
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Genet., 15 June 2021
Sec. Genomic Assay Technology
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.706540
This article is a correction to:
Sequence and Structure Characteristics of 22 Deletion Breakpoints in Intron 44 of the DMD Gene Based on Long-Read Sequencing
A Corrigendum on
Sequence and Structure Characteristics of 22 Deletion Breakpoints in Intron 44 of the DMD Gene Based on Long-Read Sequencing
by Geng, C., Tong, Y., Zhang, S., Ling, C., Wu, X., Wang, D., et al. (2021). Front. Genet. 12:638220. doi: 10.3389/fgene.2021.638220
In the original article, there was a mistake in Figure 1 as published. Supplementary Figure 1 of the article was carelessly displayed again as Figure 1. The corrected Figure 1 appears below.
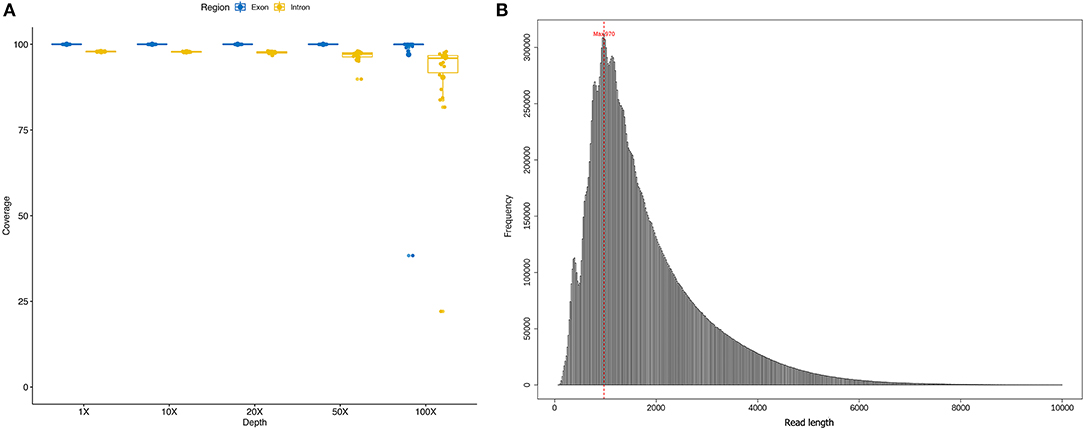
Figure 1. (A) coverage ratio in different depth level of each sample; (B) the length of sequencing reads of each sample.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: Duchenne and Becker muscular dystrophy, DMD gene, copy number variations, long-read sequencing, NHEJ, MMEJ
Citation: Geng C, Tong Y, Zhang S, Ling C, Wu X, Wang D and Dai Y (2021) Corrigendum: Sequence and Structure Characteristics of 22 Deletion Breakpoints in Intron 44 of the DMD Gene Based on Long-Read Sequencing. Front. Genet. 12:706540. doi: 10.3389/fgene.2021.706540
Received: 07 May 2021; Accepted: 24 May 2021;
Published: 15 June 2021.
Edited and reviewed by: Jiannis Ragoussis, McGill University, Canada
Copyright © 2021 Geng, Tong, Zhang, Ling, Wu, Wang and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Dai, cHVtY2hkeUBwdW1jaC5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.