, T. J., Wang, Y.-H., Zweigenbaum, J., and Khan, I. A. (2015). Authentication of True Cinnamon (Cinnamon Verum) Utilising Direct Analysis in Real Time (DART)-QToF-MS. Food Additives & Contaminants: A 32 (1), 1–8. doi:10.1080/19440049.2014.981763
Axtell, J., Jd, A., Van Scoyoc, S., and Pj, C. (1979). “Current Status of Protein Quality Improvement in Grain Sorghum,” in International Symposium on Seed Protein Improvement in Cereals and Grain Legumes, Neuherberg (Germany, FR), 4 Sep 1978 (Vienna, Austria: International Atomic Energy Agency).
Baboota, R. K., Bishnoi, M., Ambalam, P., Kondepudi, K. K., Sarma, S. M., Boparai, R. K., et al. (2013). Functional Food Ingredients for the Management of Obesity and Associated Co-morbidities - A Review. J. Funct. Foods 5 (3), 997–1012. doi:10.1016/j.jff.2013.04.014
Bagchi, D., Swaroop, A., and Bagchi, M. (2015). Genomics, Proteomics and Metabolomics in Nutraceuticals and Functional Foods. John Wiley & Sons.
Ballali, S., and Lanciai, F. (2012). Functional Food and Diabetes: a Natural Way in Diabetes Prevention? Int. J. Food Sci. Nutr. 63 (Suppl. 1), 51–61. doi:10.3109/09637486.2011.637487
Balyan, H. S., Gupta, P. K., Kumar, S., Dhariwal, R., Jaiswal, V., Tyagi, S., et al. (2013). Genetic Improvement of Grain Protein Content and Other Health‐related Constituents of Wheat Grain. Plant Breed 132 (5), 446–457.
Bandyopadhyay, T., Jaiswal, V., and Prasad, M. (2017). “Nutrition Potential of Foxtail Millet in Comparison to Other Millets and Major Cereals,” in The Foxtail Millet Genome (Springer), 123–135. doi:10.1007/978-3-319-65617-5_10
Basu, A., Schell, J., and Scofield, R. H. (2018). Dietary Fruits and Arthritis. Food Funct. 9 (1), 70–77. doi:10.1039/c7fo01435j
Ben Ayed, D. J., and Hanana, M. (2021). Artificial Intelligence to Improve the Food and Agriculture Sector. J. Food Qual. 2021.
Bennett, D. J., and Kirby, G. W. (1968). Constitution and Biosynthesis of Capsaicin. J. Chem. Soc. C, 442–446. doi:10.1039/j39680000442
Bera, S. K., Kamdar, J. H., Kasundra, S. V., Dash, P., Maurya, A. K., Jasani, M. D., et al. (2018). Improving Oil Quality by Altering Levels of Fatty Acids through Marker-Assisted Selection of Ahfad2 Alleles in Peanut (Arachis hypogaea L.). Euphytica 214, 1–15. doi:10.1007/s10681-018-2241-0
Beyer, P., Al-Babili, S., Ye, X., Lucca, P., Schaub, P., Welsch, R., et al. (2002). Golden Rice: Introducing the β-Carotene Biosynthesis Pathway into Rice Endosperm by Genetic Engineering to Defeat Vitamin A Deficiency. J. Nutr. 132 (3), 506S–510S. doi:10.1093/jn/132.3.506s
Bhatnagar, M., Prasad, K., Bhatnagar-Mathur, P., Lakshmi Narasu, M., Waliyar, F., and Sharma, K. K. (2010). An Efficient Method for the Production of Marker-free Transgenic Plants of Peanut (Arachis hypogaea L.). Plant Cel Rep 29 (5), 495–502. doi:10.1007/s00299-010-0838-4
Butelli, E., Licciardello, C., Zhang, Y., Liu, J., Mackay, S., Bailey, P., et al. (2012). Retrotransposons Control Fruit-specific, Cold-dependent Accumulation of Anthocyanins in Blood Oranges. Plant Cell 24 (3), 1242–1255. doi:10.1105/tpc.111.095232
Butt, M. S., and Sultan, M. T. (2011). Ginger and its Health Claims: Molecular Aspects. Crit. Rev. Food Sci. Nutr. 51 (5), 383–393. doi:10.1080/10408391003624848
Cai, S., Yu, G., Chen, X., Huang, Y., Jiang, X., Zhang, G., et al. (2013). Grain Protein Content Variation and its Association Analysis in Barley. BMC Plant Biol. 13 (1), 35–11. doi:10.1186/1471-2229-13-35
Cao, H., Liu, R., Zhang, J., Liu, Z., Fan, S., Yang, G., et al. (2021). Improving Sulforaphane Content in Transgenic Broccoli Plants by Overexpressing MAM1, FMO GS–OX2, and Myrosinase. PCTOC 146, 1–11. doi:10.1007/s11240-021-02079-2
Capozzi, F., and Bordoni, A. (2013). Foodomics: a New Comprehensive Approach to Food and Nutrition. Genes Nutr. 8 (1), 1–4. doi:10.1007/s12263-012-0310-x
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, Precise Modification of the Tomato Genome. Genome Biol. 16 (1), 1–15.
Čertík, M., Klempová, T., Guothová, L., Mihálik, D., and Kraic, J. (2013). Biotechnology for the Functional Improvement of Cereal‐based Materials Enriched with PUFA and Pigments. Eur. J. Lip. Sci. Tech. 115 (11), 1247–1256.
Chakraborty, P. (2018). Herbal Genomics as Tools for Dissecting New Metabolic Pathways of Unexplored Medicinal Plants and Drug Discovery. Biochimie Open 6, 9–16. doi:10.1016/j.biopen.2017.12.003
Chakraborty, S., Chakraborty, N., Agrawal, L., Ghosh, S., Narula, K., Shekhar, S., et al. (2010). Next-generation Protein-Rich Potato Expressing the Seed Protein geneAmA1is a Result of Proteome Rebalancing in Transgenic Tuber. Proc. Natl. Acad. Sci. USA 107 (41), 17533–17538. doi:10.1073/pnas.1006265107
Chen, X., Long, H., Gao, P., Deng, G., Pan, Z., Liang, J., et al. (2014). Transcriptome Assembly and Analysis of Tibetan Hulless Barley (Hordeum Vulgare L. Var. Nudum) Developing Grains, with Emphasis on Quality Properties. PLoS ONE 9 (5), e98144. doi:10.1371/journal.pone.0098144
Chen, Z., Young, T. E., Ling, J., Chang, S.-C., and Gallie, D. R. (2003). Increasing Vitamin C Content of Plants through Enhanced Ascorbate Recycling. Proc. Natl. Acad. Sci. 100 (6), 3525–3530. doi:10.1073/pnas.0635176100
Cheng, L., Zheng, W., Li, M., Huang, J., Bao, S., Xu, Q., et al. (2020). Citrus Fruits Are Rich in Flavonoids for Immunoregulation and Potential Targeting ACE2. Preprints, 2020020313.
Chiu, L.-W., Zhou, X., Burke, S., Wu, X., Prior, R. L., and Li, L. (2010). The Purple Cauliflower Arises from Activation of a MYB Transcription Factor. Plant Physiol. 154 (3), 1470–1480. doi:10.1104/pp.110.164160
Chongtham, N., Bisht, M. S., and Haorongbam, S. (2011). Nutritional Properties of Bamboo Shoots: Potential and Prospects for Utilization as a Health Food. Compr. Rev. Food Sci. Food Saf. 10 (3), 153–168. doi:10.1111/j.1541-4337.2011.00147.x
Ciura, J., Szeliga, M., Grzesik, M., and Tyrka, M. (2017). Next-generation Sequencing of Representational Difference Analysis Products for Identification of Genes Involved in Diosgenin Biosynthesis in Fenugreek (Trigonella Foenum-graecum). Planta 245 (5), 977–991. doi:10.1007/s00425-017-2657-0
Colasuonno, P., Lozito, M. L., Marcotuli, I., Nigro, D., Giancaspro, A., Mangini, G., et al. (2017). The Carotenoid Biosynthetic and Catabolic Genes in Wheat and Their Association with Yellow Pigments. BMC Genomics 18 (1), 122–218. doi:10.1186/s12864-016-3395-6
Cruet-Burgos, C., Cox, S., Ioerger, B. P., Perumal, R., Hu, Z., Herald, T. J., et al. (2020). Advancing Provitamin A Biofortification in Sorghum: Genome-wide Association Studies of Grain Carotenoids in Global Germplasm. Plant Genome 13, e20013. doi:10.1002/tpg2.20013
de Alencar Figueiredo, L. F., Sine, B., Chantereau, J., Mestres, C., Fliedel, G., Rami, J.-F., et al. (2010). Variability of Grain Quality in Sorghum: Association with Polymorphism in Sh2, Bt2, SssI, Ae1, Wx and O2. Theor. Appl. Genet. 121 (6), 1171–1185. doi:10.1007/s00122-010-1380-z
Deery, D. M., and Jones, H. G. (2021). Field Phenomics: Will it Enable Crop Improvement? Plant Phenomics 2021, 1–16.
Derosa, G., Maffioli, P., and Sahebkar, A. (2016). Piperine and its Role in Chronic Diseases. Adv. Exp. Med. Biol. 928, 173–184. doi:10.1007/978-3-319-41334-1_8
Descalsota, G. I. L., Swamy, B. P. M., Zaw, H., Inabangan-Asilo, M. A., Amparado, A., Mauleon, R., et al. (2018). Genome-wide Association Mapping in a rice MAGIC Plus Population Detects QTLs and Genes Useful for Biofortification. Front. Plant Sci. 9, 1347. doi:10.3389/fpls.2018.01347
Descalsota-Empleo, G. I., Noraziyah, A. A. S., Navea, I. P., Chung, C., Dwiyanti, M. S., Labios, R. J. D., et al. (2019). Genetic Dissection of Grain Nutritional Traits and Leaf Blight Resistance in rice. Genes 10 (1), 30. doi:10.3390/genes10010030
Devi, I., Ansari, N., and Kumar, V. (2008). “Biofortification of Safflower Oil with Gamma Linolenic Acid through Transgenic Approach Using delta-6-desaturase Gene from Borago Officinalis,” in Safflower: unexploited potential and world adaptability. 7th International Safflower Conference, Wagga Wagga, New South Wales, Australia, 3-6 November, 2008 (Bendigo, VIC: Agri-MC Marketing and Communication), 1–5.
Diapari, M., Sindhu, A., Bett, K., Deokar, A., Warkentin, T. D., and Tar’an, B. (2014). Genetic Diversity and Association Mapping of Iron and Zinc Concentrations in Chickpea (Cicer Arietinum L.). Genome 57 (8), 459–468. doi:10.1139/gen-2014-0108
Distelfeld, A., Uauy, C., Fahima, T., and Dubcovsky, J. (2006). Physical Map of the Wheat High‐grain Protein Content geneGpc‐B1and Development of a High‐throughput Molecular Marker. New Phytol. 169 (4), 753–763. doi:10.1111/j.1469-8137.2005.01627.x
Dubock, A. (2019). Golden Rice: To Combat Vitamin A Deficiency for Public Health. Vitamin A 1–21. doi:10.5772/intechopen.84445
Ducreux, L. J., Morris, W. L., Hedley, P. E., Shepherd, T., Davies, H. V., Millam, S., et al. (2005). Metabolic Engineering of High Carotenoid Potato Tubers Containing Enhanced Levels of Beta-Carotene and Lutein. J. Exp. Bot. 56 (409), 81–89. doi:10.1093/jxb/eri016
Edmunds, S. J., Roy, N. C., Davy, M., Cooney, J. M., Barnett, M. P., Zhu, S., et al. (2012). Effects of Kiwifruit Extracts on Colonic Gene and Protein Expression Levels in IL-10 Gene-Deficient Mice. Br. J. Nutr. 108 (1), 113–129. doi:10.1017/S0007114511005241
El Khoury, D., Cuda, C., Luhovyy, B., and Anderson, G. (2012). Beta Glucan: Health Benefits in Obesity and Metabolic Syndrome. J. Nutr. Metab. doi:10.1155/2012/851362
El Nagar, M. (2018). Genetic Engineering to Improve β-carotene Content in Pepper. Ann. Agric. Sci. Moshtohor 56, 119–126. doi:10.21608/assjm.2018.65121
Fang, H. Y., Chen, S. B., Guo, D. J., Pan, S. Y., and Yu, Z. L. (2011). Proteomic Identification of Differentially Expressed Proteins in Curcumin-Treated MCF-7 Cells. Phytomedicine 18 (8-9), 697–703. doi:10.1016/j.phymed.2010.11.012
FAO (2019). The State of Food and Agriculture. Availableat: http://www.fao.org/state-of-food-agriculture/2019/en/(Accessed September, 2020).
Fernandez, M. G. S., Hamblin, M. T., Li, L., Rooney, W. L., Tuinstra, M. R., and Kresovich, S. (2008). Quantitative Trait Loci Analysis of Endosperm Color and Carotenoid Content in Sorghum Grain. Crop Sci. 48 (5), 1732–1743. doi:10.2135/cropsci2007.12.0684
Fokunang, C. N., Ndikum, V., Tabi, O. Y., Jiofack, R. B., Ngameni, B., Guedje, N. M., et al. (2011). Traditional Medicine: Past, Present and Future Research and Development Prospects and Integration in the National Health System of Cameroon. Afr. J. Tradit Complement. Altern. Med. 8 (3), 284–295. doi:10.4314/ajtcam.v8i3.65276
Galpaz, N., Wang, Q., Menda, N., Zamir, D., and Hirschberg, J. (2008). Abscisic Acid Deficiency in the Tomato Mutant High-Pigment 3 Leading to Increased Plastid Number and Higher Fruit Lycopene Content. Plant J. 53 (5), 717–730. doi:10.1111/j.1365-313x.2007.03362.x
Gantait, S., Debnath, S., and Nasim Ali, M. (2014). Genomic Profile of the Plants with Pharmaceutical Value. 3 Biotech. 4 (6), 563–578. doi:10.1007/s13205-014-0218-9
Gazal, A., Dar, Z. A., Zaffar, G., Lone, A. A., Abidi, I., Shabir, A., et al. (2014). Trends in Breeding Oat for Nutritional Grain Quality - an Overview. Jans 6 (2), 904–912. doi:10.31018/jans.v6i2.552
Ghosh, B., Chandra, I., and Chatterjee, S. (2015). Fenugreek (Trigonella Foenum-graecum L.) and its Necessity. Fire J. Eng. Technol. 1 (1), 66–67.
Gibbon, B. C., and Larkins, B. A. (2005). Molecular Genetic Approaches to Developing Quality Protein maize. Trends Genet. 21 (4), 227–233. doi:10.1016/j.tig.2005.02.009
Gibon, Y., Usadel, B., Blaesing, O. E., Kamlage, B., Hoehne, M., Trethewey, R., et al. (2006). Integration of Metabolite with Transcript and Enzyme Activity Profiling during Diurnal Cycles in Arabidopsis Rosettes. Genome Biol. 7 (8), R76–R23. doi:10.1186/gb-2006-7-8-R76
Gil-Humanes, J., Pistón, F., Altamirano-Fortoul, R., Real, A., Comino, I., Sousa, C., et al. (2014). Reduced-gliadin Wheat Bread: an Alternative to the Gluten-free Diet for Consumers Suffering Gluten-Related Pathologies. PLoS ONE 9 (3), e90898. doi:10.1371/journal.pone.0090898
González, M., Salazar, E., Castillo, J., Morales, P., Mura-Jornet, I., Maldonado, J., et al. (2016). Genetic Structure Based on EST-SSR: a Putative Tool for Fruit Color Selection in Japanese Plum (Prunus Salicina L.) Breeding Programs. Mol. Breed. 36 (6), 68. doi:10.1007/s11032-016-0491-x
González-Molina, E., Domínguez-Perles, R., Moreno, D. A., and García-Viguera, C. (2010). Natural Bioactive Compounds of Citrus limon for Food and Health. J. Pharm. Biomed. Anal. 51 (2), 327–345. doi:10.1016/j.jpba.2009.07.027
Gonzali, S., Mazzucato, A., and Perata, P. (2009). Purple as a Tomato: towards High Anthocyanin Tomatoes. Trends Plant Sci. 14 (5), 237–241. doi:10.1016/j.tplants.2009.02.001
Gordeeva, E., Badaeva, E., Yudina, R., Shchukina, L., Shoeva, O., and Khlestkina, E. (2019). Marker-Assisted Development of a Blue-Grained Substitution Line Carrying the Thinopyrum Ponticum Chromosome 4Th(4D) in the Spring Bread Wheat Saratovskaya 29 Background. Agronomy 9, 723. doi:10.3390/agronomy9110723
Gordeeva, E., Shamanin, V., Shoeva, O., Kukoeva, T., Morgounov, A., and Khlestkina, E. (2020). The Strategy for Marker-Assisted Breeding of Anthocyanin-Rich Spring Bread Wheat (Triticum aestivum L.) Cultivars in Western Siberia. Agronomy 10, 1603. doi:10.3390/agronomy10101603
Gorgani, L., Mohammadi, M., Najafpour, G. D., and Nikzad, M. (2017). Piperine-The Bioactive Compound of Black Pepper: From Isolation to Medicinal Formulations. Compr. Rev. Food Sci. Food Saf. 16 (1), 124–140. doi:10.1111/1541-4337.12246
Govindaraj, M., Rai, K. N., Cherian, B., Pfeiffer, W. H., Kanatti, A., and Shivade, H. (2019). Breeding Biofortified Pearl Millet Varieties and Hybrids to Enhance Millet Markets for Human Nutrition. Agriculture 9 (5), 106.
Guizani, N. (2013). Date palm Fruits as Functional Foods. Int. J. Nutr. Pharmacol. Neurol. Dis. 3 (3), 161. doi:10.4103/2231-0738.114830
Gul, K., Tak, A., Singh, A. K., Singh, P., Yousuf, B., and Wani, A. A. (2015). Chemistry, Encapsulation, and Health Benefits of β-carotene - A Review. Cogent Food Agric. 1, 1018696. doi:10.1080/23311932.2015.1018696
Gürbüz Çolak, N., Eken, N. T., Ülger, M., Frary, A., and Doğanlar, S. (2020). Mapping of Quantitative Trait Loci for Antioxidant Molecules in Tomato Fruit: Carotenoids, Vitamins C and E, Glutathione and Phenolic Acids. Plant Sci. 292, 110393. doi:10.1016/j.plantsci.2019.110393
Hamid, S. B., and Hamid, A. F. A. (2019). Roles of Nutraceuticals and Functional Food in Prevention of Cardiovascular Disease. Nutraceutical and Functional Foods in Disease Prevention. Hershey, PA: IGI Global, 126–165. doi:10.4018/978-1-5225-3267-5.ch005
Han, Z., Zhang, J., Cai, S., Chen, X., Quan, X., and Zhang, G. (2018). Association Mapping for Total Polyphenol Content, Total Flavonoid Content and Antioxidant Activity in Barley. BMC Genomics 19 (1), 81–10. doi:10.1186/s12864-018-4483-6
Hannan, J. M. A., Ali, L., Rokeya, B., Khaleque, J., Akhter, M., Flatt, P. R., et al. (2007). Soluble Dietary Fibre Fraction ofTrigonella Foenum-Graecum(fenugreek) Seed Improves Glucose Homeostasis in Animal Models of Type 1 and Type 2 Diabetes by Delaying Carbohydrate Digestion and Absorption, and Enhancing Insulin Action. Br. J. Nutr. 97 (3), 514–521. doi:10.1017/s0007114507657869
Hao, D. C., and Xiao, P. G. (2015). Genomics and Evolution in Traditional Medicinal Plants: Road to a Healthier Life. Evol. Bioinform Online 11, 197–212. doi:10.4137/EBO.S31326
Hasler, C. M. (1998). Functional Foods: Their Role in Disease Prevention and Health Promotion. Food Technology-Champaign then Chicago 52, 63–147.
Hellwege, E. M., Czapla, S., Jahnke, A., Willmitzer, L., and Heyer, A. G. (2000). Transgenic Potato (Solanum tuberosum) Tubers Synthesize the Full Spectrum of Inulin Molecules Naturally Occurring in globe Artichoke (Cynara Scolymus) Roots. Proc. Natl. Acad. Sci. 97 (15), 8699–8704. doi:10.1073/pnas.150043797
Hill, J. O., and Peters, J. C. (2002). Biomarkers and Functional Foods for Obesity and Diabetes. Br. J. Nutr. 88 (S2), S213–S218. doi:10.1079/bjn2002685
Hindu, V., Palacios-Rojas, N., Babu, R., Suwarno, W. B., Rashid, Z., Usha, R., et al. (2018). Identification and Validation of Genomic Regions Influencing Kernel Zinc and Iron in maize. Theor. Appl. Genet. 131, 1443–1457. doi:10.1007/s00122-018-3089-3
Hu, L., Hao, C., Fan, R., Wu, B., Tan, L., and Wu, H. (2015). De Novo assembly and Characterization of Fruit Transcriptome in Black Pepper (Piper Nigrum). PLoS ONE 10 (6), e0129822. doi:10.1371/journal.pone.0129822
Huang, S., Frizzi, A., Florida, C. A., Kruger, D. E., and Luethy, M. H. (2006). High Lysine and High Tryptophan Transgenic Maize Resulting from the Reduction of Both 19- and 22-kD α-zeins. Plant Mol. Biol. 61 (3), 525–535. doi:10.1007/s11103-006-0027-6
Hussain, B., Lucas, S. J., Ozturk, L., and Budak, H. (2017). Mapping QTLs Conferring Salt Tolerance and Micronutrient Concentrations at Seedling Stagein Wheat. Sci. Rep. 7 (1), 15662–15714. doi:10.1038/s41598-017-15726-6
Iorizzo, M., Ellison, S., Senalik, D., Zeng, P., Satapoomin, P., Huang, J., et al. (2016). A High-Quality Carrot Genome Assembly Provides New Insights into Carotenoid Accumulation and Asterid Genome Evolution. Nat. Genet. 48 (6), 657–666. doi:10.1038/ng.3565
Jacobo-Valenzuela, N., Maróstica-Junior, M. R., Zazueta-Morales, J. d. J., and Gallegos-Infante, J. A. (2011). Physicochemical, Technological Properties, and Health-Benefits of Cucurbita Moschata Duchense vs. Cehualca. Food Res. Int. 44 (9), 2587–2593. doi:10.1016/j.foodres.2011.04.039
Jia, D., Fan, L., Shen, J., Qin, S., Li, F., and Yuan, Y. (2019). Genetic Transformation of the Astaxanthin Biosynthetic Genes Bkt and crtR-B into Apple Tree to Increase Photooxidation Resistance. Scientia Horticulturae 243, 428–433. doi:10.1016/j.scienta.2018.08.058
Jin, L., Xiao, P., Lu, Y., Shao, Y., Shen, Y., and Bao, J. (2009). Quantitative Trait Loci for Brown rice Color, Phenolics, Flavonoid Contents, and Antioxidant Capacity in rice Grain. Cereal Chem. J. 86 (6), 609–615. doi:10.1094/cchem-86-6-0609
Jin, M., Zhang, X., Zhao, M., Deng, M., Du, Y., Zhou, Y., et al. (2017). Integrated Genomics-Based Mapping Reveals the Genetics Underlying maize Flavonoid Biosynthesis. BMC Plant Biol. 17 (1), 17. doi:10.1186/s12870-017-0972-z
Jin, S., Sun, X., Wu, F., Su, Y., Li, Y., Song, S., et al. (2021). Lidar Sheds New Light on Plant Phenomics for Plant Breeding and Management: Recent Advances and Future Prospects. ISPRS J. Photogrammetry Remote Sensing 171, 202–223. doi:10.1016/j.isprsjprs.2020.11.006
Jourdan, M., Gagné, S., Dubois-Laurent, C., Maghraoui, M., Huet, S., Suel, A., et al. (2015). Carotenoid Content and Root Color of Cultivated Carrot: a Candidate-Gene Association Study Using an Original Broad Unstructured Population. PLoS One 10 (1), e0116674. doi:10.1371/journal.pone.0116674
Joy, P. P., Anjana, R., Rashida-Rajuva, A., and Ratheesh, A. (2018). “Fruits as a Functional Food,” in State-of-the-art Technologies in Food Science: Human Health, Emerging Issues and Specialty Topics (Boca Raton, FL: CRC Press), 111–168.
Karkute, S. G., Singh, A. K., Gupta, O. P., Singh, P. M., and Singh, B. (2017). CRISPR/Cas9 Mediated Genome Engineering for Improvement of Horticultural Crops. Front. Plant Sci. 8, 1635. doi:10.3389/fpls.2017.01635
Karmakar, A., Bhattacharya, S., Sengupta, S., Ali, N., Sarkar, S. N., Datta, K., et al. (2020). RNAi-Mediated Silencing of ITPK Gene Reduces Phytic Acid Content, Alters Transcripts of Phytic Acid Biosynthetic Genes, and Modulates mineral Distribution in rice Seeds. Rice Sci. 27, 315–328. doi:10.1016/j.rsci.2020.05.007
Kartikey, K., Singh, G., Sah, D., Singh, R. B., Singh, A. K., Takahashi, T., et al. (2019). “Functional Food Security for Osteoporosis, Carcinogenesis, Atherosclerosis and Brain Degeneration,” in The Role of Functional Food Security in Global Health (Elsevier), 639–651. doi:10.1016/b978-0-12-813148-0.00037-2
Kato, H., Takahashi, S., and Saito, K. (2011). Omics and Integrated Omics for the Promotion of Food and Nutrition Science. J. Traditional Complement. Med. 1 (1), 25–30. doi:10.1016/s2225-4110(16)30053-0
Kawatra, P., and Rajagopalan, R. (2015). Cinnamon: Mystic powers of a Minute Ingredient. Pharmacognosy Res. 7 (Suppl. 1), S1–S6. doi:10.4103/0974-8490.157990
Keilwagen, J., Lehnert, H., Berner, T., Budahn, H., Nothnagel, T., Ulrich, D., et al. (2017). The Terpene Synthase Gene Family of Carrot (Daucus Carota L.): Identification of QTLs and Candidate Genes Associated with Terpenoid Volatile Compounds. Front. Plant Sci. 8, 1930. doi:10.3389/fpls.2017.01930
Khalil, A. A., Rahman, U. u., Khan, M. R., Sahar, A., Mehmood, T., and Khan, M. (2017). Essential Oil Eugenol: Sources, Extraction Techniques and Nutraceutical Perspectives. RSC Adv. 7, 32669–32681. doi:10.1039/c7ra04803c
Khan, M. A., Nagoo, S., Naseer, S., Nehvi, F., and Zargar, S. M. (2011). Induced Mutation as a Tool for Improving Corm Multiplication in Saffron (Crocus Sativus L.). J. Phytol. 3 (7).
Khazaei, H., Podder, R., Caron, C. T., Kundu, S. S., Diapari, M., Vandenberg, A., et al. (2017). Marker-Trait Association Analysis of Iron and Zinc Concentration in Lentil (Lens Culinaris Medik.) Seeds. Plant Genome 10 (2). doi:10.3835/plantgenome2017.02.0007
Kidd, P. (2011). Astaxanthin, Cell Membrane Nutrient with Diverse Clinical Benefits and Anti-aging Potential. Altern. Med. Rev. 16, 355–364.
Kim, I.-S., Yang, M., Lee, O.-H., and Kang, S.-N. (2011). The Antioxidant Activity and the Bioactive Compound Content of Stevia rebaudiana Water Extracts. LWT - Food Sci. Technology 44 (5), 1328–1332. doi:10.1016/j.lwt.2010.12.003
Kim, S.-E., Lee, C.-J., Ji, C. Y., Kim, H. S., Park, S.-U., Lim, Y.-H., et al. (2019). Transgenic Sweetpotato Plants Overexpressing Tocopherol Cyclase Display Enhanced α-tocopherol Content and Abiotic Stress Tolerance. Plant Physiol. Biochem. 144, 436–444. doi:10.1016/j.plaphy.2019.09.046
Kim, S. J., Li, M., Jeong, C. W., Bae, H. B., Kwak, S. H., Lee, S. H., et al. (2014). Epigallocatechin-3-gallate, a green tea Catechin, Protects the Heart against Regional Ischemia-Reperfusion Injuries through Activation of RISK Survival Pathways in Rats. Arch. Pharm. Res. 37 (8), 1079–1085. doi:10.1007/s12272-013-0309-x
Kimura, E., Abe, T., Murata, K., Kimura, T., Otoki, Y., Yoshida, T., et al. (2018). Identification of OsGGR2, a Second Geranylgeranyl Reductase Involved in α-tocopherol Synthesis in rice. Sci. Rep. 8, 1870–1878. doi:10.1038/s41598-018-19527-3
Knox, R. E., Pozniak, C. J., Clarke, F. R., Clarke, J. M., Houshmand, S., and Singh, A. K. (2009). Chromosomal Location of the Cadmium Uptake Gene (Cdu1) in Durum Wheat. Genome 52 (9), 741–747. doi:10.1139/g09-042
Kole, C., Muthamilarasan, M., Henry, R., Edwards, D., Sharma, R., Abberton, M., et al. (2015). Application of Genomics-Assisted Breeding for Generation of Climate Resilient Crops: Progress and Prospects. Front. Plant Sci. 6, 563. doi:10.3389/fpls.2015.00563
Kotla, A., Phuke, R., Hariprasanna, K., Mehtre, S. P., Rathore, A., Gorthy, S., et al. (2019). Identification of QTLs and Candidate Genes for High Grain Fe and Zn Concentration in Sorghum [Sorghum Bicolor (L.)Moench]. J. Cereal Sci. 90, 102850. doi:10.1016/j.jcs.2019.102850
Kumar, A., Kujur, A., Singh, P. P., and Prakash, B. (2019). Nanoencapsulated Plant-Based Bioactive Formulation against Food-Borne Molds and Aflatoxin B1 Contamination: Preparation, Characterization and Stability Evaluation in the Food System. Food Chem. 287, 139–150. doi:10.1016/j.foodchem.2019.02.045
Kumar, A., Kumar, S., Bains, S., Vaidya, V., Singh, B., Kaur, R., et al. (2016). De Novo transcriptome Analysis Revealed Genes Involved in Flavonoid and Vitamin C Biosynthesis in Phyllanthus Emblica (L.). Front. Plant Sci. 7, 1610. doi:10.3389/fpls.2016.01610
Kumar, A., Mirza, N., Charan, T., Sharma, N., and Gaur, V. S. (2014). Isolation, Characterization and Immunolocalization of a Seed Dominant CaM from finger Millet (Eleusine Coracana L. Gartn.) for Studying its Functional Role in Differential Accumulation of Calcium in Developing Grains. Appl. Biochem. Biotechnol. 172 (6), 2955–2973. doi:10.1007/s12010-013-0714-0
Kumar, S., Hash, C., Nepolean, T., Mahendrakar, M., Satyavathi, C., Singh, G., et al. (2018). Mapping Grain Iron and Zinc Content Quantitative Trait Loci in an Iniadi-Derived Immortal Population of Pearl Millet. Genes 9 (5), 248. doi:10.3390/genes9050248
Kyriakoudi, A., Ordoudi, S., Roldán-Medina, M., and Tsimidou, M. (2015). Saffron, a Functional Spice. Austin. J. Nutri. Food Sci. 3 (1), 1059.
Lapitan, N. L. V., Hess, A., Cooper, B., Botha, A.-M., Badillo, D., Iyer, H., et al. (2009). Differentially Expressed Genes during Malting and Correlation with Malting Quality Phenotypes in Barley (Hordeum Vulgare L.). Theor. Appl. Genet. 118 (5), 937–952. doi:10.1007/s00122-008-0951-8
Laribi, B., Kouki, K., M'Hamdi, M., and Bettaieb, T. (2015). Coriander (Coriandrum Sativum L.) and its Bioactive Constituents. Fitoterapia 103, 9–26. doi:10.1016/j.fitote.2015.03.012
Lau, F. C., Shukitt-Hale, B., and Joseph, J. A. (2005). The Beneficial Effects of Fruit Polyphenols on Brain Aging. Neurobiol. Aging 26 (1), 128–132. doi:10.1016/j.neurobiolaging.2005.08.007
Lee, K.-G., and Shibamoto, T. (2001). Antioxidant property of aroma extract isolated from clove buds [Syzygium aromaticum (L.) Merr. et Perry]. Food Chem. 74 (4), 443–448. doi:10.1016/s0308-8146(01)00161-3
Li, H., Liu, H., Han, Y., Wu, X., Teng, W., Liu, G., et al. (2010). Identification of QTL Underlying Vitamin E Contents in Soybean Seed Among Multiple Environments. Theor. Appl. Genet. 120 (7), 1405–1413. doi:10.1007/s00122-010-1264-2
Li, M.-W., Muñoz, N. B., Wong, C.-F., Wong, F.-L., Wong, K.-S., Wong, J. W.-H., et al. (2016). QTLs Regulating the Contents of Antioxidants, Phenolics, and Flavonoids in Soybean Seeds Share a Common Genomic Region. Front. Plant Sci. 7, 854. doi:10.3389/fpls.2016.00854
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018). Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-mediated Multiplex Genome Editing. Front. Plant Sci. 9, 559. doi:10.3389/fpls.2018.00559
Liang, Q., Wang, K., Liu, X., Riaz, B., Jiang, L., Wan, X., et al. (2019). Improved Folate Accumulation in Genetically Modified maize and Wheat. J. Exp. Bot. 70 (5), 1539–1551. doi:10.1093/jxb/ery453
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J. Genet. Genomics 41 (2), 63–68. doi:10.1016/j.jgg.2013.12.001
Liao, P., Chen, X., Wang, M., Bach, T. J., and Chye, M.-L. (2018). Improved Fruit α-tocopherol, Carotenoid, Squalene and Phytosterol Contents through Manipulation of Brassica Juncea 3-HYDROXY-3-METHYLGLUTARYL-COA SYNTHASE1 in Transgenic Tomato. Plant Biotechnol. J. 16, 784–796. doi:10.1111/pbi.12828
Lipkie, T. E., De Moura, F. F., Zhao, Z.-Y., Albertsen, M. C., Che, P., Glassman, K., et al. (2013). Bioaccessibility of Carotenoids from Transgenic Provitamin A Biofortified Sorghum. J. Agric. Food Chem. 61 (24), 5764–5771. doi:10.1021/jf305361s
Liu, S., Li, W., Wu, Y., Chen, C., and Lei, J. (2013). De Novo transcriptome Assembly in Chili Pepper (Capsicum Frutescens) to Identify Genes Involved in the Biosynthesis of Capsaicinoids. PLoS ONE 8 (1), e48156. doi:10.1371/journal.pone.0048156
Lobo, V., Patil, A., Phatak, A., and Chandra, N. (2010). Free Radicals, Antioxidants and Functional Foods: Impact on Human Health. Phcog Rev. 4 (8), 118. doi:10.4103/0973-7847.70902
Lowe, R., Shirley, N., Bleackley, M., Dolan, S., and Shafee, T. (2017). Transcriptomics Technologies. Plos Comput. Biol. 13 (5), e1005457. doi:10.1371/journal.pcbi.1005457
Lucca, P., Hurrell, R., and Potrykus, I. (2002). Fighting Iron Deficiency Anemia with Iron-Rich rice. J. Am. Coll. Nutr. 21 (Suppl. 3), 184S–190S. doi:10.1080/07315724.2002.10719264
Luo, H., Guo, J., Yu, B., Chen, W., Zhang, H., Zhou, X., et al. (2021). Construction of ddRADseq-Based High-Density Genetic Map and Identification of Quantitative Trait Loci for Trans-resveratrol Content in Peanut Seeds. Front. Plant Sci. 12, 438. doi:10.3389/fpls.2021.644402
Ma, C., Ma, B., He, J., Hao, Q., Lu, X., and Wang, L. (2011). Regulation of Carotenoid Content in Tomato by Silencing of Lycopene β/ε-Cyclase Genes. Plant Mol. Biol. Rep. 29 (1), 117–124. doi:10.1007/s11105-010-0211-3
Mamati, G. E., Liang, Y., and Lu, J. (2006). Expression of Basic Genes Involved in tea Polyphenol Synthesis in Relation to Accumulation of Catechins and Total tea Polyphenols. J. Sci. Food Agric. 86 (3), 459–464. doi:10.1002/jsfa.2368
Martín-Lagos, R. A., Serrano, M. F. O., and Lopez, M. D. R. (1995). Determination of Organic sulphur Compounds in Garlic Extracts by Gas Chromatography and Mass Spectrometry. Food Chem. 53 (1), 91–93. doi:10.1016/0308-8146(95)95792-5
Masibo, M., and He, Q. (2008). Major Mango Polyphenols and Their Potential Significance to Human Health. Compr. Rev. Food Sci. Food Saf. 7 (4), 309–319. doi:10.1111/j.1541-4337.2008.00047.x
Mattivi, F., Guzzon, R., Vrhovsek, U., Stefanini, M., and Velasco, R. (2006). Metabolite Profiling of Grape: Flavonols and Anthocyanins. J. Agric. Food Chem. 54 (20), 7692–7702. doi:10.1021/jf061538c
Mazza, G., and Francis, F. J. (1995). Anthocyanins in Grapes and Grape Products. Crit. Rev. Food Sci. Nutr. 35 (4), 341–371. doi:10.1080/10408399509527704
McGuire, A. L., Gabriel, S., Tishkoff, S. A., Wonkam, A., Chakravarti, A., Furlong, E. E. M., et al. (2020). The Road Ahead in Genetics and Genomics. Nat. Rev. Genet. 21 (10), 581–596. doi:10.1038/s41576-020-0272-6
Melnyk, J. P., Wang, S., and Marcone, M. F. (2010). Chemical and Biological Properties of the World’s Most Expensive Spice: Saffron. Int. Food Res. 43 (8), 1981–1989.
Mertz, E. T., Bates, L. S., and Nelson, O. E. (1964). Mutant Gene that Changes Protein Composition and Increases Lysine Content of maize Endosperm. Science 145 (3629), 279–280. doi:10.1126/science.145.3629.279
Minamikawa, M. F., Nonaka, K., Kaminuma, E., Kajiya-Kanegae, H., Onogi, A., Goto, S., et al. (2017). Genome-wide Association Study and Genomic Prediction in Citrus: Potential of Genomics-Assisted Breeding for Fruit Quality Traits. Sci. Rep. 7 (1), 4721–4813. doi:10.1038/s41598-017-05100-x
Mirza, N., Taj, G., Arora, S., and Kumar, A. (2014). Transcriptional Expression Analysis of Genes Involved in Regulation of Calcium Translocation and Storage in finger Millet (Eleusine Coracana L. Gartn.). Gene 550 (2), 171–179. doi:10.1016/j.gene.2014.08.005
Mishiba, K.-I., Nishida, K., Inoue, N., Fujiwara, T., Teranishi, S., Iwata, Y., et al. (2020). Genetic Engineering of Eggplant Accumulating β-carotene in Fruit. Plant Cell Rep. 39, 1029–1039. doi:10.1007/s00299-020-02546-8
Mohan, L., Sukriti, D., and Bhattacharyya, P. (2016). Development of a High Yielding Variety, Jor Lab L-8 of Lemongrass (Cymbopogon Flexuosus L.). Ann. Agri Bio Res 21 (1), 22–23.
Mohanty, S., and Singhal, K. (2018). “Functional Foods as Personalised Nutrition: Definitions and Genomic Insights,” in Functional Food and Human Health (Springer), 513–535. doi:10.1007/978-981-13-1123-9_22
Mondal, S., Phadke, R. R., and Badigannavar, A. M. (2015). Genetic Variability for Total Phenolics, Flavonoids and Antioxidant Activity of Testaless Seeds of a Peanut Recombinant Inbred Line Population and Identification of Their Controlling QTLs. Euphytica 204 (2), 311–321. doi:10.1007/s10681-014-1324-9
Moraes, N. V., Lermen, F. H., and Echeveste, M. E. S. (2021). A Systematic Literature Review on Food Waste/loss Prevention and Minimization Methods. J. Environ. Manage. 286, 112268. doi:10.1016/j.jenvman.2021.112268
Mugode, L., Ha, B., Kaunda, A., Sikombe, T., Phiri, S., Mutale, R., et al. (2014). Carotenoid Retention of Biofortified Provitamin A maize (Zea mays L.) after Zambian Traditional Methods of Milling, Cooking and Storage. J. Agric. Food Chem. 62 (27), 6317–6325. doi:10.1021/jf501233f
Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Choudhary, M., Saha, S., Bhat, J. S., et al. (2014). Development of β-Carotene Rich Maize Hybrids through Marker-Assisted Introgression of β-carotene Hydroxylase Allele. PLoS One 9 (12), e113583. doi:10.1371/journal.pone.0113583
Nadiya, F., Anjali, N., Thomas, J., Gangaprasad, A., and Sabu, K. K. (2017). Transcriptome Profiling of Elettaria Cardamomum (L.) Maton (Small Cardamom). Genomics Data 11, 102–103. doi:10.1016/j.gdata.2016.12.013
Naidoo, D., Slavětínská, L. P., Aremu, A. O., Gruz, J., Biba, O., Doležal, K., et al. (2018). Metabolite Profiling and Isolation of Biologically Active Compounds fromScadoxus Puniceus, a Highly Traded South African Medicinal Plant. Phytotherapy Res. 32 (4), 625–630. doi:10.1002/ptr.6000
Nayak, S. N., Hebbal, V., Bharati, P., Nadaf, H. L., Naidu, G. K., and Bhat, R. S. (2020). Profiling of Nutraceuticals and Proximates in Peanut Genotypes Differing for Seed Coat Color and Seed Size. Front. Nutr. 7, 45. doi:10.3389/fnut.2020.00045
Newell, M. A., Asoro, F. G., Scott, M. P., White, P. J., Beavis, W. D., and Jannink, J.-L. (2012). Genome-wide Association Study for Oat (Avena Sativa L.) Beta-Glucan Concentration Using Germplasm of Worldwide Origin. Theor. Appl. Genet. 125 (8), 1687–1696. doi:10.1007/s00122-012-1945-0
Niazian, M., and Niedbała, G. (2020). Machine Learning for Plant Breeding and Biotechnology. Agriculture 10 (10), 436. doi:10.3390/agriculture10100436
Ogita, S., Uefuji, H., Yamaguchi, Y., Koizumi, N., and Sano, H. (2003). Producing Decaffeinated Coffee Plants. Nature 423 (6942), 823. doi:10.1038/423823a
Palou, A., Picó, C., and Bonet, M. L. (2004). Food Safety and Functional Foods in the European Union: Obesity as a Paradigmatic Example for Novel Food Development. Nutr. Rev. 62 (Suppl. l_2), S169–S181. doi:10.1111/j.1753-4887.2004.tb00089.x
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated Efficient and Heritable Targeted Mutagenesis in Tomato Plants in the First and Later Generations. Sci. Rep. 6 (1), 24765–24769. doi:10.1038/srep24765
Pandey, M. K., Wang, M. L., Qiao, L., Feng, S., Khera, P., Wang, H., et al. (2014). Identification of QTLs Associated with Oil Content and Mapping FAD2 Genes and Their Relative Contribution to Oil Quality in Peanut (Arachis hypogaea L.). BMC Genet. 15 (1), 133–214. doi:10.1186/s12863-014-0133-4
Pang, Y., Abeysinghe, I. S. B., He, J., He, X., Huhman, D., Mewan, K. M., et al. (2013). Functional Characterization of Proanthocyanidin Pathway Enzymes from Tea and Their Application for Metabolic Engineering. Plant Physiol. 161 (3), 1103–1116. doi:10.1104/pp.112.212050
Park, C., Dwiyanti, M. S., Nagano, A. J., Liu, B., Yamada, T., and Abe, J. (2019). Identification of Quantitative Trait Loci for Increased α-tocopherol Biosynthesis in Wild Soybean Using a High-Density Genetic Map. BMC Plant Biol. 19, 510–515. doi:10.1186/s12870-019-2117-z
Park, M. Y., Yi, N. R., Lee, H. Y., Kim, S. T., Kim, M., Park, J.-H., et al. (2002). Generation of Chlorsulfuron-Resistant Transgenic Garlic Plants (Allium Sativum L.) by Particle Bombardment. Mol. Breed. 9 (3), 171–181. doi:10.1023/a:1019702705996
Pattnaik, M., Pandey, P., Martin, G. J. O., Mishra, H. N., and Ashokkumar, M. (2021). Innovative Technologies for Extraction and Microencapsulation of Bioactives from Plant-Based Food Waste and Their Applications in Functional Food Development. Foods 10 (2), 279. doi:10.3390/foods10020279
Pazhamala, L. T., Kudapa, H., Weckwerth, W., Millar, A. H., and Varshney, R. K. (2021). Systems Biology for Crop Improvement. Plant Genome (TSI), 1–23. doi:10.1002/tpg2.20098
Pellati, F., Bruni, R., Righi, D., Grandini, A., Tognolini, M., Pio Prencipe, F., et al. (2013). Metabolite Profiling of Polyphenols in a Terminalia Chebula Retzius Ayurvedic Decoction and Evaluation of its Chemopreventive Activity. J. Ethnopharmacology 147 (2), 277–285. doi:10.1016/j.jep.2013.02.025
Pérez-Díaz, R., Madrid-Espinoza, J., Salinas-Cornejo, J., González-Villanueva, E., and Ruiz-Lara, S. (2016). Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis Vinífera. Front. Plant Sci. 7, 1166. doi:10.3389/fpls.2016.01166
Prakash, B., Kujur, A., Yadav, A., Kumar, A., Singh, P. P., and Dubey, N. K. (2018). Nanoencapsulation: An Efficient Technology to Boost the Antimicrobial Potential of Plant Essential Oils in Food System. Food Control 89, 1–11. doi:10.1016/j.foodcont.2018.01.018
Pu, Z.-e., Yu, M., He, Q.-y., Chen, G.-y., Wang, J.-r., Liu, Y.-x., et al. (2014). Quantitative Trait Loci Associated with Micronutrient Concentrations in Two Recombinant Inbred Wheat Lines. J. Integr. Agric. 13 (11), 2322–2329. doi:10.1016/s2095-3119(13)60640-1
Qi, X., Xie, S., Liu, Y., Yi, F., and Yu, J. (2013). Genome-wide Annotation of Genes and Noncoding RNAs of Foxtail Millet in Response to Simulated Drought Stress by Deep Sequencing. Plant Mol. Biol. 83 (4-5), 459–473. doi:10.1007/s11103-013-0104-6
Qi, Z., Han, X., Hou, M., Xin, D., Wang, Z., Zhu, R., et al. (2014). QTL Analysis of Soybean Oil Content under 17 Environments. Can. J. Plant Sci. 94 (2), 245–261. doi:10.4141/cjps2013-219
Qu, C., Zhao, H., Fu, F., Zhang, K., Yuan, J., Liu, L., et al. (2016). Molecular Mapping and QTL for Expression Profiles of Flavonoid Genes in Brassica Napus. Front. Plant Sci. 7, 1691. doi:10.3389/fpls.2016.01691
Rai, A., Yamazaki, M., Takahashi, H., Nakamura, M., Kojoma, M., Suzuki, H., et al. (2016). RNA-seq Transcriptome Analysis of Panax Japonicus, and its Comparison with Other Panax Species to Identify Potential Genes Involved in the Saponins Biosynthesis. Front. Plant Sci. 7, 481. doi:10.3389/fpls.2016.00481
Rao, A. G., Hassan, M., and Hemple, C. (1994). Structure-function Validation of High Lysine Analogs of α-hordothionin Designed by Protein Modeling. Protein Eng. Des. Sel. 7 (12), 1485–1493. doi:10.1093/protein/7.12.1485
Raza, Q., Riaz, A., Sabar, M., Atif, R. M., and Bashir, K. (2019). Meta-analysis of Grain Iron and Zinc Associated QTLs Identified Hotspot Chromosomal Regions and Positional Candidate Genes for Breeding Biofortified rice. Plant Sci. 288, 110214. doi:10.1016/j.plantsci.2019.110214
Regina, A., Kosar-Hashemi, B., Ling, S., Li, Z., Rahman, S., and Morell, M. (2010). Control of Starch Branching in Barley Defined through Differential RNAi Suppression of Starch Branching Enzyme IIa and IIb. J. Exp. Bot. 61 (5), 1469–1482. doi:10.1093/jxb/erq011
Rhodes, D. H., Hoffmann, L., Rooney, W. L., Ramu, P., Morris, G. P., and Kresovich, S. (2014). Genome-wide Association Study of Grain Polyphenol Concentrations in Global Sorghum [Sorghum Bicolor (L.) Moench] Germplasm. J. Agric. Food Chem. 62 (45), 10916–10927. doi:10.1021/jf503651t
Riccardi, G., Capaldo, B., and Vaccaro, O. (2005). Functional Foods in the Management of Obesity and Type 2 Diabetes. Curr. Opin. Clin. Nutr. Metabol. Care 8 (6), 630–635. doi:10.1097/01.mco.0000171126.98783.0c
Richards, C., Hurst, B., Messner, R., and O'Connor, G. (2021). The Paradoxes of Food Waste Reduction in the Horticultural Supply Chain. Ind. Mark. Manag. 93, 82–491. doi:10.1016/j.indmarman.2020.12.002
Roesler, K., Shintani, D., Savage, L., Boddupalli, S., and Ohlrogge, J. (1997). Targeting of the Arabidopsis Homomeric Acetyl-Coenzyme A Carboxylase to Plastids of Rapeseeds. Plant Physiol. 113 (1), 75–81. doi:10.1104/pp.113.1.75
Sagare, D. B., Shetti, P., Surender, M., and Reddy, S. (2019). Marker-assisted Backcross Breeding for Enhancing β-carotene of QPM Inbreds. Mol. Breed. 39, 1–12. doi:10.1007/s11032-019-0939-x
Saito, K. (2018). Development of Plant Metabolomics and Medicinal Plant Genomics. Yakugaku Zasshi 138 (1), 1–18. doi:10.1248/yakushi.17-00193
Santhosha, S. G., Jamuna, P., and Prabhavathi, S. N. (2013). Bioactive Components of Garlic and Their Physiological Role in Health Maintenance: A Review. Food Biosci. 3, 59–74. doi:10.1016/j.fbio.2013.07.001
Sayeed, M. A., Bracci, M., Lazzarini, R., Tomasetti, M., Amati, M., Lucarini, G., et al. (2017). Use of Potential Dietary Phytochemicals to Target miRNA: Promising Option for Breast Cancer Prevention and Treatment? J. Funct. Foods 28, 177–193. doi:10.1016/j.jff.2016.11.008
Schijlen, E., Ric de Vos, C. H., Jonker, H., Van Den Broeck, H., Molthoff, J., Van Tunen, A., et al. (2006). Pathway Engineering for Healthy Phytochemicals Leading to the Production of Novel Flavonoids in Tomato Fruit. Plant Biotechnol. J 4 (4), 433–444. doi:10.1111/j.1467-7652.2006.00192.x
Sedeek, K. E. M., Mahas, A., and Mahfouz, M. (2019). Plant Genome Engineering for Targeted Improvement of Crop Traits. Front. Plant Sci. 10, 114. doi:10.3389/fpls.2019.00114
Shahidi, F., and Ambigaipalan, P. (2015). Phenolics and Polyphenolics in Foods, Beverages and Spices: Antioxidant Activity and Health Effects - A Review. J. Funct. Foods 18, 820–897. doi:10.1016/j.jff.2015.06.018
Shan, Q., Zhang, Y., Chen, K., Zhang, K., and Gao, C. (2015). Creation of Fragrant rice by Targeted Knockout of theOsBADH2gene Using TALEN Technology. Plant Biotechnol. J. 13 (6), 791–800. doi:10.1111/pbi.12312
Shao, Y., Jin, L., Zhang, G., Lu, Y., Shen, Y., and Bao, J. (2011). Association Mapping of Grain Color, Phenolic Content, Flavonoid Content and Antioxidant Capacity in Dehulled rice. Theor. Appl. Genet. 122 (5), 1005–1016. doi:10.1007/s00122-010-1505-4
Sharma, H. C., Crouch, J. H., Sharma, K. K., Seetharama, N., and Hash, C. T. (2002). Applications of Biotechnology for Crop Improvement: Prospects and Constraints. Plant Sci. 163 (3), 381–395. doi:10.1016/s0168-9452(02)00133-4
Sharma, K., Kumar, V., Kaur, J., Tanwar, B., Goyal, A., Sharma, R., et al. (2019). Health Effects, Sources, Utilization and Safety of Tannins: A Critical Review. Toxin Rev., 1–13. doi:10.1080/15569543.2019.1662813
Sharma, P., Gaur, V. K., Sirohi, R., Varjani, S., Hyoun Kim, S., and Wong, J. W. C. (2021). Sustainable Processing of Food Waste for Production of Bio-Based Products for Circular Bioeconomy. Bioresour. Technology 325, 124684. doi:10.1016/j.biortech.2021.124684
Shasidhar, Y., Vishwakarma, M. K., Pandey, M. K., Janila, P., Variath, M. T., Manohar, S. S., et al. (2017). Molecular Mapping of Oil Content and Fatty Acids Using Dense Genetic Maps in Groundnut (Arachis hypogaea L.). Front. Plant Sci. 8, 794. doi:10.3389/fpls.2017.00794
Shetti, P., Sagare, D. B., Surender, M., and Reddy, S. S. (2020). Development of Lysine and Tryptophan Rich maize (Zea mays) Inbreds Employing Marker Assisted Backcross Breeding. Plant Gene 23, 100236. doi:10.1016/j.plgene.2020.100236
Shewmaker, C. K., Sheehy, J. A., Daley, M., Colburn, S., and Ke, D. Y. (1999). Seed-specific Overexpression of Phytoene Synthase: Increase in Carotenoids and Other Metabolic Effects. Plant J. 20 (4), 401–412. doi:10.1046/j.1365-313x.1999.00611.x
Shi, J., Arunasalam, K., Yeung, D., Kakuda, Y., Mittal, G., and Jiang, Y. (2004). Saponins from Edible Legumes: Chemistry, Processing, and Health Benefits. J. Med. Food 7, 67–78. doi:10.1089/109662004322984734
Shirgurkar, M. V., Naik, V. B., von Arnold, S., Nadgauda, R. S., and Clapham, D. (2006). An Efficient Protocol for Genetic Transformation and Shoot Regeneration of Turmeric (Curcuma Longa L.) via Particle Bombardment. Plant Cel Rep 25 (2), 112–116. doi:10.1007/s00299-005-0033-1
Shu, X., and Rasmussen, S. r. K. (2014). Quantification of Amylose, Amylopectin, and β-Glucan in Search for Genes Controlling the Three Major Quality Traits in Barley by Genome-wide Association Studies. Front. Plant Sci. 5, 197. doi:10.3389/fpls.2014.00197
Sies, H., and Stahl, W. (1995). Vitamins E and C, Beta-Carotene, and Other Carotenoids as Antioxidants. AM. J. Clin. Nutr. 62, 1315S–1321S. doi:10.1093/ajcn/62.6.1315s
Singh, B., Singh, J. P., Singh, N., and Kaur, A. (2017). Saponins in Pulses and Their Health Promoting Activities: A Review. Food Chem. 233, 540–549. doi:10.1016/j.foodchem.2017.04.161
Smith, S. A., King, R. E., and Min, D. B. (2007). Oxidative and thermal Stabilities of Genetically Modified High Oleic sunflower Oil. Food Chem. 102 (4), 1208–1213. doi:10.1016/j.foodchem.2006.06.058
Sowbhagya, H. B. (2013). Chemistry, Technology, and Nutraceutical Functions of Cumin (Cuminum Cyminum L): an Overview. Crit. Rev. Food Sci. Nutr. 53 (1), 1–10. doi:10.1080/10408398.2010.500223
Stark, A. H., Crawford, M. A., and Reifen, R. (2008). Update on Alpha-Linolenic Acid. Nutr. Rev. 66, 326–332. doi:10.1111/j.1753-4887.2008.00040.x
Street, R. A., Prinsloo, G., and Mcgaw, L. J. (2017). “Alkaloids Potential Health Benefits and Toxicity,” in Utilisation of Bioactive Compounds from Agricultural and Food Production Waste (Boca Raton, FL: CRC Press), 60–85.
Sui, M., Jing, Y., Li, H., Zhan, Y., Luo, J., Teng, W., et al. (2020). Identification of Loci and Candidate Genes Analyses for Tocopherol Concentration of Soybean Seed. Front. Plant Sci. 11, 539460. doi:10.3389/fpls.2020.539460
Sun, P., Cheng, C., Lin, Y., Zhu, Q., Lin, J., and Lai, Z. (2017a). Combined Small RNA and Degradome Sequencing Reveals Complex microRNA Regulation of Catechin Biosynthesis in tea (Camellia Sinensis). PLoS One 12 (2), e0171173. doi:10.1371/journal.pone.0171173
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017b). Generation of High-Amylose rice through CRISPR/Cas9-mediated Targeted Mutagenesis of Starch Branching Enzymes. Front. Plant Sci. 8, 298. doi:10.3389/fpls.2017.00298
Thavarajah, D., Thavarajah, P., and Gupta, D. S. (2014). Pulses Biofortification in Genomic Era: Multidisciplinary Opportunities and Challenges. Legumes in the Omic Era, 207–220. doi:10.1007/978-1-4614-8370-0_10
Thies, F., Mills, L. M., Moir, S., and Masson, L. F. (2017). Cardiovascular Benefits of Lycopene: Fantasy or Reality? Proc. Nutr. Soc. 76, 122–129. doi:10.1017/s0029665116000744
Tian, J., Bryksa, B. C., and Yada, R. Y. (2016). Feeding the World into the Future - Food and Nutrition Security: the Role of Food Science and Technology. Front. Life Sci. 9 (3), 155–166. doi:10.1080/21553769.2016.1174958
Tian, Y. S., Wang, B., Peng, R. H., Xu, J., Li, T., Fu, X. Y., et al. (2019). Enhancing Carotenoid Biosynthesis in rice Endosperm by Metabolic Engineering. Plant Biotechnol. J. 17, 849–851. doi:10.1111/pbi.13059
Tong, H., and Nikoloski, Z. (2021). Machine Learning Approaches for Crop Improvement: Leveraging Phenotypic and Genotypic Big Data. J. Plant Physiol. 257, 153354. doi:10.1016/j.jplph.2020.153354
Tran, H. T. M., Ramaraj, T., Furtado, A., Lee, L. S., and Henry, R. J. (2018). Use of a Draft Genome of Coffee (C Offea Arabica ) to Identify SNP S Associated with Caffeine Content. Plant Biotechnol. J. 16, 1756–1766. doi:10.1111/pbi.12912
Trijatmiko, K. R., Dueñas, C., Tsakirpaloglou, N., Torrizo, L., Arines, F. M., Adeva, C., et al. (2016). Biofortified Indica rice Attains Iron and Zinc Nutrition Dietary Targets in the Field. Sci. Rep. 6 (1), 19792–19813. doi:10.1038/srep19792
Tripathi, S., Sangwan, R. S., Mishra, B., Jadaun, J. S., and Sangwan, N. S. (2020). Berry Transcriptome: Insights into a Novel Resource to Understand Development Dependent Secondary Metabolism in Withania Somnifera (Ashwagandha). Physiol. Plantarum 168 (1), 148–173. doi:10.1111/ppl.12943
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., and Dubcovsky, J. (2006). A NAC Gene Regulating Senescence Improves Grain Protein, Zinc, and Iron Content in Wheat. Science 314 (5803), 1298–1301. doi:10.1126/science.1133649
Van Eenennaam, A. L., Lincoln, K., Durrett, T. P., Valentin, H. E., Shewmaker, C. K., Thorne, G. M., et al. (2003). Engineering Vitamin E Content: from Arabidopsis Mutant to Soy Oil. Plant Cell 15 (12), 3007–3019. doi:10.1105/tpc.015875
Varshney, R., Graner, A., and Sorrells, M. (2005). Genomics-assisted Breeding for Crop Improvement. Trends Plant Sci. 10 (12), 621–630. doi:10.1016/j.tplants.2005.10.004
Varshney, R. K., Nayak, S. N., May, G. D., and Jackson, S. A. (2009). Next-generation Sequencing Technologies and Their Implications for Crop Genetics and Breeding. Trends Biotechnol. 27 (9), 522–530. doi:10.1016/j.tibtech.2009.05.006
Vasconcelos, M., Datta, K., Oliva, N., Khalekuzzaman, M., Torrizo, L., Krishnan, S., et al. (2003). Enhanced Iron and Zinc Accumulation in Transgenic rice with the Ferritin Gene. Plant Sci. 164 (3), 371–378. doi:10.1016/s0168-9452(02)00421-1
Vega-Gálvez, A., Miranda, M., Aranda, M., Henriquez, K., Vergara, J., Tabilo-Munizaga, G., et al. (2011). Effect of High Hydrostatic Pressure on Functional Properties and Quality Characteristics of Aloe Vera Gel (Aloe Barbadensis Miller). Food Chem. 129 (3), 1060–1065. doi:10.1016/j.foodchem.2011.05.074
Velmurugan, B., Mani, A., and Nagini, S. (2005). Combination of S-Allylcysteine and Lycopene Induces Apoptosis by Modulating Bcl-2, Bax, Bim and Caspases during Experimental Gastric Carcinogenesis. Eur. J. Cancer Prev. 14 (4), 387–393. doi:10.1097/00008469-200508000-00012
Vetriventhan, M., Azevedo, V. C. R., Upadhyaya, H. D., Nirmalakumari, A., Kane-Potaka, J., Anitha, S., et al. (2020). Genetic and Genomic Resources, and Breeding for Accelerating Improvement of Small Millets: Current Status and Future Interventions. Nucleus 63, 217–239. doi:10.1007/s13237-020-00322-3
Vinoth, A., and Ravindhran, R. (2017). Biofortification in Millets: a Sustainable Approach for Nutritional Security. Front. Plant Sci. 8, 29. doi:10.3389/fpls.2017.00029
Wang, C., Qiao, A., Fang, X., Sun, L., Gao, P., Davis, A. R., et al. (2019a). Fine Mapping of Lycopene Content and Flesh Color Related Gene and Development of Molecular Marker-Assisted Selection for Flesh Color in Watermelon (Citrullus lanatus). Front. Plant Sci. 10, 1240. doi:10.3389/fpls.2019.01240
Wang, C., Zeng, J., Li, Y., Hu, W., Chen, L., Miao, Y., et al. (2014). Enrichment of Provitamin A Content in Wheat (Triticum aestivum L.) by Introduction of the Bacterial Carotenoid Biosynthetic Genes CrtB and CrtI. J. Exp. Bot. 65 (9), 2545–2556. doi:10.1093/jxb/eru138
Wang, M., Amakye, W. K., Guo, L., Gong, C., Zhao, Y., Yao, M., et al. (2019b). Walnut‐Derived Peptide PW5 Ameliorates Cognitive Impairments and Alters Gut Microbiota in APP/PS1 Transgenic Mice. Mol. Nutr. Food Res. 63, 1900326. doi:10.1002/mnfr.201900326
Wang, Q., Reddy, V. A., Panicker, D., Mao, H.-Z., Kumar, N., Rajan, C., et al. (2016). Metabolic Engineering of Terpene Biosynthesis in Plants Using a Trichome-specific Transcription factorMsYABBY5from Spearmint (Mentha Spicata). Plant Biotechnol. J. 14 (7), 1619–1632. doi:10.1111/pbi.12525
Wei, Y., Bergenholm, D., Gossing, M., Siewers, V., and Nielsen, J. (2018). Expression of cocoa Genes in Saccharomyces cerevisiae Improves cocoa Butter Production. Microb. Cel Fact 17 (1), 11. doi:10.1186/s12934-018-0866-2
Welch, R. M., and Graham, R. D. (2004). Breeding for Micronutrients in Staple Food Crops from a Human Nutrition Perspective. J. Exp. Bot. 55 (396), 353–364. doi:10.1093/jxb/erh064
White, J., Pacey-Miller, T., Crawford, A., Cordeiro, G., Barbary, D., Bundock, P., et al. (2006). Abundant Transcripts of Malting Barley Identified by Serial Analysis of Gene Expression (SAGE). Plant Biotechnol. J. 4 (3), 289–301. doi:10.1111/j.1467-7652.2006.00181.x
Wu, H.-x., Jia, H.-m., Ma, X.-w., Wang, S.-b., Yao, Q.-s., Xu, W.-t., et al. (2014). Transcriptome and Proteomic Analysis of Mango (Mangifera Indica Linn) Fruits. J. Proteomics 105, 19–30. doi:10.1016/j.jprot.2014.03.030
Wu, Q., Chen, Z., Sun, W., Deng, T., and Chen, M. (2016). De Novo Sequencing of the Leaf Transcriptome Reveals Complex Light-Responsive Regulatory Networks in Camellia Sinensis Cv. Baijiguan. Front. Plant Sci. 7, 332. doi:10.3389/fpls.2016.00332
Wu, Y., Li, X., Xiang, W., Zhu, C., Lin, Z., Wu, Y., et al. (2012). Presence of Tannins in Sorghum Grains Is Conditioned by Different Natural Alleles of Tannin1. Proc. Natl. Acad. Sci. 109 (26), 10281–10286. doi:10.1073/pnas.1201700109
Xu, L.-Y., Wang, L.-Y., Wei, K., Tan, L.-Q., Su, J.-J., and Cheng, H. (2018). High-density SNP Linkage Map Construction and QTL Mapping for Flavonoid-Related Traits in a tea Plant (Camellia Sinensis) Using 2b-RAD Sequencing. BMC Genomics 19, 955. doi:10.1186/s12864-018-5291-8
Yadav, M., Sehrawat, N., Singh, M., Upadhyay, S. K., Aggarwal, D., and Sharma, A. K. (2020). Cardioprotective and Hepatoprotective Potential of Citrus Flavonoid Naringin: Current Status and Future Perspectives for Health Benefits. Ajbls 9, 1–5. doi:10.5530/ajbls.2020.9.1
Yang, R., Piao, Z., Wan, C., Lee, G., Ruan, X., and Bai, J. (2020). Breeding for Three-Line Japonica Hybrid rice Combinations with High Resistant Starch Content Using Molecular Marker-Assisted Selection. Breed. Sci. 70, 409–414. doi:10.1270/jsbbs.20005
Yang, R., Yan, Z., Wang, Q., Li, X., and Feng, F. (2018). Marker-assisted Backcrossing of lcyE for Enhancement of proA in Sweet Corn. Euphytica 214, 1–12. doi:10.1007/s10681-018-2212-5
Yao, L. H., Jiang, Y. M., Shi, J., Datta, N., Singanusong, R., Chen, S. S., et al. (2004). Flavonoids in Food and Their Health Benefits. Plant Foods Hum. Nutr. 59, 113–122. doi:10.1007/s11130-004-0049-7
Ye, X., Al-Babili, S., Klöti, A., Zhang, J., Lucca, P., Beyer, P., et al. (2000). Engineering the Provitamin A (-Carotene) Biosynthetic Pathway into (Carotenoid-free) Rice Endosperm. Science 287 (5451), 303–305. doi:10.1126/science.287.5451.303
Yousuf, B., Gul, K., Wani, A. A., and Singh, P. (2016). Health Benefits of Anthocyanins and Their Encapsulation for Potential Use in Food Systems: a Review. Crit. Rev. Food Sci. Nutr. 56, 2223–2230. doi:10.1080/10408398.2013.805316
Yu, B., Lydiate, D. J., Young, L. W., Schäfer, U. A., and Hannoufa, A. (2008). Enhancing the Carotenoid Content of Brassica Napus Seeds by Downregulating Lycopene Epsilon Cyclase. Transgenic Res. 17 (4), 573–585. doi:10.1007/s11248-007-9131-x
Yusuf, M. A., and Sarin, N. B. (2007). Antioxidant Value Addition in Human Diets: Genetic Transformation of Brassica Juncea with γ-TMT Gene for Increased α-tocopherol Content. Transgenic Res. 16 (1), 109–113. doi:10.1007/s11248-006-9028-0
Zenda, T., Liu, S., Dong, A., and Duan, H. (2021). Advances in Cereal Crop Genomics for Resilience under Climate Change. Life 11 (6), 502. doi:10.3390/life11060502
Zhang, L., Su, W., Tao, R., Zhang, W., Chen, J., Wu, P., et al. (2017). RNA Sequencing Provides Insights into the Evolution of Lettuce and the Regulation of Flavonoid Biosynthesis. Nat. Commun. 8 (1), 2264–2312. doi:10.1038/s41467-017-02445-9
Zhang, M., Pinson, S. R. M., Tarpley, L., Huang, X.-Y., Lahner, B., Yakubova, E., et al. (2014). Mapping and Validation of Quantitative Trait Loci Associated with Concentrations of 16 Elements in Unmilled rice Grain. Theor. Appl. Genet. 127 (1), 137–165. doi:10.1007/s00122-013-2207-5
Zhang, P., Guo, Z., Ullah, S., Melagraki, G., Afantitis, A., and Lynch, I. (2021). Nanotechnology and Artificial Intelligence to Enable Sustainable and Precision Agriculture. Nat. Plants 7 (7), 864–876. doi:10.1038/s41477-021-00946-6
Zhao, Y., Sun, H. Y., Wang, Y. Y., Pu, Y. Y., Kong, F. M., and Li, S. S. (2013). QTL Mapping for the Color, Carotenoids and Polyphenol Oxidase Activity of Flour in Recombinant Inbred Lines of Wheat. Austr. J. Crop Sci. 7 (3), 328.
Zhao, Z.-y., Glassman, K., Sewalt, V., Wang, N., Miller, M., Chang, S., et al. (2003). “Nutritionally Improved Transgenic Sorghum,” in Plant Biotechnology 2002 and beyond (Springer), 413–416. doi:10.1007/978-94-017-2679-5_85
Zhao, Z.-y. (2007). “The Africa Biofortified Sorghum Project- Applying Biotechnology to Develop Nutritionally Improved Sorghum for Africa,” in Biotechnology and Sustainable Agriculture 2006 and beyond (Springer), 273–277. doi:10.1007/978-1-4020-6635-1_41
Zheng, X., Tang, Y., Ye, J., Pan, Z., Tan, M., Xie, Z., et al. (2018). SLAF-based Construction of a High-Density Genetic Map and its Application in QTL Mapping of Carotenoids Content in Citrus Fruit. J. Agric. Food Chem. 67 (3), 994–1002. doi:10.1021/acs.jafc.8b05176
Zhou, S., Kremling, K. A., Bandillo, N., Richter, A., Zhang, Y. K., Ahern, K. R., et al. (2019). Metabolome-scale Genome-wide Association Studies Reveal Chemical Diversity and Genetic Control of maize Specialized Metabolites. Plant Cell 31 (5), 937–955. doi:10.1105/tpc.18.00772
Keywords: nutrition, genomics, transgene, functional foods, nutraceuticals
Citation: Nayak SN, Aravind B, Malavalli SS, Sukanth BS, Poornima R, Bharati P, Hefferon K, Kole C and Puppala N (2021) Omics Technologies to Enhance Plant Based Functional Foods: An Overview. Front. Genet. 12:742095. doi: 10.3389/fgene.2021.742095
Received: 15 July 2021; Accepted: 13 October 2021;
Published: 08 November 2021.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Mehanathan Muthamilarasan, University of Hyderabad, IndiaSeetha Anitha, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India
Salej Sood, Indian Council of Agricultural Research (ICAR), India
Copyright © 2021 Nayak, Aravind, Malavalli, Sukanth, Poornima, Bharati, Hefferon, Kole and Puppala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naveen Puppala, bnB1cHBhbGFAbm1zdS5lZHU=; Chittaranjan Kole, Y2tvbGVvcmdAZ21haWwuY29t
 Spurthi N. Nayak
Spurthi N. Nayak B. Aravind
B. Aravind Sachin S. Malavalli
Sachin S. Malavalli B. S. Sukanth
B. S. Sukanth R. Poornima
R. Poornima Pushpa Bharati
Pushpa Bharati Kathleen Hefferon
Kathleen Hefferon Chittaranjan Kole4*
Chittaranjan Kole4* Naveen Puppala
Naveen Puppala