- 1Jiangsu Key Laboratory for Bioresources of Saline Soils, Jiangsu Synthetic Innovation Center for Coastal Bio-agriculture, School of Wetlands, Yancheng Teachers University, Yancheng, China
- 2College of Forestry, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Fujian Colleges and Universities Engineering Research Institute for Conservation and Utilization of Natural Bioresources, Fuzhou, China
Euscaphis konishii is an evergreen plant that is widely planted as an industrial crop in Southern China. It produces red fruits with abundant secondary metabolites, giving E. konishii high medicinal and ornamental value. Auxin signaling mediated by members of the AUXIN RESPONSE FACTOR (ARF) and auxin/indole-3-acetic acid (Aux/IAA) protein families plays important roles during plant growth and development. Aux/IAA and ARF genes have been described in many plants but have not yet been described in E. konishii. In this study, we identified 34 EkIAA and 29 EkARF proteins encoded by the E. konishii genome through database searching using HMMER. We also performed a bioinformatic characterization of EkIAA and EkARF genes, including their phylogenetic relationships, gene structures, chromosomal distribution, and cis-element analysis, as well as conserved motifs in the proteins. Our results suggest that EkIAA and EkARF genes have been relatively conserved over evolutionary history. Furthermore, we conducted expression and co-expression analyses of EkIAA and EkARF genes in leaves, branches, and fruits, which identified a subset of seven EkARF genes as potential regulators of triterpenoids and anthocyanin biosynthesis. RT-qPCR, yeast one-hybrid, and transient expression analyses showed that EkARF5.1 can directly interact with auxin response elements and regulate downstream gene expression. Our results may pave the way to elucidating the function of EkIAA and EkARF gene families in E. konishii, laying a foundation for further research on high-yielding industrial products and E. konishii breeding.
Introduction
Plant secondary metabolites are not only important to plant development, but are also significant as nutritional resources for humans, as sources for color and odorants, and as potential materials for drug discovery (Huang et al., 2020a; Bing et al., 2021). Secondary metabolites include terpenes, phenolic compounds, and alkaloids, which play essential roles as food additives and in medicine, cosmetics, skincare, and industrial chemicals. The biosynthesis of plant secondary metabolites is very complex and is regulated by various environmental signals and development cues. Because of their critical roles, much attention is being paid to the biosynthesis and regulatory mechanism of secondary metabolites.
The phytohormone auxin regulates a wide range of processes in plant growth and development, including vascular differentiation, lateral root formation, apical dominance, and fruit development (Liscum and Reed, 2002; Aloni et al., 2006). The auxin/indole-3-acetic acid (Aux/IAA) and AUXIN RESPONSE FACTOR (ARF) proteins are critical players of auxin signal transduction (Hagen, 2015). ARFs are transcription factors consisting of a DNA-binding domain (DBD), a middle transcriptional regulatory region (MR), and a dimerization domain at their C termini (CTD), with the MR serving as an activation or repressor domain (Tiwari et al., 2003; Guilfoyle and Hagen, 2007). ARF DBDs bind to auxin response elements (AuxREs) located in the promoter region of auxin-responsive genes to activate or repress their transcription, depending on the type of MR (Tiwari et al., 2003; Guilfoyle and Hagen, 2007). Aux/IAA proteins comprise four domains, denoted domains I, II, III, and IV (Tiwari et al., 2001). Located at the N terminus, domain I is characterized by the LxLxLx motif and is mainly responsible for repression of gene expression. Domain II mediates the degradation of the protein via the degron sequence (GWPPV) by the 26S proteasome (Hagen and Guilfoyle, 2002; Tiwari et al., 2004). Domains III and IV at the C terminus interact with the CTD of ARFs to form homo- and heterodimers, resulting in the transcriptional induction or repression of downstream auxin-responsive genes (Ulmasov et al., 1999; Guilfoyle, 2015). The interaction between Aux/IAA and ARF proteins also depends on auxin concentration. When auxin levels are low, ARFs bind to AuxREs, but transcription is repressed through their interaction with Aux/IAAs. With increasing auxin levels, Aux/IAAs are degraded via the 26S proteasome, thus alleviating the repressive effect of Aux/IAAs on ARFs (Gray et al., 2001; Winkler et al., 2017; Roosjen et al., 2018). Both Aux/IAA and ARF proteins are encoded by large gene families with distinct expression patterns and regulatory mechanisms, contributing to the complexity of auxin signaling (Luo et al., 2018).
Aux/IAA and ARF family members play critical and extensive roles during the entire plant life cycle. During embryogenesis, ARF5 is involved in organ formation in Arabidopsis (Arabidopsis thaliana) (Hardtke and Berleth, 1998). ARF19 and ARF7 redundantly regulate lateral root initiation (Wilmoth et al., 2005), while ARF1 and ARF2 control floral organ senescence and abscission (Ellis et al., 2005). Most of the Aux/IAA family members described in Arabidopsis, such as IAA3, IAA14, IAA28, and IAA19, affect the growth and development of lateral roots (Goh et al., 2012; Luo et al., 2018). Aux/IAAs and ARFs also play important roles in regulating fruit development. ARF8 was reported to play a negative role during fertilization and fruit initiation in Arabidopsis (Goetz et al., 2006). SlARF10 and SIARF6A are involved in chlorophyll and sugar accumulation during tomato (Solanum lycopersicum) fruit development (Yuan et al., 2018a; Yuan et al., 2019). Auxin signaling also interacts with other phytohormone signaling pathways to control fruit development. For example, the tomato gibberellic acid (GA) repressor SlDELLA interacts with SlARF7/SlIAA9 to regulate fruit initiation (Hu et al., 2018). Likewise, SlIAA3 contributes to differential growth by integrating auxin and ethylene signaling (Chaabouni et al., 2009). The accumulation of secondary metabolites is an important biological process during fruit ripening, which includes fruit color formation and biosynthesis of volatile odorants. In apple (Malus domestica), MdARF13 interacts with MdIAA121 to regulate anthocyanin biosynthesis (Wang et al., 2018). Overexpression of MdIAA26 boosts anthocyanin biosynthesis in apple and Arabidopsis seedlings (Wang et al., 2020). In addition to anthocyanins, auxin induces the accumulation of flavonols by promoting the expression of the gene encoding a key biosynthetic enzyme (Lewis et al., 2011). Auxin signaling also participates in the accumulation of glucosinolates, a class of important plant defense metabolites (Mitreiter and Gigolashvili, 2021). Although auxin plays critical roles in secondary metabolite biosynthesis, how auxin signaling synergistically regulates the contents of multiple secondary metabolites is unclear.
Euscaphis plants belong to the Staphyleaceae family and are deciduous shrubs or small trees widely distributed in East Asia, from Japan to Southern China (Zhang et al., 2012). These plants constitute an important industrial crop due to the medicinal compounds and other industrially desirable products extracted from their fruits, leaves, and roots, such as triterpenes, phenolic acid, and flavonoids (Liang et al., 2018). In China, E. konishii is cultivated as an ornamental plant due to its beautiful red-winged pericarp and also has a long history of use as a medicinal plant to cure colds and allergies (Yuan et al., 2018b). The potential medical applications of Euscaphis were recently supported by data indicating that total phenolic and methanolic extracts of Euscaphis plants can mitigate liver fibrosis in mice and inhibit hepatic stem cell proliferation (Lee et al., 2009; Huang et al., 2020b), prompting the expansion of E. konishii cultivation (Sun et al., 2019). Therefore, E. konishii is an economically useful crop for the production of medicinal compounds. However, the molecular mechanisms governing the biosynthesis of these compounds in this species are poorly understood, which limits the breeding of improved E. konishii varieties. Although the Aux/IAA and ARF families have been systematically identified and characterized in many plants, including Arabidopsis (Remington et al., 2004; Okushima et al., 2005), rice (Oryza sativa) (Jain et al., 2006; Wang et al., 2007), poplar (Populus trichocarpa) (Kalluri et al., 2007), maize (Zea mays) (Wang et al., 2010; Xing et al., 2011), soybean (Glycine max) (Van Ha et al., 2013; Singh and Jain, 2015), and pepper (Capsicum annuum) (Yu et al., 2017; Waseem et al., 2018), their functions in E. konishii are not clear. Given the importance of Aux/IAA and ARF proteins in plant development, we undertook a comprehensive survey of the Aux/IAA and ARF gene families in E. konishii to better understand the biosynthetic pathways of medicinal secondary metabolites. In this study, we identified 34 Aux/IAA and 29 ARF genes in E. konishii and analyzed their sequence features, phylogenetic relationships, cis-elements, and co-expression profiles. We also explored the function of EkARFs in the context of phenolic and anthocyanin biosynthesis. Our results may have uncovered a potential role for auxin in the biosynthesis of secondary products, which may provide useful information for breeding E. konishii with a high content of medicine compounds.
Materials and Methods
Identification of IAA and ARF Genes
Hidden Markov model (HMM) logos of Aux/IAA (PF02309) and ARF (PF06507) proteins were downloaded from the Pfam database (Finn et al., 2014) and used to scan the E. konishii predicted proteome (Sun et al., 2021) with the HMMER software package (Finn et al., 2011). The resulting Aux/IAA and ARF candidates were further used to generate HMM logos for EkIAAs and EkARFs using hmm-build from the HMMER suite (Finn et al., 2011), before scanning the E. konishii proteome again. Proteins with an E-value lower than 0.01 were retained, and the presence of conserved ARF or IAA domains was confirmed using the Conserved Domains Database (Marchler-Bauer et al., 2011), Pfam (Finn et al., 2014), and the Simple Modular Architecture Research Tool (Letunic and Bork, 2018). The proteins meeting all of the above criteria were used for further study. The number of amino acids, the predicted molecular weight, and the theoretical isoelectric point (pI) were determined using the ExPASy server (http://web.expasy.org/protparam/) (Gasteiger et al., 2003).
Gene Structure and Motif Analysis
TBtools (Chen et al., 2020) was employed to illustrate the exon/intron structures of all EkIAA and EkARF genes. Conserved protein motifs in their encoded proteins were predicted by the MEME program (parameters: number of maximum patterns, 10; maximum width, 50) (http://memesuite.org/tools/meme) (Bailey et al., 2006).
Multiple Sequence Alignment and Phylogenetic Analysis
The protein sequences for the 29 ARFs and 34 IAAs from Arabidopsis were obtained from published references (https://www.arabidopsis.org), and the protein sequences of 14 ARFs and 11 IAAs from Amborella trichopoda were identified following the same method described for E. konishii. Full-length protein sequences for all IAAs and ARFs identified in E. konishii, Arabidopsis, and A. trichopoda were used for phylogenetic analysis. The phylogenetic tree was built with the maximum likelihood method on the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at/) (Nguyen et al., 2015).
Analysis of Cis-acting Elements in the EkIAA and EkARF Promoters
The upstream sequences (2 kb) of EkIAA and EkARF genes were extracted via TBtools (Chen et al., 2020) and then submitted to the PlantCARE database (Lescot et al., 2002) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify cis-elements.
EkIAA and EkARF Chromosomal Location and Duplication Event Analysis
The chromosomal distribution and location of all EkIAA and EkARF genes were acquired from the E. konishii genome annotation file. Colinear circles for EkIAA and EkARF genes were drawn with TBtools (Chen et al., 2020). Duplication events were confirmed on the basis of coverage (>70% of the entire gene body) and similarity (70%) of the two aligned sequences (Gu et al., 2002) and were considered tandem duplication pairs if they were located within 100 kb (Mehan et al., 2004). Genes located in duplicated regions with 70% similarity were identified as segmental duplications (Mehan et al., 2004). Ka/Ks values were calculated with TBtools (Chen et al., 2020).
Expression Analysis of IAA and ARF Genes by Transcriptome Deep Sequencing (RNA-Seq)
The RNA-seq data for three development stages (green, turning, and red fruit) and three tissues (red-winged pericarp, branch, and leaf) were downloaded from the National Center for Biotechnological Information (NCBI) Sequence Read Archive (SRA) under the accession numbers PRJNA548305 and PRJNA548305, respectively. The RNA-seq reads were mapped to the E. konishii reference genome via Salmon algorithm, and the transcripts per million reads (TPM) for AUX/IAA and ARF genes were extracted for further analysis. The heatmaps were drawn using TBtools (Chen et al., 2020).
Correlation Analysis
To study the effect of auxin on the regulation of anthocyanin and terpenoid biosynthesis in E. konishii, a comprehensive correlation analysis was first performed using the correlation test in R between anthocyanin contents and the expression levels of anthocyanin biosynthetic genes. Biosynthetic genes whose expression was positively correlated with anthocyanin contents were selected for further correlation analysis between their expression levels and those of EkARF genes. Pearson’s correlation coefficients (r, p-value < 0.05) were used to define five correlation levels: no correlation (|r| ≤ 0.2), weak correlation (0.21 ≤ |r| ≤ 0.35), moderate correlation (0.36 ≤ |r| ≤ 0.67), strong correlation (0.68 ≤ |r| ≤ 0.90), and very strong correlation (0.91 ≤ |r| ≤ 1), with r > 0 indicating positive correlations and r < 0 negative correlations (Prion and Haerling, 2014).
RT-qPCR Verification of EkARF5.1 Gene Expression
Fruits at the green stage, turning stage, and red stage from E. konishii were harvested as materials for RT-qPCR. Total RNA was extracted using the RNAprep Pure kit (Tiangen, China), and then 1 µg of total RNA per sample was subjected to reverse transcription using the PrimeScript RT Reagent Kit (Takara, China) with gDNA Eraser (Takara, China). The specific primers for EkARF5.1 (F: 5′-GCAACCTCCAACTCAAGAGC-3′, R: 5′-GACGCCTCACACCCACTAAT-3′) were designed by Primer 5 software and synthetized by Sangon Biotech (Shanghai, China). UBC23 (F: 5′-AGCCACATAATCTCCGTGTAAG-3′, R: 5′-GCTGACCATGTTCGAGTAGTT-3′) was used as an internal reference (Yuan et al., 2018b). The reaction mixture consisted of 10 µl of 2× GoTaq qPCR Master Mix (Promega, United States), 0.4 µl of each gene-specific primer, 1 µl of cDNA (10× dilution), 0.4 µl of dye, and 7.8 µl of nuclease-free water. The reaction conditions were as follows: 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative gene expression levels were calculated by the comparative ΔCt method. Three biological replications were assessed per sample.
DNA Binding and Transactivation Assay
The full-length EkARF5.1 coding sequence was cloned into the pGBKT7 vector to generate BD-EkARF5.1, which was then introduced into yeast strain Y2HGold. The resulting colonies were grown on synthetic defined (SD) medium lacking Trp and His for 2 days to observe the transcriptional activation activity of EkARF5.1, using empty pGBKT7 as a negative control.
To test the binding of EkARF5.1 to AuxREs, seven repeats of the AuxRE element (TGTCTC) were inserted into the multiple cloning site of the pAbAi vector to generate the 7×TGTCTC-pAbAi vector, which was integrated into the Y1HGold genome to construct the bait reporter strain. The AD-EkARF5.1 clone was generated by subcloning the full-length EkARF5.1 coding sequence into the pGADT7 vector. AD-EkARF5.1 was then transformed into the bait reporter strain. The transformants were spotted onto SD medium lacking Leu or the same medium containing 100 ng/ml of the antibiotic aureobasidin A (AbA) and allowed to grow for 48 h to analyze binding activity.
Constructs consisting of the β-GLUCURONIDASE (GUS) reporter 7×TGTCTC:GUS (cloned in pMDC164) and 35S:EkARF5.1 (cloned in pDMC32) were introduced into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 for infiltration in Nicotiana benthamiana leaf epidermal cells. Agrobacterium harboring 7×TGTCTC:GUS and 35S:EkARF5.1 were infiltrated into the abaxial side of N. benthamiana leaves with a syringe as described previously (Liu et al., 2014). N. benthamiana leaves were stained for GUS activity 3 days after infiltration.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: The E. konishii chromosome-level genome assembly and annotation data (Accession No. PRJCA005268/GWHBCHS00000000) were available from National Genomics Data Center at BioProject/GWH (https://bigd.big.ac.cn/gwhc).
Results
Genome-wide Identification of EkIAA and EkARF Genes in the E. konishii Genome
To identify EkIAA and EkARF genes, we searched the E. konishii genome using HMMER v3 in two rounds (see Materials and Methods for details). We analyzed the resulting protein sequences using the Conserved Domains Database, Simple Modular Architecture Research Tool, and Pfam database, which resulted in 34 Aux/IAA and 29 ARF candidate proteins (Figure 1; Supplementary Table S1). We numbered the E. konishii IAA and ARF genes based on their homologs in Arabidopsis (Remington et al., 2004; Okushima et al., 2005); the full list is provided in Supplementary Table S1, along with their gene IDs, their coding sequences, genomic DNA and protein sequences, the lengths of the coding and protein sequences, and the predicted molecular weights and isoelectric points (pI) of the proteins.
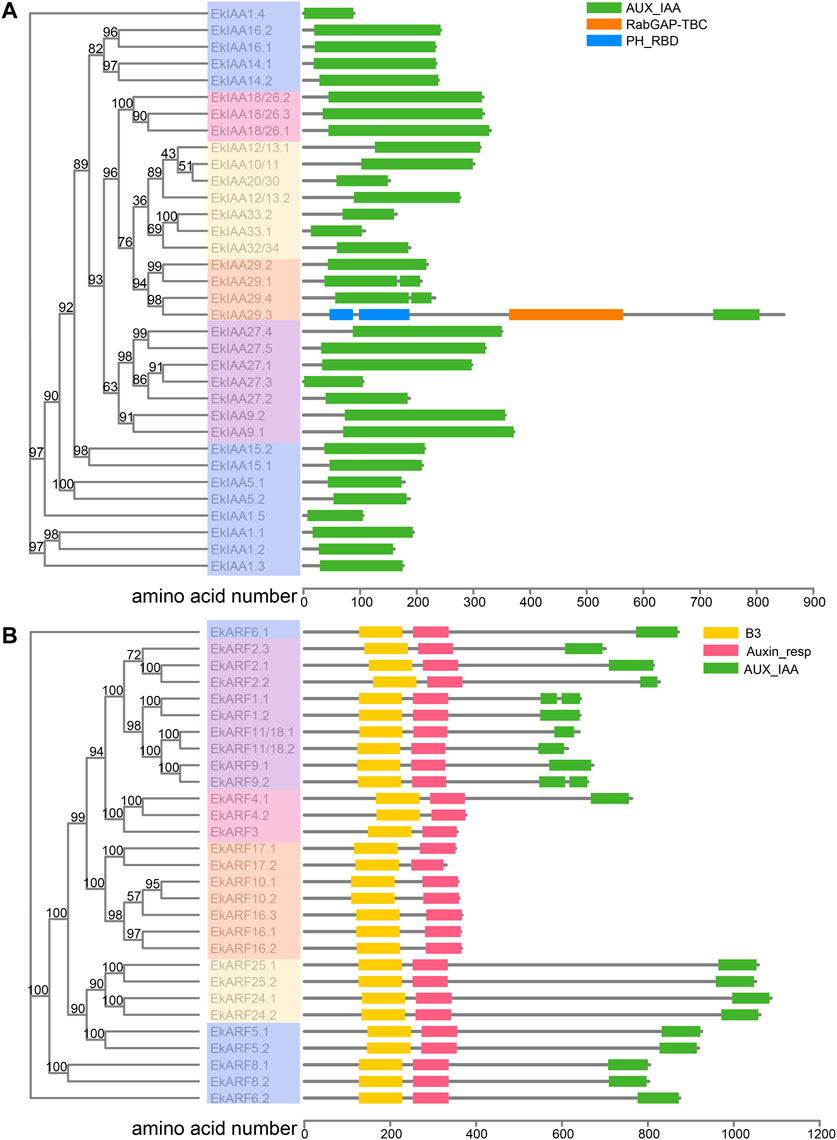
FIGURE 1. Phylogenetic relationships and protein domain maps of E. konishii IAA (A) and ARF (B) proteins. Left, phylogenetic relationship of 34 IAAs and 29 ARFs from E. konishii. Different colors represent different groups. Right, IAA and ARF protein domain composition. Bootstrap values are shown close to branch nodes.
We observed a broad variation in the lengths and biochemical properties of EkIAA and EkARF proteins. EkIAA proteins ranged from 91 (EkIAA1.4) to 849 (EkIAA29.3) amino acids, with predicted molecular weights from 10.5 to 97.2 kDa (Supplementary Table S1). The predicted pI values of EkIAA proteins varied from 4.7 (EkIAA32/34) to 9.3 (EkIAA33.2). Similarly, EkARF proteins ranged in length from 432 (EkARF17.2) to 1,110 (EkARF24.1) amino acids with predicted molecular weights from 48.2 to 122.7 kDa (Supplementary Table S1). The predicted pI values of EkARF proteins varied from 5.41 (EkARF5.1) to 8.42 (EkARF10.2) (Supplementary Table S1).
Phylogenetic Analysis of EkIAAs and EkARFs
To better understand their evolutionary history, we subjected all IAAs and ARFs identified in the model plant Arabidopsis, the early angiosperm A. trichopoda, and E. konishii to phylogenetic analysis with the MEGA-X software package (Kumar et al., 2018) and the IQ-TREE web server (Nguyen et al., 2015). In both protein families, individual members clustered into five branches, indicating that IAAs and ARFs are highly differentiated (Figure 2A), as previously reported in Arabidopsis (Remington et al., 2004) and poplar (Kalluri et al., 2007). EkIAA proteins were equally divided among groups Ⅰ, Ⅲ, and Ⅴ (Figure 2A). EkIAA5 and EkIAA15 from group I appeared to have undergone gene duplication, while group I had no clear E. konishii orthologs for Arabidopsis IAA6 or IAA19 (Figure 2A). As illustrated by the size of groups Ⅱ and Ⅳ, EkIAA genes have undergone gene duplication, especially IAA27 and IAA29 (Figure 2A). As with IAAs, the phylogenetic analysis of ARFs also divided the proteins into five groups (Figure 2B), as previously reported in Arabidopsis (Okushima et al., 2005). Group I consisted of six EkARF and five Arabidopsis ARF members, the latter having been reported to exhibit transcriptional activation activity (Guilfoyle and Hagen, 2007). Of note, ARF5, ARF6, and ARF8 all showed gene duplication in the E. konishii genome, while orthologs for ARF7 and ARF19 appeared to be lacking. Some, but not all, group II members showed signs of gene duplication (Figure 2B), for example, ARF1 and ARF9. Notably, IAAs and ARFs displayed the same distribution across groups in Arabidopsis and E. konishii, with the exception of group Ⅴ ARFs, which indicates that the IAA and ARF gene families in E. konishii are likely conserved. Within each group, several EkIAA and EkARF members had experienced duplication, with the exception of group Ⅴ IAA members (Figure 2A). In addition, we noted the absence of clear orthologs for several IAAs and ARFs in several groups (Figure 2), indicative of their independent evolution in E. konishii since the divergence from Arabidopsis.
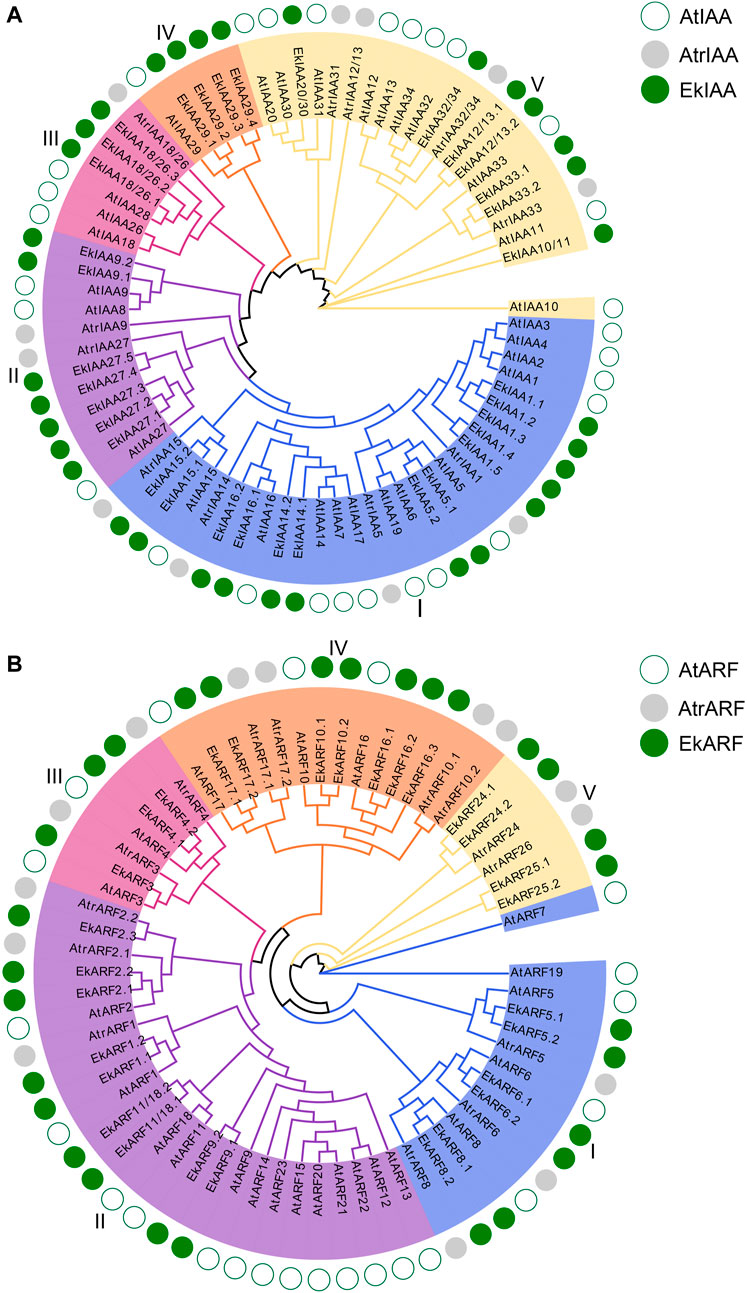
FIGURE 2. Phylogenetic trees of EkIAAs (A) and EkARFs (B) from Arabidopsis, Amborella trichopoda, and E. konishii. Different groups have different colors. The green open circles, gray circles, and green solid circles represent Arabidopsis, A. trichopoda, and E. konishii, respectively.
Chromosomal Location and Gene Duplication Events of EkIAA and EkARF Genes
We determined the genomic positions of all EkIAA and EkARF genes along the linkage groups (LGs) of the E. konishii genome. Both groups of genes were unevenly distributed in the E. konishii LGs (Figure 3). For example, LG02 alone harbored eight EkIAA genes, whereas no EkIAA gene mapped to LG07 or LG11 (Figure 3). Several EkIAA genes clustered in close proximity on LG01, LG2, LG03, and LG08 (Figure 3). The pattern for the EkARF genes was similar: six EkARF genes mapped to LG03, five to LG05, three each to LG06 and LG07, one each to LG09 and LG12, and none to LG02 (Figure 3).
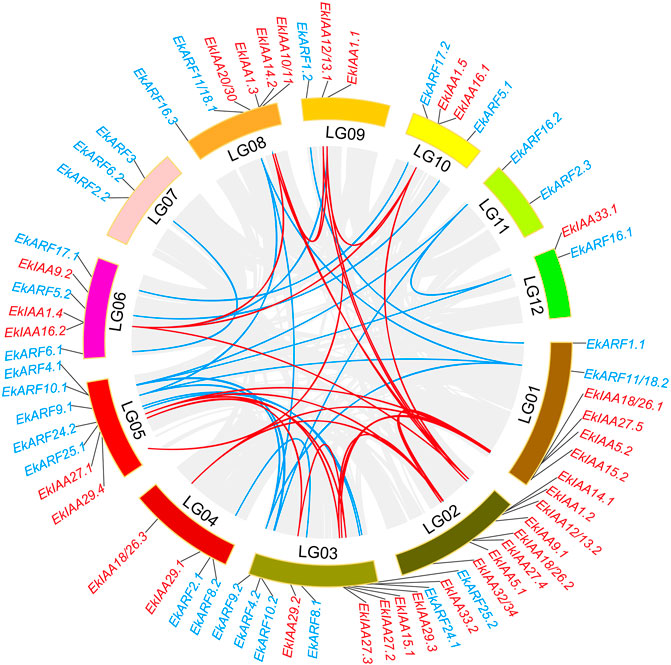
FIGURE 3. Chromosomal locations and segmental duplicated genes for 34 EkIAA and 29 EkARF genes. Linkage group numbers (LG01–LG12) are shown at the bottom. Gray lines show all synteny blocks in the E. konishii genome, red lines show segmental duplication of IAA genes, and blue lines show segmental duplication of ARF genes.
During genome evolution, gene duplication and neofunctionalization are driven by tandem and segmental duplication (Cannon et al., 2004; Freeling, 2009). To elucidate the expansion of the EkIAA and EkARF gene families in E. konishii, we studied their segmental and tandem duplications. We identified 20 instances of segmental duplication events, involving 11 EkIAA and 20 EkARF genes, but no tandem duplication events in either gene family (Figure 3). For the 10 pairs of segmental duplicated EkIAA genes and the 10 pairs of segmental duplicated EkARF genes identified above, we calculated the ratios of nonsynonymous to synonymous substitutions (Ka/Ks) to evaluate their molecular evolutionary rates (Supplementary Table S2). The ratios for all duplicated pairs were less than 1 (Supplementary Table S2), suggesting that duplicated pairs of genes underwent purifying selection during evolution, thus raising the possibility that the biochemical characteristics of these EkIAAs and EkIAAs may not have changed very much since the initial duplication event.
Analysis of Conserved Motifs and Gene Structure
Protein motifs are critical for protein function and structure maintenance (Smith-Gill, 1991). Accordingly, we looked for conserved functional motifs in the predicted EkIAA and EkARF proteins with the MEME web server tool. We identified four domains conserved in EkIAA proteins (motifs 1–4), corresponding to IAA domains Ⅳ, Ⅲ, Ⅱ, and Ⅰ, respectively (Figure 4). Of the 34 EkIAA proteins, 23 (61.8%) contained all four conserved domains (domains I–IV) (Figure 4). Some EkIAA proteins lost one or more domains: For example, EkIAA1.3, EkIAA29.1, EkIAA29.2, EkIAA29.3, and EkIAA29.4 lack domain I; EkIAA20/30, EkIAA32/34, EkIAA33.1, and EkIAA33.2 lack domain II; EkIAA1.5 lacks domains I and IV; EkIAA27.2 lacks domains III and IV; and EkIAA1.4 and EkIAA27.3 have only domain I (Figure 4).
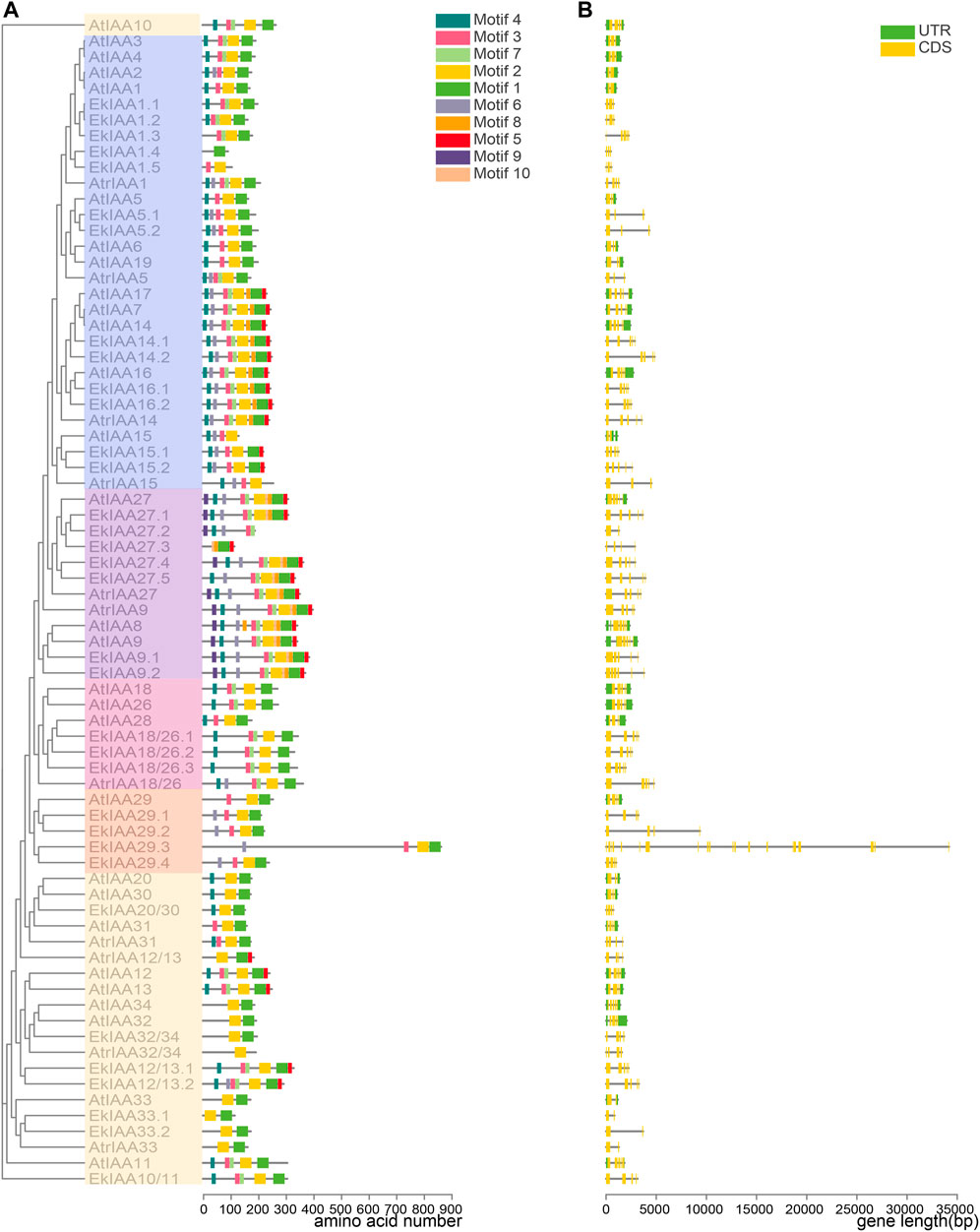
FIGURE 4. Gene structures of EkIAAs and conserved motifs in EkIAAs. (A) EkIAA phylogenetic tree and corresponding EkIAA gene structures. (B) Conserved protein motifs within EkIAAs. The phylogenetic tree was generated using the IQ-tree web server. Gene structures of EkIAAs were predicted with TBtools. The conserved motifs were analyzed using the MEME web server.
To understand the evolution of EkIAA genes, we examined their exon-intron structures (Figure 4A). Most EkIAA genes consisted of five exons and four introns, as in Arabidopsis, although introns in EkIAA genes were larger than those in their Arabidopsis counterparts (Figure 4). Three EkIAA genes (EkIAA27.2, EkIAA33.1, and EkIAA33.2) comprised only two exons and one intron, with another six EkIAA genes (EkIAA1.1, EkIAA1.2, EkIAA1.4, EkIAA1.5, EkIAA5.1, and EkIAA5.2) having three exons and two introns. Six EkIAA genes (EkIAA20/30, EkIAA27.3, EkIAA29.1, EkIAA29.2, EkIAA29.4, and EkIAA32/34) had four exons and three introns. EkIAA9.2 had six exons and five introns, while EkIAA29.3 had by far the most exons (20) and 19 introns (Figure 4). As the presence of conserved domains in the EkIAA proteins and the EkIAA gene structure are similar to those in their Arabidopsis orthologs (Remington et al., 2004), these results suggested that the EkIAA family in E. konishii is conserved.
We analyzed the conserved motifs and gene structure of EkARF proteins and EkARF genes, respectively (Supplementary Figure S1). EkARF proteins belonging to the same clade in the phylogenetic tree had the same functional motifs (Supplementary Figure S1). The DNA binding domain was represented by motifs 1, 9, and 10, while motifs 3, 6, and 8 matched the variable middle transcriptional regulatory region (MR). Motifs 7 and 5 formed part of the C-terminal dimerization domain (CTD) (Supplementary Figure S1). Of the 29 EkARFs, 21 (72.4%) contained all three functional domains, with only eight EkARFs (EkARF3, EkARF10.1, EkARF10.2, EkARF16.1, EkARF16.2, EkARF16.3, EkARF17.1, and EkARF17.2) lacking the CTD (Supplementary Figure S1). This variation in functional protein motifs may reflect mutations or deletions in the gene structure. Most EkARF genes contained 14 exons and 13 introns (Supplementary Figure S1). However, EkARF17.1 consisted of only two exons and one intron, three EkARF genes (EkARF16.1, EkARF16.2, and EkARF16.3) were composed of three exons and two introns, EkARF17.2 had five exons and four introns, and EkARF3 contained 11 exons and 10 introns, which is consistent with the observed variation in protein domains (Supplementary Figure S1). Overall, EkIAA and EkARF genes appeared to be relatively conserved during evolution, but those derived from segmental duplication have experienced some structural divergence.
Cis-element Analysis of EkIAA and EkARF Promoters
To explore the transcriptional regulation of EkARF and EkIAA genes and predict their functions, we analyzed the cis-regulatory elements in their promoters. We extracted 2,000 bp of upstream sequence, which we submitted to the PlantCARE online tool (Lescot et al., 2002). We then counted the number of phytohormone-, environment-, and flavonoid-responsive elements and noted their locations (Figure 5). cis-elements in EkIAA and EkARF promoters exhibited a similar pattern (Figure 5). Indeed, phytohormone-responsive and environmental stress–related cis-elements were present in all promoters of the EkIAA and EkARF gene family in E. konishii.
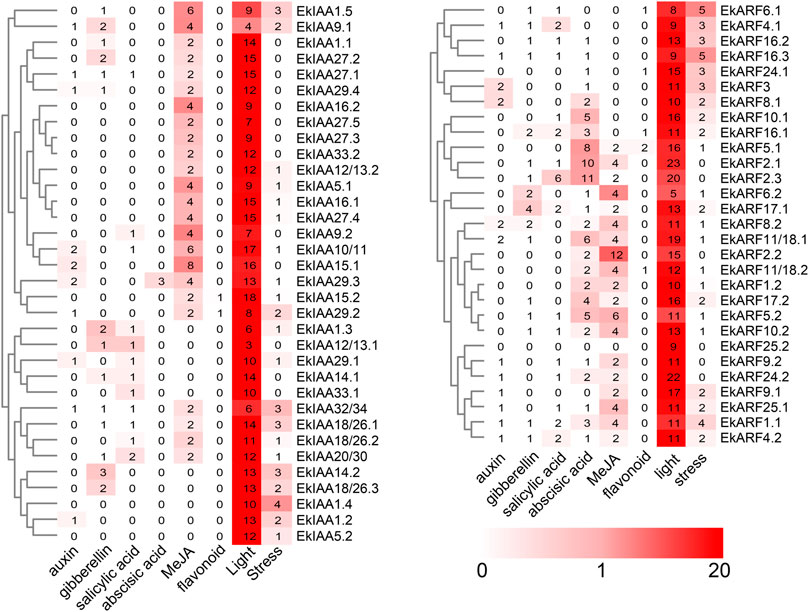
FIGURE 5. Predicted cis-elements in the EkIAA and EkARF promoters. The promoter sequences (−2,000 bp) of 34 EkIAA and 29 EkARF genes were analyzed by PlantCARE. The color bar indicates the number of cis-elements.
Expression Patterns of EkIAA and EkARF Genes in E. konishii
We then used publicly available RNA-seq datasets to analyze the expression patterns of EkIAA and EkARF genes at four fruit developmental stages, including green, turning red fruit, and red-winged pericarp stages, as well as in branches and leaves (Yang et al., 2020); the results are summarized as heatmaps in Figure 6. EkIAA gene family members showed varying expression patterns. Most EkIAA genes were highly expressed in green fruits, with the exception of EkIAA15.2, EkIAA33.1, EkIAA33.2, and EkIAA29.3, of which the first two were expressed specifically in branches, whereas the latter two genes were specifically expressed in red-winged pericarp (Figure 6A). EkIAA27.5 and EkIAA32/34 were highly expressed during the red fruit stage. EkARF genes were highly expressed during the fruit maturation stage (Figure 6B), of which EkARF1.2, EkARF2.2, EkARF4.2, EkARF5.1, EkARF5.2, EkARF16.3, and EkARF17.1 showed high expression levels in the red fruit stage. Most EkIAA and EkARF genes were highly expressed in fruits, hinting at their potential involvement in fruit maturation, including the accumulation of associated secondary metabolites.
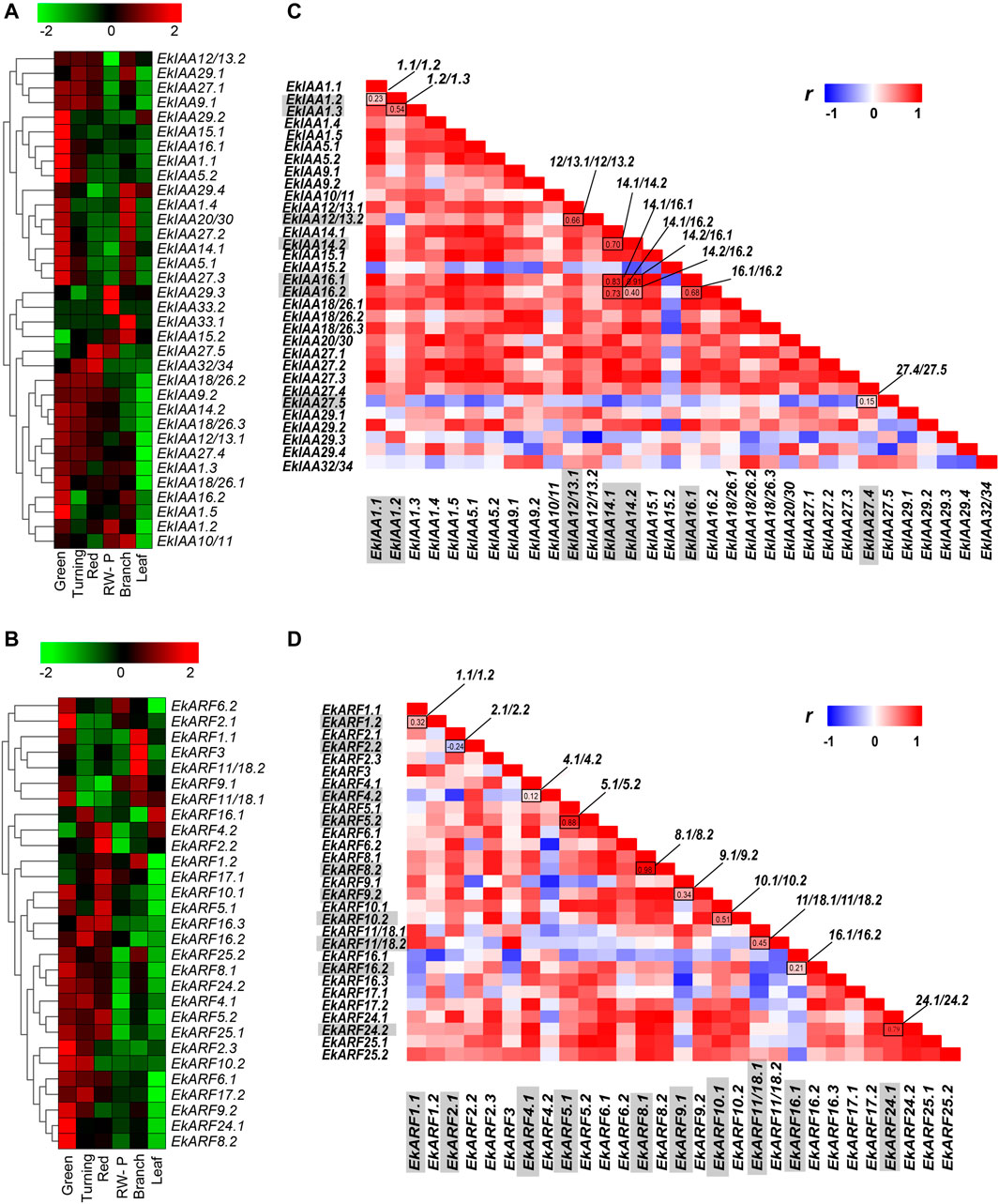
FIGURE 6. Heatmap representation of the expression of EkIAA (A) and EkARF (B) genes during fruit developmental stages (green, turning, red fruit, and red-winged pericarp) in branches and leaves. The color bar indicates Log2-normalized transcripts per million reads (TPM). RW-P, red-winged pericarp.
To assess the extent of functional diversification within these two families, we focused on duplicated gene pairs (10 EkIAA pairs and 10 EkARF pairs) and calculated the Pearson’s correlation coefficients of their expression profiles. Several duplicated gene pairs did in fact exhibit differential expression across the samples tested. Six EkIAA gene pairs (EkIAA12/13.1/-12/13.2, EkIAA14.1/-14.2, EkIAA14.1/-16.1, EkIAA14.1/-16.2, EkIAA14.2/-16.1, and EkIAA16.1/-16.2) and three EkARF gene pairs (EkARF5.1/-5.2, EkARF8.1/-8.2, and EkARF24.1/-24.2) showed similar expression patterns within the pairs, as evidenced by their high correlation coefficients (Figure 6B). We also identified one EkIAA pair and one EkARF pair with distinct expression levels between duplicated copies: EkIAA27.5 (expressed at high levels in all samples) and the duplicated copy EkIAA27.4 (expressed at relatively low levels), with a correlation coefficient of 0.15 (Figure 6C); and EkARF11/18.1 (expressed at low levels in all tissues) and EkARF18.2 (highly expressed in branches), with a correlation coefficient of 0.45. (Figure 6D). These results suggested that the functions of duplicated genes may have diverged following the initial duplication event.
EkARFs May Regulate the Biosynthesis of Terpenoids and Anthocyanins
Medicinal compounds such as triterpenes, phenolic acids, and flavonoids have been isolated from Euscaphis fruits, leaves, and roots (Liang et al., 2018). The accumulation of anthocyanin and terpenoid secondary metabolites coincides with E. konishii fruit maturation (Yuan et al., 2018b; Liang et al., 2019). Generally, genes with similar expression patterns may have related roles as they belong to the same regulatory pathway or are regulated by the same upstream factors. Thus, co-expression detected from our correlation analyses may provide cues as to gene regulation or function. Because anthocyanin contents, and the key anthocyanin biosynthetic genes, were previously well characterized during Euscaphis fruit development (Yuan et al., 2018b), we performed a correlation analysis between anthocyanin levels and the expression estimates for anthocyanin biosynthesis genes, EkIAA, and EkARF genes. We determined that anthocyanin contents were positively and strongly correlated with the expression of five genes encoding key enzymes (CHALCONE SYNTHASE8 [CHS8], CHALCONE ISOMERASE2 [CHI2], FLAVANONE 3-HYDROXYLASE1 [F3H1], F3H2, and F3H3) and very strongly correlated with that of another eight genes (CHS2, CHI3, F3H4, FLAVONOID 3′-HYDROXYLASE [F3′H], DIHYDROFLAVONOL 4-REDUCTASE1 [DFR1], LEUCOANTHOCYANIDIN REDUCTASE [LAR], FLAVONOL 3-O-GLUCOSYLTRANSFERASE3 [UFGT3], and UFGT7) (Figure 7A). Because ARF proteins regulate the expression of their target genes by binding to their cognate cis-elements in promoters, we looked for EkARF genes co-expressed with the anthocyanin biosynthetic genes listed above (Figure 7A), leading to the identification of seven such genes (EkARF1.2, EkARF2.2, EkARF4.2, EkARF5.1, EkARF5.2, EkARF16.3, and EkARF17.1).
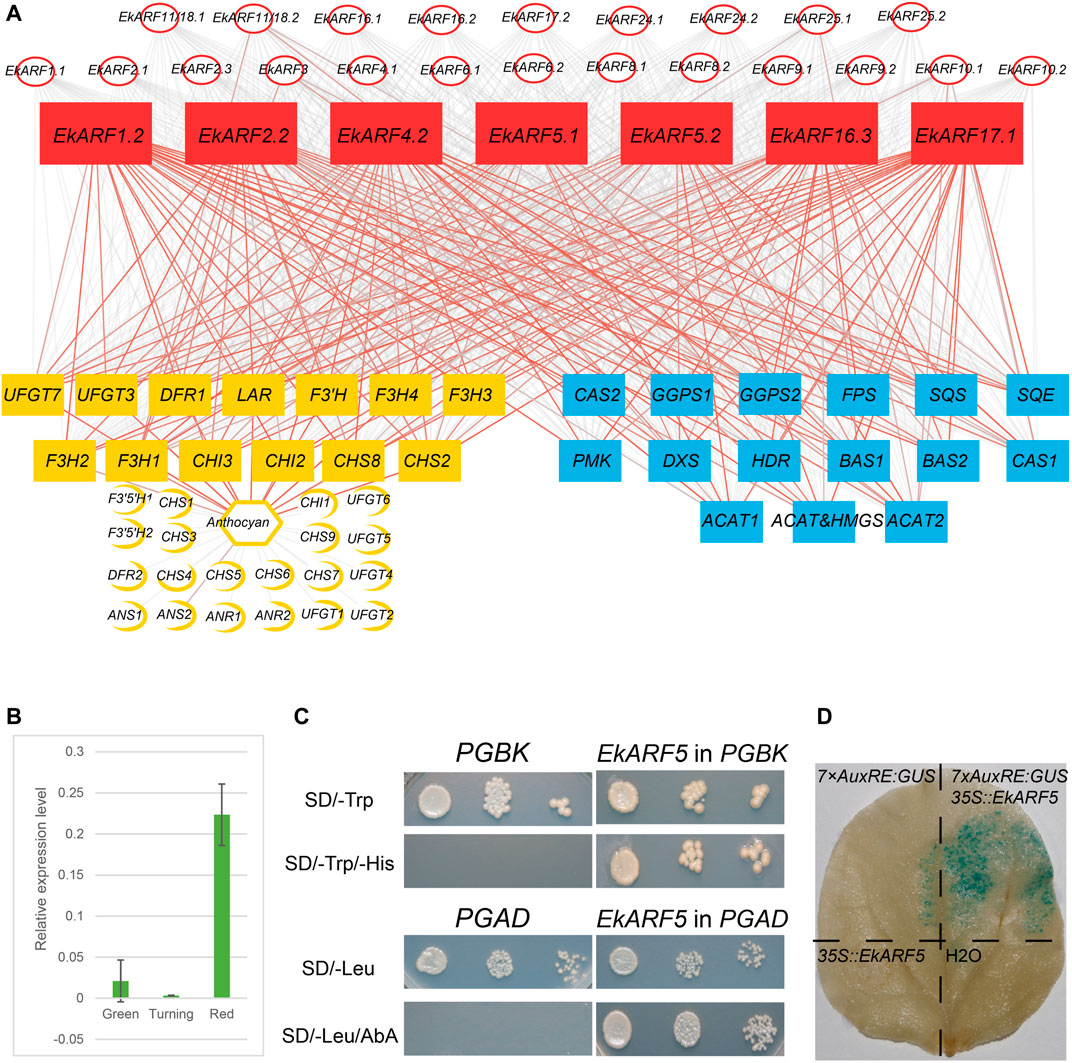
FIGURE 7. Correlation analysis between anthocyanin contents and EkARF expression, and triterpenoid contents and EkARF expression. (A) Red, EkARFs; yellow, anthocyanins; blue, triterpenoids. The gray lines indicate weak correlation, and red lines indicate strong correlation. (B) RT-qPCR results. (C) Yeast one-hybrid analysis results. (D) Results from GUS staining.
Triterpenoids accumulate to high levels in Euscaphis fruits and are important raw materials for natural products, food additives, and chemical products. Genes encoding the enzymes involved in the biosynthesis of triterpenoids have been described in the Euscaphis genome (Huang et al., 2019; Liang et al., 2019), prompting us to test for correlations between their expression patterns and those of ARFs genes (Figure 7A). This analysis highlighted seven EkARF genes (EkARF1.2, EkARF2.2, EkARF4.2, EkARF5.1, EkARF5.2, EkARF16.3, and EkARF17.1) whose expression was positively and strongly correlated with genes involved in triterpenoid accumulation (Figure 7). These results suggested that ARF genes contribute to secondary metabolite biosynthesis in Euscaphis.
ARF proteins are transcription factors that can bind to AuxREs (TGTCTC) to regulate the expression of their target genes. We noticed at least one AuxRE either upstream, downstream, or within intronic regions of anthocyanin and triterpenoid biosynthetic genes (Supplementary Table S3). To assess the role of ARFs in anthocyanin and triterpenoid biosynthesis, we selected EkARF5.1 for further characterization. RT-qPCR showed that EkARF5.1 is highly expressed during the red fruit stage, which is consistent with the RNA-seq data (Figure 7B). We fused EkARF5.1 to the GAL4 activation domain (AD) (AD-EkARF5.1) and introduced the resulting construct into yeast strain Y1HGold carrying a reporter consisting of seven copies of the AuxRE sequence driving the expression of Aureobasidin Resistance 1, conferring resistance to the antibiotic aureobasidin A (AbA). Whereas yeast colonies carrying AD or AD-EkARF5.1 grew on synthetic defined medium (Figure 7C), only yeast cells harboring the AD-EkARF5.1 construct survived growth on AbA-containing medium (Figure 7C), supporting the notion that EkARF5.1 binds to the AuxRE. We then tested the transactivation activity of EkARF5.1 by fusing full-length EkARF5.1 to the GAL4 DNA binding domain (BD) to generate the BD-EkARF5.1 fusion protein; the resulting encoding construct was introduced into yeast strain Y2HGold, which harbors the His3 gene driven by a GAL4-responsive promoter (Figure 7C). Only yeast cells carrying the BD-EkARF5.1 construct survived on synthetic medium lacking histidine, unlike yeast cells carrying the empty GAL4 DB vector (Figure 7C). These results indicated that EkARF5.1 has transactivation activity in yeast cells. Finally, we tested EkARF5.1 in N. benthamiana leaf epidermal cells by co-infiltrating a construct overexpressing EkARF5.1 and a β-GLUCURONIDASE (GUS) reporter construct whose expression is driven by seven copies of the AuxRE. We detected GUS activity in plant cells only when EkARF5 was co-expressed (Figure 7D). These results demonstrate that EkARF5.1 may play a role during anthocyanin and triterpenoid biosynthesis.
Discussion
Plant secondary metabolites consist of various bioactive compounds with applications in medicine and industry. The biosynthesis of plant secondary metabolites is regulated not only by plant growth and development signaling, but also by environmental stress cues. In China, E. konishii is widely planted as a medicinal and ornamental plant, but the regulatory mechanism of secondary metabolite biosynthesis is poorly understood, which limits genetic improvement and development of agronomic management techniques. In this study, we identified 34 Aux/IAA genes and 29 ARF coding sequences that map to some, but not all, linkage groups that constitute the E. konishii genome. Co-expression analysis suggested that seven EkARF genes may regulate anthocyanin and triterpenoid biosynthesis in E. konishii. Our data improve the understanding of the EkIAA and EkARF gene families and may provide valuable information on their biological functions in the context of secondary metabolite biosynthesis.
Auxin is an essential plant hormone, and Aux/IAAs and ARFs are key components of the signaling transduction process. Most of the current knowledge on Aux/IAA and ARF function, gene expression, and regulation has been obtained from studies in annual herbaceous plants such as Arabidopsis, rice, and tomato (Luo et al., 2018), while much less is known about IAAs and ARFs in longer-lived species, such as the evergreen shrub E. konishii. In this study, we identified 34 Aux/IAA and 29 ARF family members in the E. konishii genome (Figure 1; Supplementary Table S1), which was comparable to the numbers in other species, such as Arabidopsis (29 Aux/IAAs and 23 ARFs) (Remington et al., 2004), poplar (35 Aux/IAAs and 39 ARFs) (Kalluri et al., 2007), maize (31 Aux/IAAs and 31 ARFs) (Wang et al., 2010), and rice (31 Aux/IAAs and 25 ARFs) (Jain et al., 2006). EkIAA and EkARF genes clustered into five groups, as previously reported in Arabidopsis (Remington et al., 2004). Most EkIAA and EkARF genes within the same phylogenetic group shared similar exon-intron structures and the same arrangement of functional motifs in their encoded proteins (Figure 4), likely reflecting the gene duplication events that have shaped the expansion of the EkIAA and EkARF gene families in the E. konishii genome. Ka/Ks values of homologous genes further showed that duplicated genes underwent purifying selection (Supplementary Table S1). These results indicated that the two gene families are evolutionarily conserved with those from other plant species and may thus exhibit the same function and biochemical characteristics in E. konishii. However, we also identified six EkIAAs (EkIAA1.3, EkIAA29.1, EkIAA29.2, EkIAA29.3, and EkIAA29.4) that lack domain I (Figure 4), which are not expected to repress their downstream targets because domain I can repress the expression of target genes when in close proximity to the promoter (Hagen and Guilfoyle, 2002). Therefore, the E. konishii genome encodes conserved EkIAA and EkARF gene family members, although some members exhibit domain loss, possibly having arisen from unknown segmental duplication events, which will increase the complexity of auxin regulation.
The phytohormone auxin plays critical roles during plant growth. During fruit development, auxin also induces fruit set and growth, whereas it represses fruit maturation and ripening (Pattison et al., 2014). During tomato fruit maturation, the SIARF2 expression level increases in response to stimulation by ethylene, suggesting that auxin may repress fruit ripening (Pattison et al., 2014). Although gene structure and their encoded protein motifs were conserved in EkIAA and EkARF gene families, a subset of EkIAA and EkARF genes showed high expression during fruit maturation and ripening (Figure 6). Seven EkARFs were highly expressed during fruit maturation, of which EkARF1.2, EkARF2.2, EkARF4.2, EkARF16.3, and EkARF17.1 encode proteins containing a proline/serine/threonine-rich domain that acts as a transcriptional repressor (Supplementary Figure S1) (Tiwari et al., 2003). These results suggested that E. konishii fruit maturation and ripening may be similar to tomato. Interestingly, the two ARF5 homologs EkARF5.1 and EkARF5.2 contained a glutamate-rich domain that functions as a transcriptional activator domain; their encoding genes were highly expressed during E. konishii fruit maturation and ripening (Supplementary Figure S1) (Tiwari et al., 2003; Guilfoyle and Hagen, 2007), indicating that auxin signaling may play distinct roles during E. konishii fruit maturation and ripening processes. In Arabidopsis, ARF5 affects meristem development (Dastidar et al., 2019), while we showed here that EkARF5 is highly expressed during fruit maturation and ripening, likely reflecting changes in the promoter region associated with E. konishii and suggesting that the EkARF family underwent subfunctionalization during its evolutionary history. The high accumulation of secondary metabolites is a main feature of E. konishii fruit maturation and ripening (Yuan et al., 2018b). Co-expression analysis further revealed that EkARF5.1 and EkARF5.2 expression is positively and strongly correlated with that of anthocyanin and triterpenoid biosynthetic genes (Figure 7A). We confirmed that EkARF5.1 is highly expressed during fruit maturation and ripening stages and that EkARF5.1 can directly bind to AuxREs located within the promoter regions of anthocyanin and triterpenoid biosynthetic genes to activate their transcription in yeast and plant cells (Figures 7C,D). These results strongly suggest that EkARF5.1 and EkARF5.2 may be positive regulators of secondary metabolite biosynthesis, although the exact mechanisms by which they regulate anthocyanin biosynthesis require further study.
E. konishii fruit maturation and ripening involve pericarp splitting, pericarp overturn, and the accumulation of secondary metabolites (Yuan et al., 2018b; Liang et al., 2019; Huang et al., 2019; Sun et al., 2021). Pericarp overturn, the process in which the pericarp morphological changes after pericarp splitting (Supplementary Figure S2), is the main difference between E. konishii fruits and those of grape (Vitis vinifera) and tomato (Yuan et al., 2018b; Liang et al., 2019; Sun et al., 2021), which and may contribute to its survival and enable expansion to new environments (Sun et al., 2021). Pericarp overturn may be the result of uneven cell growth between epicarp and endocarp (Sun et al., 2021), which is associated with cell growth or differentiation (Ding et al., 2011). Auxin asymmetric distribution mediated by development and environmental cues results in uneven cell growth, thus regulating plant growth and response to environmental changes (Ding et al., 2011). Therefore, the highly expressed EkARF5s are probably involved in pericarp overturn in the last stage of E. konishii fruit maturation (Figure 6B; Figure 7B). It has been reported that auxin or its signaling is involved in biosynthesis of secondary metabolites such as anthocyanin, flavonols, and glucosinolates (Lewis et al., 2011; Wang et al., 2018; Wang et al., 2020). Given that pericarp split, pericarp overturn, and secondary metabolites are coupled (Yuan et al., 2018b), they may be regulated by similar or identical molecular mechanisms. In our study, we identified AuxREs in the promoters of anthocyanin and triterpenoid biosynthetic genes (Supplementary Table S3), to which EkARF5 can bind in yeast and plant cells, suggesting that EkARF5-mediated auxin signaling may regulate multiple signaling pathways in E. konishii fruit maturation.
Conclusion
We comprehensively analyzed the Aux/IAA and ARF gene families in E. konishii, which are evolutionarily well conserved. Expression and co-expression analyses showed that EkARF5 may play critical roles during the regulation of secondary metabolite biosynthesis. This study provides the basis for uncovering the regulatory mechanisms necessary to boost the production of industrial products and breed new E. konishii varieties with high economic output.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: The E. konishii chromosome-level genome assembly and annotation data (Accession No. GWHBCHS00000000) were available from National Genomics Data Center (https://ngdc.cncb.ac.cn/gwh/Assembly/reviewer/DsVzSvoOmVHEsqvwRYFCzmaHLYVsIsbpuBGJNDyDMGIWICZvIzpChREHnNqERbWc).
Author Contributions
BL conceived the project. JZ carried out all analyses together with BL. JZ wrote the manuscript draft, and BL and X-XZ revised the manuscript. LL, QY, BH, and QW helped with the experiments. BL and S-QZ acquired the funding. All authors have read and approved the manuscript.
Funding
This work was supported by startup funds for scientific research of Yancheng Teachers University (72672166005C), Funds from Fujian Agriculture and Forestry University (KFA17295A, CXZX2019044G, 11899170128), and by the project in Fujian Province (2020N5004 and Min(2018)TG14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.737293/full#supplementary-material
Abbreviations
AbA, antibiotic aureobasidin A; AD, activation domain; ARF, auxin response factor; Aux/IAA, auxin/indole-3-acetic acid; AuxRE, auxin response element; BD, binding domain; CTD, C-terminal dimerization domain; DBD, DNA-binding domain; GA, gibberellic acid; HMM, Hidden Markov model; LG, linkage group; Ka, number of nonsynonymous substitutions per nonsynonymous site; Ks, number of synonymous substitutions per synonymous site; MR, middle region; pI, isoelectric point; SD, synthetic defined.
References
Aloni, R., Aloni, E., Langhans, M., and Ullrich, C. I. (2006). Role of Cytokinin and Auxin in Shaping Root Architecture: Regulating Vascular Differentiation, Lateral Root Initiation, Root Apical Dominance and Root Gravitropism. Ann. Bot. 97 (5), 883–893. doi:10.1093/aob/mcl027
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: Discovering and Analyzing DNA and Protein Sequence Motifs. Nucleic Acids Res. 34 (Web Server issue), W369–W373. doi:10.1093/nar/gkl198
Bing, H., Liua, H., Han, X., Cui, P., and Xub, L-A. (2021). Multi-Omics Analysis of Ginkgo Biloba Preliminarily Reveals the Co-Regulatory Mechanism between Stilbenes and Flavonoids. Ind. Crops Prod. 167, 113434. doi:10.1016/j.indcrop.2021.113434
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis T. BMC Plant Biol. 4, 10. doi:10.1186/1471-2229-4-10
Chaabouni, S., Jones, B., Delalande, C., Wang, H., Li, Z., Mila, I., et al. (2009). Sl-IAA3, a Tomato Aux/IAA at the Crossroads of Auxin and Ethylene Signalling Involved in Differential Growth. J. Exp. Bot. 60 (4), 1349–1362. doi:10.1093/jxb/erp009
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Dastidar, M. G., Scarpa, A., Mägele, I., Ruiz-Duarte, P., von Born, P., Bald, L., et al. (2019). ARF5/MONOPTEROS Directly Regulates miR390 Expression in the Arabidopsis T Primary Root Meristem. Plant Direct 3 (2), e00116. doi:10.1002/pld3.116
Ding, Z., Galván-Ampudia, C. S., Demarsy, E., Łangowski, Ł., Kleine-Vehn, J., Fan, Y., et al. (2011). Light-Mediated Polarization of the PIN3 Auxin Transporter for the Phototropic Response in Arabidopsis. Nat. Cel Biol 13 (4), 447–452. doi:10.1038/ncb2208
Ellis, C. M., Nagpal, P., Young, J. C., Hagen, G., Guilfoyle, T. J., and Reed, J. W. (2005). AUXIN RESPONSE FACTOR1andAUXIN RESPONSE FACTOR2regulate Senescence and floral Organ Abscission inArabidopsis Thaliana. Development 132 (20), 4563–4574. doi:10.1242/dev.02012
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the Protein Families Database. Nucl. Acids Res. 42 (Database issue), D222–D230. doi:10.1093/nar/gkt1223
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 39 (Web Server issue), W29–W37. doi:10.1093/nar/gkr367
Freeling, M. (2009). Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition. Annu. Rev. Plant Biol. 60, 433–453. doi:10.1146/annurev.arplant.043008.092122
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: The Proteomics Server for In-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 31 (13), 3784–3788. doi:10.1093/nar/gkg563
Goetz, M., Vivian-Smith, A., Johnson, S. D., and Koltunow, A. M. (2006). AUXIN RESPONSE FACTOR8Is a Negative Regulator of Fruit Initiation inArabidopsis. Plant Cell 18 (8), 1873–1886. doi:10.1105/tpc.105.037192
Goh, T., Kasahara, H., Mimura, T., Kamiya, Y., and Fukaki, H. (2012). Multiple AUX/IAA-ARF Modules Regulate Lateral Root Formation: The Role of Arabidopsis SHY2/IAA3-Mediated Auxin Signalling. Phil. Trans. R. Soc. B 367 (1595), 1461–1468. doi:10.1098/rstb.2011.0232
Gray, W. M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin Regulates SCFTIR1-Dependent Degradation of AUX/IAA Proteins. Nature 414 (6861), 271–276. doi:10.1038/35104500
Gu, Z., Cavalcanti, A., Chen, F.-C., Bouman, P., and Li, W.-H. (2002). Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 19 (3), 256–262. doi:10.1093/oxfordjournals.molbev.a004079
Guilfoyle, T. J., and Hagen, G. (2007). Auxin Response Factors. Curr. Opin. Plant Biol. 10 (5), 453–460. doi:10.1016/j.pbi.2007.08.014
Guilfoyle, T. J. (2015). The PB1 Domain in Auxin Response Factor and Aux/IAA Proteins: A Versatile Protein Interaction Module in the Auxin Response. Plant Cell 27 (1), 33–43. doi:10.1105/tpc.114.132753
Hagen, G., and Guilfoyle, T. (2002). Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors. Plant Mol. Biol. 49 (3-4), 373–385. doi:10.1007/978-94-010-0377-3_9
Hardtke, C. S., and Berleth, T. (1998). The Arabidopsis Gene MONOPTEROS Encodes a Transcription Factor Mediating Embryo Axis Formation and Vascular Development. Embo j 17 (5), 1405–1411. doi:10.1093/emboj/17.5.1405
Hu, J., Israeli, A., Ori, N., and Sun, T.-P. (2018). The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 30 (8), 1710–1728. doi:10.1105/tpc.18.00363
Huang, B., Rong, H., Ye, Y., Ni, Z., Xu, M., Zhang, W., et al. (2020). Transcriptomic Analysis of Flower Color Variation in the Ornamental Crabapple (Malus spp.) Half-Sib Family through Illumina and PacBio Sequel Sequencing. Plant Physiol. Biochem. 149, 27–35. doi:10.1016/j.plaphy.2020.01.033
Huang, W., Ding, H., Chen, L-Y., Ni, L., Ruan, Y-F., Zou, X-X., et al. (2019). Protective Effect of the Total Triterpenes of Euscaphis Konishii Hayata Pericarp on Bacillus Calmette-Guérin Plus Lipopolysaccharide-Induced Liver Injury. Evid. Based Complement. Alternat Med. 2019, 1806021. doi:10.1155/2019/1806021
Huang, W., Zheng, Y., Feng, H., Ni, L., Ruan, Y.-F., Zou, X.-X., et al. (2020). Total Phenolic Extract of Euscaphis Konishii Hayata Pericarp Attenuates Carbon Tetrachloride (CCl4)-Induced Liver Fibrosis in Mice. Biomed. Pharmacother. 125, 109932. doi:10.1016/j.biopha.2020.109932
Jain, M., Kaur, N., Garg, R., Thakur, J. K., Tyagi, A. K., and Khurana, J. P. (2006). Structure and Expression Analysis of Early Auxin-Responsive Aux/IAA Gene Family in Rice (Oryza Sativa). Funct. Integr. Genomics 6 (1), 47–59. doi:10.1007/s10142-005-0005-0
Kalluri, U. C., DiFazio, S. P., Brunner, A. M., and Tuskan, G. A. (2007). Genome-Wide Analysis of Aux/IAA and ARF Gene Families in Populus trichocarpa. BMC Plant Biol. 7, 59. doi:10.1186/1471-2229-7-59
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Lee, M. K., Lee, K. Y., Jeon, H. Y., Sung, S. H., and Kim, Y. C. (2009). Antifibrotic Activity of Triterpenoids from the Aerial Parts ofEuscaphis Japonicaon Hepatic Stellate Cells. J. Enzyme Inhib. Med. Chem. 24 (6), 1276–1279. doi:10.3109/14756360902829709
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2018). 20 Years of the SMART Protein Domain Annotation Resource. Nucleic Acids Res. 46 (D1), D493–d496. doi:10.1093/nar/gkx922
Lewis, D. R., Ramirez, M. V., Miller, N. D., Vallabhaneni, P., Ray, W. K., Helm, R. F., et al. (2011). Auxin and Ethylene Induce Flavonol Accumulation through Distinct Transcriptional Networks. Plant Physiol. 156 (1), 144–164. doi:10.1104/pp.111.172502
Liang, W., Ni, L., Carballar-Lejarazú, R., Zou, X., Sun, W., Wu, L., et al. (2019). Comparative Transcriptome Among Euscaphis Konishii Hayata Tissues and Analysis of Genes Involved in Flavonoid Biosynthesis and Accumulation. BMC Genomics 20 (1), 24. doi:10.1186/s12864-018-5354-x
Liang, W., Zou, X., Carballar-Lejarazú, R., Wu, L., Sun, W., Yuan, X., et al. (2018). Selection and Evaluation of Reference Genes for qRT-PCR Analysis in Euscaphis Konishii Hayata Based on Transcriptome Data. Plant Methods 14, 42. doi:10.1186/s13007-018-0311-x
Liscum, E., and Reed, J. W. (2002). Genetics of Aux/IAA and ARF Action in Plant Growth and Development. Plant Mol. Biol. 49 (3-4), 387–400. doi:10.1007/978-94-010-0377-3_10
Liu, B., Wang, L., Zhang, J., Li, J., Zheng, H., Chen, J., et al. (2014). WUSCHEL-Related Homeobox Genes in Populus Tomentosa: Diversified Expression Patterns and a Functional Similarity in Adventitious Root Formation. BMC Genomics 15, 296. doi:10.1186/1471-2164-15-296
Luo, J., Zhou, J. J., and Zhang, J. Z. (2018). Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 19 (1), 259. doi:10.3390/ijms19010259
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: A Conserved Domain Database for the Functional Annotation of Proteins. Nucleic Acids Res. 39 (Database issue), D225–D229. doi:10.1093/nar/gkq1189
Mehan, M. R., Freimer, N. B., and Ophoff, R. A. (2004). A Genome-Wide Survey of Segmental Duplications that Mediate Common Human Genetic Variation of Chromosomal Architecture. Hum. Genomics 1 (5), 335–344. doi:10.1186/1479-7364-1-5-335
Mitreiter, S., and Gigolashvili, T. (2021). Regulation of Glucosinolate Biosynthesis. J. Exp. Bot. 72 (1), 70–91. doi:10.1093/jxb/eraa479
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi:10.1093/molbev/msu300
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional Genomic Analysis of theAUXIN RESPONSE FACTORGene Family Members inArabidopsis Thaliana: Unique and Overlapping Functions ofARF7andARF19. Plant Cell 17 (2), 444–463. doi:10.1105/tpc.104.028316
Pattison, R. J., Csukasi, F., and Catalá, C. (2014). Mechanisms Regulating Auxin Action during Fruit Development. Physiol. Plantarum 151 (1), 62–72. doi:10.1111/ppl.12142
Prion, S., and Haerling, K. A. (2014). Making Sense of Methods and Measurement: Pearson Product-Moment Correlation Coefficient. Clin. Simulation Nurs. 10 (11), 587–588. doi:10.1016/j.ecns.2014.07.010
Remington, D. L., Vision, T. J., Guilfoyle, T. J., and Reed, J. W. (2004). Contrasting Modes of Diversification in the Aux/IAA and ARF Gene Families. Plant Physiol. 135 (3), 1738–1752. doi:10.1104/pp.104.039669
Roosjen, M., Paque, S., and Weijers, D. (2018). Auxin Response Factors: Output Control in Auxin Biology. J. Exp. Bot. 69 (2), 179–188. doi:10.1093/jxb/erx237
Singh, V. K., and Jain, M. (2015). Genome-Wide Survey and Comprehensive Expression Profiling of Aux/IAA Gene Family in Chickpea and Soybean. Front. Plant Sci. 6, 918. doi:10.3389/fpls.2015.00918
Smith-Gill, S. J. (1991). Protein-Protein Interactions: Structural Motifs and Molecular Recognition. Curr. Opin. Biotechnol. 2 (4), 568–575. doi:10.1016/0958-1669(91)90082-g
Sun, W. H., Li, Z., Xiang, S., Ni, L., Zhang, D., Chen, D-Q., et al. (2021). The Euscaphis Japonica Genome and the Evolution of Malvids. Plant J. doi:10.1111/tpj.15518
Sun, W., Yuan, X., Liu, Z.-J., Lan, S., Tsai, W.-C., and Zou, S.-Q. (2019). Multivariate Analysis Reveals Phenotypic Diversity of Euscaphis Japonica Population. PLoS One 14 (7), e0219046. doi:10.1371/journal.pone.0219046
Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2004). Aux/IAA Proteins Contain a Potent Transcriptional Repression Domain. Plant Cell 16 (2), 533–543. doi:10.1105/tpc.017384
Tiwari, S. B., Hagen, G., and Guilfoyle, T. (2003). The Roles of Auxin Response Factor Domains in Auxin-Responsive Transcription. Plant Cell 15 (2), 533–543. doi:10.1105/tpc.008417
Tiwari, S. B., Wang, X. J., Hagen, G., and Guilfoyle, T. J. (2001). AUX/IAA Proteins Are Active Repressors, and Their Stability and Activity Are Modulated by Auxin. Plant Cell 13 (12), 2809–2822. doi:10.1105/tpc.13.12.2809
Ulmasov, T., Hagen, G., and Guilfoyle, T. J. (1999). Activation and Repression of Transcription by Auxin-Response Factors. Proc. Natl. Acad. Sci. 96 (10), 5844–5849. doi:10.1073/pnas.96.10.5844
Van Ha, C., Le, D. T., Nishiyama, R., Watanabe, Y., Sulieman, S., Tran, U. T., et al. (2013). The Auxin Response Factor Transcription Factor Family in Soybean: Genome-Wide Identification and Expression Analyses during Development and Water Stress. DNA Res. 20 (5), 511–524. doi:10.1093/dnares/dst027
Wang, C.-K., Han, P.-L., Zhao, Y.-W., Ji, X.-L., Yu, J.-Q., You, C.-X., et al. (2020). Auxin Regulates Anthocyanin Biosynthesis through the Auxin Repressor Protein MdIAA26. Biochem. Biophysical Res. Commun. 533 (4), 717–722. doi:10.1016/j.bbrc.2020.09.065
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-Wide Analysis of the Auxin Response Factors (ARF) Gene Family in rice (Oryza Sativa). Gene 394 (1-2), 13–24. doi:10.1016/j.gene.2007.01.006
Wang, Y.-C., Wang, N., Xu, H.-F., Jiang, S.-H., Fang, H.-C., Su, M.-Y., et al. (2018). Auxin Regulates Anthocyanin Biosynthesis through the Aux/IAA-ARF Signaling Pathway in Apple. Hortic. Res. 5, 59. doi:10.1038/s41438-018-0068-4
Wang, Y., Deng, D., Bian, Y., Lv, Y., and Xie, Q. (2010). Genome-Wide Analysis of Primary Auxin-Responsive Aux/IAA Gene Family in maize (Zea mays. L). Mol. Biol. Rep. 37 (8), 3991–4001. doi:10.1007/s11033-010-0058-6
Waseem, M., Ahmad, F., Habib, S., and Li, Z. (2018). Genome-Wide Identification of the Auxin/indole-3-Acetic Acid (Aux/IAA) Gene Family in Pepper, its Characterisation, and Comprehensive Expression Profiling under Environmental and Phytohormones Stress. Sci. Rep. 8 (1), 12008. doi:10.1038/s41598-018-30468-9
Wilmoth, J. C., Wang, S., Tiwari, S. B., Joshi, A. D., Hagen, G., Guilfoyle, T. J., et al. (2005). NPH4/ARF7 and ARF19 Promote Leaf Expansion and Auxin-Induced Lateral Root Formation. Plant J. 43 (1), 118–130. doi:10.1111/j.1365-313x.2005.02432.x
Winkler, M., Niemeyer, M., Hellmuth, A., Janitza, P., Christ, G., Samodelov, S. L., et al. (2017). Variation in Auxin Sensing Guides AUX/IAA Transcriptional Repressor Ubiquitylation and Destruction. Nat. Commun. 8, 15706. doi:10.1038/ncomms15706
Xing, H., Pudake, R. N., Guo, G., Xing, G., Hu, Z., Zhang, Y., et al. (2011). Genome-Wide Identification and Expression Profiling of Auxin Response Factor (ARF) Gene Family in maize. BMC Genomics 12, 178. doi:10.1186/1471-2164-12-178
Yang, C. L., Yuan, X. Y., Zhang, J., Sun, W. H., Liu, Z. J., and Zou, S. Q. (2020). Comprehensive Transcriptome Analysis of Reference Genes for Fruit Development of Euscaphis Konishii. PeerJ 8, e8474. doi:10.7717/peerj.8474
Yu, C., Zhan, Y., Feng, X., Huang, Z. A., and Sun, C. (2017). Identification and Expression Profiling of the Auxin Response Factors in Capsicum Annuum L. Under Abiotic Stress and Hormone Treatments. Int. J. Mol. Sci. 18 (12), 2719. doi:10.3390/ijms18122719
Yuan, X., Sun, W., Zou, X., Liu, B., Huang, W., Chen, Z., et al. (2018). Sequencing of Euscaphis Konishii Endocarp Transcriptome Points to Molecular Mechanisms of Endocarp Coloration. Int. J. Mol. Sci. 19 (10), 3209. doi:10.3390/ijms19103209
Yuan, Y., Mei, L., Wu, M., Wei, W., Shan, W., Gong, Z., et al. (2018). SlARF10, an Auxin Response Factor, Is Involved in Chlorophyll and Sugar Accumulation during Tomato Fruit Development. J. Exp. Bot. 69 (22), 5507–5518. doi:10.1093/jxb/ery328
Yuan, Y., Xu, X., Gong, Z., Tang, Y., Wu, M., Yan, F., et al. (2019). Auxin Response Factor 6A Regulates Photosynthesis, Sugar Accumulation, and Fruit Development in Tomato. Hortic. Res. 6, 85. doi:10.1038/s41438-019-0167-x
Keywords: Euscaphis konishii, Aux/IAA, ARF, triterpenoids, anthocyanin, fruit development
Citation: Liu B, Zhu J, Lin L, Yang Q, Hu B, Wang Q, Zou X-X and Zou S-Q (2022) Genome-Wide Identification and Co-Expression Analysis of ARF and IAA Family Genes in Euscaphis konishii: Potential Regulators of Triterpenoids and Anthocyanin Biosynthesis. Front. Genet. 12:737293. doi: 10.3389/fgene.2021.737293
Received: 06 July 2021; Accepted: 18 November 2021;
Published: 05 January 2022.
Edited by:
Sreepriya Pramod, Altria, United StatesReviewed by:
Hui Song, Qingdao Agricultural University, ChinaYulong Ren, Institute of Crop Sciences (CAAS), China
Yourong Chai, Southwest University, China
Xiaojiao Han, Chinese Academy of Forestry, China
Copyright © 2022 Liu, Zhu, Lin, Yang, Hu, Wang, Zou and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bobin Liu, bGl1YmJAeWN0dS5lZHUuY24=; Xiao-Xing Zou, enh4MDI5OUBmYWZ1LmVkdS5jbg==
 Bobin Liu
Bobin Liu Juanli Zhu2,3
Juanli Zhu2,3