
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 11 August 2021
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.698983
Ulcerative colitis (UC) is a complex inflammatory bowel disorder that can induce colonic and rectal dysfunction. Mesalazine, a first-line medicine, is routinely prescribed for UC treatment. However, the pharmacological targets of mesalazine against UC are not detailed in current publications. In the current study, a transcriptomics strategy was applied to reveal the therapeutic targets and molecular mechanisms of mesalazine for treating dextran sulfate sodium (DSS)-induced UC in mice. Compared with the UC group, a total of 1,663 differentially expressed genes were identified in mesalazine-treated mice, of which 262 were upregulated and 1,401 were downregulated. GO and KEGG enrichment analyses indicated that the protective actions of mesalazine for treating UC were related to the functional regulation of immune inflammatory response, such as the regulation of T cells, white blood cells, and cytokine receptor pathways. In addition, ingenuity pathway analysis of the gene network further revealed the inhibitory action of mesalazine on C–C motif chemokine ligands (CCL11 and CCL21) and C–X–C motif chemokine ligands (CXCL3 and CXCR2). Taken together, the current transcriptomic findings revealed anti-UC pharmacological targets, including the newly discovered biotargets CCL11, CCL21, CXCL3, and CXCR2, of mesalazine against DSS-induced intestinal inflammation.
Ulcerative colitis (UC), a refractory enteritis disease, is characterized by mucosal inflammation and enterocyte lesions present in the gastrointestinal tract (Guan, 2019). It is reported that the prevalence and burden of UC are increasing worldwide, including in China (Ordás et al., 2012). Genetic factors, environmental factors, and dietetic alterations may be crucial risk indicators for UC (Eisenstein, 2018). If left untreated, chronic UC in Asian patients may result in an elevated risk of developing colorectal cancer when compared with that of the general population (Bopanna et al., 2017). The pathological etiology of UC is multifaceted, including impaired barrier function in the mucus layer, lamina propria lesion, gut microbiota imbalance, and neutrophilic immune response (Kobayashi et al., 2020). Early and precise diagnosis of UC using endoscopy, CT scan, blood biomarker determination, fecal calprotectin testing, and bowel ultrasonography is critical to efficacious therapy (Conrad et al., 2014). Treat-to-target, a countermeasure used for treating chronic disorders, may be achieved by determining pathological targets and mitigating UC (Ungaro et al., 2019). Mesalazine, a precursor of 5-aminosalicylic acid, can be used clinically for mild to moderate UC, and is characterized by a good safety profile and low tolerance (Sehgal et al., 2018). Mesalazine is clinically used a the first-line treatment for patients with UC based on its high effectiveness and safety (Kato et al., 2018). However, or new mesalazine anti-UC targets remain minimally reported in preclinical studies. Transcriptomics, a next-generation sequencing technology, is an emerging and rapid method that can identify genome-wide annotation of DNA functions and perform detailed comparative genomic studies in humans and mice (Breschi et al., 2017). Interestingly, transcriptomic strategies have been used to unravel the detailed targets of pathological liver cancer (Lai et al., 2020), perfluorooctanesulfonate-induced immunotoxicity (Li et al., 2020a), glutathione against cleft lip (Li et al., 2021b), and vitamin C against hepatotoxicity (Li et al., 2021a). Therefore, in the present study, we utilized transcriptomic analysis to reveal detailed and novel biotargets of mesalazine against UC in vivo.
Mature male C57BL/6J mice, aged around 7 weeks, were purchased from STA Lab Animal Co., Ltd. (Changsha, China). All mice were adaptively maintained for approximately 7 days, and then 3% DSS solution was provided freely in drinking water for UC induction. Meanwhile, DSS-exposed mice were treated with 0.4 g/kg mesalazine for 10 days. In addition, mice treated with DSS-free solution were used as the control group. At the end of the experiment, all mice were euthanized via cervical dislocation, and colorectal tissue samples were immediately isolated and snap-frozen with liquid nitrogen before being used for transcriptomics analysis (Su et al., 2019).
Total RNA (n = 5 from each treatment) was extracted using TRIzol reagent, following the manufacturer’s instructions. RNA samples with a RIN number >8.0, were used for library construction. Briefly, poly(A)-mRNA was enriched from ten-micrograms of total RNA using poly T oligo-attached magnetic beads. After purification, the mRNA was fragmented and subjected to cDNA library construction using an Illumina mRNA Seq sample preparation kit. Paired-end sequencing (150 bp each) was performed on an Illumina HiSeq 4000, following the manufacturer’s instructions.
The adaptor sequences and sequencing primers were trimmed. Low-quality reads with q quality scores lower than 20 were removed. The clean sequence reads were mapped to the UCSC reference genome1 using the HISAT package. StringTie and edgeR were used to determine the expression levels of all the transcripts. Genes with a lLog2 (fold change) > 1 and with statistical significance (q-value < 0.05) were defined as differentially expressed genes (DEGs). The DEGs were subjected to Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, and ingenuity pathway analysis (IPA) to examine the effects of mesalazine on DSS-induced colitis.
The colon samples were homogenized, and the supernatants were used for the measurement of inflammatory cytokines using ELISA kits (Elisa Biotech, Shanghai, China). The experimental procedures were performed as described previously (Xu et al., 2019).
The colon samples from different groups were fixed with 4% paraformaldehyde prior to being cut into 4–5 μm thick sections using a microtome. Thereafter, the sections were dewaxed using xylene and were stained with hematoxylin and eosin dyes for histopathological examination (Wu et al., 2019; Li et al., 2020b).
In this study, we performed comparative transcriptomic analysis to understand the molecular basis of DSS-induced colitis and to delineate the molecular mechanism underlying the immunosuppressive effect of mesalazine. In the first part of the study, we attempted to understand the detailed mechanism underlying DSS-induced colitis in a mouse model. It has been well reported that the major cause of UC results from abnormal immune responses to antigens derived from the intestinal microbiota (Podolsky, 2002; Macdonald and Monteleone, 2005). Our results showed that DSS could lead to a large number of differentially expressed genes. The results of GO and pathway analyses further highlighted that the induction of inflammatory and immune responses is mainly through alterations of cytokine-cytokine receptor interaction, IL-17 signaling pathway, leukocyte transendothelial migration, Toll-like receptor signaling pathway, and intestinal immune network for IgA production. In addition, IBD was highlighted in the analysis, suggesting that the model was valid. Cytokine-cytokine receptor interactions play an important role in immune regulation (Wu et al., 2005). It is involved in the T-cell receptor signaling pathway (Qian et al., 2019) and has been reported to be associated with autoimmune disorders (Liu et al., 2013; Xing et al., 2020). Many autoimmune and inflammatory diseases can result from excessive cytokine production or responsiveness. The IL-17 signaling pathway is the founding member of a novel family of inflammatory cytokines (Amatya et al., 2017). IL-17, the hallmark cytokine of the newly defined T helper 17 [T(H)17] cell subset, plays a crucial role in the inflammatory pathology of autoimmune diseases (Gaffen, 2009). The role of the IL-17 family in most common autoimmune diseases, such as psoriasis, IBD, and rheumatoid arthritis, has been extensively characterized (Zhang et al., 2015). Our funding is concordant with the previous IL-17R knockout mice study that IL-17 signaling plays a critical role in the development of TNBS-induced colitis and may represent a target for therapeutic intervention for IBD (Zhang et al., 2006). The toll-like receptor signaling pathway has been recognized as a key repressor of inflammatory signaling and plays an essential role in the pathogenesis of autoimmune diseases (Chen et al., 2016; Shamilov and Aneskievich, 2018). It is a primary sensor of both innate and adaptive immune systems through its modulation of numerous genes that function in host defense, including inflammatory cytokines, chemokines, and antigen presenting molecules (Liu et al., 2006). It has been reported that Toll-like receptor (TLR) signaling-related protein (TLR4) was detected in the colon tissue of DSS model mice (Bai et al., 2021). Furthermore, macrophages in the intestinal mucosa can rapidly induce TLR-mediated inflammatory responses (Wang et al., 2021). Our results provide a better understanding of the immunopathogenesis of UC and may help to identify novel targets for more potent interventions.
To understand the molecular mechanism underlying DSS-induced colitis and the immunosuppressive effects of mesalazine on DSS-induced colitis, a comparative transcriptomic analysis was conducted, followed by bioinformatic analysis. Deep sequencing of RNA libraries derived from control and treatment groups each generated at least 43 million quality-trimmed clean reads (Supplementary Table 1); the read utilization ratio was over 97%. A total of 110 Gb quality-trimmed bases were obtained from transcriptome sequencing (Supplementary Table 1). Over 90% of the sequencing reads could be mapped to the exonic regions of the mouse reference genome (Supplementary Table 2). In the comparative transcriptomic analysis, a total of 1,627 differentially expressed genes (DEGs), including 1,326 upregulated and 301 downregulated genes, were identified in the DSS-induced colitis mice as compared to the control group mice (Figure 1A and Supplementary Table 3). The DEGs were then subjected to GO and KEGG enrichment analyses to understand the alteration of biological functions and signaling pathways in the DSS-induced colitis model. GO results suggested that numerous biological processes, cellular components, and molecular functions were altered in the DSS-induced colitis model. In the analysis of biological processes, we found the activation of numerous inflammatory processes (Figure 1B and Table 1). The KEGG pathway analysis further highlighted the alteration of pathways related to immune and inflammatory responses, including cytokine-cytokine receptor interaction, inflammatory bowel disease (IBD), IL-17 signaling pathway, leukocyte transendothelial migration, Toll-like receptor signaling pathway, and intestinal immune network for IgA production (Figure 1C and Table 2).
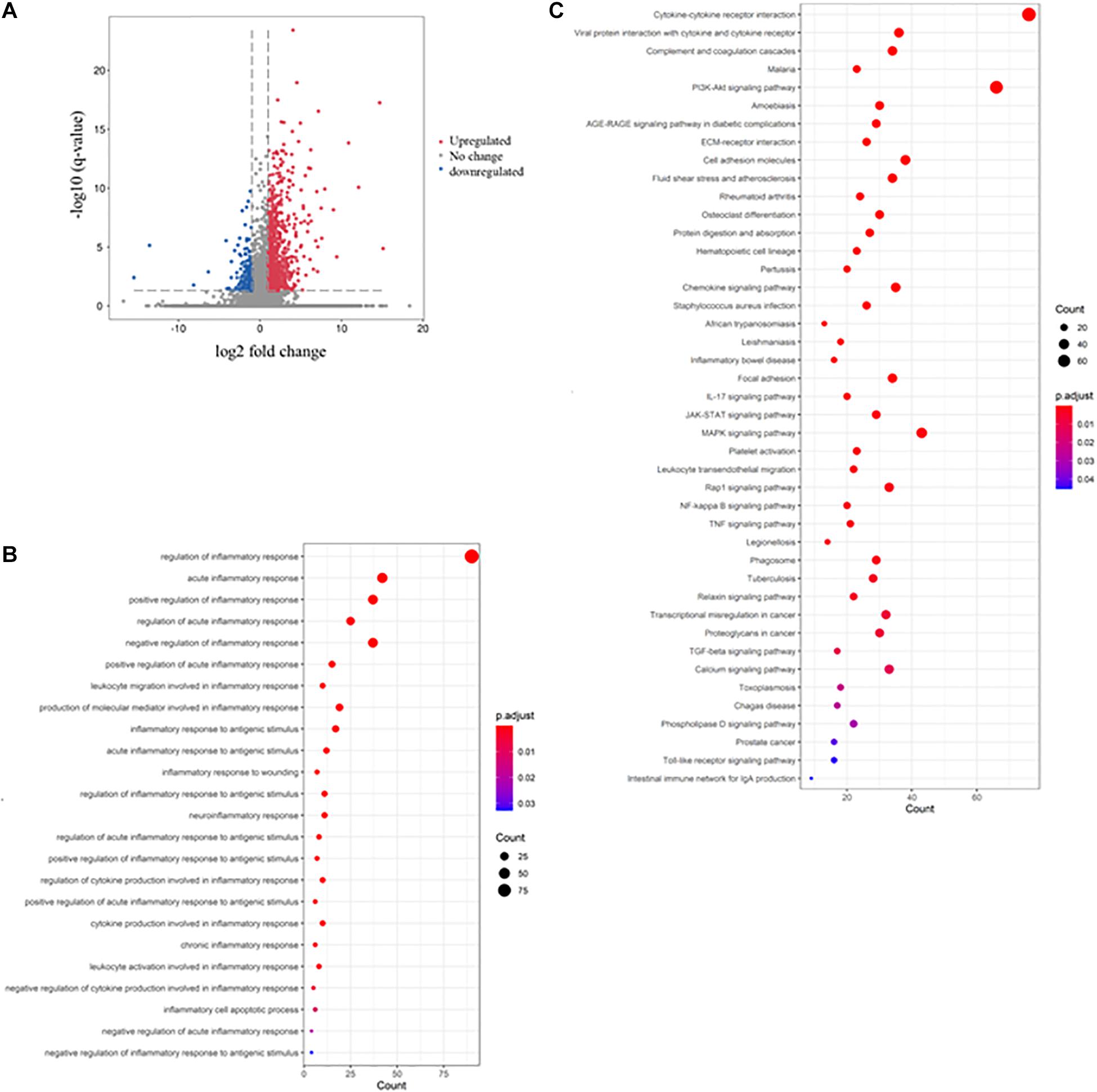
Figure 1. Dextran sulfate sodium (DSS) induced colitis through the induction of inflammatory response. The volcano plot shows the differential expression of genes in the gut of DSS-induced colitis mice. The genes with | log2 (fold change: DSS/Ctrl)| > 1 and -log B and H corrected p-value > 1.3 were considered differentially expressed genes (DEGs). The red dot represents upregulated genes, the green dot represents downregulated genes, and the gray dot represents genes with no significant change. (B) The rich factor plot shows the alteration of biological processes related to immunity in the gut of DSS-induced colitis mice. The size of the dot represents the number of DEGs. The color intensity of the dots represents the significance of the biology processes. (C) The rich factor plot showed the alteration of cell signaling pathways related to immunity in the gut of DSS-induced colitis mice. The size of the dot represents the number of DEGs. The color intensity of the dots represents the significance of the signaling pathways.
A similar approach was used to determine the immunosuppressive effect of mesalazine on DSS-induced colitis. When we compared the DSS group and DSS + mesalazine group, we identified 1,663 DEGs, of which 262 were upregulated and 1,401 were downregulated (Figure 2A and Supplementary Table 4). In the bioinformatic analysis, we primarily focused on the GO terms and pathways related to immune responses. The results of GO analysis showed that low-dose mesalazine treatment altered different immune processes. This could be due to the regulation of different immune cell types, such as T cells, B cells, natural killer cells, and leukocytes (Figure 2B and Table 3). This results in the regulation of cytokine functions such as cytokine biosynthetic process, cytokine production, cytokine secretion, and cytokine binding (Figure 2B and Table 3), leading to alterations in the immune response of the gut system (Figure 2B and Table 3). The results of KEGG pathway analysis further highlighted the immune-responsive pathways such as ECM-receptor interaction, T-cell receptor signaling pathway, TNF signaling pathway, cytokine-cytokine receptor interaction, chemokine signaling pathway, IL-17 signaling pathway, Th1, Th2, and Th17 cell differentiation, IBD, Hippo signaling pathway, and TGF-beta signaling pathway (Figure 2C and Table 4).
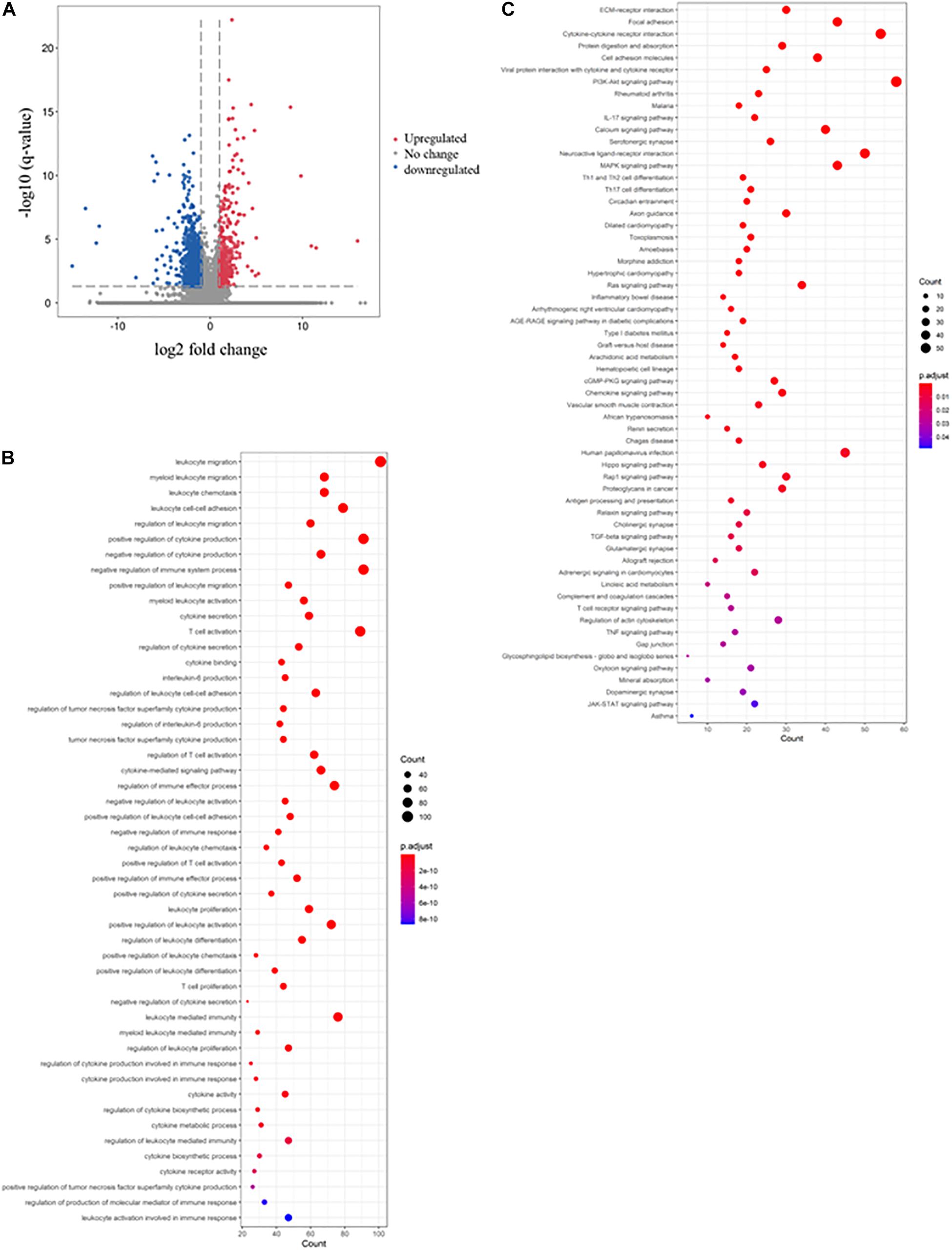
Figure 2. Immunosuppressive effects of mesalazine on DSS-induced colitis. The volcano plot shows the differential expression of genes in the mesalazine-treated colitis mice. The genes with | log2 (fold change: mesalazine + DSS/DSS)| > 1 and -log B and H corrected p-value > 1.3 were considered DEGs. The red dot represents upregulated genes, the green dot represents downregulated genes, and the gray dot represents genes with no significant change. (B) The rich factor plot shows the alteration of biological processes related to immunity in the gut of mesalazine-treated mice with colitis. The size of the dot represents the number of DEGs. The color intensity of the dots represents the significance of the biology processes. (C) The rich factor plot showed the alteration of cell signaling pathways related to immunity in the gut of mesalazine-treated colitis mice. The size of the dots represents the number of DEGs. The color intensity of the dots represents the significance of the signaling pathways.
The DEGs were then subjected to IPA to determine the immunosuppressive effect of mesalazine on the treatment of UC. The results of canonical pathway analysis highlighted the substantial alteration of a large number of cell signaling pathways related to immune and inflammatory responses (p < 0.05) (Table 5). They included the regulation of interleukin signaling, such as IL-6, IL-8, IL-15, and IL-17 signaling pathways (Table 5). Furthermore, the results highlighted T cell differentiation, including Th1, Th2, and T helper cells (Table 5). In addition to the T cells, we also observed the regulation of B cells by mesalazine treatment (Table 5). Finally, we conducted IPA gene network analysis to delineate the detailed molecular mechanisms underlying the immunosuppressive responses caused by mesalazine. Our results showed that mesalazine regulated a large number of C–C motif chemokine ligands (CCL5, CCL7, CCL11, and CL21) and C-X-C motif chemokine ligands (CXCL3, CXCL6, CXCL9, and CXCL11) in the extracellular space (Figure 3). This led to the modulation of a group of transmembrane receptors, including interleukin receptors (IL2RB, IL12RB1, and IL12RB2), T-cell surface glycoprotein (CD3E, CD3G, CD28, and CD247), and chemokine receptors (CXCR2 and CXCR3) (Figure 3). The regulation of these receptors has been reported to regulate different enzymes (PDE1A, PDE1B, PDE1C, and MYH10) and kinases (ITK, LCK NOX1, and MAP3K8) involved in immune responses (Figure 3). More importantly, mesalazine treatment reversed the suppressive effect of immune responsive genes, including chemokine (C–X–C motif) ligand (CXCL3), chemokine (C–C motif) ligand (CCL11 and CCL21), chemokine receptor (CXCR2), and colony-stimulating factor (CSF3) in the DSS-induced colitis model (Table 6). Taken together, our results suggest that mesalazine treatment could reverse the DSS-induced immune response through its immunosuppressive effect.
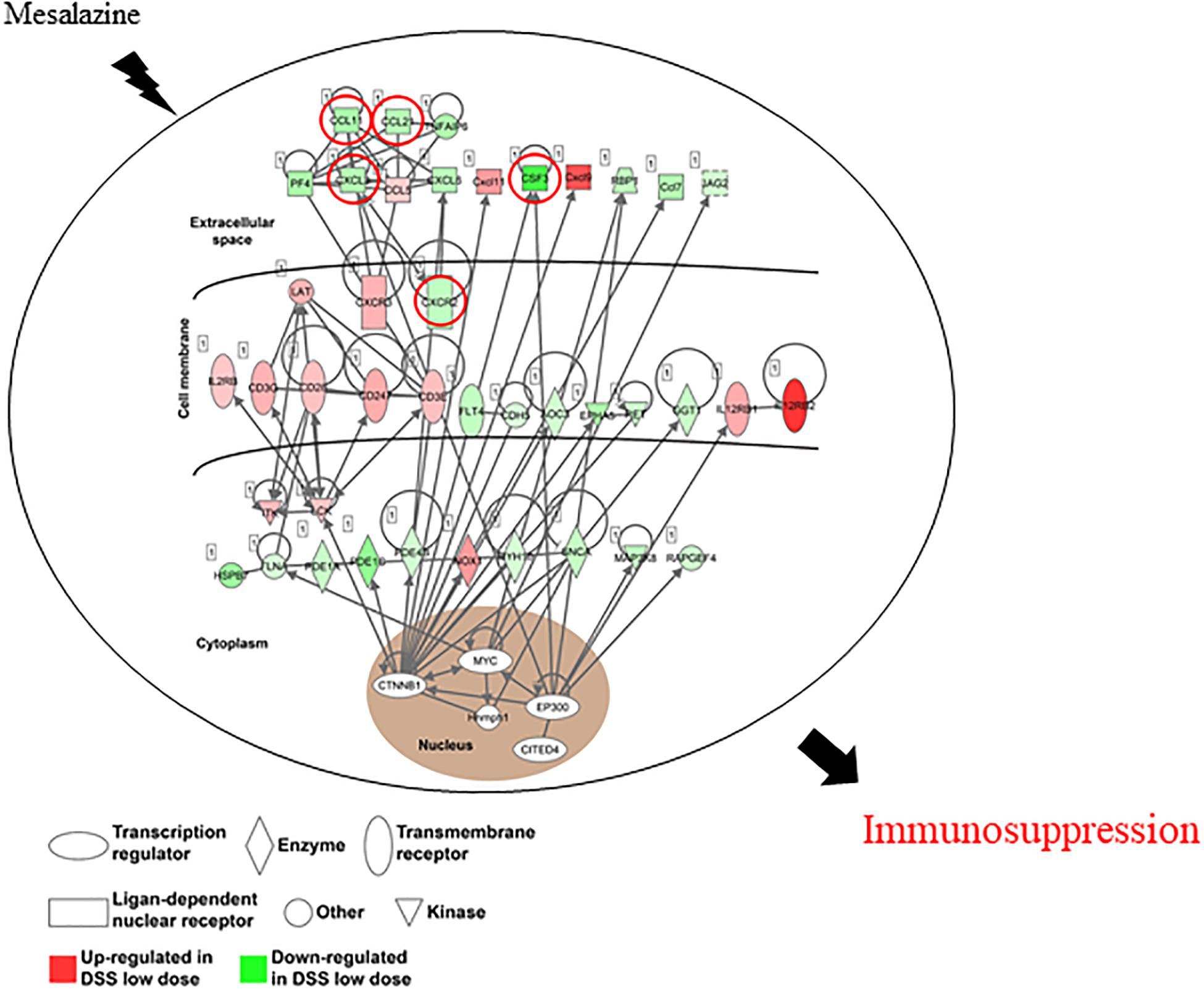
Figure 3. The gene network involved in the immunosuppressive effect of mesalazine on DSS-induced colitis. Ingenuity pathway analysis shows the contribution of different molecules in the immunosuppressive effect of mesalazine. The red symbol represents upregulated genes; the green shape represents downregulated genes.
In the later part of this study, we aimed to determine the molecular pathways underlying the immunosuppressive effect of mesalazine on DSS-induced colitis. Therefore, we focused on immune-related biological processes and pathways in bioinformatic analysis. Our results show that mesalazine treatment can alter different immune cells, including T cells, B cells, and natural killer cells, leading to the control of autoimmune responses (Zitti and Bryceson, 2018; Meffre and O’Connor, 2019; Wing et al., 2019). T cells are reported to contribute to the pathogenesis of chronic intestinal inflammation and are considered targets for UC treatment due to their role in the autoimmune system (Eri et al., 2012). For example, a study using a T cell transfer colitis model demonstrated that CD4(+) T helper cells strongly contributed to the pathogenesis of IBD (Maschmeyer et al., 2021). In addition, imbalanced CD4+ T cells have been reported as predisposing factors for colitis (Pandit et al., 2021). Furthermore, mesalazine treatment mediated cytokine biosynthesis and secretion. As mentioned before, cytokines play an important role in the autoimmune response. More importantly, our gene network analysis further highlighted the suppression of C–C motif chemokine ligands (CCL11 and CCL21) and C–X–C motif chemokine ligands (CXCL3 and CXCR2) by mesalazine treatment. All of these ligands were increased in the DSS-induced model. CCL11, a prototypical Th2 chemokine, is associated with enrichment in Th2 CD4+ T cells (Chao et al., 2014) and is involved in eosinophil recruitment (Hornig et al., 2016). CCL11 expression levels correlated with the expression levels of different ILs. Reduced expression of CCL11 has been reported to alleviate allergic symptoms and allergic inflammatory responses by reducing serum cytokine levels (Lei et al., 2020). It has been reported that CD4+ CD25+ T cells inhibit the development of colitis induced by both Th1 and Th2 cells in a mouse model (Xu et al., 2003). The Th1/Th2 balance of peripheral CD4-positive T cells is considered a biomarker for patients with refractory ulcerative colitis (Nakase et al., 2009). CCL21, a ligand of the receptor CCR7, contributes to the balance of immunity and tolerance through thymocyte development, secondary lymphoid organogenesis, high affinity antibody responses, and regulatory and memory T-cell function (Förster et al., 2008; Comerford et al., 2013). It has been shown that the loss of CCL21 responsiveness in the normal development of the memory T-cell effector function does not hold for autoimmune diseases (Christopherson et al., 2003). A mouse colitis study demonstrated that CCL21 suppression could decrease damage induced by ulcerative colitis (Zhang et al., 2014), suggesting that CCL21 might be a therapeutic target for UC treatment.
To determine the pharmacological effects of mesalazine against colitis, the structure of the colon tissues was studied. Our results showed that the length of the colon was shortened and colon tissues became fragile in the colitis mouse model, and ulcers were formed, including mucosal abscess and congestion. Mesalazine intervention improved pathological changes in mice with colitis. Histomorphological observations using H&E staining showed that the colon tissue of the colitis mice changed in morphology and showed alterations, such as crypt abscess, decreased density in the colon, and inflammatory infiltration. Mesalazine treatment mitigated mucosal structural changes and reduced inflammatory infiltration (Figure 4A). ELISA data showed that the expression levels of TNF-α, IL-1α, IL-6, and IL-8 in colitis mice were significantly reduced compared with those in control samples. Interestingly, the elevation of inflammatory cytokines in mesalazine-treated colitis mice was decreased (Figure 4B).
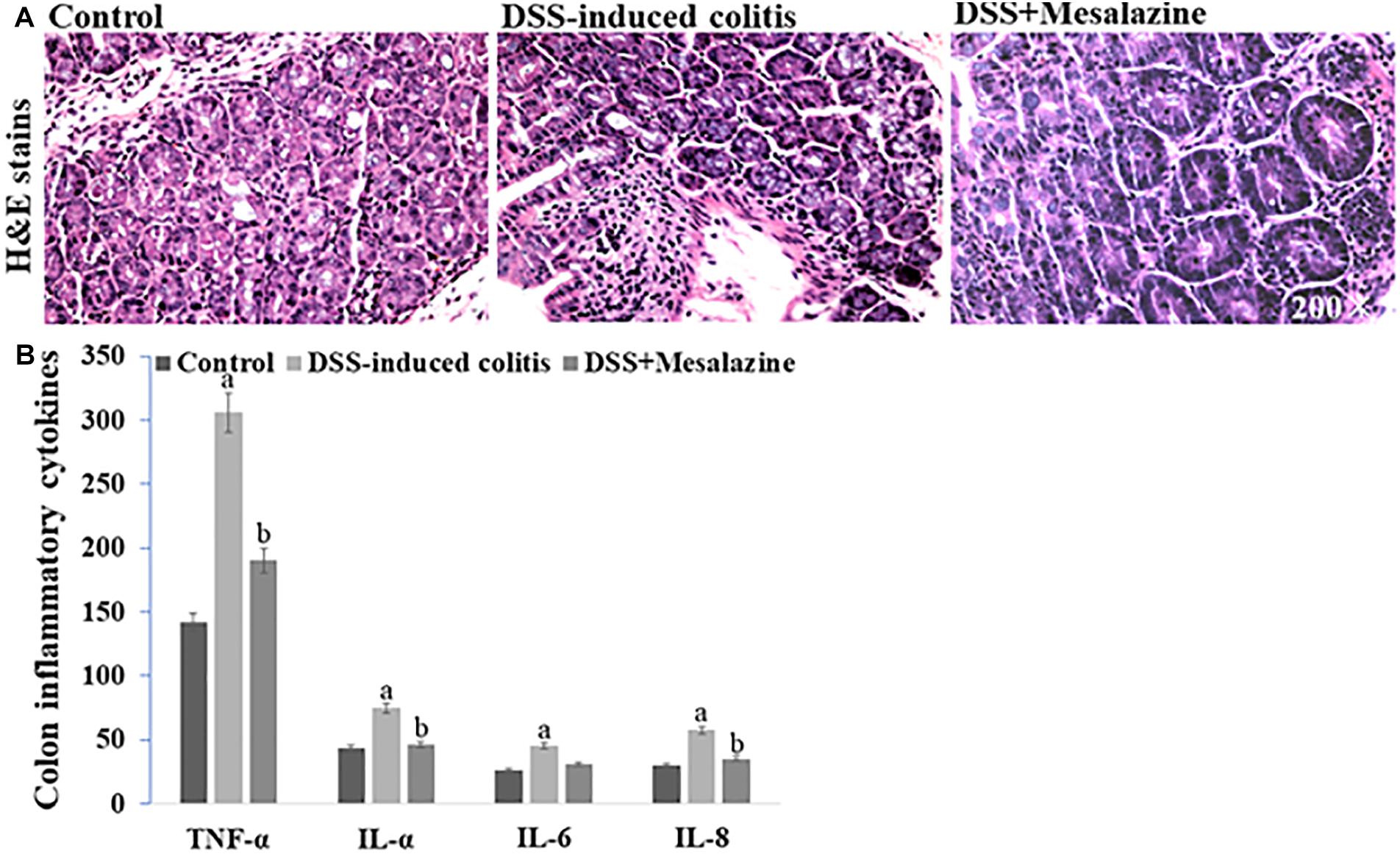
Figure 4. In vivo effects of mesalazine treatment on DSS-induced colitis. (A) Hematoxylin and eosin staining show the structural changes including visible crypt abscess, inflammatory infiltration, and cytoclasis in DSS-induced colitis. These alterations were reversed by mesalazine treatment. (B) Mesalazine treatment relieved DSS-induced inflammatory cytokines including TNF-α, IL-1α, IL-6, and IL-8 in the colon samples via its anti-inflammation action.
CXCL3, a small cytokine, controls the migration and adhesion of monocytes through its interaction with the cell surface chemokine receptor CXCR2 (Smith et al., 2005). CXCR2 is a functional receptor for GRO-family chemokines involved in monocyte recruitment and inhibits inflammation. CXCR2 has been found to be significantly increased in colonic mucosal tissues of patients with active UC (Zhu et al., 2020). It has been reported that CXCL3 plays a role in controlling intestinal inflammation and gut mucosal healing, and is considered a key prognostic parameter in the management of IBD (Stronati et al., 2019). Therefore, the suppressive effect of mesalazine on these C–C motif chemokine ligands and C–X–C motif chemokine ligands suggests a possible strategy for the treatment of UC, and a better understanding of the molecular mechanism underlying the immunosuppressive effect of mesalazine has important implications for developing novel anti-UC drugs directed against the CXC chemokine signaling system. However, the newly identified markers need to be further confirmed and investigated using a human model.
The data presented in the study are deposited in the BioProject repository, accession number PRJNA732984.
The animal study was reviewed and approved by West China School of Basic Medical Sciences and Forensic Medicine of Sichuan University.
RL and LZ conceived and designed the experiments and wrote the manuscript. RL, LC, and QW performed the experiments. RL analysed the data. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.698983/full#supplementary-material
Amatya, N., Garg, A. V., and Gaffen, S. L. (2017). IL-17 signaling: the Yin and the Yang. Trends Immunol. 38, 310–322. doi: 10.1016/j.it.2017.01.006
Bai, B., Wu, F., Ying, K., Xu, Y., Shan, L., Lv, Y., et al. (2021). Therapeutic effects of dihydroartemisinin in multiple stages of colitis-associated colorectal cancer. Theranostics 11, 6225–6239. doi: 10.7150/thno.55939
Bopanna, S., Ananthakrishnan, A. N., Kedia, S., Yajnik, V., and Ahuja, V. (2017). Risk of colorectal cancer in Asian patients with ulcerative colitis: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2, 269–276. doi: 10.1016/s2468-1253(17)30004-3
Breschi, A., Gingeras, T. R., and Guigó, R. (2017). Comparative transcriptomics in human and mouse. Nat. Rev. Genet. 18, 425–440. doi: 10.1038/nrg.2017.19
Chao, G. Y., Wallis, R. H., Marandi, L., Ning, T., Sarmiento, J., Paterson, A. D., et al. (2014). Iddm30 controls pancreatic expression of Ccl11 (Eotaxin) and the Th1/Th2 balance within the insulitic lesions. J. Immunol. 192, 3645–3653. doi: 10.4049/jimmunol.1302383
Chen, J. Q., Szodoray, P., and Zeher, M. (2016). Toll-like receptor pathways in autoimmune diseases. Clin. Rev. Allergy Immunol. 50, 1–17. doi: 10.1007/s12016-015-8473-z
Christopherson, K. W. II, Hood, A. F., Travers, J. B., Ramsey, H., and Hromas, R. A. (2003). Endothelial induction of the T-cell chemokine CCL21 in T-cell autoimmune diseases. Blood 101, 801–806. doi: 10.1182/blood-2002-05-1586
Comerford, I., Harata-Lee, Y., Bunting, M. D., Gregor, C., Kara, E. E., and McColl, S. R. (2013). A myriad of functions and complex regulation of the CCR7/CCL19/CCL21 chemokine axis in the adaptive immune system. Cytokine Growth Fact. Rev. 24, 269–283. doi: 10.1016/j.cytogfr.2013.03.001
Conrad, K., Roggenbuck, D., and Laass, M. W. (2014). Diagnosis and classification of ulcerative colitis. Autoimmun. Rev. 13, 463–466. doi: 10.1016/j.autrev.2014.01.028
Eri, R., McGuckin, M. A., and Wadley, R. (2012). T cell transfer model of colitis: a great tool to assess the contribution of T cells in chronic intestinal inflammation. Methods Mol. Biol. 844, 261–275. doi: 10.1007/978-1-61779-527-5_19
Förster, R., Davalos-Misslitz, A. C., and Rot, A. (2008). CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8, 362–371. doi: 10.1038/nri2297
Gaffen, S. L. (2009). Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9, 556–567. doi: 10.1038/nri2586
Guan, Q. (2019). A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J. Immunol. Res. 2019:7247238.
Hornig, M., Gottschalk, G., Peterson, D. L., Knox, K. K., Schultz, A. F., Eddy, M. L., et al. (2016). Cytokine network analysis of cerebrospinal fluid in myalgic encephalomyelitis/chronic fatigue syndrome. Mol. Psychiatry 21, 261–269. doi: 10.1038/mp.2015.29
Kato, S., Ishibashi, A., Kani, K., and Yakabi, K. (2018). Optimized management of ulcerative Proctitis: when and how to use Mesalazine suppository. Digestion 97, 59–63. doi: 10.1159/000484224
Kobayashi, T., Siegmund, B., Le Berre, C., Wei, S. C., Ferrante, M., Shen, B., et al. (2020). Ulcerative colitis. Nat. Rev. Dis. Primers 6:74.
Lai, K. P., Cheung, A., Ho, C. H., Tam, N. Y., Li, J. W., Lin, X., et al. (2020). Transcriptomic analysis reveals the oncogenic role of S6K1 in hepatocellular carcinoma. J. Cancer 11, 2645–2655. doi: 10.7150/jca.40726
Lei, H., Sun, Y., and Quan, S. (2020). IL-37 relieves allergic inflammation by inhibiting the CCL11 signaling pathway in a mouse model of allergic rhinitis. Exp. Ther. Med. 20, 3114–3121.
Li, R., Guo, C., Lin, X., Chan, T. F., Lai, K. P., and Chen, J. (2020a). Integrative omics analyses uncover the mechanism underlying the immunotoxicity of perfluorooctanesulfonate in human lymphocytes. Chemosphere 256:127062. doi: 10.1016/j.chemosphere.2020.127062
Li, R., Guo, C., Lin, X., Chan, T., Su, M., Zhang, Z., et al. (2021a). Integrative Omics analysis reveals the protective role of vitamin C on Perfluorooctanoic acid-induced Hepatoxicity. J. Adv. Res. doi: 10.1016/j.jare.2021.04.003
Li, R., Guo, C., Tse, W. K. F., Su, M., Zhang, X., and Lai, K. P. (2020b). Metabolomic analysis reveals metabolic alterations of human peripheral blood lymphocytes by perfluorooctanoic acid. Chemosphere 239:124810. doi: 10.1016/j.chemosphere.2019.124810
Li, R., Huang, C., Ho, J. C. H., Leung, C. C. T., Kong, R. Y. C., Li, Y., et al. (2021b). The use of glutathione to reduce oxidative stress status and its potential for modifying the extracellular matrix organization in cleft lip. Free Radic. Biol. Med. 164, 130–138. doi: 10.1016/j.freeradbiomed.2020.12.455
Liu, G., Jiang, Y., Chen, X., Zhang, R., Ma, G., Feng, R., et al. (2013). Measles contributes to rheumatoid arthritis: evidence from pathway and network analyses of genome-wide association studies. PLoS One 8:75951. doi: 10.1371/journal.pone.0075951
Liu, H., Komai-Koma, M., Xu, D., and Liew, F. Y. (2006). Toll-like receptor 2 signaling modulates the functions of CD4+ CD25+ regulatory T cells. Proc. Natl. Acad. Sci. U.S.A. 103, 7048–7053. doi: 10.1073/pnas.0601554103
Macdonald, T. T., and Monteleone, G. (2005). Immunity, inflammation, and allergy in the gut. Science 307, 1920–1925. doi: 10.1126/science.1106442
Maschmeyer, P., Zimmermann, J., and Kühl, A. A. (2021). Murine T-cell transfer colitis as a model for inflammatory bowel disease. Methods Mol. Biol. 2285, 349–373. doi: 10.1007/978-1-0716-1311-5_26
Meffre, E., and O’Connor, K. C. (2019). Impaired B-cell tolerance checkpoints promote the development of autoimmune diseases and pathogenic autoantibodies. Immunol. Rev. 292, 90–101. doi: 10.1111/imr.12821
Nakase, H., Mikami, S., and Chiba, T. (2009). Alteration of CXCR4 expression and Th1/Th2 balance of peripheral CD4-positive T cells can be a biomarker for leukocytapheresis therapy for patients with refractory ulcerative colitis. Inflamm. Bowel Dis. 15, 963–964. doi: 10.1002/ibd.20755
Ordás, I., Eckmann, L., Talamini, M., Baumgart, D. C., and Sandborn, W. J. (2012). Ulcerative colitis. Lancet 380, 1606–1619.
Pandit, M., Acharya, S., Gu, Y., Seo, S. U., Kweon, M. N., Kang, B., et al. (2021). Geranylgeranyl pyrophosphate amplifies Treg differentiation via increased IL-2 expression to ameliorate DSS-induced colitis. Eur. J. Immunol. 51, 1461–1472. doi: 10.1002/eji.202048991
Qian, Z., Zhang, Z., and Wang, Y. (2019). T cell receptor signaling pathway and cytokine-cytokine receptor interaction affect the rehabilitation process after respiratory syncytial virus infection. PeerJ 7:7089.
Sehgal, P., Colombel, J. F., Aboubakr, A., and Narula, N. (2018). Systematic review: safety of mesalazine in ulcerative colitis. Aliment Pharmacol. Ther. 47, 1597–1609. doi: 10.1111/apt.14688
Shamilov, R., and Aneskievich, B. J. (2018). TNIP1 in autoimmune diseases: regulation of Toll-like receptor signaling. J. Immunol. Res. 2018:3491269.
Smith, D. F., Galkina, E., Ley, K., and Huo, Y. (2005). GRO family chemokines are specialized for monocyte arrest from flow. Am. J. Physiol. Heart Circ. Physiol. 289, 1976–1984.
Stronati, L., Palone, F., Negroni, A., Colantoni, E., Mancuso, A. B., Cucchiara, S., et al. (2019). Dipotassium Glycyrrhizate improves intestinal mucosal healing by modulating extracellular matrix remodeling genes and restoring epithelial barrier functions. Front. Immunol. 10:939. doi: 10.3389/fimmu.2019.00939
Su, M., Liang, X., Xu, X., Wu, X., and Yang, B. (2019). Hepatoprotective benefits of vitamin C against perfluorooctane sulfonate-induced liver damage in mice through suppressing inflammatory reaction and ER stress. Environ. Toxicol. Pharmacol. 65, 60–65. doi: 10.1016/j.etap.2018.12.004
Ungaro, R., Colombel, J. F., Lissoos, T., and Peyrin-Biroulet, L. A. (2019). Treat-to-target update in ulcerative colitis: a systematic review. Am. J. Gastroenterol. 114, 874–883. doi: 10.14309/ajg.0000000000000183
Wang, L., Zhang, N., Han, D., Su, P., Chen, B., Zhao, W., et al. (2021). MTDH promotes intestinal inflammation by positively regulating TLR signaling. J. Crohns. Colitis. 2021:jjab086.
Wing, J. B., Tanaka, A., and Sakaguchi, S. (2019). Human FOXP3+ regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity 50, 302–316. doi: 10.1016/j.immuni.2019.01.020
Wu, K., Wei, P., Liu, M., Liang, X., and Su, M. (2019). To reveal pharmacological targets and molecular mechanisms of curcumol against interstitial cystitis. J. Adv. Res. 20, 43–50. doi: 10.1016/j.jare.2019.05.003
Wu, S., Gessner, R., von Stackelberg, A., Kirchner, R., Henze, G., and Seeger, K. (2005). Cytokine/cytokine receptor gene expression in childhood acute lymphoblastic leukemia: correlation of expression and clinical outcome at first disease recurrence. Cancer 103, 1054–1063. doi: 10.1002/cncr.20869
Xing, Y., Lin, Q., Tong, Y., Zhou, W., Huang, J., Wang, Y., et al. (2020). Abnormal Neutrophil transcriptional signature may predict newly diagnosed latent autoimmune diabetes in adults of South China. Front. Endocrinol. 11:581902. doi: 10.3389/fendo.2020.581902
Xu, D., Liu, H., Komai-Koma, M., Campbell, C., McSharry, C., Alexander, J., et al. (2003). CD4+CD25+ regulatory T cells suppress differentiation and functions of Th1 and Th2 cells, Leishmania major infection, and colitis in mice. J. Immunol. 170, 394–399. doi: 10.4049/jimmunol.170.1.394
Xu, X., Guo, C., Liang, X., Li, R., and Chen, J. (2019). Potential biomarker of fibroblast growth factor 21 in valproic acid-treated livers. Biofactors 45, 740–749. doi: 10.1002/biof.1519
Zhang, H., Bernuzzi, F., Lleo, A., Ma, X., and Invernizzi, P. (2015). Therapeutic potential of IL-17-mediated signaling pathway in autoimmune liver diseases. Med. Inflamm. 2015:436450.
Zhang, H., Zhang, X., Ding, X., Cao, W., Qu, L., and Zhou, G. (2014). Effect of secondary lymphoid tissue chemokine suppression on experimental ulcerative colitis in mice. Genet. Mol. Res. 13, 3337–3345. doi: 10.4238/2014.april.29.12
Zhang, Z., Zheng, M., Bindas, J., Schwarzenberger, P., and Kolls, J. K. (2006). Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm. Bowel. Dis. 12, 382–388. doi: 10.1097/01.mib.0000218764.06959.91
Zhu, F., He, H., Fan, L., Ma, C., Xu, Z., Xue, Y., et al. (2020). Blockade of CXCR2 suppresses proinflammatory activities of neutrophils in ulcerative colitis. Am. J. Transl. Res. 12, 5237–5251.
Keywords: ulcerative colitis, mesalazine, transcriptomics, differentially expressed genes, pharmacological targets
Citation: Li R, Cheng L, Wang Q and Zhou L (2021) Comparative Transcriptomic Analysis Reveals the Immunosuppressive Targets of Mesalazine in Dextran Sulfate Sodium-Induced Ulcerative Colitis. Front. Genet. 12:698983. doi: 10.3389/fgene.2021.698983
Received: 22 April 2021; Accepted: 03 June 2021;
Published: 11 August 2021.
Edited by:
Cesar Wong, Hong Kong Polytechnic University, ChinaReviewed by:
Alvin Ma, Hong Kong Polytechnic University, ChinaCopyright © 2021 Li, Cheng, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liming Zhou, c2N1X3pob3VsbUBzaW5hLmNvbQ==; emhvdTEwOEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.