- Department of Obstetrics and Gynecology, Zhongnan Hospital of Wuhan University, Wuhan, China
Background: Inhibin A (INHBA), a member of the TGF-β superfamily, has been shown to be differentially expressed in various cancer types and is associated with prognosis. However, its role in cervical cancer remains unclear.
Methods: We aimed to demonstrate the relationship between INHBA expression and pan-cancer using The Cancer Genome Atlas (TCGA) database. Next, we validated INHBA expression in cervical cancer using the Gene Expression Omnibus (GEO) database, including GSE7803, GSE63514, and GSE9750 datasets. Enrichment analysis of INHBA was performed using the R package “clusterProfiler.” We analyzed the association between immune infiltration level and INHBA expression in cervical cancer using the single-sample gene set enrichment analysis (ssGSEA) method by the R package GSVA. We explored the association between INHBA expression and prognosis using the R package “survival”.
Results: Pan-cancer data analysis showed that INHBA expression was elevated in 19 tumor types, including cervical cancer. We further confirmed that INHBA expression was higher in cervical cancer samples from GEO database and cervical cancer cell lines than in normal cervical cells. Survival prognosis analysis indicated that higher INHBA expression was significantly associated with reduced Overall Survival (p = 0.001), disease Specific Survival (p = 0.006), and Progression Free Interval (p = 0.001) in cervical cancer and poorer prognosis in other tumors. GSEA and infiltration analysis showed that INHBA expression was significantly associated with tumor progression and some types of immune infiltrating cells.
Conclusion: INHBA was highly expressed in cervical cancer and was significantly associated with poor prognosis. Meanwhile, it was correlated with immune cell infiltration and could be used as a promising prognostic target for cervical cancer.
1 Introduction
Although the introduction of prophylactic vaccination against human papillomavirus (HPV) and screening would substantially reduce the incidence of cervical cancer, it remains the most common gynecologic malignancy worldwide Matsuo et al. (2017). The global mortality rate from cervical cancer is approximately 54% Kiran et al. (2018). In 2018, an estimated 570 000 cases of cervical cancer were diagnosed, and 311 000 women died from the disease Canfell et al. (2020). Persistent infection with high-risk types of HPV (hrHPV) is a necessary cause of cervical cancer; however, alterations in tumor-suppressor genes and/or oncogenes may also be necessary for cervical cancer progression Babion et al. (2018). The combination of early detection via screening and effective treatment with surgery, chemotherapy, and radiotherapy has meant that early stage cervical cancer can be successfully treated, with 5-years overall survival (OS) rates as high as 90% Potikanond et al. (2017). However, metastatic cervical cancer is virtually incurable, mainly due to limited treatment options, with 5-years OS rates below 10% Wallbillich et al. (2020). Thus, better prognostic biomarkers for cervical cancer development are urgently required to increase patient survival.
Inhibin A (INHBA), a member of the transforming growth factor-β superfamily, is located in 7p14.1, and encodes the βA-subunit of the activins/inhibins Tournier et al. (2014). In mammals, prolonged upregulation of INHBA has been associated with cardiac remodeling and failure Dogra et al. (2017). INHBA also plays a crucial role in follicle activation in females Bellingham et al. (2013) and encodes for the major regulator of FSH secretion in adult males d’Aurora et al. (2015). Previous studies have identified an association between INHBA expression and invasion or poor survival in cancer Singh et al. (2018); Lee et al. (2015); Seder et al. (2009); Chen et al. (2019); Okano et al. (2013); Antsiferova et al. (2017). The possible mechanism of INHBA contribution to tumorigenesis lies in its interaction with other receptor subunits or other molecular partners depending on their relative expression levels. Liu et al. found that INHBA expression level is associated with cancer aggressiveness and may be a potential diagnostic marker of invasive breast cancer Liu et al. (2018). Moreover, INHBA has been demonstrated to be a significantly mutated driver candidate gene in endometrial cancer Kadara et al. (2017). Although it has been proven that INHBA is important for tumor development, no study has explored the precise function and mechanism of INHBA in cervical cancer.
In this study, we comprehensively evaluated the difference in INHBA expression between normal and tumor tissues using RNA-seq data from The Cancer Genome Atlas (TCGA) database as well as its association with patient prognosis. Furthermore, to reveal its potential functions, we performed gene set enrichment analysis (GSEA) on the low and high expression groups of INHBA. Finally, we examined the correlation between INHBA expression and immune cell infiltration levels to explore the possible mechanism by which INHBA induces tumor occurrence and progression.
2 Materials and Methods
2.1 Collection of INHBA Expression Data From TCGA Database
INHBA expression data with clinical information (including 13 normal and 306 cervical cancer tissues) were obtained from TCGA public database (cancergenome.nih.gov). Level 3 HTSeq-fragments per kilobase per million (FPKM) samples were computed with an HTseq tool and subsequently transformed to transcripts per million (TPM) units. We also downloaded publicly available transcript data from TCGA and Genotype-Tissue Expression (GTEx) database, which was uniformly managed by the Toil process from UCSC Xena (https://xenabrowser.net/datapages/). Cervical cancer microarray data were obtained from the Gene Expression Omnibus (GEO) database, including GSE7803 (Platform: GPL96), GSE63514 (Platform: GPL570), and GSE9750 (Platform: GPL96) datasets.
2.2 qRT-PCR
We selected an RNAsimple Total RNA Kit (TIANGEN, Beijing, China) to extract total RNA from cervical cancer cell lines. The Servicebio®RT First Strand cDNA Synthesis Kit was used for qRT-PCR (Servicebio, Wuhan, China). Next, we performed real-time PCR using TB Green® Premix Ex TaqTM (Takara, Japan). The primers used were as follows: human INHBA -Forward: 5′-ATCATCACGTTTGCCGAGTCA-3′; human INHBA -Reverse: 5′-GAAGAGGCGGATGGTGACTTT-3′; human GAPDH-Forward: 5′-GGAGTCCACTGGCGTCTTCA-3′; human GAPDH-Reverse, 5′-GTCATGAGTCCTTCCACGATACC-3′.
2.3 Correlation and INHBA-Related Gene Enrichment Analysis
Functional networks and gene connectivity data were extracted using the STRING database (https://www.string-db.org/, version 11.5). The parameters used were as follows: minimum required interaction score [“medium confidence (0.400)”], meaning of network edges (“evidence”), max number of interactors to show (“no more than 10 interactors” in first shell), and active interaction sources (“all”).
The correlation between expression levels of INHBA and other mRNAs in cervical cancer was determined using TCGA data, and the Pearson correlation coefficient was calculated for all correlation analyses. The top100 genes most significantly positively correlated with INHBA were selected for enrichment analysis to reflect their functional roles. Gene Ontology (GO) term enrichment analysis was performed using the R package clusterProfiler (v3.14.3). For KEGG pathway analysis, the R package clusterProfiler (v3.14.3) was employed. GSEA between high- and low-INHBA groups was carried out using the R package clusterProfiler (version 3.14.3). The process was repeated 1,000 times for each analysis and c2.cp.v7.2.symbols.gmt in MSigDB Collections were used as a reference gene collection. For gene sets to be considered significantly enriched, the false discovery rate (FDR) q-value needed to be smaller than 0.25 and P adjust
2.4 Survival Prognosis Analysis
We obtained patient survival data from TCGA and performed survival analysis using R packages, including package survival (version 3.2.10) and survminer (version 0.4.9). We selected 50% as the threshold, and the cohorts were divided into low-and high-expression groups. The relationships between INHBA expression and patient prognosis, including Overall Survival (OS), disease Specific Survival (DSS), and Progression Free Interval (PFI), were investigated.
2.5 Immune Cell Infiltration Analysis
Immune infiltration analysis of cervical cancer was performed using the single-sample gene set enrichment analysis (ssGSEA) method with the R package GSVA (version 1.34.0). To explore the correlation between INHBA and the infiltration levels of immune cells and the association of immune cell infiltration with the different expression groups of INHBA, Spearman’s rank correlation and Wilcoxon rank sum test were used. Statistical significance was set at p
3 Results
3.1 INHBA Expression Analysis in Pan-Cancer and Cervical Cancer Cell Lines
We performed pan-cancer analyses using the Mann-Whitney U test (Wilcoxon rank sum test) to compare INHBA expression in normal tissues and tumor samples using RNA sequencing data obtained from TCGA and GTEx databases (Figure 1A). INHBA expression was significantly higher in 19 tumor types, including BLCA (P
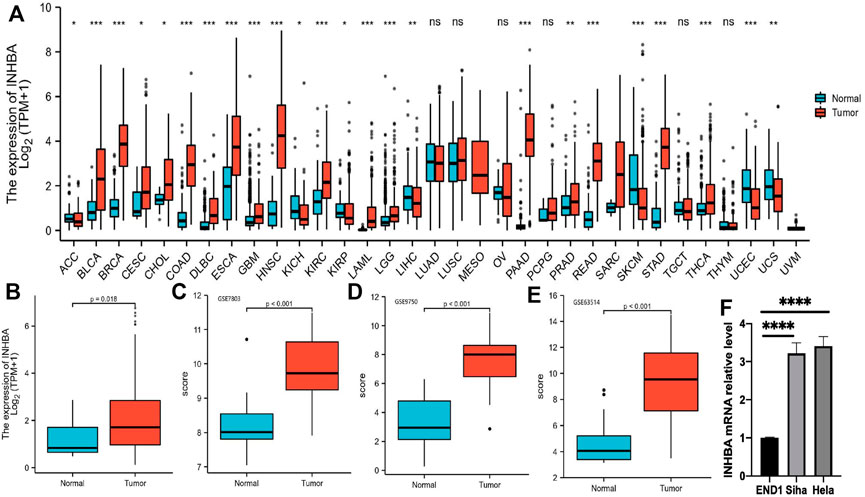
FIGURE 1. INHBA expression level analysis in pan-cancer. (A) INHBA expression in normal and tumor tissues in TCGA and GTEx data. (B) Boxplot representation of INHBA expression in cervical cancer tissue. (C–E) INHBA expression in normal cervical tissues and cervical cancer epithelial component from GSE7803, GSE9750 and GSE63514. (F) qRT-PCR analysis of INHBA expression in the indicated cell lines.
3.2 Correlation and INHBA-Related Gene Enrichment Analysis
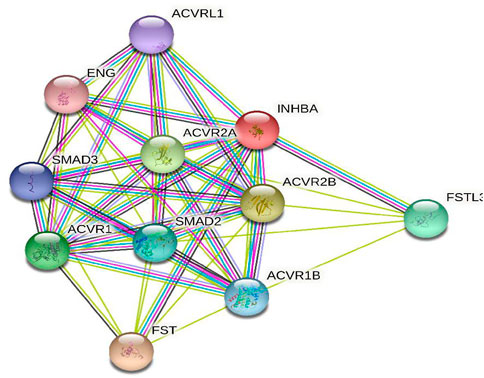
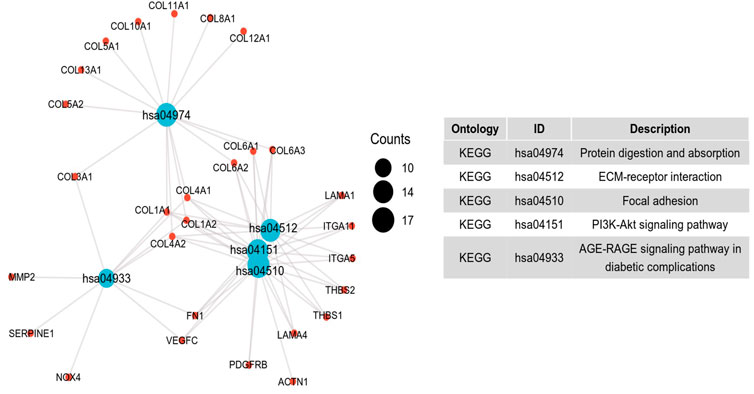
To further investigate the potential molecular mechanism of INHBA involvement in cervical cancer development and progression, the STRING tool was utilized. Based on the STRING tool, a database of known and predicted protein-protein interactions, we obtained the top 10 INHBA -binding proteins. Figure 2 demonstrates the interaction network of these proteins. We downloaded the expression data from the TCGA cancer browser website to investigate the functional enrichment information and pathways involved. Next, we selected the top100 genes that were most positively correlated with INHBA using the R stats package (Supplementary Table S1). As shown in Figure 3, the GO enrichment analysis of related genes revealed a significant enrichment of GO terms associated with extracellular matrix, such as extracellular matrix organization, collagen-containing extracellular matrix, extracellular matrix structural constituent, and others. The KEGG data analysis indicated that the “PI3K-Akt signaling pathway” might be related to the involvement of INHBA in tumor pathogenesis (Figure 4). Further, we performed GSEA to identify INHBA -related signaling pathways, and a total of 1069 pathways were enriched. There were 536 datasets that showed significant differential enrichment in the INHBA high expression phenotype. We selected the top nine datasets with a high normalized enrichment score (NES) (Figure 5). The results revealed that ECM receptor interaction and focal adhesion were significantly enriched in the KEGG pathway. In addition, extracellular matrix organization, degradation of the extracellular matrix, NON-integrin membrane ECM interactions, ECM proteoglycans, collagen formation, and collagen degradation were significantly enriched in Reactome pathway analysis (Supplementary Table S2).
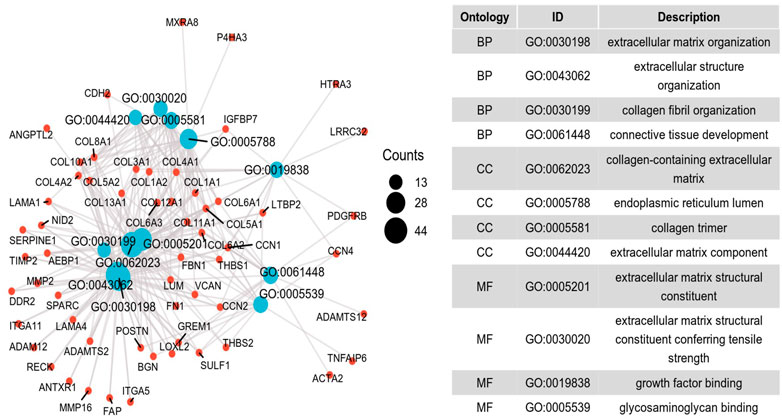
FIGURE 3. Significant Gene Ontology terms of the top 100 genes most positively associated with INHBA, including BP (biological processes), MF (molecular function) and CC (cell component).
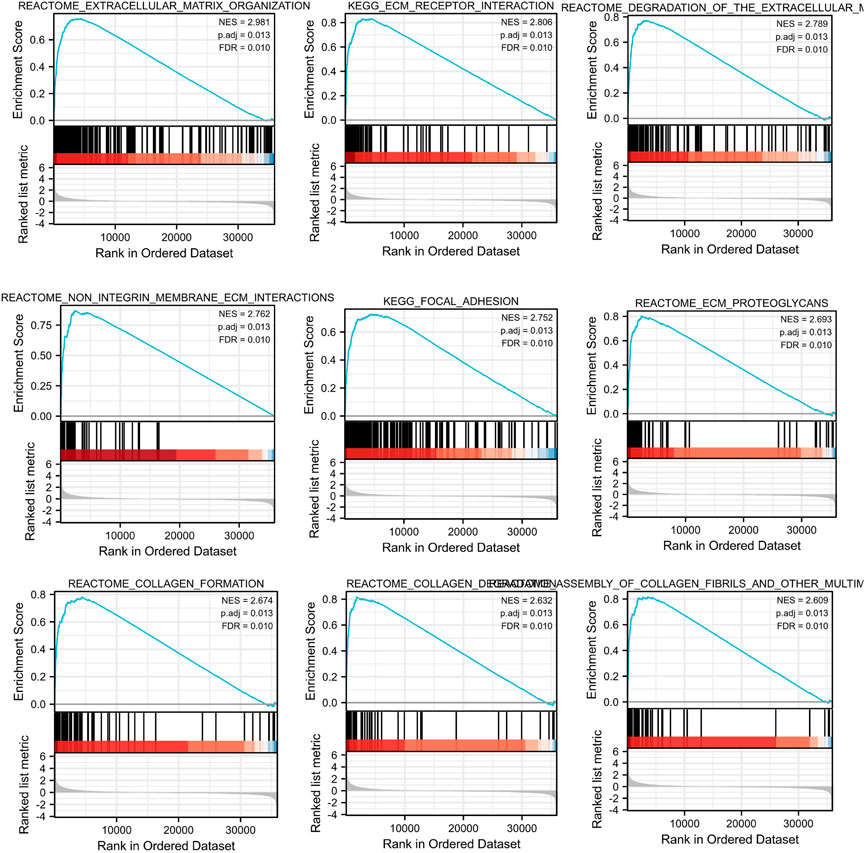
FIGURE 5. Top 9 significant results most positively associated with INHBA, including Reactome pathways and KEGG pathways.
3.3 Association Between INHBA Expression and Cancer Patients Survival Prognosis
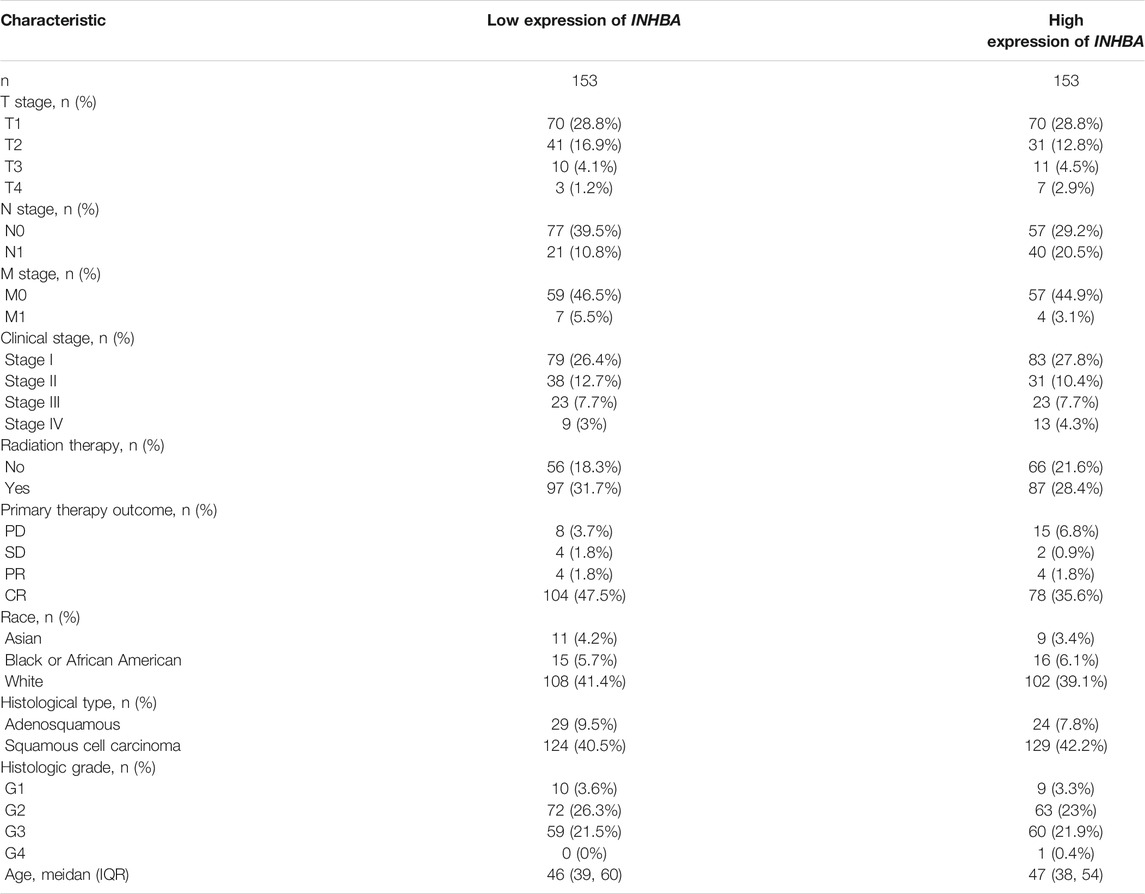
The characteristics of cervical cancer patients are listed in Table 1, in which clinical data and mRNA expression profiles from 306 patients with cervical cancer were collected from the TCGA database. Mean INHBA expression was defined as the cutoff to classify patients into high expression (n = 153) and low expression groups (n = 153). To identify the prognostic value of markers for cervical cancer, we evaluated the correlation between high expression of INHBA and patients’ survival time using Kaplan-Meier analysis with Cox regression, including OS, DSS, and PFI. The results demonstrated that higher INHBA expression was associated with poorer prognosis for OS (p = 0.001, HR = 2.30, 95% CI: 1.41–3.76) (Figure 6A), DSS (p = 0.006, HR = 2.18, 95% CI: 1.25–3.81) (Figure 6B), and PFI (p = 0.001, HR = 2.26, 95% CI: 1.39–3.67) (Figure 6C) in cervical cancer. Furthermore, we placed INHBA in the broader context of cancer by performing several pan-cancer analyses. We found that higher INHBA expression was associated with poorer OS in patients with HNSC (p = 0.001, HR = 1.59, 95% CI: 1.21–2.09) (Figure 6D) and STAD (p = 0.031, HR = 1.42, 95% CI: 1.02–1.98) (Figure 6E). There was a trend toward decreased DSS with increasing INHBA expression in BRCA (p = 0.048, HR = 1.54, 95% CI: 1.00–2.38) (Figure 6F) and HNSC (p = 0.002, HR = 1.73, 95% CI: 1.21–2.47) (Figure 6G). Moreover, upon analysis of the PFI data of patients with BRCA (p = 0.02, HR = 1.47, 95% CI: 1.06–2.04) (Figure 6H), DLBC (p = 0.041, HR = 0.2, 95% CI: 0.04–0.93) (Figure 6I), and HNSC (p = 0.03, HR = 1.37, 95% CI: 1.03–1.83) (Figure 6J), high INHBA expression was found to be correlated with poor prognosis.
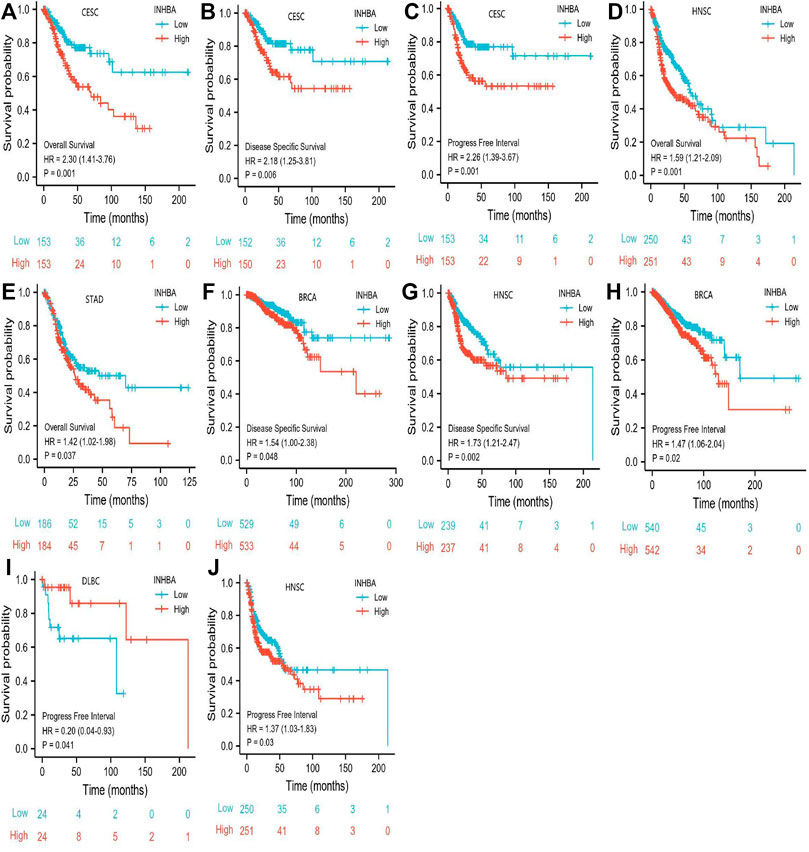
FIGURE 6. Association between INHBA expression and cancer survival prognosis. (A) CESC INHBA OS Survival. (B) CESC INHBA DSS Survival. (C) CESC INHBA PFI Survival. (D) HNSC INHBA OS Survival. (E) STAD INHBA OS Survival. (F) BRCA INHBA DSS Survival. (G) HNSC INHBA DSS Survival. (H) BRCA INHBA PFI Survival. (I) DLBC INHBA PFI Survival. (J) HNSC INHBA PFI Survival.
3.4 The Correlation Between INHBA Expression and Immune Cell Infiltration
ssGSEA with Spearman’s rank correlation was employed to measure the correlation between INHBA expression and infiltration levels of 24 immune cell types (Figure 7). The results showed that INHBA expression was correlated with several subsets of myeloid cells, including macrophages and mast cells, natural killer (NK) cells, neutrophils, Th2 cells, and eosinophils. Our study indicated that the expression of INHBA was positively associated with these immune cell types (Figures 8A–F, P
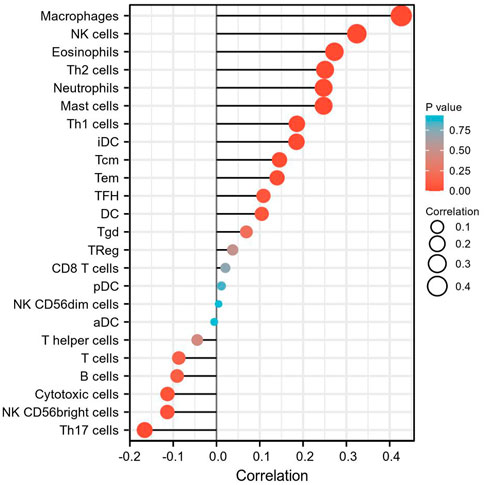
FIGURE 7. The correlation between INHBA expression level and 24 immune cell types. The size of dots indicate the absolute value of Spearman r.
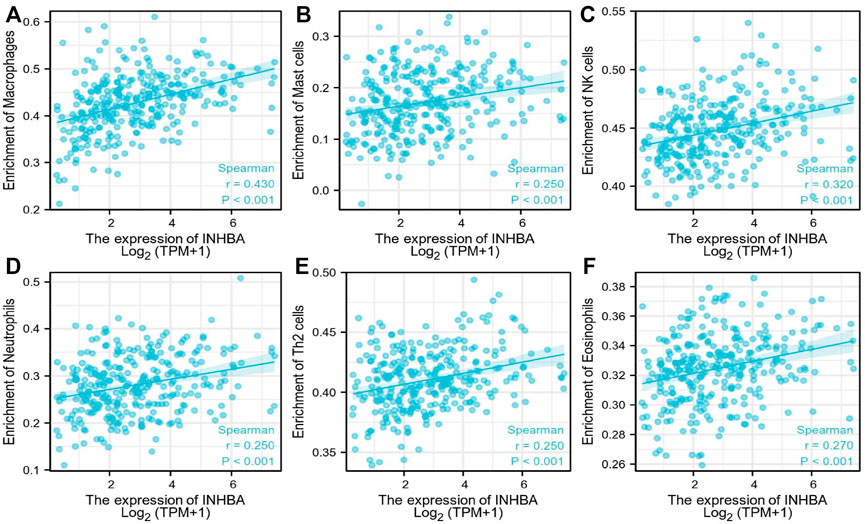
FIGURE 8. Immune cells postively correlated with INHBA expression. (A) Correlation between INHBA expression and Macrophages. (B) Correlation between INHBA expression and Mast cells. (C) Correlation between INHBA expression and NK cells. (D) Correlation between INHBA expression and Neutrophils. (E) Correlation between INHBA expression and Th2 cells. (F) Correlation between INHBA expression and Eosinophils.
4 Disscusion
Despite advances in screening, diagnosis, and treatment, cervical cancer remains a leading cause of cancer-related deaths worldwide. Therefore, accurate biomarkers are needed for the early detection and monitoring of disease progression. Previous studies have proven that INHBA is significantly increased in various tumors and it may be a potential prognostic biomarker for predicting the survival outcome of cancer patients Lyu et al. (2018); Li et al., 2020. While the role of INHBA has been comprehensively studied in many cancer types, its function in cervical cancer remains insufficiently understood. In this study, we attempted to address the role of INHBA in cervical cancer for the first time.
INHBA encodes a subunit of activin and inhibin, members of the TGFβ superfamily, which play context-dependent roles in cancer progression Li et al., 2020. Previous studies have shown that INHBA expression is upregulated in lung cancer and esophageal adenocarcinoma and has been shown to promote cell proliferation Liang et al. (2012); Dai et al. (2018). Furthermore, upregulated INHBA has also been implicated in promoting cancer stem cell and metastatic properties Hadadi et al. (2020). We also found that INHBA was highly overexpressed in most cancer types, including cervical cancer tissues, compared to that in adjacent normal tissues. In our study, we found that high expression of INHBA correlated with poor OS, DSS, and PFI in cervical cancer patients. Identical results were also observed at the pan-cancer level. High expression of the protein in tumors was associated with significantly shorter survival rates than that of patients whose tumors expressed lower levels of INHBA in BRCA, DLBC, HNSC, and STAD. Based on these findings, INHBA can be considered as a potential therapeutic target. Kalli et al. demonstrated that knocking down INHBA levels delayed primary breast tumor growth and suppressed the formation of lung metastases in vivo Kalli et al. (2019).
However, in some malignancies, INHBA serves as a tumor suppressor gene. Since INHBA/activins proteins are multifunctional ligands and their superfamily member, TGF-β, is closely involved in angiogenesis, INHBA may also play a role in tumor angiogenesis. For example, INHBA substantially inhibits tumor angiogenesis in gastric cancer in vivo Kaneda et al. (2011) and neuroblastoma models Liang et al. (2012). To the best of our knowledge, inhibition of angiogenesis potentially prevents tumor growth and metastasis to other organs.
To further investigate the function of INHBA in detail, we performed functional annotation based on the enrichment analysis. Among the first ten top-ranked primary INHBA interactors in the STRING protein-protein interaction (PPI) network, most genes were involved in the TGF-β signaling pathway. Previous studies have also demonstrated that INHBA plays a role in the regulation of cancer cell growth, proliferation, and survival via the TGF-β signaling pathway Chen et al. (2019); Yu et al. (2021); Basu et al. (2015). GO and GSEA analysis revealed that INHBA -related genes were associated with extracellular matrix, collagen formation and degradation, the misregulation of which is a key factor in epithelial to mesenchymal transition (EMT) Burke et al. (2015). Basu et al. Basu et al. (2015) also indicated that INHBA expression contributes to EMT in both normal and ovarian cancer cells. Several studies have shown that INHBA controls cell proliferation and apoptosis through the PI3K/Akt pathway Tsai et al. (2019); Loomans and Andl (2014), which is consistent with our KEGG pathway analysis.
The tumor microenvironment is a complex assembly of tumor, immune, stromal, and extracellular components, and it has emerged as an important component that contributes to tumor progression and metastasis Lee et al. (2019). Tumor-associated macrophages are considered essential components of the tumor microenvironment and play critical roles in the regulation of tumor progression Zhu et al. (2017). According to our research, there was a significant positive correlation between INHBA expression and macrophage infiltration. Additionally, INHBA has been shown to affect macrophage polarization in vitro Hreha et al. (2020). We hypothesized that INHBA may influence the tumor microenvironment by regulating macrophage polarization, which in turn affects tumor cells. Based on our research, mast cells, NK cells, neutrophils, Th2 cells, and eosinophils were also positively correlated with INHBA expression. However, the role of INHBA in the immune system is not fully understood, with only a handful of publications providing evidence for its involvement in T cell biology and neutrophils Locci et al. (2016); Sideras et al. (2013). Thus, a more comprehensive survey of the relationship between INHBA and immune infiltration is warranted.
In conclusion, we found that INHBA was overexpressed in cervical cancer and was significantly related to poor prognosis. INHBA may be involved in tumor progression and metastasis. Moreover, all the results indicated that INHBA is likely to play a key role in macrophage polarization and immune cell infiltration. Therefore, it could be used as a potential prognostic target for cervical cancer.
Although our study has uncovered some new facts, it also has the following limitations. First, our study is based on bioinformatics analyses, and the data are from public databases; therefore, it lacks the verification of clinical data of our hospital. Second, further validation studies with a long-term follow-up and larger cohorts of patients are needed to definitively validate INHBA as a prognostic predictor. Finally, further in-depth studies are required to address the relationship between INHBA gene expression and immune infiltration in more detail.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA data portal (https://portal.gdc.cancer.gov/), UCSC Xena (https://xenabrowser.net/datapages), and GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Author Contributions
KZ and WZ contributed to the study conception and design. Material preparation, data collection and analysis were performed by KZ, and ZM. YY and KZ contributed to the literature search. The first draft of the manuscript was written by KZ and all authors commented on previous versions of the manuscript. WZ and YY reviewed the paper and gave suggestions on the revision of the article. All authors read and approved the final manuscript.
Funding
The research was supported by the project of improving the ability of diagnosis and treatment of difficult diseases. Project No.: ZLYNXM202019.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.705512/full#supplementary-material
References
Antsiferova, M., Piwko‐Czuchra, A., Cangkrama, M., Wietecha, M., Sahin, D., Birkner, K., et al. (2017). Activin Promotes Skin Carcinogenesis by Attraction and Reprogramming of Macrophages. EMBO Mol. Med. 9, 27–45. doi:10.15252/emmm.201606493
Babion, I., Snoek, B., Novianti, P., Jaspers, A., van Trommel, N. V., Heideman, D., et al. (2018). Triage of High-Risk Hpv-Positive Women in Population-Based Screening by Mirna Expression Analysis in Cervical Scrapes; a Feasibility Study. Clin. Epigenetics 10, 1. doi:10.1186/s13148-018-0509-9
Basu, M., Bhattacharya, R., Ray, U., Mukhopadhyay, S., Chatterjee, U., and Roy, S. (2015). Invasion of Ovarian Cancer Cells Is Induced Bypitx2-Mediated Activation of Tgf-β and Activin-A. Mol. Cancer 14, 162. doi:10.1186/s12943-015-0433-y
Bellingham, M., Amezaga, M. R., Mandon-Pépin, B., Speers, C. J. B., Kyle, C. E., Evans, N. P., et al. (2013). Exposure to Chemical Cocktails before or after conception - the Effect of Timing on Ovarian Development. Mol. Cell Endocrinol. 376, 156–172. doi:10.1016/j.mce.2013.06.016
Burke, S. L., Hammell, M., and Ambros, V. (2015). Robust Distal Tip Cell Pathfinding in the Face of Temperature Stress Is Ensured by Two Conserved Micrornas in caenorhabditis Elegans. Genetics 200, 1201–1218. doi:10.1534/genetics.115.179184
Canfell, K., Kim, J. J., Brisson, M., Keane, A., Simms, K. T., Caruana, M., et al. (2020). Mortality Impact of Achieving Who Cervical Cancer Elimination Targets: a Comparative Modelling Analysis in 78 Low-Income and Lower-Middle-Income Countries. The Lancet 395, 591–603. doi:10.1016/s0140-6736(20)30157-4
Chen, Z. L., Qin, L., Peng, X. B., Hu, Y., and Liu, B. (2019). INHBA Gene Silencing Inhibits Gastric Cancer Cell Migration and Invasion by Impeding Activation of the TGF‐β Signaling Pathway Signaling Pathway. J. Cel Physiol 234, 18065–18074. doi:10.1002/jcp.28439
Dai, Y., Wang, Q., Gonzalez Lopez, A., Anders, M., Malfertheiner, P., Vieth, M., et al. (2018). Genome-Wide Analysis of Barrett's Adenocarcinoma. A First Step towards Identifying Patients at Risk and Developing Therapeutic Paths. Translational Oncol. 11, 116–124. doi:10.1016/j.tranon.2017.10.003
d’Aurora, M., Ferlin, A., Nicola, M. D., Garolla, A., Toni, L. D., Franchi, S., et al. (2015). Deregulation of Sertoli and Leydig Cells Function in Patients with Klinefelter Syndrome as Evidenced by Testis Transcriptome Analysis. BMC Genomics 16, 156. doi:10.1186/s12864-015-1356-0
Dogra, D., Ahuja, S., Kim, H.-T., Rasouli, S. J., Stainier, D., and Reischauer, S. (2017). Opposite Effects of Activin Type 2 Receptor Ligands on Cardiomyocyte Proliferation during Development and Repair. Nat. Commun. 8, 1. doi:10.1038/s41467-017-01950-1
Hadadi, E., Taylor, W., mei Li, X., Aslan, Y., Villote, M., Rivière, J., et al. (2020). Chronic Circadian Disruption Modulates Breast Cancer Stemness and Immune Microenvironment to Drive Metastasis in Mice. Nat. Commun. 11, 3193. doi:10.1038/s41467-020-16890-6
Hreha, T. N., Collins, C., Daugherty, A. L., Griffith, J. M., Hruska, K., and Hunstad, D. (2020). Androgen-influenced Polarization of Activin A-Producing Macrophages Accompanies post-pyelonephritic Renal Scarring. Front. Immunol. 11, 1641. doi:10.3389/fimmu.2020.01641
Kadara, H., Choi, M., Zhang, J., Parra, E., Rodriguez-Canales, J., Gaffney, S. G., et al. (2017). Erratum: Whole-Exome Sequencing and Immune Profiling of Early-Stage Lung Adenocarcinoma with Fully Annotated Clinical Follow-Up. Ann. Oncol. 29, 75. doi:10.1093/annonc/mdw436
Kalli, M., Mpekris, F., Wong, C., Panagi, M., Ozturk, S., Thiagalingam, S., et al. (2019). Activin a Signaling Regulates Il13rα2 Expression to Promote Breast Cancer Metastasis. Front. Oncol. 9, 32. doi:10.3389/fonc.2019.00032
Kaneda, H., Arao, T., Matsumoto, K., de Velasco, M. A., Tamura, M. D., Aomatsu, K., et al. (2011). Activin a Inhibits Vascular Endothelial Cell Growth and Suppresses Tumour Angiogenesis in Gastric Cancer. Br. J. Cancer 105, 1210–1217. doi:10.1038/bjc.2011.348
Kiran, S., Dar, A., Singh, S. K., Lee, K. Y., and Dutta, A. (2018). The Deubiquitinase USP46 Is Essential for Proliferation and Tumor Growth of HPV-Transformed Cancers. Mol. Cel 72, 823–835. e5. doi:10.1016/j.molcel.2018.09.019
Lee, H.-Y., Li, C., Huang, C.-N., ming Li, W., Yeh, H., Ke, H.-L., et al. (2015). Inhba Overexpression Indicates Poor Prognosis in Urothelial Carcinoma of Urinary Bladder and Upper Tract. J. Surg. Oncol. 111, 414. doi:10.1002/jso.23836
Lee, Y.-C., Kurtova, A., Xiao, J., Nikolos, F., Hayashi, K., Tramel, Z., et al. (2019). Collagen-rich Airway Smooth Muscle Cells Are a Metastatic Niche for Tumor Colonization in the Lung. Nat. Commun. 10, 1. doi:10.1038/s41467-019-09878-4
Li, X., Yu, W., Liang, C., Xu, Y., Zhang, M., Ding, X., et al. (2020). Inhba Is a Prognostic Predictor for Patients with colon Adenocarcinoma. BMC Cancer 20, 305. doi:10.1186/s12885-020-06743-2
Liang, H., Cheung, L. W. T., Li, J., Ju, Z., Yu, S., Stemke-Hale, K., et al. (2012). Whole-exome Sequencing Combined with Functional Genomics Reveals Novel Candidate Driver Cancer Genes in Endometrial Cancer. Genome Res. 22, 2120–2129. doi:10.1101/gr.137596.112
Liu, Y., Pandey, P. R., Sharma, S., Xing, F., Wu, K., Chittiboyina, A., et al. (2018). Id2 and Gjb2 Promote Early-Stage Breast Cancer Progression by Regulating Cancer Stemness. Breast Cancer Res. Treat. 175, 77–90. doi:10.1007/s10549-018-05126-3
Locci, M., Wu, J. E., Arumemi, F., Mikulski, Z., Dahlberg, C., Miller, A. T., et al. (2016). Activin a Programs the Differentiation of Human Tfh Cells. Nat. Immunol. 17, 976–984. doi:10.1038/ni.3494
Loomans, H., and Andl, C. (2014). Intertwining of Activin A and TGFβ Signaling: Dual Roles in Cancer Progression and Cancer Cell Invasion Signaling: Dual Roles in Cancer Progression and Cancer Cell Invasion. Cancers 7, 70–91. doi:10.3390/cancers7010070
Lyu, S., Jiang, C., Xu, R., Huang, Y., and Yan, S. (2018). Inhba Upregulation Correlates with Poorer Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Cmar 10, 1585–1596. doi:10.2147/cmar.s160186
Matsuo, K., Shimada, M., Aoki, Y., Sakamoto, M., Takeshima, N., Fujiwara, H., et al. (2017). Comparison of Adjuvant Therapy for Node-Positive Clinical Stage Ib-Iib Cervical Cancer: Systemic Chemotherapy versus Pelvic Irradiation. Int. J. Cancer 141, 1042. doi:10.1002/ijc.30793
Okano, M., Yamamoto, H., Ohkuma, H., Kano, Y., Kim, H., Nishikawa, S., et al. (2013). Significance of INHBA Expression in Human Colorectal Cancer. Oncol. Rep. 30, 2903–2908. doi:10.3892/or.2013.2761
Potikanond, S., Sookkhee, S., Takuathung, M. N., Mungkornasawakul, P., Wikan, N., Smith, D. R., et al. (2017). Kaempferia Parviflora Extract Exhibits Anti-cancer Activity against Hela Cervical Cancer Cells. Front. Pharmacol. 8, 630. doi:10.3389/fphar.2017.00630
Seder, C. W., Hartojo, W., Lin, L., Silvers, A. L., Wang, Z., Thomas, D. G., et al. (2009). Upregulated Inhba Expression May Promote Cell Proliferation and Is Associated with Poor Survival in Lung Adenocarcinoma. Neoplasia 11 (4), 388–396. doi:10.1593/neo.81582
Sideras, P., Apostolou, E., Stavropoulos, A., Sountoulidis, A., Gavriil, A., Apostolidou, A., et al. (2013). Activin, Neutrophils, and Inflammation: Just Coincidence? Semin. Immunopathol 35, 481–499. doi:10.1007/s00281-013-0365-9
Singh, P., Jenkins, L. M., Horst, B., Alers, V., Pradhan, S., Kaur, P., et al. (2018). Inhibin Is a Novel Paracrine Factor for Tumor Angiogenesis and Metastasis. Cancer Res. 78, 2978–2989. doi:10.1158/0008-5472.can-17-2316
Tournier, I., Marlin, R., Walton, K., Charbonnier, F., Coutant, S., Théry, J. C., et al. (2014). Germline Mutations of Inhibins in Early‐Onset Ovarian Epithelial Tumors. Hum. Mutat. 35, 294–297. doi:10.1002/humu.22489
Tsai, C.-N., Tsai, C.-L., Yi, J.-S., Kao, H., Huang, Y., Wang, C.-I., et al. (2019). Activin a Regulates the Epidermal Growth Factor Receptor Promoter by Activating the Pi3k/sp1 Pathway in Oral Squamous Cell Carcinoma Cells. Scientific Rep. 9, 1. doi:10.1038/s41598-019-41396-7
Wallbillich, J. J., Tran, P. M., Bai, S., Tran, L. K., Sharma, A. K., Ghamande, S. A., et al. (2020). Identification of a Transcriptomic Signature with Excellent Survival Prediction for Squamous Cell Carcinoma of the Cervix. Am. J. Cancer Res. 10, 1534–1547.
Yu, Y., Wang, W., Lu, W., Chen, W., and Shang, A. (2021). Inhibin β-A (INHBA) Induces Epithelial-Mesenchymal Transition and Accelerates the Motility of Breast Cancer Cells by Activating the TGF-β Signaling Pathway. Bioengineered 12, 4681–4696. doi:10.1080/21655979.2021.1957754
Zhu, Y., Herndon, J. M., Sojka, D. K., Kim, K.-W., Knolhoff, B. L., Zuo, C., et al. (2017). Tissue-resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 47, 323–338. e6. doi:10.1016/j.immuni.2017.07.014
Glossary
BLCA Bladder urothelial carcinoma
BRCA Breast invasive carcinoma
CESC Cervical squamous cell carcinoma and endocervical adenocarcinoma
CHOL Cholangiocarcinoma
COAD Colon adenocarcinoma
DLBC Lymphoid Neoplasm Diffuse Large B-cell Lymphoma
DSS disease Specific Survival
ESCA Esophageal carcinoma; GBM: Glioblastoma multiforme
GEO Gene Expression Omnibus
GSEA gene-set enrichment analysis
GO Gene Ontology
KEGG Kyoto Encyclopedia of genes and genomes
HNSC Head and neck squamous cell carcinoma
INHBA Inhibin A
LAML Acute myeloid leukemia
LGG Lower grade glioma
LIHC Lever hepatocellular carcinoma
LUAD Lung adenocarcinoma
LUSC Lung squamous cell carcinoma
KICH Kidney chromophobe
KIRC Kidney renal clear cell carcinoma
KIRP Kidney renal papillary cell carcinoma
OS Overall Survival
OV Ovarian serous cystadenocarcinoma
PAAD Pancreatic adenocarcinoma
PCPG Pheochromocytoma and paraganglioma
PFI Progress Free Interval
PRAD Prostate adenocarcinoma
READ Rectum adenocarcinoma
SKCM Skin cutaneous melanoma
STAD Stomach adenocarcinoma
TCGA The Cancer Genome Atlas
TGCT Testicular germ cell tumor
THCA Thyroid carcinoma
THYM Thymoma
UCEC Uterine corpus endometrial carcinoma
UCS Uterine carcinosarcoma
Keywords: INHBA, biomarker, cervical cancer, prognosis, immune infiltration
Citation: Zhao K, Yi Y, Ma Z and Zhang W (2022) INHBA is a Prognostic Biomarker and Correlated With Immune Cell Infiltration in Cervical Cancer. Front. Genet. 12:705512. doi: 10.3389/fgene.2021.705512
Received: 05 May 2021; Accepted: 06 December 2021;
Published: 04 January 2022.
Edited by:
Nicholas Anagnou, Biomedical Research Foundation of the Academy of Athens (BRFAA), GreeceReviewed by:
Olga Vadimovna Kurmyshkina, Petrozavodsk State University, RussiaMusalula Sinkala, University of Cape Town, South Africa
Copyright © 2022 Zhao, Yi, Ma and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhang, em4wMDI2NDZAd2h1LmVkdS5jbg==
 Kaidi Zhao
Kaidi Zhao Yuexiong Yi
Yuexiong Yi