
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 16 June 2021
Sec. Epigenomics and Epigenetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.680633
This article is part of the Research Topic Role of Epigenetic Regulators in the Initiation, Progression, and Metastasis of Cancer View all 15 articles
Epigenetic dysregulation has been implicated in a variety of pathological processes including carcinogenesis. A major group of enzymes that influence epigenetic modifications are lysine demethylases (KDMs) also known as “erasers” which remove methyl groups on lysine (K) amino acids of histones. Numerous studies have implicated aberrant lysine demethylase activity in a variety of cancers, including melanoma. This review will focus on the structure, classification and functions of KDMs in normal biology and the current knowledge of how KDMs are deregulated in cancer pathogenesis, emphasizing our interest in melanoma. We highlight the current knowledge gaps of KDMs in melanoma pathobiology and describe opportunities to increases our understanding of their importance in this disease. We summarize the progress of several pre-clinical compounds that inhibit KDMs and represent promising candidates for further investigation in oncology.
There are numerous genetic alterations which promote carcinogenesis which include mutations of certain genes and chromosomes. Epigenetics is defined as heritable functional changes in the genome which do not involve a change in the DNA sequence (Dupont et al., 2009). The Greek prefix “epi” denotes “over, outside of, or around” implying additional factors that may influence traditional genetic inheritance patterns. Epigenetics is essential in normal development and biology but dysregulation has been implicated as a key impetus of carcinogenesis and resistance. All cells contain genetic information in the form of DNA which is wound around proteins called histones (Kornberg, 1974). The DNA is assembled into units called nucleosomes which form a complex consisting of histones and DNA known as chromatin (Kornberg, 1974). Chromatin is organized into compact, transcriptionally inactive regions called heterochromatin, usually around the periphery of the nucleus and loosely arranged, transcriptionally active chromatin called euchromatin (Elgin, 1996). This formation has an important role in the regulation of gene expression as well as controlling the transcription, replication, recombination and repairing of DNA (Elgin, 1996). Various epigenetic changes can affect chromatin structure and hence gene expression. One of the most well studied epigenetic changes involves methylation which may occur at the DNA or histone level (Tost, 2010). DNA methylation involves the transfer of methyl groups by DNA methyltransferases (DNMTs) to individual nucleotide bases, altering gene expression (Bilian Jin and Robertson, 2011). The family of DNMT enzymes adds methyl groups while removal is mediated by the ten eleven translocation (TET) family (Zhang et al., 2010).
The other major class of methylation is histone methylation which is a post-translational modification (PTM) of histone tails. Histone methylation involves the addition of (via writer enzymes) or removal of (via eraser enzymes) methyl groups, typically on lysine (K) or arginine (R) amino acids of histone type 1–4 tails (Figure 1). Lysine methyltransferases (KMTs) drive the addition of methyl groups, whereas lysine demethylases (KDMs) are responsible for the removal of methyl groups. Depending on the type of histone modifications, the consequence might induce either an open or closed chromatin state which regulates gene expression. KDMs rarely act in isolation as enzymatic erasers but are typically members of large epigenetic complexes, consisting of other enzymes and transcription factors. This is suggestive of a scaffolding protein role for KDMs in addition to a catalytic one. The most well investigated histone lysine methylation sites include H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20 (Figure 1) that will either compact or open chromatin depending on lysine position and the number of methyl groups. Hence this review will focus on what is currently known about KDMs in normal biology and cancers including melanoma. A summary of the progress of current KDM inhibitors is also discussed.
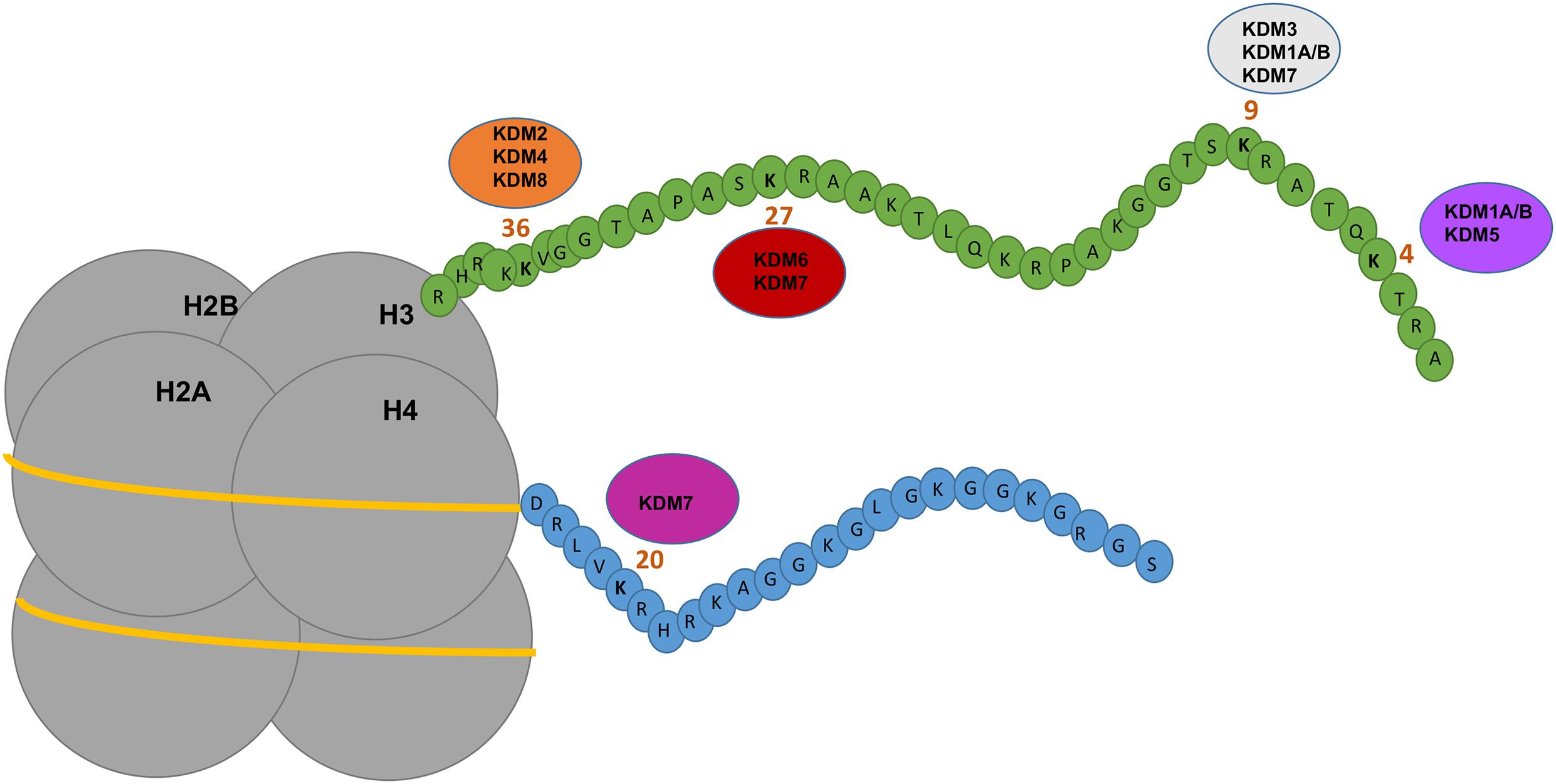
Figure 1. Depicting the mechanism of action of KDMs on histones. Nucleosomes consist of four subunits of histones which contain amino acid tails that are modified by epigenetic regulators including KDMs. KDM1A/B demethylates H3K9me1/H3K9me2 and H3K4me1/me2 inducing gene repression or activation. KDM2 and KDM4 demethylases target H3K36me1/me2/me3 inducing gene activation. KDM3 demethylases target H3K9me1/me2 inducing gene activation. KDM5 demethylases targets H3K4me2/me3 inducing gene repression. KDM6 and KDM7 demethylases target H3K27me2/me3 inducing gene activation. KDM7 demethylases target H4K20me1 and H3K9me2/me3. KDM8 demethylase target H3K36me3 inducing gene activation. Figure is adapted from Verde et al. (2017).
Lysine demethylases are critical for normal development but numerous studies have implicated the dysregulation of several KDMs in cancers. This is likely due to the ability of KDMs to govern major changes in the transcriptional networks of hundreds of genes with potential roles in all major hallmarks of cancer. The roles of each KDM in normal biology and cancer is described below and Table 1 summarizes studies in cancer.
Lysine demethylases are classified into two groups according to the catalytic mechanisms of demethylation- amine oxidase or jumonji C domain containing KDMs. Class I is the amine-oxidase lysine specific demethylases 1 and 2 (LSD1 and 2), also known as KDM1A and KDM1B. KDM1A and KDM1B use Flavin adenine dinucleotide (FAD) as a substrate to generate an imine intermediate which is hydrolyzed to produce the demethylated lysine residue (Walport et al., 2012). The amines-oxidase like (AOL) catalytic domain at the C-terminal in KDM1A and KDM1B consists of two folded subdomains. The FAD and substrate binding regions are structurally related to the superfamily of monoamine oxidases (MAO). In particular MAO-A and MAO-B- enzymes which catalyze the oxidation of monoamine (contain one amino group) neurotransmitters which include serotonin and dopamine (Shi et al., 2004; Willmann et al., 2012). The N-terminal of KDM1A contains the SWIRM domain which is named after the SWI3, RSC8 and MOIRA proteins and is essential for protein stability and interactions with histone tails.
KDM1A was the first histone demethylase identified (Shi et al., 2004). Functionally, KDM1A predominantly catalyzes the removal of methyl groups from mono and di-methylated lysine residues at H3K4 inducing gene repression (Walport et al., 2012). An early in vivo study showed that KDM1A demethylation of K1096 in the DNA methyl transferase enzyme DNMT1 was essential for the gastrulation stage of murine embryogenesis (Wang et al., 2009). Genetic ablation of KDM1A in a knockout mouse model was embryonic lethal (Wang et al., 2007, 2009).
Interestingly, KDM1A can demethylate non-histone proteins in osteosarcoma cells such as the tumor suppressor p53 which has an important role in regulation of pro-apoptotic genes. Another example showed KDM1A can demethylate K185 of transcription factor E2F1 which has an important role in regulating the cell cycle and tumor suppressor genes in lung cancer cells (Huang et al., 2007; Wang et al., 2009; Kontaki and Talianidis, 2010; Xie et al., 2011). A study found that overexpression of KDM1A induces E2F1 signaling via histone demethylation and promotes cell proliferation in oral cancer and that inhibition of KDM1A reduces E2F1 signaling, implying an oncogenic role of KDM1A in oral cancer (Narayanan et al., 2015).
The other KDM1 family member, KDM1B has been reported to catalyze the removal of methyl groups at H3K4 and this enzyme is essential for oocyte development (Ciccone et al., 2009). KDM1B knockout mice exhibit embryonic lethality (Ciccone et al., 2009), highlighting its importance in normal biology.
Class II KDMs are the demethylases that contain a Jumonji C domain (JMjC KDMs “Jumonji” meaning cruciform in Japanese). This includes the KDM2-6 subfamilies which consists of 20 enzymes that are grouped into five subfamilies: KDM2, KDM3, KDM4, KDM5, and KDM6 (Klose et al., 2006). The JMjC KDMs catalyze the removal of methyl groups from mono, di and trimethylated lysines at various sites and use dioxygen and 2-oxoglutarate (2OG) as substrates with Fe (II) as an essential cofactor (Walport et al., 2012). Unlike KDM1A/1B the JMjC KDMs are able to remove methyl groups from trimethylated lysine sites since the mechanism doesn’t require a formation of an imine. The catalytic domain of the JMjC KDMs is structurally related to the superfamily of 2OG- dependent oxygenases which play an important role in fatty acid metabolism, protein biosynthesis and nucleic acid repair/modification (Loenarz and Schofield, 2008).
The KDM2 subfamily consists of two demethylases KDM2A and KDM2B. Both KDM2A and KDM2B have been reported to be oncogenic (Pedersen and Helin, 2010). KDM2A has been reported to be upregulated and induces proliferation in lung, gastric and breast cancer. KDM2A overexpression was found to increase cell proliferation and invasion through activation of ERK1/2 signaling in lung cancer as well as being associated with poor prognosis in lung cancer patients (Wagner et al., 2013). KDM2A overexpression also promoted cell growth and migration in gastric cancer by downregulation of tumor suppressor gene programmed cell death 4 (PDCD4) (Huang et al., 2015). In breast cancer, KDM2A is highly expressed in myoepithelial cells which have been reported to have anti-tumor properties. KDM2A ablation in these cells induced increased invasion and migration via downregulation of MMP proteins and repression of EF21 signaling (Rizwani et al., 2014).
KDM2B is overexpressed in numerous cancers. Knockdown of KDM2B reduced cell growth in gastric cancer in vitro and in vivo and induced autophagy- a process in which cells remove damaged cell components (Zhao et al., 2017). KDM2B overexpression induces transformation of hematopoietic progenitor cells in acute myeloid leukemia whereas reduction of KDM2B inhibited Hox9/Meis1 induced leukemic transformation (He et al., 2011). Overexpression of KDM2B is observed in ovarian cancer and when knocked down in vitro and in vivo reduced cell proliferation, migration and tumor growth (Kuang et al., 2017). Pancreatic cancers with increased KDM2B promoted tumor formation in cooperation with the oncogene Kras in an in vivo model (Tzatsos et al., 2013). Another study showed that KDM2B overexpression was associated with poor prognosis in glioma and KDM2B knockdown inhibited cell proliferation and induced cell cycle arrest (Wang Y. et al., 2018).
The KDM3 subfamily consists of three demethylases; KDM3A, KDM3B, and JMJD1C. It has been reported that KDM3A is important in spermatogenesis and male KDM3A knockout mice are infertile (Pedersen and Helin, 2010). The function of KDM3B and JMJD1C is largely unknown.
The KDM4 subfamily consists of five demethylases- KDM4A, KDM4B, KDM4C, KDM4D, and KDM4E. KDM4A, KDM4B, KDM4C, and KDM4D have been reported to be important in oncogenesis, but the function of KDM4E is unknown (Pedersen and Helin, 2010). KDM4A overexpression was found to stimulate the AR, inducing the expression of prostate specific antigen, implicated in the progression of prostate cancer (Kim et al., 2016). KDM4A is overexpressed in breast cancer and KDM4A knockdown inhibited cell proliferation, migration and invasion (Li et al., 2012). Lung cancers with increased KDM4A are associated with poor prognosis (Xu et al., 2016).
KDM4B overexpression promoted DNA damage in breast cancer cells which was significantly reduced upon pharmacological inhibition of KDM4B, by induction of apoptosis in triple negative breast cancers deficient in the tumor suppressor gene, PTEN (Wang W. et al., 2018; Xiang et al., 2019). KDM4B overexpression also promoted proliferation, growth and glucose uptake in colorectal cancer cells whereas KDM4B knockdown inhibited tumor growth significantly in an in vivo model (Li et al., 2020).
KDM4C overexpression is associated with poor prognosis in prostate cancer and can co-regulate transcriptional activation of the AR. KDM4C knockdown significantly reduced proliferation, colony formation, AR transcriptional activity in prostate cancer cells and inhibited tumor growth of a prostate cancer model in zebrafish (Lin et al., 2019).
The KDM5 subfamily consists of four demethylases- KDM5A, KDM5B, KDM5C, and KDM5D. This group of demethylases can remove methyl groups from di and trimethylated lysines on H3K4. The KDM5 subfamily has been reported to play an important role in development, identified in drosophila melanogaster as Lid (Little Imaginal Disks) protein due to the phenotype visible in mutant larvae (Gildea et al., 2000). This protein was classified as a H3K4 histone demethylase which had all the domains of the human JARID1 family. The Lid protein binds to drosophila Myc (dMyC)—a transcription factor that is important in cell cycle progression, cell proliferation and apoptosis, and is frequently dysregulated in cancer (Secombe and Eisenman, 2007; Li et al., 2010).
KDM5A was initially identified as a binding partner of retinoblastoma protein (pRB) (Defeo-Jones et al., 1991). pRB is a tumor suppressor that inhibits cell cycle progression, preventing cell growth and promoting senescence. Embryonic fibroblasts isolated from KDM5A knockout mouse revealed an important role for KDM5A in mitochondrial function (Varaljai et al., 2015) and cell differentiation (Benevolenskaya et al., 2005; Lopez-Bigas et al., 2008).
KDM5B/JARID1B/PLU1 was initially identified in a study targeting genes regulated by the tyrosine kinase HER2 (Lu et al., 1999). A later study found that KDM5B had H3K4 histone demethylase activity and inhibited the expression of tumor suppressor genes BRAC1 and CAV1 (Yamane et al., 2007). In addition, KDM5B has been reported to be overexpressed in numerous cancers and has been identified as a potential oncogene.
KDM5D/JARID1D is the least well investigated demethylases from the KDM5 subfamily, but has been implicated in prostate cancer progression (Perinchery et al., 2000).
The KDM6 subfamily consists of three demethylases- KDM6A, KDM6B, and UTY. KDM6A also known as UTX can remove methyl groups from di and trimethylated lysines on H3K27- a mark associated with suppression of gene transcription (Pedersen and Helin, 2010). KDM6A was initially identified as playing an important role in embryonic development and cell differentiation (Hong et al., 2007). KDM6A together with methyltransferases MLL2 (KMT2D) and MLL3 (KMT2C) is an important component of the COMPASS complex also known as the ASCOM complex which mediates the transcriptional activation of genes via H3K4 trimethylation and H3K27me2/3 demethylation (Shilatifard, 2008; Van der Meulen et al., 2014; Ford and Dingwall, 2015). The COMPASS complex can also promote histone acetylation a demethylation independent activity, by interaction with histone acetyltransferase p300 (CBP) as well as chromatin remodeling via the SWI/SNF complex and transcriptional elongation by interacting with transcription elongation factors (Figure 2; Wang et al., 2017; Schulz et al., 2019). KDM6A therefore has dual roles in activation of gene expression. Not only does it remove suppressive marks on H3K27 but also activates genes by H3K4 trimethylation and H3K27 acetylation, highlighting the complex interplay between histone erasers and writers to remodel chromatin in a highly orchestrated fashion.
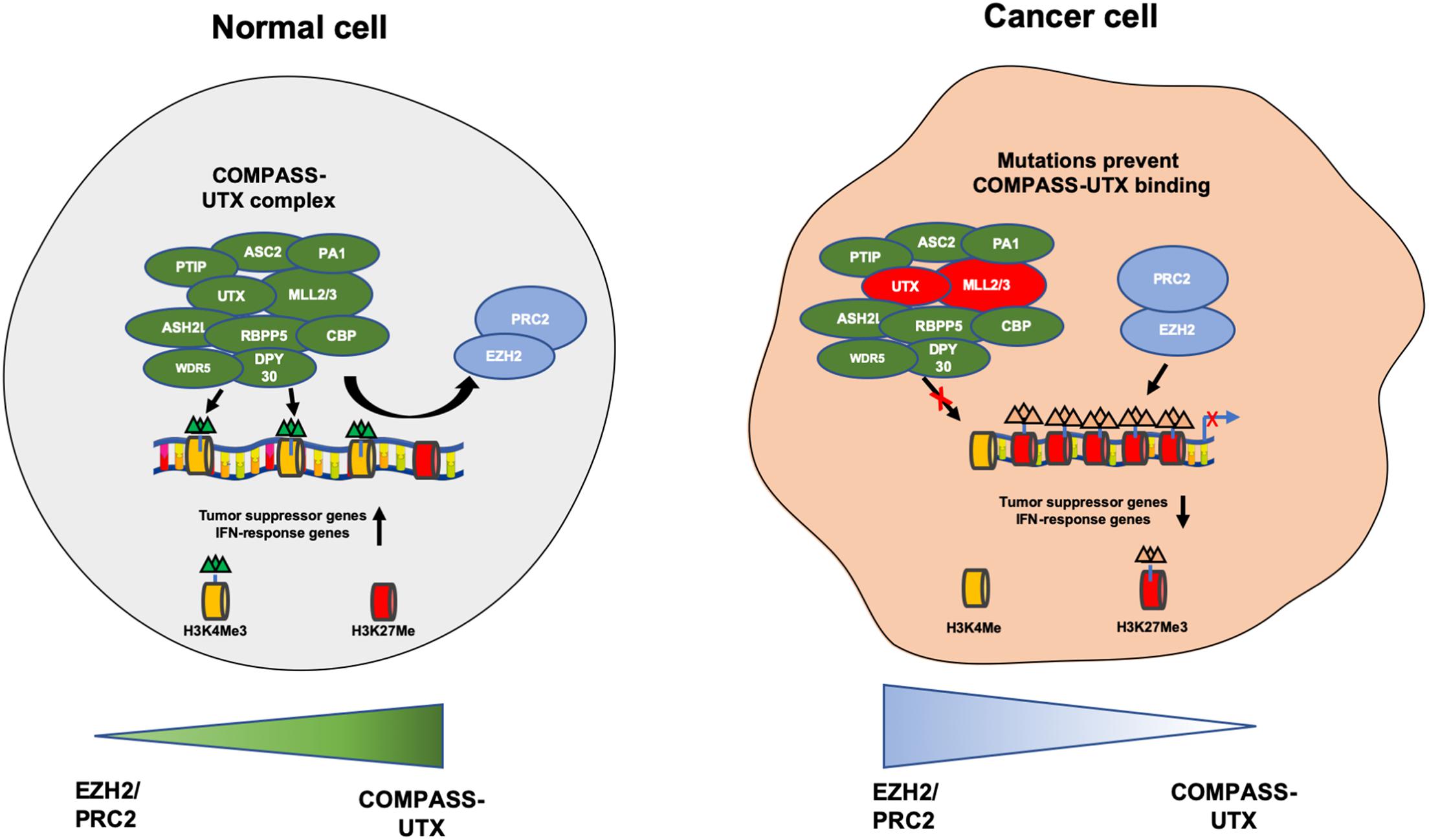
Figure 2. Depicting the COMPASS complex and PCR2 complex that contribute to the open and closed chromatin states. In a normal healthy cell the COMPASS complex removes methyl groups from H3K27me3 and adds methyl groups to H3K4me and prevents PRC2 complex from adding methyl groups to H3K27me. This induces transcriptional activation of tumor suppressive and IFN-response genes. In the context of cancer UTX (KDM6A) and MLL2/3 are frequently mutated in the COMPASS complex, causing a loss of expression. This prevents UTX from binding to the compass complex and no H3K27me3 demethylation occurs, enabling the PRC2 complex to add methyl groups on H3K27me and remove methyl groups from H3K4me, inducing transcriptional repression of tumor suppressor and IFN response genes, promoting cancer cell growth.
KDM6A exhibits another methylation independent role by directly interacting with DNA binding transcription factors, including nuclear receptors such as estrogen and retinoic acid receptors (Cho et al., 2007; Rocha-Viegas et al., 2014; Xie et al., 2017). KDM6A has been found to interact with retinoblastoma binding proteins including RBBP5 in vivo, potentially having an influence on the regulation of cell cycle and cell differentiation by RB family proteins (Shpargel et al., 2012; Van der Meulen et al., 2014).
KDM6A has been reported to be inactivated by mutations in 70% of non-invasive bladder cancer causing a loss of KDM6A expression (Ler et al., 2017). This is suggestive of a tumor suppressor role in this cancer. Studies have suggested loss of KDM6A may amplify PRC2 complex mediated gene repression and dependency in bladder cancer cells that can be sensitized to EZH2 inhibitors (Atala, 2017). Loss of KDM6A expression has also been associated with other cancers such as multiple myeloma (MM) in which KDM6A mutations lead to low KDM6A expression, resulting in increased proliferation, adhesion and tumorigenicity. Loss of KDM6A also sensitized MM cells to EZH2 inhibitors GSK343 and GSK126, inducing cell death and decreased proliferation (Ezponda et al., 2017). Squamous cell, metastatic pancreatic cancer in females was also associated with loss of KDM6A expression in a knockout mouse model. This was attributed to deregulation of the COMPASS complex and activation of oncogenes MYC and RUNX3 (Zhu et al., 2014). The cells also had increased sensitivity to bromodomains and extra-terminal (BET) inhibitors that target a type of epigenetic “reader” protein. Collectively this data suggests opportunities to indirectly target KDMs by studying the rich network of histone eraser or reader proteins that KDMs interact with.
In contrast to loss of KDM6A its presence may also be associated with other cancer types such as cervical cancer, where it appeared necessary for HPVE7 expressing cells to survive and de-repress the cell cycle DNA replication inhibitor p21 (Soto et al., 2017). In addition, KDM6A has been shown to support the oncogenic function of the estrogen receptor in breast cancer (Kim et al., 2017; Taube et al., 2017; Xie et al., 2017). This suggests KDM6A may operate in a cell type dependant manner and further investigation is required to resolve the tumor suppressive vs oncogenic role in different types of cancer.
KDM6B also known as JMJD3 has been reported to induce expression of oncogenes of the RAS/RAF MAP kinase signaling pathway and is expressed on activated macrophages, purported to play a role in inflammation (Agger et al., 2009).
Initial studies showed that KDM6B expression was important in the progression and prognosis of colon cancer (Pereira et al., 2011; Tokunaga et al., 2016). KDM6B has been reported to be overexpressed in prostate cancer with its expression increasing incrementally as the diseases progresses (Daures et al., 2016). In addition, KDM6B was found to induce epithelial to mesenchymal transition (EMT) and metastasis in clear cell renal carcinoma via activation of EMT factor Slug (Li et al., 2015). KDM6B has been reported to be overexpressed in diffuse large B-cell lymphoma (DLBCL) and is associated with poor survival. When DLBCL cells were treated with a small molecule KDM6 inhibitor (GSK-J4) not only was KDM6B expression inhibited, the cells were sensitized to chemotherapy agents (Mathur et al., 2017). KDM6B is highly expressed in multiple myeloma cells and when KDM6B was knocked down, growth and survival of these cells was inhibited (Ohguchi et al., 2017). Recent studies found that KDM6B overexpression promoted ovarian cancer cell migration and invasion via modulation of transforming growth factor-β1 (TGF-β1) (Pinton et al., 2018; Liang et al., 2019). In contrast, KDM6B overexpression induced cell apoptosis in non-small cell lung cancer (NSCLC) via translocation of FOXO1 (Ma et al., 2015) indicating that the role of KDM6B may be cell type dependent.
KDM7A/JHDM1D can remove methyl groups from di and trimethylated lysines on histone H3K4 as well as methyl groups from H3K9 (Klose et al., 2006; Wen et al., 2010). KDM7A has been reported to play an important role in the regulation of neural differentiation in particular the regions of the brain (Huang et al., 2010; Tsukada et al., 2010) but is not well studied in cancer.
The most recently identified KDM is KDM8 also known as JMJD5 and demethylates H3K36me2 inducing gene activation. KDM8 was initially identified to play an important role in embryogenesis and stem cell renewal (Oh and Janknecht, 2012; Zhu et al., 2014) and can promote carcinogenesis. A study showed KDM8 overexpression induced the expression of cell cycle promoter gene cyclin D1, promoting cell proliferation in a breast cancer cells model in vitro and conversely, KDM8 knockdown inhibited cell growth (Hsia et al., 2010). In addition, KDM8 overexpression induces activation of AR transcriptional activity and promotes cell growth in prostate cancer in vitro and in vivo (Wang et al., 2019).
Lysine demethylases have been implicated in a variety of cancers (summarized in Table 1) however, very few KDMs have been investigated in melanoma. We analyzed the percentage of mutation rates and types found in KDMs in the melanoma cohort obtained from the cancer genome atlas (TCGA) (Figure 3) and in a second independent dataset known as the Australian Melanoma Genome Project (AMGP) (Figure 4). We found that KDM1B and KDM5B exhibited the highest mutation rates in melanoma in the TCGA dataset. The majority of KDMs with the exception of KDM6C are upregulated in this dataset further emphasizing their importance in melanogenesis (Figure 3). KDM2B and KDM4C contained the highest number of alterations in the AMGP, however, this dataset does not include mRNA expression. The current knowledge and relevance of KDMs in melanoma will be discussed in the following section.
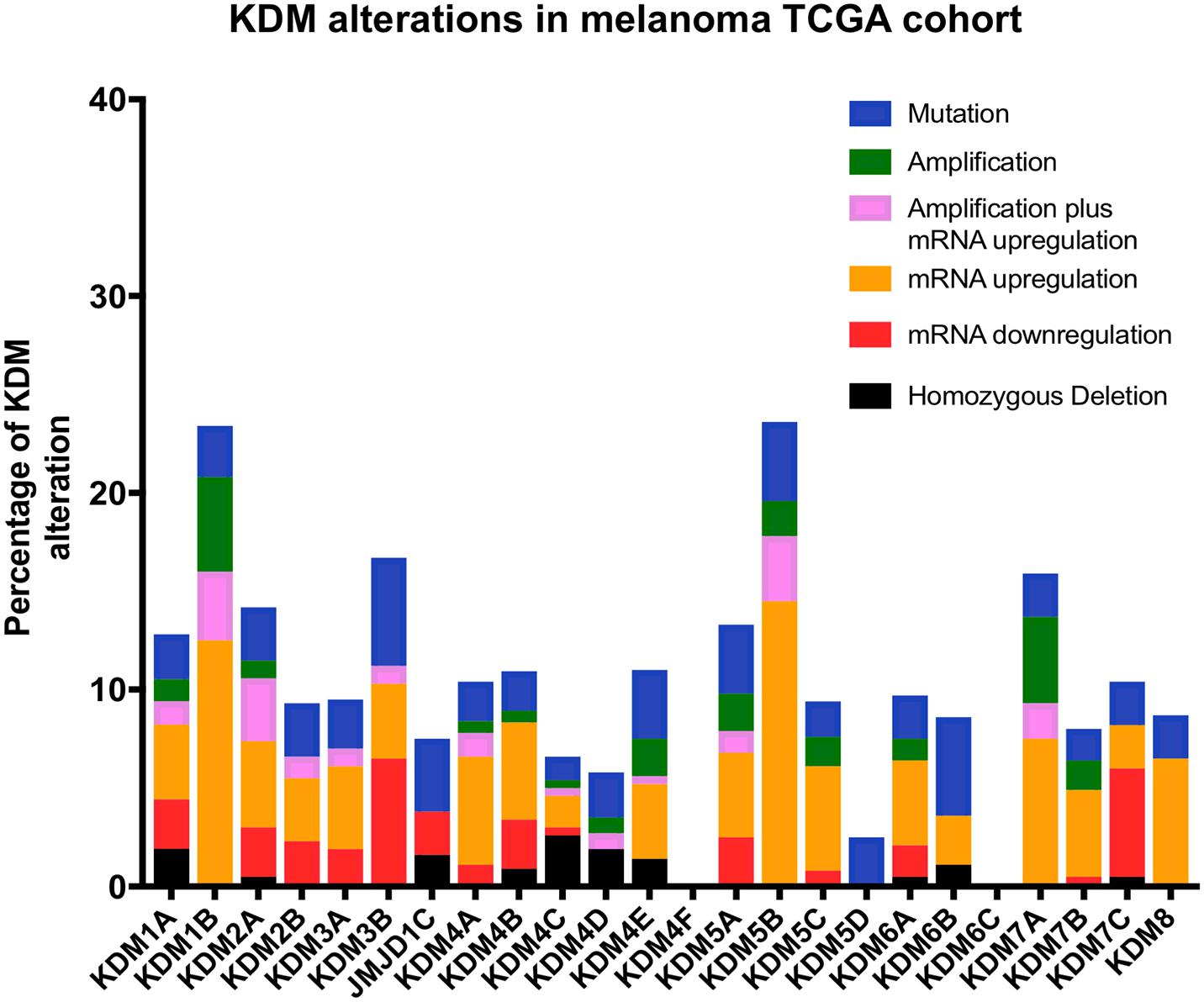
Figure 3. Bar graph showing the percentage and type of each KDM mutations obtained from the cancer genome atlas (TCGA) database. The data was obtained from a total of 472 patients with skin cutaneous melanoma (SKCM).
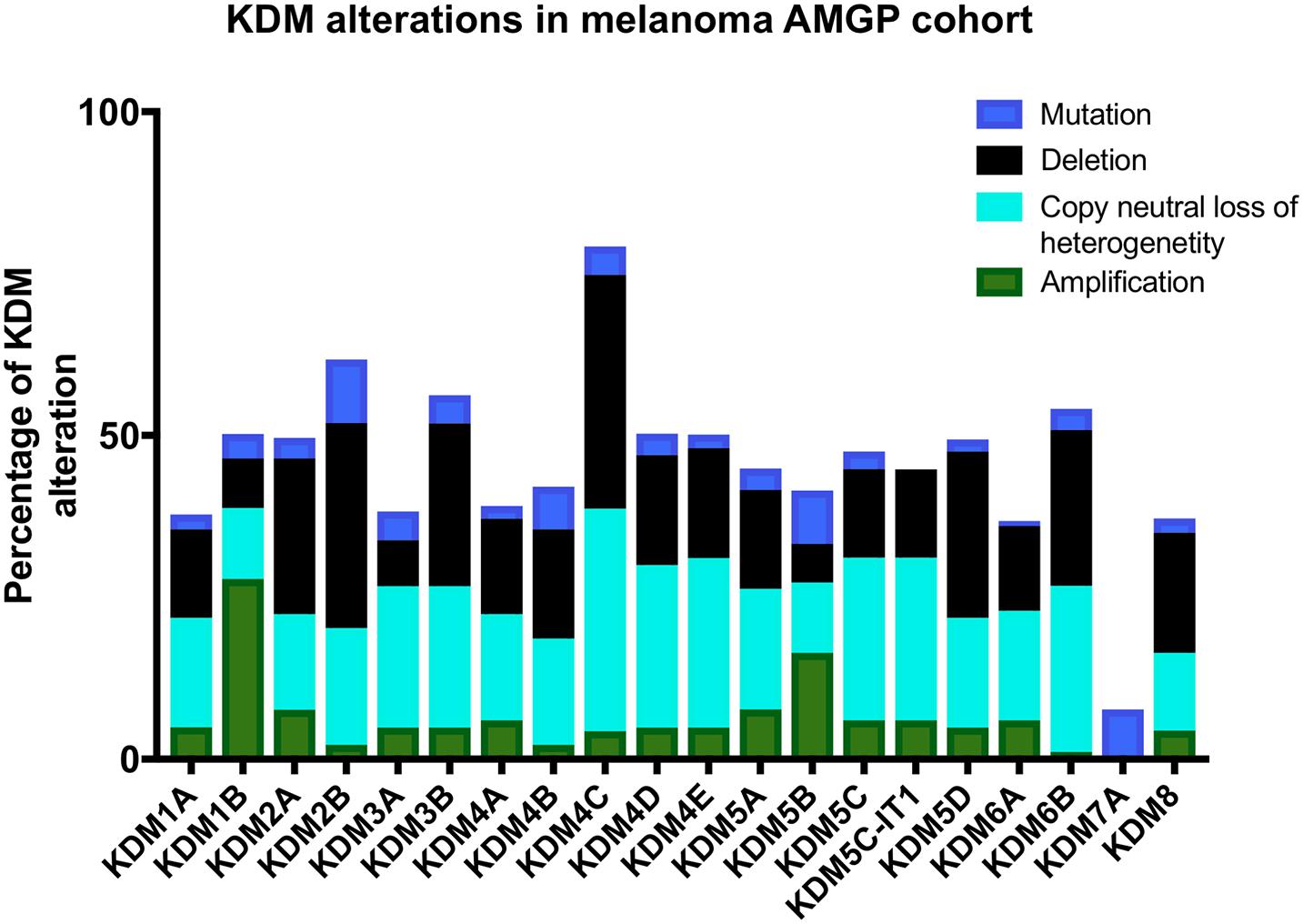
Figure 4. Bar graph showing the percentage of each and type of KDM alterations in 183 melanoma patients from the Australian melanoma genome project (AMGP).
Melanoma is the most deadly type of skin cancer and is the most commonly diagnosed cancer in young Australians (AIHW, 2017). Analysis of two independent cohorts showed that KDM alterations are frequent in melanoma (Figures 3, 4) and thus warrant further investigation into their mechanistic role.
The most extensively studied KDM in melanoma is KDM5B. Studies found that KDM5B expression is higher in melanocytic nevi compared to advanced and metastatic melanomas (Roesch et al., 2005, 2010). High KDM5B expression was associated with a slow cycling population of melanoma cells that prolonged growth and self-renewal. An effect was observed on Notch signaling in that KDM5B suppressed the Notch ligand Jagged 1, causing less notch cleavage and a decrease in the expression of Notch target genes. The study suggests that KDM5B may have an important role in the maintenance of stem like progenitors that seed tumor progression and metastasis in melanoma.
KDM6B was found to be essential for melanoma tumor growth and metastasis (Park et al., 2016). KDM6B could activate NF-kB and bone morphogenic protein signaling promoting melanoma growth and progression.
Another recent study showed that H3K9 demethylases (which include KDM3B) can disable senescence, allowing ras/braf mutant melanoma development and progression. This was reversed when treated with H3K9 inhibitors in vitro and in vivo (Yu et al., 2018). The main role of KDM6A/B may be as an antagonist of EZH2 which has been implicated in the growth and progression in melanoma. Early studies found that elevated EZH2 expression was associated with poor survival (Bachmann et al., 2006; McHugh et al., 2007). A melanoma EZH2 mouse model showed that melanocyte specific loss of EZH2, or treatment with an EZH2 inhibitor, abolished the spread of metastatic melanoma (Zingg et al., 2015). EZH2 was able to induce resistance to immunotherapy treatments anti-CTLA-4 and IL-2 in a melanoma mouse model and when inactivated, reversed this resistance (Zingg et al., 2017). A recent study also found that EZH2 induced loss of primary cilia, enhanced Wnt signaling and promoted melanoma metastasis (Zingg et al., 2018). Collectively these studies show that EZH2 has an important role in the progression of melanoma, therefore its only known antagonist, KDM6A/B, is likely to be of equal importance and warrants exploration. It is also unclear what role the COMPASS complex which contains KDM6A may have in melanoma progression. Importantly it is not clear what role these complexes may have in determining the sensitivity or resistance to inhibitors of EZH2 and KDM6A/B.
There is a striking and unexplained predominance for males to be diagnosed and die from cancer, including melanoma, compared to females (Joosse et al., 2013; Clocchiatti et al., 2016; Enninga et al., 2017). It is postulated that this is due to particular X-linked genes that escape X-inactivation also known as “escape from X-inactivation tumor suppressor” (EXITS) genes (Dunford et al., 2017). This means that females express two copies of EXITS genes compared to males, effectively doubling the amount of tumor suppressive function. One of these KDMS identified in this category was KDM6A. Our recent study showed that KDM6A had a significant prognostic effect in female melanoma patients inducing better overall survival (Emran et al., 2020). Our TCGA gene set enrichment analysis (GSEA) suggests that high KDM6A level associated with upregulation of several immune related pathways like IFN-gamma which may help anti-tumor immunity and survival advantage in female melanoma patients compared to male (Emran et al., 2020).
Other studies support KDM6A in having sex-specific roles in normal biology and cancer. An early study showed that KDM6A expression was significantly higher in the brains and organs of female mice compared to male mice (Xu et al., 2008). In the context of cancer, a study showed that KDM6A had a gender-specific, tumor suppressive effect in T-cell acute lymphoblastic leukemia (T-ALL). The study showed that KDM6A is frequently mutated in male T-ALL patients and KDM6A expression is significantly lower compared to females T-ALL patients (Van der Meulen et al., 2015). In addition, loss of KDM6A expression in female mice induced poorer survival by downregulation of T-ALL associated tumor suppressor genes and upregulation of T-ALL associated oncogenes (Van der Meulen et al., 2015). Loss of KDM6A expression in female mice exhibited a squamous-like, malignant phenotype via activation of oncogenes MYC and RUNX3 in a pancreatic cancer mouse model (Andricovich et al., 2018).
UTY also known as KDM6C is the male equivalent of KDM6A/UTX and is expressed on the Y chromosome but importantly, has significantly reduced H3K27me3 demethylation activity compared to KDM6A, due to the substitution of important amino acids in the Jumonji C domain (Walport et al., 2014).
A study in pancreatic cancer found that concurrent loss of UTY and KDM6A in male patients was associated with a more malignant phenotype and poorer and prognosis (Andricovich et al., 2018). In addition, male mice that expressed UTY and female mice with heterozygous KDM6A expression exhibited a less aggressive pancreatic cancer phenotype, while male and female mice with no UTY or KDM6A exhibited a more aggressive malignant phenotype (Andricovich et al., 2018). Hence this study suggests that KDM6A and UTY have tumor-suppressive roles in pancreatic cancer and that sex-specific mechanism should be investigated in melanoma and other cancers.
Numerous KDM inhibitors are showing promise in targeting KDMs in certain types of cancers in both preclinical and clinical studies. A summary of inhibitors is provided below and in Table 2, grouped by KDM family.
There have been numerous irreversible and reversible inhibitors that have been developed and tested preclinically and in clinical trials that target the KDM1 or LSD family. These include tranylcypromine derived KDM1A inhibitors that have comprehensively described elsewhere (Fang et al., 2019) and summarized below.
ORY-1001 has been reported to induce expression of differentiation markers in mixed lineage leukemia (MLL) cells as well as reducing tumor growth in an acute myeloid leukemia (AML) mouse model and possesses good oral bioavailability (Harris et al., 2012; Maes et al., 2013, 2015). ORY-1001 has also been shown to suppress growth in lung cancer in vitro (Lu et al., 2018). ORY-1001 is currently being tested in phase I and II clinical trials in patients with relapsed and/or refractory AML and small cell lung cancer (SCLC) (Register ECT, 2018).
Another selective KDM1A inhibitor is GSK2879552, tested in phase I clinical trials in SCLC. A study found that GSK2879552 promotes differentiation in AML cells and when SCLC cells were treated, proliferation was reduced in vitro. In mouse models of AML and SCLC, GSK2879552 prolonged survival (Dhanak and Jackson, 2014; Mohammed et al., 2014). However, clinical trials in patients with AML and SCLC and were terminated due to adverse side effects (Table 2).
Initially, the KDM1A inhibitor known as INCB059872 treatment was shown to induce cell differentiation in progenitor cells in AML (Johnston et al., 2020), prostate cancer and Ewing sarcoma (Fang et al., 2019). However, INCB059872 was tested in a clinical trial in patients with Ewing sarcoma and terminated due to recruitment issues, this inhibitor in conjunction with an anti-PD-1 checkpoint inhibitor is currently being tested in patients with colorectal and lung cancer (Table 2).
Another strategy involves dual inhibition of both histone deacetylases (HDAC) and KDM1A with 4SC-202 which targets HDAC1, 2, 3, and KDM1A and has been shown to inhibit the stem-related properties of cancer cells reducing their viability (Henning et al., 2010). A phase I clinical study has been conducted for 4SC-202 in patients with advanced leukemia, found to possess anti-cancer activity and to be well-tolerated in patients (von Tresckow et al., 2014, 2019). Another study found that treatment with 4SC-202 significantly increased the survival of mice that had AML tumors without any toxicity effects (Fiskus et al., 2014). Currently, there are clinical trials being undertaken that are testing the effect of 4SC-202 in patients with melanoma, merkel cell carcinoma and gastrointestinal cancer (Table 2).
SP-2577 reversibly inhibits KDM1A demethylation and has been recently found to promote anti tumor immunity in mutated ovarian cancer cells in vitro and has also been found to inhibit growth in Ewing Sarcoma xenografts (Salarius Pharmaceuticals, Inc., 2020; Soldi et al., 2020). Currently SP-2577 is being tested in patients with Ewing sarcoma, ovarian and endometrial cancers (Table 2).
The reversible KDM1A inhibitor CC-90011 has been found to induce cellular differentiation exhibits anti-tumor efficacy in vitro and in vivo in AML and SCLC (Kanouni et al., 2020). CC-90011 was also tested in patients with hematological cancers, showing robust anti-cancer properties and good tolerability in patients (Hollebecque et al., 2021). Currently, CC-90011 is being tested in clinical trials in patients with prostate and SCLC (Table 2).
The most prominent KDM4 inhibitor is caffeic acid, a naturally occurring compound found in various sources including eucalyptus globus (Santos et al., 2011). Caffeic acid has been reported to mainly target KDM4C (Nielsen et al., 2012) and displays potent anti-cancer activity against esophageal cancer in vitro and in vivo (Hirose et al., 1998). Currently, caffeic acid is being tested in a clinical trial in patients with esophageal cancer (Table 2).
The most advanced preclinical KDM4 inhibitor is JIB-04 which targets KDM4A, KDM4B, and KDM4E and can inhibit growth and reduce tumor burden in non-small cell lung cancer (NSCLC) and breast cancer in vitro and in vivo (Wang et al., 2013). In addition, JIB-04 treatment reduced colony formation, growth and migration in vitro and reduced tumorigenic activity in a colorectal cancer model in vivo. The mechanism has been attributed to downregulating genes of the Wnt signaling pathway which are essential for promoting carcinogenesis (Kim et al., 2018). JIB-04 is yet to be tested in clinical trials.
Another KDM4 inhibitor in the toolbox is ML324 which targets KDM4B and KDM4E (Rai et al., 2010). A study reported that ML324 treatment reduced tumor volume and growth in a triple negative breast cancer mouse model (Wang W. et al., 2018) and also inhibits proliferation in prostate cancer in vitro and in vivo (Carter et al., 2017). ML324 is yet to be tested in clinical trials.
The most advanced KDM5 inhibitor is EPT-103182. This small molecule compound targets KDM5B which has been shown to have an anti-proliferative effect in hematological and solid cancer cell lines as well as inhibiting tumor growth in a dose-dependent manner in xenograft models (Hancock et al., 2015; Maes et al., 2015). This inhibitor has yet to be tested in clinical trials.
Another recently identified KDM5 inhibitor is PBIT, shown to specially target and inhibit KDM5B (Sayegh et al., 2013). In the context of cancer, PBIT treatment inhibited proliferation of breast cancer by derepression and upregulation of the tumor suppressor HEXIMI in vitro (Montano et al., 2019). PBIT is yet to be tested in clinical trials.
KDOAM-25 has been shown to target and inhibit KDM5A-C demethylases (a pan-KDM5 family inhibitor), but especially KDM5B (Tumber et al., 2017). KDOAM-25 treatment has been shown to reduce proliferation and growth in breast cancer and multiple myeloma which highly expressed KDM5B (Tumber et al., 2017; Montano et al., 2019). KDOAM-25 is yet to be tested in clinical trials.
Other inhibitors which target JMJC domain containing KDMs are known as GSK-J4 that targets KDM6A and KDM6B, shown to reduce the production of pro-inflammatory cytokines by human macrophages, although there have been questions in regards to its specificity (Kruidenier et al., 2012; Heinemann et al., 2014). Over the last few years, numerous preclinical studies have demonstrated that GSK-J4 could be a potential therapeutic for certain cancers. GSK-J4 treatment was initially shown to be effective in inhibiting cell growth and inducing cell cycle arrest and apoptosis in primary human T-cell acute lymphoblastic leukemia (T-ALL) lines (Ntziachristos et al., 2014). A recent study found that GSK-J4 treatment inhibited cell proliferation and colony forming ability of acute myeloid leukemia (AML) cell lines and inhibited tumor growth in an AML xenograft mouse model (Li et al., 2018).
Other studies have shown that GSK-J4 treatment inhibited proliferation of castration-resistant prostate cancer cells by inhibiting AR–driven transcription and can also inhibit proliferation in glioma cells in a dose-dependent manner in vitro (Morozov et al., 2017; Sui et al., 2017). A study also showed that GSK-J4 suppressed the ability of breast cancer and ovarian cancer stem cells to proliferate and grow (Sakaki et al., 2015; Yan et al., 2017). GSK-J4 in combination with an anti-diabetic drug metformin induced cell death and inhibited cell growth in non-small cell lung cancer (NSCLC) cell lines (Watarai et al., 2016) and another study found GSK-J4 treatment sensitized diffuse large B-lymphoma cells to chemotherapy drugs (Mathur et al., 2017). GSK-J4 inhibited cell growth and upregulated apoptosis markers in neuroblastoma cell lines and inhibited tumor growth in an in vivo neuroblastoma model (Lochmann et al., 2018). GSK-J4 is yet to be tested in clinical trials.
Dysregulation of lysine methyl demethylases due to genetic changes or aberrant signaling are associated with a number of different cancers. The amine oxidase group of KDMs appear to mediate their effects by action on the H3K4 and H3K9 histone marks but in addition by interactions with the P53 tumor suppressor and E2F transcription factors regulating cell division.
The 2-oxoglutarate dependent oxygenases are the largest group of KDMs and mediate demethylation of histones at several important activating or repressive marks. Their effects in cancers range from tumor suppression to promotion and growth of cancers. Additional roles in immune responses against cancer have been revealed for KDM5C and KDM6A in survival studies on human melanoma patients and may be linked to higher expression from X linked chromosomes. Several KDMs are part of protein complexes like the COMPASS complex that contains KDM6A as well as methyl transferases MLL2 and MLL3 that regulate gene transcription in certain cancers.
The role of KDMs in cancer have identified them as potential therapeutic targets and a wide range of pharmacological agents have been developed. Given the complex interactions of KDMs with other epigenetic regulators it is not surprising that drugs targeting KDMs are yet to enter clinical practice and this remains a focus of future research.
GP-M, AE, and JT: design and structure of review. GP-M and AE: figures. GP-M: tables. GP-M, AE, PH, and JT: editing the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Australian National Health and Medical Research Council (NHMRC) program grant 633004, Cancer Council New South Wales (CCNSW) research grant RG 18-05, and the Cancer Council NSW grant RG 21-07.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank all the patients involved in the TCGA and AGMP cohort and also the people who obtained and made the data accessible for this review.
Agger, K., Cloos, P. A., Rudkjaer, L., Williams, K., Andersen, G., Christensen, J., et al. (2009). The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 23, 1171–1176. doi: 10.1101/gad.510809
Amente, S., Milazzo, G., Sorrentino, M. C., Ambrosio, S., Di Palo, G., Lania, L., et al. (2015). Lysine-specific demethylase (LSD1/KDM1A) and MYCN cooperatively repress tumor suppressor genes in neuroblastoma. Oncotarget 6, 14572–14583. doi: 10.18632/oncotarget.3990
Andricovich, J., Perkail, S., Kai, Y., Casasanta, N., Peng, W., and Tzatsos, A. (2018). Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33, 512–526.e8. doi: 10.1016/j.ccell.2018.02.003
Atala, A. (2017). Re: loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. J. Urol. 198, 984–985. doi: 10.1016/j.juro.2017.08.009
Bachmann, I. M., Halvorsen, O. J., Collett, K., Stefansson, I. M., Straume, O., Haukaas, S. A., et al. (2006). EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 24, 268–273. doi: 10.1200/JCO.2005.01.5180
Benevolenskaya, E. V., Murray, H. L., Branton, P., Young, R. A., and Kaelin, W. G. Jr. (2005). Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18, 623–635. doi: 10.1016/j.molcel.2005.05.012
Berry, W. L., Shin, S., Lightfoot, S. A., and Janknecht, R. (2012). Oncogenic features of the JMJD2A histone demethylase in breast cancer. Int. J. Oncol. 41, 1701–1706. doi: 10.3892/ijo.2012.1618
Bilian Jin, Y. L., and Robertson, K. D. (2011). DNA methylation: superior or subordinate in the epigenetic hierachy? Genes Cancer 2, 607–617.
Carter, D. M., Specker, E., Przygodda, J., Neuenschwander, M., von Kries, J. P., Heinemann, U., et al. (2017). Identification of a novel benzimidazole pyrazolone scaffold that inhibits KDM4 lysine demethylases and reduces proliferation of prostate cancer cells. SLAS Discov. 22, 801–812. doi: 10.1177/2472555217699157
Catchpole, S., Spencer-Dene, B., Hall, D., Santangelo, S., Rosewell, I., Guenatri, M., et al. (2011). PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int. J. Oncol. 38, 1267–1277. doi: 10.3892/ijo.2011.956
Chen, L., Vasilatos, S. N., Qin, Y., Katz, T. A., Cao, C., Wu, H., et al. (2017). Functional characterization of lysine-specific demethylase 2 (LSD2/KDM1B) in breast cancer progression. Oncotarget 8, 81737–81753. doi: 10.18632/oncotarget.19387
Cho, Y. W., Hong, T., Hong, S., Guo, H., Yu, H., Kim, D., et al. (2007). PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406. doi: 10.1074/jbc.M701574200
Ciccone, D. N., Su, H., Hevi, S., Gay, F., Lei, H., Bajko, J., et al. (2009). KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418. doi: 10.1038/nature08315
Clocchiatti, A., Cora, E., Zhang, Y., and Dotto, G. P. (2016). Sexual dimorphism in cancer. Nat. Rev. Cancer 16, 330–339. doi: 10.1038/nrc.2016.30
Daures, M., Ngollo, M., Judes, G., Rifai, K., Kemeny, J. L., Penault-Llorca, F., et al. (2016). The JMJD3 histone demethylase and the EZH2 histone methyltransferase in prostate cancer. OMICS 20, 123–125. doi: 10.1089/omi.2015.0113
Defeo-Jones, D., Huang, P. S., Jones, R. E., Haskell, K. M., Vuocolo, G. A., Hanobik, M. G., et al. (1991). Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature 352, 251–254. doi: 10.1038/352251a0
Dhanak, D., and Jackson, P. (2014). Development and classes of epigenetic drugs for cancer. Biochem. Biophys. Res. Commun. 455, 58–69. doi: 10.1016/j.bbrc.2014.07.006
Dong, R., Yao, R., Du, J., Wang, S., and Fan, Z. (2013). Depletion of histone demethylase KDM2A enhanced the adipogenic and chondrogenic differentiation potentials of stem cells from apical papilla. Exp. Cell Res. 319, 2874–2882. doi: 10.1016/j.yexcr.2013.07.008
Dunford, A., Weinstock, D. M., Savova, V., Schumacher, S. E., Cleary, J. P., Yoda, A., et al. (2017). Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 49, 10–16. doi: 10.1038/ng.3726
Dupont, C., Armant, D. R., and Brenner, C. A. (2009). Epigenetics: definition, mechanisms and clinical perspective. Semin. Reprod. Med. 27, 351–357. doi: 10.1055/s-0029-1237423
Elgin, S. C. (1996). Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev. 6, 193–202. doi: 10.1016/s0959-437x(96)80050-5
Emran, A. A., Nsengimana, J., Punnia-Moorthy, G., Schmitz, U., Gallagher, S. J., Newton-Bishop, J., et al. (2020). Study of the female sex survival advantage in melanoma-A focus on X-linked epigenetic regulators and immune responses in two cohorts. Cancers (Basel) 12:2082. doi: 10.3390/cancers12082082
Enninga, E. A. L., Moser, J. C., Weaver, A. L., Markovic, S. N., Brewer, J. D., Leontovich, A. A., et al. (2017). Survival of cutaneous melanoma based on sex, age, and stage in the United States, 1992–2011. Cancer Med. 6, 2203–2212. doi: 10.1002/cam4.1152
Ezponda, T., Dupere-Richer, D., Will, C. M., Small, E. C., Varghese, N., Patel, T., et al. (2017). UTX/KDM6A loss enhances the malignant phenotype of multiple myeloma and sensitizes cells to EZH2 inhibition. Cell Rep. 21, 628–640. doi: 10.1016/j.celrep.2017.09.078
Fang, Y., Liao, G., and Yu, B. (2019). LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J. Hematol. Oncol. 12:129. doi: 10.1186/s13045-019-0811-9
Feng, T., Wang, Y., Lang, Y., and Zhang, Y. (2017). KDM5A promotes proliferation and EMT in ovarian cancer and closely correlates with PTX resistance. Mol. Med. Rep. 16, 3573–3580. doi: 10.3892/mmr.2017.6960
Fiskus, W., Sharma, S., Shah, B., Portier, B. P., Devaraj, S. G., Liu, K., et al. (2014). Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia 28, 2155–2164. doi: 10.1038/leu.2014.119
Ford, D. J., and Dingwall, A. K. (2015). The cancer COMPASS: navigating the functions of MLL complexes in cancer. Cancer Genet. 208, 178–191. doi: 10.1016/j.cancergen.2015.01.005
Gildea, J. J., Lopez, R., and Shearn, A. (2000). A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156, 645–663.
Hancock, R. L., Dunne, K., Walport, L. J., Flashman, E., and Kawamura, A. (2015). Epigenetic regulation by histone demethylases in hypoxia. Epigenomics 7, 791–811. doi: 10.2217/epi.15.24
Harris, W. J., Huang, X., Lynch, J. T., Spencer, G. J., Hitchin, J. R., Li, Y., et al. (2012). The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21, 473–487. doi: 10.1016/j.ccr.2012.03.014
He, J., Nguyen, A. T., and Zhang, Y. (2011). KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 117, 3869–3880. doi: 10.1182/blood-2010-10-312736
Heinemann, B., Nielsen, J. M., Hudlebusch, H. R., Lees, M. J., Larsen, D. V., Boesen, T., et al. (2014). Inhibition of demethylases by GSK-J1/J4. Nature 514, E1–E2. doi: 10.1038/nature13688
Henning, S. W., Doblhofer, R., Kohlhof, H., Jankowsky, R., Maier, T., Beckers, T., et al. (2010). Preclinical characterization of 4SC-202, a novel isotype specific HDAC inhibitor. EJC Suppl. 8:61.
Hirose, M., Takesada, Y., Tanaka, H., Tamano, S., Kato, T., and Shirai, T. (1998). Carcinogenicity of antioxidants BHA, caffeic acid, sesamol, 4-methoxyphenol and catechol at low doses, either alone or in combination, and modulation of their effects in a rat medium-term multi-organ carcinogenesis model. Carcinogenesis 19, 207–212. doi: 10.1093/carcin/19.1.207
Hollebecque, A., Salvagni, S., Plummer, R., Isambert, N., Niccoli, P., Capdevila, J., et al. (2021). Phase I study of lysine-specific demethylase 1 inhibitor, CC-90011, in patients with advanced solid tumors and relapsed/refractory non-hodgkin lymphoma. Clin. Cancer Res. 27, 438–446. doi: 10.1158/1078-0432.CCR-20-2380
Hong, S., Cho, Y. W., Yu, L. R., Yu, H., Veenstra, T. D., and Ge, K. (2007). Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U. S. A. 104, 18439–18444. doi: 10.1073/pnas.0707292104
Hou, J., Wu, J., Dombkowski, A., Zhang, K., Holowatyj, A., Boerner, J. L., et al. (2012). Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am. J. Transl. Res. 4, 247–256.
Hsia, D. A., Tepper, C. G., Pochampalli, M. R., Hsia, E. Y., Izumiya, C., Huerta, S. B., et al. (2010). KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 107, 9671–9676. doi: 10.1073/pnas.1000401107
Huang, C., Xiang, Y., Wang, Y., Li, X., Xu, L., Zhu, Z., et al. (2010). Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 20, 154–165. doi: 10.1038/cr.2010.5
Huang, J., Sengupta, R., Espejo, A. B., Lee, M. G., Dorsey, J. A., Richter, M., et al. (2007). p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108. doi: 10.1038/nature06092
Huang, Y., Liu, Y., Yu, L., Chen, J., Hou, J., Cui, L., et al. (2015). Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol. 36, 271–278. doi: 10.1007/s13277-014-2630-5
Johnston, G., Ramsey, H. E., Liu, Q., Wang, J., Stengel, K. R., Sampathi, S., et al. (2020). Nascent transcript and single-cell RNA-seq analysis defines the mechanism of action of the LSD1 inhibitor INCB059872 in myeloid leukemia. Gene 752:144758. doi: 10.1016/j.gene.2020.144758
Joosse, A., Collette, S., Suciu, S., Nijsten, T., Patel, P. M., Keilholz, U., et al. (2013). Sex is an independent prognostic indicator for survival and relapse/progression-free survival in metastasized stage III to IV melanoma: a pooled analysis of five European organisation for research and treatment of cancer randomized controlled trials. J. Clin. Oncol. 31, 2337–2346. doi: 10.1200/JCO.2012.44.5031
Kanouni, T., Severin, C., Cho, R. W., Yuen, N. Y., Xu, J., Shi, L., et al. (2020). Discovery of CC-90011: a potent and selective reversible inhibitor of lysine specific demethylase 1 (LSD1). J. Med. Chem. 63, 14522–14529. doi: 10.1021/acs.jmedchem.0c00978
Katz, T. A., Vasilatos, S. N., Harrington, E., Oesterreich, S., Davidson, N. E., and Huang, Y. (2014). Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast Cancer Res. Treat. 146, 99–108. doi: 10.1007/s10549-014-3012-9
Kawazu, M., Saso, K., Tong, K. I., McQuire, T., Goto, K., Son, D. O., et al. (2011). Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 6:e17830. doi: 10.1371/journal.pone.0017830
Kim, J. H., Sharma, A., Dhar, S. S., Lee, S. H., Gu, B., and Chan, C. H. (2017). UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 77:2553. doi: 10.1158/0008-5472.CAN-17-0639
Kim, J. Y., Kim, K. B., Eom, G. H., Choe, N., Kee, H. J., Son, H. J., et al. (2012). KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol. Cell Biol. 32, 2917–2933. doi: 10.1128/MCB.00133-12
Kim, M. S., Cho, H. I., Yoon, H. J., Ahn, Y. H., Park, E. J., Jin, Y. H., et al. (2018). JIB-04, a small molecule histone demethylase inhibitor, selectively targets colorectal cancer stem cells by inhibiting the wnt/beta-catenin signaling pathway. Sci. Rep. 8:6611. doi: 10.1038/s41598-018-24903-0
Kim, T. D., Jin, F., Shin, S., Oh, S., Lightfoot, S. A., Grande, J. P., et al. (2016). Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1. J. Clin. Invest. 126, 706–720. doi: 10.1172/JCI78132
Klose, R. J., Kallin, E. M., and Zhang, Y. (2006). JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727. doi: 10.1038/nrg1945
Komura, K., Jeong, S. H., Hinohara, K., Qu, F., Wang, X., Hiraki, M., et al. (2016). Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc. Natl. Acad. Sci. U. S. A. 113, 6259–6264. doi: 10.1073/pnas.1600420113
Komura, K., Yoshikawa, Y., Shimamura, T., Chakraborty, G., Gerke, T. A., Hinohara, K., et al. (2018). ATR inhibition controls aggressive prostate tumors deficient in Y-linked histone demethylase KDM5D. J. Clin. Invest. 128, 2979–2995. doi: 10.1172/JCI96769
Kong, Y., Zou, S., Yang, F., Xu, X., Bu, W., Jia, J., et al. (2016). RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting KDM2A. Cancer Lett. 381, 138–148. doi: 10.1016/j.canlet.2016.07.038
Kontaki, H., and Talianidis, I. (2010). Lysine methylation regulates E2F1-induced cell death. Mol. Cell 39, 152–160. doi: 10.1016/j.molcel.2010.06.006
Kornberg, R. D. (1974). Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871. doi: 10.1126/science.184.4139.868
Kruidenier, L., Chung, C. W., Cheng, Z., Liddle, J., Che, K., Joberty, G., et al. (2012). A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408. doi: 10.1038/nature11262
Kuang, Y., Lu, F., Guo, J., Xu, H., Wang, Q., Xu, C., et al. (2017). Histone demethylase KDM2B upregulates histone methyltransferase EZH2 expression and contributes to the progression of ovarian cancer in vitro and in vivo. Onco. Targets Ther. 10, 3131–3144. doi: 10.2147/OTT.S134784
Lee, K. H., Hong, S., Kang, M., Jeong, C. W., Ku, J. H., Kim, H. H., et al. (2018). Histone demethylase KDM7A controls androgen receptor activity and tumor growth in prostate cancer. Int. J. Cancer 143, 2849–2861. doi: 10.1002/ijc.31843
Ler, L. D., Ghosh, S., Chai, X., Thike, A. A., Heng, H. L., Siew, E. Y., et al. (2017). Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci. Transl. Med. 9:eaai8312. doi: 10.1126/scitranslmed.aai8312
Li, B. X., Luo, C. L., Li, H., Yang, P., Zhang, M. C., Xu, H. M., et al. (2012). Effects of siRNA-mediated knockdown of jumonji domain containing 2A on proliferation, migration and invasion of the human breast cancer cell line MCF-7. Exp. Ther. Med. 4, 755–761. doi: 10.3892/etm.2012.662
Li, H., Lan, J., Wang, G., Guo, K., Han, C., Li, X., et al. (2020). KDM4B facilitates colorectal cancer growth and glucose metabolism by stimulating TRAF6-mediated AKT activation. J. Exp. Clin. Cancer Res. 39:12. doi: 10.1186/s13046-020-1522-3
Li, L., Greer, C., Eisenman, R. N., and Secombe, J. (2010). Essential functions of the histone demethylase lid. PLoS Genet. 6:e1001221. doi: 10.1371/journal.pgen.1001221
Li, N., Dhar, S. S., Chen, T. Y., Kan, P. Y., Wei, Y., Kim, J. H., et al. (2016). JARID1D is a suppressor and prognostic marker of prostate cancer invasion and metastasis. Cancer Res. 76, 831–843. doi: 10.1158/0008-5472.CAN-15-0906
Li, Q., Hou, L., Ding, G., Li, Y., Wang, J., Qian, B., et al. (2015). KDM6B induces epithelial-mesenchymal transition and enhances clear cell renal cell carcinoma metastasis through the activation of SLUG. Int. J. Clin. Exp. Pathol. 8, 6334–6344.
Li, Y., Zhang, M., Sheng, M., Zhang, P., Chen, Z., Xing, W., et al. (2018). Therapeutic potential of GSK-J4, a histone demethylase KDM6B/JMJD3 inhibitor, for acute myeloid leukemia. J. Cancer Res. Clin. Oncol. 144, 1065–1077. doi: 10.1007/s00432-018-2631-7
Liang, S., Yao, Q., Wei, D., Liu, M., Geng, F., Wang, Q., et al. (2019). KDM6B promotes ovarian cancer cell migration and invasion by induced transforming growth factor-beta1 expression. J. Cell Biochem. 120, 493–506. doi: 10.1002/jcb.27405
Lin, C. Y., Wang, B. J., Chen, B. C., Tseng, J. C., Jiang, S. S., Tsai, K. K., et al. (2019). Histone demethylase KDM4C stimulates the proliferation of prostate cancer cells via activation of AKT and c-Myc. Cancers (Basel) 11:1785. doi: 10.3390/cancers11111785
Lochmann, T. L., Powell, K. M., Ham, J., Floros, K. V., Heisey, D. A. R., Kurupi, R. I. J., et al. (2018). Targeted inhibition of histone H3K27 demethylation is effective in high-risk neuroblastoma. Sci. Transl. Med. 10:eaao4680. doi: 10.1126/scitranslmed.aao4680
Loenarz, C., and Schofield, C. J. (2008). Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 4, 152–156. doi: 10.1038/nchembio0308-152
Lopez-Bigas, N., Kisiel, T. A., Dewaal, D. C., Holmes, K. B., Volkert, T. L., Gupta, S., et al. (2008). Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell 31, 520–530. doi: 10.1016/j.molcel.2008.08.004
Lu, P. J., Sundquist, K., Baeckstrom, D., Poulsom, R., Hanby, A., Meier-Ewert, S., et al. (1999). A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274, 15633–15645.
Lu, Z., Guo, Y., Zhang, X., Li, J., Li, L., Zhang, S., et al. (2018). ORY-1001 suppresses cell growth and induces apoptosis in lung cancer through triggering hk2 mediated warburg effect. Front. Pharmacol. 9:1411. doi: 10.3389/fphar.2018.01411
Ma, J., Wang, N., Zhang, Y., Wang, C., Ge, T., Jin, H., et al. (2015). KDM6B elicits cell apoptosis by promoting nuclear translocation of FOXO1 in non-small cell lung cancer. Cell Physiol. Biochem. 37, 201–213. doi: 10.1159/000430345
Maes, T., Carceller, E., Salas, J., Ortega, A., and Buesa, C. (2015). Advances in the development of histone lysine demethylase inhibitors. Curr. Opin. Pharmacol. 23, 52–60. doi: 10.1016/j.coph.2015.05.009
Maes, T., Tirapu, I., Mascaro, C., Ortega, A., Estiarte, A., Valls, N., et al. (2013). Preclinical characterization of a potent and selective inhibitor of the histone demethylase KDM1A for MLL leukemia. J. Clin. Oncol. 31:e13543.
Mathur, R., Sehgal, L., Havranek, O., Kohrer, S., Khashab, T., Jain, N., et al. (2017). Inhibition of demethylase KDM6B sensitizes diffuse large B-cell lymphoma to chemotherapeutic drugs. Haematologica 102, 373–380. doi: 10.3324/haematol.2016.144964
McHugh, J. B., Fullen, D. R., Ma, L., Kleer, C. G., and Su, L. D. (2007). Expression of polycomb group protein EZH2 in nevi and melanoma. J. Cutan. Pathol. 34, 597–600. doi: 10.1111/j.1600-0560.2006.00678.x
Mohammed, H., Smitheman, K., Van Aller, G., Cusan, M., Kamat, S., Liu, Y., et al. (2014). Novel anti-tumor activity of targeted LSD1 inhibiton by GSK2879552. Eur. J. Cancer 50:72.
Montano, M. M., Yeh, I. J., Chen, Y., Hernandez, C., Kiselar, J. G., de la Fuente, M., et al. (2019). Inhibition of the histone demethylase, KDM5B, directly induces re-expression of tumor suppressor protein HEXIM1 in cancer cells. Breast Cancer Res. 21:138. doi: 10.1186/s13058-019-1228-7
Morozov, V. M., Li, Y., Clowers, M. M., and Ishov, A. M. (2017). Inhibitor of H3K27 demethylase JMJD3/UTX GSK-J4 is a potential therapeutic option for castration resistant prostate cancer. Oncotarget 8, 62131–62142. doi: 10.18632/oncotarget.19100
Narayanan, S. P., Singh, S., Gupta, A., Yadav, S., Singh, S. R., and Shukla, S. (2015). Integrated genomic analyses identify KDM1A’s role in cell proliferation via modulating E2F signaling activity and associate with poor clinical outcome in oral cancer. Cancer Lett. 367, 162–172. doi: 10.1016/j.canlet.2015.07.022
Nielsen, A. L., Kristensen, L. H., Stephansen, K. B., Kristensen, J. B., Helgstrand, C., Lees, M., et al. (2012). Identification of catechols as histone-lysine demethylase inhibitors. FEBS Lett. 586, 1190–1194. doi: 10.1016/j.febslet.2012.03.001
Niu, X., Zhang, T., Liao, L., Zhou, L., Lindner, D. J., Zhou, M., et al. (2012). The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene 31, 776–786. doi: 10.1038/onc.2011.266
Ntziachristos, P., Tsirigos, A., Welstead, G. G., Trimarchi, T., Bakogianni, S., Xu, L., et al. (2014). Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514, 513–517. doi: 10.1038/nature13605
Oh, S., and Janknecht, R. (2012). Histone demethylase JMJD5 is essential for embryonic development. Biochem. Biophys. Res. Commun. 420, 61–65. doi: 10.1016/j.bbrc.2012.02.115
Ohguchi, H., Harada, T., Sagawa, M., Kikuchi, S., Tai, Y. T., Richardson, P. G., et al. (2017). KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia 31, 2661–2669. doi: 10.1038/leu.2017.141
Osawa, T., Muramatsu, M., Wang, F., Tsuchida, R., Kodama, T., Minami, T., et al. (2011). Increased expression of histone demethylase JHDM1D under nutrient starvation suppresses tumor growth via down-regulating angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 108, 20725–20729. doi: 10.1073/pnas.1108462109
Park, W. Y., Hong, B. J., Lee, J., Choi, C., and Kim, M. Y. (2016). H3K27 Demethylase JMJD3 employs the NF-kappaB and BMP signaling pathways to modulate the tumor microenvironment and promote melanoma progression and metastasis. Cancer Res. 76, 161–170. doi: 10.1158/0008-5472.CAN-15-0536
Pedersen, M. T., and Helin, K. (2010). Histone demethylases in development and disease. Trends Cell Biol. 20, 662–671. doi: 10.1016/j.tcb.2010.08.011
Pereira, F., Barbachano, A., Silva, J., Bonilla, F., Campbell, M. J., Munoz, A., et al. (2011). KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum. Mol. Genet. 20, 4655–4665. doi: 10.1093/hmg/ddr399
Perillo, B., Ombra, M. N., Bertoni, A., Cuozzo, C., Sacchetti, S., Sasso, A., et al. (2008). DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206. doi: 10.1126/science.1147674
Perinchery, G., Sasaki, M., Angan, A., Kumar, V., Carroll, P., and Dahiya, R. (2000). Deletion of Y-chromosome specific genes in human prostate cancer. J. Urol. 163, 1339–1342.
Pinton, G., Nilsson, S., and Moro, L. (2018). Targeting estrogen receptor beta (ERbeta) for treatment of ovarian cancer: importance of KDM6B and SIRT1 for ERbeta expression and functionality. Oncogenesis 7:15. doi: 10.1038/s41389-018-0027-9
Qiu, M. T., Fan, Q., Zhu, Z., Kwan, S. Y., Chen, L., Chen, J. H., et al. (2015). KDM4B and KDM4A promote endometrial cancer progression by regulating androgen receptor, c-myc, and p27kip1. Oncotarget 6, 31702–31720. doi: 10.18632/oncotarget.5165
Rai, G., Kawamura, A., Tumber, A., Liang, Y., Vogel, J. L., Arbuckle, J. H., et al. (2010). Discovery of ML324, a JMJD2 Demethylase Inhibitor with Demonstrated Antiviral Activity. Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD: National Center for Biotechnology Information.
Ramadoss, S., Guo, G., and Wang, C. Y. (2017a). Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non-histone protein p53. Oncogene 36, 47–59. doi: 10.1038/onc.2016.174
Ramadoss, S., Sen, S., Ramachandran, I., Roy, S., Chaudhuri, G., and Farias-Eisner, R. (2017b). Lysine-specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene 36, 6508. doi: 10.1038/onc.2017.331
Register ECT (2018). A Pilot Study To Assess the Safety, Tolerability, Dose Finding and Efficacy of ORY-1001 in Combination with Azacitidine in Older Patients with AML in First Line Therapy. Available online at: www.clinicaltrialsregister.eu/ORY-1001
Rizwani, W., Schaal, C., Kunigal, S., Coppola, D., and Chellappan, S. (2014). Mammalian lysine histone demethylase KDM2A regulates E2F1-mediated gene transcription in breast cancer cells. PLoS One 9:e100888. doi: 10.1371/journal.pone.0100888
Rocha-Viegas, L., Villa, R., Gutierrez, A., Iriondo, O., Shiekhattar, R., and Di Croce, L. (2014). Role of UTX in retinoic acid receptor-mediated gene regulation in leukemia. Mol. Cell Biol. 34, 3765–3775. doi: 10.1128/MCB.00839-14
Roesch, A., Becker, B., Meyer, S., Hafner, C., Wild, P. J., Landthaler, M., et al. (2005). Overexpression and hyperphosphorylation of retinoblastoma protein in the progression of malignant melanoma. Mod. Pathol. 18, 565–572. doi: 10.1038/modpathol.3800324
Roesch, A., Fukunaga-Kalabis, M., Schmidt, E. C., Zabierowski, S. E., Brafford, P. A., Vultur, A., et al. (2010). A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141, 583–594. doi: 10.1016/j.cell.2010.04.020
Sakaki, H., Okada, M., Kuramoto, K., Takeda, H., Watarai, H., Suzuki, S., et al. (2015). GSKJ4, a selective jumonji H3K27 demethylase inhibitor, effectively targets ovarian cancer stem cells. Anticancer Res. 35, 6607–6614.
Salarius Pharmaceuticals, Inc. (2020). Ewing Sarcoma. available online at: http://investors.salariuspharma.com
Santos, S. A., Freire, C. S., Domingues, M. R., Silvestre, A. J., and Pascoal Neto, C. (2011). Characterization of phenolic components in polar extracts of Eucalyptus globulus Labill. bark by high-performance liquid chromatography-mass spectrometry. J. Agric. Food Chem. 59, 9386–9393. doi: 10.1021/jf201801q
Sayegh, J., Cao, J., Zou, M. R., Morales, A., Blair, L. P., Norcia, M., et al. (2013). Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. J. Biol. Chem. 288, 9408–9417. doi: 10.1074/jbc.M112.419861
Schulte, J. H., Lim, S., Schramm, A., Friedrichs, N., Koster, J., Versteeg, R., et al. (2009). Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 69, 2065–2071. doi: 10.1158/0008-5472.CAN-08-1735
Schulz, W. A., Lang, A., Koch, J., and Greife, A. (2019). The histone demethylase UTX/KDM6A in cancer: progress and puzzles. Int. J. Cancer 145, 614–620. doi: 10.1002/ijc.32116
Sechler, M., Parrish, J. K., Birks, D. K., and Jedlicka, P. (2017). The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing Sarcoma cell migration and metastasis. Oncogene 36, 4150–4160. doi: 10.1038/onc.2017.44
Secombe, J., and Eisenman, R. N. (2007). The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle 6, 1324–1328. doi: 10.4161/cc.6.11.4269
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. doi: 10.1016/j.cell.2004.12.012
Shilatifard, A. (2008). Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20, 341–348. doi: 10.1016/j.ceb.2008.03.019
Shin, S., and Janknecht, R. (2007). Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem. Biophys. Res. Commun. 359, 742–746. doi: 10.1016/j.bbrc.2007.05.179
Shpargel, K. B., Sengoku, T., Yokoyama, S., and Magnuson, T. (2012). UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8:e1002964. doi: 10.1371/journal.pgen.1002964
Smith, J. A., White, E. A., Sowa, M. E., Powell, M. L., Ottinger, M., Harper, J. W., et al. (2010). Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U. S. A. 107, 3752–3757. doi: 10.1073/pnas.0914818107
Soldi, R., Ghosh Halder, T., Weston, A., Thode, T., Drenner, K., Lewis, R., et al. (2020). The novel reversible LSD1 inhibitor SP-2577 promotes anti-tumor immunity in SWItch/Sucrose-NonFermentable (SWI/SNF) complex mutated ovarian cancer. PLoS One 15:e0235705. doi: 10.1371/journal.pone.0235705 PubMed ; PubMed Central PMCID: PMCPMC7351179 he is an inventor on the patent for SP-2577 compound. Dr. Soldi declares ownership of stocks from Salarius Pharmaceuticals. This does not alter our adherence to PLoS One policies on sharing data and materials. The other authors have declared that no other competing interests exist
Soto, D. R., Barton, C., Munger, K., and McLaughlin-Drubin, M. E. (2017). KDM6A addiction of cervical carcinoma cell lines is triggered by E7 and mediated by p21CIP1 suppression of replication stress. PLoS Pathog. 13:e1006661. doi: 10.1371/journal.ppat.1006661
Sui, A., Xu, Y., Li, Y., Hu, Q., Wang, Z., Zhang, H., et al. (2017). The pharmacological role of histone demethylase JMJD3 inhibitor GSK-J4 on glioma cells. Oncotarget 8, 68591–68598. doi: 10.18632/oncotarget.19793
Taube, J. H., Sphyris, N., Johnson, K. S., Reisenauer, K. N., Nesbit, T. A., Joseph, R., et al. (2017). The H3K27me3-demethylase KDM6A is suppressed in breast cancer stem-like cells, and enables the resolution of bivalency during the mesenchymal-epithelial transition. Oncotarget 8, 65548–65565. doi: 10.18632/oncotarget.19214
Terashima, M., Ishimura, A., Wanna-Udom, S., and Suzuki, T. (2017). Epigenetic regulation of epithelial-mesenchymal transition by KDM6A histone demethylase in lung cancer cells. Biochem. Biophys. Res. Commun. 490, 1407–1413. doi: 10.1016/j.bbrc.2017.07.048
Tokunaga, R., Sakamoto, Y., Nakagawa, S., Miyake, K., Izumi, D., Kosumi, K., et al. (2016). The prognostic significance of histone lysine demethylase JMJD3/KDM6B in colorectal cancer. Ann. Surg. Oncol. 23, 678–685. doi: 10.1245/s10434-015-4879-3
Tost, J. (2010). DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 44, 71–81. doi: 10.1007/s12033-009-9216-2
Tsukada, Y., Ishitani, T., and Nakayama, K. I. (2010). KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 24, 432–437. doi: 10.1101/gad.1864410
Tumber, A., Nuzzi, A., Hookway, E. S., Hatch, S. B., Velupillai, S., Johansson, C., et al. (2017). Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem. Biol. 24, 371–380. doi: 10.1016/j.chembiol.2017.02.006
Tzatsos, A., Paskaleva, P., Ferrari, F., Deshpande, V., Stoykova, S., Contino, G., et al. (2013). KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J. Clin. Invest. 123, 727–739. doi: 10.1172/JCI64535
Van der Meulen, J., Sanghvi, V., Mavrakis, K., Durinck, K., Fang, F., Matthijssens, F., et al. (2015). The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125, 13–21. doi: 10.1182/blood-2014-05-577270
Van der Meulen, J., Speleman, F., and Van Vlierberghe, P. (2014). The H3K27me3 demethylase UTX in normal development and disease. Epigenetics 9, 658–668. doi: 10.4161/epi.28298
van Zutven, L. J., Onen, E., Velthuizen, S. C., van Drunen, E., von Bergh, A. R., van den Heuvel-Eibrink, M. M., et al. (2006). Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer 45, 437–446. doi: 10.1002/gcc.20308
Varaljai, R., Islam, A. B., Beshiri, M. L., Rehman, J., Lopez-Bigas, N., and Benevolenskaya, E. V. (2015). Increased mitochondrial function downstream from KDM5A histone demethylase rescues differentiation in pRB-deficient cells. Genes Dev. 29, 1817–1834. doi: 10.1101/gad.264036.115
Verde, G., Querol-Panos, J., Cebria-Costa, J. P., Pascual-Reguant, L., Serra-Bardenys, G., Iturbide, A., et al. (2017). Lysine-specific histone demethylases contribute to cellular differentiation and carcinogenesis. Epigenomes 1:4. doi: 10.3390/epigenomes1010004
von Tresckow, B., Goebeler, M., Sayehli, C., Gundermann, S., Eichenauer, D., Aulitzky, W., et al. (2014). First-in-man study of 4sc-202, a novel oral HDAC inhibitor in aptients with advanced hematoloigcal malignancies (Topas Study). Haematologica 99, 711–712.
von Tresckow, B., Sayehli, C., Aulitzky, W. E., Goebeler, M. E., Schwab, M., Braz, E., et al. (2019). Phase I study of domatinostat (4SC-202), a class I histone deacetylase inhibitor in patients with advanced hematological malignancies. Eur. J. Haematol. 102, 163–173. doi: 10.1111/ejh.13188
Wagner, K. W., Alam, H., Dhar, S. S., Giri, U., Li, N., Wei, Y., et al. (2013). KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J. Clin. Invest. 123, 5231–5246. doi: 10.1172/JCI68642
Walport, L. J., Hopkinson, R. J., and Schofield, C. J. (2012). Mechanisms of human histone and nucleic acid demethylases. Curr. Opin. Chem. Biol. 16, 525–534. doi: 10.1016/j.cbpa.2012.09.015
Walport, L. J., Hopkinson, R. J., Vollmar, M., Madden, S. K., Gileadi, C., Oppermann, U., et al. (2014). Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J. Biol. Chem. 289, 18302–18313. doi: 10.1074/jbc.M114.555052
Wang, H. J., Pochampalli, M., Wang, L. Y., Zou, J. X., Li, P. S., Hsu, S. C., et al. (2019). KDM8/JMJD5 as a dual coactivator of AR and PKM2 integrates AR/EZH2 network and tumor metabolism in CRPC. Oncogene 38, 17–32. doi: 10.1038/s41388-018-0414-x
Wang, J., Hevi, S., Kurash, J. K., Lei, H., Gay, F., Bajko, J., et al. (2009). The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129. doi: 10.1038/ng.268
Wang, J., Scully, K., Zhu, X., Cai, L., Zhang, J., Prefontaine, G. G., et al. (2007). Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887. doi: 10.1038/nature05671
Wang, L., Chang, J., Varghese, D., Dellinger, M., Kumar, S., Best, A. M., et al. (2013). A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun. 4:2035. doi: 10.1038/ncomms3035
Wang, S. P., Tang, Z., Chen, C. W., Shimada, M., Koche, R. P., Wang, L. H., et al. (2017). A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol. Cell 67, 308–321.e6. doi: 10.1016/j.molcel.2017.06.028
Wang, W., Oguz, G., Lee, P. L., Bao, Y., Wang, P., Terp, M. G., et al. (2018). KDM4B-regulated unfolded protein response as a therapeutic vulnerability in PTEN-deficient breast cancer. J. Exp. Med. 215, 2833–2849. doi: 10.1084/jem.20180439
Wang, Y., Zang, J., Zhang, D., Sun, Z., Qiu, B., and Wang, X. (2018). KDM2B overexpression correlates with poor prognosis and regulates glioma cell growth. Onco. Targets Ther. 11, 201–209. doi: 10.2147/OTT.S149833
Watarai, H., Okada, M., Kuramoto, K., Takeda, H., Sakaki, H., Suzuki, S., et al. (2016). Impact of H3K27 demethylase inhibitor GSKJ4 on NSCLC cells alone and in combination with metformin. Anticancer Res. 36, 6083–6092. doi: 10.21873/anticanres.11198
Wen, H., Li, J., Song, T., Lu, M., Kan, P. Y., Lee, M. G., et al. (2010). Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 285, 9322–9326. doi: 10.1074/jbc.C109.097667
Willmann, D., Lim, S., Wetzel, S., Metzger, E., Jandausch, A., Wilk, W., et al. (2012). Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. Int. J. Cancer 131, 2704–2709. doi: 10.1002/ijc.27555
Wilson, S., Fan, L., Sahgal, N., Qi, J., and Filipp, F. V. (2017). The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells. Oncotarget 8, 30328–30343. doi: 10.18632/oncotarget.15681
Wissmann, M., Yin, N., Muller, J. M., Greschik, H., Fodor, B. D., Jenuwein, T., et al. (2007). Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9, 347–353. doi: 10.1038/ncb1546
Xiang, Y., Yan, K., Zheng, Q., Ke, H., Cheng, J., Xiong, W., et al. (2019). Histone demethylase KDM4B promotes DNA damage by activating long interspersed nuclear element-1. Cancer Res. 79, 86–98. doi: 10.1158/0008-5472.CAN-18-1310
Xiang, Y., Zhu, Z., Han, G., Ye, X., Xu, B., Peng, Z., et al. (2007). JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 104, 19226–19231. doi: 10.1073/pnas.0700735104
Xie, G., Liu, X., Zhang, Y., Li, W., Liu, S., Chen, Z., et al. (2017). UTX promotes hormonally responsive breast carcinogenesis through feed-forward transcription regulation with estrogen receptor. Oncogene 36, 5497–5511. doi: 10.1038/onc.2017.157
Xie, Q., Bai, Y., Wu, J., Sun, Y., Wang, Y., Zhang, Y., et al. (2011). Methylation-mediated regulation of E2F1 in DNA damage-induced cell death. J. Recept. Signal. Transduct. Res. 31, 139–146. doi: 10.3109/10799893.2011.552914
Xu, J., Deng, X., Watkins, R., and Disteche, C. M. (2008). Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 28, 4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008
Xu, W., Jiang, K., Shen, M., Chen, Y., and Huang, H. Y. (2016). Jumonji domain containing 2A predicts prognosis and regulates cell growth in lung cancer depending on miR-150. Oncol. Rep. 35, 352–358. doi: 10.3892/or.2015.4349
Yamamoto, K., Tateishi, K., Kudo, Y., Sato, T., Yamamoto, S., Miyabayashi, K., et al. (2014). Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPalpha. Carcinogenesis 35, 2404–2414. doi: 10.1093/carcin/bgu136
Yamane, K., Tateishi, K., Klose, R. J., Fang, J., Fabrizio, L. A., Erdjument-Bromage, H., et al. (2007). PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25, 801–812. doi: 10.1016/j.molcel.2007.03.001
Yan, N., Xu, L., Wu, X., Zhang, L., Fei, X., Cao, Y., et al. (2017). GSKJ4, an H3K27me3 demethylase inhibitor, effectively suppresses the breast cancer stem cells. Exp. Cell Res. 359, 405–414. doi: 10.1016/j.yexcr.2017.08.024
Yan, Q., Bartz, S., Mao, M., Li, L., and Kaelin, W. G. Jr. (2007). The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol. Cell Biol. 27, 2092–2102. doi: 10.1128/MCB.01514-06
Yu, Y., Schleich, K., Yue, B., Ji, S., Lohneis, P., Kemper, K., et al. (2018). Targeting the senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell 33:785. doi: 10.1016/j.ccell.2018.03.009
Zhang, H., Zhang, X., Clark, E., Mulcahey, M., Huang, S., and Shi, Y. G. (2010). TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 20, 1390–1393. doi: 10.1038/cr.2010.156
Zhao, E., Tang, C., Jiang, X., Weng, X., Zhong, X., Zhang, D., et al. (2017). Inhibition of cell proliferation and induction of autophagy by KDM2B/FBXL10 knockdown in gastric cancer cells. Cell Signal. 36, 222–229. doi: 10.1016/j.cellsig.2017.05.011
Zhu, H., Hu, S., and Baker, J. (2014). JMJD5 regulates cell cycle and pluripotency in human embryonic stem cells. Stem Cells 32, 2098–2110. doi: 10.1002/stem.1724
Zingg, D., Arenas-Ramirez, N., Sahin, D., Rosalia, R. A., Antunes, A. T., Haeusel, J., et al. (2017). The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep. 20, 854–867. doi: 10.1016/j.celrep.2017.07.007
Zingg, D., Debbache, J., Pena-Hernandez, R., Antunes, A. T., Schaefer, S. M., Cheng, P. F., et al. (2018). EZH2-mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer Cell 34, 69–84.e14. doi: 10.1016/j.ccell.2018.06.001
Keywords: epigenetics, histones, lysine demethylases, small molecule inhibitors, cancer, melanoma
Citation: Punnia-Moorthy G, Hersey P, Emran AA and Tiffen J (2021) Lysine Demethylases: Promising Drug Targets in Melanoma and Other Cancers. Front. Genet. 12:680633. doi: 10.3389/fgene.2021.680633
Received: 15 March 2021; Accepted: 24 May 2021;
Published: 16 June 2021.
Edited by:
Ritu Kulshreshtha, Indian Institute of Technology Delhi, IndiaReviewed by:
Silvio Zaina, University of Guanajuato, MexicoCopyright © 2021 Punnia-Moorthy, Hersey, Emran and Tiffen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessamy Tiffen, ai50aWZmZW5AY2VudGVuYXJ5Lm9yZy5hdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.