
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 10 September 2021
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.703688
This article is part of the Research TopicGenomics in Flower Development: From ‘Omics’ to Functional CharacterizationView all 11 articles
 Yuanyuan Jiang1,2†
Yuanyuan Jiang1,2† Yicun Liu3†
Yicun Liu3† Yongshun Gao2,4,5
Yongshun Gao2,4,5 Jiangrong Peng2
Jiangrong Peng2 Wenbing Su2,6
Wenbing Su2,6 Yuan Yuan1
Yuan Yuan1 Xianghui Yang2
Xianghui Yang2 Chongbin Zhao2
Chongbin Zhao2 Man Wang2
Man Wang2 Shunquan Lin2*
Shunquan Lin2* Ze Peng2*
Ze Peng2* Fangfang Xie2*
Fangfang Xie2*Flowering is an integral part of the life cycle of flowering plants, which is essential for plant survival and crop production. Most woody fruit trees such as apples and pears bloom in spring, but loquat blooms in autumn and winter. Gibberellin (GA) plays a key role in the regulation of plant flower formation. In this study, we sprayed loquat plants with exogenous GA3, which resulted in vigorous vegetative growth rather than floral bud formation. We then performed a comprehensive RNA-seq analysis on GA3-treated and control-treated leaves and buds over three time periods to observe the effects of exogenous GA3 application on floral initiation and development. The results showed that 111 differentially expressed genes (DEGs) and 563 DEGs were down-regulated, and 151 DEGs and 506 DEGs were up-regulated in buds and leaves, respectively, upon treatment with GA3. Among those that are homologs of the DELLA-mediated GA signal pathway genes, some may be involved in the positive regulation of flower development, including EjWRKY75, EjFT, EjSOC1, EjAGL24, EjSPL, EjLFY, EjFUL, and EjAP1; while some may be involved in the negative regulation of flower development, including EjDELLA, EjMYC3, EjWRKY12, and EjWRKY13. Finally, by analyzing the co-expression of DEGs and key floral genes EjSOC1s, EjLFYs, EjFULs, EjAP1s, 330 candidate genes that may be involved in the regulation of loquat flowering were screened. These genes belong to 74 gene families, including Cyclin_C, Histone, Kinesin, Lipase_GDSL, MYB, P450, Pkinase, Tubulin, and ZF-HD_dimer gene families. These findings provide new insights into the regulation mechanism of loquat flowering.
The floral transformation of plants is affected by various endogenous and exogenous factors, forming a very sophisticated and complex regulatory network. It can accurately respond to internal and external signals and integrate them together to ensure that plants bloom at a favorable time and reproduce successfully. Plants can accurately sense photoperiod changes and adjust flowering time (Shim et al., 2017); in addition, gibberellin (GA), temperature, vernalization and age signals can also affect plant flowering (Moon et al., 2005; Amasino, 2010; Song et al., 2013; Teotia and Tang, 2015). These signals are not independent. In Arabidopsis, they integrate related signals to regulate plant flower formation through integrators FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), LEAFY (LFY), etc. (Blazquez et al., 1998; Kardailsky et al., 1999; Lee et al., 2000).
In Arabidopsis, the GA signal mainly regulates the flower formation of plants through the interaction with the photoperiod signal and regulates the expression of FT under the conditions of LD and SD (Osnato et al., 2012; Song et al., 2012). In leaves, MYB-type transcription factor ASYMMETRIC LEAVES positively regulates the expression of GA biosynthesis gene GA20ox1 (Song et al., 2012). AS1 forms a complex with CO protein and regulates FT expression by directly binding to the FT promoter (Song et al., 2012). As a central inhibitor of the GA signaling pathway, DELLA has been proven to interact with many transcription factors in leaves and stem tips and regulate their activities, thereby regulating plant flowering (Bao et al., 2020). For example, under long-day conditions, DELLA directly binds to the CCT domain of the CO protein and sequentially separates CO from the binding of the FT promoter, thereby down-regulating the expression of FT (Wang et al., 2016; Xu et al., 2016). In addition, DELLA can inhibit the interaction between CO and Nuclear factor Y (NF-Y) subunit B (NF-YB), and DELLA can also regulate the expression of FT through the interaction between PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and MYC3 (de Lucas et al., 2008; Xu et al., 2016; Bao et al., 2019, 2020).
In addition, GA can directly promote flowering by up-regulating flowering integrons LFY, SOC1 and AGAMOUS-LIKE 24 (AGL24) independently of the photoperiod pathway (Blazquez and Weigel, 2000; Moon et al., 2003; Hisamatsu and King, 2008; Liu et al., 2008). Hou et al. (2014) found that GA signal can regulate the expression of SOC1 through epigenetic modification mediated by NF-Y complex. Under short-day conditions, the promoter activity of LFY gradually increased during vegetative growth, and GA enhanced the promoter activity and accelerated plant flowering (Blazquez et al., 1998). GA mainly regulates miR159 by inhibiting the expression of DELLA protein, thereby regulating the transcription of downstream MYB33, ultimately regulating the expression of LFY, and regulating the floral transformation of plants (Blazquez and Weigel, 2000; Gocal et al., 2001; Achard et al., 2009; Davis, 2009). DELLA may recruit different SPLs to target various downstream target genes, so that GA can play a role in different development environments (Bao et al., 2020). In addition, GA signal can regulate the expression of WRKY12, WRKY13 and WRKY75 genes through DELLA protein to regulate plant flower formation (Li et al., 2016; Zhang et al., 2018). In summary, the role of GA signal in the regulation of plant flower formation is very important and complex, but there are relatively few studies on gibberellin-mediated flower formation in woody fruit trees.
Loquat (Eriobotrya japonica Lindl.) is an evergreen fruit tree, which belongs to the Maloideae subfamily of the Rosaceae family which is mainly planted in subtropical regions. In Rosaceae, the flowering transition time and flowering time usually occur in different years (flower buds differentiate in summer and autumn, and the flower buds bloom in the spring of the second year after dormancy), including apples, pears, and strawberries (Kurokura et al., 2013). However, the flowering transformation and flowering of loquat occur in the same year. Flower bud differentiation generally occurs from July to September, with flowering occurring between October and January of the same year (Lin, 2007). The phenomenon of autumn flowering and spring harvest of loquat is very unique among woody fruit trees. In spring, the selection of fresh fruit varieties is greatly reduced. Therefore, the market demand for fresh loquat fruits in spring is relatively high and the price is relatively high. However, in cold winters (especially extreme weather events), the newly opened loquat flowers or young fruits are very susceptible to freezing damage (Peng et al., 2021), resulting in a reduction in loquat production or even no harvest.
Recent research results show that although the start time of loquat flower bud differentiation is similar to that of apples and pears, it occurs from June to July; the difference is that the development of loquat flower buds is continuous and does not undergo dormancy, and it blooms in autumn and winter (Jiang et al., 2019c). In addition, after treating loquat plants with the exogenous hormone GA3, the plants are in vigorous vegetative growth and cannot form flowers, and genes such as EjSOC1s, EjAP1s and EjLFYs are strongly inhibited (Jiang et al., 2019a,b,c). These studies show that gibberellin can regulate the flowering of loquat by regulating the expression of genes such as EjSOC1s, EjAP1s and EjLFYs. In Arabidopsis, SOC1, AP1 and LFY genes are all downstream of the floral regulation network, and GA regulates their expression through the inhibition or activation of other transcription factors mediated by DELLAs (Bao et al., 2020). However, the mechanism of gibberellin regulating loquat flowering is not clear. Transcriptome sequencing technology is based on exogenous sequencing technology to quickly and comprehensively understand the difference level of transcripts. The application of transcriptome sequencing technology has accelerated the gene expression profile analysis and gene identification of many plant species. In this study, transcriptome analysis was performed on the materials of the GA3 treatment and the control group to screen the key genes related to the regulation of loquat flowering, in order to analyze the regulation mechanism of loquat flowering.
The loquat tissue materials involved in the experiment were taken from 12-year-old “JieFangZhong” loquat plants in the loquat plant germplasm resource nursery of South China Agricultural University (Guangzhou, China, N23°09′N, 113°20′E). The experimental plants have entered the flowering and fruiting age for several years and have grown well. Loquat trees are planted in the loquat germplasm resource nursery and grow under natural conditions.
The trees were sprayed with an aqueous solution containing 0.1% (v/v) phosphoric acid and 0.025% (v/v) Triton X-100 as a surfactant and 300 mg L–1 GA3 (Dingguo Biotechnology Co., Ltd., Guangzhou, China). Spray the control plants with a solution containing only 0.1% (v/v) phosphoric acid and 0.025% (v/v) Triton X-100. The experimental treatment method was: spraying all leaves and top buds (soaked, the leaves began to drip) every 2 weeks, from May 18th to August 10th. The differentiation time of loquat flower buds is from the end of June to the beginning of July (Jiang et al., 2019c). Accordingly, the sampling time points for the GA3 treatment group and the control group were set as: May 25, June 29, and August 17. Mature leaves and apical buds were used in this experiment (randomly mixed with tissue samples with the same maturity in different directions and different heights of the plant, as a biological repeat). Samples of the treatment group and the control group were taken at the same time. The sample was placed in a clean centrifuge tube that has been marked, immediately frozen and stored in liquid nitrogen, and then stored in an −80°C refrigerator until use. Three independent biological replicates (the biological replicates were from separate plants) were performed for each treatment.
Total RNA were extracted following the manufacturer of the RNA Prep Pure Plant Kit (TIANGEN, China). Their purity and integrity were checked and assessed using the NanoPhotometer® spectrophotometer (IMPLEN, CA, United States) and RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, United States), respectively. Subsequently, total 1 μg RNA of each sample was used as input material for the RNA sample preparations. mRNA was purified using poly-T oligo-attached magnetic beads. First-strand and second-strand cDNA was synthesized according to the manufacturer of M-MuLV Reverse Transcriptase (RNase H–), and DNA Polymerase I and RNase H–, respectively. cDNA fragments of 250∼300 bp in length were selected and purified with AMPure XP system (Beckman Coulter, Beverly, MA, United States). Besides, the library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated.
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. Clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. Q20, Q30, GC content, and sequence duplication levels in the clean data were calculated (Supplementary Table 1). All the downstream analyses were based on the clean data with high quality. Raw reads of the RNA-seq data are uploaded to Sequence Read Archive (SRA) database of NCBI with Bioproject ID number PRJNA729650.
Reference genome and gene model annotation files were downloaded from genome website (Su et al., 2021). The building of index of the reference genome, and the alignment between clean reads and reference genome all using Hisat2 (version 2.0.5). FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) of each gene was calculated by featureCounts (version 1.5.0) (Florea et al., 2013).
DEGs (Differentially expressed genes) were defined by DESeq2 R package (version 1.16.1) with an adjusted P-value < 0.05. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (Benjamini and Hochberg, 1995). GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways enrichment analysis of DEGs was implemented by the clusterProfiler R package. Volcano plots, Venn diagrams and heatmaps were drawed by TBtools (Chen C. et al., 2020). WGCNA (weighted gene co-expression network analysis) was performed in R with the WGCNA package (Langfelder and Horvath, 2008) and visualized the networks by Cytoscape (version 3.8.2) (Shannon et al., 2003).
At 10–20 days after treating loquat plants with 300 mg L–1 GA3, vigorous vegetative growth was observed, and the stems grew rapidly. In the beginning of September when obvious inflorescence could be observed in the control group, plants in the GA3 treatment group were still in vigorous vegetative growth (Figures 1A,B).
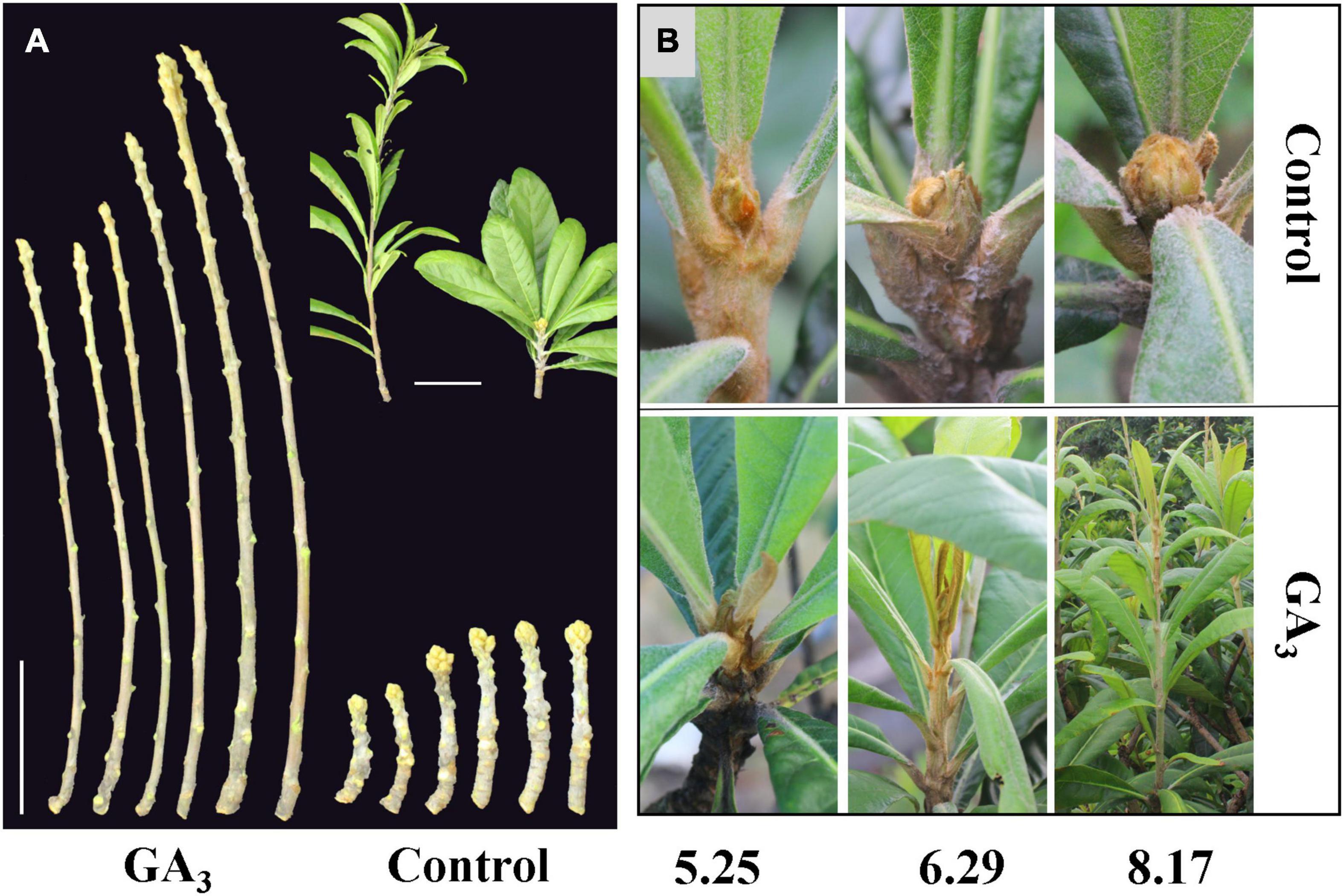
Figure 1. The phenotype of loquat in the GA3 treatment group and the control group. (A) GA3 treatment promotes the growth of stems, bars = 10 cm; (B) Phenotypes of samples used for transcriptome sequencing at different stages.
Transcriptome sequencing results showed that 41.68–56.71 Mb clean reads were obtained from the 18 samples in the control group; 41.39–61.024 Mb clean reads were obtained from the 18 samples in the GA3 treatment group. The GC content of the GA3 treatment group and the control group was similar, ranging from 45.59 to 47.08% (Supplementary Table 1). For 36 samples, 96.70% of the bases had a quality score greater than 20, and Q30 ≥ 91.39%. The sequencing data are of high quality and can meet the requirements of subsequent analysis.
Compared with the control group, there were 1,901 down-regulated DEGs and 1,268 up-regulated DEGs in the buds 7 days after GA3 treatment on May 25th (Figure 2 and Supplementary Tables 2-1, 2); 1,867 down-regulated DEGs and 2,003 up-regulated DEGs in the buds on June 29th (Figure 2 and Supplementary Tables 2-3, 4); 5,172 down-regulated DEGs and 5,383 up-regulated DEGs in the buds on August 17th (Figure 2 and Supplementary Tables 2-5, 6). In leaves, compared with the control group, there were 7,052 down-regulated DEGs and 7,079 up-regulated DEGs in the leaves 7 days after GA3 treatment on May 25th (Figure 2 and Supplementary Tables 3-1, 2); 4,137 down-regulated DEGs and 4,097 up-regulated DEGs in the leaves on June 29th (Figure 2 and Supplementary Tables 3-3, 4); 3,207 down-regulated DEGs and 4,012 up-regulated DEGs in the leaves on August 17th (Figure 2 and Supplementary Tables 3-5, 6).
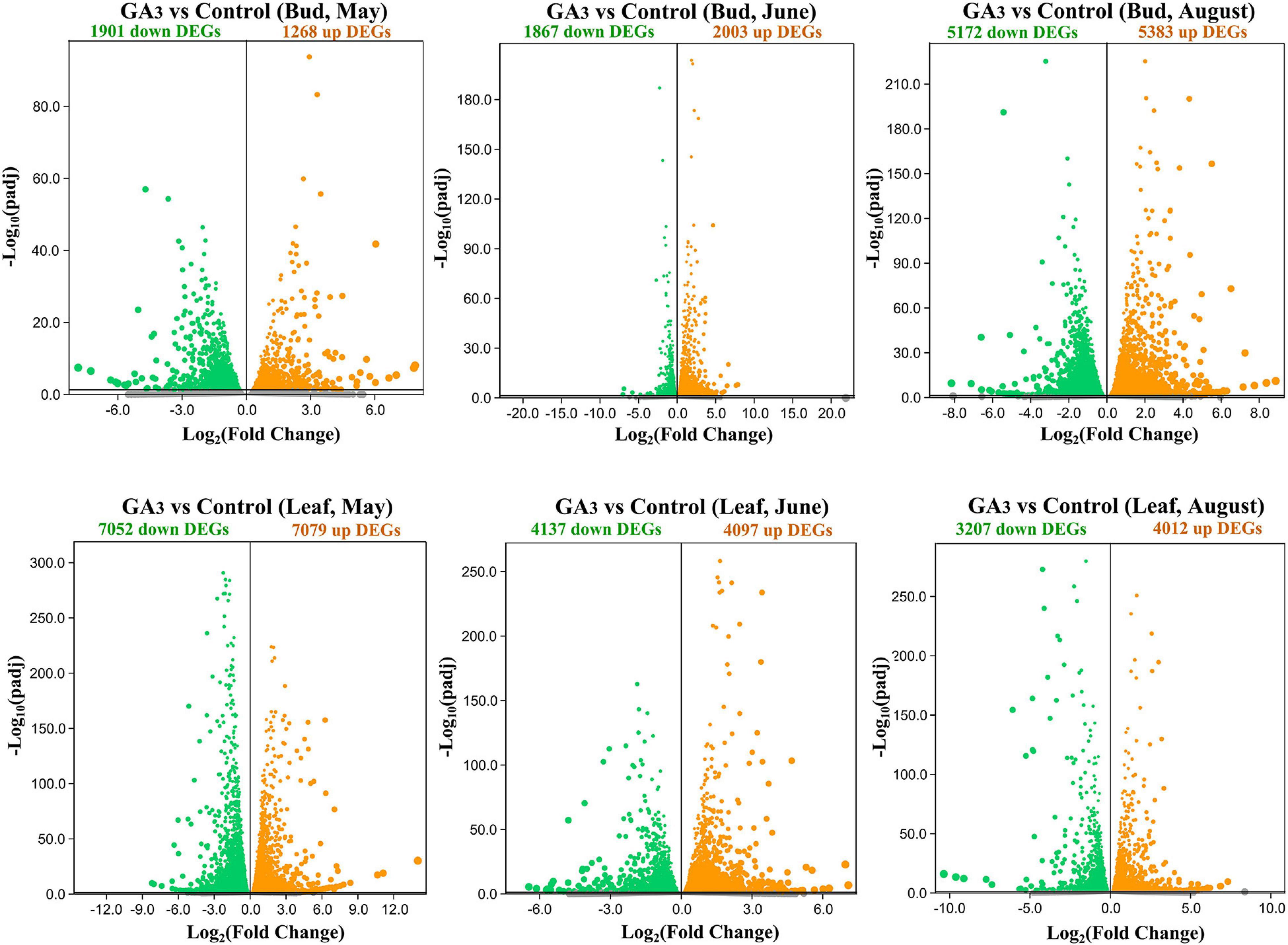
Figure 2. Volcano map of DEGs in the buds and leaves between the GA3 treatment group and the control group. Orange dots indicate up-regulated DEGs, and green dots indicate down-regulated DEGs, and gray dots indicate genes that were not differential expressed.
The results showed that compared with the control group, the number of DEGs in the apical buds of the GA3 treatment group increased rapidly from June 29th to August 17th (Figure 2). It implies that during this period, GA3 regulates the flower bud differentiation of loquat by up-regulating or down-regulating the expressions of a large number of flowering-related genes. In comparison, highly number of DEGs were expressed in May in leaves before the bud differentiation of loquat (Figure 2).
In order to explore how loquat responds to GA3 signals, GO and KEGG enrichment analysis were performed on DEGs the compare between GA3 treatment and control of buds and leaves. GO enrichment showed that DEGs of buds mainly involved in biological signal binding and catalytic activity, such as heme binding, tetrapyrrole binding, hydrolase activity, acting on glycosyl bonds, etc. (Figure 3 and Supplementary Tables 4-1, 2, 3). In leaves, DEGs mainly involved in metabolism, transcriptional activity, and biosynthesis, such as peptide metabolic process, amide biosynthetic process, peptide biosynthetic process, nucleic acid binding transcription factor activity, transcription factor activity (sequence-specific DNA binding), etc. (Supplementary Figure 1 and Supplementary Tables 4-4, 5, 6).
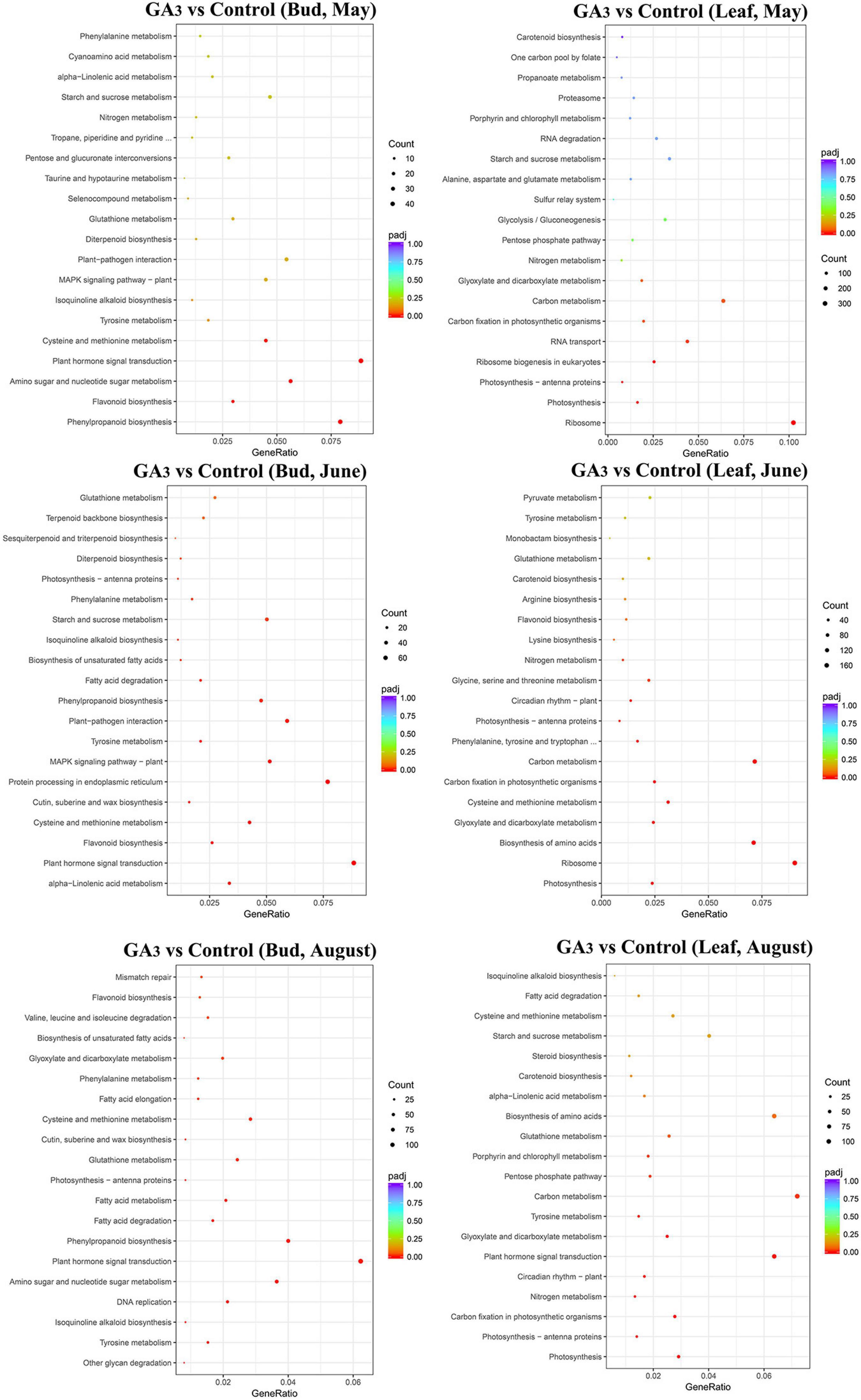
Figure 3. KEGG analysis of DEGs in different tissues of GA3 treatment group and control group at different periods.
The KEGG annotation shows that the 841 DEGs in the buds 7 days after GA3 treatment were enriched in 110 KEGG pathways, among which plant hormone signal transduction (47 genes, 5.59%), phenylpropanoid biosynthesis (42 genes, 4.99%), and amino acids biosynthesis (35 genes, 4.16%) were significantly enriched pathways (Figure 3 and Supplementary Table 5-1). Total of 1,278 DEGs in the buds on June 29th were enriched in 115 KEGG pathways, among which plant hormone signal transduction (70 genes, 5.48%), protein processing in endoplasmic reticulum (61 genes, 4.77%), and plant-pathogen interaction (47 genes, 3.68%) were significantly enriched pathways (Figure 3 and Supplementary Table 5-2). Total of 3,327 DEGs in the buds on August 17th were enriched in 120 KEGG pathways, among which carbon metabolism (125 genes, 3.76%), ribosome (125 genes, 3.76%), and plant hormone signal transduction (124 genes, 3.73%) were significantly enriched pathways (Figure 3 and Supplementary Table 5-3).
In leaves, KEGG annotation shows that the 4,627 DEGs in the leaves on May 25th were enriched in 120 KEGG pathways, among which ribosome (302 genes, 6.53%), carbon metabolism (189 genes, 4.08%), biosynthesis of amino acids (153, 3.31%), and plant hormone signal transduction (137 genes, 2.96%) were significantly enriched pathways (Figure 3 and Supplementary Table 5-4). A total of 3,059 DEGs in the leaves on June 29th were enriched in 120 KEGG pathways, among which ribosome (171 genes, 5.59%), carbon metabolism (136 genes, 4.45%), biosynthesis of amino acids (135 genes, 4.41%), and plant hormone signal transduction (89 genes, 2.91%) were significantly enriched pathways (Figure 3 and Supplementary Table 5-5). 2,395 DEGs in the leaves on August 17th were enriched in 119 KEGG pathways, among which carbon metabolism (105 genes, 4.38%), plant hormone signal transduction (93 genes, 3.88%), biosynthesis of amino acids (93 genes, 3.88%), and ribosome (84 genes, 3.51%) were significantly enriched pathways (Figure 3 and Supplementary Table 5-6).
The results showed that DEGs were mainly enriched in the plant hormone signal transduction pathway after GA3 treatment, which also indicated that after GA3 treatment, loquats mainly responded to GA3 signals through these DEGs, and ultimately participated in the regulation of loquat flower bud differentiation.
In order to further understand the expression patterns of genes related to flower bud differentiation, we performed a cluster analysis on the selected DEGs. It was revealed that after GA3 treatment, 111 DEGs were down-regulated in the three stages of buds (Figures 4A,C and Supplementary Table 6-1) and 151 DEGs were up-regulated (Figures 4B,D and Supplementary Table 6-2). In addition, we found in the leaves that 563 DEGs were down-regulated in the three stages after GA3 treatment (Figure 4A and Supplementary Table 6-3), and 506 DEGs were up-regulated (Figure 4B and Supplementary Table 6-4).

Figure 4. DEGs analysis of loquat leaves and buds transcriptome datasets. (A) Venn diagrams of DEGs that are down-regulated in leaves and buds at different stages; (B) Venn diagrams of DEGs that are up-regulated in leaves and buds at different stages; (C) The expression patterns of 111 down-regulated DEGs in buds; (D) The expression patterns of 151 up-regulated DEGs in buds. a, b, and c respectively represent the material on May 25th, June 29th, and August 17th after GA3 treatment; d, e, and f represent the control group materials on May 25th, June 29th, and August 17th, respectively.
Through the analysis of all down-regulated and up-regulated DEGs in buds with annotation results, it was found that 44 down-regulated DEGs belonged to 27 gene families (Supplementary Figure 2A); while the 58 up-regulated DEGs belonged to 28 gene families (Supplementary Figure 2B) including AP2, bZIP, F-box, MYB, WRKY and other gene family genes. The flower buds of loquat cannot differentiate after GA3 treatment, which also implies that the down-regulated DEGs after treatment are possibly positive-regulatory genes involved in loquat flowering, and these up-regulated DEGs may be negative-regulatory genes for loquat flowering.
In Arabidopsis thaliana, the GA pathway genes involved in the regulation of flower formation mainly include DELLA, CO, MYC3, WRKY75, WRKY12, WRKY13, SOCl, SPL, FT, AGL24, LFY, FUL, AP1, etc. (Bao et al., 2020). Thirty-three homologous genes in loquat were obtained through sequence alignment by blast the transcripts of loquat and coding sequence of Arabidopsis thaliana genome (Figure 5 and Supplementary Table 7). Based on the expression patterns of these homologous genes in the GA3 treatment group and the control group, thirteen candidate genes, including EjWRKY75 (Eri011414), EjFT (Eri036481), EjSOC1 (Eri012338, Eri023104), EjAGL24 (Eri026753), EjSPL (Eri001949, Eri003494), EjLFY (Eri007397, Eri022269), EjFUL (Eri009416, Eri033768), EjAP1 (Eri000407, Eri030184), were highly expressed in control group than GA3 treatment group in buds, and may be involved in the positive regulation of flower development. Besides, eleven candidate genes, including EjDELLA (Eri016831, Eri029753, Eri030473, Eri031405, Eri038200, Eri038624), EjMYC3 (Eri010051, Eri034229), EjWRKY12 (Eri034804), EjWRKY13 (Eri012544, Eri035481), were highly expressed in GA3 treatment group than control group in buds, and may be involved in the negative regulation of flower development. However, we found that the expressions of EjCOs in leaves did not decrease but increased after GA3 treatment. It suggests that the EjCOs in loquat may be mainly regulated by photoperiod, rather than regulating loquat flower development by responding to GA3 signals.
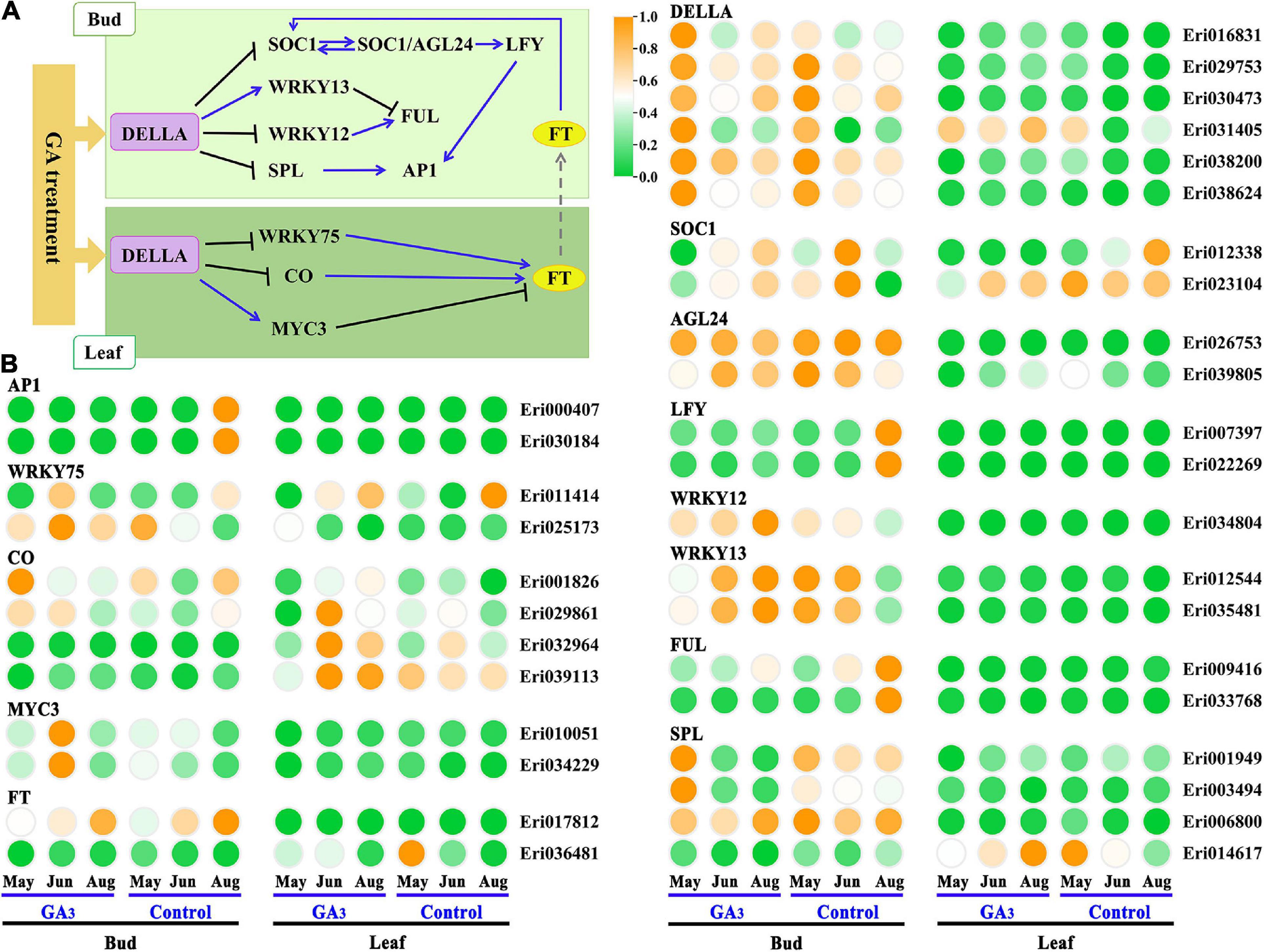
Figure 5. Expression analysis of homologous genes from DELLA-mediated GA signal regulatory pathway in loquat. (A) DELLA-mediated GA signal regulation pathway in Arabidopsis. (B) The expression pattern of homologous genes from the GA pathway in loquat leaves and buds.
EjSOC1s play an active role in the flowering process of loquat, EjAP1s and EjLFYs can be used as marker genes for loquat flowering regulation (Jiang et al., 2019c). Our experimental results also further confirmed this conclusion. In Arabidopsis, SOC1 or LFY activates the expressions of floral meristem identity genes LFY, AP1, and FUL to initiate flower bud differentiation (Blazquez et al., 1998; Blazquez and Weigel, 2000).
In order to further understand the regulatory network of loquat flowering genes and the regulatory relationship of flowering-related genes, WGCNA analysis was carried out to investigate the co-expression networks of DEGs in the transcriptome data, in which all the co-expressed genes were connected to each other with varying association strengths. DEGs are clustered into 27 modules according to the expression patterns (Figure 6A), interestingly, EjSOC1-2 (Eri023104), EjLFY-1 (Eri007397), EjLFY-2 (Eri022269), EjFUL-1 (Eri009416), EjFUL-2 (Eri033768), EjAP1-1 (Eri000407) and EjAP1-2 (Eri030184) were assigned to the brown module, except for EjSOC1-1 (Eri012338) in the yellow module (weight < 0.1). Further analysis of the seven genes in the brown module revealed that 330 genes were co-expressed with them (Figure 6B and Supplementary Table 8), belonging to 74 gene families, including Cyclin_C, Histone, Kinesin, Lipase_GDSL, MYB, P450, Pkinase, Tubulin, ZF-HD_dimer gene family (Figure 6C). It showed that their expression patterns are similar to that of EjSOC1-2, EjLFYs, EjFULs, EjAP1s, and they are all inhibited by GA3 treatment. It implied that these genes may play a positive regulatory role in the flowering of loquat. Therefore, these data provide new directions and useful candidate genes for research on loquat flowering regulation.
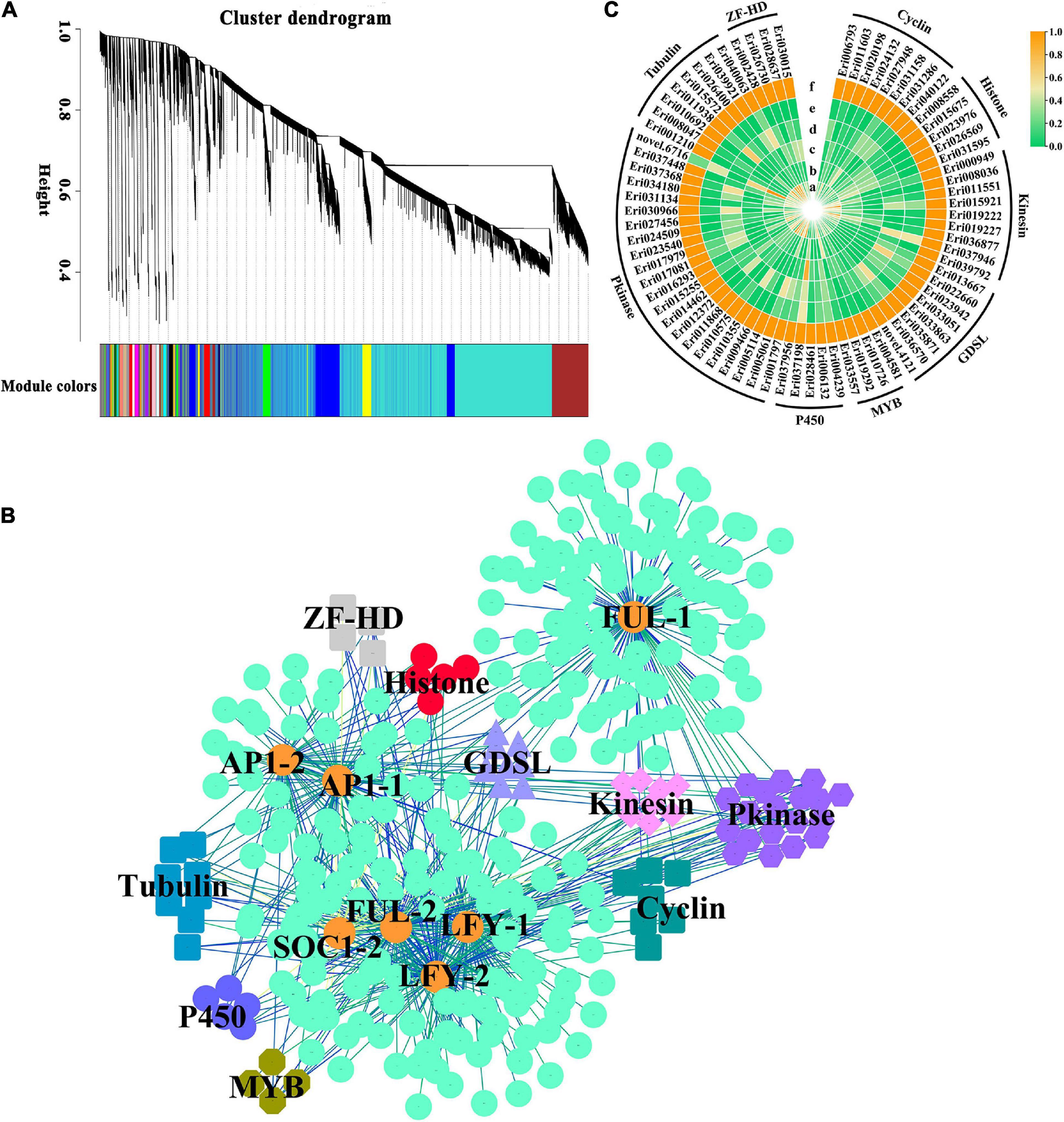
Figure 6. WGCNA co-expression analysis of homolog genes from DELLA-mediated GA signal regulatory pathway in loquat. (A) Module trait relationships in the GA3 treatment group and the control group. (B) The co-expression network connecting the key genes in DELLA-mediated GA signal regulatory pathway with candidate genes in brown module. Expression associations between homolog genes (orange circle) from GA pathway and candidate genes (other shapes except orange circle) are indicated with colored lines according to weight. (C) The expression pattern of candidate genes in loquat buds.
The Rosaceae family contains many fruit crop species, such as apple, pear, peach, strawberry and loquat. However, the unique flowering time of autumn flowering and spring fruit ripening of cultivated loquat suggests that it may have a flowering regulation system different from other woody plants in Rosaceae family during the evolutionary process. Although the flowering habit of loquat is very special, there are few reports about it. So far, several flowering-related genes have been cloned from cultivated loquat, including EjAP1, EjAP3, EjFT, EjLFY, EjSOC1, EjSVP, EjSPL, EjFRI and EjTFL1 (Esumi et al., 2005; Liu et al., 2013, 2017; Reig et al., 2017; Jiang et al., 2019a,b,c; Chen W. et al., 2020). EdCO, EdGI, EdFT and EdFD have been cloned from wild loquat Eriobotrya deflexa Nakai forma koshunensis (Zhang L. et al., 2016; Zhang et al., 2019). However, the loquat floral regulation network is still unclear. This study enriched the loquat floral regulation network resources, which can provide an important reference for further analysis of the molecular mechanism of loquat flowering, and also provide a theoretical basis for the later research on the floral regulation of Rosaceae.
The two hormone systems “gibberellin” and “florigen” (FT) play a key role in crop yield and quality (Eshed and Lippman, 2019). Previous studies have shown that GA3 treatment can inhibit the flower bud induction of nectarine, sweet cherry, mango, citrus and apple (García-Pallas et al., 2001; Lenahan et al., 2006; Nakagawa et al., 2012; Goldberg-Moeller et al., 2013; Zhang S. et al., 2016). The miFT in mangoes can regulate the flowering by responding to GA signals (Nakagawa et al., 2012); in citrus (Citrus reticulata Blanco × Citrus temple Hort. ex Y. Tanaka), FT, AP1 and a few flower-organ-identity genes are inhibited by GA, but GA promotes the expression of LFY (Goldberg-Moeller et al., 2013); The expression levels of MdSPLs, MdFT, MdSOC1 and MdAP1 genes in apples are all inhibited by the GA3 treatment (Zhang S. et al., 2016). In this study, we also found similar conclusions to that from these woody fruit trees. For example, GA3 treatment inhibited the flowering of loquat, and the expressions of genes such as EjFT, EjSOC1, EjSPL, and EjAP1 were inhibited. But different from the expression in citrus, the expression of EjLFY in loquat was inhibited by GA3. Our results also showed that some genes are different from the expression patterns in model plants, such as EjMYC3, which mainly plays a regulatory role in Arabidopsis leaves, while the difference in expression of EjMYC3 in loquat after GA3 treatment mainly occurs in buds. There is little difference in expression in leaves, suggesting that it mainly plays a role in regulating flowering in buds. In addition, we found that the expressions of several EjCO genes did not decrease but increased after GA3 treatment. We speculated that it may be mainly through response to photoperiod signals to regulate flower formation, rather than GA signals.
In this study, by analyzing all the down-regulated and up-regulated DEGs in the apical buds of different periods, some candidate genes that may participate in loquat flower bud differentiation were screened, including AP2, bZIP, F-box, MYB, WRKY and other gene families (Supplementary Figure 2). In addition, through WGCNA analysis, candidate genes co-expressed with EjSOC1-2, EjLFYs, EjFULs, EjAP1s, and possibly involved in loquat flower bud differentiation, including Cyclin_C, Histone, Kinesin, Lipase_GDSL, MYB, P450, Pkinase, Tubulin, ZF- HD_dimer gene family. The discovery of these candidate genes has brought great convenience to the subsequent study of loquat flower formation. Future work will focus on verifying and analyzing the functions and mechanisms of these candidate genes in the formation of loquat flowers.
Referring to the DELLA-mediated GA signal regulation network diagram in Arabidopsis (Figure 6A), we constructed a hypothetical model for loquat flowering regulation network based on the expression patterns of related homologous genes in loquat (Figure 7). As follows: In leaves, GA3 promotes the expression of DELLA family genes (Eri031405) to inhibit the expression of EjWRKY75 (Eri011414), and then EjWRKY75 down-regulates the expression of EjFT (Eri036481), and finally the EjFT protein is transported to SAMs to take effect. In SAMs, GA3 promotes the expression of DELLA family genes (Eri016831, Eri029753, Eri030473, Eri038200, and Eri038624). On the one hand, DELLA family genes inhibit the expression of EjFUL (Eri009416, Eri033768) by promoting the expression of EjWRKY12 (Eri034804) and EjWRKY13 (Eri012544, Eri035481). On the other hand, DELLA family genes inhibited the expression of flower-specific genes EjLFY (Eri007397, Eri022269) and EjAP1 (Eri000407, Eri030184) by inhibiting the expression of EjSOC1 (Eri012338, Eri023104), EjAGL24 (Eri026753) and EjSPL (Eri001949, Eri003494). Finally, the flower development of loquat was inhibited.
The original contributions presented in the study are publicly available in NCBI using accession number PRJNA729650.
YJ, YL, and FX designed the research. YJ and FX mainly performed the research. YG, JP, WS, YY, XY, CZ, MW, and SL contributed reagents, materials, and analysis tools. YJ, FX, and ZP wrote the manuscript. ZP and FX revised and approved the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Natural Science Foundation of Guangdong Province (2021A1515011048), Young Innovative Talents Project in Ordinary Colleges and Universities in Guangdong Province (2020KQNCX075), University-level scientific research project of Shaoguan University (SZ2019ZK11), Shaoguan City Science and Technology Planning Project (Social Development Direction-Supporting Scientific Research Workers Project) (200811094530739), and Collaborative innovation project from Fujian Provincial People’s Government and Chinese Academy of Agricultural Sciences (KXXYJBG2021006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past collaboration with one of the author YJ.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.703688/full#supplementary-material
Achard, P., Gusti, A., Cheminant, S., Alioua, M., Dhondt, S., Coppens, F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193. doi: 10.1016/j.cub.2009.05.059
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Bao, S., Hua, C., Huang, G., Cheng, P., Gong, X., Shen, L., et al. (2019). Molecular basis of natural variation in photoperiodic flowering responses. Dev Cell. 50, 90–101.e3. doi: 10.1016/j.devcel.2019.05.018
Bao, S., Hua, C., Shen, L., and Yu, H. (2020). New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62, 118–131. doi: 10.1111/jipb.12892
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Blazquez, M. A., Green, R., Nilsson, O., Sussman, M. R., and Weigel, D. (1998). Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell. 10, 791–800.
Blazquez, M. A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. doi: 10.1038/35009125
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Wang, P., Wang, D., Shi, M., Xia, Y., He, Q., et al. (2020). EjFRI, FRIGIDA (FRI) ortholog from Eriobotrya japonica, delays flowering in Arabidopsis. Int J Mol Sci. 21:1087. doi: 10.3390/ijms21031087
Davis, S. J. (2009). Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 32, 1201–1210. doi: 10.1111/j.1365-3040.2009.01968.x
de Lucas, M., Daviere, J. M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Eshed, Y., and Lippman, Z. B. (2019). Revolutions in agriculture chart a coursresee for targeted breeding of old and new crops. Science 366:eaax0025. doi: 10.1126/science.aax0025
Esumi, T., Tao, R., and Yonemori, K. (2005). Isolation of LEAFY and TERMINAL FLOWER 1 homologues from six fruit tree species in the subfamily Maloideae of the Rosaceae. Sex. Plant Reprod. 17, 277–287. doi: 10.1007/s00497-004-0239-3
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res. 2:188. doi: 10.12688/f1000research.2-188.v2
García-Pallas, I., Val, J., and Blanco, A. (2001). The inhibition of flower bud differentiation in ‘Crimson Gold’ nectarine with GA3 as an alternative to hand thinning. Sci. Hortic. 90, 265–278. doi: 10.1016/S0304-4238(01)00229-1
Gocal, G. F., Sheldon, C. C., Gubler, F., Moritz, T., Bagnall, D. J., MacMillan, C. P., et al. (2001). GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 127, 1682–1693.
Goldberg-Moeller, R., Shalom, L., Shlizerman, L., Samuels, S., Zur, N., Ophir, R., et al. (2013). Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in Citrus buds. Plant Sci. 198, 46–57. doi: 10.1016/j.plantsci.2012.09.012
Hisamatsu, T., and King, R. W. (2008). The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J. Exp. Bot. 59, 3821–3829. doi: 10.1093/jxb/ern232
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., and Yu, H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. doi: 10.1038/ncomms5601
Jiang, Y., Peng, J., Wang, M., Su, W., Gan, X., Jing, Y., et al. (2019a). The Role of EjSPL3, EjSPL4, EjSPL5, and EjSPL9 in regulating flowering in loquat (Eriobotrya japonica Lindl.). Int. J. Mol. Sci. 21:248. doi: 10.3390/ijms21010248
Jiang, Y., Peng, J., Zhang, Z., Lin, S., Lin, S., and Yang, X. (2019b). The role of EjSVPs in flower initiation in Eriobotrya japonica. Int J Mol Sci. 20:5933. doi: 10.3390/ijms20235933
Jiang, Y., Peng, J., Zhu, Y., Su, W., Zhang, L., Jing, Y., et al. (2019c). The role of EjSOC1s in flower initiation in Eriobotrya japonica. Front. Plant Sci. 10:253. doi: 10.3389/fpls.2019.00253
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965.
Kurokura, T., Mimida, N., Battey, N. H., and Hytonen, T. (2013). The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 64, 4131–4141. doi: 10.1093/jxb/ert233
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9:559. doi: 10.1186/1471-2105-9-559
Lee, H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. doi: 10.1101/gad.813600
Lenahan, O. M., Whiting, M. D., and Elfving, D. C. (2006). Gibberellic acid inhibits floral bud induction and improves ‘Bing’ sweet cherry fruit quality. Hortscience 41, 654–659.
Li, W., Wang, H., and Yu, D. (2016). Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant. 9, 1492–1503. doi: 10.1016/j.molp.2016.08.003
Lin, S. (2007). World loquat production and research with special reference to China. Proc. Sec. Int. Symp. Loquat. 750, 37–43. doi: 10.17660/ActaHortic.2007.750.2
Liu, C., Chen, H., Er, H. L., Soo, H. M., Kumar, P. P., Han, J. H., et al. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–1491. doi: 10.1242/dev.020255
Liu, Y., Zhao, Q., Meng, N., Song, H., Li, C., Hu, G., et al. (2017). Over-expression of EjLFY-1 leads to an early flowering habit in strawberry (Fragaria x ananassa) and its asexual progeny. Front. Plant Sci. 8:496. doi: 10.3389/fpls.2017.00496
Liu, Y. X., Song, H. W., Liu, Z. L., Hu, G. B., and Lin, S. Q. (2013). Molecular characterization of loquat EjAP1 gene in relation to flowering. Plant Growth Regul. 70, 287–296. doi: 10.1007/s10725-013-9800-0
Moon, J., Lee, H., Kim, M., and Lee, I. (2005). Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46, 292–299. doi: 10.1093/pcp/pci024
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623.
Nakagawa, M., Honsho, C., Kanzaki, S., Shimizu, K., and Utsunomiya, N. (2012). Isolation and expression analysis of FLOWERING LOCUS T-like and gibberellin metabolism genes in biennial-bearing mango trees. Sci. Hortic. 139, 108–117. doi: 10.1016/j.scienta.2012.03.005
Osnato, M., Castillejo, C., Matias-Hernandez, L., and Pelaz, S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3:808. doi: 10.1038/ncomms1810
Peng, J., Li, W., Yuan, Y., Han, Z., Cao, Y., Shahid, M. Q., et al. (2021). Removal of the Main Inflorescence to Induce Reflowering of Loquat. Hortic. Plant J. in press. doi: 10.1016/j.hpj.2021.03.009
Reig, C., Gil-Munoz, F., Vera-Sirera, F., Garcia-Lorca, A., Martinez-Fuentes, A., Mesejo, C., et al. (2017). Bud sprouting and floral induction and expression of FT in loquat [Eriobotrya japonica (Thunb.) Lindl.]. Planta. 246, 915–925. doi: 10.1007/s00425-017-2740-6
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: cONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Song, Y. H., Lee, I., Lee, S. Y., Imaizumi, T., and Hong, J. C. (2012). CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J. 69, 332–342. doi: 10.1111/j.1365-313X.2011.04793.x
Su, W., Jing, Y., Lin, S., Yue, Z., Yang, X., Xu, J., et al. (2021). Polyploidy underlies co-option and diversification of biosynthetic triterpene pathways in the apple tribe. Proc. Natl. Acad. Sci. U. S. A. 118:e2101767118. doi: 10.1073/pnas.2101767118
Teotia, S., and Tang, G. (2015). To bloom or not to bloom: role of microRNAs in plant flowering. Mol. Plant. 8, 359–377. doi: 10.1016/j.molp.2014.12.018
Wang, H., Pan, J., Li, Y., Lou, D., Hu, Y., and Yu, D. (2016). The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol. 172, 479–488. doi: 10.1104/pp.16.00891
Xu, F., Li, T., Xu, P. B., Li, L., Du, S. S., Lian, H. L., et al. (2016). DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 590, 541–549. doi: 10.1002/1873-3468.12076
Zhang, L., Chen, L., and Yu, D. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Zhang, L., Jiang, Y., Zhu, Y., Su, W., Long, T., Huang, T., et al. (2019). Functional characterization of GI and CO homologs from Eriobotrya deflexa Nakai forma koshunensis. Plant Cell Rep. 38, 533–543. doi: 10.1007/s00299-019-02384-3
Zhang, L., Yu, H., Lin, S., and Gao, Y. (2016). Molecular characterization of FT and FD homologs from Eriobotrya deflexa Nakai forma koshunensis. Front. Plant Sci. 7:8. doi: 10.3389/fpls.2016.00008
Zhang, S., Zhang, D., Fan, S., Du, L., Shen, Y., Xing, L., et al. (2016). Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ apple (Malus domestica Borkh.). Plant Physiol. Biochem. 107, 178–186. doi: 10.1016/j.plaphy.2016.06.005
Keywords: GA3, flowering, RNA-seq, co-expression, loquat
Citation: Jiang Y, Liu Y, Gao Y, Peng J, Su W, Yuan Y, Yang X, Zhao C, Wang M, Lin S, Peng Z and Xie F (2021) Gibberellin Induced Transcriptome Profiles Reveal Gene Regulation of Loquat Flowering. Front. Genet. 12:703688. doi: 10.3389/fgene.2021.703688
Received: 30 April 2021; Accepted: 24 August 2021;
Published: 10 September 2021.
Edited by:
Yunpeng Cao, Central South University Forestry and Technology, ChinaReviewed by:
Zhongqi Fan, Fujian Agriculture and Forestry University, ChinaCopyright © 2021 Jiang, Liu, Gao, Peng, Su, Yuan, Yang, Zhao, Wang, Lin, Peng and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shunquan Lin, bG9xdWF0QHNjYXUuZWR1LmNu; Ze Peng, cGVuZ3plMTk5MDEyMUBnbWFpbC5jb20=; Fangfang Xie, eGllZmFuZ2ZhbmcyMDIwMTJAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.