- 1College of Agriculture, Shanxi Agricultural University, Taiyuan, China
- 2College of Life Sciences, Hebei Agricultural University, Baoding, China
- 3Center for Agricultural Genetic Resources Research, Shanxi Agricultural University, Taiyuan, China
Calmodulin-binding proteins belong to the IQ67 domain (IQD) gene family and play essential roles in plant development and stress responses. However, the role of IQD gene family in potato (Solanum tuberosum L.) is yet to be known. In the present study, 23 StIQDs were identified in the potato genome and named StIQD1 to StIQD23. They were unevenly distributed on 10 of the 12 chromosomes. Phylogenetic analysis divided the IQDs into four subfamilies (IQD I–IV). StIQDs found in three of the four subfamilies. Synteny analysis confirmed that potato and tomato shared a close evolutionary relationship. Besides, RNA-Seq data analysis revealed that the expression of 19 of the 23 StIQDs was detected in at least one of the 12 tissues, and some of which showed a tissue-specific pattern. Quantitative reverse transcriptase–polymerase chain reaction results further confirmed that 14 StIQDs responded differently to various abiotic stresses, including drought, extreme temperature, and CaCl2 treatment, suggesting their significance in stress response. This study presents a comprehensive overview of the potato IQD gene family and lays a foundation for further analysis of the StIQDs functions in plant development and stress response.
Introduction
Potato (Solanum tuberosum L.) is an annual Solanaceae crop that is widely cultivated worldwide (Xu et al., 2011). It is the fourth most food-cultivated crop worldwide after wheat, rice, and maize (Zhang et al., 2017). Its tubers are an important source of starch, protein, antioxidants, and vitamins and are also used for vegetative propagation (Burlingame et al., 2009).
There are numerous signaling pathways and complex networks in plants participating in plants’ response to stress (Mulligan et al., 1997; Lamers et al., 2020). Among them, calcium (Ca2+) signaling plays a major role in many biological functions (Schulz et al., 2013; Aldon et al., 2018). It utilized intracellular Ca2+ as a second messenger. The Ca2+-binding proteins act as Ca2+ sensors, thereby causing plants to respond to various biological processes by regulating the concentration of Ca2+ in the cytoplasm instantaneously (Bouché et al., 2005). There are four major classes of Ca2+ sensors in plants. They include calmodulins (CaMs), CaM-like proteins, calcineurin B–like proteins, and Ca2+-dependent protein kinases (Ranty et al., 2006). Calmodulins are among the most dominant proteins in Ca2+ sensors. Their role in Ca2+ signal transduction has been widely studied (Bouché et al., 2005). CaMs lack enzymatic activity but can interact and regulate the activity of the target proteins. The CaM-mediated signal can only play its role in regulating the physiological functions of cells through its downstream target protein CaM-binding proteins (CaMBPs) (Perochon et al., 2011; Kudla et al., 2018). Studies postulate that the IQ67 motif contains proteins, known as IQ67 domain (IQD) protein families. They are common representatives of CaMBPs (Abel et al., 2013; Bürstenbinder et al., 2017a). The IQD family proteins have two common features. It contains a conserved domain of 67 amino acid residues (the IQD) and a conserved exon–intron boundary that interrupts codons 16 and 17 via a phase-0 intron (Abel et al., 2005). To date, IQD gene families have been comprehensively analyzed in numerous plants. For instance, 33 IQD-like genes have been identified in Arabidopsis, 29 in rice, 26 in maize, 23 in Brachypodium, and 40 in poplar (Abel et al., 2005; Filiz et al., 2013; Ma et al., 2014; Cai et al., 2016).
The functions of IQD family genes have also been widely studied. In Arabidopsis, they regulate cell function, shape, and growth. Overexpression of AtIQD1 could enhance the accumulation of glucosinolate and strengthen resistance to herbivores (Levy et al., 2005; Bürstenbinder et al., 2017a). The IQD12/SUN of tomato regulates multiple tomato developmental processes, especially the morphological development of the leaves and fruit shape (Xiao et al., 2008; Wu et al., 2011). IQDs have been found to participate in abiotic stress responses. For example, 12 IQD members participate in poplar’s response to methyl jasmonate (MeJA) and Polyethylene glycol (PEG) treatment (Ma et al., 2014). In maize, the ZmIQD genes play crucial roles in response to drought stress (Cai et al., 2016). These reports indicate that the IQD genes can regulate plant development and response to stress or hormonal changes. However, IQD members and functions in potato are yet to be identified. In this study, the IQD gene family from the whole potato genome was identified. Detailed information about the IQDs regarding their chromosomal locations, synteny, and the phylogenetic relationship was also retrieved. In addition, expression levels of the IQDs in different tissues were analyzed using RNA-Seq data. Genes responsible for six abiotic stresses were further identified by quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR). This study provides a theoretical basis for further studies on the function of the IQD gene family in potato.
Materials and Methods
Identification of IQD Genes in Potato
The protein sequences of Arabidopsis thaliana IQD family members (AtIQDs) were downloaded from the TAIR database.1 The protein sequences of potato (version 4.03) were downloaded from PGSC.2 First, the AtIQD sequences were utilized as query, and BLASTP program was used to search against the potato proteins for the identification of the potato StIQD family. The sequences were selected as candidate StIQD if their E-value was < 0.001. Second, the candidate sequences were further confirmed by using the HMMER software3 with the Pfam IQD motif (PF00612) and SMART (Simple Modular Architecture Research Tool) online tool (Letunic et al., 2021).
Analysis of Sequences and Motifs
The physiochemical data [length, molecular weight, and theoretical isoelectric points (pIs)] of StIQD protein sequences were analyzed by the ProtParam server of Expasy.4 For the gene structure of StIQD analysis, the sequences of DNA and their corresponding coding sequences were analyzed by using GSDS (Hu et al., 2015). The conserved domains of StIQD gene family members in potato were analyzed by MEME online tool (Bailey et al., 2015), and the maximum number of motifs to identify was set to 10. In addition, the position of putative CaM-binding sites of each StIQD protein was predicted by the CaM Target Database (Yap et al., 2000).
Chromosomal Location and Gene Synteny
The chromosomal location of StIQD genes was based on genome annotation information in PGSC database and drafted by Mapgene2chrom online tool.5 For the synteny analysis of genes, the genome and genome annotation file of potato (version 4.03), tomato (version SL3.0) and Arabidopsis (version TAIR 10) were used, and the MCScan of python version6 (Python-version) was used to analyze the synteny.
Phylogenetic Analysis
Protein sequences of the potato, tomato, Arabidopsis, maize, and rice were aligned using ClustalX2 (Larkin et al., 2007), and the multiple sequence alignment result was used for phylogenetic analysis. For the phylogenetic analysis, the FastTree 2 software was used by using the maximum likelihood method, and the bootstrap value was set as 1,000 (Price et al., 2010).
RNA-Seq Data Analysis
The published transcriptome data (NCBI SRA database accession no. SRA030516) of different tissues of potato were used to analyze StIQDs expression patterns (Xu et al., 2011). For the expression analysis, normalized gene expression values FPKM (fragments per kilobase of exon per million fragments mapped) were first transformed using log2 (FPKM + 1) and then plotted by using ggplot2 in R (Wickham, 2011). The MeV7 was used to conduct the coexpression analysis. The gene expression levels were used and clustered using the k-means method. For the coexpressed genes, the agriGO (Tian et al., 2017) was used for GO enrichment analysis.
Plant Materials and Stress Treatments
The potato cultivar Desiree sterile tissue cultured seedlings for 15 days were transferred to perlite substrate and were initially irrigated with standard Hoagland culture medium for another 15 days. The culture temperature was set at 25°C, and the light cycle was set at 16 h/8 h (light/dark). Seedlings with consistent growth were selected and washed with 1/4 Hoagland culture medium for three times. The seedlings were treated with 150 mmol/L NaCl, 50 mmol/L CaCl2, 20% PEG6000, 4 and 35°C for 24 h, respectively. Each treatment was repeated for three times, and the same volume of double steaming water was used as the control. The leaves and root tissues of each treatment were quickly treated with liquid nitrogen and then stored at −80°C.
RNA Extraction and qRT-PCR Analysis
Total RNA was isolated from the collected samples using Trizol Reagents (Invitrogen, United States). After digestion with DNase I, the quantified of the RNA was evaluated by using NanoDrop 2000. Approximately 4 μg of total RNA was reverse transcribed by M-MLV reverse transcriptase (Promega, United States). The qRT-PCR was performed using SYBR Green (TaKaRa, Japan) and BioRad Real-Time System CFX96TM C1000 thermal cycler (Bio-Rad, Hercules, CA, United States). The PCR conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s. Three replicates were performed for each sample. The 2–ΔΔCt method was used to calculate the relative gene expression levels, and the potato housekeeping EF1α gene was used as reference (Livak and Schmittgen, 2001). The primer sequences used in this study were listed in Supplementary Table 1.
cis-Element Analysis
The putative promoters of IQDs, composed of 2-kb upstream of transcriptional start site, were acquired on the base of the annotation of potato genome. The PLANT CARE online software was used to analyze the cis-acting elements of promoters (Lescot et al., 2002).
Protein–Protein Interaction Network Analysis
For the protein–protein interaction (PPI) predicted analysis, all of the StIQD proteins sequences were first searched against the STRING database version 11.0 (Szklarczyk et al., 2019). The “confidence score” of STRING was used, and a confidence score of > 0.7 (high confidence) between proteins was used. The interaction networks of protein-protein generated by STRING were used to show the relationship of proteins with StIQDs.
Results
Genome-Wide Identification of IQD Gene Family
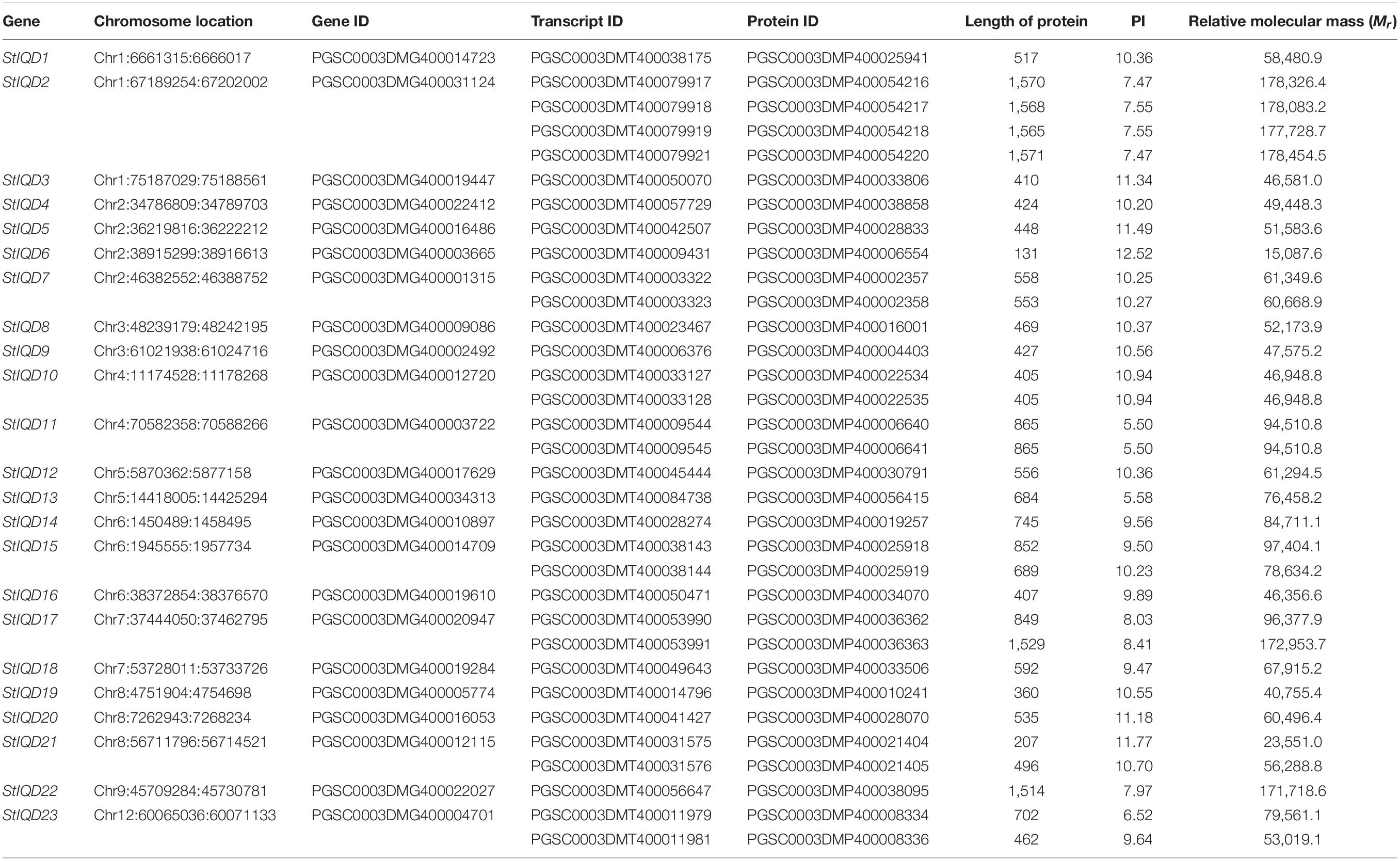
The IQD motif (PF00612) was used as the query sequence to search for similar potato protein sequence in the HMMER program. The search was performed to identify the IQD family members in potato. The SMART tool was then used to confirm whether the candidates contained the IQ domain. Consequently, 23 IQD genes that comprised 33 transcripts were identified and named as StIQD1 to StIQD23 according to the position of their corresponding genes on chromosomes 1–12 (Table 1, Supplementary Table 2, and Supplementary Figure 1).
The physical parameters of each IQD protein were subsequently calculated using the ExPASy server. As shown in Table 1, the full-length protein sequences of the StIQD ranged between 131 (StIQD6) and 1,571 amino acids (StIQD2). Their molecular weights ranged between 15,087.6 and 178,454.5 Da. Most of the StIQDs had relatively high pIs (9.38 ± 1.89), with only two having less than 7. This finding was in agreement with that of IQDs identified in other plants, such as Arabidopsis and maize (Levy et al., 2005; Cai et al., 2016).
Conserved Motifs and Structural Analysis of Potato IQD Genes
The MEME suite was used to identify the conserved domains of the 23 StIQD protein sequences. Motifs I and VI were the most common in StIQD protein sequences, occurring 13 and 10 times, respectively. In addition, we found that the 23 StIQD sequences contained either Motif I or Motif VI. However, the motifs did not occur together in a single StIQD sequence. The motif I contained the IQ67 motif (LQXXXRXXXXR), whereas motif VI contained the relaxed version of the IQ67 motif (IQXXXRGXXXR) (Figure 1). Prediction of the CaM-binding sites of each StIQD revealed that all StIQD proteins contained at least one string of high-scoring regions that indicated the location of the putative CaM interaction sites (Table 2). Evolutionary relationships of the StIQDs based on the 23 StIQD protein sequences revealed that they could be roughly categorized into two subfamilies: subfamilies I and II (Figure 2). The 23 StIQDs formed seven sister pairs with high bootstrap support (≥ 89%). The structural diversity of StIQD genes analyzed using their cDNA and genome sequences revealed that they had between 2 (StIQD6) and 38 (StIQD2, StIQD17, and StIQD22) exons. However, exons in subfamily I were more than those in subfamily II. Genes with the maximum number of exon cluster together. These results demonstrated the conservative nature of the motifs and the structural diversity of the genes.
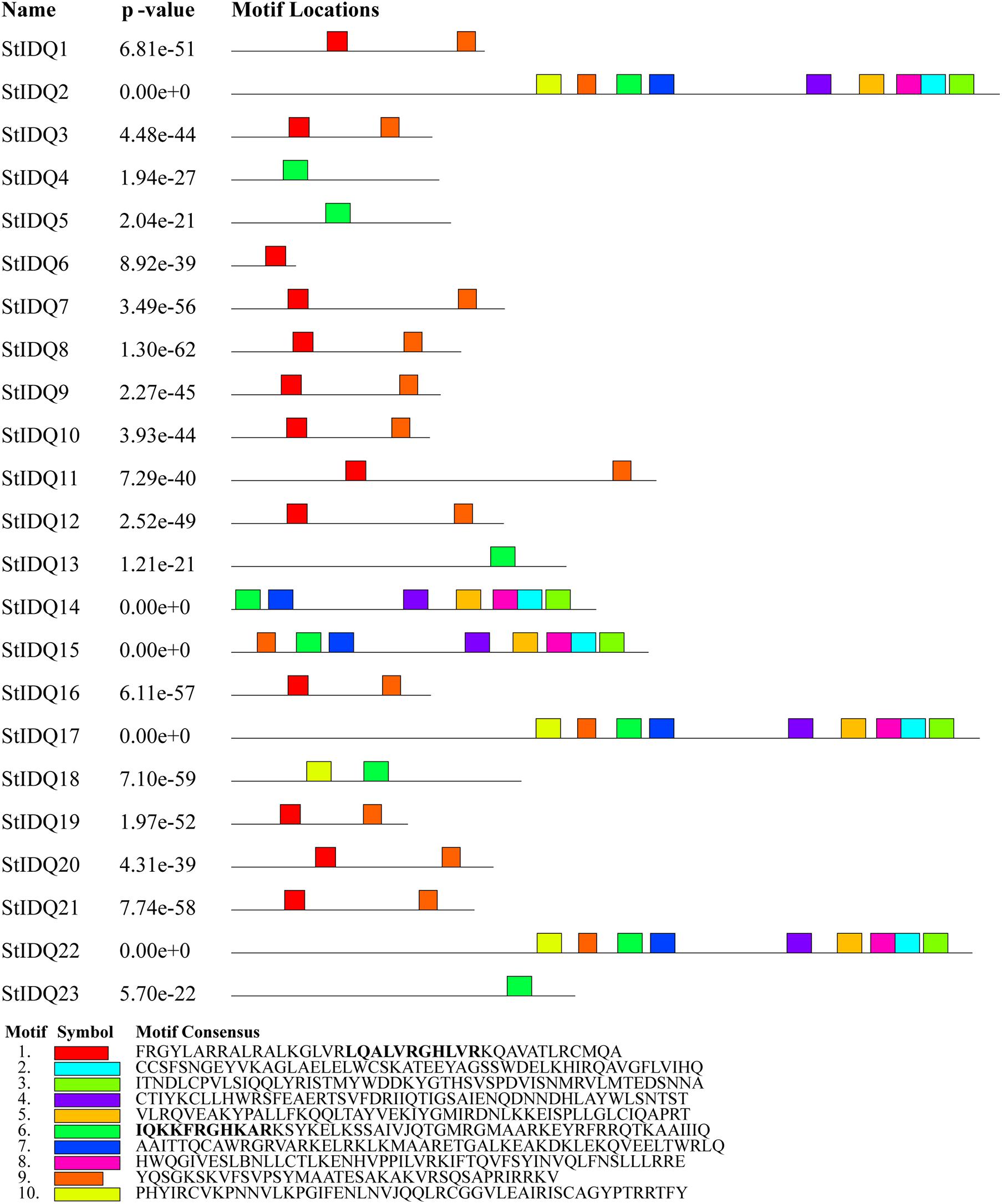
Figure 1. The motifs in IQD proteins of Solanum tuberosum L. (StIQD). Each motif was represented by different colored box. The sequence of each motif is listed below with the same color.
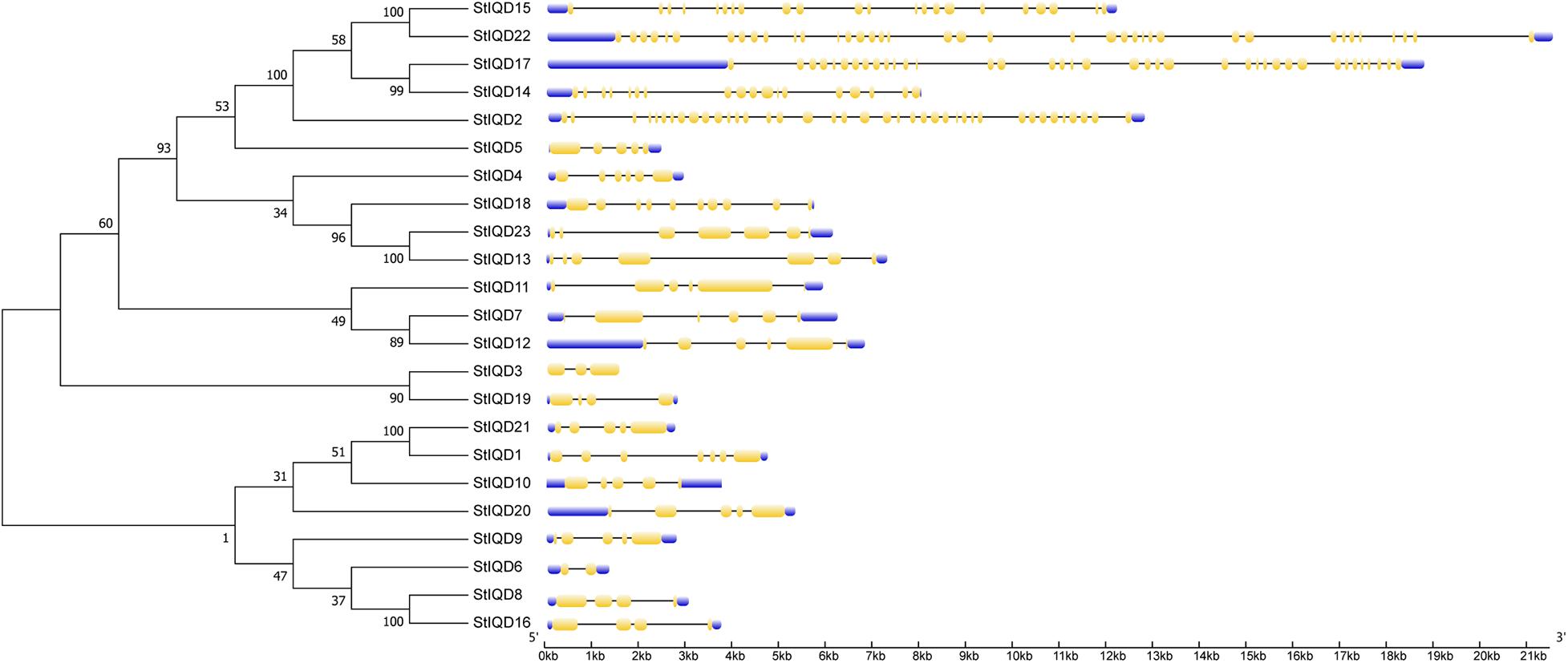
Figure 2. Phylogenetic analysis and exon–intron organization of StIQD. Maximum likelihood trees of full-length amino acid sequences encoded by StIQD (left) and exon–intron organization of StIQD (right). Exons: yellow boxes; introns: black lines; Untranslated regions: blue box.
Synteny Analysis of Potato With Arabidopsis and Tomato
The synteny blocks of potato with tomato and potato with Arabidopsis were analyzed further based on the total number of genes in potato, tomato, and Arabidopsis. There were 105 synteny blocks containing 18,988 pairs of synteny genes between the potato and tomato (Figure 3). The blocks contained nearly 50% of the total genes of the two genomes. Moreover, potato and Arabidopsis had 377 synteny blocks containing 4,872 pairs of syntenic genes. The blocks contained 10.6 and 15.6% of the total genes of potato and Arabidopsis, respectively. These results confirmed the evolutionary relationship results that found potato to be more closely related to the tomato (Xu et al., 2011). Further analysis of 5 pairs of synteny genes between potato and tomato and 16 pairs between potato and Arabidopsis revealed that three StIQDs (PGSC0003DMT400006376, PGSC0003DMT400041427, and PGSC0003DMT400045444) of potato showed synteny with those of Arabidopsis and tomato. These findings demonstrated the evolutionary conservatism and the important function of these genes. Interestingly, we found PGSC0003DMT400006376 showed synteny with AT1G17480.1 and AT1G72670.1, respectively, suggesting the possibility of a gene replication event.
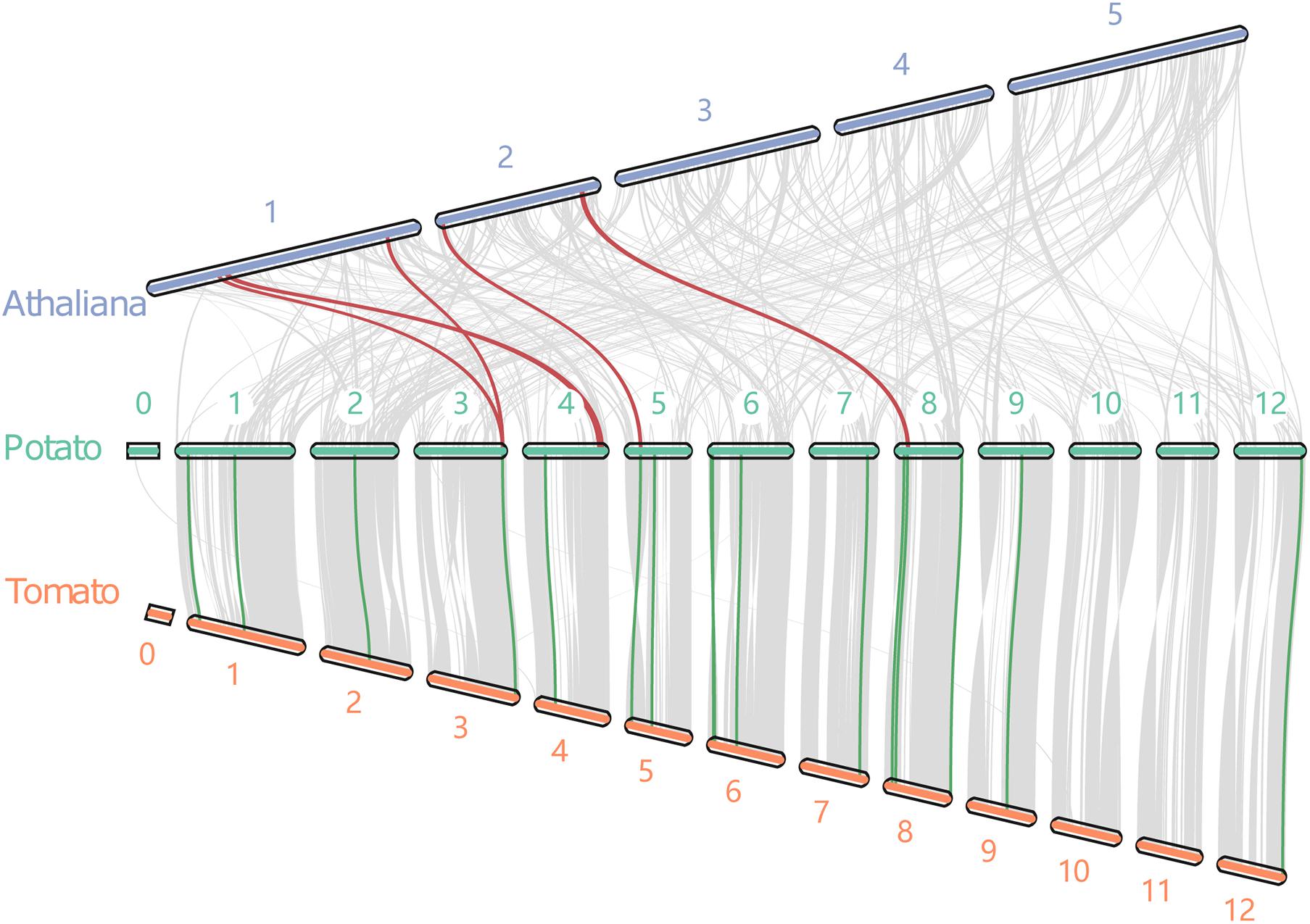
Figure 3. The collinear relationship between potato, tomato and Arabidopsis. Red and blue lines indicate the collinear gene pairs of potato and Arabidopsis, and potato and tomato, respectively.
Phylogenetic Analysis of Potato IQD Genes
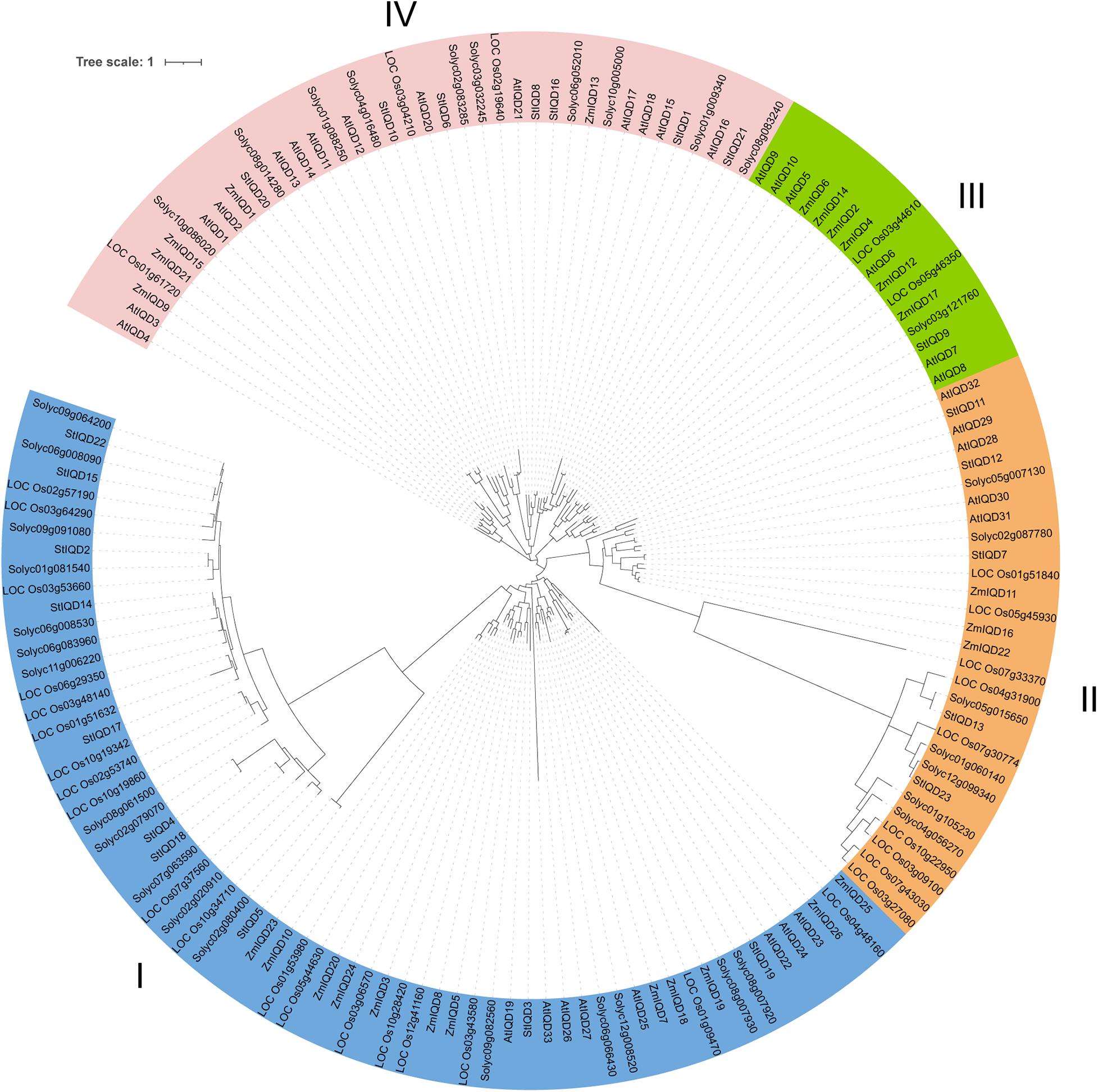
The protein sequences of the IQDs from Arabidopsis, tomato, maize, and rice were analyzed using the maximum likelihood (bootstrap = 1,000) method to understand the phylogenetic relationship of StIQD in plants. The analyzed IQDs clustered into four distinct classes (I–IV), with the potato StIQDs clustering in all four classes (Figure 4). Class I had most of the IQDs (66), whereas class III had the least IQD members (16). All the IQD family genes of dicotyledons were more closely conserved than monocots. While, in the combined phylogenetic trees, classes I and IV contained one ortholog pair, respectively, formed by Arabidopsis and rice orthologs AtIQD20/LOC_Os03g04210, and potato and rice orthologs Solyc02g020910/LOC_Os10g34710. The IQD genes of potato were more closely related to these of tomato. These results were consistent with the evolutionary relationships among the five species.
Expression Profiling of StIQD in Different Tissues
The expression levels of the StIQDs in different tissues were analyzed using the publicly available RNA-Seq data to reveal the roles of StIQDs in potato (Xu et al., 2011). Among the 23 StIQD genes, 19 expressed at least one tissue (FPKM > 1). Nineteen were divided into two major groups (Figure 5). The first group was composed of StIQD7, StIQD11, and StIQD20. The three exhibited relatively high expression levels in most tissues. The remaining 16 StIQD genes formed the second group. The group contained eight high-expression genes (StIQD2, StIQD8, StIQD12, StIQD13, StIQD17, StIQD18, StIQD22, and StIQD23) and eight low-expression genes (StIQD1, StIQD4, StIQD9, StIQD10, StIQD15, StIQD16, StIQD19, and StIQD21). In addition, we found that the tissues could also be divided into two clusters: the tissues of vegetative tissues (leaves, shoots, roots, and petioles) and the tissues of reproductive tissues (tubers, stamens, stolons, petals, sepals, flowers, callus, carpels). In addition, we found some of the StIQD genes exhibited a tissue-specific expression. For instance, the average expression level of StIQD20 in reproductive tissues is nearly three times (2.75-fold change) that in vegetative tissues. While the expression level of StIQD8 in vegetative tissue was higher, more than six times (6.47-fold change) of that in reproductive tissue. These results indicated that the StIQD genes were tissue-specific. Furthermore, in order to reveal the molecular network of IQD family in cellular metabolism, we classified the detected genes into 15 coexpression modules for 12 different tissues, and 19 expressed IQDs belong to 7 modules (Supplementary Figure 2). Then, the coexpression genes were annotated using GO enrichment method. We found the signal and response to stress-related terms were enriched in the coexpressed modules, such as “signal transduction,” “single organism signaling,” “cellular response to stress,” “cellular response to stimulus,” “response to oxidative stress,” and “response to hormone.” These results suggest that the IQDs and their coexpressed genes may play an important role in the signal transduction and stress response.
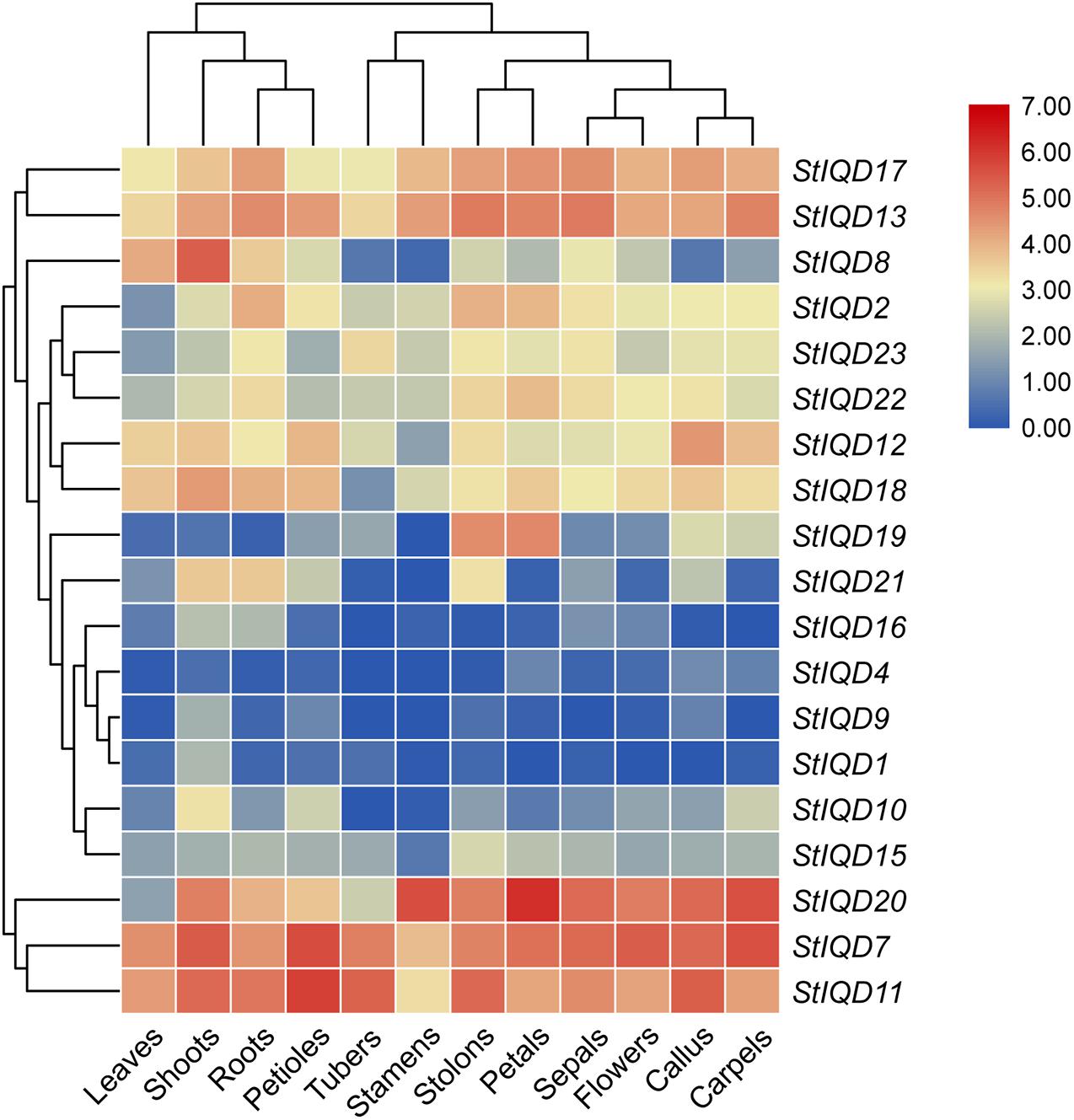
Figure 5. Hierarchical clustering of expression values of StIQD in 12 tissues. Red indicates high expression level; blue, low expression level.
Identification of the StIQD Genes Related to Abiotic Stresses
Heat, cold, PEG, NaCl, and CaCl2 stresses were induced to explore the role of StIQD genes in abiotic stresses. Fifteen StIQD genes were cloned and analyzed by qRT-PCR. The expression of 14 of the 15 genes was either induced or suppressed under a specific stressor in specific tissues (Figure 6). Most of the genes were up-regulated in leaves but down-regulated in the roots. StIQD6 and StIQD23 participated in almost all stress responses. Notably, the expression of StIQD6 increased nearly 30 times under heat stress (35°C). These findings suggested that StIQD6 and StIQD23 played crucial roles in response to various stresses. Seven StIQD genes (StIQD3, StIQD6, StIQD7, StIQD11, StIQD15, StIQD17, StIQD20, and StIQD21) were down-regulated in the roots under PEG stress. Among them, StIQD7, StIQD15, StIQD17, and StIQD21 responded only to PEG stress, indicating they played a negative role in response to PEG. StIQD15 was up-regulated (2.41-fold change) in the leaf but down-regulated (0.15-fold change) in the root under PEG stress, indicating the diverse role of StIQD15 in response to PEG stress. Besides, eight StIQD genes (StIQD3, StIQD5, StIQD6, StIQD13, StIQD17, StIQD20, StIQD22, and StIQD23) responded to calcium stimulation. The majority of the eight were up-regulated. StIQD20 was up-regulated (2.75-fold change) in the root, whereas it was down-regulated (0.42-fold change) in the leaf. These results demonstrated that StIQD genes play crucial roles in multiple stresses and exhibit different responses in various organizations.
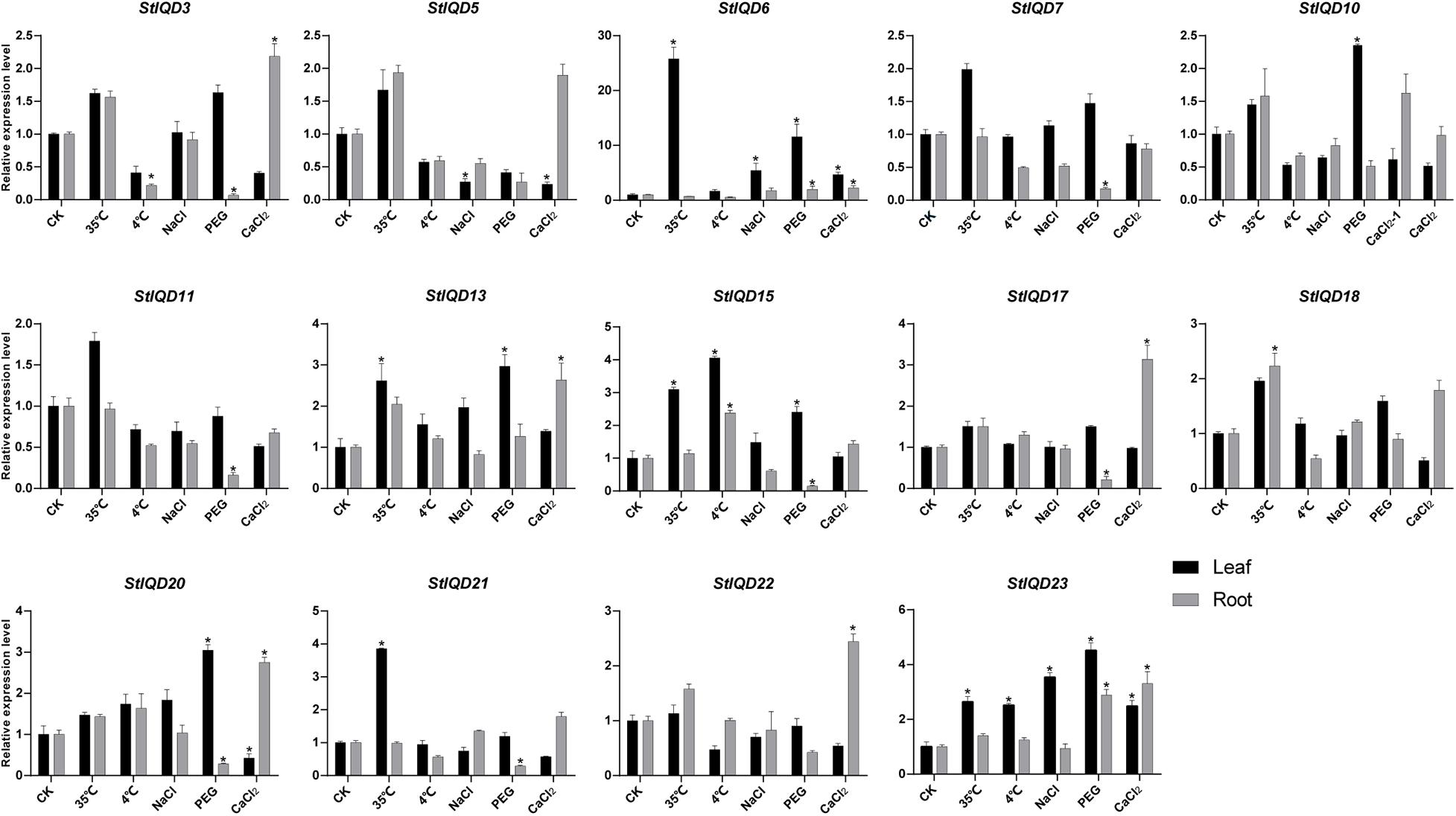
Figure 6. qRT-PCR analysis of StIQDs genes in response to heat, cold, NaCl, CaCl2, and PEG. Error bars represent standard error of the mean (n = 3). *P < 0.05.
Analysis of cis-Acting Elements of SOD Gene Promoters
The cis-acting elements play an important role in cell regulatory network. To explore the potential roles of the potato IQD genes under environmental stresses, the IQD gene promoters coupled with 2-kb genomic sequences upstream of the transcriptional start site were analyzed by PlantCARE online tool. The IQD family genes possessed a variety of elements, including the hormone-related responsive elements: ABA (ABRE), MeJA, and salicylic acid; the abiotic stress-related responsive elements: MBS (Drought) and ARE (anaerobic induction); and the light response elements: AE-box, Box 4, G-box, GATA motif, and GT1 motif (Figure 7). Among them, we found 13 of 23 IQD family genes contained MBS elements. For instance, StIQD6, which contained two MBS elements could be up-regulated in four abiotic stresses (heat, NaCl, PEG, and CaCl2), suggesting the two MBS elements may contribute to the stress response. Further, 10 of the above 13 IQDs were analyzed by qRT-PCR and found that 8 (StIQD3, StIQD6, StIQD7, StIQD11, StIQD13, StIQD15, StIQD20, StIQD23) of which could be responsive to the PEG stress (Figure 6). These results suggested that IQD family genes play important roles in abiotic stresses and might be related to hormone stimuli.
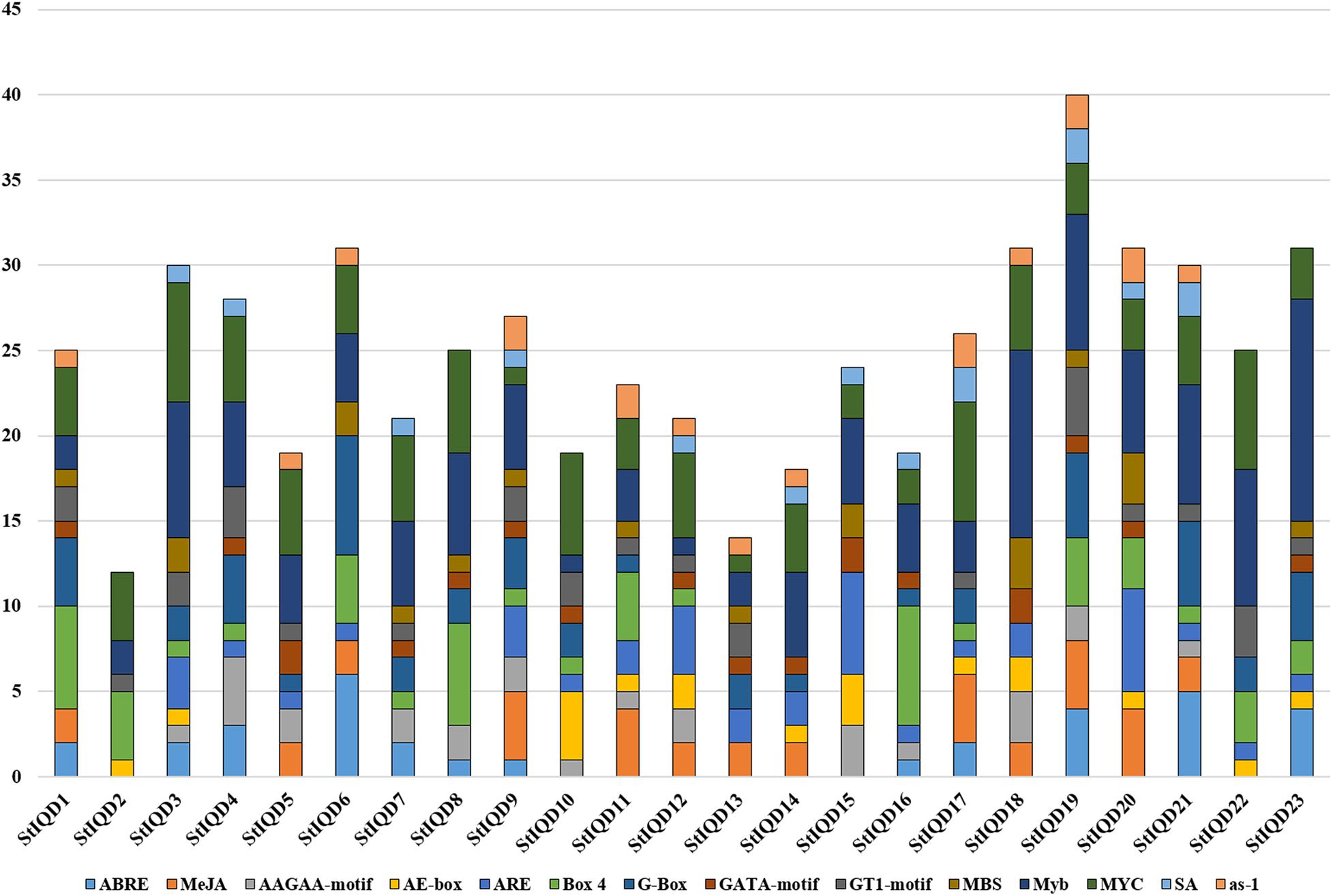
Figure 7. The cis-acting elements of putative IQD promoters predicted by PlantCARE. The different cis-elements are present with different colors.
Protein–Protein Interaction Analysis
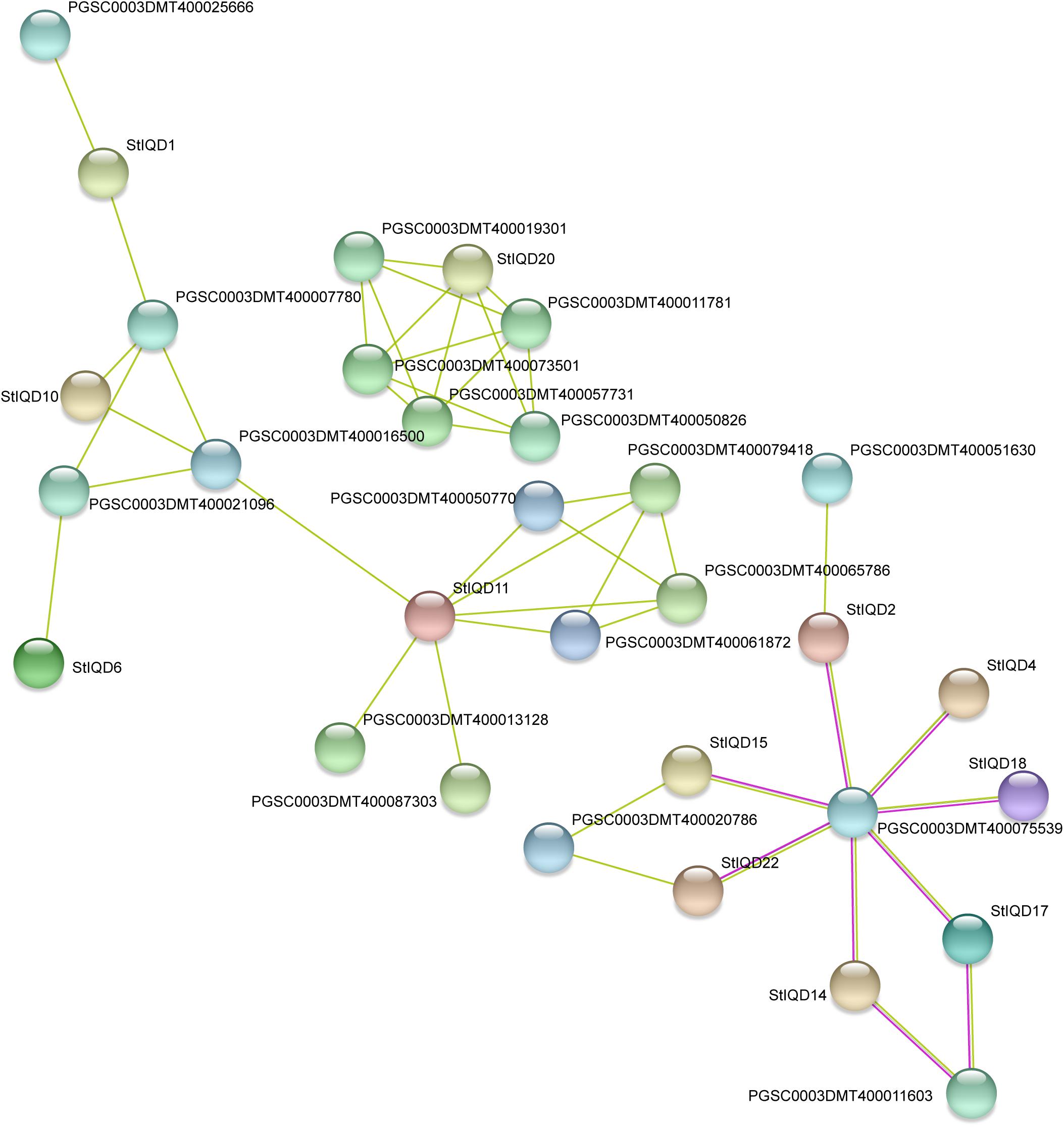
In order to identify the function of IQD genes, the PPI networks of IQD protein were built by using the STRING database. The results showed that the PPI networks can be divided into three groups and contained 12 StIQDs (Figure 8). StIQD11 was predicted to interact with most proteins (six proteins), in which two encoded the amine oxidase, which play important roles in plant development and stress response. StIQD20 was predicted to interact with the five proteins, in which two belong to the NB-LRR family proteins (PGSC0003DMT400019301 and PGSC0003DMT400050826). This indicates that StIQDs may be involved in abiotic stress responses. In addition, we found the gene PGSC0003DMT400075539, which encodes an exocyst complex component and implicated in tethering secretory vesicles to the plasma membrane (Heider and Munson, 2012), was predicted to interact with seven StIQDs, and these IQDs contained the myosin-related motif, such as the “Myosin_head” and “Myosin_N,” indicating these StIQDs may participate in the vesicle transport. These results suggest that the StIQDs may interact with these proteins to take part in potato development.
Discussion
Plants evolve many specific gene families to adapt to environmental changes because of the specificity of plant growth. IQD genes are among the specific gene families that are widespread in photosynthetic plants (Abel et al., 2013). Previous studies postulate that IQD participate in various crucial biological processes in plants (Bürstenbinder et al., 2017a,b). Herein, bioinformatics analyses were employed to analyze the IQDs gene family of potato at the whole-genome level.
There were 23 non-redundant IQD genes identified in the potato genome. This number was not significantly different from that of Arabidopsis, which has 33 IQD genes (Abel et al., 2005). Although the proteins all belong to the StIQDs, the length, molecular weight, and pI of proteins showed a significant difference. For instance, the StIQD6 contained only 131 amino acid residues and contained only 1 IQD domain, whereas the StIDQ2 contained 1,571 amino acid residues and contained not only IQD domains but also the myosin-related domains (Myosin_N, MYSc, and DIL). The diversity of gene sequence length leads to the diversity of gene function. For an amino acid, the pI is the average of pKa values for the amine and the carboxyl group, which are the pH value at which the molecule carries no electrical charge (Moldoveanu and David, 2017). We found the lowest pI of StIQD was 5.5 (StIQD11), whereas the highest pI was 12.52 (StIQD6), indicating the complex properties of StIQDs.
The IQDs genes of potato were divided into two classes: the conventional IQ67 motif (LQXXXRXXXXR) and the more relaxed version IQ67 motif (IQXXXRGXXXR). In Arabidopsis, four IQ67 motifs have been identified. They include [IQXXXRGXXXR or (ILV) QXXXRXXXX (R, K)], the 1-5-10 motif [(FILVW)X3(FILV)X4(FILVW)], and the 1-8-14 motif [(FILVW)X6(FAILVW)X5(FILVW)] (Abel et al., 2005). The conventional IQ67 mediates calmodulin retention in a Ca2+-independent manner. Besides, the 1-5-10 motif and the 1-8-14 motif function in a Ca2+-dependent manner (Rhoads and Friedberg, 1997; Choi et al., 2002). Nonetheless, the difference between the conventional IQ67 motif and the relaxed version IQ67 motif has not been reported. Among of the 33 StIQD proteins encoded by 23 StIQDs, only one transcript of each gene predicting the calmodulin-binding site motif was consistent with the IQ67. These findings suggest that the alternative splicing events of genes lead to the diversity pattern of calmodulin-binding sites, which may be related to the functional diversity of the StIQD.
A comparative genomic analysis of plant IQD members from two dicotyledonous (Arabidopsis and tomato) and two monocotyledonous (maize and rice) plants was performed to explore the phylogenetic relationship of the IQDs. The phylogenetic analysis categorized the IQDs from the five plants into four distinct classes. IQD proteins in the same class were closer to each other than proteins from the same species in different classes. Nonetheless, the closeness was more between members of one family in the same class than others. Class IV did not contain IQD gene from Arabidopsis and maize. This finding suggests that the IQDs in this class evolve after species differentiation and may play a significant role in species generation. Two orthologous IQD pairs between rice and tomato (three) and rice and potato (one) were identified. These results indicated that the IQDs possibly exist before the dicotyledon–monocotyledon division. Synteny analysis is an important method for identity phylogenetic character (Tang et al., 2008). In this study, we found nearly 50% potato and tomato genes showed synteny. However, only 10.6% of potato genes and 15.6% of Arabidopsis genes showed synteny. These findings suggest that potatoes are more closely related to tomatoes.
Previous studies postulated that the IQD genes play an important role in plant development and stress response (Tang et al., 2008; Sugiyama et al., 2017; Yang et al., 2019). In this study, we found the expression of the StIQD had a close relationship with the type of organization that vegetative tissue and reproductive tissue were clearly divided into two classes. The tissue-specific expression patterns of IQD genes have also been reported in other plants such as poplar, tomato, and soybean (Huang et al., 2013; Feng et al., 2014; Ma et al., 2014). StIQD20 was highly expressed in the reproductive tissues but low in the vegetative tissues. In contrast, StIQD5 was higher in vegetative tissues but lower in reproductive tissues. The homolog gene of StIQD20 in Arabidopsis, AtIQD13, regulates xylem cell-wall deposition (Sugiyama et al., 2017). The differential function of this homolog pair is attributed to species specificity. In addition, 9 of 14 StIQDs could respond to at least two abiotic stresses among the five tested, indicating the StIQDs play important roles in plant response to stress. In Populus leaves, many IQD genes were up-regulated by PEG stress. Similarly, the BrIQD5 of Chinese cabbage is also significantly induced by PEG (Ma et al., 2014; Yuan et al., 2019). Herein, most of the studied StIQDs responded to PEG treatment. However, the regulation pattern was different in the leaf and roots; some were up-regulated in the leaf but down-regulated in the root. The differential expression was attributed to the different ways roots and leaves respond to drought stress. Hence, the specific mechanism should be studied further. Most StIQDs responded to heat stress than cold stress. Notably, StIQD6 was up-regulated by > 25-fold after heat treatment, suggesting the crucial roles of StIQDs in heat stress than in freezing stress. Ca2+ is a pivotal cytosolic second messenger, which plays a prominent role in plant growth and development, and the CaM has been extensively studied (Ng et al., 2001). When exposed to CaCl2, we found eight StIQDs were differently expressed, and most of which were up-regulated in leaves and roots. This result suggests that these StIQDs interact with CaM to transmit calcium signals at the protein level and respond to Ca2+ stress at the transcription level.
In conclusion, 23 IQD genes were identified in potato. These genes had diverse exon/intron structures, motif distribution, and phylogenetic relationships that potentially contributed to their diverse functions. RNA-Seq data analyses revealed that some of the StIQDs were expressed in a tissue-specific pattern and thus were strongly related to tissue development. qRT-PCR results further confirmed that StIQDs responded differently to various abiotic stresses, including drought, extreme temperature, and CaCl2 treatment. This study provides baseline information for further studies on IQD proteins in potato.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
CM, YL, and XD performed the experiments, analyzed the data, and wrote the manuscript. CM and RF conceived the research and revised the manuscript. All the authors read and approved the final manuscript.
Funding
This research was supported by the Key Research and Development Project of Shanxi Province (201903D221076), the Applied Basic Research Program of Shanxi Academy of Agricultural Sciences (YGC2019FZ1), and the Doctor Foundation of Institute of Crop Science, Shanxi Academy of Agricultural Sciences (ZB1901).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.693936/full#supplementary-material
Supplementary Figure 1 | The chromosome location of the IQD gene family in potato.
Supplementary Figure 2 | The coexpression gene module of expressed genes in 12 tissues.
Supplementary Table 1 | Primers used in this study.
Supplementary Table 2 | The sequences (gene, corresponding coding sequence, proteins) of StIQDs.
Footnotes
- ^ http://www.arabidopsis.org
- ^ http://solanaceae.plantbiology.msu.edu
- ^ http://hmmer.org/
- ^ https://web.expasy.org/protparam/
- ^ http://mg2c.iask.in/mg2c_v2.1/
- ^ https://github.com/tanghaibao/jcvi/wiki/MCscan
- ^ http://mev.tm4.org/
References
Abel, S., Bürstenbinder, K., and Müller, J. (2013). The emerging function of IQD proteins as scaffolds in cellular signaling and trafficking. Plant Signal. Behav. 6:e24369. doi: 10.4161/psb.24369
Abel, S., Savchenko, T., and Levy, M. (2005). Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol. Biol. 5:72.
Aldon, D., Mbengue, M., Mazars, C., and Galaud, J. P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 3:665. doi: 10.3390/ijms19030665
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49.
Bouché, N., Yellin, A., Snedden, W. A., and Fromm, H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev Plant Biol. 56, 435–466. doi: 10.1146/annurev.arplant.56.032604.144224
Burlingame, B., Mouillé, B., and Charrondière, R. (2009). Nutrients, bioactive non-nutrients and anti-nutrients in potatoes. J. Food Compost. Anal. 6, 494–502. doi: 10.1016/j.jfca.2009.09.001
Bürstenbinder, K., Möller, B., Plötner, R., Stamm, G., Hause, G., Mitra, D., et al. (2017a). The IQD family of calmodulin-binding proteins links calcium signaling to microtubules, membrane subdomains, and the nucleus. Plant Physiol. 3:1692. doi: 10.1104/pp.16.01743
Bürstenbinder, K., Mitra, D., and Quegwer, J. (2017b). Functions of IQD proteins as hubs in cellular calcium and auxin signaling: a toolbox for shape formation and tissue-specification in plants? Plant Signal. Behav. 12, e1331198. doi: 10.1080/15592324.2017.1331198.
Cai, R., Zhang, C., Zhao, Y., Zhu, K., Wang, Y., Jiang, H., et al. (2016). Genome-wide analysis of the IQD gene family in maize. Mol. Genet. Genomics 2, 543–558. doi: 10.1007/s00438-015-1122-7
Choi, J. Y., Lee, S. H., Park, C. Y., Heo, W. D., Kim, J. C., Kim, M. C., et al. (2002). Identification of calmodulin isoform-specific binding peptides from a phage-displayed random 22-mer peptide library. J. Biol. Chem. 24, 21630–21638. doi: 10.1074/jbc.m110803200
Feng, L., Chen, Z., Ma, H., Chen, X., Li, Y., Wang, Y., et al. (2014). The IQD gene family in soybean: structure, phylogeny, evolution and expression. PLoS One 10:e110896. doi: 10.1371/journal.pone.0110896
Filiz, E., Tombuloglu, H., and Ozyigit, I. I. (2013). Genome wide analysis of IQ67 domain (IQD) gene families in Brachypodium distachyon. Plant Omics 6, 425–432.
Heider, M. R., and Munson, M. (2012). Exorcising the exocyst complex. Traffic 13, 898–907. doi: 10.1111/j.1600-0854.2012.01353.x
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, Z., Van Houten, J., Gonzalez, G., Xiao, H., and van der Knaap, E. (2013). Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genomics 3-4, 111–129. doi: 10.1007/s00438-013-0733-0
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytol. 2, 414–431. doi: 10.1111/nph.14966
Lamers, J., van der Meer, T., and Testerink, C. (2020). How plants sense and respond to stressful environments. Plant physiol. 4:1624. doi: 10.1104/pp.19.01464
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Peer, Y. V. D., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Rese. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460.
Levy, M., Wang, Q., Kaspi, R., Parrella, M. P., and Abel, S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 1, 79–96. doi: 10.1111/j.1365-313x.2005.02435.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 4, 402–408. doi: 10.1006/meth.2001.1262
Ma, H., Feng, L., Chen, Z., Chen, X., Zhao, H., and Xiang, Y. (2014). Genome-wide identification and expression analysis of the IQD gene family in Populus trichocarpa. Plant Sci. 229, 96–110. doi: 10.1016/j.plantsci.2014.08.017
Moldoveanu, S. C., and David, V. (2017). “Chapter 5–Properties of analytes and matrices determining HPLC selection,” in Selection of the HPLC Method in Chemical Analysis, eds S. C. Moldoveanu and V. David (Boston, MA: Elsevier), 189–230. doi: 10.1016/B978-0-12-803684-6.00005-6
Mulligan, R. M., Chory, J., and Ecker, J. R. (1997). Signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 7:2793.
Ng, C. K. Y., McAinsh, M. R., Gray, J. E., Hunt, L., Leckie, C. P., Mills, L., et al. (2001). Calcium-based signalling systems in guard cells. New Phytol. 1, 109–120. doi: 10.1046/j.1469-8137.2001.00152.x
Perochon, A., Aldon, D., Galaud, J. P., and Ranty, B. (2011). Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 12, 2048–2053. doi: 10.1016/j.biochi.2011.07.012
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 - approximately maximum-likelihood trees for large alignments. PloS one 5:e9490. doi: 10.1371/journal.pone.0009490
Ranty, B., Aldon, D., and Galaud, J. P. (2006). Plant calmodulins and calmodulin-related proteins: multifaceted relays to decode calcium signals. Plant Signal. Behav. 3, 96–104. doi: 10.4161/psb.1.3.2998
Rhoads, A. R., and Friedberg, F. (1997). Sequence motifs for calmodulin recognition. FASEB J. 5, 331–340. doi: 10.1096/fasebj.11.5.9141499
Schulz, P., Herde, M., and Romeis, T. (2013). Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 2:523. doi: 10.1104/pp.113.222539
Sugiyama, Y., Wakazaki, M., Toyooka, K., Fukuda, H., and Oda, Y. A. (2017). A novel plasma membrane-anchored protein regulates xylem cell-wall deposition through microtubule-dependent lateral inhibition of Rho GTPase domains. Curr. Biol. 16, 2522–2528. doi: 10.1016/j.cub.2017.06.059
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613.
Tang, H., Bowers, J. E., Wang, X., Ming, R., Alam, M., and Paterson, A. H. (2008). Synteny and collinearity in plant genomes. Science 5875:486. doi: 10.1126/science.1153917
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2. 0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129.
Wickham, H. (2011). “ggplot2,”. in Wiley Interdisciplinary Reviews: Computational Statistics 3, eds J. E. Gentle and D. W. Scott (Hoboken, NJ: Wiley), 180–185. doi: 10.1002/wics.147
Wu, S., Xiao, H., Cabrera, A., Meulia, T., and van der Knaap, E. (2011). SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 157, 1175–1186. doi: 10.1104/pp.111.181065
Xiao, H., Jiang, N., Schaffner, E., Stockinger, E. J., and van der Knaap, E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 5869, 1527–1530. doi: 10.1126/science.1153040
Xu, X., Pan, S., Cheng, S., Zhang, B., Mu, D., Ni, P., et al. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Yang, X., Kirungu, J. N., Magwanga, R. O., Xu, Y., Pu, L., Zhou, Z., et al. (2019). Knockdown of GhIQD31 and GhIQD32 increases drought and salt stress sensitivity in Gossypium hirsutum. Plant Physiol. Biochem. 144, 166–177. doi: 10.1016/j.plaphy.2019.09.027
Yap, K. L., Kim, J., Truong, K., Sherman, M., Yuan, T., and Ikura, M. (2000). Calmodulin target database. J. Struct. Funct. Genomics 1, 8–14.
Yuan, J., Liu, T., Yu, Z., Li, Y., Ren, H., Hou, X., et al. (2019). Genome-wide analysis of the Chinese cabbage IQD gene family and the response of BrIQD5 in drought resistance. Plant Mol. Biol. 6, 603–620. doi: 10.1007/s11103-019-00839-5
Keywords: potato, IQD gene family, phylogenetic analysis, abiotic stress, expression profile
Citation: Mei C, Liu Y, Dong X, Song Q, Wang H, Shi H and Feng R (2021) Genome-Wide Identification and Characterization of the Potato IQD Family During Development and Stress. Front. Genet. 12:693936. doi: 10.3389/fgene.2021.693936
Received: 12 April 2021; Accepted: 16 June 2021;
Published: 27 July 2021.
Edited by:
Madhav P. Nepal, South Dakota State University, United StatesReviewed by:
Ertugrul Filiz, Duzce University, TurkeyDawei Xue, Hangzhou Normal University, China
Mehanathan Muthamilarasan, University of Hyderabad, India
Fuyou Fu, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2021 Mei, Liu, Dong, Song, Wang, Shi and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruiyun Feng, RmVuZ3J1aXl1bjE5NzBAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Chao Mei1†
Chao Mei1† Yuwei Liu
Yuwei Liu Ruiyun Feng
Ruiyun Feng