- 1Department of Systematics, Biodiversity and Evolution of Plants, Albrecht-von-Haller Institute for Plant Sciences, University of Goettingen, Goettingen, Germany
- 2Georg-August University School of Science, University of Goettingen, Goettingen, Germany
- 3Evolutionary Biology Center, Uppsala University, Uppsala, Sweden
- 4Taxonomy & Evolutionary Biology, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany
Polyploidy plays a major role in plant evolution. The establishment of new polyploids is often a consequence of a single or few successful polyploidization events occurring within a species’ evolutionary trajectory. New polyploid lineages can play different roles in plant diversification and go through several evolutionary stages influenced by biotic and abiotic constraints and characterized by extensive genetic changes. The study of such changes has been crucial for understanding polyploid evolution. Here, we use the multiploid-species Paspalum intermedium to study population-level genetic and morphological variation and ecological differentiation in polyploids. Using flow cytometry, amplified fragment length polymorphism (AFLP) genetic markers, environmental variables, and morphological data, we assessed variations in ploidy, reproductive modes, and the genetic composition in 35 natural populations of P. intermedium along a latitudinal gradient in South America. Our analyses show that apomictic auto-tetraploids are of multiple independent origin. While overall genetic variation was higher in diploids, both diploids and tetraploids showed significant variation within and among populations. The spatial distribution of genetic variation provides evidence for a primary origin of the contact zone between diploids and tetraploids and further supports the hypothesis of geographic displacement between cytotypes. In addition, a strong link between the ecological differentiation of cytotypes and spatial distribution of genetic variation was observed. Overall, the results indicate that polyploidization in P. intermedium is a recurrent phenomenon associated to a shift in reproductive mode and that multiple polyploid lineages from genetically divergent diploids contributed to the successful establishment of local polyploid populations and dispersal into new environments.
Introduction
Polyploidization in plants is a recurring and pivotal evolutionary phenomenon that brings both short- and long-term benefits for plant diversification (Werth et al., 1985; Soltis et al., 2010; Symonds et al., 2010). Comparative genomic studies show that approximately 15% of plant speciation events resulted from polyploidy (Wood et al., 2009) and that polyploidy is substantially associated to higher plant diversity (Symonds et al., 2010; Jiao et al., 2011). A crucial step of polyploidization, the formation of unreduced gametes, occurs at a noticeable rate of ∼0.5% per gamete (Ramsey and Schemske, 1998; Wood et al., 2009). Yet, the occurrence of new polyploids in natural populations is unequivocally lower than expected from such values (Suda and Herben, 2013), implying that polyploidization events are not always inherently beneficial. Polyploidization can either act as an instantaneous mechanism for divergence and speciation because of reproductive isolation (see Soltis et al., 2009) or as a step toward extinction due to competitive exclusion by the majority cytotype [i.e., minority cytotype disadvantage (Levin, 1975; Parisod et al., 2010)]. Therefore, mechanisms that help newly arisen polyploids to overcome competition, survive, and establish themselves devoid of reproductive isolation are key to the success of polyploids.
Apomixis (asexual reproduction via seeds) is a reproductive strategy almost exclusively coupled with polyploidy. Since apomictic plants skip meiosis and fertilization and form clonal euploid seed embryos, apomixis can be particularly beneficial during the establishment of new polyploids by shielding triploids from the negative effects of unbalanced chromosomal segregation, low density, and parental introgression, thus aiding new polyploids to overcome reproductive exclusion (Hojsgaard, 2018). Thus, apomixis provides reproductive assurance to polyploids (Levin, 1975) and enhances a plant’s ability to colonize new habitats reinforcing founder events (Baker, 1955). Since meiosis is bypassed during offspring formation, apomixis maintains heterozygosity levels and may even counteract genetic drift during plant dispersals (Paun et al., 2006; Cosendai et al., 2013). Furthermore, additional genetic variation among clonal individuals in apomictic populations comes from mutation accumulation and residual sexuality (Hörandl and Paun, 2007; Hojsgaard and Hörandl, 2015). Such variations provide polyploids with the genetic novelty needed to adapt to environments and expand their range. Many apomictic polyploid complexes display a pattern called geographical parthenogenesis, whereby sexual cytotypes occupy restricted geographical areas while apomicts are more widespread, often into higher latitudes (Brochmann et al., 2004; Suda et al., 2004; Hörandl, 2006).
The observed ecological amplitude or tolerance to wider, sometimes extreme, environmental conditions in apomictic polyploids compared to their sexual parents is usually explained through two main hypotheses: The General Purpose Genotype hypothesis (GPG) (Baker, 1967; Lynch, 1984) explains that one fit genotype with higher tolerance to a broader ecological setting may colonize different habitats, while the Frozen Niche Variation (FNV) hypothesis assumes that single genotypes can freeze a portion of the genetic variation of the sexual progenitors and therefore adapt to restricted ecological setups, thus efficiently partitioning underutilized resources by the ancestors (Vrijenhoek, 1984, 1994). Although these two concepts are mutually exclusive, recent studies suggest that mechanisms of inter-clonal selection acting upon natural asexual populations to acquire their ecological breadth and clonal divergence support both hypotheses (Vrijenhoek and Parker, 2009).
Cytotype coexistence implies the possibility of spontaneous exchanges of genetic information between ploidy levels. Inter-cytotype gene flow will result in patterns of genetic admixture, like observed in different polyploid apomictic complexes (Petit et al., 1999; Paun et al., 2006; Zozomová-Lihová et al., 2015). In general, gene flow is mostly unidirectional as it mainly takes place from diploids to polyploids, but cases of genetic introgression from polyploids to diploids have also been observed (Bretagnolle and Thompson, 1996; Van Dijk and Bakx-Schotman, 1997; Ramsey and Schemske, 1998). Recurrent polyploidization events from genetically divergent diploids are common in either sexual (e.g., Soltis et al., 2004) or apomictic polyploids (e.g., Sharbel and Mitchell-Olds, 2001) and can create genetically distinct populations or introduce novel genetic material in an established population to be recombined and assorted among polyploid individuals during sexual reproduction.
Studies of extant genetic variation in polyploid populations compared to their putative diploid parents not only provide evidence for the dynamic formation and fate of polyploids in natural populations (Soltis and Soltis, 2000); they also shed light on their recent evolutionary history and mechanisms of genetic and ecological differentiation (Fox et al., 2020).
Paspalum intermedium Munro ex Morong is a grass species of the sub-family Panicoideae with two major cytotypes: self-sterile sexual diploids (2n = 2x = 20) and self-fertile apomictic autotetraploids (2n = 4x = 40) (Norrmann et al., 1989). Despite the species occurring in a wide range of ecological and climatic gradients in sub-tropical South America (Zuloaga et al., 2012), a recent study revealed a significant shift in niche-optima between diploids and tetraploids and a dynamic competition and displacement between cytotypes in an ecotone where populations are segregated in allopatry, sympatry, and parapatry (Karunarathne et al., 2018). Therefore, P. intermedium makes an ideal non-model plant system to examine extant levels of genetic variation and associated traits to better understand the formation and dynamics of polyploids, the main drivers of habitat adaptation, niche divergence, and cytotype coexistence and to get a glimpse of the recent evolutionary history of the complex.
In the present study, using flow cytometry, amplified fragment length polymorphism (AFLP) markers, and ecological data, we endeavor 1) to assess extant levels of genetic variability in P. intermedium populations; 2) to evaluate the differences in population structure between cytotypes with divergent reproductive modes; 3) to examine the genetic composition of sympatric, parapatric, and allopatric populations in the contact zone; and 4) to search for associations between genetic variability parameters and distribution patterns which may support alternative ecological hypothesis and population attributes of different cytotypes that may help to clarify the recent evolutionary history of the species.
Materials and Methods
Sampling and Cytotyping
Sampling was done covering the core and peripheral distribution of P. intermedium (i.e., Pampas, Mesopotamia, and Gran Chaco of Argentina). Leaf materials were collected in silica gel from 35 populations and between 20 and 30 individuals from each population. Flow cytometry ploidy evaluations data are from our previous analyses published in Karunarathne et al. (2018). A total of 915 individuals classified as diploids (2n = 2x = 20) or tetraploids (2n = 4x = 40) and representing 24 pure tetraploid populations, nine pure diploid populations, and four mix ploidy populations were used here to analyze the population genetic structure (see Figure 1 and Table 1).
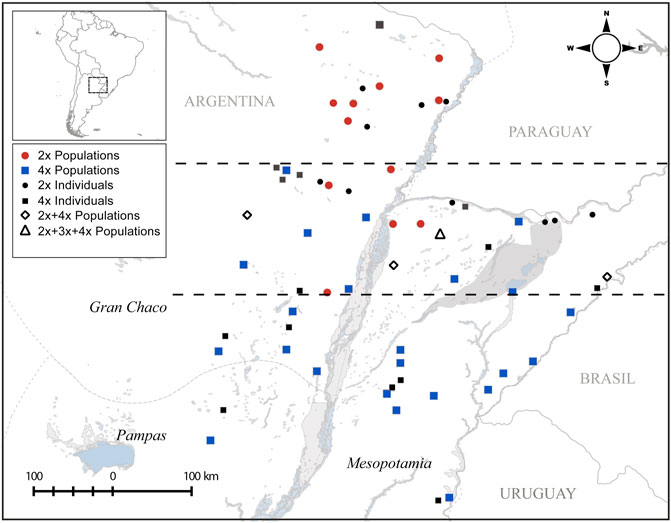
FIGURE 1. Collection locations of studied P. intermedium populations and their ploidy levels (adopted from Karunarathne et al., 2018). The dashed lines demarcate the contact zone of cytotypes in the middle separating the north and south diploid and tetraploid distribution zones, respectively.
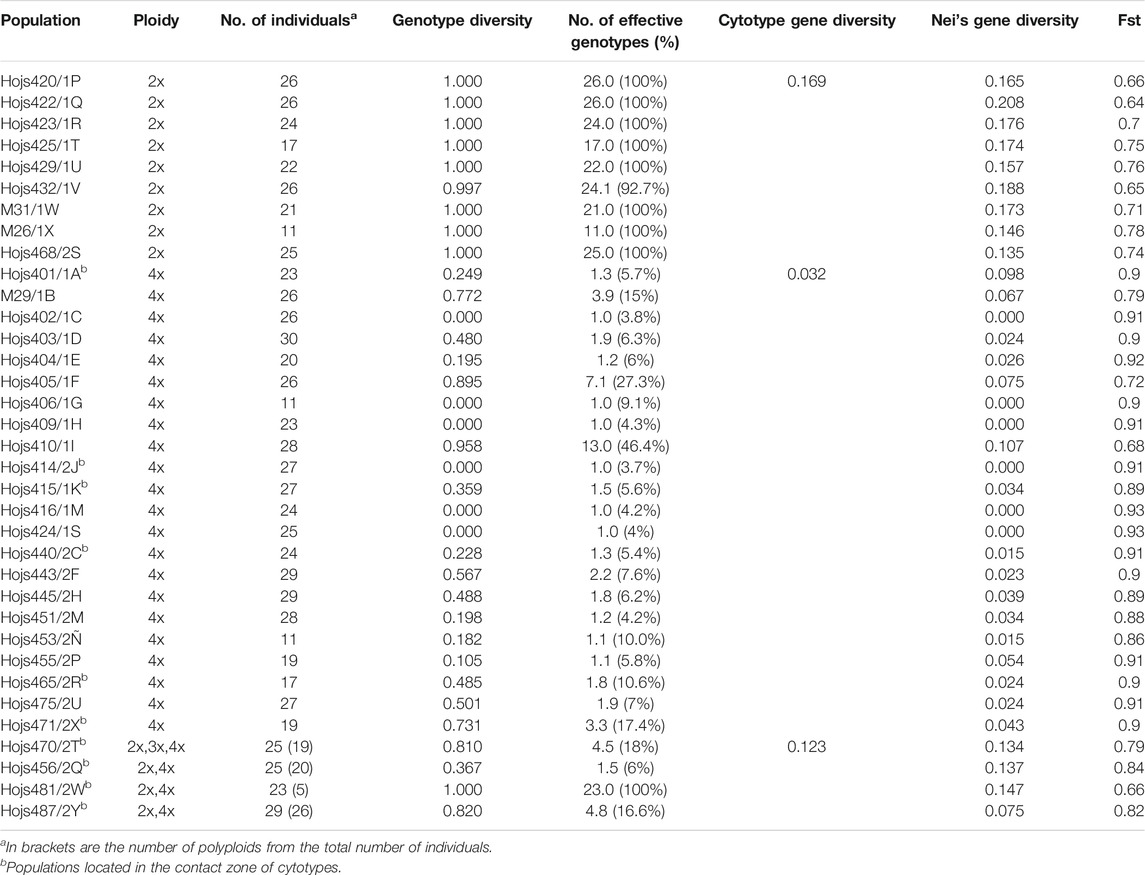
TABLE 1. Populations of P. intermedium with different ploidy levels, assessed in the present study for genetic variation. Nei’s gene diversity, genotype diversity, and the number of effective genotypes were calculated as in Nei’s (1987) with amplified fragment length polymorphism (AFLP) markers. The cytotype gene diversity was calculated for each population and averaged to diploid, tetraploid, and mixed-ploidy populations.
gDNA Extraction and Amplified Fragment Length Polymorphism
The genome DNA of all the collected samples were extracted using QIAGEN mini plant DNA extraction kit (QIAGEN GmbH, Hilden, Germany), from 50 mg of silica-dried leaf materials. The DNA concentrations were quantified and used for AFLP analysis.
For AFLP, we followed the methodology described by Vos et al. (1995) with a modification skipping the pre-selective amplification of the digested fragments. Further, selective primer combinations with four additional bases (instead of three) in one of the primers were used, though not in all combinations (see Supplementary note one for primer details). The three primer combinations used were EcoR I- ACA-5′ FAM/Mse I- GAAC, EcoR I - AATG/Mse I -AAC-5′ HEX, and EcoR I - AGA/Mse I -ACA-5’ TAMRA. This modification yielded consistent DNA fragments, reducing the laboratory time substantially. The reproducibility of PCRs was checked with 10 duplicate samples with each primer pair.
For restriction digestion and ligation, approximately 500 ng of genome DNA of each sample was digested overnight with EcoR I (5 Units) and Mse I (1 Unit) (New England Biolabs, Frankfurt, Germany), and T4 DNA ligase (Promega Corporation, Mannheim, Germany) (1 Unit) with EcoR adapter pair (5 pmol) and Mse adapter pair (50 pmol) with the presence of NaCl (0.05 M) and BSA (0.05 mg/ml) in 1X ligase buffer (Promega Corporation, Mannheim, Germany). The direct selective amplification reaction mixture consisted of 1X PCR buffer (10x NH4 reaction buffer; Bioline GmbH, Luckenwalde, Germany), 2.5 mM MgCl2 (Bioline GmbH, Luckenwalde, Germany), 0.2 mM dNTPs (Promega Corporation, Mannheim, Germany), 4 pmol of each EcoR and Mse primers, 1 Unit Taq polymerase (BioTaq–Bioline GmbH, Luckenwalde, Germany), and ca. 80 ng of digested DNA in 25 μl final volume. The PCR was done in a Thermal Cycler (BioRad T100; Bio-Rad Laboratories GmbH, Munich, Germany) with the following program. Denatured at 94°C, 2 min, 9 × (94°C, 1 s; 65°C, 30 s, 1°C/cycle; 72°C, 2 min), 23 × (94°C, 1 s; 56°C, 30 s; 72°C, 2 min), 60°C, 30 min. Amplified fragments were analyzed in an ABI 3130xl Genetic Analyzer (Applied Biosystems Inc. Foster City, CA, United States) with the 500 ROX size standard (Applied Biosystems Inc. Foster City, CA, United States). A total of 887 individual fingerprints were retained after the initial analysis of all the individuals (48 atypical fingerprints were removed). Genotyping and binary presence–absence matrices were assembled in GeneMarker 2.6.0 (Softgenetics, State College, PA, United States), with a threshold of 75 RFU for scoring bands of size range of 100–510 bp (small fragments between 50 and 100 were not considered due to the possibility of non-homologous fragments; Vekemans et al., 2002). All the peaks were checked in the panel editor eliminating non-reproducible bands by comparing against replicated samples. Reproducibility of the data was checked using the error rate with 30 duplicate samples, where the similarity of the scoring (i.e., presence–absence of fragments) was cross-checked. The error rate was less than 0.1%, indicating the high reliability of the data.
Population Genetic Analyses
The combined binary matrix of the three primer combinations was analyzed with the R-script AFLPDAT (Ehrich, 2007) to calculate diversity indices (i.e., Nei’s gene diversity for each population) and genotype diversity (using Nei’s formula for haplotype diversity; Masatoshi Nei, 1987). The binary matrix was assembled into an individual genotype data object with the R package ADEGENET (which includes a method that can handle clonal data and allows for analyses of mixed-ploidy data sets with a correction for allele copy-number ambiguity in polyploids) (Jombart, 2008), which was used in the rest of the genetic analyses in the R environment (R Core Team, 2016). A Neighbor-Joining tree was constructed using the Prevosti’s Distance Coefficient (a measurement over all the loci of the proportion of unshared alleles) with a bootstrap analysis of 1,000 sample size. The R package POPPR 2.7.1 (Kamvar et al., 2014) was used for the distance matrix and the bootstrapping. Principal coordinate analysis (PCoA) was performed based on pairwise Euclidian distance used in the DAPC (Discriminant Analysis of Principal Component) function of the ADEGENET R package. Analysis of Molecular Variance (AMOVA) was calculated on the discrete dissimilarity matrix with 1,000 permutations. For AMOVA, both ploidy and populations were used as different strata to calculate between ploidy and within- and among-population molecular variance.
Bayesian model-based clustering implemented in the “find.clusters” function of R package ADEGENET was used to determine the potential number of clusters that can describe the data best. Here, BICs (Bayesian Information Criteria) are calculated using k-means algorithm (also Ripley’s K-function, where the sum of squares from points to the assigned cluster centers is minimized; Baddeley and Turner, 2005), and the resulting BIC values are plotted against the increasing number of k (clusters). Ideally, the number of clusters where the BIC value starts to increase is taken as the best cluster solution. In our case, the BIC value did not increase (Supplementary Figure S1A). Therefore, according to the plot, any number of clusters more than two and less than 15 will describe the data. Considering the number of groups observed in the NJ tree, k = 3 was taken as the number of clusters, thus making it a biologically meaningful number of clusters. An assignment of three clusters (k = 3) in the STRUCTURE analysis (Figure 2A) best explains the variation. The assignment of k = 2 did not noticeably change the admixture values in tetraploids. Clusters were unstable with higher k (>3) indicating admixture in almost all individuals. This was also tested with ad hoc statistic ΔK based on the rate of change in the log probability of data between successive K values described by Evanno et al. (2005) (Figure S1B). Plotting the genetic clusters was performed using the R package LEA (Frichot and Francois, 2014).
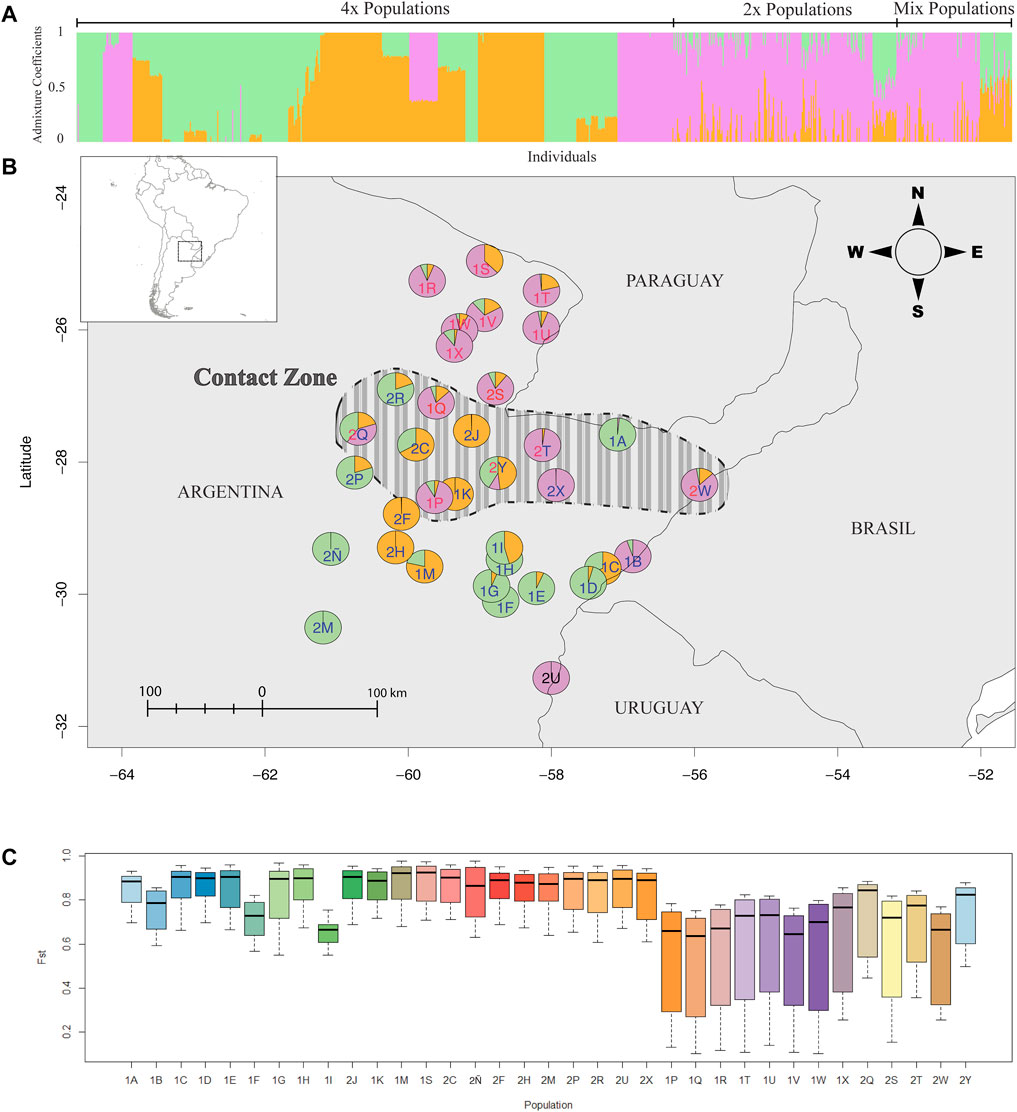
FIGURE 2. Genetic clusters in P. intermedium inferred from AFLPs. (A) Bayesian clustering of all the individuals at K = 3. Vertical bars represent the individuals with the proportion of the admixture (i.e., admixture coefficients) in different colors. (B) Admixture coefficients of populations plotted on the map indicating the collection location. Pie chart colors follow the different clusters as in (A) The color of the population labels indicates the ploidy level of each population (red: diploids, blue: tetraploid, red and blue: mixed populations). The dashed-line area on the map shows the putative zone of contact between parapatric and sympatric diploids and tetraploids. (C) Boxplots of the Fst values of each population showing their genetic divergence (color scheme only represents populations, and the population order follows the same as 4A).
Isolation by Distance and Environment
A Mantel test was performed using pairwise Euclidean distance (R package VEGAN; Oksanen et al., 2016) to calculate the geographical isolation of each cytotype and population, based on genetic data. The R package MPMCORRELOGRAM (Matesanz et al., 2011) was used to visualize the geographic isolation based on distance intervals. The R package LEA (Frichot and Francois, 2014) was used to plot the genetic admixture on the map.
We also performed environmental association analysis of genetic diversity among populations using bioclimatic data. Bioclimatic data were downloaded from www.chelsa-climate.org (Karger et al., 2017), and point data were extracted for each population. A regression analysis of Nei’s genetic diversity and each bioclimatic variable indicated eight variables showing significant correlation (p < 0.05 for all). A forward selection with 95% collinearity was used to sequentially remove intercorrelated variables, and only four variables were retained (Bio1: annual mean temperature, Bio3: isothermality, Bio6: min temperature of the coldest month, Bio15: precipitation seasonality). An environmental distance matrix was obtained using these variables, and a regression analysis of the environmental distance and the genetic diversity of populations was performed to detect isolation by environment.
Morphological Analysis
During plant material collections, at least three plants were collected from all the studied populations for morphological assessment, and the collected vouchers were deposited in several herbaria in Argentina (MNES, CTES, BAA, and SI) and Germany (B and GOET). Overall, 42 morphological characters (18 quantitative, 24 descriptive/categorical: Table S1) of both diploids and tetraploids were studied from the voucher specimens to discern any morphological variation among cytotypes and populations. A principal coordinate analysis (PCoA) was performed to identify morphological clusters among diploid and tetraploid populations. Similarity percentage analysis (SIMPER) was performed using the software PAST 3.23 (Hammer et al., 2001) to assess the amount of contribution of each morphological character to the observed morphological variation in P. intermedium individuals.
Results
Genetic Diversity in Cytotypes Across Populations
In the combined binary matrix, a total of 189 fragments were scored; 84 were from the EcoR-Mse (ACA- GAAC), 58 from EcoR-Mse (AATG- AAC), and 44 from EcoR-Mse (AGA- ACA). Out of this, 66.2% (61.6% in 2x, 70.8% in 4x) of fragments were polymorphic. The number of bands per individual ranged from 84 to 91. Cytotype-specific fragments were overall significantly higher in tetraploids (n = 41) than in diploids (n = 34) (p = 0.02).
Diversity analysis (Nei’s gene diversity) showed that all diploids have significantly higher values (paired t-test p-value < 0.001) in both genotype and gene diversity (Table 1). The effective number of genotypes was maximum in diploid populations (100%), indicating that all individuals were genetically distinct, whereas in tetraploids, the number of effective genotypes varied from 3.7 to 100% (averaging 8.05%; Table 1). Interestingly, mixed-ploidy populations showed variable levels of genotype diversity (between 6 and 100%, averaging 35.2%) which was significantly correlated to the number of diploid individuals in the population (Table 1; Pearson correlations r2 = 0.91, p < 0.01). One such population (Hojs481/2W: 24-diploids, 6-tetraploids; Table 1) harbored five non-clonal tetraploid individuals, suggesting either independent polyploidization events or (more likely) formation of recombinant polyploids by sexuality. The highest number of effective genotypes observed among tetraploid populations was 13 (46.4%), while six populations showed high Fst values (≥0.9) and a single effective genotype (Nei’s diversity = 0.0) and hence were classified as pure clonal populations (Table 1). Grouping populations by cytotype and geographic occurrence within the zone of contact between cytotypes (Figure 1) revealed that tetraploid individuals and populations in the contact zone were genetically more diverse, both in genotype and gene diversity as well as in the effective number of genotypes compared to those out of the contact zone (Table 1, 2).
When taking the complete dataset, AMOVA revealed that half of the genetic variation (50.18%) is found within populations, the rest accounting for among-population variation (ϕ = 0.4102, p = 0.001). When ‘cytotype’ was assigned as the preferred hierarchy, within-cytotype genetic variation was 36% (ϕ = 0.572, p = 0.001). Among-population variation increased to 64.8% in tetraploids when cytotypes were analyzed separately, while the values for diploids did not change noticeably (36%).
Genetic Clusters and Population Structure Variation
Three major clusters with strong bootstrap support (>90%) were observed in the unrooted NJ tree (Figure 3), and five (sub-)clusters were resolved with variable bootstrap values (<70%). Pure diploid and tetraploid populations and mixed-ploidy populations were grouped in all three clusters and interspersed among each other (Figure 3), suggesting that tetraploids were formed independently from different diploids within each cluster. In only a few diploid populations, all individuals from the same population were grouped together in a single cluster (e.g., M26/1X, Hojs429/1U, and Hojs468/2S; see Figure 3). In most cases, individuals from the same diploid population formed small groups interspersed among clusters (e.g., Hojs420/1P, Hojs422/1Q, Hojs423/1R, Hojs425/1T, Hojs432/1V, and M31/1W). Individuals from two of such populations were found in two clusters (populations Hojs420/1P and Hojs425/1T), while individuals from three diploid populations were found in three clusters (populations Hojs422/1Q, Hojs423/1R, and M31/1W). Such wide genetic representation observed among individuals of single populations indicates either an ancestral origin or regular gene flow among diploid populations. A Mantel test of genetic variation in diploid populations showed isolation by distance supporting the former hypothesis (r2 = 0.163, p = 0.031). Contrary to diploid populations, all but three tetraploid populations (populations M29/1B, Hojs405/1F, and Hojs410/1I) formed compact groups within each cluster (Figure 3). Two out of four mixed-ploidy populations (Hojs481/2W and Hojs487/2Y; Figure 3) formed a consistent single group, while individuals from the other two mixed-ploidy populations formed small, separate groups within each cluster (in populations Hojs470/2T and Hojs456/2Q; Figure 3). Mantel test for tetraploids exhibited genetic variation strongly correlated to distance among populations (r2 = 0.3040, p = 0.001). Furthermore, when the overall distance found among 4x populations and genetic clusters of the studied individuals were plotted against distance intervals (Mantel correlogram; Matesanz et al., 2011), almost all groups (except for two) indicated significant isolation (filled points in Supplementary Figure S2, p < 0.05). This is expected in apomictic species where random mating—even within populations—is limited as apomixis imposes substantial reproductive isolation among individuals (see Discussion for details). Our IBE analysis showed that the observed genetic diversity among all populations shows a strong correlation to the environmental distance showing substantial isolation by environment (F-statistic: 6.605, adjust R-squared: 0.1415, p-value: 0.0148, Supplementary Figure S4). This observation corroborates with our previous claim of the ecological divergence of cytotypes.
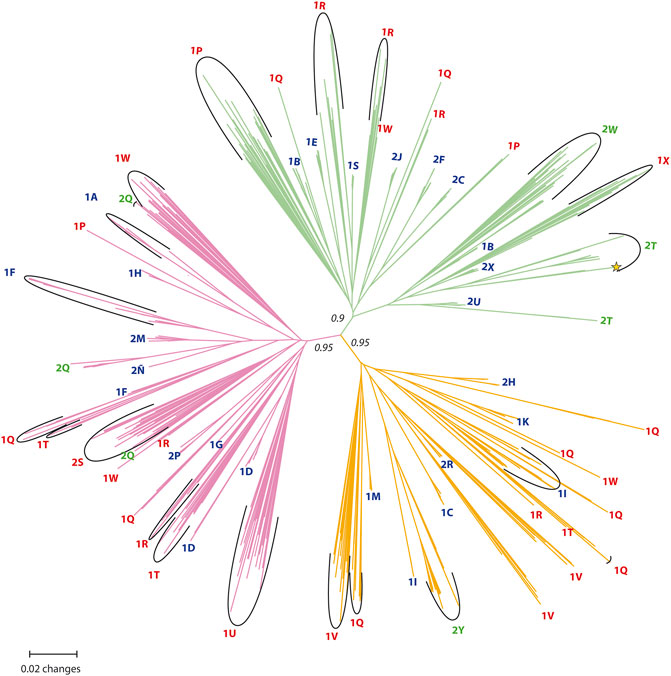
FIGURE 3. The unrooted Neighbor-Joining (NJ) tree constructed using the Prevosti’s Distance Coefficient among the amplified fragment length polymorphisms (AFLPs) of all the studied individuals of P. intermedium with a bootstrap analysis of 1,000 sample size. The bootstrap values are shown only for the main branches. The tip labels show the respective population (red: diploid populations, blue: tetraploid populations, green: mixed-ploidy populations, *: triploid individual).
The PCoA revealed higher genetic similarity among diploid populations compared to tetraploid ones (Figure 4). Despite highly clonal populations, tetraploids revealed a wider dispersion along both PCoA axes and a continuous genetic variation (Figure 4). Interestingly, the tetraploid population located in the north of the distribution (Hojs424/1S; see Figure 1) as well as the mixed-ploidy populations Hojs470/2T and the Hojs481/2W clustered together with diploids. The third mixed-ploidy population (Hojs456/2Q) clustered in between the diploids and most tetraploids, while the fourth mixed population (Hojs487/2Y) fell in the middle of the genetic dispersion of tetraploids (Figure 4).
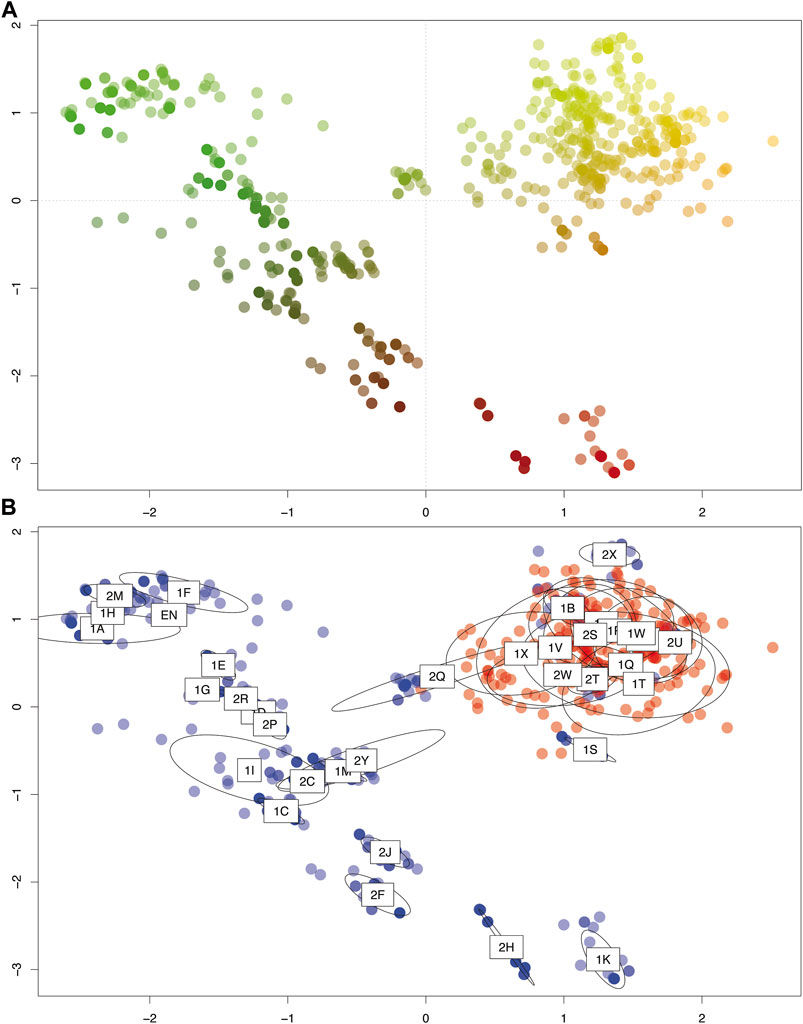
FIGURE 4. Principle coordinate analysis (PCoA) of the studied populations of P. intermedium based on pairwise Euclidian distance used in the DAPC (Discriminant Analysis of Principal Component) function of the ADEGENET R package. The first two axes represent 37 and 21% of total variation, respectively. (A) Genetic similarity among all the individuals depicted by RGY color scheme (i.e., Red, Green, and Yellow distinct groups, and the shades of each color show how much related they are to each pure RGY group). (B) Genetic variation between cytotypes (red: diploids, blue: tetraploids) and populations (labels indicating population codes are mapped on the center of ellipses representing the 95% dispersion of individuals from each population).
Bayesian clustering revealed three major clusters (k = 3) wherein most pure diploid and tetraploid populations were segregated each in individual clusters, while a few pure populations were clustered together with all mixed-ploidy populations in a third cluster (Figure 2A). Cluster analysis also showed high levels of admixture among diploid and mixed-ploidy populations (Figure 2A), including several tetraploid populations close to the contact zone (Figures 2A,B). Supporting the PCoA, the two mixed-ploidy populations (Hojs456/2Q and Hojs487/2Y) falling out of the diploid clustering (Figure 4, and Figure 2B) harbored the greatest amounts of admixture compared to all the other populations. Noticeably, geographically distant tetraploid populations like Hojs451/2M, Hojs453/2Ñ (EN), and Hojs475/2U (Figure 2B) showed minimum or no admixture. The Fst genetic divergence values of each population revealed substantially high levels of genetic divergence among most tetraploid populations compared to diploid ones, except for populations M29/1B, Hojs405/1F, and Hojs410/1I whose values were high but more similar to those from diploids (Fst = 0.79, Fst = 0.72, and Fst = 0.68, respectively) (Figure 2C).
Morphological Variation
All 42 morphological characters were measured in 64 individuals, representing three reference diploids and 61 tetraploids. A K-means clustering with K = 3 (based on the number of clusters observed in the genetic variation) showed that the morphology of individuals makes three clusters with substantial overlap among clusters (Supplementary Figure S3). This observation proved clearer when the variation was plotted on PCoA ordination plot showing a continuum of character variation among individuals with no correspondence to populations, genetic clusters, or geographic distribution. The SIMPER analysis showed that qualitative (descriptive/categorical) characters contributed the most to the observed morphological variation, although quantitative characters were also adding to the variability. Inflorescence and anthecium shape contributed 45.6 and 18.0%, respectively, to the variation observed among all the individuals (Supplementary Table S1). Among the quantitative traits, the height of the plant and number of racemes were also recognized as substantial discriminating characters of the morphological variation (each contributing 3.8%, and 2.8% respectively; Supplementary Table S1). Analyses of populations with respect to their geographic location (i.e., North, Central, and South of the species distribution) did not expose any affiliation of morphological variation to geography or climatic gradients and rather indicated a wide range of gradual character variations (Supplementary Figure S3).
Discussion
In plants, the data on genetic composition and population structure variation are a much-needed complement to information about biological traits and are essential information for understanding the recent evolutionary history of any species. In multiploid taxa with cytotypes showing alternative reproductive modes (other than sexuality) and restricted gene flow (e.g., clonal propagation and apomixis), such studies take a new dimension for understanding natural variation and the evolutionary fate of cytotypes within polyploid complexes. The present analysis revises local and regional standing genetic variation in P. intermedium populations and associated morphological and reproductive traits. In combination with data from species distribution modeling (Karunarathne et al., 2018), the present study sheds light on the evolutionary aspects of P. intermedium life history, suggesting that intraspecific variation is segregated within cytotypes and that recurrent formation of tetraploids allows a range expansion of polyploid cytotypes into new habitats, thus creating genetically divergent lineages and promoting ecologically differentiation.
Cytotype Genetic Divergence Is Explained by Differential Rates of Sexuality
The results show that fully sexual diploids have relatively few private fragments (17.9%), suggesting that regular gene flow occurs between diploid individuals among populations and there is minimal isolation by distance. Despite the noticeable number of cytotype-specific fragments among genotypes (21.6%), a considerable proportion of fragments (33.8%) were shared between diploids and tetraploids. Since tetraploids in P. intermedium are autopolyploids (Norrmann et al., 1989), the observation of ca. 34.2% of unique fragments in polyploids of mixed-ploidy populations (expectedly having a relatively recent origin, see Discussion below) indicates that those fragments are a consequence of post-polyploidization mutations, likely involving genomic rearrangements. Similar cases of rapid sequence rearrangements (indels), genomic and regulatory alterations during formation, and stabilization of allopolyploids and autopolyploids have already been reported in Paspalum spp (Calderini et al., 2006; Martelotto et al., 2007; Siena et al., 2016) and other plant systems (Akiyama et al., 2005; Conner et al., 2008) and are responsible for evolutionary novelty (e.g., Ainouche et al., 2003; Hegarty and Hiscock, 2005; Paun et al., 2006). The levels of genetic variation observed in mixed-ploidy populations of P. intermedium (Table 1) point toward regular gene flow between ploidies, either through de novo formation of polyploids (2x → 4x) or through backcrosses (2x × 4x). The finding of only one triploid individual among the four mixed populations (<1% of all individuals) instead of an expectedly higher number of triploids from backcrosses suggests that 1) there is likely an interploidy barrier between cytotypes (either pre- or post-zygotic) and 2) that rare triploids are more likely to arise from diploids through an unreduced gamete, thus providing support to the hypothesis of de novo formation of polyploids via a triploid bridge, as postulated for other Paspalum species (Quarin, 1992) and recently discussed as a pivotal step enhancing polyploid establishment in nature (Hojsgaard, 2018). The fact that the triploid individual is genetically more related to the diploid individuals in that population than to the coexisting tetraploids further supports this hypothesis and matches the above statement, indicating a probably recent origin for the mixed-ploidy populations.
In pure tetraploid populations, the data show low within-population genetic variation in concordance with the observed low genotype diversity (Table 1), a result consistent with the lack or reduced sexuality inherently associated to high proportions of apomictic reproduction in P. intermedium (Karunarathne et al., 2018; 2020b). Studies alike have also shown a reduction in the number of effective genotypes in populations of polyploid apomicts, even when the overall level of genetic diversity is maintained (e.g., Hamston et al., 2018). In P. intermedium, the most significant contribution to genetic diversity comes from the variation found among populations, holding an average proportion of private fragments (21.6%). Our results—and particularly the neighbor-joining analysis—further indicate that the observed among population variation in genotype diversity is due to independent polyploid founder events, plus the posterior sporadic occurrence of sexuality. Low proportions of sexual reproduction (<16%) had been detected through embryology on ovules and flow cytometry analysis on seeds of individuals from all tetraploid apomictic populations included here (Karunarathne et al., 2020b). Since apomixis can impose a within-population reproductive isolation among individuals, the rate of (residual) sexuality in such populations has a higher impact on gene flow analysis than geographic distance. The regular occurrence of low rates of sexuality is central to the adaptation and evolution of apomictic populations in agamic complexes (Hojsgaard and Hörandl, 2019). We previously showed that P. intermedium tetraploids can have residual reproduction within populations as high as 20% at the seed stage and up to 40% at the embryo-sac stage depending on the environmental stress on the plant (Karunarathne et al., 2020b). Varying proportions of residual sexuality and facultative apomixis have been often reported in other species from genera like Boechera spp. (Carman et al., 2019), Hieracium spp. (Hand et al., 2015), Hypericum spp. (Barcaccia et al., 2006), Paspalum spp. (Daurelio et al., 2004), Potentilla spp. (Nyléhn et al., 2003), Pilosella spp. (Krahulcová et al., 2014), Ranunculus spp. (Cosendai and Hörandl, 2010; Cosendai et al., 2013; Pellino et al., 2013), and Taraxacum spp. (Vijverberg et al., 2004; Majeský et al., 2012), among others. Strict clonality has been rarely found in nature, obligate apomictic triploids of Taraxacum officinale being perhaps one of the few examples (Van der Hulst et al., 2000). The genetic evidence in P. intermedium supports this view and points to residual sexuality playing a critical role in maintaining moderate levels of genotypic diversity despite restricted genetic diversity in apomictic populations. Such genotypic diversity supports the range expansions of polyploid genotypes with greater ecological tolerance into new geographic areas (Karunarathne et al., 2018). Since clonal tetraploid populations are distinct to each other and each population exhibits either single or multiple genotypes rather one common genotype and occupies regions differing ecologically, the data endorse the Frozen Niche Variation model (instead of a General-Purpose Genotype model) of genotype dispersal and habitat occupation for the apomictic populations of P. intermedium. The fact that genetic variation is significantly associated to environmental differences further supports this inference. The observed positive correlation between Nei’s population level genetic diversity to environmental distance suggests that genetically different clones/cytotypes are being selected to occupy environmentally different areas.
Genetic Admixture Shows Multiple Polyploid Lineages of Independent Origins
Panmictic (sexual) populations are expected to hold high levels of genetic admixture facilitated by regular random gene flow between individuals (Suda et al., 2007). The study of the spatial distribution of standing genetic variation in natural populations is relevant to identify the relative levels of genetic admixture (Cruse-Sanders and Hamrick, 2004; Matesanz et al., 2011) and to get insights into the mechanisms responsible for it (Ng et al., 2004). Variations in admixture levels can be attributed to an array of factors, historical or current, which interfere the rates of gene flow between individuals directly (e.g., lack of pollinator specificity, pre- or postzygotic isolation) or indirectly (e.g., spatial isolation, phenology) and can provide useful information on key aspects of species life history, such as dispersal, genetic drift, and selection processes.
In the present study, our results from 35 geographically widespread P. intermedium populations point to the occurrence of distinct levels of genetic admixture among populations depending upon the ploidy, the occurrence of multiple cytotypes, and the geographic location of the population (Table 1, 2; Figure 2). In populations of P. intermedium holding only a diploid cytotype, the variability observed in the Neighbor-Joining tree displays diploid individuals from each population being widespread within and (in some cases) among the three main genetic clusters of our dataset (Figure 3). Diploid populations like Hojs422/1Q, Hojs423/1R, and M31/1W are good examples and a clear evidence of high levels of genetic admixture in diploids, either through gene flow among individuals and/or migrants between populations. The discriminant analysis provides additional support by showing that all diploids are grouped together, and by the lack of populations isolated by distance (Figure 4). Similar levels of admixture are expected for sexual populations and had already been observed between sexual diploids in other agamic complexes (e.g., Paun et al., 2006; Cosendai et al., 2013).
In populations of P. intermedium with only one tetraploid cytotype, the situation is contrastingly different. Tetraploids show low admixture and genetic similarity to different diploid genetic clusters (Figure 2, 3). Unlike diploids, tetraploid individuals from single populations are grouped together, in a few cases interspersed with a few diploids in between (population Hojs410/1I; Figure 3) or diploid–tetraploid individuals from other populations (Hojs405/1F; Figure 3). This is expected in asexual lineages where genotypic diversity is constrained by clonality. Our results on ancestry point to the occurrence of at least 16 independent polyploidization events sharing high genetic similarity with 12 diploid populations (Table 3; Figure 3). In a few cases, it was possible to link the origin of a tetraploid population to one (or two) diploid parental individuals (e.g., in the orange cluster, the population Hojs445/2H likely originated from individuals Hojs422/1Q 16/17; Figure 3). A few diploid populations hold individuals sharing alleles with populations of more than one cluster and are putatively involved in the origin of several tetraploid populations (e.g., diploid population Hojs423/1R likely originated tetraploid populations Hojs404/1E, Hojs406/1G, Hojs424/1S, and Hojs414/2J; Table 3). Multiple origins of polyploidy are commonly observed in different plant species with diverse distribution patterns and life histories and are considered an important evolutionary mechanism for diversification (Fehrer et al., 2005; Lo et al., 2009).
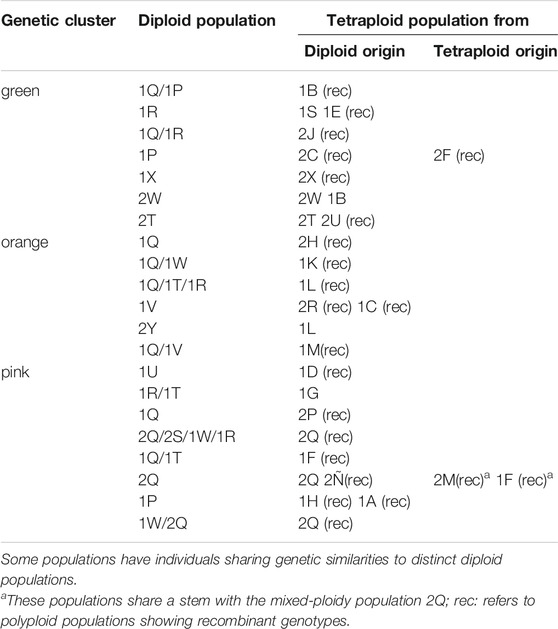
TABLE 3. Population origin inferred from Bayesian analysis and Neighbor-Joining tree based on genetic distance.
While sexual diploids in P. intermedium show no genetic or cytological (discussed above) evidence of introgression from tetraploids, diploids can inject genetic variability into a tetraploid population through recurrent formation of new polyploids. This is very likely the case in all four mixed-ploidy populations that show a higher number of genotypes compared to pure polyploid populations and intermediate fixation indices (Fst values, Table 1; see below). All other populations show little or no genotype variability. Variability in these pure polyploid populations can either be generated through post-polyploidization mutations like genome rearrangements (Birchler, 2012; Mandáková and Lysak, 2018) or by genetic reshuffling through sexual reproduction. As discussed above, most apomicts are facultative and exhibit low levels of functional sexuality, including P. intermedium (Karunarathne et al., 2018; 2020b). All but six populations represented by a single clone show evidence of genotype diversification by sexual recombination in local populations, and four tetraploid populations are likely originated from polyploid migrants (populations Hojs405/1F, Hojs416/1M, Hojs443/2F, and Hojs453/2Ñ; Table 1, 3). The tetraploid populations most diverse in terms of genotype variability and number of effective genotypes (M29/1B, Hojs405/1F, and Hojs410/1I; Table 1 and Figure 2) show fixation indices alike those found in diploids populations, suggesting lower genetic differentiation. A genome-wide analysis of variation will provide more details on the demography of these cytotypes and the species. Nevertheless, since apomixis is the dominant reproductive mechanism in these tetraploid genotypes, they capture and fix a fraction of the genetic variability present in diploid parentals, including transgressive variation, which explains the wider distribution of tetraploid populations observed in our discriminant analysis (Figure 4) despite their genetic relatedness to different diploid populations (Figure 3).
Admixture in tetraploid apomictic populations is low, and the feeble indication of gene flow in some populations (e.g., Hojs406/1G, Hojs404/1E, and Hojs403/1D) further supports the evidence of genetic isolation by distance among these populations and significant spatial dissociation (p = 0.001) (see Supplementary Figure S2). While strong isolation between distant sexual polyploids can lead to directional selection favoring local adaptations (Parisod et al., 2010), in polyploid apomictic P. intermedium individuals, isolation by distance between populations is likely reinforced by restrictions to gene flow imposed by the reproductive mode, which might favor stronger population differentiation at regional scales. Overall, the geographical distribution of genetic clusters identified using Bayesian and discriminant analyses and previous evidence of intra- and inter-population cytotype dynamics indicate that selection is acting at distinct geographic scales to sift genotypes better adapted to harsher environmental conditions in southern areas of the distribution. More specific studies including transplantation experiments and exploitation of next-generation approaches (see e.g., Yang et al., 2016) will help in clarifying these points.
The Case of the Mixed-Ploidy Populations in the Contact Zone
Contact zones represent geographic areas where two or more conspecific individuals of different ploidies come into contact, and therefore, they are excellent locations representing natural laboratories to study the specifics of biological interactions and evolutionary transitions (Lexer and van Loo, 2006). The most relevant mechanisms to be elucidated in a zone of contact between cytotypes are those enhancing differentiation and reinforcing reproductive isolation. Depending on historical aspects, contact zones are of primary or secondary origin (Petit et al., 1999). In the first case, new polyploids emerge within a parental (often diploid) population, while in the latter, allopatric cytotypes (often diploids and polyploids) come into contact after migration or range expansion of populations. Both primary and secondary contact zones are found in natural populations of many polyploid complexes, like in the Knautia arvensis agg. (Kolár et al., 2009), in Aster amellus (Castro et al., 2012), or in Galium valdepilosum (Kolař et al., 2014). In P. intermedium, our current genetic analysis shows that all but one mixed ploidy population form a consistent genetic group with conspecific diploids indicating a primary origin (Hojs470/2T, Hojs481/2W and Hojs487/2Y; Figure 3, Figure 4). Previous evidence of polyploid origin, reproductive biology, and cytotype frequencies (Norrmann et al., 1989; Karunarathne et al., 2018, 2020b), including the occurrence of a triploid plant in population Hojs470/2T, point to the occurrence of autopolyploidization events via a triploid bridge and further suggest a primary origin for the contact zones. The fourth mixed ploidy population (Hojs456/2Q) shows a more complex pattern in our Neighbor-Joining tree whereby the diploids exhibit high genetic similarity to pure diploid populations M31/1W, Hojs423/1R, and Hojs468/2S, suggesting gene flow between diploids rather than a possible secondary contact with conspecific tetraploid migrants. While inter-cytotype crosses are commonly observed in secondary contact zones in sexual plants (e.g. Zozomová-Lihová et al., 2015), they are much less frequent among sexual–apomictic polyploid complexes mainly because egg cells in apomictic ovules do not receive pollen, and yet, apomicts’ pollen can exert sizeable reproductive interference on sexuals (Hersh et al., 2016). Studies in natural populations from different Paspalum species depict a lack of interploidy hybrids, even when they are easily produced under experimental conditions (e.g., Hojsgaard et al., 2008, 2013; Karunarathne et al., 2020a). The lack of triploids, the occurrence of higher number of effective genotypes, and the levels of admixture among tetraploids from mixed-ploidy populations compared to those in pure populations of P. intermedium suggest either that tetraploids are regularly formed de novo in these contact zones or that the levels of sexuality in tetraploids are higher and recombinant genotypes are produced at higher frequencies than in pure populations (population Hojs481/2W, for instance, contains five different tetraploid genotypes; Table 1). These observations plus the existence of competition and ecological displacement between diploid and tetraploid cytotypes (Karunarathne et al., 2018) and the fact that five out of six tetraploid clonal populations occur southern from the zone of contact between cytotypes suggest that many tetraploid populations have probably originated at the contact zone and eventually spread to the south of the distribution range. In addition, the southernmost tetraploid populations show close genetic affinities to mixed-ploidy populations in the contact zone (population Hojs475/2U to mixed population Hojs470/2T, Hojs451/2M, Hojs453/2Ñ, and Hojs405/1F to Hojs456/2Q, M29/1B to Hojs481/2W, and population Hojs410/1I to mixed population Hojs487/2Y; Figure 3, and Figure 2), which supports the view of tetraploids’ dispersal to the south.
A Continuum of Morphological Variation Supports Independent Origins of Polyploid Lineages
The analysis of morphological variation found in 42 quantitative and qualitative characters displays the formation of arbitrary groups with substantial overlap among the apomictic populations of P. intermedium but lacking correspondence to either genetic clusters or geographical distribution. The variation rather exhibits a continuum of morphological character distribution among all the populations that is rooted in the variation observed for three representatives of the diploid cytotype (Figure S3). While lack of morphologically discrete cytotype groups can be expected in grass species (e.g., Amirouche and Misset, 2007; Pimentel and Sahuquillo, 2008), or in other families (e.g., in the Rosaceae; Bigl et al., 2019), cases of significant morphological differentiation between cytotypes have been observed in many other plant groups (e.g., Segraves et al., 1999; Kao and Parker, 2010), including one species from the genus Paspalum (Quarin and Hanna, 1980). Such morphological variation is mostly due to nucleotypic effects (i.e., effects associated with the DNA content in nuclei or the number of monoploid genomes) or to confound phenomena like hybridity (in allopolyploids) or ecological differentiation (Ramsey and Schemske, 2002). Polyploidization can cause immediate divergence in phenotypic traits important to establishment of neopolyploids without affecting significantly genotypic variation (Oswald and Nuismer, 2011). In P. intermedium, we observe rather the opposite, no clear morphological divergence but genetically segregant groups among tetraploid populations. Despite tetraploids showing distinct ecological preferences to diploids and occupying differentiated niches (Karunarathne et al., 2018), the multiple polyploidization events from diverse diploid gene pools detected here create morphologically overlapping groups among tetraploids from distant geographical regions. The occurrence of selfing and apomixis in these polyploid lineages further contribute to keep intrapopulation morphological variability low, as observed in other polyploid apomictic complexes like those of the subfamily Malodieae (Campbell and Dickinson, 1990) or in the Ranunculus auricomus aggregate (Hodač et al., 2014). The fact that tetraploid apomicts in P. intermedium showing overlapping morphology and evidence of single, independent founder events for each of the apomictic populations studied here (with indication of gene flow among populations as discussed above), provides support to the view that the polyploid complex is evolutionarily young. Alternatively, the paucity in genotypic variation despite the observed rates of residual sexuality might be explained by a high formation/extinction rate of polyploid lineages.
Conclusions
The present study on genetic variation in 35 natural populations of P. intermedium shows that apomictic autotetraploids are of multiple independent origins, indicating recurrent polyploidization and subsequent genotype diversification through facultative apomixis. The substantial among-population genetic variation and genetic similarity analyses further point to polyploidization events from genetically distinct diploid populations. The contact zone existing between diploids and tetraploids is primary in origin where tetraploids are persistently formed from diploids. Such polyploids might become established or outcompeted by coexisting diploid parentals, and yet, because of wider ecological tolerance, some tetraploids manage to disperse southward and establish new polyploid populations, clonal or mostly clonal. Apomictic populations display reduced genotype and genetic variability, strict geographic isolation, and high genetic differentiation which support a recent evolutionary origin for the polyploid complex.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
DH conceived and designed the project, PK and DH conducted the fieldwork, PK performed the lab work and data generation. PK and DH performed statistical analyses and wrote the manuscript.
Funding
This work was supported by the DFG in the framework of DFG-MINCyT-CONICET international cooperation (HO5462-1/1 to D.H.).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank Ana Honfi, Eric Martínez, Carlos Acuña, and various students for their help and support during sampling and/or cultivation of plant materials. Curators of herbaria MNES, CTES, BAA, SI, B, GOET, HUH, and PE are appreciated for allowing us to study the collections. We place our gratitude to Alexandra Dolynska for her assistance with AFLP procedures and Silvia Friedrich for taking care of the plants in climate chambers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.736088/full#supplementary-material
References
Ainouche, M. L., Baumel, A., Salmon, A., and Yannic, G. (2003). Hybridization, Polyploidy and Speciation in Spartina (Poaceae). New Phytol. 161, 165–172. doi:10.1046/j.1469-8137.2003.00926.x
Akiyama, Y., Hanna, W. W., and Ozias-Akins, P. (2005). High-resolution Physical Mapping Reveals that the Apospory-specific Genomic Region (ASGR) in Cenchrus Ciliaris Is Located on a Heterochromatic and Hemizygous Region of a Single Chromosome. Theor. Appl. Genet. 111, 1042–1051. doi:10.1007/s00122-005-0020-5
Amirouche, N., and Misset, M.-T. (2007). Morphological Variation and Distribution of Cytotypes in the Diploid-Tetraploid Complex of the Genus Dactylis L. (Poaceae) from Algeria. Plant Syst. Evol. 264, 157–174. doi:10.1007/s00606-006-0502-1
Baddeley, A., and Turner, R. (2005). Spatstat: An R Package for Analysizing Spatial Point Patterns. J. Stat. Softw. 12, 1–42. doi:10.18637/jss.v012.i06
Baker, H. G. (1955). Self-compatibility and Establishment after '"Long-Distance" Dispersal. Evolution 9, 347–349. doi:10.2307/240565610.1111/j.1558-5646.1955.tb01544.x
Baker, H. G. (1967). Support for Baker's Law-As a Rule. Evolution 21, 853–856. doi:10.1111/j.1558-5646.1967.tb03440.x
Barcaccia, G., Arzenton, F., Sharbel, T. F., Varotto, S., Parrini, P., and Lucchin, M. (2006). Genetic Diversity and Reproductive Biology in Ecotypes of the Facultative Apomict Hypericum perforatum L. Heredity 96, 322–334. doi:10.1038/sj.hdy.6800808
Bigl, K., Paule, J., and Dobeš, C. (2019). The Morphometrics of Autopolyploidy: Insignificant Differentiation Among Sexual-Apomictic Cytotypes. AoB Plants 11, 1–11. doi:10.1093/aobpla/plz028
Birchler, J. A. (2012). “Genetic Consequences of Polyploidy in Plants,” in “Genetic Consequences of Polyploidy in Plants,” in Polyploidy And Genome Evolution. Editors P. S. Soltis, and D. E. Soltis (Berlin, Heidelberg: Springer Berlin Heidelberg), 21–32. doi:10.1007/978-3-642-31442-1_2
Bretagnolle, F., and Thompson, J. D. (1996). An Experimental Study of Ecological Differences in winter Growth between Sympatric Diploid and Autotetraploid Dactylis Glomerata. J. Ecol. 84, 343–351. doi:10.2307/2261197
Brochmann, C., Brysting, A. K., Alsos, I. G., Borgen, L., Grundt, H. H., Scheen, A.-C., et al. (2004). Polyploidy in Arctic Plants. Biol. J. Linn. Soc. 82, 521–536. doi:10.1111/j.1095-8312.2004.00337.x
Calderini, O., Chang, S. B., De Jong, H., Busti, A., Paolocci, F., Arcioni, S., et al. (2006). Molecular Cytogenetics and DNA Sequence analysis of an Apomixis-Linked BAC in Paspalum simplex Reveal a Non pericentromere Location and Partial Microcolinearity With Rice. Theor. Appl. Genet. 112, 1179–1191. doi:10.1007/s00122-006-0220-7
Campbell, C. S., and Dickinson, T. A. (1990). Apomixis, Patterns of Morphological Variation, and Species Concepts in Subfam. Maloideae (Rosaceae). Syst. Bot. 15, 124. doi:10.2307/2419022
Carman, J. G., Mateo de Arias, M., Gao, L., Zhao, X., Kowallis, B. M., Sherwood, D. A., et al. (2019). Apospory and Diplospory in Diploid Boechera (Brassicaceae) May Facilitate Speciation by Recombination-Driven Apomixis-To-Sex Reversals. Front. Plant Sci. 10, 1–14. doi:10.3389/fpls.2019.00724
Castro, S., Loureiro, J., Procházka, T., and Münzbergová, Z. (2012). Cytotype Distribution at a Diploid-Hexaploid Contact Zone in Aster Amellus (Asteraceae). Ann. Bot. 110, 1047–1055. doi:10.1093/aob/mcs177
Conner, J. A., Goel, S., Gunawan, G., Cordonnier-Pratt, M.-M., Johnson, V. E., Liang, C., et al. (2008). Sequence Analysis of Bacterial Artificial Chromosome Clones from the Apospory-specific Genomic Region ofPennisetumandCenchrus. Plant Physiol. 147, 1396–1411. doi:10.1104/pp.108.119081
Cosendai, A.-C., and Hörandl, E. (2010). Cytotype Stability, Facultative Apomixis and Geographical Parthenogenesis in Ranunculus Kuepferi (Ranunculaceae). Ann. Bot. 105, 457–470. doi:10.1093/aob/mcp304
Cosendai, A.-C., Wagner, J., Ladinig, U., Rosche, C., and Hörandl, E. (2013). Geographical Parthenogenesis and Population Genetic Structure in the alpine Species Ranunculus Kuepferi (Ranunculaceae). Heredity 110, 560–569. doi:10.1038/hdy.2013.1
Cruse-Sanders, J. M., and Hamrick, J. L. (2004). Spatial and Genetic Structure within Populations of Wild American Ginseng (Panax Quinquefolius L., Araliaceae). J. Hered. 95, 309–321. doi:10.1093/jhered/esh046
Daurelio, L. D., Espinoza, F., Quarin, C. L., and Pessino, S. C. (2004). Genetic Diversity in Sexual Diploid and Apomictic Tetraploid Populations of Paspalum Notatum Situated in Sympatry or Allopatry. Plant Syst. Evol. 244, 189–199. doi:10.1007/s00606-003-0070-6
Ehrich, D. (2007). Documentation for AFLPdat A Collection of R Functions for Handling AFLP Data. Mol. Ecol. Notes 6, 603–604. doi:10.1007/s12144-015-9363-5
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 14, 2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Fehrer, J., Šimek, R., Krahulcová, A., Chrtek, J. Í., and Bräutigam, E. (2005). “Evolution, Hybridisation, and Clonal Distribution Central European Mountain Range” in Plant Species-Level Systematics: New Perspectives on Pattern & Process. Editors F. T. Bakker, L. W. Chatrou, B. Gravendeel, and P. B. Pelser (Ruggell, Liechtenstein: Gantner Verlag), 175–201.
Fox, D. T., Soltis, D. E., Soltis, P. S., Ashman, T.-L., and Van de Peer, Y. (2020). Polyploidy: A Biological Force from Cells to Ecosystems. Trends Cel Biol. 30, 688–694. doi:10.1016/j.tcb.2020.06.006
Frichot, E., and Francois, O. (2014). LEA: An R Package for Landscape and Ecological Association Studies.
Hammer, Ø., Harper, D. A. T., and Ryan, P. D. (2001). PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 4, 1352–1357.
Hamston, T. J., de Vere, N., King, R. A., Pellicer, J., Fay, M. F., Cresswell, J. E., et al. (2018). Apomixis and Hybridization Drives Reticulate Evolution and Phyletic Differentiation in Sorbus l.: Implications for Conservation. Front. Plant Sci. 9, 1–13. doi:10.3389/fpls.2018.01796
Hand, M. L., Vít, P., Krahulcová, A., Johnson, S. D., Oelkers, K., Siddons, H., et al. (2015). Evolution of Apomixis Loci in Pilosella and Hieracium (Asteraceae) Inferred from the Conservation of Apomixis-Linked Markers in Natural and Experimental Populations. Heredity 114, 17–26. doi:10.1038/hdy.2014.61
Hegarty, M. J., and Hiscock, S. J. (2005). Hybrid Speciation in Plants: New Insights from Molecular Studies. New Phytol. 165, 411–423. doi:10.1111/j.1469-8137.2004.01253.x
Hersh, E., Grimm, J., and Whitton, J. (2016). Attack of the Clones: Reproductive Interference between Sexuals and Asexuals in the Crepis Agamic Complex. Ecol. Evol. 6, 6473–6483. doi:10.1002/ece3.2353
Hodač, L., Scheben, A. P., Hojsgaard, D., Paun, O., and Hörandl, E. (2014). ITS Polymorphisms Shed Light on Hybrid Evolution in Apomictic Plants: a Case Study on the Ranunculus Auricomus Complex. PLoS One 9, e103003. doi:10.1371/journal.pone.0103003
Hojsgaard, D., and Hörandl, E. (2015). A Little Bit of Sex Matters for Genome Evolution in Asexual Plants. Front. Plant Sci. 6, 82. doi:10.3389/fpls.2015.00082
Hojsgaard, D. H., Martínez, E. J., and Quarin, C. L. (2013). Competition between Meiotic and Apomictic Pathways during Ovule and Seed Development Results in Clonality. New Phytol. 197, 336–347. doi:10.1111/j.1469-8137.2012.04381.x
Hojsgaard, D., and Hörandl, E. (2019). The Rise of Apomixis in Natural Plant Populations. Front. Plant Sci. 10. doi:10.3389/fpls.2019.00358
Hojsgaard, D., Schegg, E., Valls, J. F. M., Martínez, E. J., and Quarin, C. L. (2008). Sexuality, Apomixis, Ploidy Levels, and Genomic Relationships Among Four Paspalum Species of the Subgenus Anachyris (Poaceae). Flora - Morphol. Distribution, Funct. Ecol. Plants 203, 535–547. doi:10.1016/j.flora.2007.09.005
Hojsgaard, D. (2018). Transient Activation of Apomixis in Sexual Neotriploids May Retain Genomically Altered States and Enhance Polyploid Establishment. Front. Plant Sci. 9, 230. doi:10.3389/fpls.2018.00230
Hörandl, E., and Paun, O. (2007). “Patterns and Sources of Genetic Diversity in Apomictic Plants: Implications for Evolutionary Potentials,” in Apomixis: Evolution, Mechanisms, and Perspectives. Editors E. Hörandl, U. Grossniklaus, P. van Dijk, and T. F. Sharbel (Rugell, Liechtenstein: A.R.G Gantner Verlag K. G.), 169–194.
Hörandl, E. (2006). The Complex Causality of Geographical Parthenogenesis. New Phytol. 171, 525–538. doi:10.1111/j.1469-8137.2006.01769.x
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral Polyploidy in Seed Plants and Angiosperms. Nature 473, 97–100. doi:10.1038/nature09916
Jombart, T. (2008). Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 24, 1403–1405. doi:10.1093/bioinformatics/btn129
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, And/or Sexual Reproduction, 2, e281. doi:10.7717/peerj.281
Kao, R. H., and Parker, I. M. (2010). Coexisting Cytotypes ofArnica Cordifolia: Morphological Differentiation and Local‐Scale Distribution. Int. J. Plant Sci. 171, 81–89. doi:10.1086/647924
Karger, D. N., Conrad, O., Böhner, J., Kawohl, T., Kreft, H., Soria-Auza, R. W., et al. (2017). Climatologies at High Resolution for the Earth's Land Surface Areas. Sci. Data 4, 1–20. doi:10.1038/sdata.2017.122
Karunarathne, P., Feduzka, C., and Diego, H. (2020a). Ecological Setup, Ploidy Diversity, and Reproductive Biology of Paspalum Modestum, a Promising Wetland Forage Grass from South america. Genet. Mol. Biol. 43, 1–12. doi:10.1590/1678-4685-gmb-2019-0101
Karunarathne, P., Reutemann, A. V., Schedler, M., Glücksberg, A., Martínez, E. J., Honfi, A. I., et al. (2020b). Sexual Modulation in a Polyploid Grass: a Reproductive Contest between Environmentally Inducible Sexual and Genetically Dominant Apomictic Pathways. Sci. Rep. 10, 1–14. doi:10.1038/s41598-020-64982-6
Karunarathne, P., Schedler, M., Martínez, E. J., Honfi, A. I., Novichkova, A., and Hojsgaard, D. (2018). Intraspecific Ecological Niche Divergence and Reproductive Shifts foster Cytotype Displacement and Provide Ecological Opportunity to Polyploids. Ann. Bot. 121, 1183–1196. doi:10.1093/aob/mcy004
Kolař, F., Lučanova, M., Koutecky, P., Dortova, M., Knotek, A., and Suda, J. (2014). Spatio-ecological Segregation of Diploid and Tetraploid Cytotypes of Galium Valdepilosum in central Europe. Preslia 86, 155–178.
Kolář, F., Štech, M., Trávníček, P., Rauchová, J., Urfus, T., Vít, P., et al. (2009). Towards Resolving the Knautia Arvensis Agg. (Dipsacaceae) Puzzle: Primary and Secondary Contact Zones and Ploidy Segregation at Landscape and Microgeographic Scales. Ann. Bot. 103, 963–974. doi:10.1093/aob/mcp016
Krahulcová, A., Rotreklová, O., and Krahulec, F. (2014). The Detection, Rate and Manifestation of Residual Sexuality in Apomictic Populations of Pilosella (Asteraceae, Lactuceae). Folia Geobot 49, 239–258. doi:10.1007/s12224-013-9166-0
Levin, D. A. (1975). Minority Cytotype Exclusion in Local Plant Populations. Taxon 24, 35–43. doi:10.2307/1218997
Lexer, C., and van Loo, M. (2006). Contact Zones: Natural Labs for Studying Evolutionary Transitions. Curr. Biol. 16, R407–R409. doi:10.1016/j.cub.2006.05.007
Lo, E. Y. Y., Stefanović, S., and Dickinson, T. A. (2009). Population Genetic Structure of Diploid Sexual and Polyploid Apomictic Hawthorns (Crataegus; Rosaceae) in the Pacific Northwest. Mol. Ecol. 18, 1145–1160. doi:10.1111/j.1365-294X.2009.04091.x
Lynch, M. (1984). Destabilizing Hybridization, General-Purpose Genotypes and Geographic Parthenogenesis. Q. Rev. Biol. 59, 257–290. doi:10.1086/413902
Majeský, Ľ., Vašut, R. J., Kitner, M., and Trávníček, B. (2012). The Pattern of Genetic Variability in Apomictic Clones of Taraxacum officinale Indicates the Alternation of Asexual and Sexual Histories of Apomicts. PLoS One 7, e41868. doi:10.1371/journal.pone.0041868
Mandáková, T., and Lysak, M. A. (2018). Post-polyploid Diploidization and Diversification through Dysploid Changes. Curr. Opin. Plant Biol. 42, 55–65. doi:10.1016/j.pbi.2018.03.001
Martelotto, L. G., Ortiz, J. P. A., Stein, J., Espinoza, F., Quarin, C. L., and Pessino, S. C. (2007). Genome Rearrangements Derived from Autopolyploidization in Paspalum Sp. Plant Sci. 172, 970–977. doi:10.1016/j.plantsci.2007.02.001
Matesanz, S., Gimeno, T. E., de la Cruz, M., Escudero, A., and Valladares, F. (2011). Competition May Explain the fine-scale Spatial Patterns and Genetic Structure of Two Co-occurring Plant Congeners. J. Ecol. 99, 838–848. doi:10.1111/j.1365-2745.2011.01812.x
Ng, K. K. S., Lee, S. L., and Koh, C. L. (2004). Spatial Structure and Genetic Diversity of Two Tropical Tree Species with Contrasting Breeding Systems and Different Ploidy Levels. Mol. Ecol. 13, 657–669. doi:10.1046/j.1365-294X.2004.02094.x
Norrmann, G. A., Quarin, C. L., and Burson, B. L. (1989). Cytogenetics and Reproductive Behavior of Different Chromosome Races in Six Paspalum Species. J. Hered. 80, 24–28. doi:10.1093/oxfordjournals.jhered.a110783
Nyléhn, J., Hamre, E., and Nordal, I. (2003). Facultative Apomixis and Hybridization in Arctic Potentilla Section Niveae (Rosaceae) from Svalbard. Bot. J. Linn. Soc. 142, 373–381. doi:10.1046/j.1095-8339.2003.00190.x
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2016). Vegan: Community Ecology Package. R Version 3.2.4. Community Ecol. Packag. Version 2. Available at: https://cran.r-project.org/package=vegan.
Oswald, B. P., and Nuismer, S. L. (2011). A Unified Model of Autopolyploid Establishment and Evolution. The Am. Naturalist 178, 687–700. doi:10.1086/662673.A
Parisod, C., Holderegger, R., and Brochmann, C. (2010). Evolutionary Consequences of Autopolyploidy. New Phytol. 186, 5–17. doi:10.1111/j.1469-8137.2009.03142.x
Paun, O., Stuessy, T. F., and Hörandl, E. (2006). The Role of Hybridization, Polyploidization and Glaciation in the Origin and Evolution of the Apomictic Ranunculus Cassubicus Complex. New Phytol. 171, 223–236. doi:10.1111/j.1469-8137.2006.01738.x
Pellino, M., Hojsgaard, D., Schmutzer, T., Scholz, U., Hörandl, E., Vogel, H., et al. (2013). Asexual Genome Evolution in the apomicticRanunculus Auricomuscomplex: Examining the Effects of Hybridization and Mutation Accumulation. Mol. Ecol. 22, 5908–5921. doi:10.1111/mec.12533
Petit, C., Bretagnolle, F., and Felber, F. (1999). Evolutionary Consequences of Diploid-Polyploid Hybrid Zones in Wild Species. Trends Ecol. Evol. 14, 306–311. doi:10.1016/s0169-5347(99)01608-0
Pimentel, M., and Sahuquillo, E. (2008). Relationships between the Close Congeners Anthoxanthum Odoratum and A. Alpinum (Poaceae, Pooideae) Assessed by Morphological and Molecular Methods. Botan J. Linn. Soc. 156, 237–252. doi:10.1111/j.1095-8339.2007.00738.x
Quarin, C. L., and Hanna, W. W. (1980). Effect of Three Ploidy Levels on Meiosis and Mode of Reproduction in Paspalum Hexastachyum1. Crop Sci. 20, 69–75. doi:10.2135/cropsci1980.0011183x002000010016x
Quarin, C. L. (1992). The Nature of Apomixis and its Origin in Panicoid Grasses. Apomixis Newslt. Available at: http://ci.nii.ac.jp/naid/10004268589/en/(Accessed May 17, 2017). 5, 8–15.
R Core Team (2016). R: A Language and Environment for Statistical Computing. Available at: http://www.r-project.org.
Ramsey, J., and Schemske, D. W. (2002). Neopolyploidy in Flowering Plants. Annu. Rev. Ecol. Syst. 33, 589–639. doi:10.1146/annurev.ecolsys.33.010802.150437
Ramsey, J., and Schemske, D. W. (1998). Pathways, Mechanisms, and Rates of Polyploid Formation in Flowering Plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi:10.1146/annurev.ecolsys.29.1.467
Segraves, K. A., Thompson, J. N., Soltis, P. S., and Soltis, D. E. (1999). Multiple Origins of Polyploidy and the Geographic Structure of Heuchera Grossulariifolia. Mol. Ecol. 8, 253–262. doi:10.1046/j.1365-294X.1999.00562.x
Sharbel, T. F., and Mitchell-Olds, T. (2001). Recurrent Polyploid Origins and Chloroplast Phylogeography in the Arabis Holboellii Complex (Brassicaceae). Heredity 87, 59–68. doi:10.1046/j.1365-2540.2001.00908.x
Siena, L. A., Ortiz, J. P. A., Calderini, O., Paolocci, F., Cáceres, M. E., Kaushal, P., et al. (2016). An Apomixis-linked ORC3-like Peudogene is Associated With Silencing of its Functional Homolog in Apomictic Paspalum simplex. J. Exp. Bot. 67, 1965–1978.
Soltis, D. E., Albert, V. A., Leebens-Mack, J., Bell, C. D., Paterson, A. H., Zheng, C., et al. (2009). Polyploidy and Angiosperm Diversification. Am. J. Bot. 96, 336–348. doi:10.3732/ajb.0800079
Soltis, D. E., Buggs, R. J. A., Doyle, J. J., and Soltis, P. S. (2010). What We Still Don't Know about Polyploidy. Taxon 59, 1387–1403. doi:10.1002/tax.595006
Soltis, D. E., Soltis, P. S., and Tate, J. A. (2004). Advances in the Study of Polyploidy since Plant Speciation. New Phytol. 161, 173–191. doi:10.1046/j.1469-8137.2003.00948.x
Soltis, P. S., and Soltis, D. E. (2000). The Role of Genetic and Genomic Attributes in the success of Polyploids. Proc. Natl. Acad. Sci. 97, 7051–7057. doi:10.1073/pnas.97.13.7051
Suda, J., and Herben, T. (2013). Ploidy Frequencies in Plants with Ploidy Heterogeneity: Fitting a General Gametic Model to Empirical Population Data. Proc. R. Soc. B. 280, 20122387. doi:10.1098/rspb.2012.2387
Suda, J., Malcová, R., Abazid, D., Banaš, M., Procházka, F., Šída, O., et al. (2004). Cytotype Distribution in Empetrum (Ericaceae) at Various Spatial Scales in the Czech Republic. Folia Geobot 39, 161–171. Available at: http://www.springerlink.com/index/7436K131TR4711J6.pdfdoi:10.1007/bf02805244
Suda, J., Weiss-Schneeweiss, H., Tribsch, A., Schneeweiss, G. M., Travnicek, P., and Schönswetter, P. (2007). Complex Distribution Patterns of Di-, Tetra-, and Hexaploid Cytotypes in the European High Mountain Plant Senecio Carniolicus (Asteraceae). Am. J. Bot. 94, 1391–1401. doi:10.3732/ajb.94.8.1391
Symonds, V. V., Soltis, P. S., and Soltis, D. E. (2010). Dynamics of Polyploid Formation Intragopogon(asteraceae): Recurrent Formation, Gene Flow, and Population Structure. Evolution (N. Y). 64, 1984–2003. doi:10.1111/j.1558-5646.2010.00978.x
Van der Hulst, R. G. M., Mes, T. H. M., Den Nijs, J. C. M., and Bachmann, K. (2000). Amplified Fragment Length Polymorphism (AFLP) Markers Reveal that Population Structure of Triploid Dandelions (Taraxacum officinale) Exhibits Both Clonality and Recombination. Mol. Ecol. 9, 1–8. doi:10.1046/j.1365-294x.2000.00704.x
Van Dijk, P., and Bakx‐Schotman, T. (1997). Chloroplast DNA Phylogeography and Cytotype Geography in Autopolyploid Plantago media. Mol. Ecol. 6, 345–352. doi:10.1046/j.1365-294X.1997.00199.x
Vekemans, X., Beauwens, T., Lemaire, M., and Roldán-Ruiz, I. (2002). Data from Amplified Fragment Length Polymorphism (AFLP) Markers Show Indication of Size Homoplasy and of a Relationship between Degree of Homoplasy and Fragment Size. Mol. Ecol. 11, 139–151. doi:10.1046/j.0962-1083.2001.01415.x
Vijverberg, K., Van Der Hulst, R. G. M., Lindhout, P., and Van Dijk, P. J. (2004). A Genetic Linkage Map of the Diplosporous Chromosomal Region in Taraxacum officinale (Common Dandelion; Asteraceae). Theor. Appl. Genet. 108, 725–732. doi:10.1007/s00122-003-1474-y
Vos, P., Hogers, R., Bleeker, M., Reijans, M., Lee, T. v. d., Hornes, M., et al. (1995). AFLP: A New Technique for DNA Fingerprinting. Nucl. Acids Res. 23, 4407–4414. doi:10.1093/nar/23.21.4407
Vrijenhoek, R. C. (1984). “Ecological Differentiation Among Clones: the Frozen Niche Variation Model,” in Proceedings in Life Sciences. Editors K. Wöhrmann, and V. Loeschcke (Berlin, Heidelberg: Springer Berlin Heidelberg), 217–231. doi:10.1007/978-3-642-69646-6_18
Vrijenhoek, R. C., and Parker, E. D. (2009). “Geographical Parthenogenesis: General Purpose Genotypes and Frozen Niche Variation,” in In Lost Sex: The Evolutionary Biology Of Parthenogenesis. Editors I. Schön, K. Martens, and P. van Dijk (New York: Springer), 99–131. doi:10.1007/978-90-481-2770-2_6
Vrijenhoek, R. C. (1994). Unisexual Fish: Model Systems for Studying Ecology and Evolution. Annu. Rev. Ecol. Syst. 25, 71–96. doi:10.1146/annurev.es.25.110194.000443
Werth, C. R., Guttman, S. I., and Eshbaugh, W. H. (1985). Electrophoretic Evidence of Reticulate Evolution in the Appalachian asplenium Complex. Syst. Bot. 10, 184–192. doi:10.2307/2418344
Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., and Rieseberg, L. H. (2009). The Frequency of Polyploid Speciation in Vascular Plants. Proc. Natl. Acad. Sci. 106, 13875–13879. doi:10.1073/pnas.0811575106
Yang, A.-H., Wei, N., Fritsch, P. W., and Yao, X.-H. (2016). AFLP Genome Scanning Reveals Divergent Selection in Natural Populations of Liriodendron Chinense (Magnoliaceae) along a Latitudinal Transect. Front. Plant Sci. 7, 698. doi:10.3389/fpls.2016.00698
Zozomová-Lihová, J., Malánová-Krásná, I., Vít, P., Urfus, T., Senko, D., Svitok, M., et al. (2015). Cytotype Distribution Patterns, Ecological Differentiation, and Genetic Structure in a Diploid-Tetraploid Contact Zone of Cardamine Amara. Am. J. Bot. 102, 1380–1395. doi:10.3732/ajb.1500052
Keywords: apomixis, residual sexuality, Paspalum intermedium, polyploid establishment, contact zone
Citation: Karunarathne P and Hojsgaard D (2021) Single Independent Autopolyploidization Events From Distinct Diploid Gene Pools and Residual Sexuality Support Range Expansion of Locally Adapted Tetraploid Genotypes in a South American Grass. Front. Genet. 12:736088. doi: 10.3389/fgene.2021.736088
Received: 04 July 2021; Accepted: 07 September 2021;
Published: 04 October 2021.
Edited by:
Genlou Sun, Saint Mary’s University, CanadaReviewed by:
Matthias Hartmann, Institute of Botany (ASCR), CzechiaNaazneen Khan, University of Kentucky, United States
Copyright © 2021 Karunarathne and Hojsgaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piyal Karunarathne, cGl5YWwua2FydW5hcmF0aG5lQGViYy51dS5zZQ==
 Piyal Karunarathne
Piyal Karunarathne Diego Hojsgaard
Diego Hojsgaard