- 1Department of Cardiac Surgery, Beijing Anzhen Hospital, Capital Medical University, Beijing, China
- 2Key Laboratory of Remodeling-Related Cardiovascular Diseases, Ministry of Education, Beijing Institute of Heart, Lung and Vascular Diseases, Beijing, China
Calcific aortic valve disease (CAVD) is the most common structural heart disease, and the morbidity is increased with elderly population. Several microRNAs (miRNAs) have been identified to play crucial roles in CAVD, and numerous miRNAs are still waiting to be explored. In this study, the miRNA expression signature in CAVD was analyzed unbiasedly by miRNA-sequencing, and we found that, compared with the normal control valves, 152 miRNAs were upregulated and 186 miRNAs were downregulated in calcified aortic valves. The functions of these differentially expressed miRNAs were associated with cell differentiation, apoptosis, adhesion and immune response processes. Among downregulated miRNAs, the expression level of miR-139-5p was negatively correlated with the osteogenic gene RUNX2, and miR-139-5p was also downregulated during the osteogenic differentiation of primary human aortic valve interstitial cells (VICs). Subsequent functional studies revealed that miR-139-5p overexpression inhibited the osteogenic differentiation of VICs by negatively modulating the expression of pro-osteogenic gene FZD4 and CTNNB1. In conclusion, these results suggest that miR-139-5p plays an important role in osteogenic differentiation of VICs via the Wnt/β-Catenin pathway, which may further provide a new therapeutic target for CAVD.
Introduction
Calcific aortic valve disease (CAVD), which results in aortic valve stenosis, affects 25% of the population >65 years of age (Coffey et al., 2014; Lindman et al., 2016). In developing countries, CAVD represents a major cause for surgical valve replacement. CAVD confers a high clinical and economic burden, because no effective pharmacological therapy exists (Hutcheson et al., 2014). The disease progression is rapid and, although initially indolent, it results in heart failure and premature death if left untreated. Owing to its high morbidity and mortality, a strong incentive exists to identify the key molecular drivers contributing to the development of this disease, which could provide a new target for clinical treatment in its earlier stages before cardiac damage.
CAVD is marked by inflammatory infiltration, fibrotic extracellular matrix (ECM) synthesis by activated valve interstitial cells (VICs), increased leaflet thickening and stiffness and calcific mineral deposition (Peeters et al., 2018). The resultant pathological remodeling ultimately impairs valve movement and obstructs blood flow across the narrowing valve orifice. In healthy valves, VICs comprise mostly quiescent fibroblasts that maintain valvular homeostasis and physiological leaflet mechanical properties. Under pathological stimuli, VICs undergo differentiation toward osteoblastic phenotypes as a result of newly acquired expression of cytoskeletal or osteogenic genes (RUNX2, ALPP, BMP2, BGLAP, etc.) (Rutkovskiy et al., 2017). However, the pathogenic mechanisms that trigger their maladaptive differentiation in vivo are unclear.
MicroRNA (miRNA) is a type of small RNA that is generally 22–25 bases in length (Lagos-Quintana et al., 2001). miRNA binds to the 3′-untranslated regions (UTR) of the target mRNA in the cytoplasm by sequence complementarity, which inhibits the translation of the target mRNA or causes the degradation of the target mRNA (Miranda et al., 2006; Sun and Lai, 2013). Compared with normal human aortic valves, the miRNA expression profiles in calcified aortic valve tissues have changed (Wang et al., 2017). Several miRNAs [miR-204-5p (Yu et al., 2018; Song et al., 2019), miR-143 (Fiedler et al., 2019), miR-125b (Ohukainen et al., 2015), miR-34a (Toshima et al., 2020), etc.] have been reported to participate in the osteogenic differentiation of VICs and valve calcification.
In this study, to identify the key miRNAs during aortic valve calcification, we explored the miRNA expression signature in CAVD by miRNA-sequencing and combined analyzed with a published transcriptome data (https://cics.bwh.harvard.edu/multiomics_databases) (Schlotter et al., 2018). Among the differentially expressed miRNAs (DEMs), miR-139-5p was downregulated miRNA in calcified aortic valves and primary human aortic VICs. Subsequent functional studies revealed that miR-139-5p overexpression results in inhibition of VICs osteogenic differentiation by negatively modulating the expression of pro-osteogenic gene FZD4 and CTNNB1. Therefore, our study provided new insights into the function of miR-139-5p in the pathogenesis of CAVD.
Materials and Methods
Clinical Samples
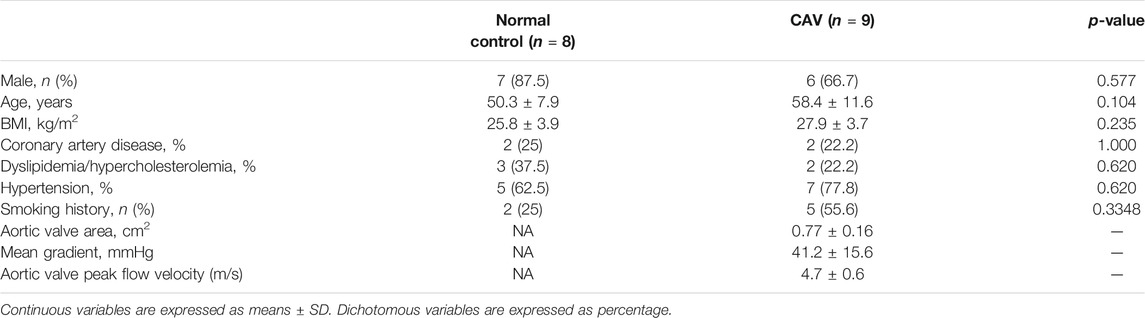
A total of nine calcified aortic valves (CAVs) (exclusion criteria: rheumatic aortic valvulopathy, infective endocarditis, congenital valve disease, bicuspid aortic valve) from patients with CAVD and 8 normal aortic valves (CONs) from patients with heart transplantation were obtained at the Department of Cardiovascular Surgery, Anzhen Hospital, affiliated to Capital Medical University. The clinical characteristics of all samples are listed in Table 1. All the studies involving human samples complied with the Declaration of Helsinki and were approved by the Ethics Committee of Anzhen Hospital, affiliated to Capital Medical University. Written informed consent was obtained from the patients before surgery.
RNA Extraction and miRNA-Sequencing
Total RNAs were extracted from aortic valves and cells using TRIzol Reagent (Life technologies) as described previously (Han et al., 2021). RNA from five calcified aortic valves and 3 normal aortic valves were used for miRNA-sequencing. After using Agilent 2100 Bio analyzer (Agilent RNA 6000 Nano Kit) to do the total RNA sample quality control, cDNA library was constructed and sequenced on DNBseq platform. DEMs were identified by comparing the gene-level fragments kilobase of exon model per million mapped reads (FKPM) between two groups and selected by using the criteria with at least a twofold change of FPKM and false discovery rate (FDR) value <0.001. The target genes of DEMs were predicted with RNAhybrid, miRanda and TargetScan database and further function analysis by gene ontology (GO) biological process (BP) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/). All the raw data can be accessed in the GEO database (GSE171208).
Real-Time PCR
For miRNAs, the isolated total RNAs were reverse transcribed into complementary cDNAs and qRT-PCR analysis using Bulge-Loop miRNA qRT-PCR kit (Ribobio, C10211-2 Guangzhou, China) and the Bulge-Loop primer of hsa-miR-374c-3p (Ribobio, miRA1000644), hsa-miR-181a-2-3p (Ribobio, miRA1000331), hsa-miR-490-5p (Ribobio, miRA1000817), hsa-miR-204-5p (Ribobio, miRA0000265), hsa-miR-1180-3p (Ribobio, miRA1000016), hsa-miR-149-5p (Ribobio, miRA1000346), hsa-miR-139-5p (Ribobio, miRA1000053), and U6 (Ribobio, miRAN0002-1-100). U6 was used as an internal control.
For mRNAs, the isolated total RNAs were reversely transcribed into complementary cDNAs using GoScript™ Reverse Transcriptase Kit (Promega, A5001) and then real-time PCR analysis was performed using SYBR Green PCR Master Mix Reagent Kit (TaKaRa), with GAPDH used as an internal control. Data were presented as values calculated by the 2ΔΔt method. The sequences of primers are as follows:
ALPP: 5′-GTGAACCGCAACTGGTACTC-3′, 5′-GAGCTGCGTAGCGATGTCC-3′; RUNX2: 5′-TGGTTACTGTCATGGCGGGTA-3′, 5′-TCTCAGATCGTTGAACCTTGCTA-3′; BMP3: 5′-TGACATCGCTAACCAAGTCTGA-3′, 5′-TGAGGGTCCATGCAGAAAGAT-3′; BGLAP: 5′-CACTCCTCGCCCTATTGGC-3′, 5′-CCCTCCTGCTTGGACACAAAG-3′; CTNNB1: 5′-AGCTTCCAGACACGCTATCAT-3′, 5′-CGGTACAACGAGCTGTTTCTAC-3’; FZD4: 5′-GTGTCACTCTGTGGGAACCAA-3′, 5′-GGCTGTATAAGCCAGCATCAT-3’; GAPDH: 5′-GGAGCGAGATCCCTCCAAAAT-3′, 5′-GGCTGTTGTCATACTTCTCATGG-3′.
VECs and VICs Isolation, Culture, and Osteogenic Differentiation
Primary valve endothelial cells (VECs) and VICs were isolated from normal aortic valve leaflets according to a previously reported protocol (Li et al., 2017a). After enzymatic digestion of the aortic valve leaflets with 1 mg/ml collagenase (Type I) at 37°C for 30 min, the endothelial cells were removed from valve leaflets by cell scraper and collected in 1 × PBS buffer. After centrifugation for 5 min, VECs were resuspended in ECM with 10% heat-inactivated FBS and 1% penicillin/streptomycin and cultured in 37°C humidified atmospheres at 5% CO2. Then, the remaining left leaflets were minced and further digested with collagenase (Type I) at 37°C for 1–2 h. After enzymatic digestion, the cell suspension was filtered with a 70 μm cell strainer and VICs were cultured in standard DMEM with 10% heat-inactivated FBS and 1% penicillin/streptomycin in a 37°C humidified atmosphere at 5% CO2.
VECs and VICs were identified by immunofluorescence staining. After fixation with 4% paraformaldehyde and blocking with 5% bovine serum albumin, primary antibody [rabbit polyclonal antibody to CD31 (Abcam, ab28364, 1:100 diluted) or rabbit polyclonal antibody to Periostin (Santa Cruz, sc-67233, 1:50 diluted)] was incubated at 4°C overnight and then a second antibody [Alexa Fluor 555 donkey anti-rabbit (Life Technologies, A31572, 1:500 diluted) or Alexa Fluor 488 donkey anti-rabbit (Life Technologies, A21206, 1:500 diluted)] was incubated for 1 h at room temperature. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Abcam).
Calcification of VICs was induced by osteogenic medium [OM, 50 μM ascorbic acid phosphate (Sigma), 100 nM dexamethasone (Sigma) and 10 μM β-glycerophosphate (Sigma)]. VICs were cultured in OM for 14 days with a replacement every 2–3 days. Alizarin red staining was used to access the matrix calcium deposition after fixation with 10% formalin.
Transfection of VICs With miRNA Mimics
VICs were transfected at a confluency of 70–80% in a 12-well plate after seeding. micrONTM hsa-miR-139-5p mimic (50 nM) or negative controls (micrONTM miRNA mimic NC #22, Ribobio) were transfected using Lipofectamine 3000 according to the manufacturer’s protocol (Invitrogen). The transfection efficiency was examined by real-time PCR after 48 h of transfection. During the osteogenic differentiation induction of VICs, miRNA mimics were added at the same time of OM replacement.
Statistical Analysis
Data processing was performed using SPSS21.0 software. The measurement data were expressed as mean ± standard deviation and the unpaired t-test was used for comparison between two groups and one-way ANOVA was used for comparison in three groups. Spearman’s correlation test was used for correlation analysis. p < 0.05 was considered to have a statistically significant different between groups.
Results
miRNA Expression Profile was Altered During Aortic Valve Calcification
To explore the DEMs, total RNAs from three control aortic valves and five calcified aortic valves were analyzed by genome-wide miRNA expression profiling using DNBseq sequencing platform (Figure 1A). The average alignment ratio of the sample comparison genome (Homo_sapiens_9606.NCBI.GCF_000001405.38_GRCh38.p12. v1904) was 93.48%. A total of 1,572 miRNAs were detected. The boxplots in Figure 1B showed comparable read count distribution across all eight samples. Based on the analysis of Pearson correlation coefficient of overall miRNA expression levels between every two samples, the eight samples could be divided into two groups, which is consistent with the original groups (Figure 1C). Then, significant DEMs were identified as those with a change fold >2.0 and FDR <0.001. Compared with the control group, 152 miRNAs were upregulated and 186 miRNAs were downregulated in calcified aortic valves (Figure 1D, Supplementary Excel S1). The expression patterns of the DEMs were shown in the hierarchical clustering heatmap (Figure 1E).
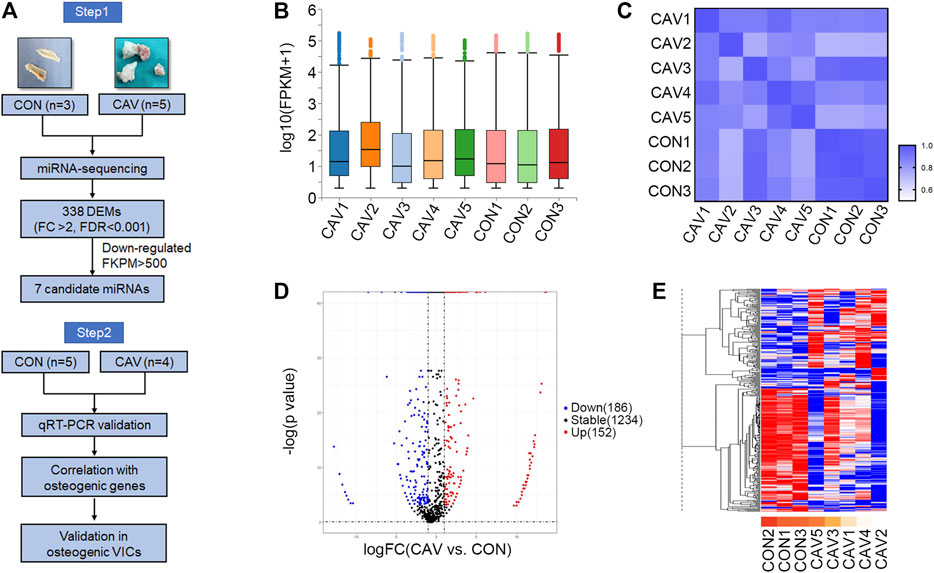
FIGURE 1. miRNA expression profile was altered during aortic valve calcification. (A) Schematic description of the workflow illustrating the two-stage-approach involving independent samples for discovery and validation. (B) Overall count distribution of miRNAs in each sample in the RNA-sequencing experiment. (C) Pearson correlation coefficient of overall miRNA expression levels between every two samples. (D) Volcano plot revealing miRNA-sequencing results comparing CON (n = 3 biologically independent samples) versus CAV (n = 5 biologically independent samples). Individual miRNAs are displayed by the p-value and the corresponding fold change. (E) Heatmap of the 338 differentially expressed miRNAs. Red color indicated higher expression level; blue color indicated lower expression level. CON, control; CAV, calcified aortic valves.
Pathway Analysis of the Predict Target Genes of Differentially Expressed miRNAs
Identification the function enrichment of predicted targets may help to understand the biological role of these DEMs. We used RNAhybrid, miRanda and TargetScan database to predict the target mRNAs for these DEMs. Through GO BP enrichment analysis, we found that the target genes of DEMs were associated with cell differentiation, apoptosis process, cell adhesion, oxidation–reduction process and immune response (Figure 2A). Through the KEGG pathway enrichment analysis, we found that the target genes of DEMs were significantly enriched in a few signaling pathways, especially the phosphatidylinositol 3ʹ-kinase (PI3K)–protein kinase B (AKT) signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, and cytokine–cytokine receptor interaction pathway (Figure 2B).
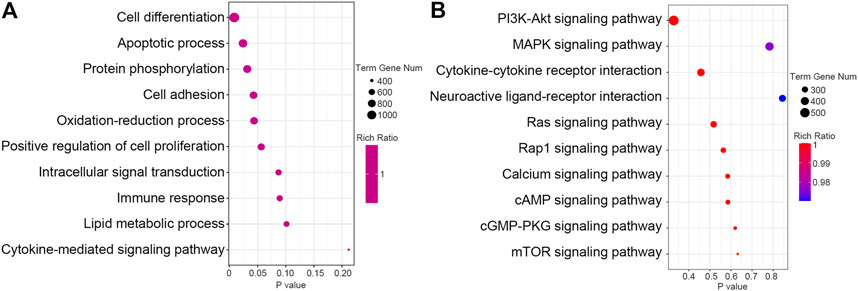
FIGURE 2. Pathway analysis of the predict target genes of differential expressed miRNAs. (A) The GO biological process (BP) function of the predict target genes of differential expressed miRNAs was analyzed and the top 10 terms were listed. (B) KEGG pathway of the predict target genes of differential expressed miRNAs was analyzed and the top 10 pathways were listed.
Dysregulated Signaling Pathway in Calcified Aortic Valves
Then, we further analyzed the altered signaling pathway in a previous published data (https://cics.bwh.harvard.edu/multiomics_databases) (Schlotter et al., 2018), which compared the transcriptome of three non-calcified aortic valves and three calcified aortic valves. Differentially expressed genes (DEGs) were identified as those with a change fold >2.0. Compared with the non-calcified group, 1,235 genes were upregulated and 544 genes were downregulated in calcified aortic valves and the hierarchical clustering heatmap is shown in Supplementary Figure S1A. Through GO BP enrichment analysis, we found that the upregulated genes were associated with immune response, chemokine-mediated signaling pathway, T-cell activation, positive regulation of extracellular signal-regulated kinase (ERK)1/2 cascade and extracellular matrix disassembly (Supplementary Figure S1B). The downregulated genes were associated with negative regulation of canonical Wnt signaling pathway, muscle contraction, negative regulation of cell proliferation and ossification (Supplementary Figure S1C). Through the KEGG pathway enrichment analysis, the upregulated DEGs were significantly enriched in cytokine–cytokine receptor interaction, osteoclast differentiation, cell adhesion molecules and PI3K–AKT signaling pathway (Supplementary Figure S1D). The downregulated DEGs were significantly enriched in protein digestion and absorption, Wnt signaling pathway, TGF-beta signaling pathway and Hippo signaling pathway (Supplementary Figure S1E). These results have shown that the function enrichment of DEMs in our miRNA-seq had some similarities with the function enrichment of DEGs in RNA-seq.
The Verification of Downregulated Candidate miRNAs in Calcified Aortic Valves
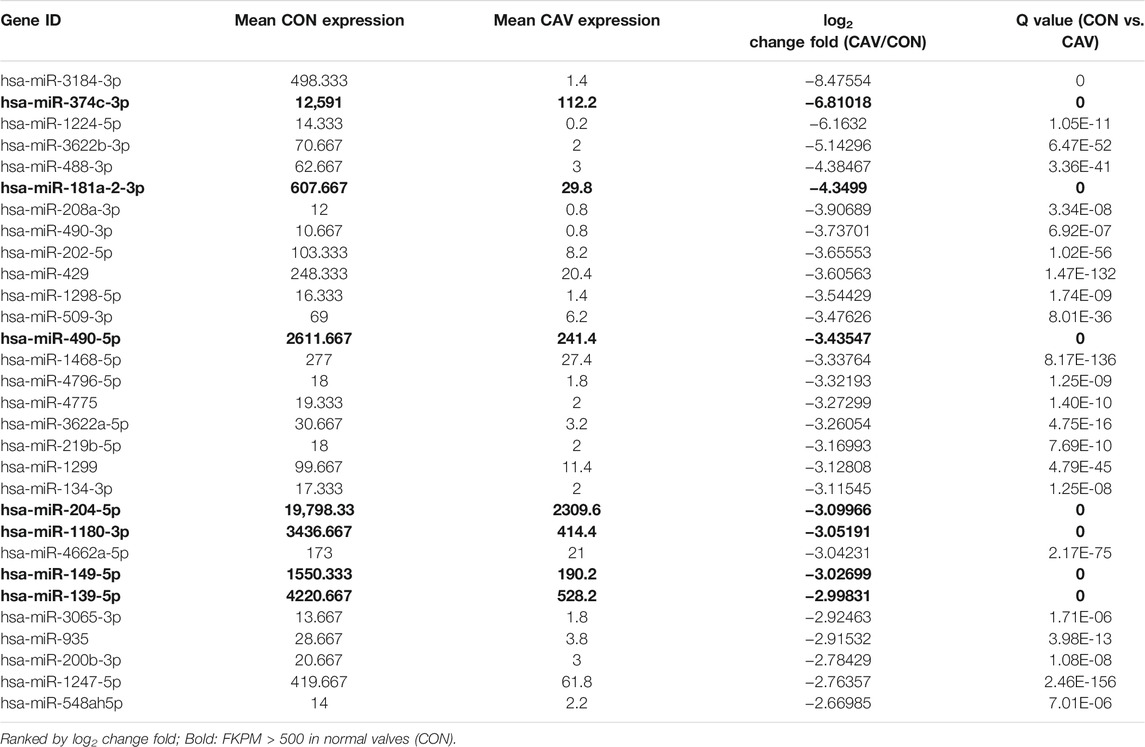
We selected candidate miRNAs from the downregulated miRNAs for validation by real-time PCR. Among the top 30 miRNAs (ranked by change fold), there were 7 miRNAs (hsa-miR-374c-3p, hsa-miR-181a-2-3p, hsa-miR-490-5p, hsa-miR-204-5p, hsa-miR-1180-3p, hsa-miR-149-5p and hsa-miR-139-5p) highly expressed in normal valves (FKPM > 500) (Table 2). Then, we validated the expression of these miRNAs in another five normal valves and four calcific valves. The expression levels of osteogenic genes ALPP, RUNX2, BMP3, BGLAP, FZD4 and CTNNB1 were higher in calcific valves than normal valves (Figure 3A and Supplementary Figure S2). Compared to the normal valves, six miRNAs (hsa-miR-374c-3p, hsa-miR-181a-2-3p, hsa-miR-490-5p, hsa-miR-204-5p, hsa-miR-149-5p, and hsa-miR-139-5p) were significantly downregulated in calcified aortic valves, while the expression levels of hsa-miR-1180-3p were not different between two groups (Figures 3B,C). Because the expression level of hsa-miR-374c-3p in valves was lower than the other five miRNAs, we removed it from the candidate miRNAs. Among the five candidate miRNAs, it was previously reported that the expression of miR-204-5p was decreased in calcified aortic valves and inhibited the osteogenic differentiation of VICs and aortic valve calcification (Yu et al., 2018; Song et al., 2019). Then, we calculated the correlation of five candidate miRNAs with the osteogenic genes RUNX2. miR-204-5p, miR-149-5p, and miR-139-5p exhibited significant negative correlations with osteogenic genes RUNX2 (Figure 3D). These data indicated that, besides miR-204-5p, miR-149-5p and miR-139-5p may also be the key miRNAs in the development of CAVD.
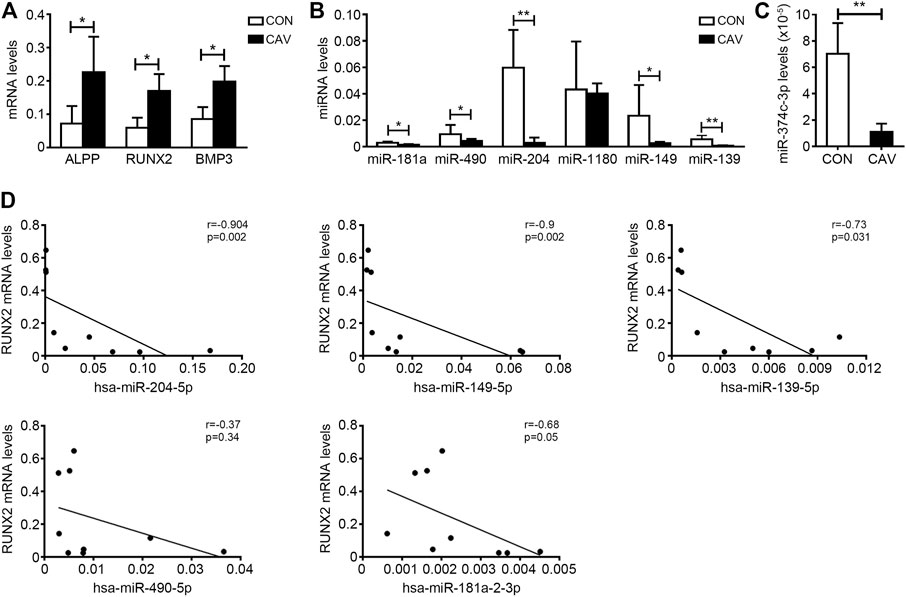
FIGURE 3. The verification of downregulated candidate miRNAs in calcified aortic valves. (A) The expression levels of osteogenic genes ALPP, RUNX2 and BMP3 in valves from CON patients (n = 5 biologically independent samples) and CAV patients (n = 4 biologically independent samples) were accessed by qRT-PCR and normalized with GAPDH. (B,C) The expression levels of hsa-miR-181a-2-3p (miR-181a), hsa-miR-490-5p (miR-490), hsa-miR-204-5p (miR-204), hsa-miR-1180-3p (miR-1180), hsa-miR-149-5p (miR-149), hsa-miR-139-5p (miR-139) (B) and hsa-miR-374c-3p (C) in valves from CON patients (n = 5 biologically independent samples) and CAV patients (n = 4 biologically independent samples) were accessed by qRT-PCR and normalized with U6. (D) Spearman’s correlation test was used to analyze the correlation of the expression of hsa-miR-204-5p, hsa-miR-149-5p, hsa-miR-139-5p, hsa-miR-181a-2-3p and hsa-miR-490-5p with the expression levels of RUNX2 in aortic valves. Unpaired Student’s t-test, *p < 0.05, **p < 0.01. CON, control; CAV, calcified aortic valves.
Both miR-149-5p and miR-139-5p Were Downregulated in Osteogenically Differentiated VICs
To confirm the cell location of miR-149-5p and miR-139-5p, we cultured the primary VICs and VECs and compared the expression levels of miR-149-5p and miR-139-5p in VICs and VECs (Figure 4A and Supplementary Figure S3). Both the expression levels of miR-149-5p and miR-139-5p were higher in VICs than that in VECs (Figure 4B). Then, we induced the osteogenic differentiation of VICs by condition medium (Figure 4C) and compared the expression levels of miR-149-5p and miR-139-5p in primary VICs and osteogenically differentiated VICs. We found that miR-149-5p and miR-139-5p decreased 4.66-fold and 2.79-fold, respectively, in osteogenically differentiated VICs (Figure 4D).

FIGURE 4. Both miR-149-5p and miR-139-5p were downregulated in osteogenic differentiated VICs significantly. (A) Primary cultured VICs and VECs were immunofluorescence stained with Periostin (green) and CD31 (red), respectively and the nucleus was stained with DAPI (blue) (the scale bar = 50 μm). (B) The expression levels of hsa-miR-149-5p (miR-149) and hsa-miR-139-5p (miR-139) in VECs and VICs were accessed by qRT-PCR and normalized with U6 (n = 4 samples from independent wells in each group). (C) Alizarin red staining was used to access the matrix calcium deposition in VICs at 15 days after OM induction. (D) The expression levels of hsa-miR-149-5p and hsa-miR-139-5p in control and osteogenic differentiated VICs were accessed by qRT-PCR and normalized with U6 (n = 4 samples from independent wells in each group). Unpaired Student’s t-test, *p < 0.05, **p < 0.01.
miR-139-5p Overexpression Could Inhibit the Osteogenic Differentiation of VICs
It was reported that miR-139-5p could repress the osteogenesis of mesenchymal stem cells via targeting Wnt/β-Catenin signaling pathway (Long et al., 2017). For the importance of Wnt/β-Catenin signaling pathway to osteogenesis of VICs and aortic valve calcification (Khan et al., 2020; Albanese et al., 2017), we focused on the effects of miR-139-5p on osteogenesis the VICs in further study. miR-139-5p mimics were transfected to VICs during osteogenic differentiation, the NC mimics as negative control. The expression of miR-139-5p in VICs transfected with miR-139-5p mimics was higher than that in NC mimics transfected VICs at 15 days after osteogenic induction, indicating that the miR-139-5p mimics were successfully transfected into VICs (Figure 5A). Alizarin Red staining showed that calcium deposition in VICs with the miR-139-5p mimics was less than that in VICs with NC mimics (Figure 5B). At the same time, the expression levels of osteogenic genes BGLAP, ALPP and RUNX2 in VICs were also decreased by miR-139-5p overexpression (Figures 5C–E). FZD4 and CTNNB1, the key genes of Wnt/β-Catenin pathway, were the reported target genes of miR-139-5p (Long et al., 2017). We found that the expression levels of FZD4 and CTNNB1 were increased in osteogenically differentiated VICs, while they were repressed by miR-139-5p overexpression (Figures 5F,G). Therefore, these data demonstrated that miR-139-5p could inhibit the osteogenic differentiation of VICs by targeting the Wnt/β-Catenin pathway.
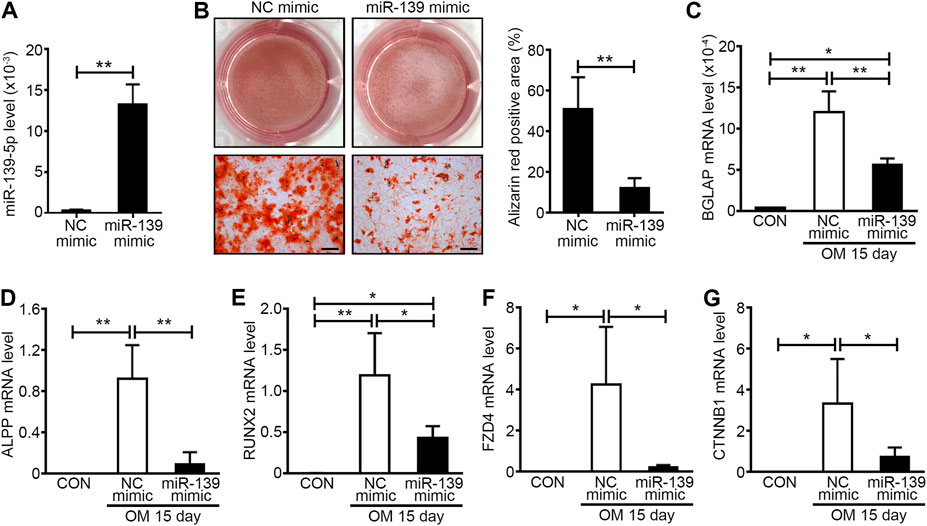
FIGURE 5. miR-139-5p overexpression could inhibit the osteogenic differentiation of VICs. (A) The expression levels of miR-139-5p in negative control (NC) mimics and miR-139-5p mimics (miR-139 mimic) transfected VICs were accessed by qRT-PCR and normalized with U6 (n = 4 samples from independent wells in each group). Unpaired Student’s t-test, **p < 0.01. (B) Alizarin red staining was used to access the matrix calcium deposition in NC mimics and miR-139-5p mimics transfected VICs at 15 days after OM induction and the right graph was the percentage of Alizarin red positive area per field (n = 3 wells in each group and three to four fields per well were counted, the scale bar = 200 μm). Unpaired Student’s t-test, **p < 0.01. (C–E) The expression levels of BGLAP (C), ALPP (D) and RUNX2 (E) in NC mimics and miR-139-5p mimics transfected VICs were accessed by qRT-PCR and normalized with GAPDH (n = 4 samples from independent wells in each group). (F,G) The expression levels of FZD4 (F) and CTNNB1 (G) in NC mimics and miR-139-5p mimics transfected VICs were accessed by qRT-PCR and normalized with GAPDH (n = 4 samples from independent wells in each group). One-way ANOVA, *p < 0.05, **p < 0.01.
Discussion
miRNAs participate in the pathology of various cardiovascular diseases. By miRNA-seq, we found that the miRNA profile was altered in calcified aortic valves. Among the downregulated miRNAs, miR-139-5p could inhibit the osteogenic differentiation of VICs by target Wnt/β-Catenin pathway associated genes FZD4 and CTNNB1.
Several studies have investigated the miRNA profile alternation in CAVD by quantitative RT-PCR based screening or microarray in the past decades. However, the detection throughput and sensitivity of those methods are relatively low and only hundreds of miRNAs can be detected. In one study, only 92 miRNAs were identified as DEMs with a change fold >2.0. Among the 92 DEMs, 53 miRNAs were downregulated and 39 were upregulated in aortic tissue from CAVD patients (Wang et al., 2017). In another study, 373 human miRNAs were examined by a quantitative RT-PCR based screening and found only 41 DEMs (Fiedler et al., 2019). In our study, miRNA-sequencing was used to the detect the DEMs in calcific aortic valves. Compared to microarray and RT-PCR-based screening, this method has the characteristics of high throughput and high sensitivity, which allows us to detect more miRNAs with higher accuracy. As a result, a total of 1,572 miRNAs were detected and there were 338 DEMs with a change fold >2.0 and FDR <0.001. Among the DEMs, 152 miRNAs were upregulated and 186 miRNAs were downregulated in calcified aortic valves, which indicated that more DEMs were found in this study. The significantly downregulated miRNAs found in the previous study, such as miR-125b (Ohukainen et al., 2015), miRNA-126 and Let-7 family (let-7a, let-7c, let-7d, let-7e and let-7f) (Wang et al., 2017), were also downregulated in our study. Other reported upregulated miRNAs, such as miR-34a (Toshima et al., 2020), miR-133a, miR-143 and miR-21 (Fiedler et al., 2019), were also upregulated in our study. Therefore, the method used in this study could contribute to fully understand miRNA profile alteration and the discovery of new functional targets in aortic valve calcification.
The pathology of calcific valve disease is an active and multifaceted condition involving lipoprotein deposition, inflammatory cell (including macrophages, T lymphocytes and mast cells) infiltration and cytokine secretion, osteogenic differentiation and apoptosis of VICs and ECM remodeling (Rutkovskiy et al., 2017; Peeters et al., 2018). In this study, by the functional and pathway enrichment analysis of the target genes of DEMs and DEGs, we found that both the DEMs and DEGs were associated with cell differentiation, apoptosis, adhesion and immune response and the associated pathways included the Wnt signaling pathway, ERK1/2 pathway, PI3K–AKT pathway, MAPK pathway and cytokine–cytokine receptor interaction pathway. Activation of the Wnt, PI3K–AKT and MAPK pathway promoted the osteogenic differentiation of VICs (Fang et al., 2014; Poggio et al., 2014; Xie et al., 2020). These results demonstrated that the DEMs in the miRNA-seq were indeed involved in the process of aortic valve calcification.
Since miRNA always acted as a negative regulator of transcription and there were more downregulated miRNAs than upregulated miRNAs and more upregulated mRNAs than downregulated mRNAs in calcified valves, we choose seven miRNAs from the downregulated miRNAs as candidates, which were abundant in normal valves. After validation in calcific valves and VICs and calculating the correlation with the osteogenic genes, we finally selected hsa-miR-139-5p from seven candidate miRNAs to verify its effect on VICs. The calcium deposition and the expression levels of BGLAP, ALPP and RUNX2 were repressed by miR-139-5p overexpression, but these genes were not in the predicted target genes list. So, the effect of miR-139-5p on the osteogenic differentiation of VICs was mediated indirectly. The role of miR-139-5p in cell differentiation has been frequently reported. Mouse 3T3-L1 preadipocyte differentiation can be suppressed by miR-139-5p (Mi et al., 2015a). As a negative regulator in myogenesis, miR-139-5p has a great influence in myoblast differentiation by blocking the Wnt1-mediated Wnt/β-catenin signaling pathway (Mi et al., 2015b). In another study, miR-139-5p was able to repress the osteogenesis of mesenchymal stem cells via targeting Wnt/β-Catenin signaling pathway during the repair of bone (Long et al., 2017). FZD4 (encodes Frizzled receptor isoforms 4) and CTNNB1 (encodes β-Catenin), the reported target genes of miR-139-5p, are key molecular drivers in the Wnt/β-Catenin signaling pathway. Frizzled receptor 4, which is the receptor of Wnt family ligands, mediates the intracellular signaling activation with the lipoprotein-related peptide 5/6 (LRP5/6) co-receptors (Ahn et al., 2011). The typical Wnt/β-catenin signaling pathway activates the transcriptional activity of osteogenic genes (RUNX2, ALPP, etc.) by nuclear translocation of β-catenin (Calvisi et al., 2005; Park et al., 2011). LRP5 deficiency could prevent the calcification in the aortic valve under hypercholesterolemia condition (Rajamannan, 2011). In this study, we found that FZD4 and CTNNB1 were increased in osteogenic VICs and miR-139-5p could inhibit the expression the FZD4 and CTNNB1. These data demonstrated that miR-139-5p could inhibit the osteogenic differentiation of VICs by targeting the Wnt/β-Catenin pathway.
In this study, we also found that miR-149-5p was downregulated in calcified valves and osteogenically differentiated VICs. In the previous study, interleukin-6 (IL-6) was reported as the target gene of miR-149-5p (Kong et al., 2018). The expression level of IL-6 was increased in calcified human aortic valves (Li et al., 2017b). Recombinant IL-6 can increase the expression of RUNX2 and osteopontin to promote the osteogenic differentiation of VICs; when IL-6 was inhibited by siRNA, the osteogenic differentiation of VICs was blocked in vitro (El Husseini et al., 2014; Grim et al., 2020). However, there is still no evidence for the function of miR-149-5p in osteogenic differentiation of VICs and CAVD and it could be explored in a future study.
In summary, our study fully described the changes in miRNA expression profile and its related pathological processes during aortic valve calcification by miRNA-seq. At the same time, we discovered that miR-139-5p plays an inhibiting role in the process of osteogenic differentiation, which may provide a new therapeutic target for aortic valve calcification.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/, GSE171208.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Anzhen Hospital, affiliated to Capital Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HZ and CZ initiated and managed the project. FZ and CZ designed and conducted major experiments. FZ collected the valves samples, extracted the total RNA from valves, and validated the RNA-sequencing results. NC and YH performed cell culture and transfection experiments. FZ written the first draft, and HZ and CZ interpreted the results and edited the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81871758, 81861128025) and Beijing Hospitals Authority Youth Programme (QML20190604).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.722564/full#supplementary-material
References
Ahn, V. E., Chu, M. L.-H., Choi, H.-J., Tran, D., Abo, A., and Weis, W. I. (2011). Structural Basis of Wnt Signaling Inhibition by Dickkopf Binding to LRP5/6. Develop. Cel. 21 (5), 862–873. doi:10.1016/j.devcel.2011.09.003
Albanese, I., Yu, B., Al-Kindi, H., Barratt, B., Ott, L., Al-Refai, M., et al. (2017). Role of Noncanonical Wnt Signaling Pathway in Human Aortic Valve Calcification. Atvb 37 (3), 543–552. doi:10.1161/atvbaha.116.308394
Calvisi, D. F., Conner, E. A., Ladu, S., Lemmer, E. R., Factor, V. M., and Thorgeirsson, S. S. (2005). Activation of the Canonical Wnt/β-Catenin Pathway Confers Growth Advantages in C-Myc/E2F1 Transgenic Mouse Model of Liver Cancer. J. Hepatol. 42 (6), 842–849. doi:10.1016/j.jhep.2005.01.029
Coffey, S., Cox, B., and Williams, M. J. (2014). The Prevalence, Incidence, Progression, and Risks of Aortic Valve Sclerosis: a Systematic Review and Meta-Analysis. J. Am. Coll. Cardiol. 63 (25 Pt A), 2852–2861. doi:10.1016/j.jacc.2014.04.018
El Husseini, D., Boulanger, M.-C., Mahmut, A., Bouchareb, R., Laflamme, M.-H., Fournier, D., et al. (2014). P2Y2 Receptor Represses IL-6 Expression by Valve Interstitial Cells through Akt: Implication for Calcific Aortic Valve Disease. J. Mol. Cell. Cardiol. 72, 146–156. doi:10.1016/j.yjmcc.2014.02.014
Fang, M., Alfieri, C. M., Hulin, A., Conway, S. J., and Yutzey, K. E. (2014). Loss of β-Catenin Promotes Chondrogenic Differentiation of Aortic Valve Interstitial Cells. Atvb 34 (12), 2601–2608. doi:10.1161/atvbaha.114.304579
Fiedler, J., Park, D.-H., Hobuss, L., Anaraki, P. K., Pfanne, A., Just, A., et al. (2019). Identification of miR-143 as a Major Contributor for Human Stenotic Aortic Valve Disease. J. Cardiovasc. Trans. Res. 12 (5), 447–458. doi:10.1007/s12265-019-09880-7
Grim, J. C., Aguado, B. A., Vogt, B. J., Batan, D., Andrichik, C. L., Schroeder, M. E., et al. (2020). Secreted Factors from Proinflammatory Macrophages Promote an Osteoblast-like Phenotype in Valvular Interstitial Cells. Arterioscler Thromb. Vasc. Biol. 40 (11), e296–e308. doi:10.1161/ATVBAHA.120.315261
Han, Y., Zhang, J., Huang, S., Cheng, N., Zhang, C., Li, Y., et al. (2021). MicroRNA-223-3p Inhibits Vascular Calcification and the Osteogenic Switch of Vascular Smooth Muscle Cells. J. Biol. Chem. 296, 100483. doi:10.1016/j.jbc.2021.100483
Hutcheson, J. D., Aikawa, E., and Merryman, W. D. (2014). Potential Drug Targets for Calcific Aortic Valve Disease. Nat. Rev. Cardiol. 11 (4), 218–231. doi:10.1038/nrcardio.2014.1
Khan, K., Yu, B., Kiwan, C., Shalal, Y., Filimon, S., Cipro, M., et al. (2020). The Role of Wnt/β-Catenin Pathway Mediators in Aortic Valve Stenosis. Front. Cel Dev. Biol. 8, 862. doi:10.3389/fcell.2020.00862
Kong, Y.-G., Cui, M., Chen, S.-M., Xu, Y., Xu, Y., and Tao, Z.-Z. (2018). LncRNA-LINC00460 Facilitates Nasopharyngeal Carcinoma Tumorigenesis through Sponging miR-149-5p to Up-Regulate IL6. Gene 639, 77–84. doi:10.1016/j.gene.2017.10.006
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of Novel Genes Coding for Small Expressed RNAs. Science 294 (5543), 853–858. doi:10.1126/science.1064921
Li, F., Yao, Q., Ao, L., Cleveland, J. C., Dong, N., Fullerton, D. A., et al. (2017). Klotho Suppresses High Phosphate-Induced Osteogenic Responses in Human Aortic Valve Interstitial Cells through Inhibition of Sox9. J. Mol. Med. 95 (7), 739–751. doi:10.1007/s00109-017-1527-3
Li, G., Qiao, W., Zhang, W., Li, F., Shi, J., and Dong, N. (2017). The Shift of Macrophages toward M1 Phenotype Promotes Aortic Valvular Calcification. J. Thorac. Cardiovasc. Surg. 153 (6), 1318–1327. doi:10.1016/j.jtcvs.2017.01.052
Lindman, B. R., Clavel, M.-A., Mathieu, P., Iung, B., Lancellotti, P., Otto, C. M., et al. (2016). Calcific Aortic Stenosis. Nat. Rev. Dis. Primers 2, 16006. doi:10.1038/nrdp.2016.6
Long, H., Sun, B., Cheng, L., Zhao, S., Zhu, Y., Zhao, R., et al. (2017). miR-139-5p Represses BMSC Osteogenesis via Targeting Wnt/β-Catenin Signaling Pathway. DNA Cel. Biol. 36 (8), 715–724. doi:10.1089/dna.2017.3657
Mi, L., Chen, Y., Zheng, X., Li, Y., Zhang, Q., Mo, D., et al. (2015). MicroRNA-139-5p Suppresses 3T3-L1 Preadipocyte Differentiation through Notch and IRS1/PI3K/Akt Insulin Signaling Pathways. J. Cel. Biochem. 116 (7), 1195–1204. doi:10.1002/jcb.25065
Mi, L., Li, Y., Zhang, Q., Zhao, C., Peng, Y., Yang, G., et al. (2015). MicroRNA-139-5p Regulates C2C12 Cell Myogenesis through Blocking Wnt/β-Catenin Signaling Pathway. Biochem. Cel Biol. 93 (1), 8–15. doi:10.1139/bcb-2014-0079
Miranda, K. C., Huynh, T., Tay, Y., Ang, Y.-S., Tam, W.-L., Thomson, A. M., et al. (2006). A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes. Cell 126 (6), 1203–1217. doi:10.1016/j.cell.2006.07.031
Ohukainen, P., Syväranta, S., Näpänkangas, J., Rajamäki, K., Taskinen, P., Peltonen, T., et al. (2015). MicroRNA-125b and Chemokine CCL4 Expression Are Associated with Calcific Aortic Valve Disease. Ann. Med. 47 (5), 423–429. doi:10.3109/07853890.2015.1059955
Park, K.-H., Kang, J. W., Lee, E.-M., Kim, J. S., Rhee, Y. H., Kim, M., et al. (2011). Melatonin Promotes Osteoblastic Differentiation through the BMP/ERK/Wnt Signaling Pathways. J. pineal Res. 51 (2), 187–194. doi:10.1111/j.1600-079x.2011.00875.x
Peeters, F. E. C. M., Meex, S. J. R., Dweck, M. R., Aikawa, E., Crijns, H. J. G. M., Schurgers, L. J., et al. (2018). Calcific Aortic Valve Stenosis: Hard Disease in the Heart. Eur. Heart J. 39 (28), 2618–2624. doi:10.1093/eurheartj/ehx653
Poggio, P., Branchetti, E., Grau, J. B., Lai, E. K., Gorman, R. C., Gorman, J. H., et al. (2014). Osteopontin-CD44v6 Interaction Mediates Calcium Deposition via Phospho-Akt in Valve Interstitial Cells from Patients with Noncalcified Aortic Valve Sclerosis. Atvb 34 (9), 2086–2094. doi:10.1161/atvbaha.113.303017
Rajamannan, N. M. (2011). The Role of Lrp5/6 in Cardiac Valve Disease: Experimental Hypercholesterolemia in the ApoE−/−/Lrp5−/− Mice. J. Cel. Biochem. 112 (10), 2987–2991. doi:10.1002/jcb.23221
Rutkovskiy, A., Malashicheva, A., Sullivan, G., Bogdanova, M., Kostareva, A., Stensløkken, K. O., et al. (2017). Valve Interstitial Cells: The Key to Understanding the Pathophysiology of Heart Valve Calcification. J. Am. Heart Assoc. 6 (9). doi:10.1161/JAHA.117.006339
Schlotter, F., Halu, A., Goto, S., Blaser, M. C., Body, S. C., Lee, L. H., et al. (2018). Spatiotemporal Multi-Omics Mapping Generates a Molecular Atlas of the Aortic Valve and Reveals Networks Driving Disease. Circulation 138 (4), 377–393. doi:10.1161/circulationaha.117.032291
Song, R., Zhai, Y., Ao, L., Fullerton, D. A., and Meng, X. (2019). MicroRNA-204 Deficiency in Human Aortic Valves Elevates Valvular Osteogenic Activity. Int. J. Mol. Sci. 21 (1). doi:10.3390/ijms21010076
Sun, K., and Lai, E. C. (2013). Adult-specific Functions of Animal microRNAs. Nat. Rev. Genet. 14 (8), 535–548. doi:10.1038/nrg3471
Toshima, T., Watanabe, T., Narumi, T., Otaki, Y., Shishido, T., Aono, T., et al. (2020). Therapeutic Inhibition of microRNA-34a Ameliorates Aortic Valve Calcification via Modulation of Notch1-Runx2 Signalling. Cardiovasc. Res. 116 (5), 983–994. doi:10.1093/cvr/cvz210
Wang, H., Shi, J., Li, B., Zhou, Q., Kong, X., and Bei, Y. (2017). MicroRNA Expression Signature in Human Calcific Aortic Valve Disease. Biomed. Res. Int. 2017, 4820275. doi:10.1155/2017/4820275
Xie, F., Li, F., Li, R., Liu, Z., Shi, J., Zhang, C., et al. (2020). Inhibition of PP2A Enhances the Osteogenic Differentiation of Human Aortic Valvular Interstitial Cells via ERK and P38 MAPK Pathways. Life Sci. 257, 118086. doi:10.1016/j.lfs.2020.118086
Keywords: calcific aortic valve disease, miR-139-5p, aortic valve interstitial cells, osteogenesis, Wnt/b-catenin
Citation: Zhang F, Cheng N, Han Y, Zhang C and Zhang H (2021) miRNA Expression Profiling Uncovers a Role of miR-139-5p in Regulating the Calcification of Human Aortic Valve Interstitial Cells. Front. Genet. 12:722564. doi: 10.3389/fgene.2021.722564
Received: 11 June 2021; Accepted: 27 September 2021;
Published: 22 October 2021.
Edited by:
King-Hwa Ling, Universiti Putra Malaysia, MalaysiaReviewed by:
Kenji Miki, Massachusetts General Hospital, United StatesJun Jie Tan, Universiti Sains Malaysia (USM), Malaysia
Copyright © 2021 Zhang, Cheng, Han, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Congcong Zhang, zcc_anzhen@163.com; Haibo Zhang, zhanghb2318@163.com
 Fan Zhang1
Fan Zhang1 Congcong Zhang
Congcong Zhang