
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 04 May 2021
Sec. Nutrigenomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.675780
DNA methylation plays an important role in the maintenance of genomic stability. Ten-eleven translocation proteins (TETs) are a family of iron (Fe2+) and α-KG -dependent dioxygenases that regulate DNA methylation levels by oxidizing 5-methylcystosine (5mC) to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). These oxidized methylcytosines promote passive demethylation upon DNA replication, or active DNA demethylation, by triggering base excision repair and replacement of 5fC and 5caC with an unmethylated cytosine. Several studies over the last decade have shown that loss of TET function leads to DNA hypermethylation and increased genomic instability. Vitamin C, a cofactor of TET enzymes, increases 5hmC formation and promotes DNA demethylation, suggesting that this essential vitamin, in addition to its antioxidant properties, can also directly influence genomic stability. This review will highlight the functional role of DNA methylation, TET activity and vitamin C, in the crosstalk between DNA methylation and DNA repair.
The protective role of vitamin C in cancer progression has historically been attributed to its antioxidant activity and the prevention of DNA damage induced by oxidative stress (Padayatty and Levine, 2016). Vitamin C also enhances the activity of a large family of iron (Fe2+) and α-ketoglutarate-dependent dioxygenases (α-KGDDs), which include epigenetic regulators of DNA methylation known to play important roles in the maintenance of genomic stability. Ten-Eleven Translocation proteins (TETs) are a subfamily of α-KGDDs that promote DNA demethylation in the genome (Tahiliani et al., 2009). Loss of function in TET proteins and altered levels of DNA methylation are hallmarks of cancer (Figueroa et al., 2010a, b; Akalin et al., 2012) that drive genomic instability and malignant transformation (An et al., 2015; Ko et al., 2015). Recent studies have shown that vitamin C, by enhancing TET activity, can directly influence DNA methylation levels that in turn alter chromatin structure, and the expression of tumor suppressors and DNA repair enzymes. Vitamin C deficiency has been widely reported in cancer patients (Mayland et al., 2005; Huijskens et al., 2016) and accelerates cancer progression in disease models (Agathocleous et al., 2017). In addition to its potential role in the prevention of cancer, vitamin C added to cell culture media can improve the quality of stem cells and reprogrammed cells for use in stem cell therapies and regenerative medicine by maintaining genomic integrity (Li et al., 2009; Wang et al., 2011; Chen J. et al., 2013; Gustafson et al., 2015; Hore et al., 2016). Through its ability to promote DNA demethylation, vitamin C has clinical application in the treatment of cancer and especially hematological malignancies where mutation in TET enzymes and aberrant DNA methylation are frequently observed. While several recent reviews have underscored the role of vitamin C as an anticancer agent (Das et al., 2021), modulator of immune responses (Yue and Rao, 2020), and in the reprogramming of stem cells (Lee Chong et al., 2019), herein we highlight these activities within a context of DNA damage, repair, and genomic stability. Understanding how DNA methylation levels and TET activity influence genomic stability provides the context in which vitamin C, as a co-factor of TET enzymes, can play a pivotal role as an epigenetic regulator of DNA damage and repair.
DNA methylation directly regulates essential biological functions such as gene expression, chromatin organization, DNA imprinting and X-chromosome inactivation that in combination instruct embryonic development and cellular differentiation (Guo et al., 2014). DNA methylation also plays direct and indirect roles in maintaining genomic stability. Both hypomethylation and hypermethylation of DNA is associated with increased genomic instability that can lead to malignant transformation. Under normal, steady-state conditions, up to 80% of cytosines, in the context of CpG dinucleotides, are methylated in the mammalian genome, reviewed in Law and Jacobsen (2010). DNA methylation is conventionally known as a repressive epigenetic mark, and the majority of methylated cytosines are concentrated in heterochromatic regions, ensuring that chromatin remains closed and genes silenced when not required to be actively transcribed by the cell. DNA methylation also directly silences gene expression by hypermethylation of CpG-rich regions known as CpG islands (CGIs) in promoter regions that alter the ability to recruit transcription factors (Curradi et al., 2002). This is a key mechanism by which DNA methylation regulates gene expression, as approximately 70% of human gene promoters have associated CGIs (Saxonov et al., 2006; Deaton and Bird, 2011). Furthermore, almost 50% of the human genome consists of long terminal repeats (LTRs), short or long interspersed nuclear elements (SINES or LINES), and other endogenous retroviruses that are enriched for CpGs and silenced into heterochromatic regions by both DNA methylation and repressive histone modifications (Kondo and Issa, 2003; Pehrsson et al., 2019). DNA hypomethylation at repetitive regions in the genome can reduce the formation of heterochromatin, leading to transposition of DNA and the aberrant expression of oncogenes that can drive tumorigenesis (Lopez-Moyado et al., 2019). DNA hypermethylation can also promote genomic instability, by silencing the expression of DNA repair genes, or by inhibiting the recruitment of DNA repair proteins (Toffolatti et al., 2014; Tsuboi et al., 2020). Methylated cytosines are also intrinsically more mutagenic than unmethylated cytosines (Poulos et al., 2017; Kusmartsev et al., 2020). Balancing the activity of writers and erasers of DNA methylation ensures that epigenetic information is interpreted and inherited correctly, but also protects cells from acquiring permanent changes to the genetic code.
DNA methyltransferases (DNMT1, 3A and 3B) catalyze the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to the 5′ position of cytosine residues generating 5-methylcytosine (5mC) in the genome. DNMT1 is essential for the maintenance of methylation marks during DNA replication, whereas DNMT3A/B are responsible for de novo synthesis and their activity is independent of the cell cycle (Bestor et al., 1988; Okano et al., 1999). DNMT1 preferentially recognizes hemi-methylated DNA so that DNA methylation patterns are inherited upon DNA replication (Bashtrykov et al., 2012), whereas DNMT3A and B show an equivalent affinity for both hemimethylated and unmethylated DNA (Kareta et al., 2006).
TET proteins (TET1-3) are a sub-family of α-KGDDs that promote DNA demethylation by catalyzing the iterative oxidation of 5mC to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Tahiliani et al., 2009; He et al., 2011; Ito et al., 2011). 5hmC is a stable modification that constitutes 5–10% of the total level of 5mC in embryonic stem cells (ESCs) (Tahiliani et al., 2009) but this frequency can vary widely in different adult tissues, with 5hmC present at 40% the level of 5mC in Purkinje cells of the brain (Kriaucionis and Heintz, 2009) compared to 1% in immune cells (Ko et al., 2010). The presence of 5hmC in the genome causes the passive loss of DNA methylation upon DNA replication, given that DNMT1 is unable to recognize hemi-methylated 5hmC sites (Otani et al., 2013). 5fC and 5caC levels are rare modifications in the genome and constitute approximately 2 or 0.5% of the total level of 5hmC in wild-type mouse ESCs, respectively (He et al., 2011; Ito et al., 2011).
The low abundance of 5fC and 5caC in DNA is attributed to their removal and replacement with an unmethylated cytosine by active DNA demethylation via base excision repair (BER). The conversion of 5hmC to 5-hydroxymethyluracil (5hmU) by cytidine deaminase (AID or APOBEC) is one proposed mechanism by which thymine or uracil DNA glycosylases (TDG or SMUG1, respectively) excise oxidized mCs (Cortellino et al., 2011). Subsequently it was shown that TDG most likely targets 5fC:G and 5caC:G mismatches for removal, given that TDG-deficient ESCs accumulate up to 10-fold higher levels of 5fC and 5caC in their genome (He et al., 2011; Shen et al., 2013; Song et al., 2013), and TDG targets 5caC:G and 5fC:G with higher affinity than T:G with no activity toward 5hmC:G (Maiti and Drohat, 2011; Zhang et al., 2012). Other components of the DNA damage and BER machinery have been identified as specific readers of 5fC or 5caC, including p53, TDG, PARP, GADD45 and NEIL1/2 (Spruijt et al., 2013), and depletion studies of these factors in cells have shown that their activity is required to prevent DNA hypermethylation (Cortellino et al., 2011; Ciccarone et al., 2012; Li et al., 2015; Schomacher et al., 2016; Tovy et al., 2017). TET enzymes have also been shown to oxidize thymine to generate 5hmU directly, leading to 5hmU:A mismatches (Pfaffeneder et al., 2014; Olinski et al., 2016). Protein readers of 5hmU:G and 5hmU:A also include chromatin regulators and DNA repair enzymes involved in BER (Pfaffeneder et al., 2014), and the repair of 5hmU:A by long-patch BER or non-canonical mismatch repair could lead to the indirect removal of adjacent 5mC in the genome (Metivier et al., 2008; Santos et al., 2013; Grin and Ishchenko, 2016; Spada et al., 2020). In primed embryonic murine pluripotent stems cells (PSCs) and the developing zygote, other non-oxidative and cell-cycle independent mechanisms of DNA demethylation have also been reported that may utilize deamination of mC by AID and DNA repair processes (Santos et al., 2013; Amouroux et al., 2016; Spada et al., 2020), however, the majority of active DNA demethylation in mPSCs was shown to be driven by oxidation of 5mC (Spada et al., 2020). These studies provide further evidence that TET-oxidized bases and DNA repair mechanisms can work together, directly or indirectly, to mediate DNA demethylation in the genome.
Studies into the genomic distribution of 5hmC, 5fC, and 5caC revealed their enrichment in correlation with active gene expression at transcriptional start sites and gene bodies (Ficz et al., 2011; Williams et al., 2011; Wu H. et al., 2011; Raiber et al., 2012; Song et al., 2013; Neri et al., 2015), enhancers (Lu et al., 2014; Rasmussen et al., 2015), the edges of large “canyons” of low DNA methylation in the genome (Jeong et al., 2014), and at the boundaries of topologically associated domains (TADs) marked by the insulator protein CCCTF binding factor (CTCF) (Song et al., 2013; Nanan et al., 2019). Several laboratories have developed methods to sequence the genomic distribution of 5hmU at base resolution (Yu et al., 2015; Bullard et al., 2017; Kawasaki et al., 2017), however, the patterning of this modification in the genome of mammalian cells has not yet been described.
Differences in the level of DNA methylation and oxidized 5mC, or their aberrant distribution across the genome, can influence how cells interpret these epigenetic cues. Both hypomethylation and hypermethylation of the DNA is associated with increased genomic instability that can lead to malignant transformation (Figure 1). Dysregulation of DNA methylation patterning can occur by several mechanisms including defective or decreased activity of DNMT or TET DNA demethylases that leads to hypomethylation and/or hypermethylation. DNA methylation changes, in combination with altered expression or activity of cytosine deaminases, and other readers or repair enzymes recruited at 5mC, 5hmC, 5fC, or 5caC modified cytosines, can accelerate mutational processes that drive cancer progression.
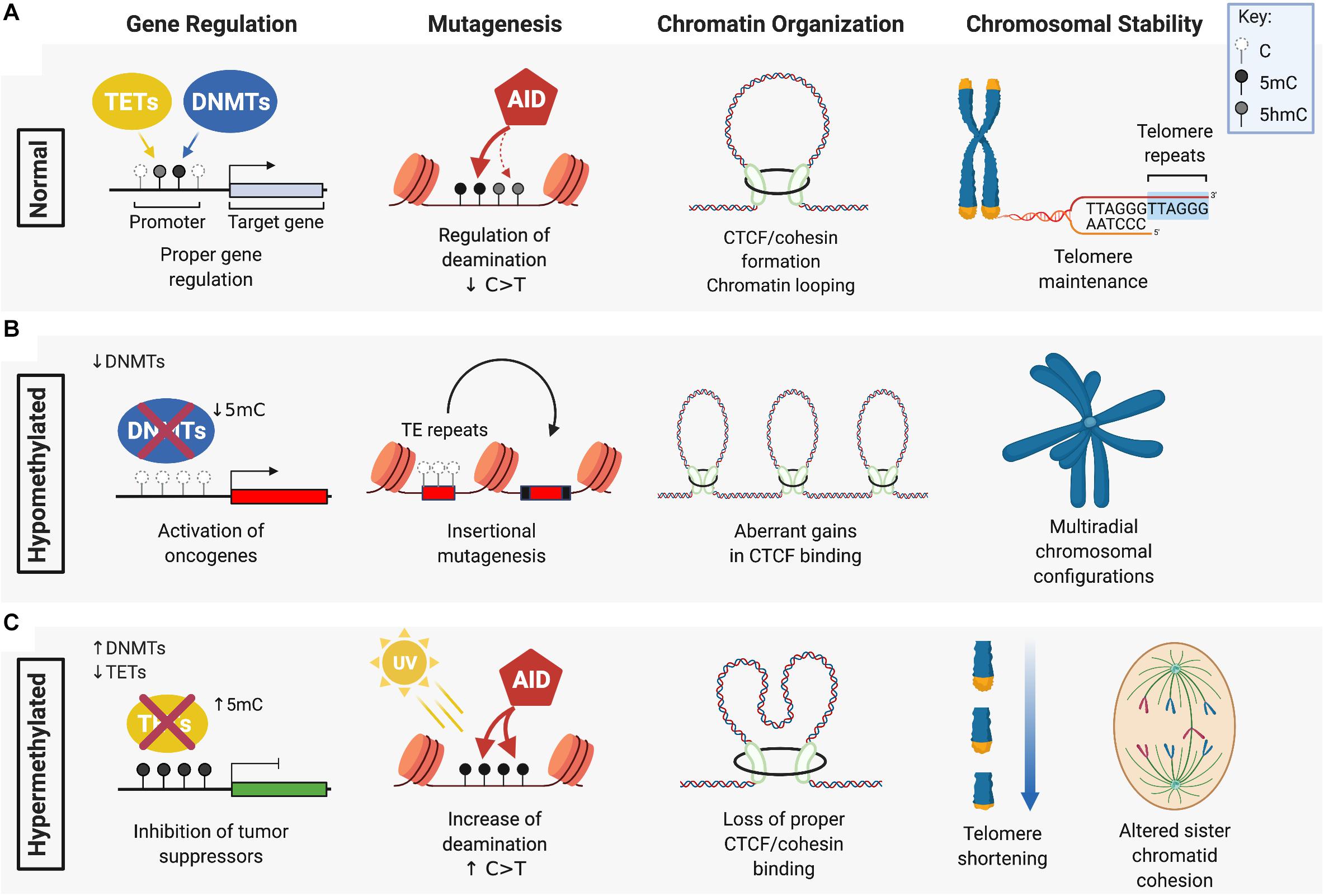
Figure 1. The role of DNMTs and TETs in DNA methylation maintenance and genomic stability. (A) DNA methyltransferases (DMNTs) regulate the methylation of cytosine residues (white) in CpG islands typically found in the promoter region of target genes, while Ten-Eleven Translocation (TET) enzymes promote the demethylation of 5-methylcytosines (5mC) (black). Together, these enzymes regulate normal DNA methylation patterns and genomic stability by ensuring: proper silencing and activation of gene expression; cytosine deamination by AID or APOBEC (not shown) that preferentially target 5mC over C/5hmC (gray) for C > T transition; CTCF/cohesin-guided boundaries between enhancers and promoters of genes; and maintenance of telomeres. (B) DNA hypomethylation, caused by a loss of function of DNMTs, can lead to: aberrant oncogene activation by removal of promoter silencing; upregulated activity of transposable elements (TE) that drive insertional mutagenesis; gains in CTCF binding that exacerbate oncogene expression; and loss of heterochromatin at peri-centromeric regions causing defective chromosome segregation and multi-radial configurations (specifically seen in ICF patients with DNMT3B mutation). (C) DNA hypermethylation is associated with: aberrant silencing of tumor suppressors from increased DNMT activity or loss of TET function; an increased frequency of C > T transitions due to enhanced mutagenicity and deamination of 5mC; loss of CTCF/cohesin binding leading to gene mis-regulation; and telomere shortening and altered sister chromatid cohesion. Figure created with BioRender.com.
Defects in DNMT expression and function can cause global hypomethylation that correlates with genomic instability and tumor progression due to loss of silencing at the loci of oncogenes (Soes et al., 2014; Kushwaha et al., 2016; Sheaffer et al., 2016). One of the most frequently observed upregulated oncogenes in cancer that exhibits a hypomethylated gene locus and is known to cause genomic instability is the transcription factor c-Myc. Large genomic amplifications induced by c-Myc overexpression are attributed to replicative stress caused by unscheduled DNA replication and increased oxidative stress leading to DNA breaks (Dominguez-Sola et al., 2007; Kuzyk and Mai, 2014). While c-Myc DNA hypomethylation is observed in many different cancers, other examples of hypomethylated oncogenic loci are cancer-specific, such as the melanoma-associated antigens (MAGE) gene that is hypomethylated specifically in gastric cancer and colorectal cancer (Honda et al., 2004; Kim et al., 2006) or synuclein γ (SNCG) in breast and ovarian cancers and solid tumors of the liver, gastric and (Gupta et al., 2003; Liu et al., 2005). Overexpression of these oncogenes promote, proliferation, metastasis, disruption of mitotic checkpoints, enhanced transcriptional activity and accelerated rates of chromosomal instability.
Hypomethylated repetitive DNA sequences, including CG repeats in CGIs and in transposable elements (TEs), are commonly found in cancers that could be caused by loss of function in DNA methyltransferase activity. Hypomorphic activity of Dnmt1 in mice induces thymic lymphomas with recurring insertions of a transposable element within the Notch gene leading to its oncogenic activation (Howard et al., 2008). LINE-1 and Alu elements are retrotransposons that constitute over 30% of the human genome and are normally inhibited by DNA methylation yet become frequently hypomethylated in cancer (Daskalos et al., 2009; Pehrsson et al., 2019). Increased LINE-1 and Alu activity causes genomic instability by promoting deletions, chromosome breaks, and translocations (Mandal et al., 2013; Bakshi et al., 2016).
DNA hypomethylation is also observed at microsatellite and pericentromeric heterochromatic regions in chromosomes of DNMT3B-mutant disease. DNMT3B loss of function mutations have been described in ∼60% of patients with an autosomal recessive disorder known as immunodeficiency, centromeric instability and facial anomaly (ICF) syndrome (Hansen et al., 1999; Okano et al., 1999; Xu et al., 1999). ICF is a rare disease showing symptoms in early childhood of recurrent gastrointestinal and pulmonary infections as a result of agammaglobulinemia (Ehrlich et al., 2008; Hagleitner et al., 2008). ICF patients display evident decondensation of heterochromatin caused by DNA hypomethylation at pericentromeric regions in chromosomes 1, 9, and 16, causing upregulated gene expression at these loci, chromosome breaks, and rearrangements in radial structures that are detectable in stimulated lymphocytes and increase the risk of hematological malignancies (Brown et al., 1995; Kaya et al., 2011; Gossling et al., 2017).
Cancer cells on average display up to 30% genome-wide losses in DNA methylation (Ehrlich and Lacey, 2013; Baylin and Jones, 2016) which could significantly impact three-dimensional chromatin architecture and genomic stability by altering oncogene activity and activation of TEs. CTCF, in cooperation with cohesin proteins, regulates long-range looping interactions between enhancers and promoters (Dixon et al., 2012) and preferentially binds at hypomethylated CpGs of cis-regulatory insulator regions of the genome (Nanavaty et al., 2020). DNA hypomethylation could allow for aberrant gains in CTCF binding, creating stronger insulation at oncogenic super-enhancers and an increased frequency of tandem duplications (Gong et al., 2018). Mis-regulated gene expression via stronger TAD boundaries could also cooperate with oncogenic transcription factors to exacerbate genomic instability (Fang et al., 2020). Thus, hypomethylated states may confer an advantage of cancer cells by the increase and maintenance of elevated oncogenic expression during tumor development (Kang et al., 2015).
Hypermethylation of the genome is associated with aberrant silencing of many tumor suppressors that could drive or accelerate carcinogenesis. DNA hypermethylation can be caused by a decrease in the activity of DNA demethylases such as the TET proteins leading to the silencing of DNA repair enzymes and tumor suppressors. Silencing due to aberrant DNA hypermethylation at imprinted loci (Rainier et al., 1993; Moulton et al., 1994; Steenman et al., 1994; Plass and Soloway, 2002), senescence genes (Kamb et al., 1994; Zhao et al., 2016; Xie et al., 2019) and lineage specific transcription factors, that normally slow proliferation and drive differentiation, reviewed in (Suelves et al., 2016; Pfeifer, 2018), may cause a buildup in unchecked DNA damage and DNA replication errors.
Methylated cytosines are approximately fivefold more likely to undergo mutagenesis than unmethylated cytosines (Poulos et al., 2017). This could be due to a greater tendency for 5mC to spontaneously deaminate compared to unmethylated cytosines (Supek et al., 2014; Blokzijl et al., 2016). Furthermore, C > T and G > A mutations disproportionally affect CpG dinucleotides. In one study, approximately 18% of C > T and G > A missense and nonsense mutations in genes of inherited human diseases occurred within CpGs, a frequency 10-fold higher would have been expected by chance alone (Cooper et al., 2010). Cytosine deaminases (AID and APOBEC1-3) also exhibit greater activity toward 5mC than 5hmC (Nabel et al., 2012; Rangam et al., 2012), and 5mC deamination generates T:G mismatches that can potentially recruit error-prone mismatch repair (MMR) complexes (Cortellino et al., 2011). Importantly, C > T mutation rates are 50% lower for C or 5hmC compared to 5mC (Tomkova et al., 2016). Given that C > T transition mutations are the most frequent, age-associated mutation signature in cancer (Alexandrov et al., 2013; Kandoth et al., 2013) loss of TET activity could mimic a premature aging phenotype with regard to the frequency at which C > T mutations accumulate. Distinct mutational signatures are also caused by aberrant deaminase activity, with AID/APOBEC-driven signatures dominated by high levels of C > T transitions in specific cancers (Alexandrov et al., 2020). Upregulated oncogenic activity of AID/APOBEC deaminases, in combination with elevated levels of 5mC in the genome, could synergistically accelerate malignant transformation.
Studies in ESCs have shown that loss of TET function causes decreased 5hmC and DNA hypermethylation in telomeric regions, that leads to telomere shortening, reduced telomere recombination and chromosome segregation defects (Lu et al., 2014; Kafer et al., 2016; Yang et al., 2016). Sub-telomeres are also hypermethylated in TET depleted ESCs, which may further impede telomere elongation by recombination (Yang et al., 2016). DNA hypermethylation caused by loss of TET activity increases nucleosome occupancy, and subsequently, loss of CTCF binding and a block in the downstream recruitment of the cohesin complex, reducing the formation of CTCF/cohesion-mediated chromatin loops thereby downregulating the expression of neighboring genes (Wiehle et al., 2019; Nanavaty et al., 2020). Loss of CTCF itself can also induce DNA hypermethylation at CTCF binding sites and the fusion of TAD boundaries that establish oncogenic expression patterns and increased cancer progression (Akdemir et al., 2020; Damaschke et al., 2020). TETs also regulate chromosomal architecture by protecting large undermethylated genomic regions known as DNA methylation “canyons” from becoming hypermethylated (Wiehle et al., 2016; Zhang X. et al., 2016). Loss of Dnmt3a or Tet2 causes canyon edges to collapse and become hypermethylated, suggesting that de novo methyltransferase activity and Tet2-mediated hydroxylation of 5mC work together to maintain 5hmC and hypomethylation at these loci (Jeong et al., 2014; Zhang X. et al., 2016).
Studies using ESCs with a triple knockout of all three TET enzymes revealed an increase of 5mC signal across all chromosomes by whole-genome bisulfite sequencing (Lu et al., 2014) and an increased frequency in telomere loss and chromosomal fusion (Yang et al., 2016). Genomic instability has also been observed in Tet2/3 double knockout hematopoietic stem and progenitor cells (HSPCs) and immature myeloid cells, which show impairment of DNA repair responses, lower homologous recombination (HR) or non-homologous end-joining (NHEJ) gene expression and spontaneously accumulate DNA double strand breaks (DSBs) marked by increased phosphorylated histone H2A.X (γH2AX) in comparison to WT cells (An et al., 2015). These studies, in combination with numerous reports of TET deficiency in other solid tumors (Haffner et al., 2011; Kudo et al., 2012; Yang et al., 2013; Munari et al., 2016) demonstrate the loss of 5hmC by defective TET enzymatic activity as a hallmark of cancer that may drive genomic instability.
DNA methylation and oxidized mCs regulate the susceptibility of cells to genomic instability by also actively participating in sensing and repair processes upon DNA damage. C > T transition mutations could lead to losses in methylation at otherwise silenced gene loci that accumulate over our lifetime due to spontaneous deamination and exposure to ionizing radiation or chemical carcinogens (Baylin and Jones, 2016). UV irradiation primarily induces C > T transitions in the epidermis, with cells exposed to UVA biased toward mutations of CC > TT dipyrimidines more frequently than by UVB, and with a greater propensity for 5mC than C (Martinez-Fernandez et al., 2017; Ikehata, 2018). UV irradiation can also induce the expression of DNMT1 in human dermal fibroblasts leading to DNA hypermethylation at specific gene loci, such as TIMP2 (Kim et al., 2018).
Reactive oxygen species (ROS) are estimated to create up to 50,000 DNA lesions in a single human cell, which can lead to potentially oncogenic mutations if not repaired (Rossi et al., 2007). ROS can influence DNA methylation levels by the oxidation of guanosine to 8-oxo-2′-deoxyguanosine (8-oxo-dG) or by hydroxyl radicals that via abstraction of a hydrogen from the methyl group of mC leads to the formation of 5hmC (Madugundu et al., 2014). The presence of 8-oxo-dG can lead to DNA hypomethylation, by preventing the adjacent cytosine in a Cp8-oxo-dG dinucleotide from being recognized by DNMTs, thereby promoting passive DNA demethylation. Furthermore, 8-oxo-dG DNA glycosylase (OGG1) recruited to these lesions interacts with TET1 that can trigger active DNA demethylation by BER (Zhou et al., 2016). ROS can also induce G quadruplex formation as well as R-loops (DNA-RNA hybrids that are enriched at CpG islands), which can cause genomic instability at active transcriptional sites and delay the removal and repair of these ROS-induced damaged bases (Tan et al., 2020). Further direct evidence of the involvement of TET enzymes in DNA repair come from studies in ESCs, where genotoxic insults such as aphidicolin treatment to induce double strand breaks (DSBs) were shown to induce 5hmC, but not 5mC, foci that co-localized with γH2AX marks, and the repair proteins 53BP1 and RAD51 (Kafer et al., 2016). Together these studies illustrate how TETs can act as the bridge between DNA methylation regulation and DNA repair. The ability of TET proteins to influence transcription and DNA repair at oxidized mCs implies a dual role in the regulation of gene expression and DNA damage responses.
The strong association between aberrant DNA methylation and tumorigenesis is evidenced by the frequent occurrence of mutations, and altered expression levels, of DNMT and TET enzymes in solid tumors and hematological malignancies. DNMT1 and DNMT3B mutations or amplifications are more common in cancers of epithelial tissues of the breast, ovaries, skin, bladder, lung and colon (reviewed in Zhang et al., 2020), whereas loss-of-function DNMT3A mutations are the most frequent lesions identified in acute myeloid leukemia (AML) where they are found on average in 30% of patients (Ley et al., 2010; Cancer Genome Atlas Research, 2013; Brunetti et al., 2017). The mutation hotspot encoding arginine 882 (R882) accounts for ∼65% of all DNTM3A variants and blocks its methyltransferase activity (Russler-Germain et al., 2014; Nguyen et al., 2019; Gao et al., 2020) leading to genome-wide DNA hypomethylation with focal increases in promoter CGI DNA methylation that are hallmarks of AML (Figueroa et al., 2010b; Spencer et al., 2017). DNMT3A mutations also predispose patients with pre-malignant clonal hematopoiesis to transformation and drive relapse in AML patients (Jaiswal et al., 2014; Buscarlet et al., 2017). The inability of DNMT3A R882 mutants to sense and repair DNA torsional stress results in increased mutagenesis and resistance to anthracycline (Guryanova et al., 2016) and demonstrates how the role of these epigenetic regulators in DNA damage-sensing, in combination with altered genomic methylation, can drive cancer progression.
Unopposed DNA methyltransferase activity due to loss of TET function can cause DNA hypermethylation, and decreased TET1-3 expression or reduced 5hmC levels in the genome are common across multiple solid tumors (Haffner et al., 2011; Kudo et al., 2012; Yang et al., 2013, 2015) and blood cell malignancies (Ko et al., 2010; Lemonnier et al., 2018; Zhang et al., 2018). Similar to the lineage bias observed for mutation of DNMTs, TET1 and TET3 are more frequently mutated in carcinomas (Li L. et al., 2016; Lee et al., 2020), whereas TET2 deletions and missense mutations induce loss of its catalytic function and are observed at much higher frequency, in ∼10-30% of patients with either clonal hematopoiesis ((Busque et al., 2012; Jaiswal et al., 2014; Bowman et al., 2018), myeloid (Abdel-Wahab et al., 2009; Delhommeau et al., 2009; Langemeijer et al., 2009), or lymphoid (Quivoron et al., 2011; Lemonnier et al., 2012) malignancies.
DNMT3A and TET2 mutation are independently associated with an adverse outcome and poor prognosis in intermediate risk AML (Kosmider et al., 2009; Ribeiro et al., 2012). Furthermore, the prevalence of DNMT3A and TET2 mutation in hematological malignancies, and their early emergence from within the HSPC compartment to drive transformation (Delhommeau et al., 2009; Quivoron et al., 2011; Welch et al., 2012; Papaemmanuil et al., 2013), emphasizes the importance of DNA methylation in the pre-malignant regulation of stem cell self-renewal and blood cell lineage differentiation. Hypermethylated CpGs that should normally be targeted by TET2 for demethylation could suffer increased C > T transition rates, based on the higher mutagenicity of 5mC compared to unmethylated cytosines or 5hmC (Poulos et al., 2017). Cells from myelodysplastic (MDS) and AML patients with TET2 mutation harbor more non-synonymous somatic mutations than TET2 wild-type patients, and HSPCs of Tet2 knockout mice also exhibit increased mutation rates (Pan et al., 2017). Whether CGI hypermethylation in clonal hematopoiesis patients increases susceptibility to additional mutations that drive transformation is not yet known. Both DNMT3A and TET2 have been shown to prevent DNA hypermethylation of canyons and promoter CGIs, suggesting that de novo methylation and 5hmC formation by DNMT3A and TET2 work simultaneously in HSPCs to prevent specific tumor suppressor gene loci from becoming dysregulated, and their potential cooperation at these sites requires further study.
The recent discovery that vitamin C can act as an epigenetic regulator by enhancing the activity of α-KGDDs such as the TET proteins has transformed our understanding of the role of vitamin C in biology (Figure 2A). Removal of histone and DNA methylation is essential for the efficient epigenetic reprogramming and generation of induced pluripotent stem cells (iPSCs) from somatic cells. Studies using ESCs and reprogramming fibroblasts were the first to show that vitamin C, in a TET-dependent manner, could increase 5hmC, 5fC, and 5caC production, triggering a global DNA hypomethylation that improved the quality of cells in culture and enhance the formation of iPSCs (Blaschke et al., 2013; Chen Q. et al., 2013; Minor et al., 2013; Yin et al., 2013). ESCs express high levels of TET1 and TET2 (Koh et al., 2011; Dawlaty et al., 2013), and 100 μM vitamin C is sufficient to increase 5hmC by up to ∼4-fold above basal levels in ESCs within 24hrs of treatment, with even larger effects on the levels of 5fC (10-fold increase) and 5caC (20-fold increase) (Yin et al., 2013). A study using human colorectal cancer cells also reported significantly elevated levels of 5hmU (up 18.5-fold increase) in response to vitamin C treatment (Modrzejewska et al., 2016). In HSPCs and human leukemia cell lines, treatment with low or high doses of vitamin C also cause 2–4 fold increases in 5hmC formation and genome-wide DNA hypomethylation (Liu et al., 2016; Cimmino et al., 2017; Mingay et al., 2018) similar to what has been observed in ESCs (Chung et al., 2010). Subsequent studies of vitamin C treatment in other tissue specific stem cells (He et al., 2015; Wulansari et al., 2017) and carcinoma cells of the kidney, bladder, lung, colon and breast, amongst others (Ge et al., 2018; Peng et al., 2018; Sant et al., 2018) all reported similar increases in 5hmC formation and/or DNA hypomethylation.
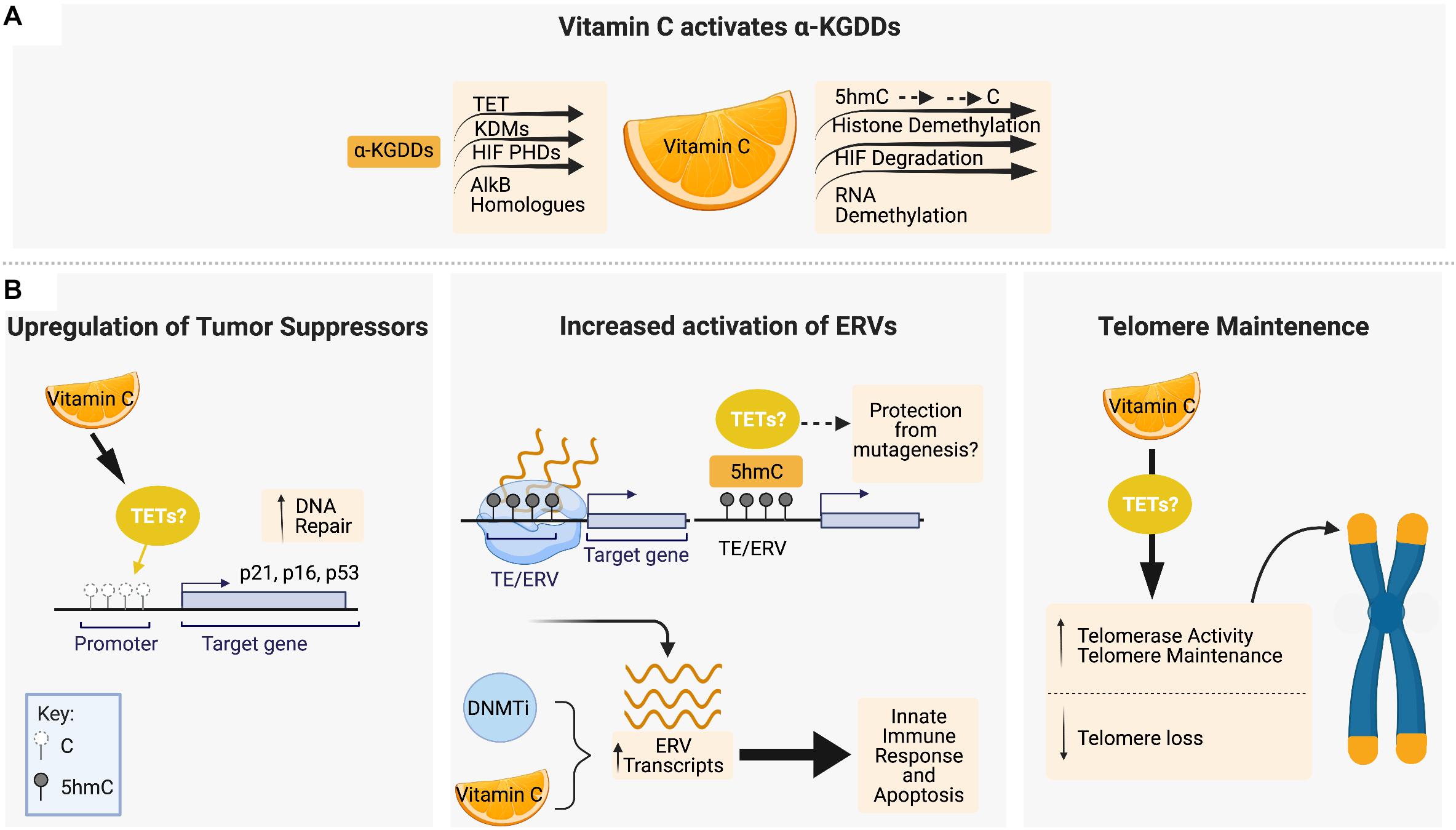
Figure 2. Vitamin C maintains genomic stability through interactions with epigenetic regulators, tumor suppressor upregulation and telomere maintenance. (A) Vitamin C serves as a cofactor for members of the α-ketoglutarate-dependent dioxygenases (α-KGDD) family such as TETs, KDMs, HIF PHDs, and ALKBHs, which can work to maintain genomic stability through the demethylation of 5mC residues, histone demethylation, HIF-1α degradation, and RNA demethylation, respectively. (B) Vitamin C supplementation has been shown to promote DNA demethylation in the promotor regions of tumor suppressor loci encoding p16, p21, and p53. Vitamin C promotes DNA demethylation and 5hmC formation at transposable elements (TEs) in the genome, leading to upregulated expression of endogenous retroviral genes (ERVs) in combination with DNA methyltransferase inhibitors (DNMTis). In cancer cells, ERV upregulation initiates an innate immune response and apoptotic cell death which is enhanced by vitamin C. Vitamin C treatment has also been shown to increase telomerase activity and the expression of genes that protect telomere integrity and decrease the rate of telomere loss. Figure created with BioRender.com.
The effect of vitamin C treatment on cell viability and genomic stability under steady-state conditions or in response to DNA damage can vary depending on developmental stage or normal versus malignant cell context. Vitamin C treatment can promote survival and maintain proliferation while protecting stem cells from DNA damage and senescence, whereas in cancer cells, the reactivation of tumor suppressors can reduce cell viability and alter sensitivity to therapeutic agents. Given that TET enzymes play a primary role as tumor suppressors, the ability to enhance 5hmC formation and DNA hypomethylation by vitamin C is of great interest in the maintenance of genomic stability, and cancer prevention and treatment (Figure 2B).
CpG islands in the promoters of DNA damage response genes, such as the tumor suppressor p16INK4a and p21, are frequently hypermethylated in cancer (Roman-Gomez et al., 2002; Zhao et al., 2016; Ocker et al., 2019). The p16 and p21 proteins belong to a family of cyclin–dependent kinase (CDK) inhibitors, also known as CDKN2A and CDKN1A, respectively, that bind and inactivate CDKs to block cell cycle progression. Reactivation of their expression by vitamin C has been shown to induce senescence, apoptosis and halt proliferation of cancer cells. Vitamin C treatment of human skin and colon cancer cells increases 5hmC formation and reduces 5mC in the promoter CGIs of p16INK4a and p21 that correlates with upregulated expression (Lin et al., 2014; Gerecke et al., 2018). Vitamin C also upregulates p53 and p21 protein expression in other cancer cells, such as oral squamous cell (Zhou et al., 2020) and hepatocellular carcinoma (Lv et al., 2018) causing growth arrest and apoptosis, however, in these studies the effect on 5hmC/5mC levels at these gene loci was not measured.
The studies in cancer cells on the effect of vitamin C in regulating p16INK4a and p21 expression seem at odds with the observation in ESCs, iPSCs and tissue specific stem cell cultures in which vitamin C treatment silences the p16INK4a/p19ARF locus and is associated with reduced expression of p53 and p21 (He et al., 2008; Li et al., 2009; Esteban et al., 2010; Li Y. et al., 2016; Zhang P. et al., 2016). Furthermore, TET activity at promoter CGIs in stem cells may be counteracted by vitamin C-mediated co-activation of the H3K36 demethylase JHDM1b (KDM2B), which removes gene body H3K36me2/3 marks leading to p16INK4a/p19ARF gene silencing (He et al., 2008; Tzatsos et al., 2009). These different responses highlight the context specific effect of vitamin C in the regulation of senescence in normal stem cells compared to cancer cells. Overexpression of the reprogramming factors such as OCT4, SOX2, KLF4, and MYC increase ROS production by up to threefold compared to controls which could induce oxidative damage and trigger the premature senescence of non-cancerous stem cells (Krishnamurthy et al., 2004; Li et al., 2009). Vitamin C antioxidant effects can mitigate the high levels of ROS induced during reprogramming to prevent oxidative damage, thereby blocking ROS-induced senescence mechanisms (Krishnamurthy et al., 2004; Li et al., 2009).
Vitamin C treatment also induces the expression of a TET2-dependent gene expression signature in human leukemia cell lines and primary murine HSPCs involved in BER such as GADD45, PARP, and DNA glycosylases (Cimmino et al., 2017). The ability of vitamin C to drive increased BER activity could be a direct consequence of the increased need to actively remove and replace TET catalyzed oxidation products of mC in the DNA.
DNA methyltransferase inhibitors (DNMTis), such as 5-azacytidine and decitabine, are cytidine analogs used for the treatment of hematological malignancies by inducing global DNA hypomethylation (Hackanson et al., 2005; Shadduck et al., 2007; Santos et al., 2010). Restoring TET function by vitamin C administration, in combination with DNMTi therapy, may help erase DNA hypermethylation at tumor suppressor loci to promote differentiation and cell death. In addition, studies in a variety of cancers cells have shown that DNA hypomethylation by DNMTis causes the increased expression of endogenous retroviruses (ERVs) that mimic a viral infection and trigger an innate immune response leading to apoptosis (Chiappinelli et al., 2015; Roulois et al., 2015; Liu et al., 2016). Vitamin C was shown to synergize with DNMTi treatment in a TET2-dependent manner to increase 5hmC, and drive DNA hypomethylation to further increase ERV expression and enhance apoptosis of leukemia and solid tumor cell lines (Liu et al., 2016). Interestingly, ERVs in ESCs treated with vitamin C have been shown to acquire and retain high levels of 5hmC that do not become demethylated for several days, in contrast to the rapid and dynamic hydroxymethylation and demethylation induced at CGIs in promoters and TSS sites of hypermethylated pluripotency and blastocyst-like genes (Blaschke et al., 2013). This distinction may be a key factor in how vitamin C treatment, by promoting TET activity, can induce the expression of ERVs while preventing genomic instability that would be caused by a lack of 5mC or 5hmC at these retrotransposable elements (REs). REs are one of the two classes of TEs, which unlike the cut and paste mechanisms of DNA transposons, insert themselves by duplicating elements into a new genomic location via an RNA intermediate (Friedli and Trono, 2015; Chuong et al., 2016). In ESCs, TET2 has been shown to be recruited to ERV loci by an ERV RNA-binding protein Paraspeckle Component -1 (PSPC1) to regulate the expression of adjacent genes during embryonic development (Guallar et al., 2018). Maintenance of 5hmC levels at ERVs by vitamin C may therefore reduce the risk of insertional mutagenesis while exploiting these cis-regulatory sequences for transcriptional control of neighboring genes.
Human telomeres shorten by 20–200 bp per cell division (Harley et al., 1990) and telomere length in circulating blood cells has been used as a biomarker of human aging (von Zglinicki and Martin-Ruiz, 2005). Higher intake of antioxidants by multivitamin supplementation or high vegetable intake is associated with increased telomere length in human studies (Xu et al., 2009; Marcon et al., 2012; Tsoukalas et al., 2019), however, direct evidence that vitamin C supplementation alone can suppress telomere attrition in vivo requires further study. Using in vitro cellular models derived from human iPSCs, vitamin C treatment has been shown to increase telomerase activity and the expression of genes encoding telomerase-related RNA and protein components that protect telomere stability (Wei et al., 2012; Kim et al., 2013). Patients with Werner Syndrome (WS) harbor a mutation in the WRN gene that leads to loss of telomere maintenance, premature aging and increased cancer rates (Burtner and Kennedy, 2010; Zhang et al., 2015). Vitamin C treatment of a human WS mesenchymal stem cell model was shown to slow down telomere loss and downregulate the senescence-inducing p16Ink4a protein (Zhang et al., 2013, 2015). In ESCs, the protection and maintenance of telomere length by vitamin C has not yet been correlated directly to its role as a TET coactivator; however, given that mouse ESCs with TET deficiency exhibit shorter telomeres, chromosomal instability, sub-telomere DNA hypermethylation and reduced telomere recombination (Lu et al., 2014; Kafer et al., 2016; Lazzerini-Denchi and Sfeir, 2016; Yang et al., 2016), vitamin C most likely plays a direct role in TET-mediated telomere maintenance.
Vitamin C can participate as a cofactor to enhance and maintain the activity of numerous other α-KGDD family members, such as histone lysine demethylases (KDMs), hypoxia inducible factor (HIF) prolyl hydroxylases, and AlkB homologs (ALKBHs), in addition to TET proteins, that in combination can influence genomic stability.
Vitamin C is required for the optimal activity and demethylation capacity of multiple KDMs that hydroxylate and remove mono-, di-, or trimethyl-lysines in histones (Klose et al., 2006; Tsukada et al., 2006). During somatic cell reprogramming, vitamin C-mediated removal of histone modifications such as H3K4me3 and H3K36me3, potentially by KDM5 and KDM2, respectively, can turn off the expression of genes, while demethylation of H3K9me2/me3 by members of the KDM3/4/7 family can remodel heterochromatin regions that can facilitate DNA hypomethylation at these otherwise silenced loci (Li et al., 2009; Tzatsos et al., 2009; Esteban et al., 2010; Huang et al., 2019). Histone modifications and KDMs have a wide variety of roles in regulating genomic stability that could potentially be influenced by vitamin C with different biological outcomes in either stem cells or cancer cells. Suppression of H3K9me2/me3 demethylase activity such as KDM3A (JMJD1A), KDM4C (JMJD2C), and KDM7C (PHF2) in tissue specific stem cell cultures and cancer cells can lead to premature senescence, increased DNA damage and genomic instability (Huang et al., 2019; Pappa et al., 2019; Fan et al., 2020). Vitamin C can coactivate KDM5B/C to demethylate H3K4me3, and KDM5B (JARID1B) has been shown to increase DNA DSB repair by recruiting factors Ku70 and BRCA1 in osteosarcoma (U2OS) cells (Li et al., 2014). A deficiency in KDM5B was shown to disengage the DNA repair process, promote spontaneous DNA damage, activate p53 signaling, and sensitize cells to genotoxic insults (Li et al., 2014). Demethylation of H3K4me3 by KDM5C (JARID1C) leads to heterochromatin formation, and renal cell carcinoma patients with JARID1C mutations exhibit genome-wide DNA hypomethylation, increased genomic rearrangements, and an overall worse prognosis (Rondinelli et al., 2015). In these settings, vitamin C could enhance KDM activity to prevent genomic instability. However, other KDM activity, such as overexpression of KDM2B (JHDM1B) and KDM4A (JHDM3A) that target H3K36 for demethylation could promote increased DNA damage through decreased HR repair (Pfister et al., 2014; Staberg et al., 2018).
HIF-1 and HIF-2 are transcription factors induced in low oxygen (hypoxic) environments typical of tumor niches, and are repressed in normoxic conditions by prolyl hydroxylases (PHDs) (Rankin and Giaccia, 2016). Vitamin C has been shown to function as a direct cofactor of HIF PHDs (Osipyants et al., 2018) that catalyze the hydroxylation of HIF-1α (Appelhoff et al., 2004). HIF-1α is involved in microsatellite instability and mismatch repair deficiency in a colon cancer model (Koshiji et al., 2005) and induces transcriptional changes leading to the downregulation of several DNA damage repair genes in an oral squamous cell carcinoma model (Nakamura et al., 2018). Treatment with vitamin C could promote the degradation of HIF-1α by stimulating HIF prolyl hydroxylases that would restore DNA repair activity and remove HIF-1 α mediated protection from genomic instability (Knowles et al., 2003; Zhao et al., 2015). HIF stabilization is also associated with therapeutic resistance to DNA damage. Hypoxic mouse stromal cells are shown to be more resistant to irradiation than the same cells under normoxic conditions in a HIF-1 dependent manner (Calvo-Asensio et al., 2018), and a lung cancer model expressing a HIF-1-stabilizing micro RNA exhibits a hypoxic phenotype and increased radioresistance (Grosso et al., 2013). Inhibition of leukemia cell growth by vitamin C treatment also correlates with the downregulation of HIF-1α mRNA expression (Kawada et al., 2013). Vitamin C could therefore target the PHD family of α-KGDDs to maintain genomic stability and counteract hypoxic tumor conditions to influence disease progression or sensitivity to DNA damage.
In mammalian cells, homologues of the E. coli DNA dealkylation enzyme AlkB act as RNA demethylases, catalyzing the oxidative demethylation of N6-methyladenosine (m6A) (Fedeles et al., 2015). Nine mammalian homologues have been identified, ALKBH1-8 and the fat mass and obesity associated protein (FTO), with each displaying unique roles in genomic stability (Fedeles et al., 2015). ALKBH5 and FTO are the most well characterized of the AlkB homologs and their silencing or deletion reduces the expression of a number of HR and other repair genes in glioblastoma stem cells (Kowalski-Chauvel et al., 2020) and osteoblasts (Zhang et al., 2019), increasing their susceptibility to genotoxic damage. Interestingly, ALKBH2 and ALKBH3 have been shown biochemically to act as DNA repair enzymes that oxidize 5mC to generate 5hmC, 5fC, and 5caC, similarly to TET enzymes (Bian et al., 2019). However, in certain cancer cell models, ALKBH activity may promote survival or resistance to chemotherapy. ALKBH2 knockdown sensitize lung cancer cells to cisplatin (Wu S.S. et al., 2011) and glioblastoma cells to temozolomide (Johannessen et al., 2013) and ALKBH8-knockout in mouse embryonic fibroblasts elevates ROS levels that could promote DNA damage (Endres et al., 2015). While vitamin C was shown to stimulate oxoglutarate turnover in E. coli AlkB (Welford et al., 2003), studies on the role of vitamin C in the regulation of mammalian ALKBH proteins are lacking. Given the recent confirmation that vitamin C binds directly to FTO (Wang et al., 2020) and the diverse roles of m6A in the regulation of gene expression in cancer (He et al., 2019) the role of vitamin C in m6A demethylation and the implication on genomic stability and cancer therapy requires further investigation.
Isocitrate dehydrogenases (IDHs) are citric acid cycle enzymes responsible for making α-KG, and as such are inextricably linked with α-KGDD functional activity. Mutually exclusive IDH1 or IDH2 mutations are common in myeloid malignancies and gliomas, and cause the production of 2-hydroxyglutarate (2HG), an oncometabolite that impairs α-KGDD function by acting as a competitive inhibitor of α-KG binding (Dang et al., 2010; Han et al., 2020). 2HG inhibition in IDH-mutant AML patients impairs TET2 function that leads to loss of 5hmC and DNA hypermethylation (Figueroa et al., 2010a; Lu et al., 2012; Sasaki et al., 2012). 2HG can also cause H3K9 hypermethylation due to decreased KDM4B activity, which can block the recruitment of repair factors at DSB sites in the genome and reduce HR repair efficiency (Sulkowski et al., 2020). Vitamin C treatment, however, has been shown to reduce proliferation in an IDH-mutant leukemia model that was associated with demethylation at the loci of myeloid differentiating factors (Mingay et al., 2018), suggesting that it could potentially override the effect of 2-HG to activate α-KGDDs such as the TET demethylases.
Vitamin C is an essential dietary micronutrient for humans, whereas other mammals, including mice, can synthesize vitamin C from glucose via the liver enzyme L-gulonolactone oxidase (GULO). GULO catalyzes the last step of ascorbate biosynthesis but is mutated and non-functional in humans (Linster and Van Schaftingen, 2007). Vitamin C is water soluble and optimal physiological plasma concentrations of approximately ∼70–80 μM can be sustained by the daily intake of 200 mg in the diet (Lindblad et al., 2013; Padayatty and Levine, 2016). Vitamin C is most well-known for its role in the prevention of scurvy, a disease caused by prolonged periods of low dietary vitamin C intake (<10 mg/day) that reduces plasma levels to below 11.4 μM, leading to insufficient collagen production that manifests with symptoms ranging from fever, confusion and depression, to internal bleeding (Prinzo, 1999; Schleicher et al., 2009; Padayatty and Levine, 2016).
While scurvy is now seen as a rare modern-day disease, patients with cancer are often markedly vitamin C-deficient (Mayland et al., 2005; Huijskens et al., 2016; Liu et al., 2016), and restoring or maintaining physiological levels has been shown to slow malignant cell growth (Campbell et al., 2015; Liu et al., 2016; Agathocleous et al., 2017). Plasma concentrations of ascorbate can differ up to ten-fold from person to person (Khaw et al., 2001) and in the United States, it is estimated that more than 7% of the population (>20 million people) are deficient in vitamin C (Schleicher et al., 2009). A study in the United Kingdom found that 25–46% of low income population and smokers exhibit deficient or depleted vitamin C plasma levels (Mosdol et al., 2008). Mild vitamin C deficiency may be underreported owing to its non-specific symptoms such as fatigue, irritability, dull aching pains, and weight loss (Prinzo, 1999; Schleicher et al., 2009). All-cause mortality decreases when vitamin C serum levels rise above 60 μM (Goyal et al., 2013; Wang et al., 2018), while in cancer patients, low serum vitamin C levels of <20–30 μM are frequently observed (Waldo and Zipf, 1955; Anthony and Schorah, 1982; Mayland et al., 2005; Carr et al., 2020). A study of leukocytes isolated from colon cancer patients showed decreased levels of 5mC and 5hmC that correlated with blood plasma ascorbate below 20 μM, while expression of TET genes was not significantly changed (Starczak et al., 2018). 5hmC/5mC ratios in the DNA of peripheral blood cells could potentially act as a biomarker of vitamin C status or bioavailability.
Vitamin C is water soluble and transported across cellular membranes by sodium-dependent vitamin C transporters (SVCTs) and facilitative glucose transporters (GLUTs). SVCTs transport vitamin C directly, whereas GLUTs transport the oxidized form of vitamin C, dehydroascorbate (DHA), which is reduced to vitamin C inside cells by glutathione (GSH) (Figure 3A). Plasma levels of vitamin C are tightly controlled by two sodium-dependent vitamin C transporters (SVCTs), with SVCT1 being responsible for gastrointestinal absorption and renal reabsorption, and SVCT2, playing a primary role in whole body cellular uptake (Lindblad et al., 2013). Mouse models with genetic inactivation of the Gulo locus or Svct1/2 knockout have been used to model cell-intrinsic or systemic dietary deficiency in vitamin C (Maeda et al., 2000; Sotiriou et al., 2002; Corpe et al., 2010). Supplementation of ascorbate in the drinking water at 3.3 g/L is sufficient to maintain normal 80 μM concentrations in the plasma of Gulo knockout mice; however, 0.33 g/L will reduce plasma concentrations to 30 μM (Schleicher et al., 2009; Kim et al., 2012). This model of dietary deficiency in mice causes hematopoietic defects that mimic the effect of TET2 deficiency, including pre-leukemic HSPC expansion and loss of 5hmC in the genome (Beamer et al., 2000; Sotiriou et al., 2002). Importantly, these effects could be reversed upon increased dietary vitamin C administration (Agathocleous et al., 2017). Svct2 knockout in bone marrow cells expressing the AML oncogene Flt3ITD has also been shown to accelerate leukemia progression in mice and exacerbate 5hmC loss in Tet2 deficient HSCs, suggesting that vitamin C depletion could further impair the activity of other TET proteins in pre-leukemic cells (Agathocleous et al., 2017). Severe deficiency of both Tet2 and Tet3, is associated with >90% loss of 5hmC in HSPCs and causes the spontaneous accumulation of DSBs, marked by elevated and persistent yH2AX foci and rapid leukemia progression (An et al., 2015). Taken together, these findings suggest that vitamin C, by maintaining TET activity in HSPCs, provides protection from leukemia transformation.
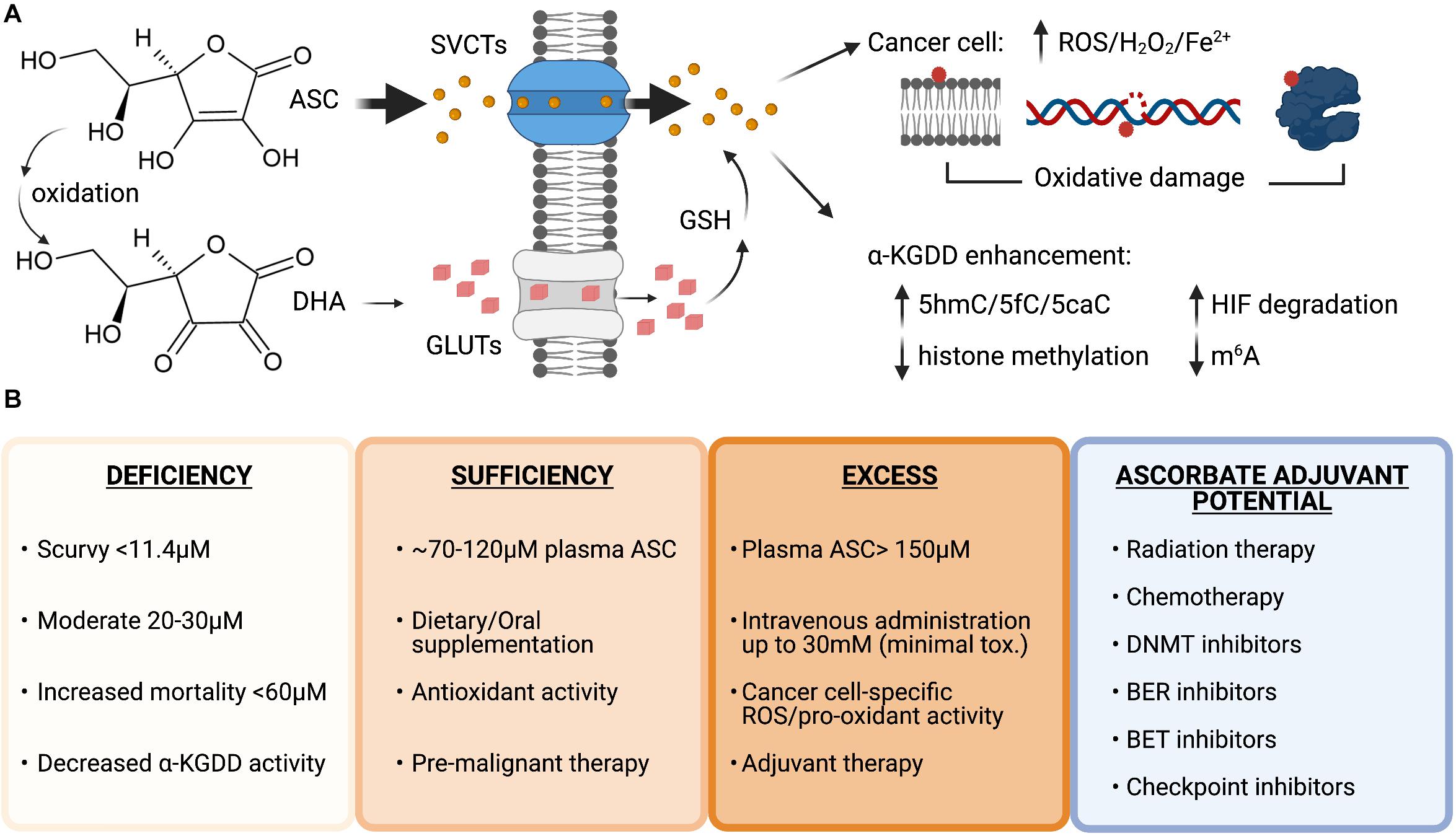
Figure 3. Vitamin C uptake and therapeutic potential as an anti-cancer agent. (A) Structures of ascorbate (ASC) and dehydroascorbate (DHA). ASC primarily enters cells through sodium-dependent vitamin C transporters (SVCTs) and can be transported in its oxidized form as DHA via glucose transporters (GLUTs) then reduced back to ASC by the antioxidant glutathione (GSH) once inside the cell. ASC can enhance the activity of α-ketoglutarate dependent dioxygenases (α-KGDD), and high doses can act as a pro-oxidant, creating increased levels of reactive oxygen species (ROS), H2O2 and increased redox active iron levels (Fe2+) that cause lipid, DNA, and protein oxidation in cancer cells. (B) Plasma ascorbate levels correlate with disease states, overall health, and therapeutic potential. Ascorbate deficiency is associated with increased all-cause mortality and lower α-KGDD activity. Oral supplementation can restore plasma levels to normal and allow vitamin C antioxidant properties to provide benefit as a pre-malignant therapy potentially lowering genomic instability and preventing transformation. High-dose intravenous administration of ASC can be used to generate millimolar (mM) plasma concentrations and generate ROS and pro-oxidant damage to cancer cells. Vitamin C has currently shown efficacy as an adjuvant for many cancer therapies in pre-clinical and clinical trials in combination with standard chemotherapy, DNA hypomethylating agents and targeted inhibitors: Carboplatin and Paclitaxel, Azacitidine, poly-adenosine diphosphate-ribose polymerase (PARP) inhibitors (Olaparib), immune checkpoint inhibitors (anti-PD1 and anti-CTLA4), and bromodomain and extraterminal domain (BET) inhibitors (JQ1). Figure created with BioRender.com.
Vitamin C supplementation is being explored as an adjuvant to many existing cancer therapies (Figure 3B). As cancer patients frequently present with decreased plasma vitamin C levels (Waldo and Zipf, 1955; Anthony and Schorah, 1982; Mayland et al., 2005; Carr et al., 2020) that are further decreased by chemotherapy (Mousseau et al., 2005; Carr et al., 2020), vitamin C treatment to restore normal amounts in the circulation as an adjuvant for standard chemotherapy could be of therapeutic benefit. The vast majority of patients are TET2 haploinsufficient (Abdel-Wahab et al., 2009; Delhommeau et al., 2009; Kosmider et al., 2009; Langemeijer et al., 2009), suggesting that enhancing residual TET2 activity, or restoring the activity of functionally defective mutant TET2 proteins, could be a particularly viable therapeutic strategy for TET2 deficient hematological malignancies. A recent study showed that oral supplementation of 500 mg/day of vitamin C in combination with DNMTi treatment in patients with myeloid malignancies was able to raise deficient vitamin C plasma levels to ∼100 μM concentration, increase 5hmC/5mC ratios and decrease global 5mC levels that were elevated at baseline in the peripheral blood cells (Gillberg et al., 2019). While previous clinical studies in advanced cancer patients given similar oral doses did not show any benefit (Creagan et al., 1979; Moertel et al., 1985), long-term oral vitamin C supplementation may have important clinical implications for pre-malignant and low-risk patients with mutation in specific epigenetic regulators such as TET2 and should be investigated further.
Maximal plasma concentrations from oral vitamin C doses above 500 mg/day do not exceed 150 μM, due to homeostatic down-regulation of vitamin C transporters in enterocytes and kidney cells, leading to reduced absorption and increased urinary excretion (Young et al., 2015). High-dose vitamin C (up to 100 g) administered by intravenous (IV) infusion can bypass the limited oral bioavailability without reported toxicity, and raise plasma concentrations to high millimolar levels that remain above 100 μM for up to 6 h (Padayatty et al., 2004). An intriguing effect of high-dose vitamin C treatment in cancer cells is its ability to create ROS and act as a pro-oxidant.
Vitamin C in the form of ascorbate, under normal growth conditions, behaves as an anti-oxidant that donates electrons to quench damage-inducing free radicals, becoming a relatively stable ascorbate radical while to protecting cells from lipid, protein, and DNA oxidation (Padayatty et al., 2003). In contrast to the effect vitamin C has in reprogramming and stem cell cultures, which is to reduce ROS, increase proliferation and protect cells from apoptosis, vitamin C treatment of cancer cells correlates with an increase in ROS-mediated oxidative stress and reduced viability (Chen et al., 2008; Noguera et al., 2017; Ghanem et al., 2020; Wu et al., 2020; Zhou et al., 2020). Moreover, it has been demonstrated that while diverse cancer cell types are sensitive to low- and sub-millimolar doses of vitamin C, even with high millimolar doses, limited cytotoxicity is observed in normal tissue (Chen et al., 2008; Noguera et al., 2017). This points to a unique synthetic lethality of cancer cells to high-dose vitamin C treatment that can be exploited for cancer therapy. In a Burkitt’s lymphoma model, toxicity to ascorbate was associated with ROS-induced peroxide formation that could be abrogated by catalase treatment (Chen et al., 2005). Follow up work by the same group showed elevated ascorbate radical and peroxide levels in vivo in the microenvironment of three separate models where vitamin C reduced tumor burden in mice (Chen et al., 2008). Another study showed mitochondrial hyperpolarization associated with high-dose ascorbate and ROS production (Ghanem et al., 2020).
The pro-oxidant effects of vitamin C in these studies has been attributed to excess uptake and formation of redox active iron. In a series of reactions, ascorbate can reduce ferric (Fe3+) iron to ferrous (Fe2+), which can then be returned to the ferric state after reducing O2 to superoxide. Superoxide reacts with itself and protons to generate hydrogen peroxide and O2, and the hydrogen peroxide can be reduced by Fe2+ in a Fenton reaction to form hydroxide radicals that can create the pro-oxidant killing effects observed in cancer cells (Du et al., 2012). Another possibility is that high-dose vitamin C can exhaust the primary cellular antioxidant GSH when taken up in higher amounts by more metabolically active cells, thus rendering cancer cells more vulnerable to oxidative stress. In this capacity, high-dose vitamin C was shown to be selectively toxic to KRAS or BRAF mutant colorectal cancer cells due to increased cellular uptake of oxidized vitamin C (DHA) through upregulated GLUT transporters, which led to GSH depletion and lethal levels of ROS (Yun et al., 2015).
Multiple cellular and animal models and recent clinical trials have shown that high-dose vitamin C treatment can reduce cancer cell viability and improve treatment outcome in combination therapies. In patients with ovarian cancer, high-dose intravenous vitamin C (IVC) administered in combination with carboplatin and paclitaxel enhanced chemosensitivity and reduced adverse side effects of chemotherapy (Ma et al., 2014). Glioblastoma patients receiving IVC in combination with radiation therapy and temozolomide have also shown improved survival compared to previous studies (Schoenfeld et al., 2017). Another notable adjuvant use of vitamin C was with PARP inhibitor treatment for a small group of patients with HR-deficient advanced metastatic cancers (primarily BRCA mutations) where partial or complete responses were observed in all eight patients (Demiray, 2020).
High-dose parenteral vitamin C treatment can be modeled in mice via IP administration, where 4 g/Kg delivered IP can induce mM concentrations in plasma similar to pharmacokinetic studies in patients treated with high-dose IVC (Padayatty et al., 2004; Yun et al., 2015). Vitamin C treatment of Tet2-deficient mouse HSPCs and patient-derived AML leads to increased 5hmC formation, DNA hypomethylation and a block in aberrant self-renewal in vitro with suppression of disease progression in vivo (Cimmino et al., 2017). High-dose vitamin C also enhances immune checkpoint inhibitor therapy, that in animals models was shown to be curative when used to treat tumors with high mutational burdens such as mismatch repair–deficient (MMRd) or microsatellite instable (MSI) breast and colorectal mouse tumors that otherwise would be unresponsive to single immune checkpoint inhibitors (Magri et al., 2020). MSI cancers exhibit microsatellite DNA hypermethylation and accumulate 5mC mutations in CpG in the absence MMR that has been attributed to methylation-associated repair deficiencies (Ling et al., 2010; Poulos et al., 2017). While a role for TET mediated hypomethylation in microsatellite stability was not assessed, this may be one mechanism by which vitamin C treatment could play a role in MMR-deficient cancer treatment. Importantly the therapeutic efficacy of vitamin C in combination with checkpoint inhibitors was dependent on a functional immune system, suggesting that tumor cell-intrinsic and immune microenvironmental effects of vitamin C can work together to slow tumor growth.
Multiple studies continue to identify new potential combination cancer treatment strategies with vitamin C. In cellular and preclinical studies, high-dose vitamin C enhances the sensitivity of hematological cell malignancies to arsenic trioxide (Huijskens et al., 2016; Noguera et al., 2017) and increases chemosensitivity and radiosensitivity of various solid tumor cells including ovarian (Ma et al., 2014), pancreatic (Du et al., 2015) glioblastoma and non-small cell lung carcinoma cells (Schoenfeld et al., 2017). Vitamin C has also been shown to sensitize melanoma cells to bromodomain inhibitors (Mustafi et al., 2018). These studies argued in favor of combination therapies based primarily on the mechanism of oxidative stress generated by high-dose ascorbate, that could increase cancer cell susceptibility to standard therapies by exacerbating DNA damage. The effect of vitamin C as a co-factor of α-KGDDs and specifically as a regulator of DNA demethylation via increased TET function were not investigated. However, it has been shown that TET-mediated DNA oxidation induced by vitamin C can create a synthetic lethality, where AML cells being forced to undergo active DNA demethylation renders them hypersensitive to PARP inhibition (Cimmino et al., 2017). In this study, vitamin C treatment increases the expression of genes involved in BER that may be a response to the increased oxidation of 5mC and in combination with the PARP inhibitor, Olaparib, enhances the killing of human AML cells greater than either agent alone (Cimmino et al., 2017). Future studies on the use of vitamin C as a therapeutic agent would benefit from the inclusion of additional correlative studies such as DNA methylation changes, oxidized mC formation or even histone and RNA methylation levels in order to fully appreciate the cell-intrinsic or microenvironmental epigenetic biomarkers of its anti-cancer efficacy.
Given the numerous roles for vitamin C in biological processes that maintain and influence genomic stability, it is no surprise that vitamin C continues to be explored for potential anticancer activity. From the studies by Cameron and Pauling in the 1970s (Cameron and Pauling, 1976, 1978) to the present day, there is an ever-expanding volume of research attempting to elucidate the true and, more relevantly, meaningful roles of vitamin C in disease etiology, treatment, and prevention. But important consideration must be given to robust data collection and interpretation, especially where clinical samples are involved (Lykkesfeldt, 2020). Considering vitamin C’s accessibility and prevalence as a dietary nutrient, synthesizing meaningful conclusions on the role of vitamin C in cancer prevention by oral supplementation will require large population studies. However, vitamin C’s role as an anti-tumor agent that can restore DNA damage and repair signaling processes at high doses appears to go beyond just regulating the oxidation state of the cell. Vitamin C has also been shown to influence many enzymes within the Fe2+ and α-KGDD superfamily that can work together to maintain genomic stability beyond the direct effect of vitamin C in TET-mediated modulation of cytosine modification and turnover in the genome. The role of vitamin C as an epigenetic regulator will have context specific effects that depend on the cancer cell lineage. The strong association of DNA hypermethylation coupled with the frequent mutation of TET2 in hematological malignancies suggest that vitamin C treatment could be a targeted therapy for these patients. Future studies into how vitamin C can maintain the function of the diverse α-KGDDs known to have direct and indirect roles in the maintenance of genomic stability will allow us to fully understand the effect of this essential vitamin in the etiology, prevention, and treatment of cancer.
JB and TL contributed equally in the writing and organization of the manuscript. SM contributed to the writing of the manuscript. LC supervised and edited the manuscript. All the authors contributed to the article and approved the submitted version.
This research was supported by the St. Baldrick’s Cancer Research Foundation (ID# 703429), The Woman’s Cancer Association of University of Miami, and The Dresner MDS foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abdel-Wahab, O., Mullally, A., Hedvat, C., Garcia-Manero, G., Patel, J., Wadleigh, M., et al. (2009). Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114, 144–147. doi: 10.1182/blood-2009-03-210039
Agathocleous, M., Meacham, C. E., Burgess, R. J., Piskounova, E., Zhao, Z., Crane, G. M., et al. (2017). Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481. doi: 10.1038/nature23876
Akalin, A., Garrett-Bakelman, F. E., Kormaksson, M., Busuttil, J., Zhang, L., Khrebtukova, I., et al. (2012). Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 8:e1002781. doi: 10.1371/journal.pgen.1002781
Akdemir, K. C., Le, V. T., Chandran, S., Li, Y., Verhaak, R. G., Beroukhim, R., et al. (2020). Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat. Genet. 52, 294–305. doi: 10.1038/s41588-019-0564-y
Alexandrov, L. B., Kim, J., Haradhvala, N. J., Huang, M. N., Tian, Ng, A. W., et al. (2020). The repertoire of mutational signatures in human cancer. Nature 578, 94–101. doi: 10.1038/s41586-020-1943-3
Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Aparicio, S. A., Behjati, S., Biankin, A. V., et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. doi: 10.1038/nature12477
Amouroux, R., Nashun, B., Shirane, K., Nakagawa, S., Hill, P. W., D’Souza, Z., et al. (2016). De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233. doi: 10.1038/ncb3296
An, J., Gonzalez-Avalos, E., Chawla, A., Jeong, M., Lopez-Moyado, I. F., Li, W., et al. (2015). Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 6:10071. doi: 10.1038/ncomms10071
Anthony, H. M., and Schorah, C. J. (1982). Severe hypovitaminosis C in lung-cancer patients: the utilization of vitamin C in surgical repair and lymphocyte-related host resistance. Br. J. Cancer 46, 354–367. doi: 10.1038/bjc.1982.211
Appelhoff, R. J., Tian, Y. M., Raval, R. R., Turley, H., Harris, A. L., Pugh, C. W., et al. (2004). Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279, 38458–38465. doi: 10.1074/jbc.M406026200
Bakshi, A., Herke, S. W., Batzer, M. A., and Kim, J. (2016). DNA methylation variation of human-specific Alu repeats. Epigenetics 11, 163–173. doi: 10.1080/15592294.2015.1130518
Bashtrykov, P., Jankevicius, G., Smarandache, A., Jurkowska, R. Z., Ragozin, S., and Jeltsch, A. (2012). Specificity of Dnmt1 for methylation of hemimethylated CpG sites resides in its catalytic domain. Chem. Biol. 19, 572–578. doi: 10.1016/j.chembiol.2012.03.010
Baylin, S. B., and Jones, P. A. (2016). Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 8:a019505. doi: 10.1101/cshperspect.a019505
Beamer, W. G., Rosen, C. J., Bronson, R. T., Gu, W., Donahue, L. R., Baylink, D. J., et al. (2000). Spontaneous fracture (sfx): a mouse genetic model of defective peripubertal bone formation. Bone 27, 619–626.
Bestor, T., Laudano, A., Mattaliano, R., and Ingram, V. (1988). Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203, 971–983.
Bian, K., Lenz, S. A. P., Tang, Q., Chen, F., Qi, R., Jost, M., et al. (2019). DNA repair enzymes ALKBH2, ALKBH3, and AlkB oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in vitro. Nucleic Acids Res. 47, 5522–5529. doi: 10.1093/nar/gkz395
Blaschke, K., Ebata, K. T., Karimi, M. M., Zepeda-Martinez, J. A., Goyal, P., Mahapatra, S., et al. (2013). Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226. doi: 10.1038/nature12362
Blokzijl, F., de Ligt, J., Jager, M., Sasselli, V., Roerink, S., Sasaki, N., et al. (2016). Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264. doi: 10.1038/nature19768
Bowman, R. L., Busque, L., and Levine, R. L. (2018). Clonal Hematopoiesis and Evolution to Hematopoietic Malignancies. Cell Stem Cell 22, 157–170. doi: 10.1016/j.stem.2018.01.011
Brown, D. C., Grace, E., Sumner, A. T., Edmunds, A. T., and Ellis, P. M. (1995). ICF syndrome (immunodeficiency, centromeric instability and facial anomalies): investigation of heterochromatin abnormalities and review of clinical outcome. Hum. Genet. 96, 411–416. doi: 10.1007/BF00191798
Brunetti, L., Gundry, M. C., and Goodell, M. A. (2017). DNMT3A in Leukemia. Cold Spring Harb. Perspect. Med. 7:a030320. doi: 10.1101/cshperspect.a030320
Bullard, W., Kieft, R., and Sabatini, R. (2017). A method for the efficient and selective identification of 5-hydroxymethyluracil in genomic DNA. Biol. Methods Protoc. 2:bw006. doi: 10.1093/biomethods/bpw006
Burtner, C. R., and Kennedy, B. K. (2010). Progeria syndromes and ageing: what is the connection? Nat. Rev. Mol. Cell Biol. 11, 567–578. doi: 10.1038/nrm2944
Buscarlet, M., Provost, S., Zada, Y. F., Barhdadi, A., Bourgoin, V., Lepine, G., et al. (2017). DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130, 753–762. doi: 10.1182/blood-2017-04-777029
Busque, L., Patel, J. P., Figueroa, M. E., Vasanthakumar, A., Provost, S., Hamilou, Z., et al. (2012). Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44, 1179–1181. doi: 10.1038/ng.2413
Calvo-Asensio, I., Dillon, E. T., Lowndes, N. F., and Ceredig, R. (2018). The Transcription Factor Hif-1 Enhances the Radio-Resistance of Mouse MSCs. Front. Physiol. 9:439. doi: 10.3389/fphys.2018.00439
Cameron, E., and Pauling, L. (1976). Supplemental ascorbate in the supportive treatment of cancer: Prolongation of survival times in terminal human cancer. Proc. Natl. Acad. Sci. U S A. 73, 3685–3689. doi: 10.1073/pnas.73.10.3685
Cameron, E., and Pauling, L. (1978). Supplemental ascorbate in the supportive treatment of cancer: reevaluation of prolongation of survival times in terminal human cancer. Proc. Natl. Acad. Sci. U S A. 75, 4538–4542. doi: 10.1073/pnas.75.9.4538
Campbell, E. J., Vissers, M. C., Bozonet, S., Dyer, A., Robinson, B. A., and Dachs, G. U. (2015). Restoring physiological levels of ascorbate slows tumor growth and moderates HIF-1 pathway activity in Gulo(-/-) mice. Cancer Med. 4, 303–314. doi: 10.1002/cam4.349
Cancer Genome Atlas Research, N. (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074. doi: 10.1056/NEJMoa1301689
Carr, A. C., Spencer, E., Das, A., Meijer, N., Lauren, C., MacPherson, S., et al. (2020). Patients Undergoing Myeloablative Chemotherapy and Hematopoietic Stem Cell Transplantation Exhibit Depleted Vitamin C Status in Association with Febrile Neutropenia. Nutrients 12:nu12061879. doi: 10.3390/nu12061879
Chen, J., Guo, L., Zhang, L., Wu, H., Yang, J., Liu, H., et al. (2013). Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 45, 1504–1509. doi: 10.1038/ng.2807
Chen, Q., Chen, Y., Bian, C., Fujiki, R., and Yu, X. (2013). TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564. doi: 10.1038/nature11742
Chen, Q., Espey, M. G., Krishna, M. C., Mitchell, J. B., Corpe, C. P., Buettner, G. R., et al. (2005). Pharmacologic ascorbic acid concentrations selectively kill cancer cells: action as a pro-drug to deliver hydrogen peroxide to tissues. Proc. Natl. Acad. Sci. U S A. 102, 13604–13609. doi: 10.1073/pnas.0506390102
Chen, Q., Espey, M. G., Sun, A. Y., Pooput, C., Kirk, K. L., Krishna, M. C., et al. (2008). Pharmacologic doses of ascorbate act as a prooxidant and decrease growth of aggressive tumor xenografts in mice. Proc. Natl. Acad. Sci. U S A. 105, 11105–11109. doi: 10.1073/pnas.0804226105
Chiappinelli, K. B., Strissel, P. L., Desrichard, A., Li, H., Henke, C., Akman, B., et al. (2015). Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 162, 974–986. doi: 10.1016/j.cell.2015.07.011
Chung, T. L., Brena, R. M., Kolle, G., Grimmond, S. M., Berman, B. P., Laird, P. W., et al. (2010). Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells 28, 1848–1855. doi: 10.1002/stem.493
Chuong, E. B., Elde, N. C., and Feschotte, C. (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. doi: 10.1126/science.aad5497
Ciccarone, F., Klinger, F. G., Catizone, A., Calabrese, R., Zampieri, M., Bacalini, M. G., et al. (2012). Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS One 7:e46927. doi: 10.1371/journal.pone.0046927
Cimmino, L., Dolgalev, I., Wang, Y., Yoshimi, A., Martin, G. H., Wang, J., et al. (2017). Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 170, 1079–1095e1020. doi: 10.1016/j.cell.2017.07.032
Cooper, D. N., Mort, M., Stenson, P. D., Ball, E. V., and Chuzhanova, N. A. (2010). Methylation-mediated deamination of 5-methylcytosine appears to give rise to mutations causing human inherited disease in CpNpG trinucleotides, as well as in CpG dinucleotides. Hum. Genomics 4, 406–410. doi: 10.1186/1479-7364-4-6-406
Corpe, C. P., Tu, H., Eck, P., Wang, J., Faulhaber-Walter, R., Schnermann, J., et al. (2010). Vitamin C transporter Slc23a1 links renal reabsorption, vitamin C tissue accumulation, and perinatal survival in mice. J. Clin. Invest. 120, 1069–1083. doi: 10.1172/JCI39191
Cortellino, S., Xu, J., Sannai, M., Moore, R., Caretti, E., Cigliano, A., et al. (2011). Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79. doi: 10.1016/j.cell.2011.06.020
Creagan, E. T., Moertel, C. G., O’Fallon, J. R., Schutt, A. J., O’Connell, M. J., Rubin, J., et al. (1979). Failure of high-dose vitamin C (ascorbic acid) therapy to benefit patients with advanced cancer. A controlled trial. N. Engl. J. Med. 301, 687–690. doi: 10.1056/NEJM197909273011303
Curradi, M., Izzo, A., Badaracco, G., and Landsberger, N. (2002). Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell Biol. 22, 3157–3173. doi: 10.1128/mcb.22.9.3157-3173.2002
Damaschke, N. A., Gawdzik, J., Avilla, M., Yang, B., Svaren, J., Roopra, A., et al. (2020). CTCF loss mediates unique DNA hypermethylation landscapes in human cancers. Clin. Epigenet. 12:80. doi: 10.1186/s13148-020-00869-7
Dang, L., Jin, S., and Su, S. M. (2010). IDH mutations in glioma and acute myeloid leukemia. Trends Mol. Med. 16, 387–397. doi: 10.1016/j.molmed.2010.07.002
Das, A. B., Smith-Diaz, C. C., and Vissers, M. C. M. (2021). Emerging epigenetic therapeutics for myeloid leukemia: modulating demethylase activity with ascorbate. Haematologica 106, 14–25. doi: 10.3324/haematol.2020.259283
Daskalos, A., Nikolaidis, G., Xinarianos, G., Savvari, P., Cassidy, A., Zakopoulou, R., et al. (2009). Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int. J. Cancer 124, 81–87. doi: 10.1002/ijc.23849
Dawlaty, M. M., Breiling, A., Le, T., Raddatz, G., Barrasa, M. I., Cheng, A. W., et al. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323. doi: 10.1016/j.devcel.2012.12.015
Deaton, A. M., and Bird, A. (2011). CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022. doi: 10.1101/gad.2037511
Delhommeau, F., Dupont, S., Della Valle, V., James, C., Trannoy, S., Masse, A., et al. (2009). Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360, 2289–2301. doi: 10.1056/NEJMoa0810069
Demiray, M. (2020). Combinatorial Therapy of High Dose Vitamin C and PARP Inhibitors in DNA Repair Deficiency: A Series of 8 Patients. Integr. Cancer Ther. 19:1534735420969812. doi: 10.1177/1534735420969812
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Dominguez-Sola, D., Ying, C. Y., Grandori, C., Ruggiero, L., Chen, B., Li, M., et al. (2007). Non-transcriptional control of DNA replication by c-Myc. Nature 448, 445–451. doi: 10.1038/nature05953
Du, J., Cieslak, J. A. III, Welsh, J. L., Sibenaller, Z. A., Allen, B. G., Wagner, B. A., et al. (2015). Pharmacological Ascorbate Radiosensitizes Pancreatic Cancer. Cancer Res. 75, 3314–3326. doi: 10.1158/0008-5472.CAN-14-1707
Du, J., Cullen, J. J., and Buettner, G. R. (2012). Ascorbic acid: chemistry, biology and the treatment of cancer. Biochim. Biophys. Acta 1826, 443–457. doi: 10.1016/j.bbcan.2012.06.003
Ehrlich, M., and Lacey, M. (2013). DNA hypomethylation and hemimethylation in cancer. Adv. Exp. Med. Biol. 754, 31–56. doi: 10.1007/978-1-4419-9967-2_2
Ehrlich, M., Sanchez, C., Shao, C., Nishiyama, R., Kehrl, J., Kuick, R., et al. (2008). ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity 41, 253–271. doi: 10.1080/08916930802024202
Endres, L., Begley, U., Clark, R., Gu, C., Dziergowska, A., Malkiewicz, A., et al. (2015). Alkbh8 Regulates Selenocysteine-Protein Expression to Protect against Reactive Oxygen Species Damage. PLoS One 10:e0131335. doi: 10.1371/journal.pone.0131335
Esteban, M. A., Wang, T., Qin, B., Yang, J., Qin, D., Cai, J., et al. (2010). Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6, 71–79. doi: 10.1016/j.stem.2009.12.001
Fan, L., Xu, S., Zhang, F., Cui, X., Fazli, L., Gleave, M., et al. (2020). Histone demethylase JMJD1A promotes expression of DNA repair factors and radio-resistance of prostate cancer cells. Cell Death Dis. 11:214. doi: 10.1038/s41419-020-2405-4
Fang, C., Wang, Z., Han, C., Safgren, S. L., Helmin, K. A., Adelman, E. R., et al. (2020). Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation. Genome Biol. 21:247. doi: 10.1186/s13059-020-02152-7
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D., and Essigmann, J. M. (2015). The AlkB Family of Fe(II)/alpha-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J. Biol. Chem. 290, 20734–20742. doi: 10.1074/jbc.R115.656462
Ficz, G., Branco, M. R., Seisenberger, S., Santos, F., Krueger, F., Hore, T. A., et al. (2011). Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402. doi: 10.1038/nature10008
Figueroa, M. E., Abdel-Wahab, O., Lu, C., Ward, P. S., Patel, J., Shih, A., et al. (2010a). Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567. doi: 10.1016/j.ccr.2010.11.015
Figueroa, M. E., Lugthart, S., Li, Y., Erpelinck-Verschueren, C., Deng, X., Christos, P. J., et al. (2010b). DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17, 13–27. doi: 10.1016/j.ccr.2009.11.020
Friedli, M., and Trono, D. (2015). The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell Dev. Biol. 31, 429–451. doi: 10.1146/annurev-cellbio-100814-125514
Gao, L., Emperle, M., Guo, Y., Grimm, S. A., Ren, W., Adam, S., et al. (2020). Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 11:3355. doi: 10.1038/s41467-020-17109-4
Ge, G., Peng, D., Xu, Z., Guan, B., Xin, Z., He, Q., et al. (2018). Restoration of 5-hydroxymethylcytosine by ascorbate blocks kidney tumour growth. EMBO Rep. 19:201745401. doi: 10.15252/embr.201745401
Gerecke, C., Schumacher, F., Edlich, A., Wetzel, A., Yealland, G., Neubert, L. K., et al. (2018). Vitamin C promotes decitabine or azacytidine induced DNA hydroxymethylation and subsequent reactivation of the epigenetically silenced tumour suppressor CDKN1A in colon cancer cells. Oncotarget 9, 32822–32840. doi: 10.18632/oncotarget.25999
Ghanem, A., Melzer, A. M., Zaal, E., Neises, L., Baltissen, D., Matar, O., et al. (2020). Ascorbate kills breast cancer cells by rewiring metabolism via redox imbalance and energy crisis. Free Radic. Biol. Med. 163, 196–209. doi: 10.1016/j.freeradbiomed.2020.12.012
Gillberg, L., Orskov, A. D., Nasif, A., Ohtani, H., Madaj, Z., Hansen, J. W., et al. (2019). Oral vitamin C supplementation to patients with myeloid cancer on azacitidine treatment: Normalization of plasma vitamin C induces epigenetic changes. Clin. Epigenet. 11:143. doi: 10.1186/s13148-019-0739-5
Gong, Y., Lazaris, C., Sakellaropoulos, T., Lozano, A., Kambadur, P., Ntziachristos, P., et al. (2018). Stratification of TAD boundaries reveals preferential insulation of super-enhancers by strong boundaries. Nat. Commun. 9:542. doi: 10.1038/s41467-018-03017-1
Gossling, K. L., Schipp, C., Fischer, U., Babor, F., Koch, G., Schuster, F. R., et al. (2017). Hematopoietic Stem Cell Transplantation in an Infant with Immunodeficiency, Centromeric Instability, and Facial Anomaly Syndrome. Front. Immunol. 8:773. doi: 10.3389/fimmu.2017.00773
Goyal, A., Terry, M. B., and Siegel, A. B. (2013). Serum antioxidant nutrients, vitamin A, and mortality in U.S. Adults. Cancer Epidemiol. Biomark. Prev. 22, 2202–2211. doi: 10.1158/1055-9965.EPI-13-0381
Grin, I., and Ishchenko, A. A. (2016). An interplay of the base excision repair and mismatch repair pathways in active DNA demethylation. Nucleic Acids Res. 44, 3713–3727. doi: 10.1093/nar/gkw059
Grosso, S., Doyen, J., Parks, S. K., Bertero, T., Paye, A., Cardinaud, B., et al. (2013). MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 4:e544. doi: 10.1038/cddis.2013.71
Guallar, D., Bi, X., Pardavila, J. A., Huang, X., Saenz, C., Shi, X., et al. (2018). RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 50, 443–451. doi: 10.1038/s41588-018-0060-9
Guo, H., Zhu, P., Yan, L., Li, R., Hu, B., Lian, Y., et al. (2014). The DNA methylation landscape of human early embryos. Nature 511, 606–610. doi: 10.1038/nature13544
Gupta, A., Godwin, A. K., Vanderveer, L., Lu, A., and Liu, J. (2003). Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 63, 664–673.
Guryanova, O. A., Shank, K., Spitzer, B., Luciani, L., Koche, R. P., Garrett-Bakelman, F. E., et al. (2016). DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat. Med. 22, 1488–1495. doi: 10.1038/nm.4210
Gustafson, C. B., Yang, C., Dickson, K. M., Shao, H., Van Booven, D., Harbour, J. W., et al. (2015). Epigenetic reprogramming of melanoma cells by vitamin C treatment. Clin. Epigenet. 7:51. doi: 10.1186/s13148-015-0087-z
Hackanson, B., Robbel, C., Wijermans, P., and Lubbert, M. (2005). In vivo effects of decitabine in myelodysplasia and acute myeloid leukemia: review of cytogenetic and molecular studies. Ann. Hematol. 84(Suppl. 1), 32–38. doi: 10.1007/s00277-005-0004-1
Haffner, M. C., Chaux, A., Meeker, A. K., Esopi, D. M., Gerber, J., Pellakuru, L. G., et al. (2011). Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2, 627–637.
Hagleitner, M. M., Lankester, A., Maraschio, P., Hulten, M., Fryns, J. P., Schuetz, C., et al. (2008). Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome). J. Med. Genet. 45, 93–99. doi: 10.1136/jmg.2007.053397
Han, S., Liu, Y., Cai, S. J., Qian, M., Ding, J., Larion, M., et al. (2020). IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br. J. Cancer 122, 1580–1589. doi: 10.1038/s41416-020-0814-x
Hansen, R. S., Wijmenga, C., Luo, P., Stanek, A. M., Canfield, T. K., Weemaes, C. M., et al. (1999). The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. U S A. 96, 14412–14417. doi: 10.1073/pnas.96.25.14412
Harley, C. B., Futcher, A. B., and Greider, C. W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460. doi: 10.1038/345458a0
He, J., Kallin, E. M., Tsukada, Y., and Zhang, Y. (2008). The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat. Struct. Mol. Biol. 15, 1169–1175. doi: 10.1038/nsmb.1499
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18:176. doi: 10.1186/s12943-019-1109-9
He, X. B., Kim, M., Kim, S. Y., Yi, S. H., Rhee, Y. H., Kim, T., et al. (2015). Vitamin C facilitates dopamine neuron differentiation in fetal midbrain through TET1- and JMJD3-dependent epigenetic control manner. Stem Cells 33, 1320–1332. doi: 10.1002/stem.1932
He, Y. F., Li, B. Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. doi: 10.1126/science.1210944
Honda, T., Tamura, G., Waki, T., Kawata, S., Terashima, M., Nishizuka, S., et al. (2004). Demethylation of MAGE promoters during gastric cancer progression. Br. J. Cancer 90, 838–843. doi: 10.1038/sj.bjc.6601600
Hore, T. A., von Meyenn, F., Ravichandran, M., Bachman, M., Ficz, G., Oxley, D., et al. (2016). Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naive pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. U S A. 113, 12202–12207. doi: 10.1073/pnas.1608679113
Howard, G., Eiges, R., Gaudet, F., Jaenisch, R., and Eden, A. (2008). Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27, 404–408. doi: 10.1038/sj.onc.1210631
Huang, B., Wang, B., Yuk-Wai Lee, W., Pong, U. K., Leung, K. T., Li, X., et al. (2019). KDM3A and KDM4C Regulate Mesenchymal Stromal Cell Senescence and Bone Aging via Condensin-mediated Heterochromatin Reorganization. iScience 21, 375–390. doi: 10.1016/j.isci.2019.10.041
Huijskens, M. J., Wodzig, W. K., Walczak, M., Germeraad, W. T., and Bos, G. M. (2016). Ascorbic acid serum levels are reduced in patients with hematological malignancies. Results Immunol. 6, 8–10. doi: 10.1016/j.rinim.2016.01.001
Ikehata, H. (2018). Mechanistic considerations on the wavelength-dependent variations of UVR genotoxicity and mutagenesis in skin: the discrimination of UVA-signature from UV-signature mutation. Photochem. Photobiol. Sci. 17, 1861–1871. doi: 10.1039/c7pp00360a
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi: 10.1126/science.1210597
Jaiswal, S., Fontanillas, P., Flannick, J., Manning, A., Grauman, P. V., Mar, B. G., et al. (2014). Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498. doi: 10.1056/NEJMoa1408617
Jeong, M., Sun, D., Luo, M., Huang, Y., Challen, G. A., Rodriguez, B., et al. (2014). Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 46, 17–23. doi: 10.1038/ng.2836
Johannessen, T. C., Prestegarden, L., Grudic, A., Hegi, M. E., Tysnes, B. B., and Bjerkvig, R. (2013). The DNA repair protein ALKBH2 mediates temozolomide resistance in human glioblastoma cells. Neuro Oncol. 15, 269–278. doi: 10.1093/neuonc/nos301
Kafer, G. R., Li, X., Horii, T., Suetake, I., Tajima, S., Hatada, I., et al. (2016). 5-Hydroxymethylcytosine Marks Sites of DNA Damage and Promotes Genome Stability. Cell Rep. 14, 1283–1292. doi: 10.1016/j.celrep.2016.01.035
Kamb, A., Gruis, N. A., Weaver-Feldhaus, J., Liu, Q., Harshman, K., Tavtigian, S. V., et al. (1994). A cell cycle regulator potentially involved in genesis of many tumor types. Science 264, 436–440. doi: 10.1126/science.8153634
Kandoth, C., McLellan, M. D., Vandin, F., Ye, K., Niu, B., Lu, C., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. doi: 10.1038/nature12634
Kang, J. Y., Song, S. H., Yun, J., Jeon, M. S., Kim, H. P., Han, S. W., et al. (2015). Disruption of CTCF/cohesin-mediated high-order chromatin structures by DNA methylation downregulates PTGS2 expression. Oncogene 34, 5677–5684. doi: 10.1038/onc.2015.17
Kareta, M. S., Botello, Z. M., Ennis, J. J., Chou, C., and Chedin, F. (2006). Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 281, 25893–25902. doi: 10.1074/jbc.M603140200
Kawada, H., Kaneko, M., Sawanobori, M., Uno, T., Matsuzawa, H., Nakamura, Y., et al. (2013). High concentrations of L-ascorbic acid specifically inhibit the growth of human leukemic cells via downregulation of HIF-1alpha transcription. PLoS One 8:e62717. doi: 10.1371/journal.pone.0062717
Kawasaki, F., Beraldi, D., Hardisty, R. E., McInroy, G. R., van Delft, P., and Balasubramanian, S. (2017). Genome-wide mapping of 5-hydroxymethyluracil in the eukaryote parasite Leishmania. Genome Biol. 18:23. doi: 10.1186/s13059-017-1150-1
Kaya, N., Al-Muhsen, S., Al-Saud, B., Al-Bakheet, A., Colak, D., Al-Ghonaium, A., et al. (2011). ICF syndrome in Saudi Arabia: immunological, cytogenetic and molecular analysis. J. Clin. Immunol. 31, 245–252. doi: 10.1007/s10875-010-9488-0
Khaw, K. T., Bingham, S., Welch, A., Luben, R., Wareham, N., Oakes, S., et al. (2001). Relation between plasma ascorbic acid and mortality in men and women in EPIC-Norfolk prospective study: a prospective population study. European Prospective Investigation into Cancer and Nutrition. Lancet 357, 657–663. doi: 10.1016/s0140-6736(00)04128-3
Kim, H. Y., Lee, D. H., Shin, M. H., Shin, H. S., Kim, M. K., and Chung, J. H. (2018). UV-induced DNA methyltransferase 1 promotes hypermethylation of tissue inhibitor of metalloproteinase 2 in the human skin. J. Dermatol. Sci. 91, 19–27. doi: 10.1016/j.jdermsci.2018.03.009
Kim, H., Bae, S., Yu, Y., Kim, Y., Kim, H. R., Hwang, Y. I., et al. (2012). The analysis of vitamin C concentration in organs of gulo(-/-) mice upon vitamin C withdrawal. Immune Netw. 12, 18–26. doi: 10.4110/in.2012.12.1.18
Kim, K. H., Choi, J. S., Kim, I. J., Ku, J. L., and Park, J. G. (2006). Promoter hypomethylation and reactivation of MAGE-A1 and MAGE-A3 genes in colorectal cancer cell lines and cancer tissues. World J. Gastroenterol. 12, 5651–5657. doi: 10.3748/wjg.v12.i35.5651
Kim, Y. Y., Ku, S. Y., Huh, Y., Liu, H. C., Kim, S. H., Choi, Y. M., et al. (2013). Anti-aging effects of vitamin C on human pluripotent stem cell-derived cardiomyocytes. Age 35, 1545–1557. doi: 10.1007/s11357-012-9457-z
Klose, R. J., Kallin, E. M., and Zhang, Y. (2006). JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727. doi: 10.1038/nrg1945
Knowles, H. J., Raval, R. R., Harris, A. L., and Ratcliffe, P. J. (2003). Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 63, 1764–1768.
Ko, M., An, J., Pastor, W. A., Koralov, S. B., Rajewsky, K., and Rao, A. (2015). TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol. Rev. 263, 6–21. doi: 10.1111/imr.12239
Ko, M., Huang, Y., Jankowska, A. M., Pape, U. J., Tahiliani, M., Bandukwala, H. S., et al. (2010). Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843.
Koh, K. P., Yabuuchi, A., Rao, S., Huang, Y., Cunniff, K., Nardone, J., et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. doi: 10.1016/j.stem.2011.01.008
Kondo, Y., and Issa, J. P. (2003). Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 278, 27658–27662. doi: 10.1074/jbc.M304072200
Koshiji, M., To, K. K., Hammer, S., Kumamoto, K., Harris, A. L., Modrich, P., et al. (2005). HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol. Cell 17, 793–803. doi: 10.1016/j.molcel.2005.02.015
Kosmider, O., Gelsi-Boyer, V., Ciudad, M., Racoeur, C., Jooste, V., Vey, N., et al. (2009). TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica 94, 1676–1681. doi: 10.3324/haematol.2009.011205
Kowalski-Chauvel, A., Lacore, M. G., Arnauduc, F., Delmas, C., Toulas, C., Cohen-Jonathan-Moyal, E., et al. (2020). The m6A RNA Demethylase ALKBH5 Promotes Radioresistance and Invasion Capability of Glioma Stem Cells. Cancers 13:13010040. doi: 10.3390/cancers13010040
Kriaucionis, S., and Heintz, N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. doi: 10.1126/science.1169786
Krishnamurthy, J., Torrice, C., Ramsey, M. R., Kovalev, G. I., Al-Regaiey, K., Su, L., et al. (2004). Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 114, 1299–1307. doi: 10.1172/JCI22475
Kudo, Y., Tateishi, K., Yamamoto, K., Yamamoto, S., Asaoka, Y., Ijichi, H., et al. (2012). Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 103, 670–676. doi: 10.1111/j.1349-7006.2012.02213.x
Kushwaha, G., Dozmorov, M., Wren, J. D., Qiu, J., Shi, H., and Xu, D. (2016). Hypomethylation coordinates antagonistically with hypermethylation in cancer development: a case study of leukemia. Hum. Genomics 10(Suppl. 2):18. doi: 10.1186/s40246-016-0071-5
Kusmartsev, V., Drozdz, M., Schuster-Bockler, B., and Warnecke, T. (2020). Cytosine Methylation Affects the Mutability of Neighboring Nucleotides in Germline and Soma. Genetics 214, 809–823. doi: 10.1534/genetics.120.303028
Kuzyk, A., and Mai, S. (2014). c-MYC-induced genomic instability. Cold Spring Harb. Perspect. Med. 4:a014373. doi: 10.1101/cshperspect.a014373
Langemeijer, S. M., Kuiper, R. P., Berends, M., Knops, R., Aslanyan, M. G., Massop, M., et al. (2009). Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 41, 838–842. doi: 10.1038/ng.391
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Lazzerini-Denchi, E., and Sfeir, A. (2016). Stop pulling my strings - what telomeres taught us about the DNA damage response. Nat. Rev. Mol. Cell Biol. 17, 364–378. doi: 10.1038/nrm.2016.43
Lee Chong, T., Ahearn, E. L., and Cimmino, L. (2019). Reprogramming the Epigenome With Vitamin C. Front. Cell Dev. Biol. 7:128. doi: 10.3389/fcell.2019.00128
Lee, C. J., Ahn, H., Jeong, D., Pak, M., Moon, J. H., and Kim, S. (2020). Impact of mutations in DNA methylation modification genes on genome-wide methylation landscapes and downstream gene activations in pan-cancer. BMC Med. Genomics 13(Suppl. 3):27. doi: 10.1186/s12920-020-0659-4
Lemonnier, F., Couronne, L., Parrens, M., Jais, J. P., Travert, M., Lamant, L., et al. (2012). Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120, 1466–1469. doi: 10.1182/blood-2012-02-408542
Lemonnier, F., Poullot, E., Dupuy, A., Couronne, L., Martin, N., Scourzic, L., et al. (2018). Loss of 5-hydroxymethylcytosine is a frequent event in peripheral T-cell lymphomas. Haematologica 103, 115–118e. doi: 10.3324/haematol.2017.167973
Ley, T. J., Ding, L., Walter, M. J., McLellan, M. D., Lamprecht, T., Larson, D. E., et al. (2010). DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433. doi: 10.1056/NEJMoa1005143
Li, H., Collado, M., Villasante, A., Strati, K., Ortega, S., Canamero, M., et al. (2009). The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139. doi: 10.1038/nature08290
Li, L., Li, C., Mao, H., Du, Z., Chan, W. Y., Murray, P., et al. (2016). Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Sci. Rep. 6:26591. doi: 10.1038/srep26591
Li, X., Liu, L., Yang, S., Song, N., Zhou, X., Gao, J., et al. (2014). Histone demethylase KDM5B is a key regulator of genome stability. Proc. Natl. Acad. Sci. U S A. 111, 7096–7101. doi: 10.1073/pnas.1324036111
Li, Y., Zhang, W., Chang, L., Han, Y., Sun, L., Gong, X., et al. (2016). Vitamin C alleviates aging defects in a stem cell model for Werner syndrome. Protein Cell 7, 478–488. doi: 10.1007/s13238-016-0278-1
Li, Z., Gu, T. P., Weber, A. R., Shen, J. Z., Li, B. Z., Xie, Z. G., et al. (2015). Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 43, 3986–3997. doi: 10.1093/nar/gkv283
Lin, J. R., Qin, H. H., Wu, W. Y., He, S. J., and Xu, J. H. (2014). Vitamin C protects against UV irradiation-induced apoptosis through reactivating silenced tumor suppressor genes p21 and p16 in a Tet-dependent DNA demethylation manner in human skin cancer cells. Cancer Biother. Radiopharm. 29, 257–264. doi: 10.1089/cbr.2014.1647
Lindblad, M., Tveden-Nyborg, P., and Lykkesfeldt, J. (2013). Regulation of vitamin C homeostasis during deficiency. Nutrients 5, 2860–2879. doi: 10.3390/nu5082860
Ling, Z. Q., Tanaka, A., Li, P., Nakayama, T., Fujiyama, Y., Hattori, T., et al. (2010). Microsatellite instability with promoter methylation and silencing of hMLH1 can regionally occur during progression of gastric carcinoma. Cancer Lett. 297, 244–251. doi: 10.1016/j.canlet.2010.05.017
Linster, C. L., and Van Schaftingen, E. (2007). Vitamin C. Biosynthesis, recycling and degradation in mammals. FEBS J. 274, 1–22. doi: 10.1111/j.1742-4658.2006.05607.x
Liu, H., Liu, W., Wu, Y., Zhou, Y., Xue, R., Luo, C., et al. (2005). Loss of epigenetic control of synuclein-gamma gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 65, 7635–7643. doi: 10.1158/0008-5472.CAN-05-1089
Liu, M., Ohtani, H., Zhou, W., Orskov, A. D., Charlet, J., Zhang, Y. W., et al. (2016). Vitamin C increases viral mimicry induced by 5-aza-2’-deoxycytidine. Proc. Natl. Acad. Sci. U S A. 113, 10238–10244. doi: 10.1073/pnas.1612262113
Lopez-Moyado, I. F., Tsagaratou, A., Yuita, H., Seo, H., Delatte, B., Heinz, S., et al. (2019). Paradoxical association of TET loss of function with genome-wide DNA hypomethylation. Proc. Natl. Acad. Sci. U S A. 116, 16933–16942. doi: 10.1073/pnas.1903059116
Lu, C., Ward, P. S., Kapoor, G. S., Rohle, D., Turcan, S., Abdel-Wahab, O., et al. (2012). IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478. doi: 10.1038/nature10860
Lu, F., Liu, Y., Jiang, L., Yamaguchi, S., and Zhang, Y. (2014). Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 28, 2103–2119. doi: 10.1101/gad.248005.114
Lv, H., Wang, C., Fang, T., Li, T., Lv, G., Han, Q., et al. (2018). Vitamin C preferentially kills cancer stem cells in hepatocellular carcinoma via SVCT-2. NPJ Precis. Oncol. 2:1. doi: 10.1038/s41698-017-0044-8
Lykkesfeldt, J. (2020). On the effect of vitamin C intake on human health: How to (mis)interprete the clinical evidence. Redox. Biol. 34:101532. doi: 10.1016/j.redox.2020.101532
Ma, Y., Chapman, J., Levine, M., Polireddy, K., Drisko, J., and Chen, Q. (2014). High-dose parenteral ascorbate enhanced chemosensitivity of ovarian cancer and reduced toxicity of chemotherapy. Sci. Transl. Med. 6:222ra218. doi: 10.1126/scitranslmed.3007154
Madugundu, G. S., Cadet, J., and Wagner, J. R. (2014). Hydroxyl-radical-induced oxidation of 5-methylcytosine in isolated and cellular DNA. Nucleic Acids Res. 42, 7450–7460. doi: 10.1093/nar/gku334
Maeda, N., Hagihara, H., Nakata, Y., Hiller, S., Wilder, J., and Reddick, R. (2000). Aortic wall damage in mice unable to synthesize ascorbic acid. Proc. Natl. Acad. Sci. U S A. 97, 841–846.
Magri, A., Germano, G., Lorenzato, A., Lamba, S., Chila, R., Montone, M., et al. (2020). High-dose vitamin C enhances cancer immunotherapy. Sci. Transl. Med. 12:aay8707. doi: 10.1126/scitranslmed.aay8707
Maiti, A., and Drohat, A. C. (2011). Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338. doi: 10.1074/jbc.C111.284620
Mandal, P. K., Ewing, A. D., Hancks, D. C., and Kazazian, H. H. Jr. (2013). Enrichment of processed pseudogene transcripts in L1-ribonucleoprotein particles. Hum. Mol. Genet. 22, 3730–3748. doi: 10.1093/hmg/ddt225
Marcon, F., Siniscalchi, E., Crebelli, R., Saieva, C., Sera, F., Fortini, P., et al. (2012). Diet-related telomere shortening and chromosome stability. Mutagenesis 27, 49–57. doi: 10.1093/mutage/ger056
Martinez-Fernandez, L., Banyasz, A., Esposito, L., Markovitsi, D., and Improta, R. (2017). UV-induced damage to DNA: effect of cytosine methylation on pyrimidine dimerization. Signal Transduct. Target Ther. 2:17021. doi: 10.1038/sigtrans.2017.21
Mayland, C. R., Bennett, M. I., and Allan, K. (2005). Vitamin C deficiency in cancer patients. Palliat. Med. 19, 17–20. doi: 10.1191/0269216305pm970oa
Metivier, R., Gallais, R., Tiffoche, C., Le Peron, C., Jurkowska, R. Z., Carmouche, R. P., et al. (2008). Cyclical DNA methylation of a transcriptionally active promoter. Nature 452, 45–50. doi: 10.1038/nature06544
Mingay, M., Chaturvedi, A., Bilenky, M., Cao, Q., Jackson, L., Hui, T., et al. (2018). Vitamin C-induced epigenomic remodelling in IDH1 mutant acute myeloid leukaemia. Leukemia 32, 11–20. doi: 10.1038/leu.2017.171
Minor, E. A., Court, B. L., Young, J. I., and Wang, G. (2013). Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 288, 13669–13674. doi: 10.1074/jbc.C113.464800
Modrzejewska, M., Gawronski, M., Skonieczna, M., Zarakowska, E., Starczak, M., Foksinski, M., et al. (2016). Vitamin C enhances substantially formation of 5-hydroxymethyluracil in cellular DNA. Free Radic. Biol. Med. 101, 378–383. doi: 10.1016/j.freeradbiomed.2016.10.535
Moertel, C. G., Fleming, T. R., Creagan, E. T., Rubin, J., O’Connell, M. J., and Ames, M. M. (1985). High-dose vitamin C versus placebo in the treatment of patients with advanced cancer who have had no prior chemotherapy. A randomized double-blind comparison. N. Engl. J. Med. 312, 137–141. doi: 10.1056/NEJM198501173120301
Mosdol, A., Erens, B., and Brunner, E. J. (2008). Estimated prevalence and predictors of vitamin C deficiency within UK’s low-income population. J. Public Health 30, 456–460. doi: 10.1093/pubmed/fdn076
Moulton, T., Crenshaw, T., Hao, Y., Moosikasuwan, J., Lin, N., Dembitzer, F., et al. (1994). Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat. Genet. 7, 440–447. doi: 10.1038/ng0794-440
Mousseau, M., Faure, H., Hininger, I., Bayet-Robert, M., and Favier, A. (2005). Leukocyte 8-oxo-7,8-dihydro-2’-deoxyguanosine and comet assay in epirubicin-treated patients. Free Radic. Res. 39, 837–843. doi: 10.1080/10715760500042860
Munari, E., Chaux, A., Vaghasia, A. M., Taheri, D., Karram, S., Bezerra, S. M., et al. (2016). Global 5-Hydroxymethylcytosine Levels Are Profoundly Reduced in Multiple Genitourinary Malignancies. PLoS One 11:e0146302. doi: 10.1371/journal.pone.0146302
Mustafi, S., Camarena, V., Volmar, C. H., Huff, T. C., Sant, D. W., Brothers, S. P., et al. (2018). Vitamin C Sensitizes Melanoma to BET Inhibitors. Cancer Res. 78, 572–583. doi: 10.1158/0008-5472.CAN-17-2040
Nabel, C. S., Jia, H., Ye, Y., Shen, L., Goldschmidt, H. L., Stivers, J. T., et al. (2012). AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 8, 751–758. doi: 10.1038/nchembio.1042
Nakamura, H., Bono, H., Hiyama, K., Kawamoto, T., Kato, Y., Nakanishi, T., et al. (2018). Differentiated embryo chondrocyte plays a crucial role in DNA damage response via transcriptional regulation under hypoxic conditions. PLoS One 13:e0192136. doi: 10.1371/journal.pone.0192136
Nanan, K. K., Sturgill, D. M., Prigge, M. F., Thenoz, M., Dillman, A. A., Mandler, M. D., et al. (2019). TET-Catalyzed 5-Carboxylcytosine Promotes CTCF Binding to Suboptimal Sequences Genome-wide. iScience 19, 326–339. doi: 10.1016/j.isci.2019.07.041
Nanavaty, V., Abrash, E. W., Hong, C., Park, S., Fink, E. E., Li, Z., et al. (2020). DNA Methylation Regulates Alternative Polyadenylation via CTCF and the Cohesin Complex. Mol. Cell 78, 752–764e756. doi: 10.1016/j.molcel.2020.03.024
Neri, F., Incarnato, D., Krepelova, A., Rapelli, S., Anselmi, F., Parlato, C., et al. (2015). Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics. Cell Rep. 10, 674–683. doi: 10.1016/j.celrep.2015.01.008
Nguyen, T. V., Yao, S., Wang, Y., Rolfe, A., Selvaraj, A., Darman, R., et al. (2019). The R882H DNMT3A hot spot mutation stabilizes the formation of large DNMT3A oligomers with low DNA methyltransferase activity. J. Biol. Chem. 294, 16966–16977. doi: 10.1074/jbc.RA119.010126
Noguera, N. I., Pelosi, E., Angelini, D. F., Piredda, M. L., Guerrera, G., Piras, E., et al. (2017). High-dose ascorbate and arsenic trioxide selectively kill acute myeloid leukemia and acute promyelocytic leukemia blasts in vitro. Oncotarget 8, 32550–32565. doi: 10.18632/oncotarget.15925
Ocker, M., Bitar, S. A., Monteiro, A. C., Gali-Muhtasib, H., and Schneider-Stock, R. (2019). Epigenetic Regulation of p21(cip1/waf1) in Human Cancer. Cancers 11:11091343. doi: 10.3390/cancers11091343
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. doi: 10.1016/s0092-8674(00)81656-6
Olinski, R., Starczak, M., and Gackowski, D. (2016). Enigmatic 5-hydroxymethyluracil: Oxidatively modified base, epigenetic mark or both? Mutat. Res. Rev. Mutat. Res. 767, 59–66. doi: 10.1016/j.mrrev.2016.02.001
Osipyants, A. I., Poloznikov, A. A., Smirnova, N. A., Hushpulian, D. M., Khristichenko, A. Y., Chubar, T. A., et al. (2018). L-ascorbic acid: A true substrate for HIF prolyl hydroxylase? Biochimie 147, 46–54. doi: 10.1016/j.biochi.2017.12.011
Otani, J., Kimura, H., Sharif, J., Endo, T. A., Mishima, Y., Kawakami, T., et al. (2013). Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells. PLoS One 8:e82961. doi: 10.1371/journal.pone.0082961
Padayatty, S. J., and Levine, M. (2016). Vitamin C: the known and the unknown and Goldilocks. Oral Dis. 22, 463–493. doi: 10.1111/odi.12446
Padayatty, S. J., Katz, A., Wang, Y., Eck, P., Kwon, O., Lee, J. H., et al. (2003). Vitamin C as an antioxidant: evaluation of its role in disease prevention. J. Am. Coll. Nutr. 22, 18–35. doi: 10.1080/07315724.2003.10719272
Padayatty, S. J., Sun, H., Wang, Y., Riordan, H. D., Hewitt, S. M., Katz, A., et al. (2004). Vitamin C pharmacokinetics: implications for oral and intravenous use. Ann. Intern. Med. 140, 533–537.
Pan, F., Wingo, T. S., Zhao, Z., Gao, R., Makishima, H., Qu, G., et al. (2017). Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat. Commun. 8:15102. doi: 10.1038/ncomms15102
Papaemmanuil, E., Gerstung, M., Malcovati, L., Tauro, S., Gundem, G., Van Loo, P., et al. (2013). Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627. doi: 10.1182/blood-2013-08-518886
Pappa, S., Padilla, N., Iacobucci, S., Vicioso, M., Alvarez de la Campa, E., Navarro, C., et al. (2019). PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc. Natl. Acad. Sci. U S A. 116, 19464–19473. doi: 10.1073/pnas.1903188116
Pehrsson, E. C., Choudhary, M. N. K., Sundaram, V., and Wang, T. (2019). The epigenomic landscape of transposable elements across normal human development and anatomy. Nat. Commun. 10:5640. doi: 10.1038/s41467-019-13555-x
Peng, D., Ge, G., Gong, Y., Zhan, Y., He, S., Guan, B., et al. (2018). Vitamin C increases 5-hydroxymethylcytosine level and inhibits the growth of bladder cancer. Clin. Epigenet. 10:94. doi: 10.1186/s13148-018-0527-7
Pfaffeneder, T., Spada, F., Wagner, M., Brandmayr, C., Laube, S. K., Eisen, D., et al. (2014). Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 10, 574–581. doi: 10.1038/nchembio.1532
Pfeifer, G. P. (2018). Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 19:19041166. doi: 10.3390/ijms19041166
Pfister, S. X., Ahrabi, S., Zalmas, L. P., Sarkar, S., Aymard, F., Bachrati, C. Z., et al. (2014). SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018. doi: 10.1016/j.celrep.2014.05.026
Plass, C., and Soloway, P. D. (2002). DNA methylation, imprinting and cancer. Eur. J. Hum. Genet. 10, 6–16. doi: 10.1038/sj.ejhg.5200768
Poulos, R. C., Olivier, J., and Wong, J. W. H. (2017). The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. Nucleic Acids Res. 45, 7786–7795. doi: 10.1093/nar/gkx463
Prinzo, Z. (1999). Scurvy and its prevention and control in major emergencies. Geneva: World Health Organization.
Quivoron, C., Couronne, L., Della Valle, V., Lopez, C. K., Plo, I., Wagner-Ballon, O., et al. (2011). TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event during Human Lymphomagenesis. Cancer Cell 20, 25–38. doi: 10.1016/j.ccr.2011.06.003
Raiber, E. A., Beraldi, D., Ficz, G., Burgess, H. E., Branco, M. R., Murat, P., et al. (2012). Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 13:R69. doi: 10.1186/gb-2012-13-8-r69
Rainier, S., Johnson, L. A., Dobry, C. J., Ping, A. J., Grundy, P. E., and Feinberg, A. P. (1993). Relaxation of imprinted genes in human cancer. Nature 362, 747–749. doi: 10.1038/362747a0
Rangam, G., Schmitz, K. M., Cobb, A. J., and Petersen-Mahrt, S. K. (2012). AID enzymatic activity is inversely proportional to the size of cytosine C5 orbital cloud. PLoS One 7:e43279. doi: 10.1371/journal.pone.0043279
Rankin, E. B., and Giaccia, A. J. (2016). Hypoxic control of metastasis. Science 352, 175–180. doi: 10.1126/science.aaf4405
Rasmussen, K. D., Jia, G., Johansen, J. V., Pedersen, M. T., Rapin, N., Bagger, F. O., et al. (2015). Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 29, 910–922. doi: 10.1101/gad.260174.115
Ribeiro, A. F., Pratcorona, M., Erpelinck-Verschueren, C., Rockova, V., Sanders, M., Abbas, S., et al. (2012). Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood 119, 5824–5831. doi: 10.1182/blood-2011-07-367961
Roman-Gomez, J., Castillejo, J. A., Jimenez, A., Gonzalez, M. G., Moreno, F., Rodriguez Mdel, C., et al. (2002). 5’ CpG island hypermethylation is associated with transcriptional silencing of the p21(CIP1/WAF1/SDI1) gene and confers poor prognosis in acute lymphoblastic leukemia. Blood 99, 2291–2296. doi: 10.1182/blood.v99.7.2291
Rondinelli, B., Rosano, D., Antonini, E., Frenquelli, M., Montanini, L., Huang, D., et al. (2015). Histone demethylase JARID1C inactivation triggers genomic instability in sporadic renal cancer. J. Clin. Invest. 125, 4625–4637. doi: 10.1172/JCI81040
Rossi, D. J., Bryder, D., Seita, J., Nussenzweig, A., Hoeijmakers, J., and Weissman, I. L. (2007). Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729. doi: 10.1038/nature05862
Roulois, D., Loo Yau, H., Singhania, R., Wang, Y., Danesh, A., Shen, S. Y., et al. (2015). DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 162, 961–973. doi: 10.1016/j.cell.2015.07.056
Russler-Germain, D. A., Spencer, D. H., Young, M. A., Lamprecht, T. L., Miller, C. A., Fulton, R., et al. (2014). The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25, 442–454. doi: 10.1016/j.ccr.2014.02.010
Sant, D. W., Mustafi, S., Gustafson, C. B., Chen, J., Slingerland, J. M., and Wang, G. (2018). Vitamin C promotes apoptosis in breast cancer cells by increasing TRAIL expression. Sci. Rep. 8:5306. doi: 10.1038/s41598-018-23714-7
Santos, F. P., Kantarjian, H., Garcia-Manero, G., Issa, J. P., and Ravandi, F. (2010). Decitabine in the treatment of myelodysplastic syndromes. Expert Rev. Anticancer Ther. 10, 9–22. doi: 10.1586/era.09.164
Santos, F., Peat, J., Burgess, H., Rada, C., Reik, W., and Dean, W. (2013). Active demethylation in mouse zygotes involves cytosine deamination and base excision repair. Epigenet. Chromatin 6:39. doi: 10.1186/1756-8935-6-39
Sasaki, M., Knobbe, C. B., Munger, J. C., Lind, E. F., Brenner, D., Brustle, A., et al. (2012). IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488, 656–659. doi: 10.1038/nature11323
Saxonov, S., Berg, P., and Brutlag, D. L. (2006). A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U S A. 103, 1412–1417. doi: 10.1073/pnas.0510310103
Schleicher, R. L., Carroll, M. D., Ford, E. S., and Lacher, D. A. (2009). Serum vitamin C and the prevalence of vitamin C deficiency in the United States: 2003-2004 National Health and Nutrition Examination Survey (NHANES). Am. J. Clin. Nutr. 90, 1252–1263. doi: 10.3945/ajcn.2008.27016
Schoenfeld, J. D., Sibenaller, Z. A., Mapuskar, K. A., Wagner, B. A., Cramer-Morales, K. L., Furqan, M., et al. (2017). O2(-) and H2O2-Mediated Disruption of Fe Metabolism Causes the Differential Susceptibility of NSCLC and GBM Cancer Cells to Pharmacological Ascorbate. Cancer Cell 32:268. doi: 10.1016/j.ccell.2017.07.008
Schomacher, L., Han, D., Musheev, M. U., Arab, K., Kienhofer, S., von Seggern, A., et al. (2016). Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat. Struct. Mol. Biol. 23, 116–124. doi: 10.1038/nsmb.3151
Shadduck, R. K., Latsko, J. M., Rossetti, J. M., Haq, B., and Abdulhaq, H. (2007). Recent advances in myelodysplastic syndromes. Exp. Hematol. 35(4 Suppl. 1), 137–143. doi: 10.1016/j.exphem.2007.01.022
Sheaffer, K. L., Elliott, E. N., and Kaestner, K. H. (2016). DNA Hypomethylation Contributes to Genomic Instability and Intestinal Cancer Initiation. Cancer Prev. Res. 9, 534–546. doi: 10.1158/1940-6207.CAPR-15-0349
Shen, L., Wu, H., Diep, D., Yamaguchi, S., D’Alessio, A. C., Fung, H. L., et al. (2013). Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706. doi: 10.1016/j.cell.2013.04.002
Soes, S., Daugaard, I. L., Sorensen, B. S., Carus, A., Mattheisen, M., Alsner, J., et al. (2014). Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience 1, 367–374. doi: 10.18632/oncoscience.42
Song, C. X., Szulwach, K. E., Dai, Q., Fu, Y., Mao, S. Q., Lin, L., et al. (2013). Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691. doi: 10.1016/j.cell.2013.04.001
Sotiriou, S., Gispert, S., Cheng, J., Wang, Y., Chen, A., Hoogstraten-Miller, S., et al. (2002). Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nat. Med. 8, 514–517. doi: 10.1038/nm0502-514
Spada, F., Schiffers, S., Kirchner, A., Zhang, Y., Arista, G., Kosmatchev, O., et al. (2020). Active turnover of genomic methylcytosine in pluripotent cells. Nat. Chem. Biol. 16, 1411–1419. doi: 10.1038/s41589-020-0621-y
Spencer, D. H., Russler-Germain, D. A., Ketkar, S., Helton, N. M., Lamprecht, T. L., Fulton, R. S., et al. (2017). CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression. Cell 168, 801–816e813. doi: 10.1016/j.cell.2017.01.021
Spruijt, C. G., Gnerlich, F., Smits, A. H., Pfaffeneder, T., Jansen, P. W., Bauer, C., et al. (2013). Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159. doi: 10.1016/j.cell.2013.02.004
Staberg, M., Rasmussen, R. D., Michaelsen, S. R., Pedersen, H., Jensen, K. E., Villingshoj, M., et al. (2018). Targeting glioma stem-like cell survival and chemoresistance through inhibition of lysine-specific histone demethylase KDM2B. Mol. Oncol. 12, 406–420. doi: 10.1002/1878-0261.12174
Starczak, M., Zarakowska, E., Modrzejewska, M., Dziaman, T., Szpila, A., Linowiecka, K., et al. (2018). In vivo evidence of ascorbate involvement in the generation of epigenetic DNA modifications in leukocytes from patients with colorectal carcinoma, benign adenoma and inflammatory bowel disease. J. Transl. Med. 16:204. doi: 10.1186/s12967-018-1581-9
Steenman, M. J., Rainier, S., Dobry, C. J., Grundy, P., Horon, I. L., and Feinberg, A. P. (1994). Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat. Genet. 7, 433–439. doi: 10.1038/ng0794-433
Suelves, M., Carrio, E., Nunez-Alvarez, Y., and Peinado, M. A. (2016). DNA methylation dynamics in cellular commitment and differentiation. Brief Funct. Genomics 15, 443–453. doi: 10.1093/bfgp/elw017
Sulkowski, P. L., Oeck, S., Dow, J., Economos, N. G., Mirfakhraie, L., Liu, Y., et al. (2020). Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature 582, 586–591. doi: 10.1038/s41586-020-2363-0
Supek, F., Lehner, B., Hajkova, P., and Warnecke, T. (2014). Hydroxymethylated cytosines are associated with elevated C to G transversion rates. PLoS Genet 10:e1004585. doi: 10.1371/journal.pgen.1004585
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Tan, J., Wang, X., Phoon, L., Yang, H., and Lan, L. (2020). Resolution of ROS-induced G-quadruplexes and R-loops at transcriptionally active sites is dependent on BLM helicase. FEBS Lett. 594, 1359–1367. doi: 10.1002/1873-3468.13738
Toffolatti, L., Scquizzato, E., Cavallin, S., Canal, F., Scarpa, M., Stefani, P. M., et al. (2014). MGMT promoter methylation and correlation with protein expression in primary central nervous system lymphoma. Virchows Arch. 465, 579–586. doi: 10.1007/s00428-014-1622-6
Tomkova, M., McClellan, M., Kriaucionis, S., and Schuster-Boeckler, B. (2016). 5-hydroxymethylcytosine marks regions with reduced mutation frequency in human DNA. Elife 5:17082. doi: 10.7554/eLife.17082
Tovy, A., Spiro, A., McCarthy, R., Shipony, Z., Aylon, Y., Allton, K., et al. (2017). p53 is essential for DNA methylation homeostasis in naive embryonic stem cells, and its loss promotes clonal heterogeneity. Genes Dev. 31, 959–972. doi: 10.1101/gad.299198.117
Tsoukalas, D., Fragkiadaki, P., Docea, A. O., Alegakis, A. K., Sarandi, E., Vakonaki, E., et al. (2019). Association of nutraceutical supplements with longer telomere length. Int. J. Mol. Med. 44, 218–226. doi: 10.3892/ijmm.2019.4191
Tsuboi, M., Kondo, K., Soejima, S., Kajiura, K., Kawakita, N., Toba, H., et al. (2020). Chromate exposure induces DNA hypermethylation of the mismatch repair gene MLH1 in lung cancer. Mol. Carcinog. 59, 24–31. doi: 10.1002/mc.23125
Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. doi: 10.1038/nature04433
Tzatsos, A., Pfau, R., Kampranis, S. C., and Tsichlis, P. N. (2009). Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc. Natl. Acad. Sci. U S A. 106, 2641–2646. doi: 10.1073/pnas.0813139106
von Zglinicki, T., and Martin-Ruiz, C. M. (2005). Telomeres as biomarkers for ageing and age-related diseases. Curr. Mol. Med. 5, 197–203. doi: 10.2174/1566524053586545
Wang, L., Song, C., Wang, N., Li, S., Liu, Q., Sun, Z., et al. (2020). NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 16, 1394–1402. doi: 10.1038/s41589-020-0601-2
Wang, S. M., Fan, J. H., Taylor, P. R., Lam, T. K., Dawsey, S. M., Qiao, Y. L., et al. (2018). Association of plasma vitamin C concentration to total and cause-specific mortality: a 16-year prospective study in China. J. Epidemiol. Community Health 72, 1076–1082. doi: 10.1136/jech-2018-210809
Wang, T., Chen, K., Zeng, X., Yang, J., Wu, Y., Shi, X., et al. (2011). The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9, 575–587. doi: 10.1016/j.stem.2011.10.005
Wei, F., Qu, C., Song, T., Ding, G., Fan, Z., Liu, D., et al. (2012). Vitamin C treatment promotes mesenchymal stem cell sheet formation and tissue regeneration by elevating telomerase activity. J. Cell Physiol. 227, 3216–3224. doi: 10.1002/jcp.24012
Welch, J. S., Ley, T. J., Link, D. C., Miller, C. A., Larson, D. E., Koboldt, D. C., et al. (2012). The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278. doi: 10.1016/j.cell.2012.06.023
Welford, R. W., Schlemminger, I., McNeill, L. A., Hewitson, K. S., and Schofield, C. J. (2003). The selectivity and inhibition of AlkB. J. Biol. Chem. 278, 10157–10161. doi: 10.1074/jbc.M211058200
Wiehle, L., Raddatz, G., Musch, T., Dawlaty, M. M., Jaenisch, R., Lyko, F., et al. (2016). Tet1 and Tet2 Protect DNA Methylation Canyons against Hypermethylation. Mol. Cell Biol. 36, 452–461. doi: 10.1128/MCB.00587-15
Wiehle, L., Thorn, G. J., Raddatz, G., Clarkson, C. T., Rippe, K., Lyko, F., et al. (2019). DNA (de)methylation in embryonic stem cells controls CTCF-dependent chromatin boundaries. Genome Res. 29, 750–761. doi: 10.1101/gr.239707.118
Williams, K., Christensen, J., Pedersen, M. T., Johansen, J. V., Cloos, P. A., Rappsilber, J., et al. (2011). TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348. doi: 10.1038/nature10066
Wu, H., D’Alessio, A. C., Ito, S., Xia, K., Wang, Z., Cui, K., et al. (2011). Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393. doi: 10.1038/nature09934
Wu, S. S., Xu, W., Liu, S., Chen, B., Wang, X. L., Wang, Y., et al. (2011). Down-regulation of ALKBH2 increases cisplatin sensitivity in H1299 lung cancer cells. Acta Pharmacol. Sin. 32, 393–398. doi: 10.1038/aps.2010.216
Wu, T. M., Liu, S. T., Chen, S. Y., Chen, G. S., Wu, C. C., and Huang, S. M. (2020). Mechanisms and Applications of the Anti-cancer Effect of Pharmacological Ascorbic Acid in Cervical Cancer Cells. Front. Oncol. 10:1483. doi: 10.3389/fonc.2020.01483
Wulansari, N., Kim, E. H., Sulistio, Y. A., Rhee, Y. H., Song, J. J., and Lee, S. H. (2017). Vitamin C-Induced Epigenetic Modifications in Donor NSCs Establish Midbrain Marker Expressions Critical for Cell-Based Therapy in Parkinson’s Disease. Stem Cell Rep. 9, 1192–1206. doi: 10.1016/j.stemcr.2017.08.017
Xie, W., Baylin, S. B., and Easwaran, H. (2019). DNA methylation in senescence, aging and cancer. Oncoscience 6, 291–293. doi: 10.18632/oncoscience.476
Xu, G. L., Bestor, T. H., Bourc’his, D., Hsieh, C. L., Tommerup, N., Bugge, M., et al. (1999). Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191. doi: 10.1038/46052
Xu, Q., Parks, C. G., DeRoo, L. A., Cawthon, R. M., Sandler, D. P., and Chen, H. (2009). Multivitamin use and telomere length in women. Am. J. Clin. Nutr. 89, 1857–1863. doi: 10.3945/ajcn.2008.26986
Yang, H., Liu, Y., Bai, F., Zhang, J. Y., Ma, S. H., Liu, J., et al. (2013). Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 32, 663–669. doi: 10.1038/onc.2012.67
Yang, J., Guo, R., Wang, H., Ye, X., Zhou, Z., Dan, J., et al. (2016). Tet Enzymes Regulate Telomere Maintenance and Chromosomal Stability of Mouse ESCs. Cell Rep. 15, 1809–1821. doi: 10.1016/j.celrep.2016.04.058
Yang, L., Yu, S. J., Hong, Q., Yang, Y., and Shao, Z. M. (2015). Reduced Expression of TET1, TET2, TET3 and TDG mRNAs Are Associated with Poor Prognosis of Patients with Early Breast Cancer. PLoS One 10:e0133896. doi: 10.1371/journal.pone.0133896
Yin, R., Mao, S. Q., Zhao, B., Chong, Z., Yang, Y., Zhao, C., et al. (2013). Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135, 10396–10403. doi: 10.1021/ja4028346
Young, J. I., Zuchner, S., and Wang, G. (2015). Regulation of the Epigenome by Vitamin C. Annu. Rev. Nutr. 35, 545–564. doi: 10.1146/annurev-nutr-071714-034228
Yu, M., Song, C. X., and He, C. (2015). Detection of mismatched 5-hydroxymethyluracil in DNA by selective chemical labeling. Methods 72, 16–20. doi: 10.1016/j.ymeth.2014.11.007
Yue, X., and Rao, A. (2020). TET family dioxygenases and the TET activator vitamin C in immune responses and cancer. Blood 136, 1394–1401. doi: 10.1182/blood.2019004158
Yun, J., Mullarky, E., Lu, C., Bosch, K. N., Kavalier, A., Rivera, K., et al. (2015). Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 350, 1391–1396. doi: 10.1126/science.aaa5004
Zhang, J., Yang, C., Wu, C., Cui, W., and Wang, L. (2020). DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy. Cancers 12:12082123. doi: 10.3390/cancers12082123
Zhang, L., Lu, X., Lu, J., Liang, H., Dai, Q., Xu, G. L., et al. (2012). Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 8, 328–330. doi: 10.1038/nchembio.914
Zhang, P., Li, J., Qi, Y., Zou, Y., Liu, L., Tang, X., et al. (2016). Vitamin C promotes the proliferation of human adipose-derived stem cells via p53-p21 pathway. Organogenesis 12, 143–151. doi: 10.1080/15476278.2016.1194148
Zhang, Q., Riddle, R. C., Yang, Q., Rosen, C. R., Guttridge, D. C., Dirckx, N., et al. (2019). The RNA demethylase FTO is required for maintenance of bone mass and functions to protect osteoblasts from genotoxic damage. Proc. Natl. Acad. Sci. U S A. 116, 17980–17989. doi: 10.1073/pnas.1905489116
Zhang, T. J., Zhou, J. D., Yang, D. Q., Wang, Y. X., Wen, X. M., Guo, H., et al. (2018). TET2 expression is a potential prognostic and predictive biomarker in cytogenetically normal acute myeloid leukemia. J. Cell Physiol. 233, 5838–5846. doi: 10.1002/jcp.26373
Zhang, W., Li, J., Suzuki, K., Qu, J., Wang, P., Zhou, J., et al. (2015). Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348, 1160–1163. doi: 10.1126/science.aaa1356
Zhang, W., Qu, J., Suzuki, K., Liu, G. H., and Izpisua Belmonte, J. C. (2013). Concealing cellular defects in pluripotent stem cells. Trends Cell Biol. 23, 587–592. doi: 10.1016/j.tcb.2013.07.001
Zhang, X., Su, J., Jeong, M., Ko, M., Huang, Y., Park, H. J., et al. (2016). DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 48, 1014–1023. doi: 10.1038/ng.3610
Zhao, L., Quan, Y., Wang, J., Wang, F., Zheng, Y., and Zhou, A. (2015). Vitamin C inhibit the proliferation, migration and epithelial-mesenchymal-transition of lens epithelial cells by destabilizing HIF-1alpha. Int. J. Clin. Exp. Med. 8, 15155–15163.
Zhao, R., Choi, B. Y., Lee, M. H., Bode, A. M., and Dong, Z. (2016). Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine 8, 30–39. doi: 10.1016/j.ebiom.2016.04.017
Zhou, J., Chen, C., Chen, X., Fei, Y., Jiang, L., and Wang, G. (2020). Vitamin C Promotes Apoptosis and Cell Cycle Arrest in Oral Squamous Cell Carcinoma. Front. Oncol. 10:976. doi: 10.3389/fonc.2020.00976
Keywords: DNA methylation (5mC), DNMT, 5-hydroxymethylation (5hmC), TET, genomic stability, vitamin C
Citation: Brabson JP, Leesang T, Mohammad S and Cimmino L (2021) Epigenetic Regulation of Genomic Stability by Vitamin C. Front. Genet. 12:675780. doi: 10.3389/fgene.2021.675780
Received: 03 March 2021; Accepted: 06 April 2021;
Published: 04 May 2021.
Edited by:
Fiammetta Vernì, Sapienza University of Rome, ItalyReviewed by:
Ryszard Oliński, Nicolaus Copernicus University in Toruń, PolandCopyright © 2021 Brabson, Leesang, Mohammad and Cimmino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luisa Cimmino, bHVpc2EuY2ltbWlub0BtZWQubWlhbWkuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.