
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 26 August 2021
Sec. Livestock Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.691272
This article is part of the Research Topic The Effects of Genetic Mutations in Poultry View all 4 articles
 Tong Li1†
Tong Li1† Bingjie Chen1†
Bingjie Chen1† Chengjie Wei1
Chengjie Wei1 Dan Hou1
Dan Hou1 Panpan Qin1
Panpan Qin1 Zhenzhu Jing1
Zhenzhu Jing1 Haoran Ma1
Haoran Ma1 Xinran Niu1
Xinran Niu1 Chunxiu Wang1
Chunxiu Wang1 Ruili Han1
Ruili Han1 Hong Li1
Hong Li1 Xiaojun Liu1,2
Xiaojun Liu1,2 Huifen Xu1
Huifen Xu1 Xiangtao Kang1,2
Xiangtao Kang1,2 Zhuanjian Li1,2*
Zhuanjian Li1,2*Analyzing marker-assisted breeding is an important method utilized in modern molecular breeding. Recent studies have determined that a large number of molecular markers appear to explain the impact of “lost heritability” on human height. Therefore, it is necessary to locate molecular marker sites in poultry and investigate the possible molecular mechanisms governing their effects. In this study, we found a 104-bp insertion/deletion polymorphism in the 5′UTR of the ADPRHL1 gene through resequencing. In cross-designed F2 resource groups, the indel was significantly associated with weight at 0, 2, 4, 6, and 10 weeks and a number of other traits [carcass weight (CW), semi-evisceration weight (SEW), evisceration weight (EW), claw weight (CLW), wings weight (DWW), gizzard weight (GW), pancreas weight (PW), chest muscle weight (CMW), leg weight (LW), leg muscle weight (LMW), shedding Weight (SW), liver rate (LR), and leg muscle rate (LMR)] (P < 0.05). In brief, the insertion-insertion (II) genotype was significantly associated with the greatest growth traits and meat quality traits, whereas the values associated with the insertion-deletion (ID) genotype were the lowest in the F2 reciprocal cross chickens. The mutation sites were genotyped in 4,526 individuals from 12 different chicken breeds and cross-designed F2 resource groups. The II genotype is the most important genotype in commercial broilers, and the I allele frequency observed in these breeds is relatively high. Deletion mutations tend to be fixed in commercial broilers. However, there is still considerable great potential for breeding in dual-purpose chickens and commercial laying hens. A luciferase reporter assay showed that the II genotype of the ADPRHL1 gene possessed 2.49-fold higher promoter activity than the DD genotype (P < 0.05). We hypothesized that this indel might affect the transcriptional activity of ADPRHL1, thereby affecting the growth traits of chickens. These findings may help to elucidate the function of the ADPRHL1 gene and facilitate enhanced reproduction in the chicken industry.
ADP-ribosylhydrolase like-1 (ADPRHL1, encoded by the ADPRHL1 gene) is a pseudoenzyme expressed during myocardial development in all vertebrates (Smith et al., 2020). Pseudoenzymes cannot catalyze chemical reactions, but researchers have observed that they can perform a variety of other tasks in cells (Leslie, 2013; Adrain, 2020). Accumulating evidence has shown that these pseudoenzymes play important roles in regulating inflammation and growth factor signal transduction; therefore so they play important roles in growth and development and in many diseases (Zhang et al., 2014; Adrain, 2020).
ADPRHL1 is so named because of its sequence similarity to a small group of ADP-ribosylhydrolase enzymes encoded in vertebrate genomes: ADPRH (sometimes called ARH1), ADPRHL1 (ARH2), and ADPRHL2 (ARH3). Mono-ADP-ribosylation of proteins is a post-translational modification catalyzed by ADP-ribosyltransferase and a number of bacterial toxins (Tsuge and Tsurumura, 2014). The natural protein target of ADPRH has not been fully elucidated, and while mice lacking ADPRH are viable, evidence obtained from this knockout indicates that ADPRH acts as an important tumor suppressor (Tang et al., 2013). In zebra fish embryos, the expression of ADPRH was briefly detected in the formed somatic cells, suggesting that ADP-ribosylation may be involved in skeletal muscle development (Reischauer et al., 2014). Studies on another member of the homologous gene family, ADP-ribosylhydrolase-like 2 (ADPRHL2), showed that it has a 22% amino acid sequence homology with ADPRH and can act on two distinct classes of substrates.
ADPRHL1 (Ono et al., 2006) is another member of this protein family, and the 354-amino-acid sequences of human ADPRHL1 exhibits have 46% identity to ADPRH. Interestingly, ADPRHL1 seems to lack similar enzymatic activity compared to the other two proteins (Ono et al., 2006). Pseudoenzymes similar to ADPRHL1 can be a challenge to study, but there is accumulating evidence that ADPRHL1 may be an important factor in cardiogenesis. Gene knockdown and overexpression experiments demonstrate that ADPRHL1 is essential for heart chamber outgrowth and that alteration of ADPRHL1 expression levels affects myofibril assembly. Elevated ADPRHL1 is associated with disarrayed myofibril patterns, contractile filaments with diverging orientations and prominent branches at the actin-Z-disc boundary (Smith et al., 2016). However, to date, whether ADPRHL1 plays a similar role in the growth and development of poultry is unknown.
Long sequence indels longer than 50 bp are also called structural variation (SVs), which is the main mechanism of genome evolution (Kidd et al., 2010; Depristo et al., 2011). Genomic SVs seem to be more useful than SNPs to explain the diversity of human populations (Zhuo et al., 2020). Accumulating researchers have focused on the importance of SVs for animal phenotypes and diseases. For example, there has been found that SVs are associated with a variety of complex traits in dairy cows, and may potentially contribute to improving the accuracy of genomic prediction of dairy traits (Chen et al., 2021).
However, whether ADPRHL1 gene has a similar effect in poultry and the polymorphism of ADPRHL1 gene has not been studied. Here, a novel 104-bp SV in the 5′UTR of the ADPRHL1 gene was employed as a genetic marker, and genotyped in an F2 designed full-sib resource population. The association of ADPRHL1 gene polymorphisms with chicken growth and carcass traits was analyzed. Our dataset confirms the importance of pseudoenzymes for biological growth. The mRNA levels of the ADPRHL1 gene were determined by qPCR to investigate the expression pattern of the gene and to confirm the deduced association between ADPRHL1 polymorphisms and phenotypes. Our dataset elucidates the importance of pseudoenzymes for biological growth.
All applicable international, national, and institutional guidelines for the care and use of animals were followed. All animal experiments were performed according to the Regulations of the Chinese National Research Council (1994) and approved by the Henan Agricultural University Institutional Animal Care and Use Committee (Permit Number: 11–0085).
The F2 resource population was constructed by cross-breeding Gushi chickens (GSs) and Anak broiler chickens (AKs). The F1 generation was formed through the hybridization of 24 Gushi hens with 4 Anak cocks and 12 Anak hens with 2 Gushi cocks. From this hybrid combination, individuals were selected according to phenotypic quality and heterozygosity to increase the separation of F2 traits, and a maximum of nine hens were randomly selected to mate with each rooster. F2 individuals consisted of seven families and were mated with unrelated hens (the cock was not related to the female), resulting in a total of 783 F2 chickens. The F2 population of the hybrid offspring was raised in the same environment, and free food and water were provided to the animals. At 84 days of age, each chicken was sacrificed by cervical dislocation and decapitation. The following growth characteristics were measured during this period: body weight (BW) at 0 days and 2, 4, 6, 8, 10, and 12 weeks; shank length (SL) at 0, 4, 8, and 12 weeks; and shank girth (SG), chest width (CW), body slant length (BSL), and pelvis width (PW) at 4, 8, and 12 weeks. All the detailed measuring methods were described previously (Ren et al., 2019).
Two blood samples were taken from the jugular vein during slaughter. One sample was placed in a centrifuge tube for separation of the serum and stored at −80°C. The other sample was placed in an anticoagulation tube for DNA extraction and subsequently stored at −20°C. After blood collection, 783 individuals were slaughtered to determine the carcass weights, such as carcass weight (CW), semi-evisceration weight (SEW), evisceration weight (EW), and claw weight (CLW).
To determine the presence or absence of the insertion/deletion allele at the locus of interest, DNA from 4,526 individuals was collected from blood. Samples were obtained from the F2 generation resource population, commercial broilers, dual-purpose chickens, commercial laying hens, and local Chinese chickens. The number of samples provided by each group was as follows: F2 populations (F2, n = 783), Ross 308 broilers (n = 192), breed 817 (817, n = 74), Arbor Acres broilers (AA, n = 552), Cobb (n = 192), Hubbard (n = 595), Gushi chicken (GS, n = 284), Guifei chickens (GF, n = 156), Wuhei green-eggshell (WH, n = 441), Xichuan black-bone (XC, n = 572), Changshun blue-eggshell (CS, n = 184), commercial H-line brown layers (HL, n = 392), and Lohmann laying hen (LM, n = 64).
Genomic DNA was obtained from fresh whole-blood samples in EDTA-K2 vacutainer tubes using phenol/chloroform extraction (Huang et al., 2014). DNA quality was assessed by gel electrophoresis, and DNA quantity was assessed by optical density (OD) using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). DNA samples were diluted to approximately 10 ng/μl and stored at −20°C.
The primers used to amplify the target fragments were designed by NCBI1 and synthesized by Sangon Biotech Company (Shanghai, China). The primer pairs used in this study are listed in Table 1.
DNA from each individual was subjected to indel identification and genotype analysis using ADPRHL1 primers. Sequencing was performed by Sangon Biotech Company (Shanghai, China), and the differences in the distribution of different genotypes among populations were analyzed using SPSS 24.0 software (version 24.0; Statistical Product and Service Solutions, IBM Corporation, Armonk, NY, United States). The allele frequency and genotype frequency of each mutation point were calculated using GENEPOP software,2 and the polymorphism information content (PIC), effective allele numbers (Ne), expected heterozygosity (He), and observed heterozygosity (Ho) were calculated simultaneously.
To study the expression of ADPRHL1 in different varieties, samples were taken from chickens at different stages of development for RNA experiments. A total of eight tissues (heart, liver, spleen, lung, kidney, pancreas, breast muscle, and leg muscle) were collected from GSs and AA broilers aged 1 week. Leg muscle tissues of GSs were collected at 1 day, 1 week, 6 weeks, 14 weeks, 22 weeks, and 30 weeks. Ten, 12, 14, 16, and 18 embryonic days of Gushi chicken leg muscle tissue were collected. In addition, the breast and leg tissues of broilers at embryonic ages of 10, 12, 14, 16, and 18 and at 1 day, 1 week, and 3 weeks of age were obtained from LS chickens and AA broilers, respectively.
To study the expression level of ADPRHL1 in chickens of different genotypes, samples were taken from Arbor Acres broilers and LS chickens of different genotypes at 3 weeks old for RNA experiments. Since the DD genotype was not obtained in AA broilers, the template was collected from the liver, muscle stomach, spleen, kidney, abdominal fat, duodenum, leg muscles, and hearts of these two broilers. All tissues were immediately placed in RNAwait (Solarbio Cat#SR0020) and stored at −80°C. The samples were treated with TRIzol reagent (Takara, Otsu, Japan) following the manufacturer’s recommended protocol. The RNA concentration and integrity were estimated spectrophotometrically using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and verified through electrophoresis using an agarose gel. Only samples with an OD absorption ratio (OD 260 nm/OD 280 nm) between 1.9 and 2.0 and exhibiting no signs of degradation were used for further analysis. cDNA synthesis was performed using a PrimeScript RT reagent kit with gDNA Eraser (Takara).
The qPCR reaction (10 μl) contained 5 μl of 2 × SYBR Premix ExTaq (Takara, Dalian, China), 0.5 μl of each primer, and 1 μl of cDNA (approximately 100 ng/μl). qPCR conditions were as follows: 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The reactions were performed using the QuantStudio 5 and a ProFlexTM PCR instrument. The results were analyzed using the 2–ΔΔCT method (Schmittgen, 2008). The figures were drawn using GraphPad Prism 6 (GraphPad Software Inc., 2007, San Diego, CA, United States).
The allele fragment of ADPRHL1 was amplified by polymerase chain reaction (PCR) using primers in Table 1, and cloned into a PMD18-T vector (Takara, Tokyo, Japan). To construct the luciferase reporter, the insert was released by EcoRV and KpnI digestion and was subsequently subcloned into the luciferase reporter vector pGL4-Basic (Promega, Madison, WI, United States). Two vectors, designated the pGL4-pro1-inserted allele and pGL4-pro1-deleted allele, were constructed to test the effects of the ADPRHL1 indel. The respective sequences of these vectors were obtained from 30 GS chickens mixed gene pools. The vector pRL-TK (Promega, Madison, WI, United States) was used as an internal reference to the luciferase reporter assay.
Human embryonic kidney 293T (HEK293T) cells were seeded in 96-well plates at a density of 5 × 104 cells per well, and 100 μl growth medium was added to each well. The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen) and double antibiotic (1% penicillin and streptomycin) at 37°C in a humidified 5% CO2 atmosphere.
The Bonferroni test was performed for multiple comparisons. Model I was used to determine growth traits, meat quality traits, and serum variables. With BW being taken as a variable, according to the effect of BW on growth traits, type II model body traits were used for analysis (Li et al., 2019, 2020; Ren et al., 2019).
Model I: Yijklm = μ + Gi + Sj + Hk + fl + eijklm
Model II: Yijklm = μ + Gi + Sj + Hk + fl + b (Wijklm −) + eijklm
Our model is designed based on least squares. In these models, Yijklm is the observed value, μ is the overall population mean, and Gi is the fixed effect of the genotype (i = 3), including the additive and dominant effects of the gene (additive effect values are −1, 0, and 0.1 represent II, ID, and DD genotypes, respectively, while dominant effect values 1, 1, −1, and 1 represent II, ID, and DD genotypes, respectively), Sj is the fixed effect of gender (j = 2), and Hk is hatching. The fixed effect of (k = 1, 2), fl is the fixed effect of family (l = 1, 7), eijklm stands for random error, b is the regression coefficient of BW, Wijklm is the individual slaughter weight, and is the average slaughter weight. In this study, all data are expressed as the mean ± SEM. A P-value < 0.05 was considered statistically significant, and the Bonferroni test was used for multiple comparisons.
The chicken ADPRHL1 gene is located on chromosome 1 (GenBank accession number NC_006088.5), which includes two variable spliceosomes and contains eight exons that encode a protein of 1,934 amino acids. A synteny analysis was performed to examine the context of the gene in eight different species (Figure 1). The results showed that the genomic region bracketing the ADPRHL1 gene was conserved across species and contains several common genes, including TMCO3 (ENSGALG00000016824), TFDP1 (ENSGALG0000001 6823), FAM70B (ENSGALG00000016821), ATP4B (ENSGA LG00000016822), GRTP1 (ENSGALG00000016828), LAMP1 (ENSGALG00000037697), CUL4A (ENSGALG00000016830), and PCID2 (ENSGALG00000016831).
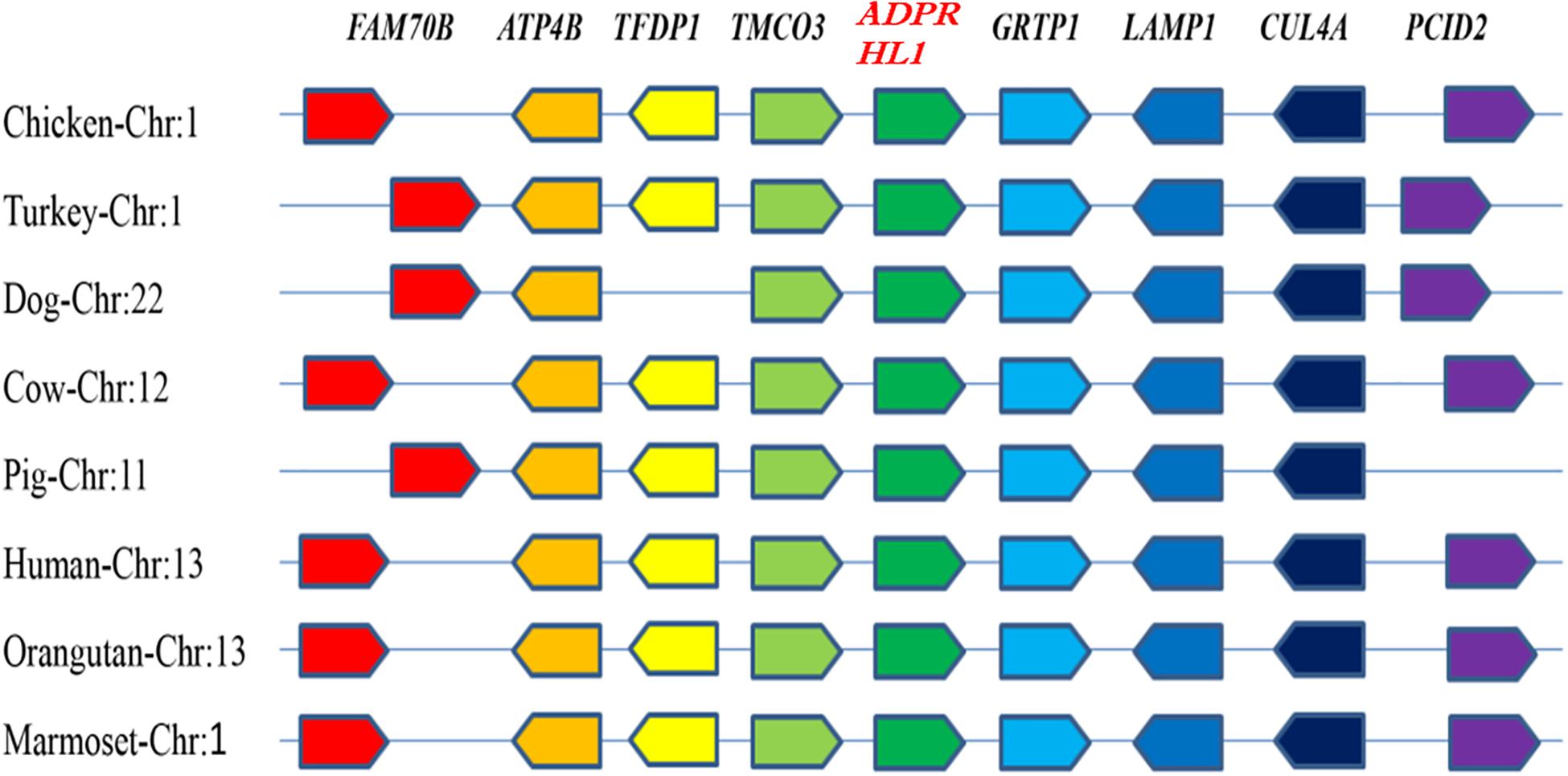
Figure 1. Syntenic analysis of ADPRHL1 genes in different species. Different colors represent different genes.
The expression of ADPRHL1 mRNA in various tissues of 1-week-old Gushi broilers and AA commercial broilers was analyzed. ADPRHL1 was expressed in all the tested tissues. Notably, higher levels of expression were observed in the myocardium and leg muscles (Figure 2). The high expression of ADPRHL1 in a variety of muscle tissues suggests a role for ADPRHL1 in muscle-muscle formation and differentiation, which is consistent with previous findings (Smith et al., 2016).
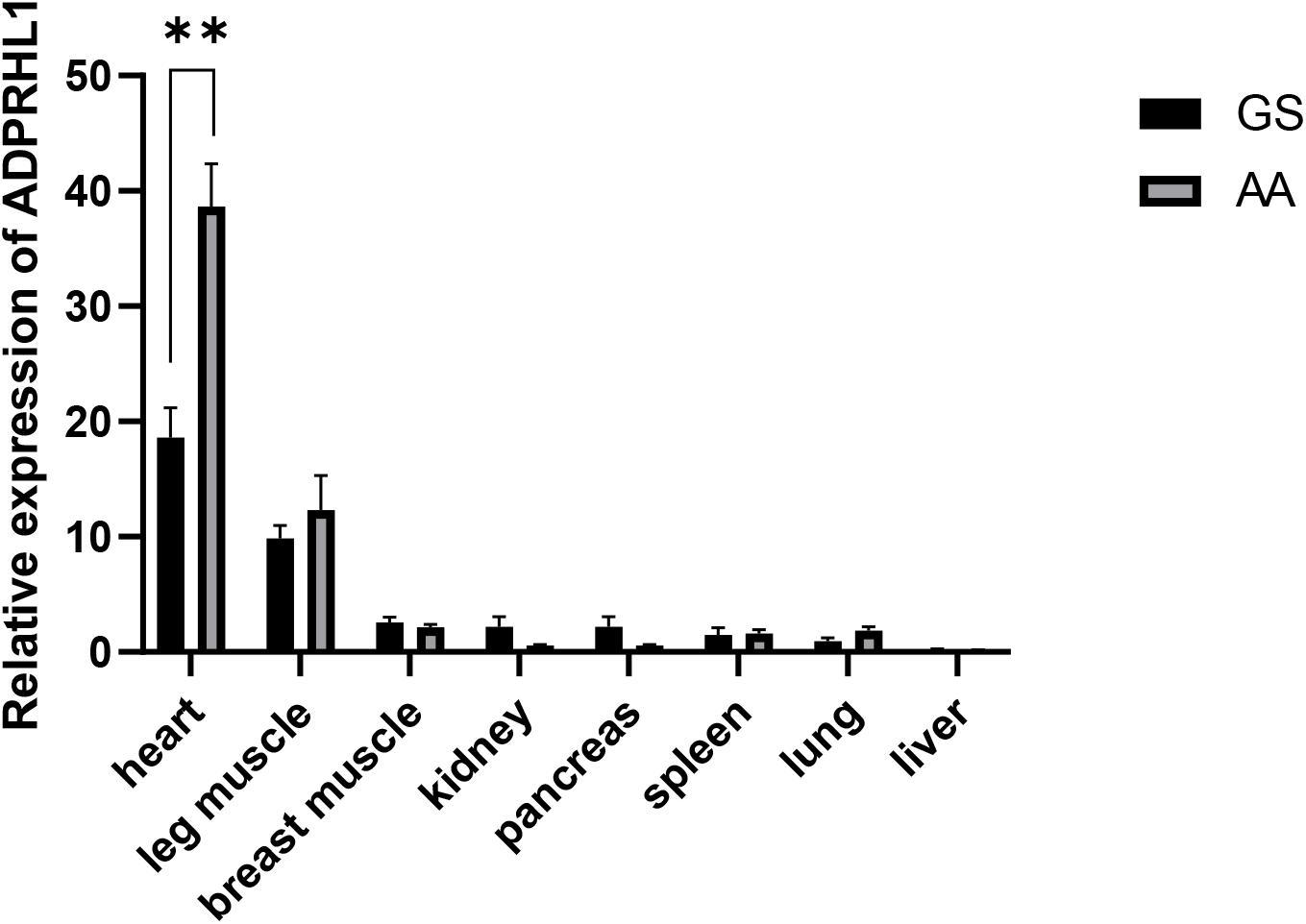
Figure 2. Relative expression patterns of ADPRHL1. Relative expression patterns of ADPRHL1 in GS chicken and AA broiler tissues at 1 week of age. GS, Gushi chicken; AA, Arbor Acres broilers. n = 8. ∗∗P < 0.01.
Expression of ADPRHL1 gene in breast and leg muscle tissues was studied at embryonic day of 10, 12, 14, 16, and 18 and at 1 day, 1 week, and 3 weeks of age in LS chickens and AA broilers. The expression of ADPRHL1 in breast muscle and leg muscle showed a trend of first increasing, then decreasing and then increasing at successive developmental stages. Specifically, the expression level was highest at approximately 16–18 days of embryonic life, decreased rapidly after birth, and then increased rapidly with developmental stages to reach its peak at 3 weeks of age (Figures 3A,B). The expression level of ADPRHL1 in AA broilers was significantly higher than that in LS broilers at 3 weeks of age (P < 0.05). Notably, the expression of ADPRHL1 in breast muscle did not change significantly at 1 day of age and 1 week of age, which was different from the expression observed in leg muscle. To examine the cause of this difference, we analyzed the expression level of ADPRHL1 in the leg muscle of GS chickens. The results showed that the expression level of ADPRHL1 in GS chicken leg muscle reached the highest level at 1 week, decreased decline from 1 to 14 weeks, and remained largely stable from 14 to 30 weeks (Figure 3C).
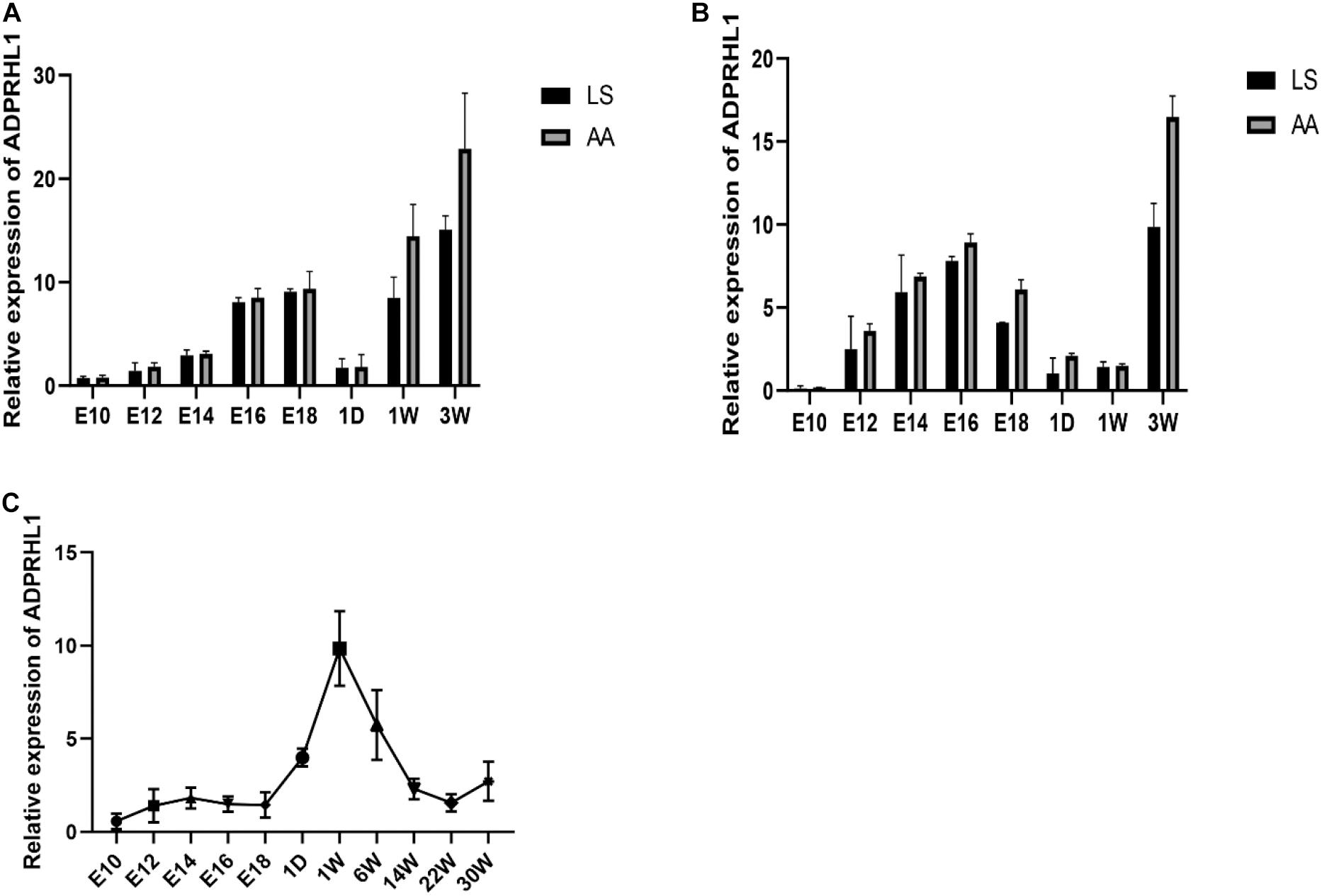
Figure 3. Relative expression patterns of ADPRHL1. (A) Relative expression patterns of ADPRHL1 in the leg muscle in LS and AA chickens tissues at 10, 12, 14, 16, and 18 embryo and 1 day, 1 week, and 3 weeks of age. LS, Lushi chicken; AA, Arbor Acres broilers. n = 6. (B) Relative expression patterns of ADPRHL1 in the breast muscle in LS and AA chickens tissues at 10, 12, 14, 16, and 18 embryonic days and 1 day, 1 week, and 3 weeks of age. LS, Lushi chicken; AA, Arbor Acres broiler; n = 6. (C) Relative expression patterns of ADPRHL1 in the leg muscle in GS chickens tissues at 10, 12, 14, 16, and 18 embryo and 1 day, 1 week, 6 weeks, 14 weeks, 22 weeks, and 30 weeks of age. GS, Gushi chicken; n = 6.
To identify the potential factors affecting the expression of ADPRHL1, the sequence mutation of ADPRHL1 gene was studied. A new 104-bp deletion (GenBank accession number NC_006088.5, Chr1:138752529–138752632) mutation was detected in the 5′UTR of the ADPRHL1 gene by whole-genome resequencing. Indel polymorphisms were genotyped by PCR amplification of the region and electrophoresis of products was performed in a 2.0% agarose gel. Three possible genotypes were identified, designated II (218 bp), ID (218 bp and 114 bp), and DD (114 bp) (Figure 4).
Table 1 details the PCR primers related to the locus. The results of qPCR showed that there were significant differences in the expression levels between ID and DD genotypes in 3-week-old Lushi chickens and 1-week-old AA broilers (no DD genotypes were found in AA broilers in our population) (P < 0.05, Figure 5).
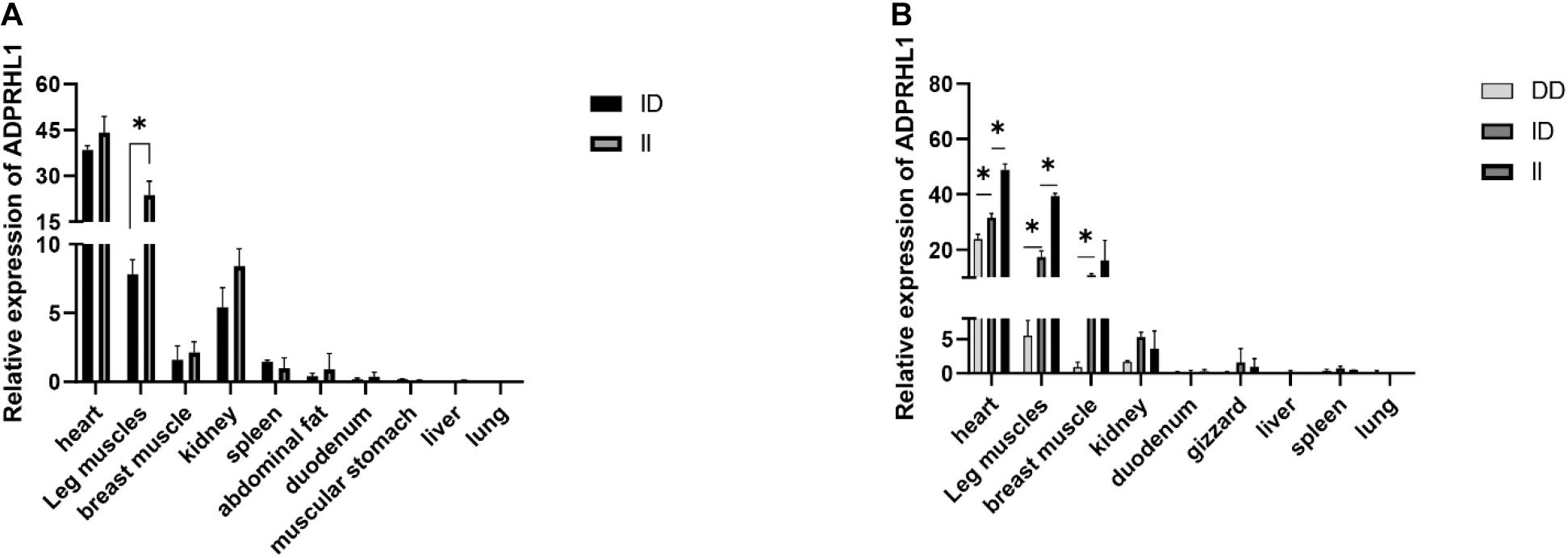
Figure 5. Relative expression patterns of ADPRHL1. (A) Expression patterns of different genotypes of ADPRHL1 in Arbor Acres broiler at 1 week of age. ID, n = 3; DD, n = 5. (B) Expression patterns of different genotypes of ADPRHL1 in Lushi chicken at 3 weeks of age. DD, n = 3; ID, n = 3; DD, n = 3. Error bars represent the SEM. ∗P < 0.05.
These results suggest that the 104-bp indel locus affects the expression of ADPRHL1 and may influence the growth traits of chickens. It was therefore important to examine the relationship between the 104-bp indel locus and growth traits in a larger chicken population.
To examine the relationship between the 104-indel locus and growth traits in chickens, allele frequencies and other genetic parameters associated with the ADPRHL1 Indel locus were calculated for 4,526 individuals in the study. This analysis included F2 generation resource populations, dual-purpose chickens (GS, GF, WH, XC, and CS), commercial laying hens (HL and LM) and commercial broilers (Ross 308, 817, Hubbard, Cobb, and AA). The frequency of genotype II in commercial broilers (Ross 308/67%, 817/70%, AA/99%, Cobb/74%, and Hubbard/77%) was notably greater than that in any other breed (F2/43%, GS/37%, GF/25%, WH/46%, XC/47%, CS/39%, HL/22%, and LM/42%) in our study. The frequency of the DD genotype was lower in all varieties, except type GF, and it was highly rare in commercial broilers (Ross 308/4%, 817/0%, AA/0%, Cobb/1%, and Hubbard/0%). Finally, the frequency of the I alleles in commercial broilers (Ross 308/81.2%, 817/85.1%, AA/99.7%, Cobb/86.7%, and Hubbard/88.4%) was higher than that in other breeds (F2/70.1%, GS/68.0%, GF/48.7%, WH/67.6%, XC/69.5%, CS/62.5%, HL/55.6%, and LM/64.8%) (Table 2).
These results suggested that genotype II was the dominant genotype and has been highly selected in commercial broilers. The genetic indices (Ho, He, Ne, and PIC) for these 13 chicken populations are presented in Table 2. Ho, He, Ne, and Pic are not only indicators to measure allelic polymorphisms and the degree of gene mutation, but also indicators of genetic variation within a population. In commercial broilers, the observed heterozygosity was in the range of 0.01–0.03 and the expected heterozygosity was in the range of 0.01–0.31. In addition, all the breeds exhibited low polymorphism except Ross 308 in commercial broilers. In the F2 generation resource population, dual-purpose chickens and commercial laying hens, the observed heterozygosity was in the range of 0.43–0.67 and the expected heterozygosity was in the range of 0.42–0.50. Moreover, the F2 generation resource population, dual-purpose chickens and commercial laying hens showed moderate polymorphism, indicating that they have greater genetic variation and selection potential. These results suggested that commercial broiler breeds (especially Arbor Acres broilers) might have experienced great pressure of artificial selection.
The results of the correlation analysis show that the 104-bp indel polymorphism was significantly associated with BW at 4, 6, and 10 weeks of age and with body size indices, including 0-week SL and 4-week BSL (P < 0.05), in addition to exhibiting a highly significant correlation with BW at 0 and 2 weeks (P < 0.01) (Table 3). The carcass traits were significantly associated with CW, SEW, EW, CLW, DWW, GW, PW, CMW, LW, LMW, and SW (P < 0.05); Moreover, there were significant associations between this 104-bp indel polymorphism and meat quality traits (Table 4). The results of the serum variable association analysis further showed that the indel was significantly associated with alanine transaminase and lactate dehydrogenase (P < 0.05) (Table 5).

Table 5. Association of the 104-bp locus with serum variables in an F2 population at 86 days of age.
To elucidate the molecular mechanisms associated with 104-bp indel at the cellular level, we attempted to identify changes in promoter activity across different genotypes. Pooled genomic DNA from 30 GS chickens was utilized as a template, and various genotypes were obtained by PCR using a pair of specific primers (Table 1). The transcriptional activity of the different genotypes was subsequently measured in HEK293T cells using a dual luciferase reporter analysis system. Genotype II exhibited a 2.49-fold higher activity than the genotype DD (P < 0.05) (Figure 6).
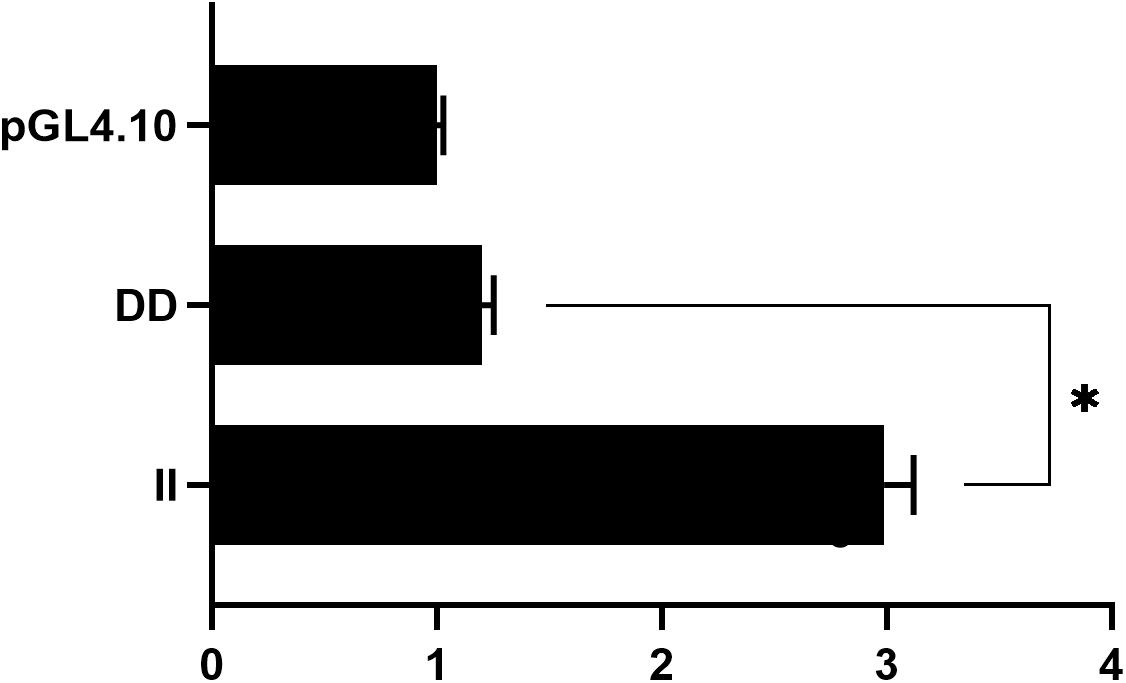
Figure 6. Relative luciferase activity detection of ADPRHL1 gene promoter. Luciferase activity in HEK293T cells transfected with recombinant plasmids containing serial promoter fragments (pGL4-II, pGL4-DD) of the ADPRHL1 promoter. The backbone vector pGL4.10 was used as a negative control; Each experiment was repeated at least five times. The data were the mean ± standard error (S.E.) of the normalized luciferase activity (∗P < 0.05).
ADPRHL1 is only widely expressed in vertebrates, suggesting that it may be a driving force in evolution (Smith et al., 2016). In addition, it was previously observed that ADPRHL1 expresses one of the mRNAs induced cardiac differentiation of human embryonic stem cells in vitro (Beqqali et al., 2006). As a pseudoenzyme-expressing gene, the specific mechanism of ADPRHL1 in biological growth and development has not been determined. In our study, we first indirectly suggested the important role of ADPRHL1 in chicken heart, leg muscle, chest muscle, and other tissues, by analyzing the expression of this gene in different tissues at different times. Notably, ADPRHL1 appears to play a role at different times in different organizations. Previous studies have suggested that ADPRHL1 plays a direct role in the modification of Z-disc and actin dynamics (Smith et al., 2020). In our study, ADPRHL1 has higher expression in the breast muscle and heart, and we surmise that it may have a positive effect on muscle development. This may be on account of Adprhl1 plays a similar role in chickens. We believe that ADPRHL1 is involved in the development of chicken embryos in the middle and late stages of development and primarily mainly functions between 0 and 6 weeks after birth.
Many studies have elucidated the effects of other 5′UTR indel mutations on animal growth and development (Glanzmann et al., 2014). For example, the 18-bp indel in the porcine SOX9 5′-UTR is of functional importance and may therefore indeed be a causative variation in SOX9 associated traits (Brenig et al., 2015). Genome SVs is a major source of genetic and phenotypic variation and has received increasing attention (Liu et al., 2021). Some researchers have made some progress through the role of SVs in evolution and the mechanism of SVs in promoting pig phenotypic variation (Du et al., 2021). This evidence supports the importance of 5′UTR for large indel. We first discovered a novel 104-bp SV in 5′UTR of the ADPRHL1 gene in whole-genome resequencing of thirty GS chickens and verified it in thirteen populations. In cross-designed F2 resource groups, the indel was significantly associated with many growth traits, carcass traits and serum variables in our study (Tables 3–5). The results indicate the importance of this 104-bp indel for the growth and development of chickens. In addition, we further studied the genetic diversity of the mutation and characterized its genetic characteristics.
Allele frequencies are a reflection of genetic diversity between populations and may indicate genetic drift or the introduction of new mutations (Jia et al., 2016). Artificial selection has an important influence on the development of varieties and commodity populations and determines the amount and distribution of genetic variation during domestication. Notably, although genotype II is dominant in commercial broiler flocks, the gap between genotypes II and DD is considerably smaller in dual-purpose and commercial laying hens. Moreover, dual-purpose chickens and commercial laying hens showed moderate polymorphism, which indicated that they possessed greater genetic variation and selection potential than commercial broilers. Notably, Ross 308 chickens also showed moderate polymorphism, which indicated that the Ross 308 chickens still had room for breeding.
Growth and development are important processes that affect different body parts of all animals in different life stages, and are affected by many factors, including genetics, nutrient absorption and environmental conditions (Hui et al., 2020; De Robertis and Gurdon, 2021; Liu et al., 2021). We found that a significant association between 104-bp indel and growth characteristics occurred in the early stage of chicken growth. Consistent with this result, the expression of the ADPRHL1 gene with genotype II was significantly higher in the leg muscle, breast muscle, and myocardium of 3-week-old LS chickens than that in those with genotypes ID and DD. Although the DD genotype was not observed in large numbers of AA broilers, the expression of the ADPRHL1 gene with genotype II was significantly higher in the leg muscle of 1-week-old AA broilers than in that of the ID chickens. There was no significant difference in the expression of these two genotypes in the breast muscle of 1-week-old AA broilers, which might be related to the low expression level of ADPRHL1 in the breast muscle of 1-week-old. Nevertheless, differences in the expression levels of ADPRHL1 with different polymorphisms in various muscle tissues demonstrate the possibility that this indel is related to muscle development. In addition to the above description, this locus was significantly associated with such traits as PW, CW, and GW (P < 0.05). These results indicate that the ADPRHL1 gene plays an important role in muscle development. Further studies should consider the role of its regulatory network and components in the development of skeletal muscle in chickens.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Henan Agricultural University.
TL performed the research, analyzed the data, and wrote the manuscript. BC, CJW, DH, and PQ analyzed the data and involved in the study design. HM, XN, ZJ, and CXW performed the statistical analysis. RH, HL, XL, HX, and XK were involved in the design of the study. ZL conceived the study and involved in its design and coordination. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by grants from the Key Science and Technology Research Project of Henan Province (202102110085), the National Natural Science Foundation of China-Henan joint grant (U1804107), the Zhongyuan Youth Talent Support Program (ZYQR202012203), and the Earmarked Fund for Modern Agro-Industry Technology Research Systems of China (No. CARS-40-K04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adrain, C. (2020). Pseudoenzymes: dead enzymes with a lively role in biology. FEBS J. 287, 4102–4105. doi: 10.1111/febs.15535
Beqqali, A., Kloots, J., Ward-van Oostwaard, D., Mummery, C., and Passier, R. (2006). Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells 24, 1956–1967. doi: 10.1634/stemcells.2006-0054
Brenig, B., Duan, Y., Xing, Y., Ding, N., and Huang, L. (2015). Porcine SOX9 gene expression is influenced by an 18 bp Indel in the 5’-untranslated region. PLoS One 10:e0139583. doi: 10.1371/journal.pone.0139583
Chen, L., Pryce, J. E., Hayes, B. J., and Daetwyler, H. D. (2021). Investigating the effect of imputed structural variants from whole-genome sequence on genome-wide association and genomic prediction in dairy cattle. Animals 11:541. doi: 10.3390/ani11020541
De Robertis, E. M., and Gurdon, J. B. (2021). A brief history of Xenopus in biology. Cold Spring Harb. Protoc. doi: 10.1101/pdb.top107615 [Epub ahead of print].
Depristo, M. A., Banks, E., Poplin, R., Garimella, K. V., and Daly, M. J. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498.
Du, H., Zheng, X., Zhao, Q., Hu, Z., Wang, H., Zhou, L., et al. (2021). Analysis of structural variants reveal novel selective regions in the genome of meishan pigs by whole genome sequencing. Front. Genet. 12:550676. doi: 10.3389/fgene.2021.550676
Glanzmann, B., Lombard, D., Carr, J., and Bardien, S. (2014). Screening of two indel polymorphisms in the 5′UTR of the DJ-1 gene in South African Parkinson’s disease patients. J. Neural Trans. 121, 135–138. doi: 10.1007/s00702-013-1094-x
Huang, Y. Z., Sun, J. J., Zhang, L. Z., Li, C. J., Womack, J. E., Li, Z. J., et al. (2014). Genome-wide DNA methylation profiles and their relationships with mRNA and the microRNA transcriptome in bovine muscle tissue (Bos taurine). Sci. Rep. 13:6546. doi: 10.1038/srep06546
Hui, Y., Zhang, Y., Wang, K., Pan, C., Chen, H., Qu, L., et al. (2020). Goat DNMT3B: an indel mutation detection, association analysis with litter size and mRNA expression in gonads. Theriogenology 147, 108–115. doi: 10.1016/j.theriogenology.2020.02.025
Jia, X., Lin, H., Nie, Q., Zhang, X., and Lamont, S. J. (2016). A short insertion mutation disrupts genesis of miR-16 and causes increased body weight in domesticated chicken. Sci. Rep. 6:36433.
Kidd, J. M., Graves, T., Newman, T. L., Fulton, R., Hayden, H. S., Malig, M., et al. (2010). A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143, 837–847. doi: 10.1016/j.cell.2010.10.027
Leslie, M. (2013). Molecular biology. ‘Dead’ enzymes show signs of life. Science 340, 25–27. doi: 10.1126/science.340.6128.25
Li, W., Jing, Z., Cheng, Y., Wang, X., Li, D., Han, R., et al. (2020). Analysis of four complete linkage sequence variants within a novel lncRNA located in a growth QTL on chromosome 1 related to growth traits in chickens. J. Anim. Sci. 5:skaa122.
Li, W., Liu, D., Tang, S., Li, D., Han, R., Tian, Y., et al. (2019). A multiallelic indel in the promoter region of the Cyclin-dependent kinase inhibitor 3 gene is significantly associated with body weight and carcass traits in chickens. Poult. Sci. 98, 556–565. doi: 10.3382/ps/pey404
Liu, S., Gao, G., Layer, R. M., Thorgaard, G. H., Wiens, G. D., Leeds, T. D., et al. (2021). Identification of high-confidence structural variants in domesticated rainbow trout using whole-genome sequencing. Front. Genet. 12:639355. doi: 10.3389/fgene.2021.639355
Ono, T., Kasamatsu, A., Oka, S., and Moss, J. (2006). The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 103, 16687–16691. doi: 10.1073/pnas.0607911103
Reischauer, S., Arnaout, R., Ramadass, R., and Stainier, D. Y. (2014). Actin binding GFP allows 4D in vivo imaging of myofilament dynamics in the zebrafish heart and the identification of Erbb2 signaling as a remodeling factor of myofibril architecture. Circ. Res. 115, 845–856. doi: 10.1161/circresaha.115.304356
Ren, T., Li, W., Liu, D., Liang, K., Wang, X., Li, H., et al. (2019). Two insertion/deletion variants in the promoter region of the QPCTL gene are significantly associated with body weight and carcass traits in chickens. Anim. Genet. 50, 279–282. doi: 10.1111/age.12741
Schmittgen, T. D. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Smith, S. J., Towers, N., Demetriou, K., and Mohun, T. J. (2020). Defective heart chamber growth and myofibrillogenesis after knockout of adprhl1 gene function by targeted disruption of the ancestral catalytic active site. PLoS One 15:e0235433. doi: 10.1371/journal.pone.0235433
Smith, S. J., Towers, N., Saldanha, J. W., Shang, C. A., Mahmood, S. R., Taylor, W. R., et al. (2016). The cardiac-restricted protein ADP-ribosylhydrolase-like 1 is essential for heart chamber outgrowth and acts on muscle Actin filament assembly. Dev. Biol. 416, 373–388. doi: 10.1016/j.ydbio.2016.05.006
Tang, Y., Wang, Y. L., Yang, L., Xu, J. X., Xiong, W., Xiao, M., et al. (2013). Inhibition of arginine ADP-ribosyltransferase 1 reduces the expression of poly(ADP-ribose) polymerase-1 in colon carcinoma. Intern. J. Mol. Med. 32, 130–136. doi: 10.3892/ijmm.2013.1370
Tsuge, H., and Tsurumura, T. (2014). Reaction mechanism of mono-ADP-ribosyltransferase based on structures of the complex of enzyme and substrate protein. Curr. Top. Microbiol. Immunol. 384, 69–87. doi: 10.1007/82_2014_415
Zhang, J., Wang, W., Chen, S., Ruo, Y., Zhong, X., and Wu, X. (2014). Bi-pseudoenzyme synergetic catalysis to generate a coreactant of peroxydisulfate for an ultrasensitive electrochemiluminescence-based cholesterol biosensor. Biosens. Bioelectron. 57, 71–76. doi: 10.1016/j.bios.2014.01.046
Keywords: chicken, ADPRHL1, muscle, pseudoenzyme, structural variation
Citation: Li T, Chen B, Wei C, Hou D, Qin P, Jing Z, Ma H, Niu X, Wang C, Han R, Li H, Liu X, Xu H, Kang X and Li Z (2021) A 104-bp Structural Variation of the ADPRHL1 Gene Is Associated With Growth Traits in Chickens. Front. Genet. 12:691272. doi: 10.3389/fgene.2021.691272
Received: 06 April 2021; Accepted: 29 July 2021;
Published: 26 August 2021.
Edited by:
Tomoyoshi Komiyama, Tokai University, JapanReviewed by:
Tatsuhiko Goto, Obihiro University of Agriculture and Veterinary Medicine, JapanCopyright © 2021 Li, Chen, Wei, Hou, Qin, Jing, Ma, Niu, Wang, Han, Li, Liu, Xu, Kang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuanjian Li, bGl6aHVhbmppYW5AMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.