- Department of Biotechnology, University of Kashmir, Srinagar, India
Multiple sclerosis (MS) is an early onset chronic neurological condition in adults characterized by inflammation, demyelination, gliosis, and axonal loss in the central nervous system. The pathological cause of MS is complex and includes both genetic and environmental factors. Non-protein-coding RNAs (ncRNAs), specifically miRNAs and lncRNAs, are important regulators of various biological processes. Over the past decade, many studies have investigated both miRNAs and lncRNAs in patients with MS. Since then, insightful knowledge has been gained in this field. Here, we review the role of miRNAs and lncRNAs in MS pathogenesis and discuss their implications for diagnosis and treatment.
Introduction
Multiple sclerosis (MS) is a chronic inflammatory demyelinating neurodegenerative disease of the central nervous system (CNS) (Thompson et al., 2018). It mainly affects young adults, with onset between the ages of 20 and 40 years, and is predominant in women (Thompson et al., 2018). The pathological hallmark of MS is the accumulation of focal plaques, which are areas of demyelination along with infiltration of immune cells found throughout the CNS (Mahad et al., 2015).
The clinical manifestations and course of MS vary and are broadly divided into three types: relapsing-remitting MS (RRMS), primary progressive MS (PPMS), and secondary progressive MS (SPMS). Almost 85% of patients typically present with RRMS, which is characterized by episodes of disability, followed by a period of recovery (Confavreux and Vukusic, 2014; Mahad et al., 2015; Thompson et al., 2018). Approximately 10–15% of patients exhibit PPMS, which is characterized by a slow progression of disease from the beginning without remission (Confavreux and Vukusic, 2014; Thompson et al., 2018). Over 10 years, roughly half of RRMS patients progress to the SPMS stage characterized by chronic inflammation, sclerosis, and brain atrophy with few or no periods of remission (Confavreux and Vukusic, 2014; Thompson et al., 2018).
The pathophysiological mechanism of MS is heterogeneous and is thought to involve complex gene-environment interactions (Mahad et al., 2015). However, the major cause of MS development is a pro-inflammatory response. Immune cells such as CD4+ and CD8+ T cells, B cells, macrophages, and other cells infiltrate the CNS through a disrupted blood-brain barrier (BBB) (Mahad et al., 2015). These cells, together with resident activated microglia and astrocytes, damage oligodendrocytes and myelin through contact-dependent mechanisms and the secretion of cytokines and chemokines (Mahad et al., 2015).
In the initial stages of MS development, CD4+ T helper type 1 (Th1) and CD4+ T helper type 17 (Th17) are autoreactive to myelin and have therefore been intensively investigated (Sospedra and Martin, 2005). Both Th1 and Th17 cells are enhanced in the CNS, cerebrospinal fluid (CSF), and the blood of MS patients, as well as in the experimental autoimmune encephalomyelitis (EAE) model of MS. Th1 cells and Th17 cells produce cytokine interferon-γ (IFN-γ) and cytokine interleukin-17 (IL-17), respectively, which initiate inflammation and neuronal cell death (Sospedra and Martin, 2005). Inhibition of Th1 or Th17 cells or expansion of anti-inflammatory Th2 cells ameliorates disease in EAE animal models (Aharoni et al., 2000; O’Connor et al., 2008; Jäger et al., 2009). In addition, CD4+ regulatory T cells (Tregs), which normally prevent damage to host cells by limiting the immune response, are decreased in the frequency and suppressive function of MS (Sospedra and Martin, 2005). In EAE animal models, Treg cells and cytokine interleukin-10 (IL-10) negatively regulate disease development. In summary, the homeostasis of pro-inflammatory cells and anti-inflammatory and cytokine activities, including TNF-α, IFN-γ, IL-17, IL-6, and IL-18, appears to be significantly dysregulated in MS.
Non-coding RNAs (ncRNAs) provide an intricate network that controls gene expression and immune system responses (Esteller, 2011; Wright and Bruford, 2011). They are tightly regulated and play critical roles in development and physiology. Therefore, their dysregulation plays an important role in the pathogenesis of MS. In this respect, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) have provided potential biomarkers and mechanistic insights into MS (Liu et al., 2014; Yang et al., 2018). miRNAs are small ncRNA molecules with lengths of 21–25 nucleotides that regulate gene expression at the post-transcriptional level by causing degradation or translational repression of target mRNAs (Qurashi et al., 2007). On the other hand, lncRNAs are a heterogeneous group of ncRNAs with a length of more than 200 nucleotides that regulate all steps of gene expression, including transcription, post-transcription, and translation (Mercer et al., 2009; Ponting et al., 2009; Chen and Carmichael, 2010; Hung and Chang, 2010; Ma et al., 2013; Beermann et al., 2016). In multiple studies, miRNAs and lncRNAs were profiled for expression in demyelinating lesions and body fluids [CSF, peripheral blood mononuclear cells (PBMCs), plasma, and whole blood] from MS patients (Yang et al., 2018; Ghaderian et al., 2020). These studies have identified a large set of miRNAs and lncRNAs that are dysregulated in MS. Some of these miRNAs or lncRNAs are dysregulated in a lineage-specific manner, in specific cell populations or during specific stages/subtypes of MS, providing new MS-specific biomarkers to predict disease progression or therapy response. Figure 1A summarizes some of the common miRNAs that have been identified as dysregulated in a variety of patient samples or studies. However, some critical issues that could lead to discrepancies in such studies need to be addressed. These include an appropriate sample number, biological sample (serum, plasma, PBMC, and blood), stage of the disease (RPMS, PPMS, and SPMS), and technical and analytical methods selected for analysis. Given the overwhelming number of ncRNAs identified in these studies, only a few have been functionally defined and are thus discussed here.
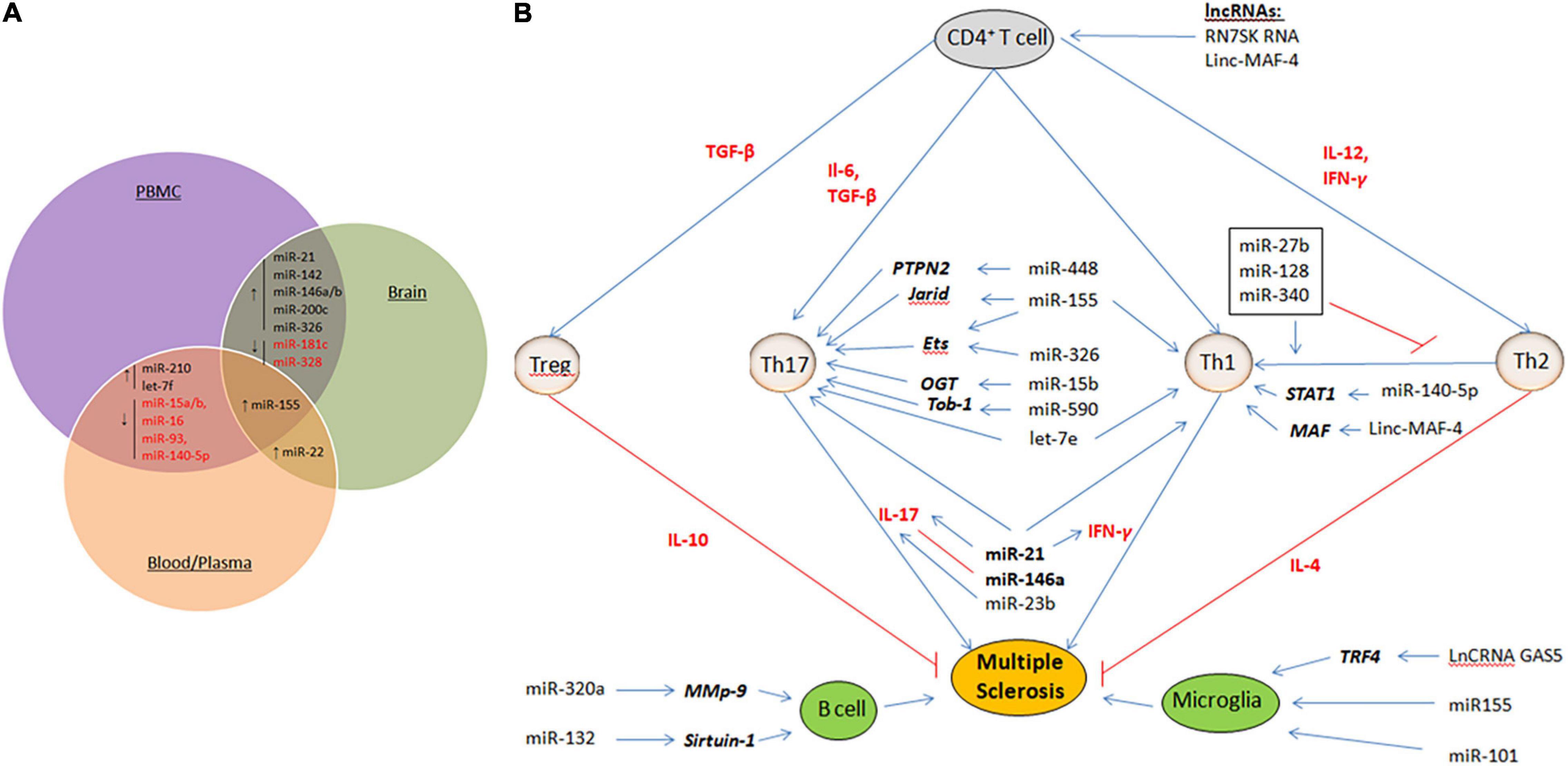
Figure 1. (A) Dysregulated miRNAs that are common in various samples (PBMC, blood/Plasma and brain tissue) of MS patients. ↑ represent up-regulated miRNAs and ↓ represent down-regulated miRNAs. (B) Mechanisms of ncRNAs in MS. Shown are the miRNAs and lncRNAs that function in the modulation of various immune cells, including upregulating activity of the proinflammatory Th1 cells and Th17 cells, B cells, and Microglia cells. Aberant expression of such ncRNAs eventually results in demyelination within the brain and spinal cord, and axonal damage. Cytokines that promote cell fate and differentiation are shown in red. Target genes of miRNAs are shown in bold italics.
miRNAs Involved in MS Pathogenesis
Multiple studies have reported a repertoire of aberrantly expressed miRNAs in both the immune and CNS cells of MS patients. Some of these miRNAs have been functionally characterized to play critical roles in MS pathogenesis. Table 1 summarizes the miRNAs and their targets, as described in the literature. While most of the miRNAs were found to be uniquely dysregulated in a particular cell type, some were dysregulated in many cell types and studies (Figures 1A,B).
Dysregulation of miRNA in the Immune Cells of Multiple Sclerosis Patients
The upregulated expression of miR-326 in Th17 cells isolated from the peripheral blood of MS patients with RRMS was related to disease severity and the production of IL-17 (Du et al., 2009). It inhibits Ets-1, a negative regulator of Th17 cell differentiation, and its overexpression increases Th17 cell number and leads to severe EAE. In contrast, miR-326 knockdown reduced the number of Th17 cells and alleviated EAE (Du et al., 2009). Similarly, miR-155 expression in T cells promotes Th17 differentiation and function by inhibiting the transcription of Ets-1 (O’Connell et al., 2010; Hu et al., 2013; Zhang et al., 2014). Similarly, miR-590 is upregulated in relapse-MS patients to promote the differentiation of Th17 cells by targeting Tob1 (Transducer of Erb-2), a member of the Tob/BTG anti-proliferative family of proteins (Liu Q. et al., 2017). In addition, miR-590 affects the pathogenicity of Th17 cells by upregulating inflammation-related molecules, such as CXCL3, CSF2, and IL-23R (Liu Q. et al., 2017). miR-448 can increase Th17 differentiation by directly inhibiting the anti-inflammatory protein tyrosine phosphatase non-receptor type 2 (PTPN2) (Wu et al., 2017). The let-7e miRNA was significantly upregulated in both experimental EAE and MS patients. Let-7e inhibition shifts the immune response to a Th2 profile and reduces disease severity, whereas let-7e overexpression increases Th1 and Th17 cells and worsens EAE (Guan et al., 2013).
Downregulation of many miRNAs has also been shown to influence Th1 or Th17 cell differentiation. For example, miR-15b targets O-linked N-acetylglucosamine transferase, which is normally required for CD4+ T cell activation and the ability to induce inflammation (Liu R. et al., 2017). However, in patients with MS, miR-15b is downregulated to inhibit Th1 or Th17 cell differentiation. In-line knockdown of miR-15b aggravated EAE, and overexpression of miR-15b alleviated EAE. Similarly, the miR-132 cluster is downregulated in CD4+ cells (Hanieh and Alzahrani, 2013). It targets Bcl-6 (B-cell lymphoma 6), a negative regulator of Th17 differentiation. Its downregulation is associated with the severity of EAE (Hanieh and Alzahrani, 2013). Similarly, under normal conditions, miR-1405p inhibits the differentiation of Th1 cells by downregulating STAT1 (signal transducer and activator of transcription) (Guan et al., 2016). However, its expression is markedly downregulated in MS, and consequently increases the development of Th1 cells and disease severity (Guan et al., 2016).
Dysregulation of miRNAs in MS is not limited to T helper cells, but has been reported in other types of T cells to influence their polarization. For example, miR-128 and miR-27b expression is elevated in naïve CD4+T cells, and miR-340 expression is increased in CD4+ memory T cells (Guerau-de-Arellano et al., 2011). These miRNAs inhibit the differentiation of Th2 cells by directly decreasing the expression of IL-4 and BMI1 (B lymphoma Mo-MLV insertion region 1 homolog), resulting in a Th2 to Th1 response shift. Interestingly, oligonucleotides against these miRNAs restore Th2 responses in patients with MS (Murugaiyan et al., 2011). In CD4+ memory T cells, miR-29b, which is induced by IFN-γ, acts in a negative feedback loop by inhibiting T-bet and IFN-γ transcription to control Th1 cell bias (Smith et al., 2012). Treg cells in MS have differential expression of 23 miRNAs when compared to healthy controls (De Santis et al., 2010). Among the significantly increased miRNAs were miR-106b and miR-25, which influenced TGF-β signaling, which is important for the development of both Th17 and Treg cells (Petrocca et al., 2008). TGF-β signaling is also increased in pro-inflammatory CD8+ T cells through the upregulation of miR-629 (Huang et al., 2016). Together, these lines of evidence have demonstrated that miRNAs through the inhibition of various target genes influence the differentiation of proinflammatory Th1 cells and Th17 cells, the development of Tregs, and the alteration of the Th2 to Th1 response in MS.
Dysregulation of miRNAs in Antigen-Presenting Cells (B Cells, Macrophages, and Dendritic Cells) of Multiple Sclerosis Patients
In addition to T cells, several important miRNAs have been identified to be differentially expressed in B lymphocytes of MS (Table 1). The expression of miR-320a is considerably decreased in B cells (Aung et al., 2015). Among many targets, miR-320a inhibits matrix metallopeptidase-9 (MMP-9) produced by activated B cells. In MS, increased MMP-9 expression and secretion in B cells due to downregulation of miR-320a disrupts the BBB and degrades myelin basic protein (Chandler et al., 1995; Asahi et al., 2001). Similarly, significantly increased expression of miR-132 in patients with MS reduces the level of Sirtuin-1 in B lymphocytes, which in turn accounts for the elevated expression of pro-inflammatory cytokines such as lymphotoxin and tumor necrosis factor (TNF-α) (Miyazaki et al., 2014). miR-17–92 inhibits the expression of Bim, a pro-apoptotic protein, and PTEN, a tumor suppressor. Therefore, downregulation of miR-17–92 results in elevated levels of Bim, which in turn suppresses the development of pro-B to pre-B cells (Ventura et al., 2008). Compared to controls, in untreated RRMS patients, miR-155 expression was significantly increased in both peripheral circulating CD14+ monocytes and active lesions CD68+ cells from perivascular (blood-derived macrophages) and parenchymal (microglia) brain regions. Upregulation of miR-155 in these cells was subsequently associated with increased proinflammatory cytokine secretion (Moore et al., 2013). miR-124 is considered a key regulator of microglial quiescence. Accordingly, in EAE, miR-124 expression is decreased in activated microglia, while overexpression of miR-124 could promote activated microglia into a phenotype resembling microglia quiescence and suppress EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway (Ponomarev et al., 2011). Together, these studies demonstrate the crucial effects of miRNAs on antigen-presenting cell-mediated mechanisms by influencing their activation and effector functions.
Dysregulation of miRNA in CNS Cells of Multiple Sclerosis Patients
Similar to immune cells, aberrant expression of miRNAs in the CNS contributes to the mechanism underlying inflammation in MS. In a study examining cell type-specific miRNA profiles using laser capture microdissection, 10 miRNAs were identified to be substantially elevated in active MS lesions (Junker et al., 2009). CD47, which is ubiquitously expressed in a variety of human cells to prevent phagocytosis, was decreased in the active lesions of MS patients. Three miRNAs, miR-155, miR-34a, and miR-326, which are upregulated in MS, can target CD47, consequently releasing macrophages from inhibitory control and promoting myelin breakdown (Junker et al., 2009). These changes occur primarily in astrocytes. In addition, miR-155 directly targets and downregulates SOCS1, a negative regulator of cytokine production in astrocytes (Moore et al., 2013; Arruda et al., 2015). miR-155 also targets the neurosteroid synthesis enzymes ARK1C1 and ARK1C2 (Noorbakhsh et al., 2011). miR-23 control of lamin B1 was shown to be important for oligodendrocyte growth and myelination in a previous study, implying that it may play a role in the pathogenesis of MS (Lin and Fu, 2009). Compared to controls, miR-219 and miR-338 are reduced in patients with MS (Dugas et al., 2010). Overexpression of miR-219 and miR-338 promotes the differentiation of oligodendroglial precursor cells (OPCs) in culture. Milbreta et al. (2019) introduced these miRNAs into the oligodendrocytes of rats and demonstrated their therapeutic potential in promoting oligodendrocyte differentiation and myelination. Based on these findings, it is clear that dysregulation of miRNAs affects an environment that promotes remyelination and axon regeneration, both of which are impaired in MS.
lncRNAs in MS
Although research into the role of lncRNAs in MS is still in its infancy, abnormal lncRNA expression has been investigated in serum, plasma, PBMCs, and blood exclusively in patients with RRMS and SPMS (Santoro et al., 2016). Like miRNA, lncRNAs can modulate the activity of various immune cells (Figure 1B). In MS patients, excess NEAT1 leads to re-localization of SFPQ (splicing factor proline/glutamine-rich) from the IL-8 promoter, resulting in transcriptional activation of IL-8 (Imamura et al., 2014). RN7SK RNA is involved in the regulation of CD4+ T lymphocytes and contributes to inflammation (Sung and Rice, 2006). The upregulation of RN7SK RNA in the 7SK snRNP complex might catalytically repress P-TEFb, a Cdk9/cyclin T1 kinase complex, which is important for the differentiation of CD4+ T cells into several sub-populations. TUG1 is upregulated in several neurodegenerative diseases, including MS. The TUG1 promoter contains conserved p53-binding and is a downstream target of p53 participating in the apoptotic pathways (Rossi et al., 2014). Sun et al. (2017) documented the upregulation of the lncRNA growth arrest-specific 5 (GAS5) in amoeboid-shaped microglial cells of MS patients. GAS5 was demonstrated to promote polarization of the M1 subgroup of microglial cells and, consequently, demyelination. Interfering with GAS5 in transplanted microglia slowed the course of EAE and promoted remyelination. GAS5 inhibits the proliferation of T cells by binding to PRC2, the polycarbonate 2 suppressor complex, and suppresses IRF4 transcription factor. Zhang et al. found that linc-MAF-4 levels were considerably higher in PBMCs from MS patients than in healthy controls. Linc-MAF-4 exacerbates MS pathogenesis by altering the Th1/Th2 ratio and by targeting MAF, a Th2 cell transcription factor required for Th2 differentiation (Zhang et al., 2017). Given these findings and the ability of lncRNAs to activate or repress gene expression at multiple levels through various mechanisms, it is conceivable that they act as critical regulators in the pathogenesis of MS. They are also promising biomarker candidates for MS diagnosis because of their stability in body fluids and cell specificity.
Conclusion and Perspectives
Multiple sclerosis is a chronic inflammatory disease of the nervous system with autoimmune manifestations. The complexity associated with its pathophysiology and clinical presentation underlies the need for specific biomarkers and therapies. Large repertoires of aberrantly expressed miRNAs and lncRNAs have been identified in the immune and CNS cells of MS patients. Although the role of some of these miRNAs or lncRNAs in mediating MS pathogenesis has been demonstrated, others need to be functionally characterized. Interestingly, several miRNAs that are dysregulated in MS meet most of the required criteria for being an ideal biomarker, such as accessibility, high specificity, and sensitivity.
Given the wide range of cells and immunological responses implicated in MS pathogenesis, as well as the numerous targetsof ncRNAs (particularly miRNAs), it is important to explore specific target genes and pathways that drive aberrant immune responses in MS. Therefore, the combination of ncRNAs and their targets may provide better signatures for developing specific biomarkers and new therapeutic interventions in MS. In this context, disease-modifying therapies may include drugs and treatment methods that modulate ncRNA expression or function. Because some of the immune abnormalities in MS have been described in other types of autoimmune diseases, it is reasonable to assume that such studies will add to our understanding of the complex regulatory networks in autoimmune disorders in general.
Author Contributions
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank all the members of the Qurashi lab for their critical reading of the manuscript.
References
Aharoni, R., Teitelbaum, D., Leitner, O., Meshorer, A., Sela, M., and Arnon, R. (2000). Specific Th2 cells accumulate in the central nervous system of mice protected against experimental autoimmune encephalomyelitis by copolymer 1. Proc. Natl. Acad. Sci. U.S.A. 97, 11472–11477. doi: 10.1073/pnas.97.21.11472
Ahmadian-Elmi, M., Bidmeshki Pour, A., Naghavian, R., Ghaedi, K., Tanhaei, S., Izadi, T., et al. (2016). miR-27a and miR-214 exert opposite regulatory roles in Th17 differentiation via mediating different signaling pathways in peripheral blood CD4+ T lymphocytes of patients with relapsing-remitting multiple sclerosis. Immunogenetics 68, 43–54. doi: 10.1007/s00251-015-0881-y
Arruda, L. C., Lorenzi, J. C., Sousa, A. P., Zanette, D. L., Palma, P. V., Panepucci, R. A., et al. (2015). Autologous hematopoietic SCT normalizes miR-16, -155 and -142-3p expression in multiple sclerosis patients. Bone Marrow Transpl. 50, 380–389.
Asahi, M., Wang, X., Mori, T., Sumii, T., Jung, J. C., Moskowitz, M. A., et al. (2001). Effects of matrix metalloproteinase-9 gene knock-out on the proteolysis of blood-brain barrier and white matter components after cerebral ischemia. J. Neurosci. 21, 7724–7732.
Aung, L. L., Mouradian, M. M., Dhib-Jalbut, S., and Balashov, K. E. (2015). MMP-9 expression is increased in B lymphocytes during multiple sclerosis exacerbation and is regulated by microRNA-320a. J. Neuroimmunol. 278, 185–189. doi: 10.1016/j.jneuroim.2014.11.004
Beermann, J., Piccoli, M. T., Viereck, J., and Thum, T. (2016). Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 96, 1297–1325. doi: 10.1152/physrev.00041.2015
Chandler, S., Coates, R., Gearing, A., Lury, J., Wells, G., and Bone, E. (1995). Matrix metalloproteinases degrade myelin basic protein. Neurosci. Lett. 201, 223–226. doi: 10.1016/0304-3940(95)12173-0
Chen, L. L., and Carmichael, G. G. (2010). Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 22, 357–364. doi: 10.1016/j.ceb.2010.03.003
Confavreux, C., and Vukusic, S. (2014). The clinical course of multiple sclerosis. Handb. Clin. Neurol. 122, 343–369. doi: 10.1016/B978-0-444-52001-2.00014-5
Cruz, L. O., Hashemifar, S. S., Wu, C. J., Cho, S., Nguyen, D. T., Lin, L. L., et al. (2017). Excessive expression of miR-27 impairs Treg-mediated immunological tolerance. J. Clin. Invest. 127, 530–542. doi: 10.1172/JCI88415
De Santis, G., Ferracin, M., Biondani, A., Caniatti, L., Rosaria Tola, M., Castellazzi, M., et al. (2010). Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J. Neuroimmunol. 226, 165–171. doi: 10.1016/j.jneuroim.2010.06.009
Du, C., Liu, C., Kang, J., Zhao, G., Ye, Z., Huang, S., et al. (2009). MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10, 1252–1259. doi: 10.1038/ni.1798
Dugas, J. C., Cuellar, T. L., Scholze, A., Ason, B., Ibrahim, A., Emery, B., et al. (2010). Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611. doi: 10.1016/j.neuron.2010.01.027
Escobar, T. M., Kanellopoulou, C., Kugler, D. G., Kilaru, G., Nguyen, C. K., Nagarajan, V., et al. (2014). miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity 40, 865–879. doi: 10.1016/j.immuni.2014.03.014
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi: 10.1038/nrg3074
Fontana, L., Pelosi, E., Greco, P., Racanicchi, S., Testa, U., Liuzzi, F., et al. (2007). MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 9, 775–787. doi: 10.1038/ncb1613
Ghaderian, S., Shomali, N., Behravesh, S., Danbaran, G. R., Hemmatzadeh, M., Aslani, S., et al. (2020). The emerging role of lncRNAs in multiple sclerosis. J. Neuroimmunol. 347:577347. doi: 10.1016/j.jneuroim.2020.577347
Guan, H., Fan, D., Mrelashvili, D., Hao, H., Singh, N. P., Singh, U. P., et al. (2013). MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur. J. Immunol. 43, 104–114. doi: 10.1002/eji.201242702
Guan, H., Singh, U. P., Rao, R., Mrelashvili, D., Sen, S., Hao, H., et al. (2016). Inverse correlation of expression of microRNA-140-5p with progression of multiple sclerosis and differentiation of encephalitogenic T helper type 1 cells. Immunology 147, 488–498. doi: 10.1111/imm.12583
Guerau-de-Arellano, M., Smith, K. M., Godlewski, J., Liu, Y., Winger, R., Lawler, S. E., et al. (2011). Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain 134, 3578–3589. doi: 10.1093/brain/awr262
Hanieh, H., and Alzahrani, A. (2013). MicroRNA-132 suppresses autoimmune encephalomyelitis by inducing cholinergic anti-inflammation: a new Ahr-based exploration. Eur. J. Immunol. 43, 2771–2782. doi: 10.1002/eji.201343486
Hu, R., Huffaker, T. B., Kagele, D. A., Runtsch, M. C., Bake, E., Chaudhuri, A. A., et al. (2013). MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J. Immunol. 190, 5972–5980. doi: 10.4049/jimmunol.1300351
Huang, Q., Xiao, B., Ma, X., Qu, M., Li, Y., Nagarkatti, P., et al. (2016). MicroRNAs associated with the pathogenesis of multiple sclerosis. J. Neuroimmunol. 29, 148–161. doi: 10.1016/j.jneuroim.2016.04.014
Hung, T., and Chang, H. Y. (2010). Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 7, 582–585. doi: 10.4161/rna.7.5.13216
Imamura, K., Imamachi, N., Akizuki, G., Kumakura, M., Kawaguchi, A., Nagata, K., et al. (2014). Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 53, 393–406. doi: 10.1016/j.molcel.2014.01.009
Ingwersen, J., Menge, T., Wingerath, B., Kaya, D., Graf, J., Prozorovski, T., et al. (2015). Natalizumab restores aberrant miRNA expression profile in multiple sclerosis and reveals a critical role for miR-20b. Ann. Clin. Transl. Neurol. 2, 43–55. doi: 10.1002/acn3.152
Jäger, A., Dardalhon, V., Sobel, R. A., Bettelli, E., and Kuchroo, V. K. (2009). Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 183, 7169–7177. doi: 10.4049/jimmunol.0901906
Junker, A., Krumbholz, M., Eisele, S., Mohan, H., Augstein, F., Bittner, R., et al. (2009). MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132, 3342–3352. doi: 10.1093/brain/awp300
Jurkin, J., Schichl, Y. M., Koeffel, R., Bauer, T., Richter, S., Konradi, S., et al. (2010). miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J. Immunol. 184, 4955–4965. doi: 10.4049/jimmunol.0903021
Lin, S. T., and Fu, Y. H. (2009). miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model Mech. 2, 178–188. doi: 10.1242/dmm.001065
Liu, Q., Gao, Q., Zhang, Y., Li, Z., and Mei, X. (2017). MicroRNA-590 promotes pathogenic Th17 cell differentiation through targeting Tob1 and is associated with multiple sclerosis. Biochem. Biophys. Res. Commun. 493, 901–908. doi: 10.1016/j.bbrc.2017.09.123
Liu, R., Ma, X., Chen, L., Yang, Y., Zeng, Y., Gao, J., et al. (2017). MicroRNA-15b Suppresses Th17 Differentiation and Is Associated with Pathogenesis of Multiple Sclerosis by Targeting. J. Immunol. 198, 2626–2639. doi: 10.4049/jimmunol.1601727
Liu, X. F., Luo, Y. B., Luo, Z. H., and Yang, H. (2014). Biomarker studies in multiple sclerosis: from proteins to noncoding RNAs. Neurochem. Res. 39, 1661–1674. doi: 10.1007/s11064-014-1386-z
Lorenzi, J. C., Brum, D. G., Zanette, D. L., de Paula Alves, Souza, A., Barbuzano, F. G., et al. (2012). miR-15a and 16-1 are downregulated in CD4+ T cells of multiple sclerosis relapsing patients. Int. J. Neurosci. 122, 466–471. doi: 10.3109/00207454.2012.678444
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 925–933. doi: 10.4161/rna.24604
Mahad, D. H., Trapp, B. D., and Lassmann, H. (2015). Pathological mechanisms in progressive multiple sclerosis. Lancet Neurol. 14, 183–193. doi: 10.1016/S1474-4422(14)70256-X
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Milbreta, U., Lin, J., Pinese, C., Ong, W., Chin, J. S., Shirahama, H., et al. (2019). Scaffold-Mediated Sustained, Non-viral Delivery of miR-219/miR-338 Promotes CNS Remyelination. Mol. Ther. 27, 411–423. doi: 10.1016/j.ymthe.2018.11.016
Miyazaki, Y., Li, R., Rezk, A., Misirliyan, H., Moore, C., Farooqi, N., et al. (2014). A novel microRNA-132-sirtuin-1 axis underlies aberrant B-cell cytokine regulation in patients with relapsing-remitting multiple sclerosis [corrected]. PLoS One. 9:e105421. doi: 10.1371/journal.pone.0105421
Moore, C. S., Rao, V. T., Durafourt, B. A., Bedell, B. J., Ludwin, S. K., Bar-Or, A., et al. (2013). miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann. Neurol. 74, 709–720.
Murugaiyan, G., Beynon, V., Mittal, A., Joller, N., and Weiner, H. L. (2011). Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 187, 2213–2221. doi: 10.4049/jimmunol.1003952
Mycko, M. P., Cichalewska, M., Cwiklinska, H., and Selmaj, K. W. (2015). miR-155-3p drives the development of autoimmune demyelination by regulation of heat shock protein 40. J. Neurosci. 35, 16504–16515. doi: 10.1523/JNEUROSCI.2830-15.2015
Noorbakhsh, F., Ellestad, K. K., Maingat, F., Warren, K. G., Han, M. H., Steinman, L., et al. (2011). Impaired neurosteroid synthesis in multiple sclerosis. Brain 134, 2703–2721. doi: 10.1093/brain/awr200
O’Connell, R. M., Kahn, D., Gibson, W. S., Round, J. L., Scholz, R. L., Chaudhuri, A. A., et al. (2010). MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619. doi: 10.1016/j.immuni.2010.09.009
O’Connor, R. A., Prendergast, C. T., Sabatos, C. A., Lau, C. W., Leech, M. D., Wraith, D. C., et al. (2008). Cutting edge: Th1 cells facilitate the entry of Th17 cells to the central nervous system during experimental autoimmune encephalomyelitis. J. Immunol. 181, 3750–3754. doi: 10.4049/jimmunol.181.6.3750
Petrocca, F., Vecchione, A., and Croce, C. M. (2008). Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 68, 8191–8194. doi: 10.1158/0008-5472.CAN-08-1768
Ponomarev, Veremeyko, T., Barteneva, N., Krichevsky, A. M., and Weiner, H. L. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 17, 64–70. doi: 10.1038/nm.2266
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Qurashi, A., Chang, S., and Peng, J. (2007). Role of microRNA pathway in mental retardation. Scient. World J. 7, 146–154. doi: 10.1100/tsw.2007.208
Rossi, S., Motta, C., Studer, V., Macchiarulo, G., Volpe, E., Barbieri, F., et al. (2014). Interleukin-1β causes excitotoxic neurodegeneration and multiple sclerosis disease progression by activating the apoptotic protein p53. Mol. Neurodegener. 9:56. doi: 10.1186/1750-1326-9-56
Santoro, M., Nociti, V., Lucchini, M., De Fino, C., Losavio, F. A., and Mirabella, M. (2016). Expression Profile of Long Non-Coding RNAs in Serum of Patients with Multiple Sclerosis. J. Mol. Neurosci. 59, 18–23. doi: 10.1007/s12031-016-0741-8
Sievers, C., Meira, M., Hoffmann, F., Fontoura, P., Kappos, L., and Lindberg, R. L. (2012). Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin. Immunol. 144, 70–79. doi: 10.1016/j.clim.2012.04.002
Smith, K. M., Guerau-de-Arellano, M., Costinean, S., Williams, J. L., Bottoni, A., Mavrikis Cox, G., et al. (2012). miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J. Immunol. 189, 1567–1576. doi: 10.4049/jimmunol.1103171
Sospedra, M., and Martin, R. (2005). Immunology of multiple sclerosis. Annu. Rev. Immunol. 23, 683–747. doi: 10.1146/annurev.immunol.23.021704.115707
Sun, D., Yu, Z., Fang, X., Liu, M., Pu, Y., Shao, Q., et al. (2017). LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 18, 1801–1816. doi: 10.15252/embr.201643668
Sun, L., Liu, B., Lin, Z., Yao, Y., Chen, Y., Li, Y., et al. (2015). MiR-320a acts as a prognostic factor and Inhibits metastasis of salivary adenoid cystic carcinoma by targeting ITGB3. Mol. Cancer 14:96. doi: 10.1186/s12943-015-0344-y
Sung, T. L., and Rice, A. P. (2006). Effects of prostratin on Cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology 3:66. doi: 10.1186/1742-4690-3-66
Thompson, A. J., Baranzini, S. E., Geurts, J., Hemmer, B., and Ciccarelli, O. (2018). Multiple sclerosis. Lancet 391, 1622–1636. doi: 10.1016/S0140-6736(18)30481-1
Ventura, A., Young, A. G., Winslow, M. M., Lintault, L., Meissner, A., Erkeland, S. J., et al. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886. doi: 10.1016/j.cell.2008.02.019
Vistbakka, J., Elovaara, I., Lehtimäki, T., and Hagman, S. (2017). Circulating microRNAs as biomarkers in progressive multiple sclerosis. Mult. Scler. 23, 403–412. doi: 10.1177/1352458516651141
Wang, B., Yang, Z., Wang, H., Cao, Z., Zhao, Y., Gong, C., et al. (2015). MicroRNA-320a inhibits proliferation and invasion of breast cancer cells by targeting RAB11A. Am. J. Cancer Res. 5, 2719–2729.
Wright, M. W., and Bruford, E. A. (2011). Naming ‘junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Hum. Genomics 5, 90–98. doi: 10.1186/1479-7364-5-2-90
Wu, R., He, Q., Chen, H., Xu, M., Zhao, N., Xiao, Y., et al. (2017). MicroRNA-448 promotes multiple sclerosis development through induction of Th17 response through targeting protein tyrosine phosphatase non-receptor type 2 (PTPN2). Biochem. Biophys. Res. Commun. 486, 759–766. doi: 10.1016/j.bbrc.2017.03.115
Yang, X., Wu, Y., Zhang, B., and Ni, B. (2018). Noncoding RNAs in multiple sclerosis. Clin. Epigenet. 10:149. doi: 10.1186/s13148-018-0586-9
Zhang, F., Liu, G., Wei, C., Gao, C., and Hao, J. (2017). Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J. 31, 519–525. doi: 10.1096/fj.201600838R
Keywords: multiple sclerosis, central nervous system, microRNA, long noncoding RNA, neurodegeneration
Citation: Yousuf A and Qurashi A (2021) Non-coding RNAs in the Pathogenesis of Multiple Sclerosis. Front. Genet. 12:717922. doi: 10.3389/fgene.2021.717922
Received: 31 May 2021; Accepted: 04 August 2021;
Published: 30 September 2021.
Edited by:
Vishal Shinde, Massachusetts Eye and Ear Infirmary and Harvard Medical School, United StatesReviewed by:
James A. Poulter, University of Leeds, United KingdomChaitanya R. Joshi, University of Texas Southwestern Medical Center, United States
Copyright © 2021 Yousuf and Qurashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abrar Qurashi, YWJyYXIucXVyYXNoaUB1b2suZWR1Lmlu
 Aadil Yousuf
Aadil Yousuf Abrar Qurashi
Abrar Qurashi